Abstract
Coronaviruses are prone to emergence into new host species most recently evidenced by SARS-CoV-2, the causative agent of the COVID-19 pandemic1. Small animal models that recapitulate SARS-CoV-2 disease are desperately needed to rapidly evaluate medical countermeasures (MCMs)2,3. SARS-CoV-2 cannot infect wildtype laboratory mice due to inefficient interactions between the viral spike (S) protein and the murine ortholog of the human receptor, ACE24. We used reverse genetics5 to remodel the interaction between S and mACE2 resulting in a recombinant virus (SARS-CoV-2 MA) that could utilize mACE2 for entry. SARS-CoV-2 MA replicated in both the upper and lower airways of both young adult and aged BALB/c mice. Importantly, disease was more severe in aged mice, and showed more clinically relevant phenotypes than those seen in HFH4-hACE2 transgenic mice. We then demonstrated the utility of this model through vaccine challenge studies in immune competent mice with native expression of mACE2. Lastly, we show that clinical candidate interferon (IFN) lambda-1a can potently inhibit SARS-CoV-2 replication in primary human airway epithelial cells in vitro, and both prophylactic and therapeutic administration diminished replication in mice. Our mouse-adapted SARS-CoV-2 model demonstrates age-related disease pathogenesis and supports the clinical use of pegylated IFN lambda-1a treatment in human COVID-19 infections6.
Zoonotic coronaviruses readily traffic into new host species, as evidenced by the emergence of SARS-CoV in 2002–2003, Middle East Respiratory Coronavirus in 2012 and SARS-CoV-2, the causative agent of the COVID-19 pandemic. SARS-CoV-2 has caused millions of infections and hundreds of thousands of deaths worldwide. As there are currently no approved vaccines and only one antiviral approved for emergency use for SARS-CoV-27 small animal model systems are vital to better understand COVID-19 disease mechanisms and to evaluate medical counter measures (MCMs) for improved global health. Mouse models not only provide key insights into the pathogenic mechanisms of CoV disease but can serve as high-throughput preclinical evaluation platforms to identify high performance antivirals and vaccines2,8. SARS-CoV-2 enters host cells through the binding of the cellular receptor angiotensin-converting enzyme 2 (ACE2). Unfortunately, standard laboratory mice do not support infection with SARS-CoV-2 due to incompatibility of the S protein to the murine ortholog (mACE2) of the human receptor, complicating model development4.
Infection of HFH4-hACE2 transgenic mice
Animal models are critical for development of MCMs to combat the COVID-19 pandemic. While laboratory mice infected with mouse adapted strains of SARS-CoV-1 and MERS-CoV have informed our understanding of viral pathogenesis and intervention strategiesSeveral animal models for SARS-CoV-2 have been reported with varying degrees of viral replication and clinical disease including hACE2 transgenic mice9–11, virally transduced hACE2 mice8,12,13, ferrets14, hamsters15,16, and non-human primates17–19. To determine the utility of our previously characterized HFH4 (FoxJ1) promoter driven hACE2-overexpressing transgenic mice as a model for SARS-CoV-2 disease, we infected with HFH4-hACE2 mice with SARS-CoV-29,20,21. The HFH4 promoter drives expression of hACE2 in ciliated cells of respiratory tract epithelium but also in in the central nervous system22,23. HFH4-hACE2 mice infected with SARS-CoV-2 lost minimal weight yet only 60% survived by 5 days post infection (dpi) (Extended Data 1A–B). Virus was detected in the lung on 2 and 5dpi (Extended Data 1C). Similar to previously reported SARS-CoV-1 and SARS-CoV-2 infection of HFH4-hACE2 mice, SARS-CoV-2 virus was detected in the brains of mice that succumbed to infection suggesting that mortality was driven by viral neuroinvasion (Extended Data 1D)9,21. Lastly, we utilized whole body plethysmography (WBP) to monitor pulmonary function in SARS-CoV-2 infected mice24. Using WBP, we evaluated several complementary metrics of pulmonary obstruction and bronchoconstriction including PenH and Rpef which remained at normal levels for the duration of these studies proving further evidence that respiratory infection was likely not a major driver of mortality (Extended Data 1F–G). While hACE2 expression driven by the HFH4 promoter facilitates SARS-CoV-2 infection of mice, the pathogenesis observed fails to accurately model the more severe disease manifestations observed in humans.
Remodeling the SARS-CoV-2 mACE2 binding interface
Rather than genetically alter the host, we next sought to remodel the SARS-CoV-2 spike receptor binding domain to facilitate efficient binding to mACE2. We compared the ACE2 contact residues in the RBDs of several group 2B coronaviruses capable of infecting mice to those of SARS-CoV-2 (Figure 1A, Extended Data 2)21,25,26. While there is variation among SARS-CoV-1, WIV1, and SHC014 at a few amino acid positions, residue 498 of SARS-CoV-2 was uniquely divergent, suggesting incompatibility of SARS-CoV-2 Q498 with mACE2. In addition, molecular modeling of the SARS-CoV-2 RBD and receptor interface predicted a loss of interaction between Q498 of the SARS-CoV-2 spike and Q42 of hACE2 compared to mACE2 (Figure 1B–C), which may diminish binding efficiency. Thus, substitution of residue Q498, and adjacent P499, with Y and T, respectively, from WIV1 and SARS-CoV-1 might restore the interaction with Q42 of mACE2, while preserving interaction with hACE2 (Figure 1D–E). Using reverse genetics22, we engineered Q498Y/P499T into the SARS-CoV-2 S gene and recovered the recombinant virus (SARS-CoV-2 MA). Importantly, SARS-CoV-2 MA replicated to slightly lower titers compared to parental wildtype (WT) virus in Vero E6 cells (Figure 1F) and human primary differentiated bronchiolar airway epithelial cells (HAE) (Figure 1G). Notably, unlike WT virus, SARS-CoV-2 MA RNA could be detected in cells expressing mACE2 by 24 hours post infection (Figure 1H).
Figure 1. Generation of mouse adapted SARS-CoV-2 MA.
(A) Amino acid (AA) table of group 2B spike receptor binding domain (RBD) hACE2 interaction residues. AA positions are numbered relative to SARS-CoV-1 (above) and SARS-CoV-2 (below). Green residues are contacts as determined by published crystal structures. AAs colored by BLOSUM62 conservation score relative to SARS-CoV-1 Urbani (red=least conserved; blue=most conserved). SARS-CoV-1 Urbani, SARS-CoV-1 MA15, WIV1, and SHC014 can utilize mACE2 as a functional receptor whereas SARS-CoV-2 cannot. Red box indicates Q498 in SARS-CoV-2 that is a hACE2 contact for both SARS-CoV-1 and SARS-CoV-2, and is uniquely divergent in SARS-CoV-2. (B) SARS-CoV-2 WT RBD and hACE2 interface (PDB: 6m0j). SARS-CoV-2 Q498 (red) interacts with Q42 (magenta) of hACE2. (C) Modelling of SARS-CoV-2 WT RBD and mACE2. SARS-CoV-2 Q498 no longer interacts with Q42 of mACE2. (D) Modelling of SARS-CoV-2 Q498Y and P499T (orange) substitutions restore interaction with Q42 of mACE2. (E) Modelling of SARS-CoV-2 Q498Y/P499T maintains interaction with Q42 of hACE2. (F–G) Single step growth curve of SARS-CoV-2 WT and SARS-CoV-2 MA in Vero E6 cells (F) and differentiated primary human bronchiolar epithelial cells (HAEs) (G). Dotted line represents limit of detection (LOD). Log transformed data analyzed by 2-way ANOVA followed by Sidak’s multiple comparisons. (F) ‘*’ denotes p=0.0053 (36 hours). (H) Non-permissive DBT-9 cells were transfected to express hACE2 or mACE2 and infected with SARS-CoV-2 WT and SARS-CoV-2 MA. Viral RNA was quantified by qRT-PCR at 24 hours post infection and scaled to empty vector transfected cells. Log transformed data analyzed by 2-way ANOVA followed by Dunnett’s multiple comparisons. ‘*’ denotes p=0.0322 (hACE2), <0.0001 (mACE2). (F-H) n=3 techical replicates for each group, representative of 2 independent experiments. Error bars represent standard deviation about the mean.
Mouse adapted SARS-CoV-2 replicates in the upper and lower airways of WT mice
After demonstrating SARS-CoV-2 MA could utilize mACE2 for entry, we sought to determine if this virus could infect young adult WT mice. While overt clinical signs of infection such as weight loss were not observed in young adult BALB/c mice infected with 105 plaque forming units (PFU) SARS-CoV-2 MA (Figure 2A), high titer virus replication was noted in lung tissue on 2dpi but was cleared by 4dpi (Figure 2B). Under identical conditions, SARS-CoV-2 WT replication was not detected. Using WBP to monitor pulmonary function as described above, we found SARS-CoV-2 MA young infected mice showed a small but significant change in PenH (Figure 2D) and a significant decrease in Rpef (Figure 2E) on 2dpi, indicative of impaired lung function. Histological analysis of SARS-CoV-2 MA infected mice revealed insterstitial congestion, epithelial damage, inflammatory infiltrate, and peribronchiolar lympocytic inflammation surrounding airways on 2dpi which were also the main target of virus replication as determined by viral antigen staining (Figure 2F). At 4dpi, histological analysis showed increased inflammation and hemorrhage in the lung (Figure 2G). Concordant with infectious titer data demonstrating virus clearance by 4dpi, viral antigen was not detected in lung tissues sections at this time (Figure 2B, Figure 2G). Similar to that observed in humans, SARS-CoV-2 MA virus was also observed in the upper airway and antigen was present in nasal turbinate epithelium on 2dpi (Figure 2C, 2H). Thus, like often seen in humans, infection of young adult mice with SARS-CoV-2 MA resulted in efficient virus replication in the upper and lower airways, limited replication in the parenchyma and was associated with mild to moderate disease.
Figure 2: SARS-CoV-2 MA replicates in young BALB/c mice.
12-week-old female BALB/c mice were mock infected (gray), or infected with 105 PFU SARS-CoV-2 WT (black) or MA (red). Data combined from two independent experiments. (A) Percent starting weight. Dotted line represents weight loss criteria for humane euthanasia. Data analyzed by mixed effects analysis followed by Dunnett’s multiple comparisons. (B–C) Viral lung (B) and nasal turbinate (C) titer. 2dpi: n=5 WT, n=10 MA; 4dpi: n=5 WT, n=9 MA. Dotted line represents limit of detection (LOD). Undetected samples are plotted at half the LOD. Log transformed data analyzed by 2-way ANOVA followed by Sidak’s multiple comparisons. ‘ND’=not determined. (B) ‘*’ denotes p=<0.0001. (D–E) Whole body plethysmography assessing pulmonary function for PenH (D) and Rpef (E). Data analyzed by 2-way ANOVA followed by Dunnett’s multiple comparisons. Error bars represent standard error about the mean. (D) ‘*’ denotes p=0.012 (2dpi), 0.0025 (3dpi), 0.0030 (4dpi), 0.0018 (6dpi). (E) ‘*’ denotes p=0.0426 (1dpi), 0.0194 (2dpi), 0.0442 (3dpi) (F–H) 200X images of lung sections from 2dpi (F) and 4dpi (G) and 100X images of nasal turbinates from 2dpi (H). Top: hematoxylin and eosin (H&E). Bottom: immunohistochemistry staining (IHC) for SARS-CoV-2 nucleocapsid protein, counterstained with hematoxylin. Scale bars=200 μm. Representative of two independent experiments.
Modeling age-related exacerbation of SARS-CoV-2 disease in mice
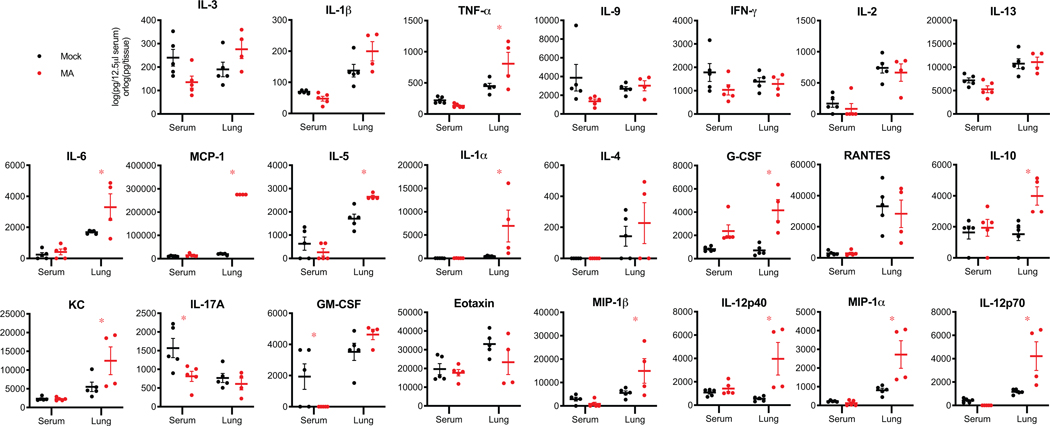
Higher morbidity and mortality rates have been consistently observed in older human populations throughout the COVID-19 pandemic27. Additionally, wildtype and mouse adapted SARS-CoV-1 shows strong age dependent disease phenotypes in humans and mice, respectively28,29. To determine if infection of aged mice would recapitulate the age-related increase in disease severity observed in humans with COVID-19, we infected 1-year-old BALB/c mice with SARS-CoV-2 MA. In contrast to young adult mice, aged BALB/c mice exhibited a transient yet significant decrease in body weight by 3–4dpi compared to mock infected mice (Figure 3A) (p-values of weights of old versus young SARS-CoV-2 MA infected mice on 3 and 4dpi are <0.0001 and 0.0040, respectively). Similar to young adult mice, aged mice had high titer replication in the lung on 2dpi, and but unlike young adult mice viral clearence was delayed as evidenced by detectable virus at 4dpi (Figure 3B). Similarly, replication in the upper airway persisted in half of the mice at 4dpi (Figure 3C). The loss of pulmonary function was more pronounced in aged animals as evidenced by significant differences in PenH and Rpef among mock and SARS-CoV-2 MA infected animals (Figure 3D–E) (PenH was statistically higher in SARS-CoV-2 MA infected old mice on 2dpi compared young mice with p-value of 0.0457. Rpef was significantly lower in old infected mice on 2 and 4dpi with p-values of 0.0264 and 0.0280, respectively). Compared to young mice, SARS-CoV-2 MA infected old mice displayed increased epithelial damage, peribronchiolar lymphocytic inflammation, hemorrhage, and edema in the lung at 2dpi and 4dpi, and viral antigen was found in both conducting airway epithelium and interstitium and nasal epithelium, with minimal antigen staining present at 4dpi, concordant with detection of viral titer (Figure 3B., Fig 3F–H). Additionally, several proinflammatory cytokines were elevated in the lung, but not serum, at 2dpi, indicative of a localized cytokine and chemokine response (Extended Data 3).
Figure 3. SARS-CoV-2 MA replicates in old BALB/c mice with minor disease.
1-year-old female BALB/c were mock infected (gray), or infected with 105 PFU SARS-CoV-2 WT (black) or MA (red). Data combined from two independent experiments. (A) Percent starting weight. Dotted line represents weight loss criteria for humane euthanasia. Data analyzed by mixed effects analysis followed by Dunnett’s multiple comparisons. ‘*’ denotes p=<0.0001 (3dpi), 0.0305 (4dpi). (B–C) Viral lung (B) and nasal turbinate (C) titer. 2dpi & 4dpi: n=5 WT, n=10 MA. Dotted line represents limit of detection (LOD). Undetected samples are plotted at half the LOD. Log transformed data analyzed by 2-way ANOVA followed by Sidak’s multiple comparisons. ‘ND’=not determined (B) ‘*’ denotes p=<0.0001. (D–E) Whole body plethysmography assessing pulmonary function for PenH (D) and Rpef (E). Data analyzed by 2-way ANOVA followed by Dunnett’s multiple comparisons. Error bars represent standard error about the mean. (D) ‘*’ denotes p=0.0014 (2dpi). (E) ‘*’ denotes p=<0.0001 (2dpi), 0.0242 (3dpi), 0.0130 (4dpi), 0.0481 (5dpi). (F–H) 200X images of lung sections from 2dpi (F) and 4dpi (G) and 100X images of nasal turbinates from 2dpi (H). Top: hematoxylin and eosin (H&E). Bottom: immunohistochemistry staining (IHC) for SARS-CoV-2 nucleocapsid protein, counterstained with hematoxylin. Scale bars=200 μm. Representative of two independent experiments.
Vectored vaccine and interferon lambda-1a efficacy against SARS-CoV-2
As demonstrated with mouse adapted strains of SARS-CoV-125, a replication competent SARS-CoV-2 MA strain facilitates the elucidation of virus and host factors that guide pathogenesis and disease severity and importantly, allows for rapid testing of intervention strategies in standard laboratory mice. Utilizing a virus replicon particle (VRP) system, we vaccinated 10-week-old BALB/c mice against SARS-CoV-2 spike (S), nucleocapsid (N), and GFP as a control, boosted after 3 weeks, and challenged 4 weeks post boost with SARS-CoV-2 MA. Unlike mice vaccinated with GFP or N, serum from S vaccinated mice 3 weeks post boost potently neutralized SARS-CoV-2 reporter virus expressing nanoluciferase (nLUC) (Figure 4A). Upon challenge with SARS-CoV-2 MA, only those vaccinated with VRP expressing S significantly diminished lung and nasal turbinate titer (Figure 4B–C).
Figure 4. Evaluation of prevention and intervention strategies against SARS-CoV-2 MA infection in mice.
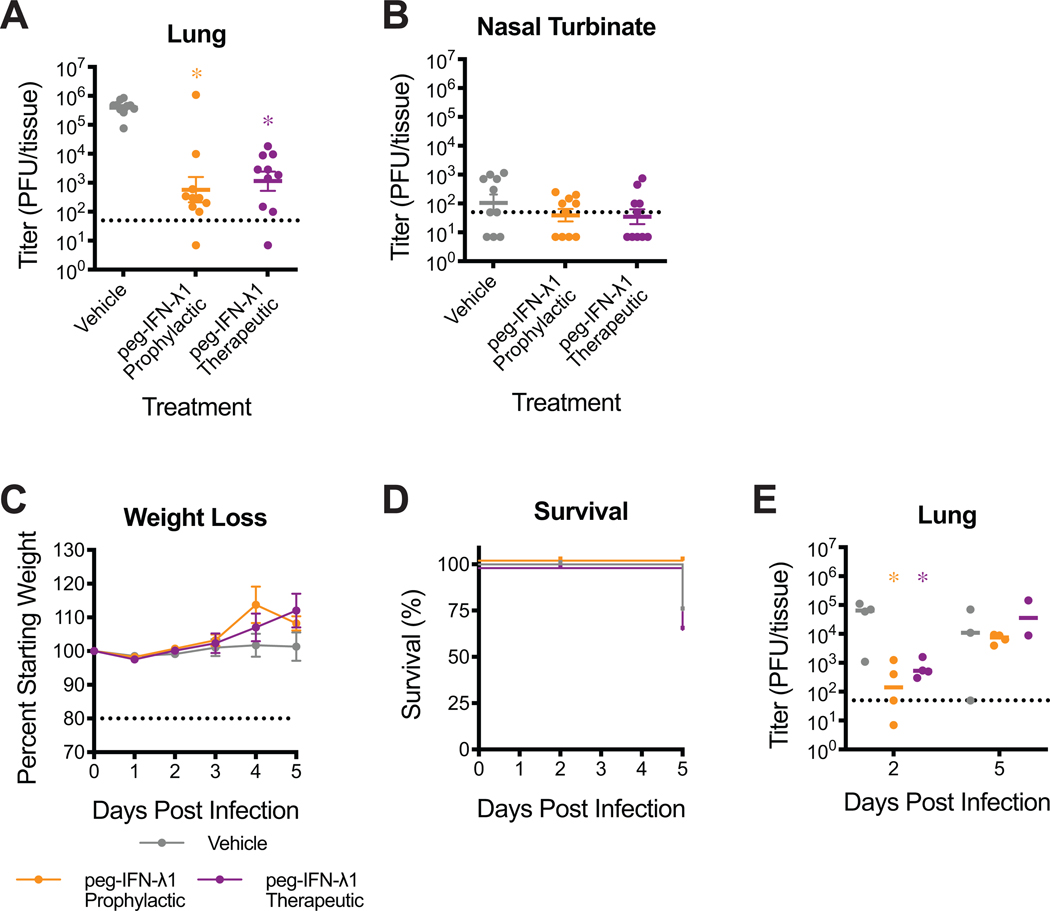
(A–C) Groups of 10-week-old female BALB/c mice were vaccinated with VRPs expressing wildtype spike (S, n=10), nucleocapsid (N, n=8), or GFP (n=4). (A) 50% inhibitory concentration (IC50) of sera from 3 weeks post boost to neutralize SARS-CoV-2 WT. Dotted line represents limit of detection (LOD). Log transformed data analyzed by 1-way ANOVA followed by Dunnett’s multiple comparisons. ‘*’ denotes p=0.0003. (B–C) Lung (B) and nasal turbinate (C) viral titer on 2dpi. Dotted line represents LOD. Undetected samples are plotted at half the LOD. Log transformed data analyzed as in (A). (B) ‘*’ denotes p=<0.0001. (C) ‘*’ denotes p=0.0360. (D) Human primary airway epithelial cells were pretreated for 24hrs with peg-IFN-λ1 followed by infection with SARS-CoV-2 WT. Infectious virus in apical washes from 48 hours post infection was titered. Remdesivir (RDV) was used as positive control. Data representative of two independent experiments in unique human donors. (E–I) 1-year-old female BALB/c mice were subcutaneously treated with vehicle or with 2μg peg-IFN-λ1 prophylactically or therapeutically and infected with SARS-CoV-2 MA. n=5 per group per timepoint. (E) Percent starting weight. Dotted line represents weight loss criteria for humane euthanasia. Data analyzed by mixed effects analysis followed by Dunnett’s multiple comparisons. (B) ‘*’ denotes p=<0.0001 (prophylactic, 2dpi), 0.0128 (therapeutic, 2dpi), 0.0042 (prophylactic, 4dpi), 0.0037 (therapeutic, 4dpi). (F–G) Viral titers in the lung (F) and nasal turbinates (G) at 2 and 4dpi. Dotted line represents LOD. Log transformed data analyzed by 2-way ANOVA followed by Dunnett’s multiple comparisons. (H–I) Whole body plethysmography assessing pulmonary function for PenH (H) and Rpef (I). Data analyzed as in (E). All error bars represent standard error about the mean. (H) ‘*’ denotes p=0.0083 (prophylactic, 2dpi), 0.0080 (therapeutic, 2dpi), 0.0029 (prophylactic, 3dpi), 0.0020 (therapeutic, 3dpi), 0.0327 (therapeutic, 4dpi). (I) ‘*’ denotes p=0.0442 (prophylactic, 1dpi), 0.0033 (therapeutic, 1dpi), 0.0048 (prophylactic, 2dpi), 0.0118 (therapeutic, 3dpi), 0.0259 (prophylactic, 4dpi), 0.0247 (therapeutic, 4dpi).
Interferon lambda is a type III interferon whose receptors are largely limited to epithelial cells, including the lungs30,31. Treatment with interferons has been employed as pan viral treatment for several viral infections, including trials for the treatment of SARS-CoV-1 and MERS-CoV infections. Pegylated human interferon lambda-1 (peg-IFN-λ1) is Phase 3-ready for hepatitis delta virus infection and has been proposed to treat COVID-19 patients32. We first sought to determine if peg-IFN-λ1 would initiate an antiviral response capable of inhibiting productive infection of primary human airway epithelial (HAE) cell cultures by SARS-CoV-2. Pretreatment of HAE with peg-IFN-λ1 provided a potent dose dependent reduction in SARS-CoV-2 infectious virus production (Fig 4D).
To determine if the in vitro antiviral effect of peg-IFN-λ1 translates to in vivo efficacy, we performed prophylactic and therapeutic efficacy studies in 1-year old BALB/c mice. We subcutaneously administered 2μg peg-IFN-λ1 18hr prior or 12hr after infection with 105 PFU SARS-CoV-2 MA. Both prophylactic and therapeutic administration of peg-IFN-λ1 significantly prevented weight loss at 2 and 3dpi (Figure 4E) and diminished SARS-CoV-2 MA replication in the lung measured at 2dpi (Figure 4F). Peg-IFN-λ1 had minimal effect on nasal turbinate titer, likely limited by low limits of detection (Figure 4G). Importantly, both prophylactic and therapeutic peg-IFN-λ1 protected mice from pulmonary dysfuction as measured by PenH and Rpef (Figure 4H–I). Peg-IFN-λ1 also reduced SARS-CoV-2 MA titer in the lung in treated young mice at 2dpi (Extended Data 3A–B). When tested in HFH4-hACE2 mice, peg-IFN-λ1 reduced lung viral titer at 2dpi, but had no effect at 5dpi (Extended Data 3C–E), possibly due to decreased potentancy by 5dpi. Altogether, these data demonstrate the utility of these models to rapidly evaluate vaccine and therapeutic drug efficacy in standard laboratory mice. In addition, we show that peg-IFN-λ1 exerts potent antiviral activity against SARS-CoV-2 in vitro and can diminish virus replication in vivo even when given therapeutically.
Discussion
Structural studies have solved the SARS-CoV and SARS-CoV-2 RBD interaction residues that bind hACE233,34. Coupling molecular modeling and reverse genetics22, we altered the SARS-CoV-2 receptor binding domain (RBD) allowing viral entry via mACE2 and highlight the precision of structure-based predictions in virus entry. Unlike parental wildtype (WT) virus, the resultant recombinant virus, SARS-CoV-2 MA, used mACE2 for infection in vitro. Importantly, recombinant SARS-CoV-2 MA did not grow more efficiently than WT virus in primary human bronchiolar epithelial cells or Vero cells, suggesting that this virus is not more fit in human cells. SARS-CoV-2 MA replicated in both the upper and lower airways of BALB/c mice, with more severe disease in aged mice, reproducing the age-related increase in disease observed in humans. SARS-CoV-2 MA infection is cleared by 4dpi in young, but not old adult mice, likely due to control by interferons and the innate immune system12,35 and is exemplified by sensitivity to peg-IFN-λ1.
SARS-CoV-2 is a pandemic pathogen of concern and experiments to intentionally alter host range was reviewed for risk potential for Potential Pandemic Pathogen Care and Oversight (P3C0) concerns prior to start of research. By current US review standards, SARS-CoV-2 MA was not considered dual use research of concern, which is limited to a subset of pathogens. Prior to the start of studies, appropriate discussion and documentation of proposed experiments and protocols were reviewed and approvals obtained by institutional and external review. We recommend that SARS-CoV-2 MA and its derivitives be maintained in a biosafety level 3 laboratory.
Importantly, we show the utility of the SARS-CoV-2 MA model for screening MCMs through vaccine challenge studies and the evaluation of a novel clinical candidate, pegylated interferon lambda-1. Peg-IFN-λ1 is a Phase 3-ready drug in clinical development for hepatitis delta virus infection. It has been given to over 3000 patients in the context of 19 clinical trials as a weekly subcutaneous injection, often for 24–48 weeks to treat patients with chronic viral hepatitis36,37. For SARS-CoV-2, peg-IFN-λ1 is a promising therapeutic for the treatment of COVID-19 infections6, blocks porcine coronavirus replication in gut cells in vitro38, and SARS-CoV-1 replication in human airway cells39. Our data, which demonstrates reduction in SARS-CoV-2 infection in primary human cells and in mice, support further multiple investigator sponsored studies currently underway to evaluate peg-IFN-λ1 for prevention and treatment of SARS-CoV-2 infection (Clinicaltrials.gov identifiers: NCT04331899, NCT04343976, NCT04344600, NCT04354259).
Although the SARS-CoV-2 MA RBD mutations may attenuate the function of select human monoclonal antibodies or vaccines in mice, this phenotype was not observed with mouse adapted strains of SARS-CoV-140–42 or alter the potent neutralizing activity of two human monoclonal antibodies that target the SARS-CoV-2 RBD43. In any event, we demonstrate that our previously described HFH4-hACE2 transgenic mouse model supports efficient SARS-CoV-2 replication and pathogenesis in vivo, as well21. Consequently, transgenic models offer an alternative replication and disease model that uses wildtype SARS-CoV-2, appropriate for evaluating therapeutic antibodies and other countermeasures that target RBD epitopes altered in SARS-CoV-2 MA.
Similar to SARS-CoV-1 infection, SARS-CoV-2 infection of HFH4-hACE2 transgenic mice results in a mild bronchiolitis in young mice and about 40% mortality, the latter phenotype associated with viral invasion of the CNS9,21,44. Recently, Bao, et al used a murine ACE2 promoter to drive overexpression of hACE2 and infected 6- to 11-month-old mice showed mild weight loss and pulmonary inflammation. The discrepancy in weight loss between the HFH4-hACE2 model presented here and the Bao et al. study may be due to differences in age of mice utilized or differences in hACE2 expression distribution. Virally delivered hACE2 overexpression models have also been reported8,12. While these systems allow for rapid studies in commercially available mice, including knockout mice, the cellular distribution of hACE2 may not faithfully recapitulate endogenously expressed ACE2 and proper SARS-CoV-2 tropism.
We note that a new SARS-CoV-2 young and aged BALB/c mouse model that captures multiple aspects of SARS-CoV-2 pathogenesis, provides a urgently needed high-throughput in vivo system to evaluate MCMs during this devastating pandemic. Since this model uses standard immune competent laboratory mice, the accessibility, ease of use, availability of reagents, cost, and utility far exceeds that of other hACE2 transgenic mice, ferret, hamster, and non-human primate models. While this model provides a critically necessary tool in COVID-19 countermeasure research, it also provides an important first step in SARS-CoV-2 serial adaptation in mice25, potentially selecting for variants that develop more severe pathogenic manifestations of acute respiratory distress syndrome (ARDS), coagulopathies, and other human disease outcomes. In addition, the SARS-CoV-2 MA model can be used to evaluate the role of host genetics and antiviral defense genes in viral pathogenesis using transgenic and knockout mice. Together, these data describe critical new animal models, each appropriate for testing of various medical countermeasures.
Material & Methods
Ethics and biosafety
The generation of recombinant SARS-CoV-2 MA was approved for use under BSL3 conditions by the University of North Carolina at Chapel Hill Institutional Review Board (UNC-CH IBC) and by a Potential Pandemic Pathogen Care and Oversight committee at the National Institute of Allergy and Infectious Diseases (NIAID). All animal work was approved by Institutional Animal Care and Use Committee at University of North Carolina at Chapel Hill according to guidelines outlined by the Association for the Assessment and Accreditation of Laboratory Animal Care and the U.S. Department of Agriculture. All work was performed with approved standard operating procedures and safety conditions for SARS-CoV-2. Our institutional BSL3 facilities have been designed to conform to the safety requirements recommended by Biosafety in Microbiological and Biomedical Laboratories (BMBL), the U.S. Department of Health and Human Services, the Public Health Service, the Centers for Disease Control and Prevention (CDC), and the National Institutes of Health (NIH). Laboratory safety plans have been submitted, and the facility has been approved for use by the UNC Department of Environmental Health and Safety (EHS) and the CDC.
SARS-CoV-2 receptor binding domain and ACE2 analysis and modelling
Group 2B coronavirus spike and ACE2 amino acid sequences were aligned using Geneious Prime (Version 2020.0.5). Accession numbers used: SARS-CoV-1 Urbani (AY278741), WIV1 (KF367457), SHC014 (KC881005), SARS-CoV-2 (MN985325.1), hACE2 (BAB40370), mACE2 (NP_081562). Protein similarity scores were calculated using BLOSUM62 matrix. Contact residues previously identified by crystal structures33,34,45. Structure modelling was performed using Modeller (Version 9.20) and visualized using PyMOL (Version 1.8.6.0).
Viruses, cells, and transfections
All viruses used were derived from an infectious clone of SARS-CoV-2, which was designed using similar strategies for SARS-CoV and MERS-CoV22,46,47. The Q498Y/P499T substitutions were generated by site directed mutagenesis using the following primers: Forward: 5’-ATA TGG TTT CTA CAC GAC TAA TGG TGT TGG TTA CCA ACC-3’, Reverse: 5’-TAG TCG TGT AGA AAC CAT ATG ATT GTA AAG GAA AGT AAC AAT TAA AAC CTT C-3’. Viruses were derived following systematic cDNA assembly of the infections clone, followed by in vitro transcription and electroporation into Vero E6 cells. Virus stocks were passaged once on Vero E6 cells and titered via plaque assay. Briefly, virus was serial diluted and inoculated onto confluent monolayers of Vero E6 cells, followed by agarose overlay. Plaques were visualized on day 2 post infection via staining with neutral red dye.
Vero E6 cells were obtained from USAMRIID in 2003 (ATCC CRL-1586) and were maintained in Dulbecco’s modified Eagle’s medium (DMEM; Gibco),5% Fetal Clone II serum (FCII, Hyclone), and 1X antibiotic/antimycotic (Gibco). DBT-9 were previously clonally derived in our lab maintained in DMEM, 10% FCII, and 1X antibiotic/antimycotic. Cells were confirmed to be netgative for mycoplasma contamination.
For Vero E6 single step growth curve, cells were infected at a multiplicity of infection (MOI) of 0.5 for 1 hour. Inoculum was removed and monolayer was washed twice with PBS and replaced with media. For HAE growth curve, cells were infected at a multiplicity of infection (MOI) of 0.5 for 2 hours. Inoculum was removed and cells were washed three times with PBS. At designated timepoints, Vero E6 supernatant was removed without replacement or HAEs were washed apically with 200μL 1X PBS for 10 minutes and stored at −80°C until titered by plaque assay as described above.
For ACE2 receptor usage, non-permissive DBT-9 cells were transfected with pcDNA3.1 empty-vector, pcDNA3.1-hACE2, or pcDNA3.1-mACE2 using lipofectamine 2000 (Invitrogen). At 24 hours post transfection, cells were infected at an MOI of 0.5 for 1 hour, removed and washed twice with PBS. At 24hrs post infection, media was removed, and total cellular RNA was collected via TRIzol (Invitrogen) and extracted using Direct-Zol RNA MiniPrep kit (Zymo Research). Viral RNA was quantified via qRT-PCR using TaqMan Fast Virus 1-Step Master Mix (Thermo Fisher Scientific) on a QuantStudio 3 (Applied Biosystems). SARS-CoV-2 RNA was quantified using US Centers of Disease Control and Prevention diagnostic N1 assay: Forward: 5’-GAC CCC AAA ATC AGC GAA AT-3’, probe: 5’-FAM-AC CCC GCA TTA CGT TTG GTG GAC C-BHQ1–3’, reveres: 5’-TCT GGT TAC TGC CAG TTG AAT CTG-3’. Host 18S rRNA was used as housekeeping control (Invitrogen, product number 4319413E). Viral RNA was analyzed using ΔΔCt and fold change over viral RNA in empty-vector transfected cells.
In vivo Infections
HFH4-hACE2 overexpressing mice were bred and maintained at University of North Carolina at Chapel Hill. BALB/c mice were obtained from Envigo (strain 047). Mice were infected with 105 PFU intranasally under ketamine/xylazine anesthesia. Body weight was monitored daily and WBP was performed as indicated. Briefly, mice were allowed to equilibrate in WBP chambers (DSI Buxco respiratory solutions, DSI Inc.) for 30 minutes prior to a 5 minute data acquisition period using FinePointe software. At indicated timepoints, a subset of mice were euthanized by isoflurane overdose and tissue samples were harvested for titer and histopathology analysis. A subset of mice for nasal turbinate histopathology were perfused with 10% phosphate buffered formalin prior to tissue collection. The right caudal lung lobe was taken for titer and stored at −80°C until homogenized in 1mL PBS and titered by plaque assay as described above. The left lung lobe was taken for histopathology and were fixed in 10% phosphate buffered formalin for 7 days before paraffin embedding and sectioning.
Histopathology and antigen staining
Lungs were fixed for 7 days in 10% phosphate buffered formalin, paraffin embedded, and sectioned at 4μm. Serial sections were stained with hematoxylin and eosin, and stained for immunohistochemistry for SARS-CoV-2 nucleocapsid using a monoclonal anti-SARS-CoV-1 nucleocapsid antibody (1:250, NB100–56576, Novus Biologicals) on deparaffinized sections on the Ventana Discovery Ultra platform (Roche). Photomicrographs were captured on an Olympus BX43 light microscope at 200X magnification with a DP27 camera using cellSens Entry software.
Vaccination studies
Non-select BSL2 Venezuelan equine encephalitis virus strain 3526 based replicon particles (VRPs) were generated to express GFP, SARS-CoV-2 spike (S), or nucleocapsid (N) as described previously5,48. Mice were vaccinated via hind footpad infection with 103 VRP in 10 μL, boosted identically at 3 weeks post prime, and bled via submandibular bleed at 3 weeks to confirm presence of neutralizing antibodies. Neutralizing antibody levels were assessed via neutralization assay using SARS-CoV-2 WT expressing nanoluciferase (nLUC) in place of ORF7a. Briefly, the ORF7a gene of SARS-CoV-2 was removed from the molecular clone and nLUC inserted downstream of the ORF7a transcription regulatory sequence. Recombinant viruses encoding nLUC (SARS-CoV-2 nLUC) were recovered, titered and serial dilutions of sera were incubated with virus for 1 hour at 37°C, then added to monolayers of Vero E6 cells. 48hrs post infection, viral infection was quantified using nLUC activity via Nano-Glo Luciferase Assay System (Promega). 50% inhibitory concentration (IC50) values were calculated from full dilution curves.
Mice were challenged 4 weeks post boost with 105 PFU intranasally under ketamine/xylazine anesthesia. Body weight was monitored daily. On day 2 post infection, mice were euthanized by isoflurane overdose and tissue samples were harvested for titer analysis as described above.
Pegylated-IFN-λ1 treatment in vitro and in vivo
Peginterferon Lambda-1a was obtained from Eiger BioPharmaceuticals by MTA in GMP prefilled syringes, 0.18 mg/syringe (0.4 mg/mL). Primary HAE cell cultures were obtained from the Tissue Procurement and Cell Culture Core Laboratory in the Marsico Lung Institute/Cystic Fibrosis Research Center at UNC. Human tracheobronchial epithelial cells provided by Dr. Scott Randell were obtained from airway specimens resected from patients undergoing surgery under University of North Carolina Institutional Review Board-approved protocols (#03–1396) by the Cystic Fibrosis Center Tissue Culture Core. Primary cells were expanded to generate passage 1 cells and passage 2 cells were plated at a density of 250,000 cells per well on Transwell-COL (12mm diameter) supports (Corning). Human airway epithelium cultures (HAE) were generated by differentiation at an air-liquid interface for 6 to 8 weeks to form well-differentiated, polarized cultures that resembled in vivo pseudostratified mucociliary epithelium49. HAEs were treated with a range of peg-IFN-λ1 doses basolaterally for 24hrs prior to infection. 1μM remdesivir was obtained from Gilead Sciences by MTA and was used as a positive control. Cultures were infected at an MOI of 0.5 for 2 hours. Inoculum was removed and culture was washed three times with PBS. At 48hrs post infection, apical washes were taken to measure viral replication via plaque assays as described above. This study was repeated in two separate human donors.
1-year or 10-week-old BALB/c, or 4- to 7-week old HFH4-hACE2 mice were subcutaneously treated with a single 2μg dose of peg-IFN-λ1 prophylactically at 18hrs prior to infection, therapeutically at 12hrs post infection50, or PBS vehicle treated, and infected with 105 PFU of SARS-CoV-2 MA intranasally under ketamine/xylazine anesthesia. 4 to 7-week-old HFH4-hACE2 mice treated as above and infected with 105 PFU of SARS-CoV-2 MA. Body weight was monitored daily. WBP was performed as indicated. On days indicated, mice were euthanized by isoflurane overdose and tissue samples were harvested for titer analysis as described above.
Data analysis and presentation
All data visualize and analyzed in Prism (version 8.4.2). Non-parametric tests were performed as described in figure legends. Figures arranged in Adobe Illustrator (version 24.1).
Data and Material availability
All relevant data is included in this article. Viral genome sequences uploaded to GenBank (SARS-CoV-2 MA: MT844088. SARS-CoV-2 nLuc: MT844089). Reagents and resources available upon request to corresponding author (rbaric@email.unc.edu) and with material transfer agreement.
Extended Data
Extended Data 1: SARS-CoV-2 infection in HFH4-hACE2 transgenic mice.
Male and female HFH4-hACE2 mice were infected with 105 PFU SARS-CoV-2 WT. (A) Percent starting weight. Dotted line represents weight loss criteria for humane euthanasia. n=2 mock and 5 SARS-CoV-2. (B) Survival. (C–D) Lung (C) and brain (D) viral titer. Dotted line represents limit of detection (LOD). Undetected samples are plotted at half the LOD. ‘x’ symbol indicates mice that succumbed to infection. 2dpi: n=2; 5dpi: n=5. (E–F) Whole body plethysmography assessing pulmonary function for PenH (E) and Rpef (F).
Extended Data 2: Group 2B coronavirus spike receptor binding domain alignment.
Amino acid positions are numbered above in reference to SARS-CoV-1, and below in reference to SARS-CoV-2. Green highlighted residues are hACE2 contacts as determined by published crystal structures.
Extended Data 3: Cytokine analysis in SARS-CoV-2 MA infected 1-year-old BALB/c mice.
Cytokine and chemokine levels in serum and lung homogenates of 1-year-old female BALB/c mice from Figures 3 at 2 and 4dpi. For each analyte, data analyzed by 2-way ANOVA followed by Sidak’s multiple comparisons. ‘*’ denotes p=0.0155 (TNF-α), 0.0189 (IL-6), <0.0001 (MCP-1), 0.0115 (IL-5), 0.0127 (IL-1α), 0.0004 (G-CSF), 0.0070 (IL-10), 0.0243 (KC), 0.0152 (IL-17A), 0.0408 (GM-CSF), 0.0261 (MIP-1β), 0.0025 (IL-12p40), 0.0015 (MIP-1α), 0.0019 (IL-12p70).
Extended Data 4: Evaluation of peg-IFN-λ1 against SARS-CoV-2 MA infection in young BALB/c and HFH4-hACE2 mice.
(A–B) 12-week-old female BALB/c mice were subcutaneously treated with vehicle or with 2μg peg-IFN-λ1 prophylactically or therapeutically and infected with SARS-CoV-2 MA. Viral titers in the lung (A) and nasal turbinates (B) at 2dpi. n=10 for each group, combined from two independent experiments. Dotted line represents limit of detection (LOD). Undetected samples plotted at half the LOD. Log transformed data analyzed by 1-way ANOVA followed by Holm-Sidak’s multiple comparisons. (A) ‘*’ denotes p=<0.0001 (prophylactic), <0.0001 (therapeutic). (C–E) 4- to 7-week old HFH4-hACE2 male and female mice were treated with peg-IFN-λ1 as done in (A–B) and infected with 105 PFU SARS-CoV-2 WT. n=8 vehicle; n=10 prohpylactic, n=7 therapeutic. (C) Percent starting weight. Dotted line represents weight loss criteria for humane euthanasia. Data analyzed by mixed effects analysis followed by Sidak’s multiple comparisons. (D) Survival. (E) Lung viral titer at 2 and 5dpi. 2dpi: n=4 vehicle, n=4 prophylactic, n=4 therapeutic: 5dpi: n=3 vehicle, n=4 prophylactic, n=2. Data analyzed by 2-way ANOVA followed by Dunnett’s multiple comparisons. ‘*’ denotes p=0.0037 (prophylactic, 2dpi), 0.0365 (therapeutic, 2dpi). All error bars represent standard error about the mean.
Acknowledgements
This project was funded in part by the National Institute of Allergy and Infectious Diseases, National Institutes of Health, Department of Health and Human Service award: 1U19 AI142759 (Antiviral Drug Discovery and Development Center awarded to R.S.B); 5R01AI132178 (partnership grant awarded to T.P.S. and R.S.B) and an animal models contract from the NIH (HHSN272201700036I). This project was supported in part by the North Carolina Policy Collaboratory at the University of North Carolina at Chapel Hill with funding from the North Carolina Coronavirus Relief Fund established and appropriated by the North Carolina General Assembly. K.H.D. is funded by an NIH NIAID T32 AI007419. D.R.M. is funded by an NIH NIAID T32 AI007151 and a Burroughs Wellcome Fund Postdoctoral Enrichment Program Award. The Marsico Lung Institute Tissue Procurement and Cell Culture Core is supported by NIH grant DK065988 and Cystic Fibrosis Foundation grant BOUCHE15RO. Animal histopathology service was performed by Dawud Hilliard and Ling Wang in the Animal Histopathology & Laboratory Medicine Core at the University of North Carolina, which is supported in part by an NCI Center Core Support Grant (5P30CA016086–41) to the UNC Lineberger Comprehensive Cancer Center.
Footnotes
Competing Interests
M.D.B and I.C.C. are employees, and J.S.G is the founder and a board member, of Eiger BioPharmaceuticals, Inc., which produces peg-IFN-λ1.
References
- 1.Li Q et al. Early Transmission Dynamics in Wuhan, China, of Novel Coronavirus-Infected Pneumonia. N Engl J Med 382, 1199–1207, doi: 10.1056/NEJMoa2001316 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Corbett KS et al. SARS-CoV-2 mRNA Vaccine Development Enabled by Prototype Pathogen Preparedness. bioRxiv, doi: 10.1101/2020.06.11.145920 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hassan AO et al. A SARS-CoV-2 Infection Model in Mice Demonstrates Protection by Neutralizing Antibodies. Cell 182, 744–753 e744, doi: 10.1016/j.cell.2020.06.011 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhou P et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature 579, 270–273, doi: 10.1038/s41586-020-2012-7 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hou YJ et al. SARS-CoV-2 Reverse Genetics Reveals a Variable Infection Gradient in the Respiratory Tract. Cell 182, 429–446 e414, doi: 10.1016/j.cell.2020.05.042 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Andreakos E & Tsiodras S. COVID-19: lambda interferon against viral load and hyperinflammation. EMBO Mol Med 12, e12465, doi: 10.15252/emmm.202012465 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Humeniuk R et al. Safety, Tolerability, and Pharmacokinetics of Remdesivir, an Antiviral for Treatment of COVID-19, in Healthy Subjects. Clin Transl Sci, doi: 10.1111/cts.12840 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hassan AO et al. A SARS-CoV-2 Infection Model in Mice Demonstrates Protection by Neutralizing Antibodies. Cell, doi: 10.1016/j.cell.2020.06.011 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jiang RD et al. Pathogenesis of SARS-CoV-2 in Transgenic Mice Expressing Human Angiotensin-Converting Enzyme 2. Cell 182, 50–58 e58, doi: 10.1016/j.cell.2020.05.027 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bao L et al. The pathogenicity of SARS-CoV-2 in hACE2 transgenic mice. Nature, doi: 10.1038/s41586-020-2312-y (2020). [DOI] [PubMed] [Google Scholar]
- 11.Sun SH et al. A Mouse Model of SARS-CoV-2 Infection and Pathogenesis. Cell Host Microbe 28, 124–133 e124, doi: 10.1016/j.chom.2020.05.020 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sun J et al. Generation of a Broadly Useful Model for COVID-19 Pathogenesis, Vaccination, and Treatment. Cell, doi: 10.1016/j.cell.2020.06.010 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Israelow B et al. Mouse model of SARS-CoV-2 reveals inflammatory role of type I interferon signaling. J Exp Med 217, doi: 10.1084/jem.20201241 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kim YI et al. Infection and Rapid Transmission of SARS-CoV-2 in Ferrets. Cell Host Microbe 27, 704–709 e702, doi: 10.1016/j.chom.2020.03.023 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Imai M et al. Syrian hamsters as a small animal model for SARS-CoV-2 infection and countermeasure development. Proc Natl Acad Sci U S A, doi: 10.1073/pnas.2009799117 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sia SF et al. Pathogenesis and transmission of SARS-CoV-2 in golden hamsters. Nature, doi: 10.1038/s41586-020-2342-5 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rockx B et al. Comparative pathogenesis of COVID-19, MERS, and SARS in a nonhuman primate model. Science 368, 1012–1015, doi: 10.1126/science.abb7314 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yu P et al. Age-related rhesus macaque models of COVID-19. Animal Model Exp Med 3, 93–97, doi: 10.1002/ame2.12108 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Munster VJ et al. Respiratory disease in rhesus macaques inoculated with SARS-CoV-2. Nature, doi: 10.1038/s41586-020-2324-7 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ostrowski LE, Hutchins JR, Zakel K & O’Neal WK Targeting expression of a transgene to the airway surface epithelium using a ciliated cell-specific promoter. Mol Ther 8, 637–645, doi: 10.1016/s1525-0016(03)00221-1 (2003). [DOI] [PubMed] [Google Scholar]
- 21.Menachery VD et al. SARS-like WIV1-CoV poised for human emergence. Proc Natl Acad Sci U S A 113, 3048–3053, doi: 10.1073/pnas.1517719113 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hou YJ et al. SARS-CoV-2 Reverse Genetics Reveals a Variable Infection Gradient in the Respiratory Tract. Cell, doi: 10.1016/j.cell.2020.05.042 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Abdi K et al. Uncovering inherent cellular plasticity of multiciliated ependyma leading to ventricular wall transformation and hydrocephalus. Nat Commun 9, 1655, doi: 10.1038/s41467-018-03812-w (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Menachery VD, Gralinski LE, Baric RS & Ferris MT New Metrics for Evaluating Viral Respiratory Pathogenesis. PLoS One 10, e0131451, doi: 10.1371/journal.pone.0131451 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Roberts A et al. A mouse-adapted SARS-coronavirus causes disease and mortality in BALB/c mice. PLoS Pathog 3, e5, doi: 10.1371/journal.ppat.0030005 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Menachery VD et al. A SARS-like cluster of circulating bat coronaviruses shows potential for human emergence. Nat Med 21, 1508–1513, doi: 10.1038/nm.3985 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang D et al. Clinical Characteristics of 138 Hospitalized Patients With 2019 Novel Coronavirus-Infected Pneumonia in Wuhan, China. JAMA, doi: 10.1001/jama.2020.1585 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.de Wit E, van Doremalen N, Falzarano D & Munster VJ SARS and MERS: recent insights into emerging coronaviruses. Nat Rev Microbiol 14, 523–534, doi: 10.1038/nrmicro.2016.81 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Roberts A et al. Aged BALB/c mice as a model for increased severity of severe acute respiratory syndrome in elderly humans. J Virol 79, 5833–5838, doi: 10.1128/JVI.79.9.5833-5838.2005 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kotenko SV et al. IFN-lambdas mediate antiviral protection through a distinct class II cytokine receptor complex. Nat Immunol 4, 69–77, doi: 10.1038/ni875 (2003). [DOI] [PubMed] [Google Scholar]
- 31.Sheppard P et al. IL-28, IL-29 and their class II cytokine receptor IL-28R. Nat Immunol 4, 63–68, doi: 10.1038/ni873 (2003). [DOI] [PubMed] [Google Scholar]
- 32.Elazar M & Glenn JS Emerging concepts for the treatment of hepatitis delta. Curr Opin Virol 24, 55–59, doi: 10.1016/j.coviro.2017.04.004 (2017). [DOI] [PubMed] [Google Scholar]
- 33.Lan J et al. Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature, doi: 10.1038/s41586-020-2180-5 (2020). [DOI] [PubMed] [Google Scholar]
- 34.Shang J et al. Structural basis of receptor recognition by SARS-CoV-2. Nature, doi: 10.1038/s41586-020-2179-y (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Felgenhauer U et al. Inhibition of SARS-CoV-2 by type I and type III interferons. J Biol Chem, doi: 10.1074/jbc.AC120.013788 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Muir AJ et al. A randomized phase 2b study of peginterferon lambda-1a for the treatment of chronic HCV infection. J Hepatol 61, 1238–1246, doi: 10.1016/j.jhep.2014.07.022 (2014). [DOI] [PubMed] [Google Scholar]
- 37.Chan HLY et al. Peginterferon lambda for the treatment of HBeAg-positive chronic hepatitis B: A randomized phase 2b study (LIRA-B). J Hepatol 64, 1011–1019, doi: 10.1016/j.jhep.2015.12.018 (2016). [DOI] [PubMed] [Google Scholar]
- 38.Li L et al. IFN-lambda preferably inhibits PEDV infection of porcine intestinal epithelial cells compared with IFN-alpha. Antiviral Res 140, 76–82, doi: 10.1016/j.antiviral.2017.01.012 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mordstein M et al. Lambda interferon renders epithelial cells of the respiratory and gastrointestinal tracts resistant to viral infections. J Virol 84, 5670–5677, doi: 10.1128/JVI.00272-10 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rockx B et al. Escape from human monoclonal antibody neutralization affects in vitro and in vivo fitness of severe acute respiratory syndrome coronavirus. J Infect Dis 201, 946–955, doi: 10.1086/651022 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sui J et al. Effects of human anti-spike protein receptor binding domain antibodies on severe acute respiratory syndrome coronavirus neutralization escape and fitness. J Virol 88, 13769–13780, doi: 10.1128/JVI.02232-14 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rockx B et al. Structural basis for potent cross-neutralizing human monoclonal antibody protection against lethal human and zoonotic severe acute respiratory syndrome coronavirus challenge. J Virol 82, 3220–3235, doi: 10.1128/JVI.02377-07 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zost SJ et al. Potently neutralizing human antibodies that block SARS-CoV-2 receptor binding and protect animals. bioRxiv, doi: 10.1101/2020.05.22.111005 (2020). [DOI] [Google Scholar]
- 44.McCray PB Jr. et al. Lethal infection of K18-hACE2 mice infected with severe acute respiratory syndrome coronavirus. J Virol 81, 813–821, doi: 10.1128/JVI.02012-06 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Li F, Li W, Farzan M & Harrison SC Structure of SARS coronavirus spike receptor-binding domain complexed with receptor. Science 309, 1864–1868, doi: 10.1126/science.1116480 (2005). [DOI] [PubMed] [Google Scholar]
- 46.Yount B et al. Reverse genetics with a full-length infectious cDNA of severe acute respiratory syndrome coronavirus. Proc Natl Acad Sci U S A 100, 12995–13000, doi: 10.1073/pnas.1735582100 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Scobey T et al. Reverse genetics with a full-length infectious cDNA of the Middle East respiratory syndrome coronavirus. Proc Natl Acad Sci U S A 110, 16157–16162, doi: 10.1073/pnas.1311542110 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Agnihothram S et al. Development of a Broadly Accessible Venezuelan Equine Encephalitis Virus Replicon Particle Vaccine Platform. J Virol 92, doi: 10.1128/JVI.00027-18 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Fulcher ML, Gabriel S, Burns KA, Yankaskas JR & Randell SH Well-differentiated human airway epithelial cell cultures. Methods Mol Med 107, 183–206, doi: 10.1385/1-59259-861-7:183 (2005). [DOI] [PubMed] [Google Scholar]
- 50.Davidson S et al. IFNlambda is a potent anti-influenza therapeutic without the inflammatory side effects of IFNalpha treatment. EMBO Mol Med 8, 1099–1112, doi: 10.15252/emmm.201606413 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]