Abstract
Plants establish their root system as a three-dimensional structure, which is then used to explore the soil to absorb resources and provide mechanical anchorage. Simplified two-dimensional growth systems, such as agar plates, have been used to study various aspects of plant root biology. However, it remains challenging to study the more realistic three-dimensional structure and function of roots hidden in opaque soil. Here, we optimized X-ray computer tomography (CT)-based visualization of an intact root system by using Toyoura sand, a standard silica sand used in geotechnology research, as a growth substrate. Distinct X-ray attenuation densities of root tissue and Toyoura sand enabled clear image segmentation of the CT data. Sorghum grew especially vigorously in Toyoura sand and it could be used as a model for analyzing root structure optimization in response to mechanical obstacles. The use of Toyoura sand has the potential to link plant root biology and geotechnology applications.
Keywords: root system architecture, Sorghum bicolor, Toyoura sand, X-ray computer tomography
The spatial arrangement of plant roots (root system architecture: RSA) is shaped by innate genetic programs, environmental factors, and interactions thereof to optimize root functions, such as water and nutrient uptake (reviewed by Morris et al. 2017). RSA also plays a physical role, supporting aboveground shoots against mechanical forces and preventing lodging or uprooting (Bailey et al. 2002; Jenkins 1930; Yang et al. 2017). Two conifer tree species grown in a wind tunnel developed more lateral roots on their windward and leeward sides to help counteract wind stresses (Stokes et al. 1995), suggesting an intriguing link between force sensing in the shoot and changes in RSA in the root. However, a mechanistic understanding of such adaptive structural optimization has been hampered by the difficulty of observing time-course changes in intact, three-dimensional (3D) RSA in soil. X-ray computed tomography (CT) is a promising technology that enables the non-destructive visualization of inner 3D structures based on different X-ray attenuation densities among materials (Hounsfield 1973). Although CT is extensively used in various areas such as engineering, medical, and archaeological studies, visualization using CT of plant roots growing in soil has been challenging (reviewed by Mooney et al. 2012). First, plant root diameters are often too thin (ca. 100 µm in Arabidopsis thaliana) to be distinguished by the sub-millimeter resolution of CT, although this issue could be circumvented by using larger plants with thicker roots and/or micro-CT scanners with higher resolutions. Second, the X-ray attenuation densities of roots and soil are similar and often overlap, making it difficult to perform image segmentation to distinguish roots from the background soil in the acquired CT data. Recently, various advances have been made to improve the utility of CT for root structure phenotyping in three major ways, namely by refining CT scanning parameters (Teramoto et al. 2020), by developing programs for improved image analysis (Mairhofer et al. 2012), and by optimizing root growth settings. Use of inorganic growth substrates, such as sand, silica beads, or calcined clay granules, was found effective in visualization of root structure by CT (Piñeros et al. 2016; Rogers et al. 2016; Teramoto et al. 2020). This is based on the clear difference in X-ray attenuation densities between these inorganic substrates (high) and root tissue (low). In this study, we aimed to extend the use of inorganic growth substrates in CT visualization of roots by using Toyoura sand, a standard sand with well characterized physical properties. Additionally, we mixed sand with materials having X-ray attenuation densities higher than that of sand, which enabled us to visualize root responses to hard obstacles.
Toyoura sand (Toyoura Keiseki Kogyo, Yamaguchi, Japan) is a fine quartz sand with uniformly graded particle size; with average (50% passing) diameter of 234 µm and 87% of the grains having a diameter between 180 and 300 µm (Hosono and Yoshimine 2009). We sought to compare the CT visibility of plant roots grown in standard soil with that of plants grown in Toyoura sand. To do this, we first prepared a custom columnar pot by cutting polyvinyl chloride (PVC) pipe (VU65, Japan Industrial Standards) (Figure 1A). We partitioned it into two halves using a strip of cardboard and filled each side with either Toyoura sand or organic soil mixture (Cell-baido from Latec, Miyazaki, Japan), then removed the cardboard strip. Tomato seeds (Solanum lycopersicum cv. Pritz) were water-imbibed, then sown on both sand and soil sides. Germinated seedlings were cultivated in a growth room at 22°C under continuous light (Figure 1B). We performed CT scanning (TOSCANER-32300 FPD; Toshiba ITC, Japan) with a tube voltage and current of 190 kV and 500 µA, respectively. Samples were placed on a rotating stage and imaged from 1,000 view-angles over 360°. Each image of a view was obtained from the average of three acquisitions, with an acquisition time of 166 ms each (8.3 min of X-ray exposure in total). We did not find any negative symptoms in subsequent growth after four repeated CT scans (data not shown), although any heritable effects in subsequent generations were not investigated. Final reconstructed data had a voxel resolution of ∼120 µm, and the entire imaged area was 1,024×1,024×878 voxels (approximately 12×12×10 cm3). A representative single slice of the CT data is shown in Figure 1C. In the Toyoura sand area, root tissue with low attenuation density can be seen as dark spots or filaments on a brighter background (quartz sand with high attenuation density). However, in the soil area, root tissue could not be distinguished from the soil background because of their similar attenuation density. To extract root images, we used Fiji ImageJ software (https://fiji.sc/) to crop the region of interest and convert the gray-scale image into binary by thresholding pixel brightness. Then, the 3D Viewer plugin of ImageJ (Schmid et al. 2010) was used to draw a 3D rendering of the extracted root pixels. It was confirmed that the 3D structure of the roots growing Toyoura sand area were clearly visible after such simple image processing (Figure 1D). Owing to the voxel resolution of 120 µm in our settings, we could judge the dark spots or filaments as roots when their diameters were at minimum 2–3 voxels (240–360 µm). Higher voxel resolution may enable detecting even thinner roots such as those of Arabidopsis, but as a tradeoff, it limits the size of field of view because of fixed matrix size.
Figure 1. Establishment of Toyoura sand cultivation system for CT. A: Custom columnar pot made of a cut PVC tube and a punched end cap. B: Growth of tomato seedlings in a pot filled with Toyoura sand (left side) and organic soil mix (right side). C: Reconstructed single slice image of CT data, showing Toyoura sand in lighter color (left side) and organic soil mix in darker color (right side). Primary roots (asterisks) and lateral roots (arrowheads) are clearly visible in Toyoura sand area. D: 3D rendering of segmented tomato root tissue. Brightness is inverted from the original CT image, so Toyoura sand is shown in darker color (left side) and organic soil mix is shown in lighter color (right side). E: Growth of Arabidopsis (Col-0 accession) 14 days after sowing on organic soil mix (left) and Toyoura sand (right). F: Growth of sorghum 22 days after sowing on Toyoura sand (left) and organic soil mix (right). G–J: 3D rendering of root CT image from various species. G: Radish, 44 days after sowing. Note poorer hypocotyl thickening in Toyoura sand area (left side). H: Turnip, 44 days after sowing. I: Pea, 7 days after sowing. J: Rice, 90 days after sowing. Scale bars: 1 cm.
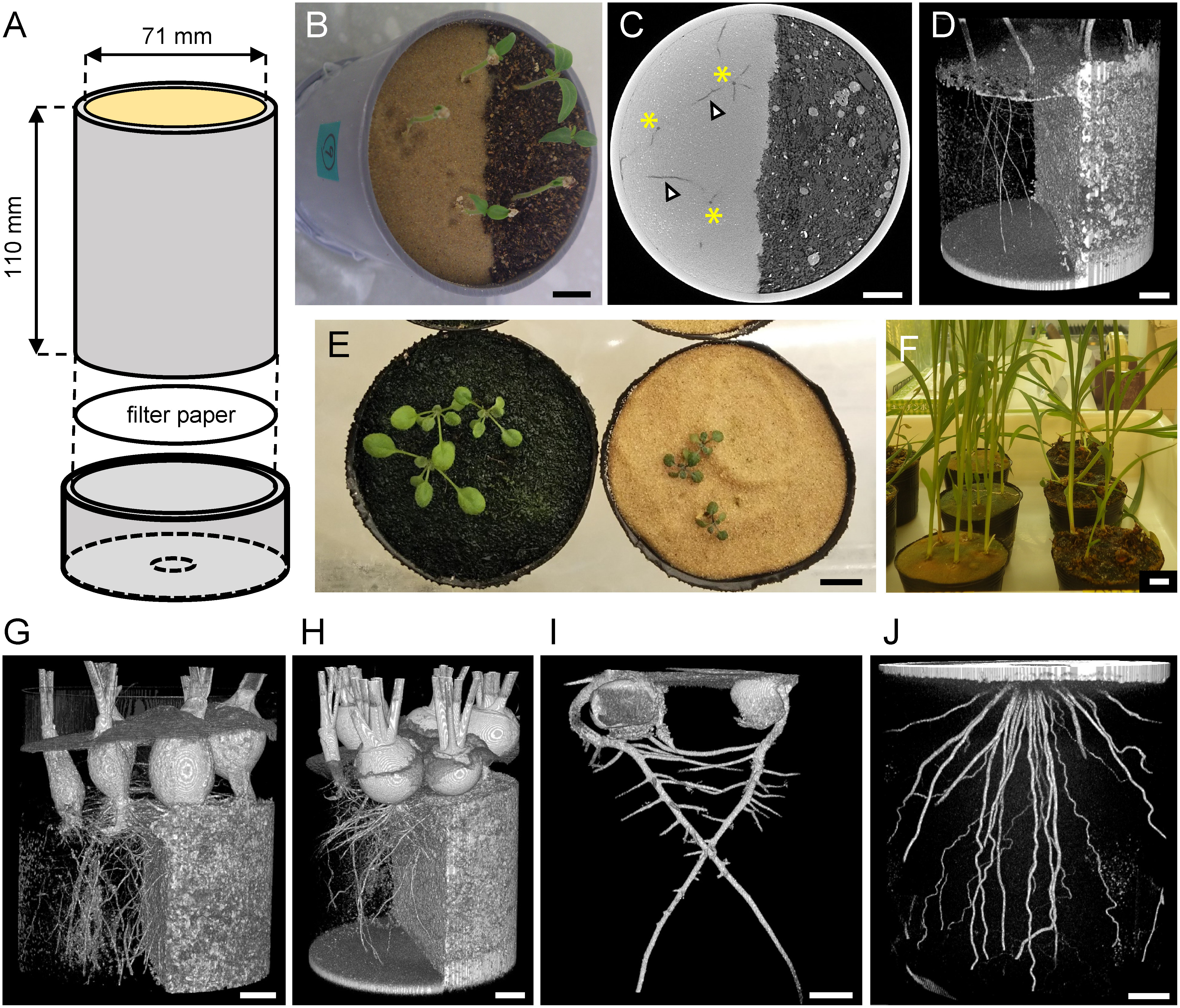
To extend the utility of Toyoura sand in plant biology, we cultivated various other species on Toyoura sand and compared their growth with that of standard soil-grown plants. Tested plants were Arabidopsis (Arabidopsis thaliana: accessions Columbia-0, Landsberg erecta, Wassilewskija, C24, Cape Verdi Islands), radish (Raphanus sativus var. sativus cv. ‘Comet’), daikon radish (R. sativus var. longipinnatus cv. ‘Utsugi Gensuke’), and turnip (Brassica rapa ssp. rapa cv. ‘Shin-Kanamachi’) from Brassicaceae; pea (Pisum sativum cv. ‘Tsurunashi Snap’) and soybean (Glycine max cv. ‘Fukuyutaka’) from Fabaceae; carrot (Daucus carota ssp. sativus cv. ‘Kuroda Gosun’) from Apiaceae; and rice (Oryza sativa cv. ‘Taichung 65’), maize (Zea mays cv. ‘Canberra 90’), and sorghum (Sorghum bicolor cv. ‘Metre Sorgo’) from Poaceae. A general tendency was observed that the plant growth was to some extent poorer on Toyoura sand than on soil, even if well-fertilized with Hoagland’s hydroponics solution (Hoagland and Arnon 1950). Arabidopsis is one of the most important model plants, about which there is extensive knowledge of its root development, but its growth was severely inhibited on Toyoura sand (Figure 1E). Although we tested several laboratory strains, Arabidopsis seeds collected from world-wide natural populations are available from stock centers (such as ABRC: https://abrc.osu.edu/), and some of these strains were native to sandy habitats (coastal dunes). It would be interesting to test whether these sand dune ecotypes could thrive better on Toyoura sand. Nevertheless, the other plant species grew sufficiently well in Toyoura sand to perform CT scanning. We could obtain clear 3D renderings of diverse root structures grown in Toyoura sand (Figure 1G–J).
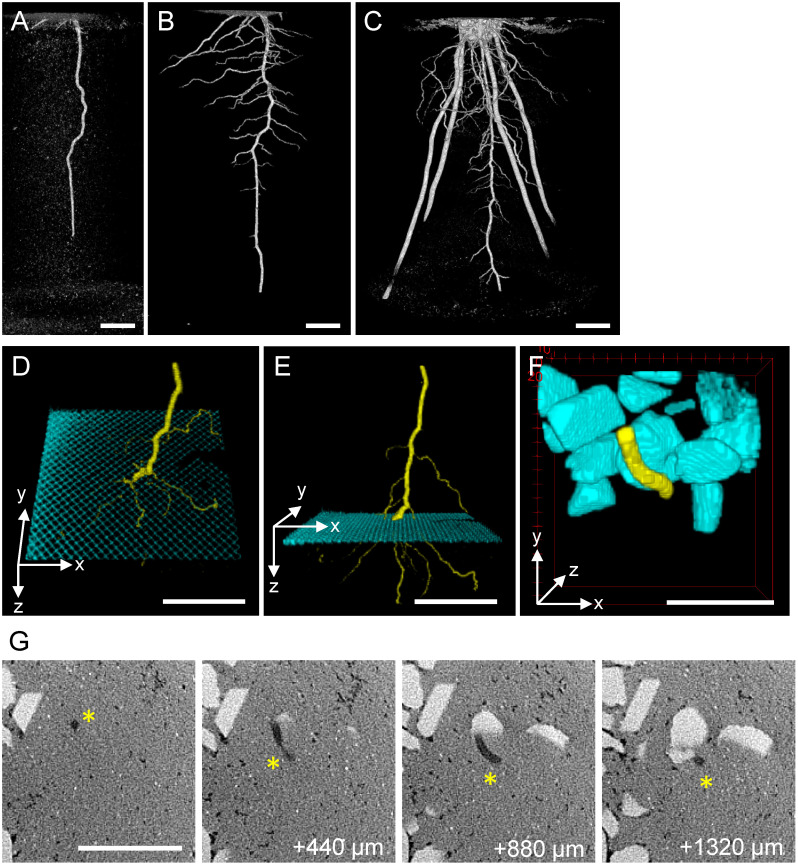
Sorghum (cv. ‘Metre Sorgo’ from Takii Seeds, Kyoto, Japan) grew vigorously on Toyoura sand and even outperformed soil-grown controls under our cultivation conditions (Figure 1F). We used X-ray CT scanning to visualize the time course of sorghum roots grown in Toyoura sand. Seven days after germination, a single strand of embryonic primary root grew straight down (Figure 2A). Fourteen days after germination, lateral roots branched out from the primary root and grew near horizontally (Figure 2B). Thirty-five days after germination, thick crown roots derived from the basal stem node grew obliquely to form a typical fibrous root system (Figure 2C). We noted that the bulk density of Toyoura sand affected the growth pattern of sorghum roots. Loosely packed sand (1.4 g cm−3) permitted deep and straight penetration of primary roots, while densely packed sand (1.6 g cm−3) inhibited primary root elongation and caused root waving (Kotaro Iwashita and Jun Otani, unpublished). Thus, Toyoura sand could also be used to study root responses to soil compaction (Correa et al. 2019; Rogers et al. 2016).
Figure 2. CT visualization of sorghum root growth in Toyoura sand. A–C: Time-course development of sorghum root system. A: 7 days after sowing. B: 14 days after sowing. C: 35 days after sowing. Panels A and B are from an identical plant. D–E: Passage of sorghum root (yellow) through metal mesh (cyan) buried in Toyoura sand, viewed from two angles. F: Detour growth of sorghum root (yellow) interfered by calcite gravels (cyan) mixed in Toyoura sand. G: Selected slices of the original CT data used to depict F. Four depth series (440 µm interval) of the same XY area, showing Toyoura sand as light-gray background, calcite gravels as whitish polygons, and root tissues as dark-gray filaments (asterisks). Scale bars: 1 cm.
Clear separation of root and Toyoura sand led us to add a third material that has X-ray attenuation density distinct from those of plant tissue and silica. We buried stainless-steel mesh (0.65 mm aperture) or calcite gravels (approximately 3–10 mm diameter) inside Toyoura sand and cultivated sorghum seedlings to test how root growth is affected when encountering these hard obstacles. After CT scanning, we separately segmented root tissues and stainless-steel mesh (or calcite gravels), assigned different channels, and merged them to draw 3D volume rendering images. We observed that the sorghum root passed through stainless-steel mesh by branching out multiple fine lateral roots (Figure 2D, E). However, we could not determine the root structure inside the mesh pore because of too high X-ray attenuation by stainless steel, known as CT metal artifact (Katsura et al. 2018). Further optimization of experimental design and data analysis are needed. When sorghum root growth is interfered by calcite gravels, we observed curvature of root growth path to flexibly detour the obstacle, instead of pushing it aside (Figure 2F). Four of original slices (440 µm interval) are shown in Figure 2G, which clearly detected light-gray background of Toyoura sand, whitish polygonal calcite, and dark-gray filamentous root that changed its growth path within a millimeter depth. It will be interesting to manipulate the candidate genes involved in touch sensing or root growth angle (such as Nakagawa et al. 2007; Uga et al. 2013; Wang et al. 2018) and assess how this root structure optimization is controlled.
Here, we established a simple method for visualizing plant roots grown in Toyoura sand by CT. Toyoura sand has been widely used in Japan as the standard sand for geotechnical engineering research (Hosono and Yoshimine 2009). Plant root-inspired design of novel building foundation piles is emerging as a new interest in the field of geotechnical engineering (Frost et al. 2017). The use of Toyoura sand in plant research thus has a potential to facilitate the interdisciplinary transfer of knowledge from plant root biology to geotechnology applications. For example, load testing of living root system grown in Toyoura sand will reveal the mechanical difference between plant roots and artificial foundation piles.
Acknowledgments
We are grateful to X-EARTH Center, Kumamoto University for providing access to the CT facility. Kotaro Iwashita, Takahiro Sato, and Chihiro Furumizu assisted sorghum cultivation, CT scanning, and shared unpublished data. Hidehiko Sunohara provided rice seeds. PVC pots were fabricated by using machine tools facility at Creative Engineering & Design Education Center, Kumamoto University. This research has been funded by KAKENHI Grant Number, 18H05487 to S.S.
References
- Bailey PH, Currey JD, Fitter AH (2002) The role of root system architecture and root hairs in promoting anchorage against uprooting forces in Allium cepa and root mutants of Arabidopsis thaliana. J Exp Bot 53: 333–340 [DOI] [PubMed] [Google Scholar]
- Correa J, Postma JA, Watt M, Wojciechowski T (2019) Soil compaction and the architectural plasticity of root systems. J Exp Bot 70: 6019–6034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frost JD, Martinez A, Mallett SD, Roozbahani MM, DeJong JT (2017) Intesection of modern soil mechanics with ants and roots. Proceedings of Geotechnical Frontiers 2017: Geotechnical Materials, Modeling, and Testing (GSP 280). Orlando, Florida, pp 900–909
- Hoagland DR, Arnon DI (1950) The water-culture method for growing plants without soil. Calif Agric Exp Stn 434: 1–32 [Google Scholar]
- Hosono Y, Yoshimine M (2009) Toyoura-suna no ryudo-bunpu. 64th Japan Society of Civil Engineers Annual Meeting 64: 335–336 (in Japanese)
- Hounsfield GN (1973) Computerized transverse axial scanning (tomography): Part 1. Description of system. Br J Radiol 46: 1016–1022 [DOI] [PubMed] [Google Scholar]
- Jenkins MT (1930) Heritable characters of maize: XXXIV—Rootless. J Hered 21: 79–80 [Google Scholar]
- Katsura M, Sato J, Akahane M, Kunimatsu A, Abe O (2018) Current and novel techniques for metal artifact reduction at CT: Practical guide for radiologists. Radiographics 38: 450–461 [DOI] [PubMed] [Google Scholar]
- Mairhofer S, Zappala S, Tracy SR, Sturrock C, Bennett M, Mooney SJ, Pridmore T (2012) RooTrak: Automated recovery of three-dimensional plant root architecture in soil from X-ray microcomputed tomography images using visual tracking. Plant Physiol 158: 561–569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mooney SJ, Pridmore TP, Helliwell J, Bennett MJ (2012) Developing X-ray computed tomography to non-invasively image 3-D root systems architecture in soil. Plant Soil 352: 1–22 [Google Scholar]
- Morris EC, Griffiths M, Golebiowska A, Mairhofer S, Burr-Hersey J, Goh T, von Wangenheim D, Atkinson B, Sturrock CJ, Lynch JP, et al. (2017) Shaping 3D root system architecture. Curr Biol 27: R919–R930 [DOI] [PubMed] [Google Scholar]
- Nakagawa Y, Katagiri T, Shinozaki K, Qi Z, Tatsumi H, Furuichi T, Kishigami A, Sokabe M, Kojima I, Sato S, et al. (2007) Arabidopsis plasma membrane protein crucial for Ca2+ influx and touch sensing in roots. Proc Natl Acad Sci USA 104: 3639–3644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piñeros MA, Larson BG, Shaff JE, Schneider DJ, Falcão AX, Yuan L, Clark RT, Craft EJ, Davis TW, Pradier PL, et al. (2016) Evolving technologies for growing, imaging and analyzing 3D root system architecture of crop plants. J Integr Plant Biol 58: 230–241 [DOI] [PubMed] [Google Scholar]
- Rogers ED, Monaenkova D, Mijar M, Nori A, Goldman DI, Benfey PN (2016) X-ray computed tomography reveals the response of root system architecture to soil texture. Plant Physiol 171: 2028–2040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid B, Schindelin J, Cardona A, Longair M, Heisenberg M (2010) A high-level 3D visualization API for Java and ImageJ. BMC Bioinformatics 11: 274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stokes A, Fitter AH, Courts MP (1995) Responses of young trees to wind and shading: Effects on root architecture. J Exp Bot 46: 1139–1146 [Google Scholar]
- Teramoto S, Takayasu S, Kitomi Y, Arai-Sanoh Y, Tanabata T, Uga Y (2020) High-throughput three-dimensional visualization of root system architecture of rice using X-ray computed tomography. Plant Methods 16: 66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uga Y, Sugimoto K, Ogawa S, Rane J, Ishitani M, Hara N, Kitomi Y, Inukai Y, Ono K, Kanno N, et al. (2013) Control of root system architecture by DEEPER ROOTING 1 increases rice yield under drought conditions. Nat Genet 45: 1097–1102 [DOI] [PubMed] [Google Scholar]
- Wang L, Guo M, Li Y, Ruan W, Mo X, Wu Z, Sturrock CJ, Yu H, Lu C, Peng J, et al. (2018) LARGE ROOT ANGLE1, encoding OsPIN2, is involved in root system architecture in rice. J Exp Bot 69: 385–397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang M, Défossez P, Danjon F, Dupont S, Fourcaud T (2017) Which root architectural elements contribute the best to anchorage of Pinus species?: Insights from in silico experiments. Plant Soil 411: 275–291 [Google Scholar]