Abstract
Autoactivation of two-component systems (TCSs) can increase the sensitivity to signals but inherently cause a delayed response. Here, we describe a unique negative feedback mechanism enabling the global NtrB/NtrC regulator to rapidly respond to nitrogen starvation over the course of histidine utilization (hut) in Pseudomonas fluorescens. NtrBC directly activates transcription of hut genes, but overexpression will produce excess ammonium leading to NtrBC inactivation. To prevent this from occurring, the histidine-responsive repressor HutC fine-tunes ntrBC autoactivation: HutC and NtrC bind to the same operator site in the ntrBC promoter. This newly discovered low-affinity binding site shows little sequence similarity with the consensus sequence that HutC recognizes for substrate-specific induction of hut operons. A combination of genetic and transcriptomic analysis indicated that both ntrBC and hut promoter activities cannot be stably maintained in the ΔhutC background when histidine fluctuates at high concentrations. Moreover, the global carbon regulator CbrA/CbrB is involved in directly activating hut transcription while de-repressing hut translation via the CbrAB-CrcYZ-Crc/Hfq regulatory cascade. Together, our data reveal that the local transcription factor HutC plays a crucial role in governing NtrBC to maintain carbon/nitrogen homeostasis through the complex interactions between two TCSs (NtrBC and CbrAB) at the hut promoter.
INTRODUCTION
All living organisms have evolved the ability to recognize internal and external environmental stimuli and produce appropriate physiological responses. In bacteria, signal transduction mediated by protein phosphorylation is predominantly carried out by two-component systems (TCSs) consisting of a sensor kinase (SK) and a response regulator (RR) (1–3). SK responds to the presence of a stimulatory ligand and regulates phosphorylation of its cognate RR. Most RRs possess a transcriptional regulator output domain and determine the expression levels of downstream target genes. Interestingly, many phosphorylated RRs activate their own expression (4,5). This phenomenon (known as autoactivation or positive autoregulation) is widespread for transcription factors in bacteria. Autoactivation can increase the sensitivity to signals and exert history-dependent hysteretic responses (6–8). However, both experimental and theoretical analysis indicated that autoactivation causes an inherent delayed response as more time is required for protein synthesis (9,10). A slow response is potentially disadvantageous for bacteria living in ever-changing environmental conditions, i.e. nutrient fluctuation. To overcome the fitness cost of TCS autoactivation, a coupled negative feedback loop is predicted to be necessary but only a few mechanisms have been uncovered thus far, including autorepression at a secondary weak binding site (11–13).
A typical example of TCS autoactivation concerns the NtrBC nitrogen (N) regulator, the first TCS described in enteric bacteria (14,15). Under N-excess conditions, there is very little NtrB and NtrC in the cell. When N becomes limiting, intracellular glutamine levels decline relative to α-ketoglutarate. This causes the NtrB kinase to catalyze NtrC phosphorylation (NtrC∼P) coupled with PII uridylation. The NtrC∼P then activates σ54-RNA polymerase holoenzyme leading to expression of a LysR-type transcriptional regulator NAC (plus other genes and the ntrBC operon itself). NAC in turn activates the expression of many N catabolic pathways from a σ70 promoter (16), including histidine utilization (hut) genes. Histidine is a good source of nutrient (17), but its utilization poses a significant challenge as it delivers excess nitrogen over carbon. Overspeed of the hut catabolism would lead to the buildup of intracellular ammonium and subsequent inactivation of the NtrBC system (18,19). If that happens, a transition period occurs to enable cells to physiologically shift back to N-limiting status and then ‘jump start’ the NtrBC system (20). Extra time will be required to produce NtrBC and then NAC proteins to sufficient levels for hut activation. Therefore, expression of hut enzymes must be tightly regulated to circumvent the potential metabolic shifts associated with ntrBC autoactivation.
Historically, the hut pathway has been a model for studying gene regulation, particularly the coordination of C/N metabolism (20). Early studies in enteric bacteria led to the concept of catabolite repression, which explains the inhibitory effects of glucose on the utilization of alternative C sources, such as histidine and lactose (21). In summary, hut transcription is activated by the catabolite-activating protein (CAP) charged with cAMP and the aforementioned NtrBC/NAC cascade when histidine is used as a source of C and N, respectively (20). However, this well-defined paradigm does not hold for many non-enteric bacteria, including those of the closely related genus Pseudomonas (22). The Pseudomonas genome does contain homologues of CAP and NAC, but their involvement (and also the role of cAMP) in hut activation has been eliminated (23–25).
Pseudomonads are metabolically versatile but use succinate as one of the most preferred C sources. Succinate-induced carbon catabolite repression (CCR) of hut enzymes was first reported in 1960s in various Pseudomonas species, but a molecular explanation is still lacking (22,26). Recent progress showed that CCR in Pseudomonas occurs at the post-transcriptional level with the Crc/Hfq protein complex as the principal mediator (27–29). In the presence of succinate, Crc/Hfq represses the expression of catabolic genes through specific binding to target mRNAs. When succinate is consumed, Crc/Hfq is sequestrated by ncRNAs (CrcY and CrcZ) whose expression is activated by the CbrAB two-component system. However, previous work with P. putida suggested that the crc gene was not involved in the CCR control of hut enzymes (30).
Expression of hut genes is shown to be controlled by substrate-specific induction in addition to the general induction mediated by global regulators. Like in enteric bacteria, histidine-induced expression of hut in Pseudomonas is negatively regulated by the HutC repressor, which is a representative member of the GntR/HutC family of transcriptional regulators (23,31). It possesses a N-terminal winged helix-turn-helix (wHTH) DNA-binding domain and a C-terminal substrate-binding domain. HutC binds to operator sites (Phut) of hut promoters, and the repression is relieved by urocanate (the first intermediate of the histidine degradation pathway). In regard to positive regulation of hut, previous gene deletion analysis showed that cbrAB and ntrBC are functionally required for bacterial growth on histidine, but it remains unclear how they mediate the regulatory effects (18).
We hereby describe the mechanisms of hut activation mediated by the CbrAB and NtrBC systems in P. fluorescens SBW25. Our study began with genetic characterization of the hut promoters and in vitro protein-DNA interactions using purified CbrBHis6 and NtrCHis6 proteins. Results consistently indicated that both CbrAB and NtrBC activate hut expression in a direct manner. Furthermore, we present empirical evidence showing that CbrAB activates hut transcription in response to C-limitation and de-represses hut translation through the CbrAB-CrcYZ-Crc/Hfq regulatory cascade. When growing on histidine as a source of sole N but alternative C (i.e. a minimal salt medium supplemented with succinate and histidine), the CbrAB-mediated promoter activity is weak, and the global nitrogen regulator NtrBC plays the dominant role in activating hut transcription. Subsequent genetic and biochemical analysis led to an unexpected finding that HutC and NtrC target the same operator DNA (Pntr) in the PntrBC promoter. This implies HutC represents a transcription factor that can recognize two distinct DNA-binding motifs (Phut and Pntr) that share little sequence similarity. Finally, experiments were designed to test the hypothesis that the histidine-responsive repressor HutC fine-tunes ntrBC expression, acting as an essential negative feedback loop for maintaining C/N homeostasis during bacterial growth on histidine at high concentrations.
MATERIALS AND METHODS
Bacterial strains and culture conditions
P. fluorescens SBW25 is a plant growth-promoting bacterium originally isolated from the phyllosphere of sugar beet grown at the University of Oxford farm, Wytham, Oxford, UK (32). Wild-type SBW25 and its derived mutants were routinely grown in lysogeny broth (LB) medium at 28°C. When growth was examined in the M9 minimal salt medium, succinate and histidine were added at the final concentration of 20 and 10 mM, respectively (except where otherwise specified). To ensure that strains being compared were physiologically equal, bacterial cells in overnight LB culture were washed twice and then subjected to starvation in M9 salt solution at 28°C for 2 h. When necessary, antibiotics were added at the following concentrations (μg/ml): ampicillin (Ap), 100; tetracycline (Tc), 15; spectinomycin (Sp), 100; kanamycin (Km), 50; gentamicin (Gm), 25 and nitrofurantoin (NF), 100. Bacterial strains and plasmids used in this study are listed in Supplementary Table S1.
Strain construction
Escherichia coli DH5αλpir was used for general gene cloning and subsequent conjugation into Pseudomonas. Standard DNA recombination techniques were used following manufacturers’ recommendations. The splicing by overlapping extension PCR (SOE-PCR) strategy was adopted for introducing mutations into the probe DNAs and also for the construction of Pseudomonas mutant strains (33). Briefly, two pairs of oligonucleotide primers were designed to amplify two DNA fragments of similar sizes flanking the target region. Complementary sequences carrying the desired mutation were incorporated into the primers, hence the two PCR products were joined via an additional PCR. A summary of the oligonucleotide primers is provided in Supplementary Table S2. As a general practice, the PCR product was cloned into plasmid vector pCR8/GW/TOPO from Invitrogen (Auckland, New Zealand), and sequence identity was confirmed using Sanger's method of DNA sequencing. The resulting recombinant plasmid can be used as the template for preparing DNA probes using PCR with a biotin-labeled primer. To construct mutant strains, the DNA insert was sub-cloned into an integration suicide vector pUIC3 (34), and the recombinant plasmid was mobilized into Pseudomonas by conjugation with the help of pRK2013. Allelic exchange mutants were selected using a modified procedure of D-cycloserine enrichment (23).
Assays for gene expression
To construct a transcriptional lacZ fusion, an error-free DNA fragment in pCR8/GW/TOPO was subcloned into either pUIC3 (34) or pUC18-mini-Tn7T-Gm-lacZ (35). The obtained pUIC3 recombinant plasmid was mobilized into Pseudomonas and integrated into the target locus of the chromosome via a homologous recombination event of insertion-duplication. In contrast, the recombinant plasmid of pUC18-mini-Tn7T-Gm-lacZ was electroporated into Pseudomonas together with the helper plasmid pUX-BF13 (35,36). The mini-Tn7 element containing the lacZ reporter fusion was integrated into a unique chromosome site located downstream of glmS. Gene expression at the translational level was estimated using lacZ fusion cloned in pXY2, a modified version of pUC18-mini-Tn7T-Gm-lacZ (37). β-Galactosidase activity was assayed using 4-methylumbelliferyl-β-d-galactoside (4MUG) as the enzymatic substrate. The fluorescent product, 7-hydroxy-4-methylcoumarin (4MU), was detected at 460 nm with an excitation wavelength of 365 nm using a Synergy 2 plate reader (Bio-Tek). The enzyme activity was expressed as the amount of 4MU (μM) produced per minute per cell (A600). Statistical analyses were performed using tools in GraphPad Prism v7.
EMSA and DNase I footprinting assays
The coding regions of CbrB and NtrC were amplified by PCR from the genomic DNA of P. fluorescens SBW25 with the integration of His6 tag at the N-terminal. The PCR product was first cloned into pCR8/GW/TOPO, and the error-free DNA fragment was subcloned into the protein expression vector pTrc99A at the NcoI and HindIII sites. The recombinant plasmids were then transformed into E. coli BL21(DE3). Protein expression was induced by 1 mM IPTG (isopropyl-β-d-thiogalactopyranoside) and it was subsequently purified using the Talon metal affinity resin (Clontech Laboratories Inc.) according to the manufacturer's protocol. Of note, HfqHis6 purification involved an additional washing step with a high concentration of imidazole (70 mM) whereby the effects of residual HfqE.coli were eliminated (27).
Probe DNAs were prepared by PCR using a biotinylated primer. The single-stranded RNA probes were synthesized by IDT (Integrated DNA Technologies, Inc.) with a 5’-biotin modification. The protein–DNA/RNA interactions were examined in vitro as previously described (27,31). The protein–RNA interaction was assayed in a 20 μl reaction containing 0.1 μM biotin-labeled probe and varying concentrations of HfqHis6 in addition to 10 mM HEPES (pH 7.9), 35 mM KCl and 2 mM MgCl2. Yeast tRNA (1 μg) was added as a control agent for nonspecific binding. For EMSA with a DNA probe, the 20 μl reaction was composed of 20 nM probe, 1 μg salmon sperm DNA, 10 mM HEPES, 50 mM KCl, 5 mM MgCl2 and 1 mM dithiothreitol (DTT) (pH 7.5). After 30 min of incubation at room temperature, DNA or RNA probes were subject to 6% polyacrylamide gel electrophoresis (PAGE) in half-strength Tris–borate–EDTA (TBE) buffer at low temperatures (∼4°C). Probes in the gel were then transferred to a positively charged Whatman Nytran SuPerCharge nylon membrane (Sigma-Aldrich) and were heat immobilized at 80°C for 30 min. Finally, the Pierce's LightShift Chemiluminescent EMSA kit (Thermo Fisher Scientific) was used for probe detection, and the image was visualized with a LAS-4000 luminescent imager equipped with the ImageQuant LAS 4000 software (Fujifilm). Equilibrium dissociation constant (Kd) was calculated by plotting the fractions of bound DNA against protein concentrations in GraphPad Prism v7.
DNase I analyses were performed under the same condition described for EMSA but in a 50 μl reaction containing 2 μM probe. After 30 min incubation, the reaction mixture was mixed with 50 μl co-factor solution (5 mM CaCl2 and 10 mM MgCl2), and then treated with 0.02 unit of DNase I (Invitrogen) for 5 min at room temperature. Next, 100 μl stop buffer containing 200 mM NaCl, 20 mM EDTA and 1% (w/v) SDS was added to terminate the enzymatic reaction. The DNAs were then subjected to purification with 1:1 phenol–chloroform, and precipitation with three volumes of ethanol with the addition of 1 μl glycogen (20 mg/ml), 1/10 volume of 3 M sodium acetate (pH 5.2). After centrifugation, DNAs were dissolved in 8 μl loading buffer containing 95% (v/v) formamide, 0.05% (w/v) bromophenol blue and 20 mM EDTA. DNAs were denatured by heat treatment (95°C, 10 min) before they were loaded onto a 6% urea–polyacrylamide gel (21 by 40 cm) in 1× TBE buffer using the Sequi-Gen GT electrophoresis system (Bio-Rad Laboratories Pty). The DNAs were then transferred from the gel to a positively charged nylon membrane by contact blotting and detected using the LightShift Chemiluminescent EMSA kit as described above for EMSA. The DNA sequence ladder was obtained by the G + A chemical sequencing reaction with the same biotin-labeled probe, and it was included in the gel to identify the protein-protected DNA regions (38).
RNA-seq analysis
Transcriptomes of the ΔcbrB and ΔntrC mutants were performed in parallel with wild-type P. fluorescens SBW25 and the derived ΔhutC mutant as described previously (31). Cells were harvested from bacteria grown in mid-exponential phase in M9 minimal salt medium supplemented with succinate (20 mM) and histidine (10 mM) as the sources of C and N. Total RNAs were prepared using the Promega SV Total RNA Isolation System (Thermo Fisher Scientific, Auckland, New Zealand). Quality of the RNA samples was determined using an Agilent 2100 Bioanalyzer (Agilent Technologies, Inc.), and sequencing was performed on an Illumina HiSeq 4000 platform using the services provided by Novogene Technology Co. Ltd. (Beijing, China). Data of the obtained 150-bp paired-end reads were processed using Geneious 9.0.5 (Biomatters Ltd, Auckland, New Zealand). Reads were mapped to the reference genome of P. fluorescens SBW25 (NC_012660.1). Next, reads per kilobase per million (RPKM) and transcripts per million (TPM) were subsequently calculated for each assembly, and differentially expressed genes were determined based on transcript comparison normalized using the Median of Gene Expression Ratios method.
RESULTS
Genetic identification of NtrC and CbrB target sites in the PhutU promoter
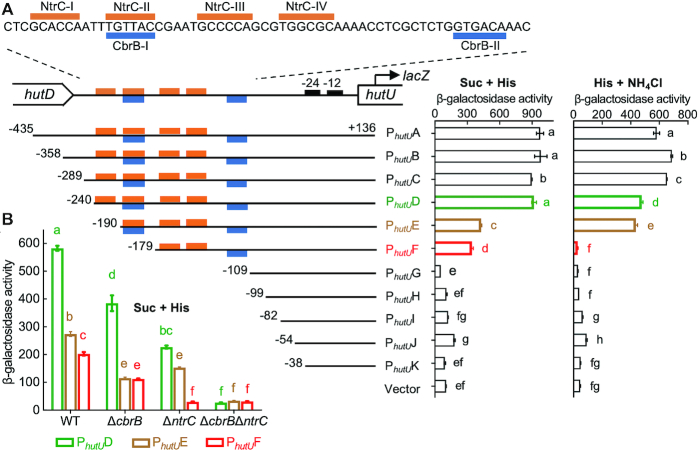
The hut genes of P. fluorescens SBW25 are organized in three transcriptional units, but only one promoter (PhutU) was involved in the CbrAB- and NtrBC-mediated positive regulation (Supplementary Figure S1). Interrogation of the PhutU sequence identified two overlapping DNA regions, which are potentially targeted by NtrC and CbrB (Figure 1A). To map the PhutU promoter, eleven PhutU variants with variable 5′-ends were fused to a promoterless lacZ reporter gene, and then integrated into the mini-Tn7 site of wild-type SBW25 (Figure 1A). β-Galactosidase activities were first examined for cells grown in minimal medium with histidine as the sole C source (Figure 1A). Results indicate that the 11 nt sequence harboring the CbrB-I site (PhutUE versus PhutUF) was functionally required for PhutU expression.
Figure 1.
Genetic analysis of PhutU promoter showing the direct regulatory roles of cbrAB and ntrBC. (A) Putative NtrC and CbrB target sites are indicated by orange and blue bars, respectively. β-Galactosidase activities were measured for wild-type SBW25 containing chromosomally integrated lacZ fusions to eleven PhutU variants (named PhutUA to PhutUK). The empty mini-Tn7 element was included as a negative control. Bacteria were grown in minimal salt medium supplemented with succinate (Suc) + histidine (His) or histidine + NH4Cl. (B) Expression of three PhutU-lacZ variants was assessed in wild-type (WT) and three mutant backgrounds (ΔcbrB, ΔntrC, ΔcbrB ΔntrC) in minimal medium supplemented with succinate and histidine (Suc + His). Data are means and standard errors of four independent cultures. Bars that are not connected by the same letter (shown above each) are significantly different (P < 0.05) by Tukey's HSD.
Next, we measured promoter activities in minimal medium of succinate + histidine (Figure 1A). In line with our expectations, PhutU expression was reduced by half with the deletion of NtrC-I (PhutUE), and the promoter activity ceased completely when the second NtrC target site (NtrC-III/IV) was further deleted (PhutUG). A small but significant reduction was noted between PhutUE and PhutUF with the deletion of the NtrC-II (or CbrB-I) half site (Figure 1A). This result was initially surprising, as NtrC-II would have negligible effects on NtrC-mediated PhutU activation in the absence of the primary NtrC-I half site. However, the data was consistent with the functional involvement of CbrB in activating PhutU when histidine is the sole N source (Supplementary Figure S1A). The role of CbrB was further confirmed by comparing the expression levels of PhutUD, PhutUE and PhutUF in mutants devoid of cbrB and/or ntrC (Figure 1B). Significant difference between PhutUE and PhutUF was detected in the wild-type but not in the ΔcbrB mutant background (with remaining activity in ΔcbrB attributable to the second NtrC binding site NtrC-III/IV, as PhutUF expression was abolished in the ΔntrC background). Together, the genetic data implicate that NtrBC and CbrAB coordinate PhutU expression in a direct manner.
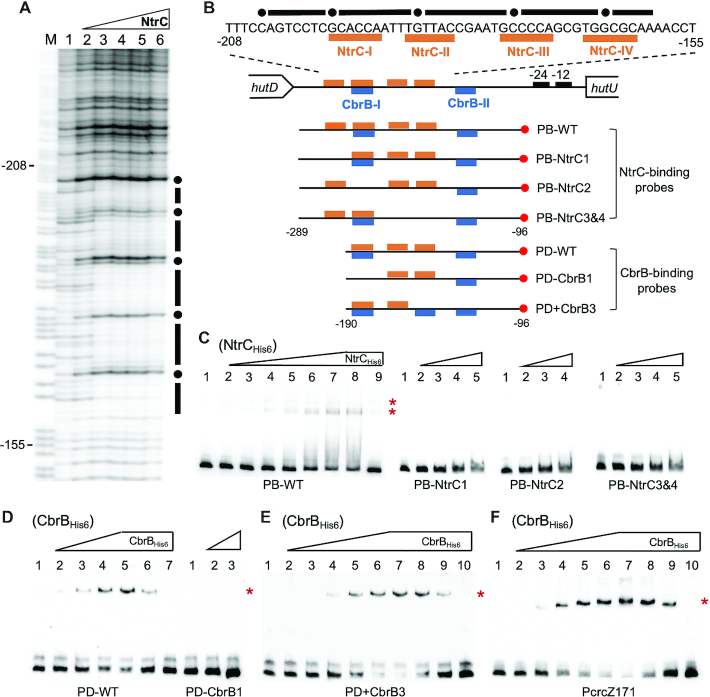
NtrC and CbrB bind in vitro with PhutU promoter
To determine the direct interactions between NtrC and PhutU, His6-tagged NtrC from P. fluorescens SBW25 was first subjected to DNase I footprinting analysis with a biotin-labeled PhutU probe (PB-WT). Results showed that a 44-bp DNA region was protected from DNase I digestion, and it contains the two NtrC-binding sites genetically identified above (Figure 2A and B). Next, EMSA was conducted with PB-WT and three mutant probes carrying substitutions of NtrC-I, NtrC-II and NtrC-III/IV sites, respectively (Figure 2C). A significant shift of the wild-type PB-WT probe was observed in the presence of increasing concentrations of NtrCHis6. However, DNA retardation was not observed for the three mutant probes (Figure 2C). The data thus suggest that NtrC-I, NtrC-II and NtrC-III/IV sites are all functionally required for stable binding of NtrCHis6 to the PhutU promoter.
Figure 2.
In vitro protein–DNA interactions showing the direct roles of CbrAB and NtrBC in PhutU expression. (A) DNase I footprinting was performed using purified NtrCHis6 and a 194 bp biotin-labeled DNA probe PB-WT. Lane M is G+A marker; Lane 1–6, NtrCHis6 added at an increasing concentration of 0, 0.2, 0.6, 1.6, 3.2 and 5 μM respectively. The protected region is indicated by black bars with hypersensitive residues being marked with filled circles. (B) The hutD - hutU intergenic region showing NtrC-protected DNA sequence and locations of the probe DNAs. The biotin-labeled 3′-ends are marked with red circles. (C) EMSA with NtrCHis6. For wild-type probe PB-WT, NtrC was added at 0, 1.25, 2.5, 5, 10, 15, 20, 25 and 25 μM from lane 1 to 9. In lane 9, the unlabeled probe was added at 60-fold molar excess. For the three mutant probes, NtrC was added at 0, 5, 15, 25 and 37.5 μM from lane 1 to 5. (D) EMSA with CbrBHis6 and PhutU probes. For PD-WT, CbrB was added at 0, 7.83, 11.75, 15.66, 19.58, 19.58 and 19.58 μM from lanes 1 to 7. In lanes 6 and 7, unlabeled probe (competitor DNA) was added at 10- and 60-fold molar excess, respectively. For PD-CbrB1, CbrB was added at 0, 15.66 and 19.58 μM from lane 1 to 3. (E) EMSA with the PD+CbrB3 probe. CbrB was added at 0, 1.31, 3.92, 7.83, 11.75, 15.66, 19.58, 19.58, 19.58, 19.58 μM from lane 1 to 10. Competitor DNA (unlabeled PD+CbrB3) was added at 10-, 20- and 60-fold molar excess in lanes 8–10. (F) EMSA of CbrBHis6 with PcrcZ171 probe. CbrB was added in lanes 1–10 at 0, 1.31, 3.92, 6.53, 9.14, 11.75, 14.36, 14.36, 14.36, 14.36 μM, respectively. Lanes 8–10 contained unlabeled probe at 10-, 20- and 60-fold molar excess. Of note, the NtrC-I, NtrC-II/CbrB-I, NtrC-III and NtrC-IV sites were altered to random sequences of TTCAAG, AACCGT, TAAAGC and AACCGT, respectively. The CbrB-III artificial site ‘GTAACA’ was introduced into the PD+CbrB3 probe sequence at the original NtrC-IV site. Asterisks on right-hand side of each gel denote positions of the shifted bands.
Next, we performed EMSA with purified CbrBHis6 from P. fluorescens SBW25 (Figure 2D). A significant shift of the wild-type PhutU probe PD-WT was observed in the presence of increasing concentrations of CbrBHis6 (lanes 2–5). DNA retardation was eventually abolished with the addition of competitor DNAs from low to high concentrations (lanes 6–7). More importantly, DNA retardation was not observed with the mutant probe PD-CbrB1 lacking the CbrB-I half site (Figure 2D). The data thus confirmed the direct interaction between CbrB and PhutU. Furthermore, EMSA was performed with a PcrcZ probe (PcrcZ171), and significant shifts were observed when CbrBHis6 was added at an increasing concentration from lanes 2 to 7 (Figure 2F). PcrcZ is a well-characterized promoter under the direct control of CbrB in Pseudomonas, including P. fluorescens SBW25 (27). CrcZ is also the key mediator for CbrAB to control hut expression at the post-transcriptional level (see details below). CbrB appears to bind PcrcZ with a relatively higher affinity than the PhutU promoter (Supplementary Figure S2). The precise modes of CbrB interaction with DNA is currently poorly understood. It was thus interesting to note that PcrcZ promoters from different Pseudomonas species contain a third highly conserved CbrB half site (CbrB-III) (Supplementary Figure S3), which is absent in PhutU promoters (Supplementary Figure S4). This suggested that CbrB has evolved a low-affinity interaction with PhutU promoter due to the lack of this CbrB-III site. To test this hypothesis, we performed EMSA with a mutant probe containing an introduced CbrB-III site in PhutU (PD+CbrB3). Indeed, CbrB-binding was increased though the difference was not significant from the estimated Kd values (Figure 2E, Supplementary Figure S2). DNA retardation was eventually abolished by adding competitor DNA at an increasing concentration from lanes 8 to 10 (Figure 2E). Together, the EMSA data confirmed the results of genetic analysis that NtrC and CbrB directly activate hut transcription.
hut expression is subject to CCR control mediated by the CbrAB-CrcYZ-Crc/Hfq cascade
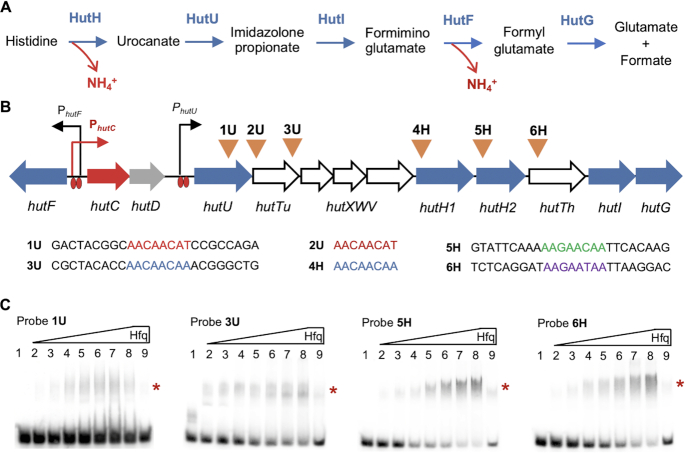
The hut locus contains six putative Crc/Hfq-binding sites (AAnAAnAA) with four types of sequence variants (Figure 3B). They are present in hutTh and hutTu genes (encoding the histidine- and urocanate-specific transporters), and also in hutH2 (histidase) and hutU (urocanase) responsible for the enzymatic breakdown of histidine and urocanate, respectively. This suggests that CCR occurs for histidine (and urocanate) utilization by targeting substrate uptake and the first two enzymes of the histidine catabolic pathway. To test the predicted molecular interactions in vitro, we performed EMSAs with HfqHis6 and four oligoribonucleotide probes (25 nt) centred on the putative target sequence (Figure 3C). A significant shift of biotin-labeled RNA was observed for all four probes (1U, 3U, 5H and 6H) in the presence of increasing concentrations of HfqHis6 (lanes 2–8). RNA retardation was nearly eliminated with the addition of excess unlabeled competitor RNA, i.e. the same but unlabeled probe (lane 9). The data thus confirm that Hfq specifically targets the hutU-G transcript (Figure 3B).
Figure 3.
Specific interactions between Hfq and its target hut mRNAs. (A) Histidine is sequentially broken down by the following five enzymes: HutH, histidine ammonia lyase or histidase; HutU, urocanase; HutI, imidazolone propionate (IPA) amidohydrolase; HutF, formiminoglutamate (FIGLU) iminohydrolase; HutG, formylglutamate (FG) amidohydrolase. (B) Locations of the six putative Crc/Hfq-binding sites are indicated by inverted triangles above the hut genes. The four oligoribonucleotide probes (1U, 3U, 5H and 6H) are 25 nt in length centred by the predicted Crc/Hfq-binding sequences. Histidine- and urocanate-induced expression of the three hut operons (hutF, hutCD and hutU-G) is mediated by HutC targeting operator sites located in the front of hutU and the hutF-C intergenic region. hutTu and hutTh encode the high-affinity transporter for urocanate and histidine, respectively. hutXWV encodes a high-affinity ABC-type transporter. The hut locus contains two hutH homologues, but hutH1 was not required for bacterial growth on histidine. The function of hutD remains unknown. (C) EMSA was performed using purified HfqHis6 and each of the four RNA probes labeled with biotin at the 5′-ends. HfqHis6 was added at 0, 55, 110, 220, 330, 440, 550, 660 and 660 nM in lanes 1–9, respectively. A 200-fold molar excess of the same unlabeled probe was added in lane 9 as a specific competitor for RNA binding. Asterisks on right-hand side of each gel denote positions of the shifted bands.
Subsequent assays of urocanate consumption, gene expression and growth kinetics (summarised in Supplementary Figures S5–S9) consistently revealed the regulatory interplay between CbrAB and NtrBC. Especially in the ‘succinate + histidine’ medium, bacteria face an obvious physiological dilemma in regard to hut enzymes. hut expression ought to be maximized as histidine is the sole N source. However, the physiological demand for C is low due to the presence of succinate, and hence hut expression ought to be repressed. Our data showed that under this nutrient condition CbrAB weakly activates hut transcription while de-repressing its expression at the translational level through the CrcYZ-Crc/Hfq cascade; NtrBC plays the dominant role in activating hut transcription. However, an analogous negative feedback loop was lacking with regard to homeostatic ntrBC regulation of hut.
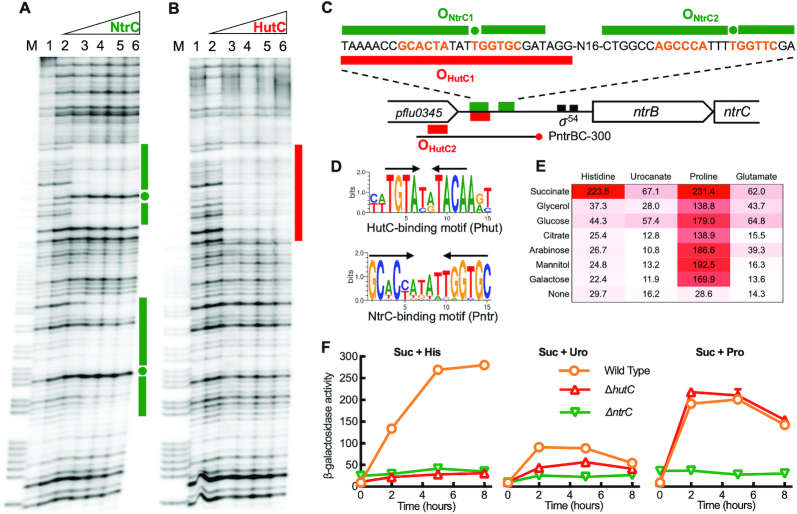
Identifying HutC as a direct regulator for ntrBC expression
ntrB and ntrC are overlapped by four nucleotides (ATGA), and the promoter region contains a typical σ54-binding site and two NtrC-binding motifs (designated Pntr) (Figure 4C). These suggest that ntrBC are co-transcribed and subject to autoactivation. Results of DNase I footprinting showed that NtrCHis6 has a strong binding affinity with ONtrC1 (Figure 4A). This is consistent with the prediction that ONtrC1 is the primary target site for ntrBC autoactivation. A relatively weak binding affinity was noted for ONtrC2 (Figure 4A).
Figure 4.
Biochemical and genetic characterization of the ntrBC promoter. (A) Identification of NtrC target sites by DNase I footprinting. Lane M, G+A marker; lanes 1–6, NtrCHis6 added at an increasing concentration from 0, 0.07, 0.2, 0.54, 1.1 to 1.7 μM. NtrC-protected regions are indicated by green bars, and hypersensitive residues are marked with filled circles. (B) DNase I analysis showing specific interactions between HutCHis6 and PntrBC promoter. Lane M, G+A marker; lanes 1–6, HutCHis6 added at an increasing amount from 0, 1.16, 2.32, 4.64, 7.54 to 10.44 μM. HutC protected region is marked by a red bar on the right side of the gel image. (C) A schematic map of the PntrBC promoter showing DNA sequences that were protected by NtrC and HutC from DNase I cleavage. The footprinting assays were performed by using the PntrBC-300 probe with a biotin-labeled 3′-end. (D) Sequence logos were generated from separate comparative analysis of PhutU and PntrBC promoter regions across 30 Pseudomonas species. Inverted repeats are marked with arrows. (E) A heat map showing variation in PntrBC promoter activities under different combinations of carbon and nitrogen sources. Bacteria were grown in minimal salt media supplemented with one of the seven C substrates (20 mM) and an amino acid (10 mM) as the sole N source, as indicated in the figure. β-Galactosidase activity was measured at 0, 2, 5 and 8 h after inoculation, but only mean values at 5 h are shown here for clarity. (F) Levels of PntrBC expression in wild type, ΔhutC and ΔntrC backgrounds. Bacteria were grown on succinate (20 mM) plus histidine, urocanate or proline (10 mM). Data are means and standard errors of six independent cultures.
Interestingly, the ntrBC locus possesses a typical HutC binding motif (designated Phut), which is located at the 3′-end of pflu0345 upstream of the ntrBC operon (Figure 4C). This Phut site (OHutC2) was shown to have negligible effects on ntrBC expression (31). However, in the process of OHutC2 functional analysis we surprisingly found that HutC was bound to an unexpected DNA region containing the dominant Pntr site (ONtrC1) for NtrC interaction (Figure 4B). The Phut and Pntr sites share little sequence similarity (Figure 4D). The HutCHis6-protected region was 3 bp longer than that of NtrCHis6, and it did not show the hypersensitive bands observed with NtrCHis6, highlighting the structural difference when the same probe DNAs (PntrBC-300) were bound with NtrCHis6 and HutCHis6 (Figure 4A and B). Furthermore, we showed that the protein-DNA interaction between HutCHis6 and the PntrBC-300 probe was disrupted by urocanate (Supplementary Figure S10A). Substitution of the Pntr site with random sequence eliminated the HutCHis6 binding activity (Supplementary Figure S10B). The binding affinity of HutC with a probe DNA containing the Pntr site (PntrBC-196) was measured to be 922.6 nM (Supplementary Figure S11), which is relatively much lower than the Kd with PhutU promoter (44.6 nM) (31). Together, the in vitro data indicate that HutC is capable of binding to the primary NtrC operator site (ONtrC1), and thus, it has the potential to directly inhibit ntrBC transcription.
HutC is functionally required for ntrBC expression
To test the hypothesis that HutC fine-tunes ntrBC expression, we first compared PntrBC-lacZ promoter activities in wild type and two isogenic mutants ΔhutC and ΔntrC in succinate + histidine medium (Figure 4F). PntrBC was not expressed in ΔntrC background, which is consistent with the predicted role of NtrC in activating PntrBC. However, PntrBC expression was also abolished in the ΔhutC background. This result was initially surprising as it cannot be explained by the known function of HutC as a transcriptional repressor.
Next, we systematically assessed PntrBC activities when the wild-type strain was grown on seven different carbon sources (succinate, glycerol, glucose, citrate, arabinose, mannitol and galactose) and five nitrogen sources (histidine, urocanate, proline, glutamate and NH4Cl). Results indicate that PntrBC was not expressed in NH4Cl-containing media (data not shown). PntrBC was expressed at high levels on proline regardless of the carbon sources (Figure 4E). When histidine was the sole N source, only succinate can greatly enhance PntrBC expression. Despite the fact that histidine breakdown generates one more ammonium than urocanate, PntrBC was generally expressed at higher levels on histidine than on urocanate (Figure 4E).
With this knowledge, we proceeded to compare PntrBC expression in wild type and ΔhutC by growing bacteria on succinate-containing media with histidine, urocanate or proline as the sole N source (Figure 4F). In the ΔhutC background, PntrBC was not expressed on histidine, but it was expressed on urocanate albeit at reduced levels when compared with wild type. More importantly, PntrBC was expressed normally in ΔhutC when bacteria were grown on succinate + proline (Figure 4F). From these datasets we conclude that the direct HutC/PntrBC interaction is not involved in the transcriptional activation of PntrBC. When growing on succinate + histidine, hutC is truly required for PntrBC activation, but the influence most likely be exerted indirectly through effects on the NtrC activator.
HutC acts as a governor of histidine catabolism
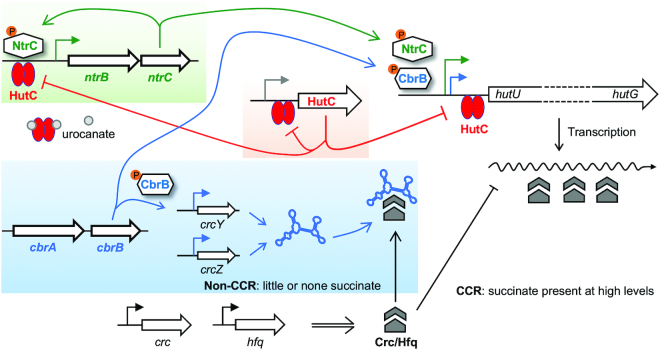
Available data led to a governor's model which posits that the low-affinity HutC/PntrBC interaction functions as a negative feedback loop for ntrBC autoactivation, setting an upper bound to the level of hut pathway activity when histidine fluctuates at high concentrations (Figure 5). Hence, the local transcription factor HutC plays a previously unrecognised coordinating role in controlling histidine catabolic rate, ensuring that bacterial cells maintain a N-starved physiological condition that is essentially required for NtrC functionality.
Figure 5.
C/N homeostatic regulation of hut genes by CbrAB, NtrBC and HutC. When histidine is utilized as a N source, transcription of hut genes is predominantly activated by NtrBC whose expression is autoactivated and involves repression by HutC as a negative feedback loop. HutC thus coordinates the expression of hut genes, hutC itself and the NtrBC activator in a histidine concentration-dependent manner. Utilization of histidine as a C source is subject to succinate-induced CCR control. In succinate-deplete media, CbrAB activates hut transcription while de-repressing the translation of hut mRNA mediated by the Crc/Hfq complex, which is sequestrated by the CbrAB-activated ncRNAs (CrcY and CrcZ). Of note, the CbrA sensor kinase can potentially detect histidine availability (52,54).
To further test the governor model, we first investigated how PntrBC responds to fluctuating C/N ratios in extracellular nutrients. Bacteria were grown in minimal media with varying concentrations of succinate relative to histidine or urocanate (Supplementary Figure S12). The data revealed a significant positive correlation between PntrBC activities and C/N ratios (Spearman's rho = 0.976, P < 0.0001 for histidine; rho = 0.891, P = 0.0011 for urocanate). Next, we tested PntrBC expression in response to intracellular dynamics of hut catabolism. Results described above show that PntrBC expression was abolished in ΔhutC cells grown on succinate (20 mM) and histidine (10 mM). The governor model posits that NtrC-inactivation was caused by over-expression of hut genes and the consequent build-up of excess N. Therefore, if we introduce a cbrB mutation into ΔhutC background, hut expression will be greatly reduced as a result of strong CCR in addition to the lack of CbrB-mediated weak promoter activity; and consequently, PntrBC expression should be restored in the double deletion mutant (ΔcbrB ΔhutC). Indeed, PntrBC expression was detected in ΔcbrB ΔhutC background from five hours after inoculation (Figure 6A).
Figure 6.
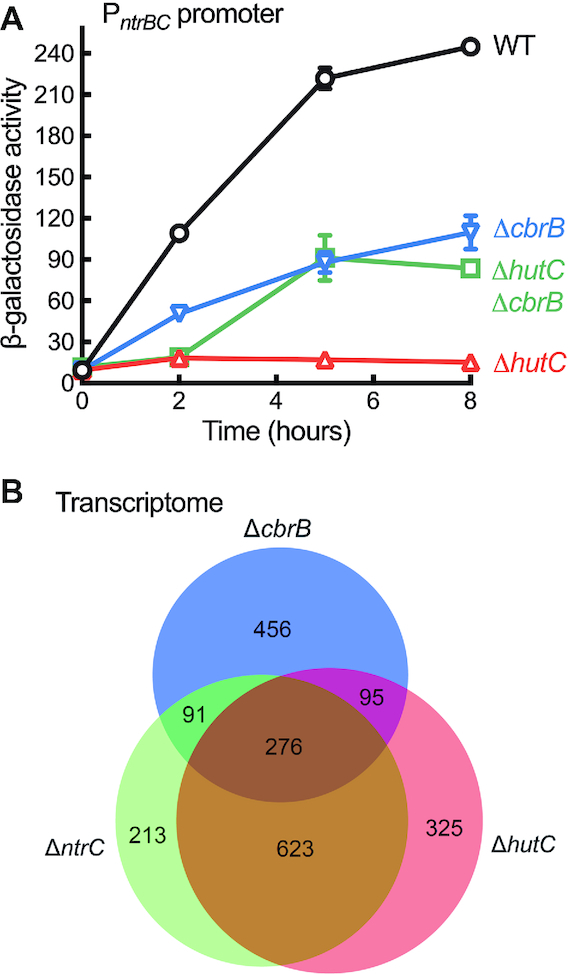
Role of hutC in determining PntrBC promoter activities and the global profiles of gene expression. (A) PntrBC-lacZ expression in wild type (WT) and three isogenic mutants devoid of hutC and/or cbrB. Data are means and standard errors of three independent cultures on succinate (20 mM) and histidine (10 mM). Two-way ANOVA revealed significant differences between genotypes (F3,8 = 678.6, P < 0.0001). (B) A Venn diagram showing the numbers of DEGs in mutants ΔcbrB, ΔntrC and ΔhutC. Total RNA was prepared from three biological replicates of bacterial cells exponentially grown on succinate (20 mM) and histidine (10 mM). Expression was detected by RNA-seq analysis for ∼97% of the predicted genes in the SBW25 genome.
Given that ntrBC is not expressed in ΔhutC for cells grown on succinate (20 mM) and histidine (10 mM), we would expect a large set of genes in the NtrC regulon to be affected by hutC deletion. This has been evidenced by a comparative transcriptome (RNA-seq) analysis of three mutants (ΔntrC, ΔhutC and ΔcbrB) relative to wild type. The ΔntrC and ΔhutC mutants shared a total of 899 differentially expressed genes (DEGs), which represents 68.16% DEGs detected for the ΔhutC mutant (Figure 6B and Supplementary Figure S13). These include genes involved in nitrogen metabolism, siderophore production and the synthesis of exopolysaccharides and flagella (Supplementary dataset 1).
Finally, we measured PntrBC and PhutU activities by growing wild type and ΔhutC mutant in minimal salt medium supplemented with 20 mM succinate and varying concentrations of histidine. The model posits that PntrBC ought to be expressed at constant high levels in wild type, and the expression cannot be maintained in ΔhutC, particularly in histidine-replete environments. As for PhutU, it ought to display a typical pattern of concentration-dependant induction along with the increase of histidine abundance. In the ΔhutC background, PhutU should be fully expressed at low histidine concentrations as we would normally expect for a substrate-specific transcriptional repressor, but the expression cannot be maintained in histidine-replete environments without HutC fine-tuning the PntrBC promoter activities. The results shown in Figure 7 are fully consistent with these predictions.
Figure 7.
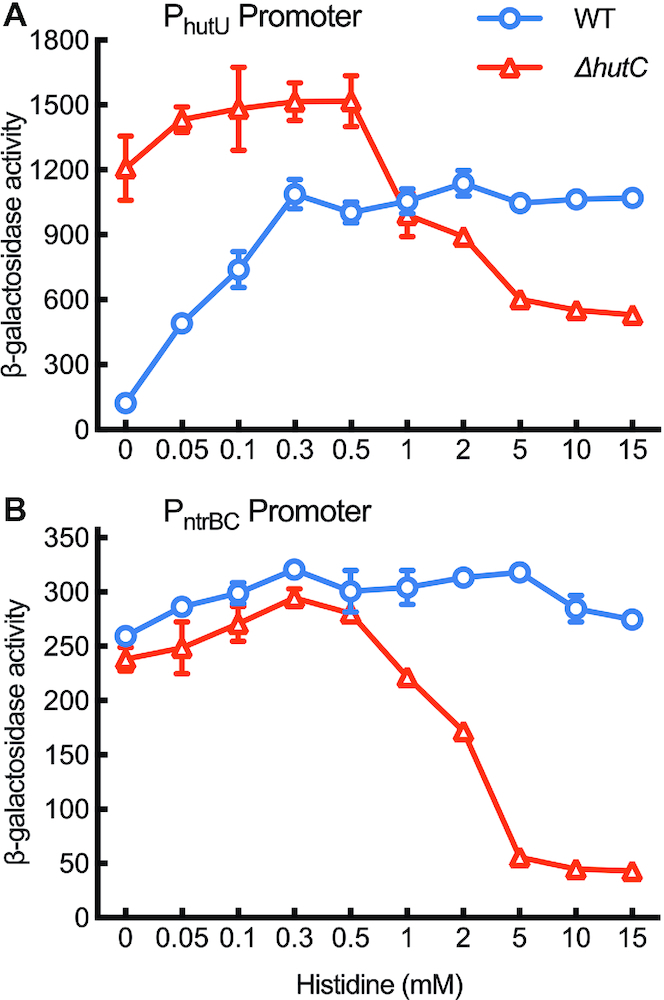
Role of hutC in maintaining PhutU and PntrBC expression in histidine-replete environments. Wild-type and ΔhutC mutant cells were N-starved for 2 hours in minimal salt medium supplemented with 20 mM succinate (A600 = ∼0.1). Bacterial growth started with the addition of histidine at varying concentrations shown in x-axis in a nonlinear scale. β-Galactosidase activities were measured at 5 hours after addition of histidine. Data are means and standard errors of three independent cultures. (A) PhutU promoter activities. Two-way ANOVA revealed a significant interaction between genotype and medium (F9,40 = 99.92, P < 0.0001). Differences between wild type and ΔhutC were highly significant (P < 0.001) in all mediums except 1 mM histidine. (B) PntrBC promoter activities. Two-way ANOVA revealed a significant interaction between genotype and medium (F9,40 = 141.3, P < 0.0001). Multiple t-tests show that differences between genotypes were highly significant (P < 0.001) when histidine was added at 1 mM or above.
DISCUSSION
Cells, like engines, are complex machines. Proper function of an engine requires the activity of individual parts to be maintained within acceptable bounds. A critical device in this regard is the governor - an element that measures and regulates the speed of an engine, protecting it from damage due to excessive rotational speed. While parallels in the design of machines and living systems are not uncommon (39), the governor-mode of regulatory control has rarely been observed in living cells (40,41). Here, we describe a new function for the HutC repressor whose behaviour is analogous to a governor. More specifically, HutC detects histidine availability, and meanwhile regulates the rate of hut catabolism through directly targeting the ntrBC promoter DNA. This negative feedback loop can help prevent the expression levels of ntrBC and hut genes from exceeding their critical upper limits.
HutC is a typical substrate-specific transcriptional regulator, controlling the expression of hut genes for histidine uptake and breakdown (23,31,42). HutC de-repression involves a positive feedback loop, which means that the activity of hut enzymes scales as a function of histidine availability (Figure 5). This is problematic in two accounts: first, excessive intracellular ammonium is poisonous to the cell; and second, ammonium sends a ‘N-replete’ signal to the NtrBC system leading to its inactivation. This physiological challenge is more serious for Pseudomonas strains as they use the 5-step hut pathway producing an additional ammonium compared with the four-step pathway adopted by most enteric bacteria (Figure 3A) (16).
Available evidence suggest that at least four distinct mechanisms are involved in the speed control of hut catabolism in Pseudomonas. First, hutD (a gene co-transcribed with hutC encoding a hypothetical protein) has an implicated function to prevent the accumulation of hut intermediates (i.e. formylglutamate) (43). Deletion of hutD resulted in hut expression at higher levels and caused a significant fitness reduction for bacterial growth on histidine (23). Secondly, at the present study we revealed that NtrBC activates hut expression in a direct manner. This is different from enteric bacteria wherein NtrBC indirectly activates hut genes through the NAC regulator (20). The direct control mode is thus likely evolved in Pseudomonas as a strategy to improve the response time to the N status of the cell.
The third potential mechanism involves ntrBC autorepression. Like many other TCSs, the ntrBC system is subject to autoactivation (4,14), and it has been well established that such a positive feedback loop is associated with a delayed response (44,45). Hence, P. fluorescens must have evolved additional mechanisms to overcome the fitness cost of autoactivation ensuring that ntrBC is expressed at optimal levels. In E. coli, ntrBC (also called glnLG) is located downstream of the glnA gene encoding glutamine synthetase. NtrC∼P activates ntrBC expression from a strong σ54-dependent promoter (glnAp2), while repressing the expression of two weak σ70-dependent promoters (glnAp1 and glnLp) (15). Moreover, the CAP-cAMP complex plays an inhibitory role in the NtrBC-mediated glnAp2 activity (46). The complex interactions between NtrC∼P and CAP-cAMP allow the integration of both C and N nutrient signals into ntrBC expression. Interestingly, Atkinson et al. identified a low-affinity NtrC-binding site in the glnAp2 promoter region, constituting a ‘governor’ responsible for setting the upper boundary of the promoter activity when the intracellular NtrC∼P is high (41). A coupled autorepression has been described in other positively autoregulated TCSs, such as PhoB/PhoR (47). Gao and Stock (13) has recently demonstrated that a high-affinity activation site of phoBR allows a fast response at low transcription factor (TF) levels, while the low-affinity site enables repression at high TF levels and ensures that expression of the TF does not exceed the optimal levels. In the present study, we identified a low-affinity NtrC-binding site in the PntrBC promoter of P. fluorescens SBW25 (Figure 4A) and revealed similar patterns of strong and weak NtrC-binding sites in the ntrBC loci of Pseudomonas spp. including P. aeruginosa and P. putida (Supplementary Figure S14). Therefore, autorepression at the low-affinity site is most likely a mechanism also adopted by Pseudomonas for preventing ntrBC over-expression. Such an autorepression mechanism is seemingly crucial for NtrBC to play its general role as a master nitrogen regulator, but it appears to be insufficient for maintaining C/N homeostasis for the utilization of histidine. To prevent NtrBC from ‘stalling’, the hut catabolic rate signal ought to be integrated into the transcriptional control of ntrBC.
The fourth mechanism concerns HutC acting as an ideal governor of hut catabolism. HutC allows the NtrBC system to directly sense histidine availability in addition to the general C/N status of the cell. A key piece of evidence leading to the governor's model was the specific binding of HutC with the PntrBC promoter DNA as revealed by DNase I footprinting (Figure 4B). Given that hutC expression is subject to autoregulation, the low-affinity HutC-PntrBC interaction is functionally significant only when histidine is present at high concentrations (and HutC is consequently abundant within the cell). When there is little or no histidine in the environment, the HutC-PntrBC interaction will be negligible and produce no significant effects on the expression of other nitrogen catabolic genes in the NtrBC regulon. This was evidenced by PntrBC expression in succinate + proline medium (Figure 4F).
It is interesting to note that the 28-bp HutC-protected region contains the highly conserved NtrC-binding site (Pntr) but shows little sequence similarity with the HutC consensus sequence (Phut) previously identified from the alignment of hut promoters (31). This finding is seemingly against the general concept that each TF recognizes a single consensus DNA sequence (or binding-site motif) (48). However, recently it has been found that some eukaryotic TFs have evolved the ability to recognize multiple DNA binding-site motifs (49). Eukaryotic TFs can potentially read the shape of the DNA molecule (not just the base sequence) as a major source of information for specific site recognition (50). For prokaryotic TFs, they typically recognize longer DNA sequences, which are considered sufficient to ensure specificity in small genomes (48). Given that TFs usually display some degrees of low-affinity DNA binding, it is highly possible that prokaryotic TFs, such as HutC, may have evolved specific binding to distinct DNA sites conferring new functions to be integrated into a sophisticated regulatory network (51).
Finally, we have resolved the CCR mechanism for histidine utilization in P. fluorescens SBW25. CbrAB is directly involved in activating hut transcription while indirectly de-repressing hut translation through the CbrAB-CrcYZ-Crc/Hfq cascade in response to the availability of C sources. Unlike ntrBC, expression of cbrA/cbrB and their modes of action remain largely rudimentary (52). cbrB is likely transcribed from its own promoter (18,19), whereas the expression of cbrA is translationally coupled with the preceding gene called cbrX (53). There has been no evidence suggesting that NtrBC is directly involved in cbrAB expression (and vice versa). Therefore, the C/N homeostasis is mainly maintained through the interplay between CbrB∼P and NtrC∼P at the operator sites of the PhutU promoter. More importantly, various feedback loops are involved in fine-tuning the CbrAB- and NtrBC-mediated hut activation (Figure 5), which includes HutC acting as a governor for the positively autoregulated NtrBC system.
However, it appears that CbrAB is not subject to direct control by the HutC governor and one may wonder why. A possible explanation resides in the unique domain structure of the CbrA sensor kinase. Its C-terminal autokinase domain is connected to a putative transporter in the sodium/solute symporter family (SSSF, TC 2A.21). In a previous work, we genetically characterised the cbrA gene of P. fluorescens SBW25, and the results suggested that the N-terminal SSSF domain of CbrA plays a dual role in histidine uptake and sensing (52). Interestingly, the histidine-specific transport activities have recently been demonstrated for CbrA in P. putida KT2440 (54). The transporter domain is not essentially required for phosphoryl transfer between CbrA and CbrB, but it is likely involved in modulating the CbrA autokinase activity (54). If CbrA is capable of sensing histidine availability by itself, it then makes sense that HutC is not involved in fine-tuning the expression of CbrA and CbrB. Together, the new modes of gene regulation revealed in this study highlight the functioning principles that are commonly found in living systems and artificial machines, and have important practical implications in synthetic biology (55).
DATA AVAILABILITY
The RNA-seq data have been deposited in NCBI’s Gene Expression Omnibus with accession number GSE160152.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Paul Rainey for his mentorship, Jacob Malone and Tim Cooper for helpful discussions, and Kiran Jayan, Yunhao Liu and Jonathan Gauntlett for technical assistance. N.N. was a recipient of a Massey University Doctoral Scholarship. X.-X.Z. acknowledges support from the Massey University Research Foundation (MURF) and MBIE Catalyst Fund, New Zealand (project no. 92846082).
Contributor Information
Naran Naren, School of Natural and Computational Sciences, Massey University at Albany, Auckland 0745, New Zealand.
Xue-Xian Zhang, School of Natural and Computational Sciences, Massey University at Albany, Auckland 0745, New Zealand.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Massey University Research Foundation (MURF); New Zealand Ministry for Business, Innovation and Employment (MBIE) Catalyst Fund [92846082 to X.-X.Z.]. Funding for open access charge: Massey University Research Foundation.
Conflict of interest statement. None declared.
REFERENCES
- 1. Stock A.M., Robinson V.L., Goudreau P.N.. Two-component signal transduction. Annu. Rev. Biochem. 2000; 69:183–215. [DOI] [PubMed] [Google Scholar]
- 2. Zschiedrich C.P., Keidel V., Szurmant H.. Molecular mechanisms of two-component signal transduction. J. Mol. Biol. 2016; 428:3752–3775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Igo M.M., Slauch J.M., Silhavy T.J.. Signal transduction in bacteria: kinases that control gene expression. New Biol. 1990; 2:5–9. [PubMed] [Google Scholar]
- 4. Groisman E.A. Feedback control of two-component regulatory systems. Annu. Rev. Microbiol. 2016; 70:103–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Goulian M. Two-component signaling circuit structure and properties. Curr. Opin. Microbiol. 2010; 13:184–189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bourret R.B. Learning from adversity?. J. Bacteriol. 2017; 199:e00420–00417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Mitrophanov A.Y., Groisman E.A.. Positive feedback in cellular control systems. Bioessays. 2008; 30:542–555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Gao R., Godfrey K.A., Sufian M.A., Stock A.M.. Counterbalancing regulation in response memory of a positively autoregulated two-component system. J. Bacteriol. 2017; 199:e00390-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Hermsen R., Erickson D.W., Hwa T.. Speed, sensitivity, and bistability in auto-activating signaling circuits. PLoS Comput. Biol. 2011; 7:e1002265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Maeda Y.T., Sano M.. Regulatory dynamics of synthetic gene networks with positive feedback. J. Mol. Biol. 2006; 359:1107–1124. [DOI] [PubMed] [Google Scholar]
- 11. Stein B.J., Fiebig A., Crosson S.. Feedback control of a two-component signaling system by an Fe-S-binding receiver domain. mBio. 2020; 11:e03383-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Ray J.C., Igoshin O.A.. Adaptable functionality of transcriptional feedback in bacterial two-component systems. PLoS Comput. Biol. 2010; 6:e1000676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Gao R., Stock A.M.. Overcoming the cost of positive autoregulation by accelerating the response with a coupled negative feedback. Cell Rep. 2018; 24:3061–3071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Ninfa A.J., Magasanik B.. Covalent modification of the glnG product, NRI, by the glnL product, NRII, regulates the transcription of the glnALG operon in Escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 1986; 83:5909–5913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Schumacher J., Behrends V., Pan Z.S., Brown D.R., Heydenreich F., Lewis M.R., Bennett M.H., Razzaghi B., Komorowski M., Barahona M.et al.. Nitrogen and carbon status are integrated at the transcriptional level by the nitrogen regulator NtrC in vivo. Mbio. 2013; 4:e00881-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Bender R.A. A NAC for regulating metabolism: the nitrogen assimilation control protein (NAC) from Klebsiella pneumoniae. J. Bacteriol. 2010; 192:4801–4811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Lonergan Z.R., Palmer L.D., Skaar E.P.. Histidine utilization is a critical determinant of Acinetobacter pathogenesis. Infect. Immun. 2020; 88:e00118-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Zhang X.X., Rainey P.B.. Dual involvement of CbrAB and NtrBC in the regulation of histidine utilization in Pseudomonas fluorescens SBW25. Genetics. 2008; 178:185–195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Nishijyo T., Haas D., Itoh Y.. The CbrA-CbrB two-component regulatory system controls the utilization of multiple carbon and nitrogen sources in Pseudomonas aeruginosa. Mol. Microbiol. 2001; 40:917–931. [DOI] [PubMed] [Google Scholar]
- 20. Bender R.A. Regulation of the histidine utilization (hut) system in bacteria. Microbiol. Mol. Biol. Rev. 2012; 76:565–584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Magasanik B. Catabolite repression. Cold Spring Harb. Symp. Quant. Biol. 1961; 26:249–256. [DOI] [PubMed] [Google Scholar]
- 22. Rojo F. Carbon catabolite repression in Pseudomonas: optimizing metabolic versatility and interactions with the environment. FEMS Microbiol. Rev. 2010; 34:658–684. [DOI] [PubMed] [Google Scholar]
- 23. Zhang X.X., Rainey P.B.. Genetic analysis of the histidine utilization (hut) genes in Pseudomonas fluorescens SBW25. Genetics. 2007; 176:2165–2176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Phillips A.T., Mulfinger L.M.. Cyclic adenosine 3',5'-monophosphate levels in Pseudomonas putida and Pseudomonas aeruginosa during induction and carbon catabolite repression of histidase synthesis. J. Bacteriol. 1981; 145:1286–1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Suh S.J., Runyen-Janecky L.J., Maleniak T.C., Hager P., MacGregor C.H., Zielinski-Mozny N.A., Phibbs P.V., West S.E.H.. Effect of vfr mutation on global gene expression and catabolite repression control of Pseudomonas aeruginosa. Microbiology. 2002; 148:1561–1569. [DOI] [PubMed] [Google Scholar]
- 26. Hug D.H., Roth D., Hunter J.. Regulation of histidine catabolism by succinate in Pseudomonas putida. J. Bacteriol. 1968; 96:396–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Liu Y.H., Gokhale C.S., Rainey P.B., Zhang X.X.. Unravelling the complexity and redundancy of carbon catabolic repression in Pseudomonas fluorescens SBW25. Mol. Microbiol. 2017; 105:589–605. [DOI] [PubMed] [Google Scholar]
- 28. Sonnleitner E., Abdou L., Haas D.. Small RNA as global regulator of carbon catabolite repression in Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. U.S.A. 2009; 106:21866–21871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Moreno R., Marzi S., Romby P., Rojo F.. The Crc global regulator binds to an unpaired A-rich motif at the Pseudomonas putida alkS mRNA coding sequence and inhibits translation initiation. Nucleic. Acids. Res. 2009; 37:7678–7690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Hester K.L., Lehman J., Najar F., Song L., Roe B.A., MacGregor C.H., Hager P.W., Phibbs P.V., Sokatch J.R.. Crc is involved in catabolite repression control of the bkd operons of Pseudomonas putida and Pseudomonas aeruginosa. J. Bacteriol. 2000; 182:1144–1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Naren N., Zhang X.X.. Global regulatory roles of the histidine-responsive transcriptional repressor HutC in Pseudomonas fluorescens SBW25. J. Bacteriol. 2020; 202:e00792-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Silby M.W., Cerdeno-Tarraga A.M., Vernikos G.S., Giddens S.R., Jackson R.W., Preston G.M., Zhang X.X., Moon C.D., Gehrig S.M., Godfrey S.A.et al.. Genomic and genetic analyses of diversity and plant interactions of Pseudomonas fluorescens. Genome Biol. 2009; 10:R51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Horton R.M., Hunt H.D., Ho S.N., Pullen J.K., Pease L.R.. Engineering hybrid genes without the use of restriction enzymes: gene splicing by overlap extension. Gene. 1989; 77:61–68. [DOI] [PubMed] [Google Scholar]
- 34. Rainey P.B. Adaptation of Pseudomonas fluorescens to the plant rhizosphere. Environ. Microbiol. 1999; 1:243–257. [DOI] [PubMed] [Google Scholar]
- 35. Choi K.H., Gaynor J.B., White K.G., Lopez C., Bosio C.M., Karkhoff-Schweizer R.R., Schweizer H.P.. A Tn7-based broad-range bacterial cloning and expression system. Nat. Methods. 2005; 2:443–448. [DOI] [PubMed] [Google Scholar]
- 36. Bao Y., Lies D.P., Fu H., Roberts G.P.. An improved Tn7-based system for the single-copy insertion of cloned genes into chromosomes of Gram-negative bacteria. Gene. 1991; 109:167–168. [DOI] [PubMed] [Google Scholar]
- 37. Liu Y., Rainey P.B., Zhang X.X.. Mini-Tn7 vectors for studying post-transcriptional gene expression in Pseudomonas. J. Microbiol. Methods. 2014; 107:182–185. [DOI] [PubMed] [Google Scholar]
- 38. Maxam A.M., Gilbert W.. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980; 65:499–560. [DOI] [PubMed] [Google Scholar]
- 39. Alberts B. The cell as a collection of protein machines: preparing the next generation of molecular biologists. Cell. 1998; 92:291–294. [DOI] [PubMed] [Google Scholar]
- 40. Noakes T.D., Peltonen J.E., Rusko H.K.. Evidence that a central governor regulates exercise performance during acute hypoxia and hyperoxia. J. Exp. Biol. 2001; 204:3225–3234. [DOI] [PubMed] [Google Scholar]
- 41. Atkinson M.R., Pattaramanon N., Ninfa A.J.. Governor of the glnAp2 promoter of Escherichia coli. Mol. Microbiol. 2002; 46:1247–1257. [DOI] [PubMed] [Google Scholar]
- 42. Allison S.L., Phillips A.T.. Nucleotide sequence of the gene encoding the repressor for the histidine utilization genes of Pseudomonas putida. J. Bacteriol. 1990; 172:5470–5476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Gerth M.L., Liu Y., Jiao W., Zhang X.X., Baker E.N., Lott J.S., Rainey P.B., Johnston J.M.. Crystal structure of a bicupin protein HutD involved in histidine utilization in Pseudomonas. Proteins. 2017; 85:1580–1588. [DOI] [PubMed] [Google Scholar]
- 44. Gao R., Stock A.M.. Quantitative kinetic analyses of shutting off a two-component system. mBio. 2017; 8:e00412-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Mitrophanov A.Y., Hadley T.J., Groisman E.A.. Positive autoregulation shapes response timing and intensity in two-component signal transduction systems. J. Mol. Biol. 2010; 401:671–680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Mao X.J., Huo Y.X., Buck M., Kolb A., Wang Y.P.. Interplay between CRP-cAMP and PII-Ntr systems forms novel regulatory network between carbon metabolism and nitrogen assimilation in Escherichia coli. Nucleic Acids Res. 2007; 35:1432–1440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Diniz M.M., Goulart C.L., Barbosa L.C., Farache J., Lery L.M., Pacheco A.B., Bisch P.M., von Kruger W.M.. Fine-tuning control of phoBR expression in Vibrio cholerae by binding of phoB to multiple pho boxes. J. Bacteriol. 2011; 193:6929–6938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Todeschini A.L., Georges A., Veitia R.A.. Transcription factors: specific DNA binding and specific gene regulation. Trends Genet. 2014; 30:211–219. [DOI] [PubMed] [Google Scholar]
- 49. Siggers T., Gordan R.. Protein-DNA binding: complexities and multi-protein codes. Nucleic. Acids. Res. 2014; 42:2099–2111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Inukai S., Kock K.H., Bulyk M.L.. Transcription factor-DNA binding: beyond binding site motifs. Curr. Opin. Genet. Dev. 2017; 43:110–119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Nakagawa S., Gisselbrecht S.S., Rogers J.M., Hartl D.L., Bulyk M.L.. DNA-binding specificity changes in the evolution of forkhead transcription factors. Proc. Natl. Acad. Sci. U. S. A. 2013; 110:12349–12354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Zhang X.X., Gauntlett J.C., Oldenburg D.G., Cook G.M., Rainey P.B.. Role of the transporter-like sensor kinase CbrA in histidine uptake and signal transduction. J. Bacteriol. 2015; 197:2867–2878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Monteagudo-Cascales E., Garcia-Maurino S.M., Santero E., Canosa I.. Unraveling the role of the CbrA histidine kinase in the signal transduction of the CbrAB two-component system in Pseudomonas putida. Sci. Rep. 2019; 9:9110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Wirtz L., Eder M., Schipper K., Rohrer S., Jung H.. Transport and kinase activities of CbrA of Pseudomonas putida KT2440. Sci. Rep. 2020; 10:5400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. de Lorenzo V. Evolutionary tinkering vs. rational engineering in the times of synthetic biology. Life Sci. Soc. Policy. 2018; 14:18. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The RNA-seq data have been deposited in NCBI’s Gene Expression Omnibus with accession number GSE160152.