Hidradenitis suppurativa (HS) is a chronic inflammatory skin disorder resulting in recurrent, painful nodules, abscesses, and sinuses with predilection for intertriginous sites.1 In previous cohorts relying primarily on chart review, 30–40% of patients reported family history of disease,2 but familial risk has not been formally assessed with more meticulous and focused data collection. 676 HS patients at the University of North Carolina Chapel Hill were enrolled in our clinical registry from August 2018-December 2019, which collected detailed family history data using questionnaires and skilled interviewers. 57.5% of patients reported HS in either first- or second-degree relatives, including 49.5% with an affected first-degree relative. This suggests a possible genetic contribution to HS. This study’s aim was to quantify familial risk in patients with HS.
Sibling recurrence risk ratio (λs) is a metric of familial aggregation defined as the risk ratio of disease in the affected patient’s siblings as compared to prevalence in the general population.3 λs is calculated using all available pedigree data from a set of probands and serves as a widely used metric of familial risk calculated based on pedigree data. Larger λs indicates high risk of disease among individuals with an affected sibling. Though environmental factors may also influence disease risk, λs quantifies potential genetic contributions of a disease. Parent-offspring recurrence risk (λo) is calculated similarly and also approximates familial risk, though, in some instances, offspring knowledge of parental disease is less complete than for siblings.
Pedigrees from 281 consecutively recruited subjects from our registry that included affected and unaffected parents, grandparents, siblings, half-siblings, and children with HS were constructed and analyzed. Affected aunts, uncles, cousins, and grandchildren were included when available. λs was determined using the formula outlined by Olson and Cordell.4
Proband subjects were 80% female, 53.4% black, 42.6% white, and 6% other races with median age 35 years. Of 371 identified siblings, 74 are affected. Based on an overall estimated population risk of 1%, analysis of 280 pedigrees found a λs of 19.9 without significant difference by race. To avoid underestimating risk by including family members who may have been too young to develop disease, we evaluated a range of age thresholds (Figure 1A). Although we expected λs to increase as we excluded younger relatives that may not yet have developed disease, it decreased instead. This may indicate that subjects are more aware of disease in younger relatives such as their children or siblings than of older, more distant relatives. Thus, excluding younger relatives may underestimate overall risk. λo was similarly found to be 18.6, though age data for offspring was often missing and limited meaningful calculations with age cut-offs. In comparison with λs from diseases with known genetic contributions (Figure 1B), the λs of 19.9 observed in HS is relatively high, consistent with a strong genetic contribution to the etiology of HS. These findings are in concordance with a recent study of 58 twin pairs with HS. Dizygotic and monozygotic concordance were 0.08 and 0.31, respectively, with calculated heritability of 77%.5 This approximates heritability of Crohn’s disease, which is consistent with our data.
Figure 1:
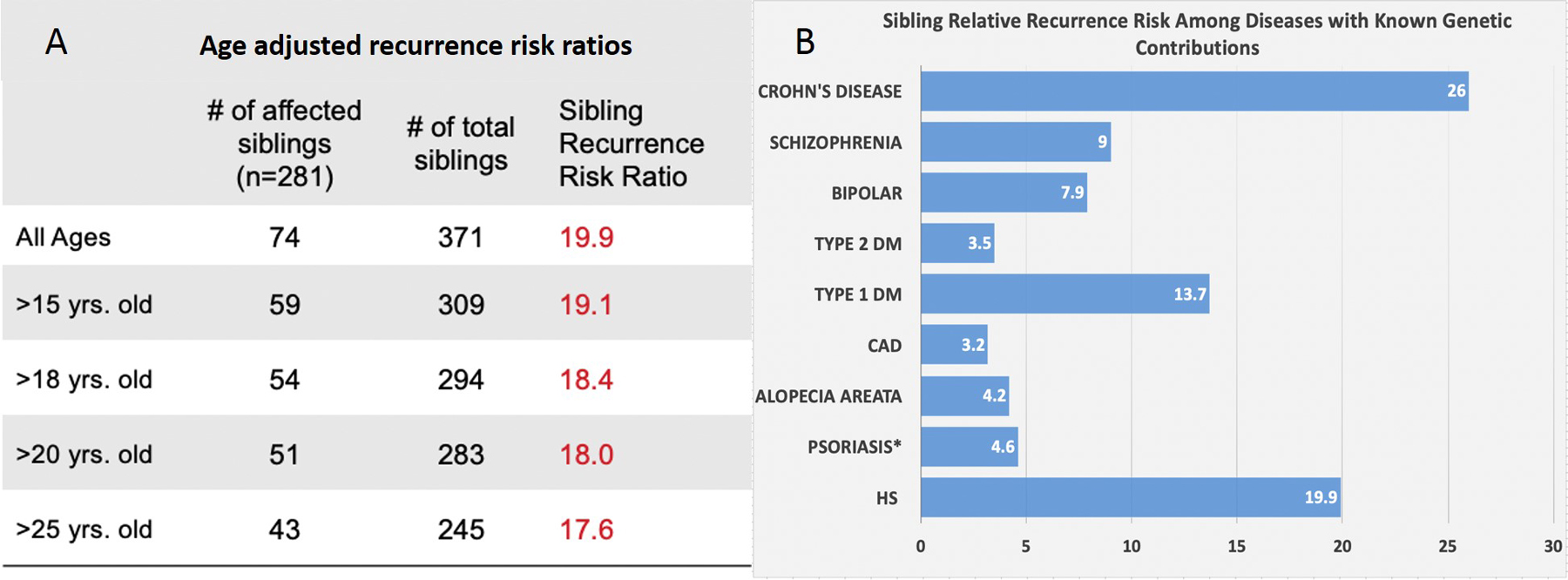
A. Age-adjusted recurrence risk ratios of hidradenitis suppurativa. B. Sibling Relative Recurrence Risk for HS compared to diseases with known genetic contributions7–9.
*λs in psoriasis of 1.6–4.6 was calculated based on a reported estimate 8–23% of first-degree relatives of psoriasis patients being affected and an assumed population prevalence of 5%8
In clinical practice, patients often inquire about disease risk for family members. Recent estimates suggest HS may affect 0.7–1% of the general population.1 Assuming 0.7–1% prevalence, λs of 19.9 indicates 13.9–19.9% of siblings and offspring of patients may be affected. While it is possible that environmental effects such as shared microbiome characteristics, smoking and obesity among related subjects could explain familial clustering, there is a notable lack of reports suggesting concordance or spread between unrelated household contacts, spouses, or sexual partners that share environments and close physical contact. The environment may influence susceptibility, but the more conspicuous clustering among related individuals suggests that genetic susceptibility plays a prominent role.
Our data are limited by lack of confirmed clinical diagnosis in relatives, but subjects reporting typical and recurrent HS lesions in family members is likely highly predictive.6 Only a single subject was adopted and lacked knowledge about family history, which did not significantly influence λs estimation. We may also underestimate familial risk if patients are unaware of their family members’ symptoms. We also did not analyze specific phenotypic or genetic factors that may influence familial risk due to the limited sample size that prevents reasonably-powered analyses of sub-categories. Despite these limitations, an estimate of general familial risk for this population is highly valuable for clinicians counseling patients on risk of disease development for children and other relatives, particularly since robust genetic data is currently unavailable.
Future research including genome-wide association studies are essential to further characterize the genetic, environmental and clinical factors that influence familial risk, and to better understand the genetic architecture of HS.
Acknowledgments
Funding Sources: Funding was provided from the National Center for Advancing Translational Sciences (NCATS), National Institutes of Health, through Grant Award Number UL1TR002489. The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH. Fellowship Funding for the project was provided by the Howard Holderness Foundation Medical Fellowship. Additional funding was provided by the University of North Carolina School of Medicine.
Footnotes
Conflicts of Interest: Christopher J. Sayed, MD has the following conflicts of interest to disclose:
Abbvie, Inc: speaker, co-investigator, and advisory board
Novartis: speaker, co-investigator
InflaRx: investigator, scientific advisory board
UCB: investigator
Chemocentryx: investigator
This study has been approved by the University of North Carolina School of Medicine Institutional Review Board, approval # 18-1209
References
- 1.Alikhan A, Sayed C, Alavi A, et al. North American Clinical Management Guidelines for Hidradenitis Suppurativa: a Publication from the United States and Canadian Hidradenitis Suppurativa Foundations. Part I: Diagnosis, Evaluation, and the use of Complementary and Procedural Management. Journal of the American Academy of Dermatology. 2019. [DOI] [PMC free article] [PubMed]
- 2.Schrader AM, Deckers IE, van der Zee HH, Boer J, Prens EP. Hidradenitis suppurativa: a retrospective study of 846 Dutch patients to identify factors associated with disease severity. Journal of the American Academy of Dermatology. 2014;71(3):460–467. [DOI] [PubMed] [Google Scholar]
- 3.Wray NR, Yang J, Goddard ME, Visscher PM. The genetic interpretation of area under the ROC curve in genomic profiling. PLoS Genet. 2010;6(2):e1000864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cordell HJ, Olson JM. Correcting for ascertainment bias of relative-risk estimates obtained using affected-sib-pair linkage data. Genet Epidemiol. 2000;18(4):307–321. [DOI] [PubMed] [Google Scholar]
- 5.van Straalen KR, Prens EP, Willemsen G, Boomsma DI, van der Zee HH. Contribution of Genetics to the Susceptibility to Hidradenitis Suppurativa in a Large, Cross-Sectional Dutch Twin Cohort. JAMA Dermatol. 2020. [DOI] [PMC free article] [PubMed]
- 6.Esmann S, Dufour DN, Jemec GB. Questionnaire-based diagnosis of hidradenitis suppurativa: specificity, sensitivity and positive predictive value of specific diagnostic questions. The British journal of dermatology. 2010;163(1):102–106. [DOI] [PubMed] [Google Scholar]
- 7.Risch N Linkage strategies for genetically complex traits. I. Multilocus models. Am J Hum Genet. 1990;46(2):222–228. [PMC free article] [PubMed] [Google Scholar]
- 8.Gupta R, Debbaneh MG, Liao W. Genetic Epidemiology of Psoriasis. Curr Dermatol Rep. 2014;3(1):61–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Petukhova L, Duvic M, Hordinsky M, et al. Genome-wide association study in alopecia areata implicates both innate and adaptive immunity. Nature. 2010;466(7302):113–117. [DOI] [PMC free article] [PubMed] [Google Scholar]


