Abstract
Human adenoviruses (HAdVs) are widespread pathogens that cause a number of partially overlapping, species-specific infections associated with respiratory, urinary, gastrointestinal, and ocular diseases. The early 3 (E3) region of adenoviruses is highly divergent between different species and it encodes a multitude of proteins with immunomodulatory functions. The study of genetic diversity in the E3 region offers a unique opportunity to gain insight into how the various HAdVs have evolutionarily adapted in response to the selection pressures exerted by host immune defenses. The objective of this review is to discuss subversion of host antiviral immune responses by HAdVs, with a focus on suppression of MHC class I antigen presentation, as a window into host-HAdV adaptation.
Keywords: Adenovirus, MHC class I antigen presentation, viral immune evasion, E3–19K protein, E3 region, E1A region, host immunity
Introduction
Human adenoviruses (HAdVs) are widespread pathogens in the world population. There are about 100 recognized HAdV types (from HAdV-1 to HAdV-100) that are classified into species A to G (http://hadvwg.gmu.edu/), based on their hemagglutination properties, DNA homology, and oncogenicity in rodents [1,2]. Members of different HAdV species are associated with distinct, yet partially overlapping, human diseases that are linked to respiratory (species B, C, and E), urinary (species B), gastrointestinal (species A, F, and G), and ocular (species D) infections. While primary HAdV infection elicits host antiviral immune responses in which cytotoxic T cells (CTLs) play an important role, these responses are usually insufficient to clear the virus. Consequently, HAdVs may often cause long-term asymptomatic infections in healthy individuals. The ability of HAdV to persist and spread in host cells presupposes that the virus is capable of subverting antiviral immune responses.
The HAdV genome contains a number of genes that encode proteins with immunomodulatory functions [3]. Some of these proteins function by downregulating the cell-surface expression of MHC class I molecules, which impairs antigen presentation to CTLs and thus protects infected cells against lysis by HAdV-specific CTLs. For example, the highly oncogenic HAdV-12 of species A (HAdV-A12) encodes proteins in the early 1A (E1A) region that specifically repress transcription of the major histocompatibility complex (MHC) class I heavy chain gene [4]. The mechanism by which MHC I gene expression is shut down is not clearly determined. The E3 region, on the other hand, is unique in that it encodes proteins dedicated at counteracting host immune defenses. The original discovery of the E3–19K protein as a modulator of host immune responses [5–7] was not only groundbreaking, but it also drove research efforts to identify and characterize immunomodulatory proteins in other human viruses. Notably, HAdVs of species A and F lack the common E3–19K protein and, instead, express proteins not found in other species. To date, these species-specific E3-encoded proteins have not been characterized and thus their functions are unknown. Finally, a number of other E3-encoded immunomodulatory proteins have been characterized: 49K, RIDα, RIDβ, 6.7K, and 14.7K. These proteins do not downregulate cell-surface MHC I, but they employ different mechanisms like inhibition of tumor necrosis factor (TNF) activities and cell-surface downregulation of Fas and tumor necrosis factor-related apoptosis-inducing ligand (TRAIL) receptors [8].
The E3 region is one of the most divergent gene regions between HAdV species. Although the reasons for this genetic variability are not fully understood, it points to an important role for the E3 proteins in the clinical diseases caused by the various HAdVs. As such, an important factor contributing to species-specific variations in HAdV diseases is likely the ability of E3 proteins to adapt to host immune defenses at the sites of infection. Another contributing factor is undoubtedly the ability of HAdVs to infect first tissue-specific cell types. The study of genetic diversity in the E3 region thus offers a unique opportunity to gain insight into how the virus has evolutionary adapted in the face of immune challenges that it encounters inside host cells. In this review, we will discuss subversion of host antiviral immune defenses by HAdVs, with a focus on the E3 region and downregulation of MHC I molecules, as an opportunity to illuminate host-HAdV adaptation. This knowledge is important for our understanding of mechanisms at play in the pathogenicity and persistence of the various HAdV species.
Adenovirus transcription units
E1A region
The early phase of HAdV infection is driven by the expression of proteins encoded in the E1A region that function as transcriptional activators of the other early transcription units [9]. Although this is a common feature of all HAdVs species, the E1A region of HAdV-A12 has also the unique ability to downregulate cell-surface MHC I levels in transformed rat and human cells [10–12]. It is thought that a 289-amino acid protein encoded in the HAdV-A12 E1A region is specifically responsible for reducing mRNA levels of the class I heavy chain gene. Evidence has been provided that HAdV-A12 E1A also represses transcription of genes that encode the transporters associated with class I antigen processing 1 (TAP1) and TAP2, as well as the proteasome-associated latent membrane proteins 2 (LMP2) and 7 (LMP7) [13]. The exact mechanism by which HAdV-A12 E1A suppresses transcription of key genes of the MHC gene region is not clear, but it may involve interference with processing of the nuclear factor kappa B (NF-κB) transcription factor [14].
E3 region
The E3 region is not essential for HAdV replication in cultured cells. However, the fact that E3 is always maintained in natural isolates suggests that the proteins encoded in this region are critical for natural infections in humans. The E3 region is one of the most divergent gene regions between HAdV species. For example, in species D, this region measures approximately 5.2 kilobases (kb), while it encompasses only 3 kb in species F [15]. Nevertheless, the proteins encoded by the E3 region exhibit mostly immunomodulatory functions.
Species C, which is one of the most studied HAdV, seems to express seven E3 gene products: 12.5K, 6.7K, 19K, 11.6K, receptor internalization and degradation α (RIDα) (formerly 10.4K), RIDβ (formerly 14.5K) and 14.7K [3,16–18]. The 19K protein is present in species B, C, D and E. The 12.5K protein is found in species A to E, while RIDα, RIDβ, and 14.7K proteins are encoded by all species [3,19]. Interestingly, the E3 region from species A encodes two proteins, 29.4K and 30.7K, that have counterparts only in species F [19]. Similarly, species B harbors the 16K gene product, with homologs found only in species D and E. Notably, HAdVs from species B and D encode two unique E3 gene products, 9K and 49K, respectively [20–23]. Characterization of the signal sequences and transmembrane segments revealed that the majority of the E3 proteins are transmembrane proteins, with the exception of the 12.5K and 14.7K proteins [19,24,25]. The immunomodulatory functions of E3-encoded proteins support evasion of host antiviral defenses, which is thought to contribute to the establishment of persistent infections by HAdVs.
E3–19K and other E3 proteins
E3–19K
The E3–19K protein was first described as a membrane glycoprotein that co-immunoprecipitated with transplantation antigens, but with no defined function [26]. E3–19K is now known to function as an immunomodulatory protein by interfering with the MHC I antigen presentation pathway [5–7]. It was established that E3–19K binds to MHC I molecules in the endoplasmic reticulum (ER) of infected cells [5,7,27–30]. Deletion studies and experiments with mutants of E3–19K identified a C-terminal di-Lysine motif as highly important for ER retrieval, defining E3–19K as an ER-resident protein[31–33]. More recently, it was found that the transmembrane domain of E3–19K harbors an ER-retention signal that also contributes to the intracellular sequestration of MHC I molecules [34]. It is thought that both the ER retrieval and retention signals of E3–19K work cooperatively to suppress MHC I transport to the cell surface. By preventing the egress of MHC I molecules to the cell surface, HAdV-infected cells become more resistant to lytic attack by CTLs [35–39]. The discovery of the MHC I-binding and ER-retrieval/-retention functions, and their effects on cell-surface expression of MHC I, provided the first evidence that a virus can counteract host antiviral T cell responses.
Mature E3–19K varies between 139 and 151 amino acids, depending on the HAdV species [40]. E3–19K is composed of a N-terminal ER-luminal domain, a transmembrane domain, and a C-terminal cytosolic tail. The ER-luminal domain contains 19 residues that are strictly conserved across Ad species[15,40]. Early structure-function studies identified Lys42, Met87, Lys91, and Tyr93 in HAdV-C2 as critical for the binding of MHC I by E3–19K [29,30,41–45]. The interaction between E3–19K and MHC I was more thoroughly understood from our determination of the x-ray crystal structure of Ad2 (species C) E3–19K bound to human leukocyte antigen-A2 (HLA-A2) [46] (Fig. 1). The structure showed that E3–19K binds at the N-terminus of the α1-helix on HLA-A2 and also contacts the C-terminus of the α2-helix and loops on the α3-domain, and β2 microgobulin (β2m) (Fig. 1). This binding mode was also seen in the x-ray structure of the HAdV-E4 E3–19K/HLA-A2 complex [47]. These structures provided critical information to understand that the conserved residues form a tightly packed hydrophobic core essential to maintains the unique tertiary fold of E3–19K (Fig. 2A), while the non-conserved residues form most contacts with HLA-A2 (Fig. 2B).
Figure 1. Structure of Ad2 E3-19K/HLA-A2 complex.
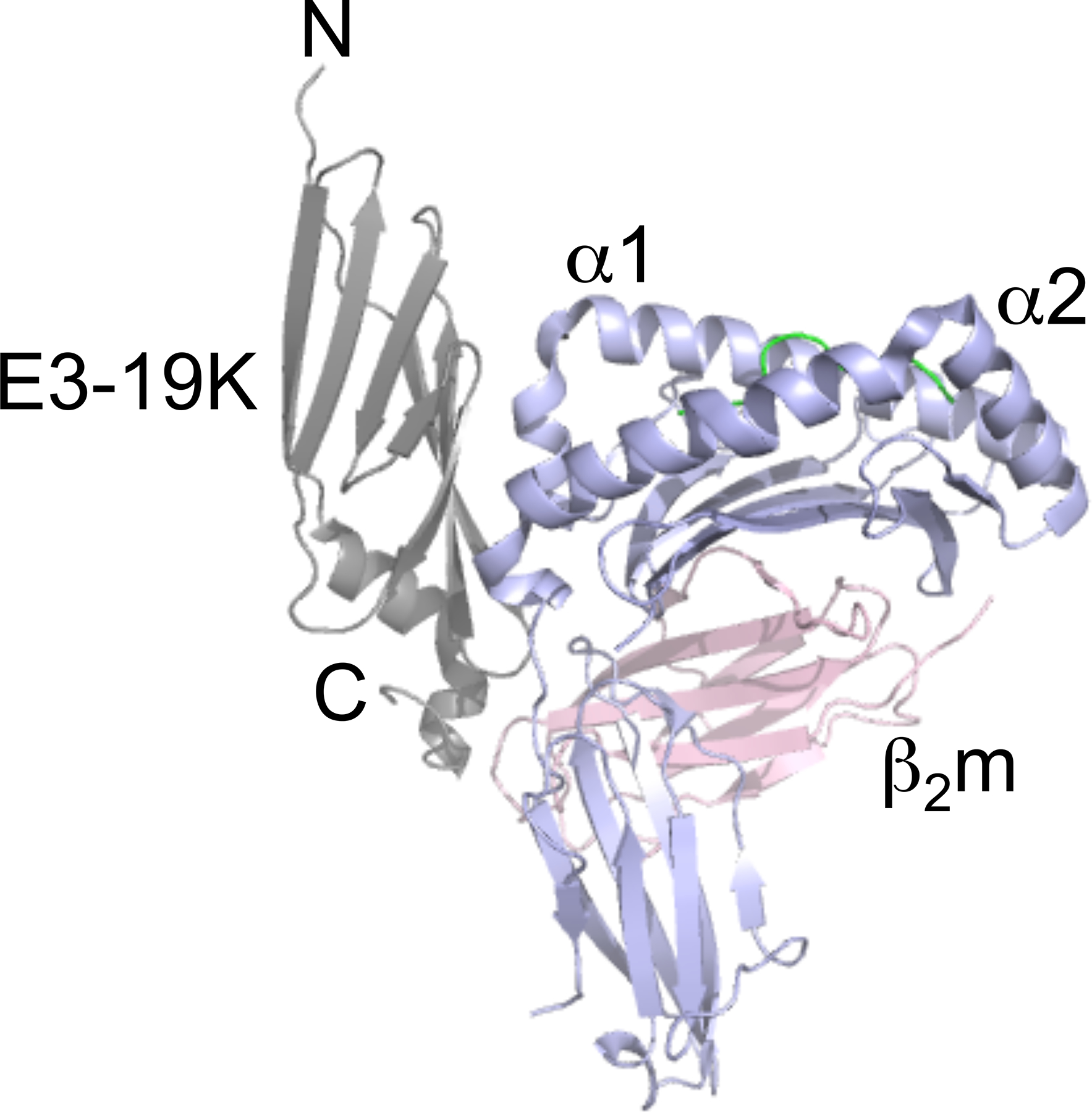
Ribbon representation of the structure of Ad2 E3–19K/HLA-A2: E3–19K, grey; HLA-A2 heavy chain (α1-, α2-, and α3-domains), purple; β2m, pink; HIV-1 Tax peptide LLFGYPVYV, green.
Figure 2. Conserved and non-conserved amino-acid residues in E3-19K.
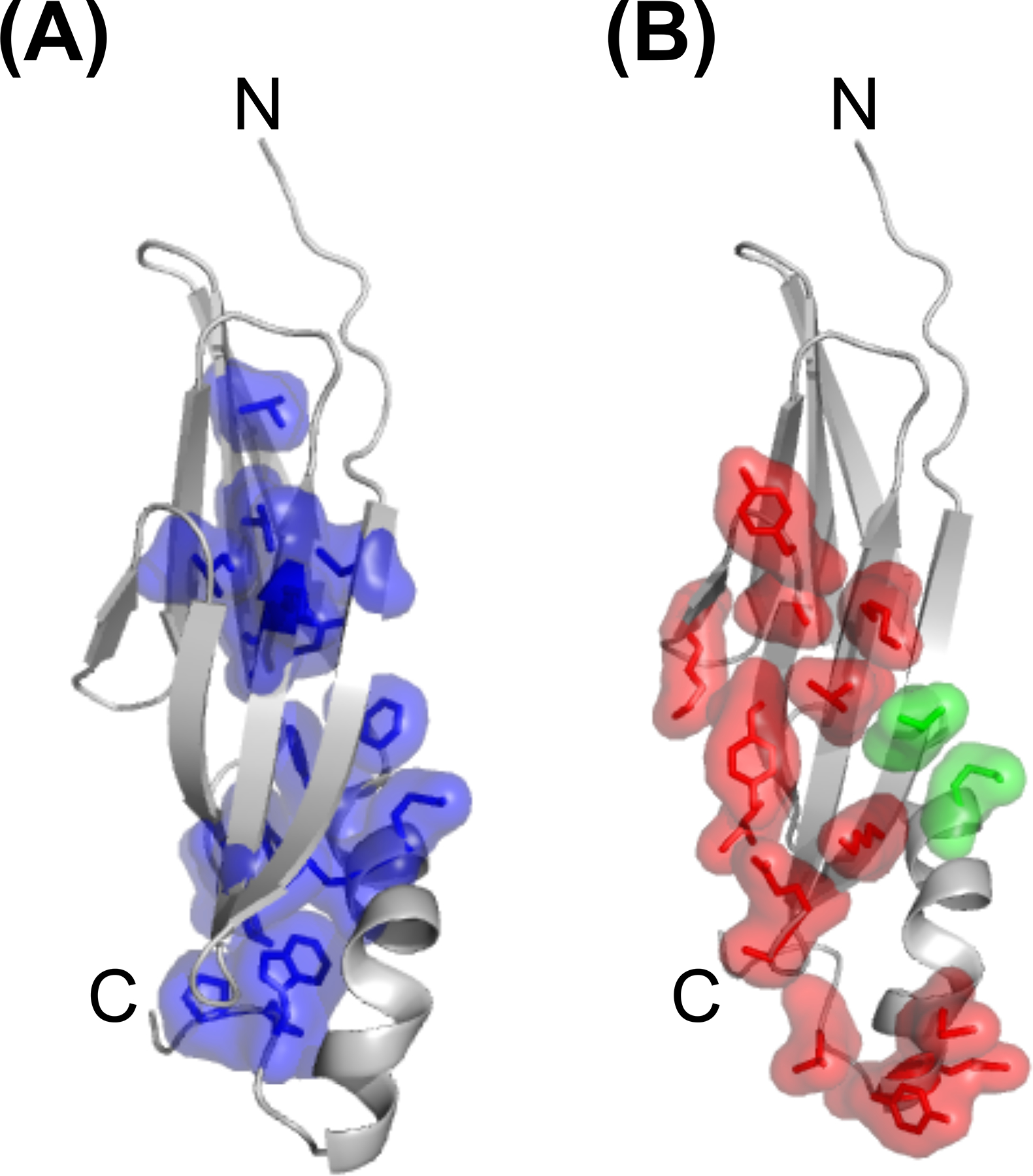
(A) Ribbon representation of liganded Ad2 E3–19K showing that the sidechains of strictly conserved amino-acid residues (purple) are largely involved in forming the hydrophobic core of the E3–19K fold. (B) Ribbon representation of liganded Ad2 E3–19K showing that the sid chains of non-conserved amino-acid residues (red) are solvent-exposed and serve primarily as contact sites with MHC I (see also Fig. 1). Residues Thr14 and Met82 (green) were shown to affect interactions with the MIC proteins [44]. The N- and C-termini of Ad2 E3–19K are labeled.
Immunoprecipitation assays with cells infected by HAdVs from species C (HAdV-C2 and HAdV-C5) or D (HAdV-D19a/-D64) indicated that E3–19K binds to MHC I molecules with different affinities, resulting in differential sensitivities of MHC I downregulation on infected cells [21,48,49]. This interaction between E3–19K and MHC I was later confirmed by an in vitro binding assay using recombinant, soluble E3–19K proteins of species B, C, D and E and HLA-A, HLA-B and HLA-C molecules [41,42]. These studies showed that E3–19K associates with HLA-A and HLA-B but, interestingly, has reduced affinity for HLA-C. The locus-specific downregulation of MHC I is thought to counteract natural killer (NK)-cell cytotoxic responses against HLA-devoid cells, as shown for the HIV-1 negative regulatory factor (Nef) immunomodulatory protein [50], although this remains to be determined.
In addition to interfering with the MHC I antigen presentation pathway, E3–19K was shown to promote intracellular sequestration of the MHC class I-related chain A (MIC A) and MIC B, which are important cell-surface ligands for NK cells to recognize target cells [51]. Alanine scanning mutagenesis of the luminal domain of E3–19K revealed that substitutions of Thr14 and Met82 compromise MIC A and MIC B downregulation with minimal effects on MHC I downregulation [44]. Interestingly, these two E3–19K residues are located outside of the MHC I-binding surface (Figs. 1 and 2B). The possibility that E3–19K interacts directly with both MHC I and the MIC proteins would provide distinct mechanisms for subverting T-cell and NK-cell effector functions. Finally, one report suggests that E3–19K also binds to TAP, which likely prevents TAP from interacting with the MHC I accessory protein tapasin, thus interfering with MHC I assembly [52].
HAdV species that lack an E3–19K protein
HAdVs of species F, which comprises HAdV-F40 and HAdV-F41, are critical pathogens and second only to rotavirus as a cause of acute gastroenteritis in children worldwide [53,54]. A striking feature of HAdVs of species F is that they lack the E3–19K protein. Instead, the E3 region of both HAdV-F40 and HAdV-F41 encodes two proteins, 19.4K and 31.6K, that are conspicuously absent in other species. To date, neither of these proteins have been characterized and no immune-evasion function has been ascribed to these two HAdVs. However, because the 19.4K and 31.6K proteins are expressed by both HAdV-F40 and HAdV-F41, and each protein shares 99% amino acid sequence homology with its counterpart[55–57], it suggests that these unique proteins likely play important roles in species F pathogenesis. Moreover, it is noteworthy that the 19.4K and 31.6K proteins have no homology with E3–19K proteins, suggesting that they may be newly acquired. Another striking feature of species F HAdVs is that they show a narrow tropism for intestinal cells. Given the clinical significance of infections associated with species F HAdVs, there is a pressing need to characterize the 19.4K and 31.6K proteins and to understand whether and how they possess immune modulatory functions. We suggest that species F HAdVs may have adapted to specifically downregulate the cell-surface expression of the MIC A and MIC B molecules. The MIC proteins are constitutively found almost exclusively on intestinal epithelial cells and are recognized by NK cells for the elimination of infected cells in the gut. It is noteworthy that the MIC proteins are the target of other human viruses that evade immune surveillance [58–62].
Much like species F, the highly oncogenic HAdV strain of species A preferentially replicates in the gastrointestinal track and also lack an E3–19K gene. Consequently, HAdV-A12 is unable to retain MHC I molecules in the ER of infected human cells [6]. However, and importantly, HAdV-A12-transformed rodent and human cells showed reduced cell-surface expression of MHC I molecules [10–12], an effect attributed to the regulation of the MHC class I heavy chain gene by the E1A region (see above). Notably, HAdV-A12 expresses the unique E3-encoded 29.4K and 30.7K proteins that share 32% and 38% similarity with the 19.4K and 31.6K proteins of species F, respectively [56]. To date, the functions of the 29.4K and 30.7K proteins are undetermined. Finally, given that HAdV-G52 was cultured from the stool of a patient with gastroenteritis, this new HAdV species deserves future attention [63,64].
49K, RIDα, RIDβ, 6.7K, and 14.7K
The E3–49K protein is uniquely encoded by species D and it is the first species-specific E3 protein that has been functionally characterized [22,23]. During HAdV infection, E3–49K undergoes proteolysis; this results in the secretion of the N-terminal part of its ectodomain, which is referred to as sec49K. Studies showed that sec49K is capable of suppressing activation and effector functions of both T cells and NK cells [23]. Notably, purified sec49K shows some tropism as it binds to lymphoid cell lines and primary leukocytes, but not to fibroblasts or epithelial cells [23]. This cell tropism is likely relevant for the immune function of sec49K in host cells. It remains to be established whether this newly identified function of E3–49K is a prerogative of the species D HAdVs associated with epidemic keratoconjunctivitis, as this would point to a role for E3–49K in the pathogenesis of this ocular disease [23].
HAdVs have also evolved several strategies to prevent apoptotic cell death. At least four E3-encoded proteins are known to contribute to anti-apoptotic functions: RIDα and RIDβ, which together form the RID complex, 6.7K, and 14.7K proteins. The RID complex is known to stimulate the lysosomal degradation of specific death receptors, such as Fas (CD95) [65–67], TRAIL-R1[68,69], and TRAIL-R2[68], thereby allowing HAdV-infected cells to escape FasL- or TRAIL-mediated apoptosis. Further studies showed that a third E3 protein, namely E3–6.7K, was required for the degradation of TRAIL-R2 [8,68]. Finally, the E3–14.7K has been shown to suppress inflammation and TNF-mediated apoptosis [40,70,71], possibly through a mechanism involving the inhibition of NF-κB transcriptional activity [71].
Conclusion and perspectives
Significant progress has been made in the last decades towards the characterization of the gene regions of various HAdVs. In particular, the E3 region has attracted much attention because of its role in the interplay between the virus and the host’s immune system. During infection, a number of E3 proteins are produced to subvert different immune functions and pathways, enabling the virus to establish long-term, possibly life-long, persistence in the host. E3–19K protein is the most studied E3-region gene product for both historical significance and its ability to downregulate MHC I molecules that prevents recognition of infected cells by CTLs. The structures of HAdV-C2 and HAdV-E4 E3–19K bound to HLA-A2 have reconciled many years of research by various laboratories on the characterization of E3–19K. The possibility that E3–19K might directly target the MIC proteins would add another layer of complexity to E3–19K. There remains much to be learnt about the functions of other E3 proteins, in particular the species-specific proteins. The recent characterization of the species D E3–49K protein is an example of a species-specific protein with an intriguing functional activity [23]. Collectively, the characterization of species-specific E3 proteins significantly expands our understanding of the role of E3 proteins in HAdV pathogenicity.
Acknowledgments
We would like to thank Dr. Lenong Li for making the figures, and Dr. Ann Tollefson for many insightful discussions over the years on a variety of topics on Adenoviruses. We are grateful to our colleagues who have provided reagents for our research, in particular Drs. H.-G. Burgert, J. Chodosh, and W. Wold. This work was supported, in whole or in part, by the US National Institute of Allergy and Infectious Diseases grants R01 AI114467, R01 AI108546, and R21 AI135917 (to MB).
Abbreviations
- HAdV
Human adenovirus
- E3
early transcription unit 3
- MHC
major histocompatibility complex
- CTL
cytotoxic T cells
- E1A
early 1A
- TNF
tumor necrosis factor
- TRAIL
tumor necrosis factor-related apoptosis-inducing ligand
- TAP
transporter associated with class I antigen processing
- β2m
beta 2 microglobulin domain
- LMP
latent membrane protein
- FasL
Fas ligand
- NF-κB
nuclear factor kappa B
- RID
receptor internalization and degradation
- ER
endoplasmic reticulum
- HLA
human leukocyte antigen
- NK
natural killer
- Nef
negative regulatory factor
- MIC
MHC class I-related chain
Footnotes
Conflict of interest
The authors declare no conflict of interest.
References
- [1].Green M, Mackey JK, Wold WSM, Rigden P. Thirty-one human adenovirus serotypes (Ad1–Ad31) form five groups (A–E) based upon DNA genome homologies. Virology 1979;93(2):481–492. [DOI] [PubMed] [Google Scholar]
- [2].Wadell G Molecular epidemiology of human adenoviruses. In: Current Topics in Microbiology and Immunology Springer Berlin Heidelberg; 1984.p. 191–220. [DOI] [PubMed] [Google Scholar]
- [3].Wold WSM, Gooding LR. Region E3 of adenovirus: A cassette of genes involved in host immunosurveillance and virus-cell interactions. Virology 1991;184(1):1–8. [DOI] [PubMed] [Google Scholar]
- [4].Vasavada R, Eager KB, Barbanti-Brodano G, Caputo A, Ricciardi RP. Adenovirus type 12 early region 1A proteins repress class I HLA expression in transformed human cells. Proceedings of the National Academy of Sciences 1986;83(14):5257– 5261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Burgert HG, Kvist S. An adenovirus type 2 glycoprotein blocks cell surface expression of human histocompatibility class I antigens. Cell 1985;41(3):987–997. [DOI] [PubMed] [Google Scholar]
- [6].Paabo S, Nilsson T, Peterson PA. Adenoviruses of subgenera B, C, D, and E modulate cell-surface expression of major histocompatibility complex class I antigens. Proceedings of the National Academy of Sciences 1986;83(24):9665–9669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Burgert H, Kvist S. The E3/19K protein of adenovirus type 2 binds to the domains of histocompatibility antigens required for CTL recognition. EMBO Journal 1987;6(7):2019–2026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Lichtenstein D, Toth K, Doronin K, Tollefson A, Wold W. Functions and mechanisms of action of the adenovirus E3 proteins. International Reviews of Immunology 2004;23(1–2):75–111. [DOI] [PubMed] [Google Scholar]
- [9].Nevins JR. Mechanism of activation of early viral transcription by the adenovirus E1A gene product. Cell 1981;26(2):213–220. [DOI] [PubMed] [Google Scholar]
- [10].Schrier P, Bernards R, Vaessen R, Houweling A, van der Eb A. Expression of class I major histocompatibility antigens switched off by highly oncogenic adenovirus 12 in transformed rat cells. Nature 1983;305(5937):771–775. [DOI] [PubMed] [Google Scholar]
- [11].Eager KB, Williams J, Breiding D, Pan S, Knowles B, Appella E, Ricciardi RP. Expression of histocompatibility antigens H-2K, -D, and -L is reduced in adenovirus-12-transformed mouse cells and is restored by interferon gamma. Proceedings of the National Academy of Sciences 1985;82(16):5525–5529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Vaessen R, Houweling A, Israel A, Kourilsky P, van der Eb A. Adenovirus E1A-mediated regulation of class I MHC expression. EMBO Journal 1986;5(2):335–341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Rotem-Yehudar R LMP-associated proteolytic activities and TAP-dependent peptide transport for class 1 MHC molecules are suppressed in cell lines transformed by the highly oncogenic adenovirus 12. Journal of Experimental Medicine 1996;183(2):499–514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Schouten G, van der Eb A, Zantema A. Downregulation of MHC class I expression due to interference with p105-NF kappa B1 processing by Ad12E1A. EMBO Journal 1995;14(7):1498–1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Burgert H, Blusch J. Immunomodulatory functions encoded by the E3 transcription unit of adenoviruses. Virus Genes 2000;21(1–2):13–25. [PubMed] [Google Scholar]
- [16].Tollefson A, Stewart A, Yei S, Saha S, Wold W. The 10,400- and 14,500-dalton proteins encoded by region E3 of adenovirus form a complex and function together to down-regulate the epidermal growth factor receptor. Journal of Virology 1991;65(6):3095–3105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Tollefson AE, Ryerse JS, Scaria A, Hermiston TW, Wold WSM. The E3–11.6-kDa adenovirus death protein (ADP) is re-quired for efficient cell death: Characterization of cells infected with adp mutants. Virology 1996;220(1):152–162. [DOI] [PubMed] [Google Scholar]
- [18].Tollefson A, Scaria A, Hermiston T, Ryerse J, Wold L, Wold W. The adenovirus death protein (E3–11.6K) is required at very late stages of infection for efficient cell lysis and release of adenovirus from infected cells. Journal of Virology 1996;70(4):2296–2306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Windheim M, Hilgendorf A, Burgert HG. Immune evasion by adenovirus E3 proteins: Exploitation of intracellular trafficking pathways. In: Current Topics in Microbiology and Immunology Springer Berlin Heidelberg; 2004.p. 29–85. [DOI] [PubMed] [Google Scholar]
- [20].Frietze KM, Campos SK, Kajon AE. No evidence of a death-like function for species B1 human adenovirus type 3 E3–9K during A549 cell line infection. BMC Research Notes 2012;5(1):429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Deryckere F, Burgert H. Early region 3 of adenovirus type 19 (subgroup D) encodes an HLA-binding protein distinct from that of subgroups B and C. Journal of Virology 1996;70(5):2838–2841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Windheim M, Burgert HG. Characterization of E3/49K, a novel, highly glycosylated E3 protein of the epidemic keratoconjunctivitis-causing adenovirus type 19a. Journal of Virology 2002;76(2):755–766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Windheim M, Southcombe JH, Kremmer E, Chaplin L, Urlaub D, Falk CS, Claus M, Mihm J, Braithwaite M, Dennehy K, Renz H, Sester M, Watzl C, Burgert HG. A unique secreted adenovirus E3 protein binds to the leukocyte common antigen CD45 and modulates leukocyte functions. Proceedings of the National Academy of Sciences 2013;110(50):E4884–E4893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Wang E, Scott M, Ricciardi R. An adenovirus mRNA which encodes a 14,700-Mr protein that maps to the last open reading frame of region E3 is expressed during infection. Journal of Virology 1988;62(4):1456–1459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Hawkins LK, Wold WSM. A 12, 500 MW protein is coded by region E3 of adenovirus. Virology 1992;188(2):486–494. [DOI] [PubMed] [Google Scholar]
- [26].Pääbo S, Weber F, Kämpe O, Schaffner W, Peterson PA. Association between transplantation antigens and a viral membrane protein synthesized from a mammalian expression vector. Cell 1983;33(2):445–453. [DOI] [PubMed] [Google Scholar]
- [27].Cox J, Yewdell J, Eisenlohr L, Johnson P, Bennink. Antigen presentation requires transport of MHC class I molecules from the endoplasmic reticulum. Science 1990;247(4943):715–718. [DOI] [PubMed] [Google Scholar]
- [28].Feuerbach D, Etteldorf S, Ebenau-Jehle C, Abastado J, Madden D, Burgert H. Identification of amino acids within the MHC molecule important for the interaction with the adenovirus protein E3/19K. The Journal of Immunology 1994;153(4):1626–1636. [PubMed] [Google Scholar]
- [29].Gabathuler R, Lévy F, Kvist S. Requirements for the association of adenovirus type 2 E3/19K wild-type and mutant proteins with HLA antigens. Journal of Virology 1990;64(8):3679–3685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Hermiston T, Tripp R, Sparer T, Gooding L, Wold W. Deletion mutation analysis of the adenovirus type 2 E3-gp19K protein: identification of sequences within the endoplasmic reticulum lumenal domain that are required for class I antigen binding and protection from adenovirus-specific cytotoxic T lymphocytes. Journal of Virology 1993;67(9):5289–5298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Pääbo S, Bhat BM, Wold WSM, Peterson PA. A short sequence in the COOH-terminus makes an adenovirus membrane glycoprotein a resident of the endoplasmic reticulum. Cell 1987;50(2):311–317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Cox JH. Retention of adenovirus E19 glycoprotein in the endoplasmic reticulum is essential to its ability to block antigen presentation. Journal of Experimental Medicine 1991;174(6):1629–1637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Nilsson T, Jackson M, Peterson PA. Short cytoplasmic sequences serve as retention signals for transmembrane proteins in the endoplasmic reticulum. Cell 1989;58(4):707–718. [DOI] [PubMed] [Google Scholar]
- [34].Sester M, Ruszics Z, Mackley E, Burgert HG. The transmembrane domain of the adenovirus E3/19K protein acts as an endoplasmic reticulum retention signal and contributes to intracellular sequestration of major histocompatibility complex class I molecules. Journal of Virology 2013;87(11):6104–6117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Flomenberg P, Piaskowski V, Truitt R, Casper J. Human adenovirus-specific CD8+ T-cell responses are not inhibited by E3–19K in the presence of gamma interferon. Journal of Virology 1996;70(9):6314–6322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Tanaka Y, Tevethia SS. Differential effect of adenovirus 2 E3/19K glycoprotein on the expression of H-2Kb and H-2Db class I antigens and H-2Kb- and H-2Db-restricted SV40-specific CTL-mediated lysis. Virology 1988;165(2):357–366. [DOI] [PubMed] [Google Scholar]
- [37].Andersson M, McMichael A, Peterson P. Reduced allorecognition of adenovirus-2 infected cells. The Journal of Immunology 1987;138(11):3960–3966. [PubMed] [Google Scholar]
- [38].Rawle F, Tollefson A, Wold W, Gooding L. Mouse anti-adenovirus cytotoxic T lymphocytes. Inhibition of lysis by E3 gp19K but not E3 14.7K. The Journal of Immunology 1989;143(6):2031–2037. [PubMed] [Google Scholar]
- [39].Burgert HG, Maryanski JL, Kvist S. “E3/19K” protein of adenovirus type 2 inhibits lysis of cytolytic T lymphocytes by blocking cell-surface expression of histocompatibility class I antigens. Proceedings of the National Academy of Sciences 1987;84(5):1356–1360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Burgert HG, Ruzsics Z, Obermeier S, Hilgendorf A, Windheim M, Elsing A. Subversion of host defense mechanisms by adenoviruses. In: Current Topics in Microbiology and Immunology Springer Berlin Heidelberg; 2002.p. 273–318. [DOI] [PubMed] [Google Scholar]
- [41].Liu H, Fu J, Bouvier M. Allele- and locus-specific recognition of class I MHC molecules by the Immunomodulatory E3–19K protein from adenovirus. The Journal of Immunology 2007;178(7):4567–4575. [DOI] [PubMed] [Google Scholar]
- [42].Fu J, Li L, Bouvier M. Adenovirus E3–19K proteins of different serotypes and subgroups have similar, yet distinct, immunomodulatory functions toward major histocompatibility class I molecules. Journal of Biological Chemistry 2011;286(20):17631–17639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Flomenberg P, Szmulewicz J, Gutierrez E, Lupatkin H. Role of the adenovirus E3–19k conserved region in binding major histocompatibility complex class I molecules. Journal of Virology 1992;66(8):4778–4783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Sester M, Koebernick K, Owen D, Ao M, Bromberg Y, May E, Stock E, Andrews L, Groh V, Spies T, Steinle A, Menz B, Burgert HG. Conserved amino acids within the adenovirus 2 E3/19K protein differentially affect downregulation of MHC class I and MICA/B proteins. The Journal of Immunology 2010;184(1):255–267. [DOI] [PubMed] [Google Scholar]
- [45].Fu J, Bouvier M. Determinants of the endoplasmic reticulum (ER) lumenal-domain of the adenovirus serotype 2 E3–19K protein for association with and ER-retention of major histocompatibility complex class I molecules. Molecular Immunology 2011;48(4):532–538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Li L, Muzahim Y, Bouvier M. Crystal structure of adenovirus E3–19K bound to HLA-A2 reveals mechanism for immunomodulation. Nature Structural & Molecular Biology 2012;19(11):1176–1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Li L, Santarsiero BD, Bouvier M. Structure of the adenovirus type 4 (species E) E3–19K/HLA-A2 complex reveals species-specific features in MHC class I recognition. The Journal of Immunology 2016;197(4):1399–1407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Severinsson L, Martens I, Peterson P. Differential association between two human MHC class I antigens and an adenoviral glycoprotein. The Journal of Immunology 1986;137(3):1003–1009. [PubMed] [Google Scholar]
- [49].Körner H, Burgert H. Down-regulation of HLA antigens by the adenovirus type 2 E3/19K protein in a T-lymphoma cell line. Journal of Virology 1994;68(3):1442–1448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Cohen GB, Gandhi RT, Davis DM, Mandelboim O, Chen BK, Strominger JL, Baltimore D. The selective downregulation of class I major histocompatibility complex proteins by HIV-1 protects HIV-infected cells from NK cells. Immunity 1999;10(6):661–671. [DOI] [PubMed] [Google Scholar]
- [51].McSharry BP, Burgert HG, Owen DP, Stanton RJ, Prod’homme V, Sester M, Koebernick K, Groh V, Spies T, Cox S, Little AM, Wang EC, Tomasec P, Wilkinson GW. Adenovirus E3/19K promotes evasion of NK cell recognition by intracellular sequestration of the NKG2D ligands major histocompatibility complex class I chain-related proteins A and B. Journal of Virology 2008;82(9):4585–4594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Bennett E, Bennink J, Yewdell J, Brodsky F. Cutting edge: Adenovirus E19 has two mechanisms for affecting class I MHC expression. The Journal of Immunology 1999;162(9):5049–5052. [PubMed] [Google Scholar]
- [53].Uhnoo I, Wadell G, Svensson L, Johansson M. Importance of enteric adenoviruses 40 and 41 in acute gastroenteritis in infants and young children. Journal of Clinical Microbiology 1984;20(3):365–372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Allard A, Girones R, Juto P, Wadell G. Polymerase chain reaction for detection of adenoviruses in stool samples. Journal of Clinical Microbiology 1990;28(12):2659–2567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Davison AJ, Telford EAR, Watson MS, McBride K, Mautner V. The DNA sequence of adenovirus type 40. Journal of Molecular Biology 1993;234(4):1308–1316. [DOI] [PubMed] [Google Scholar]
- [56].Bailey A, Mautner V. Phylogenetic relationships among adenovirus serotypes. Virology 1994;205(2):438–452. [DOI] [PubMed] [Google Scholar]
- [57].Yeh H, Pieniazek N, Pieniazek D, Luftig R. Genetic organization, size, and complete sequence of early region 3 genes of human adenovirus type 41. Journal of Virology 1996;70(4):2658–2663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Cosman D, Müllberg J, Sutherland CL, Chin W, Armitage R, Fanslow W, Kubin M, Chalupny NJ. ULBPs, novel MHC class I–related molecules, bind to CMV glycoprotein UL16 and stimulate NK cytotoxicity through the NKG2D receptor. Immunity 2001;14(2):123– 133. [DOI] [PubMed] [Google Scholar]
- [59].Wu J, Chalupny NJ, Manley TJ, Riddell SR, Cosman D, Spies T. Intracellular retention of the MHC class I-related chain B ligand of NKG2D by the human cytomegalovirus UL16 glycoprotein. The Journal of Immunology 2003;170(8):4196– 4200. [DOI] [PubMed] [Google Scholar]
- [60].Ashiru O, Bennett NJ, Boyle LH, Thomas M, Trowsdale J, Wills MR. NKG2D ligand MICA is retained in the cis-Golgi apparatus by human cytomegalovirus protein UL142. Journal of Virology 2009;83(23):12345–12354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Schneider CL, Hudson AW. The human herpesvirus-7 (HHV-7) U21 immunoevasin subverts NK-mediated cytoxicity through modulation of MICA and MICB. PLoS Pathogens 2011;7(11):e1002362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Dassa L, Seidel E, Oiknine-Djian E, Yamin R, Wolf DG, Le-Trilling VTK, Mandelboim O. The Human cytomegalovirus protein UL148A downregulates the NK cell-activating ligand MICA to avoid NK cell attack. Journal of Virology 2018;92(17):e00162–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Jones MS, Harrach B, Ganac RD, Gozum MM, Dela Cruz WP, Riedel B, Pan C, Delwart EL, Schnurr DP. New adenovirus species found in a patient presenting with gastroenteritis. Journal of Virology 2007;81(11):5978–5984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].de Jong JC, Osterhaus AD, Jones MS, Harrach B. Human adenovirus type 52: a type 41 in disguise? Journal of Virology 2008;82(7):3809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Elsing A, Burgert HG. The adenovirus E3/10.4K-14.5K proteins down-modulate the apoptosis receptor Fas/Apo-1 by inducing its internalization. Proceedings of the National Academy of Sciences 1998;95(17):10072–10077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Shisler J, Yang C, Walter B, Ware C, Gooding L. The adenovirus E3–10.4K/14.5K complex mediates loss of cell surface Fas (CD95) and resistance to Fas-induced apoptosis. Journal of Virology 1997;71(11):8299–8396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Tollefson AE, Hermiston TW, Lichtenstein DL, Colle CF, Tripp RA, Dimitrov T, Toth K, Wells CE, Doherty PC, Wold WS. Forced degradation of Fas inhibits apoptosis in adenovirus-infected cells. Nature 1998;392(6677):726–730. [DOI] [PubMed] [Google Scholar]
- [68].Benedict CA, Norris PS, Prigozy TI, Bodmer JL, Mahr JA, Garnett CT, Martinon F, Tschopp J, Gooding LR, Ware CF. Three adenovirus E3 proteins cooperate to evade apoptosis by tumor necrosis factor-related apoptosis-inducing ligand receptor-1 and −2. Journal of Biological Chemistry 2000;276(5):3270–3278. [DOI] [PubMed] [Google Scholar]
- [69].Tollefson AE, Toth K, Doronin K, Kuppuswamy M, Doronina OA, Lichtenstein DL, Hermiston TW, Smith CA, Wold WS. Inhibition of TRAIL-induced apoptosis and forced internalization of TRAIL receptor 1 by adenovirus proteins. Journal of Virology 2001;75(19):8875–8887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Horton T, Ranheim T, Aquino L, Kusher D, Saha S, Ware C, Wold WS, Gooding LR. Adenovirus E3 14.7K protein functions in the absence of other adenovirus proteins to protect transfected cells from tumor necrosis factor cytolysis. Journal of Virology 1991;65(5):2629–2639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Carmody RJ, Maguschak K, Chen YH. A novel mechanism of nuclear factor-kappaB regulation by adenoviral protein 14.7K. Immunology 2006;117(2):188–195. [DOI] [PMC free article] [PubMed] [Google Scholar]


