Abstract
Many different protein domains are conserved among numerous species, but their function remains obscure. Proteins with DUF1127 domains number >17 000 in current databases, but a biological function has not yet been assigned to any of them. They are mostly found in alpha- and gammaproteobacteria, some of them plant and animal pathogens, symbionts or species used in industrial applications. Bioinformatic analyses revealed similarity of the DUF1127 domain of bacterial proteins to the RNA binding domain of eukaryotic Smaug proteins that are involved in RNA turnover and have a role in development from Drosophila to mammals. This study demonstrates that the 71 amino acid DUF1127 protein CcaF1 from the alphaproteobacterium Rhodobacter sphaeroides participates in maturation of the CcsR sRNAs that are processed from the 3′ UTR of the ccaF mRNA and have a role in the oxidative stress defense. CcaF1 binds to many cellular RNAs of different type, several mRNAs with a function in cysteine / methionine / sulfur metabolism. It affects the stability of the CcsR RNAs and other non-coding RNAs and mRNAs. Thus, the widely distributed DUF1127 domain can mediate RNA-binding, affect stability of its binding partners and consequently modulate the bacterial transcriptome, thereby influencing different physiological processes.
Graphical Abstract
Graphical Abstract.
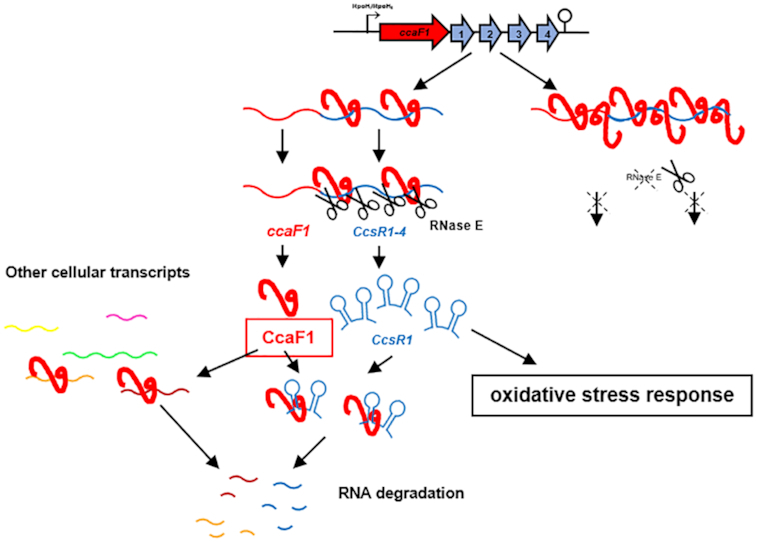
CcaF1 protein(red) binds to the ccaF1-ccsR1-4 transcript and together with RNase E contributes to maturation of the CcsR1-4 RNAs. Decreased RNase E activity or excess of CcaF1 impede this maturation. By acting on CcsR maturation CcaF1 contributes to the oxidative stress response. CcaF1 also binds to several other cellular RNAs suggesting its influence also on other physiological processes.
INTRODUCTION
Bacterial genomes typically harbor many small open reading frames that have not been annotated in the past due to their small size. More recently it has emerged that small proteins participate in a multitude of cellular processes (1,2). Although only a minor fraction of the small proteins could be analyzed up to now, they exhibit a great diversity in their mechanisms of action and their physiological functions. Important roles for small proteins in e.g. cell division, transport, spore formation and signal transduction have been unraveled (2).
This study uncovers a new function for a small protein from Rhodobacter sphaeroides and, to the best of our knowledge, represents the first functional assignment to a member of the DUF1127 proteins. Rhodobacter sphaeroides is a facultative phototrophic alphaproteobacterium living in fresh and brackish water habitats. If sufficient oxygen is available, it can perform aerobic respiration. When oxygen becomes limiting or under anaerobic conditions, ATP is produced by anoxygenic photosynthesis, anaerobic respiration or fermentation. Since the simultaneous presence of (bacterio-) chlorophylls, light and oxygen leads to the production of the harmful singlet oxygen, the formation of photosynthetic complexes is tightly controlled by redox and light signals (3–6). Furthermore, R. sphaeroides has developed a complex regulatory network consisting of proteins and sRNAs to defend against singlet oxygen stress (7–11). To date several different sRNAs that are induced by oxidative stress and have roles in oxidative/singlet oxygen stress response have been investigated in R. sphaeroides (12–17). By interacting to the mRNA for the transcriptional regulator FlhR, the four homologous sRNAs CcsR1–4 modulate the C1 metabolism and the pyruvate dehydrogenase complex in response to various stresses (18). As a consequence, the pool of the reductant glutathione is increased and aerobic electron transport, a main source of reactive oxygen species (ROS), is reduced. The CcsR1–4 sRNAs are derived from the 3′ UTR of the RSP_6037 mRNA (Figure 1A). Transcription of the RSP_6037-CcsR genes is initiated at a RpoHI/RpoHII-dependent promoter (18).
Figure 1.
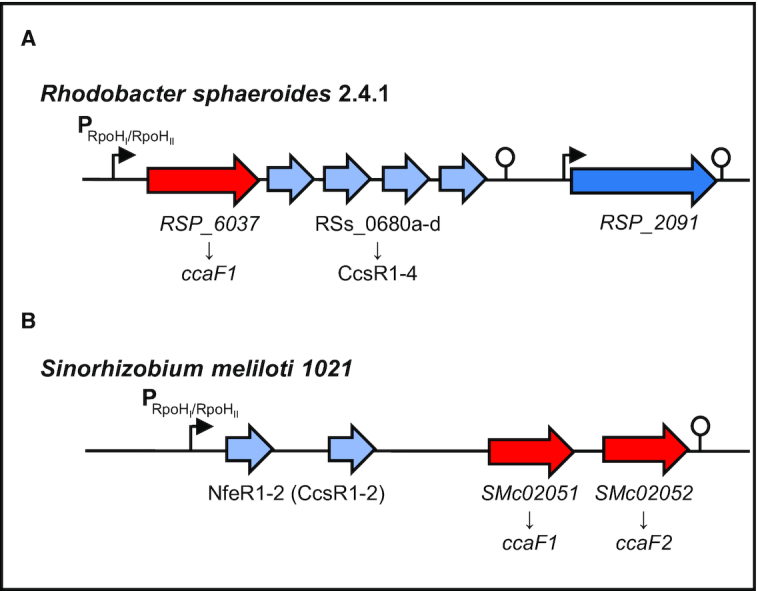
Schematic representations of the CcsR-RNA/DUF1127-protein loci in R. sphaeroides 2.4.1 and S. meliloti 1021. (A) Genomic context of the DUF1127 protein RSP_6037/CcaF1 (red) and the CcsR1–4 sRNAs (light blue) from R. sphaeroides 2.4.1. The protein–sRNA operon is preceded by a RpoHI/RpoHII promoter (black arrow) and a Rho-independent terminator structure is located at the 3′ end (modified from Billenkamp et al. (18)). (B) Corresponding locus of CcsR(NfeR)-RNAs and DUF1127 proteins in S. meliloti 1021. Open reading frames of the DUF1127 proteins are colored red, while sRNAs are colored in light blue. An RpoHI/RpoHII dependent promoter and a terminator are indicated by an arrow and a hairpin structure.
The two alternative sigma factors RpoHI and RpoHII are known to activate many genes in R. sphaeroides under a variety of stress conditions (8,19,20) and are also important for outgrowth after long stationary phase (21). Each CcsR RNA harbors two hairpin–loop structures and each loop contains a CCUCCUCCC anti-Shine Dalgarno sequence (7) that prompted Reinkensmeier and Giegerich (22) to name them ‘Cuckoo’ RNAs. RSP_6037 encodes a small protein of unknown function of 71 amino acids. Amino acids 23–62 constitute a DUF1127 domain (18). More than 17,000 bacterial protein sequences with DUF1127 domains in about 4000 bacterial species are listed in InterPro and the number is steadily increasing (23). The DUF1127 domain consists of 45–50 amino acids and often covers almost the entire protein. Alternatively, the DUF1127 domain is located at the C-terminus of slightly larger proteins with 60–75 amino acids. DUF1127 proteins are widely distributed among Alpha- and Gammaproteobacteria and mostly found in the orders Rhizobiales, Rhodobacterales, Enterobacteriales and Pseudomonales. Our previous work demonstrated that RSP_6037 influences the amounts of the CcsR RNAs, that are produced from the 3′ UTR of the RSP_6037 transcript (18).
According to Reinkensmeier and Giegerich (22) adjacency to ORFs with DUF1127 domains defines an orthologous subgroup of Cuckoo RNAs that is characterized by this genomic context. This subgroup, labeled CIN1 (conserved intergenic neighborhood 1), is present only in the Rhodobacteraceae, Brucellaceae, Rhizobiaceae and Phylobacteraceae (22). Barnett et al. (24) found that the RSP_6037 ortholog SMc02051 (47 aa) from Sinorhizobium meliloti 1021 has a RpoHI and RpoHII responsive expression as described for the CcsR locus in Rhodobacter sphaeroides (Figure 1B). Interestingly, a similar responsiveness was observed for an adjacent DUF1127-containing ORF (SMc02052). SmMc02051 also has adjacent Cuckoo RNAs with a predicted σ70-dependent promoter and is classified as a CIN1 member (25).
Here, we present major advancements in the investigation of RSP_6037 as a model for the DUF1127 domain containing ORFs that are adjacent to Cuckoo RNAs and demonstrate that this DUF1127-containing protein binds RNAs and can affect RNA stability.
MATERIALS AND METHODS
Bacterial strains and growth conditions
Rhodobacter sphaeroides strains (listed in S1 Table) were cultivated in a malate minimal-salt medium or on solid medium containing 1.6% (w/v) agar at 32°C in the dark (26). For microaerobic growth conditions (25–30 μM of dissolved oxygen) Erlenmeyer flasks filled up to 80% of the maximum volume were shaken at 140 rpm. When necessary, tetracycline (2 μg ml–1) or spectinomycin (10 μg ml−1) was added to liquid and solid growth media. Stress conditions were generated by a final concentration of 300 μM tBOOH, 1 mM H2O2 or 250 μM paraquat (O2–) or by temperature shift to 42°C under microaerobic conditions.
To culture Escherichia coli strains (listed in S1 Table), cells were continuously shaken at 180 rpm in Luria–Bertani (LB) medium at 37°C or grown on solid growth media containing 1.6% (w/v) agar. When necessary, tetracycline (20 μg ml−1) or spectinomycin (10 μg ml−1) was added to the media.
S. meliloti 1021 was cultivated similar to R. sphaeroides strains using TY medium (27).
Construction of overexpression plasmids
For construction of a plasmid for constitutive overexpression of the small DUF1127 protein CcaF1 (RSP_6037) from R. sphaeroides 2.4.1 without the sRNA cluster CcsR1–4 but with the terminator structure of this gene-locus at the 3′ end, a 310 bp fragment of the ccaF1 (RSP_6037) gene locus (primers: CcaF1_f and CcaF1int_r; Supplementary Table S2) and a 135 bp fragment of the terminator sequence (primers: CcaF1int_f and CcaF1_r; Tabel S2) were amplified by PCR using chromosomal R. sphaeroides 2.4.1 DNA as template. Both fragments have an overlapping region. By a second PCR step both fragments were fused resulting in a 445 bp fragment of the ccaF1 (RSP_6037) gene sequence and the terminator sequence. The corresponding fragment was sub-cloned into the BamHI and EcoRI sites of the pJET1.2 cloning vector (Thermo Fisher Scientific, Waltham, MA, USA) and, after digestion with the corresponding restriction enzymes, ligated into the expression vector pRK4352 (28).
The constitutive overexpression of the ccaF1 (RSKD131_0402) gene from R. sphaeroides KD131 (Primers are listed in Supplementary Table S2) was performed in the same way like for ccaF1 (RSP_6037) from R. sphaeroides 2.4.1, but using chromosomal R. sphaeroides KD131 DNA as template. The constitutive overexpression of the sRNA cluster CcsR1–4 and the whole gene locus from R. sphaeroides KD131 (primers are listed in Supplementary Table S2) was performed in the same way as described in (18), but using chromosomal R. sphaeroides KD131 DNA as template. The resulting overexpression plasmids were conjugated from E. coli S17-1 to R. sphaeroides 2.4.1 (29).
Construction of 3xFLAG-tagged CcaF1 and RSP_0557 expression strains
The R. sphaeroides RSP_6037 and RSP_0557 loci were amplified by PCR of pRK6037 and pBBR0557 (17) plasmid DNA using primers CcaF1FLAG_NT_f and CcaF1FLAG_NT_r or RSP0557FLAG_NT_f and RSP0557FLAG_NT_r (primers are listed in Supplementary Table S2). The amplified fragments start with an ATG start-codon at the 5′ end followed by the 3xFLAG sequence at the N-terminus and the ccaF1 (RSP_6037) or RSP_0557 gene. The fragment was sub-cloned into the BamHI and EcoRI sites of the pJET1.2 cloning vector (Thermo Fisher Scientific, Waltham, MA, USA) and ligated into the pRK4352 overexpression vector (28) after digestion with suitable restriction enzymes. The resulting overexpression plasmids pRKCcaF1FLAG_NT and pRK0557FLAG_NT were transferred from E. coli S17-1 to R. sphaeroides 2.4.1 by biparental conjugation (29).
Construction of a His6-MBP-TEV-CcaF1 overexpression plasmid
The Gibson assembly method was used to construct the plasmids for heterologous protein expression. The used oligonucleotides (primers are listed in Supplementary Table S2) provided overlaps of at least 20 nucleotides. Purified PCR fragments were used in equimolar concentrations and the reaction was incubated for 60 minutes at 50°C. Subsequently the reaction mixture was cooled to 4°C and transformed into chemically competent cells of strain E. coli DH5α λpir (30).
Zone of inhibition, survival assay and spot assay
Zone of inhibition assay was performed as described in Li et al. (31). The 5 mm filter-paper disks contain 5 μl of oxidative agent (200 μM paraquat or 700 μM tBOOH). The plates were incubated for 48 h at 32°C in the dark and the diameter of the zone of inhibition indicates the sensitivity of the cells against the agent.
For determination of survival rates, cultures were grown under microaerobic conditions. 300 μM tBOOH were added and after 30 min, 60 min or 90 min dilutions were plated on solid malate minimal-salt medium. The plates were incubated for 48 h at 32°C in the dark. The number of colonies of a control culture grown without the addition of any oxidative stress agents was defined as 100% survival.
To test the growth behavior of the different strains in presence of various stresses we performed a spot assay. Cultures were grown at 32°C in microaerobic condition. At an OD660 of 0.5 10 μl of different dilutions (100–10–5) were spotted on an agar plate containing the stress agents (10 μM CdCl2, 250 mM NaCl, 100 μM tBOOH). The plates were incubated for 48 h at 32°C in the dark. For heat shock, plates were incubated at 42°C over-night and afterwards at 32°C. The intensity of the spots was quantified by the 1D-Quantity One software (Bio-Rad). The control spot (100) was defined as 100% survival.
Determination of RNA half-life
The R. sphaeroides cultures of interest were incubated under the desired growth condition (see bacterial growth conditions) to an OD660 0.5. After taking sample t0, rifampicin was added to a final concentration of 0.2 mg/ml. The cells were harvested by centrifugation (10 000 rpm, 10 min, 4°C) at defined time points and RNA was isolated.
RNA isolation
For RNA isolation R. sphaeroides cultures were grown to an OD660 nm 0.5 under the different growth conditions (see bacterial growth conditions). Cells were harvested by centrifugation at 10 000 rpm for 10 min at 4°C. RNA was isolated for Northern Blot analysis, RT-PCR and RNA sequencing analysis using the hot phenol method (32) and precipitated with 1/10× vol. 3 M sodium acetate pH 4.5 and 2.5× vol. 96% ethanol. For RNA sequencing the remaining DNA was removed by TURBO-DNase treatment (Invitrogen).
Northern blot
For detection of small RNAs 7.5 μg total RNA were separated on a 10% polyacrylamide gel containing 7 M urea. Afterwards RNA was transferred to Nylon membranes (Roth) by semi-dry electroblotting. Oligodeoxynucleotides (listed in S2 Table) for detection were labeled with [γ-32P]-ATP (Hartmann Analytic) by T4 polynucleotide kinase (Fermentas; #EK0031) and were hybridized overnight. Membranes were exposed on phosphoimaging screens (Bio-Rad) and analyzed by the 1D-Quantity One software (Bio-Rad). For determination of mRNAs and precursor transcripts 10 μg total RNA were separated on a 1% (w/v) agarose 2.2 M formaldehyde gel and transferred to nylon membrane by vacuum pressure blotting. DNA fragments of the mRNAs and precursor transcripts were labeled with [α-32P]-dCTP (Hartmann Analytic) using nick translation (nick translation kit; Amersham Pharmacia Biotech). Membranes were hybridized overnight, exposed on phosphoimaging screens (Bio-Rad) and analyzed by the 1D-Quantity One software (Bio-Rad).
Co-immunoprecipitation
For co-immunoprecipitation R. sphaeroides pRKCcaF1FLAG_NT and R. sphaeroides pRKCcaF1 or R. sphaeroides pRK0557FLAG_NT and R. sphaeroides pRK0557 were grown under microaerobic conditions and harvested in exponential growth phase (culture volume of 400 ml) by centrifugation at 10 000 rpm at 4°C. Pellets were resuspended in 2 ml of cold lysis buffer (20 mM Tris pH 7.5, 150 mM KCl, 1 mM MgCl2, 1 mM DTT) and disrupted by sonication (33). Cell lysate was centrifuged for 10 min at 13 000 rpm and 4°C, followed by an ultra-centrifugation step (100 000 rpm, 1 h, 4°C). Afterwards the supernatant was mixed with 40 μl of ANTI-FLAG M2 Magnetic Beads (Sigma-Aldrich) and incubated for 2 h, at 4°C under rotation. Following five washing steps with 500 μl of lysis buffer, magnetic beads were resuspended in 500 μl of lysis buffer and RNA was isolated with phenol and chloroform–isoamyl alcohol followed by precipitation with 1/10× vol. 3 M sodium acetate pH 4.5 and 2.5× vol. 96% ethanol overnight. The precipitated CoIP RNA was treated by DNase I (Invitrogen) to remove any DNA contaminations. The isolated RNA was analyzed by RNA sequencing and RT-PCR.
Reverse transcription (RT) PCR
CoIP RNA was analyzed after DNase-treatment by a reverse transcription (RT) PCR using the One-Step Brilliant III QRT-PCR Master Mix Kit (Agilent). Each 10 μl reaction mixture contained 5 μl Master Mix (supplied), 0.1 μl DTT (100 mM, supplied), 0.5 μl Ribo-Block solution (supplied), 0.4 μl water, 1 μl of each primer (10 pmol/μl) listed in Supplementary Table S2, and 2 μl RNA (20 ng/μl). The reactions were performed in a spectrofluorometric thermal cycler (BioRad) and analyzed by BioRad CFXManager 3.0. Afterwards the RT-PCR products were separated on a 10% polyacrylamide gel and analyzed by ethidium-bromide staining.
Protein production and purification
Strain E. coli BL21 DE3 (New England Biolabs, Germany) carrying a pET24c plasmid was used for recombinant protein expression. Cells were grown in LB medium supplemented with kanamycin (50 mg/l) at 37°C under vigorous shaking until an OD600 of 0.75 was reached. Subsequently the culture was cooled for 10 min in an ice bath before protein expression was induced by addition of d-(+)-lactose-monohydrate (12.5 g/l). The culture was then incubated at 16°C under vigorous shaking for 16 h. Cells were harvested by centrifugation (5000 rpm, 10 min, 4°C), the resulting cell pellet flash frozen in liquid nitrogen and stored at –20°C until use.
For protein purification cells were resuspended in lysis buffer (50 mM Tris–HCl, 500 mM NaCl, 50 mM KCl, 10 mM MgCl2·6H2O, 20 mM imidazole, 1 mM DTT, 0.02% Tween20, pH 8) and lysed by sonication (Bandelin Sonoplus). Cell debris and intact cells were removed by centrifugation (20 000 rpm, 30 min, 4°C) and filtration. The obtained lysate was then loaded onto a 5 ml HisTrap HP (GE healthcare) column and equilibrated with lysis buffer using the ÄKTA PURE25 system. The column was then washed with 10 CV (column volume) lysis buffer. A linear gradient of 3 CV elution buffer (50 mM Tris–HCl, 500 mM NaCl, 50 mM KCl, 10 mM MgCl2·6H2O, 600 mM imidazole, 1 mM DTT, 0.02% Tween20, pH 8) from 10 to 100% was used to elute the protein, followed by 2 CV elution buffer. Elution fractions were analyzed by 12% SDS-PAGE. Samples containing the protein of interest were then combined and centrifuged (13 000 rpm, 1 min, 4°C) prior to loading 500 μl onto a Superdex 200 increase 10/300 GL SEC column equilibrated with SEC buffer (50 mM Tris-–HCl, 500 mM NaCl, 50 mM KCl, 10 mM MgCl2·6H2O, 1 mM DTT, pH 8) and connected to the ÄKTA PURE25 system. After isocratic elution with SEC buffer (0.5 ml/min) fractions containing the protein of interest were identified by 12% SDS PAGE.
100 U (w/w) of TEV protease (New England Biolabs, Germany) were applied over night at 4°C for cleavage of the fusion protein. The His6-TEV protease and the His6-MBP-tag were removed by nickel NTA agarose beads (Qiagen). Fractions containing the protein of interest were identified by 12% SDS PAGE.
Electrophoretic mobility shift assay (EMSA)
RNA was transcribed in vitro using T7 polymerase (NEB) and PCR products as template, which contain the T7 promoter region at the 5′ ends. 150 fmol of the radio-labelled RNA was denatured separately for 1 min at 95°C, cooled down for 2 min on ice and renatured for 5 min at 32°C. After these de- and renaturing steps, 5x structure buffer (25 mM MgCl2 and 300 mM KCl) and the purified protein CcaF1 in different molar ratios were added in a final volume of 10 μl. For formation of the RNA–protein complex, the samples were incubated for 30 min at 32°C. Afterwards, the reactions were mixed with 3 μl of loading dye (50% glycerol, 0.5× TBE, 0.2% bromophenol blue) and loaded onto a 6% non-denaturing polyacrylamide gel containing 0.5× TBE. Gels were pre-run in 0.5× TBE running buffer at 100 V for 60 min at room temperature before loading. Electrophoresis was performed at room temperature by applying 200 V for 4 h. Gels were dried, exposed on phosphoimaging screens (Bio-Rad) and analyzed by the 1D-Quantity One software (Bio-Rad).
Library construction, RNA sequencing and data analysis
RNA sequencing data are based on triplicates and the RNA for each triplicate stemmed from three independent cultures. After harvesting the respective aliquots total RNA was extracted followed by DNase treatment. RNA quality was checked using a 2100 Bioanalyzer with the RNA 6000 Nano kit (Agilent Technologies). The RNA integrity number (RIN) for all samples was between 2.2 and 5.1. 300 ng of total RNA were used for the preparation of a cDNA library with the NEBNext Multiplex Small RNA Library Prep kit for Illumina (NEB) in accordance with the manufacturers’ instructions with modifications: RNA was dephosphorylated at the 3′ end, phosphorylated at the 5′ end and decapped using 10 U T4-PNK ± 40 nmol ATP and 5 U RNA 5′ pyrophosphohydrolase (RppH), respectively (NEB). After each enzymatic treatment RNA was purified with the Zymo RNA Clean & Concentrator kit. The RNA fragments were ligated for cDNA synthesis to 3′ SR adapter and 5′ SR adapter diluted 1:3 with nuclease-free water before use. PCR amplification to add Illumina adaptors and indices to the cDNA was performed for 14 cycles with 1:3 diluted primer. Barcoded DNA Libraries were purified using magnetic MagSi-NGSPREP Plus beads (AMSBIO) at a 1.8 ratio of beads to sample volume. Libraries were quantified with the Qubit 3.0 fluorometer (ThermoFisher) and the library quality and size distribution were checked using a 2100 Bioanalyzer with the DNA-1000 kit (Agilent). Sequencing of pooled libraries, spiked with 10% PhiX control library, was performed in single-end mode on the NextSeq 500 platform (Illumina) with the High Output Kit v2.5 (75 Cycles). Demultiplexed FASTQ files were generated with bcl2fastq2 v2.20.0.422 (Illumina). The sequencing data are available at NCBI Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo) under the accession number GSE144523 and GSE145045. The adapter sequences were removed from the sequence reads in Fastq format. Read processing, generation of statistics, gene-wise read counting, coverage calculations and normalization were performed using READemption version 0.4.3 (34) using segemehl version 0.2.0 (35,36) for read alignments. Gene expression analysis was computed via DESeq version 1.22.1 (37). Downstream processing and statistical analysis were performed using the statistical language R (http://www.r-project.org).
Phylogenetic tree of conserved CcaF1 regions
Ninety-five CcaF1 amino acid sequences based on the CIN1 loci from Reinkensmeier and Giegerich 2015 (22) were collected from the NCBI database and aligned based on the DUF1127 domain in MEGA X (38). A phylogenetic tree was generated using the UPMGA method (39). A bootstrap consensus tree was derived from 500 replicates (40). Branches corresponding to partitions reproduced in <50% bootstrap replicates were collapsed. All positions with <95% site coverage were eliminated, i.e., fewer than 5% alignment gaps, missing data, and ambiguous bases were allowed at any position (partial deletion option). There was a total of 46 positions in the final dataset consisting of the DUF1127 domain and flanking amino acids, while further extensions were removed from analysis.
RESULTS
Comparison of different CcsR loci in Alphaproteobacteria
The genomic context of Cuckoo RNAs associated with a DUF1127-containing orthologous gene was labeled CIN1 (22). CIN1 includes the RNA family RSs0680 (now CcsR) and several members of the Rfam RNA family ar14. ar14 from Sinorhizobium meliloti 1021 includes the DUF1127-containing ORF SMc02051. The Cuckoo RNAs from this locus have been named NfeR (nodule formation efficiency RNA) and are expressed in root nodules and under salt stress, possibly from an RpoHI/HII-dependent promoter (25). Our data demonstrate that the CcsR RNAs from this CIN1 locus of S. meliloti 1021 are also induced by heat stress, but not by oxidative stress. The same result was obtained for CcsR1 from S. fredii HH103, while CcsR1 from R. capsulatus SB1003 was induced by superoxide and heat (Supplementary Figure S1). Therefore, the name CcsR (conserved CCUCCUCCC (‘cuckoo’)-motif stress-induced RNA) will be generally used for Cuckoo RNAs in CIN1 loci throughout the manuscript following the name of their first characterized example (18).
In R. sphaeroides 2.4.1 the CcsR RNAs are derived from the 3′ UTR of the RSP_6037 mRNA by RNase E-dependent processing (41). In contrast, the CcsR RNAs in S. meliloti 1021 appear to be derived from the 5′ UTR of SMc02051 (Figure 1B). This leads to two common CIN1-locus orientations that are partially exclusive to taxonomic groups (Supplementary Figure S2).
A phylogenetic tree of the conserved regions of 95 selected amino acid sequences of DUF1127-proteins from CIN1 loci correlates with a phylogenetic tree based on 16S rDNA and with the family level assignments of the organisms harboring the CcsR RNA loci (Supplementary Figure S2). The typical R. sphaeroides locus with multiple CcsR RNAs derived from the 3′ UTR of the DUF1127-containing ORF is predominant in the Rhodobacteraceae, Phylobacteraceae and most Rhizobiaceae, while the Brucellaceae typically harbor two distinct CIN1 loci, each with only one CcsR RNA. The amino acid sequences of the associated DUF1127-proteins form separate clusters in this case (Supplementary Figure S2).
In the CIN1 loci of Sinorhizobium, one DUF1127-coding ORF is directly adjacent to the CcsR-RNAs and clusters together with the DUF1127-coding ORFs of the CIN1 loci in other Alphaproteobacteria. This ORF is followed by a second DUF1127-coding ORF, which might be an extension to the CIN1 locus based on expression in S. meliloti 1021. However, the representatives of this ORF form a separate cluster in the phylogenetic tree that is distinct from the other CIN1 related DUF1127-proteins (Supplementary Figure S2).
Since the DUF1127-containing ORFs form signature ORFs for the definition of CIN1 loci, we named RSP_6037 and its orthologs ‘CcaF1’ (conserved CcsR associated factor).
In silico characterization of the CcaF proteins
The 71 aa CcaF1 protein from R. sphaeroides 2.4.1 comprises an arginine-rich DUF1127 domain in its C-terminal part. The PHYRE2 webserver predicts a structure for this CcaF1 protein, which strongly resembles (71% confidence) the RNA-binding domain of the Smaug protein of Drosophila melanogaster (Supplementary Figure S3). The Smaug protein represses translation and induces mRNA decay in Drosophila embryos (42,43). PHYRE2 also detected structural homology to SAM (sterile alpha motif) pointed domain containing proteins with up to 68% confidence. SAM pointed domains are involved in protein-protein interaction and occur in eukaryotic and some bacterial proteins (www.ebi.ac.uk/interpro/entry/ IPR013761). Interestingly, the RNA binding domain of Smaug also comprises a SAM sub-domain, which was shown to interact with RNA (44). This was the first report that SAM domains can also interact with RNA. Some residues of this small domain are conserved between the Smaug domain and CcaF1, while other residues are only conserved among the bacteria (Supplementary Figure S3). CcaF1 from R. sphaeroides was shown to influence CcsR levels (18). Hence, CcaF1 from R. sphaeroides 2.4.1 can serve as a hub for a more detailed analysis of the CcaF protein group. Due to the small sizes of CcaF proteins, characterization of this protein group also goes along with functional characterization of the DUF1127 domain.
The CcaF1 protein affects CcsR levels in trans and alters stress resistance
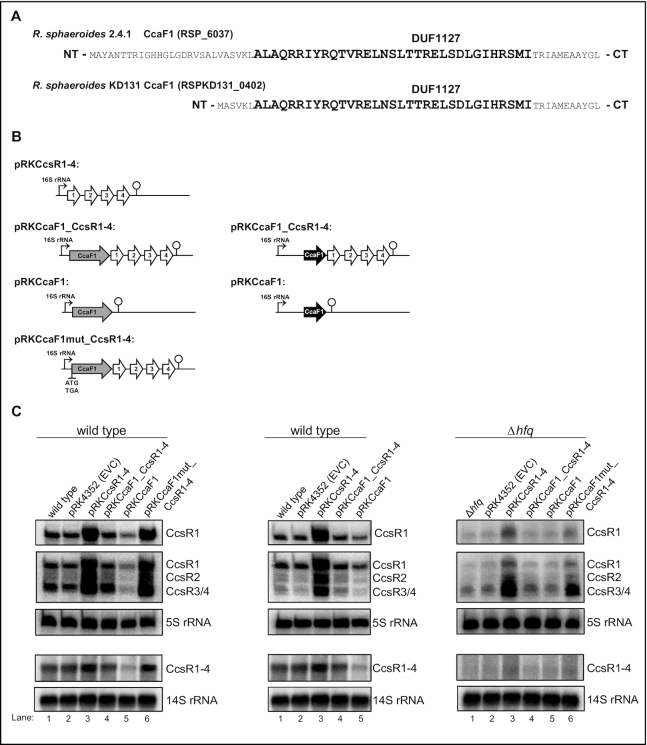
We did not achieve deletion of the chromosomal ccaF1-ccsR locus indicating that these genes are essential (18). Therefore, we chose an overexpression strategy to study the role of CcaF1. Figure 2A shows the complete amino acid sequences of CcaF1 orthologs from the R. sphaeroides strains 2.4.1 (71 aa) and KD131 (50 aa). A schematic overview of the plasmid constructs we used to study the effect of CcaF1 (RSP_6037) on the CcsR levels is shown in Figure 2B. All combinations of ccaF1 and ccsR genes were cloned under the control of the strong 16S promoter on a plasmid. Plasmid pRK4352 with 16S promoter but no cloned genes from R. sphaeroides served as control (EVC). As demonstrated previously (18), expression of the CcsR RNAs from the 16S promoter leads to a strong increase in CcsR levels, while there was no visible increase when the ccaF1 gene was co-expressed together with the ccsR genes (Figure 2C, left panel, lanes 2–4). When only the ccaF1 gene was present on the plasmid, the level of CcsR RNAs expressed from the chromosome, was clearly decreased (Figure 2C, left panel, lanes 2 and 5). This demonstrates that CcaF1 can also act on CcsR levels in trans. To verify that the effect on the CcsR level is mediated by the CcaF1 small protein and not by the ccaF1 mRNA, we exchanged the ATG start codon of the ccaF1 gene to TGA. No other in frame ATG is present in the ccaF1 gene. Expression of this mutated gene together with the CcsR RNAs from the plasmid had the same effect on CcsR levels as overexpression of the CcsR RNAs alone (Figure 2C, left panel, lanes 3 and 6) strongly supporting the assumption that CcsR levels are affected by the CcaF1 protein, not the ccaF1 mRNA. The effects of the different plasmids were identical for all individual CcsR RNAs and were also observed for the CcsR1–4 precursor. We also performed a Northern blot with a ccaF1 (RSP_6037) specific probe to confirm higher ccaF1 transcript levels in presence of plasmid pRKCcaF1(Supplementary Figure S4). This Northern also shows increased levels of the ccaF1-CcsR1–4 precursor transcripts in strains expressing ccaF1 together with CcsR1–4, but no ccaF1 specific transcript is present in those strains suggesting that longer precursors are translated to produce CcaF1.
Figure 2.
Small DUF1127 proteins affect CcsR level in R. sphaeroides. (A) Amino acid sequence of the small DUF1127 protein CcaF1 from R. sphaeroides 2.4.1 and the corresponding homologue RSPKD131_0402 from R. sphaeroides KD131. The conserved DUF1127 domain as shown in Supplementary Figure S3 is high-lighted in bold letters. (B) Schematic overview of the plasmids introduced into the wild type strain 2.4.1 or the mutant lacking the hfq gene. The ccaf1 gene is shown in light grey, the RSKD131_0402 gene in black, CcsR RNAs in white. In plasmid pRKCcaF1mut_CcsR1–4 the ATG of the ccaF1 gene was changed to TGA. (C) Northern blots of total RNA from strains containing an empty vector control (pRK4352, EVC) with just the 16S promoter, or plasmids as shown in (B). DNA fragments specific for CcsR1, CcsR2, CcsR3/CcsR4 or CcsR1–4 were used as probes. Signals for 5S RNA or 14S RNA were used as loading controls. R. sphaeroides cleaves the 23S RNA into fragments of 16S and 14S (70). The upper three panels stem from 10% denaturing polyacryamide gels, the lower two panels from 1% formaldehyde agarose gels.
Overexpression of CcsRs results in considerably slower growth (doubling time 5.4 h compared to the wild type with 3.5 h), while the doubling time of a strain having CcaF1 with CcsR1–4 together on a plasmid is only slightly increased (4.0 h) compared to that of the wild type. Overexpression of ccaF1 alone results in an extended lag phase but growth in exponential phase is not affected (doubling time of 3.4 h). All strains reached the same OD in stationary phase (Figure 3A).
Figure 3.
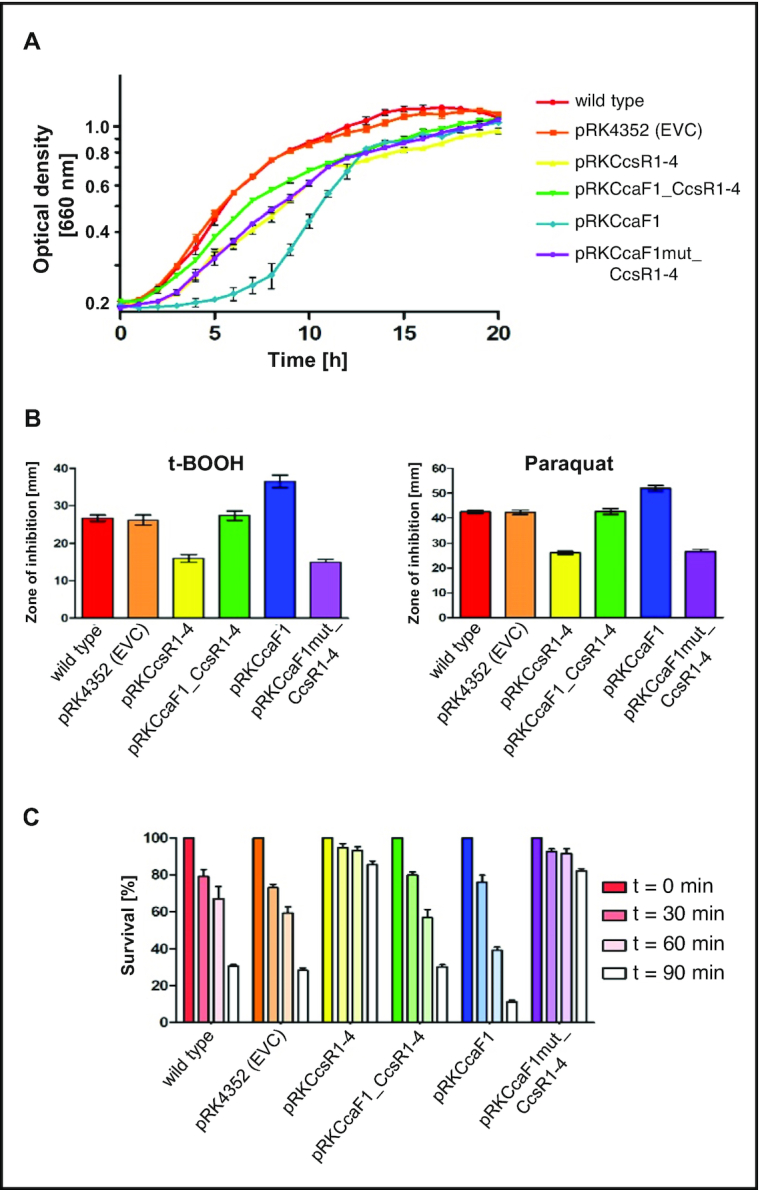
DUF1127 protein CcaF1 (RSP_6037) affects stress resistance in R. sphaeroides. (A) Growth curves of the wild type or the wild type with the empty vector (EVC) or the plasmids as shown in Figure 2B. All strains were cultivated under microaerobic conditions. The average of three independent measurements and the standard deviation are plotted. The color code for the different strains was also applied in (B) and (C). (B) Zone of inhibition assay of strains overexpressing the plasmids shown in Figure 2B in comparison to the wild type and wild type with empty vector control (pRK4352) under organic peroxide (700 mM tBOOH) and superoxide stress (300 mM paraquat). The plotted values represent the mean of at least three independent experiments and the standard deviation is indicated. (C) Survival assay of strains overexpressing the plasmids shown in Figure 2B in comparison to the wild type and wild type with empty vector control (pRK4352) under organic peroxide stress (300 mM tBOOH). The number of colonies of a control culture grown without the addition of any oxidative stress agents was defined as 100% survival. The bars represent the mean of three independent plating assays and the standard deviation is indicated.
The 71 aa CcaF1 protein of R. sphaeroides 2.4.1 consists mostly of the DUF1127 domain (bold in Figure 2A) but harbors 27 additional amino acids in the N-terminal domain and 11 additional amino acids at the C-terminus. In contrast, the R. sphaeroides strain KD131 encodes a 50 aa CcaF1 protein (RSKD131–0402) with only six additional amino acids N-terminal of the DUF1127 domain (Figure 2A). Otherwise, the amino acid sequences of the two small proteins are identical. The genomic context around the ccaF1-ccsR locus is also identical in the two strains, the overall synteny is highly conserved between these two R. sphaeroides strains. When we expressed the shorter CcaF1 protein of strain KD131 from the 16S promoter on a plasmid (pRKCcaF1) in strain R. sphaeroides 2.4.1, we saw a similar reduction in the abundance of the CcsR RNAs (Figure 2C, middle panel, lanes 2 and 5) as that observed with CcaF1 from strain 2.4.1 (Figure 2C, left panel, lanes 2 and 5). This demonstrates that the 50 aa CcaF1 from R. sphaeroides KD131 is sufficient for the effect on CcsR levels.
A previous study demonstrated lower amounts of CcsR1–4 in a strain lacking Hfq (7). To test, whether Hfq affects CcaF1 function, we also transferred the different plasmids into the hfq mutant and analyzed CcsR levels. As observed in the wild type, CcaF1 counteracted the stronger expression of CcsR1–4 when present on the plasmid together with the ccsR1–4 genes (Figure 2C, right panel, compare lanes 3 and 4). Overexpression in trans of the ccaF1 gene alone had little effect on the CcsR levels (Figure 2C, right panel, compare lanes 2 and 5). The exact role of Hfq in CcsR maturation and possibly further CcaF1-dependenten processes needs to be addresses in the future.
Increased levels of CcsR1–4 were previously shown to lead to increased resistance of R. sphaeroides to the superoxide generating paraquat and to tertiary butyl-alcohol (t-BOOH) (18). t-BOOH represents organic peroxides that are produced from cellular components due to singlet oxygen exposure. Figure 3B confirms the increase of resistance to the two chemicals when CcsR levels are increased. When CcsR1–4 are overexpressed together with the ccaF1 gene, the resistance level resembles those of the controls, but only if the ATG is not mutated. Overexpression of CcaF1 alone, resulted in decreased resistance to paraquat and t-BOOH compared to the control. This effect of overexpression of CcaF1 on the oxidative stress response was also confirmed in survival assays (Figure 3C). Thus, the effect of the different plasmid constructs on resistance reflects the CcsR amounts that were detected in the Northern blots.
In addition, we performed spot assays to test the growth behavior of the different strains in the presence of various stresses. These assays (Supplementary Figure S4) confirmed the results we obtained by t-BOOH zone of inhibition and survival assays: overexpression of CcsR1–4 (pRKCcsR1–4) resulted in significantly increased survival, while overexpression of CcaF1 (pRKCcaF1) decreased survival. The same correlation between expression and survival was observed for heat stress (42°C) or stress by CdCl2. The CcaF1-CcsR1–4 overexpression strain showed similar survival as the wild type and the control strain harboring an empty vector in presence of salt (NaCl), while the strain overexpressing CcaF1 alone showed reduced survival (Supplementary Figure S5).
RNase E and CcaF1 are involved in processing of the ccaF1-ccsR transcript and maturation of the CcsR RNAs
When the CcsR1–4 RNAs (originally designated RSs0680 a-d) were detected as photooxidative stress-induced sRNAs in a RNAseq data set, their co-transcription was already proposed based on 5′ RACE and RT-PCR (7). This strongly suggested that the individual sRNAs are generated by RNA processing.
To exclude that overexpression of ccaf1 affects CcsR levels by altering promoter activity, we applied reporter constructs that have the ccaF1 promoter including 100 nt or 200 nt upstream of the promoter transcriptionally fused to eCFP (16) and monitored fluorescence. As shown in Supplementary Figure S6Rhodobacter strains without eCFP exhibit autofluorescence, which was set to 100% relative fluorescence. Presence of the promoter:eCFP fusions elevated the fluorescence to 120–135% during exponential growth at 32°C. After heat stress, fluorescence was increased to 160–190% independently of overexpression of ccaF1 (Supplementary Figure S6). This excludes an effect of CcaF1 on the activity of the ccaF1 promoter and supports the assumption that RNA processing/degradation is responsible for the effect of CcaF1 on CcsR levels.
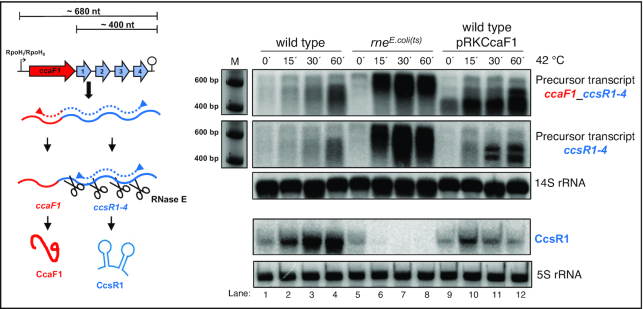
The endoribonuclease E is involved in the maturation of several sRNAs from the 3′ or 5′ UTR of mRNAs in E. coli and R. sphaeroides (45,41). Despite the different GC contents (51% for E. coli, 69% for R. sphaeroides) RNase E recognizes AU rich sequences in both organisms (41). The RNAseq data set that compares total RNA reads and RNA 5′ ends in the control strain and in a mutant that expresses a temperature sensitive RNase E from E. coli (41) reveals RNase E cleavage sites at the 5′ ends of the individual CcsR RNAs (Supplementary Figure S7). A main 5′ end is detected in the wild type at 32°C and due to induction of the RpoHI/HII-dependent promoter shows higher abundance at 42°C. In the strain expressing the temperature-sensitive RNase E the 5′ end is more abundant at 32°C compared to the wild type and this difference is more pronounced at the non-permissive temperature of 42°C. Our previous study demonstrated that RNase E cleavage is already partly impaired in the mutant at 32°C due to the different RNase E enzyme (41). RNase E-dependent 5′ ends are also detected for the individual CcsR RNAs (Supplementary Figure S7). Cleavages at the 5′ ends occur at the sequences GUUUCC (for CcsR1, nucleotides adjacent to cleavage sites in bold), CUCUUC (for CcsR2), ACUUC (for CcsR3) and ACUUC (for CcsR4). To further confirm the important role of RNase E in CcsR maturation, we performed Northern blots with probes directed against the CcsR1 RNA or against precursor transcripts (Figure 4). At time point 0 the cultures were shifted to 42°C which leads to induction of the RpoHI/HII-dependent promoter (Supplementary Figure S7) and also to inactivation of RNase E. Figure 4 demonstrates increased levels of the precursor transcripts (a precursor harbouring ccaF1 and CcsR1–4 would comprise 680 nt, a precursor including CcsR1–4 about 400 nt) as well as strongly increased CcsR1 levels after the shift to 42°C in the wild type. In the rnets mutant however, strong accumulation of precursor transcripts occurs but the level of CcsR1 is strongly decreasing, supporting a major role for RNase E in CcsR maturation.
Figure 4.
RNase E and CcaF1 are involved in CcsR maturation. Analysis of ccaF1-ccsR1–4 and ccsR1–4 precursor and CcsR1 RNA by Northern blot in R. sphaeroides 2.4.1 wild type, rnets mutant and strain 2.4.1 that overexpresses ccaF1 (pRKCcaF1). Cells were harvested at 32°C and at different time points after shift to 42°C. Total RNA was isolated and either run on a 1% formaldehyde agarose gel and, after blotting, hybridized against probes spanning the CcaF1-CcsR1–4 or the CcsR1–4 region, or on a 10% denaturing polyacryamide gel for detection of CcsR1. 5S and 14S rRNA serve as loading controls.
We also shifted strain 2.4.1 (pRKCcaF1) to 42°C, which overexpresses ccaF1. The accumulation of CcsR1 upon the temperature shift was clearly reduced compared to the wild type (Figure 4). At the same time, precursor transcripts accumulate that show however a different pattern than the precursors accumulating in the mutant with reduced RNase E activity (Figure 4). This demonstrates that not only RNase E but also CcaF1 is involved in maturation of the CcsR RNAs.
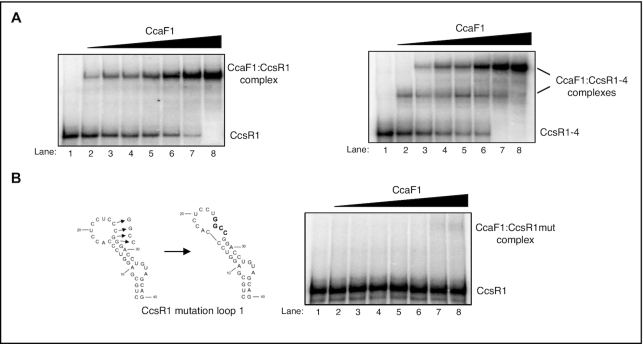
CcaF1 directly binds to CcsR RNAs
The high arginine content of the DUF1127 domain, the structural similarity to the Smaug domain and its role in CcsR maturation suggest that CcaF1 may have RNA-binding capacity. To test this hypothesis, we applied the purified protein for in vitro RNA binding assays. CcaF1 was purified as His6-MBP-fusion protein, which was cleaved by TEV protease to release CcaF1 (Supplementary Figure S8). Figure 5 shows gel retardation assays with radiolabeled CcsR1 or CcsR1–4. While addition of CcaF1 to CcsR1 leads to a single retarded band (A), two distinct bands were visible with the longer CcsR1–4 transcript. This suggests that more than one CcaF1 protein (or protein complex) can bind to CcsR1–4. Addition of an excess of unlabeled CcsR1 to the reaction abolished the retardation, indicating that the binding is specific (Supplementary Figure S9 A). When we added unlabeled RSs0827 RNA, even at high molar excess complex formation between CcsR1 and CcaF1 was not abolished (Supplementary Figure S9B). RSs0827 is the most abundant sRNA in stationary phase in R. sphaeroides (21) and was not found in the CoIP analysis with CcaF1 (see below). Thus, we selected it as unspecific competitor in our assay. Furthermore, we used a CcsR1 variant that has four nucleotides in loop 1 exchanged, which is predicted to also change the local structure. This mutant variant was not bound by CcaF1 (Figure 5B).
Figure 5.
CcaF1 binds to CcsR1 and the CcsR1–4 precursor. (A) Gel motility shift RNA-binding experiments with 150 fmol radioactively labelled CcsR1 or CcsR1–4 incubated with increasing amounts (2, 5, 10, 50, 100, 200 or 500 nM) of purified CcaF1. The reactions were run on a 6% native polyacrylamide gel. (B) Predicted structures of the 5′ hairpin in the CcsR1 RNA with and without a mutation of four nucleotides (CCGG→GGCC) in the loop (left side) and gel motility shift experiments with 150 fmol radioactively labelled, mutated CcsR1 RNA incubated with increasing amounts (2, 5, 10, 50, 100, 200 or 500 nM) of purified CcaF1 (right side). The reactions were run on a 6% native polyacrylamide gel.
Effect of CcaF1 overexpression on the transcriptome of R. sphaeroides
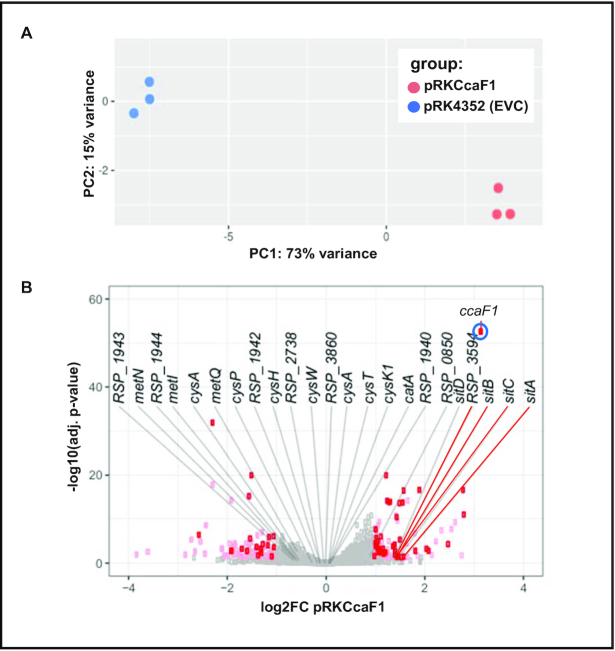
To analyze the global effect of CcaF1 on the transcriptome, we performed RNAseq on total RNA from a strain harboring plasmid pRKCcaF1 (Figure 2B) and a control strain only harboring the vector. Strains were cultivated under microaerobic conditions at 32°C. The reads for the individual genes and non-coding RNAs were compared for the two strains. PCA plots verified the high reproducibility of the technical triplicates, each from biological triplicates (Figure 6A). For few selected genes we also performed real time RT-PCR with the RNA samples used for RNAseq analysis to verify the results (not shown). Although the fold-changes showed some variation between the two methods, the direction of change was consistent for the tested RNAs.
Figure 6.
Overexpression of CcaF1 impacts the R. sphaeroides transcriptome. (A) A principal component analysis was performed as part of the downstream DESeq2 analysis. The scatterplot shows two distinct groups, each one harbors the replicates belonging to one of the individual strains used in this study. (B) Volcano plot for the comparison of the ccaF1 overexpressing strain and the wild type, based on RNAseq data. Genes with significant change in abundance are colored red (adjusted P-value ≤ 0.05, log2fold change ≤–1 or ≥1, basemean > 100) and pink (adjusted P-value ≤0.05, log2fold change ≤–1 or ≥1, basemean ≤ 100). Gray dots: adjusted P-value >0.05. The 24 RNAs with highest enrichment in the CoIP are labelled.
A volcano plot displays the strongest differences in read numbers between the strain overexpressing CcaF1 and the wild type strain (Figure 6B). In order to avoid mis-leading results due to low read numbers, we set a basemean of ≥100. Thus, we consider only genes with a normalized average read count over all samples above this threshold to dismiss potential false positive genes and to increase reliability. With this cut-off only 1487 of total 4411 genes remained in our analysis. We further chose a cut off log2fold ≤–1 or log2fold ≥1 and an adjusted P-value of ≤0.05 (Benjamini-Hochberg algorithm). The strongest fold change (marked in blue) was observed for ccaF1, which was overexpressed from the plasmid. In addition to ccaF1, 42 RNAs showed increased levels of log2fold ≥ 1.0 (Supplementary Table S3). Among these genes were RSP_3095 and RSP_3094 for a sigma-70 factor and an anti-sigma factor that are involved in the adaptation to stationary phase (46), and sitA, sitB, sitC for an Mn2+ ABC transporter. The enriched protein-encoding RNAs belong to different COG (cluster of orthologous groups) functions, 8 of them encode hypothetical proteins. Three non-coding RNAs were enriched, among them SorX (formerly RSs2461). SorX is derived from the 3′ UTR of the ompR-1 gene by RNase E cleavage and affects resistance to singlet oxygen and organic hydroperoxides by interacting with the mRNA for a polyamine transporter (16,41,47).
Nineteen RNAs showed lower levels in the overexpression strain compared to the wild type, among them one non-coding RNA of unknown function, 23S and 16S rRNAs and six mRNAs encoding hypothetical proteins (Supplementary Table S3). Three of these 19 RNAs encode cold shock proteins (RSP_3620, RSP_1952 and RSP_3621). Several tRNAs were also decreased in the overexpression strain but the P-values were above our cut-off. Likewise, in our analysis the P-values for the CcsR RNAs and for other known non-coding RNAs were too high for reliable conclusions.
The RNAseq results demonstrate that CcaF1 does not only affect CcsR levels (as demonstrated by northern blots) but several other cellular RNAs with different physiological functions. These effects may be direct, by interaction of CcaF1 with those RNAs, or indirect through other RNAs that interact with CcaF1.
Co-immunoprecipitation identifies RNA targets of CcaF1
To discriminate between direct and indirect effects of CcaF1 on RNA levels, we expressed a CcaF1 variant with an N-terminal FLAG-tag and performed co-immunoprecipitation with total RNA from R. sphaeroides. We confirmed that the tagged CcaF1 protein has the same effect on CcsR levels as the non-tagged-version (Supplementary Figure S10) and is thus functional. The total CoIP sample was analyzed on an SDS gel and by high resolution MS (LS-ESI-HRMS). Silver stain of the SDS gel shows as faint band that was confirmed as CcaF1FLAG by western blot (Supplementary Figure S11). Faint bands at higher molecular weights indicate the formation of stable multimers. By far the most abundant protein detected by MS of this sample was CcaF1, no other protein was present in similar amounts (Supplementary Table S4). Due to the high sensitivity of the MS, minor amounts of other proteins were detected. Most of these proteins are abundant in the cell and are known to interact with many proteins (GroES, GroEL) or to interact with RNA (ribosomal proteins, Rho, TufA) that was also present in the analyzed CoIP sample. Neither Hfq nor RNase E were detected in the CoIP sample, excluding their direct association with CcaF1.
The co-immunoprecipitated RNA was used for RNAseq (Rip-seq) and compared to RNAseq from total RNA and to a negative control, a CoIP with RNA from cells not expressing the FLAG-tagged CcaF1. From the RNAs identified in the CoIP, a subset was selected for further analysis using northern blot and real time RT-PCR. Supplementary Table S5 provides a quantification of the RNAs that were strongly enriched in the co-immunoprecipitation. This table considers only RNAs with >10 reads in the CoIP sample, and at least 10-fold higher read number in the CoIP sample compared to the control (CcaF1 overexpression without FLAG-tag). The strongest enrichment was observed for the mRNA of the catalase gene (catA, RSP_2779, factor 219), the sit genes (ABC Mn transporter), and for RSP_1943 (hypothetical protein) and RSP_1944 (Uroporphyrin-III-methyltransferase / siroheme synthase). Interestingly, many RNAs which were strongly enriched in the CoIP have known or predicted roles in cystein / methionine / sulfur metabolism: RSP_1944 (methyltransferase), RSP_1942 (sulfite/nitrite reductase), cysH (RSP_1941, phosphoadenosine phosphosulfate reductase), cysK1 (RSP_1109, cystein synthase), cysA, cysP, cysT, cysW (RSP_3696–3699), and RSP_3861 (ABC sulfate/ thiosulfate transporter), RSP_3860 (probable rhodanese-related sulfurtransferase), RSP_3859 (ABC sulfate/molybdate transporter), metN, metQ and metI (RSP_0129, 0130, 0132, methionine uptake transporter). The RNAseq data indicate that the adjacently located genes are transcribed into long polycistronic transcripts.
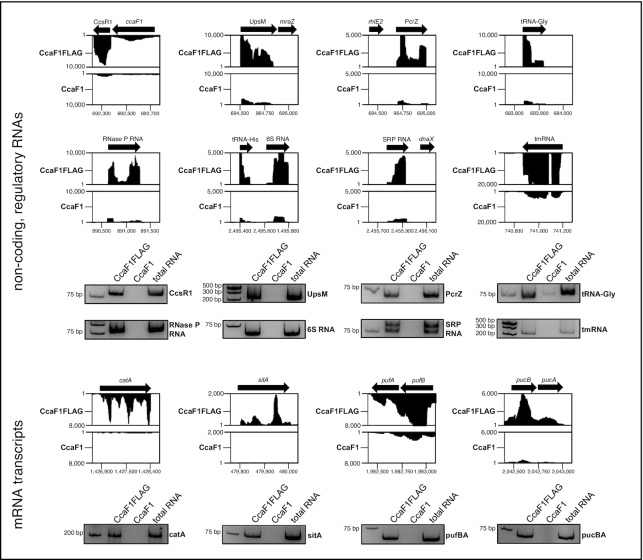
Read coverage plots for the CoIP results for selected non-coding RNAs and mRNAs are shown in Figure 7 together with results from RT-PCR. Quantitative data from real time RT-PCR from the CoIP are shown in Supplementary Figure S12. We observed accumulation of CcsR1, UpsM, and PcrZ, 6S RNA, SRP RNA, tmRNA, and tRNA Gly when the FLAG-tagged CcaF1 was present. UpsM is a highly abundant sRNA that is derived from the 5′ UTR of the dcw gene cluster mRNA by RNase E cleavage (41,48) and PcrZ has an important role in the regulation of photosynthesis genes (28,49). puf and puc genes encode pigment-binding proteins of the photosynthetic apparatus. For the small RNA UpsM (enrichment in CoIP about 12-fold, Supplementary Figure S12) we also demonstrated direct binding to CcaF1 by gel retardation (Supplementary Figure S9C). As expected from the enrichment factors in the CoIP (Supplementary Figure S12) higher amounts of CcaF1 were required for complex formation with UpsM than with CcsR1.
Figure 7.
CoIP analysis identifies targets of CcaF1. Analysis of co-immunoprecipitated RNA by RNAseq (Rip-seq) using CcaF1 with 3xFLAG-tag (CcaF1FLAG) or without 3xFLAG-tag (CcaF1, control) in exponential growth phase at 32°C and microaerobic conditions. Read coverage plots from the Integrated Genome Browser display the sequencing reads for selected RNAs. The specific non-coding RNAs and mRNA transcripts of the co-immunoprecipitation were also analyzed by reverse transcription (RT) PCR. The RT-PCR products were separated on a 10% polyacrylamide gel and analyzed by ethidium-bromide staining and are shown below the corresponding read coverage plots.
Of the RNAs with expression changes in the DESeq2 analysis upon over-expression of CcaF1, only 5 RNAs were enriched in the CoIP (S3 Table): expE1 for a hemolysin-type calcium-binding region, sitA, sitB, sitC encoding subunits of an ABC Mn2+ transporter and RSP_0850 for a hypothetical protein. This supports the view that many changes in the transcriptome are not due to a direct interaction with CcaF1. Vice versa, one could expect that a direct interaction to CcaF1 as suggested by the CoIP would result in changed levels of those RNAs in the DESeq2 analysis. As seen in Supplementary Table S5, this is only true for the five RNAs mentioned above. All other RNAs enriched in the CoIP did not reach the P-value we used for the cut-off in the DESeq2.
To further support the view that CcaF1 specifically binds to a set of RNAs we also performed CoIP analysis with the FLAG-tagged RSP_0557 protein. RSP_0557 encodes a 70 amino acid long DUF1127 protein which is not found in a CIN1 locus. The gene is under control of a RpoHI/HII-dependent promoter and is controlled by the sRNA Pos19 (17). Previous studies revealed strong increase of ccaF1 and RSP_0557 transcript levels in transition from exponential to stationary phase (46). To date no function could be assigned to the RSP_0557 protein. Supplementary Figure S13 demonstrates that some RNAs like CcsR1, UpsM and tmRNA are bound by both, CcaF1 and RSP_0557 proteins, with similar efficiency. However, other RNAs are preferentially bound by only one of the two DUF1127 proteins. These data also support the view that not only DUF1127 proteins of CIN1 loci function as RNA-binding proteins.
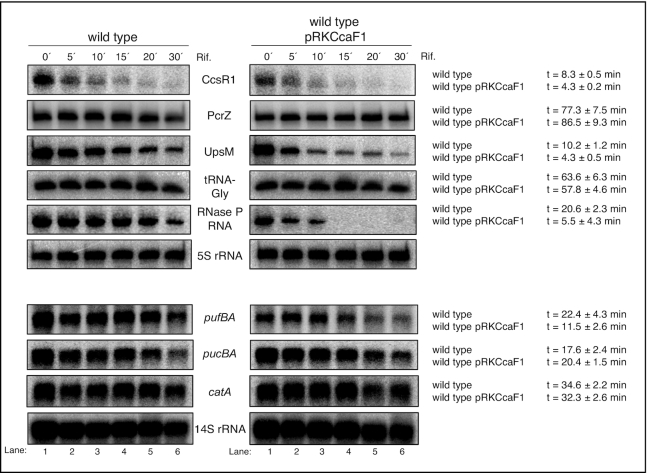
CcaF1 affects stability of some of its targets
We assumed that CcaF1 controls the amounts of its targets by either affecting their maturation, their stability, or both. To test the effect of CcaF1 on stability, we compared half-lives of selected RNAs in the wild type and the CcaF1 overexpression strain by quantifying the RNA levels after addition of rifampicin (inhibits initiation of transcription) by Northern blot analyses (Figure 8). These experiments confirmed a destabilizing effect of CcaF1 on the sRNAs CcsR1 and UpsM, the RNaseP-RNA and the pufBA mRNA. Such an effect was not observed for the sRNA PcrZ, tRNA-Gly. These RNAs turned out to be very stable and half-life determination after long time periods in the presence of rifampicin is not reliable. We also did not observe changed half-lives for pucBA, or catA, although the half-lives were in the same range as for CcsR1, which was less stable in the overexpression strain. PucBA is part of a polycistronic transcript, while catA mRNA is monocistronic. The effect of CcaF1 on the amounts of these RNAs needs further investigation.
Figure 8.
CcaF1 affects stability of some coding and non-coding RNA transcripts. Determination of RNA half-life of R. sphaeroides wild type and wild type with pRKCcaF1 for selected coding and non-coding RNA transcripts. Cells were harvested in exponential growth phase under microaerobic conditions. Samples were taken after adding rifampicin at different time points. Total RNA was isolated and either run on a 10% denaturing polyacryamide gel (upper six panels) or on a 1% formaldehyde agarose gel (lower four panels) and blotted. 5S or 14S rRNA serve as loading controls. For quantification RNA signal intensities were normalized to 5S or 14S rRNA signals. The average half-life was calculated from three independent experiments and the standard deviation is indicated.
We conclude that CcaF1 can control RNA levels by affecting the stability of the mature transcript, but that also other mechanisms, like maturation from precursor transcripts are involved.
DISCUSSION
Although numerous small open reading frames are found in bacterial genomes, the importance of small proteins was realized only about a decade ago (1,2). Numerous small proteins, mostly found in alpha- and g–ammaproteobacteria, harbor the domain of unknown function DUF1127. First evidence for the involvement of a DUF1127 protein in bacterial physiology was provided for RSP_6037 (CcaF1) that has a role in stress responses in R. sphaeroides (18). In Brucella abortus deletion of the gene for a DUF1127 protein caused a defect in fucose metabolism (50). Recently a role of DUF1127 proteins in phosphate and carbon metabolism in Agrobacterium tumefaciens was demonstrated (51), as well as a role of the Salmonella protein YjiS in virulence (52). YjiS is a DUF1127 protein with 20% identity to CcaF1. The mechanisms by which these DUF1127 proteins affect physiology remain elusive. This study identifies CcaF1 of R. sphaeroides as a new type of RNA-binding protein.
The ccaF1 gene of R. sphaeroides is co-transcribed with 4 homologous sRNAs. Our bioinformatic analysis revealed that the arrangement of sRNAs and genes for DUF1127 proteins (sRNAs in the 5′ or 3′ UTR) in CIN1 loci correlates with phylogenetic assignments by 16S rRNA. If duplicate loci occur in a family, these loci fall into two distinct clusters. Remarkably one single genus (Sulfitobacter) showed ‘Cuckoo’-RNAs adjacent to both sides of a gene for a DUF1127 protein. The high correlation between the 16S rRNA phylogenetic tree and the phylogenetic tree based on CcaF1 amino acid sequences indicates, that the DUF1127 protein coding ORFs have likely been acquired by a common ancestor and sequences co-evolved. This hypothesis is underlined by the observation, that two similar CIN1 loci, which might be a result of a locus duplication, occur in many Brucellaceae and form distinct clusters based on the CcaF1 amino acid sequence. Furthermore, occurrence of multiple highly similar ‘Cuckoo’-RNAs in Sinorhizobium on the chromosome and on plasmids, with only weak association with genes for DUF1127 proteins, shows that CIN in general and CIN1 loci in particular are subject to duplication and genomic reorganization and may result in different locus configurations.
Our data revealed a strong effect of the CcaF1 protein on CcsR levels, which did not require the N-terminal 21 amino acids. CcaF1 promotes RNase E-dependent cleavage of the ccaF1-CcsR1–4 precursor transcript. The cleavages occur adjacent to U residues, a preference that was also revealed by a previous global study mapping RNase E cleavage sites in R. sphaeroides (41). Overexpression of CcaF1 also resulted in the accumulation of precursor transcripts, that showed a pattern differing from the precursors accumulated in the rnets mutant. This suggests that CcaF1 is not solely reducing RNase E activity. It is conceivable that structural changes upon binding of CcaF1 affect cleavages of the precursor transcripts. This needs to be tested in the future. Gel retardation assays with purified CcaF1 proved that the protein can specifically bind to CcsR1 and to the CcsR1–4 precursor.
RNAseq analyses demonstrated that not only the amount of CcsR RNAs is influenced by CcaF1 but that this protein affects the levels of many RNAs including sRNAs, tRNAs, rRNAs, other non-coding RNAs like 6S RNAs or signal recognition RNA, and mRNAs. While the non-coding RNAs are mostly small and highly structured, also the amount of several large mRNAs, including polycistronic transcripts was affected. Considering the mRNAs mostly affected in the RNAseq data, there was no clear preference for certain functional groups, indicating that CcaF1 may influence multiple biological functions. Our experiments verified an effect of CcaF1 on resistance to oxidative stress, CdCl2 and heat stress.
Results from co-immunoprecipitation with tagged CcaF1 followed by RNAseq support a direct interaction between CcaF1 and many RNAs, including sRNAs, tRNAs, rRNAs, other non-coding RNAs and mRNAs. In vitro gel retardation experiments with CcaF1 and CcsR1, CcsR1–4 or UpsM confirmed that CcaF1 is a bona fide RNA binding protein. How can a small protein affect the levels of many different RNAs? It is unlikely that the small DUF1127 domain exhibits a catalytic function that degrades RNA. But binding of proteins to RNA can restrict or enhance the action of ribonucleases (53). Such a function of CcaF1 is supported by our data: the amount of CcsR RNAs is dependent on RNase E activity and CcaF1 abundance. In Drosophila, Smaug can recruit the Argonaute 1 protein to an mRNA to trigger translational repression and/or decay (54). Argonaute proteins are also encoded by many bacterial and archaeal genomes (55) and a plasmid-encoded Argonaute from Rhodobacter sphaeroides ATCC17025 was investigated in more detail (56–58). The genome of R. sphaeroides 2.4.1 does not encode such a protein excluding the involvement of Argonaute in CcaF1-dependent RNA destabilization.
Several RNA-binding proteins, some of them small proteins, have established functions as RNA chaperones in prokaryotes (59,60). The Hfq protein (77 aa in R. sphaeroides) is considered as a global regulator of sRNA-based networks. It acts as an RNA chaperone in gram-negative bacteria by stabilizing the imperfect base-pairing between trans-encoded sRNAs and their mRNA targets. The 72 aa CsrA protein of E. coli (member of the CsrA/RsmA family) recognizes an AUGGA motif in RNA loop regions. It preferentially binds to the ribosome binding site or to the start codon of mRNAs and either represses translation or regulates transcript stability. CsrA was also shown to act as a chaperone that can promote complex formation between an sRNA and its mRNA target in Bacillus subtilis (61). ProQ (about 220 aa) is an RNA chaperone of the FinO family that is commonly found in Proteobacteria (62). It binds double-stranded RNAs and prefers highly structured RNAs, mostly promoting binding of sense and anti-sense RNAs, but can also regulate trans-acting sRNAs (63). CspA is another small (68 aa in R. sphaeroides) RNA chaperone and can passively remodel RNA structures by preferentially binding to pyrimidine-rich RNA sequences (64). In Staphylococcus aureus a RIP-CHIP assay also identified sRNAs as CspA targets (65). CspA binding destabilizes secondary structures to promote translation or alter mRNA turnover. CcaF1 has only low sequence similarity (maximal 30%) to these well studied RNA-binding proteins and no obvious structural homology (based on Phyre 2).
The CcaF1 coding region which is part of the ccaF1-CcsR precursor transcript is also enriched in the CoIP. Higher amounts of CcaF1 lead to impeded maturation of the CcsR RNAs and to stabilization of the precursor RNA, strongly suggesting that processing by RNase E is negatively affected. Nevertheless, further degradation of CcsR1 is accelerated by CcaF1. Thus, binding of CcaF1 can have different effects on the stability of individual transcripts. To understand the exact mechanisms, how CcaF1 acts on the stability of its targets, future work needs to identify all RNases involved in the maturation and degradation of CcaF1 targets and to follow changes in RNA structure upon binding by CcaF1. Such structural changes may lead to sequestration as well as to exposure of RNase cleavage sites, which may cause stabilization or destabilization of transcripts.
A binding motif for the RNA-binding SAM domain of the eukaryotic Smaug protein was identified in the past (66,67). The Smaug-recognition element (SRE) consists of a stem–loop structure with the sequence CUGGC in the loop. A translational control element (TCE) of 184 nucleotides is required for translational control of nos mRNA in Drosophila. The TCE contains a pair of redundant SREs (68,69). Interestingly, the two stem-loops of the CcsR RNAs that were highly enriched in the CcaF1 CoIP, also mostly contain the CUGGC sequence. For many of the CcaF1 binding partners, pairs of stem–loop structures are predicted (not shown), but identification of CcaF1 bindings sites will require further investigation.
The CoIP data strongly suggest that another small DUF1127 protein from R. sphaeroides, the RSP_0557 protein binds to RNA. RSP_0557 is not associated with ccsR genes on the chromosome. Despite their small size and the strong similarity of the DUF1127 domains, the CcaF1 and RSP_0557 proteins show differences in their preference for RNA binding partners. It will be interesting to further elucidate the molecular basis for this binding specificity.
DATA AVAILABILITY
The sequencing data are available at NCBI Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo) under the accession number GSE144523 and GSE145045.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Kerstin Haberzettl and Andrea Weisert (Klug lab) for excellent technical assistance, Elnaz Jajin (Förstner lab) for help with mapping of the CoIP reads, Günter Lochnit (University of Giessen) for the MS analysis, Jeong Kock Lee (Sogang University, South Korea) for providing strain R. sphaeroides KD131, and Oliver Roßbach, Patrick Barth, and Kai Thormann (University of Giessen) for support of our project. We are grateful to Matthew McIntosh for corrections on the manuscript. We thank the Core Unit SysMed at the University of Würzburg for RNA-seq data generation.
Notes
Present address: Vivian Monzon, European Molecular Biology Laboratory, European Bioinformatics Institute, CB 10, 1SD Hinxton, UK.
Contributor Information
Julian Grützner, Institute of Microbiology and Molecular Biology, Justus Liebig University Giessen, IFZ, Heinrich-Buff-Ring 26–32, D-35292 Giessen, Germany.
Fabian Billenkamp, Institute of Microbiology and Molecular Biology, Justus Liebig University Giessen, IFZ, Heinrich-Buff-Ring 26–32, D-35292 Giessen, Germany; Institute of Animal Nutrition, Friedrich Loeffler Institute, Bundesalle 37, D-38116 Braunschweig, Germany.
Daniel-Timon Spanka, Institute of Microbiology and Molecular Biology, Justus Liebig University Giessen, IFZ, Heinrich-Buff-Ring 26–32, D-35292 Giessen, Germany.
Tim Rick, Institute of Microbiology and Molecular Biology, Justus Liebig University Giessen, IFZ, Heinrich-Buff-Ring 26–32, D-35292 Giessen, Germany.
Vivian Monzon, ZB MED-Information Center of Life Science, Germany.
Konrad U Förstner, ZB MED-Information Center of Life Science, Germany; Institute of Information Science, TH Köln, University of Applied Science, Gustav-Heinemann-Ufer 54, D-50968 Köln, Cologne, Germany.
Gabriele Klug, Institute of Microbiology and Molecular Biology, Justus Liebig University Giessen, IFZ, Heinrich-Buff-Ring 26–32, D-35292 Giessen, Germany.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Deutsche Forschungsgemeinschaft [Kl 561/37-1, RTG 2355]; IZKF at the University Würzburg [project Z-6]. Funding for open access charge: Deutsche Forschungsgemeinschaft/University of Giessen.
Conflict of interest statement. None declared.
REFERENCES
- 1. Hobbs E.C., Fontaine F., Yin X., Storz G.. An expanding universe of small proteins. Curr. Opin. Microbiol. 2011; 14:167–173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Storz G., Wolf Y.I., Ramamurthi K.S.. Small proteins can no longer be ignored. Annu. Rev. Biochem. 2014; 83:753–777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Braatsch S., Gomelsky M., Kuphal S., Klug G.. A single flavoprotein, AppA, integrates both redox and light signals in Rhodobacter sphaeroides. Mol. Microbiol. 2002; 3:827–569. [DOI] [PubMed] [Google Scholar]
- 4. Zeilstra-Ryalls J.H., Kaplan S.. Oxygen intervention in the regulation of gene expression: the photosynthetic bacterial paradigm. Cell. Mol. Life Sci.: CMLS. 2004; 61:417–436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Mackenzie C., Eraso J.M., Choudhary M., Roh J.H., Zeng X., Bruscella P., Puskás A., Kaplan S.. Postgenomic adventures with Rhodobacter sphaeroides. Annu. Rev. Microbiol. 2007; 61:283–307. [DOI] [PubMed] [Google Scholar]
- 6. Yin L., Bauer C.E.. Controlling the delicate balance of tetrapyrrole biosynthesis. Philos. Trans. Roy. Soc. London. B, Biol. Sci. 2013; 368:20120262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Berghoff B.A., Glaeser J., Sharma C.M., Vogel J., Klug G.. Photooxidative stress-induced and abundant small RNAs in Rhodobacter sphaeroides. Mol. Microbiol. 2009; 74:1497–1512. [DOI] [PubMed] [Google Scholar]
- 8. Nuss A.M., Glaeser J., Berghoff B.A., Klug G.. Overlapping alternative sigma factor regulons in the response to singlet oxygen in Rhodobacter sphaeroides. J. Bacteriol. 2010; 192:2613–2623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Berghoff B.A., Konzer A., Mank N.N., Looso M., Rische T., Förstner K.U., Krüger M., Klug G.. Integrative “omics”-approach discovers dynamic and regulatory features of bacterial stress responses. PLos Genet. 2013; 9:e1003576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Glaeser J., Nuss A.M., Berghoff B.A., Klug G.. Singlet oxygen stress in microorganisms. Adv. Microb. Physiol. 2011; 58:141–173. [DOI] [PubMed] [Google Scholar]
- 11. Hess W.R., Berghoff B.A., Wilde A., Steglich C., Klug G.. Riboregulators and the role of Hfq in photosynthetic bacteria. RNA Biology. 2014; 11:413–426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Adnan F., Weber L., Klug G.. The sRNA SorY confers resistance during photooxidative stress by affecting a metabolite transporter in Rhodobacter sphaeroides. RNA Biol. 2015; 12:569–577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Rui B., Shen T., Zhou H., Liu J., Chen J., Pan X., Liu H., Wu J., Zheng H., Shi Y.. A systematic investigation of Escherichia coli central carbon metabolism in response to superoxide stress. BMC Syst. Biol. 2010; 4:122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Valdivia-González M., Pérez-Donoso J.M., Vásquez C.C.. Effect of tellurite-mediated oxidative stress on the Escherichia coli glycolytic pathway. Biometals. 2012; 25:451–458. [DOI] [PubMed] [Google Scholar]
- 15. Bignucolo A., Appanna V.P., Thomas S.C., Auger C., Han S., Omri A., Appanna V.D.. Hydrogen peroxide stress provokes a metabolic reprogramming in Pseudomonas fluorescens: enhanced production of pyruvate. J. Biotechnol. 2013; 167:309–315. [DOI] [PubMed] [Google Scholar]
- 16. Peng T., Berghoff B.A., Oh J.-I., Weber L., Schirmer J., Schwarz J., Glaeser J., Klug G.. Regulation of a polyamine transporter by the conserved 3′ UTR-derived sRNA SorX confers resistance to singlet oxygen and organic hydroperoxides in Rhodobacter sphaeroides. RNA Biol. 2016; 13:988–999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Müller K.M.H., Berghoff B.A., Eisenhardt B.D., Remes B., Klug G.. Characteristics of Pos19 - a small coding RNA in the oxidative stress response of Rhodobacter sphaeroides. PLoS One. 2016; 11:e0163425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Billenkamp F., Peng T., Berghoff B.A., Klug G.. A cluster of four homologous small RNAs modulates C1 metabolism and the pyruvate dehydrogenase complex in Rhodobacter sphaeroides under various stress conditions. J. Bacteriol. 2015; 197:1839–1852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Nuss A.M., Glaeser J., Klug G.. RpoH(II) activates oxidative-stress defense systems and is controlled by RpoE in the singlet oxygen-dependent response in Rhodobacter sphaeroides. J. Bacteriol. 2009; 191:220–230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Dufour Y.S., Landick R., Donohue T.J.. Organization and evolution of the biological response to singlet oxygen stress. J. Mol. Biol. 2008; 383:713–730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Remes B., Rische-Grahl T., Müller K.M.H., Förstner K.U., Yu S.-H., Weber L., Jäger A., Peuser V., Klug G.. An RpoHI-dependent response promotes outgrowth after extended stationary phase in the alphaproteobacterium Rhodobacter sphaeroides. J. Bacteriol. 2017; 199:e00249-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Reinkensmeier J., Giegerich R.. Thermodynamic matchers for the construction of the cuckoo RNA family. RNA Biology. 2015; 12:197–207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Mitchell A.L., Attwood T.K., Babbitt P.C., Blum M., Bork P., Bridge A., Brown S.D., Chang H.-Y., El-Gebali S., Fraser M.I.et al.. InterPro in 2019: improving coverage, classification and access to protein sequence annotations. Nucleic Acids Res. 2018; 47:D351–D360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Barnett M.J., Bittner A.N., Toman C.J., Oke V., Long S.R.. Dual RpoH sigma factors and transcriptional plasticity in a symbiotic bacterium. J. Bacteriol. 2012; 194:4983–4994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Robledo M., Peregrina A., Millán V., García-Tomsig N.I., Torres-Quesada O., Mateos P.F., Becker A., Jiménez-Zurdo J.I.. A conserved α-proteobacterial small RNA contributes to osmoadaptation and symbiotic efficiency of rhizobia on legume roots. Environ. Microbiol. 2017; 19:2661–2680. [DOI] [PubMed] [Google Scholar]
- 26. Remes B., Berghoff B.A., Förstner K.U., Klug G.. Role of oxygen and the OxyR protein in the response to iron limitation in Rhodobacter sphaeroides. BMC Genomics. 2014; 15:794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Baumgardt K., Šmídová K., Rahn H., Lochnit G., Robledo M., Evguenieva-Hackenberg E.. The stress-related, rhizobial small RNA RcsR1 destabilizes the autoinducer synthase encoding mRNA sinI in Sinorhizobium meliloti. RNA Biol. 2016; 13:486–499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Mank N.N., Berghoff B.A., Hermanns Y.N., Klug G.. Regulation of bacterial photosynthesis genes by the small noncoding RNA PcrZ. PNAS. 2012; 109:16306–16311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Klug G., Drews G.. Construction of a gene bank of Rhodopseudomonas capsulata using a broad host range DNA cloning system. Arch. Microbiol. 1984; 139:319–325. [DOI] [PubMed] [Google Scholar]
- 30. Gibson D.G., Young L., Chuang R.-Y., Venter J.C., Hutchison C.A., Smith H.O.. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat. Methods. 2009; 6:343–345. [DOI] [PubMed] [Google Scholar]
- 31. Li K., Härtig E., Klug G.. Thioredoxin 2 is involved in oxidative stress defence and redox-dependent expression of photosynthesis genes in Rhodobacter capsulatus. Microbiology. 2003; 149:419–430. [DOI] [PubMed] [Google Scholar]
- 32. Damm K., Bach S., Müller K.M.H., Klug G., Burenina O.Y., Kubareva E.A., Grünweller A., Hartmann R.K.. Impact of RNA isolation protocols on RNA detection by Northern blotting. Methods Mol. Biol. 2015; 1296:29–38. [DOI] [PubMed] [Google Scholar]
- 33. Pfeiffer V., Sittka A., Tomer R., Tedin K., Brinkmann V., Vogel J.. A small non-coding RNA of the invasion gene island (SPI-1) represses outer membrane protein synthesis from the Salmonella core genome. Mol. Microbiol. 2007; 66:1174–1191. [DOI] [PubMed] [Google Scholar]
- 34. Förstner K.U., Vogel J., Sharma C.M.. READemption-a tool for the computational analysis of deep-sequencing-based transcriptome data. Bioinformatics (Oxford, England. 2014; 30:3421–3423. [DOI] [PubMed] [Google Scholar]
- 35. Hoffmann S., Otto C., Kurtz S., Sharma C.M., Khaitovich P., Vogel J., Stadler P.F., Hackermüller J.. Fast mapping of short sequences with mismatches, insertions and deletions using index structures. PLoS Comput. Biol. 2009; 5:e1000502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Hoffmann S., Otto C., Doose G., Tanzer A., Langenberger D., Christ S., Kunz M., Holdt L.M., Teupser D., Hackermüller J., Stadler P.F.. A multi-split mapping algorithm for circularRNA, splicing, trans-splicing and fusion detection. Genome Biol. 2014; 15:R34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Anders S., Huber W.. Differential expression analysis for sequence count data. Genome Biol. 2010; 11:R106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Kumar S., Stecher G., Li M., Knyaz C., Tamura K.. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018; 35:1547–1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Sneath & Sokal 1973, Sneath P.H.A, Sokal R.R.. Numerical Taxonomy: The Principles and Practice of Numerical Classification. 1973; San Francisco: Freeman; 573. [Google Scholar]
- 40. Felsenstein J. Confidence limits on phylogenies: An approach using the bootstrap. Evolution. 1985; 39:783–791. [DOI] [PubMed] [Google Scholar]
- 41. Förstner K.U., Reuscher C.M., Haberzettl K., Weber L., Klug G.. RNase E cleavage shapes the transcriptome of Rhodobacter sphaeroides and strongly impacts phototrophic growth. Life Sci. Alliance. 2018; 1:e201800080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Green J.B., Edwards T.A., Trincao J., Escalante C.R., Wharton R.P., Aggarwal A.K.. Crystallization and characterization of Smaug: a novel RNA-binding motif. Biochem. Biophys. Res. Commun. 2002; 297:1085–1088. [DOI] [PubMed] [Google Scholar]
- 43. Tadros W., Goldman A.L., Babak T., Menzies F., Vardy L., Orr-Weaver T., Hughes T.R., Westwood J.T., Smibert C.A., Lipshitz H.D.. SMAUG is a major regulator of maternal mRNA destabilization in Drosophila and its translation is activated by the PAN GU kinase. Dev. Cell. 2007; 12:143–155. [DOI] [PubMed] [Google Scholar]
- 44. Green J.B., Gardner C.D., Wharton R.P., Aggarwal A.K.. RNA recognition via the SAM domain of Smaug. Mol. Cell. 2003; 11:1537–1548. [DOI] [PubMed] [Google Scholar]
- 45. Chao Y., Li L., Girodat D., Förstner K.U., Said N., Corcoran C., Śmiga M., Papenfort K., Reinhardt R., Wieden H.-J.et al.. In vivo cleavage map Illuminates the central role of RNase E in coding and non-coding RNA pathways. Mol. Cell. 2017; 65:39–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. McIntosh M., Eisenhardt K., Remes B., Konzer A., Klug G.. Adaptation of the Alphaproteobacterium Rhodobacter sphaeroides to stationary phase. Environ. Microbiol. 2019; 21:4425–4445. [DOI] [PubMed] [Google Scholar]
- 47. Zhao Z., Peng T., Oh J.-I., Glaeser J., Weber L., Li Q., Klug G.. A response regulator of the OmpR family is part of the regulatory network controlling the oxidative stress response of Rhodobacter sphaeroides. Environ. Microbiol. Rep. 2019; 11:118–128. [DOI] [PubMed] [Google Scholar]
- 48. Weber L., Thoelken C., Volk M., Remes B., Lechner M., Klug G.. The conserved Dcw gene cluster of R. sphaeroides is preceded by an uncommonly extended 5′ leader featuring the sRNA UpsM. PLoS One. 2016; 11:e0165694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Mank N., Berghoff B.A., Klug G.. A mixed incoherent feed-forward loop contributes to the regulation of bacterial photosynthesis genes. PLoS One. 2013; 10:347–352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Budnick J.A., Sheehan L.M., Kang L., Michalak P., Caswell C.C.. Characterization of three small proteins in Brucella abortus linked to fucose utilization. J. Bacteriol. 2018; 200:e00127-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Kraus A., Weskamp M., Zierles J., Balzer M., Busch R., Eisfeld J., Lambertz J., Nowaczyk M.M., Narberhaus F.. Arginine-rich small proteins with a domain of unknown function, DUF1127, play a role in phosphate and carbon metabolism of Agrobacterium tumefaciens. J. Bacteriol. 2020; 202: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Venturini E., Svensson S.L., Maaß S., Gelhausen R., Eggenhofer F., Li L., Cain A.K., Parkhill J., Becher D., Backofen R., Barquist L., Sharma C.M., Westermann A.J., Vogel J.. A global data-driven census of Salmonella small proteins and their potential function in bacteria virulence. microLife. 2020; 1:uqaa002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Bandyra K.J., Luisi B.F.. Licensing and due process in the turnover of bacterial RNA. RNA Biology. 2013; 10:627–635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Pinder B.D., Smibert C.A.. microRNA-independent recruitment of Argonaute 1 to nanos mRNA through the Smaug RNA-binding protein. EMBO Rep. 2013; 14:80–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Makarova K.S., Wolf Y.I., van der Oost J., Koonin E.V.. Prokaryotic homologs of Argonaute proteins are predicted to function as key components of a novel system of defense against mobile genetic elements. Biol. Direct. 2009; 4:29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Olovnikov I., Chan K., Sachidanandam R., Newman D.K., Aravin A.A.. Bacterial argonaute samples the transcriptome to identify foreign DNA. Mol. Cell. 2013; 51:594–605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Miyoshi T., Ito K., Murakami R., Uchiumi T.. Structural basis for the recognition of guide RNA and target DNA heteroduplex by Argonaute. Nat. Commun. 2016; 7:11846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Liu Y., Esyunina D., Olovnikov I., Teplova M., Kulbachinskiy A., Aravin A.A., Patel D.J.. Accommodation of helical imperfections in Rhodobacter sphaeroides argonaute ternary complexes with guide RNA and target DNA. Cell Rep. 2018; 24:453–462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Holmqvist E., Vogel J.. RNA-binding proteins in bacteria. Nat. Rev. Microbiol. 2018; 16:601–615. [DOI] [PubMed] [Google Scholar]
- 60. Quendera A.P., Seixas A.F., dos Santos R.F., Santos I., Silva J.P.N., Arraiano C.M., Andrade J.M.. RNA-binding proteins driving the regulatory activity of small non-coding RNAs in bacteria. Front. Mol. Biosci. 2020; 7:78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Müller P., Gimpel M., Wildenhain T., Brantl S.. A new role for CsrA: promotion of complex formation between an sRNA and its mRNA target in Bacillus subtilis. RNA Biol. 2019; 16:972–987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Olejniczak M., Storz G.. ProQ/FinO-domain proteins: another ubiquitous family of RNA matchmakers?. Mol. Microbiol. 2017; 104:905–915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Smirnov A., Schneider C., Hör J., Vogel J.. Discovery of new RNA classes and global RNA-binding proteins. Curr. Opin. Microbiol. 2017; 39:152–160. [DOI] [PubMed] [Google Scholar]
- 64. Wang N., Yamanaka K., Inouye M.. CspI, the ninth member of the CspA family of Escherichia coli, is induced upon cold shock. J. Bacteriol. 1999; 181:1603–1609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Caballero C.J., Menendez-Gil P., Catalan-Moreno A., Vergara-Irigaray M., García B., Segura V., Irurzun N., Villanueva M., Ruiz de los Mozos I., Solano C.et al.. The regulon of the RNA chaperone CspA and its auto-regulation in Staphylococcus aureus. Nucleic Acids Res. 2018; 46:1345–1361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Dahanukar A., Walker J.A., Wharton R.P.. Smaug, a novel RNA-binding protein that operates a translational switch in Drosophila. Mol. Cell. 1999; 4:209–218. [DOI] [PubMed] [Google Scholar]
- 67. Smiber C.A., Lie Y.S., Shillinglaw W., Henzel W.J., Macdonald P.M.. Smaug, a novel and conserved protein, contributes to repression of nanos mRNA translation in vitro. RNA. 1999; 5:1535–1547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Smibert C.A., Wilson J.E., Kerr K., Macdonald P.M.. Smaug protein represses translation of unlocalized nanos mRNA in the Drosophila embryo. Genes Dev. 1996; 10:2600–2609. [DOI] [PubMed] [Google Scholar]
- 69. Dahanukar A., Wharton R.P.. The Nanos gradient in Drosophila embryos is by translational regulation. Genes Dev. 1996; 10:2610–2620. [DOI] [PubMed] [Google Scholar]
- 70. Kordes E., Jock S., Fritsch J., Bosch F., Klug G.. Cloning of a gene involved in rRNA precursor processing and 23S rRNA cleavage in Rhodobacter capsulatus. J. Bacteriol. 1994; 176:1121–1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The sequencing data are available at NCBI Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo) under the accession number GSE144523 and GSE145045.