Abstract
A large and significant portion of eukaryotic transcriptomes consists of noncoding RNAs (ncRNAs) that have minimal or no protein-coding capacity but are functional. Diverse ncRNAs, including both small RNAs and long ncRNAs (lncRNAs), play essential regulatory roles in almost all biological processes by modulating gene expression at the transcriptional and posttranscriptional levels. In this review, we summarize the current knowledge of plant small RNAs and lncRNAs, with a focus on their biogenesis, modes of action, local and systemic movement, and functions at the nexus of plant development and environmental responses. The complex connections among small RNAs, lncRNAs, and small peptides in plants are also discussed, along with the challenges of identifying and investigating new classes of ncRNAs.
Keywords: microRNA, siRNA, lncRNA, circRNA, plant development, stress response
1. INTRODUCTION
Although up to 90% of the eukaryotic genome is transcribed into RNA, only approximately 2% of transcribed RNAs give rise to protein products (Pauli et al. 2011, Rai et al. 2018). The remaining transcriptome comprises noncoding RNAs (ncRNAs) arising from what were previously considered silent regions, such as intergenic regions, repetitive sequences, transposons, and pseudogenes. Moreover, transcripts from these regions were initially regarded as transcriptional noise due to their lack of or minimal protein-coding capacity and due to poorly conserved sequences (Ariel et al. 2015, Pauli et al. 2011). However, computational analysis and experimental validation in 15 diverse flowering plant species predict approximately 40% of intergenic transcribed regions and ncRNAs to be similar in features to protein-coding or RNA genes and to likely be functional (Lloyd et al. 2018).
ncRNAs can be classified into small RNAs [18–30 nucleotides (nt)], medium-sized ncRNAs (31–200 nt), and long ncRNAs (lncRNAs) (>200 nt). The vital roles of small RNAs, particularly microRNAs (miRNAs), in diverse biological processes such as plant growth, development, and hormone and stress responses are now being elucidated in detail (Chen 2009, D’Ario et al. 2017, Martinez & Köhler 2017, Tang & Chu 2017). Studies have also shed light on the essential regulatory functions exerted by lncRNAs (Böhmdorfer & Wierzbicki 2015, Pauli et al. 2011, Zhang & Chen 2013). In this review, we summarize recent progress on plant small RNAs and lncRNAs, with an emphasis on their biogenesis, modes of action, non–cell autonomy, and conserved and diverse functions in different plant species.
2. SMALL RNAS
To date, hundreds of thousands of small RNAs have been identified in diverse plant species. Small RNAs, despite their tiny size, play important roles in myriad intracellular processes by regulating the expression of target genes at either the transcriptional or the posttranscriptional level (Chen 2009, D’Ario et al. 2017, Martinez & Köhler 2017, Tang & Chu 2017). Three major types of small RNAs are present in plants: miRNAs, transposable element (TE)-derived small interfering RNAs (siRNAs), and phased siRNAs (phasiRNAs). These small RNAs differ in terms of their precursors, biogenesis, and modes of action.
2.1. MicroRNAs
miRNAs constitute a major class of small RNAs in plants and impact various aspects of plant development and stress responses by posttranscriptionally regulating gene expression.
2.1.1. Biogenesis of microRNAs.
miRNA biogenesis is a multistep process including transcription, processing, modification, and assembly of the RNA-induced silencing complex (RISC) (Rogers & Chen 2013, Yu et al. 2017) (Figure 1). miRNAs are encoded by MIR genes, which are mainly located in intergenic regions and transcribed by RNA POLYMERASE II (Pol II) to give rise to long, single-stranded, 5′-capped and 3′-polyadenylated primary miRNAs (pri-miRNAs). An RNase III family DICER-LIKE (DCL) enzyme, usually DCL1, assisted by HYPONASTIC LEAVES 1 (HYL1) and SERRATE (SE), sequentially processes a pri-miRNA first into a precursor miRNA (pre-miRNA) and then into a short miRNA/miRNA* duplex. The miRNA/miRNA* duplex undergoes 2′-O-methylation on both 3′ terminal riboses catalyzed by the methyltransferase HUA ENHANCER 1 (HEN1). In most cases, miRNAs are stabilized by the 3′ methylation. Loss of the protective methyl group usually leads to 3′ uridylation (i.e., the addition of one to several U residues to the 3′ end) and subsequent degradation (Sanei & Chen 2015). Nevertheless, some unmethylated and uridylated miRNAs acquire the ability to trigger the generation of secondary siRNAs; examples include miR171 in Arabidopsis and miR1510 in soybean (Fei et al. 2018, Tu et al. 2015).
Figure 1.
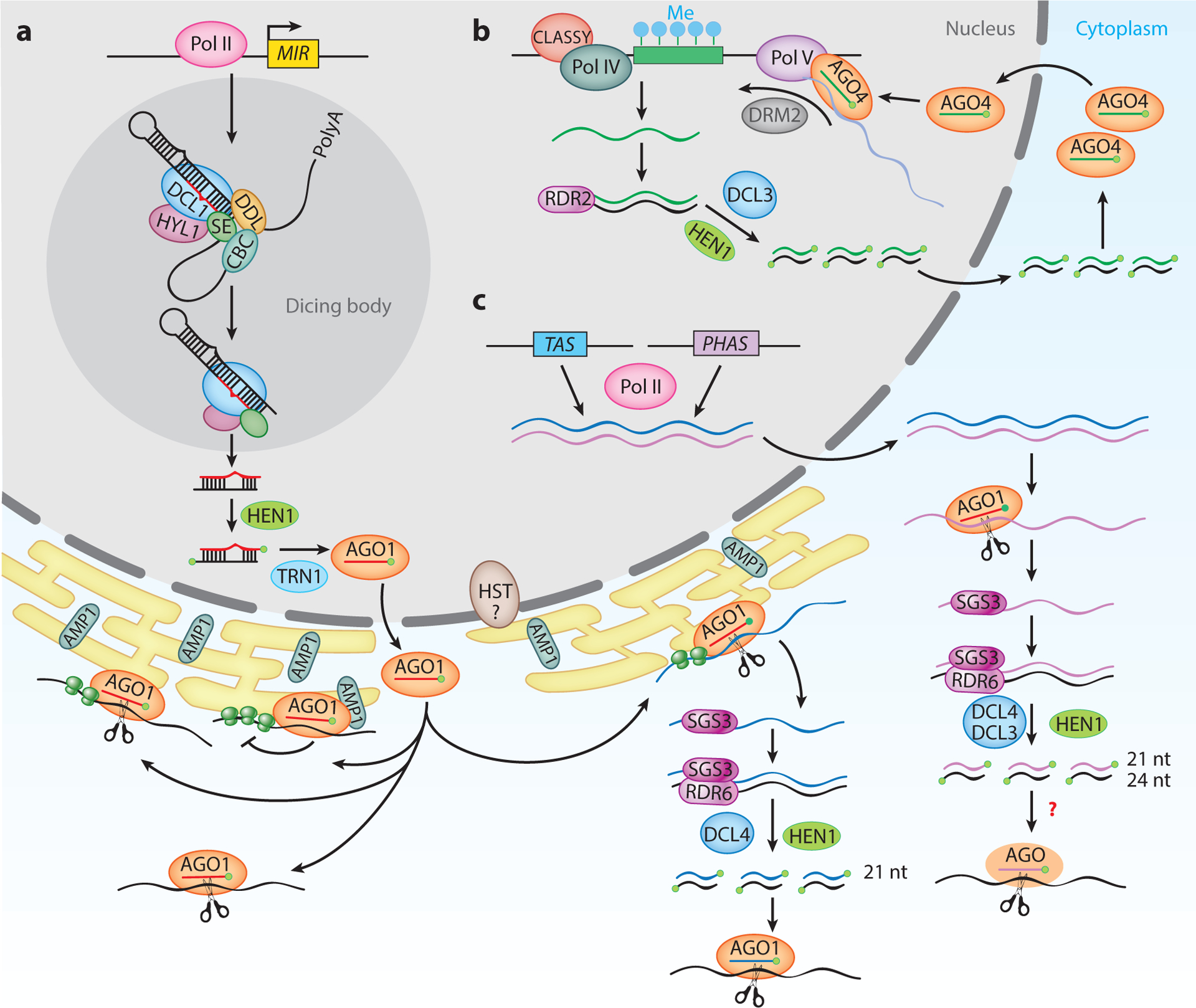
Biogenesis and modes of action of plant small RNAs. (a) A MIR gene is transcribed into a pri-miRNA, which is sequentially processed first into a pre-miRNA and then into a miRNA/miRNA* duplex. The duplex is methylated by HEN1, and the miRNA strand is loaded into AGO1 in the nucleus. The miRNA-AGO1 complex is transported to the cytoplasm and regulates target gene expression through transcript cleavage and/or translation repression. (b) Pol IV generates single-stranded siRNA precursors, which are converted into dsRNAs and processed into 24-nt siRNA duplexes.Methylated siRNAs are loaded into AGO4 in the cytoplasm and are transported to the nucleus, followed by the recruitment of these siRNA-AGO4 complexes to Pol V transcripts. The subsequent recruitment of DRM2 catalyzes DNA methylation at RdDM target loci. (c) TAS or PHAS loci are transcribed into single-stranded RNAs that are targeted by an miRNA-AGO1/7 complex. The 5′ or 3′ cleavage fragment is protected by SGS3 and converted into dsRNA by RDR6. DCL proteins process these dsRNAs into 21- or 24-nt phasiRNAs. The 21-nt tasiRNAs, which are phasiRNAs from TAS loci, are primarily loaded into AGO1 and guide transcript cleavage of their targets. Abbreviations: AGO, ARGONAUTE; AMP1, ALTERED MERISTEM PROGRAM 1; CBC, CAP-BINDING COMPLEX; DCL, DICER-LIKE; DDL, DAWDLE; DRM2, DOMAINS REARRANGED METHYLASE 2; dsRNA, double-stranded RNA; HEN1, HUA ENHANCER 1; HST, HASTY; HYL1, HYPONASTIC LEAVES 1; Me, methylated; phasiRNA, phased siRNA; Pol, RNA polymerase; pre-miRNA, precursor miRNA; pri-miRNA, primary miRNA; RdDM, RNA-directed DNA methylation; RDR2/6, RNA-DEPENDENT RNA POLYMERASE 2/6; SE, SERRATE; SGS3, SUPPRESSOR OF GENE SILENCING 3; siRNA, small interfering RNA; tasiRNA, trans-acting siRNA; TRN1, TRANSPORTIN 1.
Most mature miRNA strands are incorporated into ARGONAUTE 1 (AGO1) in the nucleus, followed by the removal of the miRNA* strand and the export of the miRNA-AGO1 complex to the cytoplasm, where miRNAs guide posttranscriptional gene silencing (Baumberger & Baulcombe 2005, Bologna et al. 2018). Beyond the core components mentioned above, many other miRNA biogenesis pathway factors have been identified (Yu et al. 2017), underscoring that the biogenesis of miRNAs is precisely controlled.
2.1.2. Modes of action of plant microRNAs.
Plant miRNAs repress target gene expression through two major modes of action: transcript cleavage and translation repression (Chen 2009, Yu et al. 2017) (Figure 1). miRNAs recognize target mRNAs via sequence complementarity and direct AGO1 to cleave the target mRNA at the phosphodiester bond corresponding to nucleotides 10 and 11 of the miRNA. miRNAs also inhibit the translation of target mRNAs with the aid of the endoplasmic reticulum (ER)-associated protein ALTERED MERISTEM PROGRAM 1 (AMP1) (Li et al. 2013). Transcript cleavage was originally thought to be the predominant mode of action for plant miRNAs due to the high degree of sequence complementarity between miRNAs and targets (Chen 2009). However, sequence complementarity is not the factor dictating the mode of action of plant miRNAs, as supported by evidence that miRNA targets with nearly perfect sequence complementarity to the corresponding miRNAs are regulated by cleavage and translation repression. For example, SQUAMOSA PROMOTER BINDING PROTEIN-LIKE 3 (SPL3) (targeted by miR156), PHABULOSA (PHB) and REVOLUTA (targeted by miR165/166), SCARECROW-LIKE PROTEIN 4 (SCL4) (targeted by miR171), APETALA 2 (AP2) (targeted by miR172), and COPPER/ZINC SUPEROXIDE DISMUTASE 1 (CSD1) and CSD2 (targeted by miR398) are subjected to both transcript cleavage and translation repression (Yu et al. 2017). Moreover, most miRNAs are enriched on membrane-bound polysomes (Li et al. 2016), suggesting that miRNA-mediated transcript cleavage and translation repression take place on the ER. Nevertheless, transcript cleavage may also occur independently of polysomes in the cytosol.
2.1.3. Functions of microRNAs in plant development.
Plant miRNAs target many transcription factors that participate in various regulatory pathways (Figure 2). A single miRNA or an miRNA family often targets multiple members of a gene family, and evolutionarily conserved miRNAs among related plant species also tend to have conserved targets. For example, members of the conserved miR156 family regulate vegetative phase transition by modulating the expression of SPL genes in diverse flowering plants (Wang 2014). miR172 regulates floral development and flowering time in Arabidopsis through the repression of AP2 genes (Aukerman & Sakai 2003, Chen 2004). miR172’s role in flowering control has been reported in a variety of plant species, including maize, barley, soybean, and rice (Tang & Chu 2017). In Arabidopsis and maize, miR164 and its target, plant-specific transcription factor NAC DOMAIN CONTAINING PROTEIN 1 (NAC1), are involved in the formation of lateral roots (Guo et al. 2005, J. Li et al. 2012). Similarly, overexpressing miR164 in potato under osmotic stress causes reduced expression of NAC262 and limits the number of lateral roots (Zhang et al. 2018).
Figure 2.
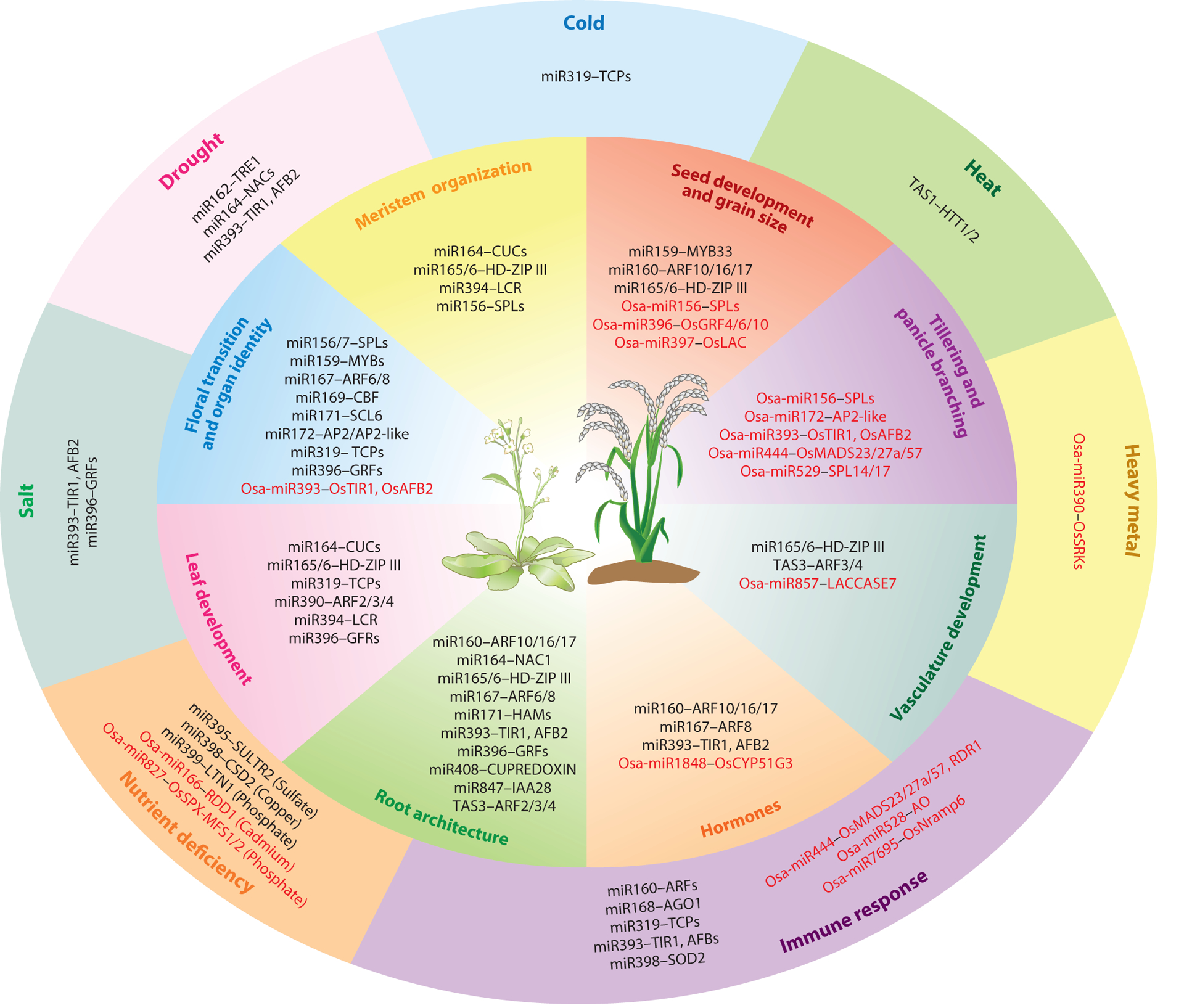
Functions of miRNAs in plant development and stress responses and an overview of the current understanding of miRNA-mediated regulation during development (inner circle) and responses to biotic and abiotic stresses (outer circle) in Arabidopsis and rice. Red font indicates miRNA-target modules that act in rice, but not in Arabidopsis.
In addition to conserved functions, distinct roles of the same miRNA-target modules have been uncovered. For instance, the miR156-SPL module regulates tillering in switchgrass, controls ear development and grain formation in maize, and participates in grain size control and panicle branching in rice (D’Ario et al. 2017, Tang & Chu 2017). Thus, miR156-SPL has a diversified regulatory function in axillary meristem initiation in monocots. Vegetative phase transition in barley requires the miR171-SCL module to activate the miR156-SPL pathway, and this appears to be a monocot-specific function of miR171-SCL (Curaba et al. 2013).
The functions of different miRNAs can also converge on one biological event. In Arabidopsis, the shoot apical meristem (SAM) is maintained by the miR165/166 and miR394 families, which restrict the expression of the HOMEODOMAIN LEUCINE ZIPPER III (HD-ZIP III) and LEAF CURLING RESPONSIVENESS (LCR) genes, respectively (D’Ario et al. 2017). miR165/166 also regulates leaf polarity together with miR390, which triggers the production of siRNAs that target several AUXIN RESPONSE FACTOR (ARF) genes (Chitwood & Timmermans 2010, Liu et al. 2009). Two miRNA families, miR156/157 and miR172, cooperate to control the juvenile-to-adult transition and flowering (Wang et al. 2009, Wu et al. 2009). In tomato, both miR156 and miR319 regulate the floral transition in response to gibberellin signaling (Silva et al. 2018). In rice, grain size is regulated by miR156, miR396, and miR397, and tillering is controlled by miR156, miR393, and miR444 (Tang & Chu 2017). Collectively, these findings demonstrate the integrated functionalities of unrelated miRNAs.
Aside from conserved miRNAs, species-specific miRNAs constitute a large proportion of plant miRNAs. miR528, a monocot-specific miRNA, is induced by nitrogen luxury conditions in maize and regulates lodging resistance by targeting the lignin biosynthesis genes ZmLACCASE 3 (ZmLAC3) and ZmLAC5 (Sun et al. 2018). In rice, the monocot-specific miR444 controls tillering (Guo et al. 2013) and participates in antiviral defense (H. Wang et al. 2016) by targeting the three MIKCC-type MADS-box genes OsMADS23, OsMADS27a, and OsMADS57; these genes repress the expression of RNA-DEPENDENT RNA POLYMERASE 1 (RDR1), a key component of the antiviral RNA silencing pathway (Garcia-Ruiz et al. 2010, H. Wang et al. 2016).
2.2. Transposable Element–Derived Small Interfering RNAs
Most endogenous siRNAs in plants are heterochromatic siRNAs derived from repeats and TEs. They are involved in transcriptional gene silencing by directing DNA methylation and/or histone methylation through a process known as RNA-directed DNA methylation (RdDM) (Du et al. 2015, Matzke et al. 2015) (Figure 1).
2.2.1. Transposable element–derived small interfering RNAs in genome stability control.
RdDM involves siRNA biogenesis from TEs and repeats as well as siRNA-guided DNA methylation at the source loci and homologous sites. The plant-specific RNA polymerase IV (Pol IV) is recruited by CLASSY chromatin remodeling factors to RdDM loci to generate single-stranded siRNA precursors (Blevins et al. 2015, S. Li et al. 2015, Zhai et al. 2015a, Zhou et al. 2018). RNA-DEPENDENT RNA POLYMERASE 2 (RDR2) converts these precursors into double-stranded RNAs (dsRNAs) that are subsequently processed by DCL3, producing 24-nt mature siRNAs that are preferentially loaded into AGO4 (Law & Jacobsen 2010). RNA polymerase V (Pol V), another plant-specific RNA polymerase, generates noncoding transcripts at the same RdDM loci, thereby recruiting the siRNA-AGO4 complex through sequence complementarity (Du et al. 2015). This activity promotes the recruitment of DNA REARRANGED METHYLASE 2 (DRM2) to trigger DNA methylation at RdDM loci (Du et al. 2015, Law & Jacobsen 2010).
The recruitment of Pol IV and Pol V to DNA is enhanced by existing repressive chromatin features such as histone H3 lysine 9 methylation and DNA methylation (Du et al. 2015). Thus, the above-described RdDM mechanism likely maintains existing heterochromatic features. Non-canonical RdDM, which involves 21–22-nt siRNAs likely generated by Pol II, may be responsible for the initiation of DNA methylation (Cuerda-Gil & Slotkin 2016).
RdDM (including noncanonical RdDM) is responsible for de novo methylation in all sequence contexts (CG, CHG, and CHH) as well as for methylation maintenance at CHH contexts in short TEs and at the edges of long TEs. In Arabidopsis, transposon derepression is commonly observed in RdDM-defective mutants, but transposition occurs only at a few loci (Ito et al. 2011). In maize, with a genome of approximately 85% TEs, methylated CHH islands often occur at the boundaries between active genes and nearby silent TEs. Loss of CHH methylation is often accompanied by CG and CHG hypomethylation at the adjacent TEs, suggesting that methylated CHH islands reinforce TE silencing by preventing the spread of active euchromatin to heterochromatin (Q. Li et al. 2015).
2.2.2. Functions of transposable element–derived small interfering RNAs in reproduction and hybridization.
Reproduction is a key period when TE-derived siRNAs monitor genome compatibility and dosage. In developing pollen, certain TEs are demethylated and reactivated in the vegetative nucleus and produce 21-, 22-, and 24-nt siRNAs that are thought to move into sperm cells (Calarco et al. 2012, Martínez et al. 2016, Slotkin et al. 2009). However, overall levels of CHH methylation are greatly reduced in sperm cells, probably owing to the low levels of expression of the RdDM machinery (Calarco et al. 2012). In the female gametophyte, the central cell and the egg cell are fertilized by sperm cells to produce the endosperm and the embryo, respectively. CHH methylation levels increase during embryogenesis (Bouyer et al. 2017, Jullien et al. 2012, Martínez et al. 2016), suggesting that the paternal genome gains CHH methylation after fertilization. The siRNAs that guide CHH methylation in the embryo may come from two sources: sperm and endosperm. The maternal genome in the endosperm is undermethylated at numerous loci, probably due to active demethylation in the central cell (Ibarra et al. 2012, Martinez & Köhler 2017). The demethylation leads to reactivation of TEs from the maternal genome in the endosperm and the production of 24-nt siRNAs. The siRNAs are thought to move into the embryo to guide DNA methylation (Martinez & Köhler 2017). In contrast, 21–22-nt siRNAs in the sperm serve as a quantitative output of paternal genome dosage in the endosperm. Such siRNAs from some loci interfere with RdDM in the endosperm, perhaps by competing with 24-nt siRNAs for Pol V–derived scaffold transcripts (Martinez et al. 2018). Thus, TE-derived siRNAs likely mediate interactions between maternal and paternal genomes. Surprisingly, a recent study identified a group of 23–24-nt meiocyte-specific siRNAs (ms-sRNAs) that were significantly enriched in genic regions rather than TEs. Moreover, unlike siRNAs in somatic cells, these ms-sRNAs are positively correlated with gene expression during reproductive development in a fashion unrelated to DNA methylation, implying a novel role of siRNAs in meiocytes (Huang et al. 2019).
Perhaps owing to the central role of TE-derived siRNAs in genome interactions, these siRNAs contribute to the transgressive phenotypes of plant hybrids, i.e., hybrid vigor and novel phenotypes that transgress the parental range and are inherited stably in subsequent generations. For example, in tomato introgression lines generated from Solanum lycopersicum (cultivated tomato) and Solanum pennellii (wild tomato), several differentially expressed siRNA loci (DSR loci) were identified at locations where the siRNA abundance was either higher or lower than in the parental lines, along with corresponding hypermethylation or hypomethylation of their target DNAs (Shivaprasad et al. 2014). In addition, RdDM mediated by siRNAs at the DSR loci seems to contribute to the paramutation-like phenotype in tomato hybrids, wherein the epigenetic modification associated with a silent allele is transferred to an active allele (Gouil & Baulcombe 2018). Similarly, nonadditive expression of siRNAs and consequent DNA methylation result in transgressive phenotypes in interspecific hybrids or allotetraploids of cotton (Song et al. 2017).
2.3. Phased Small Interfering RNAs
PhasiRNAs constitute another class of endogenous siRNAs. PhasiRNAs are generated from miRNA target transcripts and may have their own targets in trans.
2.3.1. Models of phased small interfering RNA biogenesis.
Although mRNA cleavage fragments generated by miRISCs are typically subjected to rapid degradation, a small proportion of them are further processed into secondary siRNAs in a phenomenon that is widespread and mechanistically conserved in plants (Chen 2009, Rogers & Chen 2013, Yu et al. 2017). After AGO-mediated slicing, SUPPRESSOR OF GENE SILENCING 3 (SGS3) associates with the 5′ or 3′ cleavage fragments and recruits RDR6, which converts the single-stranded cleavage fragments into dsRNAs (Figure 1). DCL proteins then dice these dsRNAs into a series of 21- or 24-nt siRNAs, termed phasiRNAs, which are arranged head to tail and are in phase relative to the miRNA cleavage sites.
Trans-acting siRNAs (tasiRNAs) are a class of DCL4-dependent 21-nt phasiRNAs generated from noncoding TAS transcripts (Chen 2009, Fei et al. 2013). In addition to TAS loci, phasiRNAs are produced from protein-coding genes, such as NUCLEOTIDE-BINDING LEUCINE-RICH REPEAT (NB-LRR) and PENTATRICOPEPTIDE REPEAT (PPR) genes in dicots and lncRNAs from PHAS (phasiRNA-generating) loci in monocots (Chen 2009, Fei et al. 2013, Yang et al. 2018). In most cases, phasiRNAs are trigged by a 22-nt miRNA with only one binding site in the target transcript—the so-called one-hit model (Chen et al. 2010, Fei et al. 2013). In contrast, the two-hit model entails two miRNA binding sites present in the target transcript, as exemplified by TAS3 (Axtell et al. 2006). Besides the number of miRNA binding sites and the length of the miRNA trigger, the following factors also influence phasiRNA biogenesis: AGO1 slicer activity, the asymmetric bulge within the miRNA/miRNA* duplex, the degree of complementarity of the miRNA/target duplex, and the position of the miRNA binding site relative to the short open reading frame (ORF) of TAS transcripts (Yu et al. 2017). Recently, TAS transcripts were found to be associated with membrane-bound polysomes (Li et al. 2016), suggesting that phasiRNA biogenesis from TAS may be initiated on the rough ER. The finding further implies a potential relationship between translation and phasiRNA biogenesis.
2.3.2. Biological functions of phased small interfering RNAs.
tasiRNAs are the best characterized phasiRNAs in terms of biological functions. In Arabidopsis, TAS1 and TAS2 are targeted by miR173 to produce tasiRNAs, some of which can target PPR transcripts to cause further production of phasiRNAs (Chen et al. 2010, Fei et al. 2013). TAS1 tasiRNAs also target the heat stress transcription factor genes HEAT-INDUCED TAS1 TARGET 1 (HTT1) and HTT2 to regulate plant thermotolerance (Li & He 2014). TAS3 tasiRNAs are triggered by the miR390-AGO7 complex (Axtell et al. 2006) and target ARF family members to regulate diverse biological processes, including embryo development, developmental transitions, leaf morphology, flower and root architecture, stress responses, and phytohormone cross talk (D’Ario et al. 2017, Xia et al. 2016). TAS4 tasiRNAs are induced by miR828 from the TAS4 locus and repress MYB genes, including MYB113, PRODUCTION OF ANTHOCYANIN PIGMENT 1 (PAP1), and PAP2, which regulate anthocyanin biogenesis (Luo et al. 2012). In contrast to TAS1–TAS4 in Arabidopsis, the biogenesis and functions of TAS5–TAS10 are ill defined. TAS5 was reported in tomato and does not exhibit all of the characteristics of a TAS locus; specifically, it resembles a protein-coding transcript, and the tasiRNAs may function in cis (F. Li et al. 2012). TAS6 from moss was found adjacent to TAS3 loci, and the production of tasiRNAs is triggered by miR156 or miR529 (Arif et al. 2012, Cho et al. 2012). Interestingly, TAS6A and TAS3A share the same primary transcript, and the two tasiRNA-generating regions are separated by only a small central intron (Cho et al. 2012), indicating linked biogenesis of TAS6A and TAS3A tasiRNAs. TAS7–TAS10 were identified in grapevine and tomato (Zhang et al. 2012, Zuo et al. 2017), but their biogenesis pathways and functions have not been described.
In dicots, NB-LRR genes exist widely in diverse plant species and represent the largest gene family that produces phasiRNAs (Fei et al. 2015, Zhai et al. 2011). NB-LRR genes are targeted by 22-nt miRNAs, such as miR2118 in Medicago truncatula and miR472 in Arabidopsis, and give rise to 21-nt DCL4-dependent phasiRNAs that may in turn regulate NB-LRR transcripts at the posttranscriptional level, perhaps both in cis and in trans (Cai et al. 2018, Zhai et al. 2011). In soybean, many NB-LRR genes are preferentially expressed in nodules and targeted by miR482, miR1507, and miR1510 (Fei et al. 2013). PPR genes are another large phasiRNA-generating gene family. In addition to being regulated by TAS1/2 tasiRNAs, some PPR transcripts are directly targeted by miRNAs such as miR7122 and miR161 (Hou et al. 2018, Xia et al. 2013). PPR phasiRNAs contribute to pathogen defense by potentially silencing Phytophthora transcripts during infection (Hou et al. 2018).
In monocots, miR2118 targets noncoding transcripts arising from PHAS loci and generates 21-nt phasiRNAs during anther development (Fei et al. 2013, Zhai et al. 2015b). In rice, 21-nt phasiRNAs derived from more than 700 PHAS loci are associated with the germline-specific AGO protein MEIOSIS ARRESTED AT LEPTOTENE 1 (MEL1), which has key functions in the development of premeiotic germ cells and the progression of meiosis (Komiya et al. 2014). In maize, mutation of OUTER CELL LAYER 4 (OCL4) leads to anther defects and male sterility, accompanied by a lack of 21-nt phasiRNAs (Zhai et al. 2015b). In addition to 21-nt phasiRNAs, a class of 24-nt meiotic phasiRNAs triggered by miR2275 and processed by DCL5 is present in both male and female reproductive organs in rice and maize, suggesting that they may be involved in male and female germinal development (Kakrana et al. 2018, Zhai et al. 2015b). The grass phasiRNAs resemble mammalian PIWI-interacting RNAs (piRNAs) in that they lack sequence conservation in related species and that they are specifically found in the germline (Kakrana et al. 2018, Patel et al. 2018).
3. PLANT LONG NONCODING RNAS
RNA transcripts longer than 200 nt that have no coding potential or lack an ORF encoding >100 amino acids are classified as lncRNAs. Numerous lncRNAs have been identified in a variety of eukaryotes, including plants. There is increasing evidence that lncRNAs are essential modulators of a wide range of biological processes and function through diverse mechanisms.
3.1. Processing and Regulatory Features of Plant Long Noncoding RNAs
lncRNAs are characterized by their wide-ranging types and origins. They arise from intergenic regions [long intergenic ncRNAs (lincRNAs)], intronic regions [intronic ncRNAs (incRNAs)], and coding regions [natural antisense transcripts (NATs)] and can be subdivided according to their processing mechanisms (Chekanova 2015). lincRNAs, incRNAs, and NATs are conventional linear lncRNAs. Circular RNAs (circRNAs) are another class of lncRNAs and mostly arise from coding regions or intronic regions. Each lncRNA type is produced via specific mechanisms and has distinct regulatory features in cis or in trans.
3.1.1. Linear long noncoding RNAs.
lincRNAs, incRNAs, and NATs are linear lncRNAs that constitute the majority of annotated lncRNAs, most of which are transcribed by Pol II. They also have typical mRNA-like features, with a 5′ m7G cap and a 3′ poly (A) tail; thus, they are processed as mRNA mimics (Wu et al. 2017). However, these molecules have a lower degree of conservation, lower abundance, more tissue-specific expression, and lower splicing efficiency than mRNAs (Ulitsky & Bartel 2013).
lncRNAs may regulate the expression of neighboring genes in cis and that of distant genes in trans (Liu et al. 2015, Yang et al. 2014). The in cis–regulatory feature of NATs was first globally implicated by a study that examined transcriptomic responses to light in Arabidopsis (H. Wang et al. 2014). This study discovered the widespread existence of NATs (approximately 70% of annotated mRNAs in Arabidopsis have NATs) and the potential roles of NATs in mediating histone modifications at the corresponding gene loci. It was subsequently reported that NAT expression is often positively correlated with that of their cognate sense genes (Zhao et al. 2018). For example, the NAT MAS regulates MADS AFFECTING FLOWERING 4 (MAF4) in cis (Zhao et al. 2018) (Figure 3). Other examples include the cis regulation of PHOSPHATE1;2 (PHO1;2) by NATpho1;2 (Jabnoune et al. 2013), CYCLING DOF FACTOR 5 (CDF5) by FLORE (Henriques et al. 2017), FLOWERING LOCUS C (FLC) by COOLAIR (Chen & Penfield 2018, Marquardt et al. 2014), and LEUCINE-RICH REPEAT RECEPTOR KINASE (LRK) by LAIR (Y. Wang et al. 2018) (Figure 3). NATs may also suppress the expression of their cognate sense genes. For example, Pol II read-through of the lncRNA SVALKA (SVK) generates a NAT transcript of C-repeat/dehydration-responsive element binding factor 1 (CBF1), and Pol II collision is thought to suppress CBF1 expression (Kindgren et al. 2018).
Figure 3.
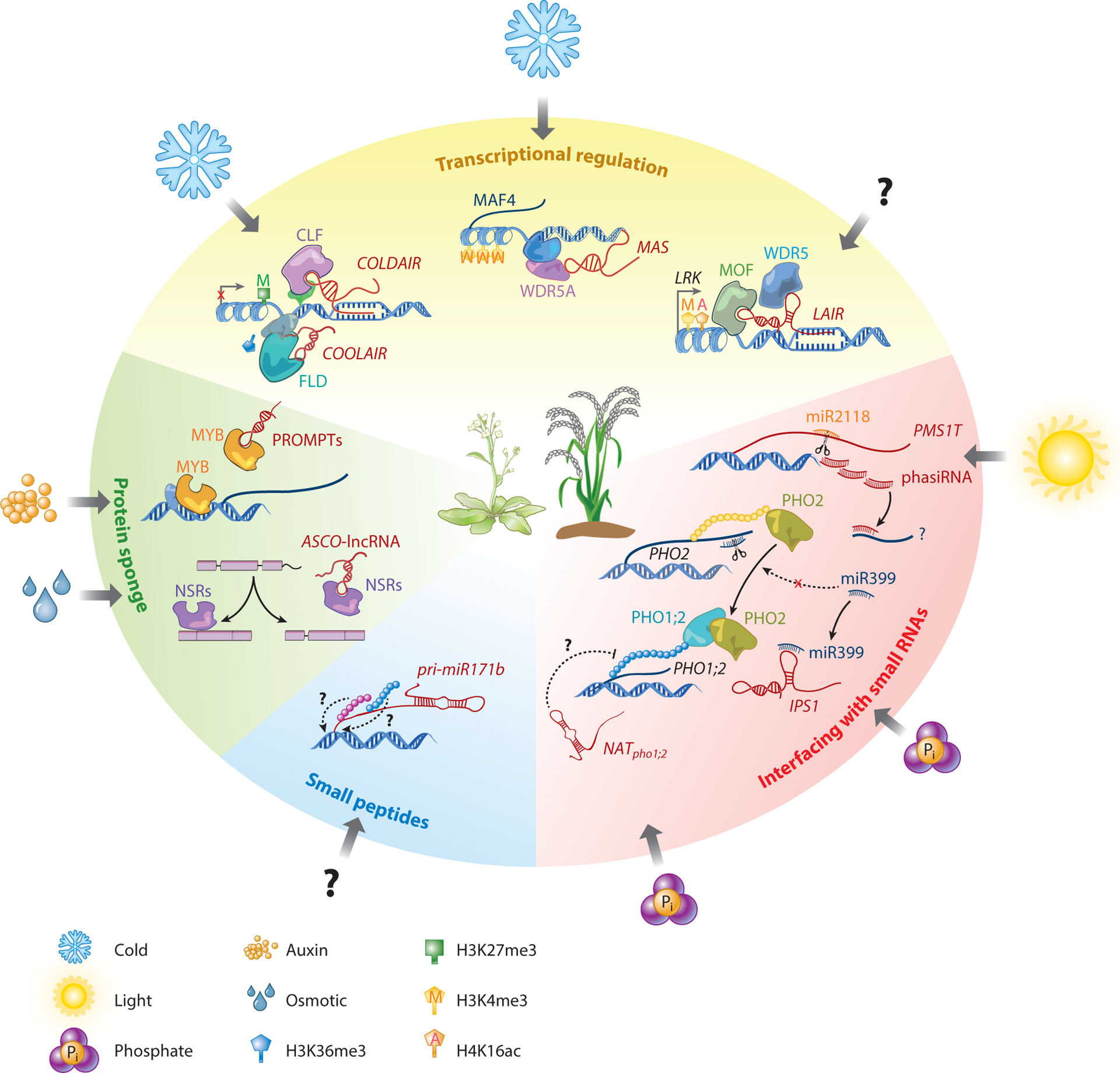
Representative models for the roles of plant long noncoding RNAs (lncRNAs). lncRNAs can serve as scaffolds, molecular mimics and sponges, and small interfering RNA precursors. They can also be translated into small peptides to regulate target genes in cis or in trans during plant development. The expression of most reported plant lncRNAs is induced by diverse environmental conditions. COLDAIR, COOLAIR, MAS, and LAIR are lncRNAs regulating mRNA transcription in cis. PMS1T is a lncRNA acting as a phasiRNA precursor. IPS1 regulates PHO2 by acting as the endogenous target mimic of miR399 and affects phosphate homeostasis together with another lncRNA, NATpho1;2. Pri-miR171b is a peptide-encoding lncRNA. PROMPTs and ASCO-lncRNA are protein-binding lncRNAs that suppress the function of target proteins.
In contrast to NAT expression, lincRNA expression is not significantly correlated with that of their neighboring genes (Y.C. Zhang et al. 2014). In fact, a number of lincRNAs function in trans in plants. For example, the lincRNA HIDDEN TREASURE 1 (HID1) modulates the transcription of PHYTOCHROME-INTERACTING FACTOR 3 (PIF3) in trans (Y. Wang et al. 2014). Nuclear alternative splicing regulators (NSRs) are trans regulated by lincRNAs in M. truncatula and Ara- bidopsis (Bardou et al. 2014, Campalans et al. 2004). The Arabidopsis lincRNA ELF18-INDUCED LONG-NONCODING RNA 1 (ELENA1) interacts with MEDIATOR SUBUNIT 19a (MED19a) in trans and affects MED19a enrichment at the PATHOGENESIS-RELATED 1 (PR1) promoter to enhance resistance to pathogens (Seo et al. 2017).
Few functional incRNAs have been identified in plants. The incRNA COLDAIR is transcribed in the sense direction from the first intron of FLC in Arabidopsis and regulates FLC transcription in cis (De Lucia et al. 2008, Kim & Sung 2013, Sung & Amasino 2004) (Figure 3). Finally, linear lincRNAs named promoter upstream transcripts (PROMPTs) are transcribed approximately 0.5–2.5 kb upstream of transcription start sites of protein-coding genes (Wu et al. 2017). PROMPTs have rapid turnover rates and regulate target genes both in cis and in trans (Song et al. 2018) (Figure 3).
Given the limited number of studies on plant lncRNAs, the above-described cis or trans regulatory preferences revealed by recent studies may not reflect general rules of lncRNA activities. Functional studies such as in situ activation/inactivation of lncRNAs will be useful for identifying their targets and uncovering their regulatory functions.
3.1.2. Circular long noncoding RNAs.
Although circRNAs are present in many species, they cannot be detected by transcriptome profiling using polyadenylated RNAs owing to their non-polyadenylated loop structures, and they were only recently identified by specific RNA sequencing approaches (X.O. Zhang et al. 2014). Most circRNAs are produced from back-splicing reactions of internal exons in pre-mRNAs and are exported to the cytoplasm (Wu et al. 2017). Other circRNAs are produced from excised intron lariats that fail to be debranched, and these circRNAs are preferentially localized in the nucleus (Wu et al. 2017).
A large number of circRNAs have been identified from Arabidopsis (Chen et al. 2017, Pan et al. 2018), rice (Oryza sativa) (Lu et al. 2015, Ye et al. 2017), tomato (S. lycopersicum) (Tan et al. 2017), sea buckthorn fruit (Hippophae rhamnoides Linn.) (Zhang et al. 2017), kiwifruit (Actinidia Lindl.) (Wang et al. 2017), barley (Hordeum vulgare) (Darbani et al. 2016), cotton (several Gossypium species) (Xiang et al. 2018, T. Zhao et al. 2017), soybean (Glycine max) (Chen et al. 2018a,W. Zhao et al. 2017), maize (Zea mays) (Chen et al. 2018b, Tang et al. 2018), and wheat (Triticum aestivum) (Y. Wang et al. 2016). circRNAs are expressed in a tissue-specific manner in different plants and exhibit a much higher degree of conservation than linear lncRNAs, but their abundance is extremely low (Chu et al. 2018, Lai et al. 2018). In animals, most circRNAs have complementary sequences such as repetitive elements in the introns flanking circularized exons; these sequences are essential for efficient exon circularization by RNA pairing across the flanking introns (Aktaş et al. 2017). In plants, however, intron pairing–driven circularization appears not to be the main mechanism of circRNA biogenesis. Instead, the production of most plant circRNAs may depend on noncanonical (non-GT/AG) splicing signals (Chu et al. 2018, Ye et al. 2017).
Few functional studies have been performed on plant circRNAs. One Arabidopsis circRNA from exon 6 of SEPALLATA 3 (SEP3) negatively regulates its parental gene in cis (Conn et al. 2017). The SEP3 exon 6 circRNA binds to its cognate DNA locus to form an R-loop, which results in transcriptional pausing and increases the abundance of the exon-skipped alternative splicing variant of SEP3, in turn driving floral homeotic phenotypes (Conn et al. 2017). Beyond this example, further studies are needed to reveal the regulatory functions of circRNAs.
3.2. Modes of Action of Long Noncoding RNAs
The modes of action of plant lncRNAs are diverse and complex. lncRNAs can act with different molecules to modulate transcription, translation, or epigenetic modification of their target genes. Interestingly, a subset of lncRNAs encodes peptides (<100 amino acids) necessary for a variety of cellular processes (Plaza et al. 2017).
3.2.1. Long noncoding RNAs partner with different molecules.
lncRNAs influence gene expression by acting as molecular scaffolds or decoys. As molecular scaffolds, lncRNAs influence gene expression by targeting regulatory factors such as chromatin remodelers to specific gene loci. As decoys, lncRNAs sequester proteins from their targets of action to regulate gene expression.
Plant lncRNAs regulate transcription through chromatin modifications, they may bind both DNA and protein, and they probably act as scaffolds. For example, COLDAIR and COOLAIR, both transcribed from the FLC locus in Arabidopsis, physically associate with FLC chromatin and recruit chromatin remodelers. This activity affects histone marks such as H3K36me3, H3K4me3, and H3K27me3 to regulate vernalization and seed dormancy (Chen & Penfield 2018, Kim & Sung 2013, Marquardt et al. 2014) (Figure 3). Similarly, the rice NAT LAIR localizes to the LRK genomic region and recruits chromatin-modifying complexes (MALES-ABSENT-ON-THE-FIRST and WD REPEAT DOMAIN 5) to increase H3K4me3 and H4K16ac levels at this gene (Y. Wang et al. 2018) (Figure 3). Although not confirmed, this mechanism may also underlie the function of the lincRNA HID1, which promotes photomorphogenesis and suppresses cotyledon greening in Arabidopsis (Y. Wang et al. 2014).
In addition to regulating transcription, some lncRNAs function as decoys that alter the behavior of target proteins. ASCO-lncRNA is an Arabidopsis lincRNA that binds NSRs, which are splicing factors, and competes with the alternative splicing targets of NSRs to modulate alternative splicing and gene expression during developmental transitions (Bardou et al. 2014) (Figure 3). Interestingly, NSRs also bind to the lncRNA ENOD40, inducing its relocalization from nuclear speckles to the cytoplasm in both Arabidopsis and M. truncatula (Campalans et al. 2004). PROMPTs can also act as decoy lncRNAs. For example, PROMPT_1281 binds MYB transcription factors to prevent them from interacting with DNA to induce target gene expression (Song et al. 2018) (Figure 3). It remains to be determined whether the decoy mechanism is universal for PROMPTs.
3.2.2. Long noncoding RNAs may encode small peptides.
Although lncRNAs by definition do not encode proteins or harbor ORFs for proteins greater than 100 amino acids, some lncRNAs are translated into small polypeptides of less than 100 amino acids (Matsumoto & Nakayama 2018). In some cases, these small peptides are necessary for a variety of cellular processes.
ENOD40 was the first lncRNA shown to encode functional peptides in Medicago sativa and soybean (Rohrig et al. 2002, Sousa et al. 2001). Two small peptides of 12 and 24 amino acids are synthesized from soybean ENOD40 RNA; both bind to the nodulin100 protein (a subunit of sucrose synthase), regulate sucrose use in nodules, and contribute to root nodule organogenesis (Rohrig et al. 2002). However, the ENOD40 RNA molecule also plays a role independent of peptides, such as binding the M. truncatula RNA BINDING PROTEIN 1 and SMALL NODULIN ACIDIC RNA-BINDING PROTEIN peptides (Campalans et al. 2004, Laporte et al. 2010). This role suggests that peptides encoded by lncRNAs are sometimes required for lncRNA function. Pri-miRNAs also encode peptides in M. truncatula and Arabidopsis (Lauressergues et al. 2015) (Figure 3). circRNAs are localized mainly in the cytosol, raising the possibility of their being translated. In animals, peptides are produced from circRNAs in a cap-independent manner and are functional (Pamudurti et al. 2017). Translation of circRNAs has not been demonstrated in plants.
Not all small ORFs present in lncRNAs encode peptides in vivo, or even if small peptides are made, they may not be functional. LAIR, discussed above, has the potential to encode short peptides, but mutations in the stop or start codons of the predicted small peptides do not affect LAIR function, indicating that LAIR more likely functions as a lncRNA (Y. Wang et al. 2018). A priori, it may not be clear whether lncRNA function is mediated by the RNA or the small peptide it encodes. It is therefore necessary to predict small ORFs when studying the function of lncRNAs and to generate transgenic plants that eliminate the ORFs to ascertain the potential functional contribution of the small peptides.
3.3. Plant Long Noncoding RNAs in Stress Responses and Development
Plants are often exposed to biotic or abiotic stresses harmful for development and survival, such as pathogen infection, extreme temperature, drought, and salt. Reproductive development is especially sensitive to certain stresses, which may cause sterility or reduced yield (Begcy & Dresselhaus 2018). To counter such stresses, plants have evolved survival strategies that involve lncRNAs, which tend to be stress responsive as well as spatially and temporally specific in expression. Therefore, lncRNAs are thought to function as effectors during stress responses.
3.3.1. Long noncoding RNAs that respond to various stresses.
Numerous plant lncRNAs are regulated by abiotic stresses. In Arabidopsis, differentially expressed lncRNAs have been identified under drought, cold, salinity, heat, and abscisic acid stresses (Di et al. 2014). lncRNAs responsive to various abiotic stresses have also been identified in other plant species (Ding et al. 2019, Pang et al. 2019, Qi et al. 2013, A. Wang et al. 2019, P. Wang et al. 2019, T.Z. Wang et al. 2015, W. Zhang et al. 2014). Biotic stress-responsive lncRNAs have been identified in wheat, Arabidopsis, grapevine, Hevea brasiliensis, and tomato (J. Wang et al. 2015, Xin et al. 2011, Xing et al. 2019, Yin et al. 2019, Zhang et al. 2013, Zhu et al. 2014). Several stress-responsive lncRNAs have been functionally analyzed, including the Arabidopsis drought- and salt-responsive lncRNA DRIR, whose elevated expression increases tolerance to drought and salt stresses (Qin et al. 2017). Tomato lncRNA16397 responsive to Phytophthora infestans infection is a NAT of GLUTAREDOXIN 22 and functions in cis to induce GLUTAREDOXIN 22 expression, resulting in enhanced pathogen resistance (Cui et al. 2017).
Some conditions that lead to stress-specific expression patterns of circRNAs in plants include dehydration stress in wheat (Y. Wang et al. 2016), chilling in tomato (Zuo et al. 2016), nutrient depletion in rice (Ye et al. 2015) and barley (Darbani et al. 2016), and Pseudomonas syringae pv. actinidiae infection in kiwifruit (Wang et al. 2017). Differential expression of circRNAs usually does not correlate with the expression of their precursor mRNAs, suggesting that circRNAs are not simply by-products of host gene expression but rather may be functional molecules in environmental and stress responses.
Functional studies of plant stress-responsive lncRNAs are at an early stage. Given that plant lncRNAs are extremely responsive to stresses in their expression and that they evolve rapidly compared with protein-coding genes, they make suitable environmental sensors or effectors to help plants adapt to changing environments.
3.3.2. Regulatory roles of long noncoding RNAs at the nexus of plant development and environmental responses.
Interestingly, lncRNAs involved in plant developmental regulation are often also regulated by environmental conditions, indicating their potential impacts on both plant development and environmental responses. The most representative examples are COLDAIR and COOLAIR, which promote flowering when plants are exposed to cold (Whittaker & Dean 2017) (Figure 3). Another example is long-day-specific male-fertility-associated RNA (LDMAR), which is highly and specifically expressed under long days and is required for normal pollen development under long-day conditions (Ding et al. 2012a). A single-nucleotide polymorphism (SNP) at the LDMAR locus increases RdDM at its promoter region, reducing LDMAR transcription specifically under long-day conditions and resulting in premature programmed cell death in developing anthers (Ding et al. 2012a,b). The photoperiod-sensitive genic male sterility 1 (Pms1) locus encodes another lncRNA, PMS1T, which is associated with photoperiod-sensitive male sterility (Fan et al. 2016) (Figure 3). An example of a salinity-responsive lncRNA is npc536, and its over-expression in Arabidopsis increases salt tolerance as well as primary and secondary root growth (Ben Amor et al. 2009). Recent studies have identified transcription factors that regulate lncRNA expression under biotic and abiotic stresses (Di et al. 2014, Nejat & Mantri 2018, Zhu et al. 2014).
The expression of stress resistance genes is often associated with reduced fitness, reflecting the inherent trade-off between stress responses and growth. The involvement of lncRNAs in both stress responses and plant growth makes them potential balancing factors in plants grown under different environmental conditions. Thus, lncRNAs may be good targets for the genetic engineering of crops aimed at increasing broad-spectrum disease resistance or stress resistance while mitigating yield loss.
4. LINKS BETWEEN LONG NONCODING RNAS AND SMALL RNAS
Long or medium-sized ncRNAs can be related to small RNAs in that they serve as precursors to siRNAs and miRNAs, as scaffolds to recruit siRNAs, or as sponges to sequester miRNAs. By definition, precursors to miRNAs and phasiRNAs lack long ORFs and are lncRNAs. So far, there has been no evidence of these lncRNAs having functions other than producing small RNAs. For example, PMS1T is a precursor to phasiRNAs. PMS1T is targeted by miR2118 to produce 21-nt phasiRNAs that preferentially accumulate in a photoperiod-sensitive male sterile mutant under long-day conditions (Fan et al. 2016). A SNP near the miR2118 recognition site leads to differential accumulation of phasiRNAs and underlies variations in fertility (Fan et al. 2016) (Figure 3). The abovementioned LDMAR locus was reported to generate siRNAs (Ding et al. 2012b), and our inspection of the locus suggested that LDMAR is likely a PHAS locus producing phasiRNAs. This implicates lncRNA-phasiRNAs as playing critical roles in sexual reproductive development in grasses (Yu et al. 2018). Pol V–generated transcripts are lncRNAs that serve as scaffolds to recruit siRNAs to chromatin (Wierzbicki et al. 2008). These relationships between lncRNAs and siRNAs are discussed in Sections 1 and 2.
A commonly observed function of lncRNAs in animals and plants is binding miRNAs as target mimics. In Arabidopsis, rice, and tomato, lncRNAs acting as potential target mimics have been identified on a large scale through RNA sequencing and bioinformatic prediction (Jiang et al. 2019, Wu et al. 2013). However, only a small number of endogenous target mimics (eTMs) are known to be functional. INDUCED BY PHOSPHATE STARVATION 1 (IPS1) was the first confirmed eTM lncRNA in plants (Franco-Zorrilla et al. 2007) (Figure 3). Arabidopsis IPS1 and its close paralog At4 are target mimics of miR399 and are involved in phosphate (Pi) accumulation; Pi-deficiency-induced long-noncoding RNA1 has a similar function in maize (Du et al. 2018, Khan et al. 2014) (Figure 3). In animals, a small number of circRNAs have been shown to sequester miRNAs to upregulate expression of their targets (Li et al. 2018). However, plant circRNAs are not known to act through miRNAs. Harboring of an miRNA binding site by a lncRNA does not necessarily impart an eTM function. The levels of the lncRNA need to be comparable to those of the miRNA for the lncRNA to be a functional eTM. In plants, miRNAs can cause the cleavage of target RNAs. Thus, a functional eTM needs to be able to bind the miRNA but also avoid cleavage, as in the case of IPS1.
Why do plants and animals employ eTMs to repress miRNA function? Other mechanisms for repressing miRNA functions include inhibiting miRNA biogenesis and enhancing miRNA turnover. We speculate that eTMs offer a faster way to repress miRNA activity than repressing miRNA abundance. miRNAs tend to have long half-lives, which makes it difficult to quickly reduce the abundance of miRNAs when necessary (such as when plants are under stress). In fact, artificial target mimic RNAs cause the degradation of cognate miRNAs (Todesco et al. 2010, Yan et al. 2012), and eTMs may thus trigger miRNA turnover in addition to blocking miRNA activity.
5. CELL-TO-CELL AND SYSTEMIC MOVEMENT OF NONCODING RNAS
5.1. Trafficking of microRNAs
Plant small RNAs can move locally between adjacent cells or over long distances, serving as functional molecules to spread silencing signals (Chitwood & Timmermans 2010, Gursanscky et al. 2011). miRNAs are better known for short-range intercellular trafficking, largely through plasmodesmata. For proper leaf patterning, miR165/166 movement from the abaxial (lower) to the adaxial (upper) leaf region results in a gradient distribution that helps establish adaxial domain–specific expression of HD-ZIP III genes (Benkovics & Timmermans 2014). In roots, miR165/166 moves from the endodermis to the vasculature to control protoxylem and metaxylem patterning by forming an expression gradient of PHB in the vasculature (Carlsbecker et al. 2010, Miyashima et al. 2011). In shoot meristems, the movement of L1 layer–expressed miR394 to the underlying L2 and L3 layers, where its target gene LCR is expressed, is essential for stem cell maintenance (Knauer et al. 2013). In maize, miR2118 is specifically expressed in the epidermis of developing anthers and moves to the subepidermal cell layers, where it targets noncoding transcripts for the biogenesis of phasiRNAs (Zhai et al. 2015b). The cell-to-cell trafficking of miRNAs probably occurs through passive diffusion through the plasmodesmata (Carlsbecker et al. 2010). However, miRNA movement is likely regulated, as the capacity for and the directionality of trafficking are different at different cell-to-cell interfaces and different from that of mobile proteins (Skopelitis et al. 2018). In addition to these examples of local cell-to-cell movement, some miRNAs spread over long distances via phloem. For example, miR399 moves from shoots to roots in response to phosphate deficiency in Arabidopsis (Lin et al. 2008, Pant et al. 2008). Similarly, the levels of miR395 and miR398 are significantly increased in phloem sap (PS) when Brassica rapa plants undergo sulfate and copper deficiency, respectively (Buhtz et al. 2008, Yoo et al. 2004), suggesting that these miRNAs function as systemic silencing signals to regulate their target genes. Under conditions of nutrient stress in Arabidopsis, miR395 and miR399 translocate from wild-type scions to hen1–1 mutant rootstocks, accompanied by reduced levels of their targets in the roots (Buhtz et al. 2010). Moreover, the shoot-to-root translocation of miR2111 in Lotus japonicus contributes to balancing bacterial infection and nodule organogenesis through repressing the symbiosis suppressor TOO MUCH LOVE (TML) (Tsikou et al. 2018).
5.2. Systemic Movement of Transposable Element–Derived Small Interfering RNAs
Almost all plant siRNAs are capable of moving locally or over long distances, as evidenced by studies using grafting and agroinfiltration strategies (Bai et al. 2011, Tamiru et al. 2018). In Arabidopsis, grafting experiments were performed combining wild type with the dcl2/3/4 triple mutant, in which the production of 21–24-nt siRNAs is compromised. The experiments show that 24-nt mobile siRNAs translocate from shoot to root across graft unions and mediate DNA methylation in recipient cells at thousands of loci associated with transgenes or endogenous TEs (Lewsey et al. 2016; Melnyk et al. 2011a,b). Furthermore, mobile siRNA-directed DNA methylation primarily occurs in non-CG contexts and largely depends on DRM1/DRM2 (Lewsey et al. 2016). However, it remains unclear whether TE-derived siRNAs also move from root to shoot and affect DNA methylation in the shoot. Although mobile siRNAs target many TE loci, they have few effects on gene expression, probably due to the low density and transposition activity of TEs in Arabidopsis.
5.3. Trafficking of Phased Small Interfering RNAs Between Cells
Similar to miRNAs and TE-derived siRNAs, phasiRNAs are also mobile, and the best example is the cell-to-cell trafficking of tasiRNA-ARFs. The biogenesis of tasiRNA-ARFs is thought to occur in the adaxial-most cell layers of leaves, where AGO7 (a gene required for tasiRNA-ARF biogenesis) and TAS3A (a gene from which tasiRNA-ARFs are generated) are expressed. But mature tasiRNA-ARFs form an adaxial-abaxial gradient to restrict ARF3 expression to the abaxial side, which ensures proper leaf patterning. Additionally, tasiRNA-ARFs are generated in the cells beneath the SAM, where AGO7 is expressed, and act non–cell autonomously within the SAM region (Chitwood et al. 2009). The intercellular mobilization of 24-nt reproductive phasiRNAs in rice and maize anthers has also been proposed, although decisive evidence is lacking (Ono et al. 2018, Zhai et al. 2015b).
5.4. Movements of Long Noncoding RNAs Through the Phloem
It was recently reported that lncRNAs are enriched in the PS and respond to imposed phosphate stress (Zhang et al. 2019). Specifically, hundreds of lncRNAs were detected in source tissues, sink tissues, and PS in cucumber. Among these PS lncRNAs are those encoding IPS1 and 24 other potential eTMs. As with the PS mRNAs, a CU-rich polypyrimidine-tract-binding motif was identified in the mobile lncRNAs (Zhang et al. 2019). These findings raise the possibility that lncRNAs are transported to distant tissues and may even act in systemic signaling. Future studies investigating long-distance RNA movement in plants will need to address how RNAs are selected for phloem access, whether they are unloaded into recipient tissues, and whether and how they function in recipient tissues. Viroid circRNAs have been reported to move cell-to-cell and long distances in plants (Ding et al. 2005, Wang & Ding 2010). circRNAs have also been found in animal exosomes, which can be released into the extracellular microenvironment (Fanale et al. 2018). Further work will elucidate whether plant circRNAs are mobile and serve as signaling molecules.
6. FUTURE PERSPECTIVES
While miRNAs have long been recognized as key components of cellular regulatory networks, siRNAs as well as lncRNAs are also emerging as regulatory molecules. However, studies of plant phasiRNAs, lncRNAs, and circRNAs are at a relatively early stage, with many outstanding questions awaiting further investigation. Thus, unraveling the complexity, biogenesis, and action of plant ncRNAs, especially lncRNAs, remains an important challenge. Furthermore, new classes of ncRNAs in plants likely await discovery. For example, novel lncRNA species that have recently been identified in mammals—such as small nucleolar RNA (snoRNA)-ended and tRNA-ended lncRNAs, which are processed by noncanonical mechanisms—have not yet been reported in plants (Wu et al. 2017). Although not discussed in this review, snoRNAs are key regulators of the transcriptome and translatome by guiding RNA methylation and/or pseudouridylation (Kiss 2002). Plant snoRNAs are poorly understood in terms of their targets and biological functions.
The functions of small RNAs, lncRNAs, and small peptides overlap in many cases, and revealing these functions during plant development or stress responses is one major challenge. The CRISPR-Cas system will enable new strategies for further identifying and analyzing components of the plant ncRNA network. Crop breeding aims to select plant varieties with disease resistance and desired growth characteristics such as high yield; however, plant growth is usually repressed by an active immune response (J. Wang et al. 2018). As molecules involved in both development and immunity, lncRNAs may be a good resource for balancing growth and immunity in crops.
ACKNOWLEDGMENTS
Support for this work is provided by Guangdong Province (2016A030308015) and the National Natural Science Foundation of China (91640202, 91740202, and 31801078).
DISCLOSURE STATEMENT
The authors are not aware of any affiliations, memberships, funding, or financial holdings that might be perceived as affecting the objectivity of this review.
LITERATURE CITED
- Aktaş T, Avşar Ilık I, Maticzka D, Bhardwaj V, Pessoa Rodrigues C, et al. 2017. DHX9 suppresses RNA processing defects originating from the Alu invasion of the human genome. Nature 544:115–19 [DOI] [PubMed] [Google Scholar]
- Ariel F, Romero-Barrios N, Jégu T, Benhamed M, Crespi M. 2015. Battles and hijacks: noncoding transcription in plants. Trends Plant Sci. 20:362–71 [DOI] [PubMed] [Google Scholar]
- Arif MA, Fattash I, Ma Z, Cho SH, Beike AK, et al. 2012. DICER-LIKE3 activity in Physcomitrella patens DICER-LIKE4 mutants causes severe developmental dysfunction and sterility. Mol. Plant 5:1281–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aukerman MJ, Sakai H. 2003. Regulation of flowering time and floral organ identity by a microRNA and its APETALA2-like target genes. Plant Cell 15:2730–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Axtell MJ, Jan C, Rajagopalan R, Bartel DP. 2006. A two-hit trigger for siRNA biogenesis in plants. Cell 127:565–77 [DOI] [PubMed] [Google Scholar]
- Bai S, Kasai A, Yamada K, Li T, Harada T. 2011. A mobile signal transported over a long distance induces systemic transcriptional gene silencing in a grafted partner. J. Exp. Bot 62:4561–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bardou F, Ariel F, Simpson CG, Romero-Barrios N, Laporte P, et al. 2014. Long noncoding RNA modulates alternative splicing regulators in Arabidopsis. Dev. Cell 30:166–76 [DOI] [PubMed] [Google Scholar]
- Baumberger N, Baulcombe DC. 2005. Arabidopsis ARGONAUTE1 is an RNA slicer that selectively recruits microRNAs and short interfering RNAs. PNAS 102:11928–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Begcy K, Dresselhaus T. 2018. Epigenetic responses to abiotic stresses during reproductive development in cereals. Plant Reprod. 31:343–55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben Amor B, Wirth S, Merchan F, Laporte P, d’Aubenton-Carafa Y, et al. 2009. Novel long non-protein coding RNAs involved in Arabidopsis differentiation and stress responses. Genome Res. 19:57–69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benkovics AH, Timmermans MC. 2014. Developmental patterning by gradients of mobile small RNAs. Curr. Opin. Genet. Dev 27:83–91 [DOI] [PubMed] [Google Scholar]
- Blevins T, Podicheti R, Mishra V, Marasco M, Wang J, et al. 2015. Identification of Pol IV and RDR2-dependent precursors of 24 nt siRNAs guiding de novo DNA methylation in Arabidopsis. eLife 4:e09591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Böhmdorfer G, Wierzbicki AT. 2015. Control of chromatin structure by long noncoding RNA. Trends Cell Biol. 25:623–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bologna NG, Iselin R, Abriata LA, Sarazin A, Pumplin N, et al. 2018. Nucleo-cytosolic shuttling of ARGONAUTE1 prompts a revised model of the plant microRNA pathway. Mol. Cell 68:709–19 [DOI] [PubMed] [Google Scholar]
- Bouyer D, Kramdi A, Kassam M, Heese M, Schnittger A, et al. 2017. DNA methylation dynamics during early plant life. Genome Biol. 18:179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buhtz A, Pieritz J, Springer F, Kehr J. 2010. Phloem small RNAs, nutrient stress responses, and systemic mobility. BMC Plant Biol. 10:64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buhtz A, Springer F, Chappell L, Baulcombe DC, Kehr J. 2008. Identification and characterization of small RNAs from the phloem of Brassica napus. Plant J. 53:739–49 [DOI] [PubMed] [Google Scholar]
- Cai Q, Liang C, Wang S, Hou Y, Gao L, et al. 2018. The disease resistance protein SNC1 represses the biogenesis of microRNAs and phased siRNAs. Nat. Commun 9:5080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calarco J, Borges F, Donoghue MA, Vanex F, Jullien P, et al. 2012. Reprogramming of DNA methylation in pollen guides epigenetic inheritance via small RNA. Cell 151:194–205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campalans A, Kondorosi A, Crespi M. 2004. Enod40, a short open reading frame–containing mRNA, induces cytoplasmic localization of a nuclear RNA binding protein in Medicago truncatula. Plant Cell 16:1047–59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlsbecker A, Lee JY, Roberts CJ, Dettmer J, Lehesranta S, et al. 2010. Cell signalling by microRNA165/6 directs gene dose-dependent root cell fate. Nature 465:316–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chekanova JA. 2015. Long non-coding RNAs and their functions in plants. Curr. Opin. Plant Biol 27:207–16 [DOI] [PubMed] [Google Scholar]
- Chen G, Cui J, Wang L, Zhu Y, Lu Z, Jin B. 2017. Genome-wide identification of circular RNAs in Arabidopsis thaliana. Front. Plant Sci 8:1678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen HM, Chen LT, Patel K, Li YH, Baulcombe DC, Wu SH. 2010. 22-nucleotide RNAs trigger secondary siRNA biogenesis in plants. PNAS 107:15269–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, Ding X, Zhang H, He T, Li Y, et al. 2018a. Comparative analysis of circular RNAs between soybean cytoplasmic male-sterile line NJCMS1A and its maintainer NJCMS1B by high-throughput sequencing. BMC Genom. 19:663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, Zhang P, Fan Y, Lu Q, Li Q, et al. 2018b. Circular RNAs mediated by transposons are associated with transcriptomic and phenotypic variation in maize. New Phytol. 217:1292–306 [DOI] [PubMed] [Google Scholar]
- Chen M, Penfield S. 2018. Feedback regulation of COOLAIR expression controls seed dormancy and flowering time. Science 360:1014–17 [DOI] [PubMed] [Google Scholar]
- Chen X 2004. A microRNA as a translational repressor of APETALA2 in Arabidopsis flower development. Science 303:2022–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X 2009. Small RNAs and their roles in plant development. Annu. Rev. Cell Dev. Biol 25:21–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chitwood DH, Nogueira FT, Howell MD, Montgomery TA, Carrington JC, Timmermans MC. 2009. Pattern formation via small RNA mobility. Genes Dev. 23:549–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chitwood DH, Timmermans MCP. 2010. Small RNAs are on the move. Nature 467:415. [DOI] [PubMed] [Google Scholar]
- Cho SH, Coruh C, Axtell MJ. 2012. miR156 and miR390 regulate tasiRNA accumulation and developmental timing in Physcomitrella patens. Plant Cell 24:4837–49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu Q, Bai P, Zhu X, Zhang X, Mao L, et al. 2018. Characteristics of plant circular RNAs. Brief. Bioinform 10.1093/bib/bby111 [DOI] [PubMed] [Google Scholar]
- Conn VM, Hugouvieux V, Nayak A, Conos SA, Capovilla G, et al. 2017. A circRNA from SEPALLATA3 regulates splicing of its cognate mRNA through R-loop formation. Nat. Plants 3:17053. [DOI] [PubMed] [Google Scholar]
- Cuerda-Gil D, Slotkin RK. 2016. Non-canonical RNA-directed DNA methylation. Nat. Plants 2:16163. [DOI] [PubMed] [Google Scholar]
- Cui J, Luan Y, Jiang N, Bao H, Meng J. 2017. Comparative transcriptome analysis between resistant and susceptible tomato allows the identification of lncRNA16397 conferring resistance to Phytophthora infestans by co-expressing glutaredoxin. Plant J. 89:577–89 [DOI] [PubMed] [Google Scholar]
- Curaba J, Talbot M, Li Z, Helliwell C. 2013. Over-expression of microRNA171 affects phase transitions and floral meristem determinancy in barley. BMC Plant Biol. 13:6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Ario M, Griffiths-Jones S, Kim M. 2017. Small RNAs: big impact on plant development. Trends Plant Sci. 22:1056–68 [DOI] [PubMed] [Google Scholar]
- Darbani B, Noeparvar S, Borg S. 2016. Identification of circular RNAs from the parental genes involved in multiple aspects of cellular metabolism in barley. Front. Plant Sci 7:776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Lucia F, Crevillen P, Jones AM, Greb T, Dean C. 2008. A PHD-polycomb Repressive Complex 2 triggers the epigenetic silencing of FLC during vernalization. PNAS 105:16831–36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di C, Yuan J, Wu Y, Li J, Lin H, et al. 2014. Characterization of stress-responsive lncRNAs in Arabidopsis thaliana by integrating expression, epigenetic and structural features. Plant J. 80:848–61 [DOI] [PubMed] [Google Scholar]
- Ding B, Itaya A, Zhong X. 2005. Viroid trafficking: a small RNA makes a big move. Curr. Opin. Plant Biol 8:606–12 [DOI] [PubMed] [Google Scholar]
- Ding J, Lu Q, Ouyang Y, Mao H, Zhang P, et al. 2012a. A long noncoding RNA regulates photoperiod-sensitive male sterility, an essential component of hybrid rice. PNAS 109:2654–59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding J, Shen J, Mao H, Xie W, Li X, Zhang Q. 2012b. RNA-directed DNA methylation is involved in regulating photoperiod-sensitive male sterility in rice. Mol. Plant 5:1210–16 [DOI] [PubMed] [Google Scholar]
- Ding Z, Tie W, Fu L, Yan Y, Liu G, et al. 2019. Strand-specific RNA-seq based identification and functional prediction of drought-responsive lncRNAs in cassava. BMC Genom. 20:214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du J, Johnson LM, Jacobsen SE, Patel DJ. 2015. DNA methylation pathways and their crosstalk with histone methylation. Nat. Rev. Mol. Cell Biol 16:519–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du Q, Wang K, Zou C, Xu C, Li WX. 2018. The PILNCR1-miR399 regulatory module is important for low phosphate tolerance in maize. Plant Physiol. 177:1743–53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan Y, Yang J, Mathioni SM, Yu J, Shen J, et al. 2016. PMS1T, producing phased small-interfering RNAs, regulates photoperiod-sensitive male sterility in rice. PNAS 113:15144–49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fanale D, Taverna S, Russo A, Bazan V. 2018. Circular RNA in exosomes. Adv. Exp. Med. Biol 1087:109–17 [DOI] [PubMed] [Google Scholar]
- Fei Q, Li P, Teng C, Meyers BC. 2015. Secondary siRNAs from Medicago NB-LRRs modulated via miRNA-target interactions and their abundances. Plant J. 83:451–65 [DOI] [PubMed] [Google Scholar]
- Fei Q, Xia R, Meyers BC. 2013. Phased, secondary, small interfering RNAs in posttranscriptional regulatory networks. Plant Cell 25:2400–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fei Q, Yu Y, Liu L, Zhang Y, Baldrich P, et al. 2018. Biogenesis of a 22-nt microRNA in Phaseoleae species by precursor-programmed uridylation. PNAS 115:8037–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franco-Zorrilla JM, Valli A, Todesco M, Mateos I, Puga MI, et al. 2007. Target mimicry provides a new mechanism for regulation of microRNA activity. Nat. Genet 39:1033–37 [DOI] [PubMed] [Google Scholar]
- Garcia-Ruiz H, Takeda A, Chapman EJ, Sullivan CM, Fahlgren N, et al. 2010. Arabidopsis RNA-dependent RNA polymerases and dicer-like proteins in antiviral defense and small interfering RNA biogenesis during Turnip Mosaic Virus infection. Plant Cell 22:481–96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gouil Q, Baulcombe DC. 2018. Paramutation-like features of multiple natural epialleles in tomato. BMC Genom. 19:203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo HS, Xie Q, Fei JF, Chua NH. 2005. MicroRNA directs mRNA cleavage of the transcription factor NAC1 to downregulate auxin signals for Arabidopsis lateral root development. Plant Cell 17:1376–86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo S, Xu Y, Liu H, Mao Z, Zhang C, et al. 2013. The interaction between OsMADS57 and OsTB1 modulates rice tillering via DWARF14. Nat. Commun 4:1566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gursanscky NR, Searle IR, Carroll BJ. 2011. Mobile microRNAs hit the target. Traffic 12:1475–82 [DOI] [PubMed] [Google Scholar]
- Henriques R, Wang H, Liu J, Boix M, Huang LF, Chua NH. 2017. The antiphasic regulatory module comprising CDF5 and its antisense RNA FLORE links the circadian clock to photoperiodic flowering. New Phytol. 216:854–67 [DOI] [PubMed] [Google Scholar]
- Hou Y, Zhai Y, Feng L, Karimi HZ, Rutter BD, et al. 2018. A Phytophthora effector suppresses trans-kingdom RNAi to promote disease susceptibility. Cell Host Microbe 25:153–65.e5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J, Wang C, Wang H, Lu P, Zheng B, et al. 2019. Meiocyte-specific and AtSPO11–1-dependent small RNAs and their association with meiotic gene expression and recombination. Plant Cell 31:444–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibarra CA, Feng X, Schoft VK, Hsieh T-F, Uzawa R, et al. 2012. Active DNA demethylation in plant companion cells reinforces transposon methylation in gametes. Science 337:1360–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito H, Gaubert H, Bucher E, Mirouze M, Vaillant I, Paszkowski J. 2011. An siRNA pathway prevents transgenerational retrotransposition in plants subjected to stress. Nature 472:115. [DOI] [PubMed] [Google Scholar]
- Jabnoune M, Secco D, Lecampion C, Robaglia C, Shu Q, Poirier Y. 2013. A rice cis-natural antisense RNA acts as a translational enhancer for its cognate mRNA and contributes to phosphate homeostasis and plant fitness. Plant Cell 25:4166–82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang N, Cui J, Shi Y, Yang G, Zhou X, et al. 2019. Tomato lncRNA23468 functions as a competing endogenous RNA to modulate NBS-LRR genes by decoying miR482b in the tomato–Phytophthora infestans interaction. Hortic. Res 6:28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jullien PE, Susaki D, Yelagandula R, Higashiyama T, Berger F. 2012. DNA methylation dynamics during sexual reproduction in Arabidopsis thaliana. Curr. Biol 22:1825–30 [DOI] [PubMed] [Google Scholar]
- Kakrana A, Mathioni SM, Huang K, Hammond R, Vandivier L, et al. 2018. Plant 24-nt reproductive phasiRNAs from intramolecular duplex mRNAs in diverse monocots. Genome Res. 28:1333–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan GA, Bouraine S, Wege S, Li Y, de Carbonnel M, et al. 2014. Coordination between zinc and phosphate homeostasis involves the transcription factor PHR1, the phosphate exporter PHO1, and its homologue PHO1;H3 in Arabidopsis. J. Exp. Bot 65:871–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim DH, Sung S. 2013. Coordination of the vernalization response through a VIN3 and FLC gene family regulatory network in Arabidopsis. Plant Cell 25:454–69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kindgren P, Ard R, Ivanov M, Marquardt S. 2018. Transcriptional read-through of the long non-coding RNA SVALKA governs plant cold acclimation. Nat. Commun 9:4561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiss T 2002. Small nucleolar RNAs: an abundant group of noncoding RNAs with diverse cellular functions. Cell 109:145–48 [DOI] [PubMed] [Google Scholar]
- Knauer S, Holt AL, Rubio-Somoza I, Tucker EJ, Hinze A, et al. 2013. A protodermal miR394 signal defines a region of stem cell competence in the Arabidopsis shoot meristem. Dev. Cell 24:125–32 [DOI] [PubMed] [Google Scholar]
- Komiya R, Ohyanagi H, Niihama M, Watanabe T, Nakano M, et al. 2014. Rice germline-specific Argonaute MEL1 protein binds to phasiRNAs generated from more than 700 lincRNAs. Plant J. 78:385–97 [DOI] [PubMed] [Google Scholar]
- Lai X, Bazin J, Webb S, Crespi M, Zubieta C, Conn SJ. 2018. CircRNAs in plants. Adv. Exp. Med. Biol 1087:329–43 [DOI] [PubMed] [Google Scholar]
- Laporte P, Satiat-Jeunemaitre B, Velasco I, Csorba T, Van de Velde W, et al. 2010. A novel RNA-binding peptide regulates the establishment of the Medicago truncatula–Sinorhizobium meliloti nitrogen-fixing symbiosis. Plant J. 62:24–38 [DOI] [PubMed] [Google Scholar]
- Lauressergues D, Couzigou JM, Clemente HS, Martinez Y, Dunand C, et al. 2015. Primary transcripts of microRNAs encode regulatory peptides. Nature 520:90–93 [DOI] [PubMed] [Google Scholar]
- Law JA, Jacobsen SE. 2010. Establishing, maintaining and modifying DNA methylation patterns in plants and animals. Nat. Rev. Genet 11:204–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewsey MG, Hardcastle TJ, Melnyk CW, Molnar A, Valli A, et al. 2016. Mobile small RNAs regulate genome-wide DNA methylation. PNAS 113:E801–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F, Orban R, Baker B. 2012. SoMART: a web server for plant miRNA, tasiRNA and target gene analysis. Plant J. 70:891–901 [DOI] [PubMed] [Google Scholar]
- Li J, Guo G, Guo W, Guo G, Tong D, et al. 2012. miRNA164-directed cleavage of ZmNAC1 confers lateral root development in maize (Zea mays L.). BMC Plant Biol. 12:220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q, Gent JI, Zynda G, Song J, Makarevitch I, et al. 2015. RNA-directed DNA methylation enforces boundaries between heterochromatin and euchromatin in the maize genome. PNAS 112:14728–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, He Y. 2014. HEAT-INDUCED TAS1 TARGET1 mediates thermotolerance via HEAT STRESS TRANSCRIPTION FACTOR A1a–directed pathways in Arabidopsis. Plant Cell 26:1764–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Le B, Ma X, Li S, You C, et al. 2016. Biogenesis of phased siRNAs on membrane-bound polysomes in Arabidopsis. eLife 5:e22750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Liu L, Zhuang X, Yu Y, Liu X, et al. 2013. MicroRNAs inhibit the translation of target mRNAs on the endoplasmic reticulum in Arabidopsis. Cell 153:562–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Vandivier LE, Tu B, Gao L, Won SY, et al. 2015. Detection of Pol IV/RDR2-dependent transcripts at the genomic scale in Arabidopsis reveals features and regulation of siRNA biogenesis. Genome Res. 25:235–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Yang L, Chen LL. 2018. The biogenesis, functions, and challenges of circular RNAs. Mol. Cell 71:428–42 [DOI] [PubMed] [Google Scholar]
- Lin SI, Chiang SF, Lin WY, Chen JW, Tseng CY, et al. 2008. Regulatory network of microRNA399 and PHO2 by systemic signaling. Plant Physiol. 147:732–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Wang H, Chua NH. 2015. Long noncoding RNA transcriptome of plants. Plant Biotechnol. J 13:319–28 [DOI] [PubMed] [Google Scholar]
- Liu Q, Yao X, Pi L, Wang H, Cui X, Huang H. 2009. The ARGONAUTE10 gene modulates shoot apical meristem maintenance and establishment of leaf polarity by repressing miR165/166 in Arabidopsis. Plant J. 58:27–40 [DOI] [PubMed] [Google Scholar]
- Lloyd JP, Tsai ZT-Y, Sowers RP, Panchy NL, Shiu S-H. 2018. A model-based approach for identifying functional intergenic transcribed regions and noncoding RNAs. Mol. Biol. Evol 35:1422–36 [DOI] [PubMed] [Google Scholar]
- Lu T, Cui L, Zhou Y, Zhu C, Fan D, et al. 2015. Transcriptome-wide investigation of circular RNAs in rice. RNA 21:2076–87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo QJ, Mittal A, Jia F, Rock CDR. 2012. An autoregulatory feedback loop involving PAP1 and TAS4 in response to sugars in Arabidopsis. Plant Mol. Biol 80:117–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marquardt S, Raitskin O, Wu Z, Liu F, Sun Q, Dean C. 2014. Functional consequences of splicing of the antisense transcript COOLAIR on FLC transcription. Mol. Cell 54:156–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez G, Köhler C. 2017. Role of small RNAs in epigenetic reprogramming during plant sexual reproduction. Curr. Opin. Plant Biol 36:22–28 [DOI] [PubMed] [Google Scholar]
- Martínez G, Panda K, Köhler C, Slotkin RK. 2016. Silencing in sperm cells is directed by RNA movement from the surrounding nurse cell. Nat. Plants 2:16030. [DOI] [PubMed] [Google Scholar]
- Martinez G, Wolff P, Wang Z, Moreno-Romero J, Santos-Gonzalez J, et al. 2018. Paternal easiRNAs regulate parental genome dosage in Arabidopsis. Nat. Genet 50:193–98 [DOI] [PubMed] [Google Scholar]
- Matsumoto A, Nakayama KI. 2018. Hidden peptides encoded by putative noncoding RNAs. Cell Struct. Funct 43:75–83 [DOI] [PubMed] [Google Scholar]
- Matzke MA, Kanno T, Matzke AJ. 2015. RNA-directed DNA methylation: the evolution of a complex epigenetic pathway in flowering plants. Annu. Rev. Plant Biol 66:243–67 [DOI] [PubMed] [Google Scholar]
- Melnyk CW, Molnar A, Bassett A, Baulcombe DC. 2011a. Mobile 24 nt small RNAs direct transcriptional gene silencing in the root meristems of Arabidopsis thaliana. Curr. Biol 21:1678–83 [DOI] [PubMed] [Google Scholar]
- Melnyk CW, Molnar A, Baulcombe DC. 2011b. Intercellular and systemic movement of RNA silencing signals. EMBO J. 30:3553–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyashima S, Koi S, Hashimoto T, Nakajima K. 2011. Non-cell-autonomous microRNA165 acts in a dose-dependent manner to regulate multiple differentiation status in the Arabidopsis root. Development 138:2303–13 [DOI] [PubMed] [Google Scholar]
- Nejat N, Mantri N. 2018. Emerging roles of long non-coding RNAs in plant response to biotic and abiotic stresses. Crit. Rev. Biotechnol 38:93–105 [DOI] [PubMed] [Google Scholar]
- Ono S, Liu H, Tsuda K, Fukai E, Tanaka K, et al. 2018. EAT1 transcription factor, a non-cell-autonomous regulator of pollen production, activates meiotic small RNA biogenesis in rice anther tapetum. PLOS Genet. 14:e1007238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pamudurti NR, Bartok O, Jens M, Ashwal-Fluss R, Stottmeister C, et al. 2017. Translation of circRNAs. Mol. Cell 66:9–21.e7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan T, Sun X, Liu Y, Li H, Deng G, et al. 2018. Heat stress alters genome-wide profiles of circular RNAs in Arabidopsis. Plant Mol. Biol 96:217–29 [DOI] [PubMed] [Google Scholar]
- Pang J, Zhang X, Ma X, Zhao J. 2019. Spatio-temporal transcriptional dynamics of maize long non-coding RNAs responsive to drought stress. Genes 10:138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pant BD, Buhtz A, Kehr J, Scheible WR. 2008. MicroRNA399 is a long-distance signal for the regulation of plant phosphate homeostasis. Plant J. 53:731–38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel P, Mathioni S, Kakrana A, Shatkay H, Meyers BC. 2018. Reproductive phasiRNAs in grasses are compositionally distinct from other classes of small RNAs. New Phytol. 220:851–64 [DOI] [PubMed] [Google Scholar]
- Pauli A, Rinn JL, Schier AF. 2011. Non-coding RNAs as regulators of embryogenesis. Nat. Rev. Genet 12:136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plaza S, Menschaert G, Payre F. 2017. In search of lost small peptides. Annu. Rev. Cell Dev. Biol 33:391–416 [DOI] [PubMed] [Google Scholar]
- Qi X, Xie S, Liu Y, Yi F, Yu J. 2013. Genome-wide annotation of genes and noncoding RNAs of foxtail millet in response to simulated drought stress by deep sequencing. Plant Mol. Biol 83:459–73 [DOI] [PubMed] [Google Scholar]
- Qin T, Zhao H, Cui P, Albesher N, Xiong L. 2017. A nucleus-localized long non-coding RNA enhances drought and salt stress tolerance. Plant Physiol. 175:1321–36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rai MI, Alam M, Lightfoot DA, Gurha P, Afzal AJ. 2018. Classification and experimental identification of plant long non-coding RNAs. Genomics. In press. 10.1016/j.ygeno.2018.04.014 [DOI] [PubMed] [Google Scholar]
- Rogers K, Chen X. 2013. Biogenesis, turnover, and mode of action of plant microRNAs. Plant Cell 25:2383–99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohrig H, Schmidt J, Miklashevichs E, Schell J, John M. 2002. Soybean ENOD40 encodes two peptides that bind to sucrose synthase. PNAS 99:1915–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanei M, Chen X. 2015. Mechanisms of microRNA turnover. Curr. Opin. Plant Biol 27:199–206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo JS, Sun HX, Park BS, Huang CH, Yeh SD, et al. 2017. ELF18-INDUCED LONG-NONCODING RNA associates with mediator to enhance expression of innate immune response genes in Arabidopsis. Plant Cell 29:1024–38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shivaprasad PV, Dunn RM, Santos BACM, Bassett A, Baulcombe DC. 2014. Extraordinary transgressive phenotypes of hybrid tomato are influenced by epigenetics and small silencing RNAs. EMBO J. 31:257–66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva GFF, Silva EM, Correa JPO, Vicente MH, Jiang N, et al. 2018. Tomato floral induction and flower development are orchestrated by the interplay between gibberellin and two unrelated microRNA-controlled modules. New Phytol. 221:1328–44 [DOI] [PubMed] [Google Scholar]
- Skopelitis DS, Hill K, Klesen S, Marco CF, von Born P, et al. 2018. Gating of miRNA movement at defined cell-cell interfaces governs their impact as positional signals. Nat. Commun 9:3107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slotkin RK, Vaughn M, Borges F, Tanurdzić M, Becker JD, et al. 2009. Epigenetic reprogramming and small RNA silencing of transposable elements in pollen. Cell 136:461–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song Q, Zhang T, Stelly DM, Chen ZJ. 2017. Epigenomic and functional analyses reveal roles of epialleles in the loss of photoperiod sensitivity during domestication of allotetraploid cottons. Genome Biol. 18:99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song Y, Xuan A, Bu C, Ci D, Tian M, Zhang D. 2018. Osmotic stress-responsive promoter upstream transcripts (PROMPTs) act as carriers of MYB transcription factors to induce the expression of target genes in Populus simonii. Plant Biotechnol. J 17:164–77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sousa C, Johansson C, Charon C, Manyani H, Sautter C, et al. 2001. Translational and structural requirements of the early nodulin gene enod40, a short-open reading frame–containing RNA, for elicitation of a cell-specific growth response in the alfalfa root cortex. Mol. Cell. Biol 21:354–66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Q, Liu X, Yang J, Liu W, Du Q, et al. 2018. microRNA528 affects lodging resistance of maize by regulating lignin biosynthesis under nitrogen-luxury conditions. Mol. Plant 11:806–14 [DOI] [PubMed] [Google Scholar]
- Sung S, Amasino RM. 2004. Vernalization in Arabidopsis thaliana is mediated by the PHD finger protein VIN3. Nature 427:159–64 [DOI] [PubMed] [Google Scholar]
- Tamiru M, Hardcastle TJ, Lewsey MG. 2018. Regulation of genome-wide DNA methylation by mobile small RNAs. New Phytol. 217:540–46 [DOI] [PubMed] [Google Scholar]
- Tan J, Zhou Z, Niu Y, Sun X, Deng Z. 2017. Identification and functional characterization of tomato circRNAs derived from genes involved in fruit pigment accumulation. Sci. Rep 7:8594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang B, Hao Z, Zhu Y, Zhang H, Li G. 2018. Genome-wide identification and functional analysis of circRNAs in Zea mays. PLOS ONE 13:e0202375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang J, Chu C. 2017. MicroRNAs in crop improvement: fine-tuners for complex traits. Nat. Plants 3:17077. [DOI] [PubMed] [Google Scholar]
- Todesco M, Rubio-Somoza I, Paz-Ares J, Weigel D. 2010. A collection of target mimics for comprehensive analysis of microRNA function in Arabidopsis thaliana. PLOS Genet. 6:e1001031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsikou D, Yan Z, Holt DB, Abel NB, Reid DE, et al. 2018. Systemic control of legume susceptibility to rhizobial infection by a mobile microRNA. Science 362:233–36 [DOI] [PubMed] [Google Scholar]
- Tu B, Liu L, Xu C, Zhai J, Li S, et al. 2015. Distinct and cooperative activities of HESO1 and URT1 nucleotidyl transferases in microRNA turnover in Arabidopsis. PLOS Genet. 11:e1005119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulitsky I, Bartel DP. 2013. lincRNAs: genomics, evolution, and mechanisms. Cell 154:26–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang A, Hu J, Gao C, Chen G, Wang B, et al. 2019. Genome-wide analysis of long non-coding RNAs unveils the regulatory roles in the heat tolerance of Chinese cabbage (Brassica rapa ssp. chinensis). Sci. Rep 9:5002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Chung PJ, Liu J, Jang IC, Kean MJ, et al. 2014. Genome-wide identification of long noncoding natural antisense transcripts and their responses to light in Arabidopsis. Genome Res. 24:444–53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Jiao X, Kong X, Humaira S, Wu Y, et al. 2016. A signaling cascade from miR444 to RDR1 in rice antiviral RNA silencing pathway. Plant Physiol. 170:2365–77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J 2014. Regulation of flowering time by the miR156-mediated age pathway. J. Exp. Bot 65:4723–30 [DOI] [PubMed] [Google Scholar]
- Wang J, Yu W, Yang Y, Li X, Chen T, et al. 2015. Genome-wide analysis of tomato long non-coding RNAs and identification as endogenous target mimic for microRNA in response to TYLCV infection. Sci. Rep 5:16946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Zhou L, Shi H, Chern M, Yu H, et al. 2018. A single transcription factor promotes both yield and immunity in rice. Science 361:1026–28 [DOI] [PubMed] [Google Scholar]
- Wang JW, Czech B, Weigel D. 2009. miR156-regulated SPL transcription factors define an endogenous flowering pathway in Arabidopsis thaliana. Cell 138:738–49 [DOI] [PubMed] [Google Scholar]
- Wang P, Dai L, Ai J, Wang Y, Ren F. 2019. Identification and functional prediction of cold-related long non-coding RNA (lncRNA) in grapevine. Sci. Rep 9:6638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang TZ, Liu M, Zhao MG, Chen R, Zhang WH. 2015. Identification and characterization of long non-coding RNAs involved in osmotic and salt stress in Medicago truncatula using genome-wide high-throughput sequencing. BMC Plant Biol. 15:131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Ding B. 2010. Viroids: small probes for exploring the vast universe of RNA trafficking in plants. J. Integr. Plant Biol 52:28–39 [DOI] [PubMed] [Google Scholar]
- Wang Y, Fan X, Lin F, He G, Terzaghi W, et al. 2014. Arabidopsis noncoding RNA mediates control of photomorphogenesis by red light. PNAS 111:10359–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Luo X, Sun F, Hu J, Zha X, et al. 2018. Overexpressing lncRNA LAIR increases grain yield and regulates neighbouring gene cluster expression in rice. Nat. Commun 9:3516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Yang M, Wei S, Qin F, Zhao H, Suo B. 2016. Identification of circular RNAs and their targets in leaves of Triticum aestivum L. under dehydration stress. Front. Plant Sci 7:2024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Liu Y, Li D, Li L, Zhang Q, et al. 2017. Identification of circular RNAs in kiwifruit and their species-specific response to bacterial canker pathogen invasion. Front. Plant Sci 8:413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whittaker C, Dean C. 2017. The FLC locus: a platform for discoveries in epigenetics and adaptation. Annu. Rev. Cell Dev. Biol 33:555–75 [DOI] [PubMed] [Google Scholar]
- Wierzbicki AT, Haag JR, Pikaard CS. 2008. Noncoding transcription by RNA polymerase Pol IVb/Pol V mediates transcriptional silencing of overlapping and adjacent genes. Cell 135:635–48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu G, Park MY, Conway SR, Wang JW, Weigel D, Poethig RS. 2009. The sequential action of miR156 and miR172 regulates developmental timing in Arabidopsis. Cell 138:750–59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H, Yang L, Chen LL. 2017. The diversity of long noncoding RNAs and their generation. Trends Genet. 33:540–52 [DOI] [PubMed] [Google Scholar]
- Wu HJ, Wang ZM, Wang M, Wang XJ. 2013. Widespread long noncoding RNAs as endogenous target mimics for microRNAs in plants. Plant Physiol. 161:1875–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia R, Meyers BC, Liu Z, Beers EP, Ye S, Liu Z. 2013. MicroRNA superfamilies descended from miR390 and their roles in secondary small interfering RNA biogenesis in eudicots. Plant Cell 25:1555–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia R, Xu J, Meyers BC. 2016. The emergence, evolution, and diversification of the miR390-TAS3-ARF pathway in land plants. Plant Cell 29:1232–47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang L, Cai C, Cheng J, Wang L, Wu C, et al. 2018. Identification of circularRNAs and their targets in Gossypium under Verticillium wilt stress based on RNA-seq. PeerJ 6:e4500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xin M, Wang Y, Yao Y, Song N, Hu Z, et al. 2011. Identification and characterization of wheat long non-protein coding RNAs responsive to powdery mildew infection and heat stress by using microarray analysis and SBS sequencing. BMC Plant Biol. 11:61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing Q, Zhang W, Liu M, Li L, Li X, et al. 2019. Genome-wide identification of long non-coding RNAs responsive to Lasiodiplodia theobromae infection in grapevine. Evol. Bioinform. Online 15:1176934319841362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan J, Gu Y, Jia X, Kang W, Pan S, et al. 2012. Effective small RNA destruction by the expression of a short tandem target mimic in Arabidopsis. Plant Cell 24:415–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L, Froberg JE, Lee JT. 2014. Long noncoding RNAs: fresh perspectives into the RNA world. Trends Biochem. Sci 39:35–43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Zhou Y, Zhang Y, Chen Y. 2018. Grass phasiRNAs and male fertility. Sci. China Life Sci 61:1–7 [DOI] [PubMed] [Google Scholar]
- Ye CY, Chen L, Liu C, Zhu QH, Fan L. 2015. Widespread noncoding circular RNAs in plants. New Phytol. 208:88–95 [DOI] [PubMed] [Google Scholar]
- Ye CY, Zhang X, Chu Q, Liu C, Yu Y, et al. 2017. Full-length sequence assembly reveals circular RNAs with diverse non-GT/AG splicing signals in rice. RNA Biol. 14:1055–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin H, Zhang X, Zhang B, Luo H, He C. 2019. Revealing the dominant long noncoding RNAs responding to the infection with Colletotrichum gloeosporioides in Hevea brasiliensis. Biol. Direct 14:7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo BC, Kragler F, Varkonyi-Gasic E, Haywood V, Archer-Evans S, et al. 2004. A systemic small RNA signaling system in plants. Plant Cell 16:1979–2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Y, Jia T, Chen X. 2017. The ‘how’ and ‘where’ of plant microRNAs. New Phytol. 216:1002–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Y, Zhou Y, Zhang Y, Chen Y. 2018. Grass phasiRNAs and male fertility. Sci. China Life Sci 61:148–54 [DOI] [PubMed] [Google Scholar]
- Zhai J, Bischof S, Wang H, Feng S, Lee T-F, et al. 2015a. A one precursor one siRNA model for Pol IV–dependent siRNA biogenesis. Cell 163:445–55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhai J, Jeong DH, De Paoli E, Park S, Rosen BD, et al. 2011. MicroRNAs as master regulators of the plant NB-LRR defense gene family via the production of phased, trans-acting siRNAs. Genes Dev. 25:2540–53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhai J, Zhang H, Arikit S, Huang K, Nan G-L, et al. 2015b.. Spatiotemporally dynamic, cell-type–dependent premeiotic and meiotic phasiRNAs in maize anthers. PNAS 112:3146–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C, Li G, Wang J, Fang J. 2012. Identification of trans-acting siRNAs and their regulatory cascades in grapevine. Bioinformatics 28:2561–68 [DOI] [PubMed] [Google Scholar]
- Zhang G, Duan A, Zhang J, He C. 2017. Genome-wide analysis of long non-coding RNAs at the mature stage of sea buckthorn (Hippophae rhamnoides Linn) fruit. Gene 596:130–36 [DOI] [PubMed] [Google Scholar]
- Zhang H, Chen X, Wang C, Xu Z, Wang Y, et al. 2013. Long non-coding genes implicated in response to stripe rust pathogen stress in wheat (Triticum aestivum L.). Mol. Biol. Rep 40:6245–53 [DOI] [PubMed] [Google Scholar]
- Zhang L, Yao L, Zhang N, Yang J, Zhu X, et al. 2018. Lateral root development in potato is mediated by Stu-mi164 regulation of NAC transcription factor. Front. Plant Sci 9:383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W, Han Z, Guo Q, Liu Y, Zheng Y, et al. 2014. Identification of maize long non-coding RNAs responsive to drought stress. PLOS ONE 9:e98958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang XO, Wang HB, Zhang Y, Lu X, Chen LL, Yang L. 2014. Complementary sequence–mediated exon circularization. Cell 159:134–47 [DOI] [PubMed] [Google Scholar]
- Zhang YC, Chen YQ. 2013. Long noncoding RNAs: new regulators in plant development. Biochem. Biophys. Res. Commun 436:111–14 [DOI] [PubMed] [Google Scholar]
- Zhang YC, Liao JY, Li ZY, Yu Y, Zhang JP, et al. 2014. Genome-wide screening and functional analysis identify a large number of long noncoding RNAs involved in the sexual reproduction of rice. Genome Biol. 15:512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Zheng Y, Ham BK, Zhang S, Fei Z, Lucas WJ. 2019. Plant lncRNAs are enriched in and move systemically through the phloem in response to phosphate deficiency. J. Integr. Plant Biol 61:492–508 [DOI] [PubMed] [Google Scholar]
- Zhao T, Wang L, Li S, Xu M, Guan X, Zhou B. 2017. Characterization of conserved circular RNA in polyploidy Gossypium species and their ancestors. FEBS Lett. 591:3660–69 [DOI] [PubMed] [Google Scholar]
- Zhao W, Cheng Y, Zhang C, You Q, Shen X, et al. 2017. Genome-wide identification and characterization of circular RNAs by high throughput sequencing in soybean. Sci. Rep 7:5636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X, Li J, Lian B, Gu H, Li Y, Qi Y. 2018. Global identification of Arabidopsis lncRNAs reveals the regulation of MAF4 by a natural antisense RNA. Nat. Commun 9:5056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou M, Palanca AMS, Law JA. 2018. Locus-specific control of the de novo DNA methylation pathway in Arabidopsis by the CLASSY family. Nat. Genet 50:865–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu QH, Stephen S, Taylor J, Helliwell CA, Wang MB. 2014. Long noncoding RNAs responsive to Fusarium oxysporum infection in Arabidopsis thaliana. New Phytol. 201:574–84 [DOI] [PubMed] [Google Scholar]
- Zuo J, Wang Q, Han C, Ju Z, Cao D, et al. 2017. SRNAome and degradome sequencing analysis reveals specific regulation of sRNA in response to chilling injury in tomato fruit. Physiol. Plant 160:142–54 [DOI] [PubMed] [Google Scholar]
- Zuo J, Wang Q, Zhu B, Luo Y, Gao L. 2016. Deciphering the roles of circRNAs on chilling injury in tomato. Biochem. Biophys. Res. Commun 479:132–38 [DOI] [PubMed] [Google Scholar]


