Abstract
Changes in the enzymatic activity of protein arginine methyltransferase (PRMT) 5 have been associated with cancer; however, the protein’s role in acute myeloid leukemia (AML) has not been fully evaluated. Here, we show that increased PRMT5 activity enhanced AML growth in vitro and in vivo while PRMT5 downregulation reduced it. In AML cells, PRMT5 interacted with Sp1 in a transcription repressor complex and silenced miR-29b preferentially via dimethylation of histone 4 arginine residue H4R3. As Sp1 is also a bona fide target of miR-29b, the miR silencing resulted in increased Sp1. This event in turn led to transcription activation of FLT3, a gene that encodes a receptor tyrosine kinase. Inhibition of PRMT5 via sh/siRNA or a first-in-class small molecule inhibitor (HLCL-61) resulted in significantly increased expression of miR-29b and consequent suppression of Sp1 and FLT3 in AML cells. As a result, significant antileukemic activity was achieved. Collectively, our data support a novel leukemogenic mechanism in AML where PRMT5 mediates both silencing and transcription of genes that participate in a “yin-yang” functional network supporting leukemia growth. As FLT3 is often mutated in AML and pharmacologic inhibition of PRMT5 appears feasible, the PRMT5-miR29b-FLT3 network should be further explored as a novel therapeutic target for AML.
Keywords: PRMT5, AML, arginine methylation, miR-29b, Sp1, FLT3
INTRODUCTION
Acute myeloid leukemia (AML) is the most common type of acute leukemia in adults.1,2,3 The incidence and mortality rate associated with AML increase with age, and the long-term overall survival achieved by younger (<60 years) and older (≥60 years) AML patients is ~30–40% and <10%, respectively.2,4 The highly heterogeneous nature of this disease (cytogenetically and clinically) complicates treatment and further highlights the significance of using biomarkers for risk-stratification and treatment guidance.5,6
Aberrant activity of epigenetic modifiers, which has been shown to play an important role in malignant transformation, represents a novel class of therapeutic targets in cancer.7,8,9,10 In AML, low-intensity treatment with DNA-hypomethylating agents has been successfully explored and been shown to have significant antileukemic effects.11,12
Recently, aberrant methylation of protein arginine residues mediated by protein arginine methyltransferase (PRMT) enzymes has been linked to cancer.13,14 PRMTs constitute a family of proteins with 11 known members that catalyze the transfer of one or two methyl groups to the guanidine nitrogen atoms of peptide arginine residues.15,16 Among these enzymes, PRMT5, having multiple substrates including histones (H3, H4, and H2A) and other, non-chromatin proteins (e.g., p53, p65, and HOXA9) 17,18,19,20 has gained attention among cancer researchers.21,22 Posttranslational histone changes induced by PRMT5 have been shown to significantly affect gene expression and ultimately induce abnormal cell growth and proliferation.23 Furthermore, overexpression of PRMT5 has been reported in hematologic and solid malignancies (mantle cell lymphoma,24,25 lung and bladder cancer, 20 gastric cancer, 26 germ cell tumors. 27).
In regard to myeloid cell malignancies, Liu et al. (2011) reported that the constitutively active tyrosine kinase (TK) mutant JAK2V617F interacts with and phosphorylates PRMT5 in myeloproliferative neoplasms. This event leads to reduced PRMT5 enzyme activity, hematopoietic progenitor expansion, and accelerated erythroid differentiation.28 To our knowledge, however, the role of PRMT5 in AML has not been fully investigated. Here we report an epigenetic-mediated proleukemic role of PRMT5 in AML through a dual function as a suppressor and activator of genes (i.e., miR-29b and FLT3, respectively), participating in a functional miR-protein network that contributes to leukemia growth.
MATERIALS AND METHODS
Cell lines and primary blasts
MV4–11 and THP-1 cells (ATCC, Manassas, VA) were maintained in RPMI 1640 medium supplemented with 10% calf serum. Blasts from AML patients were obtained from apheresis blood samples collected from patients treated at the Ohio State University (OSU) and stored in the OSU Leukemia Tissue Bank. Informed consent to use cells for investigational studies was obtained from each patient under an OSU Institutional Review Board-approved protocol. These cells were maintained in RPMI 1640 medium supplemented with 20% fetal bovine serum, 1% HEPES buffer, and 1x StemSpan cytokine cocktail CC100 (StemCell Technologies, Vancouver, BC, Canada) containing IL-3, IL-6, and SCF. All cells were incubated at 37°C with 5% CO2. For cytogenetic and molecular characteristics of cell lines and patients’ blasts, see Supplementary Table S1.
Plasmids, transient transfections and reagents
PRMT5 cDNA and shRNA were cloned, respectively, into pCDH1-MSCV-GFP-puro-cDNA (System Biosciences) and pLKO.1-Puro shRNA (Sigma) lentivector. The full-length PRMT5, PRMT5 (WT), and the catalytically inactive mutant form (G367A/R368A), PRMT5 (Mut), were cloned 29 into pBABE-puro vector (Addgene). On-target plus siRNA-SMART pools specific for PRMT5 and Sp1 and an off-target scrambled control were obtained from Dharmacon (Lafayette, CO). A locked nucleic acid (LNA)-antimiR-29b inhibitor (hsa-miR-29b mercury LNA microRNA Power inhibitor, Exiqon, Woburn, MA) was used to knockdown miR-29b, and synthetic Pre-miR™ miRNA Precursor (Ambion) was used to overexpress miR-29b. MicroRNA transfections were carried out by siPORT NeoFX transfection reagent (Life Technologies) including proper scrambled negative control for each treatment. Gene knockdown and overexpression was carried out either by electroporation using Nucleofector Kit (Amaxa, Walkersville, MD) or infection by lentivirus (System Biosciences). PKC412 (Sigma-Aldrich, M1323) and FLT3 inhibitor (Calbiochem #343020) were purchased, whereas HLCL-61 (HLCL-61) was prepared by Hongshan Lai at Ohio State University.
Colony formation assay and cell viability analysis
Clonogenic assays were prepared by plating 1×103 cell/mL in a semisolid methylcellulose medium (MethoCult, Stem Cell Technologies). Colonies were counted after 10–14 days using an inverted microscope. Growth inhibition assays were measured using a colorimetric MTS assay: 5×104 cells were plated in 100μL final volume in a 96-well plate in the presence of different concentrations of HLCL-61 for 24, 48, and 72 hours at 37°C. Afterwards, 20 μL of the CellTiter 96W AQueous One Solution Reagent, which contains a tetrazolium compound (MTS) and an electron coupling reagent (PES) (Promega, Madison WI), was added to each well. Within 1–4 hours of incubation at 37°C, the optical density at 490 and 690 nm was measured. Cell viability was calculated with respect to the control samples and a reference background wavelength at 690 nm. At least three independent experiments were performed. Growth curve assays were performed by counting live cells using trypan blue exclusion and an inverted microscope for >12 days after plating the cells in 1×105 concentrations.
THP-1 xenograft murine model
To generate a PRMT5-overexpressing AML xenograft mouse model, THP-1 cells were transduced with a PRMT5-overexpressing lentivirus marked with GFP. Positively infected cells (1×106) were injected via tail vein into ~4–6-week-old non-obese diabetic severe combined immunodeficient gamma (NSG) mice (NOD.Cg-Prkdcscid Il2rgtm1Wjl/SzJ, The Jackson Laboratory, Bar Harbor, ME). After 10 weeks, mononuclear cells isolated from blood and spleen of sacrificed animals were processed for RNA and protein isolation. Bone marrow cells were stained with Wright-Giemsa staining to visualize the leukemic blasts. Spleen and liver were fixed in 10% formalin, and the sections were stained with hematoxylin-eosin staining. Mouse survival assays were carried out similarly except by injecting 2×106 THP-1 cells, and sick mice were scored and sacrificed upon manifestation of AML signs. All animal studies were performed in accordance with OSU institutional guidelines for animal care and under protocols approved by the OSU Institutional Animal Care and Use Committee.
A detailed description for Western immunoblot and immunoprecipitation (IP), flow cytometry, PRMT5 expression in the TCGA AML cohort, chromatin immunoprecipitation (ChIP), RNA isolation and real-time PCR, luciferase reporter assay, statistical analysis, and RNA-seq analysis is included in Supplementary Data.
RESULTS
Upregulated PRMT5 contributes to leukemia growth
Following reports of PRMT5 overexpression and its positive contribution to maintaining the malignant phenotype in cancer, we investigated its effect on the leukemic phenotype and AML growth. We forced expression of PRMT5 in FLT3-ITD AML (MV4–11) and FLT3-WT AML (THP-1) cells. The FLT3 gene encodes a receptor tyrosine kinase that contributes to regulatory hematopoietic mechanisms. AML blasts often harbor FLT3 mutations [i.e., internal tandem duplication (ITD)], resulting in ligand-independent activation of the encoded protein, abnormal cell proliferation, and poor outcome. Cells were infected with PRMT5-expressing lentivirus (Lenti-PRMT5), or empty vector (Lenti-EV) control. PRMT5 expression was tested by Western blotting 48 hours later. A significant increase in proliferation and colony forming ability was observed in Lenti-PRMT5-transduced AML cell lines compared with empty vector (EV)-transduced controls (Figure 1A and 1B). Similarly, forced PRMT5 expression in primary blasts from FLT3-ITD and FLT3-WT AML patients led to significantly higher number of colony forming units (Figure 1C and 1D). Consistent with these results, we observed that ectopic expression of a catalytically inactive PRMT5 mutant yielded a colony number lower than that obtained by ectopic expression of wild-type PRMT5 (Supplementary Figure S2).
Figure 1. PRMT5 activity is important for AML cell survival and growth in vitro.
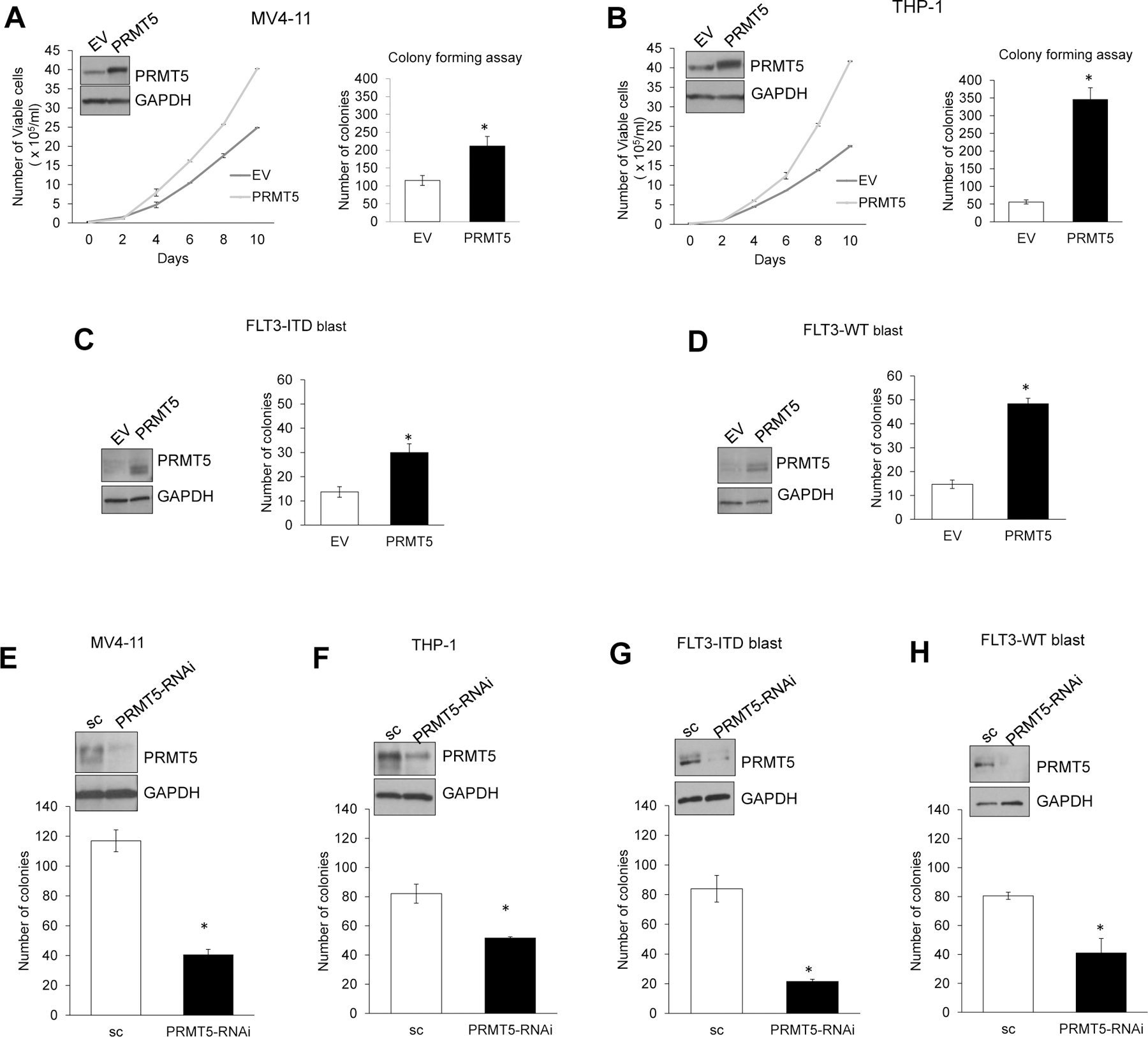
(A) Left: Growth curve assay comparing proliferation rate of MV4–11 AML cells transduced with PRMT5 or empty (EV) lentivector. Western blotting was used to confirm PRMT5 overexpression in Lenti-PRMT5 treated cells compared to Lenti-EV, while GAPDH levels served as loading control (inset). Right: Bar graph depicting colony formation assay comparing proliferation potential of AML cells transduced with Lenti-PRMT5 or empty vector control. Colonies were counted 14 days after plating. All results are from three independent experiments per treatment. (B) Growth curve and colony forming assay in THP-1 AML cell line overexpressing PRMT5. (C) and (D) Colony forming assays in AML primary blasts transduced with PRMT5 or EV lentivector. (E-H) Colony formation assay in AML cell lines and patient primary blasts transfected with either PRMT5 RNA interference oligos (siPRMT5 or shPRMT5) or scrambled control (sc). Western blotting was used to confirm sufficient downregulation of PRMT5 in presence of si/shPRMT5. * is p<0.05, and ** is p<0.005. Bars indicate SEM.
Conversely, PRMT5 depletion significantly hindered AML cell growth as shown by decreased colony forming activity. To this end, MV4–11 (FLT3-ITD) and THP-1 (FLT3-WT) cell lines and patient blasts (FLT3-ITD or FLT3-WT) were treated with either scrambled control (sc) or RNA interference (RNAi) (siRNA or shRNA) against PRMT5. Significantly fewer colonies were formed by PRMT5-depleted cells than controls (at least 1.6 fold, P=0.01) (Figure 1E–H). Stable PRMT5 knockdown resulted in 100% cell death (not shown). Comparable results were obtained in vitro on additional cell lines with different genetic backgrounds (Supplementary Figure S4).
The impact of PRMT5 on leukemia growth was then validated in vivo. NOD/SCID/Gamma (NSG) mice were injected through the tail vein with THP-1 (1×106) cells that were stably transduced with either Lenti-PRMT5 (THP-1/PRMT5) or Lenti-EV (THP-1/EV) control. After ten weeks, three mice from each group were sacrificed for analysis. Mice with THP-1/PRMT5 had significantly higher white blood cell (WBC) counts (4.2 fold, P=0.04) compared with THP-1/EV (Figure 2A). Cytospin of peripheral blood from THP-1/PRMT5 showed more circulating blast cells compared with that from THP-1/EV, which had mostly differentiated cells (Figure 2B). THP-1/PRMT5 mice had also higher extramedullary disease burden with larger spleens (3.5 fold, P=0.03) and livers (1.7 fold, P=0.03) compared with the THP-1/EV control (Figure 2C). In addition, histopathology of hematoxylin/eosin-stained sections of spleen and liver revealed that THP-1/PRMT5-engrafted mice had extensive infiltration of blasts with low myeloid maturation compared with THP-1/EV-engrafted mice (Figure 2D). Higher PRMT5 transcript levels were confirmed in RNA from bone marrow (BM), spleen, and blood from THP-1/PRMT5-engrafted animals (Figure 2E). In addition, PRMT5 protein levels and associated specific histone methylation marks, H3R8me2 and H4R3me2, were significantly higher in THP-1/PRMT5 mice than in control animals (Figure 2E). THP-1/PRMT5 mice had a shorter survival compared to THP-1/EV controls (median survival, 60 vs. 88 days, P<0.0001) (Figure 2F). Taken together, these data support a role of PRMT5 in enhancing leukemia growth in AML.
Figure 2. PRMT5 activity contributes to leukemia growth in vivo.
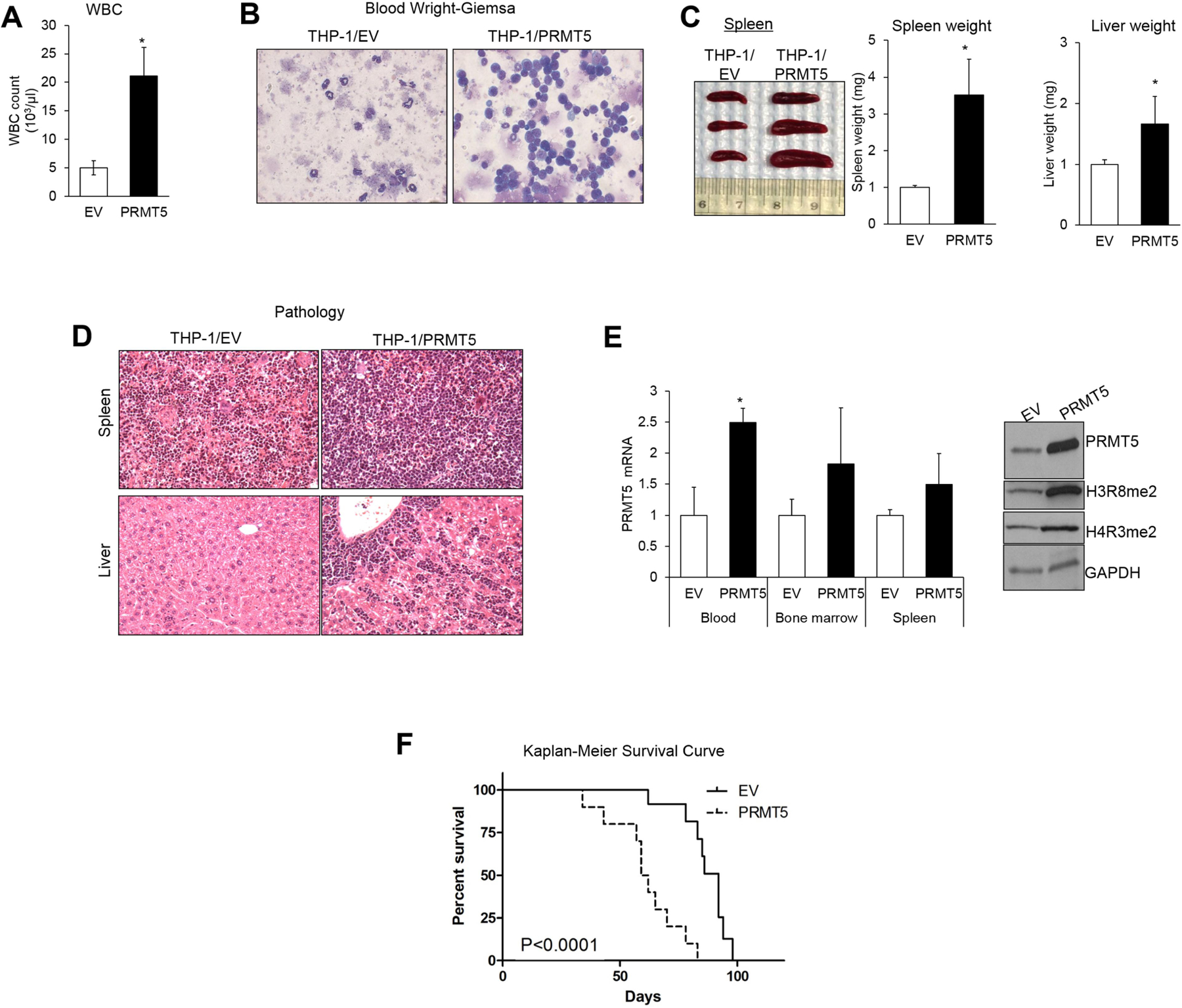
(A) White blood cell (WBC) counts in PRMT5-overexpressing mice and controls. (B) Wright-Giemsa staining of blood of THP-1/PRMT5 vs THP1-EV cell-engrafted mice. (C) Spleens harvested from mice engrafted with THP-1 cells transduced with PRMT5 or EV lentivectors. 1×106 cells were injected per mouse, and animals were euthanized after 10 weeks. Enlarged spleens and livers were in mice engrafted with THP1/PRMT5 compared to those engrafted with THP1/EV. The bar graphs represent significant increases in weight of the spleens and livers taken from PRMT5-overexpressing animals compared to negative control mice. (D) Hematoxylin-eosin staining of liver and spleen of THP-1/PRMT5 vs THP1-EV cell-engrafted mice. (E) Left: PRMT5 expression by quantitative RT-PCR in THP-1/PRMT5 vs. THP-1/EV mice. Right: Western blotting showed upregulation of PRMT5 and associated epigenetic marks, symmetrically dimethylated H3 (H3R8me2) and H4 (H4R3me2), in representative spleen samples from THP-1/PRMT5 vs THP-1/EV mice. Immunostaining was performed with anti-PRMT5 antibody followed by stripping and re-staining with anti-H3R8 and anti-H4R3 antibodies while GAPDH levels served as internal loading control. (F) Survival curve comparing survival of THP-1/PRMT5 vs THP-1/EV mice; 2×106 cells were injected in each mouse.
Antileukemic activity of PRMT5 inhibition in AML cells
To provide further insight into the role of PRMT5 as a therapeutic target, we next sought inhibition of PRMT5 enzymatic activity using a first-in-class small molecule inhibitor. On the basis of our first generation PRMT5 inhibitor, BLL1, developed using in silico modeling of the human PRMT5 catalytic site, we incorporated a PRMT5 crystal structure into a revised model. In Supplementary Figure S3A, BLL-1 is depicted as a blue stick model binding to the PRMT5 active site with the carbazole moiety overlapping with cofactor SAM and the pyridine ring competing with the substrate histone arginine side chain. To enhance potency of this drug, we replaced the pyridine with another aromatic fragment to both preserve aromatic stacking with Phe327 and Typ579 and to extend molecular interactions in the local environment. One salient feature in this environment is the presence of a Lys333 side chain available for hydrogen bonding. We selected 21 neutral aromatic fragments with potential hydrogen bonding acceptors and performed simultaneous docking of the carbazole structure modified with each of the 21 fragments using the Multiple Ligand Simultaneous Docking (MLSD) method.30–32 A methoxybenzene fragment with oxygen hydrogen bonded to the Lys333 side chain showed favorable binding as depicted in Figure S1A as an atomically colored ball-and-stick model that virtually overlapped space occupied by the first generation inhibitor BLL-1. The calculated binding energy was 1 Kcal/mol (5-fold) stronger than BLL-1 because of the additional hydrogen bond. The selective inhibitory activity of this compound against PRMT5 persisted for up to 14 days (at 4oC) in an in vitro enzyme assay (not shown).
Using an in vitro enzyme assay to measure methylase activity, HLCL-61 was determined as one of the most potent of the selected inhibitors (Supplementary Figure S3B). HLCL-61 showed no inhibitory activity against the type I (PRMT1 and PRMT4) and type II (PRMT7) PRMT family members (Supplementary Figure S3B), thus indicating its specificity for PRMT5. HLCL-61 showed effective inhibition of symmetric arginine dimethylation (me2) of histones H3 and H4 in AML samples, starting at 12 hours post-treatment and persisting after 48 hours (Figure 3A).
Figure 3. Preclinical antileukemic activities of PRMT5 inhibition in AML.
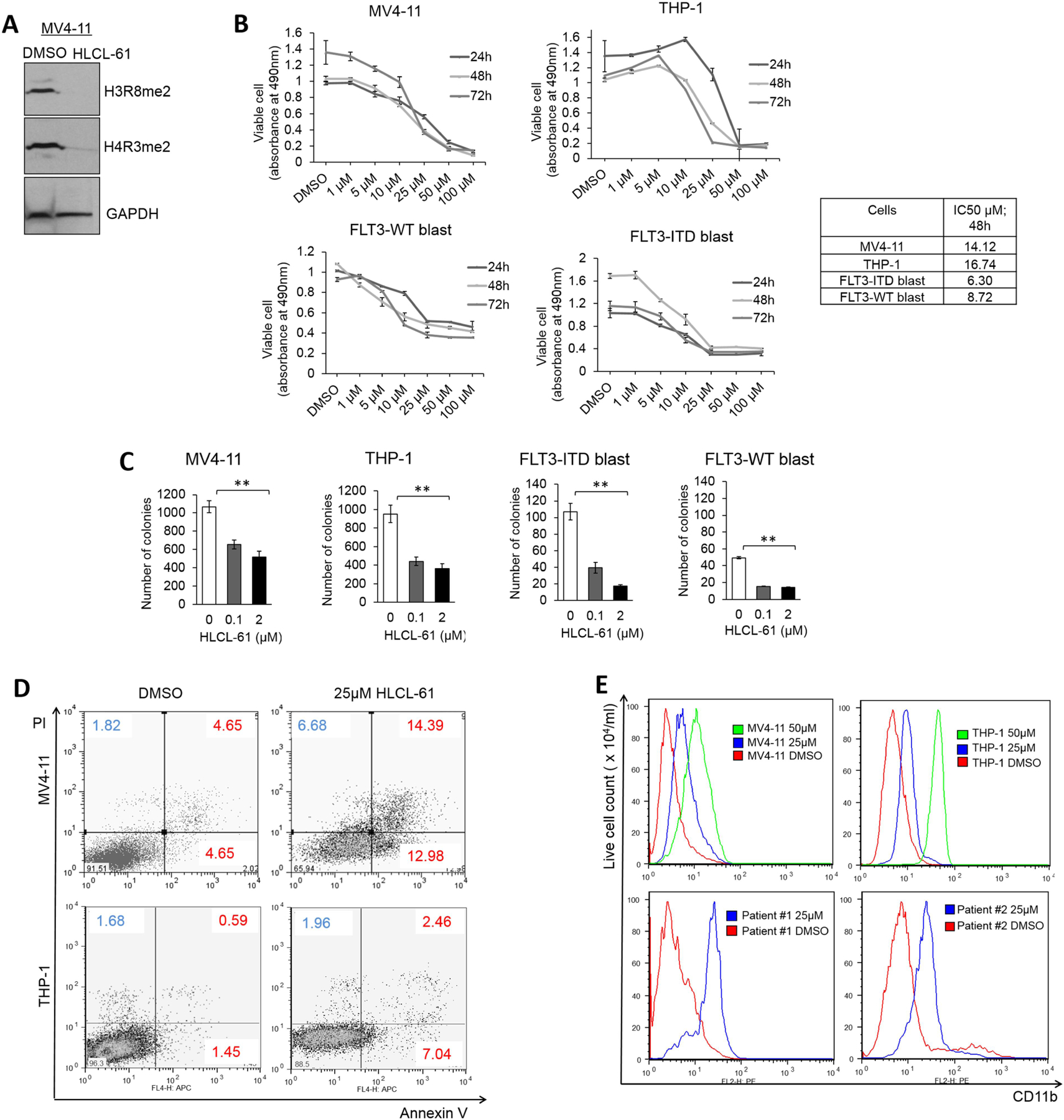
(A) Western blotting in AML cell line indicating efficient PRMT5 inhibition as shown by downregulation of symmetrically dimethylated H3 (H3R8me2) and H4 (H4R3me2) in the presence of HLCL-61. (B) MTS assay measuring the proliferation rate of AML cell lines and primary blasts from patients after 24h, 48h, and 72h incubation with different doses of PRMT5 inhibitor compound HLCL-61. Dose-dependent decrease in absorbance directly correlates with number of metabolically active live cells. IC50 values represent the concentration of the compound at which 50% cell death was achieved. (C) Colony formation assay to measure significantly reduced proliferation potential of AML cell lines and primary tumor cells in the presence of sub-lethal doses of HLCL-61. Colonies were counted 14 days after plating, and each treatment was carried out in triplicates. (D) Flow cytometry analysis of AML cell lines treated with PRMT5 inhibitor and stained for Annexin V and PI showing a dose-dependent increase in percentage of apoptotic and dead cells when compared to DMSO-treated cells after 48 hours. (E) Histogram of staining with early differentiation marker CD11b shows induction of differentiation in AML cell lines and in patient samples treated with HLCL-61 for 48 hours. The increase in CD11b expression in AML cell lines followed a dose-dependent manner.
Treatment of AML cell lines (MV4–11 and THP-1) and primary blasts with HLCL-61 also resulted in a decrease of cell viability (Figure 3B). Analysis of other AML samples with different genetic/molecular variations yielded similar results (Supplementary Figure S4A). The IC50 values of the drug with respect to inhibition of cell growth were in the low micromolar range and did not change dramatically over time (24–72 hours) for all AML cells tested (At 48 hrs treatment, IC50 = 7.21–21.46 μM for cell lines and 3.98–8.72 μM for patient blasts; Supplementary Figure S4A). Colony forming assays also demonstrated decreased clonogenic activity (>2.2 fold, as compared to DMSO-treated cells) of MV4–11, THP-1, and patient blasts (Patient #1; FLT3-ITD and Patient #2; FLT3-WT) treated with HLCL-61, even at concentrations as low as 100 nM (Figure 3C). HLCL-61 was also effective in promoting apoptosis in MV4–11 and THP-1 cells after 48 hours (Figure 3D).
To determine whether HLCL-61 could induce myeloid differentiation, we tested CD11b expression by flow cytometry on cells treated with DMSO or HLCL-61 (25 and 50 μM) for 48 hours. HLCL-61–treated AML cell lines and patient samples exhibited induction of differentiation as suggested by dose-dependent increases in the expression of CD11b (Figure 3E). These data support the potential of pharmacologic targeting of PRMT5 in AML as a novel antileukemic therapy.
Epigenetic role of PRMT5 in repression of miR-29b
Among the known substrates of PRMT5 are histones H4R3 and H3R8. It has been shown that symmetric dimethylation (me2) of H4R3 and H3R8 lead to transcription repression, whereas that of H3R8 has also been associated with transcriptional activation.33,34 Chromatin immunoprecipitation (ChIP)-Seq data collected from the mantle cell lymphoma cell lines Jeko and Mino used by our group as a prototype of malignant expression of PRMT5 revealed, among other genes, miR-29b gene regulatory elements to be >10-fold enriched for histone arginine me2 (when compared with normal IgG negative control), suggesting this locus as a potential PRMT5 target (Supplementary Figure S5). We previously reported that miR-29b silencing in AML leads to overexpression of several miR-29b target oncogenes.35,36 Therefore, we hypothesized here that the proleukemic effect of PRMT5 activation could be partly related to the impact of PRMT5 on miR-29b expression through histone arginine methylation. Consistent with our hypothesis, we detected downregulation of miR-29b in both blood and splenic mononuclear cells from THP-1/PRMT5-engrafted mice, as compared to those from THP-1/EV controls (Figure 4A). Furthermore, forced expression of PRMT5 in patient blasts resulted in suppression of miR-29b (Figure 4B), whereas increased miR-29b expression was observed in PRMT5 RNAi-treated (Figure 4C, see also RNA-seq analysis in Supplementary Figure S8) or HLCL-61–treated (Figure 4D) AML cells within 48 hours after PRMT5 inhibition.
Figure 4. Epigenetic role of PRMT5 in repression of miR-29b.
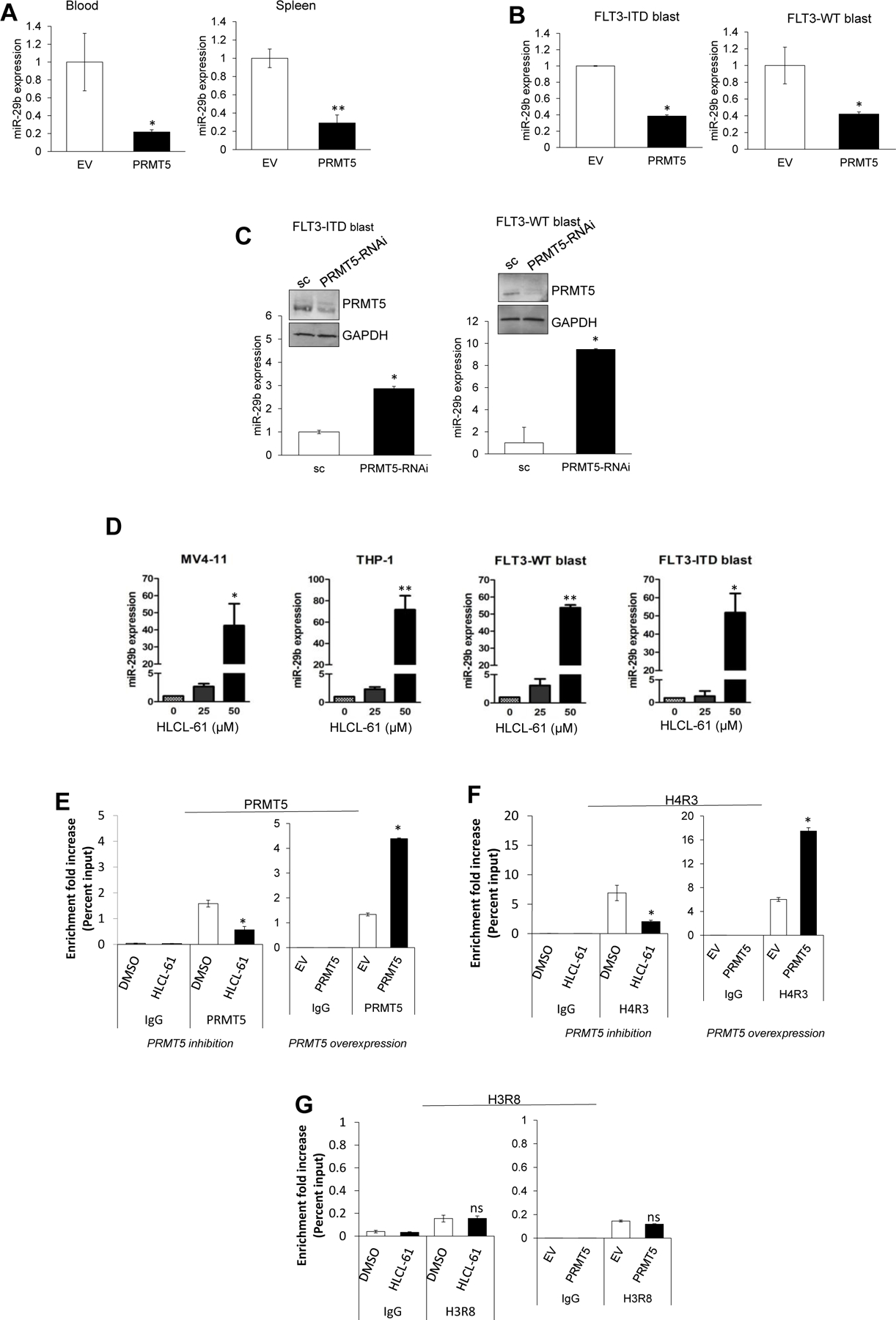
(A) qRT-PCR analysis measuring reduced miR-29b levels in blood and spleen from THP-1/PRMT5 vs THP-1/EV mice. (B) Expression of mature miR-29b following ectopic expression of PRMT5 in AML blasts. (C) Expression of miR-29b in AML blasts treated with PRMT5-siRNA as measured by quantitative PCR. (D) Time-dependent changes in miR-29b expression in AML cell lines and patient blasts after 24 hours incubation with HLCL-61. (E-G) ChIP assay to assess enrichment of PRMT5 as well as H4R3me2 and H3R8me2 (methylation marks deposited by PRMT5) on the enhancer region of miR-29b. Inhibition of PRMT5 with HLCL-61 resulted in significant decrease of H4R3me2 and PRMT5 localization onto miR-29b enhancer site. PRMT5 overexpression significantly enhanced the localization of H4R3me2 methylation mark at the miR-29b regulatory region.
To demonstrate the potential regulatory role of PRMT5 on miR-29b expression, we tested the regulatory region of miR-29b as a possible docking site for PRMT5 using ChIP assays. Enrichment of PRMT5 and the H4R3me2 methylation mark, in contrast to negative control normal IgG, were observed in the miR-29b regulatory region in AML cell lines (genomic location and primers: Supplementary Figure S7). PRMT5 inhibition by HLCL-61 and PRMT5 overexpression led to a decrease or increase, respectively, of both H4R3me2 and the presence of PRMT5 on the miR-29b regulatory element in MV4–11 cells (Figure 4E and 4F). No changes in H3R8me2 enrichment were detected (Figure 4G).
We previously reported the role of the transcription factor Sp1 and histone deacetylases (HDACs) in the transcription repression of miR-29b.37 Thus, here we tested whether PRMT5 could also be a part of this complex. Using co-immunoprecipitation (Co-IP) assays, we demonstrated the physical interaction of PRMT5 and Sp1 (Figure 5A), suggesting a possible anchoring and recruiting role of Sp1 for PRMT5 and other proteins participating in such transcriptional repressor complexes.
Figure 5. PRMT5 interacts with and significantly upregulates Sp1 protein levels.
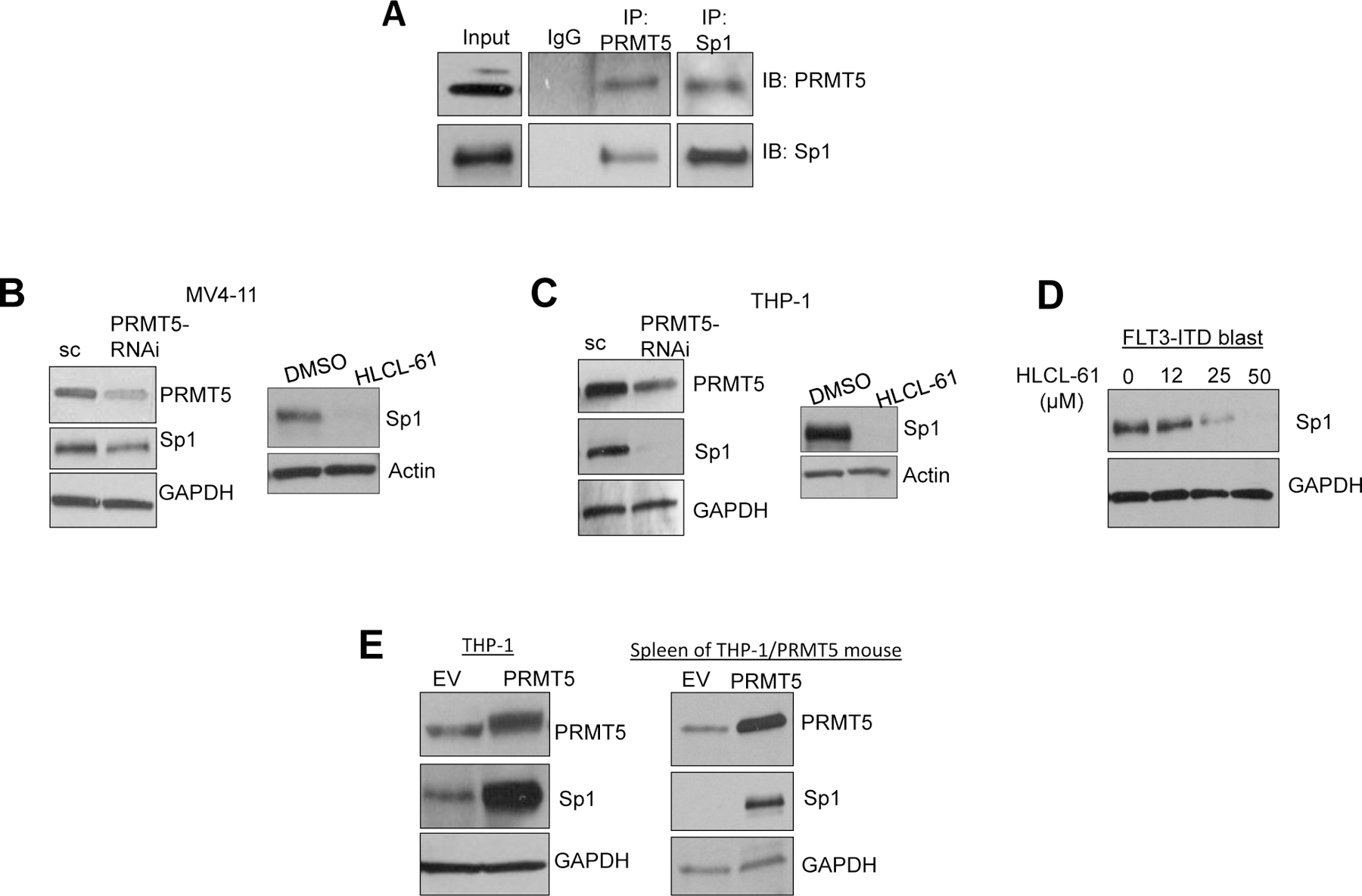
(A) Immunoprecipitation (IP) in AML cells showing physical association between PRMT5 and Sp1. Pull-downs were carried out using antibodies specific to PRMT5 and Sp1 and to normal IgG as negative control. Immunoblotting was performed with anti-PRMT5 and anti-Sp1 antibodies and was executed in a sequential manner including stripping and re-staining following each antibody. (B) and (C) Sp1 protein levels measured by Western blotting following inhibition of PRMT5 in AML cell lines. PRMT5 inhibition was carried out by HLCL-61 and si/shPRMT5; in both cases inhibition of PRMT5 resulted in significant knockdown of Sp1. Actin or GAPDH staining was used as internal loading control. (D) Downregulation of Sp1 following PRMT5 inhibition in HLCL-61–treated AML primary blasts. (E) Left: Sp1 expression by Western blotting in THP-1 cells overexpressing PRMT5 using lentivirus. Right: Sp1 expression in THP-1/PRMT5 vs THP-1/EV cells in samples from engraftment assay described in Figure 2.
We also have shown that miR-29b effectively targets Sp1 mRNA 35 (Supplementary Figure S6A and S6B) and that Sp1 participates in a miR-29b autoregulatory loop where Sp1 can repress miR-29b and indirectly upregulate its own expression.37 Therefore, we reasoned that increased PRMT5 activity should be linked to an increased Sp1 expression. In fact, PRMT5 inhibition by RNAi or HLCL-61 resulted in a significant decrease of Sp1 protein levels in AML cell lines and blasts (Figure 5B–D). Conversely, overexpression of PRMT5 in THP-1 cells resulted in significant Sp1 upregulation (Figure 5E). A similar trend was observed in vivo in THP-1/PRMT5-engrafted mice compared to THP-1/EV-engrafted controls (Figure 5E). Additionally, in a rescue experiment, Sp1 knock-down mediated an increase in miR-29b expression in cells that ectopically expressed PRMT5 (and had low miR-29b), thereby functionally linking PRMT5 activity with the expression of Sp1 and its activity on miR-29b (Supplementary Figure S6C–F). Consistently, in MV4–11 cells, an HLCL-61–dependent increase of miR-29b was attenuated by Sp1 ectopic expression (Supplementary Figure S6G). These data support a co-regulatory role of PRMT5 in transcriptional repression of the miR-29b locus, most likely via an H4R3me2 methylation event.
PRMT5 enhances FLT3 transcription in AML
Recently, it was reported that in myeloproliferative neoplasms, PRMT5 can be inhibited via phosphorylation by the constitutively active mutant of tyrosine kinase JAK2 (JAK2V617F). 28 In this context, phosphorylated PRMT5 is less active and results in enhanced expansion of hematopoietic progenitor cells and erythroid differentiation. These data suggested a direct interplay of PRMT5 with tyrosine kinases (TK) in myeloid lineage cells. JAK2 is rarely mutated in AML, but Cook et al. (2014) recently reported increased JAK/STAT activity in AML leukemia stem cells (LSC) that was associated with increased expression of the receptor tyrosine kinase (RTK) KIT and of FLT3, leading to leukemia growth. 38 Since wild type or constitutively active mutants (i.e., FLT3-ITD) of FLT3 are frequently expressed in AML, we first hypothesized a possible interplay between FLT3 and PRMT5. To address this question, we immunoprecipitated PRMT5 from FLT3-WT and FLT3-ITD AML cell lines and tested its interaction with the FLT3 protein. Figure 6A shows that, contrary to the findings with non-receptor tyrosine kinase JAK2V617F,28 we could not detect a significant physical association between FLT3 proteins and PRMT5 in AML cells.
Figure 6. PRMT5 enhances FLT3 transcription in AML.
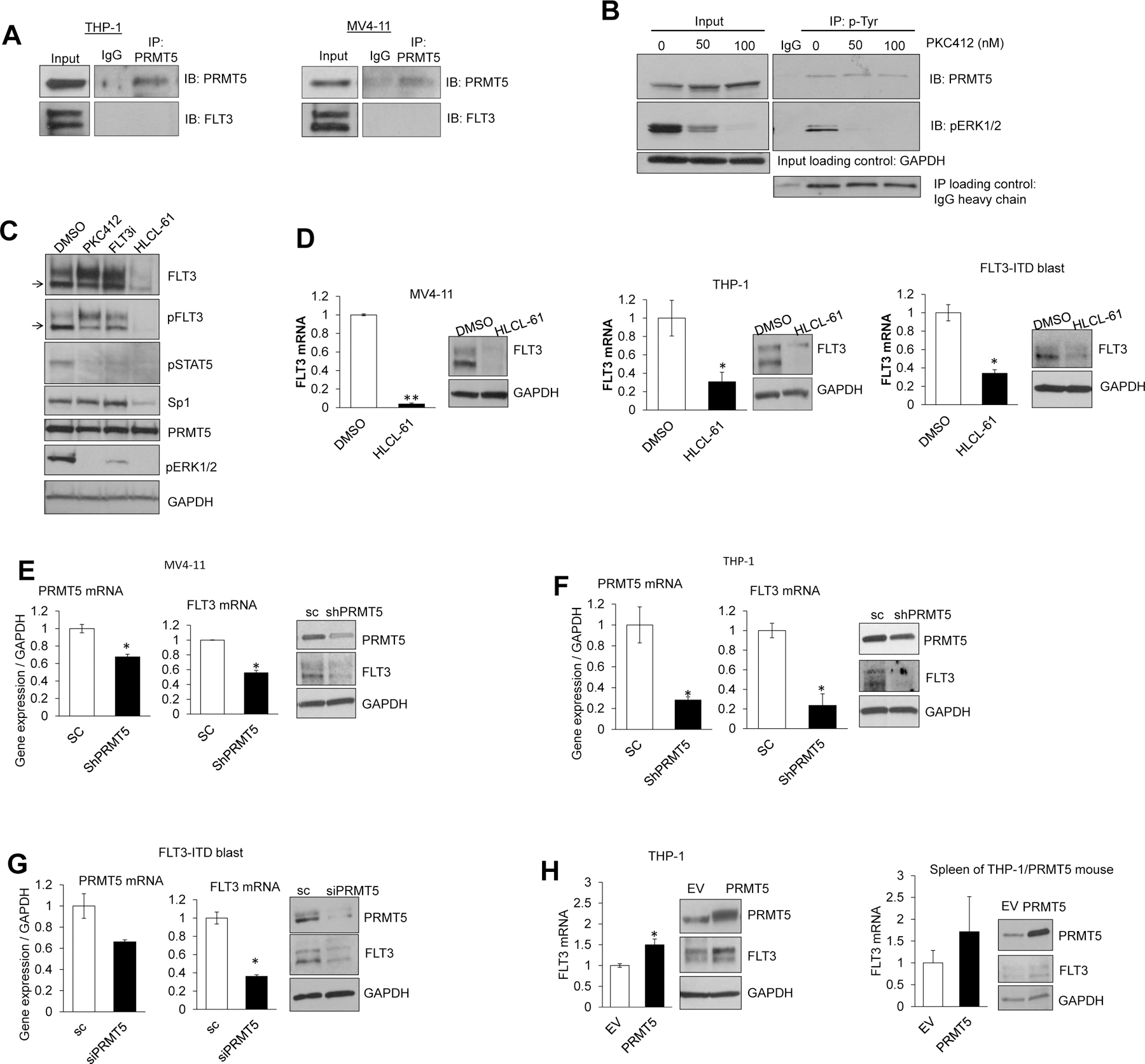
(A) Immunoprecipitation (IP) assay in FLT3-WT THP1 and FLT3-ITD MV4–11 cell lines; pulling down was performed with anti-PRMT5 and immunostaining with anti-PRMT5 and anti-FLT3. No physical association between PRMT5 and FLT3 was detected. (B) Inhibition of FLT3 kinase activity did not influence overall phosphorylated PRMT5 levels. Whole cell lysate was used to pull down phosphorylated tyrosine residues (anti-p-Tyr) in cells treated with FLT3 inhibitor. Phosphorylated ERK1/2 (p-ERK1/2) level was detected as control for p-Tyr IP and effectiveness of kinase inhibitory effects of treatment. (C) Western blotting of treatment of MV4–11 cells with PKC412, specific FLT3 inhibitor (FLT3i), and HLCL-61 for 5 hours. Levels of phosphorylated STAT5 (pSTAT5) and pERK1/2 were detected as positive control to confirm the effective FLT3 inhibition. Immunostaining was carried out sequentially after stripping the membrane following staining with anti-FLT3, anti-pFLT3, anti-Sp1, anti-PRMT5, anti-pSTAT5, anti-pERK1/2, and anti-GAPDH. (D) Treatment of AML cell lines and primary blasts with HLCL-61 led to significant suppression of FLT3 mRNA and protein levels. qRT-PCR was used to measure relative FLT3 mRNA levels and Western blotting to detect protein levels of FLT3 and GAPDH. (E-G) Small RNA interference knockdown of PRMT5 resulted in significant downregulation of FLT3 mRNA and protein expression in different AML cells. (H) Forcing PRMT5 expression in THP-1 cells resulted in FLT3 mRNA and protein upregulation.
FLT3 inhibition by either PKC412 or FLT3i (Calbiochem #343020) as assessed by Western blotting did not change the expression and/or phosphorylation levels of PRMT5 (Figure 6B) or the expression of the PRMT5 downstream target Sp1 (Figure 6C), thereby supporting that changes in FLT3 activity may have no impact on PRMT5. In contrast, treatment with HLCL-61 showed downregulation in Sp1 levels, seen only with HLCL-61 exposure (Figure 6C), further supporting our data showing that Sp1 is a downstream target of PRMT5.
Interestingly, we noted that HLCL-61 led not only to the decrease of the PRMT5 downstream target Sp1 but also to a substantial decrease of FLT3 (both WT and ITD) levels in cell lines and blasts (Figure 6D). Similarly, knock-down of PRMT5 by shRNA decreased FLT3 mRNA and protein expression (Figure 6E–G). Conversely, forced PRMT5 expression resulted in increased levels of FLT3 mRNA and protein in vitro and in vivo (Figure 6H).
We previously reported an Sp1-NFκB(p65) transactivation complex that regulates FLT3 expression in AML.37,11 Here, siRNA-mediated depletion of Sp1 resulted in significant downregulation of FLT3 expression (>2.6 fold) (Figure 7A). We have shown that PRMT5 can induce Sp1 expression through the miR-29b autoregulatory loop.37,11 Therefore, we reasoned that interfering with PRMT5 expression and activity may lead to changes in FLT3 expression. To demonstrate the effect of PRMT5 on the direct binding of Sp1 to the FLT3 promoter, we performed a ChIP assay using antibodies specific for Sp1 and primers designed to amplify predicted Sp1 binding sites (Supplementary Figure S7). HLCL-61–mediated inhibition of PRMT5 led to a significant decrease in Sp1 enrichment on the FLT3 promoter and was associated with downregulation of FLT3 expression in MV4–11 (Figure 7B). Conversely, forced expression of PRMT5 resulted in significantly enhanced binding of Sp1 to the FLT3 promoter and was accompanied by a 3-fold increase of FLT3 expression (Figure 7C). Furthermore, to support a functional role for PRMT5 in modulating FLT3 expression, the FLT3 promoter region containing the Sp1 binding site was cloned upstream of the firefly luciferase gene in the pGL4.11[luc2P] vector and transfected to THP-1 cells. Figure 7D shows that HLCL-61 treatment for 6 hours decreased the luciferase activity (4-fold) compared to DMSO-treated control.
Figure 7. PRMT5 controls FLT3 expression by modulating Sp1.
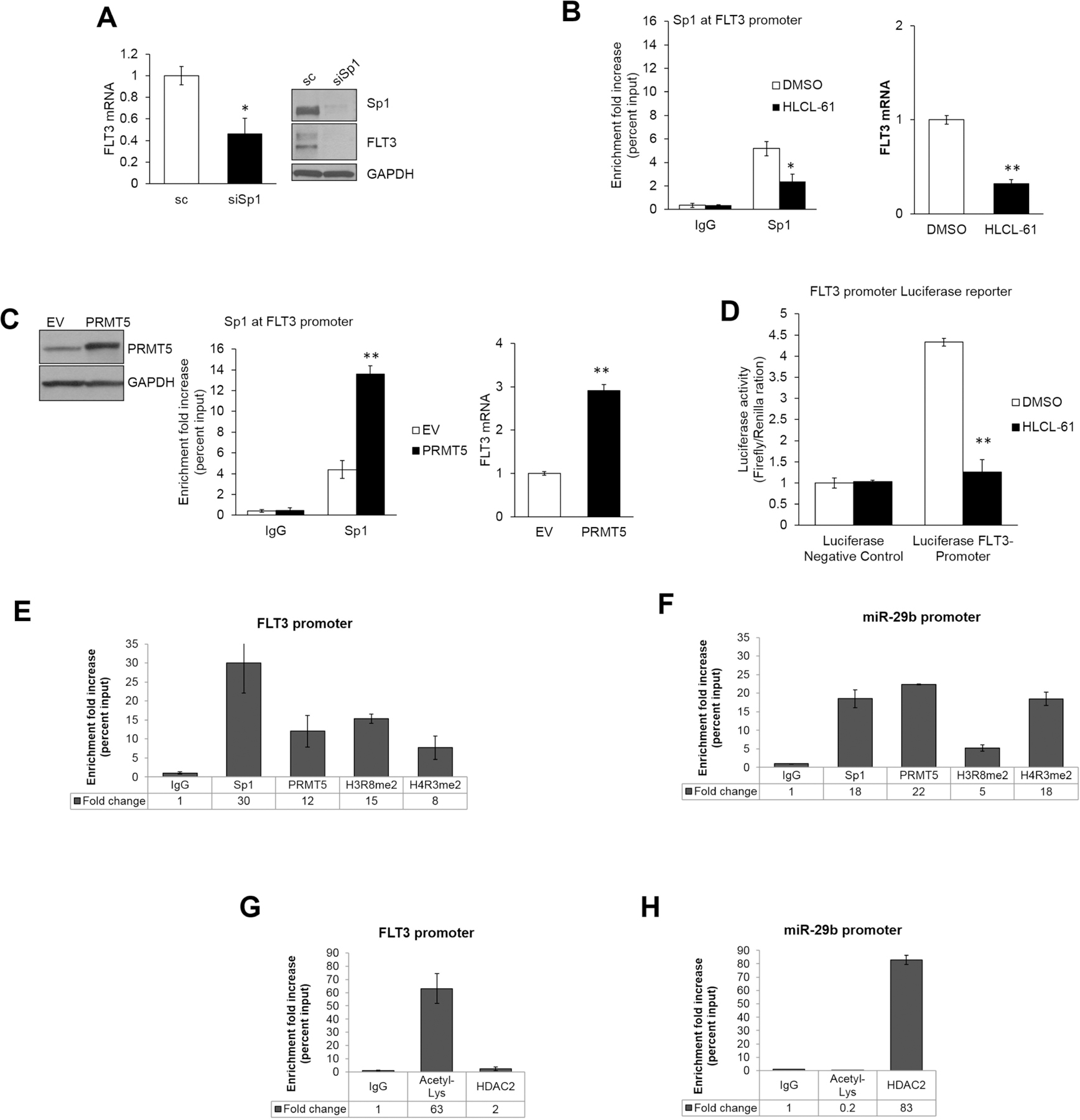
(A) FLT3 expression by qRT-PCR assay and Western blotting after transient transfection of MV4–11 AML cells with siRNA specific to Sp1 (B) Left: Chromatin immunoprecipitation (ChIP) assay to test for Sp1 enrichment on the FLT3 promoter region. Right: FLT3 transcription levels after treatment with HLCL-61 compared to vehicle treated controls. (C) Left: ChIP assays in MV4–11 cells to assess Sp1 enrichment on the FLT3 promoter upon forced expression of PRMT5 (see Western blotting). (D) Luciferase reporter assay to assess activity of the FLT3 promoter region in THP-1 cells transiently transfected with Luciferase-FLT3-promoter and Luciferase-negative-control in the presence or absence of HLCL-61. Luciferase activity measured relative to Renilla in three independent experiments. (E) ChIP assays measuring Sp1, PRMT5, H3R8me2, and H4R3me2 presence at FLT3 promoter. (F) ChIP assays measuring Sp1, PRMT5, H3R8me2, and H4R3me2 presence at miR-29b regulatory region. (G) ChIP assay to measure levels of acetylated histones and HDAC2 recruitment at FLT3 promoters. (H) ChIP assay to measure presence of acetylated histones and HDAC2 at miR-29b regulatory site.
Since we have identified a physical interaction between PRMT5 and Sp1 proteins (Figure 5A), we also tested for the direct presence of PRMT5 on the FLT3 promoter and its effect on histone methylation at this site. As expected, in addition to Sp1, enrichment of PRMT5, H3R8me2, and H4R3me2 was detected on the FLT3 promoter in MV4–11 cells when compared to the non-specific IgG negative control (Figure 7E). However, in contrast to the miR-29b promoter (Figure 7F), the levels of H3R8me2 on the FLT3 promoter were almost two-fold higher than H4R3me2. In addition, the FLT3 promoter presented high histone acetylation levels and no enrichment for HDACs (Figure 7G), which was in contrast to the hypoacetylated miR-29b regulatory region (Figure 7H). Altogether, these data support the role of PRMT5 as a two-fold gene transcription activator and repressor participating in a complex microRNA-protein regulatory network in AML.
DISCUSSION
Previous studies have shown that PRMT5 is overexpressed in a number of malignancies and is associated with enhanced tumor growth.39,40,41 Here we first demonstrated the positive contribution of PRMT5 to AML growth in vitro and in vivo. PRMT5 upregulation resulted in a survival and proliferation advantage in AML cell lines and primary blasts, along with decreased survival of mice engrafted with PRMT5-overexpressing AML cells. Consistent with these results, attenuation of PRMT5 expression and activity in AML cells significantly decreased growth and led to partial cell differentiation and eventually apoptosis. Thus, overall our work indicates a dynamic contribution of PRMT5 to leukemia growth.
The mechanisms through which PRMT5 promotes AML growth, however, appear to be quite complex and may involve methylation of both cytoplasmic and nuclear proteins. Herein, we focused on the participation of the protein in a miR-29b-Sp1–related microRNA-protein network that we previously reported to promote AML growth.37,42 We had verified that aberrant miR-29b repression is induced by a protein complex comprising Sp1, NFκB, and HDACs.37 Here, we first demonstrated that PRMT5 also participates in this repressor complex, as supported by the physical interaction of PRMT5 and Sp1 and concomitant enrichment of PRMT5 and histone H4R3me2 on miR-29b regulatory elements. PRMT5 appeared critical to miR-29b repression, as overexpression or knockdown of the protein led respectively to repression or re-expression of the microRNA. These changes in miR-29b levels could in turn mediate the effects of PRMT5 on Sp1 expression. However, a possible direct role of PRMT5 on Sp1, either at the protein level by arginine methylation or at the transcriptional level by chromatin remodeling and histone methylation, remains to be explored.
Consistent with the transcriptionally repressed or activated status, the opposite histone acetylation levels were noted for the miR-29b and FLT3 loci, with the former being mainly hypoacetylated and the latter hyperacetylated at baseline in AML cells. The notion that histone acetylation can have an impact on histone methylation and vice versa has been postulated previously, 43 but the exact mechanisms and temporal sequence of these events remain unclear. Nevertheless, the function of PRMT5 as both a gene transcription repressor and activator ultimately support a proleukemic mechanism of this protein. However, the determinants of the transcription activator or repressor properties of PRMT5 are unknown.
To this end, we demonstrated here that PRTM5 is indeed a “druggable” protein and showed that pharmacologic inhibition of PRMT5 enzyme activity results in antileukemic effects in AML. We reported here on the activity of HLCL-61, a first-in-class small molecule inhibitor of PRMT5. Treatment with this selective PRMT5 inhibitor significantly increased miR-29b expression, downregulated FLT3 levels, and affected cell survival and proliferation of AML blasts. It should be stressed, however, that HLCL-61 and other second-generation of PRMT5 inhibitors still require further optimization. Nevertheless, our results may provide a proof-of-concept for the anti-leukemia activity of PRMT5-targeting compounds.
Finally, we would like to underscore that the impact of PRMT5 inhibitors on the expression of the tyrosine kinase (TK) FLT3 is noteworthy. We showed that, while not directly interacting with FLT3 protein, PRMT5 played a pivotal role in FLT3 gene transcription. Ligand-independent activation of FLT3 occurs with the acquisition of the FLT3-ITD and confers poor prognosis to AML patients. 44,45 Nearly 25% of all AML cases harbor FLT3-ITD. The clinical use of FLT3 inhibitors induced an incomplete or transient antileukemia response in AML patients 45, which highlights the requirement for novel FLT3-aimed approaches. We have previously suggested that combining TK inhibitors with compounds that pharmacologically “shut off” FLT3 gene expression may be an efficient two-fold approach to successfully treat FLT3-ITD patients.
In conclusion, we have established a role for PRMT5 in myeloid leukemia growth through a dual complex epigenetic mechanism that silences tumor suppressor microRNAs and induces transcription of leukemogenic signaling transducers (Figure 8). To be sure, it is likely that transcription activation and expression of several other oncogenes and tumor suppressor genes are respectively affected by aberrant activity of PRMT5. Pharmacologic inhibition of PRMT5 appears biochemically feasible and results in antileukemia activity. Importantly, mining publically available data repositories 46 (Supplementary Figure S1), we found no significant difference in expression of PRMT5 among different cytogenetic/molecular subgroups of AML patients, suggesting that our data could be expanded to the distinct subsets of AML. We are working to develop more efficient PRMT5 inhibitors that can be effectively delivered in vivo and in turn be rapidly brought to the clinic to expand the armamentarium of epigenetic-targeting drugs available to AML patients.
Figure 8. Summary of PRMT5 activity and inhibition in AML.
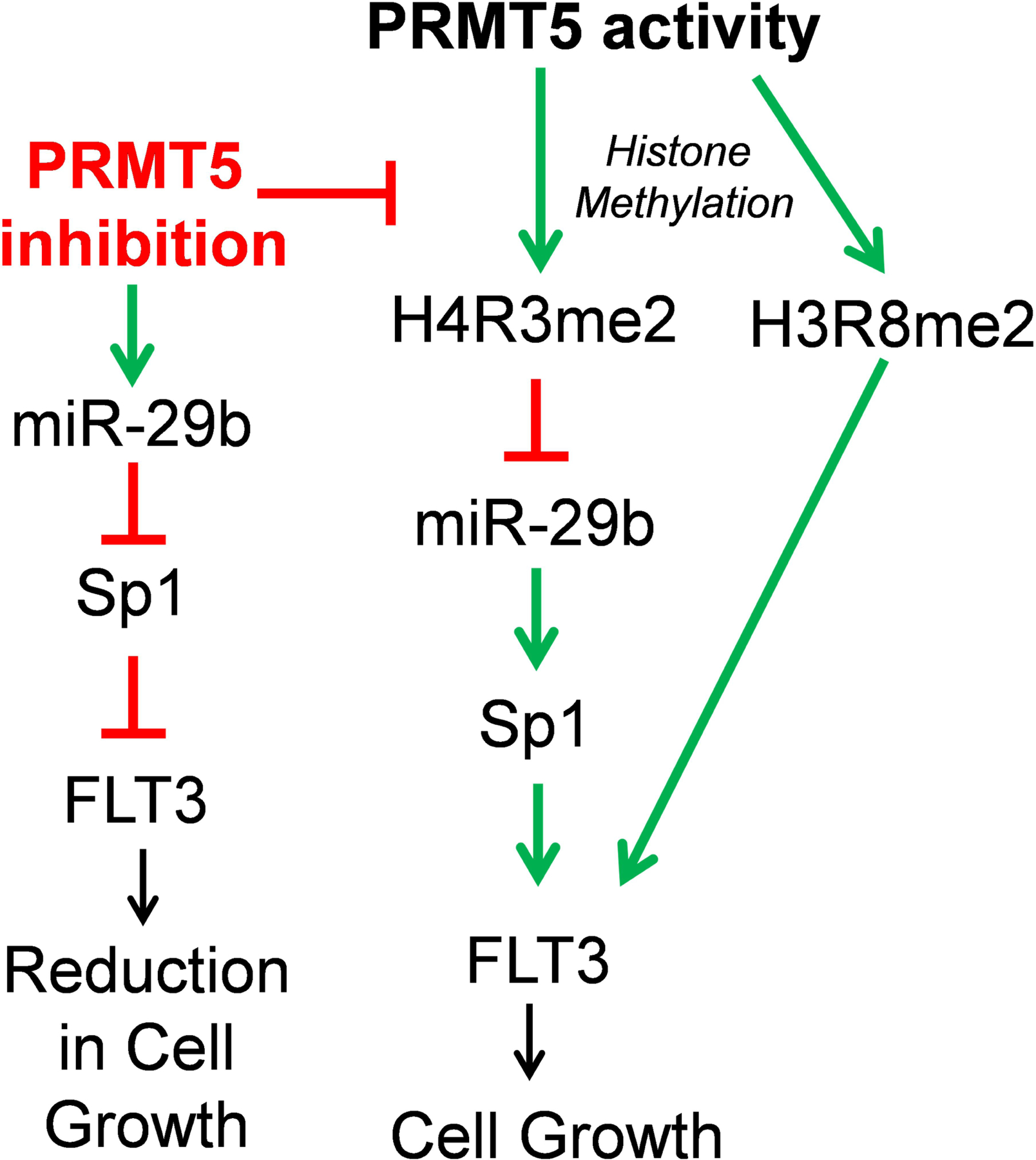
PRMT5 can suppress miR-29b expression in part by methylating histone H4R3 present at the promoter region of the miR. Low miR-29b levels leads to an increase in Sp1 protein levels and in turn elevated transcriptional activities of Sp1 at FLT3 promoter. This event ultimately can result in enhanced cell survival and growth. Our data also suggest that PRMT5 may also directly influence FLT3 expression by binding Sp1 at the promoter region of FLT3 and altering the levels of symmetrically dimethylated H3R8 mark at this locus. Inhibition of PRMT5 can block PRMT5 inhibitory effects on miR-29b expression; thus miR-29b can freely target Sp1 mRNA for translation inhibition. Low Sp1 levels leads to reduction in FLT3 expression and activity and as a result reduced cell survival and growth.
Supplementary Material
ACKNOWLEDGEMNETS
This work was supported in part by grants from the National Cancer Institute (Bethesda, Maryland, USA) (R01-CA140158, G. Marcucci), a Leukemia SPORE Grant (P50 CA140158), The Leukemia & Lymphoma Society Translational Research Program (R Baiocchi), The OSU Drug Development Institute (R Baiocchi, C Li), Friends of Jason Gould Foundation (R Baiocchi), and NIH NINDS R21 (R21NS071346, R Baiocchi, C Li).
Footnotes
ASSOCIATED CONTENT
Supporting information as noted in text.
CONFLICT OF INTEREST
No potential conflicts of interest were disclosed.
REFERENCES
- 1.Fröhling S, Scholl C, Gilliland DG, Levine RL. Genetics of myeloid malignancies: pathogenetic and clinical implications. J Clin Oncol 2005; 23: 6285–95. [DOI] [PubMed] [Google Scholar]
- 2.Estey E, Döhner H. Acute myeloid leukaemia. Lancet 2006; 368: 1894–907. [DOI] [PubMed] [Google Scholar]
- 3.McKenzie SB. Advances in understanding the biology and genetics of acute myelocytic leukemia. Clin Lab Sci 2005; 18: 28–37. [PubMed] [Google Scholar]
- 4.Dombret H Gene mutation and AML pathogenesis. Blood 2011; 118: 5366–7. [DOI] [PubMed] [Google Scholar]
- 5.Marcucci G, Haferlach T, Döhner H. Molecular genetics of adult acute myeloid leukemia: prognostic and therapeutic implications. J Clin Oncol 2011; 29: 475–86. [DOI] [PubMed] [Google Scholar]
- 6.Döhner H, Estey EH, Amadori S, Appelbaum FR, Büchner T, Burnett AK et al. Diagnosis and management of acute myeloid leukemia in adults: recommendations from an international expert panel, on behalf of the European LeukemiaNet. Blood 2010; 115: 453–74. [DOI] [PubMed] [Google Scholar]
- 7.Sharma S, Kelly TK, Jones P a. Epigenetics in cancer. Carcinogenesis 2010; 31: 27–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Oki Y, Issa JJ. Epigenetics mechanisms in AML-a target for therapy. Cancer Treatment and Research 2010; 145: 19–44. [DOI] [PubMed] [Google Scholar]
- 9.Ellis L, Atadja PW, Johnstone RW. Epigenetics in cancer: targeting chromatin modifications. Mol Cancer Ther 2009; 8: 1409–20. [DOI] [PubMed] [Google Scholar]
- 10.Marcucci G, Yan P, Maharry K, Frankhouser D, Nicolet D, Metzeler KH et al. Epigenetics meets genetics in acute myeloid leukemia: clinical impact of a novel seven-gene score. J Clin Oncol 2014; 32: 548–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Blum W, Schwind S, Tarighat SS, Geyer S, Eisfeld A-KA-K, Whitman S et al. Clinical and pharmacodynamic activity of bortezomib and decitabine in acute myeloid leukemia. Blood 2012; 119: 6025–6031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mims a, Walker a R, Huang X, Sun J, Wang H, Santhanam R et al. Increased anti-leukemic activity of decitabine via AR-42-induced upregulation of miR-29b: a novel epigenetic-targeting approach in acute myeloid leukemia. Leukemia 2013; 27: 871–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yang Y, Bedford MT. Protein arginine methyltransferases and cancer. Nat Rev Cancer 2013; 13: 37–50. [DOI] [PubMed] [Google Scholar]
- 14.Chung J, Karkhanis V, Tae S, Yan F, Smith P, Ayers LW et al. Protein arginine methyltransferase 5 (PRMT5) inhibition induces lymphoma cell death through reactivation of the retinoblastoma tumor suppressor pathway and polycomb repressor complex 2 (PRC2) silencing. J Biol Chem 2013; 288: 35534–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pal S, Sif S. Interplay Between Chromatin Remodelers and Protein Arginine Methyltransferases. J Cellular Physiology 2007; 213: 306–315. [DOI] [PubMed] [Google Scholar]
- 16.Bedford MT. Arginine methylation at a glance. J Cell Sci 2007; 120: 4243–6. [DOI] [PubMed] [Google Scholar]
- 17.Jansson M, Durant ST, Cho E-C, Sheahan S, Edelmann M, Kessler B et al. Arginine methylation regulates the p53 response. Nat Cell Biol 2008; 10: 1431–9. [DOI] [PubMed] [Google Scholar]
- 18.Guezennec X Le, Vermeulen M, Brinkman AB, Hoeijmakers WAM, Cohen A, Lasonder E et al. MBD2 / NuRD and MBD3 / NuRD , Two Distinct Complexes with Different Biochemical and Functional Properties. Mol Cell Biol 2006; 26: 843–851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bandyopadhyay S, Harris DP, Adams GN, Lause GE, McHugh A, Tillmaand EG et al. HOXA9 methylation by PRMT5 is essential for endothelial cell expression of leukocyte adhesion molecules. Mol Cell Biol 2012; 32: 1202–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wei H, Wang B, Miyagi M, She Y, Gopalan B, Huang D-B et al. PRMT5 dimethylates R30 of the p65 subunit to activate NF-κB. Proc Natl Acad Sci U S A 2013; 110: 13516–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bedford MT, Richard S. Arginine methylation an emerging regulator of protein function. Mol Cell 2005; 18: 263–72. [DOI] [PubMed] [Google Scholar]
- 22.Fay MM, Clegg JM, Uchida K a, Powers M a, Ullman KS. Enhanced Arginine Methylation of Programmed Cell Death 4 during Nutrient Deprivation Promotes Tumor Cell Viability. J Biol Chem 2014; 289: 17541–17552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Scoumanne a, Zhang J, Chen X. PRMT5 is required for cell-cycle progression and p53 tumor suppressor function. Nucleic Acids Res 2009; 37: 4965–4976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pal S, Baiocchi R a, Byrd JC, Grever MR, Jacob ST, Sif S. Low levels of miR-92b/96 induce PRMT5 translation and H3R8/H4R3 methylation in mantle cell lymphoma. EMBO J 2007; 26: 3558–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang L, Pal S, Sif S. Protein arginine methyltransferase 5 suppresses the transcription of the RB family of tumor suppressors in leukemia and lymphoma cells. Mol Cell Biol 2008; 28: 6262–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kim J, Sohn H, Yoon SY, Kim JH, Song KS, Rho S et al. Identification of Gastric Cancer − Related Genes Using a cDNA Microarray Containing Novel Expressed Sequence Tags Expressed in Gastric Cancer Cells. Clin Cancer Res 2005; 11: 473–482. [PubMed] [Google Scholar]
- 27.Eckert D, Biermann K, Nettersheim D, Gillis AJM, Steger K, Jäck H-M et al. Expression of BLIMP1/PRMT5 and concurrent histone H2A/H4 arginine 3 dimethylation in fetal germ cells, CIS/IGCNU and germ cell tumors. BMC Dev Biol 2008; 8: 106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu F, Zhao X, Perna F, Wang L, Koppikar P, Abdel-Wahab O et al. JAK2V617F-mediated phosphorylation of PRMT5 downregulates its methyltransferase activity and promotes myeloproliferation. Cancer Cell 2011; 19: 283–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pal S, Yun R, Datta A, Lacomis L, Erdjument-bromage H, Kumar J et al. mSin3A / Histone Deacetylase 2- and PRMT5-Containing Brg1 Complex Is Involved in Transcriptional Repression of the Myc Target Gene cad. Mol Cell Biol 2003; 23: 7475–7487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li H, Li C. Multiple Ligand Simultaneous Docking : Orchestrated Dancing of Ligands in Binding Sites of Protein. J Comput Chem 2010; 31: 2014–2022. [DOI] [PubMed] [Google Scholar]
- 31.Li H, Xiao H, Lin L, Jou D, Kumari V, Lin J et al. Drug Design Targeting Protein − Protein Interactions (PPIs) Using Multiple Ligand Simultaneous Docking (MLSD) and Drug Repositioning: Discovery of Raloxifene and Bazedoxifene as Novel Inhibitors of IL-6/GP130 Interface. J Med Chem 2014; 57: 632–641. [DOI] [PubMed] [Google Scholar]
- 32.Li H, Liu A, Zhao Z, Xu Y, Lin J, Jou D et al. Fragment-Based Drug Design and Drug Repositioning Using Multiple Ligand Simultaneous Docking ( MLSD ): Identifying Celecoxib and Template Compounds as Novel Inhibitors of Signal Transducer and Activator of Transcription 3 ( STAT3 ). J Med Chem 2011; 54: 5592–5596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Richard S, Morel M, Cléroux P. Arginine methylation regulates IL-2 gene expression: a role for protein arginine methyltransferase 5 (PRMT5). Biochem J 2005; 388: 379–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fabbrizio E, El Messaoudi S, Polanowska J, Paul C, Cook JR, Lee J-H et al. Negative regulation of transcription by the type II arginine methyltransferase PRMT5. EMBO Rep 2002; 3: 641–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Garzon R, Heaphy CE a, Havelange V, Fabbri M, Volinia S, Tsao T et al. MicroRNA 29b functions in acute myeloid leukemia. Blood 2009; 114: 5331–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang Y-K, Wang H, Leng Y, Li Z-L, Yang Y-F, Xiao F-J et al. Overexpression of microRNA-29b induces apoptosis of multiple myeloma cells through down regulating Mcl-1. Biochem Biophys Res Commun 2011; 414: 233–9. [DOI] [PubMed] [Google Scholar]
- 37.Liu S, Wu L-C, Pang J, Santhanam R, Schwind S, Wu Y-Z et al. Sp1/NFkappaB/HDAC/miR-29b regulatory network in KIT-driven myeloid leukemia. Cancer Cell 2010; 17: 333–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cook AM, Li L, Ho Y, Lin A, Li L, Stein A et al. Role of altered growth factor receptor-mediated JAK2 signaling in growth and maintenance of human acute myeloid leukemia stem cells. Blood 2014; 123: 2826–2837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gu Z, Gao S, Zhang F, Wang Z, Ma W, Davis RE et al. Protein arginine methyltransferase 5 is essential for growth of lung cancer cells. Biochem J 2012; 446: 235–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bao X, Zhao S, Liu T, Liu YY, Liu YY, Yang X. Overexpression of PRMT5 promotes tumor cell growth and is associated with poor disease prognosis in epithelial ovarian cancer. J Histochem Cytochem 2013; 61: 206–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nicholas C, Yang J, Peters SB, Bill M a, Baiocchi R a, Yan F et al. PRMT5 is upregulated in malignant and metastatic melanoma and regulates expression of MITF and p27(Kip1). PLoS One 2013; 8: e74710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Blum W, Garzon R, Klisovic RB, Schwind S, Walker A, Geyer S et al. Clinical response and miR-29b predictive significance in older AML patients treated with a 10-day schedule of decitabine. Proc Natl Acad Sci U S A 2010; 107: 7473–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rank G, Cerruti L, Simpson RJ, Moritz RL, Jane SM, Zhao Q. Identification of a PRMT5-dependent repressor complex linked to silencing of human fetal globin gene expression. Blood 2010; 116: 1585–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Stirewalt DL, Radich JP. The role of FLT3 in haematopoietic malignancies. Nat Rev Cancer 2003; 3: 650–65. [DOI] [PubMed] [Google Scholar]
- 45.Weisberg E, Barrett R, Liu Q, Stone R, Gray N, Griffin JD. FLT3 inhibition and mechanisms of drug resistance in mutant FLT3-positive AML. Drug Resist Updat 2009; 12: 81–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Leukemia AM. Genomic and epigenomic landscapes of adult de novo acute myeloid leukemia. N Engl J Med 2013; 368: 2059–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


