Abstract
The mechanism by which cationic polymers containing titratable amines mediate effective endosomal escape and cytosolic delivery of nucleic acids is not well understood despite the decades of research devoted to these materials. Here we utilize multiple assays investigating the endosomal escape step associated with plasmid delivery by polyethylenimine (PEI) and poly(beta-amino esters) (PBAEs) to improve the understanding of how these cationic polymers enable gene delivery. To probe the role of these materials in facilitating endosomal escape, we utilized vesicle membrane leakage and extracellular pH modulation assays to demonstrate the influence of polymer buffering capacity and effective pKa on delivery of plasmid DNA. Our results demonstrate that transfection with PBAEs is highly sensitive to the effective pKa of the overall polymer, which has broad implications for transfection. In more acidic environments, PBAE-mediated transfection was inhibited, while PEI was relatively unaffected. In neutral to basic environments, PBAEs have high buffering capacities that led to dramatically improved transfection efficacy. Cellular uptake of polymeric nanoparticles overall was unchanged as a function of pH, indicating that microenvironmental acidity was important for downstream intracellular delivery efficiency. Overall, this study motivates the use of polymer chemical characteristics, such as effective pKa values, to more efficiently evaluate new polymeric materials for enhanced intracellular delivery characteristics.
Keywords: Nanoparticle, gene delivery, poly(beta-amino ester), polyethylenimine, endosome, pKa
Graphical Abstract
Introduction:
Polycations have a long history of being used to deliver nucleic acids, due to the ability of polycationic materials to electrostatically complex with the phosphate backbone of nucleic acids to form polyplex nanoparticles. Electrostatic complexation of nucleic acids condenses them via charge neutralization to form polyplex nanoparticles small enough to be internalized to most cells via endocytosis.1, 2 Following effective internalization, traditional polycation nanoparticles must facilitate endosomal escape to achieve to effective cytosolic delivery required for functional nucleic acid delivery.3 Alternative routes to nuclear delivery include the formation of nanopores,4 trafficking through the endoplasmic reticulum5, 6 and direct cytosolic entry across the plasma membrane, similar to what occurs with electroporation.7 Lipid nanoparticles were previously demonstrated to achieve endosomal escape via temporary transient disruption of the lipid bilayer in a manner attributable to the fusogenic properties of the pH sensitive amino-lipids used;8 the mechanism of escape for polyplex nanoparticles remains much less well understood.3
Polymer-based gene delivery systems are constrained by challenges that limit their clinical uses and that often hinder transfection even in vitro. For example, poly-L-lysine (PLL) was one of the early polymers explored for its ability to complex with DNA, but it showed lackluster efficacy due to its inability to escape the endosome and successfully deliver its cargo. Polyethyleneimine (PEI), on the other hand, showed greater efficacy due to its more effective endosomal escape, but exhibited cytotoxicity due to its lack of intracellular degradation. Poly(beta-amino ester)s (PBAE), have overcome these challenges by facilitating effective delivery of genes to cells via amines that bind and encapsulate DNA into nanoparticles and hydrolyzable ester bonds that release DNA to the cytosol while also limiting potential cytotoxicity through quick degradation. Additionally, these biomaterials can be synthesized simply from a wide variety of available precursor molecular structures, which allows for design of many different polymer types with differential properties.9
The proton sponge hypothesis was originally posited as the mechanism by which PEI achieves endosomal escape10, proposing that the polycation ability to buffer hydrogen ion accumulation in endosomes triggers chloride and water influx to ultimately cause endosomal membrane disruption via polymer and osmotic swelling. While some evidence supporting this hypothesis has been observed, including chloride accumulation in endosomes containing PEI and swelling of endosomes11 this hypothesis has also been repeatedly challenged based on unclear evidence of the ability of PEI to completely buffer endosomes12 and there remains no steadfast consensus in the field for PEI’s specific mechanism of escape.3 Further supporting evidence against the ability of PEI to achieve endosomal escape via the traditional proton sponge mechanism, computational modeling of combined PEI and osmotic swelling does not appear to achieve sufficient swelling to result in endosomal membrane disruption.3 The mechanism of escape for newer cationic materials such as PBAEs are even less understood, but due to the improved gene delivery efficacy of many of their structures, motivate great interest in mechanistic research on their function.
In contrast to the traditional interpretation of the proton sponge hypothesis, a refined view put forward for endosomal escape of polycationic materials is that they achieve transient destabilization via charge driven polyplex-membrane interactions that lead to local membrane disruption and fractional escape from endosomes in contrast to full endosomal rupture.3 Both of these mechanisms could plausibly play a role in endosomal escape, along with others that have not yet been considered. By better understanding the mechanism of endosomal escape, improved rational design of next generation polymers is facilitated. Although gene therapy holds promise in a wide range of diseases, few therapeutic nanomedicines for gene delivery have made it to the clinic. Improved quantitative understanding of how biomaterial structures function can lead to new enabling technologies for gene therapy could improve medical capabilities.
Materials and Methods:
Materials
Monomers B4 (1,4-Butanediol diacrylate); S4 (4-amino-1-butanol); and E7 (1-(3-Aminopropyl)-4-methylpiperazine) were purchased from Alfa Aesar. Monomers B7 (Bisphenol A glycerolate (1 glycerol/phenol) diacrylate); and E6 (2-(3-Aminopropylamino)ethanol) were purchased from Sigma Aldrich. Branched polyethlyenimine (BPEI) 25 kDa (408727, Sigma Aldrich) and linear PEI (LPEI) 4 kDa (24885–2 Polysciences Warrington, PA, USA) were dissolved fresh on the day of use to a concentration of 10 mg/mL in 150 mM NaCl.
Cell Culture
HEK293T and B16 cells were maintained in DMEM high glucose with 10% FBS and 1% penicillin/streptomycin. GB319 glioblastoma cells were maintained in DMEM/F12 with 10% FBS and 1% antibiotic/antimycotic.13 Unless otherwise specified, all cells were cultured at 37°C in a 5% CO2 atmosphere. In the pH-dependent uptake and transfection assays, the media used was DMEM buffered with 20 mM phosphate instead of carbonate buffer, supplemented with 10% FBS and 1% penicillin/streptomycin. The pH of the medium was adjusted with HCl and NaOH to pH 5, 6, 7 or 8. Cells were exposed to the media at 37°C with atmospheric CO2 for two hours during nanoparticle incubation, after which media was changed to carbonate buffered DMEM and cells were returned to a 5% CO2 incubator.
PBAE Synthesis
Poly (beta-amino ester)s (PBAEs) were synthesized following previously described methods.13 Briefly, one diacrylate-terminated backbone monomer (B) was polymerized with one primary amine-containing sidechain monomer (S) in a neat solution by stirring for 24 hours at 90°C, forming the base polymer via Michael addition. This base polymer was dissolved in anhydrous tetrahydrofuran (THF) and mixed with one end-cap small molecule (E), then stirred at room temperature for 1 hr. The end-capped PBAE was then precipitated into diethyl ether, washed twice, and left under vacuum for 48 hours for complete removal of ether. The dry PBAE was dissolved in anhydrous DMSO at 100 mg/ml and stored at −20°C in small aliquots. The specific PBAEs used for the following studies were internally referred to as 446 or 447 for polymer formed from B4, S4 and either E6 or E7, and as 746 for polymer formed from B7, S4 and E6.
Polymer and Nanoparticle Characterization
Polymer characterization was performed using nuclear magnetic resonance (1H NMR), as described previously.14 Briefly, non-end-capped, acrylate terminated polymers and end-capped, finished polymers were sampled from reactions and washed in diethyl ether at 10X volume. They were dried for 2 h under vacuum and dissolved in CDCl3 before being analyzed using 1H NMR (Bruker 500 MHz) to determine success of reaction.
Measurement of number average molecular weight (MN), weight average molecular weight (MW), and polydispersity index (PDI) was performed using gel permeation chromatography (GPC), using a Waters system with autosampler, styragel column, and refractive index detector as described previously15 and comparing to linear polystyrene standards. The flow rate was 0.5 mL/min and the run time was 75 min per sample.
Nanoparticle characterization was performed using dynamic light scattering (DLS). After mixing polymer with DNA at the specified weight/weight ratios in 25 mM NaAc, pH 5.0 (PBAEs) and 150 mM NaCl (PEI), hydrodynamic diameters were determined using a Malvern Zetasizer NanoZS (Malvern Instruments, Marlvern, UK) with a detection angle of 173°. For characterization under different transfection pH conditions, samples were diluted at a dilution factor of 6 in 30 mM phosphate buffer with 120 mM NaCl prepared at specified pH values and hydrodynamic diameter and zeta potential were measured again.14
Nanoparticle transfection
Cells were seeded one day prior to transfection in 96-well plates at 15,000 cells/well. Weight/weight (w/w) refers to the mass ratio between polymer and plasmid. DNA. Nanoparticle N/P value was calculated as the ratio of protonatable amines in the polymer over negatively charged phosphate groups in the DNA. The ratio was calculated by determining the weight fraction of each molecule taken up by its respective moiety and multiplying the w/w by the ratio of weight fractions.
Polymers and DNA were diluted into the appropriate buffers (25 mM NaAc for PBAE, 150 mM NaCl for PEI) and mixed to achieve desired concentration and weight/weight ratio for nanoparticles. Buffers were chosen based on previously used nanoparticle formation protocols in order to maximize the transfection efficacy and reproduce the standard transfection conditions for each polymer used in the literature.10, 16, 17 BPEI 25 kDa and LPEI 4 kDa were used at a w/w ratio of 2 (6.9 N/P). PBAEs were used at either 60 w/w (446: 21.0 N/P, 447: 25.6 N/P) or 40 w/w (746: 10.4 N/P). In the pH dependent transfections, cell media was replaced with the pH-specific media immediately prior to transfection. Nanoparticles were added to the cells in their transfection media for a final specified DNA dose of 5 μg/mL in 120 μL of media per well and incubated for two hours. From prior work, this has been shown to be the optimal DNA dose for transfection, and is the standard DNA dose used in our lab.16–18 It should be noted that DNA dose can have an influence on transfection efficacy, but that high enough levels of nanoparticles can result in cytotoxicity. In standard transfections, cells were incubated at 37°C with 5% CO2, while in pH-dependent transfections, they were incubated at 37°C with atmospheric CO2 to ensure pH stability.
In all experiments, after two hours the nanoparticle containing media was aspirated and cells were washed with a dilute solution of heparin as previously described after which 100 μL of fresh medium was added.19 In pH-dependent experiments, cells were only exposed to varied pH media during the 2 hour transfection period in order to minimize the effect of media pH directly on the cells, and to ensure that only the endosomal pH varied between groups. For transfection efficacy experiments, plasmid CMV-eGFP-N1 (Addgene 2491) was used while for nanoparticle uptake experiments, nanoparticles were formed with 20% DNA covalently labeled with Cy5 as previously described.20 For initial gene expression studies, transfection efficacy was assessed by flow cytometry approximately 48 hours following transfection, while for dual uptake and expression experiments transfection efficacy was assessed at only 24 hours post nanoparticle addition.
pH Titration and Buffering Capacity
Endcapped polymer buffering capacity as a function of polymer structure was assessed by titrating 10 mg (100 μL at 100 mg/mL) of polymer dissolved in 10 mL of acidified, 100 mM NaCl from pH 4.5 to pH 8.5.21 For titrations, pH was determined using a SevenEasy pH Meter (Mettler Toledo) with pH assessed after stepwise addition of 100 mM sodium hydroxide. Effective pKa was determined as previously reported by calculating buffering capacity between each addition of sodium hydroxide.14
DNA binding affinity
DNA binding affinity was performed using a Yo-Pro-1 competition binding assay similar to as previously described but using differential pH solutions.22 Plasmid DNA and Yo-Pro-1 iodide (Thermo Fisher) were both diluted to a concentration of 2 μM in 30 mM phosphate buffer with 120 mM NaCl prepared at specified pH values and distributed (50 μL) to opaque black well plates for fluorescence measurements. A dilution series of each polymer was prepared in each specified pH solution. Fifty microliters of each polymer dilution at matched pH values were then added to respective wells containing dilute pDNA/Yo-Pro-1 for a final pDNA/Yo-Pro-1 concentration of 1 μm and polymer concentration between 200–0 w/w. After a 10 min incubation with gentle rocking, green channel fluorescence was measured using a plate reader (Biotek Synergy 2).
Vesicle leakage assay
1-Palmitoyl-2-oleoly-sn-3- glycero-phosphocholine (POPC) was purchased from Avanti Polar Lipids. The 8-aminonapthalene (ANTS) fluorophore and P-xylene-bis-pyridine (DPX) quencher pair were purchased from Thermofisher. Vesicles were prepared according to established methods to assess small molecule ANTS/DPX leakage induced by nanoparticles from vesicles.23–25 Briefly, POPC at an initial concentration of 25 mg/mL in nitrogen dried chloroform was measured using a Hamilton syringe and dried under nitrogen air flow followed by vacuum for two hours. POPC was then resuspended at 10 mg/mL in aqueous buffer containing the fluorophore quencher pair ANTS/DPX. Buffers containing ANTS/DPX were prepared to desired pH point from Millipore water with 100 mM potassium chloride and 10 mM sodium phosphate. ANTS and DPX were added to a concentration of 12.5 and 45 mM respectively. Resuspended lipid vesicles were then extruded 10 times through a 0.1 μm Nucleopore polycarbonate filter to give unilamellar vesicles of 0.1 μm diameter. Gel filtration chromatography via a Kimble Kontes FlexColumn with Sephadex G-100 resin was used to remove external ANTS/DPX from vesicles with entrapped contents. Contact with plastic containers was minimized to prevent lipid loss. Unilamellar ANTS/DPX vesicles were used on the same day of preparation.
For the vesicle leakage assay, PBAE 447 or BPEI 25 kDa polymers with or without DNA were prepared in the specified pH phosphate buffer (100mM KCl and 10 mM Na3PO4) without ANTS/DPX. Purified ANTS/DPX vesicles were mixed with varying concentrations of polymers in black well 96 well plates for a final lipid concentration of 1 mM. The mixtures were incubated for 30 minutes at 37°C prior to measuring fluorescence via plate reader assay (Synergy 2, Biotek). Triton X-100 was then added to each well to induce complete vesicle leakage and a second plate reader measurement was taken. Values shown are for individual wells normalized to their post-Triton X-100 measurement.
Data analysis, statistics and figures:
FlowJo was used for flow cytometry analysis. Prism 6 (Graphpad, La Jolla, CA) was used for all statistical analyses and curve plotting. Unless otherwise specified, statistical tests were performed with a global alpha value of 0.05 and experiments were repeated at least twice with representative results shown. Unless otherwise stated, absence of statistical significance markings where a test was stated to have been performed signified no statistical significance. Statistical significance was denoted as follows: *p<0.05; **p<0.01; ***p<0.001; ****p<0.0001.
Results and Discussion:
Polymer Synthesis and Characterization
Polymers were successfully synthesized and characterized following the procedures described in the methods. Polymer synthesis proceeded in two steps and is described in Figure S1. Polymer molecular weight characterization was performed using GPC and is shown in Figure S2. The molecular weights of PBAEs 446 and 447 were approximately the same as they were synthesized using the same 44 base polymer, whereas PBAE 746 was formed from the 74 base polymer and was smaller in molecular weight. 1H NMR spectra are shown in Figure S3 to confirm polymer identity.
Transfection with cationic polymers
Transfection efficacy was assessed using three cell lines: HEK293T human embryonic kidney cells transformed with large T-cell antigen, B16-F10 murine melanoma cells and GB319 glioblastoma cells.13 These three cell lines are representative of most rapidly dividing cell types, enabling high degrees of cellular uptake to be studied. Transfection was measured using flow cytometry after delivery of a GFP reporter plasmid. Synthesized PBAEs (Fig. 1A) were demonstrated to be highly effective for the transfection of HEK293T, B16-F10, GB319 cells in vitro (Fig. 1C–E), with transfection efficacy up to 99% and 65% respectively with the polymers selected for study here. These structures were selected because of prior utilization for delivery of plasmid DNA and mRNA26 to a diverse set of cells including in vivo to brain tumors27, as well as T-cells.28 Multiple cell lines were utilized in this study so that transfection could be probed in a more robust way. It has previously been shown that by differential PBAE structure can lead to dramatically different transfection efficacy in different cell lines, and that this phenomenon can be used as an approach for cell type selective transfection.9, 13, 29 The cell lines used for transfection in this study significantly differ in their type, as HEK293T were originally isolated from human kidney, B16-F10 are a metastatic murine melanoma cell line, and GB319 are a human glioblastoma cell line.13 The 4 kDa LPEI and 25 kDa BPEI nanoparticles were formulated with simple mixing and yielded moderate transfection in all cell lines. 4 kDa LPEI was chosen for this study as it has a similar molecular weight and physical form as a linear polymer to the PBAEs with number average molecular weight of 4–5 kDa (Fig 1B) that are also evaluated in this study. It is compared to as a control with similar macromolecular structure to the evaluated PBAEs but with similar monomeric structure to BPEI. All nanoparticles were tested at optimized doses maintaining high cell viability (Fig. 1F–H) and had similar biophysical properties (Fig. 1I, J).
Figure 1.
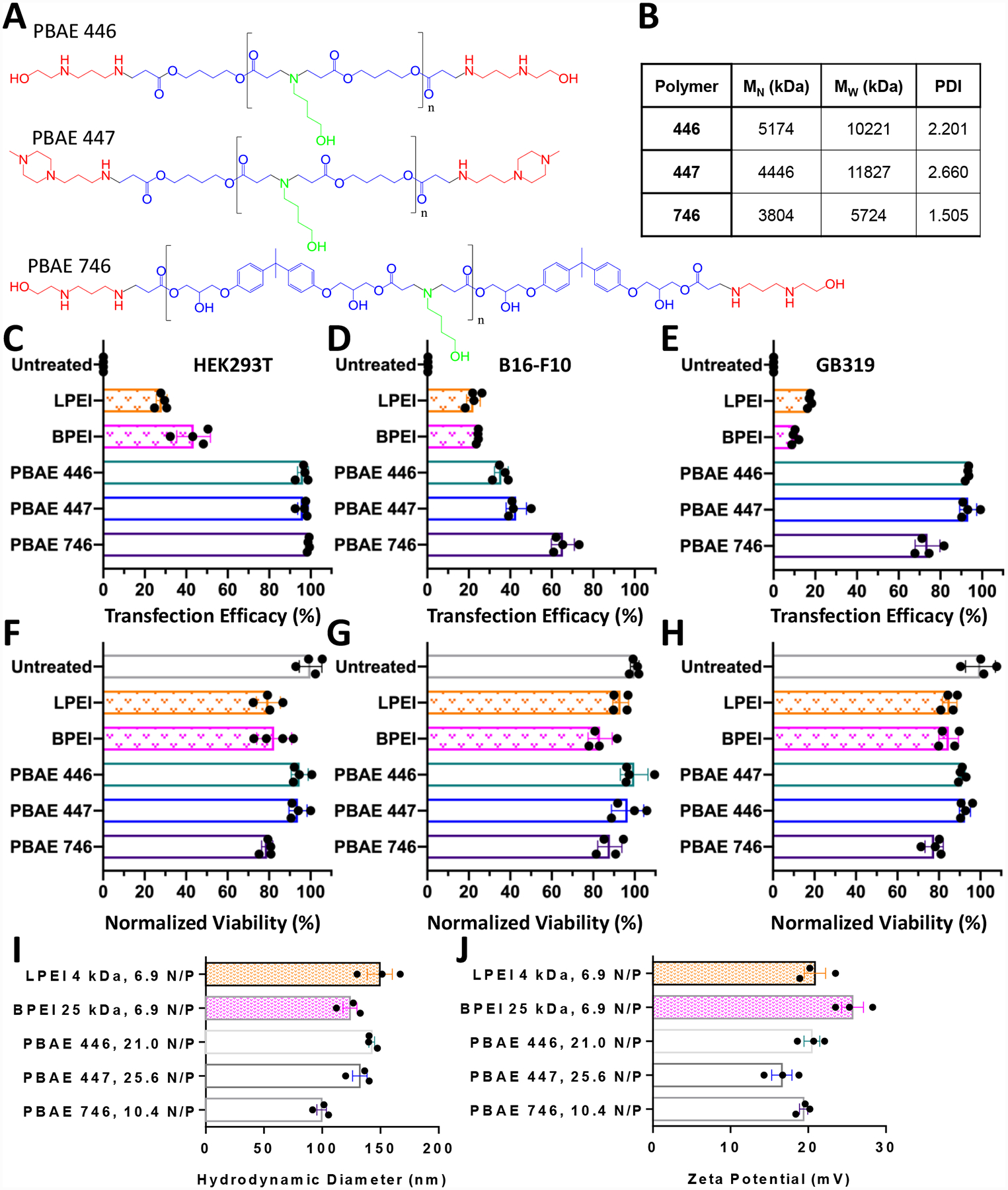
PEI and PBAEs enable transfection in mammalian cells. A) Structures of selected PBAE molecules. B) GPC data of synthesized PBAEs C) Selected PBAEs achieve up to 99% transfection efficacy in HEK293T cells, D) 65% transfection efficacy in B16-F10 cells and E) 90% transfection efficacy in GB319. F-H) Cellular viability following transfection is maintained. Transfections were performed in 10% serum conditions with a total DNA dose per well of either 450 ng (HEK293T) or 600 ng (B16-F10 or GB319 cells). I) Nanoparticle properties of hydrodynamic diameter and J) zeta potential of all materials tested. N/P ratios correspond to the following w/w values: LPEI, 2 w/w; BPEI, 2 w/w; PBAE 446, 60 w/w; PBAE 447, 60 w/w; PBAE 746, 40 w/w. Error bars show mean ± SEM of the four well replicate points shown or the mean ± SEM of three individual preparations of nanoparticles for characterization.
pH Sensitivity and Effective pKa
Following uptake and cellular internalization to the cytoplasm, early endosomes that are not recycled back out to the plasma membrane undergo transition to late-endosomes, which eventually become lysosomes, typically within 60 minutes.8 The ability of cationic polymers to protonate under acidic conditions or to buffer hydrogen ions has long been suspected as crucial to yield effective transfection; yet, buffering capacity has often shown little predictive capacity for efficacy, as more effectively-buffering materials often fail to improve gene expression.30 For this reason, we performed simple pH titration assays with the transfection materials used here demonstrating that both PEI and PBAEs are capable of buffering protons in the physiologically relevant pH range of 4.5 to 7.4 (Fig. 2A).
Figure 2.
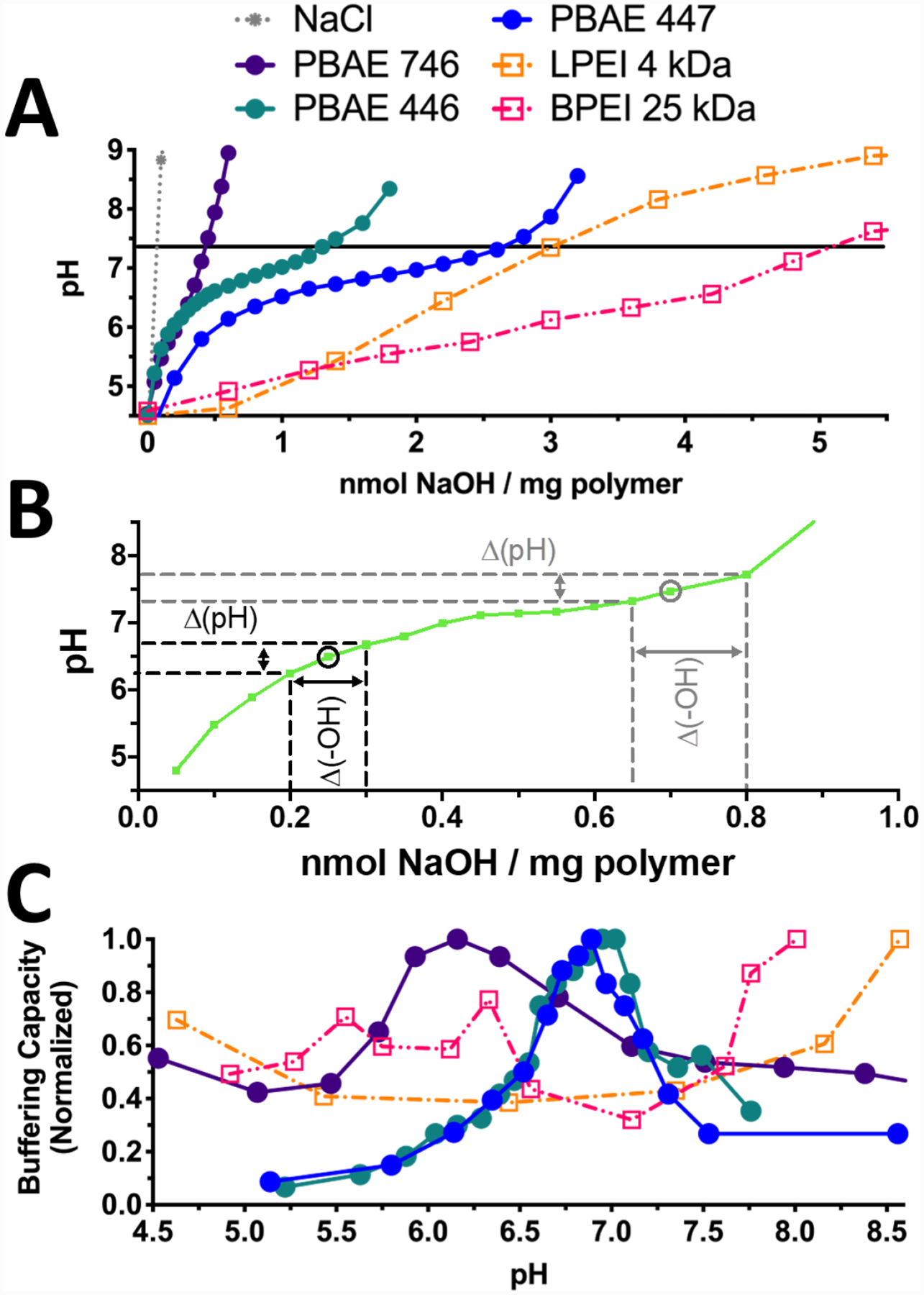
PEI and PBAE are capable of buffering acidification. A) Titration curves of PEI and PBAEs 446, 447, and 746 demonstrate lower buffering capacities of PBAEs relative to PEI on a per-mass basis. B) As shown on this representative titration curve, buffering capacity at a given pH value was calculated from the slope of each pH titration curve as Δ(-OH)/Δ(pH). C) Normalization to the maximum buffering capacity in the relevant pH range (4.5–8.5) shows that PBAEs have a clear effective pKa value between pH 6–7.4 compared to both LPEI and BPEI, which are able to buffer over a broad range and lack clear effective pKa peak.
Buffering capacity curves were generated by calculating the inverse slope of the titration curve at each pH level, given by Δ(-OH)/Δ(pH) (Fig. 2B). Effective pKa was defined as the pH at which the peak buffering capacity was observed. As PBAEs demonstrated a fairly narrow pH range for buffering, we calculated local effective pKa values for PBAEs in this pH range of 6.16, 6.95, and 7.15 respectively for PBAEs 746, 446, and 447 (Fig. 2C).14 Calculation of an effective pKa value for these polymers may prove to be similarly useful to the measurement of pKa values that has proved indispensable for ionizable lipid nanoparticle formulations.31 In contrast to PBAEs, both types of PEI studied were capable of buffering over a wide pH range and lacked a clear effective pKa peak within the acidities tested here, which matched previous work (Fig 2C).12, 21, 30, 32 The broad buffering capacity seen here is consistent with the proton sponge hypothesis, as it shows that PEI can continue to absorb protons as the endosome acidifies. Both PEI polymers seemed to trend upwards in buffering capacity past pH 8, beyond the physiologically relevant pH conditions studied here or of traditional relevance to gene delivery.
DNA Binding Affinity in PBAEs is Strongly Influenced by pH
To further investigate the effects of pH on PBAE behavior, we performed a DNA binding assay using Yo-Pro-1, a molecule that fluoresces upon binding to DNA. Because these cationic polymers compete with Yo-Pro-1 to bind DNA, polymer-DNA binding displaces Yo-Pro-1, resulting in fluorescence quenching proportional to the amount of unbound Yo-Pro-1. As a result, higher polymer to DNA amine/phosphate (N/P) ratios result in greater Yo-Pro-1 quenching, as does higher polymer charging at lower pH values. Using this assay with both w/w titration and pH modulation, we measured the influence of pH on the binding efficacy of PBAEs and BPEI to plasmid DNA at physiologically relevant N/P ratios (Fig. 3A–C). The three most disparate structures (447, 746, and BPEI) were chosen to be compared. Due to their structural and titrational similarities to PBAE 447 and BPEI, respectively, PBAE 446 and LPEI were not investigated here.
Figure 3.
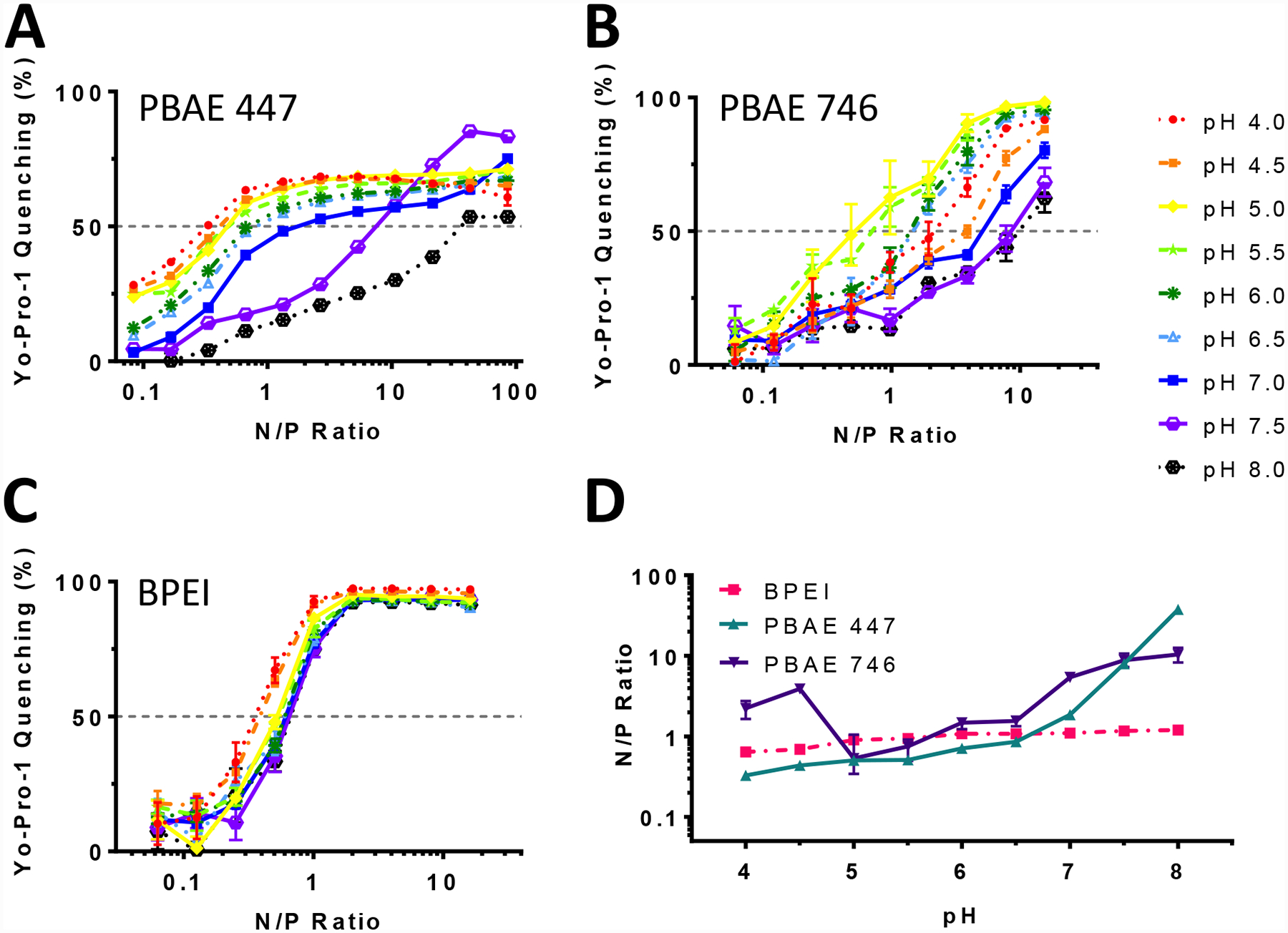
DNA binding affinity of PBAEs is influenced strongly by pH. Yo-Pro-1 fluorophore binding assay with A) PBAE 447 B) PBAE 746 and C) BPEI demonstrate the difference in pH responsiveness of these polymers in regard to DNA binding. D) The calculated effective N/P ratio to quench Yo-Pro-1 fluorescence by 50% (EQ50) at each pH. PBAE 447 binding capacity for plasmid DNA increased by 100-fold between pH 4 and pH 8, whereas BPEI’s binding capacity for DNA increased by only 2-fold. Error bars show mean ± SD of four wells.
Binding affinity was further analyzed through the EQ50, or effective polymer N/P ratio to quench Yo-Pro-1 fluorescence by 50%. These results show that while PEI’s ability to bind DNA is minimally affected by pH, PBAEs binding interactions with plasmid DNA are strongly influenced by pH (Fig. 3D). For PBAE 447 particularly, changing pH between pH 4 and pH 8 results in a 100-fold change in the degree of binding. The increase in DNA binding affinity as pH decreases can be explained by protonation. PBAE and BPEI have both been shown to form polyplexes with DNA primarily based on electrostatic interactions between cationic polymer and anionic DNA.17, 33 Thus, as moieties on the polymers are protonated, the resulting increase in positive charge allows for stronger DNA binding. Much of this change, however, occurs above pH 6.5, although it is possible that a significant increase in binding tightness would occur as a result of endosomal acidification. Notably, both BPEI and PBAE 746 show less of a pH-dependent response when compared to PBAE 447.
Vesicle leakage
To explore the role of polycation and hydrophobic effect mediated membrane disruption in PBAE-induced endosomal escape, we exposed synthetic ANTS/DPX-containing POPC vesicles to PBAE 447 and BPEI at various concentrations and pH environments (Fig. 4). These polymers were chosen as the representative PBAE and PEI polymers due to their leading efficacy in multiple at transfecting a wide variety of cells including in vivo transfection of brain tumors and T-cells (447),13, 15, 27, 28, and their frequency of use for gene delivery studies (BPEI).12, 35 The ANTS/DPX leakage assay is representative of pore formation sufficient for small molecule leakage from membranes (ANTS is 427 Da and DPX is 422 Da). Exposure to PBAE at high concentrations resulted in vesicle leakage compared to PEI. In both cases, leakage was much lower compared to the naturally occurring bee venom peptide mellitin (2846 Da), which effectively induces ~50% ANTS/DPX leakage at 5.68 μg/mL, an almost 1000-fold lower concentration (Fig 4B). At higher concentrations of ~28 μg/mL, this leakage exceeds 80%.25
Figure 4.
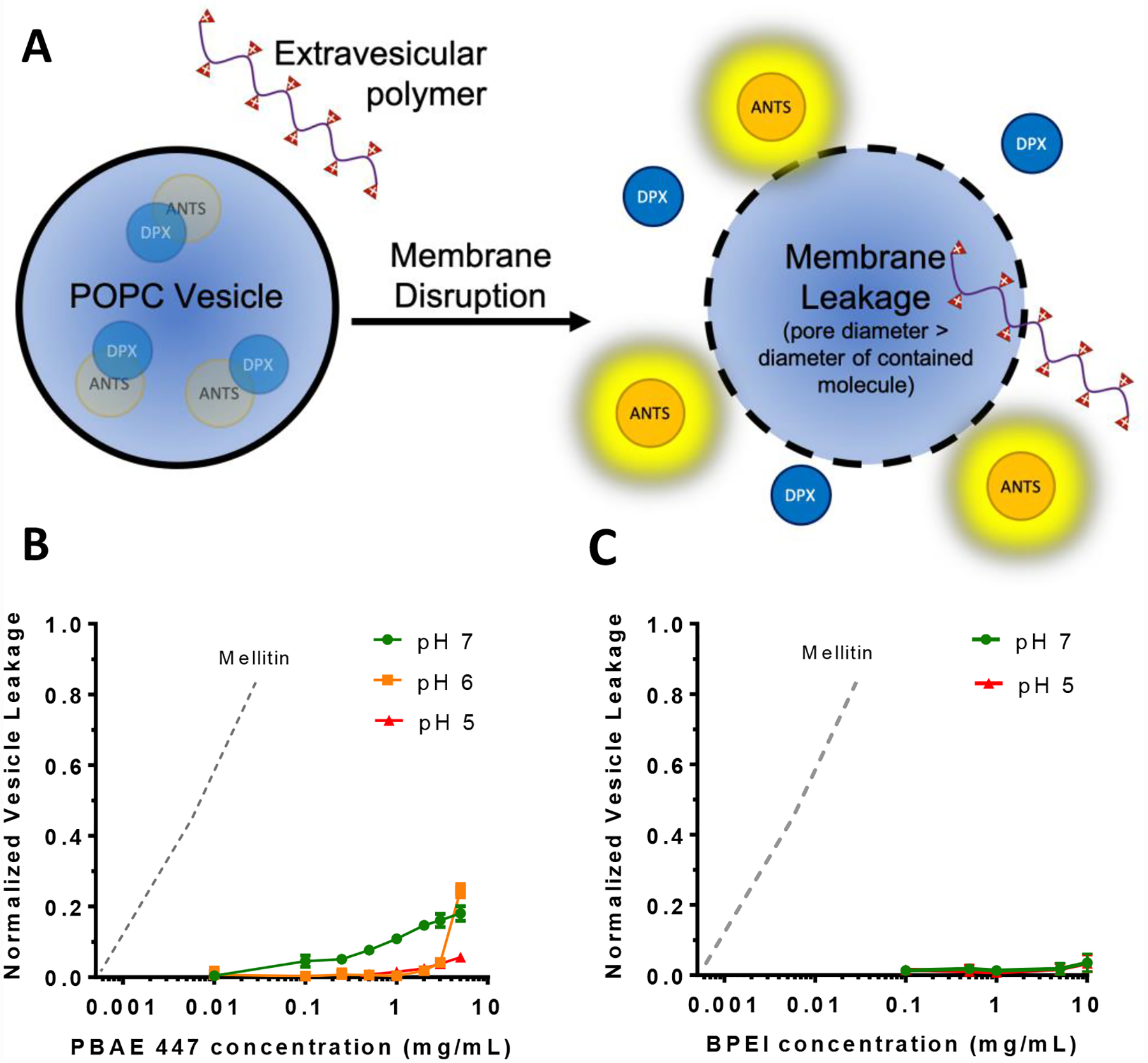
Cationic polymers do not induce efficient small molecule vesicle leakage in POPC vesicles. A) The fluorophore ANTS is quenched by close proximity quencher DPX when encapsulated in vesicles, but becomes fluorescent upon effective membrane poration. A higher fluorescence signal signifies a greater degree of leakage. B) PBAE 447 induced minor leakage of ANTS/DPX from POPC lipid vesicles at pH values of 6 and 7 but not pH 5. C) BPEI 25 kDa did not induce any meaningful leakage at either pH 5 or pH 7. Normalized vesicle leakage is reported relative to vesicles from the same prepared batch exposed to Triton X-100 to induce complete membrane leakage. The dotted line indicates vesicle leakage caused by mellitin, as approximated from the results of Wiedman et al.25
In the assay, PBAE 447 did induce up to ~25% membrane leakage, but only at high concentrations that would be inconsistent with cellular applications. In Figure 5, for example, the concentration of PBAE used was 0.3 mg/mL, which would result in less than 10% vesicle leakage (Fig. 4B). Additionally, as the local pH approaches neutral, PBAEs with tertiary amines in the backbone of the polymer become less protonated and are expected to more easily aggregate due to hydrophobic interactions, which could also minimize direct membrane disruption. As neither PBAEs nor BPEI at standard concentrations formed pores in POPC membranes large enough to induce moderate ANTS/DPX leakage, and neither of these cationic polymers induced leakage more efficiently at lower pH values, it suggests that their mode of action is not through direct charge-based membrane association and disruption. Furthermore, ANTS and DPX are both small molecules, much smaller than DNA or polymer, suggesting that compared to them, lower leakage of macromolecules is likely. It is possible, however, that the leakage that was observed with PBAE 447 under high concentration and pH could be sufficient to cause some leakage of endosomal contents that would surpass the threshold required for transfection or that this effect may work synergistically with additional polymer-mediated endosomal escape mechanisms. Cationic polymer-induced leakage experiments with a broader assortment of polymers and endosome-mimicking membranes would be interesting for future study to better elucidate these mechanisms.
Figure 5.
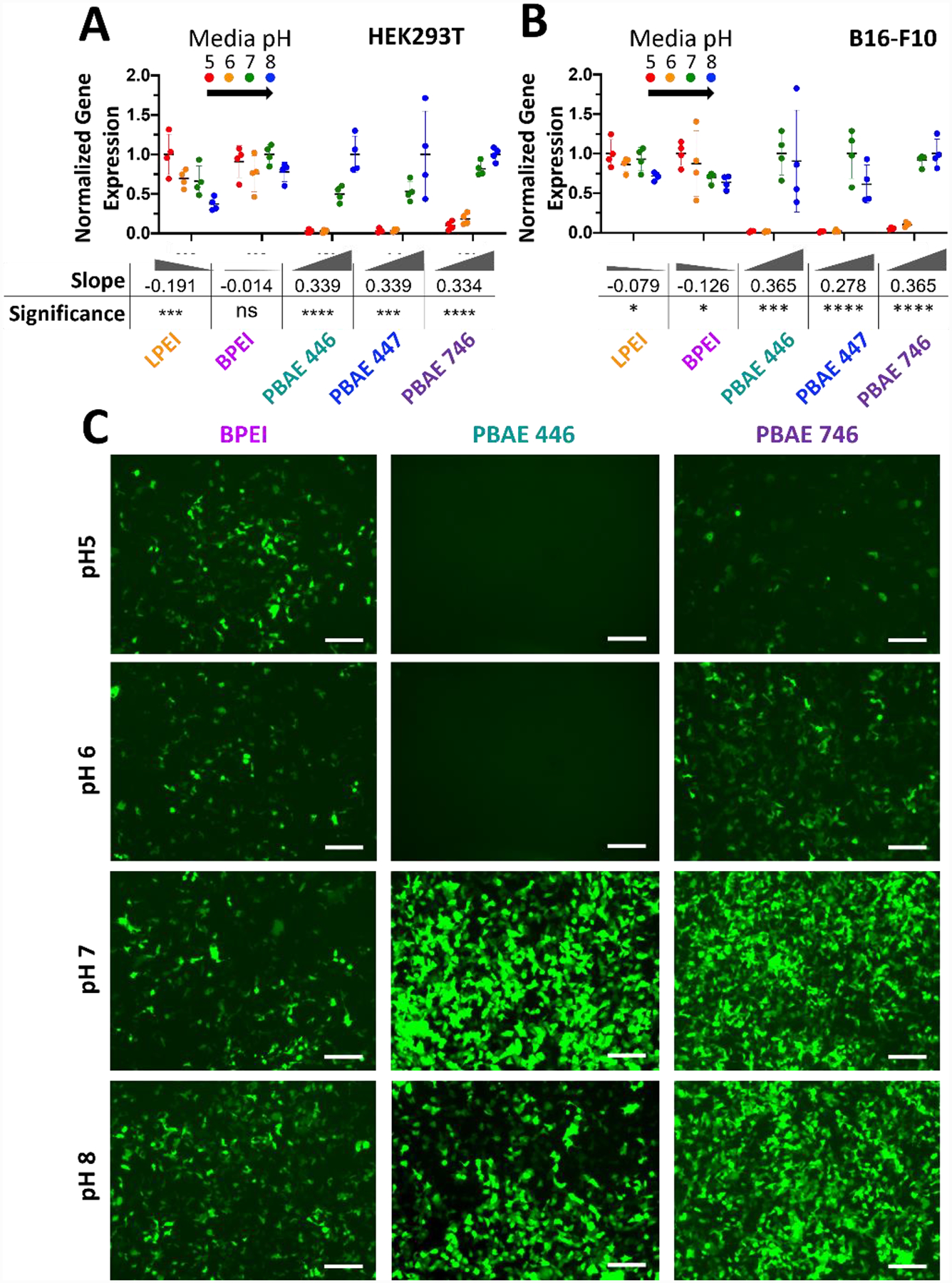
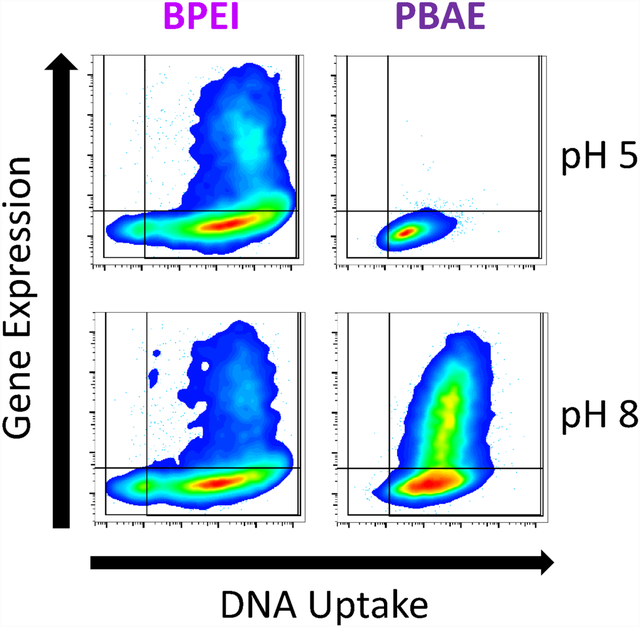
PBAE but not PEI mediated transfection is inhibited by acidic extracellular media. Modulation of extracellular media pH in A) HEK293T cells and B) B16-F10 cells set to pH 5, 6, 7 or 8 results in differences in exogenous gene expression level depending on polymer. Data points show individual well mean transfection efficacy. Error bars show mean ± SD of four wells. Slope and statistical significance were assessed by a one-way ANOVA test of linear trend across pH modulation with Dunn’s corrected multiple comparisons. C) Representative GFP fluorescence microscopy images of HEK293T cells on day two following transfection BPEI 6.9 N/P (2 w/w), 446 21.0 N/P (60 w/w) or 746 10.4 N/P (40 w/w) nanoparticles when transfections were performed with extracellular media pH set to specific values. Scale bar 50 μm.
As another limitation, this assay utilized POPC, which is found in eukaryotic lipid bilayers and is often utilized for biophysical assays as a relatively permeable lipid bilayer in contrast to membranes stabilized with cholesterol and other lipids.25 As a result, it is often the assay in which membrane leakage is most easily induced when compared to other, more biologically accurate assays, and thus lack of efficient leakage indicates that membrane leakage is less likely within the cell. If instead these cationic polymers were inducing strong POPC membrane pore formation, it would indicate that intracellular leakage is possible, but more accurate tests would be needed to confirm this. It should be noted that as these were not cell membranes, this assay has not fully eliminated potential membrane disruption as a mechanism of endosomal escape and future studies with vesicles that utilize cellular membranes or in vitro calcein studies could further elucidate polymer behavior.
Extracellular pH Influence on Transfection and Nanoparticle Uptake
To assess the influence of polymer buffering on endosomal escape steps, we performed short exposure (2-hour incubation) transfections in which the extracellular media pH was buffered to specific pH values from 5 to 8. In this manner, the fluid surrounding nanoparticles internalized during endocytosis would be pre-adjusted to a specific pH point. Our rationale was that nanoparticles internalized with the surrounding fluid at a given pH would be in equilibrium with that fluid pH at the time of internalization and only be able to undergo swelling associated with a smaller pH drop. With this assay, we quantified transfection efficacy with the tested nanoparticle formulations at 24 hours post-addition of the nanoparticles (Fig. 5A–B). We also performed nanoparticle characterization of each polymer at each transfection pH. In general, increased pH led to less charged polymers, consequently decreased nanoparticle zeta potential, and for certain more neutrally charged nanoparticle formulations, there was some particle aggregation (Fig. S4).
In both HEK293T and B16-F10 cells, both LPEI and BPEI showed a minor trend of decreased transfection with increased extracellular media pH. In all tested PBAEs, however, the opposite trend was observed where transfection efficacy increased significantly with an increased extracellular media pH of 7 or 8 and transfection was inhibited when extracellular pH was acidic. These trends were confirmed via one-way ANOVA testing of a linear trend directly observable in the number of cells expressing the reporter gene GFP at 24 hours post-transfection (Fig. 5C). Differences between PEI and PBAEs were also apparent when viewing one-dimensional flow-plots of eGFP expression (Fig. S5). One potentially contributing aspect could be an increase in hydrodynamic diameter (Fig. S4), as some previous work has indicated that larger PEI nanoparticles contribute significantly to transfection efficacy,37 although large particles can have difficulty being internalized through endocytosis. In the current studies, there does not seem to be a correlation between diameter and efficacy for PEI as pH increased. For PBAEs, there was an increase in diameter with pH, but the increase was relatively modest from pH 5–7, with particles forming only substantially bigger aggregates at pH=8. Particle size alone does not explain the dramatic increase in transfection efficacy observed by all PBAEs from pH=6 to pH=7 (Fig. 5). Nonetheless, as increasing pH was correlated with both increasing size and increasing transfection efficacy, future investigation to fully uncouple these parameters would lead to greater understanding.
To assess if the differences observed in transfection efficacy when extracellular fluid pH was modulated were attributable to DNA uptake efficacy, we co-delivered 20% Cy5-labeled plasmid DNA with the GFP coding plasmid to assess plasmid uptake and transfection efficacy in the same cells at 24 hours post-transfection. Degree of cellular uptake was minimally affected by extracellular media pH for both PEI variants and all PBAEs tested in both cell types (Fig. 6A–B). Only minor differences between nanoparticle uptake at different extracellular pH for all materials were also apparent when viewing one-dimensional flow-plots of Cy5-DNA uptake (Fig. S6). PEI nanoparticle mediated uptake of plasmid DNA decreased slightly with increasing extracellular media pH (negative slope), but by comparing two-dimensional flow-plots (Fig. 6C), it was apparent that PEI nanoparticles mediated effective uptake and transfection regardless of extracellular fluid pH.
Figure 6.
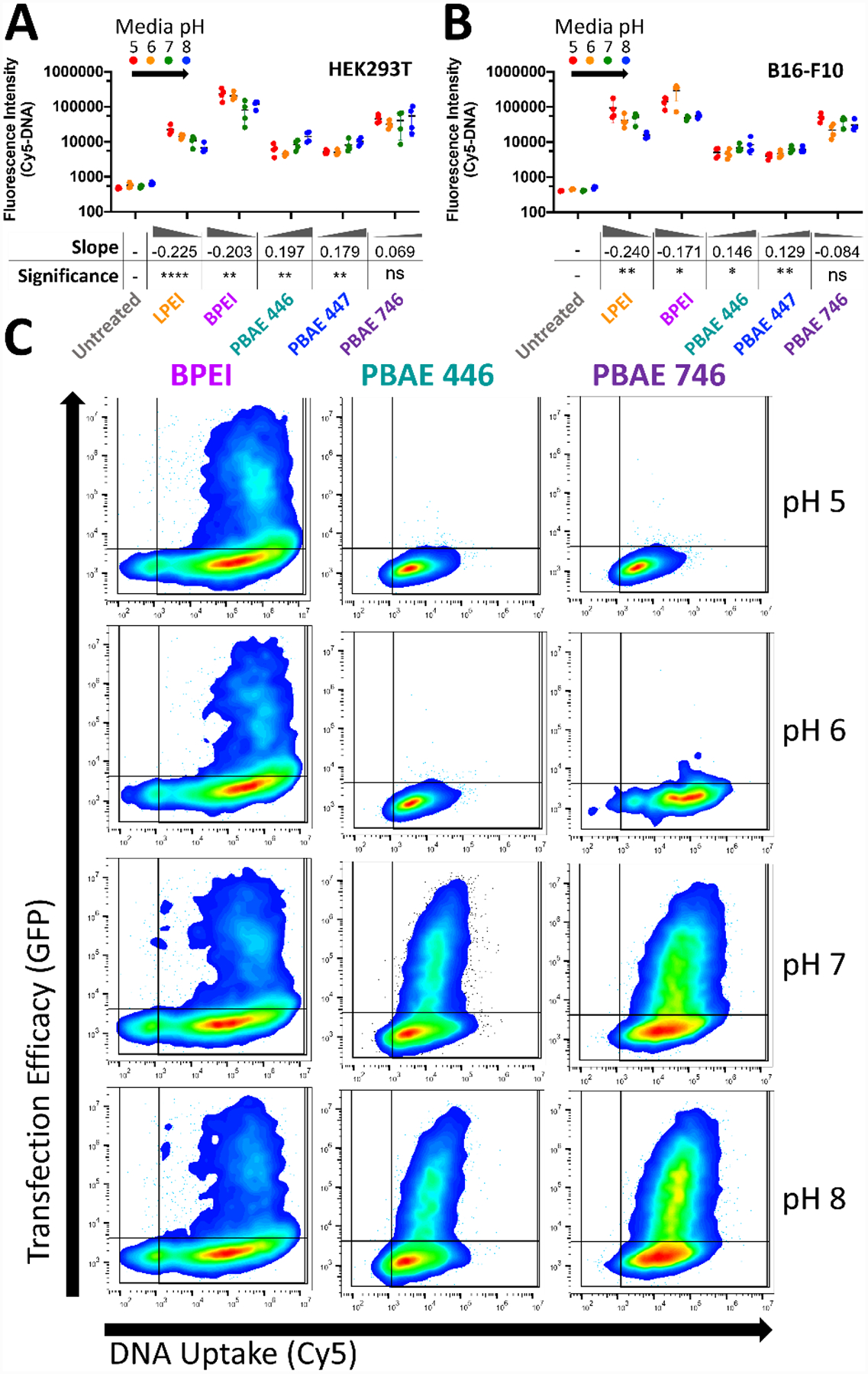
Extracellular media pH only marginally affects nanoparticle uptake and can dramatically affect transfection. Modulation of extracellular media pH in A) HEK293T cells and B) B16-F10 cells only moderately affected the degree of nanoparticle uptake. Uptake was negatively correlated with pH for PEI and positively correlated with pH for PBAEs. Points show individual well mean transfection efficacy. Error bars show mean ± SD of four wells. Slope and statistical significance were assessed by a one-way ANOVA test of linear trend across pH modulation with Dunn’s corrected multiple comparisons. C) Representative two-dimensional flow-plots of plasmid transgene expression (vertical) and plasmid DNA uptake (horizontal) with pH modulation demonstrates the lack of transfection despite effective DNA uptake for PBAEs with extracellular media of pH 5 or 6. Lines on 2D flow-plots denote gating boundary against untreated for uptake and transfection efficacy.
In contrast, PBAE nanoparticles were internalized at similar levels regardless of extracellular fluid pH (slope close to zero). These results indicate that despite similar levels of nanoparticle uptake, PBAE nanoparticles failed to enable transfection when internalized with acidic pH extracellular fluid as seen by the differences in slope of pH dependence of transfection and uptake in PBAEs. These results indicate that in acidic media (pH 5 or 6), PBAE transfection downstream of uptake is more sensitive to acidity than PEI. This effect is strongly pronounced in viewing two-dimensional flow-plots of transgene expression and plasmid DNA uptake at the level of individual cells (Fig. 6C). Specifically comparing the flow-plots of PBAE 746 at pH 6 and pH 7 which cleanly straddles the effective pKa of PBAE 746, it was highly apparent that the polymer becomes ineffective at enabling transfection below its effective pKa.
As uptake remained relatively unchanged for PBAEs at the different media acidities, the effect of pH on transfection occurred after uptake. As the volume of endosomal fluid is negligible compared to the volume of the cytosol, it is likely that the pH response is occurring within the endosome. At lower pH, a greater fraction of the protonatable amines in the PBAEs are protonated than in PEI, as shown by the differences in pKa peaks in Fig. 2C. This means that there is less buffering capability in the polymer, and thus, if PBAEs follow a proton-sponge type mechanism, there is less of a driving force to lead to endosomal escape. Alternatively, it could also indicate that PBAEs exhibit less ability to induce vesicle leakage at low pH (Fig. 4) as well as stronger DNA binding affinity at lower pH (Fig. 3). Regardless of the mechanism, these results emphasize the necessity of a more neutral environment for PBAEs at some point in the transfection timeline in order to successfully transfect cells.
Polymer degradation is another pH-dependent phenomenon that could be contributing to these results. Previous work has highlighted that gene delivery polymers should not only be stable enough extracellularly to reach the endosome, but also that they must degrade rapidly enough to release their DNA cargo.21, 38, 39 Notably the half-life of free PBAEs 446 and 447 (not in nanoparticles) were previously determined to be approximately 4–6 hrs.21 Soluble PBAEs have been shown to degrade more slowly at pH 5.1 than pH 7.4,33,38 which has been attributed to increased nucleophilic activity of unprotonated amines.21 As a result, if there is a degradation effect, PBAE nanoparticles in acidic extracellular pH may be expected to be better protected from degradation in the extracellular space and have perhaps improved transfection efficacy compared to PBAE nanoparticles in neutral extracellular pH. Ultimately, post-endosomal escape, all of the PBAE nanoparticles should degrade at the same quick rate in the neutral pH environment of the cytosol to release their DNA cargo. Further, previous work has shown that once PBAE polymer self-assembles with DNA to form a nanoparticle, the influence of degradation on transfection efficacy is lessened compared to degradation of free polymer on its own.38,39 This could be due to more effective exclusion of water by the nanoparticle, reducing ester hydrolysis, and as a result, potential differential degradation across pH likely plays an insignificant factor in the results seen here.”
To further explore this mechanism, it is important to consider the behavior of polycations when endosomal acidification is blocked. Past work has definitively shown that the transfection efficacy of PEI and PEI-containing polymers is greatly reduced under the influence of bafilomycin A1, an H+-ATPase inhibitor that blocks the endo- and lysosomal proton pump35, 40–42 and has been confirmed to prevent acidification of PEI-containing endosomes.12 If PBAEs have proton sponge-like activity, as suggested by reports in the literature18,21, it would be expected that their transfection efficacy would also decrease upon addition of a H+-ATPase inhibitor, although a transfection decrease could alternatively be explained through other charge-related mechanisms leading to endosome destabilization. Based on the overall data collected from the current studies and the literature, it is hypothesized that the buffering ability of PBAEs is an important element for their endosomal escape and transfection properties.
There are additional barriers to effective delivery of plasmid DNA between endosomal escape and reporter gene expression that may in part explain why correlation between cellular uptake and transfection remains poorly overall. First, diffusion of large plasmids or unpacked polyplexes through the cytosol likely hinders trafficking of the nucleic acid to the nucleus.44 Here we used rapidly dividing cell lines that would be expected to yield relatively efficient internalization of plasmids to the nucleus during cell division in a manner that may also be further assisted by the SV40 promoter sequence for transcription factor mediated nuclear internalization.45, 46 For PBAE NPs, nucleic acid unpacking was likewise not expected to be a large barrier given the rapid degradation of the PBAEs utilized here with ester bond half-life less than 8 hours.21, 39 As a result, it is unlikely that either of these phenomena played a part in this behavior.
Although it is difficult to quantify the relative contributions of these effects, these studies highlight the importance of considering the acidity of the extracellular medium or microenvironment of a targeted cell when transfecting with polymeric vectors, especially PBAEs. Additionally, these results could begin to hint at a crucial detail of endosomal escape, where an initially unprotonated conformation of the polymer is a necessary component for successful entry to the cytosol. The polymers without a well-defined pKa, such as BPEI, which had a significant amount of amines unprotonated even at low pH, showed fairly level transfection across pH. However, in the polymers with a clearer effective pKa, such as PBAEs, there was a much lower proportion of unprotonated amines at low pH, and subsequently a drastic shift in transfection efficacy was observed.
Even with all the studies over the past 25 years that have slowly chipped away at the mysteries of cytosolic delivery, further investigation of the precise mechanism of endosomal escape is warranted. The advent of assays that directly probe endosomal disruption in living cells using galectin-8 fluorescent protein fusions47–49 may more directly answer whether endosomal disruption occurs, but even then do not directly address cytosolic availability of the cargo molecules being delivered.
Conclusions:
In this work, multiple pH-dependent behaviors of PBAEs used for intracellular delivery have been investigated. PBAEs evaluated showed dramatically decreased ability for cell transfection when extracellular pH was made more acidic, while a small trend towards the opposite effect was shown in LPEI and BPEI. These studies highlighted that endosomal proton buffering capacity is key for PBAE-based transfection and that this is a function of both polymer structure and extracellular microenvironment. A greater sensitivity to pH was noted with PBAEs 446 and 447, which also had a narrower buffering range, when compared to PBAE 746 and BPEI, both of which showed a broader buffering peak despite drastically different buffering capacities. Other polymer characteristics, such as membrane interactions and DNA binding affinity were also affected by extracellular pH, both likely due to the protonation state of the polymer. Thus, the acidity of the extracellular medium is critically important to transfection post-cellular uptake. These findings emphasize that tuning of polymer pKa is key to ensure effective transfection in a desired pH microenvironment and has critical implications in furthering understanding of polymeric nanoparticle endosomal escape.
Supplementary Material
Acknowledgements:
The authors would like to thank Kalina Hristova and Sarah Kim of the Hristova Lab at Johns Hopkins for demonstrating the vesicle leakage assay.
Funding Sources
The authors would like to thank the following organizations for financial support: NSF Graduate Research Fellowship DGE-0707427 (DRW), Microscopy Core Grant (S10 OD016374) and the NIH (R01CA228133, R01EB022148, P41EB028239, and Wilmer Core Grant P30 EY001765). DR thanks the Hopkins Office of Undergraduate Research for the Provost’s Undergraduate Research Award (2016). JJG thanks the Bloomberg~Kimmel Institute for Cancer Immunotherapy for support.
Footnotes
Supporting information:
Figure S1. Schematic demonstrating PBAE synthesis
Figure S2. GPC characterization of PBAEs
Figure S3. 1H NMR spectra of PBAEs
Figure S4. Hydrodynamic diameter and zeta potential in varying pH
Figure S5. One-dimensional flow cytometry plots of eGFP expression
Figure S6. One-dimensional flow cytometry plots of Cy5-DNA nanoparticle uptake
References:
- 1.Duncan R; Richardson SCW, Endocytosis and intracellular trafficking as gateways for nanomedicine delivery: opportunities and challenges. Molecular pharmaceutics 2012, 9 (9), 2380–2402. [DOI] [PubMed] [Google Scholar]
- 2.Grant BD; Donaldson JG, Pathways and mechanisms of endocytic recycling. Nature reviews. Molecular cell biology 2009, 10 (9), 597–608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bus T; Traeger A; Schubert US, The great escape: how cationic polyplexes overcome the endosomal barrier. Journal of Materials Chemistry B 2018. [DOI] [PubMed] [Google Scholar]
- 4.Hong S; Leroueil PR; Janus EK; Peters JL; Kober M-M; Islam MT; Orr BG; Baker JR; Banaszak Holl MM, Interaction of polycationic polymers with supported lipid bilayers and cells: nanoscale hole formation and enhanced membrane permeability. Bioconjugate chemistry 2006, 17 (3), 728–734. [DOI] [PubMed] [Google Scholar]
- 5.Reilly MJ; Larsen JD; Sullivan MO, Polyplexes Traffic through Caveolae to the Golgi and Endoplasmic Reticulum en Route to the Nucleus. Molecular Pharmaceutics 2012, 9 (5), 1280–1290. [DOI] [PubMed] [Google Scholar]
- 6.Ross NL; Munsell EV; Sabanayagam C; Sullivan MO, Histone-targeted polyplexes avoid endosomal escape and enter the nucleus during postmitotic redistribution of ER membranes. Molecular Therapy—Nucleic Acids 2015, 4 (2), e226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tang R; Kim CS; Solfiell DJ; Rana S; Mout R; Velázquez-Delgado EM; Chompoosor A; Jeong Y; Yan B; Zhu Z-J, Direct delivery of functional proteins and enzymes to the cytosol using nanoparticle-stabilized nanocapsules. ACS nano 2013, 7 (8), 6667–6673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wittrup A; Ai A; Liu X; Hamar P; Trifonova R; Charisse K; Manoharan M; Kirchhausen T; Lieberman J, Visualizing lipid-formulated siRNA release from endosomes and target gene knockdown. Nature biotechnology 2015, 33 (8), 870–876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sunshine JC; Bishop CJ; Green JJ, Advances in polymeric and inorganic vectors for nonviral nucleic acid delivery. 2011; Vol. 2, pp 493–521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Boussif O; Lezoualc’h F; Zanta M a.; Mergny, M. D.; Scherman, D.; Demeneix, B.; Behr, J. P., A versatile vector for gene and oligonucleotide transfer into cells in culture and in vivo: polyethylenimine. Proceedings of the National Academy of Sciences of the United States of America 1995, 92 (16), 7297–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sonawane ND; Szoka FC; Verkman AS, Chloride accumulation and swelling in endosomes enhances DNA transfer by polyamine-DNA polyplexes. Journal of Biological Chemistry 2003, 278 (45), 44826–44831. [DOI] [PubMed] [Google Scholar]
- 12.Benjaminsen RV; Mattebjerg MA; Henriksen JR; Moghimi SM; Andresen TL, The possible “proton sponge” effect of polyethylenimine (PEI) does not include change in lysosomal pH. Molecular Therapy 2013, 21 (1), 149–157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tzeng SY; Guerrero-Cázares H; Martinez EE; Sunshine JC; Quiñones-Hinojosa A; Green JJ, Non-viral gene delivery nanoparticles based on poly(β-amino esters) for treatment of glioblastoma. Biomaterials 2011, 32 (23), 5402–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wilson DR; Rui Y; Siddiq K; Routkevitch D; Green JJ, Differentially branched ester amine quadpolymers with amphiphilic and pH-sensitive properties for efficient plasmid DNA delivery. Molecular pharmaceutics 2019, 16 (2), 655–668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bishop CJ; Majewski RL; Guiriba TR; Wilson DR; Bhise NS; Quiñones-Hinojosa A; Green JJ, Quantification of cellular and nuclear uptake rates of polymeric gene delivery nanoparticles and DNA plasmids via flow cytometry. Acta Biomater 2016, 37, 120–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lynn DM; Anderson DG; Putnam D; Langer R, Accelerated discovery of synthetic transfection vectors: parallel synthesis and screening of a degradable polymer library. J Am Chem Soc 2001, 123 (33), 8155–6. [DOI] [PubMed] [Google Scholar]
- 17.Anderson DG; Lynn DM; Langer R, Semi-automated synthesis and screening of a large library of degradable cationic polymers for gene delivery. Angewandte Chemie (International ed. in English) 2003, 42 (27), 3153–8. [DOI] [PubMed] [Google Scholar]
- 18.Akinc A; Lynn DM; Anderson DG; Langer R, Parallel synthesis and biophysical characterization of a degradable polymer library for gene delivery. J Am Chem Soc 2003, 125 (18), 5316–23. [DOI] [PubMed] [Google Scholar]
- 19.Wilson DR; Mosenia A; Suprenant MP; Upadhya R; Routkevitch D; Meyer RA; Quinones-Hinojosa A; Green JJ, Continuous microfluidic assembly of biodegradable poly(beta-amino ester)/DNA nanoparticles for enhanced gene delivery. J Biomed Mater Res A 2017, 105 (6), 1813–1825. [DOI] [PubMed] [Google Scholar]
- 20.Wilson DR; Routkevitch D; Rui Y; Mosenia A; Wahlin KJ; Quinones-Hinojosa A; Zack DJ; Green JJ, A Triple-Fluorophore-Labeled Nucleic Acid pH Nanosensor to Investigate Non-viral Gene Delivery. Molecular Therapy 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sunshine JC; Peng DY; Green JJ, Uptake and transfection with polymeric nanoparticles are dependent on polymer end-group structure, but largely independent of nanoparticle physical and chemical properties. Molecular pharmaceutics 2012, 9 (11), 3375–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rui Y; Wilson DR; Sanders K; Green JJ, Reducible Branched Ester-Amine Quadpolymers (rBEAQs) Codelivering Plasmid DNA and RNA Oligonucleotides Enable CRISPR/Cas9 Genome Editing. ACS Applied Materials and Interfaces 2019, 11 (11), 10472–10480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hope MJ; Bally MB; Mayer LD; Janoff AS; Cullis PR, Generation of multilamellar and unilamellar phospholipid vesicles. Chemistry and physics of lipids 1986, 40 (2–4), 89–107. [Google Scholar]
- 24.Mayer LD; Hope MJ; Cullis PR, Vesicles of variable sizes produced by a rapid extrusion procedure. Biochimica et Biophysica Acta (BBA)-Biomembranes 1986, 858 (1), 161–168. [DOI] [PubMed] [Google Scholar]
- 25.Wiedman G; Herman K; Searson P; Wimley WC; Hristova K, The electrical response of bilayers to the bee venom toxin melittin: evidence for transient bilayer permeabilization. Biochimica et Biophysica Acta (BBA)-Biomembranes 2013, 1828 (5), 1357–1364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang F; Parayath NN; Ene CI; Stephan SB; Koehne AL; Coon ME; Holland EC; Stephan MT, Genetic programming of macrophages to perform anti-tumor functions using targeted mRNA nanocarriers. Nature communications 2019, 10 (1), 1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mangraviti A; Tzeng SY; Kozielski KL; Wang Y; Jin Y; Gullotti D; Pedone M; Buaron N; Liu A; Wilson DR; Hansen SK; Rodriguez FJ; Gao G-D; DiMeco F; Brem H; Olivi A; Tyler B; Green JJ, Polymeric Nanoparticles for Nonviral Gene Therapy Extend Brain Tumor Survival in Vivo. ACS Nano 2015, 9 (2), 1236–1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Smith TT; Stephan SB; Moffett HF; McKnight LE; Ji W; Reiman D; Bonagofski E; Wohlfahrt ME; Pillai SPS; Stephan MT, In situ programming of leukaemia-specific T cells using synthetic DNA nanocarriers. Nature Nanotechnology 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kaczmarek JC; Kauffman KJ; Fenton OS; Sadtler K; Patel AK; Heartlein MW; DeRosa F; Anderson DG, Optimization of a Degradable Polymer-Lipid Nanoparticle for Potent Systemic Delivery of mRNA to the Lung Endothelium and Immune Cells. Nano Lett 2018, 18 (10), 6449–6454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Funhoff AM; van Nostrum CF; Koning GA; Schuurmans-Nieuwenbroek NME; Crommelin DJA; Hennink WE, Endosomal Escape of Polymeric Gene Delivery Complexes Is Not Always Enhanced by Polymers Buffering at Low pH. Biomacromolecules 2004, 5 (1), 32–39. [DOI] [PubMed] [Google Scholar]
- 31.Jayaraman M; Ansell SM; Mui BL; Tam YK; Chen J; Du X; Butler D; Eltepu L; Matsuda S; Narayanannair JK, Maximizing the potency of siRNA lipid nanoparticles for hepatic gene silencing in vivo. Angewandte Chemie International Edition 2012, 51 (34), 8529–8533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Choosakoonkriang S; Lobo BA; Koe GS; Koe JG; Middaugh CR, Biophysical characterization of PEI/DNA complexes. J Pharm Sci 2003, 92 (8), 1710–22. [DOI] [PubMed] [Google Scholar]
- 33.Lynn DM; Langer R, Degradable Poly(β-amino esters): Synthesis, Characterization, and Self-Assembly with Plasmid DNA. Journal of the American Chemical Society 2000, 122 (44), 10761–10768. [Google Scholar]
- 34.Mangraviti A; Tzeng SY; Gullotti D; Kozielski KL; Kim JE; Seng M; Abbadi S; Schiapparelli P; Sarabia-Estrada R; Vescovi A, Non-virally engineered human adipose mesenchymal stem cells produce BMP4, target brain tumors, and extend survival. Biomaterials 2016, 100, 53–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Akinc A; Thomas M; Klibanov AM; Langer R, Exploring polyethylenimine-mediated DNA transfection and the proton sponge hypothesis. J Gene Med 2005, 7 (5), 657–63. [DOI] [PubMed] [Google Scholar]
- 36.Wilson DR; Routkevitch D; Wahlin KJ; Zack DJ; Quinones-Hinojosa A; Green JJ In Development of a pH Sensor to Probe Endosomal Buffering of Polymeric Nanoparticles Effective for Gene Delivery, 2016; Molecular Therapy: 2016; pp S196–S196. [Google Scholar]
- 37.González-Domínguez I; Grimaldi N; Cervera L; Ventosa N; Gòdia F, Impact of physicochemical properties of DNA/PEI complexes on transient transfection of mammalian cells. N Biotechnol 2019, 49, 88–97. [DOI] [PubMed] [Google Scholar]
- 38.Green JJ; Langer R; Anderson DG, A combinatorial polymer library approach yields insight into nonviral gene delivery. Acc Chem Res 2008, 41 (6), 749–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wilson DR; Suprenant MP; Michel JH; Wang EB; Tzeng SY; Green JJ, The Role of Assembly Parameters on Polyplex Poly (Beta-Amino Ester) Nanoparticle Transfections. Biotechnology and bioengineering 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Patnaik S; Aggarwal A; Nimesh S; Goel A; Ganguli M; Saini N; Singh Y; Gupta KC, PEI-alginate nanocomposites as efficient in vitro gene transfection agents. J Control Release 2006, 114 (3), 398–409. [DOI] [PubMed] [Google Scholar]
- 41.Kim K; Ryu K; Kim TI, Cationic methylcellulose derivative with serum-compatibility and endosome buffering ability for gene delivery systems. Carbohydr Polym 2014, 110, 268–77. [DOI] [PubMed] [Google Scholar]
- 42.Kichler A; Leborgne C; Coeytaux E; Danos O, Polyethylenimine-mediated gene delivery: a mechanistic study. J Gene Med 2001, 3 (2), 135–44. [DOI] [PubMed] [Google Scholar]
- 43.Huber V; Camisaschi C; Berzi A; Ferro S; Lugini L; Triulzi T; Tuccitto A; Tagliabue E; Castelli C; Rivoltini L In Cancer acidity: An ultimate frontier of tumor immune escape and a novel target of immunomodulation, 2017; Elsevier: pp 74–89. [DOI] [PubMed] [Google Scholar]
- 44.Forrest ML; Pack DW, On the kinetics of polyplex endocytic trafficking: implications for gene delivery vector design. Molecular therapy 2002, 6 (1), 57–66. [DOI] [PubMed] [Google Scholar]
- 45.Dean DA; Strong DD; Zimmer WE, Nuclear entry of nonviral vectors. Gene therapy 2005, 12 (11), 881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Vaughan EE; Dean DA, Intracellular trafficking of plasmids during transfection is mediated by microtubules. Molecular Therapy 2006, 13 (2), 422–428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kilchrist KV; Dimobi SC; Jackson MA; Evans BC; Werfel TA; Dailing EA; Bedingfield SK; Kelly IB; Duvall CL, Gal8 Visualization of Endosome Disruption Predicts Carrier-Mediated Biologic Drug Intracellular Bioavailability. ACS nano 2019, 13 (2), 1136–1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kilchrist KV; Tierney JW; Duvall CL, Genetically encoded split luciferase biosensors to measure endosome disruption in real time in live cells. bioRxiv 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rui Y; Wilson DR; Choi J; Varanasi M; Sanders K; Karlsson J; Lim M; Green JJ, Carboxylated branched poly (β-amino ester) nanoparticles enable robust cytosolic protein delivery and CRISPR-Cas9 gene editing. Science Advances 2019, 5 (12), eaay3255. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


