Abstract
Yeast Apn2 is an AP endonuclease and DNA 3′-diesterase that belongs to the Exo III family with homology to the E. coli exonuclease III, Schizosaccharomyces pombe eth1, and human AP endonucleases APEX1 and APEX2. In the absence of Apn1, the major AP endonuclease in yeast, Apn2 can cleave the DNA backbone at an AP lesion initiating the base excision repair pathway. To study the role and relative contribution of Apn2, we took advantage of a reporter system that was previously used to delineate how uracil-derived AP sites are repaired. At this reporter, disruption of the Apn1-initiated base excision repair pathway led to a significant elevation of A:T to C:G transversions. Here we show that such highly elevated A:T to C:G transversion mutations associated with uracil residues in DNA are abolished when apn1Δ yeast cells are grown in glucose as the primary carbon source. We also show that the disruption of Apn2, either by the complete gene deletion or by the mutation of a catalytic residue, results in a similarly reduced rate of the uracil-associated mutations. Overall, our results indicate that Apn2 activity is regulated by the glucose repression pathway in yeast.
Keywords: DNA repair, AP endonuclease, Glucose repression, Uracil in DNA
Introduction
Apurinic/apyrimidic (AP) sites are one of the most common types of endogenous DNA damage and are generated by the cleavage of the N-glycosidic bond between the base and the deoxyribose moiety in DNA by DNA glycosylases (Boiteux and Jinks-Robertson 2013; Wallace 2014). Substrates of DNA N-glycosylases include oxidation-damaged bases, uracil, and spontaneous or enzymatic deamination products. After cleavage of the glycosidic bond, sugar phosphate backbone of the DNA strand adjacent to the AP site is cleaved by AP endonucleases to generate a DNA nick to initiate base excision repair (BER). Yeast contains two structurally unrelated proteins capable of incision at the AP sites, Apn1 and Apn2 (Daley et al. 2010). Apn1 belongs to the Endo IV family and comprises ~97% of both AP endonuclease and DNA 3′ diesterase activity in yeast cell extracts (Popoff et al. 1990). Apn2, which also contains AP endonuclease and DNA 3′-diesterase activity, belongs to the Exo III family with homology to the E. coli exonuclease III, Schizosaccharomyces pombe eth1, and human AP endonucleases APEX1 and APEX2 (Bennett 1999; Johnson et al. 1998; Unk et al. 2000). Endo IV family AP endonucleases are not found in most metazoan species including insects and mammals (Daley et al. 2010).
Uracil is one of the most frequent types of endogenous DNA modifications that are normally repaired by BER (Collura et al. 2012; Guillet and Boiteux 2003). One of major mechanisms underlying the appearance of uracil in DNA is through either enzymatically catalyzed or spontaneous deamination of cytosine bases (Chon et al. 2019). In various metazoan species, a family of enzymes named Alipoprotein B mRNA-editing enzyme catalytic polypeptide (APOBEC), which includes the lymphocyte-specific factor activation-induced deaminase (AID), have been shown to play a major role in deamination of cytosine in DNA resulting in U:G mis-pairs (Siriwardena et al. 2016). However, in yeast, which does not encode any APOBEC-like enzyme, deamination of cytosine does not appear to be a significant factor (Owiti et al. 2019). The spontaneous appearance of uracil in DNA can also be attributed to the eukaryotic replicative DNA polymerases that cannot distinguish between a uracil and a thymine base (Bessman et al. 1958; Warner et al. 1981). Depending on the cellular dUTP concentration, uracil is readily incorporated in place of thymine during replication and repair, resulting in a stable U:A base pair. An abasic site generated through the removal of uracil by a uracil DNA glycosylase (Ung1 in yeast; UDG in human) are either repaired in error-free manner by BER machinery or result in further mutagenic event mediated by the error-prone translesion synthesis DNA polymerases (Boiteux and Jinks-Robertson 2013). When left unrepaired, the nick left by AP endonucleases can result in DNA strand breaks leading to more detrimental genome instability events (Luke et al. 2010). Significantly, genome instability induced by uracil in DNA is the principal cause of the cytotoxicity of two major classes of anticancer drugs. The fluoropyrimidines, 5-fluorouracil (5-FU) and capecitabine are metabolized into 5-flurorode-oxyuridylate, which binds the essential enzyme thymidylate synthase (TS) to form an inhibitory complex (Longley et al. 2003). Antifolates (e.g. methotrexate) disrupt TS activity by inhibiting the enzyme dihydrofolate reductase (DHFR) and thereby interfering with the production of required co-factor tetrahydrofolate (Hagner and Joerger 2010). Because TS is the sole enzyme catalyzing the production of thymidylate (dTTP) from uracil (dUTP), in cells treated with fluoropyrimidines or antifolates, relatively lower dTTP concentration forces elevated incorporation of dUTP during replication and subsequent genome instability.
We have recently demonstrated a correlation between the elevated levels of uracil-derived mutations at highly transcribed genes and the higher densities of uracil residues in genomic DNA (Kim et al. 2007; Kim and Jinks-Robertson 2009, 2010; Owiti et al. 2018). To study how uracil in DNA are incorporated and repaired, we constructed several mutagenesis reporters designed to study the rate and the spectrum of mutations associated with active transcription in the yeast genome. One of these, the pTET-lys2-TAA reporter, contains an in-frame insertion of a TAA stop codon within the coding region of LYS2 gene. Thus, yeast cells harboring this allele are lysine auxotrophs (Lys−) and revert to lysine prototrophs (Lys+) through mutations at the TAA stop codon allowing translation read-through. It was shown that a great majority of Lys+ reversion mutations occurring at the pTET-lys2-TAA are due to the unrepaired AP lesions arising from the excision of uracil in DNA (Kim and Jinks-Robertson 2009, 2010). When the pTET-lys2-TAA was actively transcribed from the tetracycline-regulatable promoter, there was a sharp elevation in A to C or T to G transversion mutations. These mutations were dependent on the activity of translesion-synthesis (TLS) polymerases Pol zeta and Rev3, which together insert a C nucleotide opposite the AP sites (Kim et al. 2011b). And, these mutations were significantly reduced by the overexpression of dUTP pyrophosphatase Dut1 and mostly eliminated by the disruption of the uracil-specific DNA glycosylase Ung1 (Kim and Jinks-Robertson 2009, 2010). Because Dut1 performs an important function of regulating the cellular concentration of dUTP by converting dUTP to dUMP and inorganic pyrophosphate, its overexpression leads to lower dUTP pool available for incorporation into the genome (Guillet et al. 2006). And in the absence of Ung1, uracil remains in the genomic DNA without triggering base excision repair (BER) or mutagenic TLS. Overall, these data indicate that the transcription-associated A:T to C:G transversions at the pTET-lys2-TAA reporter result from the TLS-mediated bypass of AP lesions produced through uracil-excision by Ung1. The uracil-derived A:T to C:G mutations are highly elevated in strains deficient for Apn1 protein.
Here we used the previously established genetic tools to study the contribution of Apn2 AP endonuclease in the repair of uracil-derived AP lesions in yeast. Our data show that Apn2 enhances the mutations associated with uracil-derived AP sites in yeast in a manner dependent on the available carbon source. This surprising result calls for further study into how the activity of Apn2 is regulated by the presence of different carbon sources and by glucose in particular.
Materials and methods
Yeast strains and plasmids
Yeast strains used for the mutation and recombination assays were derived from YPH45 (MATa, ura3-52 ade2-101 trp1Δ1). Construction of strains containing the his4Δ::pTET-lys2-TAA allele was previously described (Kim and Jinks-Robertson 2010). See the strain list in Table S1 for more information. Gene deletions were carried out through one-step allele replacement by amplification of loxP-flanked marker cassettes (Gueldener et al. 2002). The entire APN2 ORF containing the E59A mutation was synthesized by the GeneArt Gene Synthesis service (ThermoFisher Scientific) and cloned into a pGPD2 vector (Addgene #43,972) to construct pNK231. YNK650 containing apn2 E59A allele was constructed through pop-in/pop-out method using BseRI-digested pNK231. For overexpression of APN2 gene, the APN2 ORF was amplified from the yeast genomic DNA and cloned between XbaI and XhoI sites of pGPD2 vector to construct pNK229.
Mutation/recombination rates and spectra
Mutation and recombination rates were determined using the method of the median, and 95% confidence intervals were calculated as previously described (Spell and Jinks-Robertson 2004). Two rates are considered significantly different when the confidence intervals do not overlap. Each rate was based on data obtained from 12–48 independent cultures and at least two independently derived isolates. 1 mL yeast extract-peptone (YEP) medium supplemented with 2% glycerol and 2% ethanol (YEPGE) or with 2% dextrose (YEPD) was inoculated with 250,000 cells from an overnight culture grown in the same medium. Following growth at 30 °C for 2 days in YEPD or for 3 days in YEPGE, cells were washed with water and the appropriate dilutions were plated either on synthetic, lysine-deficient medium containing 2% glucose (SCD-Lys) to select for Lys+ revertants or on SCD-Leu medium to determine the total number of cells in each culture. CAN1 forward mutation rates were determined by plating cells on SCD-Arg medium supplemented with 60 μg/mL l-canavanine sulfate (SCD-Arg+ Can; Sigma). For complementation studies in Fig. 2B, YNK103 (apn1Δ:: loxP apn2Δ::loxP-TRP1-loxP) was transformed with either the vector (pGPD2; Addgene #43,972) or pNK229 (pGPD-APN2) and cultured in synthetic, uracil-deficient medium (SC-URA) containing either 2% glucose or 2% glycerol+ 2% ethanol at 30 °C for 3 days. Then cells were washed with water and the appropriate dilutions were plated either on synthetic, lysine, uracil-deficient medium containing 2% glucose (SCD-Lys-URA) to select for Lys+ revertants or on SCD-URA medium to determine the total number of cells in each culture.
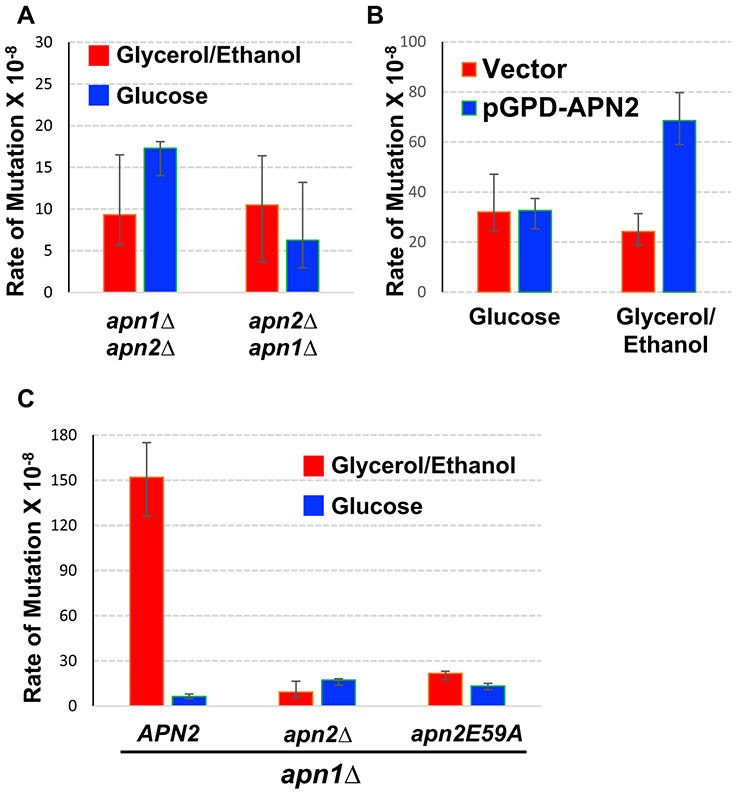
Fig. 2.
The requirement of Apn2 catalytic activity for the elevated Lys+ mutations in glycerol/ethanol. In all graphs, error bars indicate 95% confidence intervals. Two rates are considered significantly different when the error bars do not overlap. a Rates of Lys+ mutations at pTET-lys2-TAA in either apn1Δ apn2Δ (YNK103; see Table S1) or apn2Δ apn1Δ (YNK219; see Table S1). Rates were measured by fluctuation analyses in cells cultured in YEP media supplemented with either 2% glycerol+ 2% ethanol (YEPGE) or 2% glucose (YEPD). b Rates of Lys+ mutations at pTET-lys2-TAA in YNK103 (apn1Δ::loxP apn2Δ::loxP-TRP1-loxP) transformed with either the vector (pGPD2; Addgene #43,972) or pNK229 (pGPD-APN2). Rates were measured by fluctuation analyses in cells cultured in synthetic, uracil-deficient medium (SC-URA) containing either 2% glucose or 2% glycerol+ 2% ethanol as indicated. c Rates of Lys+ mutations at pTET-lys2-TAA in YNK98 (apn1Δ APN2), YNK103 (apn1Δ apn2Δ), or YNK658 (apn1Δ apn2E59A). Rates were measured by fluctuation analyses in cells cultured in YEP media supplemented with either 2% Glycerol+ 2% ethanol (YEPGE) or 2% glucose (YEPD)
To determine mutation spectra, individual colonies were used to inoculate 0.3 ml liquid YEPGE or YEPD media. After 2 or 3 days of growth at 30 °C, an appropriate fraction of each culture was plated on SCD-Lys. A single Lys+ revertant from each culture was purified on YEPD plates, and genomic DNA was prepared using a 96-well format in microtiter plates. The lys2-TAA reversion window was amplified using primers 5′-AGCTCGATGTGCCTCATGATAG-3′ and 5′-CATCACACATACCATCAAATCC-3′ and the PCR product was sent to Eurofins Genomics (Louisville, KY) for sequencing using primer 5′-TAGAGTAACCGGTGACGATG-3′. The rates of A > C and T>G were calculated by multiplying the proportion of the events by the total Lys+ mutation rate.
qRT-PCR
Total RNA was extracted using the standard hot acid phenol method and treated with DNase 1 (New England Biolabs). qRT-PCR was performed using amfiRivert cDNA synthesis Platinum Master Mix from Gendepot, Sensi-FAST SYBR No-ROX kit from Bioline, and a Biorad CFX Connect instrument. The qRT-PCR conditions were as follows: 45 °C for 10 min and 95 °C for 2 min followed by 40 cycles of 95 °C for 5 s, 60 °C for 10 s and 72 °C for 5 s. ALG9 gene was chosen as the reference gene (Teste et al. 2009). The primers used for amplification were APN2F: 5′-TTCGAATTATGGCTCACGGA-3′, APN2R: 5′-ACCAGGTTCAATTCTGTCGT-3′, ALG9F: 5′-CACGGATAGTGGCTTTGGTGAACAATTAC-3′, and ALG9R: 5′-TATGATTATCTGGCAGCAGGAAAGAACTTGGG-3′. Relative RNA levels were determined by ΔΔCq analysis as previously described (Livak and Schmittgen 2001).
Western blot analysis
Yeast whole cell extracts (WCE) prepared using NaOH lysis method were separated on a 4–20% polyacrylamide gel, and transferred using a Semi-Dry Trans-Blot Cell (BioRad). After blocking with 5% dry milk dissolved in TBS with 0.1% Tween20, HRP-conjugated anti-FLAG (Sigma, A8592) and HRP-conjugated anti-GAPDH (Invitrogen, MA5-15,738) antibodies were used for detection. The blot was developed using West-Q Pico Dura ECL Solution (GenDepot) and the ChemiDoc MP Imaging system (BioRad).
Identification of putative SUMO- or Ub-modification sites
For the identification of putative sumoylation sites and SUMO-interaction sites within Apn2 protein, we used the GPS-SUMO Webserver (Ren et al. 2009; Zhao et al. 2014). UbPred was used for the search of putative ubiquitination sites (Radivojac et al. 2010).
Results
Uracil-derived mutations at a highly transcribed gene are dependent on the growth conditions
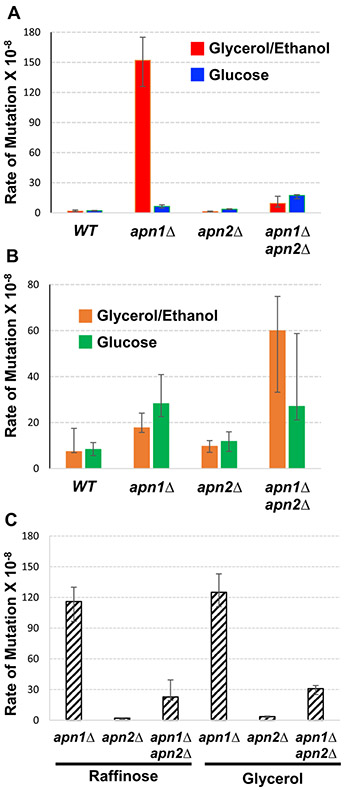
In previously reported results, the mutation rate and spectra at the pTET-lys2-TAA reporter described in “Introduction” were determined with the yeast cells cultured in rich media containing yeast extract and peptone with the addition of 2% glycerol and 2% ethanol as the primary carbon source (YEPGE). Here, we repeated the experiments by culturing the yeast cells with the same rich media containing 2% dextrose (YEPD). Surprisingly, we observed that, in an apn1Δ background, the rate of mutations at the pTET-lys2-TAA reporter when cultured in YEPD was ~ 24-fold lower than with YEPGE (Fig. 1a).
Fig. 1.
The rates of Lys+ and CanR mutations in various carbon sources. In all graphs, error bars indicate 95% confidence intervals. Two rates are considered significantly different when the error bars do not overlap. a Rates of Lys+ mutations at pTET-lys2-TAA in the indicated strain backgrounds. Rates were measured by fluctuation analyses in cells cultured in YEP media supplemented with either 2% Glycerol+ 2% ethanol (YEPGE) or 2% glucose (YEPD). b Rates of canavanine-resistant mutations at pTET-lys2-TAA in the indicated strain backgrounds. Rates were measured by fluctuation analyses in cells cultured in YEP media supplemented with either 2% glycerol+ 2% ethanol (YEPGE) or 2% glucose (YEPD). c Rates of Lys+ mutations at pTET-lys2-TAA in the indicated strain backgrounds. Rates were measured by fluctuation analyses in cells cultured in YEP media supplemented with either 2% glycerol or 2% raffinose
In a WT background, the deletion of APN2 gene, which encodes an Exo III family of AP endonuclease, did not result in a significant increase in the mutation at the pTET-lys2-TAA reporter either in YEPGE or YEPD media. However, in an apn1Δ background, the highly elevated Lys+ reversion mutation rate at the pTET-lys2-TAA reporter in cells cultured in YEPGE was reduced by a significant 16-fold reduction upon the deletion of APN2 (Fig. 1a). In addition to the reduction in the overall mutation rate, there was a significant shift in the mutation spectrum upon the deletion of APN2 in an apn1Δ background. The proportion of A:T to C:G transversions, which comprise > 90% of the mutations in apn1Δ strain, decreased to ~60% of total mutations in the apn1Δ apn2Δ strain (Table 1). Upon the deletion of APN2, the rates of T > G and A > C mutations in apn1Δ cells grown in YEPGE were reduced by 40- and 23-fold, respectively. Interestingly, this effect of APN2 deletion on the Lys+ reversion mutation rate at the pTET-lys2-TAA reporter was entirely dependent on the carbon source available. That is, in YEPD media, the rate of Lys+ reversion mutation was ~ threefold higher in an apn1Δ apn2Δ background than in an apn1Δ background (Fig. 1a). Also in YEPD media, the deletion of APN2 gene had minimal effect on the mutation spectra in apn1Δ cells with the rates of T > G and A > C mutations both increasing by less than twofold (Table 1).
Table 1.
The mutation spectra
| Glycerol/ethanol | Total rate × 10−8 (95% CI) |
Mutation type (#/total) |
Mutation rate (×10−8) |
||
|---|---|---|---|---|---|
| T > G | A > C | T > G | A > C | ||
| WTa | 1.58 (1.18–2.88) | 6/84 | 25/84 | 0.11 | 0.46 |
| apn1a | 152 (126–175) | 10/88 | 70/88 | 17.2 | 120.9 |
| apn1 apn2 | 9.33 (5.73–16.5) | 3/66 | 37/66 | 0.42 | 5.23 |
| Glucose | T > G | A > C | T > G | A > C | |
| WT | 2.21 (1.75–2.42) | 4/55 | 14/55 | 0.16 | 0.56 |
| apn1 | 6.34 (4.87–8.06) | 12/59 | 38/59 | 1.29 | 4.08 |
| apn1 apn2 | 17.3 (14–18.1) | 6/46 | 21/46 | 2.26 | 7.90 |
95% CI 95% confidence intervals
Data previously published in (Kim et al. 2011b)
Mutations at the CAN1 gene that disrupt the function of the encoded arginine transporter protein result in the resistance to the toxic arginine analog canavanine (Whelan et al. 1979). As previously reported (Hanna et al. 2004; Johnson et al. 1998), canavanine-resistance conferring mutations at the CAN1 gene (CanR) are not associated with uracil-derived AP sites. The majority of the base substitutions at CAN1 genes consists of either C:G to A:T transversions or C:G to T:A transitions. Also as previously reported, we found that the rate of CanR mutation was not affected greatly by the single deletion of APN1 or APN2 but synergistically elevated when both are knocked out (Fig. 1b). Importantly, no significant difference in the rate of CanR mutation was observed whether the carbon source provided was glycerol/ethanol or glucose. Thus, the origin of mutation at CAN1 gene is distinctly different from the uracil-derived mutation occurring at the pTET-lys2-TAA reporter under high transcription conditions. Furthermore, while the deletion of UNG1 gene encoding the uracil DNA glycosylase severely reduces the mutations at the pTET-lys2-TAA reporter, the same gene deletion was reported to elevate the rate of spontaneous mutation at CAN1 gene (Guillet and Boiteux 2003). This further indicates that the elevated mutation rate when cultured in glycerol/ethanol carbon is specific to uracil-derived AP sites.
Alternative carbon sources do not diminish the mutations at the pTET-lys2-TAA reporter
The striking difference in the rate of mutation at the pTET-lys2-TAA reporter in YEPD vs. YEPGE media led us to investigate the effect of alternate carbon sources. Raffinose is a trisaccharide made up of galactose, glucose, and fructose (Paulo et al. 2015). Raffinose can support growth by fermentation like glucose but favors mitochondrial respiration like glycerol/ethanol (Guaragnella et al. 2013). Global proteomic studies showed that two carbon sources, glucose and raffinose, result in distinctively different protein profiles (Paulo et al. 2015). For cells growing in rich media supplemented with 2% raffinose, the rate of mutation occurring at the pTET-lys2-TAA reporter in apn1Δ background was statistically the same as that in cells growing in YEPGE (Fig. 1c). And similar to the cells in YEPGE, the mutation rate was significantly decreased when APN2 was further deleted. We also tested whether the ethanol content in the YEPGE media is the decisive factor by repeating the mutation analysis in media containing only 2% glycerol. The rates of mutations at the pTET-lys2-TAA reporter were not significantly different from YEPGE in all three backgrounds tested: apn1Δ, apn2Δ, and apn1Δ apn2Δ.
The elevation of uracil-derived A:T to C:G transversions in yeast cells grown in YEPGE requires the catalytic activity of Apn2
To first confirm that Apn2 is necessary for the highly elevated mutations at the pTET-lys2-TAA reporter in apn1Δ background, we independently reconstructed the double knock-out strain by deleting APN1 gene from the apn2Δ strain. The mutation rates at the pTET-lys2-TAA reporter in this new strain (apn2Δ apn1Δ in Fig. 2a) were not significantly different from those in the original double knock-out strain (apn1Δ apn2Δ in Fig. 2a), either in YEPD or YEPGE. Similar to the mutation rate in apn1Δ apn2Δ cells, the rate of the pTET-lys2-TAA mutation in apn2Δ apn1Δ cells grown in YEPGE was ~ 14-fold lower and ~ 9-fold higher when compared to apn1Δ and apn2Δ single deletion strains, respectively (Fig. 2A). Next, we constructed a plasmid transcribing the APN2 gene from the highly active TDH3 gene promoter which is referred to as GPD promoter (Crook et al. 2011). Overexpression of APN2 from this plasmid in apn1Δ apn2Δ strain resulted in a statistically significant, ~ threefold elevation in the mutations at the pTET-lys2-TAA reporter in YEPGE but not in YPD media. However, the relative difference in the rate of mutation between apn1Δ apn2Δ strain with the control vector plasmid and the pGPD-APN2 plasmid is substantially less than the difference between apn1Δ and apn1Δ apn2Δ. This difference is likely an artifact of the overexpression system.
To determine whether the effect on the mutation rates at the pTET-lys2-TAA reporter requires the catalytic activity of Apn2 protein, we introduced an allelic mutation to change the catalytic glutamate residue E59 of Apn2 to alanine (e.g. apn2 E59A allele) (Li et al. 2019). Li et al. has demonstrated that although Apn2 E59A mutant is stably expressed and can be purified, it does not have the nucleolytic activity. In the strain expressing the catalytically inactive Apn2 E59A mutant protein in the apn1Δ background (apn1Δ apn2 E59A), the mutation rate at the pTET-lys2-TAA reporter was significantly lower than that in the apn1Δ strain and comparable to that in the apn1Δ apn2Δ strain (Fig. 2B), indicating that the catalytic activity of Apn2 endonuclease is necessary for the elevated uracil-derived A:T to C:G transversions in yeast cells grown in YEPGE. E59A mutation, however, did not affect the mutation rate in YEPD media.
Since Apn2 protein is required for the elevated mutations at the pTET-lys2-TAA reporter, the difference in the rate of mutations between YEPD and YEPGE could be due to the level of APN2 transcription in cells under these different growth conditions. Using qRT-PCR method, we quantified the level of APN2 transcripts in cell grown in YEPD or YEPGE. As shown in Table 2, either in WT or apn1Δ background, the difference in the APN2 gene expression level was not very significant. Although there was a moderately lower Apn2 transcript level in YEPD compared to YEPGE in WT background, there was no significant difference in apn1Δ background. This is consistent with the previously reported proteomics study, which showed the difference in the level of Apn2 protein in two different carbon sources was minimal with the level in raffinose being about 73% of the level in glucose (Paulo et al. 2015).
Table 2.
The rate of APN2 gene expression
| Strain background | Media | Expression fold difference relative to WT cells in YEPGE |
Expression fold difference relative to the same background cell in YEPGE |
|---|---|---|---|
| WT | YEPGE | – | – |
| WT | YPD | 0.58–0.64 | 0.58–0.64 |
| apn1Δ | YEPGE | 1.16–1.54 | – |
| apn1Δ | YPD | 1.21–1.72 | 0.91–1.29 |
| apn1Δ rfx1Δ | YEPGE | 4.13–10.75 | – |
| apn1Δ rfx1Δ | YPD | 4.73–8.72 | 0.71–1.31 |
Stress response genes YAP1, DUN1, and MSN2/MSN4 are not involved in the elevated uracil-derived mutations mediated by Apn2 in YEPGE in an apn1Δ background
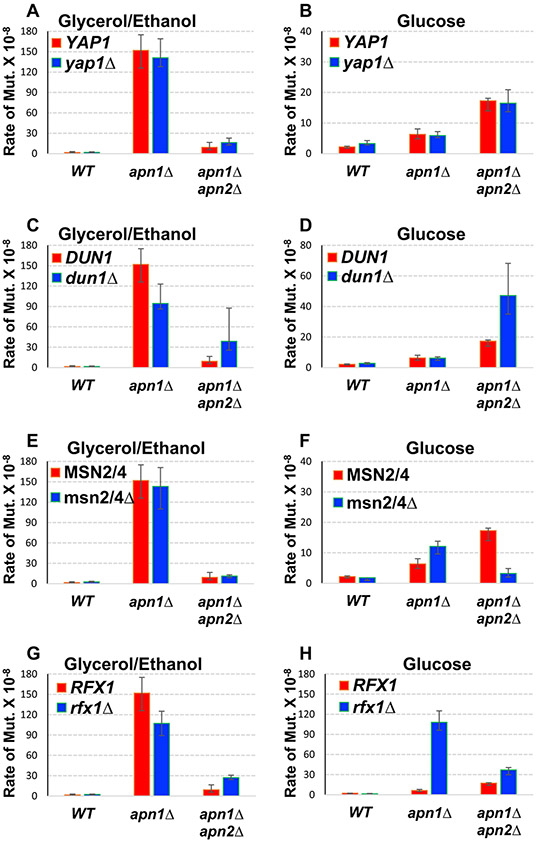
The expression and activity of many DNA repair proteins are regulated by stress response pathways. We speculated whether a stress response pathway could alter the activity of Apn2, thereby resulting in the highly elevated mutations during growth in YEPGE. We generated yeast strains defective in the stress response genes YAP1, DUN1, or MSN2/MSN4. Yap1 is a basic leucine zipper transcription factor required for oxidative stress response (Temple et al. 2005) with greater than 500 target genes (Cohen et al. 2002; Venters et al. 2011). DUN1 encodes a serine/threonine kinase that regulates the DNA damage response (Zhou and Elledge 1993). One of its best characterized targets is the transcriptional repressor Sml1, which in turn regulates the dNTP pools by transcriptional regulation of the ribonucleotide reductase genes. Msn2 and its paralog Msn4 are transcription activator proteins that bind to the stress response elements or STRE (AGGGG or CCCCT) and regulate various stress responses including DNA replication stress and nutrient starvation (Martinez-Pastor et al. 1996; Schmitt and McEntee 1996). For the rate of mutations at the pTET-lys2-TAA when in YEPGE, there were no significant changes observed upon the deletion of YAP1, DUN1, or MSN2/4 (Fig. 3a, c, e, g), indicating that none of these three stress response pathways affect the activity of Apn2 in YEPGE. When in YEPD, the rates of mutations at the pTET-lys2-TAA were slightly elevated by the deletion of DUN1 and slightly decreased by the deletion of MSN2/4 for apn1Δ apn2Δ background cells (Fig. 3d, f).
Fig. 3.
Stress response pathways and the elevated Lys+ mutations in glycerol/ethanol. In all graphs, error bars indicate 95% confidence intervals. Two rates are considered significantly different when the error bars do not overlap. a–h Rates of Lys+ mutations at pTET-lys2-TAA in the indicated strain backgrounds. Rates were measured by fluctuation analyses in cells cultured in YEP media supplemented with either 2% Glycerol + 2% ethanol (YEPGE) or 2% glucose (YEPD) as indicated above each graph. For more information about the strains used, see Table S1
The mutation rate at the pTET-lys2-TAA reporter becomes elevated upon RFX1 deletion in YEPD
Rfx1 is another DNA binding transcription factor involved in the DNA damage response. It suppresses the transcription of multiple DNA-damage inducible genes by recruiting general repressor complex Ssn6/Tup1 to the gene promoters (Zhang and Reese 2005). When we deleted RFX1 gene, the rate of mutations at the pTET-lys2-TAA reporter was not significantly changed in WT, apn1Δ, or apn1Δ apn2Δ backgrounds when in YEPGE (Fig. 3g). However, in YEPD, the rate of mutations at the pTET-lys2-TAA reporter was ~ 17-fold higher in the apn1Δ rfx1Δ strain compared to the apn1Δ strain (Fig. 3h). For WT or apn1Δ apn2Δ background, the deletion of RFX1 gene did not have any effect on the mutation rates at the pTET-lys2-TAA reporter. Rfx1 was reported to regulate the level of APN2 expression during heat-response in a high-throughput analysis (Venters et al. 2011) leading to the speculation that the elevated mutation rate we observed could be due to the alleviation of the transcriptional suppression of APN2 gene by Rfx1.
Using the qRT-PCR approach, we confirmed that the APN2 expression level is elevated by about four to tenfold in absence of Rfx1 in cells cultured in YEPD (Table 2). However, the APN2 transcription levels in rfx1Δ cells were similarly elevated when cultured in YEPGE. We also checked the level of Apn2 protein in WT, apn1Δ, and apn1Δ rfx1Δ backgrounds by western blot analysis. In three genetic backgrounds, the levels of Apn2 protein were moderately and similarly elevated when the yeast cells are grown in YEPGE compared to YEPD (Fig. 4). However, the deletion of RFX1 gene did not alter the level of Apn2 protein in either media. Overall, this data indicates that, when growing in glucose-supplemented media, Rfx1 suppresses the activity of Apn2 required for the elevated Lys+ mutations at the pTET-lys2-TAA reporter but not through the direct regulation of the level of APN2 gene transcription or translation. We speculate that Apn2 activity in glucose-supplemented media is indirectly affected by another factor under the regulation of Rfx1.
Fig. 4.
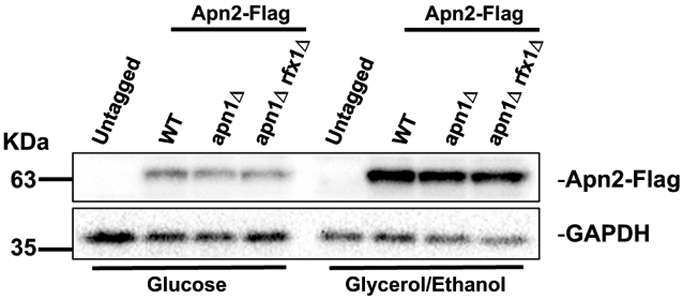
Apn2 protein level in WT, apn1Δ, and apn1Δ rfx1Δ backgrounds. Western blot analysis of yeast whole cell extracts in the indicated strain backgrounds. C-terminally flag-tagged Apn2 was detected by anti-flag antibody. Anti-GAPDH was used as loading control. The locations of relevant molecular weight marker bands are indicated at the right of western blot
Discussion
Previously, we reported that there is a transcription-associated elevation of A:T to C:G mutations at the pTET-lys2-TAA reporter located on the yeast chromosome III (Kim and Jinks-Robertson 2010; Kim et al. 2011b). Multiple lines of evidence indicate that these transversion mutations are largely due to AP lesions generated by the excision of uracil base (Kim and Jinks-Robertson 2009, 2010; Owiti et al. 2018). Briefly, the activity of uracil DNA glycosylase Ung1, which converts uracil in DNA into AP sites, is required for these mutations. Additionally, the level of cellular dUTP is an important determinant in these specific mutations. That is, overexpression of the dUTP-degrading Dut1 enzyme results in a significant decrease in A:T to C:G mutations at the pTET-lys2-TAA reporter. These mutations originate from uracil bases incorporated into DNA in place of thymine, which are then excised by Ung1 to generate AP sites. Because the specific A:T to C:G mutations occur when translesion DNA polymerases Pol zeta and Rev1 insert C nucleotides across from unrepaired AP sites during replication, they are highly elevated when BER pathway is disrupted. In yeast, when the major AP endonuclease Apn1 is absent, the repair of AP sites can also be initiated by the AP lyases Ntg1 and Ntg2 (Meadows et al. 2003). It was shown previously that Ntg1/Ntg2-associated BER is preferentially involved in repair of AP sites located on the non-transcribed DNA strand (Kim and Jinks-Robertson 2010). Those AP sites located on the transcribed DNA strand can alternatively be removed by the transcription-coupled nucleotide excision repair pathway (Kim and Jinks-Robertson 2010; Owiti et al. 2017). We have previously reported that A:T to C:G mutations at the pTET-lys2-TAA reporter are significantly elevated in apn1Δ background and further elevated when the Ntg1/Ntg2 AP lyases are absent or the alternative repair pathway of transcription-coupled nucleotide excision repair (TC-NER) is impaired.
Here we examined whether the other yeast AP endonuclease Apn2 is involved in the repair of AP sites generated by excision of uracil in DNA. Although the deletion of APN2 gene did not affect the mutations occurring at the pTET-lys2-TAA reporter in WT background, a severe decrease in the mutation rate was observed in apn1Δ background (Fig. 1a). Our data also shows that the catalytic activity of yeast Apn2 is required for the highly elevated A:T to C:G transversions at the actively transcribed pTET-lys2-TAA reporter (Fig. 2c). Apn1 and Apn2 can each initiate BER by cleavage of the phosphodiester bond adjacent to an AP lesion. Such a redundant function is shown in the rate of CanR mutations as in Fig. 1b, which are elevated in apn1Δ strain and further elevated in apn1Δ apn2Δ double mutant strain. On the other hand, the rate of uracil-derived A:T to G:C transversions at the pTET-lys2-TAA reporter are significant elevated in apn1Δ but reduced in apn1Δ apn2Δ strain (Fig. 1a), indicating the Apn2 activity involved must be a non-canonical function other than the cleavage of the DNA backbone. Recently, there have been reports of such non-canonical function of Apn2. In the absence of RNase H2-mediated ribonucleotide excision repair (RER), Top1-dependent cleavage at ribonucleotide embedded in DNA results in 2′,3′-cyclic phosphate terminated DNA ends, which leads to 2–5 bp deletion mutations at repetitive sequences (Kim et al. 2011a). Apn2 was recently shown to process these blocked 3′ ends produced during the Top1-mediated cleavage of ribonucleotides in DNA (Li et al. 2019). The ribonucleotide-associated 2–5 bp deletions are elevated by > 100-fold in absence of Apn2 but not in absence of Apn1. The same catalytic mutation (i.e. E59A) that abolishes A:T to C:G mutations at the pTET-lys2-TAA reporter here was shown to also abolish the ribonucleotide-associated small deletions in this study.
Another key finding reported here is that the highly elevated Apn2-dependent A:T to C:G transversions are not observed when the yeast cells are cultured with glucose as the primary carbon source (Fig. 1a). While dextrose is present at a sufficient concentration, yeast cells can sustain energy production and growth by fermentation even in the presence of oxygen (Pfeiffer and Morley 2014), while gene involved in the use of other carbon sources are generally suppressed in a process referred to as glucose repression (Kayikci and Nielsen 2015). Deactivation of glucose repression is required for the utilization of non-fermentable carbon sources such as glycerol (Schuller 2003). When growing in raffinose as the major carbon source, ATP production in yeast cells is primarily carried out through mitochondrial respiration and not through fermentation with no evidence of glucose repression (Guaragnella et al. 2013; Randez-Gil et al. 1998). We observed the high A:T to C:G conversion rate in yeast cells growing in media supplemented with 2% glycerol+ 2% ethanol, 2% glycerol only, or 2% raffinose only but not with 2% glucose only (Fig. 1a, c).
The suppression of Apn2 activity in glucose is not at the level of transcriptional regulation. APN2 transcripts in both WT and apn1Δ cells as measured by qRT-PCR are present in similar level whether glucose (YEPD) or glycerol/ethanol (YEPGE) is the major carbon source (Table 2) and thus cannot sufficiently account for the higher mutation rate observed in YEPGE. On the other hand, the higher Apn2 protein levels in yeast cells grown in YEPGE media compared to YEPD (Fig. 4) could partially account for the higher rate of Apn2-dependent mutations at the pTET-lys2-TAA reporter, implicating translational and/or post-translational regulation.
Either in glucose or glycerol/ethanol, the rate of Lys+ mutations at the pTET-lys2-TAA reporter was not significantly affected by disruption of the Yap1, Dun1, or Msn2/4 stress response factors (Fig. 3a-f). When RFX1 gene was deleted in apn1Δ background, the rate of Lys+ mutations at the pTET-lys2-TAA reporter in glucose was highly elevated to be similar to the rate of mutations in glycerol/ethanol (Fig. 3g, h). However, APN2 gene expression is elevated by four- to tenfold both in YEPD and YEPGE by RFX1 gene deletion (Table 2) even though there was no significant increase in the rate of mutation in an apn1Δ rfx1Δ strain compared to an apn1Δ strain in glycerol/ethanol (Fig. 3g). Furthermore, according to western blot analysis, the level of Apn2 protein is not affected by the deletion of RFX1 gene in YEPD (Fig. 4), even though the rate of mutations at the pTET-lys2-TAA reporter in YEPD was ~ 17-fold higher in apn1Δ rfx1Δ compared to apn1Δ. These results indicate that transcriptional regulation of APN2 does not sufficiently account for its non-canonical activity leading to higher uracil-associated mutations.
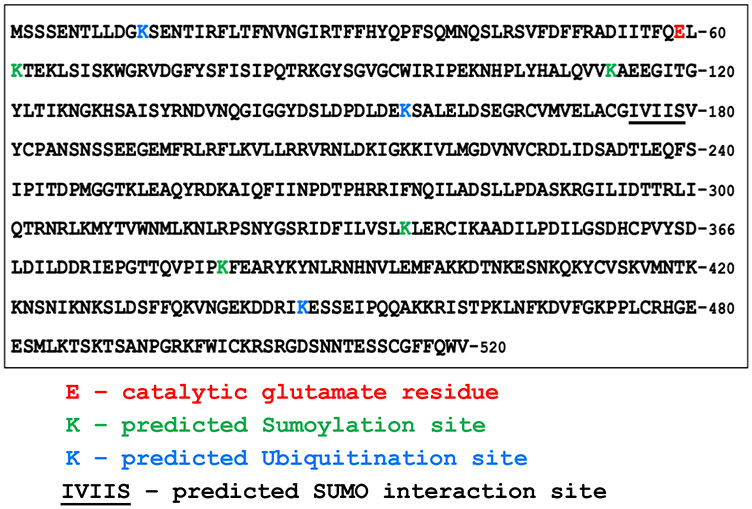
One possible way by which Apn2 activity can be regulated is through posttranslation modification. Human AP endonuclease APEX1, which is a class II endonuclease like yeast Apn2, is reported to be regulated by various types of posttranslational modifications (reviewed in (Busso et al. 2010)). For example, the cellular localization of hAPE1 is regulated by ubiquitination of the protein (Busso et al. 2009, 2010). Several putative ubiquitination sites are present in yeast Apn2 according to UbPred web tool (Radivojac et al. 2010) (Fig. 5). K12, K155, and K446 were identified as putative ubiquitination with “medium confidence” (i.e. Score range of 0.69 to 0.84, sensitivity of 0.346, and specificity of 0.950). Two additional sites identified as “low confidence” are not indicated in Fig. 5. In addition, using the GPS-SUMO webserver, four putative sumoylation sites and one sumo-interaction site were identified (Fig. 5 and Table S6). We used DISULFIND tool at Machine Learning Neural Networks Group webserver (Ceroni et al. 2006) to search for possible disulfide bonding sites and did not find any sites with positive disulfide bonding state.
Fig. 5.
Putative post-translational modification sites of Apn2. The protein sequence of S. cerevisiae Apn2 is shown in the box with the catalytic glutamate 59, putative Sumoylation sites (GPS-SUMO Webserver: http://sumosp.biocuckoo.org), and putative ubiquitination sites (UBpred: http://www.ubpred.org) indicated in red, green, and blue, respectively. Underlined residues (IVIIS) are predicted to be SUMO-interaction site (GPS-SUMO webserver: http://sumosp.biocuckoo.org)
In summary, we report here two novel findings regarding the repair of uracil-derived AP sites in the yeast genome. First, we show that, in absence of the major AP endonuclease Apn1, the catalytic activity of Apn2 contributes to elevation of uracil-derived A:T to C:G transversion mutations at an actively transcribed mutation reporter. We also show that such activity of Apn2 is suppressed when yeast cells are grown in glucose, leading to the speculation that glucose repression regulates Apn2 activity. Considering the significance of uracil as one of most prevalent types of endogenous DNA lesions, these novel findings call for future studies to determine the precise mechanism underlying such non-canonical activity of Apn2, as well as the mechanism of the glucose-dependent suppression of such activity.
Supplementary Material
Acknowledgements
We thank the members of Kim lab for discussion and critical reading of the manuscript.
Funding This work was supported by grants from National Institutes of Health R01 GM116007 and AU1875 from Welch Foundation to N.K. A.B. was supported by R01 GM116007-S1.
Footnotes
Conflict of interest The authors declare that there are no conflicts of interests.
Human/animal rights This study did not involve any human or animal subjects and followed all ethical standards of research.
Publisher’s Note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information The online version contains supplementary material available at https://doi.org/10.1007/s00294-020-01141-4.
Data availability
Yeast strains will be made available upon request.
References
- Bennett RA (1999) The Saccharomyces cerevisiae ETH1 gene, an inducible homolog of exonuclease III that provides resistance to DNA-damaging agents and limits spontaneous mutagenesis. Mol Cell Biol 19:1800–1809. 10.1128/mcb.19.3.1800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bessman MJ, Lehman IR, Adler J, Zimmerman SB, Simms ES, Kornberg A (1958) Enzymatic synthesis of deoxyribonucleic acid. Iii. The incorporation of pyrimidine and purine analogues into deoxyribonucleic acid. Proc Natl Acad Sci USA 44:633–640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boiteux S, Jinks-Robertson S (2013) DNA repair mechanisms and the bypass of DNA damage in Saccharomyces cerevisiae. Genetics 193:1025–1064. 10.1534/genetics.112.145219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busso CS, Iwakuma T, Izumi T (2009) Ubiquitination of mammalian AP endonuclease (APE1) regulated by the p53-MDM2 signaling pathway. Oncogene 28:1616–1625. 10.1038/onc.2009.5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busso CS, Lake MW, Izumi T (2010) Posttranslational modification of mammalian AP endonuclease (APE1). Cell Mol Life Sci 67:3609–3620. 10.1007/s00018-010-0487-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ceroni A, Passerini A, Vullo A, Frasconi P (2006) DISULFIND: a disulfide bonding state and cysteine connectivity prediction server. Nucleic Acids Res 34:W177–181. 10.1093/nar/gk1266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chon J, Field MS, Stover PJ (2019) Deoxyuracil in DNA and disease: genomic signal or managed situation? DNA Repair 77:36–44. 10.1016/j.dnarep.2019.02.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen BA, Pilpel Y, Mitra RD, Church GM (2002) Discrimination between paralogs using microarray analysis: application to the Yap1p and Yap2p transcriptional networks. Mol Biol Cell 13:1608–1614. 10.1091/mbc.01-10-0472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collura A, Kemp PA, Boiteux S (2012) Abasic sites linked to dUTP incorporation in DNA are a major cause of spontaneous mutations in absence of base excision repair and Rad17-Mec3-Ddc1 (9–1–1) DNA damage checkpoint clamp in Saccharomyces cerevisiae. DNA Repair 11:294–303. 10.1016/j.dnarep.2011.12.004 [DOI] [PubMed] [Google Scholar]
- Crook NC, Freeman ES, Alper HS (2011) Re-engineering multicloning sites for function and convenience. Nucleic Acids Res 39:e92. 10.1093/nar/gkr346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daley JM, Zakaria C, Ramotar D (2010) The endonuclease IV family of apurinic/apyrimidinic endonucleases. Mutat Res 705:217–227. 10.1016/j.mrrev.2010.07.003 [DOI] [PubMed] [Google Scholar]
- Guaragnella N et al. (2013) Yeast growth in raffinose results in resistance to acetic-acid induced programmed cell death mostly due to the activation of the mitochondrial retrograde pathway. Biochim Biophys Acta 1833:2765–2774. 10.1016/j.bbamcr.2013.07.017 [DOI] [PubMed] [Google Scholar]
- Gueldener U, Heinisch J, Koehler GJ, Voss D, Hegemann JH (2002) A second set of loxP marker cassettes for Cre-mediated multiple gene knockouts in budding yeast. Nucleic Acids Res 30:e23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guillet M, Boiteux S (2003) Origin of endogenous DNA abasic sites in Saccharomyces cerevisiae. Mol Cell Biol 23:8386–8394. 10.1128/mcb.23.22.8386-8394.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guillet M, Van Der Kemp PA, Boiteux S (2006) dUTPase activity is critical to maintain genetic stability in Saccharomyces cerevisiae. Nucleic Acids Res 34:2056–2066. 10.1093/nar/gkl139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagner N, Joerger M (2010) Cancer chemotherapy: targeting folic acid synthesis. Cancer Manag Res 2:293–301. 10.2147/CMR.S10043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanna M, Chow BL, Morey NJ, Jinks-Robertson S, Doetsch PW, Xiao W (2004) Involvement of two endonuclease III homologs in the base excision repair pathway for the processing of DNA alkylation damage in Saccharomyces cerevisiae. DNA Repair 3:51–59. 10.1016/j.dnarep.2003.09.005 [DOI] [PubMed] [Google Scholar]
- Johnson RE, Torres-Ramos CA, Izumi T, Mitra S, Prakash S, Prakash L (1998) Identification of APN2, the Saccharomyces cerevisiae homolog of the major human AP endonuclease HAP1, and its role in the repair of abasic sites. Genes Dev 12:3137–3143. 10.1101/gad.12.19.3137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kayikci O, Nielsen J (2015) Glucose repression in Saccharomyces cerevisiae. FEMS Yeast Res. 10.1093/femsyr/fov068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim N, Jinks-Robertson S (2009) dUTP incorporation into genomic DNA is linked to transcription in yeast. Nature 459:1150–1153. 10.1038/nature08033[pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim N, Jinks-Robertson S (2010) Abasic sites in the transcribed strand of yeast DNA are removed by transcription-coupled nucleotide excision repair. Mol Cell Biol 30:3206–3215. 10.1128/MCB.00308-10[pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim N, Abdulovic AL, Gealy R, Lippert MJ, Jinks-Robertson S (2007) Transcription-associated mutagenesis in yeast is directly proportional to the level of gene expression and influenced by the direction of DNA replication. DNA Repair 6:1285–1296. 10.1016/j.dnarep.2007.02.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim N et al. (2011a) Mutagenic processing of ribonucleotides in DNA by yeast topoisomerase I. Science 332:1561–1564. 10.1126/science.1205016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim N, Mudrak SV, Jinks-Robertson S (2011b) The dCMP transferase activity of yeast Rev1 is biologically relevant during the bypass of endogenously generated AP sites. DNA Repair 10:1262–1271. 10.1016/j.dnarep.2011.09.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F et al. (2019) Apn2 resolves blocked 3’ ends and suppresses Top1-induced mutagenesis at genomic rNMP sites. Nat Struct Mol Biol 26:155–163. 10.1038/s41594-019-0186-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)). Method Methods 25:402–408. 10.1006/meth.2001.1262 [DOI] [PubMed] [Google Scholar]
- Longley DB, Harkin DP, Johnston PG (2003) 5-fluorouracil: mechanisms of action and clinical strategies. Nat Rev Cancer 3:330–338. 10.1038/nrc1074 [DOI] [PubMed] [Google Scholar]
- Luke AM et al. (2010) Accumulation of true single strand breaks and AP sites in base excision repair deficient cells. Mutat Res 694:65–71. 10.1016/j.mrfmmm.2010.08.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Pastor MT, Marchler G, Schuller C, Marchler-Bauer A, Ruis H, Estruch F (1996) The Saccharomyces cerevisiae zinc finger proteins Msn2p and Msn4p are required for transcriptional induction through the stress response element (STRE). EMBO J 15:2227–2235 [PMC free article] [PubMed] [Google Scholar]
- Meadows KL, Song B, Doetsch PW (2003) Characterization of AP lyase activities of Saccharomyces cerevisiae Ntg1p and Ntg2p: implications for biological function. Nucleic Acids Res 31:5560–5567. 10.1093/nar/gkg749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owiti N, Lopez C, Singh S, Stephenson A, Kim N (2017) Def1 and Dst1 play distinct roles in repair of AP lesions in highly transcribed genomic regions. DNA Repair 55:31–39. 10.1016/j.dnarep.2017.05.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owiti N, Wei S, Bhagwat AS, Kim N (2018) Unscheduled DNA synthesis leads to elevated uracil residues at highly transcribed genomic loci in Saccharomyces cerevisiae. PLoS Genet 14:e1007516. 10.1371/journal.pgen.1007516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owiti N, Stokdyk K, Kim N (2019) The etiology of uracil residues in the Saccharomyces cerevisiae genomic DNA. Curr Genet 65:393–399. 10.1007/s00294-018-0895-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paulo JA, O’Connell JD, Gaun A, Gygi SP (2015) Proteome-wide quantitative multiplexed profiling of protein expression: carbon-source dependency in Saccharomyces cerevisiae. Mol Biol Cell 26:4063–4074. 10.1091/mbc.E15-07-0499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeiffer T, Morley A (2014) An evolutionary perspective on the Crabtree effect. Front Mol Biosci 1:17. 10.3389/fmolb.2014.00017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popoff SC, Spira AI, Johnson AW, Demple B (1990) Yeast structural gene (APN1) for the major apurinic endonuclease: homology to Escherichia coli endonuclease IV. Proc Natl Acad Sci USA 87:4193–4197. 10.1073/pnas.87.11.4193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radivojac P et al. (2010) Identification, analysis, and prediction of protein ubiquitination sites. Proteins 78:365–380. 10.1002/prot.22555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Randez-Gil F, Sanz P, Entian KD, Prieto JA (1998) Carbon source-dependent phosphorylation of hexokinase PII and its role in the glucose-signaling response in yeast. Mol Cell Biol 18:2940–2948. 10.1128/mcb.18.5.2940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren J et al. (2009) Systematic study of protein sumoylation: Development of a site-specific predictor of SUMOsp 2.0. Proteomics 9:3409–3412. 10.1002/pmic.200800646 [DOI] [PubMed] [Google Scholar]
- Schmitt AP, McEntee K (1996) Msn2p, a zinc finger DNA-binding protein, is the transcriptional activator of the multistress response in Saccharomyces cerevisiae. Proc Natl Acad Sci USA 93:5777–5782. 10.1073/pnas.93.12.5777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuller HJ (2003) Transcriptional control of nonfermentative metabolism in the yeast Saccharomyces cerevisiae. Curr Genet 43:139–160. 10.1007/s00294-003-0381-8 [DOI] [PubMed] [Google Scholar]
- Siriwardena SU, Chen K, Bhagwat AS (2016) Functions and malfunctions of mammalian DNA-cytosine deaminases. Chem Rev 116:12688–12710. 10.1021/acs.chemrev.6b00296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spell RM, Jinks-Robertson S (2004) Determination of mitotic recombination rates by fluctuation analysis in Saccharomyces cerevisiae. Methods Mol Biol 262:3–12. 10.1385/1-59259-761-0:003 [DOI] [PubMed] [Google Scholar]
- Temple MD, Perrone GG, Dawes IW (2005) Complex cellular responses to reactive oxygen species. Trends Cell Biol 15:319–326. 10.1016/j.tcb.2005.04.003 [DOI] [PubMed] [Google Scholar]
- Teste MA, Duquenne M, Francois JM, Parrou JL (2009) Validation of reference genes for quantitative expression analysis by real-time RT-PCR in Saccharomyces cerevisiae. BMC Mol Biol 10:99. 10.1186/1471-2199-10-99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unk I, Haracska L, Johnson RE, Prakash S, Prakash L (2000) Apurinic endonuclease activity of yeast Apn2 protein. J Biol Chem 275:22427–22434. 10.1074/jbc.M002845200 [DOI] [PubMed] [Google Scholar]
- Venters BJ et al. (2011) A comprehensive genomic binding map of gene and chromatin regulatory proteins in Saccharomyces. Mol Cell 41:480–492. 10.1016/j.molcel.2011.01.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace SS (2014) Base excision repair: a critical player in many games. DNA Repair 19:14–26. 10.1016/j.dnarep.2014.03.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warner HR, Duncan BK, Garrett C, Neuhard J (1981) Synthesis and metabolism of uracil-containing deoxyribonucleic acid in Escherichia coli. J Bacteriol 145:687–695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whelan WL, Gocke E, Manney TR (1979) The CAN1 locus of Saccharomyces cerevisiae: fine-structure analysis and forward mutation rates. Genetics 91:35–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Reese JC (2005) Molecular genetic analysis of the yeast repressor Rfx1/Crt1 reveals a novel two-step regulatory mechanism. Mol Cell Biol 25:7399–7411. 10.1128/MCB.25.17.7399-7411.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Q et al. (2014) GPS-SUMO: a tool for the prediction of sumoylation sites and SUMO-interaction motifs. Nucleic Acids Res 42:W325–330. 10.1093/nar/gku383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Z, Elledge SJ (1993) DUN1 encodes a protein kinase that controls the DNA damage response in yeast. Cell 75:1119–1127. 10.1016/0092-8674(93)90321-g [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Yeast strains will be made available upon request.