ABSTRACT
Cell imaging has entered the ‘Big Data’ era. New technologies in light microscopy and molecular biology have led to an explosion in high-content, dynamic and multidimensional imaging data. Similar to the ‘omics’ fields two decades ago, our current ability to process, visualize, integrate and mine this new generation of cell imaging data is becoming a critical bottleneck in advancing cell biology. Computation, traditionally used to quantitatively test specific hypotheses, must now also enable iterative hypothesis generation and testing by deciphering hidden biologically meaningful patterns in complex, dynamic or high-dimensional cell image data. Data science is uniquely positioned to aid in this process. In this Perspective, we survey the rapidly expanding new field of data science in cell imaging. Specifically, we highlight how data science tools are used within current image analysis pipelines, propose a computation-first approach to derive new hypotheses from cell image data, identify challenges and describe the next frontiers where we believe data science will make an impact. We also outline steps to ensure broad access to these powerful tools – democratizing infrastructure availability, developing sensitive, robust and usable tools, and promoting interdisciplinary training to both familiarize biologists with data science and expose data scientists to cell imaging.
KEY WORDS: Data science, Deep learning, Imaging, Machine learning, Microscopy
Summary: As the complexity and volume of microscopy data increases, the application of data science to cell imaging will become a critical first step to deriving biological insight.
Introduction
Microscopy provides visual access to cell appearance, organization and behavior, enabling us to discover new biology by observing cells in their basal and perturbed states. The intricate beauty of microscopy images is often engrossing. However, a digital microscopy image is a sequence of numerical values and can be interpreted not only visually, but also via mathematical analysis. Many techniques have been developed for cell biology that take advantage of the dual nature of microscopy images by using their quantitative representation to test hypotheses articulated after carefully viewing them (Ellenberg et al., 2018).
The approach of first looking and then subsequently quantifying microscopy images is becoming increasingly difficult because microscopy for cell biology now entails more – more automation for high-content image acquisition, more modes of microscopy that generate larger datasets, and more microscopes, enabling greater access to microscopy experiments by more people. Beyond generating larger and larger datasets, these advances allow us to test biological hypotheses requiring complex image data that might extend across wide spatial scales, long time-frames or many channels. Even a single complex image, such as a dense 3D mesh of actin or a spheroid of cells, can be too complicated to visually interpret. Humans have an amazing capacity to spot patterns in visual data, but the increased volume and complexity of modern cell imaging data makes visual interpretation infeasible. To draw biological conclusions from ever larger and more-complex imaging datasets, we must change how we interpret cell image data (Ouyang and Zimmer, 2017).
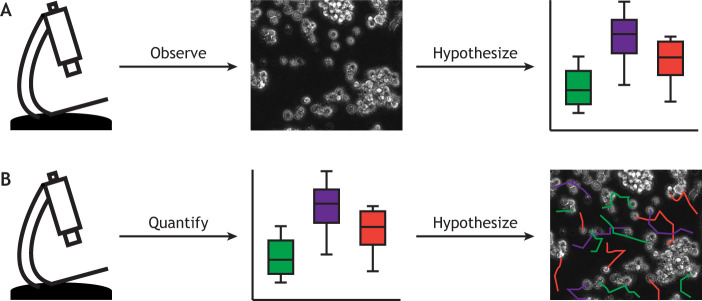
Consider the example of a recent COVID-19 drug screen with 300,000 five-channel immunofluorescence images (Heiser et al., 2020 preprint). It would not be feasible to visually assess and interpret such a large screen. Instead, a deep convolutional neural network, which is a machine-learning technique, was used to automatically extract 1024 properties from each image for statistical analysis, and the results were interpreted and visualized to communicate with other scientists and the general public. This example follows a new paradigm for drawing biological conclusions from complex or high-volume imaging data. Rather than looking and then subsequently quantifying, the order is switched, first computationally analyzing images to develop and test biological hypotheses and only then moving back to the image data to interpret the results and communicate findings (see Fig. 1). In this Perspective, we present the state of data science in cell imaging, which is currently dominated by data science-based tool building for automated quantification of routine bioimage processing. We distinguish these ‘low-level’, signal-driven, tools from ‘high-level’, biology-driven, data science, where hypotheses are raised and biological insights are derived from complex cell image data. Low-level tasks are enabling technologies to address existing questions, whereas high-level tasks, which build upon low-level tasks, open up whole new categories of currently inaccessible questions. Data science has the potential to revolutionize microscopy-based cell biology, but only if infrastructure democratization and cross-disciplinary training are advanced to enable high-level data science in cell imaging.
Fig. 1.
Image analysis workflows. (A) In a typical microscopy-heavy research project, scientists acquire and observe images, and then form and quantitatively test hypotheses based on their observations. (B) We propose that in the future it will be necessary to flip this procedure; first acquiring and quantifying images and only then interacting with the data to further form and test hypotheses.
Data science in cell biology
With the volume and complexity of imaging data increasing, we now need computation to automatically perform tasks across large datasets and to reframe complex data via pattern detection and visualization. Data science, an emerging interdisciplinary field that involves the development and application of computational tools to extract domain-specific insights from large and/or complex datasets, has already begun to supply the needed toolbox. Although the boundaries of data science remain fluid, the field combines domain knowledge with techniques from mathematics, statistics, computer science and information sciences, such as machine learning, to identify patterns hidden in data and perform statistical hypothesis testing on large data sets. The data science toolbox enables the computation-first interpretation of cell images by allowing us to iteratively alternate computational analysis with the generation of biological hypotheses and visualization of the obtained results (Wait et al., 2020).
Data science has been successfully applied to cell imaging data in multiple contexts. One prominent recent theme is the development of deep-learning inference techniques, for example inference of high-resolution images from low-resolution images or inference of cell structure directly from images (Belthangady and Royer, 2019; Eisenstein, 2020). In general, machine-learning algorithms fit generic mathematical models to data. In contrast to traditional machine learning, where models are learned from data features manually engineered by experts, deep learning enables analysis without relying on predetermined features. Instead, a hierarchy of image features is generated directly from the data, simultaneously with the model learning process. This is achieved by using ground truth annotations to train a model to map an input image to a predicted annotation, for example, mapping every pixel in a fluorescence image to its corresponding foreground or background annotation. During training, the model is automatically optimized for a given task by gradually adjusting its internal parameters according to the errors it makes, in a process called back-propagation. Deep learning has already revolutionized machine-learning-driven fields and, in microscopy, has mostly been used to improve the robustness and performance of standard bioimage-analysis tasks, such as segmentation, tracking and classification (Moen et al., 2019; Ouyang et al., 2019a; Ronneberger et al., 2015; Van Valen et al., 2016). It has also provided solutions to other, less-routine, computational tasks. For example, image restoration algorithms attempt to enhance image quality by inferring high-quality images from low-quality data (Weigert et al., 2018) using a variety of strategies, such as by taking advantage of structural redundancy in an image to reconstruct high-quality super-resolution images from under-sampled localization microscopy data (Ouyang et al., 2018), or by performing point spread function engineering for single-molecule localization (Nehme et al., 2020). Other applications include the inference of intracellular organelle localization from label-free images and the mapping of different cell microscopy modalities onto one another (Christiansen et al., 2018; Ounkomol et al., 2018), with potential applications including high-content screening (Cheng et al., 2021) and the prediction of the functional cell state, such as stages of the cell cycle or disease progression (Buggenthin et al., 2017; Eulenberg et al., 2017; Yang et al., 2020; Zaritsky et al., 2020 preprint).
A second theme of data science in cell imaging is high-content cell profiling, where the distributions of image-derived single-cell measurements, such as length, area and fluorescence brightness, are used to define fingerprints of cell populations under different experimental conditions (Perlman et al., 2004). By distilling often large image datasets into succinct fingerprints, cell profiling renders datasets accessible to biological interpretation by users. For example, CellProfiler, a popular software tool for high-content image analysis, encourages a ‘measure everything, ask questions later’ approach to image analysis (Caicedo et al., 2017; Carpenter et al., 2006; Chandrasekaran et al., 2020) by enabling users to first quickly extract and visualize a wide variety of quantitative measures before deciding which are biologically important. These image-based cell profiling ideas are now beginning to be applied to more-complex model systems, including the screening of 3D patient-derived organoids (Beck et al., 2021 preprint; Betge et al., 2019 preprint; Serra et al., 2019).
There are many other examples of the application of data science to cell imaging that are specific to particular biological subdomains. These include, for example, quantitative representations of cell shape in 2D (Bagonis et al., 2019; Chan et al., 2020 preprint; Keren et al., 2008; Pincus and Theriot, 2007) and in 3D (Driscoll et al., 2019; Elliott et al., 2015), perturbation-free inference of information flow in signaling pathways via ‘computational multiplexing’-based fluctuation analysis (Lee et al., 2015; Machacek et al., 2009), statistical-based methods for classification and characterization of protein localization patterns and intracellular organization (Boland et al., 1998; Boland and Murphy, 2001; Glory and Murphy, 2007; Ouyang et al., 2019b; Peng and Murphy, 2011), atlases for intracellular organization and their analyses (Cai et al., 2018; Heinrich et al., 2020 preprint; Thul et al., 2017; Viana et al., 2020 preprint), time-series analyses of heterogeneous dynamic molecular events (Aguet et al., 2013; Bhave et al., 2020; Goglia et al., 2020; Jacques et al., 2020 preprint; Wang et al., 2018, 2020), tracking of lineage, tissue structure and dynamics in development, morphogenesis and collective cell migration (Amat et al., 2014; Etournay et al., 2016; Hartmann et al., 2020; Keller, 2013; Zaritsky et al., 2017), graph representations of dynamic cellular processes (Gut et al., 2015), integration of single-cell omics and imaging data (Villoutreix, 2021; Yang et al., 2021), and machine learning for automated microscopy (Royer et al., 2016; Waithe et al., 2020).
The emerging use of data science tools is revolutionizing many fields, including the social sciences and business, and its impact in cell biology will likely grow. Even just a few years ago, advanced programming skills were needed to implement data science pipelines. Recently, however, user interfaces and other tools have been developed (Bannon et al., 2021; Fazeli et al., 2020; Ouyang et al., 2019a; Stringer et al., 2021; Von Chamier et al., 2020 preprint), rendering data science in cell imaging more accessible to a wide range of researchers.
Hierarchies of data processing in microscopy
The robust and versatile construction of computational pipelines for cell imaging is built on two software design concepts – modularity and abstraction. Modularity and abstraction are what make image analysis pipelines broadly useful and were arguably the key conceptual software advances that fueled the development of modern-day computing.
Building a modular pipeline requires decomposing the main image-analysis task into discrete subtasks that are as independent and generalizable as possible. For example, analysis of nuclei movement in an embryo could be decomposed into a nuclei detection problem, followed by generic object tracking, and then track analysis. The power of modularity stems from the ability to construct complex image analysis pipelines from smaller components that can be designed independently, yet function together. This promotes the reuse of successful modules in many pipelines.
Abstraction is a process that enables modular design by promoting both module reuse and simplicity, hiding algorithmic details within modules, and exposing the inner working of modules to other modules only when necessary. Abstraction enables users and tool developers to focus only on the details that are immediately relevant instead of conceptualizing the algorithm in its full complexity. For example, there exist countless proprietary microscopy file formats, each differently encoding the image and its corresponding metadata. The software Bio-Formats (Linkert et al., 2010), which is executed every time a user reads or writes image data in the image processing program Fiji, provides the abstraction that allows users to access image data without having to be aware of the exact encoding of the different file formats.
Modularity and abstraction are concepts that go hand-in-hand to enable effective problem solving with abstraction enabling modularity. For example, Fiji promotes the construction of modular image analysis pipelines via plugins. Plugins are the modular components composing these pipelines, each solving a well-defined problem and providing an abstract input–output interface. Such implementation enables straightforward reuse of the same plugin in different pipelines, switching between different components with the same interfaces, and expansion of existing pipelines.
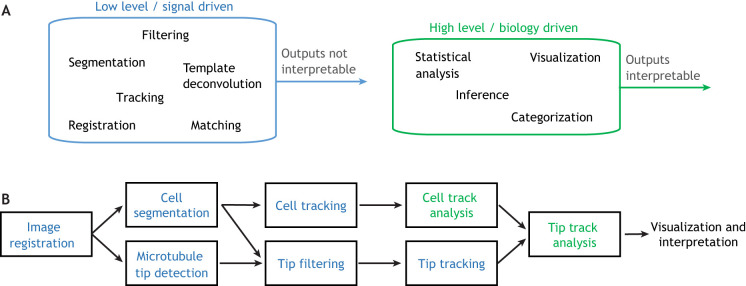
The modules that compose image analysis pipelines can be crudely partitioned into two categories, low-level (signal driven) and high-level (biology driven) (see Fig. 2). Low-level tasks are the signal-driven processing steps that take images or image-derived data and transform them into other images or sequences of numbers. Low-level tasks include image preprocessing (e.g. deconvolution, stage drift correction and tiling fields of view), detection and/or segmentation (e.g. identifying cells/intracellular organelles within an image), and tracking. It is the low-level tasks that enable the automated and complete processing of large image datasets (Danuser, 2011). Importantly, devising effective solutions for low-level tasks requires deep algorithmic knowledge, and sometimes deep understanding of the imaging and optical settings. Domain knowledge can be very helpful. For example, knowledge of the bending properties of microtubules could allow preliminarily detected microtubules that have an unrealistic bend to be excluded from further analysis. However, in most cases, deep knowledge of the biological system or question is not necessary to solve low-level tasks.
Fig. 2.
Hierarchy of image analysis tasks. (A) Image analysis pipelines can be decomposed into low-level (blue) and high-level (green) tasks, with low-level tasks generally preceding high-level tasks. (B) An example image-analysis pipeline for microtubule tracking with low-level tasks shown blue and high-level tasks shown green. Here, images are first registered, or aligned across frames, to account for microscope movement. Next, the cell is segmented, or distinguished from the background, and the microtubule tips are detected. The cell segmentation is used to filter tips by location, removing spurious detections outside the cell, and the cell segmentation and tip detections are separately tracked across frames. Finally, using information derived from a cell-tracking analysis, the tip tracks are analyzed to generate biological insight. In this example, only the track analyses are high-level tasks, since they are the only tasks whose outputs can be directly interpreted to gain biological insight.
High-level tasks are biology driven, transforming large or otherwise difficult to interpret sets of data, which are generally the outputs of low-level tasks, into information that can be directly understood to draw biological conclusions. High-level tasks include data visualization and exploration, model fitting, and statistical inference and comparisons. In contrast to most low-level tasks, high-level tasks always require deep knowledge of the particular biological domain. In order to formulate testable hypotheses, one must understand the biological process at hand and be aware of the experimental and computational techniques available to extract information hidden within the image data. Admittedly, it is currently difficult to point to specific major breakthrough discoveries in cell biology achieved by applying data science to cell imaging. However, both low- and high-level tasks carry the potential to transform the field. Biological discovery is driven by enabling technologies – data science applied to low-level tasks will open the door to addressing existing questions that were previously inaccessible due to a lack of suitable powerful methods. High-level application of data science may unlock completely new fields driven by new types of questions and new ways to discern cell imaging data.
Moving beyond tool building
Data science tools have already been extensively adapted for a variety of low level tasks, such as image enhancement (Weigert et al., 2018), segmentation (Caicedo et al., 2019; Isensee et al., 2020; Stringer et al., 2021; Van Valen et al., 2016) and tracking (Ulman et al., 2017). Indeed, most efforts in the thriving bioimage informatics community have been invested in these types of automation and tool building projects (Meijering et al., 2016). Low-level tool building is essential for advancing almost all cell-imaging-based research, but is not sufficient to answer biological questions. For example, even if all the cells in a developing zebrafish embryo are segmented and tracked, this alone does not provide biological insight. Rather, the tracks must be further visualized and analyzed with the underlying biology in mind.
Why has the bioimage analysis community so far focused on low-level analysis tasks at the expense of the high-level tasks that yield exciting biological discoveries? We believe that this focus stems from two main causes. First, low-level tasks are the most common problems encountered by any microscopist and thus draw community attention as obvious important questions worth tackling. Furthermore, they are the initial steps in any quantification. This may seem trivial; however, developing algorithms for high-level tasks is complicated by the need to first deploy an array of low-level tasks, whereas developing low-level algorithms simply requires the raw data.
Second, low-level tasks are simpler for researchers outside the field of biology to tackle, and are particularly well-suited to computer scientists. Low-level tasks are often readily formulated as abstract computational problems and developing algorithms for them does not typically require any specific ‘domain’ knowledge. In addition, a major motivation for researchers from applied computational sciences, such as computer vision, is algorithmic elegance and efficiency. Publishing and career advancement in computer science is driven by novelty in algorithm design, performance, robustness and, for some applications, usability. Utility to other fields, such as biology, is not emphasized. Moreover, the gold standard for evaluating most low-level applications is comparison with human annotation; however, there is often no correspondingly simple way of evaluating high-level algorithms whose utility is understood only in the context of a particular biological domain. Accordingly, application of data science techniques in cell imaging is heavily biased toward low-level tasks.
Building robust image analysis pipelines requires shared infrastructure
No one research lab can be expert at the full spectrum of low- and high-level tasks needed to draw robust biological conclusions from imaging data. In fact, few labs currently have the expertise and resources to take a computation-first approach to cell imaging data. To utilize the full power of modern microscopy, we must democratize access to computational analysis tools, data and training.
Although well-designed algorithms that employ modularity and abstraction enable the reuse of tools across labs, good software design alone is not enough. Moving beyond low-level tasks requires shared infrastructure to enable the joint development of algorithms and the open use of data. Such infrastructure promotes the exchange of open-source software and image-analysis toolboxes that enable an effective quantification of low-level tasks and allows developers to focus on one component of interest without the need to build a full analysis pipeline to support it. Image-analysis software, such as Fiji (Schindelin et al., 2012), CellProfiler (Carpenter et al., 2006), Icy (de Chaumont et al., 2012) and Ilastik (Berg et al., 2019), as well as open-software libraries (e.g. scikit-learn; Pedregosa et al., 2011), have so far played this role, with deep-learning-specific platforms, such as ImJoy and ZeroCostDL4Mic, beginning to be released (Haase et al., 2020; Ouyang et al., 2019a; Von Chamier et al., 2020 preprint). Support for these platforms was recently consolidated to a single online forum (https://forum.image.sc/), which is very active with frequent use by many visitors. The Bioimage Informatics Index (BII, https://biii.eu/) is a search engine that organizes the wealth of available resources by linking bioimage analysis problems to relevant tools to solve them. Another key infrastructure effort is providing open access to published data to enhance reproducibility, enable computational tool development and allow new discoveries to be made from ‘old’ data (Zaritsky, 2018). To this end, image repositories have recently received significant attention, with the planned BioImage Archive as a major example (Ellenberg et al., 2018; Williams et al., 2017). Image repositories will enable analyses of unprecedented scales of data and are critical to attracting computational researchers to the field.
Software engineers are needed to implement and maintain large-scale tools and data repositories, but these positions are expensive and currently rarely supported by governments or other funding agencies. Philanthropy efforts, such as the Chan–Zuckerberg Initiative and the Allen Institute of Cell Science, have identified this gap and now provide external support, or hire software engineers internally to produce open software. These efforts will hopefully inspire more traditional funding mechanisms to support professional engineers in building solid and shared infrastructure.
Training the next generation of data scientists in cell biology
Cell biology is inherently technology-driven and uses many different tools from biochemistry, molecular biology, microscopy and genomics. The tools of data science are in many ways no different. Effective researchers need to be able to selectively deploy technologies from other fields to forward their research, and it is becoming increasingly clear that the ability to extract quantitative information from microscopy data is essential. A modern cell biologist should be able to decompose an image analysis problem into subtasks, use existing computational tools to solve each subtask and then analyze the pipeline output. This requires basic familiarity with common image-analysis procedures for cell imaging, an ability to piece together modules using simple programming and, importantly, basic knowledge of statistics and machine learning to interpret the results of the pipeline and its limitations.
How do we train the next generation of biologists to adapt to the reality of bioimaging as a data-intensive field? With the encouragement of funding agencies, academic institutes are beginning to adjust their training programs for the ‘Big Data’ era (Barone et al., 2017; Ekmekci et al., 2016; Rubinstein and Chor, 2014; Waldrop et al., 2015). Data analysis or programming bootcamps and high-intensity basic training that last several days or weeks emerged as one of the most popular means to train inexperienced undergraduate or graduate students. However, the effectiveness of these bootcamps is questionable (Feldon et al., 2017), especially when the skills acquired during these short-format interventions are not subsequently practiced and applied. Other initiatives have focused on computational thinking, introducing the basic computer science principles of abstract, algorithmic and logical thinking to life scientists (Rubinstein and Chor, 2014), and/or full courses in developing programming skills (Ekmekci et al., 2016).
We argue that this is not enough. Experimental methods are taught, both directly in laboratory courses and indirectly through the reading of journal articles, with the background knowledge needed to understand these methods spread out among various courses. Similarly, data science and other quantitative methods can be integrated into curriculums. New, comprehensive cross-disciplinary training programs must be established to bridge the technical and cultural gaps between the disciplines. Similar to how chemistry is perceived as essential to the biology curriculum, statistics and other data science tools should also be considered a part of the modern biologist's basic training (Markowetz, 2017). These skills should be acquired early and be used continuously throughout undergraduate and graduate school, not solely in computationally focused courses (Hoffman et al., 2016). For example, when learning about microscopy, students can analyze images with Fiji and integrate results with simple python scripting. The importance of early training and continuity was supported by a recent survey (Attwood et al., 2019).
Whereas hands-on teaching of laboratory methods can require significant space and equipment, hands-on teaching of data science techniques requires only a laptop. A lack of qualified teachers can, however, be a significant challenge (Williams et al., 2019). Faculty without formal knowledge and hands-on experience in data science are asked to design and teach relevant courses. Further compounding this problem is the lack of suitable training materials and reference textbooks specifically suited for these purposes. This situation is even worse in the domain of cell imaging. Most of the textbooks and courses for quantitative thinking and/or programming aimed at biologists are focused on applications in classic ‘bioinformatics’ (omics) (Attwood et al., 2019; Cvijovic et al., 2016; Madamanchi et al., 2018; Rubinstein and Chor, 2014). Images require a different focus because of the diversity in image acquisition techniques and experiments (Gonzalez-Beltran et al., 2020), as well as their multidimensional spatial and temporal structure.
An exciting way to solve the teacher shortage is joint interdisciplinary graduate-level training that brings together students from experimental and computational sciences and introduces both biological problems and quantitative approaches to tackle them (Saunders et al., 2018; von Arnim and Missra, 2017). Another potential solution is recruiting faculty from a neighboring computational department to jointly develop with biomedical faculty, a discipline-specific data science curriculum (Marshall and Geier, 2020). Resources to facilitate cross-disciplinary teaching have also begun to sprout. Steve Royle's recent book, The Digital Cell: Cell Biology as a Data Science (Royle, 2019), is a guidebook for cell and molecular biologists on data science in cell biology, with a special focus on cell imaging. The Network of European BioImage Analysts (NEUBIAS) provides on-site and remote training in bioimage analysis for biologists. Two members of NEUBIAS, Kota Miura and Nataša Sladoje, recently published a ‘Bioimage Data Analysis Workflow’ (Miura and Sladoje, 2020), which teaches how to combine multiple image processing components to construct an effective automated image analysis pipeline suited to a specific purpose and image dataset.
We have so far focused on training biologists to do image analysis, but could we instead turn data scientists into biologists? One possible way forward is to engage computational students in the development of low-level tasks with the motivation of outperforming alternative algorithms and making tools usable for biologists. This route does not require deep domain knowledge and is premised on the hope that some students will develop a fascination with biology. Another parallel strategy is to design cross-disciplinary courses that include both biologists and data scientists. In the domain of data science for cell imaging, the curriculum could include a mix of topics, from low-level bioimage analysis to high-level inference. Similar to a course that one of us, Assaf, designed (Table S1), such a class could introduce data scientists to the amazingly complex world of cell imaging and eventually bring highly desired skills to cell biology.
What's next?
We anticipate that data science applied to cell imaging will propel cell biology forwards through these four themes.
Characterizing heterogeneity
Understanding a biological system requires considering the variability of its components rather than just population averages that mask heterogeneous phenotypes, especially since important phenotypes may be rare.
Bridging scales
Cell biological processes cross scales in space and time – molecules organize within cells, and cells organize within tissues to function. Although we have extensively studied cell biology at some specific scales, we still do not understand how information propagates between scales to enable biological function.
Integrating data across modalities
On the one hand, single-cell omics technologies provide rich information of many well-defined per-cell measurements that is missing in microscopy-based approaches. On the other hand, microscopy can provide information at the protein level, as well as the spatial and temporal context that is mostly lacking in omics. Integrating these two forms of complementary information has vast potential to transform the field (Villoutreix, 2021).
Interpretable machine learning
Machine learning and deep learning, in particular, are very effective at identifying hidden patterns in complex cell imaging data, but lack the ability to explain which biologically relevant properties are important. Developing interpretable data science approaches are absolutely necessary for mechanistic understanding.
Modern biology is becoming more and more complex, advancing toward studies with ever more physiologically relevant systems. This trend of technology-driven complexity is only expected to grow, and we, as a community, must learn to embrace and celebrate it in order to move biology forwards. The combination of more complex data with increased data volume demands infrastructure advancements. Sensitive, robust and usable tools that enable automated analysis are key to processing vast amounts of data and reproducibly analyzing complex data sets. We must train students in data science techniques that enable them to make sense of this data. Together, we can enter the era of data science in cell imaging!
Supplementary Material
Acknowledgments
We would like to thank Philippe Roudot and Dagan Segal for kindly commenting on a draft of this manuscript, as well as Yoav Ram and Natalie Elia for discussions. We would also like to thank The Company of Biologists for funding the 2020 workshop on Data Science in Cell Imaging, and all workshop participants for discussions.
Footnotes
Competing interests
The authors declare no competing or financial interests.
Funding
Our work in this area is supported by the Israeli Council for Higher Education (CHE) via the Data Science Research Center at Ben-Gurion University of the Negev, Israel (to A.Z.), the National Institutes of Health, K99GM123221 (to M.K.D.) and a pilot grant from the Lyda Hill Foundation (to M.K.D.). Deposited in PMC for release after 12 months.
Supplementary information
Supplementary information available online at https://jcs.biologists.org/lookup/doi/10.1242/jcs.254292.supplemental
References
- Aguet, F., Antonescu, C. N., Mettlen, M., Schmid, S. L. and Danuser, G. (2013). Advances in analysis of low signal-to-noise images link dynamin and AP2 to the functions of an endocytic checkpoint. Dev. Cell 26, 279-291. 10.1016/j.devcel.2013.06.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amat, F., Lemon, W., Mossing, D. P., McDole, K., Wan, Y., Branson, K., Myers, E. W. and Keller, P. J. (2014). Fast, accurate reconstruction of cell lineages from large-scale fluorescence microscopy data. Nat. Methods 11, 951-958. 10.1038/nmeth.3036 [DOI] [PubMed] [Google Scholar]
- Attwood, T. K., Blackford, S., Brazas, M. D., Davies, A. and Schneider, M. V. (2019). A global perspective on evolving bioinformatics and data science training needs. Brief. Bioinform. 20, 398-404. 10.1093/bib/bbx100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bagonis, M. M., Fusco, L., Pertz, O. and Danuser, G. (2019). Automated profiling of growth cone heterogeneity defines relations between morphology and motility. J. Cell Biol. 218, 350-379. 10.1083/jcb.201711023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bannon, D., Moen, E., Schwartz, M., Borba, E., Kudo, T., Greenwald, N., Vijayakumar, V., Chang, B., Pao, E., Osterman, E.et al. (2021). DeepCell Kiosk: scaling deep learning-enabled cellular image analysis with Kubernetes. Nat. Methods 18, 43-45. 10.1038/s41592-020-01023-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barone, L., Williams, J. and Micklos, D. (2017). Unmet needs for analyzing biological big data: a survey of 704 NSF principal investigators. PLoS Comput. Biol. 13, e1005755. 10.1371/journal.pcbi.1005755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beck, L. E., Lee, J., Cote, C., Dunagin, M. C., Salla, N., Chang, M. K., Hughes, A. J., Mornin, J. D., Gartner, Z. J. and Raj, A. (2021). Systematically quantifying morphological features reveals constraints on organoid phenotypes. bioRxiv, 2021.01.08.425947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belthangady, C. and Royer, L. A. (2019). Applications, promises, and pitfalls of deep learning for fluorescence image reconstruction. Nat. Methods 16, 1215-1225. 10.1038/s41592-019-0458-z [DOI] [PubMed] [Google Scholar]
- Berg, S., Kutra, D., Kroeger, T., Straehle, C. N., Kausler, B. X., Haubold, C., Schiegg, M., Ales, J., Beier, T., Rudy, M.et al. (2019). Ilastik: interactive machine learning for (bio)image analysis. Nat. Methods 16, 1226-1232. 10.1038/s41592-019-0582-9 [DOI] [PubMed] [Google Scholar]
- Betge, J., Rindtorff, N., Sauer, J., Rauscher, B., Dingert, C., Gaitantzi, H., Herweck, F., Miersch, T., Valentini, E., Hauber, V.et al. (2019). Multiparametric phenotyping of compound effects on patient derived organoids. bioRxiv, 660993. 10.1101/660993 [DOI] [Google Scholar]
- Bhave, M., Mino, R. E., Wang, X., Lee, J., Grossman, H. M., Lakoduk, A. M., Danuser, G., Schmid, S. L. and Mettlen, M. (2020). Functional characterization of 67 endocytic accessory proteins using multiparametric quantitative analysis of CCP dynamics. Proc. Natl. Acad. Sci. USA 117, 31591-31602. 10.1073/pnas.2020346117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boland, M. V. and Murphy, R. F. (2001). A neural network classifier capable of recognizing the patterns of all major subcellular structures in fluorescence microscope images of HeLa cells. Bioinformatics 17, 1213-1223. 10.1093/bioinformatics/17.12.1213 [DOI] [PubMed] [Google Scholar]
- Boland, M. V., Markey, M. K. and Murphy, R. F. (1998). Automated recognition of patterns characteristic of subcellular structures in fluorescence microscopy images. Cytometry 33, 366-375. [DOI] [PubMed] [Google Scholar]
- Buggenthin, F., Buettner, F., Hoppe, P. S., Endele, M., Kroiss, M., Strasser, M., Schwarzfischer, M., Loeffler, D., Kokkaliaris, K. D., Hilsenbeck, O.et al. (2017). Prospective identification of hematopoietic lineage choice by deep learning. Nat. Methods 14, 403. 10.1038/nmeth.4182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai, Y., Hossain, M. J., Hériché, J.-K., Politi, A. Z., Walther, N., Koch, B., Wachsmuth, M., Nijmeijer, B., Kueblbeck, M., Martinic-Kavur, M.et al. (2018). Experimental and computational framework for a dynamic protein atlas of human cell division. Nature 561, 411-415. 10.1038/s41586-018-0518-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caicedo, J. C., Cooper, S., Heigwer, F., Warchal, S., Qiu, P., Molnar, C., Vasilevich, A. S., Barry, J. D., Bansal, H. S., Kraus, O.et al. (2017). Data-analysis strategies for image-based cell profiling. Nat. Methods 14, 849. 10.1038/nmeth.4397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caicedo, J. C., Goodman, A., Karhohs, K. W., Cimini, B. A., Ackerman, J., Haghighi, M., Heng, C. K., Becker, T., Doan, M., McQuin, C.et al. (2019). Nucleus segmentation across imaging experiments: the 2018 Data Science Bowl. Nat. Methods 16, 1247-1253. 10.1038/s41592-019-0612-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carpenter, A. E., Jones, T. R., Lamprecht, M. R., Clarke, C., Kang, I. H., Friman, O., Guertin, D. A., Chang, J. H., Lindquist, R. A., Moffat, J.et al. (2006). CellProfiler: image analysis software for identifying and quantifying cell phenotypes. Genome Biol. 7, R100. 10.1186/gb-2006-7-10-r100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan, C. K., Hadjitheodorou, A., Tsai, T. Y.-C. and Theriot, J. A. (2020). Quantitative comparison of principal component analysis and unsupervised deep learning using variational autoencoders for shape analysis of motile cells. bioRxiv. 10.1101/2020.06.26.174474 [DOI] [Google Scholar]
- Chandrasekaran, S. N., Ceulemans, H., Boyd, J. D. and Carpenter, A. E. (2020). Image-based profiling for drug discovery: due for a machine-learning upgrade? Nat. Rev. Drug Discov. 20, 145-159. 10.1038/s41573-020-00117-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng, S., Fu, S., Kim, Y. M., Song, W., Li, Y., Xue, Y., Yi, J. and Tian, L. (2021). Single-cell cytometry via multiplexed fluorescence prediction by label-free reflectance microscopy. Sci. Adv. 7, eabe0431. 10.1126/sciadv.abe0431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christiansen, E. M., Yang, S. J., Ando, D. M., Javaherian, A., Skibinski, G., Lipnick, S., Mount, E., O'Neil, A., Shah, K., Lee, A. K.et al. (2018). In silico labeling: Predicting fluorescent labels in unlabeled images. Cell 173, 792-803.e19. 10.1016/j.cell.2018.03.040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cvijovic, M., Höfer, T., Aćimović, J., Alberghina, L., Almaas, E., Besozzi, D., Blomberg, A., Bretschneider, T., Cascante, M., Collin, O.et al. (2016). Strategies for structuring interdisciplinary education in Systems Biology: an European perspective. NPJ Syst. Biol. Appl. 2, 16011. 10.1038/npjsba.2016.11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danuser, G. (2011). Computer vision in cell biology. Cell 147, 973-978. 10.1016/j.cell.2011.11.001 [DOI] [PubMed] [Google Scholar]
- de Chaumont, F., Dallongeville, S., Chenouard, N., Herve, N., Pop, S., Provoost, T., Meas-Yedid, V., Pankajakshan, P., Lecomte, T., Le Montagner, Y.et al. (2012). Icy: an open bioimage informatics platform for extended reproducible research. Nat. Methods 9, 690-696. 10.1038/nmeth.2075 [DOI] [PubMed] [Google Scholar]
- Driscoll, M. K., Welf, E. S., Jamieson, A. R., Dean, K. M., Isogai, T., Fiolka, R. and Danuser, G. (2019). Robust and automated detection of subcellular morphological motifs in 3D microscopy images. Nat. Methods 16, 1037-1044. 10.1038/s41592-019-0539-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisenstein, M. (2020). Smart solutions for automated imaging. Nat. Methods 17, 1075-1079. 10.1038/s41592-020-00988-2 [DOI] [PubMed] [Google Scholar]
- Ekmekci, B., McAnany, C. E. and Mura, C. (2016). An introduction to programming for bioscientists: a Python-based primer. PLoS Comput. Biol. 12, e1004867. 10.1371/journal.pcbi.1004867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellenberg, J., Swedlow, J. R., Barlow, M., Cook, C. E., Sarkans, U., Patwardhan, A., Brazma, A. and Birney, E. (2018). A call for public archives for biological image data. Nat. Methods 15, 849-854. 10.1038/s41592-018-0195-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elliott, H., Fischer, R. S., Myers, K. A., Desai, R. A., Gao, L., Chen, C. S., Adelstein, R. S., Waterman, C. M. and Danuser, G. (2015). Myosin II controls cellular branching morphogenesis and migration in three dimensions by minimizing cell-surface curvature. Nat. Cell Biol. 17, 137-147. 10.1038/ncb3092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Etournay, R., Merkel, M., Popović, M., Brandl, H., Dye, N. A., Aigouy, B., Salbreux, G., Eaton, S. and Jülicher, F. (2016). TissueMiner: A multiscale analysis toolkit to quantify how cellular processes create tissue dynamics. eLife 5, e14334. 10.7554/eLife.14334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eulenberg, P., Köhler, N., Blasi, T., Filby, A., Carpenter, A. E., Rees, P., Theis, F. J. and Wolf, F. A. (2017). Reconstructing cell cycle and disease progression using deep learning. Nat. Commun. 8, 463. 10.1038/s41467-017-00623-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fazeli, E., Roy, N. H., Follain, G., Laine, R. F., von Chamier, L., Hänninen, P. E., Eriksson, J. E., Tinevez, J.-Y. and Jacquemet, G. (2020). Automated cell tracking using StarDist and TrackMate. F1000Res 9, 1279. 10.12688/f1000research.27019.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feldon, D. F., Jeong, S., Peugh, J., Roksa, J., Maahs-Fladung, C., Shenoy, A. and Oliva, M. (2017). Null effects of boot camps and short-format training for PhD students in life sciences. Proc. Natl Acad. Sci. USA 114, 9854-9858. 10.1073/pnas.1705783114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glory, E. and Murphy, R. F. (2007). Automated subcellular location determination and high-throughput microscopy. Dev. Cell 12, 7-16. 10.1016/j.devcel.2006.12.007 [DOI] [PubMed] [Google Scholar]
- Goglia, A. G., Wilson, M. Z., Jena, S. G., Silbert, J., Basta, L. P., Devenport, D. and Toettcher, J. E. (2020). A live-cell screen for altered Erk dynamics reveals principles of proliferative control. Cell Systems. 10, 240-253.e6. 10.1016/j.cels.2020.02.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez-Beltran, A. N., Masuzzo, P., Ampe, C., Bakker, G.-J., Besson, S., Eibl, R. H., Friedl, P., Gunzer, M., Kittisopikul, M., Dévédec, S. E. L.et al. (2020). Community standards for open cell migration data. GigaScience 9, giaa041. 10.1093/gigascience/giaa041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gut, G., Tadmor, M. D., Pe'er, D., Pelkmans, L. and Liberali, P. (2015). Trajectories of cell-cycle progression from fixed cell populations. Nat. Methods 12, 951. 10.1038/nmeth.3545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haase, R., Royer, L. A., Steinbach, P., Schmidt, D., Dibrov, A., Schmidt, U., Weigert, M., Maghelli, N., Tomancak, P., Jug, F.et al. (2020). CLIJ: GPU-accelerated image processing for everyone. Nat. Methods 17, 5-6. 10.1038/s41592-019-0650-1 [DOI] [PubMed] [Google Scholar]
- Hartmann, J., Wong, M., Gallo, E. and Gilmour, D. (2020). An image-based data-driven analysis of cellular architecture in a developing tissue. eLife 9, e55913. 10.7554/eLife.55913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinrich, L., Bennett, D., Ackerman, D., Park, W., Bogovic, J., Eckstein, N., Petruncio, A., Clements, J., Xu, C. S., Funke, J.et al. (2020). Automatic whole cell organelle segmentation in volumetric electron microscopy. bioRxiv, 2020.11.14.382143. 10.1101/2020.11.14.382143 [DOI] [Google Scholar]
- Heiser, K., McLean, P. F., Davis, C. T., Fogelson, B., Gordon, H. B., Jacobson, P., Hurst, B., Miller, B., Alfa, R. W., Earnshaw, B. A.et al. (2020). Identification of potential treatments for COVID-19 through artificial intelligence-enabled phenomic analysis of human cells infected with SARS-CoV-2. bioRxiv. 10.1101/2020.04.21.054387 [DOI] [Google Scholar]
- Hoffman, K., Leupen, S., Dowell, K., Kephart, K. and Leips, J. (2016). Development and assessment of modules to integrate quantitative skills in introductory biology courses. CBE—Life Sci. Educ. 15, ar14. 10.1187/cbe.15-09-0186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isensee, F., Jaeger, P. F., Kohl, S. A. A., Petersen, J. and Maier-Hein, K. H. (2020). nnU-Net: a self-configuring method for deep learning-based biomedical image segmentation. Nat. Methods 18, 203-211. 10.1038/s41592-020-01008-z [DOI] [PubMed] [Google Scholar]
- Jacques, M.-A., Dobrzynski, M., Gagliardi, P. A., Sznitman, R. and Pertz, O. (2020). CODEX, a neural network approach to explore signaling dynamics landscapes. bioRxiv. 10.1101/2020.08.05.237842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller, P. J. (2013). Imaging morphogenesis: technological advances and biological insights. Science 340, 1234168. 10.1126/science.1234168 [DOI] [PubMed] [Google Scholar]
- Keren, K., Pincus, Z., Allen, G. M., Barnhart, E. L., Marriott, G., Mogilner, A. and Theriot, J. A. (2008). Mechanism of shape determination in motile cells. Nature 453, 475-480. 10.1038/nature06952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, K., Elliott, H. L., Oak, Y., Zee, C.-T., Groisman, A., Tytell, J. D. and Danuser, G. (2015). Functional hierarchy of redundant actin assembly factors revealed by fine-grained registration of intrinsic image fluctuations. Cell Systems 1, 37-50. 10.1016/j.cels.2015.07.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linkert, M., Rueden, C. T., Allan, C., Burel, J.-M., Moore, W., Patterson, A., Loranger, B., Moore, J., Neves, C., MacDonald, D.et al. (2010). Metadata matters: access to image data in the real world. J. Cell Biol. 189, 777-782. 10.1083/jcb.201004104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machacek, M., Hodgson, L., Welch, C., Elliott, H., Pertz, O., Nalbant, P., Abell, A., Johnson, G. L., Hahn, K. M. and Danuser, G. (2009). Coordination of Rho GTPase activities during cell protrusion. Nature 461, 99-103. 10.1038/nature08242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madamanchi, A., Cardella, M. E., Glazier, J. A. and Umulis, D. M. (2018). Factors mediating learning and application of computational modeling by life scientists. In 2018 IEEE Frontiers in Education Conference (FIE), pp. 1-5: IEEE. [Google Scholar]
- Markowetz, F. (2017). All biology is computational biology. PLoS Biol. 15, e2002050. 10.1371/journal.pbio.2002050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall, B. and Geier, S. (2020). Cross-disciplinary faculty development in data science principles for classroom integration. In Proceedings of the 51st ACM Technical Symposium on Computer Science Education, pp. 1207-1213. [Google Scholar]
- Meijering, E., Carpenter, A. E., Peng, H., Hamprecht, F. A. and Olivo-Marin, J.-C. (2016). Imagining the future of bioimage analysis. Nat. Biotechnol. 34, 1250-1255. 10.1038/nbt.3722 [DOI] [PubMed] [Google Scholar]
- Miura, K. and Sladoje, N. (2020). Bioimage Data Analysis Workflows: Springer Nature. [Google Scholar]
- Moen, E., Bannon, D., Kudo, T., Graf, W., Covert, M. and Van Valen, D. (2019). Deep learning for cellular image analysis. Nat. Methods 16, 1233-1246. 10.1038/s41592-019-0403-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nehme, E., Freedman, D., Gordon, R., Ferdman, B., Weiss, L. E., Alalouf, O., Naor, T., Orange, R., Michaeli, T. and Shechtman, Y. (2020). DeepSTORM3D: dense 3D localization microscopy and PSF design by deep learning. Nat. Methods 17, 734-740. 10.1038/s41592-020-0853-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ounkomol, C., Seshamani, S., Maleckar, M. M., Collman, F. and Johnson, G. R. (2018). Label-free prediction of three-dimensional fluorescence images from transmitted-light microscopy. Nat. Methods 15, 917-920. 10.1038/s41592-018-0111-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouyang, W. and Zimmer, C. (2017). The imaging tsunami: computational opportunities and challenges. Curr. Opin. Syst. Biol. 4, 105-113. 10.1016/j.coisb.2017.07.011 [DOI] [Google Scholar]
- Ouyang, W., Aristov, A., Lelek, M., Hao, X. and Zimmer, C. (2018). Deep learning massively accelerates super-resolution localization microscopy. Nat. Biotechnol. 36, 460-468. 10.1038/nbt.4106 [DOI] [PubMed] [Google Scholar]
- Ouyang, W., Mueller, F., Hjelmare, M., Lundberg, E. and Zimmer, C. (2019a). ImJoy: an open-source computational platform for the deep learning era. Nat. Methods 16, 1199-1200. 10.1038/s41592-019-0627-0 [DOI] [PubMed] [Google Scholar]
- Ouyang, W., Winsnes, C. F., Hjelmare, M., Cesnik, A. J., Åkesson, L., Xu, H., Sullivan, D. P., Dai, S., Lan, J., Jinmo, P.et al. (2019b). Analysis of the Human Protein Atlas Image Classification competition. Nat. Methods 16, 1254-1261. 10.1038/s41592-019-0658-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedregosa, F., Varoquaux, G., Gramfort, A., Michel, V., Thirion, B., Grisel, O., Blondel, M., Prettenhofer, P., Weiss, R. and Dubourg, V. (2011). Scikit-learn: machine learning in Python. J. Mach. Learn. Res. 12, 2825-2830. [Google Scholar]
- Peng, T. and Murphy, R. F. (2011). Image-derived, three-dimensional generative models of cellular organization. Cytometry Part A 79A, 383-391. 10.1002/cyto.a.21066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perlman, Z. E., Slack, M. D., Feng, Y., Mitchison, T. J., Wu, L. F. and Altschuler, S. J. (2004). Multidimensional drug profiling by automated microscopy. Science 306, 1194-1198. 10.1126/science.1100709 [DOI] [PubMed] [Google Scholar]
- Pincus, Z. and Theriot, J. (2007). Comparison of quantitative methods for cell–shape analysis. J. Microsc. 227, 140-156. 10.1111/j.1365-2818.2007.01799.x [DOI] [PubMed] [Google Scholar]
- Ronneberger, O., Fischer, P. and Brox, T. (2015). U-net: Convolutional networks for biomedical image segmentation. In International Conference on Medical image computing and computer-assisted intervention, pp. 234-241: Springer. [Google Scholar]
- Royer, L. A., Lemon, W. C., Chhetri, R. K., Wan, Y., Coleman, M., Myers, E. W. and Keller, P. J. (2016). Adaptive light-sheet microscopy for long-term, high-resolution imaging in living organisms. Nat. Biotechnol. 34, 1267-1278. 10.1038/nbt.3708 [DOI] [PubMed] [Google Scholar]
- Royle, S. J. (2019). The Digital Cell: Cell Biology as a Data Science. Cold Spring Harbor Laboratory Press. [Google Scholar]
- Rubinstein, A. and Chor, B. (2014). Computational thinking in life science education. PLoS Comput. Biol. 10, e1003897. 10.1371/journal.pcbi.1003897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saunders, T. E., He, C. Y., Koehl, P., Ong, L. L. S. and So, P. T. C. (2018). Eleven quick tips for running an interdisciplinary short course for new graduate students. PLoS Comput. Biol. 14, e1006039. 10.1371/journal.pcbi.1006039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindelin, J., Arganda-Carreras, I., Frise, E., Kaynig, V., Longair, M., Pietzsch, T., Preibisch, S., Rueden, C., Saalfeld, S., Schmid, B.et al. (2012). Fiji: an open-source platform for biological-image analysis. Nat. Methods 9, 676-682. 10.1038/nmeth.2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serra, D., Mayr, U., Boni, A., Lukonin, I., Rempfler, M., Meylan, L. C., Stadler, M. B., Strnad, P., Papasaikas, P., Vischi, D.et al. (2019). Self-organization and symmetry breaking in intestinal organoid development. Nature 569, 66-72. 10.1038/s41586-019-1146-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stringer, C., Wang, T., Michaelos, M. and Pachitariu, M. (2021). Cellpose: a generalist algorithm for cellular segmentation. Nat. Methods 18, 100-106. 10.1038/s41592-020-01018-x [DOI] [PubMed] [Google Scholar]
- Thul, P. J., Åkesson, L., Wiking, M., Mahdessian, D., Geladaki, A., Blal, H. A., Alm, T., Asplund, A., Björk, L., Breckels, L. M.et al. (2017). A subcellular map of the human proteome. Science 356, eaal3321. 10.1126/science.aal3321 [DOI] [PubMed] [Google Scholar]
- Ulman, V., Maška, M., Magnusson, K. E. G., Ronneberger, O., Haubold, C., Harder, N., Matula, P., Matula, P., Svoboda, D., Radojevic, M.et al. (2017). An objective comparison of cell-tracking algorithms. Nat. Methods 14, 1141. 10.1038/nmeth.4473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Valen, D. A., Kudo, T., Lane, K. M., Macklin, D. N., Quach, N. T., DeFelice, M. M., Maayan, I., Tanouchi, Y., Ashley, E. A. and Covert, M. W. (2016). Deep learning automates the quantitative analysis of individual cells in live-cell imaging experiments. PLoS Comput. Biol. 12, e1005177. 10.1371/journal.pcbi.1005177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viana, M. P., Chen, J., Knijnenburg, T. A., Vasan, R., Yan, C., Arakaki, J. E., Bailey, M., Berry, B., Borensztejn, A., Brown, J. M.et al. (2020). Robust integrated intracellular organization of the human iPS cell: where, how much, and how variable? bioRxiv, 2020.12.08.415562. [Google Scholar]
- Villoutreix, P. (2021). What machine learning can do for developmental biology. Development 148, dev188474. 10.1242/dev.188474 [DOI] [PubMed] [Google Scholar]
- von Arnim, A. G. and Missra, A. (2017). Graduate training at the interface of computational and experimental biology: an outcome report from a partnership of volunteers between a University and a National Laboratory. CBE Life Sci. Educ. 16, ar61. 10.1187/cbe.17-02-0038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Von Chamier, L., Jukkala, J., Spahn, C., Lerche, M., Hernández-Pérez, S., Mattila, P., Karinou, E., Holden, S., Solak, A. C., Krull, A.et al. (2020). ZeroCostDL4Mic: an open platform to simplify access and use of Deep-Learning in Microscopy. bioRxiv. [Google Scholar]
- Wait, E. C., Reiche, M. A. and Chew, T.-L. (2020). Hypothesis-driven quantitative fluorescence microscopy - the importance of reverse-thinking in experimental design. J. Cell Sci. 133, jcs250027. 10.1242/jcs.250027 [DOI] [PubMed] [Google Scholar]
- Waithe, D., Brown, J. M., Reglinski, K., Diez-Sevilla, I., Roberts, D. and Eggeling, C. (2020). Object detection networks and augmented reality for cellular detection in fluorescence microscopy. J. Cell Biol. 219, e201903166. 10.1083/jcb.201903166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waldrop, L. D., Adolph, S. C., Diniz Behn, C. G., Braley, E., Drew, J. A., Full, R. J., Gross, L. J., Jungck, J. A., Kohler, B., Prairie, J. C.et al. (2015). Using active learning to teach concepts and methods in quantitative biology. Integr. Comp. Biol. 55, 933-948. 10.1093/icb/icv097 [DOI] [PubMed] [Google Scholar]
- Wang, C., Choi, H. J., Kim, S.-J., Desai, A., Lee, N., Kim, D., Bae, Y. and Lee, K. (2018). Deconvolution of subcellular protrusion heterogeneity and the underlying actin regulator dynamics from live cell imaging. Nat. Commun. 9, 1688. 10.1038/s41467-018-04030-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, X., Chen, Z., Mettlen, M., Noh, J., Schmid, S. L. and Danuser, G. (2020). DASC, a sensitive classifier for measuring discrete early stages in clathrin-mediated endocytosis. eLife 9, e53686. 10.7554/eLife.53686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weigert, M., Schmidt, U., Boothe, T., Müller, A., Dibrov, A., Jain, A., Wilhelm, B., Schmidt, D., Broaddus, C., Culley, S.et al. (2018). Content-aware image restoration: pushing the limits of fluorescence microscopy. Nat. Methods 15, 1090. 10.1038/s41592-018-0216-7 [DOI] [PubMed] [Google Scholar]
- Williams, E., Moore, J., Li, S. W., Rustici, G., Tarkowska, A., Chessel, A., Leo, S., Antal, B., Ferguson, R. K., Sarkans, U.et al. (2017). Image Data Resource: a bioimage data integration and publication platform. Nat. Methods 14, 775-781. 10.1038/nmeth.4326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams, J. J., Drew, J. C., Galindo-Gonzalez, S., Robic, S., Dinsdale, E., Morgan, W. R., Triplett, E. W., Burnette, J. M., III, Donovan, S. S., Fowlks, E. R.et al. (2019). Barriers to integration of bioinformatics into undergraduate life sciences education: A national study of US life sciences faculty uncover significant barriers to integrating bioinformatics into undergraduate instruction. PLoS ONE 14, e0224288. 10.1371/journal.pone.0224288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, K. D., Damodaran, K., Venkatachalapathy, S., Soylemezoglu, A. C., Shivashankar, G. and Uhler, C. (2020). Predicting cell lineages using autoencoders and optimal transport. PLoS Comput. Biol. 16, e1007828. 10.1371/journal.pcbi.1007828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, K. D., Belyaeva, A., Venkatachalapathy, S., Damodaran, K., Katcoff, A., Radhakrishnan, A., Shivashankar, G. V. and Uhler, C. (2021). Multi-domain translation between single-cell imaging and sequencing data using autoencoders. Nat. Commun. 12, 31. 10.1038/s41467-020-20249-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaritsky, A. (2018). Sharing and reusing cell image data. Mol. Biol. Cell 29, 1274-1280. 10.1091/mbc.E17-10-0606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaritsky, A., Jamieson, A. R., Welf, E. S., Nevarez, A., Cillay, J., Eskiocak, U., Cantarel, B. L. and Danuser, G. (2020). Interpretable deep learning of label-free live cell images uncovers functional hallmarks of highly-metastatic melanoma. bioRxiv. 10.1101/2020.05.15.096628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaritsky, A., Tseng, Y.-Y., Rabadán, M. A., Krishna, S., Overholtzer, M., Danuser, G. and Hall, A. (2017). Diverse roles of guanine nucleotide exchange factors in regulating collective cell migration. J. Cell Biol. 216, 1543-1556. 10.1083/jcb.201609095 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.