Abstract
This work aims to identify a new radiomics signature using imaging phenotypes and clinical variables for risk prediction of overall survival (OS) in hepatocellular carcinoma (HCC) patients treated with stereotactic body radiation therapy (SBRT). 167 patients were retrospectively analyzed with repeated nested cross-validation to mitigate overfitting issues. 56 radiomic features were extracted from pre-treatment contrast-enhanced (CE) CT images. 37 clinical factors were obtained from patients’ electronic records. Variational autoencoders (VAE) based survival models were designed for radiomics and clinical features and a convolutional neural network (CNN) survival model was used for the CECT. Finally, radiomics, clinical and raw image deep learning network (DNN) models were combined to predict the risk probability for OS. The final models yielded c-indices of 0.579 (95%CI: 0.544–0.621), 0.629 (95%CI: 0.601–0.643), 0.581 (95%CI: 0.553–0.613) and 0.650 (95%CI: 0.635–0.683) for radiomics, clinical, image input and combined models on nested cross validation scheme, respectively. Integrated gradients method was used to interpret the trained models. Our interpretability analysis of the DNN showed that the top ranked features were clinical liver function and liver exclusive of tumor radiomics features, which suggests a prominent role of side effects and toxicities in liver outside the tumor region in determining the survival rate of these patients. In summary, novel deep radiomic analysis provides improved performance for risk assessment of HCC prognosis compared with Cox survival models and may facilitate stratification of HCC patients and personalization of their treatment strategies. Liver function was found to contribute most to the OS for these HCC patients and radiomics can aid in their management.
Keywords: Hepatocellular Carcinoma (HCC), overall survival, radiomics, deep learning, variational autoencoder (VAE), convolutional neural network (CNN), computed tomography (CT)
Introduction
Liver cancer is a leading cause of cancer related deaths worldwide, with increasing incidences [1]. Hepatocellular carcinoma (HCC) is the most common type of primary liver cancer. Surgical resection and liver transplantation are used with curative intent for selected patients [2]. However, the majority of HCC patients are ineligible for surgery due to the location of the tumor or poor liver function [3]. There are several non-surgical liver-directed treatments including radiofrequency ablation (RFA), microwave ablation (MWA), trans-arterial chemoembolization (TACE) and, more recently, stereotactic body radiotherapy (SBRT). RFA/MWA can be limited by lesion size and proximity to critical organs; TACE is non-curative, with limitations such as poor neovascularization of some tumors or portal vein involvement [4]. Recently, with the development of advanced radiotherapy delivery technologies, more precise partial liver irradiation using SBRT has become an important option for HCC patients that are not suitable for resection [5, 6].
Although over 90% of tumors will be controlled by SBRT [5, 7], and the survival rate has been improved, the current survival rate is still not satisfying [8]. Radiomics, which is a field of medical image analytics, may aid in developing models to predict patient outcomes, such as overall survival and thus improving cancer management based on the prediction. Images are converted into a large number of quantitative features with subsequent datamining that relates these features to biological and clinical endpoints [9, 10]. It has been widely applied in cancer research and has shown to be able to capture distinct phenotypic differences and be associated with clinical prognosis in many cancer types [11–17]. These current studies in overall survival use mostly Cox models or random survival forests.
This study focuses on pre-SBRT arterial phase contrast-enhanced computed tomography (CECT) images, involving a relatively large dataset of 167 patients. The novelty of our work can be summarized as: (1) development of a comprehensive model based on radiomics (features from both gross tumor volume (GTV) regions and liver exclusive of the GTVs (liver-GTV)), clinical features and raw CT images; (2) novel VAE based survival model combining different sources of information; (3) investigations of correlation and contribution of clinical, radiomics, image features and miRNA data; (4) patch-based training that augmented data and improved the performance; (5) interpretability of the results, providing possible interpretation of underlying mechanism. This manuscript contributes to a better understanding of the HCC heterogeneity across patients, guidance for personalized HCC treatment planning in clinical practice and development of new methods that fuse conventional and deep learning based radiomic analyses.
Methods and Materials
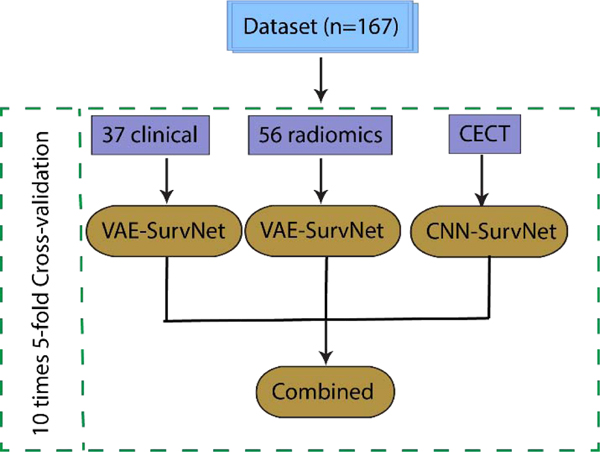
A brief description of the workflow presented in this study is shown in Fig. 1. Details are provided in the following. The detail network structures (VAE and CNN) are shown in Fig. 2.
Fig. 1.
Workflow for the radiomics survival modeling with deep learning of HCC SBRT patients. There were 167 patients enrolled in the study. Repeated 5-fold cross-validation was used. Three categories of variables – 37 clinical, 56 radiomics and raw CECT images input were integrated as a combined model.
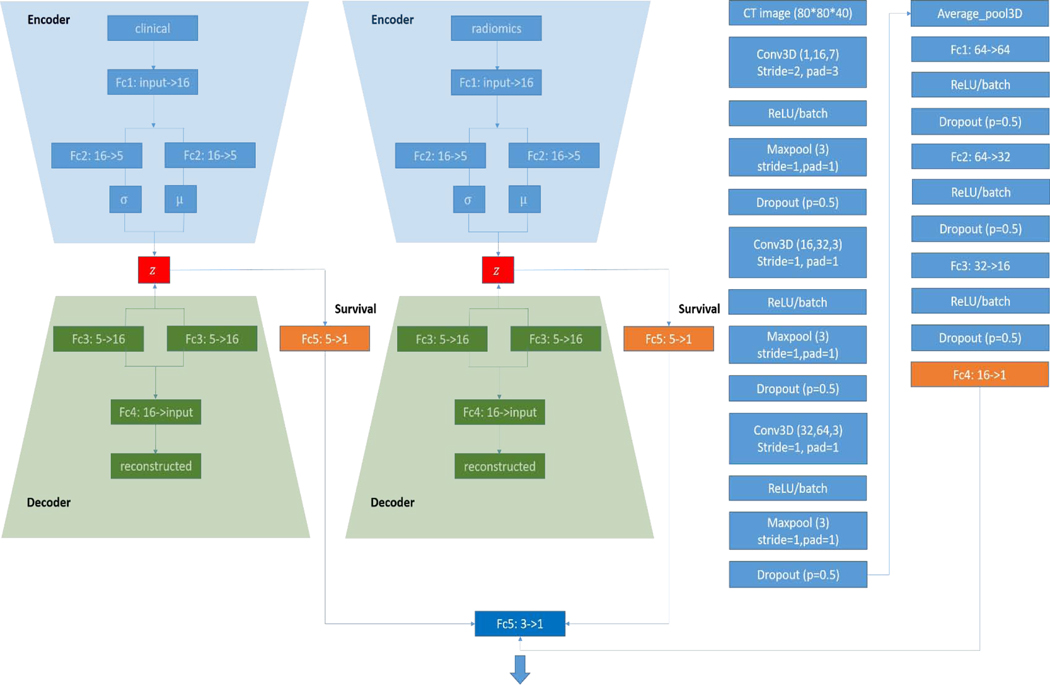
Fig. 2.
VAE-SurvNet and CNN-SurvNet structure. Clinical and radiomics features were input to the VAE SurvNet as shown in the left sub-networks. 3D CECT patches were fed into the CNN network as shown in the rightmost sub-network. FC: fully connected, Conv3D: 3D convolutional layer. Numbers in parenthesis represent the kernel dimensions
Patient cohort
After IRB approval, a HIPAA compliant retrospective analysis of HCC patients treated with SBRT was performed. A total of 303 HCC patients treated with SBRT were reviewed. Patients without: (1) contrast-enhanced CT (CECT) images; (2) gross tumor or liver contour in the database were excluded from analysis. A total of 167 HCC patients met the inclusion criteria. The overall survival (OS) events were right censored if no death until the last follow-up date (censor rate: 38.9 %).
37 clinical features, including patient basic information (e.g., gender, age, etc.), biological test results (e.g., liver function, immune cells count, etc.) and treatment (SBRT) related information (e.g., fraction number and mean liver dose) were obtained from patient records. 56 radiomics features, including seven distinct features (correlation, GLN (GLRLM), HGRE, SZE, GLN (GLSZM), ZSN, SZHGE) were extracted from both gross tumor volume (GTV) and liver-GTV regions. Due to the small sample size, though there are more radiomics features available, we limited the feature dimension via checking previously published paper on radiomic reproducibility and repeatability [18–22]. 84 miRNA features were included as well. Univariate Cox model c-index for clinical, radiomics and miRNA were investigated for these features. P-values of <0.05 were considered significant.
CT Images Acquisition and Processing
Arterial phase images and structure sets including liver, GTV, and liver-GTV from CECT were exported from an Eclipse treatment planning system (Varian Medical Systems Inc, Palo alto, CA). The contours were delineated based on the MR and CECT images by one physician and checked by at least another physician, one dosimetrist and one physicist. Eclipse was used as the workstation for the manual contouring process. The resolution of raw images ranged from 0.80 to 1.37 mm in-plane with 3 mm slice thickness. In order to extract texture features from the 3D volumes, the images were resampled to isotropic voxel sizes of 1×1×1 mm to obtain rotational invariance and also consistency across different patients. A trilinear interpolation algorithm was used for the resampling. Gray level quantization is required for the calculation of texture features (tractability). We applied the Lloyd-Max quantization. Lloyd-Max quantization is a method that tries to find a quantizer that minimizes the mean squared error (MSE) of original and new images, which can conserve most information in the images while discretizing.
Model Evaluation Metrics and Framework
Models were trained separately using clinical, radiomics and imaging data. Then, the individual models were fused and evaluated. The model fusion was implemented by concatenating the last layers of the individual models and fine-tuning the fused model. Cox proportional hazard regression [23] was applied for comparison. These models were trained and evaluated by strictly splitting data into training, validation and testing sets with a stratified 10 times 5-fold scheme, including the benchmarking methods. First, the data was split into training (75% of 4 folds), validation (25% of 4 folds) and testing fold (1 fold). The hyper-parameters were tuned on the validation data. Then, the trained model was tested on the test fold. This process was repeated 10 times to obtain the average performance and corresponding confidence intervals. The metric used was the Harrel’s c-index [24], with Kaplan-Meier plots for high and low risk groups for survival analysis. The risk groups were determined by a criterion using the median value of the outputs from the survival model. Confidence intervals were calculated by a bias-corrected and accelerated (BCa) bootstrap interval algorithm [25].
Patch-based Variational Autoencoder Survival Joint Model for Radiomics and Clinical Features
For feature selection, algorithms such as Relief-F [26], support vector machine- recursive features elimination (SVM-RFE) [27], Minimum Redundancy Maximum Relevance (mRMR) [28], etc., are available. However, these methods might cause selection bias and lead to over-optimistic results if the data are not split correctly. In addition, it is more tedious with two steps in the analysis: feature selection and subsequent model building. In comparison, the VAE-SurvNet method automatically learns a latent space to represent the important signals and train the survival model in one step efficiently.
Kingma et al. [29] introduced the Variational Autoencoders (VAEs) that resemble the naive autoencoders and variational Bayesian methods. Instead of learning a function that represents the data, variational autoencoders are able to learn a probability distribution from the data. The short coming of a pure VAE for a classification problem is that it is an unsupervised learning and the features obtained from the latent space might be irrelevant to the endpoint of interest. Thus, a supervised joint training network with a classification part was designed, which takes the latent space features as an input, goes through a fully-connected (FC) layer and outputs the risk probability. By this technique, the latent features learned by VAE are more specific to the desired task.
Specifically, the VAE consists of an encoder, which takes the input and converts it into two latent vectors (a vector of means, μ, and a vector of standard deviations, σ) that parameterize a Gaussian distribution and a decoder that reconstructs a latent space sample z back to the original space. The loss function of the VAE model is defined by two parts: (1) a reconstruction loss that measures how similar is the output comparing with the input; and (2) a regularization loss determined by Kullback-Leibler divergence (KL divergence), that measures how closely the latent variables match Gaussian distributions.
Considering the ith input sample xi, the output from the encoder is a hidden representation z, which has weights and biases θ. The encoder can be denoted as qθ(Z|x). For the decoder network, a value Z is denoted as input, and a reconstructed output x(*) is generated from some conditional distribution pφ(x|Z), which represents the decoder network. Thus, the loss function can be expressed as follows:
| #(1) |
where li is the loss for a single data point. The first term is the reconstruction loss, which encourages the network to reconstruct the input data; the second term is the KL divergence between the encoder’s distribution and the prior distribution p(z), which measures how much information is lost during the compression. This term also serves as a regularizer that prevents the network from simply copying the input and leading to overfitting.
The VAE architecture was determined by minimizing the validation loss with respect to different networks (latent space dimension and layer number). To determine the network structure, first, the classification part was ignored and only the VAE part was tuned. The number of layers and nodes in the layers were grid-searched based on the loss function. Once the VAE part was fixed, the VAE-SurvNet (including the survival part) was jointly trained by optimizing the total loss function that consists of VAE and the survival loss. A key point here is the ratio (τ) between VAE (lvae) and survival losses (lCox), which regulates the supervised and unsupervised portions, as shown in Eqn. (5). This hyper-parameter was tuned on the validation part of the training folds. Similar joint training approach was proposed in Ren et al.’s work [30].
Radiomics and clinical features are 1D vectors with 56 and 37 variables for each sample, while the CT image input is 3D matrix, which was resized to the liver size of (224, 224, 48) to be fed into the CNN network. Another important technique we used is the patch-based training, which can augment the data and improve the performance. The random crop dimension we used is (80, 80, 40), which was determined by experiments.
Neural Network based Survival Analysis
Cox proportional hazard model (CPH) is the most commonly used survival analysis method to explore the relationships between patients’ covariates and the survival time. It assumes that the log-risk of failure is a linear combination of the covariates. The hazard function is represented as the formula below:
| #(2) |
h(x) is a linear function of variables x. The weights β are tuned by optimizing the Cox partial likelihood, as shown below:
| #(3) |
Ti is the duration, Ei is the event indicator, and xi is the input feature for subject i. The risk set R(Ti) = (i: Ti ≥ t)s is the set of patients that are still at risk at time t.
However, this assumption might be too simplistic for complex relationships. To model potential nonlinearity relationship between features and the risk of failure, deep neural networks (DNNs) are used in Katzman et al.’s work, called the DeepSurv [31]. Instead of using h(x) as shown in Eqn. (2), DNN was used to estimate the log-risk function, with the output giving (xi), where β are the DNN parameters. Similarly, the objective function of the DNN is still the Cox partial likelihood:
| #(4) |
NE = 1 is the number of patients that are not censored and contribute to the log-likelihood loss calculation. The last two terms are penalties that aim to regularize the loss function, where the first penalty is an L2 norm penalty and second term is a penalty for the prediction to restrain its value not to deviate too much and cause overflow during training.
For the modeling of (xi), the original work used a pure multilayer perceptron (MLP), while in our study, a VAE architecture was applied with two advantages, (1) the latent features could be obtained; (2) jointly training partial likelihood in Eqn. (3) for survival and the VAE loss function (KL divergence and reconstruction binary cross entropy loss) makes the generated model more robust. The total loss function in the DNN is thus:
| #(5) |
τ is a weight that balances the two parts of losses, which is tuned on the training set.
Interpretation of the Deep Neural Networks
As the models become more and more complex, the interpretation becomes increasingly important, especially in the medical field. First, we did Spearman rank correlation for the three categories variables: clinical, radiomics, and raw image inputs. The average values of the second last fully connected layer of the CNN models were used as the features from raw image. Captum, a library for model interpretability, was used in this study for model visualization and interpretation [32]. The integrated gradient method was used to estimate the feature importance for clinical and radiomics features and the critical regions (voxels) for the raw image input. This method constructs a sequence of images interpolating from a baseline to the actual image and then averages the gradients across these images [33].
Results
Univariate analysis
Supplementary tables 1 and 2 show the univariate analysis of clinical and radiomics for the overall survival (OS) endpoint. Tables 1–3 show the significant clinical/radiomics/miRNA features. There are 13 significant clinical variables, 6 radiomics and 3 miRNA features.
Table 1.
Univariate results using Cox model for significant clinical variables (c-indices and corresponding p-values)
| Clinical Variables | c-index for OS | p-value for OS |
|---|---|---|
| Number of Active Liver Lesions at Time of Treatment | 0.557 | 0.009 |
| Total Number Fractions | 0.558 | 0.009 |
| Pre-RT ICGR15 | 0.568 | 0.003 |
| Na (pre-treatment) | 0.620 | 0.005 |
| Albumin (pre-treatment; g/DL) | 0.636 | 0.000 |
| Total bilirubin (pre-treatment; mg/dL) | 0.601 | 0.000 |
| MELD (baseline) | 0.574 | 0.031 |
| MELD-Na (baseline) | 0.596 | 0.009 |
| Child-Pugh (baseline) | 0.626 | 0.000 |
| ALBI Raw Score (Baseline) | 0.624 | 0.000 |
| Alkphos CTCAE Liver Toxicity Grade (pre-tx) | 0.549 | 0.004 |
| Treated previously? | 0.556 | 0.016 |
| PLATELET_pre | 0.535 | 0.028 |
Table 3.
Univariate results using Cox model for significant miRNA variables (c-indices and corresponding p-values).
| miRNA Variables | c-index for OS | p-value for OS |
|---|---|---|
| hsa-let-7i-5p | 0.689 | 0.017 |
| hsa-miR-107 | 0.621 | 0.015 |
| hsa-miR-660-5p | 0.652 | 0.046 |
Multivariate analysis and Benchmarking
For multivariate analysis, the radiomics, clinical and CT raw image individual models’ results are summarized in table 4. Proposed models were compared with the Cox model as a benchmark for time-to-event analysis. The same scheme of repeated cross validation as described previously was used for both Cox model and the proposed method. The average c-indices for the test sets are 0.554 (CI: 0.531–0.577), 0.599 (CI: 0.581–0.617) and 0.546 (CI: 0.519–0.573) for radiomics, clinical and combined models, respectively using Cox models. The average c-indexes for test sets are 0.579 (CI: 0.544–0.621, p-value: 0.0005), 0.629 (CI: 0.601–0.643, p-value: 0.149), 0.581 (CI: 0.553–0.613) and 0.650 (CI: 0.635–0.683, p-value: <0.0001) for radiomics, clinical, CT image input and combined models, respectively using DNNs. The p-values are for the comparison between corresponding deep learning models and Cox models. The Cox model cannot handle image inputs. The combined models for DNN outperformed the clinical models alone, which indicates the value of complementary information that imaging can provide. The deep learning models outperformed the Cox models significantly in all categories except the clinical model. Notice that the architecture presented here might not be the optimal structure, however, based on our experiments, the performance is not sensitive to the structure, and the goal of this work is to show the concept that VAE-SurvNet model possess predictive power, and not to find the optimal solution. The Kaplan-Meyer plots for the combined models that show stratification between high and low risk groups are shown in Fig. 3. The architecture details were shown in Fig. 2. The optimizer used is Adam. For VAE-SurvNet, the hyper-parameters τ in Eqn. (5) was tuned to be 1e-6, learning rate was 0.01, l2 penalty was 1e-4. For the CNN network, learning rate was 0.001 with no l2 penalty. The combined model, learning rate was 0.0001 for the pretrained parameters and 0.01 for the other.
Table 4.
C-indices for radiomics, clinical, raw image CNN and combined models using Cox model as a benchmark (column 2) and the proposed DNN models (column 3), as well as the corresponding Delong test p-values comparing Cox model benchmark and the proposed DNN methods.
| Cox | DNN | p-values | |
|---|---|---|---|
| Radiomics | 0.554 (0.531–0.577) | 0.579 (0.544–0.621) | 0.0005 |
| Clinical | 0.599 (0.581–0.617) | 0.629 (0.601–0.643) | 0.149 |
| Image | NA | 0.581 (0.553–0.613) | NA |
| Combined | 0.546 (0.519–0.573) | 0.650 (0.635–0.683) | <0.0001 |
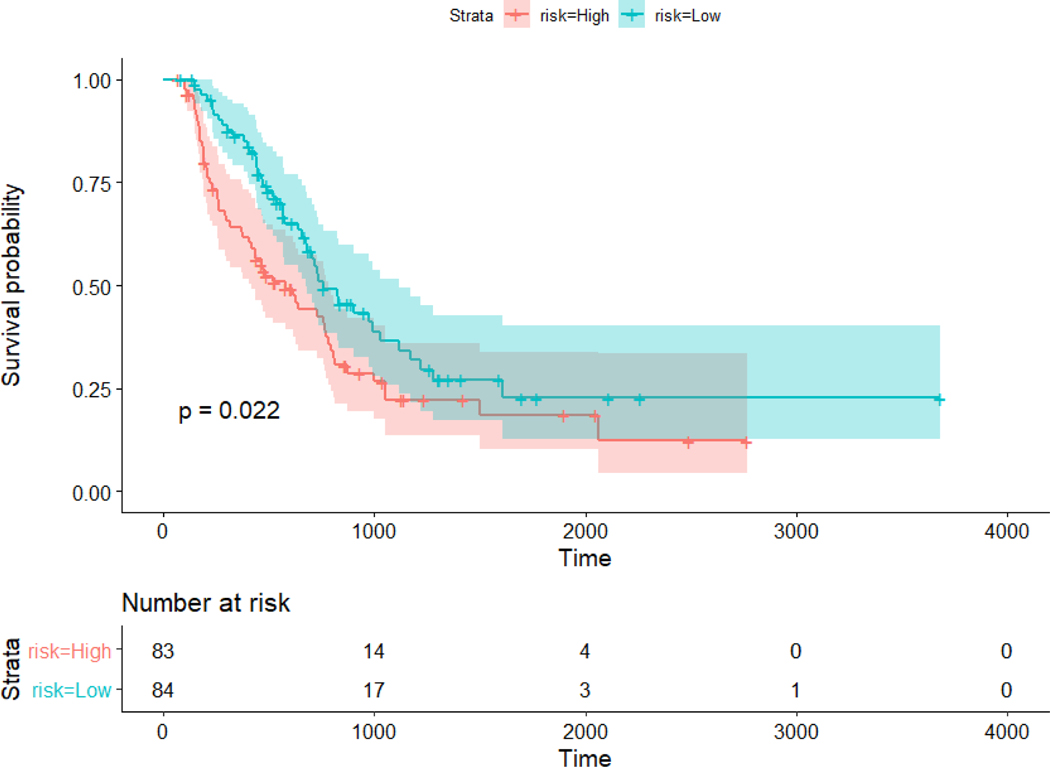
Fig. 3.
Kaplan Meier plot for high and low risk groups for overall survival. The cutoff is the median value of the output of the network for the test set. High and low risk group were stratified significantly (p-value = 0.022).
To enhance the raw image input performance, experiments of transfer learning from 3D pretrained network were carried out as well. The pretrained models are based on Resnet-18 and trained on the 3D Kinetics dataset [34, 35]. The c-indexes were calculated on test set using the repeated cross validation scheme to provide out-of-sample verification as well. The average test c-index was 0.556 (CI: 0.537–0.575), which is not better than the basic CNN structure 0.581 (CI: 0.553–0.613). This might be due to the large difference between medical images and the Kinetics dataset. So, we used the basic CNN for the image inputs. Convolutional VAE regularized training similar as discussed [30] were performed for the image input as well. The training was difficult to converge probably due to the large amount of parameters for the convolutional VAE network. In Mobadersany et al.’s work, the genomics data was integrated to the fully connected layer directly [36]. In order to compare with this method, we applied the same architecture by integrating the radiomics and clinical features to the fully connected layer of the CNN network. The result was 0.593 (CI: 0.574–0.613), which is significantly lower than the proposed methods 0.650 (CI: 0.635–0.683). The c-indexes here were also calculated on test set using the repeated cross validation scheme.
The individual models were first trained and the last layers of the three pretrained models were concatenated and fine-tuned to obtain the final results. We also tried directly training the model from scratch without the pretrained models, which is hard to converge and give good results.
Correlation analysis of clinical, radiomics, and DL-driven variables and Interpretation of models
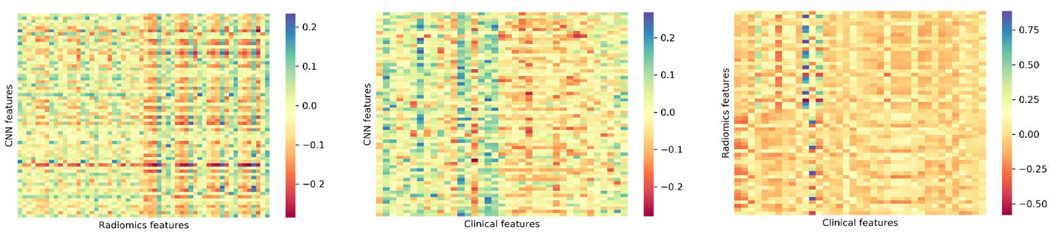
There are three categories of features: DL-driven, radiomics and clinical features. DL-driven features were extracted from the first fully connected layer (64 nodes) of the CNN model. Since there are 50 (10 times 5-fold cross-validation) trained models, the average values were calculated on the test sets. The Spearman rank correlation matrices were shown in Fig. 4 for the three categories pairwise.
Fig. 4.
Spearman rank correlation matrices for DL-driven vs. radiomics features (left), DL-driven vs. clinical features (middle) and radiomics vs. clinical features (right).
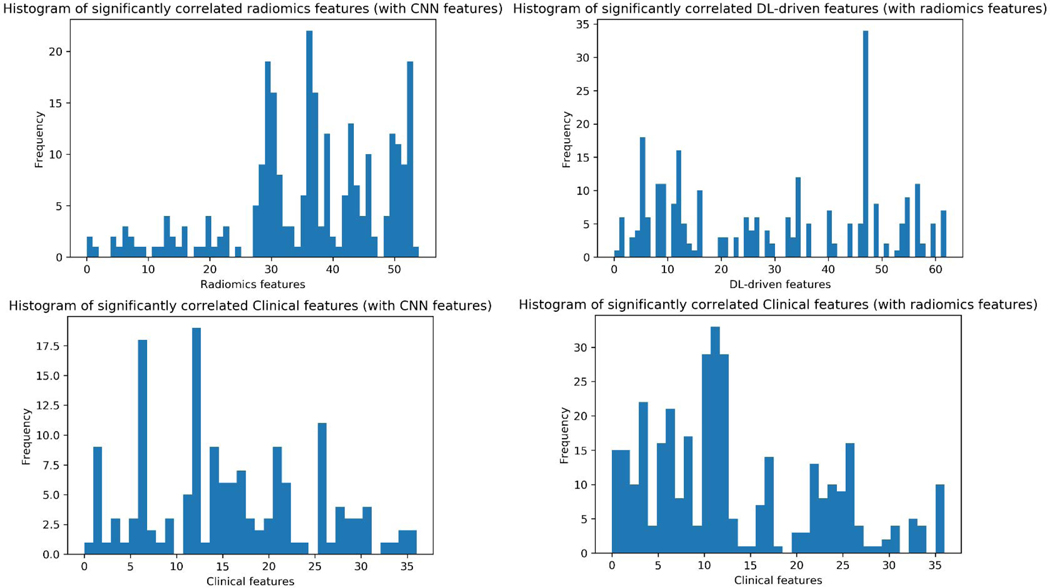
Due to the large size of the correlation matrices, we focused on the significantly correlated features. For the DL-driven vs. radiomics features, significantly correlated feature pairs were mostly between liver-GTV radiomics and DL-driven features (as shown in Fig. 4 left). The histogram in Fig. 5 top left showed the frequency of each radiomics feature that was significantly correlated with DL-driven features. The first 28 were GTV features and the last 28 were liver-GTV features. The frequency that liver-GTV features strongly correlates with DL-driven features are more than GTV features. It is consistent with the fact that the input raw images are dominant by liver regions as well as the prominent role that toxicity plays in overall survival. It also showed that the CNN models indeed learned some complex high-level features from the images that are similar to hand-crafted radiomics features. Fig. 5 top right showed the number of each DL-driven feature that was significantly correlated with radiomics features. Feature # 47 DL-driven was found to be correlated with GLN (GLSZM), ZSN and SZHGE with different gray levels from GTV regions and with correlation, SZE, GLN (GLRLM) from liver-GTV regions.
Fig. 5.
Histograms that show the frequency of each radiomics feature that significantly correlated with CNN features (top left); each CNN feature with radiomics features (top right); each clinical feature with CNN features (bottom left) and each clinical feature with radiomics features (bottom right).
The clinical features that correlated most frequently with DL-driven features were age, Total_EQD2, LIVER_GTV_Mean_Dose, ECOG_PS, Protime_with_INR and Barcelona_score, a shown in Fig. 5 bottom left. There are more significantly correlated radiomics and clinical feature pairs than DL-driven features. Based on the histogram shown in Fig. 5 bottom right, PVT, number of fractions, total EQD2, tumor volume, liver-GTV volume, liver-GTV mean dose, albumin and Barcelona score are most frequently correlated with radiomics features.
In general, the imaging features (DL-driven and radiomics features) are significantly correlated with liver function, which was represented by the clinical features (e.g., Barcelona score). In addition, the results also showed that the GTV, liver-GTV volumes are correlated with the radiomics features significantly, as discussed by Traverso et al. [37].
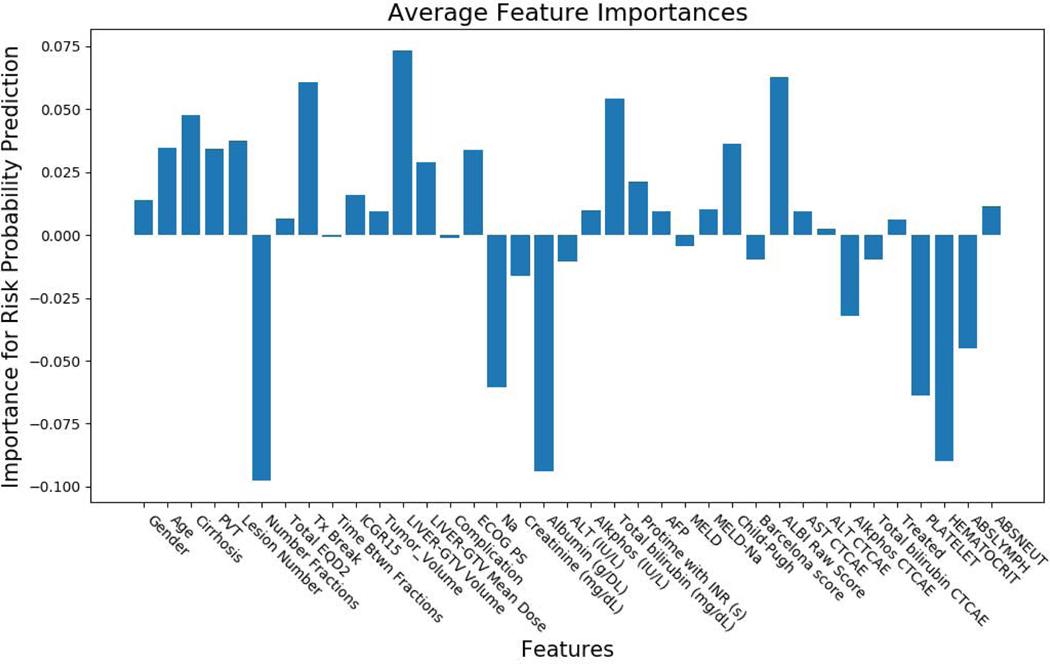
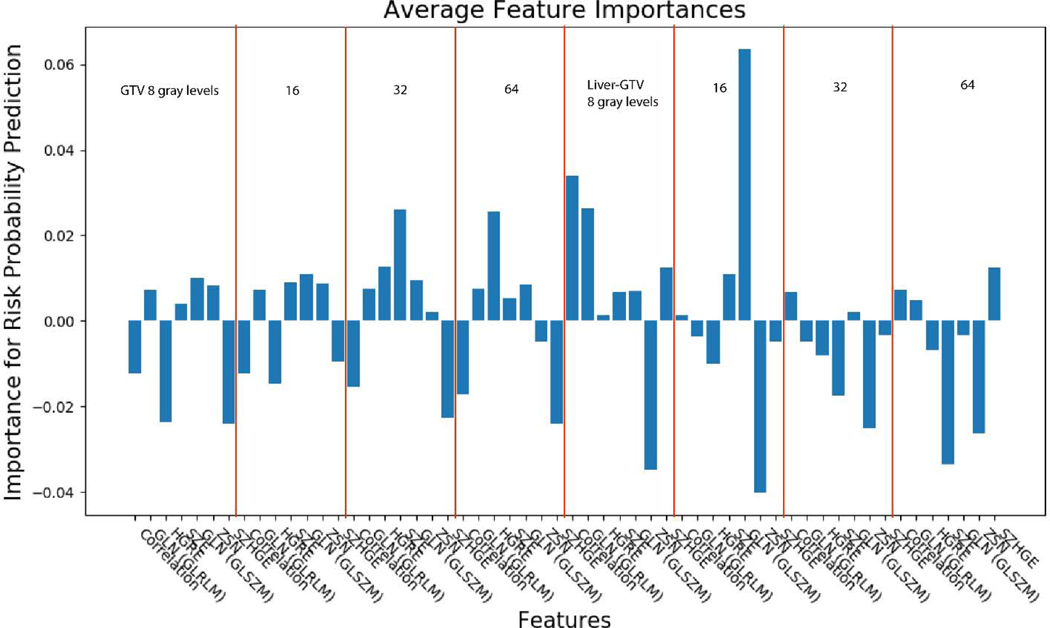
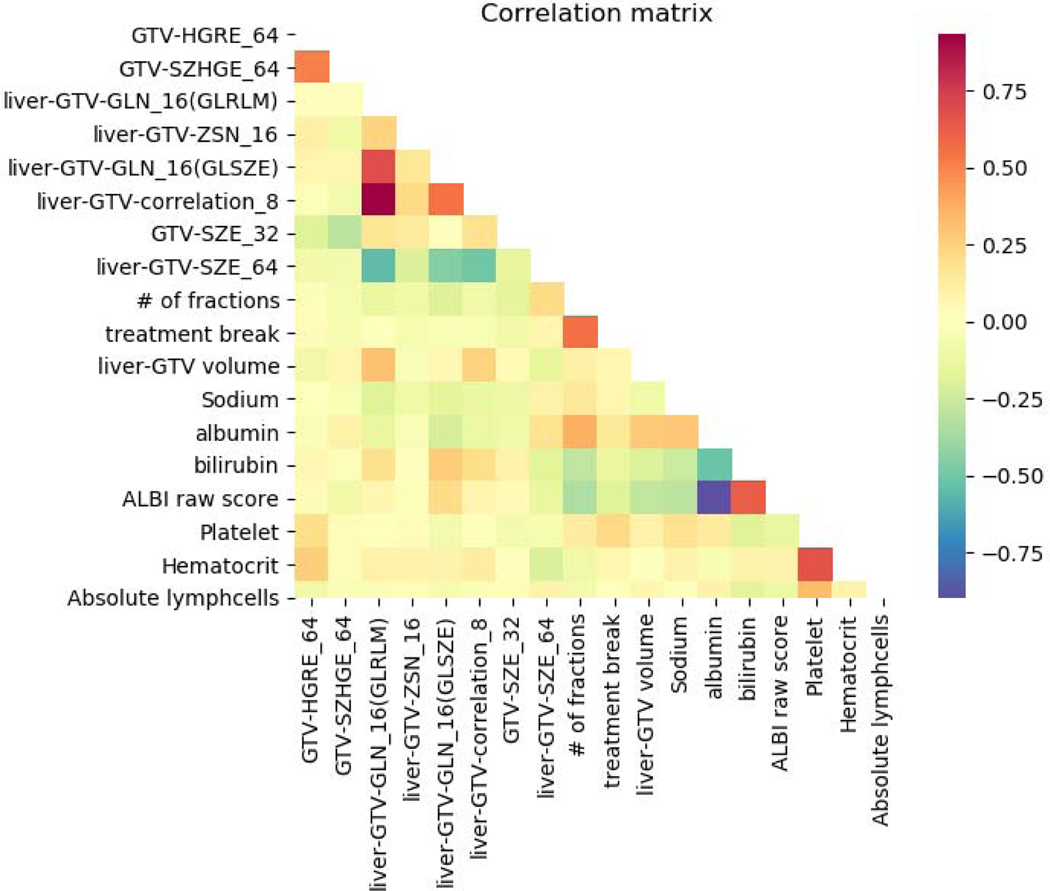
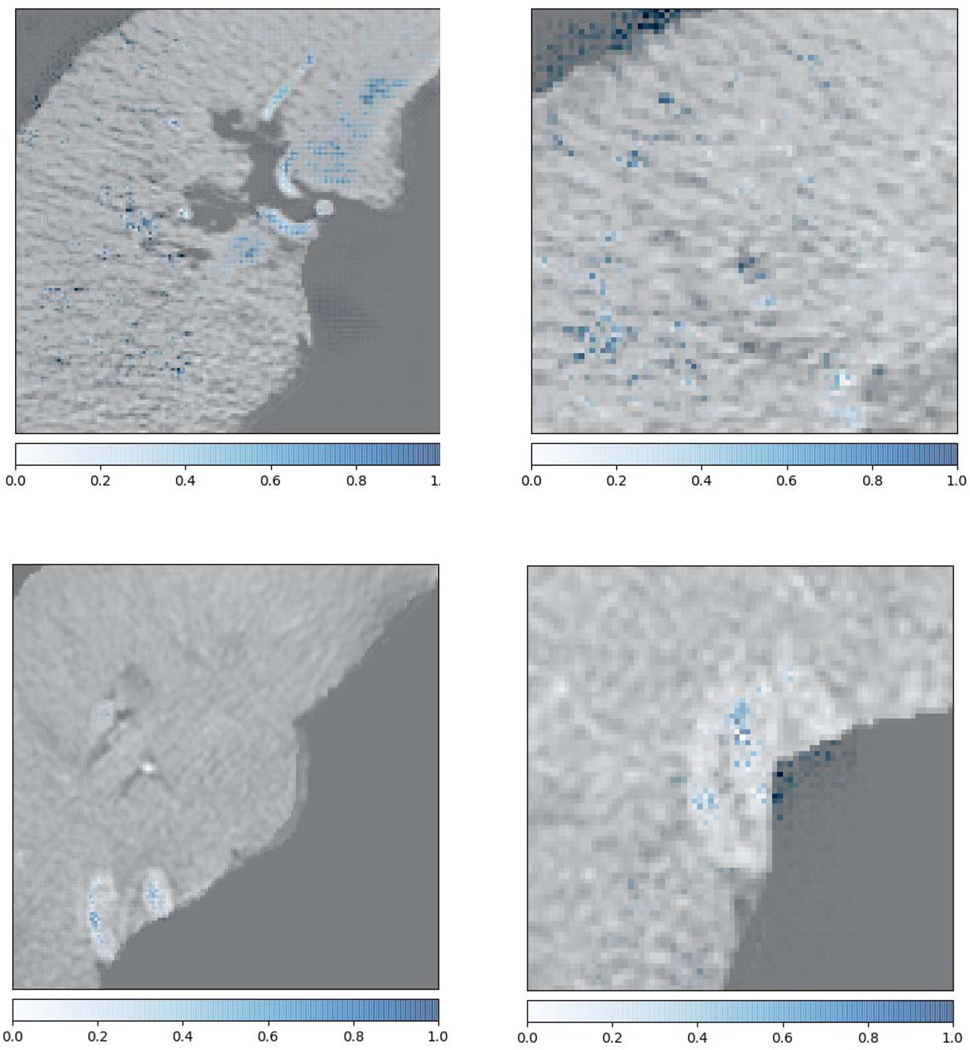
Figs. 6 and 7 show the feature importance for clinical and radiomics variables. The top ranked clinical features are total number of fractions (5), albumin (17), hematocrit (34), liver-GTV volume (11), ALBI raw score (27), platelet (33), treatment break (7), total bilirubin (20), abslymph (35) and cirrhosis (2). The top ranked radiomics features are GTV SZHGE, liver-GTV ZSN, which are ranked high for most of the gray levels (8,16,32,64). There are also several radiomics features that ranked high for particular gray level, including GTV_64_HGRE, GTV_32_SZE, liver-GTV_8_GLN (GLRLM), liver-GTV_16_GLN (GLSZM), liver-GTV_8_correlation, and liver-GTV_64_SZE. Fig. 8 shows the correlation matrix for these top ranked features. Within the radiomics features, the correlation of GTV-HGRE/SZHGE 0.512 (p-value <0.0001), liver-GTV GLN (GLRLM)/GLN (GLSZM) 0.689 (p-value: <0.0001), liver-GTV correlation/GLN (GLRLM) 0.933 (p-value <0.0001) are high. Within clinical variables, since ALBI score is calculated based on albumin and bilirubin, their correlation is intrinsically high. The correlation of albumin/number of fractions 0.378 (p-value: <0.0001), albumin/liver-GTV volume 0.276 (p-value: 0.0003), albumin/Sodium 0.295 (p-value: 0.0001) are high. More interestingly, across radiomics and clinical feature groups, correlations of liver-GTV volume/GLN (GLRLM) 0.315 (p-value: <0.0001), liver-GTV volume/correlation 0.241 (p-value: 0.002), bilirubin/liver-GTV GLN (GLSZE) 0.271 (p-value: 0.0004), bilirubin/liver-GTV SZE −0.172 (p-value: 0.026), platelet/GTV-HRGE 0.200 (p-value: 0.010), hematocrit/GTV-HGRE 0.264 (p-value: 0.0006), hematocrit/liver-GTV SZE −0.202 (p-value: 0.009) are high. Taken together, these results may suggest a prominent role of side effects and toxicities in determining the survival rate of these patients. In order to visualize the important radiomics features, Fig. 9 shows some example images with high/low liver-GTV GLN values, and the corresponding integrated gradient images that show the critical voxels for raw images CNN. It is found that the higher GLN values corresponds to higher risk, and the critical voxels distributed over the normal liver tissue. Meanwhile, lower GLN values corresponds to lower risk, and the critical pixels mostly concentrated on high intensity regions (e.g., vessels) but not through the normal tissue.
Fig. 6.
Clinical features importance using integrated gradients method for the VAE network. Gender; Age; Pre-Tx cirrhosis; Portal vein thrombosis; Number of active liver lesions at time of treatment; Total Number Fractions; Total EQD2; Tx Break; Time btwn first and final fractions (Days); Pre-RT ICGR15; Tumor volume; Liver-GTV volume (cc); Liver-GTV mean dose (Gy); Treatment-related complication; ECOG PS; Sodium; Creatinine (mg/dL); Albumin (g/DL); ALT (IU/L); Alkphos (IU/L); Total bilirubin (mg/dL); Protime with INR (s); AFP; MELD; MELD-Na; Child-Pugh; Barcelona score; ALBI Raw Score; AST CTCAE liver toxicity grade; ALT CTCAE liver toxicity grade; Alkphos CTCAE liver toxicity grade; Total bilirubin CTCAE liver toxicity grade; Treated previously; Platelet; Hematocrit; Abslymph; Absneutral.
Fig. 7.
Radiomics features importance using integrated gradients method for the VAE network. There are 7 distinct features, the red lines separate the regions (GTV, liver-GTV) and gray levels (8, 16, 32, 64). The 7 radiomics features from left to right are correlation, GLN (GLRLM), HGRE, SZE, GLN (GLSZM), ZSN, SZHGE.
Fig. 8.
The Spearman rank correlations map for the top ranked clinical and radiomics features.
Fig. 9.
Example images for low/high liver-GTV GLN (GLSZE) values. Top row shows the images for a patient with GLN 0.756, died in 264 days. Bottom row shows the images for a patient with GLN 0.154, survived a760 days (right censored). The left column shows most of the liver tissues. The right column shows the zoomed in tissues. The blue dots are the output of integrated gradients method that shows the critical pixels for the prediction of the neural network.
Subset analysis of miRNA data
Due to the small number of patients that having miRNA data, training the models for genetic information was not plausible. Instead, the correlation of miRNA and other top ranked features were investigated. Table 5 shows the correlation of the 3 significant miRNAs with the top ranked clinical and radiomics features. These three miRNA features are highly correlated with each other, with hsa-let-7i-5p and hsa-miR-10b-5p 0.459 (p-value: 0.021), hsa-let-7i-5p and hsa-miR-660–5p 0.537 (p-value: 0.006), hsa-let-7i-5p and hsa-miR-660–5p 0.511 (p-value: 0.009). hsa-let-7i-5p is significantly correlated with GTV-SZHGE_64 (−0.403, p-value: 0.046), liver-GTV-ZSN_16 (−0.452, p value: 0.023), liver-GTV-correlation_8 (−0.404, p-value: 0.045). hsa-miR-10b-5p is significantly correlated with GTV-SZHGE_64 (−0.544, p-value: 0.005), total_bilirubin (−0.423, p-value: 0.035). hsa-miR-660–5p is significantly correlated with GTV-SZHGE_64 (−0.457, p-value: 0.022), In general, it was found that the three miRNAs are significantly correlated with GLN (GLSZM), SZHGE, ZSN with different gray levels.
Table 5.
Spearman rank correlation coefficients and p-values of significant miRNA (from univariate Cox analysis) and top ranked clinical/radiomics features obtained from the Captum interpretation model. The radiomics features were named by stating the region being extracted, the name of the features and the gray level (e.g., GTV-SZHGE-64 means the SZHGE feature extracted from GTV with gray level of 64).
| miRNA | Radiomics/Clinical Variables | Correlation coefficients | p-values |
|---|---|---|---|
| hsa-let-7i-5p | GTV-SZHGE_64 | −0.403 | 0.046 |
| liver-GTV-ZSN_16 | −0.452 | 0.023 | |
| liver-GTV-correlation_8 | −0.404 | 0.045 | |
| hsa-miR-10b-5p | GTV-SZHGE_64 | −0.544 | 0.005 |
| total_bilirubin | −0.423 | 0.035 | |
| hsa-miR-660-5p | GTV-SZHGE_64 | −0.457 | 0.022 |
Discussion
Due to the challenges associated with the heterogeneity of liver cancers among different patients and the complicated etiologic factors associated with HCC, limited work to date has been done for HCC patients’ prognosis analysis. Cozzi et al. [38] conducted a retrospective study of 138 HCC patients treated with VMAT for the prediction of overall survival and local control. They applied univariate and logistic regression for clinical response and Cox regression model for survival analysis on clinical and radiomic features, which showed significant prediction performance. However, the features were extracted from non-contrast-enhanced images, which usually suffer from poor image quality, especially for liver with disease. Also linear regression methods fall short of capturing the complexity of survival analysis. Zhou et al. [18] developed a CT-based radiomics signature for preoperatively predicting the early recurrence of HCC using the LASSO algorithm. They built a radiomic and clinical combined model with AUC of 0.836 for the prediction of early recurrence. It focused on patients who underwent hepatectomy and did not consider time to event or survival analysis. Kiryu et al. [39] investigated the relationship of texture features with filtration at different filter levels and the prognosis of HCC 5-year overall survival and disease-free survival using preoperative non-contrast enhanced CT images. They showed the KM curves for OS and DFS were significantly different between patient groups dichotomized by cut-off values for all CT texture features. Bakr et al. [40] explored noninvasive biomarkers of microvascular invasion in patients with HCC (28 patients) using quantitative image features extracted from contrast-enhanced CT. Chaudhary et al. [41] conducted a deep learning study using multi-omics features (methylation, miRNA and RNA sequencing data) to identify survival subgroups of HCC. The model provides two subgroups with significant survival differences and model fit of c-index 0.68. Although, this work integrated information from different sources, it didn’t include imaging data. In addition, they used auto-encoder in an unsupervised way to obtain labels for the samples, then used SVM to predict the assigned labels, which may not be ideal, since the assigned labels tend to include some training information and may lead to biased results. Comparing with their work, our input data - radiomics, clinical, and raw imaging input are different from their gene sequencing data. In terms of methods, we used variational version of auto-encoder and directly carried out survival analysis instead of assigning labels to the samples. Mobadersanya et al. developed a model using survival CNN to integrate information from histology images and genomic biomarkers to predict time-to-event outcomes [36]. Though we both used the deepsurv network to predict time-to-event endpoints, VAE was used for dimension reduction for the clinical and radiomics input in our case, whereas the genomic data was directly fed to the fully connected layers in their model. For the visualization of the patterns in the images, integrated gradient method was used in our study, which surpasses the original heatmap method since the direction provided by the integrated gradients lead better towards the global optimum than the normal gradient which may lead to local optima that is used in basic heat map method. Additionally, the input data (CECT imaging and clinical data vs. histology imaging and genomics data) is different as well.
In general, compared to the studies above, this study assessed the prediction potential of radiomic features extracted from contrast-enhanced CT pre-treatment images, the original images, and pre-treatment clinical factors for risk assessment of overall survival using neural networks. The radiomics/raw image prediction models showed modest performance in our experiments. The possible reasons are: (1) we used a relatively strict validation framework that adopted repeated nested cross-validation; (2) Overall survival is a complex target to predict. CT images might not have sufficient predictive power; (3) the data size is too small to learn the underlying mechanism; (4) the absolute value might fluctuate for different datasets. Nonetheless, the contribution of this work comes in three ways: (1) It showed the complementary information from images that could help the clinical factors; (2) We proposed a novel VAE-Survnet that could combine multi-omics features including raw images, which outperformed the traditional Cox modeling; (3) Interpretation of the developed models by integrated gradients methods to help understand the mechanism.
The DNN based models (individuals and combined) outperformed those of the Cox based models, showing superiority of the DNN based approach in modeling non-linear, complex relationships. Although the raw imaging based individual models performed worse than the clinical models, they are still significantly better than random. One possible reason of the low predictive power of the imaging features might be the lack of good soft tissue contrast in CT, low signal to noise ratio, etc. To improve the raw image CNN model performance, different strategies were applied, such as transfer learning, it turns out the performance were all pretty similar. Thus, we used the basic CNN structure for the CT image data. We also used random crop to augment the CT image input network.
Based on the interpretation network, there are more top ranked radiomics features from liver-GTV regions than the GTV regions. These features reflect the heterogeneity in intensities in the ROIs. Feature HGRE, GLNs from both GLRLM and GLSZM, SZHGE turned out to be significant predictors for overall survival in the work by Cozzi etal [38]. Correlation was selected in the radiomiccs signature for the early recurrence prediction by Zhou et al [18]. Peritumoral HGRE was included in the early recurrence model by Shan etal [19]. The top ranked clinical features include three main categories: liver function related variables (albumin, bilirubin, ALBI), treatment related variables (total number of fractions, treatment break) and immune related variables (platelet, hematocrit). Liver function related variables were shown to be correlated with overall survival by other studies as well [42–44]. Serum sodium concentration was also found to be prognostic predictor for HCC [45–47]. Platelet count has been shown by several studies to be predictive for HCC overall survival [42, 43, 48]. Changes in lymphocytes and platelets counts, pre-treatment hematocrits, neutrophil counts were found to be the most important factors for locoregional failure in HCC [49]. The results of CNN integrated gradients method show the importance of voxels distributed in normal tissues for the survival prediction. All these results point out that the status of the normal tissue in the liver contributes to the overall survival and its exposure to unnecessary radiation may have worsened overall survival of these HCC patients.
It is of interest to investigate the relationship between imaging features and clinical/miRNA variables. Bilirubin--liver-GTV GLN, liver-GTV SZE, platelet--GTV-HGRE, hematocrit--GTV-HGRE and liver-GTV SZE, miRNA--GTV-SZHGE and liver-GTV-ZSN are highly correlated. These results showed potential ability for imaging features as non-invasive methods to investigate the tissue molecular status (e.g., miRNA), which has been found in breast cancer [50], head and neck cancer [51]. However, there are only 25 patients that have miRNA data, these findings need to be validated in larger cohort in the future. Various studies have suggested that heterogeneity of tumors is associated with genomic heterogeneity and tumoral microenvironment [52, 53], thus plays an important role in the cancer prognosis, which is found in our work as well.
The proposed methods that integrated imaging and clinical information have showed promising predictive power for the overall survival of SBRT treated HCC patients. Since we used pretreatment CECT images, which are commonly available for these SBRT treated patients clinically. This method could be used in practice to provide additional information to the physician about the individual risk of each patient. Adaptive treatment thus could be a viable option for the patient with high risk, e.g., more fractions of radiation, higher dose per fraction, etc. During the treatment planning, physician could also try to limit the organs at risk (OAR) exposure with more conservative dose due to the potential probability of retreatment later. Or simply being aware of the high risk patient groups, recommending more frequent follow-up and monitoring closely could be very beneficial for these patients’ prognosis compared to the general population. Although this work is able to provide preliminary guidance for the treatment planning based on the pre-treatment data, future work on adaptation of treatment plans (e.g., dose distribution) that customize better to the patient need to be investigated. Though we have conducted strict cross-validation to evaluate the performance, these identified biomarkers and clinical factors warrant further validation in independent, external and multi-institutional prospective studies to assess generalizability and further be applied to personalized treatment planning for HCC patients.
Conclusion
A new deep survival radiomics analysis was built based on supervised learning of imaging and clinical features for overall prognosis of liver cancer patients treated with SBRT, which showed better performance than Cox model. Interpretation of developed deep learning models suggested the importance of normal tissue status (radiomics features from liver-GTV, liver function clinical variables, and critical voxels highlighted in the normal liver regions) for the patient prognosis for overall survival, which can be used to personalize future liver cancer treatment with SBRT.
Supplementary Material
Table 2.
Univariate results using Cox model for significant radiomics variables (c-indices and corresponding p-values). There are two regions (GTV and liver-GTV) that the radiomics features were extracted from (column 1). 4 gray levels were used as the quantization levels for the extraction (column 2). The radiomics feature names were given as well (column 3).
| Regions | Gray levels | Radiomics Features | c-index for OS | p-value for OS |
|---|---|---|---|---|
| GTV | 32 | Correlation | 0.547 | 0.033 |
| Liver-GTV | 8 | ZSN | 0.586 | 0.008 |
| 16 | GLN | 0.585 | 0.014 | |
| 16 | ZSN | 0.596 | 0.015 | |
| 32 | ZSN | 0.578 | 0.047 | |
| 64 | SZHGE | 0.532 | 0.031 |
Highlights.
Novel VAE-based survival model combining radiomics, clinical features and raw CT images
Patch-based training for CNN that augmented heterogeneous data and improved overall performance
Investigations of correlation of clinical, imaging features and miRNA data
Interpretability of the DNN models: importance of different features
Prediction of the role of liver toxicity in overall survival
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Jemal A, Siegel R, Ward E, Hao Y, Xu J, Murray T, et al. Cancer statistics, 2008. CA Cancer J Clin. 2008;58:71–96. [DOI] [PubMed] [Google Scholar]
- 2.Llovet JM, Burroughs A, Bruix J. Hepatocellular carcinoma. Lancet. 2003;362:1907–17. [DOI] [PubMed] [Google Scholar]
- 3.Kwon JH, Bae SH, Kim JY, Choi BO, Jang HS, Jang JW, et al. Long-term effect of stereotactic body radiation therapy for primary hepatocellular carcinoma ineligible for local ablation therapy or surgical resection. Stereotactic radiotherapy for liver cancer. BMC Cancer. 2010;10:475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Raza A, Sood GK. Hepatocellular carcinoma review: current treatment, and evidence-based medicine. World journal of gastroenterology: WJG. 2014;20:4115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wahl DR, Stenmark MH, Tao Y, Pollom EL, Caoili EM, Lawrence TS, et al. Outcomes after stereotactic body radiotherapy or radiofrequency ablation for hepatocellular carcinoma. J Clin Oncol. 2016;34:452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schaub SK, Hartvigson PE, Lock MI, Høyer M, Brunner TB, Cardenes HR, et al. Stereotactic body radiation therapy for hepatocellular carcinoma: current trends and controversies. Technol Cancer Res Treat. 2018;17:1533033818790217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Feng M, Suresh K, Schipper MJ, Bazzi L, Ben-Josef E, Matuszak MM, et al. Individualized adaptive stereotactic body radiotherapy for liver tumors in patients at high risk for liver damage: a phase 2 clinical trial. JAMA oncology. 2018;4:40–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang C-y, Li S. Clinical characteristics and prognosis of 2887 patients with hepatocellular carcinoma: A single center 14 years experience from China. Medicine. 2019;98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Avanzo M, Wei L, Stancanello J, Vallières M, Rao A, Morin O, et al. Machine and deep learning methods for radiomics. 2020;47:e185–e202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wei L, Osman S, Hatt M, IJQNMMI El Naqa. Machine learning for radiomics-based multimodality and multiparametric modeling. 2019;63:323–38. [DOI] [PubMed] [Google Scholar]
- 11.Gillies RJ, Kinahan PE, Hricak H. Radiomics: images are more than pictures, they are data. Radiology. 2015;278:563–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vallières M, Freeman CR, Skamene SR, El Naqa I. A radiomics model from joint FDG-PET and MRI texture features for the prediction of lung metastases in soft-tissue sarcomas of the extremities. Phys Med Biol. 2015;60:5471. [DOI] [PubMed] [Google Scholar]
- 13.Tseng H-H, Wei L, Cui S, Luo Y, Ten Haken RK, El Naqa I. Machine learning and imaging informatics in oncology. Oncology. 2018:1–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Antropova N, Huynh BQ, Giger ML. A deep feature fusion methodology for breast cancer diagnosis demonstrated on three imaging modality datasets. Medical physics. 2017;44:5162–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Avanzo M, Stancanello J, El Naqa IJPM. Beyond imaging: the promise of radiomics. 2017;38:122–39. [DOI] [PubMed] [Google Scholar]
- 16.Peeken JC, Bernhofer M, Wiestler B, Goldberg T, Cremers D, Rost B, et al. Radiomics in radiooncology–challenging the medical physicist. 2018;48:27–36. [DOI] [PubMed] [Google Scholar]
- 17.Altazi BA, Fernandez DC, Zhang GG, Hawkins S, Naqvi SM, Kim Y, et al. Investigating multi-radiomic models for enhancing prediction power of cervical cancer treatment outcomes. 2018;46:180–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhou Y, He L, Huang Y, Chen S, Wu P, Ye W, et al. CT-based radiomics signature: a potential biomarker for preoperative prediction of early recurrence in hepatocellular carcinoma. Abdominal Radiology. 2017;42:1695–704. [DOI] [PubMed] [Google Scholar]
- 19.Shan Q-y, Hu H-t, Feng S-t, Peng Z-p, Chen S-l, Zhou Q, et al. CT-based peritumoral radiomics signatures to predict early recurrence in hepatocellular carcinoma after curative tumor resection or ablation. Cancer Imaging. 2019;19:11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ji G-W, Zhu F-P, Xu Q, Wang K, Wu M-Y, Tang W-W, et al. Radiomic Features at Contrast-enhanced CT Predict Recurrence in Early Stage Hepatocellular Carcinoma: A Multi-Institutional Study. Radiology. 2020:191470. [DOI] [PubMed] [Google Scholar]
- 21.Peng J, Qi X, Zhang Q, Duan Z, Xu Y, Zhang J, et al. A radiomics nomogram for preoperatively predicting prognosis of patients in hepatocellular carcinoma. Translational Cancer Research. 2018;7:936–46. [Google Scholar]
- 22.Guo D, Gu D, Wang H, Wei J, Wang Z, Hao X, et al. Radiomics analysis enables recurrence prediction for hepatocellular carcinoma after liver transplantation. Eur J Radiol. 2019;117:33–40. [DOI] [PubMed] [Google Scholar]
- 23.Cox DR. Regression models and life‐tables. Journal of the Royal Statistical Society: Series B (Methodological). 1972;34:187–202. [Google Scholar]
- 24.Harrell FE, Califf RM, Pryor DB, Lee KL, Rosati RA. Evaluating the yield of medical tests. Jama. 1982;247:2543–6. [PubMed] [Google Scholar]
- 25.Efron B. Better bootstrap confidence intervals. Journal of the American statistical Association. 1987;82:171–85. [Google Scholar]
- 26.Kira K, Rendell LA. A practical approach to feature selection. Machine Learning Proceedings 1992: Elsevier; 1992. p. 249–56. [Google Scholar]
- 27.Guyon I, Weston J, Barnhill S, Vapnik V. Gene Selection for Cancer Classification using Support Vector Machines. Machine Learning. 2002;46:389–422. doi: 10.1023/a:1012487302797. [DOI] [Google Scholar]
- 28.Ding C, Peng H. Minimum redundancy feature selection from microarray gene expression data. Journal of bioinformatics and computational biology. 2005;3:185–205. [DOI] [PubMed] [Google Scholar]
- 29.Kingma DP, Welling M. Auto-encoding variational bayes. arXiv preprint arXiv:13126114. 2013. [Google Scholar]
- 30.Ren Y, Tsai M-Y, Chen L, Wang J, Li S, Liu Y, et al. A manifold learning regularization approach to enhance 3D CT image-based lung nodule classification. 2020;15:287–95. [DOI] [PubMed] [Google Scholar]
- 31.Katzman JL, Shaham U, Cloninger A, Bates J, Jiang T, Kluger Y. DeepSurv: personalized treatment recommender system using a Cox proportional hazards deep neural network. BMC Med Res Methodol. 2018;18:24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kokhlikyan N, Miglani V, Martin M, Wang E, Reynolds J, Melnikov A, et al. PyTorch Captum. 2019. [Google Scholar]
- 33.Sundararajan M, Taly A, Yan Q. Axiomatic attribution for deep networks. arXiv preprint arXiv:170301365. 2017. [Google Scholar]
- 34.Köpüklü O, Kose N, Gunduz A, Rigoll G. Resource efficient 3d convolutional neural networks. 2019 IEEE/CVF International Conference on Computer Vision Workshop (ICCVW): IEEE; 2019. p. 1910–9. [Google Scholar]
- 35.Kay W, Carreira J, Simonyan K, Zhang B, Hillier C, Vijayanarasimhan S, et al. The kinetics human action video dataset. 2017. [Google Scholar]
- 36.Mobadersany P, Yousefi S, Amgad M, Gutman DA, Barnholtz-Sloan JS, Vega JEV, et al. Predicting cancer outcomes from histology and genomics using convolutional networks. 2018;115:E2970–E9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Traverso A, Kazmierski M, Zhovannik I, Welch M, Wee L, Jaffray D, et al. Machine learning helps identifying volume-confounding effects in radiomics. Physica Medica. 2020;71:24–30. [DOI] [PubMed] [Google Scholar]
- 38.Cozzi L, Dinapoli N, Fogliata A, Hsu W-C, Reggiori G, Lobefalo F, et al. Radiomics based analysis to predict local control and survival in hepatocellular carcinoma patients treated with volumetric modulated arc therapy. BMC Cancer. 2017;17:829. doi: 10.1186/s12885-017-3847-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kiryu S, Akai H, Nojima M, Hasegawa K, Shinkawa H, Kokudo N, et al. Impact of hepatocellular carcinoma heterogeneity on computed tomography as a prognostic indicator. Sci Rep. 2017;7:12689. doi: 10.1038/s41598-017-12688-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bakr SH, Echegaray S, Shah RP, Kamaya A, Louie J, Napel S, et al. Noninvasive radiomics signature based on quantitative analysis of computed tomography images as a surrogate for microvascular invasion in hepatocellular carcinoma: a pilot study. Journal of Medical Imaging. 2017;4:041303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chaudhary K, Poirion OB, Lu L, Garmire LX. Deep learning–based multi-omics integration robustly predicts survival in liver cancer. Clinical Cancer Research. 2018;24:1248–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hansmann J, Evers MJ, Bui JT, Lokken RP, Lipnik AJ, Gaba RC, et al. Albumin-bilirubin and platelet-albumin-bilirubin grades accurately predict overall survival in high-risk patients undergoing conventional transarterial chemoembolization for hepatocellular carcinoma. J Vasc Interv Radiol. 2017;28:1224–31. e2. [DOI] [PubMed] [Google Scholar]
- 43.Lee SK, Song MJ, Kim SH, Park M. Comparing various scoring system for predicting overall survival according to treatment modalities in hepatocellular carcinoma focused on Platelet-albumin-bilirubin (PALBI) and albumin-bilirubin (ALBI) grade: A nationwide cohort study. PLoS One. 2019;14:e0216173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kao W-Y, Su C-W, Chiou Y-Y, Chiu N-C, Liu C-A, Fang K-C, et al. Hepatocellular carcinoma: nomograms based on the albumin-bilirubin grade to assess the outcomes of radiofrequency ablation. Radiology. 2017;285:670–80. [DOI] [PubMed] [Google Scholar]
- 45.Biggins SW, Kim WR, Terrault NA, Saab S, Balan V, Schiano T, et al. Evidence-based incorporation of serum sodium concentration into MELD. Gastroenterology. 2006;130:1652–60. [DOI] [PubMed] [Google Scholar]
- 46.Huo T-I, Lin H-C, Hsia C-Y, Huang Y-H, Wu J-C, Chiang J-H, et al. The MELD-Na is an independent short-and long-term prognostic predictor for hepatocellular carcinoma: a prospective survey. Dig Liver Dis. 2008;40:882–9. [DOI] [PubMed] [Google Scholar]
- 47.Tang JY, Ohri N, Kabarriti R, Aparo S, Chuy J, Goel S, et al. Model for End-Stage Liver Disease and Sodium Velocity Predicts Overall Survival in Nonmetastatic Hepatocellular Carcinoma Patients. Canadian Journal of Gastroenterology and Hepatology. 2018;2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lai Q, Vitale A, Manzia TM, Foschi FG, Levi Sandri GB, Gambato M, et al. Platelets and Hepatocellular Cancer: Bridging the Bench to the Clinics. Cancers (Basel). 2019;11:1568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.El Naqa I, Owen D, Cuneo K, Mayo C, Lawrence T, Ten Haken R. Modeling of Locoregional Control in Hepatocellular Carcinoma after Stereotactic Body Radiation Therapy by Integrating Clinical and Immune Cell Profiles. International Journal of Radiation Oncology• Biology• Physics. 2018;102:S7. [Google Scholar]
- 50.Zhu Y, Li H, Guo W, Drukker K, Lan L, Giger ML, et al. Deciphering genomic underpinnings of quantitative MRI-based radiomic phenotypes of invasive breast carcinoma. Sci Rep. 2015;5:17787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhu Y, Mohamed AS, Lai SY, Yang S, Kanwar A, Wei L, et al. Imaging-genomic study of head and neck squamous cell carcinoma: Associations between radiomic phenotypes and genomic mechanisms via integration of The Cancer Genome Atlas and The Cancer Imaging Archive. JCO clinical cancer informatics. 2019;3:1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Aerts HJ, Velazquez ER, Leijenaar RT, Parmar C, Grossmann P, Carvalho S, et al. Decoding tumour phenotype by noninvasive imaging using a quantitative radiomics approach. Nature communications. 2014;5:1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hoshino I, Yokota H, Ishige F, Iwatate Y, Takeshita N, Nagase H, et al. Radiogenomics predicts the expression of microRNA-1246 in the serum of esophageal cancer patients. Sci Rep. 2020;10:1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.