Abstract
The transfer of electrons through protein complexes is central to cellular respiration. Exploiting proteins for charge transfer in a controllable fashion has the potential to revolutionize the integration of biological systems and electronic devices. Here we characterize the structure of an ultrastable protein filament and engineer the filament subunits to create electronically conductive nanowires under aqueous conditions. Cryoelectron microscopy was used to resolve the helical structure of gamma-prefoldin, a filamentous protein from a hyperthermophilic archaeon. Conjugation of tetraheme c3-type cytochromes along the longitudinal axis of the filament created nanowires capable of long-range electron transfer. Electrochemical transport measurements indicated networks of the nanowires capable of conducting current between electrodes at the redox potential of the cytochromes. Functionalization of these highly-engineerable nanowires with other molecules, such as redox enzymes, may be useful for bioelectronic applications.
Keywords: conductive biomaterials, electron transport, nanowires, protein engineering, protein self-assembly
Graphical Abstract
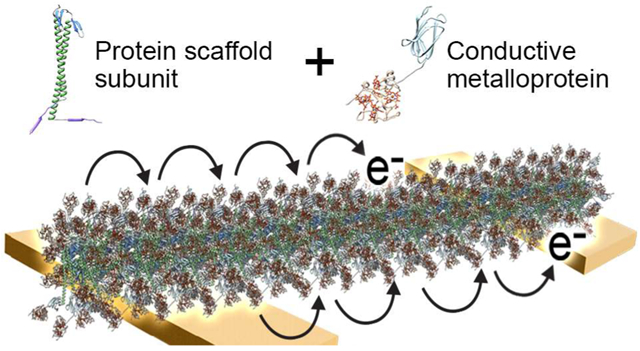
Electron transfer (ET) through protein complexes is a central process of life for the storage and use of energy in many biological systems. For example, photosynthesis requires long-range ET to convert solar energy into chemical energy. Natural bacterial biofilms have been discovered that are able to transfer electrons efficiently to extracellular acceptors, such as iron oxides.1 These biofilms are made of a network of nanowires that function as conduits for ET-related respiration and metabolism.2,3 Bacterial nanowire ET is generally described by theories of electron hopping or tunnelling along the nanowire between redox species such as cytochrome c (cytc) proteins or π-π stacked aromatic amino acids.4 While models have existed for such stacked amino acids in bacterial type 4 pili, a recent study has shown that the intensively studied Geobacter sulfurreducens nanowires are actually a polymer of hexaheme cytc proteins, and not type 4 pilin.5 Sequential electron hopping or tunnelling processes give rise to long-distance ET, which for bacterial nanowires can be several micrometers. Although naturally occurring bacterial nanowires may be useful conductive materials, these complex assemblages are difficult to engineer and repurpose, and the mechanism of their ET is not well-understood.
Exploiting the ability of proteins to self-assemble into material templates may enable replication of the ET observed in bacterial nanowires in a well-controlled and tuneable platform for bioelectronics.6,7 Advances in protein design have enabled the creation of supramolecular architectures of controllable size and symmetry,8 which can potentially be used to align cytc proteins for long-range ET to other functional materials. A recently described approach for building protein templates used engineered connector proteins to control the assembly of ultrastable filaments into geometrical structures that served as scaffolds for nanomaterials.9 The filamentous protein, called gamma-prefoldin (γPFD), is produced by Methanocaldococcus jannaschii, a hyperthermophilic archaeon.10,11 Prefoldins are molecular chaperones found in archaea and eukaryotes that generally have a jellyfish-like hexameric structure. Unusually, γPFD assembles into filaments of up to 3 μm in length composed of monomeric protein subunits, with homology modelling suggesting the subunits assemble through β-sheets with the remainder of the protein protruding as a coiled-coil domain.12 Filaments of γPFD have several advantages as scaffolds for nanomaterials including high thermal stability (Tm = 93 °C),13 inherent chaperone activity,12 and affinity for metals.9,13 Furthermore, the interface of the protein is highly malleable, which can be redesigned to impart specificity and drive filament assembly into multifaceted and ordered structures.9,14 The resulting branched assemblies of γPFD have been used to template gold nanoparticles to create conductive metallic architectures.9
Herein, we expanded the use of γPFD to position and align multi-heme cytc metalloproteins to mimic the electron-hopping ET observed in Geobacter sulfurreducens nanowires. The structure of the γPFD filament subunits was resolved by cryoelectron microscopy (cryo-EM) to a 6 Å resolution, which provided guidance for the design of proteins to attach and align cytc proteins using SpyTag-SpyCatcher chemistry. Subsequently, we demonstrated that a network of these metalloprotein nanowires conducts current under aqueous conditions through the cytc domains at their redox potential.
Results and Discussion
Cryo-EM Structural Determination of Filamentous γPFD
Application of γPFD as a template to align metalloproteins with precision requires an improved understanding of the filament structure. Cryo-EM was therefore used to determine the three-dimensional structure of recombinantly produced γPFD filaments (Figure 1a). An averaged power spectrum from approximately 17,000 overlapping cryo-EM image segments of γPFD (Figure S1) showed a meridional layer line at ~ 1/(18 Å), establishing that the rise per subunit is ~ 18 Å, and a 1-start layer line at ~ 1/(21 Å), which together define a helical symmetry unambiguously, except for an enantiomorphic ambiguity (i.e., whether the 1-start is right- or left-handed). Using the iterative helical real space reconstruction (IHRSR) approach15 and particle polishing16 a resolution was reached where the handedness of the γPFD coiled-coil was visible. Subsequently, we determined that the coiled-coil is a left-handed 1-start helix with a rise per subunit of 18.3 Å and a rotation of −48.9° along this helix (Figure 1b). In addition, a γPFD homodimer was observed in the asymmetric helical subunit, with the 2-fold axis of the homodimer perpendicular to the filament axis; hence, the filament assembly is bipolar (Figure 1c). Applying dihedral symmetry slightly improved the resolution of the cryo-EM reconstruction, enabling us to resolve that the filament termini are identical. An atomic model was built of γPFD from an initial structure obtained by homology modeling, using the αPFD subunit in PDB 2ZDI as the template.17 Residues 8–128 were built into the cryo-EM density map, but several residues at both termini could not be included due to loop flexibility. After the model building, we directly estimated the map resolution using a model:map comparison18 which yielded an estimate of 6.0 Å (Figure S2). The “gold standard” map:map Fourier shell correlation and the recently described parameter d9919 also gave a similar resolution estimate (Table S1).
Figure 1.
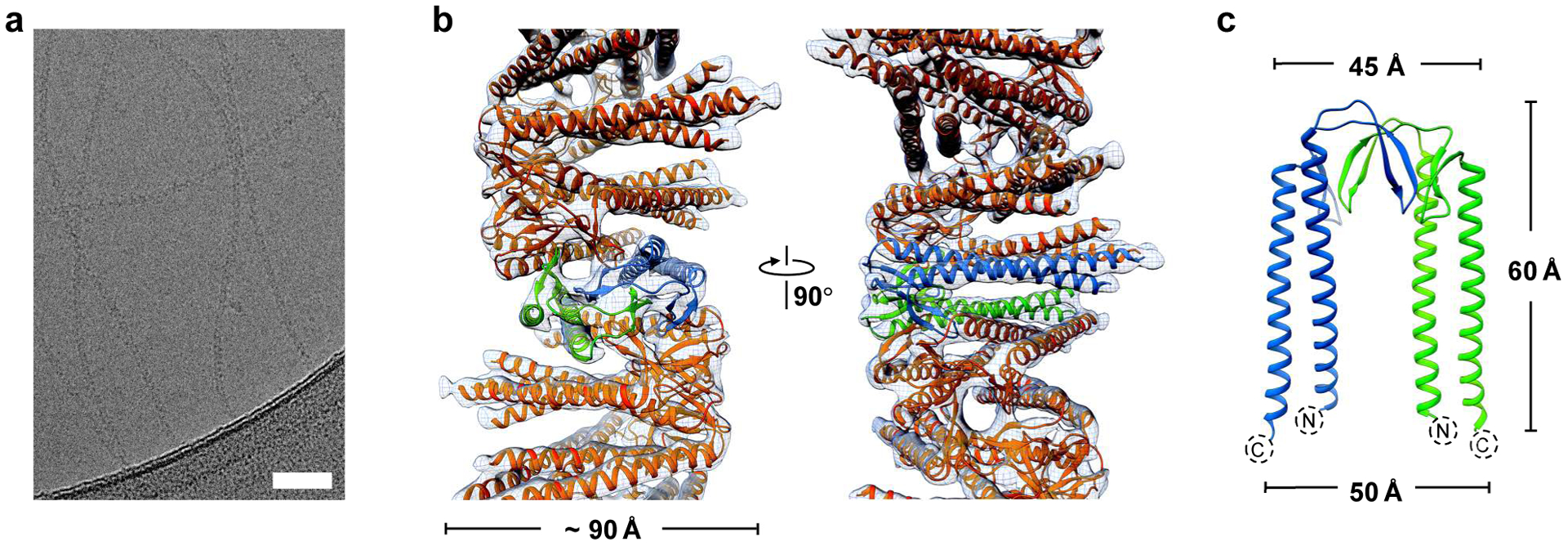
Cryo-EM reconstruction of the Methanocaldococcus jannaschii γPFD filament structure. a) Cryo-EM micrographs of γPFD filaments. Scale bars = 50 nm. b) Three-dimensional helical reconstruction of γPFD. The γPFD protein is shown in orange and two subunits in a dimer assembly are highlighted in blue and green, respectively. The map resolution was filtered to 6.0 Å. c) The γPFD dimer is shown with the two-fold axis relating the two subunits in the plane of the figure.
As the paralogous gene of αPFD, the architecture of a single γPFD subunit is structurally similar to other αPFD structures.10 The reconstituted γPFD subunit model has two long α-helices from the N- and C-terminal regions that form a ~ 60 Å long antiparallel coiled-coil, and those long helices are connected by two β hairpins (Figure 1c). A structural similarity search by DALI20 returns two archaeal αPFD structures with high percentage sequence coverage (Figure 2a). Compared with the αPFD dimers, the angle between the two coiled-coils in γPFD dimers is about 10° smaller (Figure 2b). The two archaeal αPFDs have been shown to form hetero-hexamers with βPFD subunits with a “jellyfish”-like quaternary structure (Figure 2c).17,21 The αPFD/βPFD hexamer is held together by two β-barrels, and each β-barrel contains four β-hairpins from two αPFD and two βPFD subunits. Despite γPFD and the two αPFD proteins having similar secondary structure and ~ 20% sequence identities with each other, γPFD does not assemble with either αPFD or βPFD subunit in vitro.10 Instead γPFD self-polymerizes in vitro into long filaments (Figure 1b), suggesting that this is the native assembly of γPFD.
Figure 2.
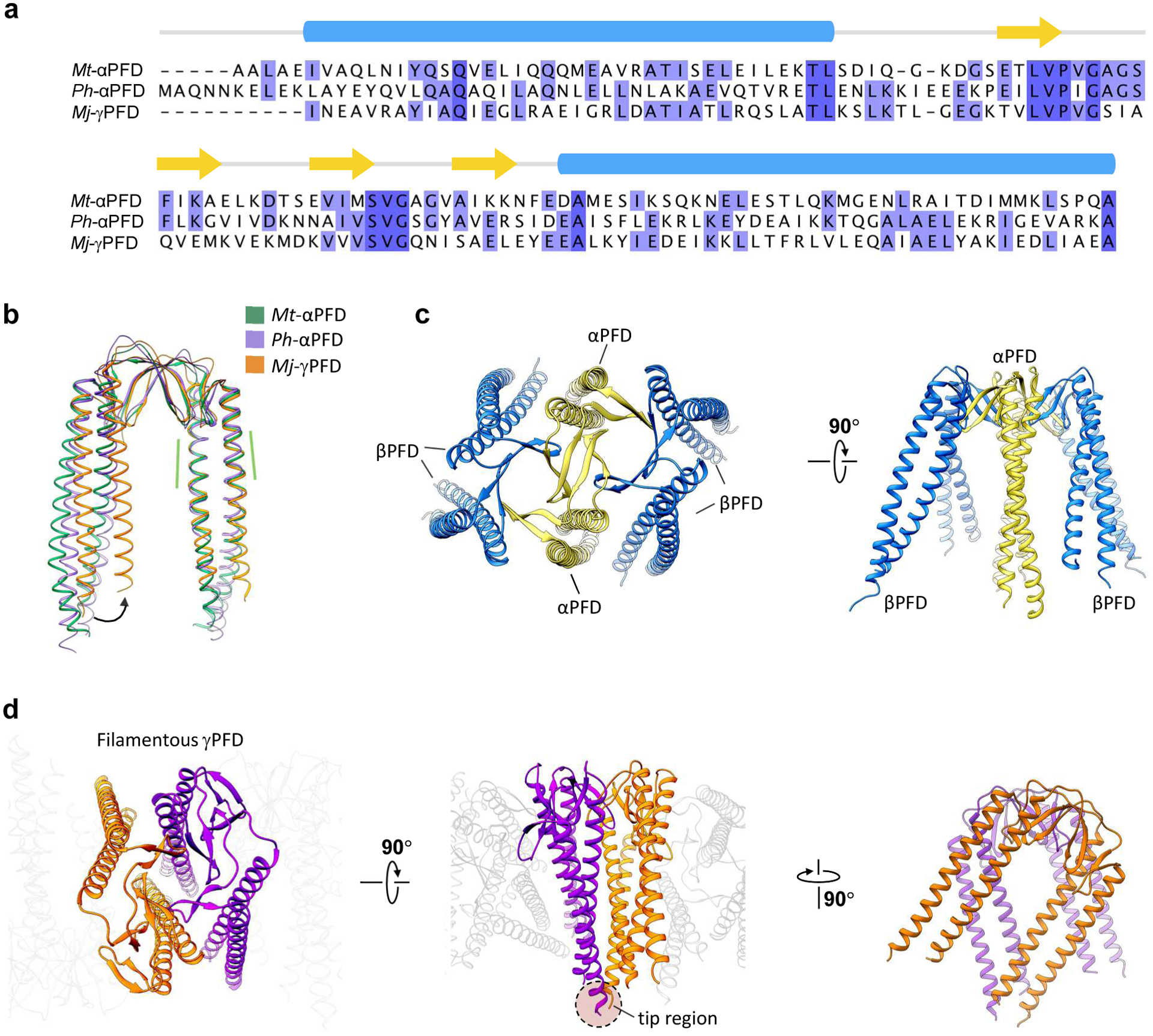
Comparison between filamentous γPFD and the αPFD-βPFD hexamer solved previously by X-ray crystallography (PDB 1FXK). a) Structure-based sequence alignment of αPFD from Methanothermobacter thermautotrophicus (Mt), αPFD from Pyrococcus horikoshii (Ph), and γPFD from Methanocaldococcus jannaschii (Mj). Only the sequences determined in the X-ray/cryo-EM structures are shown, and the corresponding secondary structure shown above the sequence (α-helical domains in blue, and β-sheets in yellow arrows). b) Structure alignment of the αPFD/γPFD dimer described in a. The region used for structural alignment is highlighted by green dashed lines. c) The “jellyfish”-like quaternary structure of the αPFD-βPFD hexamer in PDB 1FXK.21 The αPFD subunits are colored yellow, and the βPFD subunits are blue. d) The filamentous structure of the γPFD highlighting two adjacent dimers with their interactions shown from varying angles.
The buried surface area of single γPFD subunit in the β-barrel and loop region was calculated using PISA22 to be about 2,100 Å2 suggesting the β-barrel is primarily responsible for filament assembly. Strikingly, dimers of γPFD are rotated 50° along the helical axis with neighbouring dimers (Figure 2d), which contrasts with the αPFD-βPFD hexamer where the αPFD dimer is almost parallel with the βPFD dimer. As a result, the coiled-coil region of γPFD may also contribute to polymerization and the buried surface area of the coiled-coil region in a single γPFD subunit is about 620 Å2, which contributes to ~ 23% of its total buried surface. Furthermore, the helical twist brings the tips of two coiled-coils together, and extra density can be seen at the tip region that corresponds to the extra residues not built in the atomic model. It is notable that the helical filament structure determined from cryo-EM differs from the hypothetical non-helical, brush-like structure proposed in previous studies.9,12
Despite the helical arrangement of coiled-coils, filaments of γPFD have comparable chaperone activity to archaeal α/β hexameric prefoldins,10 including filaments engineered to contain only six subunits.12 The reason for the helical assembly of γPFD is unclear – presumably the filament has additional functions within M. jannaschii. Other archaeal chaperones, such as chaperonins, have been shown to form filaments in vivo and have been proposed to have cytoskeleton function for cell shape determination and division.23,24 Filaments of γPFD whose average length is sufficient to span the diameter of M. jannaschii may also have similar cytoskeleton roles.
The significant buried surface area of the γPFD coiled-coils and the extra density at the tip region observed in cryo-EM strongly suggest the distal regions of the coiled-coils are involved in filament oligomerization. A previous study demonstrated that γPFD with truncated coiled-coils was still able to assemble into filaments; however, the filaments were significantly longer and stiffer than wild-type filaments.25 It may be that intermolecular interactions of the coiled-coils sterically hinder the alignment and incorporation of incoming subunits to the helical structure of the protofilament – similar processes have been observed for the oligomerization of other filamentous proteins.26,27 The ability to tailor γPFD filament length and stiffness by truncation of its coiled-coil domain may be a useful property in a material scaffold.
Design and Fabrication of γPFD-Metalloprotein Nanowires
Exploiting the close spacing of the γPFD coiled-coils should enable precise alignment of functional molecules, such as redox proteins, along the lateral direction of the filaments. The cryo-EM resolved structure of γPFD filaments enabled us to design metalloprotein filaments to position and align cytc domains for mimicking the electron hopping mechanism of Geobacter sulfurreducens nanowires. A predicted model of the full-length γPFD monomer was created by adding the residues at both termini that could not be resolved in the cryo-EM map due to loop flexibility (Figure 3a). The tetraheme cytc 3 from Desulfovibrio vulgaris Miyazaki was chosen to mediate conductance.28 The amide bond forming SpyTag-SpyCatcher chemistry enables efficient and specific conjugation between proteins tagged with the 13-residue SpyTag peptide and counterpart proteins fused to the 12 kDa SpyCatcher domain.29,30 In the first designs, the SpyTag sequence was genetically fused to the C-terminal of γPFD, and the SpyCatcher domain genetically fused to the C-terminal of cytc3 (Figure 3b). Incubation of the cytc3-SpyCatcher with γPFD-SpyTag filaments should result in spontaneous conjugation between the SpyTag-SpyCatcher domains, completing the self-assembly of the metalloprotein nanowires. The spacing between the coiled-coil of γPFD dimers along the longitudinal axis is sufficient to accommodate the approximate 40 Å diameter of the cytc3 subunit, which when attached to γPFD-SpyTag filaments, should result in cytc3 molecules positioned adjacent to each other along the entire filament. Furthermore, in our modelling of the nanowires (Figure 3b), the abundance in electron-transporting hemes should ensure the distance between redox sites of adjacent cytc3 is less than 25 Å, the physical threshold for efficient electron hopping.31 In our model, the minimum edge-to-edge distance between the hemes of adjacent cytc3 domains on the C-terminal of γPFD can vary from 13.2 Å to 19.1 Å (Figure S3a). The varying distances of cytc3 domains are likely due to the dynamic flexibility of the γPFD coiled-coil and short linker regions that join the γPFD, SpyTag, SpyCatcher, and cytc3 domains. A greater density of cytc3 may increase conductance by staggering or spatially confining cytc3 domains in closer proximity for more efficient ET between heme molecules. To examine the effect of additional cytc3 on the nanowire for ET, a design was created with SpyTag domains at both termini of γPFD to facilitate the attachment of two cytc3 per γPFD subunit (Figure 3c). Nanowires containing one cytc3 per γPFD subunit were designated γPFD-cytc3, and nanowires containing two cytc3 per subunit were designated γPFD-[cytc3]2. In the γPFD-[cytc3]2 model, the minimum edge-to-edge distances between hemes of adjacent cytc3 domains on the C-terminal of γPFD were similar to the γPFD-cytc3 model, with distances ranging from 13.8 Å to 21.1 Å (Figure S3b). Furthermore, in the γPFD-[cytc3]2 model, the N-terminal cytc3 domains are staggered in proximity (14.9 Å to 18.7 Å) to the C-terminal cytc3 domains of subunits positioned further along the filament (Figure S3c), thereby potentially providing smaller inter-heme distances for more efficient ET.
Figure 3.
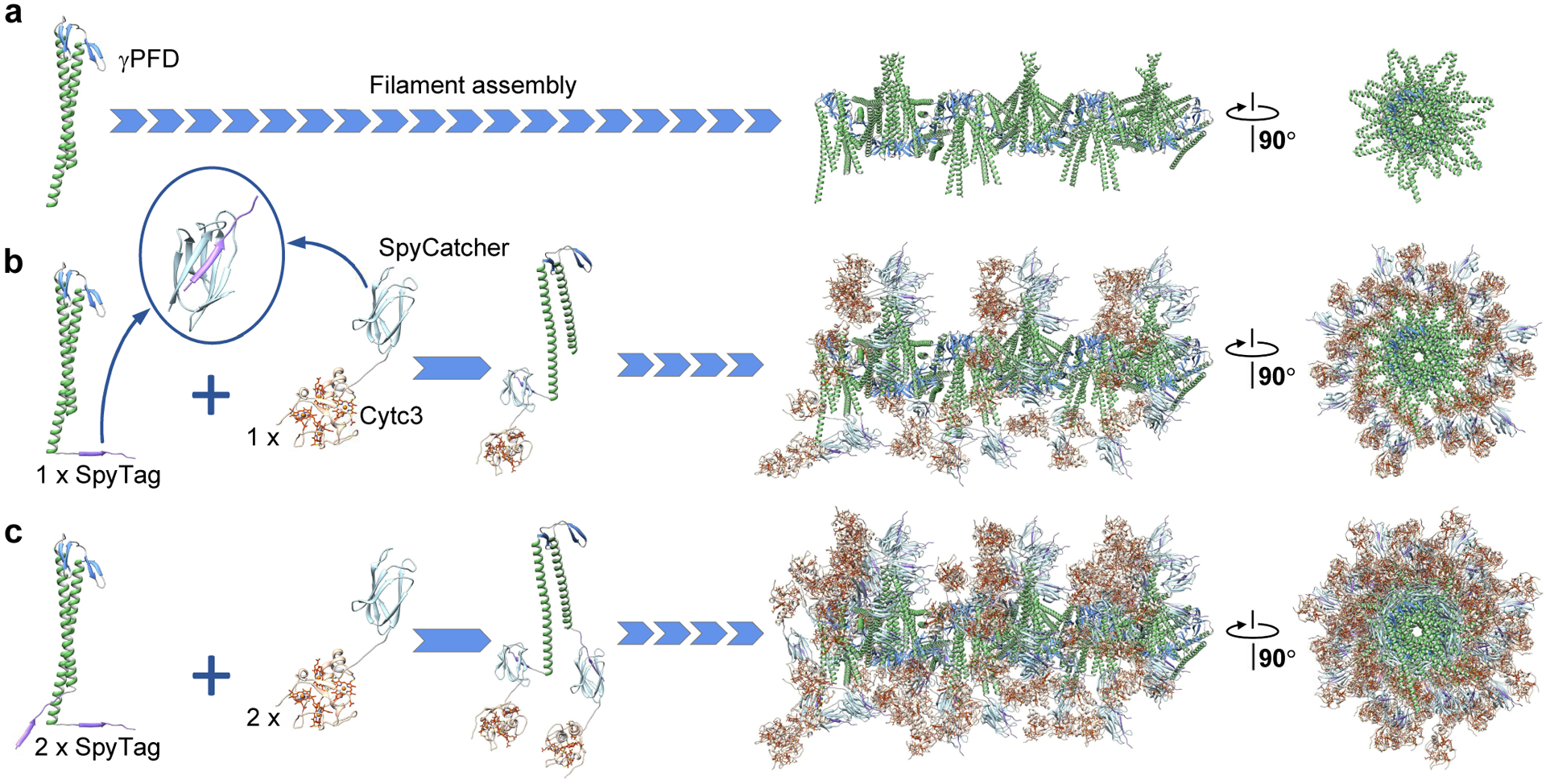
Design and assembly strategy of metalloprotein nanowires. a) Proposed structure of γPFD filaments with full-length coiled-coils. Inclusion of a SpyTag domain at b) the C-terminus or c) both termini of γPFD subunits enables covalent attachment of one or two cytc3-SpyCatcher proteins. The fused protein subunits assemble as metalloprotein nanowires.
The gene sequences of fusion proteins were chemically synthesised and cloned into plasmid DNAs for recombinant expression in E. coli. Purified subunits of γPFD containing one or two SpyTag domains were assembled into filaments at 60 °C and conjugated with cytc3-SpyCatcher subunits. The resulting nanowires containing one cytc3 per γPFD subunit were designated γPFD-cytc3, and nanowires containing two cytc3 per subunit were designated γPFD-[cytc3]2. Non-conjugated cytc3-SpyCatcher protein could be removed by attaching the nanowires to Ni-NTA through the hexa-histidine tag on the γPFD and washing away unbound proteins. Efficient conjugation of cytc3-SpyCatcher to the SpyTag domain was verified by SDS-PAGE (Figure S4). Conjugation of the 20 kDa γPFD-SpyTag with the 28 kDa cytc3-SpyCatcher produced a new band with a cumulative mass of 48 kDa in the SDS-PAGE gel.
The nanowires were imaged by negative-stain TEM and compared with γPFD-SpyTag filaments (Figure 4a). Distinct structural differences along the lateral edges of the cytc-conjugated filaments were observed for both the γPFD-cytc3 (Figure 4b) and γPFD-[cytc3]2 nanowires (Figure 4c). The helical structure of the filament should result in bound cytc3-SpyCatcher domains being symmetrically arranged around the filament (Figure 3), which is consistent with the structures imaged in TEM (Figure 4). The metalloprotein nanowires displayed Gaussian distributions of filament lengths in TEM (Figure S5), with an average length of 292 nm for the γPFD-cytc3 and 274 nm for the γPFD-[cytc3]2 (n = 130 filaments). Increased thickness of the nanowires from cytc3 conjugation was also observed by atomic force microscopy (AFM), with height measurements averaging 2.8 nm and 3.6 nm for the γPFD-cytc3 and γPFD-[cytc3]2, respectively, compared with 2.2 nm for the γPFD-SpyTag filaments (Figure S6). Cryo-EM reconstruction was also attempted for the metalloprotein nanowires; however, the conjugated cytc3-SpyCatcher domains were too structurally flexible on the filament to resolve a structure.
Figure 4.
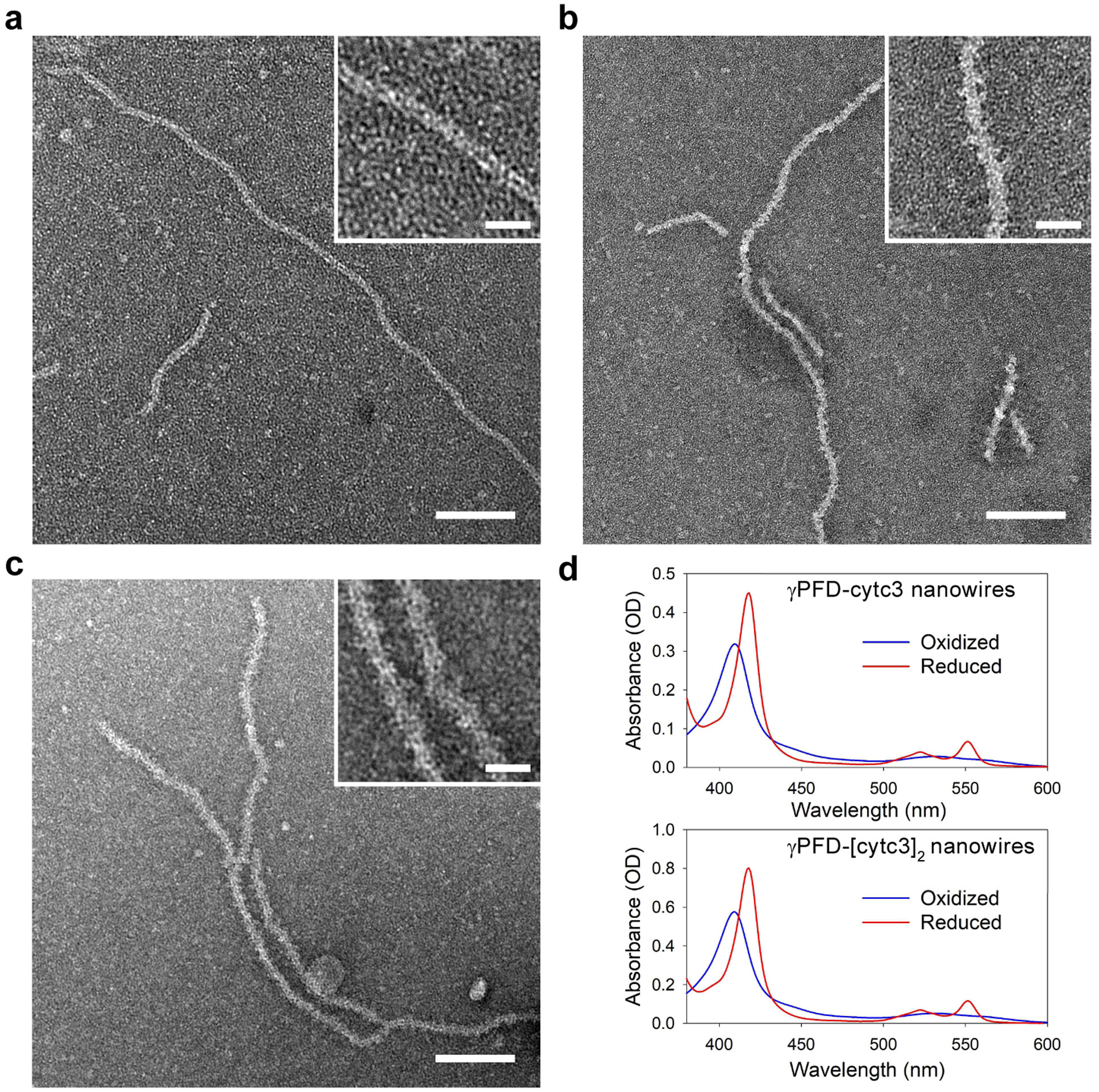
Imaging of metalloprotein nanowires. a) TEM micrographs of γPFD filaments, b) γPFD-cytc3, and c) γPFD-[cytc3]2 metalloprotein nanowires. Scale bars = 100 nm. Inserted micrographs are enlarged images of nanowires showing differences in morphology. Scale bars = 20 nm. d) Absorption spectra of the γPFD-cytc3 (top) and γPFD-[cytc3]2 (bottom) nanowires in an air oxidized or reduced state by addition of sodium borohydride.
The redox characteristics of cytc3 within the nanowires were measured by UV-Vis absorption spectroscopy (Figure 4d). In their air oxidized state, the absorption spectrum of the cytc3-containing nanowires displayed a dominant Soret peak at 409 nm, characteristic of ferric heme configurations of cytc3 when Fe(III) ions are chelated to the porphyrin molecule.28 The Soret peak shifted from 409 nm to 418 nm after addition of a reducing agent, sodium borohydride, which is representative of electron transfer to the iron core of the heme, as changes in bond energy within the porphyrin ligand introduce variations in the absorption spectrum.32 Correspondingly, α and β bands, at 522 nm and 551 nm, respectively, were observed upon reduction and are characteristic features of low energy transitions in the porphyrin under reduced state of the heme.33
Charge Transport Measurements of the Metalloprotein Nanowires
Solid-state and electrochemical ET measurements were performed on films of the metalloprotein nanowires and controls. For both measurements, nanowire samples were cast as dried films onto interdigitated electrodes separated by 6.25 μm spacing. In these film measurements, conductivity will be affected by several factors, such as the stacking density and orientation of the cytc3 and the density of the percolation network of filaments. Deposited films of γPFD-cytc3 and γPFD-[cytc3]2 nanowires were shown to exhibit long-range electronic conductivity by establishing a percolating network of filaments spanning the electrode gaps,34 with incorporation of the cytc3 shown to be critical for conductivity (Figure S7). Dry films composed of either cytc3-SpyCatcher or γPFD-cytc3 are 3.7- or 5.5- to 6.4-fold more conductive than γPFD-SpyTag alone, respectively. The greater conductivity of films composed of γPFD-cytc3 nanowires compared with films of composed of cytc3-SpyCatcher suggest that the γPFD scaffold may be aligning the cytc3 in a more efficient arrangement for multi-step charge transfer or enhancing the percolation networks of cytc3 domains, compared to those present in a randomly-deposited cytc3 film. Although the γPFD-[cytc3]2 nanowire has twice the number of cytc3 molecules aligned along the filament compared with the γPFD-cytc3 (Figure 2c), the γPFD-[cytc3]2 was only 12–16% more conductive. Cyclic voltammetry (CV) was used to measure current between a single working electrode and a Pt counter electrode with a Ag/AgCl reference to provide insight into the charge transfer process in γPFD-cytc3 nanowires. The peak current for γPFD-cytc3 films was proportional to the square root scan rate, which is indicative of a diffusion-limited redox process (Figure S8). This observation is consistent with a gradient-driven redox transport that has also been observed in Geobacter sulfurreducens biofilms, which incorporate extracellular cytc domains.3
Bipotentiostat cyclic voltammograms were used to investigate the magnitude and nature of ET through the γPFD-cytc3 nanowire films under physiologically-relevant conditions. Bipotentiostat measurements have previously been used to distinguish between redox and non-redox mediated charge transport in conductive materials.7,34,35 Briefly, in this setup (Figure 5a), the global (gate) potential of both the source and drain electrodes is swept relative to a Ag/AgCl reference electrode, while maintaining a fixed potential offset between the source and drain. The source and drain are two working electrodes which share a common counter electrode. The source-drain current, IDS, is half the difference of the separate source and drain currents at a fixed VDS offset, minus background currents at VDS = 0. For ease of viewing, the oxidation peaks during the anodic scan are provided in Figure 5, and the full anodic and cathodic bipotentiostat IDS currents are provided in Supporting Figure S9.
Figure 5.
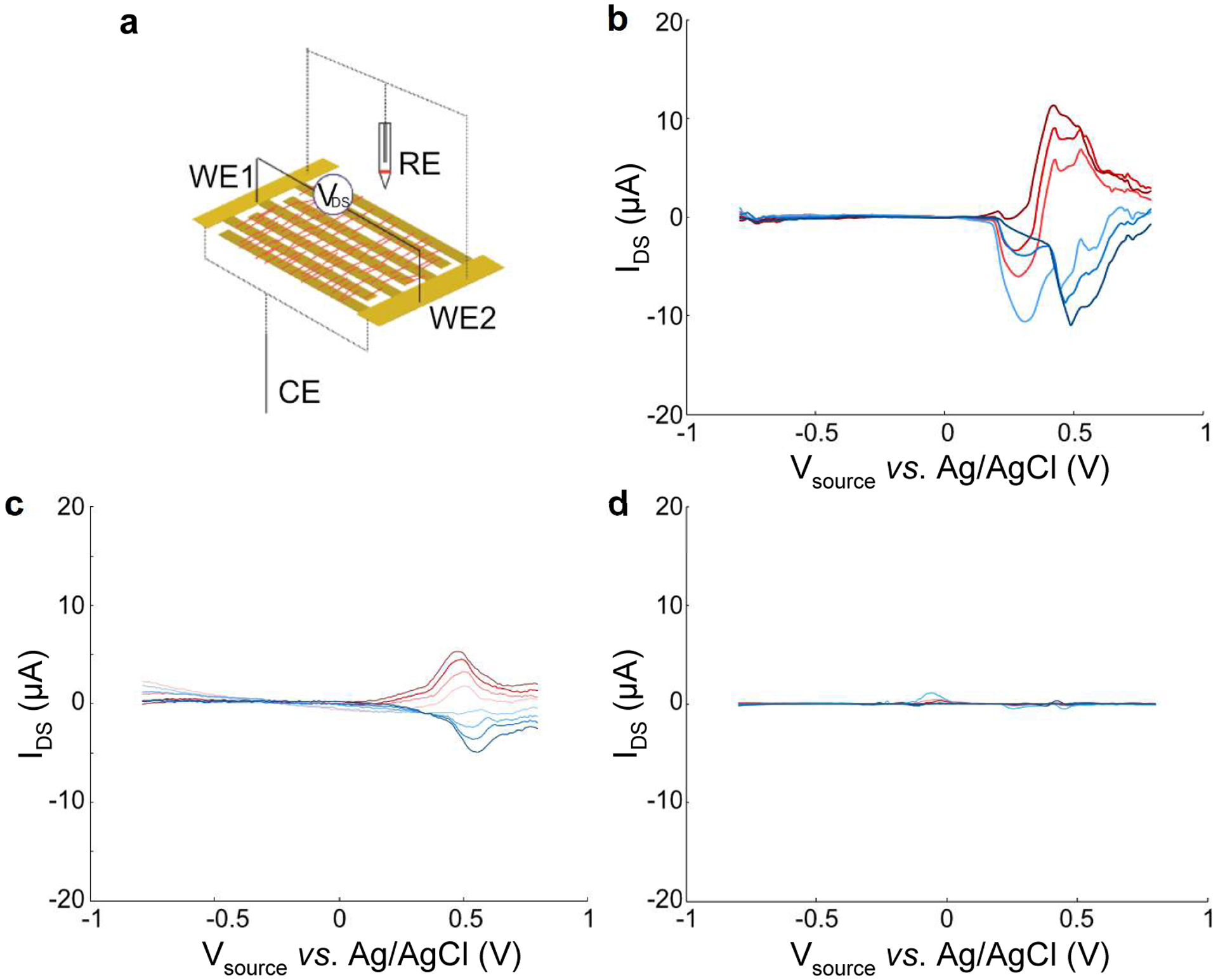
Bipotentiostat cyclic voltammogram measurements of nanowires and controls. a) Device schematic demonstrating nanowires deposited across electrodes (working electrode 1 & 2, WE1 & WE2; reference electrode, RE; counter electrode, CE). WE1 and WE2 are the “source” and “drain” electrodes, respectively. b) γPFD-cytc3 nanowire, c) cytc3-SpyCatcher, d) γPFD-SpyTag. All measurements performed at a scan rate of 50 mV/s in 0.1 M phosphate buffer, pH 8.4.
The bipotentiostat measurements show that the γPFD-cytc3 nanowire and cytc3-SpyCatcher films conduct current between the source and drain electrodes at similar redox potentials. From the individual CVs of each electrode, the redox peaks for cytc3-SpyCatcher occur around Eox = 0.6 V, Ered = −0.14 V vs. Ag/AgCl for and around. Eox = 0.5 V and Ered = −0.1 V for γPFD-cytc3 (Figure S9). As shown in Figure 5b and 5c, the bipotentiostat IDS peaks for the γPFD-cytc3 nanowire and cytc3-SpyCatcher films during the anodic scan are centered around these potentials, with the magnitude and direction of the current tracking with the magnitude and polarity of VDS. Conversely, the IDS current does not scale with VDS magnitude or polarity for the non-conducting γPFD-SpyTag film (Figure 5, Figure S9). These measurements suggest that the nanowire film is conducting current between the source and drain electrodes through the cytc3 domains. The currents through the γPFD-cytc3 films are larger than current through a film composed entirely of cytc3-SpyCatcher (Figure 5c), consistent with the solid-state measurements. Furthermore, current exchange happens at similar redox potentials, suggesting that ET in both the γPFD-cytc3 and cytc3-SpyCatcher samples occurs through the same cytc3 heme states. Small, unexpected redox peaks were also observed in the γPFD-SpyTag nanofiber films at −0.04 V and 0.42 V vs. Ag/AgCl, which may account for the small differences in redox values between the cytc3-SpyCatcher and γPFD-cytc3 systems. Although the nanowire scaffold does not appear to conduct current redox current through the source and drain electrodes, it evidently enhances current exchange between the working electrodes when coupled with cytc3. The mechanism of enhanced conduction is unclear, but it may stem from an increase in conductive bridge formation within the cytc3 network by alignment on the γPFD scaffolds, as compared to similar densities of unaligned cytc3 in the films of cytc3-SpyCatcher.
Differences in current transport between γPFD-cytc3 and control films were more apparent in solution than in solid-state measurements. These differences may be attributed to one or several of the following reasons: 1) the aqueous solution provides a more physiologically-relevant environment to support redox hopping through native conformation cytc3, 2) the use of a reference in the electrochemical setup guarantees that the measurement sweeps through the redox potential of the cytc3 domains, and 3) all films experienced partial delamination, since there was a visible reduction of material after measurements in solution, but differences in delamination were not characterized. Films composed of γPFD-SpyTag lacking cytc3 domains exhibited negligible charge transfer between the source and drain electrodes (Figure 5d), indicating that cytc3 is the primary conductive component in the γPFD-cytc3 nanowire films.
Conclusions
The present study solved the structure of γPFD to 6 Å resolution and engineered these filaments to create modular metalloprotein nanowire films capable of facilitating long-range ET. Mimicking the ET of Geobacter sulfurreducens nanowires requires a structural scaffold to align cytc metalloproteins in proximity over long distances. The ultrastable and engineerable γPFD filament was chosen as a scaffold, and its structure resolved by cryo-EM, which also provided insights into filamentous arrangements of prefoldins. As a class of molecular chaperones, prefoldins function in an ATP-independent manner to arrest protein misfolding in archaea and participate in the maturation of cytoskeletal proteins in eukaryotes.11 Except for γPFD, prefoldins assemble into hetero-hexameric complexes composed of two α-type and four β-type subunits.21 The overall hexameric quaternary structure consists of a rigid double β-barrel with six highly flexible coiled-coils extending outwards in the same direction. Previous homology modelling of γPFD using archaeal αPFD as a template supported a non-helical, brush-like structure for the filaments.9,12 The helical filament structure of γPFD determined from cryo-EM in this study was thus unexpected, and is unique amongst molecular chaperones and filamentous proteins in general.
The resolved structure of γPFD enabled the design of nanowires for the alignment of cytc3 proteins in high density. The modular SpyTag-SpyCatcher chemistry used to attach cytc3 to γPFD should enable a variety of other metalloproteins to be aligned on the filaments. Other redox proteins including iron-sulfur, zinc, or copper metalloproteins could be incorporated into γPFD filaments in place of the cytc3 to tune the electrical and semi-conductor properties of the nanowires. The ability to customize the redox properties of metalloprotein nanowires would be useful for specific bioelectronic applications or for interrogating the underlying mechanisms of charge transport in biomaterials. The distance between the hemes of the cytc3 domains was sufficient for ET, which was most likely mediated by an electron hopping mechanism. However, the flexibility of the SpyTag-SpyCatcher domains permits mobility of the cytc3 domains, which suggests that diffusion-assisted hopping mechanisms may also be occurring for conductance.4
In addition to assembly into linear nanowires, the individual γPFD-cytc3 subunits are building blocks that could potentially be used to construct conductive nanostructures or interface with other functional materials. Previously, it has been demonstrated that multiple γPFD filaments can be joined using engineered connector proteins.9 These connectors would enable the nanowires to be assembled into ordered structures, or enable attachment of functional molecules, such as redox enzymes, at specific locations – for applications in fuel-cells or biosensors. Furthermore, the interface of γPFD can be redesigned to create subunits that assemble in repeating orders to create multicomponent filaments.14 These filaments would enable a variety of metalloproteins with different redox potentials to be aligned in specific orders, potentially mimicking natural electron transport chains. The modularity and engineerability of γPFD metalloprotein nanowires hold potential for application as naturally inspired bioelectronic materials.
Methods
Cryo-EM Data Collection and Image Processing
The wild-type γPFD was expressed in E. coli and purified as described previously.9 The γPFD filament sample (4 μL at 0.2 mg mL−1) was applied to discharged lacey carbon grids and plunge frozen using a Vitrobot Mark IV (FEI). Frozen grids were imaged in a Titan Krios at 300 keV and recorded with a Falcon III camera at 1.09 Å per pixel. Micrographs were collected using a defocus range of 1.5–2.5 μm, with a total exposure time of 2.4 s (amounting to ~ 55 electrons/Å2) distributed into 24 fractions. All the micrographs were first motion corrected (ignoring the first fraction) using MotionCorr v2.136 and then used for CTF estimation by the CTFFIND3 program.37 Filament images were extracted using the e2helixboxer program within EMAN238 from the dose-weighted fractions 2–10 (amounting to ~ 20 electrons/Å2), after the images were corrected for the CTF through multiplication by the theoretical CTF. A total of 32,227 overlapping 384-px long segments (with a shift of 25 pixels, ~ 1.5 times the axial rise per subunit) were generated. The helical symmetry was determined unambiguously given the 1/(18 Å) meridional layer-line. The IHRSR method implemented in Spider15 was used to generate a ~ 7 Å reconstruction, with the volume was subsequently filtered to 10 Å as the starting reference used in Relion.16 The same micrographs and box coordinates used in the Spider reconstruction were imported into Relion. A comparable ~ 7 Å reconstruction was generated after class2D and refine3D steps, and then it was further improved to 6 Å after movie-refinement and particle polishing steps. Model:map FSC18 and d9919 were used to determine a resolution of the final volume, that was sharpened with a negative B-factor of 200.
Model Building of γPFD Filaments
The density corresponding to a γPFD dimer was segmented from the experimental filament density using UCSF Chimera.39 The initial model of γPFD was generated by homology modeling using the I-TASSER40 server using the αPFD subunit in PDB 2ZDI as the template. Then the coiled-coil part of the initial model was further optimized using CCbuilder 2.041 to maximize the real-space fitting with the cryo-EM density at the coiled-coil region. After this, the γPFD dimer was docked into the segmented map using Chimera. A filamentous model was generated using this γPFD dimer as the asymmetric unit and refined against the full cryo-EM map, using real-space refinement in PHENIX.19 To prevent model over-fitting at this resolution, protein secondary structure was restrained during the entire refinement and the coiled-coil region was refined as a rigid body. MolProbity42 was used to evaluate the quality of the filament model.
Protein Nanowire Design
UCSF Chimera software (version 1.12)39 was used to build a predicted model of the full-length γPFD monomer by adding the amino acid sequences VNEVID (residues 2–7) and QQTSEEEKAEEEENEEKAE (residues 129–147) to the N-terminus and C-terminus that were absent from the cryo-EM resolved 6 Å resolution model of the wild-type γPFD structure. The initial methionine residue was omitted due to its co-translational cleavage in E. coli. ϕ (phi) and ψ (psi) torsion angles were adjusted to −57° and −47°, respectively, to ensure an α-helical conformation of the N- and C-termini (Figure 3a).
To create hypothetical models of the γPFD monomer covalently bound to one or two cytochromes, the model of the full-length γPFD monomer as well as the available X-ray crystallography structures of the SpyCatcher-SpyTag complex (PDB ID: 4MLI)43 and of cytochrome c3 (cytc3) from Desulfovibrio vulgaris Miyazaki F (PDB ID: 2EWK)44 were joined using the UCSF Chimera software (Figure 3b). The C-terminus of cytc3 was joined to the N-terminus of the SpyCatcher domain via a GGGS linker to create a model of cytc3 fused to the SpyCatcher-SpyTag complex. Working with this new model, the N-terminus of the SpyTag from the cytc3-SpyCatcher-SpyTag model was fused to the C-terminus of the full length γPFD model via a GGGS linker to create the model of the γPFD monomer fused to a cytc3. Then, an additional cytc was added to this model, this time by fusing the C-terminus of the SpyTag domain from the cytc3-SpyCatcher-SpyTag model to the N-terminus of the γPFD subunit via a GGGS linker (Figure 3c). The models of full-length γPFD, γPFD fused to one cytc3 and γPFD fused to two cytc3 through SpyTag-SpyCatcher attachments were aligned to the resolved cryo-EM structure of the γPFD filament to create a predicted model of the filament and nanowires. In the nanowire models, torsion angles within the GGGS linkers were adjusted, where required, to eliminate clashes between cytc3 domains and Spy domains. In addition, the cytc3-SpyCatcher construct also included an N-terminal OmpA periplasmic targeting sequence. Hexa-histidine tags were included at the N-terminal of the γPFD-SpyTag and SpyTag-γPFD-SpyTag to enable IMAC techniques for protein purification. The amino acid sequences for γPFD and the designed proteins are provided in Table S2.
Recombinant Production of Designed Proteins
The gene sequences for γPFD-SpyTag, SpyTag-γPFD-SpyTag, and cytc3-SpyCatcher were chemically synthesized as gBlock Gene Fragments (Integrated DNA Technologies) and cloned into the bacterial expression vector pET19b (Novagen) via the Gibson Assembly method.45 Subsequently, the plasmids were transformed into competent BL21 T7 Express E. coli (NEB, USA), with the plasmid encoding cytc3-SpyCatcher co-transformed with pEC86, a chloramphenicol-resistant plasmid containing the heme maturation genes ccmABCDEFGH.46
Cultures of BL21 containing either the γPFD-SpyTag or SpyTag-γPFD-SpyTag plasmid were grown at 37 °C in Luria Broth supplemented with 100 μg mL−1 ampicillin. Upon reaching an optical density of 0.6 at A600, protein expression was induced for 16 h at 25 °C with the addition of 0.4 mM isopropylthiogalactoside (IPTG). Harvested cells were lysed and clarified by centrifugation for 40 min at 22 000 g. The γPFD-SpyTag and SpyTag-γPFD-SpyTag proteins were purified by immobilized metal affinity chromatography on Ni-NTA resin (Thermofisher). Eluted fractions were analysed using SDS-PAGE gels stained by QC Colloidal Coomassie G-250 stain (Bio-Rad). Fractions containing the desired pure protein were pooled, concentrated, and buffer exchanged to PBS buffer (10 mM Na2HPO4, 1.8 mM KH2PO4, 137 mM NaCl, 2.7 mM KCl, pH 7.4) using 10 kDa Amicon Ultra-15 Centrifugal Filters (Merck), followed by lyophilisation and storage at −80 °C.
Cultures containing the cytc3-SpyCatcher/pEC86 plasmids were grown at 37 °C in 2xYT media containing 100 μg mL−1 ampicillin and 34 μg mL−1 of chloramphenicol. Upon reaching an optical density of 0.6 at A600, protein expression was induced for 16 h at 25 °C with the addition of 30 μM IPTG. A periplasmic protein purification was performed on the harvested cells by resuspension in a solution containing ice-cold 30 mM Tris-HCl, 20 % w/v sucrose, 1 mM EDTA, pH 8.0, and incubated on ice for 30 min, followed by centrifugation at 10 000 g for 20 min. The supernatant was decanted off and pelleted cells were suspended in ice-cold 0.5 mM MgCl2. The cells were incubated for 30 min on ice and centrifuged to obtain a supernatant containing the periplasmic contents. The periplasmic fraction was purified by cation exchange chromatography using a HiTrap SP Sepharose FF column (GE Healthcare). Eluted fractions were analysed using SDS-PAGE gels stained by QC Colloidal Coomassie G-250 stain. Fractions containing the pure cytc3-SpyCatcher protein were pooled, concentrated, and buffer exchanged to PBS buffer using 10 kDa molecular weight cut-off Amicon Ultra-15 centrifugal filter units and stored at 4 °C.
Assembly of Metalloprotein Nanowires
Lyophilised γPFD, γPFD-SpyTag, and SpyTag-γPFD-SpyTag proteins were resuspended in 8 M guanidinium-HCl, 10 mM NaH2PO4, pH 8.0, and the protein concentrations determined by a modified Bradford assay.47 The proteins were refolded by a rapid dilution with PBS buffer to achieve a final protein concentration of 0.5 mg mL−1, and filaments assembled during a 16 h incubation at 60 °C. Covalent attachment of cytc3 to the filaments was achieved by combining an equimolar ratio of cytc3-SpyCatcher with γPFD-SpyTag and a double molar ratio of cytc3-SpyCatcher with the SpyTag-γPFD-SpyTag. Subsequently, the resulting nanowires were stored at 4 °C.
Spectrophotometric Analysis of Nanowires
Nanowire samples were diluted to a final concentration of 0.5 mg mL−1 in 20 mM NaH2PO4, 100 mM NaCl, pH 7.5, and transferred into 3 mL quartz cuvettes. Spectral absorption measurements were performed using a Lambda UV-Vis spectrometer (PerkinElmer) between wavelengths 300 to 600 nm. Sample was removed from the spectrophotometer and 0.5 mg of sodium borohydride added for the reduction of cytc3. After reduction, indicated by the colour change of the sample to bright pink, spectrophotometric absorption analysis was repeated on the samples.
Negative-Stain Electron Microscopy of Nanowires
Filament and nanowire samples for TEM were diluted to 40 μg mL−1 in PBS before deposition on Carbon Type-B, 200-mesh copper grids (Ted Pella Inc.), pre-treated with an easiGlow Glow Discharge Unit (PELCO). After a 5 min incubation at room temperature, the grids were washed with distilled water, and stained with 2 % aqueous uranyl acetate (BDH Chemicals) for 7 min. The stain was wicked off with filter paper (Whatman) and grids were left to dry before visualisation under a Tecnai G2 20 TEM (FEI). The digitized TEM images were analysed using ImageJ48 (US National Institutes of Health) to measure filament and nanowire length.
Atomic Force Microscopy of Nanowires
Sample specimen holders were prepared by mounting PELCO mica discs (Ted Pella Inc.) on AFM specimen discs (Ted Pella Inc.) using Conductive Silver Liquid (ProSciTech). After drying and overnight curing of the silver glue, the mica substrate was peeled three to four times and 10–20 μL of nanowire sample was applied. After 15 min of air-drying, the substrate was rinsed with Milli-Q water using a fine tip pipette and dried overnight. The specimen was loaded on the magnetic platform in the Dimension SPM ICON (Bruker) equipped with a SCANASYST-AIR probe (Bruker). The probe was lowered until within 1 mm above the sample surface before scanning was operated under peakforce tapping mode. Parameters were adjusted such that force was within 10–15 mV, scan rate was 0.9 Hz, while the resolution was 512 pixels per line for image dimensions of either 1 μm or 5 μm squares. Image data was processed and analysed using the NanoScope Software (Bruker).
Device Preparation for Electronic Property Characterization
Solid state and electrochemical bipotentiostat measurements were performed using interdigitated electrodes. Each electrode was comprised of 80 parallel 8 μm × 2 mm long bands with an intra-band spacing of 6.25 μm. Devices were photolithographically patterned onto Pyrex wafers with 60 nm Au and a 5 nm Ti adhesion layer deposited by electron beam evaporation (UCSD Nano3 Fabrication Foundry Services). Prior to use, devices were sonicated in washes of acetone, isopropanol, and ultrapure water. Devices were individually tested for shorts prior to sample deposition. For electrochemical measurements, 22-gauge solid core insulated wire leads were connected to the interdigitated source and drain electrodes using conductive silver epoxy (MG Materials). Exposed electrode and lead connections were sealed with waterproof silicone sealant (DAP All-Purpose Adhesive Sealant). For solid state and electrochemical charge measurements, 15 μL of sample was deposited onto each device in 5 μL intervals and allowed to dry at 4 °C. Samples were briefly rinsed with ultrapure water to remove salts prior to measurements.
Solid-State I-V Measurements of Nanowires
Solid state I-V measurements were performed with a Keithley Model 2612B Source Measure Unit. Current was monitored as a function of swept voltage from +1V to −1V under ambient conditions. Each sample was deposited onto n = 3 independent devices. Error bars are reported as the standard error.
Electrochemical Bipotentiostat Measurements of Nanowires
Bipotentiostat measurements were performed in 0.1 M phosphate buffer, pH 8.4, degassed by sparging with 20% CO2-N2 while boiling for an hour. Electrochemical measurements were performed in sealed cells with N2 flow in the headspace. For each bipotentiostat measurement, the global (gate) potential of the source and drain electrodes was swept relative to a 3.5 M KCl Ag/AgCl reference electrode, while maintaining a fixed potential offset (VDS) between the source and drain, which shared a common Pt counter electrode. The source-drain current, IDS, was taken to be half the difference of the separate source-counter and drain-counter currents at a fixed VDS offset, minus background currents at VDS = 0. Only the anodic (oxidizing) portion of each CV (−0.8 V to +0.8 V vs. Ag/AgCl) was shown in Figure 5 for clarity. The full bipotentiostat CVs (anodic and cathodic) are shown in Figure S9. Scans were performed at a scan rate of 50 mV/s.
Electron Microscopy Data
Cryo-EM structural data for the filament were deposited in the Electron Microscopy Data Bank (EMDB) and the Protein Data Bank (PDB) with the following entry codes, EMDB: EMD-21455 and PDB: 6VY1.
Supplementary Material
Acknowledgments
This work was supported by the Air Force Office of Scientific Research (FA9550-17-1-0451 to D.S.C. and D.J.G., FA9550-14-1-0350 to A.I.H.) and the National Institutes of Health GM122510 (to E.H.E.). N.L.I. also acknowledges support from a US Department of Education GAANN fellowship.
Footnotes
Supporting Information Available: Power spectrum, Fourier shell correlation between the atomic model and the cryo-EM map, proposed spatial alignment of hemes in γPFD-metalloprotein nanowires, SDS-PAGE of cytc3-SpyCatcher conjugation to γPFD-SpyTag, distribution of nanowire lengths, AFM of nanowires, solid-state conductance measurements of nanowires, cyclic voltammogram of γPFD-cytc3 films, individual cyclic voltammograms and bipotentiostat scans, table of refinement statistics for the cryo-EM model, and table of amino acid sequence of γPFD and designed proteins. This material is available free of charge via the Internet at http://pubs.acs.org
References
- (1).Reguera G; McCarthy KD; Mehta T; Nicoll JS; Tuominen MT; Lovley DR Extracellular Electron Transfer via Microbial Nanowires. Nature 2005, 435, 1098–1101. [DOI] [PubMed] [Google Scholar]
- (2).Malvankar NS; Vargas M; Nevin KP; Franks AE; Leang C; Kim B-C; Inoue K; Mester T; Covalla SF; Johnson JP; Rotello VM; Tuominen MT; Lovley DR Tunable Metallic-Like Conductivity in Microbial Nanowire Networks. Nat. Nanotechnol 2011, 6, 573–579. [DOI] [PubMed] [Google Scholar]
- (3).Snider RM; Strycharz-Glaven SM; Tsoi SD; Erickson JS; Tender LM Long-Range Electron Transport in Geobacter Sulfurreducens Biofilms Is Redox Gradient-Driven. Proc. Natl. Acad. Sci. U. S. A 2012, 109, 15467–15472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (4).Ing NL; El-Naggar MY; Hochbaum AI Going the Distance: Long-Range Conductivity in Protein and Peptide Bioelectronic Materials. J. Phys. Chem. B 2018, 122, 10403–10423. [DOI] [PubMed] [Google Scholar]
- (5).Wang F; Gu Y; O’Brien JP; Yi SM; Yalcin SE; Srikanth V; Shen C; Vu D; Ing NL; Hochbaum AI; Egelman EH; Malvankar NS Structure of Microbial Nanowires Reveals Stacked Hemes That Transport Electrons Over Micrometers. Cell 2019, 177, 361–369.e10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Altamura L; Horvath C; Rengaraj S; Rongier A; Elouarzaki K; Gondran C; Maçon ALB; Vendrely C; Bouchiat V; Fontecave M; Mariolle D; Rannou P; Le Goff A; Duraffourg N; Holzinger M; Forge V A Synthetic Redox Biofilm Made from Metalloprotein-Prion Domain Chimera Nanowires. Nat. Chem 2017, 9, 157–163. [DOI] [PubMed] [Google Scholar]
- (7).Ing NL; Spencer RK; Luong SH; Nguyen HD; Hochbaum AI Electronic Conductivity in Biomimetic α-Helical Peptide Nanofibers and Gels. ACS Nano 2018, 12, 2652–2661. [DOI] [PubMed] [Google Scholar]
- (8).Glover DJ; Clark DS Protein Calligraphy: A New Concept Begins to Take Shape. ACS Cent. Sci 2016, 2, 438–444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).Glover DJ; Giger L; Kim SS; Naik RR; Clark DS Geometrical Assembly of Ultrastable Protein Templates for Nanomaterials. Nat. Commun 2016, 7, 11771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Whitehead TA; Boonyaratanakornkit BB; Höllrigl V; Clark DS A Filamentous Molecular Chaperone of the Prefoldin Family from the Deep-Sea Hyperthermophile Methanocaldococcus Jannaschii. Protein Sci. 2007, 16, 626–634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Lim S; Glover DJ; Clark DS Prefoldins in Archaea. Adv. Exp. Med. Biol 2018, 1106, 11–23. [DOI] [PubMed] [Google Scholar]
- (12).Glover DJ; Clark DS Oligomeric Assembly Is Required for Chaperone Activity of the Filamentous γ-Prefoldin. FEBS J. 2015, 282, 2985–2997. [DOI] [PubMed] [Google Scholar]
- (13).Glover Dominic J; Giger Lars; Kim Jihyun R; Clark Douglas S. Engineering Protein Filaments with Enhanced Thermostability for Nanomaterials. Biotechnol. J 2012, 8, 228–236. [DOI] [PubMed] [Google Scholar]
- (14).Glover DJ; Lim S; Xu D; Sloan NB; Zhang Y; Clark DS Assembly of Multicomponent Protein Filaments Using Engineered Subunit Interfaces. ACS Synth. Biol 2018, 7, 2447–2456. [DOI] [PubMed] [Google Scholar]
- (15).Egelman EH A Robust Algorithm for the Reconstruction of Helical Filaments Using Single-Particle Methods. Ultramicroscopy 2000, 85, 225–234. [DOI] [PubMed] [Google Scholar]
- (16).Scheres SHW RELION: Implementation of a Bayesian Approach to Cryo-EM Structure Determination. J. Struct. Biol 2012, 180, 519–530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).Ohtaki A; Kida H; Miyata Y; Ide N; Yonezawa A; Arakawa T; Iizuka R; Noguchi K; Kita A; Odaka M; Miki K; Yohda M Structure and Molecular Dynamics Simulation of Archaeal Prefoldin: The Molecular Mechanism for Binding and Recognition of Nonnative Substrate Proteins. J. Mol. Biol 2008, 376, 1130–1141. [DOI] [PubMed] [Google Scholar]
- (18).Subramaniam S; Earl LA; Falconieri V; Milne JL; Egelman EH Resolution Advances in Cryo-EM Enable Application to Drug Discovery. Curr. Opin. Struct. Biol 2016, 41, 194–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).Afonine PV; Klaholz BP; Moriarty NW; Poon BK; Sobolev OV; Terwilliger TC; Adams PD; Urzhumtsev A New Tools for the Analysis and Validation of Cryo-EM Maps and Atomic Models. Acta Crystallogr., Sect. D: Struct. Biol 2018, 74, 814–840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (20).Holm L; Laakso LM Dali Server Update. Nucleic Acids Res. 2016, 44, W351–W355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (21).Siegert R; Leroux MR; Scheufler C; Hartl FU; Moarefi I Structure of the Molecular Chaperone Prefoldin: Unique Interaction of Multiple Coiled Coil Tentacles with Unfolded Proteins. Cell 2000, 103, 621–632. [DOI] [PubMed] [Google Scholar]
- (22).Krissinel E; Henrick K Inference of Macromolecular Assemblies from Crystalline State. J. Mol. Biol 2007, 372, 774–797. [DOI] [PubMed] [Google Scholar]
- (23).Trent JD; Kagawa HK; Yaoi T; Olle E; Zaluzec NJ Chaperonin Filaments: The Archaeal Cytoskeleton? Proc. Natl. Acad. Sci. U. S. A 1997, 94, 5383–5388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Wagstaff J; Löwe J Prokaryotic Cytoskeletons: Protein Filaments Organizing Small Cells. Nat. Rev. Microbiol 2018, 16, 187–201. [DOI] [PubMed] [Google Scholar]
- (25).Whitehead TA; Je E; Clark DS Rational Shape Engineering of the Filamentous Protein Gamma Prefoldin through Incremental Gene Truncation. Biopolymers 2009, 91, 496–503. [DOI] [PubMed] [Google Scholar]
- (26).Fitzpatrick AWP; Falcon B; He S; Murzin AG; Murshudov G; Garringer HJ; Crowther RA; Ghetti B; Goedert M; Scheres SHW Cryo-EM Structures of Tau Filaments from Alzheimer’s Disease Brain. Nature 2017, 547, 185–190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (27).Tanaka K; Takeda S; Mitsuoka K; Oda T; Kimura-Sakiyama C; Maéda Y; Narita A Structural Basis for Cofilin Binding and Actin Filament Disassembly. Nat. Commun 2018, 9, 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (28).Ozawa K; Tsapin AI; Nealson KH; Cusanovich MA; Akutsu H Expression of a Tetraheme Protein, Desulfovibrio Vulgaris Miyazaki F Cytochrome c3, in Shewanella Oneidensis MR-1. Appl. Environ. Microbiol 2000, 66, 4168–4171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (29).Zakeri B; Fierer JO; Celik E; Chittock EC; Schwarz-Linek U; Moy VT; Howarth M Peptide Tag Forming a Rapid Covalent Bond to a Protein, through Engineering a Bacterial Adhesin. Proc. Natl. Acad. Sci. U. S. A 2012, 109, E690–697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (30).Lim S; Jung GA; Muckom RJ; Glover DJ; Clark DS Engineering Bioorthogonal Protein-Polymer Hybrid Hydrogel as a Functional Protein Immobilization Platform. Chem. Commun. (Camb.) 2018, 55, 806–809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (31).Winkler JR; Gray HB Long-Range Electron Tunneling. J. Am. Chem. Soc 2014, 136, 2930–2939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (32).Wilting J; Van Buuren KJH; Braams R; Van Gelder BF The Mechanism of Reduction of Cytochrome c as Studied by Pulse Radiolysis. Biochim. Biophys. Acta, Bioenerg 1975, 376, 285–297. [DOI] [PubMed] [Google Scholar]
- (33).Gouterman M Spectra of Porphyrins. J. Mol. Spectrosc 1961, 6, 138–163. [Google Scholar]
- (34).Ing NL; Nusca TD; Hochbaum AI Geobacter Sulfurreducens Pili Support Ohmic Electronic Conduction in Aqueous Solution. Phys. Chem. Chem. Phys 2017, 19, 21791–21799. [DOI] [PubMed] [Google Scholar]
- (35).Yates MD; Golden JP; Roy J; Strycharz-Glaven SM; Tsoi S; Erickson JS; El-Naggar MY; Barton SC; Tender LM Thermally Activated Long Range Electron Transport in Living Biofilms. Phys. Chem. Chem. Phys 2015, 17, 32564–32570. [DOI] [PubMed] [Google Scholar]
- (36).Li X; Mooney P; Zheng S; Booth CR; Braunfeld MB; Gubbens S; Agard DA; Cheng Y Electron Counting and Beam-Induced Motion Correction Enable Near-Atomic-Resolution Single-Particle Cryo-EM. Nat. Methods 2013, 10, 584–590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (37).Mindell JA; Grigorieff N Accurate Determination of Local Defocus and Specimen Tilt in Electron Microscopy. J. Struct. Biol 2003, 142, 334–347. [DOI] [PubMed] [Google Scholar]
- (38).Tang G; Peng L; Baldwin PR; Mann DS; Jiang W; Rees I; Ludtke SJ EMAN2: An Extensible Image Processing Suite for Electron Microscopy. J. Struct. Biol 2007, 157, 38–46. [DOI] [PubMed] [Google Scholar]
- (39).Pettersen EF; Goddard TD; Huang CC; Couch GS; Greenblatt DM; Meng EC; Ferrin TE UCSF Chimera--A Visualization System for Exploratory Research and Analysis. J. Comput. Chem 2004, 25, 1605–1612. [DOI] [PubMed] [Google Scholar]
- (40).Yang J; Yan R; Roy A; Xu D; Poisson J; Zhang Y The I-TASSER Suite: Protein Structure and Function Prediction. Nat. Methods 2015, 12, 7–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (41).Wood CW; Woolfson DN CCBuilder 2.0: Powerful and Accessible Coiled-Coil Modeling. Protein Sci. 2018, 27, 103–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (42).Williams CJ; Headd JJ; Moriarty NW; Prisant MG; Videau LL; Deis LN; Verma V; Keedy DA; Hintze BJ; Chen VB; Jain S; Lewis SM; Arendall WB; Snoeyink J; Adams PD; Lovell SC; Richardson JS; Richardson DC MolProbity: More and Better Reference Data for Improved All-Atom Structure Validation. Protein Sci. 2018, 27, 293–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (43).Li L; Fierer JO; Rapoport TA; Howarth M Structural Analysis and Optimization of the Covalent Association between SpyCatcher and a Peptide Tag. J. Mol. Biol 2014, 426, 309–317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (44).Takayama Y; Werbeck ND; Komori H; Morita K; Ozawa K; Higuchi Y; Akutsu H Strategic Roles of Axial Histidines in Structure Formation and Redox Regulation of Tetraheme Cytochrome C3. Biochemistry 2008, 47, 9405–9415. [DOI] [PubMed] [Google Scholar]
- (45).Gibson DG; Young L; Chuang R-Y; Venter JC; Hutchison CA; Smith HO Enzymatic Assembly of DNA Molecules up to Several Hundred Kilobases. Nat. Methods 2009, 6, 343–345. [DOI] [PubMed] [Google Scholar]
- (46).Arslan E; Schulz H; Zufferey R; Künzler P; Thöny-Meyer L Overproduction of the Bradyrhizobium Japonicum C-Type Cytochrome Subunits of the Cbb3 Oxidase in Escherichia Coli. Biochem. Biophys. Res. Commun 1998, 251, 744–747. [DOI] [PubMed] [Google Scholar]
- (47).Zor T; Selinger Z Linearization of the Bradford Protein Assay Increases Its Sensitivity: Theoretical and Experimental Studies. Anal. Biochem 1996, 236, 302–308. [DOI] [PubMed] [Google Scholar]
- (48).Schneider CA; Rasband WS; Eliceiri KW NIH Image to ImageJ: 25 Years of Image Analysis. Nat. Methods 2012, 9, 671–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


