Abstract
Human primary monocytes are composed of a minor, more mature CD16+(CD14low/neg) population and a major CD16neg(CD14+) subset. The specific functions of CD16+ vs. CD16neg monocytes in steady state or inflammation remain poorly understood. In previous work we found that IL-12 is selectively produced by the CD16+ subset in response to the protozoan pathogen, Toxoplasma gondii. Here we demonstrated that this differential responsiveness correlates with the presence of an IFN-induced transcriptional signature in CD16+ monocytes already at baseline. Consistent with this observation, we found that in vitro IFN-γ-priming overcomes the defect in IL-12 response of the CD16neg subset. In contrast, pretreatment with IFN-γ had only a minor effect on IL-12p40 secretion by the CD16+ population. Moreover, inhibition of the mTOR pathway also selectively increased the IL-12 response in CD16neg but not in CD16+ monocytes. We further demonstrate that in contrast to IFN-γ, IFN-α fails to promote IL-12 production by the CD16neg subset and blocks the effect of IFN-γ-priming. Based on these observations, we propose that the acquisition of IL-12 responsiveness by peripheral blood monocyte subsets depends on extrinsic signals experienced during their developmental progression in vivo. This process can be overridden during inflammation by the opposing regulatory effects of type I and II IFN as well as the mTOR inhibition.
Introduction
The two major monocyte subsets are distinguished by distinct surface markers. Murine “classical” monocytes express high levels of Ly6C while “non-classical” monocytes are Ly6Clow (1). In humans the same populations are distinguished on the basis of their expression of the endotoxin co-receptor CD14 and the type III Fcγ receptor CD16 with CD14+CD16neg and CD14lowCD16+ phenotypically marking the classical and non-classical subsets, respectively (2). Blood monocytes constitute a dynamic population with classical monocytes continuously converting into nonclassical. Nevertheless, the peripheral blood composition is heavily skewed towards classical monocytes in humans (~85-95%) which after egress from bone marrow spend approximately 1 day in circulation (3). Most of these cells are either drawn into tissues or die, while the remaining small fraction (~1%) converts into “intermediate” (CD14+CD16low) monocytes which display a longer lifespan (4.3±0.36 days) and ultimately all differentiate into non-classical monocytes (3). Interestingly, this terminally differentiated population displays the longest lifespan (7.4±0.53 days) of all three of the monocyte subsets in blood. The factors that control the transition of classical to non-classical monocytes are still poorly understood (4-5).
Functionally, human monocytes subsets are known to display distinct cytokine responses to microbial stimulation. Although such differences can be explained in part by the preferential expression of distinct pattern recognition receptors by different subsets, other cell-intrinsic factors may play an even more important role. For example, when stimulated with LPS CD16neg monocytes secrete IL-1β but not TNF, while CD16+ monocytes produce TNF and not IL-1β (6, 7).
In a previous study, we observed a similar dichotomy in the response of human monocyte subsets to tachyzoites of the intracellular protozoan parasite Toxoplasma gondii (8). In the in vitro system studied, T. gondii-stimulated IL-12 and TNF production is triggered by live tachyzoites and not soluble parasite extracts as also observed for the CCL2 (9). However, while all monocyte subsets display a comparable rate of parasite invasion and/or uptake, only non-classical and intermediate but not classical monocytes were found to secrete significant amounts of IL-12 and TNF in response to T. gondii exposure (8). Interestingly, when we performed a preliminary genome wide microarray analysis of FACSort purified classical, intermediate and non-classical monocytes before and after stimulation with T. gondii tachyzoites, significant changes in gene expression (including several chemokines) were observed in all three subsets (K.W.T. and D.J. unpublished data). This observation argued that the failure of classical monocytes to produce IL-12 and TNF in response to T. gondii is not due to their inability to sense the parasite stimulus.
In the present study, we have further investigated the basis for differential IL-12 responsiveness of human monocytes subsets to T. gondii comparing the responses observed to that of a defined TLR agonist (R848). Our findings show that in contrast to CD16+ non-classical and intermediate monocytes, CD16neg classical monocytes require a second signal for IL-12 and TNF production in response to toxoplasma or TLR ligand exposure. This signal can be provided by IFN-γ-priming or rapamycin treatment, neither of which enhances the responsiveness of CD16+ monocytes that already exhibit an IFN-induced gene expression signature at baseline. We further observed that in contrast to IFN-γ, type I IFN does not promote IL-12 and TNF production by classical monocytes while inhibiting the stimulatory effects of IFN-γ. We propose based on these observations that the acquisition of IL-12 and TNF responsiveness by peripheral blood monocyte subsets depends on extrinsic signals experienced during their developmental progression in vivo. This process can be overridden during inflammation by the opposing regulatory effects of type I and II IFN.
Material and Methods
Healthy donors
Peripheral blood monocytes were obtained from healthy volunteers by counterflow centrifugal elutriation at the NIH Blood Bank under Institutional Review Board-approved protocols of both the National Institute of Allergy and Infectious Diseases and the Department of Transfusion Medicine. Of the 44 donors, 84% were males and 16% females; 60% were Caucasian, 34% African American and 5% Hispanic. Donor age ranged from 21-68 with a median value of 39.5.
Isolation of monocytes
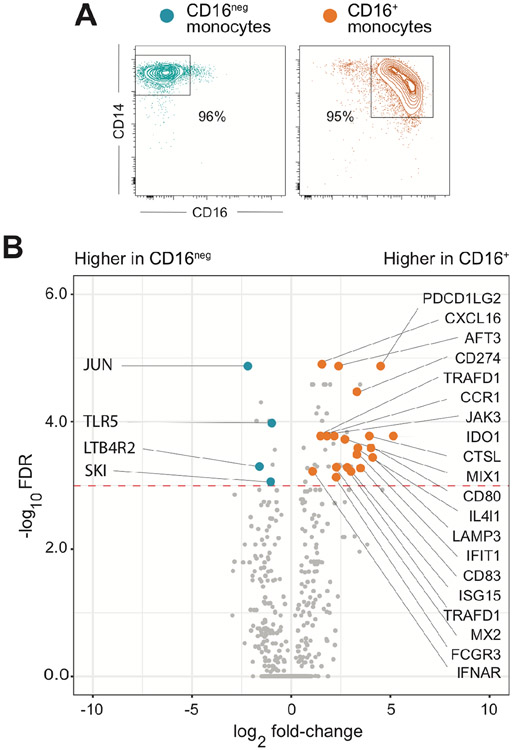
Monocytes were purified from elutriated preparations either by fluorescence-activated cell sorting (FACSorting) as previously described (8) or with MACS MicroBeads (Miltenyi Biotec, Bergisch Gladbach, Germany). To avoid variation in elutriated samples, total monocytes were isolated using CD14 MicroBeads. For monocyte subset isolation, elutriated preparations were first depleted of granulocytes and NK cells with a mixture of anti-CD15 and CD56 MicroBeads, followed by CD16+ monocytes purification with a CD16pos Monocyte Isolation Kit, and CD16neg monocytes isolation from flow throw using CD14 MicroBeads. Purity of all populations was >95% (Figure 1A).
Figure 1: CD16+ in contrast to CD16neg monocytes display an IFN-inducible gene expression signature at baseline.
(A) Representative FACS contour plots of CD16 vs. CD14 staining of MACS-purified CD16neg and CD16+ monocytes from one donor. (B) Volcano plot of mRNAs differentially expressed in purified CD16neg and CD16+ monocytes at baseline. 770 genes were analyzed using the NanoString nCounter Myeloid Innate Immunity Panel and results shown are compiled from three different donors. The red dotted line indicates the cutoff for statistical significance (p< 0.05) calculated using the Benjamini-Yekutieli FDR multiple test. The genes with increased expression in CD16+ monocytes associated with IFN-signature are indicated as orange symbols, while blue symbols depict genes that were expressed higher in the CD16neg subset.
Parasites
Toxoplasma gondii tachyzoytes of the type I strain RH88, as well as a GFP+ expressing recombinant (8), were propagated in the human foreskin fibroblast cell line Hs27 (ATCC CRL-1634). Harvested parasites (6 x 106/ml) were incubated at room temperature in medium with or without 3 μM mycalolide B (MycB; Enzo Life Sciences, Farmingdale, NY), an irreversible actin-polymerization inhibitor that blocks their ability to invade host cells.
Cell culture
Cultures were performed in RPMI 1640 medium (Life Technologies, Carlsbad, CA) supplemented with 10 % heat-inactivated FCS (HyClone, South Logan, UT), 4 mM L-glutamine, 10 mM HEPES, 1 mM sodium pyruvate, 0.1 mM MEM nonessential amino acids, 100 U/ml penicillin, 100 ug/ml streptomycin (Life Technologies), 100 μg/ml streptomycin, and 50 μM 2-ME (MilliporeSigma, Rockville, MD). Monocytes were plated at 2 x 105/0.1ml in 96-well plates (cytokine measurements) or 0.5 ml in 12-well plates (RNA analysis) and stimulated with an equal volume of untreated or MyB-treated tachyzoites (2x106/ml), or with the TLR7/8 agonist R848 (300 ng/ml; InvivoGen, San Diego, CA). Priming with IFN-γ (10ng/ml) or IFN-α2a (10ng/ml) (PeproTech, Cranbury, NJ) was performed overnight, unless otherwise specified. To inhibit the mTORC1 pathway, Rapamycin (5nM) (10) or REDD-1 inducer (10μM) (11) (Millipore Sigma, Rockville, MD) were added to monocyte cultures 1 hour before stimulation with parasites or R848.
Cell surface staining
Elutriated monocytes or MACS-purified CD16+ and CD16neg monocytes were stained with the following antibodies: anti-CD14 (clone M0Pg, BD Biosciences, San Jose, CA), anti-CD16 (clone 3G8, Biolegend, San Diego, CA), anti-HLA-DR (clone L243, Biolegend), anti-CD119 (clone GIR-208, Thermo Fisher Scientific, , Gaithersburg, MD) or anti-IFNγR2-APC (clone C38, Creative Diagnostic, Shirley, NY) and sorted or analyzed using a FACS Aria or FACS Symphony (BD Biosciences), respectively.
Intracellular staining for pSTAT1
FACSort purified monocyte populations (1 x 106) incubated in 0.1 ml in 96-well plates in serum-free medium in the presence or absence of either IFN-γ or IFN-α (10 ng/ml) at 37°C for the indicated time were fixed with BD Cytofix for 10 min at 37°C and permeabilized with BD Phosflow Perm (BD Biosciences) for 30 minutes on ice. After staining with anti-pSTAT1 Ab (clone pY701, BD Biosciences) for 1 hour at 4°C, cells were analyzed.
Cytokine measurements
Cell culture supernatants were collected after 18-24 hours of incubation and stored at −80°C. IL-12p40, IL-12p70, TNF and CCL2 were measured using Human IL-12p40 Direct ELISA kit (Life Technologies), Human TNF-α DuoSet ELISA, Human IL-12p70 Quantikine ELISA, Human CCL2/MCP-1 Quantikine ELISA (R&D Systems, Minneapolis, MN), respectively.
Western blot analysis of mTOR pathway proteins
Monocytes were lysed with Cell Lysis Buffer (Cell Signaling Technologies) as previously described (12). Protein extracts (25μg) were separated on SDS-PAGE, transferred to a PVDF membrane (both from BioRAD, Philadelphia, PA) and probed overnight at 4°C with rabbit Ab specific for mTOR (clone 7C10), phospho-mTOR Ser2448 (clone D9C2), 4E-BP1 (clone 53H11) or phospho-4E-BP1 Thr37/47 (clone 236B4) all from Cell Signaling Technology (Boston, MA). Anti-β-actin Ab (clone D6A8, Cell Signaling Technology) was added during the last hour of incubation at room temperature. After washing, the membrane was incubated with HCL-Digital Secondary Antibody for 3 hours at room temperature and chemiluminescence was measured (SuperSignal West Pico Chemiluminescent Substrate, Thermo Fisher Scientific). Band density was quantified using Image J software.
Gene expression analysis by Nanostring
Total RNA was extracted from monocytes cultured for 8 hours with or without IFN-γ by adding Trizol reagent (Life Technologies) and employing the Direct-zol RNA Miniprep kit (Zymo Research, Irvine, CA) according to the manufacturer’s instructions. For broad assessment of gene expression, we used the nCounter® Myeloid Innate Immunity Panel (NanoString Technologies, Seatttle, WA). Total RNA (170 ng) from each sample was hybridized with the probes for 770 genes. Data were processed with nSolver Analysis software (NanoString Technologies) which included assessment of quality of the runs and differential gene expression assessed. The false discovery rate (FDR) was calculated using Benjamini-Yekutieli procedure.
Quantitative RT-PCR
After 8 hours of stimulation, monocytes were collected in Trizol (Life Technologies) and RNA was isolated with a Direct-zol RNA Miniprep kit (Zymo Research). qRT-PCR assays were performed as previously described (13) with 18S rRNA used for normalization. The relative mRNA expression of genes was calculated by using the cycle threshold (ΔΔCT) method in comparison to unstimulated CD16neg population cultured in medium alone. The sequences of the specific primers are as follows:
Human ribosomal 18S, forward: 5’-GCTTAATTTGACTCAACACGGGA-3;
reverse: 5’- AGCTATCAATCTGTCAATCCTGTC-3’;
IL-12B, forward: 5’- CCCTGACATTCTGCGTTCA-3’;
reverse: 5’- AGGTCTTGTCCGTGAAGACTCTA-3’.
Cell Energy Phenotype Assay
Purified monocyte populations cultured for 3 hours in medium or with untreated or MycB-treated T. gondii tachyzoites were plated in a Seahorse XF96 Cell Culture Microplate (Agilent, Santa Clara, CA) according to the manufacturer’s protocol. Oxygen consumption rate (OCAR) and extracellular acidification rate (ECAR) were then measured in monocytes with or without metabolic stressors Carbonyl cyanide 4-(trifluoromethoxy)phenylhydrazone; (FCCP; 1.5 μM; MilliporeSigma) and Oligomycin (1 μM; MilliporeSigma) on a Seahorse XFe96 Analyzer (Agilent).
Statistical Analysis
Statistical analysis was performed by paired non-parametric Wilcoxon signed-rank test or were indicated by Student’s t tests using GraphPad Prism software version 8.0 (GraphPad).
Results
CD16+ monocytes express an IFN-inducible transcriptional signature at baseline
While primary human CD16neg (CD14+ classical) and CD16+ (both CD14+ intermediate plus CD14low non-classical) monocytes, herein referred to simply as CD16neg and CD16+, display distinct IL-12 responses after stimulation with T. gondii (8), both populations secrete comparable amounts of CCL2 (Supplemental Figure 1). To address whether the difference in IL-12 production is due to intrinsic differences among the two subsets prior to stimulation, we examined their gene expression profiles at baseline. Using a NanoString platform, we analyzed expression of genes specifically associated with the innate immune response. Of the total 770 genes assayed, the large majority (93%) were expressed at similar levels in both subsets with only 51 genes differentially expressed. Of these, 42 were increased in CD16+ and only 9 in CD16neg monocytes (Figure 1B). Importantly, closer examination of the transcripts that were significantly upregulated in CD16+ monocytes revealed that almost half of them (20) were genes previously characterized as IFN inducible (Figure 1B) indicating that the ability of human monocytes to mount an IL-12 response correlates with the presence of an IFN-inducible gene signature at steady-state. This prompted us to investigate the specific effects of both IFN-γ and type I IFN on the ability of CD16+ and CD16neg monocytes to secrete IL-12 following microbial stimulation.
The increased secretion of IL-12 triggered by IFN-γ-priming of human monocytes results largely from an effect on the CD16neg subpopulation
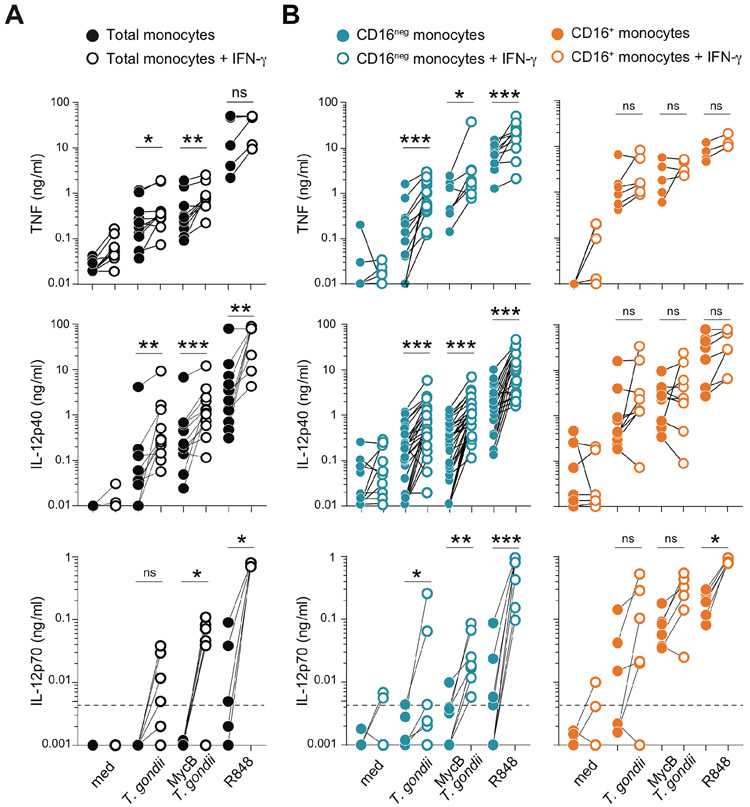
We first analyzed how IFN-γ-priming influences the ability of CD16+ and CD16neg monocytes to secrete TNF and IL-12 in response to stimulation with T. gondii or the TLR7/8 agonist R848, a known potent inducer of proinflammatory responses in monocytes. In these as well as subsequent experiments, the toxoplasma tachyzoites were either treated or not with mycalolide B (MycB), an irreversible inhibitor of actin polymerization, to assay the effect of IFN-γ-priming on the response triggered by phagocytosis vs. active invasion of the parasite. As expected, IFN-γ pretreatment of total unfractionated monocytes significantly augmented cytokine secretion in response to either toxoplasma tachyzoites or R848 (Figure 2A). Interestingly, this increase was found to be primarily due to an effect on the normally poorly responsive CD16neg subset (Figure 2B, left column). Following IFN-γ-priming, regardless of the stimulus, secretion of TNF and IL-12p40 by the normally highly responsive CD16+ monocyte population was not enhanced except for a modest increase in IL-12p70 following R848 stimulation (Figure 2B, right column). Similar results were obtained using LPS as a microbial stimulus (data not shown). A kinetic analysis revealed that for IFN-γ to enhance monocyte responsiveness it was necessary for the cytokine to be added prior to stimulation (data not shown) and that a 3-hour pre-incubation was sufficient for maximal response (Supplemental Figure 2A). Although IFN-γ can trigger cytoskeleton remodeling in human monocytes (14), the effect of cytokine pre-exposure in the case of T. gondii was not due to increased parasite invasion or the extent to which CD16neg monocytes phagocytized the MycB-treated tachyzoites (Supplemental Figure 2B). Neither did the pretreatment with IFN-γ induce expression of CD16 on CD16neg subset or covert them into macrophages as judged by morphology and gene expression (data not shown).
Figure 2: IFN-γ-priming augments T. gondii-induced IL-12 secretion by primary human monocytes is due to a selective effect on the CD16neg subset.
(A) CD14+ total monocytes and (B) MACS-purified CD16neg and CD16+ subpopulations from healthy donors were cultured overnight in the presence or absence of IFN-γ (10 ng/ml) and then left unstimulated (med) or exposed to untreated or mycalolide B (MycB) pre-treated T. gondii tachyzoites (MOI 1:1), or stimulated with R848 (300 ng/ml). After 18 hours incubation culture supernatants were collected and TNF, IL-12p40 and IL-12p70 were measured by ELISA. Pairs of connected symbols represents amount of cytokine detected in supernatants from the same donor monocytes cultured in parallel without (filled symbol) or with overnight IFN-γ-priming (open symbol) (n=14 for TNF and IL-12p40; n=6 for IL-12p70).
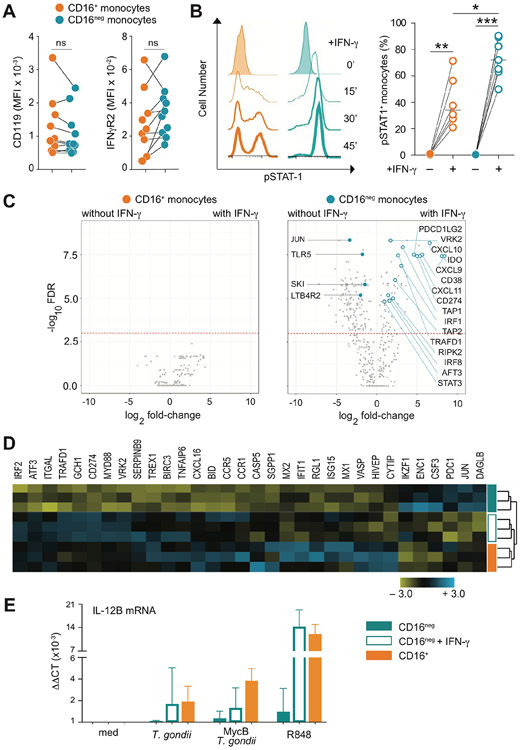
To exclude unequal expression of the IFN-γ receptor as an explanation for the distinct pattern of responsiveness, we compared the levels of the IFN-γR subunits CD119 and IFN-γR2 in both CD16+ and CD16neg monocytes populations by cell surface staining. No significant difference in expression of either receptor subunit was observed between the two subsets (Figure 3A). Interestingly, however, pSTAT1 was differentially expressed in monocyte subpopulations in response to IFN-γ-priming. Thus, as early as 15 min after IFN-γ exposure more than 70% of the CD16neg monocytes became pSTAT1+ in contrast to less than 50% in the CD16+ subset (Figure 3B). The dichotomy in pSTAT1+ expression in the CD16+ monocyte population was not due to its CD14+ heterogeneity since both the CD14lowCD16+ and CD14+CD16+ subpopulations shared the same pSTAT1 bimodal staining profile (Supplemental Figure 3A-C). Prolongation of IFN-γ exposure to 30 and 45 min did not alter the preferential expression of pSTAT1+ in the CD16neg vs. CD16+ monocytes (Figure 3B).
Figure 3: IFN-γ increases gene transcription only in CD16neg monocytes altering their mRNA expression profile to closely resemble that of the CD16pos monocytes at the baseline.
(A) Expression of IFN-γR1 (CD119) or R2 (IFN-γR2) is comparable between CD16neg and CD16+ monocytes. The graph depicts the mean fluorescence intensity (MFI) for each IFN-γR subunit on paired CD16+ vs. CD16neg populations for each donor (n=10). (B) IFN-γ response measured by pSTAT1 is more pronounced in CD16neg than CD16+ subset. Kinetics of pSTAT1+ monocytes measured by flow cytometry following stimulation with IFN-γ (10 ng/ml) for one representative donor (left) and frequencies of pSTAT1+ cells in CD16+ vs. CD16neg population at 20 min following IFN-γ exposure (right, n=6). (C) Pre-treatment with IFN-γ alters gene expression in CD16neg but not CD16+ monocytes. Volcano plots depict changes in gene expression in CD16+ (left) and CD16neg (right) monocytes cultured in the absence vs. presence of IFN-γ (10 ng/ml) assayed by the same NanoString panel used in Figure 1. The dotted line indicates the cutoff for statistical significance (p< 0.05) as calculated using a Benjamini- Yekutieli multiple test. Open blue symbols depict IFN-γ-upregulated genes known to be target of the cytokine and filled blue symbols show genes downregulated by IFN-γ, while brown dots indicate genes associated with metabolic pathways. (D) The gene expression profile of IFN-γ-primed CD16neg monocytes resembles that displayed by the CD16+ subset at the baseline. Heat-map of unsupervised clustering of CD16neg, IFN-γ-primed CD16neg and CD16+ monocytes from 3 healthy donors using 32 genes differentially expressed at baseline in CD16+ vs. CD16neg populations. (E) Expression of IL-12p40 measured by qRT-PCR in CD16neg, IFN-γ-primed CD16neg and CD16+ monocytes left unstimulated (med), exposed to either untreated or MycB T. gondii tachyzoites (MOI 1:1), or stimulated with R848 (300 ng/ml). The bar graph indicates median values ± interquartile range when compared to CD16neg monocytes culture in medium. *P < 0.05, **P < 0.01, ***P < 0.001.
We next examined the effect of IFN-γ priming on expression of the 770 genes analyzed at baseline in CD16+ and CD16neg monocyte (Figure 1). Unexpectedly, no significant changes in gene expression were detected in CD16+ monocytes as result of IFN-γ pre-treatment (Figure 3C left), while a large number of transcripts were upregulated (n=69) or downregulated (n=138) in the CD16neg population (Figure 3C right). In addition, when unsupervised clustering analysis was performed for genes differentially expressed by the CD16+ and CD16neg subsets at baseline between unprimed CD16neg vs. CD16neg monocytes pretreated with IFN-γ, IFN-γ-primed CD16neg monocytes clearly clustered with the unprimed CD16+ subset (Figure 3D). Moreover, IL-12 mRNA expression in response to either T. gondii- or R848-stimulation was also comparable in IFN-γ-primed CD16neg and CD16+ monocytes (Figure 3E). These findings indicated that CD16+ monocytes that display at baseline an interferon-inducible transcriptional signature are largely refractory to subsequent IFN-γ-priming while CD16neg monocytes which display an unbiased gene expression profile are the major subset affected by pre-treatment with the cytokine.
IFN-α-priming fails to promote IL-12 secretion by CD16neg monocytes while inhibiting the enhancing effect of IFN-γ
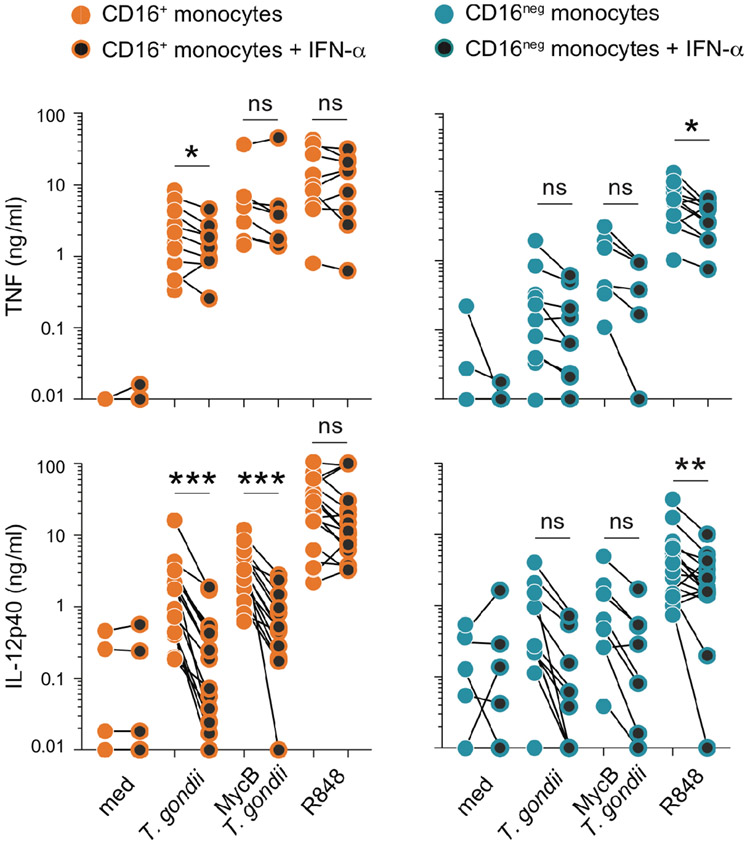
In addition to IFN-γR, all monocytes express type I interferon receptors. Nevertheless, the responses triggered by the two classes of IFNs may result in different biological effects. The role of type I interferons in regulating IL-12 production by human monocytes has been the subject of debate (15-17). Since the previous studies addressing this issue employed unfractionated monocyte populations (isolated by different methods in each case), we decided to test the effect of type I IFN (IFN-α2) addition on purified CD16+ and CD16neg monocytes prior to stimulation with T. gondii or R848. As shown in Figure 4, preincubation with IFN-α2 had an effect distinct from that induced by IFN-γ (Figure 2B), partially decreasing IL-12 secretion by CD16+ monocytes while failing to enhance cytokine production by the CD16neg subset. Nevertheless, despite this distinction, the frequency of pSTAT1+ cells following stimulation with IFN-α was very similar to that observed after IFN-γ-priming and significantly lower in CD16+ than in CD16neg monocytes (Figure 3B vs. Supplemental Figure 3D, E).
FIGURE 4: Pretreatment with type I IFN diminishes the IL-12 response of CD16+ monocytes and fails to enhance cytokine production by the CD16neg subset.
MACS-purified human primary CD16+ and CD16neg monocytes (106/ml) were cultured overnight in the presence or the absence of IFN-α (10 ng/ml) and then left untreated (med) or exposed to untreated or MycB T. gondii tachyzoites (MOI 1:1) or activated with R848 (300 ng/ml). TNF and IL-12p40 were measured by ELISA in culture supernatants collected after 18 hours of stimulation. The connected symbols represent the amount of cytokine in supernatants from monocytes from the same donor (n=6-12) cultured in parallel in the absence (orange and blue filled circles) or presence of IFN-α (black filled circles). * P < 0.05, **P < 0.01, ***P < 0.001.
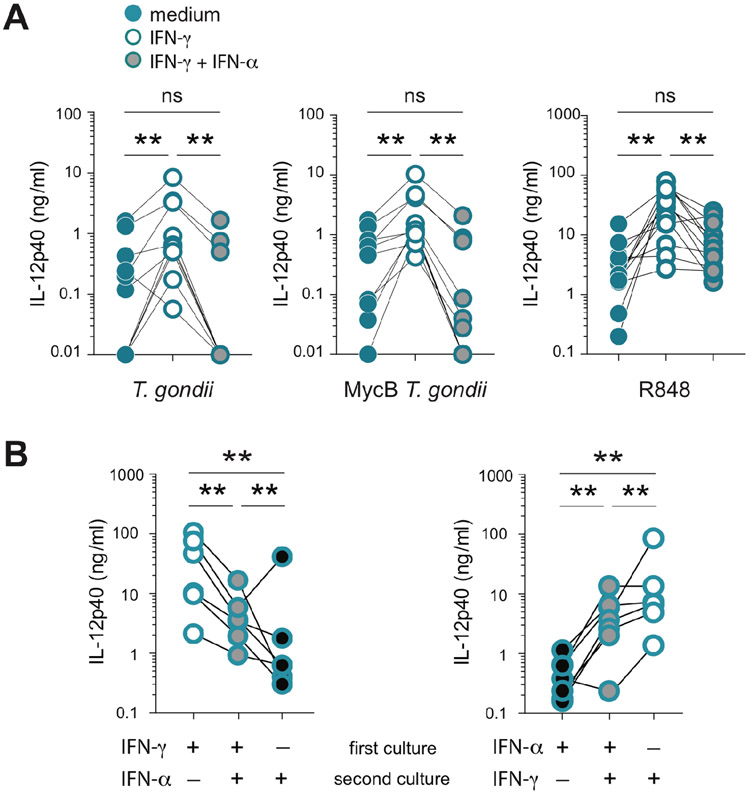
We next examined the effect of IFN-α2 on IL-12 secretion triggered by microbial stimulation of IFN-γ-primed CD16neg monocytes. In a first set of experiments, CD16neg monocytes were incubated with either IFN-γ alone or with both IFN-γ and IFN-α2 prior to stimulation. In response to either tachyzoites or R848, the IFN-γ-stimulated IL-12 secretion by CD16neg monocytes was suppressed when IFN-α2 was present during IFN-γ priming (Figure 5A). In the second assay, IFN-γ was removed after priming and CD16neg monocytes were then incubated with or without IFN-α2 before stimulation with R848. The exposure to IFN-α2 after IFN-γ priming impaired IFN-γ-induced IL-12 secretion by the CD16neg population (Figure 5B left). Nevertheless, the level of IL-12 secreted was higher than if IFN-γ was not present during priming. This intermediate phenotype was also observed when the IFN addition was reversed and CD16neg monocytes were first exposed to IFN-α2 and then IFN-γ (Figure 5B right). The pretreatment with IFN-α2 did not cause significant change in expression of CD14 or CD16 markers on either CD16neg or CD16+ monocytes (data not shown). These results demonstrated that, in addition to inhibiting IL-12-responses by CD16+ monocytes, type I interferon also dampens secretion of IL-12 by IFN-γ-primed CD16neg monocytes regardless of the temporal sequence of exposure to the two IFN species. Thus, type I and II IFN appear to have opposing effects on the IL-12 response of human monocytes.
Figure 5: The inhibitory effect of type I IFN overrides IFN-γ-mediated priming for IL-12 secretion in CD16neg monocytes.
(A) Purified human CD16neg monocytes (106/ml) were cultured overnight in the presence of medium alone (blue circles), IFN-γ (10 ng/ml) alone (white filled circles), or in the presence of both IFN-γ and IFN-α (10 ng/ml each) (gray filled circles) before stimulation with untreated (left) or MycB-treated T. gondii (middle), or R848 (right). Culture supernatants were collected 18 hours later and IL-12p40 was measured by ELISA. Connected symbols indicate the values obtained for monocytes from the same donor (n=6) treated in parallel under the 3 different conditions. (B) CD16neg monocytes (106/ml) were cultured with either IFN-γ (left panel) or IFN-α (right panel), or only in medium for 18 hours when supernatants were removed. The IFN-treated cultures were split into two wells, and medium added to one while the second was treated with the opposite IFN type from that used in the first culture. The monocytes cultured first in medium alone were treated with the same type of IFN used during second incubation. All cultures were then stimulated with R848 (300 ng/ml) in parallel for an additional 18 hours. IL-12p40 was measured by ELISA and connected symbols indicate values obtained for monocytes from the same donor (n=8) cultured in parallel under the 3 different conditions. **P <0.01.
Enhanced IL-12 production of human monocytes is metabolically influenced not at the level of glycolysis but through mTOR activity
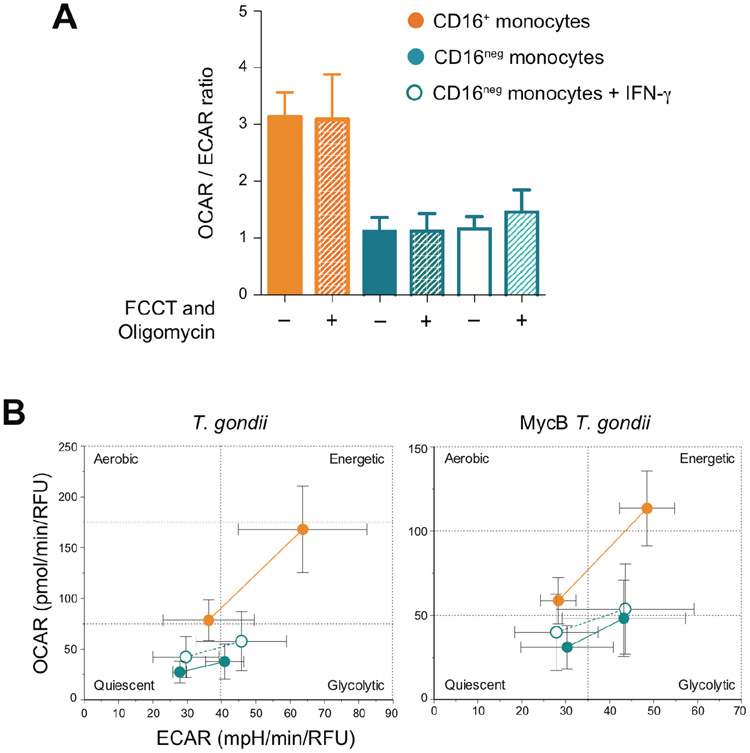
To test whether IL-12 production by human monocytes is associated with an aerobic to anaerobic metabolic shift, we measured the rates of oxidative phosphorylation (OXPHOS) and glycolysis in CD16+ and IFN-γ-primed CD16neg cells in comparison to non-primed CD16neg monocytes (Figure 6). The CD16+ subset displayed a higher OXPHOS/glycolysis ratio, but the lower ratio displayed by the CD16neg population remained unchanged after exposure to IFN-γ arguing that its enhancing effect on IL-12 production is not dependent on a glycolytic shift (Figure 6A). Moreover, the metabolic profiles of CD16+, CD16neg and IFN-γ-primed CD16neg were unaffected by the stress response or by T. gondii stimulation (Figure 6).
Figure 6: The metabolic status of CD16+ subset is distinct from that of CD16neg and IFN-γ-primed CD16neg monocytes at baseline and after T. gondii stimulation.
(A) The rate of oxygen consumption (OCAR) and extracellular acidification (ECAR) in CD16+, CD16neg and IFN- γ -pretreated CD16neg monocytes cultured in medium were measured before (solid bars) and after OXPHOS inhibition (hashed bars) as readout of a basal and a stress response, respectively. The bar graph indicates the mean values ± SD of the OCAR/ECAR ratio (n=3). (B) The monocyte populations indicated in A were infected for 3 hours with untreated (left) or MycB-treated (right) T. gondii tachyzoites and their “energy phenotype” was assayed by measuring OCAR and ECAR before and after OXPHOS inhibition. The plots depict mean ± SD of values obtained for three donors.
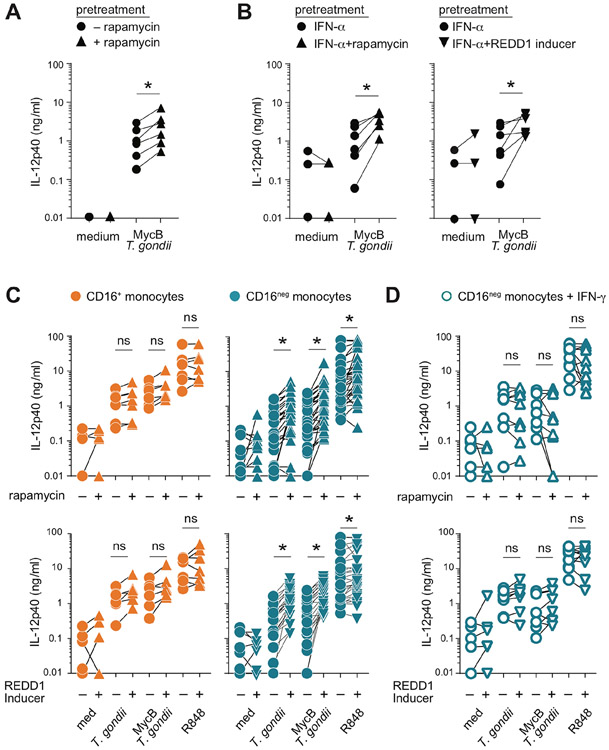
We next explored the role of the mTOR pathway which regulates the metabolism of cells according to their functional status (18-19). Previous studies have shown that IFN-γ suppresses while IFN-α stimulates mTOR activity in human macrophages (20-22). Moreover, inhibition of the mTOR complex has been shown to enhance IL-12 secretion by human PBMC (23). In agreement, with the latter observation, we found that preincubation with the mTORC1 pathway inhibitor, rapamycin increased the IL-12 response of total CD14+ monocytes to T. gondii exposure (Figure 7A). The stimulatory effect of rapamycin as well as REDD1 Inducer, a second mTORC1 inhibitor, was also observed with monocytes pre-exposed to IFN-α (Figure 7B).
Figure 7: mTOR pathway inhibition increases IL-12 secretion by human monocytes by enhancing the responsiveness of the CD16neg and not CD16+ subset.
(A) CD14+ total monocytes from healthy donors were pretreated with rapamycin (5 nM) for 1 hour prior to stimulation with MycB-treated T. gondii tachyzoites (MOI 1:1). (B-D) CD14+ total monocytes from healthy donors cultured overnight in the presence of IFN-α (10 ng/ml) (B), purified CD16+ or CD16neg (C) and IFN-γ-primed CD16neg (D) monocyte were cultured for 1 hour in the presence or absence of rapamycin (5 nM) or REDD-1 inducer (10 μM) and then left unstimulated (med) or exposed to untreated or MycB-treated T. gondii (MOI 1:1), or stimulated with R848 (300 ng/ml). Supernatants were collected after 18 hours and IL-12p40 measured by ELISA. Paired symbols indicate amount of IL-12p40 secreted by monocytes from the same donor (n=6-8) in parallel cultures treated (triangles) or not (circles) with the indicated mTOR inhibitor. *P < 0.05.
mTOR pathway inhibition promotes IL-12-secretion selectively by CD16 neg monocytes
To identify the monocyte subset(s) targeted by mTORC1 inhibition, we next examined the effect of preincubation with rapamycin or REDD1 inducer on the IL-12 response of purified CD16neg and CD16+ subsets. We first confirmed that both populations display comparable levels of the active form of mTORC1 as well as its substrate 4E-BP1 complex (Supplemental Figure 4). As previously observed with IFN-γ-priming (Figure 2B), preincubation with mTORC1 inhibitors failed to modify the response of CD16+ monocytes to either toxoplasma tachyzoites or R848 (Figure 7C). In contrast, both rapamycin and REDD1 Inducer significantly increased IL-12 production by CD16neg monocytes following the same microbial stimulation (Figure 7C). However, this enhancing effect of the two drugs was not observed with IFN-γ-primed CD16neg monocytes further demonstrating the functional similarity of the latter population with the CD16+ subset at baseline (Figure 7D). These findings demonstrated that CD16+ monocytes pre-conditioned in vivo to produce IL-12 are refractory to the enhancing effects of both IFN-γ and mTORC1 inhibition, while CD16neg monocytes that are poor responders at baseline remain susceptible to and moreover require the same regulatory signals for optimal IL-12 production.
Discussion
Blood monocytes represent a heterogenous population consisting of cells at different stages of maturation. The progression of CD16negCD14+ monocytes towards CD16+CD14low population is supported by studies in which blood monocyte were monitored in individuals after experimentally induced endotoxemia (3, 24) and by transcriptional profiling (6, 25-27). Nevertheless, the cytokine secretion profiles and specific functions of CD16neg vs. CD16+ monocytes in steady state vs. inflammation have not been formally delineated (28, 29).
The preferential expression of specific type of pattern recognition receptors such as TLR4/CD14 on CD14+CD16neg and TLR7/8 on CD14lowCD16+ monocytes has suggested their involvement in determining the selective responsiveness of these subsets to microbial stimulation (6). While the ability of human monocytes to secrete IL-12 and TNF in response to toxoplasma was previously found by us to be restricted to the CD16+ monocyte subset (8), this dichotomy is not a feature of all cytokine responses to the parasite. For example, T. gondii tachyzoites have been shown to stimulate strong CCL2 chemokine production by unfractionated CD14+ human monocytes (9) and we failed to observe any significant differences in the CCL2 response of the purified CD16neg and CD16+ subsets (Supplemental Figure 1). Moreover, production of IL-1β, associated with induction of Th17 (as opposed to Th1) responses, displays the inverse pattern to IL-12 and TNF expression with live T. gondii triggering IL-1β preferentially from CD16neg monocytes (30). These observations indicate that while toxoplasma is a potent activator of all human peripheral blood monocyte subpopulations, the type of cytokine response induced and its susceptibility to external signals is largely dictated by intrinsic differences in the individual monocyte subsets that are independent of their ability to sense the parasite.
We previously found that MycB treated parasites which can no longer actively invade host cells and become sequestered in phagosomes retain the capacity to stimulate IL-12 and TNF production by human monocytes (8). Here we extended these observations by demonstrating that MycB-tachyzoites despite their distinct mode of cell entry behave identically to live parasites in terms of the susceptibility of the cytokine response they induce to IFN-γ priming or mTOR inhibition.
In agreement with previous reports studying TNF production by human monocyte subsets (31-33) our findings demonstrate that CD16+ monocytes also display higher production of IL-12 in response to microbial stimulation. The preferential expression of IL-12 and TNF by CD16+ monocytes may be the outcome of their extended lifespan along with their prior exposure to either microbial stimulation or IFN or to both. Indirect support for this concept comes from their CD14+/low CD16+ phenotype in that GPI-anchored CD14 is known be shed after TLR stimulation (34) while CD16 (FcγRIII) is induced by IFNs (35-37). The latter hypothesis is supported by our finding that CD16+ monocytes preferentially express an IFN-induced transcriptional profile at baseline. Interestingly, all of the interferon stimulated genes (ISG) expressed by CD16+ monocytes were previously characterized by Mostafavi et al. (38) as tonically expressed ISG. The increased expression by CD16+ monocytes of some of these genes (e.g. IFITM1, IFITM2 and USP18) is also evident upon inspection of microarray data sets in previous studies comparing the different subsets (26, 27). The detection of only a subset of ISG by the CD16+ monocytes may reflect the involvement of other superimposing stimuli to which they were exposed in vivo leading to an enhanced but selective ISG profile. Consistent with this hypothesis, the frequency of CD16+ monocytes is augmented in the Th1-associated environment accompanying infection with intracellular pathogens (39-41) or autoimmune disease (29, 42, 43).
Unexpectedly, in vitro priming with IFN-γ or inhibitors of the mTOR pathway failed to augment IL-12 secretion by CD16+ monocytes in response to T. gondii tachyzoites or R848 exposure. This observation may indicate that these cells have been already primed in vivo for maximal cytokine secretion which could also activate negative feedback regulatory pathway(s). For example, activation via a PPR (e.g. TLR4) can induce expression of SOCS1 that prevents IFN induced STAT1 phosphorylation (44). Whether the fraction of CD16+ monocytes that fails to phosphorylate STAT1 upon in vitro IFN-γ stimulation (Figure 3B) reflects a recently activated population remains to be determined. The hypo-reactivity of CD16+ monocytes extends beyond cytokine responsiveness. Thus, the numbers of transcripts with altered expression after exposure to T. gondii inversely correlated with the ability of monocytes to secrete IL-12 and TNF with the lowest in CD16+CD14low, followed by CD16+CD14+ and the highest in CD16negCD14+monocytes (K.W.T. and D.J. unpublished data). Together these observations indicate that the CD16+ population represents not just a more differentiated stage but also a functionally specialized population with a prefixed and limited program.
Effector immune functions, such as cytokine production, occur at a high-energy cost and have been closely associated with a metabolic shift towards glycolysis (45). While in myeloid cells LPS stimulation simultaneously triggers decreased OXPHOS and increased glycolysis (46, 47) this may be more an exception than the rule since activation of human monocytes with other TLR ligands has been reported to enhance both OXPHOS and glycolysis (47). Moreover, glucose deprivation has been shown to promote rather than inhibit IL-12 production by monocytes (48). In agreement with previous reports of increased mitochondrial activity in CD16+ monocytes (40), we observed that these cells display a higher oxidative phosphorylation/glycolysis ratio than the CD16neg population at baseline and after microbial stimulation (Figure 6). Thus, in terms of their metabolic state CD16+ monocytes resemble memory T lymphocytes (45) and may be endowed with similar memory-like properties.
Findings from in vitro studies employing non-fractionated monocytes often reflect the properties of the dominant (~85-95%) CD16neg population. Moreover, when total monocytes are purified from peripheral blood samples, a variable and poor recovery of the CD16+ population may further bias this outcome and could explain the lack of IL-12 detection in some studies that employed unfractionated monocytes tested in the absence of IFN-γ (9). While the lack of IL-12 response by CD16neg monocytes can successfully be reversed by IFN-γ- or rapamycin-priming, neutralization of IL-10 or blocking of type I IFN receptor before and during stimulation failed to do so (data not shown), arguing that their hypo-responsiveness is not masked by either of these IL-12-suppressing cytokines.
Our findings demonstrate that CD16neg monocytes are not only less differentiated but also more functionally “plastic” since they are preferentially responsive to different environmental cues of which IFNs are important players. As shown here the reprogramming induced by IFN-γ leads to the downregulation of c-JUN mRNA (Figure 3C) that would in turn limit expression of AP-1, a transcription factor selectively associated with the CD16neg subset (26, 27). In addition, IFN-γ reprograming was found to lead to resistance to the stimulatory effect of rapamycin (Fig. 7) and on the other hand susceptibility to the inhibitory effect of type I IFN as assessed by IL-12 production (Fig. 4). Importantly, all three outcomes are properties of CD16+ monocytes at baseline. In contrast, exposure to type I IFN restrains the ability of CD16neg monocytes to secrete IL-12 that may negatively impact host protection. While strains of toxoplasma that trigger type I IFN secretion are rare (49), our results suggest that the induction of the cytokine by the parasite could represent a potential mechanism of immune evasion. Similarly, induction of type I IFN could be a means by which a concomitant viral infection (e.g. HIV) could suppress host resistance to T. gondii thereby promoting parasite growth.”
Since pSTAT1 expression in CD16neg monocytes (Figure 3B and Supplemental Figure 4) was equivalent after exposure to either IFN-α or IFN-γ despite the opposing outcomes of these cytokines on both IL-12 secretion (Figure 2 vs. 4) and CD64 expression (50), it is unlikely that the phosphorylation of STAT1 is the sole event responsible for the effects of IFN-γ priming. Previous studies have demonstrated that the cross-regulating effects of type I IFN and IFN-γ can involve downregulation of their respective receptors (51) and/or competition for STAT1 (52). Nevertheless, our finding that type I IFN inhibits IL-12 production even after preincubation with IFN-γ (Figure 5B) argues for additional regulatory mechanism(s). We tested the possibility that the inhibitory effect of type I IFN on IL-12 production is due to induction of IL-10, however no increase in expression of IL-10 mRNA in IFN-α2-primed CD16+ monocytes was observed nor did the neutralization of IL-10 before and during culture augment the production of IL-12 by IFN-α2-primed monocytes stimulated with T. gondii or R848 (data not shown). Based on the results presented here (Figure 3E) and in a previous report (16), the differential effect of IFN-γ and type I IFN on monocyte IL-12 responses is likely to stem from their ability to either promote or suppress IL-12p40 gene transcription, respectively. Alternatively, type I IFN could act by attenuating NF-κB signaling required for expression of IL-12 through direct activation of STAT3 (53) or by inducing specific epigenetic modifications (54).
At a general level the observations reported here shed new light on the functional plasticity of CD16+ versus CD16neg monocytes. Based on our findings we propose that CD16+ monocytes are poised for IL-12 and TNF expression (“ignition”) by microbial signals. However, the CD16neg subset later takes over the role as the major source of these cytokines in the response because of both their numerical dominance and higher responsiveness to external priming. This temporal distinction in the inflammatory potential of CD16neg versus CD16+ monocyte populations may be an important aspect in understanding the unique functional properties of these subsets.
Supplementary Material
Key points:
Human monocyte subsets have distinct IFN-γ priming requirements for IL-12 production
IFN-γ boosts monocyte IL-12 response by priming the less responsive CD16neg subset
IFN-α fails to enhance monocyte IL-12 production but inhibits their priming by IFN-γ
Acknowledgments:
We are grateful to Dr. Roshanak Tolouei Semnani for helpful advice throughout this study, Dr. Stephen Christensen for help with generating volcano plots and the NIAID FACS Facility for performing FACS sorting. We especially thank Drs. Raphaela Goldbach-Mansky and Bernadette Marrero for critical reading of the manuscript and valuable suggestions.
This study was funded by the Intramural Research Program of the NIAID, NIH and in part by the in part by the Brazilian National Council for Scientific and Technological Development–CNPq Fellowship (A.M.T.S.A).
Abbreviations:
- ISG
interferon stimulated gene
- FCCP
Carbonyl cyanide 4-(trifluoromethoxy)phenylhydrazone
- MycB
mycalolide B
Footnotes
Conflict of Interest:
The authors declare no conflict of interests.
References:
- 1.Ziegler-Heitbrock L, Ancuta P, Crowe S, Dalod M, Grau V, Hart DN, Leenen PJ, Liu YJ, MacPherson G, Randolph GJ, Scherberich J, Schmitz J, Shortman K, Sozzani S, Strobel H, Zembala M, Austyn JM, and Lutz MB. 2010. Nomenclature of monocytes and dendritic cells in blood. Blood 116: e74–e80. [DOI] [PubMed] [Google Scholar]
- 2.Passlick B, Flieger D, and Ziegler-Heitbrock HW. 1989. Identification and characterization of a novel monocyte subpopulation in human peripheral blood. Blood 74: 2527–2534. [PubMed] [Google Scholar]
- 3.Patel AA, Zhang Y, Fullerton JN, Boelen L, Rongvaux A, Maini AA, Bigley V, Flavell RA, Gilroy DW, Asquith B, Macallan D, and Yona S 2017. The fate and lifespan of human monocyte subsets in steady state and systemic inflammation. J. Exp. Med 214: 1913–1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gamrekelashvili J, Giagnorio R, Jussofie J, Soehnlein O, Duchene J, Briseño CG, Ramasamy SK, Krishnasamy K, Limbourg A, Häger C, Kapanadze T, Ishifune C, Hinkel R, Radtke F, Strobl LJ, Zimber-Strobl U, Napp LC, Bauersachs J, Haller H, Yasutomo K, Kupatt C, Murphy KM, Adams RH, Weber C and Limbourg FP 2016. Regulation of monocyte cell fate by blood vessels mediated by Notch signaling. Nat. Commun 7: 12597. doi: 10.1038/ncomms12597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lessard AJ, LeBel M, Egarnes B, Prefontaine P, Thériault P, Droit A, Brunet A, Rivest S, and Gosselin J 2017. Triggering of NOD2 receptor converts inflammatory Ly6Chigh into Ly6Clow Monocytes with Patrolling Properties. Cell Rep. 20: 1830–1843. doi: 10.1016/j.celrep.2017.08.009 [DOI] [PubMed] [Google Scholar]
- 6.Cros J, Cagnard N, Wollard K, Patey N, Zhang SY, Senechal B, Puel A, Biswas SK, Moshous D, Picard C, Jais JP, D'Cruz D, Casanova JL, Trouillet C, and Geissmann F, F. 2010. Human CD14dim monocytes patrol and sense nucleic acids and viruses via TLR7 and TLR8 receptors. Immunity 33: 375–386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hadadi E, Zhang B, Baidžajevas K, Yusof N, Puan KJ, Ong SM, Yeap WH, Rotzschke O, Kiss-Toth E, Wilson H, and Wong SC. 2016. Differential IL-1β secretion by monocyte subsets is regulated by Hsp27 through modulating mRNA stability. Sci. Rep 15, 6:39035. doi: 10.1038/srep39035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tosh KW, Mittereder L, Bonne-Annee S, Hieny S, Nutman TB, Singer SM, Sher A, and Jankovic D. 2016. The IL-12 response of primary human dendritic cells and monocytes to Toxoplasma gondii is stimulated by phagocytosis of live rather than host cell invasion. J. Immunol 196: 345–356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Safronova A, Araujo A, Camanzo ET, Moon TJ, Elliott MR, Beiting DP, and Yarovinsky F. 2019. Alarmin S100A11 initiates a chemokine response to the human pathogen Toxoplasma gondii. Nat. Immunol 20: 64–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li J, Kim SG, and Blenis J. 2014. Rapamycin: one drug, many effects. Cell Metabolism, 19: 373–379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kimball SR, Dang Do AN, Kutzler L, and Cavener DR, and Jefferson LS. 2008. Rapid turnover of the mTOR Complex 1 (mTORC1) repressor REDD1 and activation of mTORC1 signaling following Inhibition of protein synthesis. J. Biol. Chem 283: 3465–3475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shenderov K, Riteau N, Yip R, Mayer-Barber KD, Oland S, Hieny S, Fitzgerald P, Oberst A, Dillon CP, Green DR, Cerundolo V, and Sher A. 2014. Cutting edge: Endoplasmic reticulum stress licenses macrophages to produce mature IL-1β in response to TLR4 stimulation through a caspase-8- and TRIF-dependent pathway. J. Immunol 192, 2029–2033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kugler DG, Mittelstadt PR, Ashwell JD, Sher A, and Jankovic D. 2013. CD4+ T cells are trigger and target of the glucocorticoid response that prevents lethal immunopathology in toxoplasma infection. J. Exp. Med 210: 1919–1927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hu Y, Hu X, Boumsell L, and Ivashkiv LB. 2008. IFN-gamma and STAT1 arrest monocyte migration and modulate RAC/CDC42 pathways. J. Immunol 180: 8057–8065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hermann P, Rubio M, Nakajima T, Delespesse G, and Sarfati M. 1998. IFN-α priming of human monocytes differentially regulates gram-positive and gram-negative bacteria-induced IL-10 release and selectively enhances IL-12p70, CD80, and MHC class I expression. J. Immunol 161: 2011–2018. [PubMed] [Google Scholar]
- 16.Byrnes AA, Ma X, Cuomo P, Park L, Wah IL, Wolf SF, Zhou H, Trinchieri G, and Karp CL. 2001. Type I interferons and IL-12: convergence and cross-regulation among mediators of cellular immunity. Eur. J. Immunol 31: 2026–2034. [DOI] [PubMed] [Google Scholar]
- 17.de Paus RA, van Wengen A, Schmidt I, Visser M, Verdegaal EM, van Dissel JT, and van de Vosse E. 2013. Inhibition of the type I immune responses of human monocytes by IFNα and IFNβ. Cytokine 61, 645–655. [DOI] [PubMed] [Google Scholar]
- 18.Dazert E and Hall MN. 2011. mTOR signaling in disease. Curr. Opin. Cell Biol 23: 745–755. [DOI] [PubMed] [Google Scholar]
- 19.Peter C, Waldmann H, and Cobbold SP. 2010. mTOR signalling and metabolic regulation of T cell differentiation. Curr. Opin. Immunol 22: 655–661. [DOI] [PubMed] [Google Scholar]
- 20.Su X, Yu Y, Zhong Y, Giannopoulou EG, Hu X, Liu H, Cross JR, Rätsch G, Rice CM, and Ivashkiv L. 2015. Interferon-γ regulates cellular metabolism and mRNA translation to potentiate macrophage activation. Nat. Immunol 16: 838–849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lekmine F, Sassano A, Uddin S, Smith J, Majchrzak B, Brachmann SM, Hay N, Fish EN, and Platanias LC. 2004. Interferon-gamma engages the p70 S6 kinase to regulate phosphorylation of the 40S S6 ribosomal protein. Exp. Cell. Res 295: 173–182. [DOI] [PubMed] [Google Scholar]
- 22.Matsumoto A, Ichikawa T, Nakao K, Miyaaki H, Hirano K, Fujimito M, Akiyama M, Miuma S, Ozawa E, Shibata H, Takeshita S, Yamasaki H, Ikeda M, Kato N, and Eguchi K. 2009. Interferon-α-induced mTOR activation is an anti-hepatitis C virus signal via the phosphatidylinositol 3-kinase-Akt-independent pathway. J. Gastroenterol 45: 856–863. [DOI] [PubMed] [Google Scholar]
- 23.Turnquist HR, Cardinal J, Macedo C, Rosborough BR, Sumpter TL, Geller DA, Metes D, and Thomson AW. 2010. mTOR and GSK-3 shape the CD4+ T cell stimulatory and differentiation capacity of myeloid DCs after exposure to LPS. Blood 115: 4758–4769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tak T, van Groenendael R, Pickkers P, and Koenderman L. 2017. Monocyte subsets are differentially lost from the circulation during acute inflammation induced by human experimental endotoxemia. J. Innate Immun 9: 474–474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wong KL, Tai JJ, Wong WC, Han H, Sem X, Yeap WH, Kourilsky P, and Wong SC. 2011. Gene expression profiling reveals the defining features of the classical, intermediate and nonclassical human monocyte subsets. Blood 118, e16–31. [DOI] [PubMed] [Google Scholar]
- 26.Ingersoll MA, Spanbroek R, Lottaz C, Gautier EL, Frankenberger M, Hoffmann R, Lang R, Haniffa M, Collin M, Tacke F, Habenicht AJ, Ziegler-Heitbrock L, and Randolph GJ. 2010. Comparison of gene expression profiles between human and mouse monocyte subsets. Blood 115: e10–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schmidl C, Renner K, Peter K, Eder R, Lassmann T, Balwierz PJ, Itoh M, Nagao-Sato S, Kawaji H, Carninci P, Suzuki H, Hayashizaki Y, Andreesen R, Hume DA, Hoffmann P, Forrest AR, Kreutz MP, Edinger M, Rehli M, and FANTOM consortium 2014. Transcription and enhancer profiling in human monocyte subsets. Blood 123: e90–99. [DOI] [PubMed] [Google Scholar]
- 28.Serbina NV, Jia T, Hohl TM, and Pamer EG. 2008. Monocyte-mediated defense against microbial pathogens. Ann. Rev. Immunol 26: 421–452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Narasimhan PB, Marcovecchio P, Hamers AA, and Hedrick CC. 2019. Nonclassical monocytes in health and disease. Annu. Rev. Immunol 37, 439–456. [DOI] [PubMed] [Google Scholar]
- 30.Pandori WJ, Lima TS, Mallya S, Kao TH, Gov L, and Lodoen MB. 2019.Toxoplasma gondii activates a Syk-CARD9-NF-κB signaling axis and gasdermin D-independent release of IL-1β during infection of primary human monocytes. PLoS Pathog. 15: e1007923. doi: 10.1371/journal.ppat.1007923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Thieblemont N, Weiss L, Sadeghi HM, Estcourt C, and Haeffner-Cavaillon N. 1995. CD14low CD16high: a cytokine-producing monocyte subset which expands during human immunodeficiency virus infection. Eur. J. Immunol 25: 3418–3424. [DOI] [PubMed] [Google Scholar]
- 32.Belge KU, Dayyani F, Horelt A, Siedlar M, Frankenberger M, Frankenberger B, Espevik T, and Ziegler-Heitbrock L. 2002. The proinflammatory CD14+CD16+ DR++ monocytes are a major source of TNF. J. Immunol 168: 3536–3542. [DOI] [PubMed] [Google Scholar]
- 33.Zhao C, Tan YC, Wong WC, Sem X, Zhang H, Han H, Ong SM, Wong KL, Yeap WH, Sze SK, Kourilsky P, and Wong SC. 2010. The CD14+/lowCD16+ monocyte subset is more susceptible to spontaneous and oxidant-induced apoptosis than the CD14+CD16− subset. Cell Death & Disease 1, e95. doi: 10.1038/cddis.2010.69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shive CL, Jiang W, Anthony DD, and Lederman MM. 2015. Soluble CD14 is a nonspecific marker of monocyte activation. AIDS 29: 1263–1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Boylan MT, Crockard AD, Duddy ME, Armstrong MA, McMillan SA, and Hawkins SA. 2001. Interferon-beta1a administration results in a transient increase of serum amyloid A protein and C-reactive protein: comparison with other markers of inflammation. Immunol. Lett 75: 191–197. [DOI] [PubMed] [Google Scholar]
- 36.Aricò E, Castiello L, Urbani F, Rizza P, Panelli MC, Wang E, Marincola FM, and Belardelli F. 2011. Concomitant detection of IFNα signature and activated monocyte/dendritic cell precursors in the peripheral blood of IFNα-treated subjects at early times after repeated local cytokine treatments. J. Transl. Med 9: 67. doi: 10.1186/1479-5876-9-67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kedzierska K, Paukovics G, Handley A, Hewish M, Hocking J, Cameron PU, and Crowe SM. 2004. Interferon-γ therapy activates human monocytes for enhanced phagocytosis of Mycobacterium avium complex in HIV-infected individuals, HIV Clinical Trials 5: 80–85. [DOI] [PubMed] [Google Scholar]
- 38.Mostafavi S, Yoshida H, Moodley D, LeBoité H, Rothamel K, Raj T, Ye CJ, Chevrier N, Zhang SY, Feng T, Lee M, Casanova JL, Clark JD, Hegen M, Telliez JB, Hacohen N, De Jager PL, Regev A, Mathis D, Benoist C, and Immunological Genome Project Consortium. 2016. Parsing the Interferon Transcriptional Network and Its Disease Associations. Cell 164: 564–578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fingerle G, Pforte A, Passlick B, Blumenstein M, Ströbel M, and Ziegler-Heitbrock HW. 1993. The novel subset of CD14+/CD16+ blood monocytes is expanded in sepsis patients. Blood 82: 3170–3176. [PubMed] [Google Scholar]
- 40.Antonelli LR, Leoratti FM, Costa PA, Rocha BC, Diniz SQ, Tada MS, Pereira DB, Teixeira-Carvalho A, Golenbock DT, Gonçalves R, and Gazzinelli RT. 2014. The CD14+CD16+ inflammatory monocyte subset displays increased mitochondrial activity and effector function during acute Plasmodium vivax malaria. PLoS Pathog. 10: e1004393. doi: 10.1371/journal.ppat.1004393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Joosten SA, van Meijgaarden KE, Arend SM, Prins C, Oftung F, Korsvold GE, Kik SV, Arts RJ, van Crevel R, Netea MG, and Ottenhoff TH. 2018. Mycobacterial growth inhibition is associated with trained innate immunity. J. Clin. Invest 128: 1837–1851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Finsterbusch M, Hall P, Li A, Devi S, Westhorpe CL, Kitching AR, and Hickey MJ. 2016. Patrolling monocytes promote intravascular neutrophil activation and glomerular injury in the acutely inflamed glomerulus. Proc. Natl. Acad. Sci. USA 113: E5172–51781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhu H, Hu F, Sun X, Zhang X, Zhu L, Liu X, Li X, Xu L, Shi L, Gan Y, and Su Y. 2016. CD16+ monocyte subset was enriched and functionally exacerbated in driving T-cell activation and B-cell response in systemic lupus erythematosus. Front Immunol. 7: 512. doi: 10.3389/fimmu.2016.00512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Prêle CM, Woodward EA, Bisley J, Keith-Magee A, Nicholson SE, and Hart PH. 2008. SOCS1 regulates the IFN but not NFκB pathway in TLR-stimulated human monocytes and macrophages. J. Immunol 181: 8018–8026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Buck MD, Sowell RT,Kaech SM, and Pearce EL. 2017. Metabolic instruction of immunity. Cell 169: 570–586 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tannahill GM, Curtis AM, Adamik J, Palsson-McDermott EM, McGettrick AF, Goel G, Frezza C, Bernard NJ, Kelly B, Foley NH, Zheng L, Gardet A, Tong Z, Jany SS, Corr SC, Haneklaus M, Caffrey BE, Pierce K, Walmsley S, Beasley FC, Cummins E, Nizet V, Whyte M, Taylor CT, Lin H, Masters SL, Gottlieb E, Kelly VP, Clish C, Auron PE, Xavier RJ, and O'Neill LA. 2013. Succinate is an inflammatory signal that induces IL-1β through HIF-1α. Nature 496: 238–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lachmandas E, Boutens L, Ratter JM, Hijmans A, Hooiveld GJ, Joosten LA, Rodenburg RJ, Fransen JA, Houtkooper RH, van Crevel R, Netea MG, and Stienstra R. 2016. Microbial stimulation of different Toll-like receptor signalling pathways induces diverse metabolic programs in human monocytes. Nat. Microbiol 2, 16247. doi: 10.1038/nmicrobiol.2016.247 [DOI] [PubMed] [Google Scholar]
- 48.Kovarik JJ, Kernbauer E, Hölzl MA, Hofer J, Gualdoni GA, Schmetterer KG, Miftari F, Sobanov Y, Meshcheryakova A, Mechtcheriakova D, Witzeneder N, Greiner G, Ohradanova-Repic A, Waidhofer-Söllner P, Säemann MD, Decker T, and Zlabinger GJ, J. G 2017. Fasting metabolism modulates the interleukin-12/interleukin-10 cytokine axis. PLoS ONE 12: e0180900. doi: 10.1371/journal.pone.0180900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Melo MB, Nguyen QP, Cordeiro C, Hassan MA, Yang N, McKell R, Rosowski EE, Julien L, Butty V, Dardé ML, Ajzenberg D, Fitzgerald K, Young LH, and Saeij JP. 2013. Transcriptional analysis of murine macrophages infected with different Toxoplasma strains identifies novel regulation of host signaling pathways. PLoS Pathog. 9: e1003779. 10.1371/journal.ppat.1003779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Van Weyenbergh J, Lipinski P, Abadie A, Chabas D, Blank U, Liblau R, and Wietzerbin J. 1998. Antagonistic action of IFN-β and IFN-γ on high affinity Fcγ receptor expression in healthy controls and multiple sclerosis patients. J. Immunol 161: 1568–1574. [PubMed] [Google Scholar]
- 51.Crisler WJ, and Lenz LL. 2018. Crosstalk between type I and II interferons in regulation of myeloid cell responses during bacterial infection. Curr. Opin. Immunol 54: 35–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ho J, Pelzel C, Begitt A, Mee M, Elsheikha HM, Scott DJ, and Vinkemeier U. 2016. STAT2 is a pervasive cytokine regulator due to its inhibition of STAT1 in multiple signaling pathways. PLOS Biology 14, e2000117. doi: 10.1371/journal.pbio.2000117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ho HH, and Ivashkiv LB. 2006. Role of STAT3 in type I interferon responses. negative regulation of STAT1-dependent inflammatory gene activation. J. Biol. Chem 281: 14111–14118. [DOI] [PubMed] [Google Scholar]
- 54.Kroetz DN, Allen RM, Schaller MA, Cavallaro C, Ito T, and Kunkel SL. 2015. Type I interferon induced epigenetic regulation of macrophages suppresses innate and adaptive immunity in acute respiratory viral infection. PLoS Pathog. 11: e1005338. 10.1371/journal.ppat.1005338. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.