Abstract
Karyotype change and subsequent evolution is triggered by chromosome fusion and rearrangement events, which often occur when telomeres become dysfunctional. Telomeres protect linear chromosome ends from DNA damage responses (DDRs), and telomere dysfunction may result in genome instability. However, the complex chromosome end structures and the other possible consequences of telomere dysfunction have rarely been resolved at the nucleotide level due to the lack of the high-throughput methods needed to analyse these highly repetitive regions. Here we applied long-read sequencing technology to Caenorhabditis elegans survivor lines that emerged after telomere dysfunction. The survivors have preserved traces of DDRs in their genomes and our data revealed that variants generated by telomere dysfunction are accumulated along all chromosomes. The reconstruction of the chromosome end structures through de novo genome assemblies revealed diverse types of telomere damage processing at the nucleotide level. When telomeric repeats were totally eroded by telomere dysfunction, DDRs were mostly terminated by chromosome fusion events. We also partially reconstructed the most complex end structure and its DDR signatures, which would have been accumulated via multiple cell divisions. These finely resolved chromosome end structures suggest possible mechanisms regarding the repair processes after telomere dysfunction, providing insights into chromosome evolution in nature.
INTRODUCTION
DNA damage, such as double-strand breaks (DSBs) are the driving force for structural changes in chromosomes, and damaged telomeres caused by telomere erosion or stochastic loss can be recognised as DSBs at chromosome ends. This telomere dysfunction sometimes extends further into the chromosome, also generating structural changes in subtelomeric regions (1,2). The resulting telomere and subtelomere damage can cause chromosome end-to-end fusion or structural rearrangements at chromosome ends, leading to karyotype evolution (3–5).
Chromosome ends affected by telomere dysfunction are recognised as DSB sites and can be processed by a variety of mechanisms. In the breakage-fusion-bridge (BFB) cycle, damaged chromosome ends fuse after telomeric-repeat deletions, followed by breakage and more fusions during subsequent cell division. Some subtelomeric regions are duplicated with inverted fragments, leading to copy number doubling of those regions, making them hallmarks of the BFB cycle (3,6). Fork Stalling and Template Switching (FoSTeS) is another example, where a replicated DNA strand that is stalled by telomere dysfunction invades a different locus and continues replication to process the damaged chromosome end (2). This process adds diverse fragments from various loci in the genome, so results in odd-number or stepwise copy number variation (CNV) and either the same or the opposite directional replication.
For the repair process to terminate, the DSB sites need protection against DSB recognition. Several pathways that provide this protection include end-to-end fusion and telomerase-mediated or telomerase-independent telomere maintenance, which leave their specific signatures in the genomes. End-to-end fusion permanently conceals the damaged sites, and the fusion sites show specific features at the nucleotide level, such as comprehensive chromosome end losses and connected nonhomologous chromosomes. Telomerase is the major player in most eukaryotic cells that lengthens telomeric repeats and protects chromosome ends to stop the repair process (7). Telomerase-independent telomere maintenance mechanisms, such as alternative lengthening of telomeres (ALT), can also reconstruct the protective chromosome ends by either recombination- or replication-mediated mechanisms (8,9). Indeed, ALT is a major mechanism for lengthening and protecting chromosome end sequences in some species and acts as a backup mechanism for telomerase in various eukaryotes (10,11). For example, in the free-living nematode Caenorhabditis elegans, some rare telomerase mutant worms survive telomere dysfunction by replicating unique sequences, templates for ALT (TALTs), flanked with telomeric repeats to the dysfunctional telomeres. These TALT copies, along with the remaining trace telomeric repeats, serve as a new protective telomere sequence, so copy numbers of TALTs increase dramatically in the worms (12,13). These worms, called ALT survivors, arise within tens of generations after losing their telomerase activity. Furthermore, the survivor lines and their abnormal chromosomes can be stably maintained, so the survivor lines are a reproducible model to examine the consequences of telomere dysfunction in eukaryotes at the single nucleotide level.
Although telomere dysfunction, repair and consequent karyotype evolution have been studied in many species, the fusion sites and the new chromosome end structures at the nucleotide level are resolved rarely. It is because many molecular techniques, such as polymerase chain reaction (PCR)-based, copy number-based and short-read sequencing-based methods are insufficient for resolving the repetitive and complex structure of the chromosome ends (1,2,14,15). Furthermore, because the lack of high-throughput methods has prevented the analysis of genomic regions outside chromosome ends after telomere dysfunction events, genome instability caused by telomere dysfunction has been investigated by copy number changes, rather than genome-wide structural variation and sequence changes (16,17). Long-read sequencing technologies overcome these limitations, as the longer read length allows the resolution of highly repetitive and complex structures at the nucleotide level, and such regions can be covered by single reads. Recent technical advances have opened up opportunities to analyse genome-wide variants and chromosomal rearrangements, as well as chromosome end structures, in a single reaction in organisms with small genomes.
Here, we analysed four C. elegans ALT survivors (ALT1, ALT2, ALT3 and ALT4) using Pacific Biosciences (PacBio) long-read sequencing technology to identify genome-wide variants at the nucleotide level and resolve complex chromosome end structures after telomere dysfunction and repair. We found that ALT survivor lines accumulated thousands of variants with variable numbers, indicating that telomere dysfunction can generate genome instability. Furthermore, the C. elegans ALT survivor lines suffered from different degrees of genome instability, and DNA damage in genomic regions away from telomeric regions was accumulated only after telomere rearrangements were accumulated. Next, by reconstructing 60% of the all chromosome ends at the nucleotide level, we show that the nonhomologous chromosome fusion events after deletions in both telomeric repeats and subtelomeres were a major way to conceal the terminal DSB sites. In addition, BFB cycles were induced when fusions occurred between sister chromatids. Moreover, we show a highly complex telomere structure that was reconstructed using several subtelomeric regions as units, and at least one FoSTeS event for filling a gap between units. Finally, we show that the remaining chromosome ends were stabilised by ALT, and the TALT copies were duplicated with high accuracy and in the same direction. Resolving the DDR consequences after telomere dysfunction in the C. elegans lines studied here will shed light on how chromosome end evolution proceeds in eukaryotes.
MATERIALS AND METHODS
Strain maintenance and accessions
All worms were maintained at 20°C under standard culture conditions. ALT survivors were isolated as previously reported (12), and the trt-1(ok410) allele was used for telomerase mutation. We also used the public PacBio long-read data and genome assemblies for the CB4856, N2 and VC2010 (a descendent of N2) strains. The CB4856 genome, ASM452629v1, was obtained from the NCBI (18); the CB4856 raw read, SRR8599837, from NCBI (downsampled to 2.2 Gb, 260 000 reads using seqtk sample); the VC2010 genome: WS274 (19) was downloaded from WormBase (20,21) (ftp://ftp.wormbase.org/pub/wormbase/releases/WS274/species/c_elegans/PRJEB28388/); the VC2010 raw read, SRR7594465, was from the NCBI (downsampled to 2.2 Gb, 260,000 reads using seqtk sample); and the N2 genome, WBcel235 (ce11), was downloaded from Ensembl (used for depicting chromosome end structures of the ALT3 and ALT4 survivor lines because the long-read-based VC2010 genome assembly lacks telomeric repeats in at least one chromosome). The seqtk tool was installed from https://github.com/lh3/seqtk.
Genomic DNA preparation and whole genome sequencing
ALT survivor worms in mixed stages were collected from 10-cm NGM plates and washed three times with M9 buffer. Worms were lysed with worm lysis buffer (0.2 M NaCl, 0.1 M Tris–HCl (pH 8.5), 50 mM EDTA (pH 8.0), 0.5% SDS) with proteinase K (0.1 mg/ml) at 65°C for 2 h. One volume of phenol/chloroform/isoamyl alcohol (25:24:1) was added and mixed gently for 15 min. The aqueous phase was separated by spinning at 6000 g for 10 min at room temperature and transferred to new tubes. Genomic DNA was precipitated by adding two volumes of 100% ethanol and 0.2 M NaCl and pelleted by centrifugation at 6000 g for 15 min. DNA pellets were washed with 70% ethanol three times and resuspended in water. Macrogen performed library preparation and sequencing steps using the PacBio Single Molecule, Real-Time (SMRT) DNA sequencing technology (platform: PacBio RSII; chemistry: P6-C4).
Genome assembly and polishing
De novo genome assemblies of four ALT survivor strains were generated with ALT1 27×, ALT2 26×, ALT3 28×, ALT4 32× long reads using Canu (version 1.6, genomeSize = 100m minReadLength = 1000 -pacbio-raw filtered_subreads.fastq.gz) (22). The assemblies were corrected with PacBio raw reads to increase base quality as follows: First, PacBio raw fastq files were converted to BAM files using Picard FastqToSam (version 2.18.7, default option) (http://broadinstitute.github.io/picard/), and then the BAM files were aligned to assemblies using Pbalign (version 0.4.1, default option) from the GenomicConsensus package (https://github.com/PacificBiosciences/GenomicConsensus). The aligned BAM files were indexed and converted to SAM files using SAMtools (version 1.9, index for indexing BAM files and view for converting BAM files to SAM files, default option) and pbindex (version 1.0.6, default option) from the GenomicConsensus package (23). The assemblies were also indexed using SAMtools (version 1.9, faidx for indexing fasta files, default option) and were polished using Racon (version 1.4.3, default option) (23,24).
Genome quality assessment
Genome qualities of our ALT assemblies were assessed using Benchmarking Universal Single-Copy Orthologs (BUSCO) scores and repetitive elements. BUSCO analysis (v4.0.6) (25) was installed using bioconda (26) and conducted in the same environment using the nematoda DB (-m genome -l nematoda_odb10) (23). Repetitive elements were identified and masked by RepeatMasker (RepeatMasker -species metazoa -s) (27) with Dfam (version 3.1) (28) and Repbase (version RepBase-20170127) (29,30) libraries and tandem repeat finder (31). We estimated true repeat lengths based on the positional coverage of each repetitive element to avoid assembly errors by repetitive sequences. Raw long reads were mapped to the assemblies by minimap2 (version 2.17-r943-dirty; minimap2 -a -x map-pb) (32) and positional coverages were parsed using SAMtools (samtools depth) (23).
Identification of CNVs, genome-wide variants and translocation
We first mapped genomic long reads of ALT survivor lines to the corresponding reference genomes using minimap2 and sorted and indexed output BAM files using SAMtools (version 1.9; sort -O BAM andindex) (23,32). Then we extracted read-depth counts for every position (samtools depth -a), binned the count 100-kb intervals, and normalised the binned read depths with its interval length and the average whole genome read-depth (23). Because the rightmost interval of the chromosomes is shorter than 100 kb, we defined the two leftmost intervals and three rightmost intervals of every chromosome as subtelomeric regions.
Next, we aligned our four genome assemblies and the PD1074 genome to the CB4856 genome using the MUMmer package (version 4.0.0 beta2; nucmer –maxmatch -l 100 -c 500) and called variants between each pair of assemblies using Assemblytics (33,34). We first compared variants between ALT1 and CB4856 to variants between PD1074 and CB4856, and also compared variants between ALT2 and CB4856 to variants between PD1074 and CB4856. We found overlapped indels between each pair using BEDtools (bedtools intersect -wa -wb), and used these overlapped indels with PD1074 and CB4856 to validate our variant calling process as they came from the starting strain of ALT1 and ALT2 (35). The other variants in ALT1 and ALT2 were also compared using bedtools to find shared variants generated after telomere dysfunction and before ALT activation (35).
To assign templated insertions, we extracted inserted sequences from variant sets of our ALT lines and compared the sequences to their flanking sequences or any genomic sequences. We used 20-bp flanking sequences for ≥5- and <10-bp insertions and 100-bp flanking sequences for ≥10- and <50-bp insertions to find exact full-length matched sequences. For ≥50 bp insertions, we used BLAST to search whole genomes (makeblastdb -input_type fasta -dbtype nucl and blastn -task megablast -outfmt 6) (36).
We then extracted each nucleotide pair of left flank (−4 to +4) and right flank (−4 to +4) position pairs and compared them to test whether deletion junction sites have microhomology. We used ≥10-bp deletions rather than ≥5-bp deletions because positions of shorter deletions may be overlapped with themselves. We counted the number of matched pairs of sequences, and divided the matched number by the total number of all deletions.
To identify possible translocation blocks, we first aligned each ALT genome assembly to the corresponding reference genome (nucmer –maxmatch -l 100 -c 500), assigned only one-to-one matches (delta-filter -1), and extracted responsible one-to-one matched contigs to their reference genome (show-tiling -l 1 -g -1 -i 80.0 -v 1.0 -V 0.0) (33). We then aligned these subsets of ALT genome assemblies to the reference once again with the same commands and merged the one-to-one matched information into large chunks (mummerplot) (33). As these chunks were too fragmented to find translocation blocks, we merged ±50-kb overlapped chunks into larger blocks, and extracted >50-kb translocation blocks which have discontinuous matches compared to the reference genome.
Subtelomere and telomere analysis
We made a list of putative end-containing contigs using blastn from NCBI-BLAST (version 2.7.1+) to identify the structure of chromosome ends at the nucleotide level (36). A BLAST database of four ALT genome assemblies was made by makeblastdb from the NCBI-BLAST package (version 2.7.1+). Query sequences were canonical C. elegans telomeric-repeat sequences (six copies of TTAGGC), full-length sequences of TALT1 and TALT2, and some parts of subtelomeric sequences (∼10, 10–30, 30–50, 50–70, 70–90 kb far from telomeric repeats). Contigs that have at least 50% of the query sequence matches were used, and then we manually reviewed and selected end-containing contigs.
Local re-assembly of chromosome XL in the ALT1 survivor line
We extracted ALT1 read ids mapped to the chromosome XL ends of the reference or ALT1 genomes using SAMtools (samtools view alt1_on_cb4856.bam X:0–78000 and samtools view alt1_on_alt.bam tig00000439) (23). The mapped reads with unique read ids were extracted using seqtk (seqtk subseq), and then the FASTA reads were assembled using Canu with some different parameters (version 1.6; canu genomeSize = 200k minReadLength = 1000 corMhapSensitivity = high corMinCoverage = 0 -pacbio-raw reads_mapped_to_reference.chrXL.fa and canu genomeSize = 300k minReadLength = 1000 corMhapSensitivity = high corMinCoverage = 0 -pacbio-raw reads_mapped_to_reference.chrXL_and_ALT1.chrXL.fa) (22). Finally, we manually merged unique contigs from the two local assemblies.
RESULTS
Experimental design for analysing the consequences of telomere dysfunction
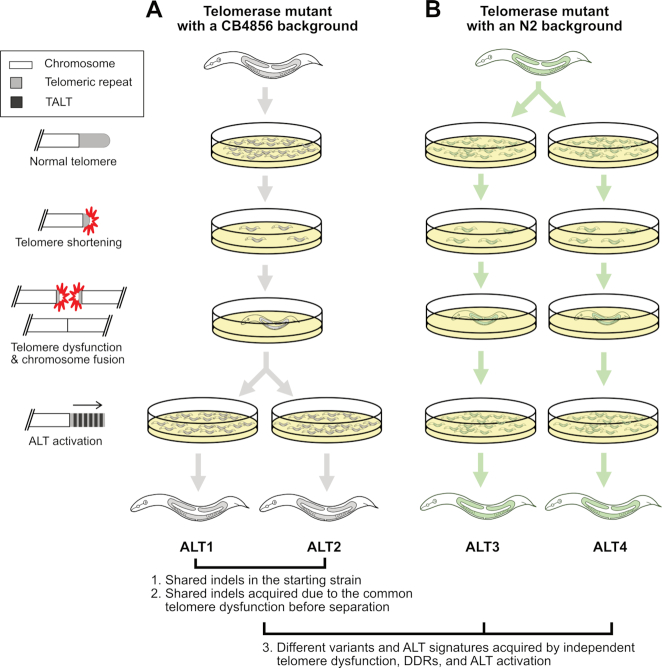
We prepared and sequenced four C. elegans ALT survivor lines using the PacBio RSII platform to examine the consequences of telomere dysfunction at the genome-wide and nucleotide levels. We first prepared two lines as internal controls because they shared known variants so we could verify our variant calling process using long read-based genome assembly. These two lines, ALT1 and ALT2, had been obtained from a common trt-1 telomerase mutant mother whose genetic background is mostly CB4856 except for some N2 background. The N2 background mix-up is due to the process of introducing the trt-1 mutation into the CB4856 strain by outcrossing. The starting trt-1 mutant strain had experienced substantial DDR events due to telomerase loss for several generations, manifested by a severely reduced brood size, then individually separated into tens of plates. A few generations after separation, two survivor lines, ALT1 and ALT2, emerged with independently acquired ALT activation (Figure 1A). Thus the two lines are expected to share both N2-type variants in the CB4856 genetic background and DDR-mediated variants that were generated during telomere dysfunction before line separation. They are also expected to contain independent DDR-mediated variants that were generated after line separation. We confirmed our variant calling process by examining how many N2-type variants are shared between variant sets of ALT1 and ALT2. We then investigated remaining non-N2-type variants to compare patterns of genomic changes generated during telomere dysfunction or just before ALT activation by checking how many non-N2-type variants were shared or independently acquired in ALT1 and ALT2 and where they are located. We used the other two lines, ALT3 and ALT4, obtained from trt-1 mutant animals in the N2 genetic background, to increase the number of replicates for independent telomere dysfunction and DDR, as they were separated before any telomere dysfunction was evoked (Figure 1B).
Figure 1.
The experimental scheme to confirm the variant calling process used in this study and to understand genomic changes after telomere dysfunction using the four Caenorhabditis elegans ALT survivor lines. (A, B) The four ALT survivor lines originated from telomerase mutant worms with two different genetic backgrounds, CB4856 and N2. The left panel represents sequential events of telomere shortening, telomere dysfunction and chromosome fusion, then ALT activation. (A) ALT1 and ALT2 originated from a telomerase mutant with a CB4856 genetic background and share some portion of N2-type variants. Their common ancestor had experienced some level of same telomere damages and were separated before new ALT-mediated chromosome end structures were stably maintained. These features gave us an opportunity to use the shared N2-type variants as internal controls to validate our genome assembly-based variant calling analysis and to investigate patterns of genomic changes generated by telomere dysfunction. (B) ALT3 and ALT4 originated from a telomerase mutant with an N2 genetic background that were separated when all telomeres were intact and were used to increase the number of replicates for understanding telomere dysfunction and ALT activation events.
Quality assessment of four ALT de novo genome assemblies
We used genome assembly to identify genomic variants between ALT lines and their respective reference genomes as our PacBio raw reads had relatively high error rates (∼5%), and first assessed qualities of our de novo genome assemblies of the 4 ALT survivor lines. We obtained a total of 26–32× long reads for each strain (N50 = 8–11 kb; Supplementary Figure S1), and most of the reads were mapped to the reference genomes with mapping quality ≥5 (86.6%, 82.4%, 89.5% and 82.6% for ALT1, ALT2, ALT3 and ALT4 survivor lines, respectively). We assembled these long reads of the four ALT survivor lines into contigs (N50: 236–395 kb; longest contig length: 1.3–4.6 Mb; Supplementary Table S1).
We then assessed and compared qualities of these four de novo genome assemblies using the Benchmarking Universal Single-Copy Orthologs (BUSCO) analysis and ratios of assembled repeat lengths to the total repeat lengths. BUSCO uses the degree of fragmentation of known single-copy ortholog genes to compare qualities of genome assemblies. Because genome assemblies with poorer qualities have shorter and more fragmented contigs, they also have lower BUSCO values. The four ALT genome assemblies had similar BUSCO values, as about 80% of the genes are complete single-copy and ∼20% are missing (Supplementary Figure S2A). Their complete single-copy BUSCO values were lower than 99% complete single-copy orthologs in reference genomes, suggesting that some ALT genomic regions may be somewhat fragmented, but still most regions were properly assembled and suitable to analyse the genomes at the contig level.
In addition, many repetitive sequences are difficult to assemble and can be missing in genome assemblies, so assembled repeat lengths can be shorter than the real repeat lengths. The real repeat length can be measured and estimated using the raw read depth as unassembled repeat reads can still be mapped to the corresponding repetitive sequences in the genome assembly. Thus we can estimate the true repeat length by mapping all raw reads to the genome assembly, measuring the raw read depth of the repetitive sequences and normalising it with the average read depth of the whole genome sequences. Following this logic, we counted the lengths of assembled repeats and also measured and estimated the total repeat lengths in the four ALT assemblies. All of these lengths were comparable to those of the reference genomes, except for the ribosomal RNA (rRNA) locus, verifying our assembly qualities (Supplementary Figure S2B and Supplementary Tables S2 and S3). The measured rRNA length of ALT1 decreased to half and that of ALT4 increased 4–5 times than those of the reference genomes.
Validation of variant calling process based on genome assembly
We then assessed our assembly-based variant calling process by testing how many internal control variants, the N2-type variants, were properly detected from the ALT1 and ALT2 genome assemblies. We called ≥5-bp insertions and deletions (indels) in ALT1 and ALT2 genome assemblies against the CB4856 genome and compared these variant sets to the N2 variant set obtained by comparing N2 and CB4856 genomes. We found that the ALT1 and ALT2 variant sets shared 506 and 391 insertions with the N2 variant set respectively, and 303 of the shared insertions were also shared by ALT1 and ALT2 lines, with identical insertion sites. We also found that the ALT1 and ALT2 variant sets shared 622 and 500 deletions with the N2 variant set respectively, and 420 of those were shared between ALT1 and ALT2 variant sets (Supplementary Table S4). Therefore, we can conclude that high portions, though not all, of shared N2-type variants in ALT1 and ALT2 were correctly called, validating our assembly-based variant calling process.
Most of indels generated by telomere dysfunction were not shared by ALT1 and ALT2
After excluding the N2-type variants from the ALT1 and ALT2 variant sets, we analysed remaining indels, which were probably produced by telomere dysfunction rather than outcrossing process, to compare degrees of genome instability before and after line separation. These two lines were separated after telomere dysfunction and before ALT activation, so shared indels would be responsible for early genome instability events after telomere dysfunction and different indels would be responsible for late genome instability events by telomere dysfunction before or after ALT activation. We found that ALT1 and ALT2 genome assemblies contained ∼6% overlapped indels only (Supplementary Table S5), suggesting that the majority of indels were acquired by the late genome instability caused by telomere dysfunction after line separation. This scarcity of shared indels implies that telomere dysfunction can generate telomere damage and also genome-wide instability, but that they might be induced at different time points, as ALT1 and ALT2 shared almost all telomere damage and chromosome fusion events (see below).
CNVs, templated insertions, simple deletions with microhomology and large translocations in the 4 ALT survivor lines
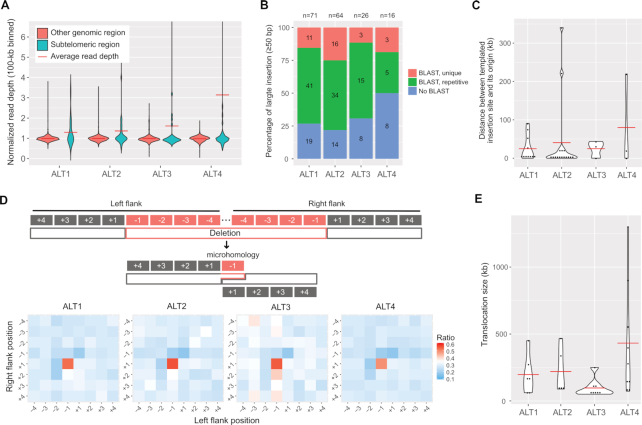
We comprehensively investigated characteristics of the variants detected by long-read sequencing data and our assembly-based indel calling process. First, we compared CNVs between subtelomeric regions and other genomic regions in the four ALT survivor lines and found that the average read depths of subtelomeric regions were greater than other genomic regions where average read depths were nearly one (Figure 2A), indicating that subtelomeric regions are susceptible to copy number changes. In contrast, indels were not enriched in the subtelomeric regions (Supplementary Figure S3A and B), so CNVs and indels may have been produced through different mechanisms.
Figure 2.
Genomic changes in the four ALT survivor lines and characteristics of CNVs, insertions, deletions and translocations. (A) Violin plots representing CNV distributions in subtelomeric regions and other genomic regions are shown. Read depths were merged in 100-kb intervals and normalised by the average whole genome depths. Subtelomeric regions were defined as 200-kb regions from each chromosomal end. (B) Ratios of templated insertions to the insertions ≥50 bp. Blue bars represent insertions with no BLAST results, green bars represent those of repetitive BLAST results, and red bars represent those of unique BLAST results. The numbers in the bars represent the actual number of large insertions, and those on top of the graph represent the total number of large insertions. (C) Size distribution of distance between a templated insertion site and its origin is shown. Red horizontal bar represents the average distance. (D) Microhomology between deletion junction sites. Top: Schematic representation of possible polymerase theta-mediated end joining (TMEJ). Typical microhomology between left flank −1 and right flank +1 positions can be achieved by TMEJ. Bottom: Heatmaps represent the ratio of the same sequences between each position pair in all ≥10-bp deletions. (E) Size distribution of >50-kb translocation. Red horizontal line represents the average size of each line.
All ALT lines contained >2000 indels (≥5 bp) of which the majority of these insertions (>97%) and deletions (>64%) were not longer than 200 bp, and the ALT4 line had the largest number of indels, 3670 indels (Supplementary Table S6). We then thoroughly examined characteristics of sequences near indels to infer possible mechanisms that generated indels, such as random insertion, which is mainly observed in nonhomologous end joining or templated insertion, which can be achieved by polymerase theta-mediated end joining (TMEJ). We first analysed whether inserted fragments had similar or same sequences near themselves to test if they were duplicated using other genomic regions as templates. We found that 12–15% of insertions had exactly full-matched sequences to their close flanking sequences (20-bp flank for 5–9 bp insertions, and 100-bp flank for 10–49 bp insertions; Supplementary Figure S3C). These small insertions, however, can have the same sequences by chance in some genomic regions, so we cannot extend search areas beyond close flanking sequences. We thus used large insertions with a size of 50 bp or longer to find their origins along whole genomes using BLAST and found that 50–80% of the large insertions had at least one BLAST result somewhere in the reference genomes (Figure 2B). Among these large templated insertions, we can calculate the distances between their original sites and insertion sites for unique templates whose insertion sites were on the same chromosomes. The templates were mainly located near the insertion sites, but some portions were located as far as several hundreds of kilobases (Figure 2C). Our data suggest that large insertions generated after telomere dysfunction would mainly be templated insertions, which is a signature of TMEJ.
Next, we examined whether deletions have microhomology in junction sites, which is another evidence of TMEJ. If polymerase theta was a major mechanism to repair DSBs, then some deleted sequences on one side and remaining sequences on the other side should have microhomology, as these sticky homologous ends can be annealed and then connected (Figure 2D). We found significant enrichment of homology in the left flank −1, right flank +1 positions (Figure 2D), which is consistent with the previous study showing that TMEJ in C. elegans mainly uses 1-bp microhomology (37).
We also had an opportunity to explore possible translocation events as our genome assemblies had sufficiently long contigs to identify translocated fragments even longer than 50 kb. We found 28 translocated fragments, 22 of which were inter-chromosomal translocations (Supplementary Table S7). Our 4 ALT lines showed at least five large translocated fragments that ranged 50–1300 kb in size, and ALT4, specifically, had remarkably larger translocated fragments than other lines, including the three fragments of which sizes were 550 kb, 900 kb and 1.3 Mb, respectively (Figure 2E).
All of these lines of evidence suggest that telomere dysfunction may not be restricted to telomeres, but lead to whole genome instability and that the genome instability might be distinct from direct telomere damage in terms of the time of action.
Subtelomere deletions in chromosome fusion and break sites
Our genome assemblies of the four ALT survivor lines gave us an opportunity to understand telomere dysfunction and end repair at the nucleotide level. We first labelled putative end-containing contigs using end-specific sequences, such as telomeric repeats, TALT copies and subtelomeric sequences. We recovered only 30 end-containing contigs (30 out of 48 ends; Supplementary Table S8) because imperfect assembly quality and the repetitive nature of subtelomeric regions restricted the full reconstruction and labelling of chromosome ends. The ALT1 assembly had nine end-containing ends out of 12 chromosome ends in the assembly, the ALT2 assembly had eight, the ALT3 assembly had seven, and the ALT4 assembly had just six ends. Among the ends, 6, 6, 6 and 2 ends in ALT1, ALT2, ALT3 and ALT4, respectively, were fused (20/30; Figure 3), and the other 3, 2, 1 and 4 ends had TALT-containing end structures with no evidence of fusion (10/30; Figures 4 and 5). Intriguingly, ALT1 and ALT2 contained fusion sites that are identical at the single nucleotide level, which validated that our assemblies are suitable for analysing chromosome end structures. In addition, this also implies that ALT1 and ALT2 were indeed separated after severe telomere dysfunction, which had led to chromosome fusion events.
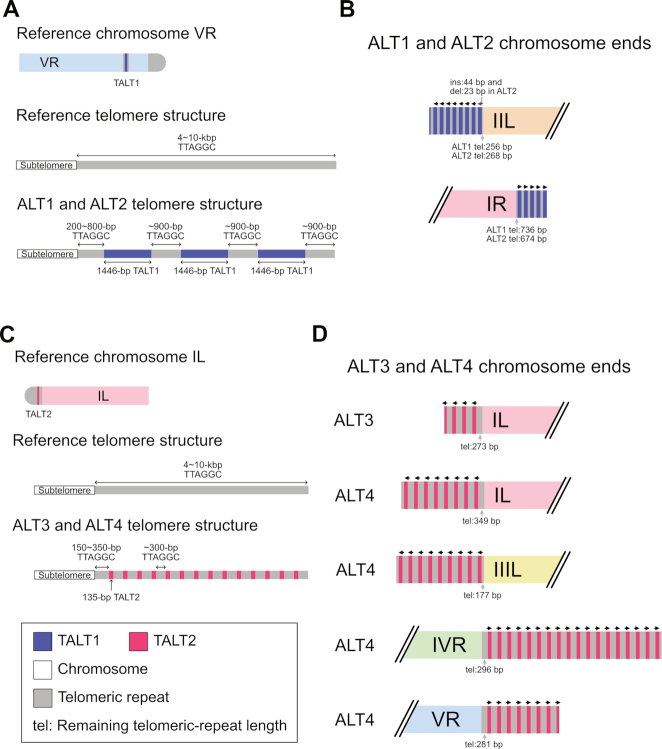
Figure 3.
A schematic representation of chromosome end structures (not to scale) is shown. (A–D) Simple fusion between nonhomologous chromosomes. (A, B) Inter-chromosomal fusion sites of the Caenorhabditis elegans ALT1 and ALT2 survivor lines. References represent CB4856 chromosomes. (A) Chromosome IL and IIR ends were fused to each other after a 50 644-bp deletion in the chromosome IL subtelomeric region and a 6387-bp deletion in the chromosome IIR subtelomeric region. (B) Chromosome VL and XR ends were fused to each other after a 156-bp deletion in the chromosome VL subtelomeric region and a 255-bp deletion in the chromosome XR subtelomeric region. (C) Inter-chromosomal fusion sites of the ALT3 survivor line. References represent N2 chromosomes. Chromosome IVL and XR ends were fused to each other after a 29,935-bp deletion in the chromosome IVL subtelomeric region and a 306-bp deletion in the chromosome XR subtelomeric region. (D) Inter-chromosomal fusion sites of the ALT4 survivor line. References represent N2 chromosomes. Chromosome IVL and VL ends were fused to each other after a 41,965-bp deletion in the chromosome IVL subtelomeric region and a 109 516-bp deletion in the chromosome VL subtelomeric region. (E–H) Complex inter-chromosomal fusion with discontinuous fragments. (E) The fusion site of chromosome IIIL and IVR ends in the Caenorhabditis elegans ALT1 survivor line contained a discontinuous fragment, which would have originated from the chromosome IVR end, but was located in the opposite direction. The origin of the discontinuous fragment was supposed to be chromosome IV:17 975 510–17 982 596 in the reference CB4856 genome. The fused chromosome IIIL end had a 1557-bp subtelomere deletion, and the chromosome IVR end had a 653-bp subtelomere deletion. References represent corresponding CB4856 chromosomes. (F) The fusion site of chromosome IIIL and IVR ends in the ALT2 survivor line contained not just the same discontinuous IVR fragment in the ALT1 survivor line, but also another discontinuous fragment, which originated from the chromosome IIIL end and was located in the same direction. The origin of the discontinuous IIIL fragment was supposed to be chromosome III:4679–8126 in the reference CB4856 genome. The fused chromosome IVR end had almost the same subtelomere deletion, but the chromosome IIIL end had a much shorter 650-bp subtelomere deletion than that of the ALT1 survivor line. References represent the corresponding CB4856 chromosomes. (G) The fusion site of chromosome IIR and VL ends in the ALT3 survivor line contained a discontinuous fragment, which would have originated from the chromosome VL end, but was located in the opposite direction. The origin of the discontinuous fragment was supposed to be chromosome IV:655–983 in the reference N2 genome. The fused chromosome IIR end had a 5282-bp subtelomere deletion, and the chromosome VL end had a 1086-bp subtelomere deletion. References represent the corresponding N2 chromosomes. (H) The fusion site of chromosome IIIL and VR ends in the ALT4 line. A discontinuous fragment would have originated from chromosome III:865–2999 in the reference N2 genome and was located in the opposite direction. The fused chromosome IIIL end had a 1640-bp subtelomere deletion, and the chromosome VR end had a 1249-bp subtelomere deletion. References represent corresponding N2 chromosomes.
Figure 4.
Schematic representation of chromosome end structures that were reconstructed by one-way replication of TALT (Template for ALT) and telomeric repeats (not to scale). All subtelomeres were intact and short telomeric repeats remained at the junctions between a subtelomeric region and a TALT copy. (A) The origin of the TALT1 (upper), typical telomere structure in the reference genome (middle) and new telomere structure of the Caenorhabditis elegans ALT1 and ALT2 survivor lines (lower). The original TALT1 (blue bar) is located in the right arm of chromosome V and consists of a 1446-bp genomic sequence flanked by short telomeric repeats (grey bars) (B) Assembled and confirmed TALT1-mediated chromosome end structures in the ALT1 and ALT2 lines. (C) The origin of the TALT2 (upper), typical telomere structure in the reference genome (middle) and new telomere structure of the ALT3 and ALT4 survivor lines (lower). The original TALT2 (red bar) is located in the far left subtelomeric region of chromosome I and consists of a 135-bp genomic sequence flanked by short telomeric repeats (gray bars). (D) Assembled and confirmed TALT2-mediated chromosome end structures in the ALT3 and ALT4 survivor lines.
Figure 5.
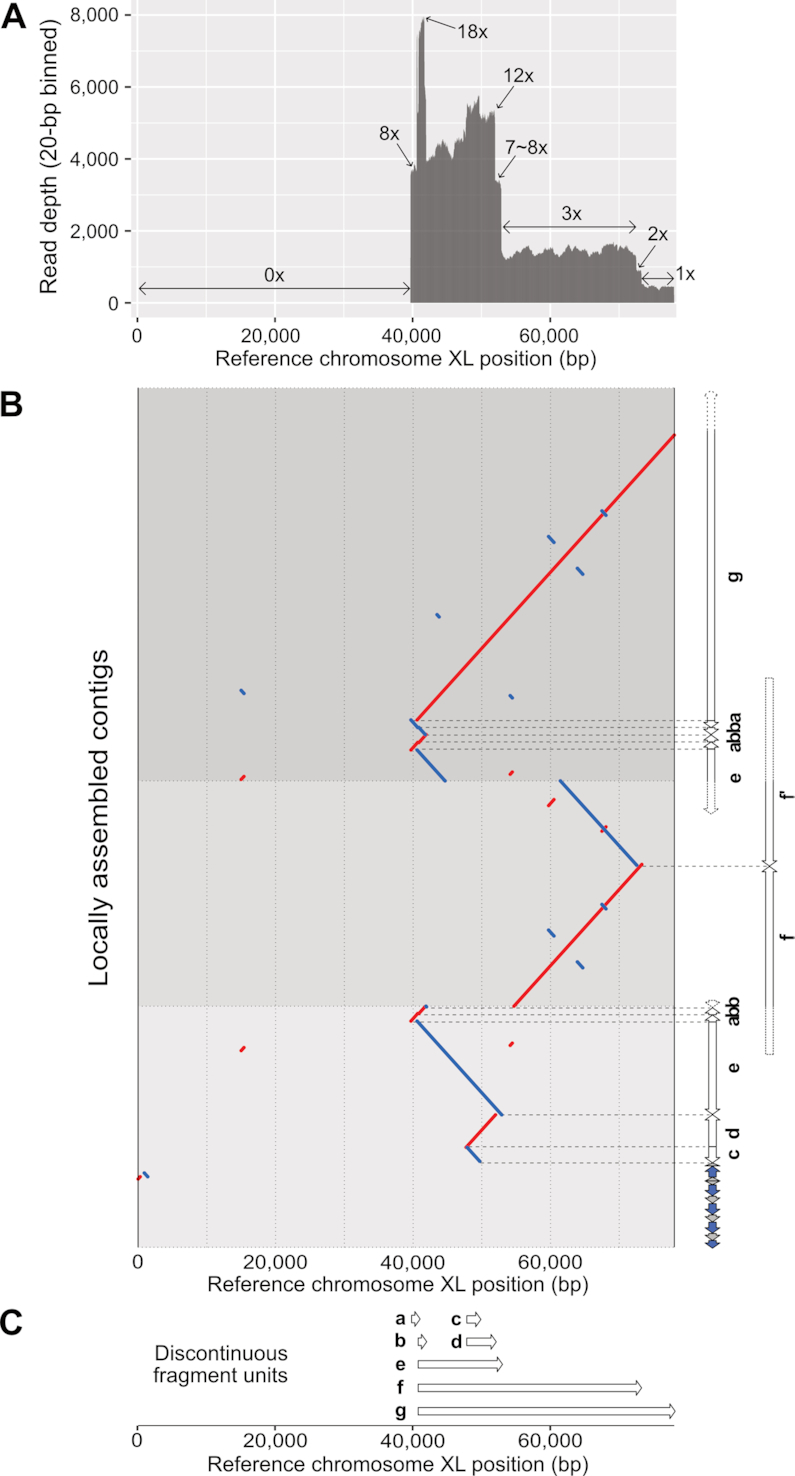
Complex chromosomal rearrangement events found in the chromosome XL end of the Caenorhabditis elegans ALT1 survivor line. (A) Complex and stepwise read-depth changes in the chromosome XL end of the ALT1. The x-axis represents reference CB4856 chromosome XL positions and the y-axis represents average read depths of raw ALT1 reads (20-bp binned). (B) Dot plot representing alignment between the reference chromosome XL end (X:0–78 000) and locally, manually re-assembled three contigs of the ALT survivor line. Each contig was aligned to the reference CB4856 genome, and units were identified and classified based on their alignments (white arrows, units a–f). Different background colours represent different contigs. Red: forward strand matches; blue: reverse strand matches; horizontal grey dots: boundaries of each contig; horizontal dashed grey lines: boundaries of each unit; blue arrows: TALT; gray arrows: telomeric repeats. (C) The list of discontinuous fragment units and genomic positions where they originated based on the reference CB4856 genome. White arrows are placed as their reference positions (a: 39 683–40 727, b: 40 833–41 912, c: 47 882–49 705, d: 47 420–51 990, e: 40 591–52 899, f: from 40 kb to 72 825, g: from 40 591 to the end of chromosome X).
Next, we resolved end-loss patterns and fusion sites in the end-containing contigs at the nucleotide level. All 30 ends exhibited telomeric-repeat deletions and 21 out of 30 contigs exhibited additional subtelomere deletion (Figure 3) in which the subtelomere deletion sizes varied from 156 to 109 516 bp (<1 kb: 8 contigs, 38%; 1–7 kb: 7 contigs, 33%; >10 kb: 6 contigs, 29%, Figures 3 and 5 and Supplementary Figure S4A). Interestingly, in all chromosome fusion sites, every chromosome end had deletions in telomeric repeats and even in subtelomeric sequences (20/20) and was concealed by fusion between nonhomologous chromosomes (Figure 3 and Supplementary Tables S9 and S10). Specifically, ALT1 and ALT2 lines, which were separated after the same telomere dysfunction and DDR, also shared the same inter-chromosomal fusion sites between chromosome IIR and IL (reference position: chromosome IIR 15 803 621 and chromosome IL 51 076; Figure 3A) and chromosome XR and VL (reference position: chromosome XR 18 070 421 and chromosome VL 3053; Figure 3B). Independently generated ALT3 and ALT4 lines had an independent and distinct fusion pattern in chromosome IVL, where the chromosome IVL was fused with chromosome XR in ALT3 (reference position: chromosome IVL 30 078 and chromosome XR 17 718 493; Figure 3C), but with chromosome VL in ALT4 (chromosome IVL: 42 114 and chromosome VL 109 843; Figure 3D).
Twelve out of 20 fused ends exhibited simple fusion between two subtelomere-deleted nonhomologous chromosomes, but the other eight fused ends had discontinuous fragments, such as partial inverted duplication (Figure 3E–H and Supplementary Table S10). The chromosome IVR and IIIL fusion site in ALT1 contained an additional inverted duplication of the subtelomeric region in chromosome IVR, and this inverted fragment had short, ∼150 bp-long, telomeric repeats (chromosome IVR 17 981 794, inverted chromosome IVR fragment [17 982 596–17 975 510] and IIIL 4679) (Figure 3E). ALT2 shared a similar chromosome fusion between the two chromosomes, but one additional discontinuous fragment from chromosome IIIL was inserted in the same direction in the fused chromosome IIIL (chromosome VR 17 981 794, inverted chromosome IVR fragment [17 982 596–17 975 510], chromosome IIIL fragment [4679–8126] and chromosome IIIL 3771) (Figure 3F).
The discontinuous fragments were found only in six out of 21 subtelomere-deleted ends, not in the ends with intact subtelomere (zero out of nine ends). Their sizes varied from 333 to 7110 bp, but most of them were 2–7 kb (Supplementary Figure S4B). The fragments were aligned to their subtelomere origins >99.9% in length and >98% identity, indicating that BFB cycles, which produce duplicated subtelomeric regions, may have inserted the fragments in the fusion sites, rather than microhomology-mediated and error-prone replication (Figure 3E–H and Supplementary Table S11). In addition, short inserted sequences, a signature of nonhomologous end joining, were found in fusion sites of chromosome IIR and chromosome VL in ALT3, as well as in the inverted duplicated fragment of chromosome IVL in ALT1 and ALT2, further supporting the possibility that BFB cycles generated discontinuous fragments (Supplementary Table S10).
One-way replication of TALTs in ALT-mediated chromosome ends
The 10 end-containing contigs that did not exhibit inter-chromosomal fusion had new chromosome end structures constituted by TALT copies and short telomeric repeats, which suggests a possible role of telomeric repeats in TALT replication (Figures 4 and 5). Subtelomere deletion was not observed in nine out of 10 contigs, and their telomeric repeats were shortened, but still remained in 200–800 bp, which is one tenth of the estimated telomeric-repeat length of 4–10 kb (38). The new chromosome ends of ALT1 and ALT2 were composed of 1.4-kb TALT1 (Figure 4A and B) and those of ALT3 and ALT4 were composed of 135-bp TALT2 (Figure 4C and D), as previously reported (12), and these TALT copies were flanked with similar-length canonical telomeric repeats (∼900 bp for TALT1 and ∼300 bp for TALT2; Figure 4B and D). This unit structure of the new chromosome ends, each TALT with flanking telomeric repeats at its both ends, was identical to that of the original TALTs, which also have flanking canonical or degenerated telomeric repeats and are located in internal chromosomal regions (Figure 4A and C). Moreover, the direction of all these TALT copies and their flanking canonical and degenerated telomeric repeats was always the same, except only one TALT on the XL end of ALT1: from the internal chromosomal region to the end (9/10 at the very first TALT of ends, 71/72 at all the TALTs found in end contigs; Figure 4B and D).
Complex chromosomal rearrangements in chromosome XL of ALT1
In this study, all the subtelomere-deleted ends in the identified chromosome ends underwent inter-chromosomal fusion, and all telomeric-repeat-remaining ends resulted in TALT-mediated telomeres. One exception was the chromosome XL of ALT1, which has extreme rearrangements among the end-containing contigs. The chromosome XL of ALT1 had a subtelomere deletion, but was covered with TALT copies and telomeric repeats. This chromosome was also the only end that has an inverted TALT copy and telomeric repeats at the beginning of the TALT-mediated telomere. We used the ALT1 reads mapped to chromosome XL of the reference genome to understand the cryptic end structure in detail and to avoid errors from possible misassembly and low contiguity in the ALT1 assembly. Their mapping pattern showed that 40-kb subtelomere deletion occurred and the read depth changed 18×, 8×, 12×, 8×, 3×, 2× and 1× along the 30-kb region of the remaining subtelomere (Figure 5A). This complex read-depth change and odd-number CNV are representative characteristics of FoSTeS, and the current state-of-the-art assemblers have limited ability to deal with such complex structures, so we locally and manually re-assembled the subtelomere structure using the reads mapped on chromosome XL of ALT1 and the reference genomes.
We partially, but more finely, assembled the structure and confirmed that high read-depth regions in the reference chromosome XL were resolved as repetitively duplicated fragments in several contigs (Figure 5B), and that the end can be partitioned into some repetitive units, which have unique start and end positions (Figure 5C, single arrows a–g). The start and end positions of the repetitive units overlapped with the regions of complex read-depth changes, suggesting that the repetitive units were used for replication and were subsequently recovered in our locally assembled subtelomere (Supplementary Table S12). The repetitive units were mainly replicated by inverted duplication, but when the unit a and b were connected as a-b-b or a-b-b-a, duplications were not in an inverted manner, but in the same direction, which cannot be generated by the BFB cycle. In addition, units a and b had 66-bp homology sequences at the connected region (Supplementary Figure S5A–C), and two copies of unit f, denoted as f and f′ that were connected in the opposite direction, also contained 110-bp homology sequences in their junction site (Supplementary Figure S5D-F). This subsequent length of homology sequences is another characteristic of FoSTeS; therefore, we speculate that at least one or two FoSTeS events may have occurred and partially contributed to reconstructing this subtelomere. In contrast, other duplications contained very short homology or insertions at the junction sites, suggesting that nonhomologous end joining was mainly used to connect remaining units.
DISCUSSION
Chromosome ends are susceptible to karyotype evolution. Telomeres provide a protective structure at chromosome ends but can suffer from gradual telomeric-repeat shortening or stochastic telomere deletion and following DDR may fuse the ends resulting in a novel karyotype. For example, the haploid chromosome number of humans contains one fewer than that of their close relative apes (39–41). Some wild mice exhibit a 20%–50% reduction in haploid chromosome numbers compared to that of the laboratory standard mouse (42–44), and these karyotype reduction events are likely the consequences of chromosome fusions between two acrocentric chromosomes (45). However, because their ancestral genome sequences were not available, it was difficult to elucidate how telomere dysfunction was terminated, for example, which DDR was involved or how fusions occurred. In this report, we used ALT survivor lines in C. elegans as a model to dissect the consequences of telomere dysfunction and karyotype evolution. Our ALT survivor lines had overcome telomere dysfunction using several DDR and ALT mechanisms, such as reconstructing TALT-mediated telomeres and chromosome fusion events, which result in fewer chromosome numbers than their ancestral lines (12). Because their ancestral lines were fully sequenced, including the chromosome ends (18,19), we applied a long-read sequencing technology to the ALT survivor lines to understand the molecular consequences of telomere dysfunction by comparing whole genome sequences of the ancestral and ALT survivor lines. We independently assembled genomes of two different ALT survivor lines that share common fusion and breakage events to validate our long-read sequencing-based methodology. Their genome assemblies share the same fusion and breakage sites at the single nucleotide level, suggesting that even genome assemblies with moderate sequencing depths can resolve genomic changes after karyotype evolution.
The high-resolution maps drawn using long-read sequencing of ALT survivor lines enabled a detailed examination of chromosomes at the single nucleotide level. Analysis of CNVs and genome-wide variants generated after telomere dysfunction suggested that DNA damage caused by telomere dysfunction is not limited to the chromosome ends, but also spreads to other chromosomal regions to generate subtle genome-wide instability. In addition, de novo genome assemblies of four ALT survivor lines depicted chromosome fusion and breakage sites and new end structures at the nucleotide level and allowed us to model these sites as specific types of traces that DDR leave behind. Thus, our long-read sequenced ALT survivor lines can serve as reproducible resources to investigate telomere dysfunction and DDR, and can model karyotype evolution events at the nucleotide level.
Telomere dysfunction causes telomere damage and genome instability at different time points
Several types of variants generated after telomere dysfunction were widespread throughout the genome of C. elegans ALT survivor lines, indicating that telomere dysfunction can generate a subtle level of genome instability in addition to telomere-specific DNA damage. Intriguingly, ALT1 and ALT2, which had commonly experienced severe telomere damage events, including chromosome fusions, shared only a small portion of indels generated after telomere dysfunction. In other words, DNA damage in other genomic regions away from telomeric regions was accumulated only after telomere rearrangements accumulated. Our finding that there is a distinct time lapse between telomere rearrangements and chromosomal damage caused by telomere dysfunction is both unprecedented and unexpected, partly because genome instability generated by telomere dysfunction has rarely been studied at the genome-wide and nucleotide levels as well as in a time series.
This time lapse phenomenon, unless only specific to ALT1 and ALT2 survivors, suggests that either it may require some time for DNA damage caused by telomere dysfunction to spread from telomeric regions to other genomic regions, or that genome-wide accumulation of indels may start just before ALT activation as ALT turn-on needs adaptation to DNA damage. In yeasts, severe telomere dysfunction results in delayed cell cycle and cellular senescence, and this cellular senescence can be overcome by adaptation to DNA damage that allows cells to bypass the cell cycle arrest and increases mutation accumulation (46). Although we cannot directly test these hypotheses because our experimental design does not contain any other lines that were separated with tight time intervals, the increased mutation rates in our lines may represent the adaptation to DNA damage, similar to the yeast, as these lines were separated before serious cellular senescence.
TMEJ is a main repair mechanism for the genome-wide DSBs generated by telomere dysfunction
The genome-wide DSBs are thought to be repaired mainly by TMEJ because they have known TMEJ signatures. TMEJ repairs DSBs by replication fork stalling, but this repair is error-prone, resulting in short-length indel generation (37). In addition to these short-length indels, TMEJ exhibits other characteristics, including microhomology between two DSB ends to connect them and templated insertions, which replicate other genomic sequences into the deletion sites (37). All of these signatures were found in the majority of our indels; therefore, the genome instability and DSBs generated after telomere dysfunction were mainly repaired by TMEJ in our ALT lines.
ALT lines suffered from different degrees of telomere dysfunction
Degrees of genome instability were different among the ALT lines. In particular, ALT4 exhibited a higher number of indels (Supplementary Table S6) and also larger translocation fragments (Figure 2E), which implies that this line may have suffered a higher level of genome-wide DSBs. How telomere dysfunction can cause genome instability and why different ALT survivor lines suffered from different degrees of DSBs still remain elusive. Variations in the number of rRNA were also observed in the ALT survivor lines (Supplementary Table S3). Although the total estimated lengths of other repetitive elements in the ALT survivor lines were still comparable to those of reference genomes, estimated rRNA lengths were reduced in ALT1 and increased in ALT4. These change are likely due to the characteristic of C. elegans genome where the rRNA genes are clustered near the end of the chromosome IR (47,48). The susceptibility of subtelomeric regions to the telomere dysfunction may result in fluctuation in rRNA copy numbers. The copy number may therefore be reduced through DNA damage and nonhomologous end joining and may be increased by microhomology-mediated replication mechanisms, such as break-induced replication. We cannot resolve the possible mechanisms because the long-read sequencing technology we used in this study still has limitations, such as the ∼10-kb read length and ∼5% error rate. The exact structure of rRNA clusters can be resolved by error-free and ultra-long sequencing technologies, as human centromeres of chromosomes 6 and X were resolved by merging the two state-of-the-art technologies (49,50); Oxford Nanopore Technologies ultra-long reads that have >1-Mb read length and PacBio high-fidelity (HiFi) reads that have 1% or less error rate (51,52).
All simple fusion sites exhibited total telomere erosion and subtelomere-subtelomere fusion
Our de novo genome assemblies of four C. elegans ALT survivor lines revealed possible repair or terminating mechanisms of DSBs at the chromosome ends. Substantial contiguity of our genome assemblies allowed us to identify chromosome end structures at the nucleotide level, but unfortunately, only 30 out of 48 chromosome ends were revealed. This may be due to the limitation of our end-searching process that listed the chromosome end by BLAST of telomeric repeats, the TALT sequence or subtelomeric sequences to the assemblies. The repetitive nature of subtelomeric regions and our end-searching process resulted in huge BLAST outputs, which restricted accurate detection of the chromosome ends. Moreover, the repetitiveness also inhibited the de novo genome assembly process, thus some chromosome ends that were partially assembled and had no unique sequences, would not be included in our search process. For example, chromosome IIIR, which was not found in any of the four lines, and chromosome XL, which was not found in three of the lines, may have no searchable unique sequences. If the contiguity can be further increased by increasing the readdepth or read length, and by reducing the read error rate, our end-searching process may retain more chromosome ends by BLAST using a unique sequence further inside the subtelomeric regions.
The simplest terminating mechanisms are simple fusion sites without any BFB cycle (Figure 6A). All of these six fusion sites involved subtelomere deletion at all 12 chromosome ends and only occurred between nonhomologous chromosomes (Figure 3A-D). This situation may arise from the holocentric chromosomes in C. elegans (53). Fusion between monocentric nonhomologous chromosomes may generate dicentric chromosomes, which leads to chromosome breakage and new DSB sites after segregation errors during cell division. In contrast, holocentric nonhomologous chromosomes do not suffer from chromosome fusion followed by segregation, because their whole chromosomal regions can act as centromeres, resulting in a stable fusion chromosome.
Figure 6.
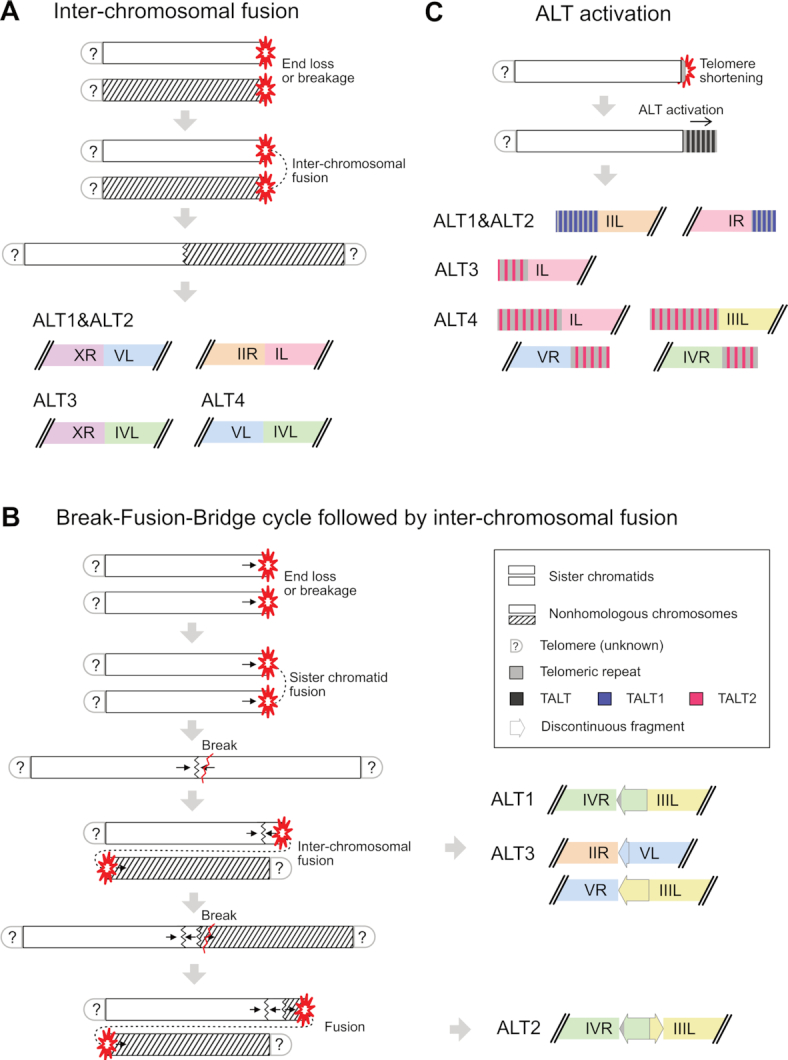
Model for chromosome end repair or reconstruction processes after telomere dysfunction. (A) Simple inter-chromosomal fusion between subtelomere-deleted chromosome ends. (B) Break-Fusion-Bridge (BFB) cycle followed by inter-chromosomal fusion. Black arrows represent the orientation of the broken, discontinuous fragments. (C) ALT-mediated chromosome end reconstruction. Short remaining telomeric repeats may facilitate the replication of TALTs and telomeric repeats to the new ends.
Our results show that all simple fusion sites underwent subtelomere-subtelomere fusions, but are different from a previous study, where ∼40% of fusion sites had telomere-subtelomere fusion (1). One possible explanation for this discrepancy is that the previous study used a PCR-based method that checked whether the nearest subtelomeric regions still remained after the fusion events. The authors directly amplified some fusion sites, but could not amplify the others because of the repetitive nature of subtelomeric regions. Thus, they moved primers far from telomeric repeats and reasoned that if the possible nearest primers work, then subtelomere deletion did not occur. This elegant approach worked well, but the repetitive nature of subtelomeric regions may have limited the detection of short, ∼100-bp subtelomere deletions. Indeed, all PCR-amplified fusion sites were identified as subtelomere-subtelomere fusion sites. Another possible reason for the discrepancy between our results and the previous report is that different methods of generating chromosome fusion lines may give different results. The previous study used brood size reduction as a signature of chromosome fusion, but we passed more generations after brood size reduction to turn on ALT mechanisms (12). Thus our ALT survivor lines had suffered from telomere dysfunction for more generations, possibly resulting in more subtelomere deletion events. In addition, the previous study used a double mutant harbouring mutations in both telomerase and a major component of the canonical nonhomologous end joining genes, while we used a telomerase single mutant worm. A third possibility is that the remaining chromosome ends that we could not assemble may have telomere-subtelomere fusions, but this is unlikely because almost all fusion sites are assembled in our ALT survivor lines, which have karyotypes of 2n = 6–8 (Supplementary Figure S6).
BFB cycles worked after sister chromatid fusion
We successfully assembled four other fusion sites that contained discontinuous fragments, which remained between nonhomologous chromosomes, suggesting that a BFB cycle between sister chromatids had occurred (Figure 3E-H). These fragments had >97% identity with the original, intact sister chromatids and four out of five fragments were inverted at the end of the sister chromatid. This suggests that the sister chromatids were fused and broken, leaving an inverted, high-identity fragment at the end (Figure 3E, G and H). Unlike nonhomologous chromosome fusion, sister chromatid fusion causes a chromosome segregation problem during the cell cycle, so the fusion chromosome must be broken, generating a discontinuous fragment.
Among the four BFB sites, three were stabilised after nonhomologous chromosome fusion events, but the BFB site in the ALT2 genome had an additional fusion event, leaving two discontinuous fragments between nonhomologous chromosomes (Figures 6B and 3F). We can speculate on the fusion and breakage events at this site based on the same breakpoints and different orientations of discontinuous fragments: the chromosome IVR fragment was inverted, but the chromosome IIIL fragment was duplicated in the same orientation. The two sister chromatids of chromosome IV were likely fused and broken, then the broken end fused with chromosome III in the common ancestor of ALT1 and ALT2. After this nonhomologous chromosome fusion, ALT1 and ALT2 would have followed different paths. The chromosome IV end in ALT1 was stabilised, but in ALT2, chromosome IV may have suffered from additional breakage and fusion, leaving the two discontinuous fragments in their different orientations (Figure 6B). The common chromosome IV fragment had 150-bp telomeric repeats, implying that telomere-subtelomere fusion occurred in the common ancestor of ALT1 and ALT2, between the two sister chromatids of chromosome IV.
Intriguingly, the sizes of the discontinuous fragments were short and varied from just 0.3 to 7.1 kb. This suggests several possibilities, either that chromosome breakage leaves only short fragments, or that the breakage ends with various sizes of fragments that can be formed, but are then subsequently shortened. In the former case, the short fragments in our assemblies may be left by chance, or by an unknown mechanism. For example, chromosome cohesion near the fusion site may physically inhibit the breakage. In the latter case, long fragments remained after breakage, but were further deleted as other subtelomere deletion events occurred. Indeed, dicentric chromosomes have been reported as more likely to be broken near the telomere after chromosome fusion events in surviving human and yeast cells (16,54). It is consistent with our data that showed that CNVs were enriched in subtelomeric regions (Figure 2A) and that discontinuous BFB blocks generated by breakage between sister chromatids fusion were actually subtelomeric fragments (Figure 3E-H). We speculate that in C. elegans, like other eukaryotes, regions near the telomere may be more susceptible to the chromosome breakage, or that worms that have breakage only near the telomeres can survive. This is one of the limitations of our ALT models as they show only final stable snapshots, rather than the whole intermediate processes.
New chromosome end structures stabilised by TALT replication
All the other ends without nonhomologous chromosome fusion contained tandem replicated copies of TALT and the canonical telomeric repeats (Figure 6C). Almost all of these chromosome ends, except chromosome XL in ALT1, had remaining short telomeric repeats and no subtelomere deletion. We speculate that these telomeric repeats may partially protect the ends from chromosome fusion and may act as a seed for TALT replication. TALT1 and TALT2 found in internal chromosome locations can be replicated to the ends that are already flanked with canonical or degenerated telomeric repeats, so the homology between telomeric repeats in TALTs and at the chromosome ends may facilitate the TALT replication.
The exceptional chromosome XL in ALT1 had a more complex structure than the other chromosome ends, including a discontinuous fragment and inversion of TALT at the beginning (Figure 5). Our local re-assembly of this region revealed that one or two probable FoSTeS events occurred, between units a and b and between units f and f′, as these pairs of units contained >50-bp-long homology sequences. We also found that ∼7 distinct discontinuous fragment units were replicated and reconstructed the chromosome end via unknown mechanisms. Nonhomologous end joining is another candidate of the replication process, as some connected units contained very short homology or inserted sequences at junction sites. Some characteristic inverted structures, such as units (a-b)-(b-a) are likely to be generated by the BFB cycle. Although we observed these signatures of candidate replication processes, we still cannot fully resolve the exact structure of XL as these units are too similar and long to be distinguished by our error-prone long reads.
The structure of ALT1 chromosome XL is too complex, so it is unlikely that this structure was established through a single replication process during a single cell division. Instead, we suggest that FoSTeS and BFB could have been induced through multiple cell divisions, resulting in repetitive complex rearrangements. Then this structure was stabilised by ALT, but possibly through a different ALT initiation process, as its TALT initiation site seemed to be in a subtelomeric region (end of unit c), and one copy of TALT and the telomeric repeats were invertedly replicated. It suggests that TALT initiation would have occurred without the original telomeric repeats, which needs further investigation.
Here, we have described the molecular consequences of telomere dysfunction and the termination processes that are retained in the ALT survivor lines of the C. elegans telomerase mutants. We clearly show that the effect of telomere dysfunction is not limited to chromosome ends, but is extended through the chromosomes. Our results strongly suggest that subtelomere deletion acts as a major source of chromosome fusion, and that even short telomeric repeats are easily repaired by an alternative telomere capture mechanism. We describe a variety of mechanisms to fix or terminate the catastrophic effects caused by telomere dysfunction in C. elegans, but it is likely that other mechanisms exist in other models. Our findings also contribute to the further understanding of karyotype evolution in nature.
DATA AVAILABILITY
Our genome assemblies and raw PacBio reads were submitted to the NCBI BioProject database (https://www.ncbi.nlm.nih.gov/bioproject) under accession number PRJNA671840.
Supplementary Material
ACKNOWLEDGEMENTS
Author contributions: Conceptualisation, J.K., E.K. and J.L.; Methodology, E.K., J.K. and C.K.; Formal Analysis, E.K. and J.K.; Investigation, E.K. and J.K.; Writing-Original Draft, J.K. and E.K.; Writing-Review & Editing, J.K., E.K. and J.L.; Funding Acqusition, J.L.; Supervision, J.L.. The authors would like to sincerely thank the anonymous reviewers for their invaluable comments.
Contributor Information
Eunkyeong Kim, Department of Biological Sciences, Seoul National University, Gwanak-ro 1, Gwanak-gu, Seoul 08826, Korea; Institute of Molecular Biology and Genetics, Seoul National University, Seoul 08826, Korea.
Jun Kim, Department of Biological Sciences, Seoul National University, Gwanak-ro 1, Gwanak-gu, Seoul 08826, Korea; Research Institute of Basic Sciences, Seoul National University, Seoul 08826, Korea.
Chuna Kim, Aging Research Center, Korea Research Institute of Bioscience and Biotechnology, Gwahak-ro 125, Daejeon 34141, Korea.
Junho Lee, Department of Biological Sciences, Seoul National University, Gwanak-ro 1, Gwanak-gu, Seoul 08826, Korea; Institute of Molecular Biology and Genetics, Seoul National University, Seoul 08826, Korea; Research Institute of Basic Sciences, Seoul National University, Seoul 08826, Korea.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Research Foundation of Korea grant funded by the Korean government (MEST) [2020R1A2C3003352]; National Research Foundation of Korea grant funded by the Korean government (MEST) [2019R1A6A1A10073437 to J.K.]; KRIBB Research Initiative Program and the National Research Foundation of Korea grant funded by the Korean government (MEST) [2020R1C1C101220611 to C.K.]. Funding for open access charge: Seoul National University.
Conflict of interest statement. None declared.
REFERENCES
- 1. Lowden M.R., Meier B., Lee T.W.-S., Hall J., Ahmed S.. End joining at Caenorhabditis elegans telomeres. Genetics. 2008; 180:741–754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Lowden M.R., Flibotte S., Moerman D.G., Ahmed S.. DNA synthesis generates terminal duplications that seal end-to-end chromosome fusions. Science. 2011; 332:468–471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Artandi S.E., Chang S., Lee S.-L., Alson S., Gottlieb G.J., Chin L., DePinho R.A.. Telomere dysfunction promotes non-reciprocal translocations and epithelial cancers in mice. Nature. 2000; 406:641–645. [DOI] [PubMed] [Google Scholar]
- 4. Hackett J.A., Feldser D.M., Greider C.W.. Telomere dysfunction increases mutation rate and genomic instability. Cell. 2001; 106:275–286. [DOI] [PubMed] [Google Scholar]
- 5. Kim C., Sung S., Kim J., Lee J.. Repair and reconstruction of telomeric and subtelomeric regions and genesis of new telomeres: implications for chromosome evolution. Bioessays. 2020; 42:1900177. [DOI] [PubMed] [Google Scholar]
- 6. McClintock B. The behavior in successive nuclear divisions of a chromosome broken at meiosis. PNAS. 1939; 25:405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Shay J., Bacchetti S.. A survey of telomerase activity in human cancer. Eur. J. Cancer. 1997; 33:787–791. [DOI] [PubMed] [Google Scholar]
- 8. Chen Q., Ijpma A., Greider C.W.. Two survivor pathways that allow growth in the absence of telomerase are generated by distinct telomere recombination events. Mol. Cell. Biol. 2001; 21:1819–1827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Dilley R.L., Verma P., Cho N.W., Winters H.D., Wondisford A.R., Greenberg R.A.. Break-induced telomere synthesis underlies alternative telomere maintenance. Nature. 2016; 539:54–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Bryan T.M., Englezou A., Dalla-Pozza L., Dunham M.A., Reddel R.R.. Evidence for an alternative mechanism for maintaining telomere length in human tumors and tumor-derived cell lines. Nat. Med. 1997; 3:1271. [DOI] [PubMed] [Google Scholar]
- 11. Mason J.M., Randall T.A., Frydrychova R.C.. Telomerase lost?. Chromosoma. 2016; 125:65–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Seo B., Kim C., Hills M., Sung S., Kim H., Kim E., Lim D.S., Oh H.-S., Choi R.M.J., Chun J.. Telomere maintenance through recruitment of internal genomic regions. Nat. Commun. 2015; 6:8189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Kim C., Sung S., Lee J.. Worm. 2016; 5:Taylor & Francis; e1146856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Gisselsson D., Jonson T., Petersén Å., Strömbeck B., Dal Cin P., Höglund M., Mitelman F., Mertens F., Mandahl N.. Telomere dysfunction triggers extensive DNA fragmentation and evolution of complex chromosome abnormalities in human malignant tumors. Proc. Natl. Acad. Sci. U.S.A. 2001; 98:12683–12688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Heacock M., Spangler E., Riha K., Puizina J., Shippen D.E.. Molecular analysis of telomere fusions in Arabidopsis: multiple pathways for chromosome end-joining. EMBO J. 2004; 23:2304–2313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Maciejowski J., Chatzipli A., Dananberg A., Chu K., Toufektchan E., Klimczak L.J., Gordenin D.A., Campbell P.J., de Lange T.. APOBEC3-dependent kataegis and TREX1-driven chromothripsis during telomere crisis. Nat. Genet. 2020; 52:884–890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Maser R.S., Choudhury B., Campbell P.J., Feng B., Wong K.-K., Protopopov A., O’Neil J., Gutierrez A., Ivanova E., Perna I.. Chromosomally unstable mouse tumours have genomic alterations similar to diverse human cancers. Nature. 2007; 447:966–971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kim C., Kim J., Kim S., Cook D.E., Evans K.S., Andersen E.C., Lee J.. Long-read sequencing reveals intra-species tolerance of substantial structural variations and new subtelomere formation in C. elegans. Genome Res. 2019; 29:1023–1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Yoshimura J., Ichikawa K., Shoura M.J., Artiles K.L., Gabdank I., Wahba L., Smith C.L., Edgley M.L., Rougvie A.E., Fire A.Z.. Recompleting the Caenorhabditis elegans genome. Genome Res. 2019; 29:1009–1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Stein L., Sternberg P., Durbin R., Thierry-Mieg J., Spieth J.. WormBase: network access to the genome and biology of Caenorhabditis elegans. Nucleic Acids Res. 2001; 29:82–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Harris T.W., Arnaboldi V., Cain S., Chan J., Chen W.J., Cho J., Davis P., Gao S., Grove C.A., Kishore R.. WormBase: a modern model organism information resource. Nucleic Acids Res. 2020; 48:D762–D767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Koren S., Walenz B.P., Berlin K., Miller J.R., Bergman N.H., Phillippy A.M.. Canu: scalable and accurate long-read assembly via adaptive k-mer weighting and repeat separation. Genome Res. 2017; 27:722–736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., Marth G., Abecasis G., Durbin R.. The sequence alignment/map format and SAMtools. Bioinformatics. 2009; 25:2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Vaser R., Sović I., Nagarajan N., Šikić M.. Fast and accurate de novo genome assembly from long uncorrected reads. Genome Res. 2017; 27:737–746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Simão F.A., Waterhouse R.M., Ioannidis P., Kriventseva E.V., Zdobnov E.M.. BUSCO: assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics. 2015; 31:3210–3212. [DOI] [PubMed] [Google Scholar]
- 26. Grüning B., Dale R., Sjödin A., Chapman B.A., Rowe J., Tomkins-Tinch C.H., Valieris R., Köster J.. Bioconda: sustainable and comprehensive software distribution for the life sciences. Nat. Methods. 2018; 15:475–476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Smit A., Hubley R., Green P.. 2015; RepeatMasker Open-4.0. 2013-2015http://www.repeatmasker.org.
- 28. Hubley R., Finn R.D., Clements J., Eddy S.R., Jones T.A., Bao W., Smit A.F., Wheeler T.J.. The Dfam database of repetitive DNA families. Nucleic Acids Res. 2016; 44:D81–D89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Jurka J. Repeats in genomic DNA: mining and meaning. Curr. Opin. Struct. Biol. 1998; 8:333–337. [DOI] [PubMed] [Google Scholar]
- 30. Bao W., Kojima K.K., Kohany O.. Repbase update, a database of repetitive elements in eukaryotic genomes. Mobile Dna. 2015; 6:11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Benson G. Tandem repeats finder: a program to analyze DNA sequences. Nucleic Acids Res. 1999; 27:573–580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Li H. Minimap2: pairwise alignment for nucleotide sequences. Bioinformatics. 2018; 34:3094–3100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Marçais G., Delcher A.L., Phillippy A.M., Coston R., Salzberg S.L., Zimin A.. MUMmer4: a fast and versatile genome alignment system. PLoS Comput. Biol. 2018; 14:e1005944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Nattestad M., Schatz M.C.. Assemblytics: a web analytics tool for the detection of variants from an assembly. Bioinformatics. 2016; 32:3021–3023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Quinlan A.R., Hall I.M.. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010; 26:841–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Altschul S.F., Gish W., Miller W., Myers E.W., Lipman D.J.. Basic local alignment search tool. J. Mol. Biol. 1990; 215:403–410. [DOI] [PubMed] [Google Scholar]
- 37. Roerink S.F., van Schendel R., Tijsterman M.. Polymerase theta-mediated end joining of replication-associated DNA breaks in C. elegans. Genome Res. 2014; 24:954–962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Wicky C., Villeneuve A.M., Lauper N., Codourey L., Tobler H., Müller F.. Telomeric repeats (TTAGGC)n are sufficient for chromosome capping function in Caenorhabditis elegans. Proc. Natl. Acad. Sci. U.S.A. 1996; 93:8983–8988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Turleau C., Roubin M., Klein M.. Ann. Genet. 1972; 15:79–84. [PubMed] [Google Scholar]
- 40. Turleau C., De Grouchy J.. New observations on the human and chimpanzee karyotypes. Humangenetik. 1973; 20:151–157. [DOI] [PubMed] [Google Scholar]
- 41. Ijdo J., Baldini A., Ward D., Reeders S., Wells R.. Origin of human chromosome 2: an ancestral telomere-telomere fusion. Proc. Natl. Acad. Sci. U.S.A. 1991; 88:9051–9055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Adolph S., Klein J.. Robertsonian variation in Mus musculus from central Europe, Spain, and Scotland. J. Hered. 1981; 72:219–221. [DOI] [PubMed] [Google Scholar]
- 43. Britton-Davidian J., Catalan J., da Graça Ramalhinho M., Ganem G., Auffray J.-C., Capela R., Biscoito M., Searle J.B., da Luz Mathias M.. Rapid chromosomal evolution in island mice. Nature. 2000; 403:158–158. [DOI] [PubMed] [Google Scholar]
- 44. PIáLEK J., Hauffe H.C., Searle J.B.. Chromosomal variation in the house mouse. Biol. J. Linn. Soc. 2005; 84:535–563. [Google Scholar]
- 45. Sánchez-Guillén R., Capilla L., Reig-Viader R., Martínez-Plana M., Pardo-Camacho C., Andrés-Nieto M., Ventura J., Ruiz-Herrera A.. On the origin of Robertsonian fusions in nature: evidence of telomere shortening in wild house mice. J. Evol. Biol. 2015; 28:241–249. [DOI] [PubMed] [Google Scholar]
- 46. Coutelier H., Xu Z., Morisse M.C., Lhuillier-Akakpo M., Pelet S., Charvin G., Dubrana K., Teixeira M.T.. Adaptation to DNA damage checkpoint in senescent telomerase-negative cells promotes genome instability. Genes Dev. 2018; 32:1499–1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Consortium*, C.e.S. Genome sequence of the nematode C. elegans: a platform for investigating biology. Science. 1998; 282:2012–2018. [DOI] [PubMed] [Google Scholar]
- 48. Stricklin S.L., Griffiths-Jones S., Eddy S.R.. C. elegans noncoding RNA genes. WormBook. 2005; 25:1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Miga K.H., Koren S., Rhie A., Vollger M.R., Gershman A., Bzikadze A., Brooks S., Howe E., Porubsky D., Logsdon G.A.. Telomere-to-telomere assembly of a complete human X chromosome. Nature. 2020; 585:79–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Bzikadze A.V., Pevzner P.A.. Automated assembly of centromeres from ultra-long error-prone reads. Nat. Biotechnol. 2020; 38:1309–1316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Jain M., Koren S., Miga K.H., Quick J., Rand A.C., Sasani T.A., Tyson J.R., Beggs A.D., Dilthey A.T., Fiddes I.T.. Nanopore sequencing and assembly of a human genome with ultra-long reads. Nat. Biotechnol. 2018; 36:338–345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Wenger A.M., Peluso P., Rowell W.J., Chang P.-C., Hall R.J., Concepcion G.T., Ebler J., Fungtammasan A., Kolesnikov A., Olson N.D.. Accurate circular consensus long-read sequencing improves variant detection and assembly of a human genome. Nat. Biotechnol. 2019; 37:1155–1162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Albertson D.G., Thomson J.N.. The kinetochores of Caenorhabditis elegans. Chromosoma. 1982; 86:409–428. [DOI] [PubMed] [Google Scholar]
- 54. Pobiega S., Marcand S.. Dicentric breakage at telomere fusions. Genes Dev. 2010; 24:720–733. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Our genome assemblies and raw PacBio reads were submitted to the NCBI BioProject database (https://www.ncbi.nlm.nih.gov/bioproject) under accession number PRJNA671840.