ABSTRACT
Rostro-caudal patterning of vertebrates depends on the temporally progressive activation of HOX genes within axial stem cells that fuel axial embryo elongation. Whether the pace of sequential activation of HOX genes, the 'HOX clock', is controlled by intrinsic chromatin-based timing mechanisms or by temporal changes in extrinsic cues remains unclear. Here, we studied HOX clock pacing in human pluripotent stem cell-derived axial progenitors differentiating into diverse spinal cord motor neuron subtypes. We show that the progressive activation of caudal HOX genes is controlled by a dynamic increase in FGF signaling. Blocking the FGF pathway stalled induction of HOX genes, while a precocious increase of FGF, alone or with GDF11 ligand, accelerated the HOX clock. Cells differentiated under accelerated HOX induction generated appropriate posterior motor neuron subtypes found along the human embryonic spinal cord. The pacing of the HOX clock is thus dynamically regulated by exposure to secreted cues. Its manipulation by extrinsic factors provides synchronized access to multiple human neuronal subtypes of distinct rostro-caudal identities for basic and translational applications.
This article has an associated ‘The people behind the papers’ interview.
KEY WORDS: Axial, HOX genes, Human, Motor neurons, Pluripotent stem cells, Spinal cord
Highlighted Article: Control by extrinsic factors of the sequential activation of HOX genes in axial progenitors is revealed by motor neuron subtype specification in human pluripotent stem cells.
INTRODUCTION
The patterning of bilaterian body is orchestrated by the differential expression of HOX transcription factors along their rostro-caudal axis. In vertebrates, HOX proteins are encoded by 39 genes, organized into four genomic clusters (HOXA, HOXB, HOXC and HOXD), that display anterior boundaries of expression co-linear to their chromosomal location. Genes located in the 3′ region of the clusters are expressed more anteriorly than their 5′ neighbors. The regionalized expression of HOX genes in post-occipital regions is initiated by their sequential, collinear, activation in axial progenitors, a dynamic cell population that fuels rostro-caudal extension of the embryo (Cambray and Wilson, 2002, 2007; Deschamps and Duboule, 2017; Forlani et al., 2003; Gouti et al., 2017; Henrique et al., 2015; Kondoh et al., 2016; Wymeersch et al., 2019). As axial progenitors contribute to progressively more caudal mesodermal and neuroectodermal structures, the 3′ to 5′ sequence of HOX gene activation is translated into a co-linear spatial pattern of expression in the progenies (Deschamps and Duboule, 2017; Henrique et al., 2015). Hence, the coupling between rostro-caudal extension of the body axis and the sequential activation of Hox genes over time, the HOX clock, is a central element of axial patterning. However, the mechanisms pacing the clock in axial progenitors are still unclear. Although extrinsic factors such as retinoic acid (RA), Wnt, fibroblast growth factors (FGFs) or growth differentiation factors (GDFs) control HOX patterns of expression, cell-intrinsic changes from a transcriptionally repressive to a permissive chromatin state occur within HOX complexes and correlate with the HOX temporal sequence of induction (Bel-Vialar et al., 2002; Deschamps and Duboule, 2017; Liu et al., 2001; Mazzoni et al., 2013; Narendra et al., 2015; Neijts et al., 2017; Noordermeer et al., 2014; Philippidou and Dasen, 2013; Soshnikova and Duboule, 2009). Whether the progressive opening of chromatin along the complexes serves as an internal timer actuated by extrinsic cues or whether sequences of secreted factors that activate progressively more caudal HOX genes largely defines the tempo of the induction remains unclear (Bel-Vialar et al., 2002; Del Corral and Storey, 2004; Deschamps and Duboule, 2017; Ebisuya and Briscoe, 2018; Lippmann et al., 2015; Mazzoni et al., 2013; Wymeersch et al., 2019). Inability to distinguish between these two hypotheses might underlie our limited ability to finely tune the rostro-caudal identity of caudal cell types derived from pluripotent stem cells [PSCs: embryonic stem cells (ESCs) and induced pluripotent stem cells (iPSCs); Diaz-Cuadros et al., 2020; Du et al., 2015; Duval et al., 2019; Faustino Martins et al., 2020; Frith et al., 2018; Gouti et al., 2014; Li et al., 2005; Lippmann et al., 2015; Matsuda et al., 2020; Maury et al., 2015; Ogura et al., 2018; Peljto et al., 2010; Sasai et al., 2014; Verrier et al., 2018]. The two models imply distinct strategies for in vitro differentiation: the ‘intrinsic model' predicts that the specification of posterior identities will require a precise synchronization between the timing of differentiation and the internal HOX timer. Alternatively, the ‘extrinsic model' implies that exposing axial progenitors to the relevant extrinsic cues should control the HOX clock to generate progenies of defined rostro-caudal identities. To approach this question and its consequences for cell engineering, we aimed to generate axial progenitors from human pluripotent stem cells (hPSCs) to study the mechanisms pacing the HOX clock during their differentiation into spinal motor neurons (MNs), a cell type that relies on a precise HOX code to acquire appropriate rostro-caudal subtype identities. Here, we report the generation of functional axial progenitors from hPSCs demonstrated by their ability to generate spinal MN subtypes located along the rostro-caudal axis of human embryos. As they become older, hPSC-derived axial progenitors generated MN subtypes born later in development and located more caudally in embryos, showing that they undergo a temporal shift in their rostro-caudal potential. Transcriptomic analysis indicated that this shift is linked to a temporal activation of HOX genes paralleled by a graded increase in FGF signaling. Blocking the FGF pathway stalled HOX temporal activation which resulted in the specification of early-born anterior MNs from late progenitors. In contrast, FGF precocious increase accelerated the temporal sequence of HOX activation and promoted the generation of more caudal, thoracic, MN subtypes. Combining FGFs with GDF11, another extrinsic factor, further accelerated the clock, favoring the specification of late-born lumbar MNs. Overall, our results indicate that extrinsic factors function as key pacers of the HOX clock tempo. Manipulations of the pacing pathways allow the efficient and synchronous engineering of human cell types of defined rostro-caudal identities for basic and translation applications.
RESULTS
HOX expression profiles and motor neuron subtypes in human embryonic spinal cord
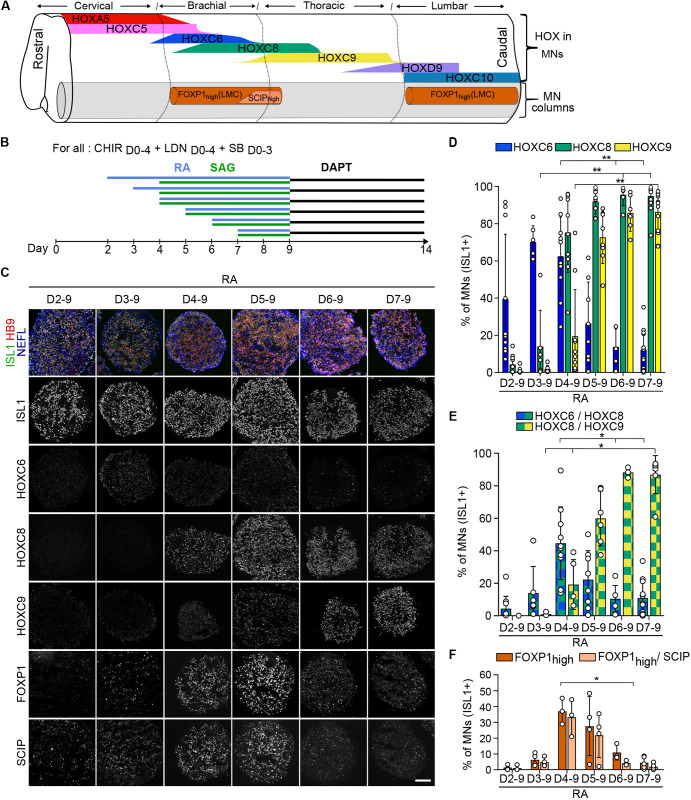
The differential expression of Hox transcription factors along the rostro-caudal axis of the vertebrate spinal cord controls the formation of molecularly and functionally distinct neuronal populations, which therefore represent a functional product of Hox gene regulation. In particular, the Hox code orchestrates the acquisition of subtype-specific features in mouse and chick spinal motor neuron (MNs), controlling the formation of the distinct locomotor circuits (Dasen, 2017; Philippidou and Dasen, 2013). Whether this spinal HOX code and associated MN subtypes is conserved in human remained unknown, thus preventing faithful assessment of HOX regulation and its link with cell ‘rostro-caudal' identity during hPSC differentiation. We therefore mapped on human embryos, at 6.3 and 7.5 weeks of development, seven HOX transcription factors that display spinal collinear expression patterns and instruct MN subtype specification in mouse and chick (Philippidou and Dasen, 2013) (Fig. 1A, Fig. S1). As in animal models, human MNs expressed ISL1 or HB9 all along the spinal cord (Fig. 1A, circles in Fig. S1) (Amoroso et al., 2013). Within MNs, HOX displayed rostro-caudal patterns resembling those of mouse and chick (Dasen et al., 2003; Liu et al., 2001): cervical MNs expressed HOXA/C5, while brachial MNs expressed HOXC6, thoracic MNs expressed HOXC9 and lumbar MNs expressed HOXC10. Caudal brachial MNs co-expressed HOXC6 and HOXC8, and anterior thoracic MNs expressed HOXC8 and HOXC9. HOXD9 labeled caudal thoracic MNs as well as anterior lumbar MNs, which also expressed HOXC10 (Fig. 1A, Fig. S1). In amniotes, this Hox code instructs the formation of distinct motor columns that innervate common muscle groups, and motor pools that innervate a single muscle (Philippidou and Dasen, 2013). To be able to assess in vitro whether changes in HOX expression have functional consequences, we mapped columnar and pool markers within HOX expression domains (Fig. 1A, Figs S1 and S2). As in mouse and chick, MNs expressing high levels of FOXP1 (FOXP1high) were observed at brachio-thoracic (HOXC6 and HOXC8/HOXC9) and lumbar (HOXC10) levels, where they formed a lateral motor column (LMC) matching the location of limb-innervating MNs (Amoroso et al., 2013; Routal and Pal, 1999) (Fig. 1A, Figs S1 and S2). Within the FOXP1high LMC, MNs expressing the transcription factor SCIP (POU3F1) together with HOXC8 were observed in the caudal brachial spinal cord (Helmbacher et al., 2003). In addition, SCIP/HOXC8/HOXC9-positive MNs were detected in the anterior thoracic region, which might correspond to hand-innervating MNs (Fig. S2) (Bell et al., 2017; Mendelsohn et al., 2017).
Fig. 1.
Human axial progenitors generate progressively more caudal motor neuron subtypes. (A) Schematic summary of data in Figs S1, S2. MNs are defined by the expression of ISL1 or HB9 (gray) are organized in motor columns in spinal ventral horns. HOX expression profiles within MNs and localization of MNs expressing high levels of FOXP1 and SCIP are represented as observed in the human embryonic spinal cord at 6.3 and 7.5 weeks of gestation. Changes in shapes indicate an increase or decrease in the number of MNs expressing a given marker. FOXP1high MNs are observed selectively in lateral motor columns (LMCs) of the brachio-thoracic and lumbar spinal cord. SCIPhigh MNs are a subset of LMC MNs in the caudal brachial spinal cord. (B) Differentiation conditions used in C to F in which the time of exposure to retinoic acid (RA)/SAG (an agonist of the sonic hedgehog pathway) is modulated. (C) Immunostaining for ISL1/HB9 (MNs), NEFL (neurons), HOX transcription factors, FOXP1 and SCIP on cryostat sections of hESC-derived EBs on day 14 of differentiation. The later RA is applied, the more caudal the MNs are. FOXP1 and SCIP MNs are mostly generated when RA is applied at day 4 and day 5, further defining the rostro-caudal identity of the MNs within HOXC8+ conditions. Scale bar: 100 µm. (D-F) Proportion of MNs (ISL1+ cells) expressing the indicated markers. Data are mean±s.d. Each circle is an independent biological replicate. (D) n=6-13, (E) n=3-13, (F) n=3 or 4. *P≤0.05, **P≤0.01 (ANOVA with Kruskal-Wallis post-hoc test).
Overall, HOX transcription factors are regionally expressed along the rostro-caudal axis of the human spinal cord. Within the HOX domains, distinct MN subtypes, identifiable by specific combination of transcription factors, are generated at stereotyped positions. These data provided readouts to assess the mechanisms regulating HOX expression in axial progenitors and their impact on cell type specification during hPSC differentiation.
Temporal regulation of HOX genes and FGF pathway in hPSC-derived axial progenitors
We first sought to test whether axial progenitors (i.e. progenitors producing derivatives along the anteroposterior axis; Cambray and Wilson, 2002, 2007; Forlani et al., 2003; Kondoh and Takemoto, 2012) could be generated from hPSCs using MN subtype specification as a readout. We have previously reported sequences of extrinsic cues leading to the efficient generation of spinal MNs from hPSCs through a putative axial progenitor stage (Maury et al., 2015). Exposure of free-floating embryoid bodies (EBs) to a WNT pathway agonist (CHIR99021, CHIR) and inhibitors of BMP and TGFβ pathways (LDN193189 and SB431542) generated progenitors expressing CDX2, which encodes a transcription factor expressed in axial progenitors and a regulator of caudal HOX gene induction (Bel-Vialar et al., 2002; Bialecka et al., 2010; Gouti et al., 2014; Maury et al., 2015; Mazzoni et al., 2013; Neijts et al., 2017). Then, human spinal MN progenitors (83% of the cells on average) were generated upon exposure of these progenitors to: (1) retinoic acid (RA), which was previously shown to guide axial progenitors toward a neural fate; and (2) an agonist of the sonic hedgehog pathway (SAG), which induces a ventral MN progenitor fate (Briscoe and Novitch, 2008; Maury et al., 2015; Ribes et al., 2009). Finally, inhibition of Notch signaling on day 9 (D9) promoted the differentiation of MN progenitors in post-mitotic MNs (Maury et al., 2015). We thus used MN subtype markers, including HOX transcription factors, to assess the rostro-caudal identity of MNs produced under the previously reported conditions (Fig. 1B) (Maury et al., 2015). First, staining for HB9, ISL1 and the pan-neuronal marker neurofilament light chain (NEFL), together with quantification of ISL1+ cells, confirmed the efficient generation of spinal MNs as previously shown (Fig. 1C, Fig. S3A). Analysis of HOX expression showed that exposing cells to RA from D2 followed by SAG on D4 gave rise to some HOXC6 MNs corresponding to anterior brachial location in vivo. This identity was acquired by most MNs when RA was added 1 day later, on D3 (70.1% of HOXC6+ and 13.9% HOXC8+ MNs) (Fig. 1D,E). Addition of RA/SAG from D4 generated MNs with a caudal brachial identity (44.6% HOXC6/C8+) from which 37.1% expressed a high level of FOXP1, which identifies limb-innervating MNs in the spinal cord (Fig. 1D-F, Fig. S3B). Overall, these results suggested that the specification of more caudal MN subtypes was dependent on either: (1) the duration of exposure to RA, with a shorter RA exposure promoting caudalization; or (2) the time-point at which the progenitors received RA, as previously suggested (Bel-Vialar et al., 2002; Del Corral and Storey, 2004; Lippmann et al., 2015). To distinguish between these two possibilities, D3 progenitors that gave rise mostly to HOXC6 MNs received shorter RA treatments (D3-5 or D3-8 versus D3-9). These two conditions failed to promote more caudal identities (Fig. S3C-E). Instead, HOXC6 expression was lost in the shortest RA treatment (48 h, D3-5, Fig. S3D), indicating a potential role for RA in HOXC6 MNs specification as previously demonstrated in chick (Liu et al., 2001). These results indicate that the day at which progenitors are exposed to RA/SAG is the main trigger of caudalization (Fig. S3C-E). Further delaying RA/SAG then induced even more caudal MN subtypes. Treatment of cells with RA/SAG on D5 generated MNs with an anterior thoracic identity (59.8% HOXC8/C9+) (Fig. 1B-E) from which 27.6% MNs acquired a FOXP1+ limb-innervating identity (Fig. 1F). Accordingly, HOXC9/FOXP1/SCIP-positive MNs, which are located in the human anterior thoracic spinal cord and might correspond to hand-innervating MNs, were observed almost exclusively in this condition (Fig. 1F, Fig. S3G). Even later treatment, on D6 or D7, specified MNs acquiring a mid-thoracic identity, as demonstrated by the expression of HOXC9 and the loss of FOXP1high MNs (Fig. 1B-F, Fig. S3F,G). The progressive caudalization of MN identity upon incremental delays of RA/SAG addition was confirmed with another hESC line and two iPSC lines (Fig. S4A,B). Interestingly, for one iPSC line, we had to increase CHIR concentration from 3 to 4 µM to observe this caudalization, suggesting line-to-line differences in WNT pathway integration (Fig. S4A-E). Indeed, and in agreement with the lack of caudalization, upon 3 µM of CHIR, this iPSC line exhibited a decreased induction of CDX2 at D3, a transcription factor induced in axial progenitors by WNT and FGF signaling pathways, and required for caudal HOX gene induction (Fig. S4F) (Amin et al., 2016; Bel-Vialar et al., 2002; Mazzoni et al., 2013; Nordström et al., 2006; Young et al., 2009).
Overall, WNT activation combined with TGF-β/BMP pathway inhibition converts hPSC into cells that can generate progenies of distinct rostro-caudal identities, a hallmark of axial progenitors. The time window between the initial exposure to WNT agonist and later exposure to RA defines HOX expression patterns in the generated progenies.
Temporal induction of caudal HOX genes and FGF target genes in axial progenitors
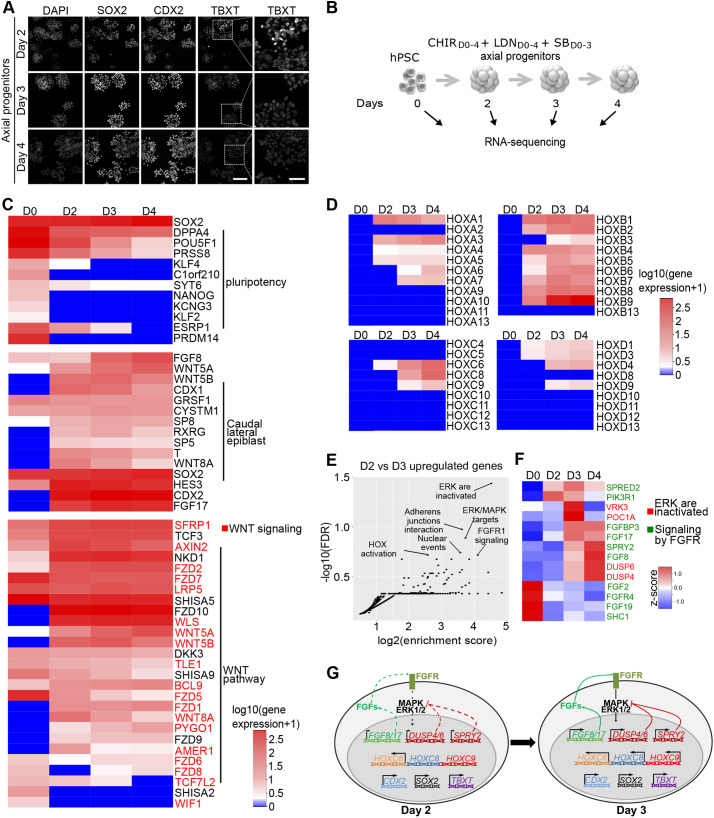
We then aimed at defining more precisely the identity of these hPSC derived-axial progenitors and the molecular changes paralleling their temporal switch in rostro-caudal potential, including HOX gene induction patterns. We first wondered whether the progenitors generated between D2 and D4 share a molecular identity with other vertebrate axial progenitors. In mouse, axial progenitor activity is carried in part by cells located in the caudal lateral epiblast (CLE) and the node-primitive streak border, which express Sox2, Nkx1.2, Cdx2 and TbxT (Brachyury) (Albors et al., 2018; Amin et al., 2016; Cambray and Wilson, 2002, 2007; Edri et al., 2019; Forlani et al., 2003; Gouti et al., 2014, 2017; Henrique et al., 2015; Koch et al., 2017; Kondoh et al., 2016; Mathis and Nicolas, 2003; Tzouanacou et al., 2009; Wymeersch et al., 2016, 2019). Single-cell lineage-tracing experiments showed that some of these axial progenitors are dual-fated neuromesodermal progenitors (NMPs) that generate progenies both in the spinal cord and paraxial mesoderm (Tzouanacou et al., 2009). The relative levels of Sox2 and TbxT transcription factors correlate with a preponderant differentiation of the cells toward the mesodermal (higher TbxT) or neural lineages (higher Sox2). We thus monitored the relative level of expression of SOX2, TBXT and NKX1.2 transcripts in D2, D3 and D4 progenitors in comparison with both hESCs and hiPSCs (Fig. S5A,B). The three markers were present from D2 to D4. SOX2 displayed levels similar to those in hPSCs, while NKX1.2 and TBXT were highly induced (Fig. S5A,B). We then stained for SOX2, TBXT and CDX2 (Fig. 2A, Fig. S5C). They were all co-expressed during the timeframe, yet TBXT progressively decreased over time to be weakly expressed by D4. This decrease in TBXT expression likely indicates a progressive transition towards preneural plate progenitors, which lose TbxT expression and are committed to the neural lineage (Del Corral and Storey, 2004; Delfino-Machín et al., 2005; Olivera-Martinez et al., 2014; Wymeersch et al., 2016).
Fig. 2.
Temporal transcriptomic analysis of hPSC-derived axial progenitors. (A) Immunostaining for axial progenitor markers on hESC-derived progenitors at day 2, day 3 and day 4 of differentiation. Scale bars: 40 µm. (B) Experimental design of RNA-seq experiment to profile the transcriptome of hESC (SA001)-derived axial progenitors. n=2 per sample. (C) Heatmap of gene expression [log10(gene expression +1)] for pluripotency genes, most common mouse NMPs markers and Wnt pathway-related genes. Genes in red are part of the ‘Formation β-catenin:TCF transactivating complex' annotation found enriched in reactome pathway analysis in Fig. S3E,F. (D) Heatmap showing temporal transcriptional changes [log10(gene expression +1)] of all HOX genes. (E) Functional enrichment analysis (Reactome pathway) of the 232 genes upregulated twofold (P<0.05) between D3 and D2: y axis, FDR (false discovery rate); x-axis, enrichment score calculated for a given Reactome pathway. (F) Heatmap based on z-score of the genes associated with the annotations ‘ERK/MAPK target' (red labels) or ‘signaling by FGFR' (green labels) in reactome analysis in 2E. (G) Schematic representation of the transcriptional and immunostaining analysis of day 2 and day 3 progenitors.
Whole-transcriptome analysis confirmed that, beside these markers, D2, D3 and D4 hPSC-derived progenitors shared many similarities with mouse axial progenitors found in the caudal epiblast (Fig. 2, Fig. S5 and Tables S1, S2). Functional enrichment analysis of genes increased in progenitors in comparison with hESCs indicated an activation of HOX genes and the WNT pathway in D3 and D4 progenitors, accompanied by a decrease of key pluripotency markers, such as NANOG or KLF2 and KLF4, as observed in CLE cells (Fig. 2C, Fig. S5D-F) (Del Corral and Storey, 2004; Deschamps and Duboule, 2017; Olivera-Martinez and Storey, 2007; Olivera-Martinez et al., 2014; Wymeersch et al., 2019). More strikingly, the most common genes defining mouse CLE cells (Edri et al., 2019; Gouti et al., 2017; Olivera-Martinez et al., 2014; Wymeersch et al., 2019) were present from D2 to D4 (Fig. 2C, Table S1), and D2 and D3 progenitors expressed 122 out of 142 genes defining mouse E8.5 and E9.5 NMPs in comparison with mesodermal progenitors (RPM≥1.0, Fig. S5G, Table S4). Out of these 122 genes, the CLE genes CDX1, FGF17, RXRG, SP8, WNT5A, WNT8A and FST all belonged to the 50 most enriched genes in D2 and D3 progenitors compared with hESCs (Fig. S5H, Tables S1 and S4). There was also a strong overlap with genes found in recently reported hPSC-derived axial progenitors (Fig. S5J, Table S3) (Frith et al., 2018). Overall, the transcriptomic signature of the progenitors, together with the co-expression of SOX2, TBXT and CDX2, demonstrate the WNT-induced specification of human axial progenitors resembling mouse CLE progenitors (in which NMPs reside) (Edri et al., 2019; Gouti et al., 2014, 2017; Henrique et al., 2015; Koch et al., 2017; Liu et al., 2001; Wymeersch et al., 2016). However, the degree of similarity with the different states of mouse axial progenitors, including early and late NMPs and whether individual cells are bipotent, would require additional analysis and experiments. Furthermore, the progressive decrease of TBXT indicates a transition toward more neural potent progenitors, as observed in the mouse anterior CLE and in regions anterior to the node where pre-neural axial progenitors are located (Del Corral et al., 2003; Diez del Corral et al., 2002; Forlani et al., 2003; Wymeersch et al., 2016).
Next, we aimed to define the molecular changes paralleling the temporal change in rostro-caudal potential of the progenitors. A comparative analysis of axial progenitor transcriptomes indicated that D2 progenitors already activated most HOXB genes (except HOXB13), which are also activated early in development in mouse and chick and are not differentially expressed along the spinal cord (Dasen et al., 2003; Denans et al., 2015; Deschamps and van Nes, 2005) (Fig. 2D). Similarly, members of the HOXA, HOXC and HOXD complexes expressed early in development with anterior borders of expression in the hindbrain or anterior spinal cord were already activated at D2: HOXA4 and HOXA5 were expressed at low levels (Fig. 2D, Table S1) whereas HOXC4 and HOXC5 were below the detection level but detectable using real time PCR (see Fig. 3B,C). HOX genes regionally expressed along the brachial and thoracic spinal cord (Fig. 1A, Fig. S1) then displayed a temporal co-linear activation (Fig. 2D). At D2, the progenitors expressed low levels of HOXC6 that increased by D3; HOXC8 was expressed at higher levels than HOXC9 on D3 (Fig. 2D). The expression of HOXC8 and HOXC9 further increased by D4 (by 3.8- and 3.4-fold, respectively). In agreement, this progenitor state will give rise to HOXC8 and some HOXC9 MNs when exposed to RA/SAG (Fig. 1C,D). Non-expressed HOX genes corresponded to the 10 to 13 paralog groups – the latest and most caudally expressed genes (Gaunt, 1991; Izpisúa-Belmonte et al., 1991; Philippidou and Dasen, 2013; Fig. 1A, Fig. S1). Hence, WNT activation induced a temporal co-linear activation of HOX genes, which prefigured the generation of MNs of progressively more caudal identities upon incremental delays of RA/SAG treatment (Fig. 1).
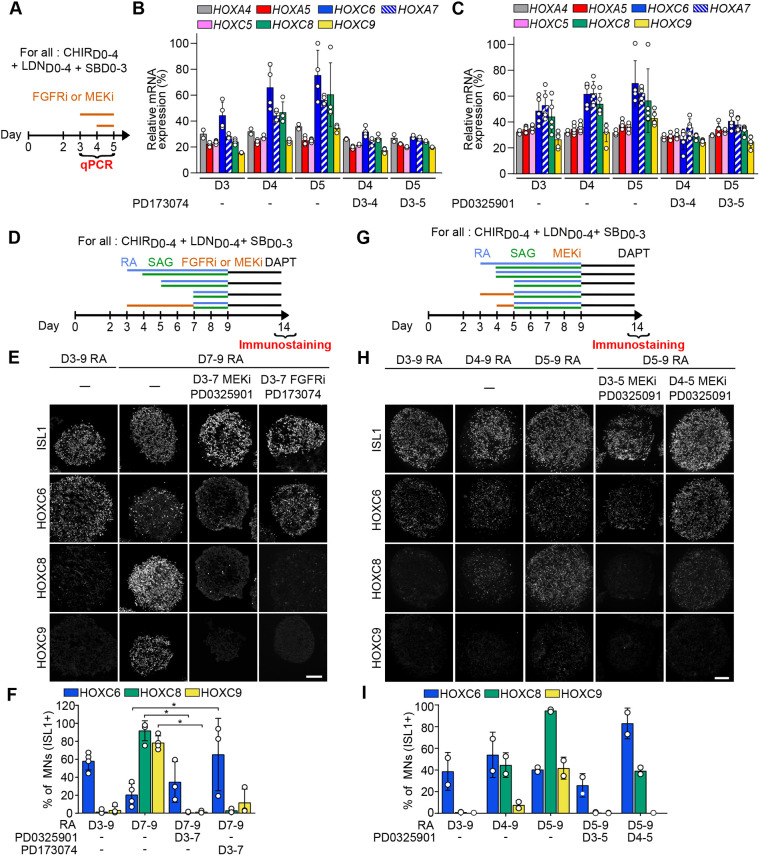
Fig. 3.
FGF and MEK pathway inhibitors stall temporal induction of HOX genes in axial progenitors and prevent specification of caudal motor neuron subtypes. (A) Differentiation conditions. PD173074 (FGFR1-3 inhibitor) or PD032590 (MEK1/2 inhibitor) were added on day 3 up to day 5 and hESC-derived progenitors were collected from day 3 to 5 for qPCR analysis. (B,C) Real-time PCR analysis of HOX mRNAs in day 3, 4 and 5 progenitors. Data represent the expression of the different genes relative to the highest expressed gene for all time points and conditions (HOXC6). MEK1/2 and FGFR inhibitors, applied from day 3, prevent the temporal increase in caudal HOX expression. (D,G) Differentiation conditions. PD173074 (FGFR1-3 inhibitor; FGFRi) or PD032590 (MEK1/2 inhibitor; MEKi) was added on day 3 up to day 7 (D) or from day 3 or day 4 until day 5 (G) and hESC-derived MNs were collected at day 14 for immunostaining analysis. Retinoic acid (RA) and SAG (an agonist of the sonic hedgehog pathway) were added at the indicated time points, between day 3 and day 7. (E,H) Immunostaining for ISL1 (MNs) and HOX transcription factors on cryostat sections of embryoid bodies on day 14 of differentiation, according to conditions presented in D for E and in G for H. MEK and FGFR inhibitors prevent the specification of HOXC8+ and HOXC9+ MNs. Instead, HOXC6+ MNs are generated. Scale bars: 100 µm. (F,I) Quantification of HOXC6, HOXC8 and HOXC9 MNs on day 14 of differentiation, according to conditions presented in D for F and in G for I. Data are mean±s.d. Each circle is an independent biological replicate: (B) n=4, (C) n=5, (F) n=3 or 4 and (I) n=2. *P≤0.05 (ANOVA with Kruskal-Wallis post-hoc test).
To define the pathways activated in parallel to the sequential induction of HOX genes, we performed functional enrichment analysis on the upregulated genes between D2 and D3 when brachial and thoracic spinal HOX genes start to be induced [log2(FC)≥1.0, P-value≤0.05, 232 genes]. We detected a significant enrichment for target genes associated with an activation of the mitogen-activated protein kinase (MAPK) pathways (Fig. 2E). Indeed, typical target genes of the MAPK ERK1/2 pathway (such as DUSP4, DUSP6, SPRED2 and SPRY2) gradually increased from D2 to D4 (Fig. 2E,F), which was confirmed by real-time PCR in hiPSC-derived progenitors (Fig. S5K). MAPKs are classical mediators of FGF signaling (Lunn et al., 2007). In agreement, we observed a temporal increase of FGF8 and FGF17 expression, two secreted FGF ligands, previously described to increase over time in the caudal epiblast of chick and mouse embryos (Fig. 2F,G) (Liu et al., 2001; Wymeersch et al., 2019).
Hence, the sequential co-linear activation of HOX genes within axial progenitors is paralleled by an increase in FGF ligands and MAPK target genes, suggesting a temporal increase in FGF pathway activity. As FGFs modulate the expression of caudal Hox genes in other systems (Bel-Vialar et al., 2002; Dasen et al., 2003; Liu et al., 2001), we hypothesized that graded paracrine or autocrine FGF signaling might be triggering the sequential induction of HOX genes.
FGF signaling is necessary for HOX sequential activation and caudal MN specification
To test whether endogenous FGF signaling was necessary for the temporal sequence of HOX gene induction and caudal MN subtype specification, we first exposed progenitors to PD173074, a selective inhibitor of FGFR1/3, and monitored the impact on HOXA and HOXC genes, which undergo temporal activation in axial progenitors (Fig. 3A,B, Fig. S6A). Inhibition of FGFR1/3 for 24 h blocked the temporal increase of HOXC6, HOXA7, HOXC8 and HOXC9 but did not impact the expression of anteriorly expressed HOXA4, HOXA5 and HOXC5, which are not temporally activated in this time-frame (Fig. 3A,B). We then tested whether this effect was mediated by MEK1/2, a downstream effector of the FGFR pathway. Exposing D3 progenitors to PD0325901, a selective MEK1/2 inhibitor, for 24 h or 48 h also blocked the temporal increase of HOXC6, HOXA7, HOXC8 and HOXC9 (Fig. 3C, Fig. S6A). MEK inhibition even induced a drop in expression of all HOX genes, including HOXA4, HOXA5 and HOXC5, that was less pronounced at 48 h than at 24 h (Fig. 3C, Fig. S6B). This result suggests a stronger impact of MEK inhibition on HOX genes, potentially reflecting a more pronounced and rapid inhibition of pathway activity when targeting downstream effectors.
To determine the functional consequences of the stalled HOX temporal induction, we monitored MN subtype specification after FGFR1/3 and MEK1/2 inhibition (Fig. 3D-I, Fig. S6). First, the efficiency of MN generation was not impacted by the two inhibitors in the different tested conditions, even though we collected a reduced number of EBs in the D3-D7 PD173074 condition (Fig. S6D,F). Exposure to RA/SAG on D7 normally leads to the specification of HOXC8 and C9 MNs (Figs 1B-D and 3D,F,H). Addition of PD173074 or PD0325901 from D3 to D7 followed by RA/SAG at D7 fully prevented the generation of HOXC8/9 MNs (Fig. 3E,F). Instead, HOXC6 MNs were specified, an identity normally obtained when D3 progenitors are exposed to RA (Figs 1B,C and 3D-F). A similar result was obtained with only a 48 h inhibition from D3 to D5 followed by RA on D5, which also gave rise to HOXC8/9 MNs in absence of inhibitor (Fig. 3E,G-I, Fig. S6B,C). These results agreed with the view that FGF pathway inhibition stalled the progression of the HOX clock, resulting in the specification of MNs normally obtained from younger progenitors. To further verify this hypothesis, we inhibited the pathway 1 day later, at D4, followed by RA on D5. In contrast to D3 inhibition, this later inhibition did not prevent HOXC8 MN specification but blocked selectively HOXC9 MNs specification (Figs 1B and 3G-I). Altogether, these results demonstrate that autocrine and/or paracrine FGF signaling is necessary for HOX temporal induction within axial progenitors and the subsequent specification of caudal brachial and anterior thoracic MNs.
FGF level regulates the pace of the HOX clock and MN subtype specification
Our transcriptomic analysis indicated a temporal increase in FGF ligand gene expression, suggesting that an increase in FGF concentration and/or duration of exposure might be pacing the HOX clock. To test whether modulating the level of environmental FGF was sufficient to modify HOX induction, we exposed early D3 progenitors to RA/SAG, together with FGF2 or FGF8 at different concentrations and for different durations. In all conditions that received FGF2, more caudal MN identities were induced without the need for delaying RA addition (Fig. 4A-D, Fig. S7). In addition, the extent of caudalization varied with FGF2 concentration, with increasing concentrations promoting more caudal identities: caudal brachial HOXC8+ MNs were induced at 15 ng/ml (68.2%, 8.6-fold increase) and HOXC9+/HOXC6− thoracic MNs at 60 ng/ml (58.9%, 825-fold increase), while 120 ng/ml further reduced HOXC6+ MNs (Fig. 4A-D, Fig. S7A-G). Similar results were obtained with FGF8 (Fig. S7B). FGFs acted directly and rapidly on axial progenitors, as addition of 120 ng/ml FGF2 for 24 h or 48 h induced MNs of a caudal brachial or mid-thoracic identity (after 24h, 49.7% of the MNs were HOXC6+, 79.4% HOXC8+, 14.7% HOXC9+ and 47.9% FOXP1high/SCIP+. After 48 h, 7.5% were HOXC6+, 76.4% HOXC9+ and 13.2% FOXP1high/SCIP+; Fig. 4A-C, S7C-I).
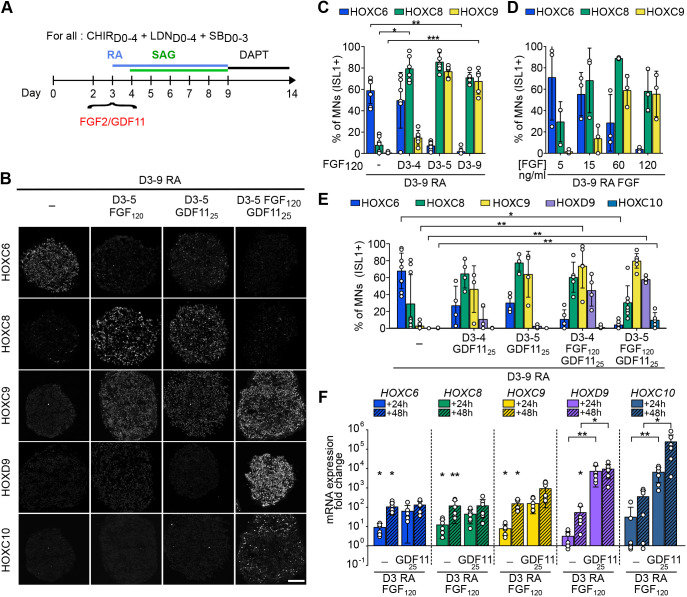
Fig. 4.
Dynamic pacing of HOX induction in axial progenitors by changes in extrinsic FGF2 and GDF11 levels. (A) Differentiation conditions. Extrinsic cues, FGF2, GDF11 or FGF2+GDF11 were added on day 3 of differentiation at various concentrations or for different durations. (B) Immunostaining for HOX proteins on cryostat sections of hESC-derived EBs on day 14 of differentiation. FGF2, GDF11 and FGF2/GDF11 induce more caudal MN subtypes. Scale bar: 100 µm. (C-E) Proportion of MNs (ISL1+ cells) expressing the indicated markers. The effect of the duration of FGF2 treatment (C), FGF2 concentration (D) and duration of GDF11 or FGF2+GDF11 (E) were monitored. (F) Real-time quantitative PCR analysis of the expression of HOX genes regionally expressed in human MNs in vivo. HOX mRNAs were monitored at day 4 (24 h post-treatment) and day 5 (48 h post-treatment) upon addition of FGF2 or GDF11, or a combination of FGF2 (120 ng/ml) and GDF11 (25 ng/ml). Data are expressed as fold changes to their respective control [day 4 retinoic acid (RA) 24 h or day 5 RA 48 h]. Asterisks above the horizontal lines indicate significance of statistical comparison between the indicated conditions; asterisks above the histogram bars are for statistical comparison with the control. Data are mean±s.d. Each circle is an independent biological replicate: (C) n=6 or 7, (D) n=3, (E) n=3-9, (F) n=6. *P≤0.05, **P≤0.01, ***P≤0.001 (ANOVA with Kruskal-Wallis post-hoc test).
To determine whether this caudalizing effect was linked to an accelerated induction of brachial and thoracic HOX genes, we performed real-time PCR analysis 24 h and 48 h post-FGF2 treatment. We indeed observed an earlier increase of HOXC8, HOXC9 and HOXD9 mRNA expression compared with RA/SAG controls (Fig. 4F).
Hence, a precocious rise in FGF signaling accelerates the induction of caudal HOX genes in early axial progenitors, resulting in the specification of more caudal cell types within the same differentiation timeline (14 days). These results demonstrate that the levels or duration of FGF signaling dynamically regulate the pace of HOX co-linear activation in axial progenitors.
FGF and GDF11 synergize to further accelerate the HOX clock
FGF2 accelerated HOX induction to generate MNs up to the mid-thoracic level, suggesting that early axial progenitors might lack competency to generate the most caudal segments. Alternatively, other extrinsic factors might be required to further accelerate the induction of HOX genes and promote more caudal identities. GDF11 is a member of the TGFβ family implicated in the control of axial elongation and MN subtype specification, and is required for the expression of the most 5′ HOX gene starting from the 10 paralogs in vivo and in vitro in late NMP-like cells (Aires et al., 2019; Gaunt et al., 2013; Lippmann et al., 2015; Liu, 2006; Liu et al., 2001; McPherron et al., 1999; Peljto et al., 2010). We exposed D3 progenitors to a combination of RA and GDF11 (25 ng/ml; Liu et al., 2001) for 24 h or 48 h. A 24 h GDF11 treatment induced caudal brachial and anterior thoracic MNs (64.7% HOXC8+ and 46.4% HOXC9+), the latter increasing after a 48 h treatment (Fig. 4A,B,E, Fig. S8A,B). However, as with FGF2, none of the more caudal identities was observed. In chick, exposure of spinal cord explants to combination of FGF2 and GDF11 promoted more caudal MNs than the two factors separately (Liu et al., 2001). We thus tested whether this combination might accelerate the HOX clock and promote caudal thoracic or lumbar identities. 24 h after exposure of D3 axial progenitors to FGF2 and GDF11, HOXC9, HOXD9 and HOXC10 mRNAs were strongly induced (respectively 65.4-, 2774.77- and 3329.44-fold compared with controls) and further increased after 48 h (Fig. 4F). In agreement, a 24 h (D3-D4) treatment combined with RA led to the specification of caudal thoracic MNs (73.9% HOXC9+ and 45% HOXD9+) whereas lumbar HOXC10+ MNs were observed after a 48 h FGF/GDF11 treatment (10.1% HOXC10+, 58% HOXD9+) (Fig. 4B,E, Fig. S8B-F). Importantly, adding FGF2/GDF11 prior RA (FGF/GDF11 at D3 followed by RA D5) largely increased the generation of lumbar MNs (81% HOXC10+, 89% HOXD9+) (Fig. S8G-I). This result suggests that RA antagonized the caudalizing activity of FGF2/GDF11, potentially by repressing FGF8 gene expression as previously shown (Del Corral and Storey, 2004; Del Corral et al., 2003) and/or by promoting a differentiation stage at which HOX expression is stalled. Hence, a combination of FGF2 and GDF11 further accelerates the HOX clock in young D3 progenitors to induce late and caudally expressed HOX10 genes, resulting in the generation of lumbar MNs.
DISCUSSION
In this study, using axial progenitors derived from hPSCs, we demonstrate that the parameters of exposure to two extrinsic factors, FGFs and GDF11, dynamically regulate the speed at which the HOX clock proceeds. Extrinsic control of the clock results in the synchronous specification of progenies of distinct rostro-caudal identities, which were born at different stages of embryogenesis. These results support the view that the pace of the HOX clock is largely regulated by extrinsic factors rather than by an intrinsic timer.
To reach these conclusions, we used 3D, embryoid body-based differentiation, of hPSC to selectively generate axial progenitors, an identity demonstrated by the co-expression of SOX2, TBXT and CDX2, as observed in mouse, chick and human embryos, complemented by detailed transcriptomic analysis and, more importantly, by functional analyses showing their ability to generate cell types found along the rostro-caudal axis of the human embryonic spinal cord (Figs 1, 2 and Figs S1-S5). Axial progenitors feed axial elongation in the embryo by generating diverse neural and mesodermal derivatives. They appear to form a heterogeneous dynamic population of progenitors comprising progenitors differentiating into diverse neural and mesodermal progenies (lateral, axial and paraxial), bi-fated NMPs that generate both spinal and paraxial mesoderm, as well as mono-fated progenitors that give rise either to mesodermal or spinal progenies (Albors et al., 2018; Forlani et al., 2003; Henrique et al., 2015; Tzouanacou et al., 2009; Wymeersch et al., 2016, 2019). Defining the precise degree of similarity between the reported hPSC-derived axial progenitors with these distinct states or with previously reported in vitro-generated axial progenitors and, in particular, whether individual progenitors are mono-fated or bi-fated NMPs will require further investigations (Wymeersch et al., 2021).
With efficient access to axial progenitors, we demonstrated that modulating the concentration, duration and combination of the caudalizing factors FGFs and GDF11 controls the speed at which the temporal activation of HOX genes occurs. The pace of the HOX clock is thus dynamically controlled by exposure parameters to extrinsic cues. Hence, the sequential changes in chromatin structure occurring along HOX complexes during their activation might be a consequence of changes in extrinsic signals rather than in the main mechanism intrinsically controlling the timing of HOX induction. (Bel-Vialar et al., 2002; Del Corral and Storey, 2004; Kimelman and Martin, 2012; Kmita and Duboule, 2003; Lippmann et al., 2015; Mazzoni et al., 2013; Narendra et al., 2015; Neijts and Deschamps, 2017; Noordermeer et al., 2011, 2014; Soshnikova and Duboule, 2009; Tschopp et al., 2009). As the sequence of activation from 3′ to 5′ genes was maintained under the different experimental conditions, the extrinsic cues seem to modulate the speed but not the directionality at which a repressive chromatin state is cleared from HOX clusters. This clearance could be temporally progressive or, alternatively, individual extrinsic factors might induce domain-wide, saltatory remodeling of repressive chromatin marks (up to HOXC9 in presence of FGFs, up to HOXD9/HOXC10 or more caudally in presence of FGF and GDF11) (Kmita and Duboule, 2003; Mazzoni et al., 2013; Narendra et al., 2016; Soshnikova and Duboule, 2009). Individual HOX genes within these transcriptionally permissive domains might be activated in a 3′ to 5′ gene direction due to their reliance on different combinations of transcription factors or differential strength of enhancer-promoter interactions (Kmita and Duboule, 2003; Mazzoni et al., 2013; Neijts and Deschamps, 2017). Determining the changes in histones marks, chromatin structure and the binding of signaling effectors within HOX complexes should help determine how extrinsic cues signal to the genome to control rostro-caudal patterning.
Dynamic environmental control of Hox expression was already suggested by grafting experiments in chick (Ensini et al., 1998; McGrew et al., 2008). In particular, it was demonstrated that heterochronic grafting of ‘old' axial progenitors to a ‘younger' caudal stem zone reverted their HOX profile to the ‘young' one (McGrew et al., 2008). Although our results might provide a potential molecular basis for environmental changes in HOX expression in caudal progenitors, we did not observe a reversion of the HOX profile upon FGF pathway inhibition but a stalled activation (Fig. 3E-I). This observation suggests that the FGF pathway regulates the tempo at which HOX activation proceeds rather than the maintenance of the HOX profile.
Importantly, the pacing of the HOX clock by secreted factors might ensure a community effect to synchronize HOX expression between neighboring progenitors, allowing the emergence of expression domains at tissue level (Durston, 2019). Furthermore, FGFs and GDFs are also implicated in embryo axial elongation (Aires et al., 2019; Boulet and Capecchi, 2012; Jurberg et al., 2013; Mallo et al., 2009; McPherron et al., 1999). A common mechanism to control rostro-caudal extension of the body axis together with Hox gene induction would be a parsimonious way to couple morphogenesis and patterning (Denans et al., 2015; Young et al., 2009). Hence, determining the mechanisms controlling the temporality of FGF and GDF expression onset, levels and duration should provide a better understanding of post-occipital tissues development and evolution.
In a bioengineering perspective, the extrinsic control of the HOX clock provides a means with which to manipulate HOX expression for the synchronous and efficient engineering of neuronal subtypes found at distinct rostro-caudal identities. In particular, we provide evidence for the controlled generation of putative hand- and leg-controlling MNs (Mendelsohn et al., 2017). Importantly, MN subtypes display differential vulnerabilities in MN diseases. The targeted differentiations in MN subtypes reported here should thus help in the more accurate study of these incurable diseases or allow the access of more controlled sources of cells for putative cell therapy approaches (Abati et al., 2019; An et al., 2019; Baloh et al., 2018; Nijssen et al., 2017; Ragagnin et al., 2019; Sances et al., 2016; Steinbeck and Studer, 2015; Tung et al., 2019). Considering the role of HOX transcription factors in instructing cell diversity in other derivatives of axial progenitors, such as control over their expression, might have broader applications for cell engineering beside spinal cell types (Deschamps and Duboule, 2017; Frith et al., 2018; Helmbacher et al., 2003; Iimura et al., 2009). More broadly, the temporal generation of distinct types of neurons or glia from the same progenitor domain is a widely used strategy to increase cell diversity in the nervous system (Dias et al., 2014; Kohwi and Doe, 2013; Oberst et al., 2019a; Rossi et al., 2017). Extrinsic cues play important roles in the unfolding of these temporal sequences (Kawaguchi, 2019; Oberst et al., 2019a,b; Syed et al., 2017; Tiberi et al., 2012). Extrinsic manipulation of the temporality of these lineages might help improve the generation of early and late-born cells for both basic research, disease modeling and cell therapy.
MATERIALS AND METHODS
Human embryonic spinal cord histology
Human fetal embryos at 6.3 weeks of gestation were obtained from pregnant women referred to the Department of Gynecology and Obstetrics at the Antoine Béclère hospital (Clamart, France) for legally induced abortions in the first trimester of pregnancy, as previously described (Lambrot et al., 2006). All women provided written informed consent for scientific use of the fetal tissues. None of the abortions was due to fetal abnormality. The fetal age was determined by measuring the length of limbs and feet (Evtouchenko et al., 1996). The project was approved by the local Medical Ethics Committee and by the French Biomedicine Agency (reference number PFS 12-002). Alternatively, human embryonic spinal cords (n=2) at stage 7.5 weeks of gestation were collected in accordance with the national guidelines of the USA (National Institutes of Health, US Food and Drug Administration) and the State of New York and under Columbia University institutionally approved ethical guidelines relating to anonymous tissue. The material was obtained after elective abortions and was classified on the basis of external morphology according to the Carnegie stages. Gestational age was determined by the date of the last menstrual period of the patient or by ultrasound, if the ultrasound estimated date differed by 1 week (as indicated by the obstetrician). In all cases, the spinal cord was removed as intactly as possible before fixation with fresh, ice-cold 4% PFA for 1.5 h, washed abundantly with PBS and then cryoprotected overnight in 30% sucrose. Post-fixation, the cord was measured and cut into anatomical sections to accommodate embedding in OCT compound (Leica) and stored at −80°C before cutting on a cryostat. Sections (16 µm) were cut along the full length of the cord.
Human pluripotent stem cell lines
Human SA001 embryonic stem cell (ESC) line (male, RRID: CVCL_B347) was obtained from Cellectis and H9 ESC line (female, RRID: CVCL_9773) was obtained from Wicell. Both cell lines were used according to the French current legislation for hESC (Agency of Biomedicine, authorization number AFSB1530532S). The induced pluripotent stem cell (iPSC) line WTSIi002 (male, RRID: CVCL_AH30, alternative name HPSI0913i-eika_2) was obtained from the European Bank for Pluripotent Stem Cells (EBISC). WTC-mEGFP-Safe harbor locus (AAVS1)-cl6 produced by the Allen Cell Institute was obtained from Coriell (AICS-0036-006, male, RRID: CVCL_JM19). Experiments with iPSCs were approved by relevant ethic committees (declaration DC-2015-2559). All PSC lines were cultured at 37°C on Matrigel (Corning) in mTSER1 medium (Stem Cell Technologies) and amplified using EDTA (Life Technologies) clump-based passaging. They were tested for potential mycoplasma contamination every other week (MycoAlertTM Mycoplasma Detection Kit, Lonza, LT07-118). No contamination was detected during the study. PSCs were thawed in presence of Y-27632 (10 μM, Stemgent or Stem Cell Technologies) and the culture medium was changed every day.
Human pluripotent stem cell differentiation
Human PSC embryoid body-based differentiation was performed as previously described (Maury et al., 2015). hPSC were dissociated with cold Accutase (Life Technologies) for 3-5 min at 37°C and resuspended in differentiation medium N2B27 [Advanced DMEM F12, Neurobasal vol:vol (Life Technologies)], supplemented with N2 (Life Technologies), B27 without Vitamin A (Life Technologies), penicillin/streptomycin 1%, β-mercaptoethanol 0.1% (Life Technologies), Y-27632 (10 μM, Stemgent or Stem Cell Technologies), CHIR-99021 (3 µM or 4 µM Selleckchem), SB431542 (20 μM, Selleckchem) and LDN 193189 (0.1 μM, Selleckchem). Cells were seeded in ultra-low attachment 6 well plates (Corning) (2×105 cells ml−1) to form embryoid bodies (EBs). All conditions of differentiation received the same medium, without Y-27632, at day 2 and at day 3, but SB431542 was removed at day 3. The differentiation then proceeded according to the schematics presented in Fig. 1B, Fig. 3A,D,G, Fig. 4A. SAG (smoothened agonist, Merck millipore), FGF2 (recombinant human FGF basic, Peprotech), RA (Sigma-Aldrich), GDF11 (recombinant human/murine/rat GDF11, Peprotech), PD0325901 (Selleckchem), PD173074 (Selleckchem) and DAPT (Stemgent or Stem Cell Technologies) were added at indicated time points. For concentrations used, see Table S5. Media were changed every other day unless specified.
Embryoid body processing for immunostaining
EBs were collected, rinsed with PBS then fixed with 4% PFA for 5 min at 4°C and rinsed with PBS three times for 5 min. EBs were cryoprotected with 30% sucrose and embedded in OCT (Leica) prior to sectioning with a cryostat. Alternatively, day 2, 3 and 4 progenitors were plated on Matrigel (Corning, diluted according to the manufacturer's recommendation) -coated coverslips and allowed to adhere between 30 and 60 min prior to fixation with 4% PFA for 5 min at 4°C.
Immunostaining
All immunostainings were performed as follows: cells or sections were incubated with a saturation solution (PBS, 10% FBS, 0.2% Triton) for 10 min. Primary antibodies (Table S5) were diluted in a staining solution (PBS, 2% FBS, 0.2% Triton) and incubated overnight at 4°C in a humidified chamber. After four PBS washes (10 min each), secondary antibodies (Alexa488, Alexa555 and Alexa647, Life Technologies, 1:1000) were added for 1 h at room temperature. After three PBS washes, DAPI was added to cells (Invitrogen, 1:3000) for 5 min. Cells or slices were then mounted in Fluoromount (Sigma-Aldrich or Cliniscience).
Image acquisition
Samples were visualized and imaged using either a Zeiss LSM 880 Confocal Laser Scanning Microscope controlled by Zen black software (Zeiss), a confocal microscope TCS SP5 II (Leica) or a DM6000 microscope (Leica) equipped with CoolSNAP EZ CDD camera, controlled by MetaMorph software. Alternatively, images were acquired using the automated microscope Cell Discoverer 7 (Zeiss), equipped with an Axiocam 506m camera, with Zen black software (Zeiss).
Quantitative RT-PCR analysis
Total RNA was extracted (RNeasy Plus Mini Kit, Qiagen) and cDNA synthesized using SuperScript III (Invitrogen). Quantitative real-time PCR was performed using a 7900HT fast real-time PCR system (Applied Biosystems) with Sybr Green PCR Master Mix (Applied Biosystems) or performed using QuantStudio 5 Real-Time PCR System (Thermofisher Scientific) and a mix with qPCR Brilliant II SYBR MM with low ROX (Agilent). Primers are listed in Table S5. All expression data were normalized to cyclophilin A mRNA (ΔCt). All analyses were performed with three technical replicates per plate. In Fig. 3 and Fig. S5, mRNA expression levels are represented as a percentage of the highest expressed gene (ΔCtmax) among all the conditions [% relative to max gene expression=(ΔCtmax/ΔCt)×100]. In Fig. 4F, expression levels are represented as a percentage relative to the maximal expression level of each gene (ΔCtgeneMax) among all conditions [% relative to max gene expression=(ΔCtgeneMax/ΔCt)×100]. In Fig. S4, data are presented as relative expression levels determined by calculating 2−ΔΔCt.
Transcriptomic analysis
hESCs were exposed to CHIR-99021 together with LDN193189 and SB431542 while forming EBs. Progenitors were collected on days 2, 3 and 4 of differentiation, and processed for total RNA preparation. For each of the eight samples, total RNA was reverse transcribed using the Ion AmpliSeq Transcriptome Human Gene Expression kit (Thermofisher Scientific). The cDNA libraries were amplified and barcoded using the Ion AmpliSeq Transcriptome Human Gene Expression core panel and Ion Xpress Barcode Adapter (Thermofisher Scientific). The amplicons were quantified using Agilent High Sensitivity DNA kit before the samples were pooled in sets of eight. Emulsion PCR and enrichment was performed on the Ion OT2 system Instrument using the Ion PI Hi-Q OT2 200 kit (Thermofisher Scientific). Samples were loaded on an Ion PI v3 Chip and sequenced on the Ion Proton System using Ion PI Hi-Q sequencing 200 kit chemistry (200 bp read length; Thermofisher Scientific). The Ion Proton reads (FASTQ files) were imported into the RNA-seq pipeline of Partek Flow software (v6 Partek) using hg19 as a reference genome. The number of reads per sample ranged from 7.5 million to 12 million reads. To determine genes that are differentially expressed between groups, mapped reads were quantified using Partek E/M algorithm normalized by the Total count/sample (the resulting counts represent the gene expression levels on reads/millions for over 20,800 different genes present in the AmpliSeq Human Gene Expression panel). Genes with an average of reads ≤1.0 in all the time points were excluded from the analysis. The evaluation of the differential expression between two conditions was performed using the EdgeR package under R. Pathway enrichment analyses were performed on upregulated genes (FC⩾2.0, P-value<0.05) between two time points by interrogating Reactome database. Hierarchical clustering was performed using Ward's method with the ward.d2 algorithm. The Euclidean distance is represented. Significant enrichments were calculated using a hyper-geometrical test and Benjamini-Hochberg correction for multiple comparisons. The enrichment score was calculated as described by Wang et al. (2017).
The normalized transcriptomic data are provided in Table S1. The GEO accession number for the raw data is GSE153519. Lists of genes differentially expressed between time points are available in Table S2. Lists of common genes between D3 enriched genes and NMPs-like genes have been identified by Frith et al. (2018) and are available in Table S3. Expression of the genes in our model that have been identified in NMP cells at E8.5 and E9.5 by Gouti et al. (2017) are listed in Table S4.
Quantification and statistical analysis
All statistics were computed using Graphpad Prism software. One-way analysis of variance (ANOVA) with a Kruskal-Wallis post-hoc analysis was performed following normality tests provided by Prism. Number of samples (n), dispersion measures and P-values are indicated in figure legends. In all figures, n are independent differentiations started from independent newly thawed hPSCs vials. For each condition at least four independent EB sections were imaged in which all the cells were quantified by automated image analysis (four EBs were quantified in one replicate of the FGFRi 3-7 conditions; in all the other experiments 6 to 9 EBs were quantified). The images were exported and saved as TIFF with Fiji if needed (Schindelin et al., 2012). Quantitative analyses on images were performed using the CellProfiler software (Carpenter et al., 2006) (Broad Institute open source at www.cellprofiler.org). DAPI-stained nuclei were segmented into primary objects using the CellProfiler segmentation pipeline and the nuclear mask was used to define objects on the target channels. The threshold for defining positive nuclei for a given target was obtained using EB sections negative for the target of interest. All images across conditions were then automatically analyzed in batch to ensure unbiased analysis. The percentage of colocalization between markers (ISL1/HOXC6/C8, ISL1/C8/C9, ISL1/C9/C10) was obtained after co-immunostaining for the markers of interest followed by quantification in Cell profiler. The analyses of the FOXP1 and SCIP immunostaining intensities were performed by combining CellProfiler with the software FCS express 7 (DeNovo Software). Nuclei were segmented into primary objects as described above, and FOXP1 and SCIP fluorescence intensities were calculated for each primary object. Fluorescence intensity plots for FOXP1 and SCIP were then generated by FCS express 7 software to visualize the intensity levels of the different markers for each individual cell and determine the percentage of cells above a given threshold. Cell profiler pipelines for quantification are available upon request. For real-time PCR analysis, one-way analysis of variance (ANOVA) with a Kruskal-Wallis post-hoc analysis and correction of multiple comparisons by controlling the FDR (Benjamini and Hochberg) was performed following normality tests provided by Prism. Number of samples (n), dispersion measures and P-values are indicated in figure legends.
Supplementary Material
Acknowledgements
We thank Frédéric Causeret for providing sections of mouse embryos; Susan Morton, Thomas Jessell and Jeremy Dasen for the generous gift of antibodies; and F. Causeret, P. Gilardi, A. Rebsam, F. Giudicelli and E. Mazzoni for critical reading of the manuscript. Differentiation were performed at the ‘Cell engineering facility' of the Institut du Fer à Moulin (IFM) and at I-STEM. Imaging experiments were carried out at the Imaging facility of the Institut du Fer à Moulin (IFM) and at I-STEM.
Footnotes
Competing interests
The authors declare no competing or financial interests.
Author contributions
Conceptualization: C.M., S.N.; Methodology: V.M., C.V., R.R., S.G., V.R., S.N.; Software: S.G.; Validation: V.M., C.V., R.R., A.T., M.D., C.M., S.N.; Formal analysis: V.M., C.V., R.R., S.G., M.J., M.D., S.N.; Investigation: V.M., C.V., R.R., M.J., A.T., L.L., M.W.M., G.C.; Resources: N.N., V.R.-F., S.N.; Data curation: R.R., M.J.; Writing - original draft: V.M., S.N.; Writing - review & editing: V.M., C.V., R.R., S.G., N.N., M.J., A.T., L.L., M.W.M., G.C., M.D., V.R.-F., H.W., V.R., C.M., S.N.; Visualization: V.M., C.V., R.R., S.G., M.W.M., G.C., M.D., S.N.; Supervision: H.W., C.M., S.N.; Project administration: H.W., C.M., S.N.; Funding acquisition: S.N.,C.M.,H.W.
Funding
This project was supported by grants from AVENIR/ATIP, from the Association Française contre les Myopathies (AFM) and from Laboratoire d'Excellence (Labex) Biopsy and Revive (ANR-10-LABX-73 and 11-LABX-0035, respectively), and from the Agence Nationale de la Recherche SYMASYM (ANR-18-CE16-0021-03) and Atomy (ANR-19-CE16-0002-02). V.M. received fellowships from Domaine d'Investissement Majeur (DIM) ‘Biothérapies' of the region Ile-de-France and from the Fondation pour la Recherche Médicale (FRM). C.V. was supported by a PhD fellowship of the Ministère de la Recherche and the Fondation pour la Recherche Médicale (FRM). V.R. and S.N.'s salaries are funded by the Institut National de la Santé et de la Recherche Médicale (INSERM). H.W., M.W.M. and G.C. were supported by Project ALS and by the National Institutes of Health (R01NS109217). Deposited in PMC for release after 12 months.
Data availability
The normalized transcriptomic data are provided in Table S1. FASTQ files are available in GEO under accession number GSE153519.
Supplementary information
Supplementary information available online at https://dev.biologists.org/lookup/doi/10.1242/dev.194514.supplemental
Peer review history
The peer review history is available online at https://dev.biologists.org/lookup/doi/10.1242/dev.194514.reviewer-comments.pdf
References
- Abati, E., Bresolin, N., Comi, G. and Corti, S. (2019). Advances, challenges, and perspectives in translational stem cell therapy for amyotrophic lateral sclerosis. Mol. Neurobiol. 56, 6703-6715. 10.1007/s12035-019-1554-x [DOI] [PubMed] [Google Scholar]
- Aires, R., de Lemos, L., Nóvoa, A., Jurberg, A. D., Mascrez, B., Duboule, D. and Mallo, M. (2019). Tail bud progenitor activity relies on a network comprising Gdf11, Lin28, and Hox13 genes. Dev. Cell 48, 383-395.e8. 10.1016/j.devcel.2018.12.004 [DOI] [PubMed] [Google Scholar]
- Albors, A., Halley, P. A. and Storey, K. G. (2018). Lineage tracing of axial progenitors using Nkx1-2CreER T2 mice defines their trunk and tail contributions. Development 145, dev164319. 10.1242/dev.164319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amin, S., Neijts, R., Simmini, S., van Rooijen, C., Tan, S. C., Kester, L., van Oudenaarden, A., Creyghton, M. P. and Deschamps, J. (2016). Cdx and T Brachyury co-activate growth signaling in the embryonic axial progenitor niche. Cell Rep. 17, 3165-3177. 10.1016/j.celrep.2016.11.069 [DOI] [PubMed] [Google Scholar]
- Amoroso, M. W., Croft, G. F., Williams, D. J., O'Keeffe, S., Carrasco, M. A., Davis, A. R., Roybon, L., Oakley, D. H., Maniatis, T., Henderson, C. E.et al. (2013). Accelerated high-yield generation of limb-innervating motor neurons from human stem cells. J. Neurosci. 33, 574-586. 10.1523/JNEUROSCI.0906-12.2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- An, D., Fujiki, R., Iannitelli, D. E., Smerdon, J. W., Maity, S., Rose, M. F., Gelber, A., Wanaselja, E. K., Yagudayeva, I., Lee, J. Y.et al. (2019). Stem cell-derived cranial and spinal motor neurons reveal proteostatic differences between ALS resistant and sensitive motor neurons. eLife 8, e44423. 10.7554/eLife.44423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baloh, R. H., Glass, J. D. and Svendsen, C. N. (2018). Stem cell transplantation for amyotrophic lateral sclerosis. Curr. Opin. Neurol. 31, 655-661. 10.1097/WCO.0000000000000598 [DOI] [PubMed] [Google Scholar]
- Bel-Vialar, S., Itasaki, N. and Krumlauf, R. (2002). Initiating Hox gene expression: in the early chick neural tube differential sensitivity to FGF and RA signaling subdivides the HoxB genes in two distinct groups. Development 129, 5103-5115. [DOI] [PubMed] [Google Scholar]
- Bell, S. W., Brown, M. J. C. and Hems, T. J. (2017). Refinement of myotome values in the upper limb: evidence from brachial plexus injuries. Surgeon 15, 1-6. 10.1016/j.surge.2015.08.004 [DOI] [PubMed] [Google Scholar]
- Bialecka, M., Wilson, V. and Deschamps, J. (2010). Cdx mutant axial progenitor cells are rescued by grafting to a wild type environment. Dev. Biol. 347, 228-234. 10.1016/j.ydbio.2010.08.032 [DOI] [PubMed] [Google Scholar]
- Boulet, A. M. and Capecchi, M. R. (2012). Signaling by FGF4 and FGF8 is required for axial elongation of the mouse embryo. Dev. Biol. 371, 235-245. 10.1016/j.ydbio.2012.08.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briscoe, J. and Novitch, B. G. (2008). Regulatory pathways linking progenitor patterning, cell fates and neurogenesis in the ventral neural tube. Philos. Trans. R. Soc. B Biol. Sci. 363, 57-70. 10.1098/rstb.2006.2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cambray, N. and Wilson, V. (2002). Axial progenitors with extensive potency are localised to the mouse chordoneural hinge. Development 129, 4855-4866. [DOI] [PubMed] [Google Scholar]
- Cambray, N. and Wilson, V. (2007). Two distinct sources for a population of maturing axial progenitors. Development 134, 2829-2840. 10.1242/dev.02877 [DOI] [PubMed] [Google Scholar]
- Carpenter, A. E., Jones, T. R., Lamprecht, M. R., Clarke, C., Kang, I. H., Friman, O., Guertin, D. A., Chang, J. H., Lindquist, R. A., Moffat, J.et al. (2006). CellProfiler: Image analysis software for identifying and quantifying cell phenotypes. Genome Biol. 7, R100. 10.1186/gb-2006-7-10-r100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dasen, J. S. (2017). Master or servant? emerging roles for motor neuron subtypes in the construction and evolution of locomotor circuits. Curr. Opin. Neurobiol. 42, 25-32. 10.1016/j.conb.2016.11.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dasen, J. S., Liu, J.-P. and Jessell, T. M. (2003). Motor neuron columnar fate imposed by sequential phases of Hox-c activity. Nature 425, 926-933. 10.1038/nature02051 [DOI] [PubMed] [Google Scholar]
- Del Corral, R. D. and Storey, K. G. (2004). Opposing FGF and retinoid pathways: A signalling switch that controls differentiation and patterning onset in the extending vertebrate body axis. BioEssays 26, 857-869. 10.1002/bies.20080 [DOI] [PubMed] [Google Scholar]
- Del Corral, R. D., Olivera-Martinez, I., Goriely, A., Gale, E., Maden, M. and Storey, K. (2003). Opposing FGF and retinoid pathways control ventral neural pattern, neuronal differentiation, and segmentation during body axis extension. Neuron 40, 65-79. 10.1016/S0896-6273(03)00565-8 [DOI] [PubMed] [Google Scholar]
- Delfino-Machín, M., Lunn, J. S., Breitkreuz, D. N., Akai, J. and Storey, K. G. (2005). Specification and maintenance of the spinal cord stem zone. Development 132, 4273-4283. 10.1242/dev.02009 [DOI] [PubMed] [Google Scholar]
- Denans, N., Iimura, T. and Pourquié, O. (2015). Hox genes control vertebrate body elongation by collinear Wnt repression. eLife 4, e04379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deschamps, J. and Duboule, D. (2017). Embryonic timing, axial stem cells, chromatin dynamics, and the Hox clock. Genes Dev. 31, 1406-1416. 10.1101/gad.303123.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deschamps, J. and van Nes, J. (2005). Developmental regulation of the Hox genes during axial morphogenesis in the mouse. Development 132, 2931-2942. 10.1242/dev.01897 [DOI] [PubMed] [Google Scholar]
- Dias, J. M., Alekseenko, Z., Applequist, J. M. and Ericson, J. (2014). Tgfβ signaling regulates temporal neurogenesis and potency of neural stem cells in the CNS. Neuron 84, 927-939. 10.1016/j.neuron.2014.10.033 [DOI] [PubMed] [Google Scholar]
- Diaz-Cuadros, M., Wagner, D. E., Budjan, C., Hubaud, A., Tarazona, O. A., Donelly, S., Michaut, A., Al Tanoury, Z., Yoshioka-Kobayashi, K., Niino, Y.et al. (2020). In vitro characterization of the human segmentation clock. Nature 580, 113-118. 10.1038/s41586-019-1885-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diez del Corral, R., Breitkreuz, D. N. and Storey, K. G. (2002). Onset of neuronal differentiation is regulated by paraxial mesoderm and requires attenuation of FGF signalling. Development 129, 1681-1691. [DOI] [PubMed] [Google Scholar]
- Du, Z.-W., Chen, H., Liu, H., Lu, J., Qian, K., Huang, C. T.-L., Zhong, X., Fan, F. and Zhang, S.-C. (2015). Generation and expansion of highly pure motor neuron progenitors from human pluripotent stem cells. Nat. Commun. 6, 6626. 10.1038/ncomms7626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durston, A. J. (2019). Some questions and answers about the role of Hox temporal collinearity in vertebrate axial patterning. Front. Cell Dev. Biol. 7. 10.3389/fcell.2019.00257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duval, N., Vaslin, C., Barata, T. C., Frarma, Y., Contremoulins, V., Baudin, X., Nedelec, S. and Ribes, V. C. (2019). Bmp4 patterns smad activity and generates stereotyped cell fate organization in spinal organoids. Development 146, dev175430. 10.1242/dev.175430 [DOI] [PubMed] [Google Scholar]
- Ebisuya, M. and Briscoe, J. (2018). What does time mean in development? Development 145, dev164368. 10.1242/dev.164368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edri, S., Hayward, P., Jawaid, W. and Arias, A. M. (2019). Neuro-mesodermal progenitors (NMPs): a comparative study between pluripotent stem cells and embryo-derived populations. Development 146, dev180190. 10.1242/dev.180190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ensini, M., Tsuchida, T. N., Belting, H. G. and Jessell, T. M. (1998). The control of rostrocaudal pattern in the developing spinal cord: specification of motor neuron subtype identity is initiated by signals from paraxial mesoderm. Development 125, 969-982. [DOI] [PubMed] [Google Scholar]
- Evtouchenko, L., Studer, L., Spenger, C., Dreher, E. and Seiler, R. W. (1996). A mathematical model for the estimation of human embryonic and fetal age. Cell Transplant. 5, 453-464. 10.1177/096368979600500404 [DOI] [PubMed] [Google Scholar]
- Faustino Martins, J.-M., Fischer, C., Urzi, A., Vidal, R., Kunz, S., Ruffault, P.-L., Kabuss, L., Hube, I., Gazzerro, E., Birchmeier, C.et al. (2020). Self-organizing 3D human trunk neuromuscular organoids. Cell Stem Cell 26, 172-186.e6. 10.1016/j.stem.2019.12.007 [DOI] [PubMed] [Google Scholar]
- Forlani, S., Lawson, K. A. and Deschamps, J. (2003). Acquisition of Hox codes during gastrulation and axial elongation in the mouse embryo. Development 130, 3807-3819. 10.1242/dev.00573 [DOI] [PubMed] [Google Scholar]
- Frith, T. J. R., Granata, I., Wind, M., Stout, E., Thompson, O., Neumann, K., Stavish, D., Heath, P. R., Ortmann, D., Hackland, J. O. S.et al. (2018). Human axial progenitors generate trunk neural crest cells in vitro. eLife 7, e35786. 10.7554/eLife.35786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaunt, S. J. (1991). Expression patterns of mouse hox genes: clues to an understanding of developmental and evolutionary strategies. BioEssays 13, 505-513. 10.1002/bies.950131004 [DOI] [PubMed] [Google Scholar]
- Gaunt, S. J., George, M. and Paul, Y.-L. (2013). Direct activation of a mouse Hoxd11 axial expression enhancer by Gdf11/Smad signalling. Dev. Biol. 383, 52-60. 10.1016/j.ydbio.2013.08.025 [DOI] [PubMed] [Google Scholar]
- Gouti, M., Tsakiridis, A., Wymeersch, F. J., Huang, Y., Kleinjung, J., Wilson, V. and Briscoe, J. (2014). In vitro generation of neuromesodermal progenitors reveals distinct roles for wnt signalling in the specification of spinal cord and paraxial mesoderm identity. PLoS Biol. 12, e1001937. 10.1371/journal.pbio.1001937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gouti, M., Delile, J., Stamataki, D., Wymeersch, F. J., Huang, Y., Kleinjung, J., Wilson, V. and Briscoe, J. (2017). A gene regulatory network balances neural and mesoderm specification during vertebrate trunk development. Dev. Cell 41, 243-261.e7. 10.1016/j.devcel.2017.04.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helmbacher, F., Dessaud, E., Arber, S., DeLapeyrière, O., Henderson, C. E., Klein, R. and Maina, F. (2003). Met signaling is required for recruitment of motor neurons to PEA3-positive motor pools. Neuron 39, 767-777. 10.1016/S0896-6273(03)00493-8 [DOI] [PubMed] [Google Scholar]
- Henrique, D., Abranches, E., Verrier, L. and Storey, K. G. (2015). Neuromesodermal progenitors and the making of the spinal cord. Development 142, 2864-2875. 10.1242/dev.119768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iimura, T., Denans, N. and Pourquié, O. (2009). Chapter 7 establishment of Hox vertebral identities in the embryonic spine precursors. Curr. Top. Dev. Biol. 88, 201-234. 10.1016/S0070-2153(09)88007-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izpisúa-Belmonte, J. C., Falkenstein, H., Dollé, P., Renucci, A. and Duboule, D. (1991). Murine genes related to the Drosophila AbdB homeotic genes are sequentially expressed during development of the posterior part of the body. EMBO J. 10, 2279-2289. 10.1002/j.1460-2075.1991.tb07764.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jurberg, A. D., Aires, R., Varela-Lasheras, I., Nóvoa, A. and Mallo, M. (2013). Switching axial progenitors from producing trunk to tail tissues in vertebrate embryos. Dev. Cell 25, 451-462. 10.1016/j.devcel.2013.05.009 [DOI] [PubMed] [Google Scholar]
- Kawaguchi, A. (2019). Temporal patterning of neocortical progenitor cells: how do they know the right time? Neurosci. Res. 138, 3-11. 10.1016/j.neures.2018.09.004 [DOI] [PubMed] [Google Scholar]
- Kimelman, D. and Martin, B. L. (2012). Anterior-posterior patterning in early development: three strategies. Wiley Interdiscip. Rev. Dev. Biol. 1, 253-266. 10.1002/wdev.25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kmita, M. and Duboule, D. (2003). Organizing axes in time and space; 25 years of colinear tinkering. Science 301, 331-333. 10.1126/science.1085753 [DOI] [PubMed] [Google Scholar]
- Koch, F., Scholze, M., Wittler, L., Schifferl, D., Sudheer, S., Grote, P., Timmermann, B., Macura, K. and Herrmann, B. G. (2017). Antagonistic activities of Sox2 and brachyury control the fate choice of neuro-mesodermal progenitors. Dev. Cell 42, 514-526.e7. 10.1016/j.devcel.2017.07.021 [DOI] [PubMed] [Google Scholar]
- Kohwi, M. and Doe, C. Q. (2013). Temporal fate specification and neural progenitor competence during development. Nat. Rev. Neurosci. 14, 823-838. 10.1038/nrn3618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondoh, H. and Takemoto, T. (2012). Axial stem cells deriving both posterior neural and mesodermal tissues during gastrulation. Curr. Opin. Genet. Dev. 22, 374-380. 10.1016/j.gde.2012.03.006 [DOI] [PubMed] [Google Scholar]
- Kondoh, H., Takada, S. and Takemoto, T. (2016). Axial level-dependent molecular and cellular mechanisms underlying the genesis of the embryonic neural plate. Dev. Growth Differ. 58, 427-436. 10.1111/dgd.12295 [DOI] [PubMed] [Google Scholar]
- Lambrot, R., Coffigny, H., Pairault, C., Donnadieu, A.-C., Frydman, R., Habert, R. and Rouiller-Fabre, V. (2006). Use of organ culture to study the human fetal testis development: Effect of retinoic acid. J. Clin. Endocrinol. Metab. 91, 2696-2703. 10.1210/jc.2005-2113 [DOI] [PubMed] [Google Scholar]
- Li, X.-J., Du, Z.-W., Zarnowska, E. D., Pankratz, M., Hansen, L. O., Pearce, R. A. and Zhang, S.-C. (2005). Specification of motoneurons from human embryonic stem cells. Nat. Biotechnol. 23, 215-221. 10.1038/nbt1063 [DOI] [PubMed] [Google Scholar]
- Lippmann, E. S., E.williams, C., Ruhl, D. A., Estevez-Silva, M. C., Chapman, E. R., Coon, J. J. and Ashton, R. S. (2015). Deterministic HOX patterning in human pluripotent stem cell-derived neuroectoderm. Stem Cell Rep. 4, 632-644. 10.1016/j.stemcr.2015.02.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, J.-P. (2006). The function of growth/differentiation factor 11 (Gdf11) in rostrocaudal patterning of the developing spinal cord. Development 133, 2865-2874. 10.1242/dev.02478 [DOI] [PubMed] [Google Scholar]
- Liu, J.-P., Laufer, E. and Jessell, T. M. (2001). Assigning the positional identity of spinal motor neurons: Rostrocaudal patterning of Hox-c expression by FGFs, Gdf11, and retinoids. Neuron 32, 997-1012. 10.1016/S0896-6273(01)00544-X [DOI] [PubMed] [Google Scholar]
- Lunn, J. S., Fishwick, K. J., Halley, P. A. and Storey, K. G. (2007). A spatial and temporal map of FGF/Erk1/2 activity and response repertoires in the early chick embryo. Dev. Biol. 302, 536-552. 10.1016/j.ydbio.2006.10.014 [DOI] [PubMed] [Google Scholar]
- Mallo, M., Vinagre, T. and Carapuço, M. (2009). The road to the vertebral formula. Int. J. Dev. Biol. 53, 1469-1481. 10.1387/ijdb.072276mm [DOI] [PubMed] [Google Scholar]
- Mathis, L. and Nicolas, J.-F. (2003). Progressive restriction of cell fates in relation to neuroepithelial cell mingling in the mouse cerebellum. Dev. Biol. 258, 20-31. 10.1016/S0012-1606(03)00098-8 [DOI] [PubMed] [Google Scholar]
- Matsuda, M., Yamanaka, Y., Uemura, M., Osawa, M., Saito, M. K., Nagahashi, A., Nishio, M., Guo, L., Ikegawa, S., Sakurai, S.et al. (2020). Recapitulating the human segmentation clock with pluripotent stem cells. Nature 580, 124-129. 10.1038/s41586-020-2144-9 [DOI] [PubMed] [Google Scholar]
- Maury, Y., Côme, J., Piskorowski, R. A., Salah-Mohellibi, N., Chevaleyre, V., Peschanski, M., Martinat, C. and Nedelec, S. (2015). Combinatorial analysis of developmental cues efficiently converts human pluripotent stem cells into multiple neuronal subtypes. Nat. Biotechnol. 33, 89-96. 10.1038/nbt.3049 [DOI] [PubMed] [Google Scholar]
- Mazzoni, E. O., Mahony, S., Peljto, M., Patel, T., Thornton, S. R., McCuine, S., Reeder, C., Boyer, L. A., Young, R. A., Gifford, D. K.et al. (2013). Saltatory remodeling of Hox chromatin in response to rostrocaudal patterning signals. Nat. Neurosci. 16, 1191-1198. 10.1038/nn.3490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGrew, M. J., Sherman, A., Lillico, S. G., Ellard, F. M., Radcliffe, P. A., Gilhooley, H. J., Mitrophanous, K. A., Cambray, N., Wilson, V. and Sang, H. (2008). Localised axial progenitor cell populations in the avian tail bud are not committed to a posterior Hox identity. Development 135, 2289-2299. 10.1242/dev.022020 [DOI] [PubMed] [Google Scholar]
- McPherron, A. C., Lawler, A. M. and Lee, S.-J. (1999). Regulation of anterior/posterior patterning of the axial skeleton by growth/differentiation factor 11. Nat. Genet. 22, 260-264. 10.1038/10320 [DOI] [PubMed] [Google Scholar]
- Mendelsohn, A. I., Dasen, J. S. and Jessell, T. M. (2017). Divergent Hox coding and evasion of retinoid signaling specifies motor neurons innervating digit muscles. Neuron 93, 792-805.e4. 10.1016/j.neuron.2017.01.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narendra, V., Rocha, P. P., An, D., Raviram, R., Skok, J. A., Mazzoni, E. O. and Reinberg, D. (2015). CTCF establishes discrete functional chromatin domains at the Hox clusters during differentiation. Science 347, 1017-1021. 10.1126/science.1262088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narendra, V., Bulajić, M., Dekker, J., Mazzoni, E. O. and Reinberg, D. (2016). CTCF-mediated topological boundaries during development foster appropriate gene regulation. Genes Dev. 30, 2657-2662. 10.1101/gad.288324.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neijts, R. and Deschamps, J. (2017). At the base of colinear Hox gene expression: cis-features and trans-factors orchestrating the initial phase of Hox cluster activation. Dev. Biol. 428, 293-299. 10.1016/j.ydbio.2017.02.009 [DOI] [PubMed] [Google Scholar]
- Neijts, R., Amin, S., van Rooijen, C. and Deschamps, J. (2017). Cdx is crucial for the timing mechanism driving colinear Hox activation and defines a trunk segment in the Hox cluster topology. Dev. Biol. 422, 146-154. 10.1016/j.ydbio.2016.12.024 [DOI] [PubMed] [Google Scholar]
- Nijssen, J., Comley, L. H. and Hedlund, E. (2017). Motor neuron vulnerability and resistance in amyotrophic lateral sclerosis. Acta Neuropathol. 133, 863-885. 10.1007/s00401-017-1708-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noordermeer, D., Leleu, M., Splinter, E., Rougemont, J., De Laat, W. and Duboule, D. (2011). The dynamic architecture of Hox gene clusters. Science 334, 222-225. 10.1126/science.1207194 [DOI] [PubMed] [Google Scholar]
- Noordermeer, D., Leleu, M., Schorderet, P., Joye, E., Chabaud, F. and Duboule, D. (2014). Temporal dynamics and developmental memory of 3D chromatin architecture at Hox gene loci. eLife 3, e02557. 10.7554/eLife.02557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nordström, U., Maier, E., Jessell, T. M. and Edlund, T. (2006). An early role for Wnt signaling in specifying neural patterns of Cdx and Hox gene expression and motor neuron subtype identity. PLoS Biol. 4, 1438-1452. 10.1371/journal.pbio.0040252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oberst, P., Fièvre, S., Baumann, N., Concetti, C., Bartolini, G. and Jabaudon, D. (2019a). Temporal plasticity of apical progenitors in the developing mouse neocortex. Nature 573, 370-374. 10.1038/s41586-019-1515-6 [DOI] [PubMed] [Google Scholar]
- Oberst, P., Agirman, G. and Jabaudon, D. (2019b). Principles of progenitor temporal patterning in the developing invertebrate and vertebrate nervous system. Curr. Opin. Neurobiol. 56, 185-193. 10.1016/j.conb.2019.03.004 [DOI] [PubMed] [Google Scholar]
- Ogura, T., Sakaguchi, H., Miyamoto, S. and Takahashi, J. (2018). Three-dimensional induction of dorsal, intermediate and ventral spinal cord tissues from human pluripotent stem cells. Development 145, dev162214. 10.1242/dev.162214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olivera-Martinez, I. and Storey, K. G. (2007). Wnt signals provide a timing mechanism for the FGF-retinoid differentiation switch during vertebrate body axis extension. Development 134, 2125-2135. 10.1242/dev.000216 [DOI] [PubMed] [Google Scholar]
- Olivera-Martinez, I., Schurch, N., Li, R. A., Song, J., Halley, P. A., Das, R. M., Burt, D. W., Barton, G. J. and Storey, K. G. (2014). Major transcriptome re-organisation and abrupt changes in signalling, cell cycle and chromatin regulation at neural differentiation in vivo. Development 141, 3266-3276. 10.1242/dev.112623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peljto, M., Dasen, J. S., Mazzoni, E. O., Jessell, T. M. and Wichterle, H. (2010). Functional diversity of ESC-derived motor neuron subtypes revealed through intraspinal transplantation. Cell Stem Cell 7, 355-366. 10.1016/j.stem.2010.07.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philippidou, P. and Dasen, J. S. (2013). Hox genes: choreographers in neural development, architects of circuit organization. Neuron 80, 12-34. 10.1016/j.neuron.2013.09.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ragagnin, A. M. G., Shadfar, S., Vidal, M., Jamali, M. S. and Atkin, J. D. (2019). Motor neuron susceptibility in ALS/FTD. Front. Neurosci. 13, 532. 10.3389/fnins.2019.00532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribes, V., Le Roux, I., Rhinn, M., Schuhbaur, B. and Dolle, P. (2009). Early mouse caudal development relies on crosstalk between retinoic acid, Shh and Fgf signalling pathways. Development 136, 665-676. 10.1242/dev.016204 [DOI] [PubMed] [Google Scholar]
- Rossi, A. M., Fernandes, V. M. and Desplan, C. (2017). Timing temporal transitions during brain development. Curr. Opin. Neurobiol. 42, 84-92. 10.1016/j.conb.2016.11.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Routal, R. V. and Pal, G. P. (1999). A study of motoneuron groups and motor columns of the human spinal cord. J. Anat. 195, 211-224. 10.1046/j.1469-7580.1999.19520211.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sances, S., Bruijn, L. I., Chandran, S., Eggan, K., Ho, R., Klim, J. R., Livesey, M. R., Lowry, E., Macklis, J. D., Rushton, D.et al. (2016). Modeling ALS with motor neurons derived from human induced pluripotent stem cells. Nat. Neurosci. 19, 542-553. 10.1038/nn.4273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasai, N., Kutejova, E. and Briscoe, J. (2014). Integration of signals along orthogonal axes of the vertebrate neural tube controls progenitor competence and increases cell diversity. PLoS Biol. 12, e1001907. 10.1371/journal.pbio.1001907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindelin, J., Arganda-Carreras, I., Frise, E., Kaynig, V., Longair, M., Pietzsch, T., Preibisch, S., Rueden, C., Saalfeld, S., Schmid, B.et al. (2012). Fiji: An open-source platform for biological-image analysis. Nat. Methods 9, 676-682. 10.1038/nmeth.2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soshnikova, N. and Duboule, D. (2009). Epigenetic temporal control of mouse hox genes in vivo. Science 324, 1321-1323. 10.1126/science.1171468 [DOI] [PubMed] [Google Scholar]
- Steinbeck, J. A. and Studer, L. (2015). Moving stem cells to the clinic: potential and limitations for brain repair. Neuron 86, 187-206. 10.1016/j.neuron.2015.03.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Syed, M. H., Mark, B. and Doe, C. Q. (2017). Playing well with others: extrinsic cues regulate neural progenitor temporal identity to generate neuronal diversity. Trends Genet. 33, 933-942. 10.1016/j.tig.2017.08.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiberi, L., Vanderhaeghen, P. and van den Ameele, J. (2012). Cortical neurogenesis and morphogens: diversity of cues, sources and functions. Curr. Opin. Cell Biol. 24, 269-276. 10.1016/j.ceb.2012.01.010 [DOI] [PubMed] [Google Scholar]
- Tschopp, P., Tarchini, B., Spitz, F., Zakany, J. and Duboule, D. (2009). Uncoupling time and space in the collinear regulation of Hox genes. PLoS Genet. 5, e1000398. 10.1371/journal.pgen.1000398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tung, Y.-T., Peng, K.-C., Chen, Y.-C., Yen, Y.-P., Chang, M., Thams, S. and Chen, J.-A. (2019). Mir-17∼92 confers motor neuron subtype differential resistance to ALS-associated degeneration. Cell Stem Cell 25, 193-209.e7. 10.1016/j.stem.2019.04.016 [DOI] [PubMed] [Google Scholar]
- Tzouanacou, E., Wegener, A., Wymeersch, F. J., Wilson, V. and Nicolas, J.-F. (2009). Redefining the progression of lineage segregations during mammalian embryogenesis by clonal analysis. Dev. Cell 17, 365-376. 10.1016/j.devcel.2009.08.002 [DOI] [PubMed] [Google Scholar]
- Verrier, L., Davidson, L., Gierlinski, M., Dady, A. and Storey, K. G. (2018). Neural differentiation, selection and transcriptomic profiling of human neuromesodermal progenitor-like cells in vitro. Development145, dev166215. 10.1242/dev.166215 [DOI] [PMC free article] [PubMed]
- Wang, J., Vasaikar, S., Shi, Z., Greer, M. and Zhang, B. (2017). WebGestalt 2017: A more comprehensive, powerful, flexible and interactive gene set enrichment analysis toolkit. Nucleic Acids Res. 45, W130-W137. 10.1093/nar/gkx356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wymeersch, F. J., Huang, Y., Blin, G., Cambray, N., Wilkie, R., Wong, F. C. K. and Wilson, V. (2016). Position-dependent plasticity of distinct progenitor types in the primitive streak. eLife 5, e10042. 10.7554/eLife.10042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wymeersch, F. J., Skylaki, S., Huang, Y., Watson, J. A., Economou, C., Marek-Johnston, C., Tomlinson, S. R. and Wilson, V. (2019). Transcriptionally dynamic progenitor populations organised around a stable niche drive axial patterning. Development 146, dev168161. 10.1242/dev.168161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wymeersch F. J., Wilson V. and Tsakiridis A. (2021). Understanding axial progenitor biology in vivo and in vitro. Development148, dev180612. 10.1242/dev.180612 [DOI] [PubMed]
- Young, T., Rowland, J. E., van de Ven, C., Bialecka, M., Novoa, A., Carapuco, M., van Nes, J., de Graaff, W., Duluc, I., Freund, J.-N.et al. (2009). Cdx and Hox genes differentially regulate posterior axial growth in mammalian embryos. Dev. Cell 17, 516-526. 10.1016/j.devcel.2009.08.010 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.