To the Editor,
With the increasing spread of COVID-19 pandemic around the world, there have been 223,983 whole genome sequences of SARS-CoV-2 submitted in GISAID database as of November 2, 2020. This wealth of sequences can be useful in probing variations in the viral genome which can potentially affect its transmissibility and virulence. The SARS-CoV-2 is a RNA virus constituting six major open reading frames (ORF) that encodes structural and non-structural proteins. Sixteen non-structural proteins (nsp 1–16) are encoded by ORF1a and 1b while the accessory genes are encoded by ORF3a, ORF6, ORF7a and b, and ORF8 (Shimamoto et al., 2015; Zhou et al., 2020). From a comparative standpoint, RNA viruses (like influenza and HIV) tend to incorporate nucleotide variations due to the lack of proof reading activity of RNA polymerase enzyme. Logically, this can bring about high mutation rate, however SARS-CoV viruses has evolved with a proof reading region, the nsp14, that keeps a check on rapid mutational changes in its genome (Denison et al., 2011). Numerically, this has been exemplified from reports which have suggested 12,000 mutations in SARS CoV2 mutations till September 2020 (Callaway, 2020).
Pakistan is a populous country with 403,311 positive cases and 166 deaths as of December 1, 2020 (https://covid.gov.pk/stats/pakistan). Therefore, following genomic surveillance for SARS-CoV-2 is imperative. As of October 16, 2020, only 14 whole genome sequences of SARS-CoV-2 has been reported from Pakistan. Nevertheless, studying these sequences with respect to its divergence from worldwide sequences, is crucial to get a lead on the possible genetic variants of SARS-CoV-2 which might be circulating in Pakistani population.
All the 14 whole genome sequences of SARS-CoV-2 reported from Pakistan till October 16, 2020 were downloaded from GISAID database (https://www.epicov.org/epi3/frontend#efa72), and the first sequence SARS-CoV-2 from Wuhan was used as reference sequence (Accession number: NC_045512.2). The multiple sequence alignment was performed using Clustal X (Larkin et al., 2007). Visualization of alignment followed by mutational analysis was performed using Jalview (Waterhouse et al., 2009). Phylogenetic analysis of 14 Pakistani sequence isolates of SARS-CoV-2, was performed using Galaxy server. For phylogenetic analysis multiple sequence alignment was performed using MAFFT followed by Maximum Likelihood tree construction using IQTree available on Galaxy server (Nguyen et al., 2015). The visualization and editing of tree was performed using FigTree (http://tree.bio.ed.ac.uk/software/figtree/). The variations in the amino acid sequences has been compared with the SARS-CoV-2 sequences reported around the world on COVIDCG database (Chen et al., 2020) as of October 16, 2020. The effect of mutations on stability of protein was studied through I-Mutant 3.0. It predicts the effect of mutation on stability of protein by estimation of Gibbs free change (ΔG) (difference of energy (DDG) between native and mutated protein). The effect of mutations on the stability of protein is classified either increasing the stability (DDG > 0.5Kcal/mol), decreasing the stability (DDG < -0.5 Kcal/mol) or neutral impact (−0.5 ≤DDG ≤0.5 Kcal/mol) on protein structure (Capriotti et al., 2005).
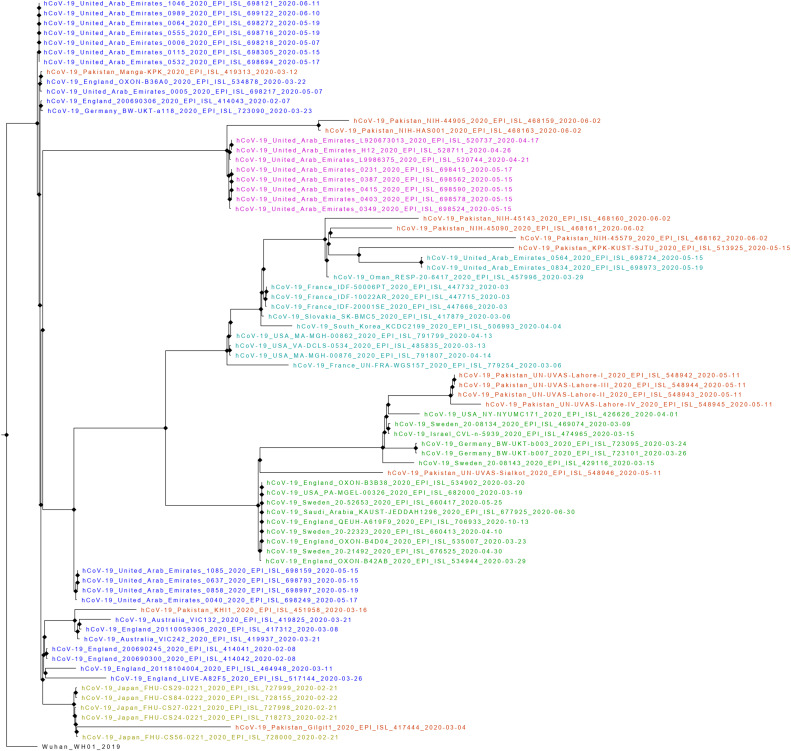
Phylogenetic analysis revealed that the SARS-CoV-2 sequences correspond to GH, S, O, GR and L clades circulating in Pakistan. Initial sequences (March 2020) from Pakistan revealed the presence of L and O clades. The L clade sequences appeared to be closely related to strains reported from United Kingdom and United Arab Emirates, while O clade sequences were closely related to SARS-CoV-2 sequences from Japan. The samples collected in May 2020 belongs to GR clade that is closely related to isolates from USA, Sweden and Germany. The GH and S clade were observed in June 2020 sequences that appear to be closely related to strains reported from United Arab Emirates (Fig. S1).
Phylogenetic analysis revealed that the SARS-CoV-2 sequences correspond to GH, S, O, GR and L clades circulating in Pakistan. Initial sequences (March 2020) from Pakistan revealed the presence of L and O clades. The L clade sequences appeared to be closely related to strains reported from United Kingdom and United Arab Emirates, while O clade sequences were closely related to SARS-CoV-2 sequences from Japan. The samples collected in May 2020 belongs to GR clade that is closely related to isolates from USA, Sweden and Germany. The GH and S clade were observed in June, 2020 sequences that appear to be closely related to strains reported from United Arab Emirates (Fig. S1).
In total, 28 amino acid variations in the structural and nonstructural proteins of SARS-CoV-2 have been identified from patient isolates across Pakistan. There are 07 non-structural genes (nsp1, 7–11, 14–16) in ORF1ab that have been found to be conserved in Pakistani isolates. The amino acid changes have been observed in nsp2, 3, 4, 5,6, 12, and 13 (Fig. 1 ).
Fig. 1.
Multiple sequence alignment of Pakistani SARA-CoV-2 sequences with the reference Wuhan-1 strain. (A) nsp2; (B) nsp3; (C) nsp4; (D) nsp5; (E) nsp6; (F) nsp13; (G) Spike; (H) ORF3; (I) ORF8; (J) Nucleocapsid; (K) ORF10. The amino acid variations are shown with white color.
In nsp2, three changes (R207C, V378I, D448N) have been observed in the sequences collected from March and one change (L450F) have been observed in the samples from June. Interestingly, these changes have not been observed in any of the sequences reported around the world. Nsp2 is an important viral protein that along with nsp8, is involved in viral replication (Angeletti et al., 2020). Hence, any change in this gene may impair viral replication and therefore requires further investigation.
In nsp3, three changes (L944S, T1246I, K1305N, and Q2702H) have been detected in the isolates from May 2020. The T1246I variant has been reported in only 0.2% sequences from different countries (Table 1 ). The K1305N has been observed in only 0.1% of the Asian sequences and 0.5% in European isolates. The Q2702H have been observed from 5 sequences recovered in May and June 2020. Comparatively worldwide, this mutation has been reported in less than 1% of the isolates. Another mutation that has been observed in nsp3 is T2016K which has been reported earlier by Ghanchi et al. 2020 (Ghanchi et al., 2020). This mutation has been reported in 15% of the isolates from Asia and found to be prevailing in October 2020 isolates as well. Functionally, the nsp3 plays its part in immunosuppression of innate immune responses of host (Lei et al., 2018). Hence, the changes in nsp3 can result in enhanced viral capability to evade innate immune defenses.
Table 1.
Details of mutations reported in Pakistani Sequences in comparison with the worldwide reported sequences and effect of mutations on stability of protein. DDG > 0.5Kcal/mol = protein stability increases, DDG < -0.5 Kcal/mol = protein stability decreases.
| Sample ID | Gene | Mutation | Worldwide Prevalence | DDG (KJ/mol) | Effect on Protein Stability |
|---|---|---|---|---|---|
| EPI_ISL_417444 | nsp2 | R207C V378I |
−0.82654688 −0.47956396 |
Decrease | |
| EPI_ISL_451958 | nsp2 | D448N | −0.91305103 | Decrease | |
| EPI_ISL_45579 | nsp2 | L450F | −1.6603629 | Decrease | |
| EPI_ISL_548942 EPI_ISL_548943 EPI_ISL_548944 EPI_ISL_548945 |
nsp3 | L944S |
−0.61244224 −0.16539386 |
Decrease | |
| T1246I | Asia = 0.2% Europe = 2.5% South America = 7.6% USA = 0.1% Canada = 0.2% Africa = 9.5% |
||||
| K1305N | Asia = 0.1% Europe = 0.6% Africa = 0.04% |
||||
| EPI_ISL_513925 EPI_ISL_45143 EPI_ISL_45090 EPI_ISL_45579 |
nsp3 | Q2702H | Asia = 0.5% Europe = 0.9% Canada = 0.2% Africa = 1.7% |
−1.4611661 | Decrease |
| EPI_ISL_417444 | nsp4 | P2965L | 0.433947 | Increase | |
| EPI_ISL_548942 EPI_ISL_548943 EPI_ISL_548944 EPI_ISL_548945 |
nsp5 | G3278S | Asia = 0.2% Europe = 6.4% South America = 7.6% Canada = 0.4% Africa = 9.6% |
−0.39907465 | Decrease |
| EPI_ISL_468160 | nsp5 | N3491K | −1.3556076 | Decrease | |
| EPI_ISL_417444 EPI_ISL_513925 EPI_ISL_45143 EPI_ISL_45090 EPI_ISL_45579 |
nsp6 | L3606F | Asia = 21.0% Europe = 9.6% South America = 2.6% USA = 3.3% Canada = 6.2% Africa = 5.1% |
−1.2923268 | Decrease |
| EPI_ISL_468159 EPI_ISL_468163 |
nsp6 | M3655I | Asia = 0.6% Europe = 0.8% Africa = 1.1% |
−0.74222727 | Decrease |
| EPI_ISL_548942 EPI_ISL_548943 EPI_ISL_548944 EPI_ISL_548945 EPI_ISL_513925 EPI_ISL_468161 EPI_ISL_548946 EPI_ISL_468160 EPI_ISL_468162 |
nsp12 | P4715L | Asia = 62.7% Europe = 90% South America = 94% USA = 89% Canada = 85% Africa = 91% |
||
| EPI_ISL_513925 EPI_ISL_417444 EPI_ISL_419313 |
nsp13 | A5561T | −1. 8,647,808 | Decrease | |
| EPI_ISL_548946 | S | N74K | 0.82838446 | Increase | |
| EPI_ISL_548946 EPI_ISL_468160 EPI_ISL_468161 EPI_ISL_548942 EPI_ISL_548943 EPI_ISL_548944 EPI_ISL_548945 EPI_ISL_513925 EPI_ISL_468162 |
S | D614G | Asia = 62.4% Europe = 87.9% South America = 94.3% USA = 89.2% Canada = 82.2% Africa = 95% |
−1.4867818 | |
| EPI_ISL_468159 EPI_ISL_468163 |
S | D830A | −0.59789075 | Decrease | |
| EPI_ISL_513925 EPI_ISL_468161 EPI_ISL_468160 EPI_ISL_468162 |
ORF3a | Q57H | Asia = 25.2% Europe = 10.8% South America = 12.0% USA = 62.0% Canada = 36.6% Africa = 9.3% |
−0.91817829 | Decrease |
| EPI_ISL_468160 | ORF3a | F105S | −0.94734895 | Decrease | |
| EPI_ISL_468161 | ORF8 | W45L | Europe = 0.1% USA = 0.2% |
−0.57599897 | Decrease |
| EPI_ISL_468159 EPI_ISL_468163 |
ORF8 | L84S | Asia = 8.3% Europe = 2.2% South America = 3.7% USA = 8.8% Canada = 12.8% Africa = 4.2% |
−1.9053577 | Decrease |
| E92K | Asia = 0.4% Africa = 1.7% |
−0.75628271 | |||
| EPI_ISL_468159 EPI_ISL_468163 EPI_ISL_468162 |
N | S202N | Asia = 2.7% Europe = 0.1% South America = 0.0% USA = 0.3% Africa = 4.2% |
0.6542265 | Increase |
| EPI_ISL_548942 EPI_ISL_548943 EPI_ISL_548944 EPI_ISL_548945 EPI_ISL_548946 |
N | R203K G204R |
Asia = 29.3% Europe = 46.7% South America = 62.7% USA = 12.9% Canada = 16.9% Africa = 53.7% |
−1.0587625 | Decrease |
| EPI_ISL_513925 | N | S327L | Asia = 0.1% Europe = 0.1% South America = 0.1% USA = 0.1% Africa = 0.3% |
0.55558303 | Increase |
| EPI_ISL_513925 | ORF10 | V30L | Asia = 0.2% Europe = 6.9% USA = 0.1% Africa = 0.2% |
−1.4376902 | Decrease |
In nsp4, only one novel change (P2965L) has been observed in one isolate. This change has not been found in any of the worldwide reported sequences to our knowledge.
In nsp5, the G3278S and N3491K changes have been observed from the isolates of May and June, respectively. The N3491K appears to be a novel mutation while G3278 has been reported with high prevalence rate (9.6%) in the initial months of pandemic and has not been observed after August 2020. The nsp5 is also known as 3C like protease, is involved in viral replication (Macchiagodena et al., 2020) and also interferes with interferon signaling (Zhu et al., 2017a; Zhu et al., 2017b). Hence, potentially any mutation can impact viral replication capability.
In nsp6, the L3606F change has been present in 5 isolates from March, and June 2020. This mutation has been shown in 21% of isolates from Asia while in 9.6% of isolates from Europe. The nsp6 protein of other coronaviruses has been reported to interfere cellular autophagy signaling by affecting the PI3K3C3 and ATG5 proteins thereby inducing autophagosome formation but blocking its maturation (Benvenuto et al., 2020; Gassen et al., 2019; Yang and Shen, 2020).
Among the structural proteins of SARS-CoV-2, the spike protein (S) is the outermost protein that is involved in entry of virus into the host cell. The D614G change in the S protein is observed in 9 Pakistani isolates from May and June 2020. The D614G mutation has been considered to increase SARS-CoV-2 infectious capability (Korber et al., 2020) by increasing the transmissibility of virus. The D830A is another novel change observed in 2 Pakistani isolates from June 2020. This change is important as D830 is present near the TMPRSS2 binding site and may have an effect on viral fusion.
In ORF3a, the important change has been observed at position Q57H from the isolates of May and June 2020. It has been reported that this mutation is prevalent worldwide and it has an impact on protein structure (Banoun, 2020). The Q57H is present in 62% of the isolates from USA, 25% of isolates from Asia and 10.8% of isolates from Europe. ORF3a and ORF8 may have a role in host immune responses (Banoun, 2020).
Three changes (W45L, L84S, and E92K) has been observed in ORF8 from the isolates of May and June 2020. The L84S has been present in Canada (12.8% of isolates), USA (8.8% of isolates) and Asia (8.3% of isolates). The E92K has been reported in less than 0.5% of worldwide isolates and not observed after July 2020. The ORF8 is important in downregulating the MHC-I molecules thus protecting the affected cells from cytotoxic T-cell killing (Banoun, 2020).
In the N gene the important mutations have been S202N, R203K, G204R, S327L observed in Pakistani isolates. In comparison with the worldwide reported sequences, it is observed that in Asian and African sequences the S202N is present in 2.7% and 4.2% of isolates, respectively but it has disappeared after July 2020. The R203K and G204R are the changes that co-exist in the isolates while observing the worldwide reported sequences. These two mutations have also been among one of the prevalent changes observed in SARS-CoV-2 genome present in 62% of isolates in America, 53% of isolates in Africa, 46% of isolates in Europe and 29% of isolates in Asia. The N protein -contribute to viral genome assembly and serve as viral suppressor of RNAi to antagonize the host immune defense system (Liu et al., 2020).
In ORF10, only one change the V30L have been observed in one of the isolate from Pakistan. The V30L have not been observed in sequences from USA and Asia after August, 2020 while in European sequences this change is still appearing. Since ORF10 is a novel protein to SARS-CoV-2, not much data available about the role of this protein in viral pathogenesis (Cagliani et al., 2020).
By analyzing the impact of mutations on the stability of protein structure through I-Mutant, all the reported variations have been suggesting a decreasing stability of protein structure with the exception of P2965L, N75K, S202N, and S327L variations which suggest an impression of increasing the stability of nsp4, S, and N protein respectively (Table1).
Pakistan is now experiencing second wave of COVID-19 insurgence. Therefore, more SARS-CoV-2 sequences are required for effective genomic surveillance of SARS-CoV-2 and for identifying sequence divergence from Pakistan. The novel mutations in the nsp2, nsp4, nsp5, nsp13, S, and ORF3a should be further evaluated in more SARS-CoV-2 genomes from Pakistan. The novel mutation in nsp4 (P2965L) and similar variants that have been reported around the world should be further investigated for its impact on protein stability as it may affect the viral counter measures in developing efficient vaccines and therapeutic solutions.
Data availability
The annotated genomes of SARS-CoV-2 from Pakistan and the sequences used for phylogenetic analysis has been retrieved from the global initiative on sharing all influenza data (GISAID) (https://www.gisaid.org/). A full list of accession number along with the acknowledgment table is provided as supplementary file 1.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
The authors declare the following financial interests/personal relationships which may be considered as potential competing interests:
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.meegid.2021.104752.
Appendix A. Supplementary data
Supplementary material
Fig. S1.
Maximum-Likelihood phylogenetic tree of 14 SARS-CoV-2 whole genome sequences from Pakistan. The Pakistani sequences are highlighted in red color. The L clade is highlighted as blue, S clade as purple, GH clade as sea green, GR clade as green, and O clade as yellow color.
References
- Angeletti S., et al. COVID-2019: the role of the nsp2 and nsp3 in its pathogenesis. J. Med. Virol. 2020;92(6):584–588. doi: 10.1002/jmv.25719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banoun H. 2020. Evolution of Sars-Cov-2: Update September 2020. SSRN. [Google Scholar]
- Benvenuto D., et al. Evolutionary analysis of SARS-CoV-2: how mutation of non-structural protein 6 (NSP6) could affect viral autophagy. J. Inf. Secur. 2020;81(1):e24–e27. doi: 10.1016/j.jinf.2020.03.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cagliani R., et al. Coding potential and sequence conservation of SARS-CoV-2 and related animal viruses. Infect. Genet. Evol. 2020;83:104353. doi: 10.1016/j.meegid.2020.104353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callaway E. The coronavirus is mutating - does it matter? Nature. 2020;585(7824):174–177. doi: 10.1038/d41586-020-02544-6. [DOI] [PubMed] [Google Scholar]
- Capriotti E., Fariselli P., Casadio R. I-Mutant2.0: predicting stability changes upon mutation from the protein sequence or structure. Nucleic Acids Res. 2005;33 doi: 10.1093/nar/gki375. (Web Server issue): p. W306–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen A.T., et al. COVID-19 CG: Tracking SARS-CoV-2 mutations by locations and dates of interest. bioRxiv. 2020 doi: 10.7554/eLife.63409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denison M.R., et al. Coronaviruses: an RNA proofreading machine regulates replication fidelity and diversity. RNA Biol. 2011;8(2):270–279. doi: 10.4161/rna.8.2.15013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gassen N.C., et al. SKP2 attenuates autophagy through Beclin1-ubiquitination and its inhibition reduces MERS-coronavirus infection. Nat. Commun. 2019;10(1):5770. doi: 10.1038/s41467-019-13659-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghanchi N.K., et al. 2020. SARS-CoV-2 genome analysis of strains in Pakistan reveals GH, S and L clade strains at the start of the pandemic. 2020.08.04.234153. [Google Scholar]
- Korber B., et al. Tracking changes in SARS-CoV-2 spike: evidence that D614G increases infectivity of the COVID-19 virus. Cell. 2020;182(4):812–827. doi: 10.1016/j.cell.2020.06.043. (e19) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larkin M.A., et al. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23(21):2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- Lei J., Kusov Y., Hilgenfeld R. Nsp3 of coronaviruses: structures and functions of a large multi-domain protein. Antivir. Res. 2018;149:58–74. doi: 10.1016/j.antiviral.2017.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q., et al. 2020. Ongoing natural selection drives the evolution of SARS-CoV-2 genomes. 2020.09.07.20189860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macchiagodena M., Pagliai M., Procacci P. Identification of potential binders of the main protease 3CL(pro) of the COVID-19 via structure-based ligand design and molecular modeling. Chem. Phys. Lett. 2020;750:137489. doi: 10.1016/j.cplett.2020.137489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen L.T., et al. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 2015;32(1):268–274. doi: 10.1093/molbev/msu300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimamoto Y., et al. Fused-ring structure of decahydroisoquinolin as a novel scaffold for SARS 3CL protease inhibitors. Bioorg. Med. Chem. 2015;23(4):876–890. doi: 10.1016/j.bmc.2014.12.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterhouse A.M., et al. Jalview version 2--a multiple sequence alignment editor and analysis workbench. Bioinformatics. 2009;25(9):1189–1191. doi: 10.1093/bioinformatics/btp033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang N., Shen H.M. Targeting the endocytic pathway and autophagy process as a novel therapeutic strategy in COVID-19. Int. J. Biol. Sci. 2020;16(10):1724–1731. doi: 10.7150/ijbs.45498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou P., et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579(7798):270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu X., et al. Porcine deltacoronavirus nsp5 inhibits interferon-beta production through the cleavage of NEMO. Virology. 2017;502:33–38. doi: 10.1016/j.virol.2016.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu, X., et al., Porcine Deltacoronavirus nsp5 Antagonizes Type I Interferon Signaling by Cleaving STAT2. J. Virol., 2017b. 91(10). [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material
Data Availability Statement
The annotated genomes of SARS-CoV-2 from Pakistan and the sequences used for phylogenetic analysis has been retrieved from the global initiative on sharing all influenza data (GISAID) (https://www.gisaid.org/). A full list of accession number along with the acknowledgment table is provided as supplementary file 1.