Abstract
Background:
In contrast to most other common diseases, few genetic variants have been identified that impact risk of cirrhosis. We aimed to identify new genetic variants that predispose to cirrhosis, to test whether such variants, aggregated into a polygenic score, enable genomic risk stratification and to test whether alcohol intake or body mass index interact with polygenic predisposition.
Methods:
A multi-trait genome-wide association study (GWAS) combining cirrhosis and alanine aminotransferase (ALT) levels, performed in five discovery studies (UK Biobank, Vanderbilt BioVU, the Atherosclerosis Risk in Communities study, and two case-control studies containing 4,829 individuals with cirrhosis and 72,705 controls and 362,539 individuals with ALT levels). Identified variants were replicated in three studies (Partners HealthCare Biobank, FinnGen and Biobank Japan containing 3,554 individuals with cirrhosis and 343,826 controls). A polygenic score was tested in Partners HealthCare Biobank
Results:
Five previously-reported and seven newly-identified genetic variants were associated with cirrhosis in both the discovery studies multi-trait GWAS (p< 5×10−8) and the replication studies (p< 0.05), including a missense variant in the APOE gene and a noncoding variant near EFN1A. These 12 variants were used to generate a polygenic score. Among Partners HealthCare Biobank participants, high polygenic score -- defined as the top quintile of the distribution -- was associated with significantly increased risk of cirrhosis (OR 2.26, p<0.001) and related comorbidities when compared to the lowest quintile. Risk was even more pronounced among those with extreme polygenic risk (top 1% of the distribution, OR 3.16, p< 0.001). The impact of extreme polygenic risk was substantially more pronounced in those with elevated alcohol consumption or body mass index. Modeled as risk by age 75 years, probability of cirrhosis with extreme polygenic risk was 13.7%, 20.1%, and 48.2% among individuals with no or modest, moderate, and increased alcohol consumption, respectively (p-interaction< 0.001). Similarly, probability among those with extreme polygenic risk was 6.5%, 10.3%, and 19.5% among individuals with normal weight, overweight, and obesity, respectively (p-interaction< 0.001).
Conclusions:
Twelve independent genetic variants, 7 of which are newly-identified in this study, conferred risk for cirrhosis. Aggregated into a polygenic score, these variants identified a subset of the population at substantially increased risk who are most susceptible to the hepatotoxic effects of excess alcohol consumption or obesity.
Keywords: Cirrhosis, chronic liver disease, genome-wide association study, genetics
Short summary
Using a multi-trait statistical genetics approach, twelve independent common genetic variants that contribute to cirrhosis risk were identified. A polygenic score composed of these variants may have utility in identifying individuals at high risk of progression to cirrhosis.
Introduction
Cirrhosis of the liver is the eleventh leading cause of mortality worldwide.1 While environmental factors such as excessive alcohol consumption, chronic viral hepatitis, and obesity contribute to the development of cirrhosis, genetic factors also play an important role. In a study of twins, up to half of the observed variation in liver fibrosis was attributed to genetic factors.2 Pinpointing genetic variants that predispose to cirrhosis may have implications for our understanding of disease biology, for clinical risk stratification, and for developing targeted therapies for the treatment of liver fibrosis and cirrhosis.
Genome-wide association studies enable systematic assessment of the impact of common genetic variants on risk of a given disease, typically by comparing frequency in affected cases versus unaffected controls.3 This approach has been highly successful in identifying robust and reproducible associations across a range of important diseases, including more than 161 variants associated with coronary artery disease4 and 243 for type 2 diabetes.5 In contrast to most other common diseases, genome-wide association studies to date for cirrhosis have cumulatively identified fewer than ten genetic loci.6,7 Approaches to increase this yield include increasing sample size (the largest prior study to date included eighteen hundred affected cases7 versus sixty-five thousand in an analogous study of coronary artery disease8) and increasing statistical power by considering multiple correlated traits (e.g., cirrhosis and alanine aminotransferase levels) together.9,10
Whether lifestyle or anthropometric factors, such as alcohol consumption or body mass index, modify the inherited risk of cirrhosis remains largely unexplored. For other common diseases, such as coronary artery disease, type 2 diabetes and dementia, a healthy lifestyle was shown to offset increased genetic risk, but relative risk reductions were similar among those with low versus high genetic risk.11–13 By contrast, adiposity and insulin resistance have been shown to have a significantly greater relative risk impact on genetic predisposition to liver fat.14,15 Prior studies have suggested the obesity may amplify the association of the genetic variants with alanine aminotransferase (ALT) levels and cirrhosis.14,16,17 If lifestyle factors such as obesity and alcohol modify risk of cirrhosis, interventions to target these factors may substantially mitigate inherited predisposition to cirrhosis.
In this study, a case-control cohort of cirrhosis was assembled and a multi-trait genome-wide association study was conducted to identify new genetic variants impacting cirrhosis. This approach leveraged the known correlation between circulating ALT levels and cirrhosis risk.18 Next, a polygenic score that integrated information from all identified genetic variants was constructed and tested against risk of cirrhosis and related traits. Finally, the interaction of inherited polygenic risk with lifestyle and anthropometric factors -- alcohol consumption and obesity -- was examined.
Methods
Study Populations and Outcomes
Individuals with cirrhosis were identified in two prospective cohorts (UK Biobank and Atherosclerosis Risk in Communities study), a medical center-based cohort (Vanderbilt BioVU) and two case-control studies from previously published genome wide association studies of alcohol-related cirrhosis.7 Definitions of cirrhosis used in each of the five cohorts are provided (Supp. Tables 1 and 2). Cases of cirrhosis caused by primary biliary cholangitis (ICD K74.3) and primary sclerosing cholangitis (ICD K83.0) were excluded, as these autoimmune disorders are directed against the biliary (and not hepatic) parenchyma.19,20 To validate our ICD-coded definition, we manually reviewed pathology reports from two hundred patients who met our ICD-coded cirrhosis definition and underwent liver biopsy in Partners Biobank. 92% of patients who met the ICD-coded definition had cirrhosis on biopsy.
Across these five studies, 4,829 individuals with cirrhosis and 72,705 controls were identified. ALT levels were measured in 362,539 individuals in UK Biobank using a Beckman Coulter enzymatic assay. ALT levels were inverse normal transformed prior to analysis as inverse normal transformation increases power to detect genetic associations relative to a non-normally distributed quantitative trait.21
Multi-trait Genome Wide Association Study
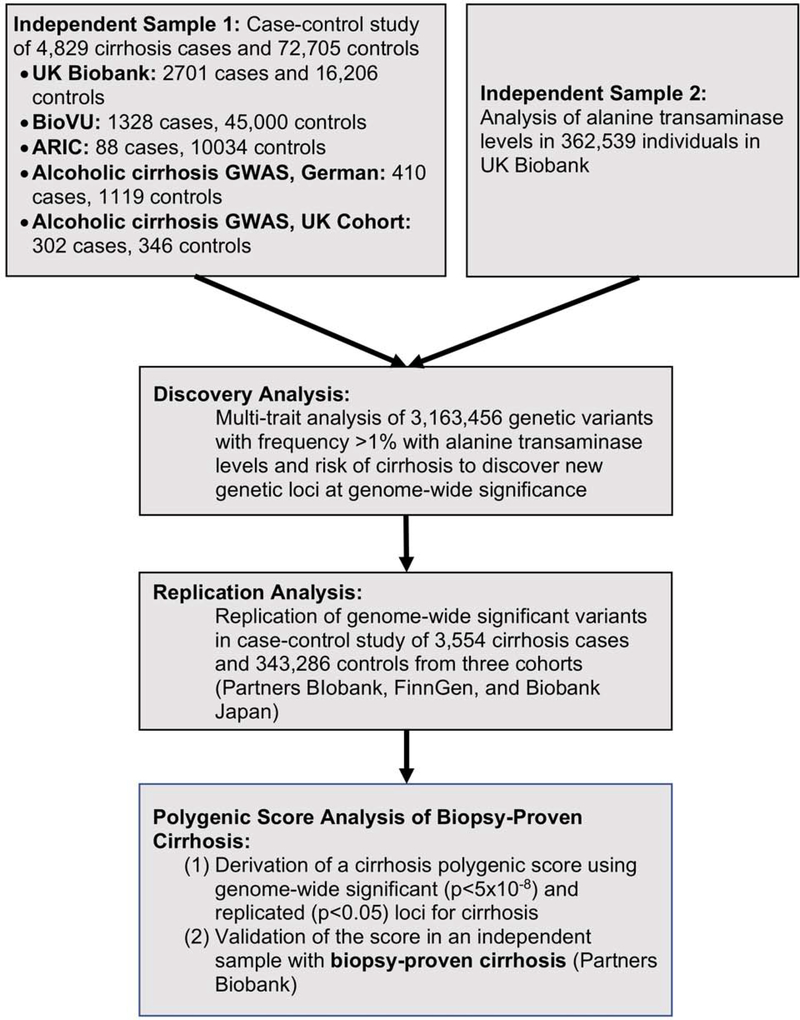
A multi-trait genome-wide association study was conducted to identify new genetic variants associated with cirrhosis. Study design is shown in Figure 1. Multi-trait analysis can provide a substantial increase in statistical power when two outcomes are correlated.9,10 ALT levels are a commonly used marker of hepatocyte injury.22 Individuals with chronic liver disease often have elevated ALT levels, and genetic variants which predispose to chronic liver disease are known to also increase ALT levels.23 Furthermore, when compared to other potential cirrhosis biomarkers, including aspartate aminotransferase (AST) levels, the Fibrosis-4 index and the AST to platelet ratio index (APRI), ALT more strongly associated with known cirrhosis variants (Supp. Table 3). ALT levels were therefore jointly analyzed with cirrhosis to increase our power to discover new variants associated with cirrhosis.
Figure 1.
Study design.
ARIC, Atherosclerosis Risk in Communities
In the UK Biobank, genotyping was performed using either the UK BiLEVE Axiom array or the UK Biobank Axiom array as previously reported.24 Phasing and imputation were performed centrally, by UK Biobank, using the Haplotype Reference Consortium and a reference panel of UK 10K merged with the 1000 Genomes phase 3 panel. One individual of each related pair of individuals (defined as second-degree relatives or closer), individuals whose genetic sex did not match self-reported sex and individuals with an excess of missing genotype calls or more heterozygosity than expected were excluded from analysis. For ARIC, genotyping was performed using the Affymetrix 6.0 array.25 Variants were imputed to the Haplotype Reference Consortium reference panel using the Michigan Imputation Server. For BioVU, genotyping was performed using the Illumina Multi-Ethnic Genotyping Array and variants were imputed to the Haplotype Reference Consortium.26 Ancestry of participants in each cohort is provided (Supp. Table 4).
For ALT levels, a genome-wide association study was performed in UK Biobank using linear regression with adjustment for age, sex and genetic ancestry, as quantified by the first ten principal components. For cirrhosis, a genome wide association study in each cohort was performed using logistic regression with adjustment for age, sex and ten principal components of ancestry. PLINK2 was used for all analyses.27 To combine estimates across cohorts, inverse variance fixed effects meta-analysis, as implemented by METAL, was used.28
The association of genetic variants with ALT levels and cirrhosis risk was then jointly analyzed using the Multi-trait Analysis of GWAS (MTAG) statistical genetics method.9 While genome-wide association studies typically analyze a single trait at a time, MTAG allows for the joint analysis of multiple traits. When two traits are correlated, such as ALT levels and cirrhosis risk, analysis of both traits can substantially increase power. For each variant, a multi-trait p-value for the association of the variant with cirrhosis is calculated. Variants were considered to be associated with cirrhosis in the discovery study if the p-value reached standard genome wide significance, less than p<5×10−8. We used an imputation quality threshold of <0.3 to exclude low quality imputed genetic variants, as previously performed.29 LD-score regression was used to test for the presence of population stratification.30
For genetic variants that associated with cirrhosis at genome-wide significance in the multi-trait discovery analysis, their association with cirrhosis was tested in independent replication studies composed of 3,554 cirrhosis cases and 343,286 controls from three cohorts - Partners BIobank, FinnGen, and Biobank Japan. In each cohort, estimates of the association of each variant with cirrhosis were deriving using logistic regression with adjustment for age, sex and principal components of ancestry. Estimates of the association of each variant with cirrhosis were pooled across cohorts using inverse variance-weighted fixed effects meta-analysis. Cirrhosis definitions in each cohort are provided (Supp. Tables 1 and 2). A p-value less than 0.05 was considered statistically significant in this replication study. In a sensitivity analysis, we also examined the association of identified variants with a more stringent definition of cirrhosis, restricted to individuals carrying ICD codes K70.3 (alcoholic cirrhosis) and K74.6 (other and unspecified cirrhosis of liver).
Polygenic Score Analysis
A polygenic score using the twelve genetic variants that were associated with cirrhosis at genome wide significance in the multi-trait discovery study and associated with cirrhosis in the independent replication sample was derived. For each individual in Partners Biobank and UK Biobank, the polygenic score was calculated by taking a weighted sum of cirrhosis-associated alleles, as previously performed.11 Specifically, for each individual, the number of alleles of twelve independent genetic variants, identified as associated with cirrhosis in the multi-trait analysis, was multiplied by the magnitude of association (natural log of the odds ratio observed in the logistic regression analysis) of each variant with cirrhosis. The polygenic score was then calculated for each individual as the sum of the weighted alleles. The polygenic score was standardized by ancestry to correct for differences in the score among individuals of differing genetic ancestry. To standardize the distribution of the polygenic score by ancestry, raw polygenic score values were corrected for genetic ancestry, creating an ancestry-adjusted polygenic score based on the residual of a linear regression that included the first 5 principal components of ancestry as previously described.31 To further account for any confounding due to a relationship between genetic ancestry and disease phenotypes, five principal components of ancestry were additionally included as covariates in the regression models.
The relationship of this polygenic score to cirrhosis and liver-related outcomes was tested in an independent cohort of 30,469 Partners HealthCare Biobank participants, none of whom were included in the derivation of the polygenic score (Supp. Table 5). This cohort, composed of patients of European ancestry in the Partners HealthCare System, included 1137 individuals with cirrhosis -- 339 individuals with biopsy-confirmed cirrhosis and an additional 798 individuals with cirrhosis ascertained through ICD codes. In Partners Biobank, we analyzed all-cause cirrhosis as assessed by ICD codes or biopsy confirmation. Sensitivity analyses were performed using only the subset of these individuals for whom cirrhosis was confirmed on biopsy. As in prior analyses11,32, individuals were classified as being at low, intermediate and high polygenic risk of cirrhosis if they were in the 0 to 19th percentile of the polygenic score, 20th to 79th percentile and 80th to 99th percentile respectively. The association of intermediate and high polygenic risk with cirrhosis, esophageal varices, hepatocellular carcinoma, liver-related death and all-cause death was tested. A Bonferroni-adjusted p-value of less than 0.01 (0.05/5) was considered significant for this analysis. Whether high polygenic risk predicted both alcohol-associated cirrhosis (cirrhosis among men reporting twenty-one drinks or more per week and women reporting fourteen drinks or more per week) and obesity-associated cirrhosis (cirrhosis among individuals with a body mass index greater than or equal to 30 kg/m2 who consumed less than twenty-one drinks per week for men and less than fourteen drinks per week for women) was tested, with alcohol and non-alcohol associated cirrhosis defined based on current clinical guidelines.33 Alcohol consumption in UK Biobank and Partners Biobank was ascertained in participants through a study questionnaire completed at time of enrollment.
Extreme polygenic risk (top 1%) analysis
Prior studies have documented a sharp increase in risk among those in the top 1% of a polygenic score distribution across a range of important diseases, in some cases conferring risk equivalent to that of rare, large-effect mutations.31 To determine if this observation was similarly true for cirrhosis, individuals in the top percentile of the cirrhosis polygenic score distribution were compared to the remainder of the score distribution. The risk conferred by ‘extreme’ polygenic risk was directly compared to the risk of cirrhosis for individuals who were carriers of known large-effect coding variants that predispose to cirrhosis in HFE, SERPINA1 and ATP7B that cause hereditary hemochromatosis, alpha-1-antitrypsin deficiency and Wilson’s disease respectively in UK Biobank (Supp Table 6). In a sensitivity analysis, we validated the association of the polygenic score in a subset of 339 individuals with biopsy-confirmed cirrhosis as compared to 12,708 controls free of known liver disease and with liver enzyme levels within the normal reference range (ALT less than 56 units per liter, AST levels less than 40 units per liter and alkaline phosphatase levels less than 147 units per liter).
Genotype-environment interaction analyses -- alcohol consumption and body mass index
Both alcohol consumption and obesity are strong risk factors for cirrhosis, but widespread heterogeneity in rates of cirrhosis have been described, even among those with similar alcohol consumption or body-mass index. Previous studies for diseases such as congestive heart failure have suggested that environmental factors preferentially ‘unmask’ inherited predisposition.34,35 To understand if a similar pattern might hold true for liver disease independent of genetic risk, we analyzed the interaction of polygenic risk with alcohol consumption or body mass index to confer risk for cirrhosis in both UK Biobank and in Partners HealthCare Biobank. Fine and Gray models with age as the underlying time variable were used to estimate cumulative risk of cirrhosis by age 75 years while accounting for both censoring from loss of follow up and the competing risk of death in each cohort. For body mass index, risk of cirrhosis among individuals with a body mass index less than 25 kg/m2, 25-<35 kg/m2 or 35 or more kg/m2 was examined. For alcohol consumption, risk of cirrhosis among individuals who consumed 14 or less drinks per week, 14 to 20 drinks per week or 21 drinks or more per week was examined.36 A formal interaction of polygenic risk with body mass index or alcohol consumption was tested by including an interaction term between the polygenic risk score as a continuous variable and either body mass or alcohol consumption as continuous variables within the Fine and Gray regression model with sex and ten principal components of ancestry as covariates. Tests for interaction were performed using continuous variables to both increase power and to prevent choice of quantiles of the underlying variable from influencing the result. To estimate cumulative risks at age 75 using clinically relevant categories, the subgroup of polygenic risk category and body mass index/alcohol consumption category was included in the model, as well as sex and ten principal components of ancestry as covariates with age as the underlying time variable. Proportional hazards assumption was examined through visual inspection of the Kaplan-Meier plot. The median age at cirrhosis diagnosis was 63 years.
Full summary statistics from this study are available at broad.cvdi.org. All statistical analysis was performed using R version 3.5.
Results
Multi-trait analysis identifies seven newly-identified loci for cirrhosis
A conventional single-trait GWAS for cirrhosis was first performed, testing the association of 3.1 million common (allele frequency > 1%) genetic variants with disease among 4,829 cases versus 72,705 controls (Supp Table 1). This analysis identified four variants previously associated with cirrhosis, near the genes PNPLA3, TM6SF2, HSD17B13 and SERPINA1, p < 5 × 10−8 for each (Figure 2A).23,37,38 Next, a second single-trait GWAS study was performed for concentrations of ALT. This analysis identified 139 associated variants, including in the PNPLA3, TM6SF2, HSD17B13, SERPINA1 and MARC1 gene regions (Figure 2B).39 As expected, across these 3.1 million common genetic variants, the effect of a variant on cirrhosis status was correlated with its effect on circulating ALT levels (linkage disequilibrium Pearson’s correlation coefficient = 0.48; p < 0.001).
Figure 2.
Discovery analysis of 4,829 cirrhosis cases, 72,705 controls free of cirrhosis and 362,539 individuals with ALT level measurements.
A total of 3.1 million snps are considered in this analysis. The red red horizontal line indicates the threshold for genome-wide significance (P = 5 × 10−8).
Having established a correlation between genomic determinants of cirrhosis and ALT levels, the recently developed multi-trait association GWAS algorithm was applied to integrate information into a single test of association with cirrhosis for each variant. This approach identified fifteen genomic variants associated with cirrhosis, including replication of five variants previously identified in prior studies (Figure 2C, Table 2).23,37–39 Reassuringly, no evidence of test statistic inflation -- as can occur due to inadequate adjustment for genetic ancestry -- was observed (intercept of 1.05 in an LD-score regression analysis).30 The five previously-identified variants included three that significantly increased risk of cirrhosis -- PNPLA3 p.Ile148Met (adjusted odds ratio [OR] 1.63, p=2.1 × 10−290), TM6SF2 p.Glu167Lys (OR 1.44, p=2.8 × 10−83) and SERPINA1 p.Gly366Lys (OR 2.03, p=7.8 × 10−56) -- and two that conferred protection: a splice-disrupting variant in HSD17B13 rs6834314 (OR 0.85; p=2.8 × 10−54) and MARC1 p.Ala165Thr (OR 0.91, p=1.2 × 10−43). Although it did not reach genome-wide significance, the previously reported variant rs641738 in MBOAT7 (membrane bound O-acyltransferase domain containing 7) also associated with cirrhosis (OR 1.11; p=9×10−5) in this analysis.7 In contrast, a variant near the GCKR (glucokinase regulatory protein) gene previously associated with non-alcoholic fatty liver disease was not associated with cirrhosis in this study (OR 0.98, p=0.42).40 A recent report identified an additional variant (rs15052) near the HNRNPUL1 gene associated with alcoholic cirrhosis.41 Although this variant did not reach genome-wide significance in the current analysis, our analysis of cirrhosis provides additional evidence in support of this association -- odds ratio 1.09; 95% CI 1.04 to 1.14; p = 0.0003.
Table 2.
Results of a multi-trait genome-wide association study jointly analyzing ALT levels with cirrhosis.
| Variant | Chr | Position | EA | RA | EAF, % | Consequence | ALT Effect (95% CI) | Cirrhosis OR (95% CI) | Multi-trait Cirrhosis P-value |
|---|---|---|---|---|---|---|---|---|---|
| Newly-identified | |||||||||
| rs12904 | 1 | 155106697 | A | G | 40 | Downstream, EFNA1 | −0.029 (−0.033, −0.024) | 0.90 (0.86, 0.95) | 1.7×10−37 |
| rs888655 | 5 | 72917439 | G | A | 26 | Upstream, ARHGEF28 | −0.016 (−0.021, −0.011) | 0.93 (0.88, 0.98) | 6.3×10−11 |
| rs9398804 | 6 | 126703390 | A | T | 45 | Intron, CENPW | −0.012 (−0.016, −0.008) | 0.93 (0.89, 0.98) | 3.8×10−8 |
| rs7029757 | 9 | 132566666 | A | G | 9 | Intron, TOR1B | −0.027 (−0.034, −0.019) | 0.85 (0.77, 0.93) | 1.4×10−12 |
| rs1799992 | 11 | 118957246 | C | T | 40 | Intron, HMBS | 0.013 (0.008, 0.017) | 1.23 (1.08, 1.43) | 1.1×10−8 |
| rs429358 | 19 | 45411941 | C | T | 15 | Missense, APOE C130R | −0.038 (−0.044, −0.032) | 0.85 (0.79, 0.92) | 7.0×10−36 |
| rs1883711 | 20 | 39179822 | C | G | 4 | Upstream, MAFB | 0.048 (0.036, 0.061) | 1.21 (1.05, 1.40) | 7.7×10−14 |
| Known | |||||||||
| rs2642438 | 1 | 220970028 | A | G | 29 | Missense, MARC1 A165T | −0.034 (−0.039, −0.029) | 0.91 (0.86, 0.96) | 1.2×10−43 |
| rs6834314 | 4 | 88213808 | G | A | 26 | Intron, HSD17B13 | −0.038 (−0.043, −0.033) | 0.85 (0.80, 0.91) | 2.8×10−54 |
| rs28929474 | 14 | 94844947 | T | C | 2 | Missense, SERPINA1 E366K | 0.124 (0.108, 0.140) | 2.03 (2.33, 1.77) | 7.8×10−56 |
| rs58542926 | 19 | 19379549 | T | C | 7 | Missense, TM6SF2 E167K | 0.081 (0.072, 0.089) | 1.44 (1.32, 1.56) | 2.8×10−83 |
| rs738409 | 22 | 44324727 | G | C | 23 | Missense, PNPLA3 I48M | 0.096 (0.091, 0.101) | 1.63 (1.54, 1.72) | 2.1×10−290 |
ALT: alanine aminotransferase, EAF: Effect allele frequency in European Non-Finnish ancenstry in gnomAD database.
Beyond the five previously-known variants ten variants were associated with cirrhosis at genome-wide significance in the multi-trait discovery analysis (Supp. Table 7). In the independent replication studies, all ten of these variants demonstrated directionally consistent associations with cirrhosis (binomial p=0.002). Seven of the ten variants were significantly associated with cirrhosis in the replication studies (p < 0.05 for each; Table 2, Supp. Table 7). Associations with cirrhosis were similar in a sensitivity analysis that used a more stringent ICD-coding based definition of cirrhosis, with no evidence of heterogeneity for any variant (p-heterogeneity > 0.05 for each, Supp. Table 8).
The newly-identified variants included the p.Cys130Arg missense variant in APOE, which was associated with a 15% lower odds of cirrhosis in the multi-trait genome wide analysis (p=7.0 × 10−36). A non-coding variant downstream of the EFNA1 gene, encoding the ephrin A1 receptor expressed on hepatocytes, associated with lower ALT levels and lower cirrhosis risk (OR 0.90, p =1.7 × 10−37). To understand the functional consequences of these variants, we (1) examined the effect of the variants on APOE and EFNA1 expression in all tissues and liver tissue using the Genotype Expression Dataset, (2) examined the effect of the variants on APOE and EFNA1 splicing in all tissues and liver tissue using the Genotype Expression Dataset, (3) examined the predicted consequences of their associated DNA sequence change on protein structure five in silico prediction algorithms (LRT score, MutationTaster, PolyPhen-2 HumDiv, PolyPhen-2 HumVar and Sorting Intolerant From Tolerant (SIFT)) as previously performed42 and (4) conducted a phenome-wide association study on 31 disease phenotypes in UK Biobank.43
APOE Cys130Arg did not effect APOE expression in liver tissue or in any other tissue in the Genotype Tissue Expression dataset and did not affect APOE splicing in liver tissue or other tissue.44 It was predicted to have a neutral effect on APOE structure by all five of the in silico prediction algorithms. Although this variant has not previously been identified to relate to liver disease, it is one of two missense variants that comprise the well-studied APOε4 variant. Prior work has linked APOε4 to a substantially increased risk of Alzheimer’s disease45 and coronary artery disease46 and prior functional work has demonstrated that this variant modified APOE structure to enhance binding to very low density lipoprotein (VLDL) particles. Similar to prior analyses, APOε4 carrier status was associated with increased risk of dementia and coronary artery disease in UK Biobank. However, APOε4 was associated with protection from cirrhosis or fatty liver. As compared to those who did not carry the APOε4 variant, participants who carried two APOε4 alleles at 19% lower odds of cirrhosis and 48% lower odds of fatty liver (p trend = 0.002 and 4.8×10−5 respectively, Supp. Table 9). In a phenome wide association study of 31 different diseases, this variant also associated with lower risk of gallstones (OR 0.93, p=1.4×10−5).”
EFNA1 rs12904 is a non-coding variant located in the 3’ untranslated region of EFNA1. It is not predicted to result in a change in the protein sequence of EFNA1. In the Genotype Tissue Expression dataset, it is predicted to reduce expression of EFNA1 across a range of tissue types, including gastrointestinal tissue, neural tissue and vascular tissue.44 This variant is a splice quantitative trait locus for EFNA1 in the liver in the Genotype Tissue Expression dataset, meaning that carriers of the protective minor allele have reduced excision of an intron of EFNA1 study (208 individuals with liver tissue, p=2.8 × 10−18).44 These findings suggest that this variant reduces expression of EFNA1 and damages excision of an intron from the protein. In a phenome wide association study of 31 different diseases, this variant also associated with a lower risk of gout (OR 0.92, p=1.5×10−6).
Population attributable fraction for individual identified variants ranged from 0.8% to 13.8% (Supp. Table 10). Total population attributable fraction for elevated genetic risk for cirrhosis was 55%, indicating that 55% of cirrhosis cases could be averted if all individuals in the population were at low polygenic risk of cirrhosis (bottom 20% of the polygenic score, Supp. Table 10).
High polygenic risk predisposes to liver disease in two cohorts
Having identified twelve genetic variants that independently confer risk of cirrhosis, a polygenic score was next constructed to integrate information from each of these sites of DNA variation into a single number for each participant. The association of the polygenic score with liver disease was tested in UK Biobank. Individuals at low (<20th percentile), intermediate (20th to 79th percentile) and high polygenic risk (≥80th percentile) were identified. High polygenic risk (defined as the top 20% of the population) was associated with an elevated risk of cirrhosis (OR 2.12, p=3.3×10−40) and hepatocellular carcinoma (OR 2.13, p=1.5×10−5) relative to individuals at low polygenic risk (Supp Figure 1). Importantly, the magnitude of risk conferred appeared similar across physician-coded etiologic assessment of cirrhosis, including alcohol-associated cirrhosis (OR 1.94, p=8.9×10−7) and obesity-associated cirrhosis (OR 2.61, p=5.5×10−11). Consistent with this effect on cirrhosis, a two-fold risk gradient in risk of esophageal varices (OR 2.37 p=2.6 × 10−14) and a two-fold risk in hepatocellular carcinoma (OR 2.13, p=1.5×10−5) was observed.
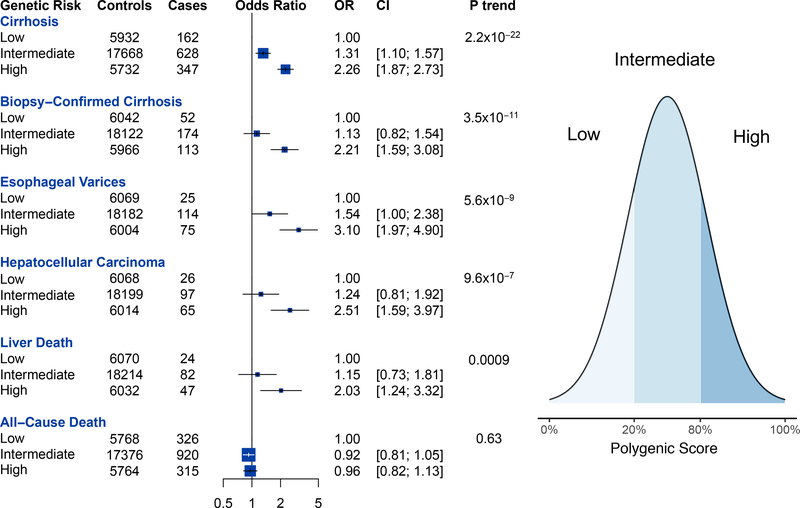
Next, the association of the polygenic score with liver disease in an independent testing dataset was examined. This cohort, composed of patients of European ancestry in the Partners HealthCare System, included 1137 individuals with cirrhosis ascertained through ICD codes of whom 339 individuals had cirrhosis confirmed on biopsy. Individuals at low (<20th percentile), intermediate (20th to 80th percentile) and high polygenic risk (>80th percentile) were identified. Individuals at high polygenic risk were at two-fold risk of cirrhosis (OR 2.26, p=2.2 × 10−22), three-fold risk of esophageal varices (OR 3.10, p=5.6×10−9) and 2.5 fold risk of hepatocellular carcinoma (OR 2.51, p=9.6 × 10−7, Figure 3). High polygenic risk was also associated with elevated risk of biopsy-confirmed cirrhosis (OR 2.21, p=3.5×10−11). Per standard deviation increase, the polygenic score was associated with elevated risks of cirrhosis (OR 1.32, p=1.1×10−22), biopsy-confirmed cirrhosis (OR 1.39, p=1.2×10−10), esophageal varices (OR 1.45, p=5.6×10−9), hepatocellular carcinoma (OR 1.39, p= 9.6×10−7) and death from liver disease (OR 1.29, p=0.0009) but was not associated with all-cause death (OR 0.99, p=0.63). Individuals in Partners Biobank at high polygenic risk had an 8.8% cumulative risk of cirrhosis at age 75 versus a 3.2% cumulative risk for individuals at low polygenic risk at age 75.
Figure 3.
Association of polygenic risk for cirrhosis with liver disease and death in the Partners HealthCare Biobank.
Estimates were derived using logistic regression with adjustment for age, sex and five principal components of ancestry. Low polygenic risk was defined as the 0th to 19th percentile of the polygenic score. Intermediate polygenic risk was defined as the 20th to 79th percentile. High polygenic risk was defined as the 80th to 100th percentile.
In an exploratory analysis involving 1802 black participants of the Partners Biobank, individuals at intermediate polygenic risk were at elevated risk of cirrhosis compared to those at low risk (OR 3.63 CI 1.55, 8.5, p=0.003). A nonsignificant trend towards higher cirrhosis risk was observed in the high polygenic risk category (OR 2.44 CI 0.92, 6.48, p=0.072, Supp. Table 11). The association of the polygenic score was largely consistent across individuals stratified by hepatitis B or hepatitis C status (Supp. Table 12). For individuals with hepatitis B, high polygenic risk (top 20 percent of the polygenic score) was associated with five-fold risk of cirrhosis (OR 4.73 CI 1.12, 20, p=0.03). For individuals with hepatitis C, high polygenic risk was associated with two-fold risk of cirrhosis (OR 2.20 CI 1.20, 4.05, p=0.01). A polygenic score composed of only the seven newly-identified variants independently associated with cirrhosis in Partners Biobank (OR 1.12 CI 1.05, 1.18 per SD of the score, p=0.0003, Supp. Table 13). The polygenic score continued to be associated with cirrhosis risk when 1389 individuals with non-cirrhotic liver disease (fatty liver, primary biliary cholangitis, primary sclerosing cholangitis or autoimmune hepatitis) were excluded and when they were controlled for as a binary variable (Supp. Table 14).
Extreme polygenic risk can identify individuals at markedly elevated risk for cirrhosis
Polygenic scores for other diseases have noted even more pronounced risk when examining extreme tails of the score distribution. Within the Partners HealthCare Biobank, extreme polygenic score -- defined as the top 1% of the score distribution -- was associated with a more than three-fold risk of cirrhosis (OR 3.16, p= 3×10−7) and a six-fold risk of biopsy-confirmed cirrhosis (OR 6.12, p=7.9×10−11 Figure 4). In order to put the clinical importance of a very high polygenic score in context of other known genetic risk factors, the association of extreme polygenic score to cirrhosis was directly compared to known rare large-effect genetic variants. Such participants included homozygous carriers of the HFE p.Cys282Tyr variant causal for hemochromatosis, homozygous carriers of SERPINA1 p.Glu366Lys causal for alpha-1-antitrypsin deficiency (Z/Z genotype) or ATP7B loss-of-function variants causal for Wilson’s disease. In aggregate, 1.6% of patients with fibrosis or cirrhosis harbored any of these three genetic risk factors versus 0.5% of controls -- OR 2.98, p = 6×10−5 (Figure 4). When 19 individuals who were homozygous for rs28929474 (SERPINA1 E366K) were excluded from analysis, individuals at extreme polygenic risk continued to be at highly elevated risk of biopsy-diagnosed cirrhosis (OR 5.96, p=5×10−10).
Figure 4.
Association of extreme polygenic risk or rare large-effect mutations with cirrhosis. HFE, SERPINA1 or ATP7B refer to HFE p.Cys282Tyr, SERPINA1 p.Glu366Lys (Z/Z genotype) and ATP7B loss-of-function mutations in the homozygous state.
Estimates were derived using logistic regression with adjustment for age, sex and five principal components of ancestry. OR, odds ratio
Alcohol consumption and body mass index amplify risk of liver disease conferred by extreme polygenic risk in two cohorts
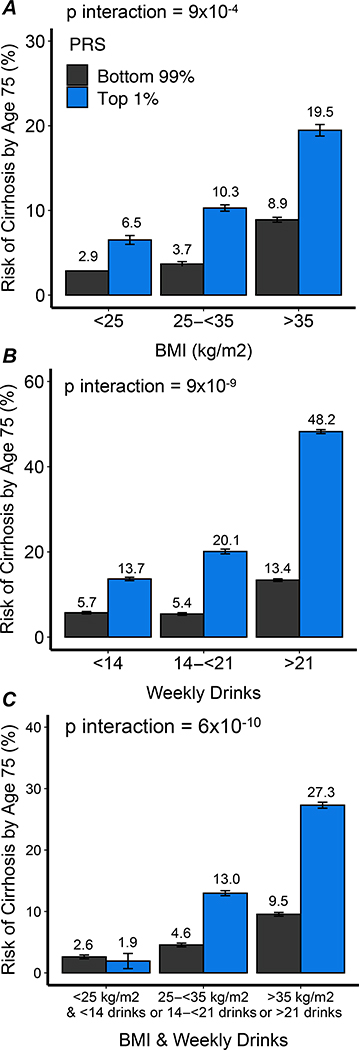
To examine the interplay of polygenic risk and environmental factors in determining risk of cirrhosis, Partners HealthCare Biobank were next stratified according to alcohol consumption and weight. Alcohol consumption and body mass index did not differ by polygenic predisposition to cirrhosis (Supp. Tables 15 and 16). Among individuals at low and high polygenic risk in Partners Biobank, 70% and 72% consumed less than 14 drinks, respectively, while 5% and 5% consumed more than 21 drinks per week, respectively. Among individuals at low and high polygenic risk in Partners Biobank, 38% and 38% had a BMI of less than 25 kg/m2, respectively, while 11% and 11% had a BMI of 35 kg/m2 or more, respectively. However, the impact of polygenic risk on cirrhosis was substantially more pronounced in those with either of these risk factors, with an overall p-interaction = 9×10−9 for alcohol consumption and an overall p-interaction =0.0009 for elevated BMI. When modeled as absolute probability of cirrhosis by age 75 years using a Fine and Gray model, probability of cirrhosis with high polygenic risk was 13.7%, 20.1%, and 48.2% among individuals with minimal or modest, moderate, and increased alcohol consumption, respectively, in Partners HealthCare Biobank (Figure 5). Similarly, probability among those with high polygenic risk was 6.5%, 10.3%, and 19.5% among individuals with normal weight, overweight, and obesity, respectively, in Partners HealthCare Biobank respectively. For those individuals with high polygenic risk who both maintained normal weight and avoided excess alcohol consumption, probability of cirrhosis was similar to those with normal polygenic risk -- 2.6% versus 1.9% respectively. Individuals at extreme polygenic risk who were obese or consumed alcohol in excess had an estimated probability of cirrhosis ten-fold higher than those who maintained normal bodyweight and alcohol consumption, 27.3% versus 1.9% respectively. A similar interaction by alcohol consumption and body mass index was observed among UK Biobank participants (Supp. Figure 2). An interaction by alcohol consumption and body mass index was observed among both male and female participants in UK Biobank (Supp. Figure 3).
Figure 5.
Association of alcohol consumption and body weight with cumulative risk of cirrhosis among individuals at extreme and non-extreme polygenic risk of cirrhosis. A. Cumulative risk of cirrhosis by body mass index. B. Cumulative risk of cirrhosis by alcohol consumption. C. Cumulative risk of cirrhosis by body-mass index and alcohol consumption.
Extreme polygenic risk was defined as the top percentile of the polygenic score. BMI, body-mass index. PRS, polygenic risk score.
Discussion
In this study, increased sample size and a new statistical genetics method were used to identify twelve independent genetic variants that impact risk of all-cause cirrhosis, including newly-identified variants in the APOE and EFNA1 gene regions. A polygenic score, which integrated risk information from each of these twelve variants was constructed. Individuals at extreme polygenic risk were strongly predisposed to liver disease and liver cancer. Alcohol consumption and increased body mass index amplified the association of polygenic risk with cirrhosis. Individuals at extreme polygenic risk who were obese or consumed alcohol in excess had an estimated probability of cirrhosis ten-fold higher than those who maintained normal bodyweight and alcohol consumption, 27.3% versus 2.6% respectively.
The identification of genetic variants associated with cirrhosis may have implications for understanding of disease biology and potential therapeutic strategies. Of note, the recently reported protective association of MARC1 p.Ala165Thr with cirrhosis was confirmed and extended in this study.39 Among the newly-identified variants, a common variant in a noncoding region of the genome downstream of the Ephrin A1 (EFNA1) gene was associated with a reduced risk of cirrhosis. Ephrin A1 is a transmembrane receptor highly expressed by hepatocytes, known to be upregulated in cirrhosis and liver cancer.47 Within the liver, ephrin A1 has been reported to induce expression of hepatocyte growth and promote angiogenesis.47 An antisense oligonucleotide targeting ephrin A1 inhibits hepatocellular carcinoma cell lines.47 Interestingly APOε4 carriers, at elevated risk of both Alzheimer’s disease and coronary artery disease, were found to be at significantly lower risk of cirrhosis. This finding suggests that APOE, like PNPLA3 and TM6SF2, has opposing effects on risk of coronary artery disease and liver disease.48
From a methodologic perspective, the multi-trait approach applied in the current study substantially improved power to detect new genetic variants associated with cirrhosis. By jointly analyzing ALT levels and cirrhosis, the number of variants identified in the current study increased from four to twelve. Prior analyses of polygenic scores for prediction of cirrhosis have largely focused on one to four variants but have shown that the combined effects of PNPLA3 p.I48M and TM6SF2 p.E147K predict progression to cirrhosis.49,50 In an analysis of two hundred non-alcoholic fatty liver disease cases and two hundred controls, a polygenic score predicted the prevalence of fatty liver with an odds ratio of 3 for the highest category of the score.50 We extend this prior work by analyzing the combined effects of twelve variants at a large scale with more than thirty thousand participants of the Partners Biobank replication cohort. Development of genome-wide polygenic scores -- which integrate information from even more variants -- may further enhance risk stratification for cirrhosis. Previous familial studies have shown that one in five first-degree relatives of probands with cirrhosis may have advanced fibrosis.51 Whether multi-trait polygenic scores could be utilized in these families to dissect out the first-degree relatives with a higher risk of progression to cirrhosis warrants further study..
Individuals at extreme polygenic risk were at markedly-elevated risk of cirrhosis and liver-related death. However, this excess risk of liver disease was amplified by both body mass index and by alcohol consumption, with significant interactions by both risk factors. Individuals at extreme polygenic risk who consumed alcohol in moderation had a significantly lower risk of developing cirrhosis by age 75 compared to those who consumed alcohol in excess (13.7% risk versus 48.2% risk). These findings may help account for widespread variability in rates of cirrhosis, even among individuals who consume similarly high amounts of alcohol.52
Similarly, individuals at extreme polygenic risk who maintained normal body weight had a three-fold lower risk of developing end-stage liver disease by age 75 compared to those who were obese. These findings are consistent with the observation that genetic variants predisposing to nonalcoholic fatty liver disease have greater effects on liver fat with increasing body mass index.14 They suggest that individuals at elevated polygenic risk of cirrhosis may be able to offset increased genetic risk through weight loss, and suggest that interventions to promote weight loss, including pharmacotherapy or bariatric surgery, may have increased efficacy in mitigating excess risk of liver disease among individuals at high genetic risk of cirrhosis.14
One important limitation of this study is that, among the cirrhosis cases analyzed in both UK Biobank and Partners Biobank, hepatitis B or hepatitis C was not systematically assessed. Additional research is needed to determine if the polygenic score developed in this study predicts cirrhosis among individuals exposed to hepatitis B or hepatitis C. Similarly, the current score was derived and validated in populations of predominantly European ancestry. Further research is required to determine the performance of the score in non-European populations. The association of the score with liver-related mortality but not all-cause mortality suggests that further analyses are needed to understand the relationship between genetic predisposition to cirrhosis and non-liver related mortality.53 Although we did not observe a significant association between a polygenic score for cirrhosis and all-cause mortality, our analysis may have been underpowered to detect an association and warrants further exploration in larger datasets.
In conclusion, we performed a multi-trait genome wide association study and identified seven newly-identified loci for cirrhosis. We observed a ten-fold increase in the risk of cirrhosis due to combined effects of alcohol and obesity among those who are genetically susceptible to develop cirrhosis. These data suggest that polygenic scores may have use in stratifying patients at risk for progressive liver disease and suggest clear lifestyle guidance to minimize alcohol use and maintain normal weight among those with the highest genetic risk for developing cirrhosis.
Supplementary Material
Table 1.
Baseline characteristics of analyzed cohorts.
| Discovery Studies | Replication Studies | |||||||
|---|---|---|---|---|---|---|---|---|
| UK Biobank, n=405,569 | BioVU, n=46,328 | ARIC, n=10,122 | Buch et al. German cohort7, n=1490 | Buch et al. UK Cohort7, n=648 | Partners Biobank, n=30,469 | Biobank Japan54, n=212,453 | FinGen55, n=94918 | |
| Data analyzed | Individual-level data | Individual-level data | Individual-level data | Summary-level association statistics | Summary-level association statistics | Individual-level data | Summary-level association statistics | Summary-level association statistics |
| Mean Age, yrs (SD) | 57 (8) | 60 (20) | 54 (6) | 45 (17) | 51 (18) | 59 (17) | 62 (14) | 58 (17) |
| Female sex | 218,376 (54%) | 25545 | 5501 (54%) | 119 (8%) | 176 (27%) | 16,103 (53%) | 99,747 (47%) | 54,262 (57%) |
| Mean BMI, kg/m2 (SD) | 27 (5) | 28 (14) | 28 (5) | 25 (7) | 25 (5) | 28 (6) | 23 (4) | 27 (4) |
| Type 2 Diabetes, n (%) | 20458 (5%) | 8795 (20%) | 909 (9%) | 142 (10%) | 0 (0%) | 2537 (8%) | 40,250 (19%) | 11,006 (12%) |
| Weekly alcoholic drinks | 8 (10) | NA | 3 (4) | ≥ 30 drinks | ≥ 30 drinks | 4 (7) | NA | NA |
| Cirrhosis | 2701 (1%) | 1,328 (3%) | 88 (1%) | 410 (28%) | 302 (47%) | 1137 (4%) | 2184 (1%) | 233 (0.2%) |
NA, not available.
What you need to know.
Background and context:
Whether common genetic variation influences risk of cirrhosis is unclear.
New findings:
Using a multi-trait statistical genetics approach, twelve independent common DNA sequence variants, including seven novel variants, were found to associate with cirrhosis risk. When aggregated into a polygenic risk score, these variants identified a subset of the population at several-fold increased risk who are most susceptible to the hepatotoxic effects of obesity or excess alcohol consumption.
Limitations:
Study participants were predominantly of European ancestry. Further research is required to determine the performance of the polygenic score in non-European populations.
Impact:
Polygenic scores of common variants may be useful for identifying patients at highly elevated risk of liver disease and identifying patients who may substantially benefit from weight loss and alcohol consumption reduction.
Acknowledgements:
This research has been conducted using the UK Biobank resource, application 7089. Funding support was provided by grants 1K08HG010155 (to A.V.K.) and 5UM1HG008895 (to S.K., A.V.K.) from the National Human Genome Research Institute, a Hassenfeld Scholar Award from Massachusetts General Hospital (to A.V.K.), a Merkin Institute Fellowship from the Broad Institute of MIT and Harvard (to A.V.K.), and a sponsored research agreement from IBM Research (to A.V.K.).
Competing financial interests: CAE has served as a consultant or received honoraria from Acceleron Pharma, Deerfield Management, Navitor Pharma, Nference, Novartis and Verve Therapeutics. JH, CN and AZ are employees of Color Genomics. SK is an employee of Verve Therapeutics and has received a research grant from Bayer Healthcare; and consulting fees from Merck, Novartis, Sanofi, AstraZeneca, Alnylam Pharmaceuticals, Leerink Partners, Noble Insights, MedGenome, Aegerion Pharmaceuticals, Regeneron Pharmaceuticals, Quest Diagnostics, Color Genomics, Genomics PLC, and Eli Lilly and Company; and holds equity in San Therapeutics, Catabasis Pharmaceuticals, Verve Therapeutics and Maze Therapeutics. A.V.K. has served as a consultant to Sanofi, Medicines Company, Maze Pharmaceuticals, Navitor Pharmaceuticals, Verve Therapeutics, Amgen, and Color Genomics; received speaking fees from Illumina, the Novartis Institute for Biomedical Research; received sponsored research agreements from the Novartis Institute for Biomedical Research and IBM Research, and reports a patent related to a genetic risk predictor (20190017119). All other authors have reported that they have no relationships relevant to the contents of this paper to disclose.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.GBD 2017 Mortality Collaborators. Global, regional, and national age-sex-specific mortality and life expectancy, 1950–2017: a systematic analysis for the Global Burden of Disease Study 2017. Lancet. 2018;392(10159):1684–1735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Loomba R, Schork N, Chen C-H, et al. Heritability of Hepatic Fibrosis and Steatosis Based on a Prospective Twin Study. Gastroenterology. 2015;149(7):1784–1793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pearson TA, Manolio TA. How to interpret a genome-wide association study. JAMA. 2008;299(11):1335–1344. [DOI] [PubMed] [Google Scholar]
- 4.van der Harst P, Verweij N. Identification of 64 Novel Genetic Loci Provides an Expanded View on the Genetic Architecture of Coronary Artery Disease. Circ Res. 2018;122(3):433–443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mahajan A, Taliun D, Thurner M, et al. Fine-mapping type 2 diabetes loci to single-variant resolution using high-density imputation and islet-specific epigenome maps. Nat Genet. 2018;50(11):1505–1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen VL, Chen Y, Du X, Handelman SK, Speliotes EK. Genetic variants that associate with cirrhosis have pleiotropic effects on human traits. Liver Int. December 2019. doi: 10.1111/liv.14321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Buch S, Stickel F, Trépo E, et al. A genome-wide association study confirms PNPLA3 and identifies TM6SF2 and MBOAT7 as risk loci for alcohol-related cirrhosis. Nat Genet. 2015;47(12):1443–1448. [DOI] [PubMed] [Google Scholar]
- 8.Klarin D, Zhu QM, Emdin C, et al. Genetic analysis in UK Biobank links insulin resistance and transendothelial migration pathways to coronary artery disease. Nat Genet. 2017;49(9):1392–1397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Turley P, Walters RK, Maghzian O, et al. Multi-trait analysis of genome-wide association summary statistics using MTAG. Nat Genet. 2018;50(2):229–237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Craig JE, Han X, Qassim A, et al. Multitrait analysis of glaucoma identifies new risk loci and enables polygenic prediction of disease susceptibility and progression. Nat Genet. January 2020:1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Khera AV, Emdin C, Drake I, et al. Genetic Risk, Adherence to a Healthy Lifestyle, and Coronary Disease. N Engl J Med. 2016;375(24):2349–2358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lourida I, Hannon E, Littlejohns TJ, et al. Association of Lifestyle and Genetic Risk With Incidence of Dementia. JAMA. July 2019. doi: 10.1001/jama.2019.9879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nettleton JA, Hivert M-F, Lemaitre RN, et al. Meta-analysis investigating associations between healthy diet and fasting glucose and insulin levels and modification by loci associated with glucose homeostasis in data from 15 cohorts. Am J Epidemiol. 2013;177(2):103–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Stender S, Kozlitina J, Nordestgaard BG, Tybjærg-Hansen A, Hobbs HH, Cohen JC. Adiposity amplifies the genetic risk of fatty liver disease conferred by multiple loci. Nat Genet. 2017;49(6):842–847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Barata L, Feitosa MF, Bielak LF, et al. Insulin Resistance Exacerbates Genetic Predisposition to Nonalcoholic Fatty Liver Disease in Individuals Without Diabetes. Hepatol Commun. 2019;3(7):894–907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Romeo S, Sentinelli F, Dash S, et al. Morbid obesity exposes the association between PNPLA3 I148M (rs738409) and indices of hepatic injury in individuals of European descent. Int J Obes. 2010;34(1):190–194. [DOI] [PubMed] [Google Scholar]
- 17.del Giudice EM, Grandone A, Cirillo G, et al. The association of PNPLA3 variants with liver enzymes in childhood obesity is driven by the interaction with abdominal fat. PLoS One. 2011;6(11):e27933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Alexander M, Loomis AK, van der Lei J, et al. Risks and clinical predictors of cirrhosis and hepatocellular carcinoma diagnoses in adults with diagnosed NAFLD: real-world study of 18 million patients in four European cohorts. BMC Med. 2019;17(1):95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Qiu F, Tang R, Zuo X, et al. A genome-wide association study identifies six novel risk loci for primary biliary cholangitis. Nat Commun. 2017;8:14828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ji S-G, Juran BD, Mucha S, et al. Genome-wide association study of primary sclerosing cholangitis identifies new risk loci and quantifies the genetic relationship with inflammatory bowel disease. Nat Genet. 2017;49(2):269–273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McCaw ZR, Lane JM, Saxena R, Redline S, Lin X. Operating characteristics of the rank-based inverse normal transformation for quantitative trait analysis in genome-wide association studies. Biometrics. December 2019. doi: 10.1111/biom.13214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kim WR, Flamm SL, Di Bisceglie AM, Bodenheimer HC, Public Policy Committee of the American Association for the Study of Liver Disease. Serum activity of alanine aminotransferase (ALT) as an indicator of health and disease. Hepatology (Baltimore, Md). 2008;47(4):1363–1370. http://doi.wiley.com/10.1002/hep.22109. [DOI] [PubMed] [Google Scholar]
- 23.Abul-Husn NS, Cheng X, Li AH, et al. A Protein-Truncating HSD17B13 Variant and Protection from Chronic Liver Disease. N Engl J Med. 2018;378(12):1096–1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bycroft C, Freeman C, Petkova D, et al. The UK Biobank resource with deep phenotyping and genomic data. Nature. 2018;562(7726):203–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Psaty BM, O’Donnell CJ, Gudnason V, et al. Cohorts for Heart and Aging Research in Genomic Epidemiology (CHARGE) Consortium: Design of prospective meta-analyses of genome-wide association studies from 5 cohorts. Circ Cardiovasc Genet. 2009;2(1):73–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Roden DM, Pulley JM, Basford MA, et al. Development of a large-scale de-identified DNA biobank to enable personalized medicine. Clin Pharmacol Ther. 2008;84(3):362–369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chang CC, Chow CC, Tellier LC, Vattikuti S, Purcell SM, Lee JJ. Second-generation PLINK: rising to the challenge of larger and richer datasets. Gigascience. 2015;4(1):7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Willer CJ, Li Y, Abecasis GR. METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics. 2010;26(17):2190–2191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Emdin C, Khera AV, Chaffin M, et al. Analysis of predicted loss-of-function variants in UK Biobank identifies variants protective for disease. Nat Commun. 2018;9(1):1613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bulik-Sullivan BK, Loh P-R, Finucane HK, et al. LD Score regression distinguishes confounding from polygenicity in genome-wide association studies. Nat Genet. 2015;47(3):291–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Khera AV, Chaffin M, Aragam KG, et al. Genome-wide polygenic scores for common diseases identify individuals with risk equivalent to monogenic mutations. Nat Genet. 2018;50(9):1219–1224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mega JL, Stitziel NO, Smith JG, et al. Genetic risk, coronary heart disease events, and the clinical benefit of statin therapy: an analysis of primary and secondary prevention trials. Lancet. 2015;385(9984):2264–2271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chalasani N, Younossi Z, Lavine JE, et al. The diagnosis and management of nonalcoholic fatty liver disease: Practice guidance from the American Association for the Study of Liver Diseases. Hepatology. 2018;67(1):328–357. [DOI] [PubMed] [Google Scholar]
- 34.Ware JS, Amor-Salamanca A, Tayal U, et al. Genetic Etiology for Alcohol-Induced Cardiac Toxicity. J Am Coll Cardiol. 2018;71(20):2293–2302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Qi Q, Chu AY, Kang JH, et al. Sugar-sweetened beverages and genetic risk of obesity. N Engl J Med. 2012;367(15):1387–1396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Karlson E, Boutin N, Hoffnagle A, Allen N. Building the partners healthcare biobank at partners personalized medicine: informed consent, return of research results, recruitment lessons and operational considerations. Journal of personalized medicine. 2016;6(1):2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kozlitina J, Smagris E, Stender S, et al. Exome-wide association study identifies a TM6SF2 variant that confers susceptibility to nonalcoholic fatty liver disease. Nat Genet. 2014;46(4):352–356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Romeo S, Kozlitina J, Xing C, et al. Genetic variation in PNPLA3 confers susceptibility to nonalcoholic fatty liver disease. Nat Genet. 2008;40(12):1461–1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Emdin C, Haas M, Khera AV, et al. A missense variant in Mitochondrial Amidoxime Reducing Component 1 gene and protection against liver disease. bioRxiv. 2019;5:594523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Speliotes EK, Yerges-Armstrong LM, Wu J, et al. Genome-wide association analysis identifies variants associated with nonalcoholic fatty liver disease that have distinct effects on metabolic traits. PLoS Genet. 2011;7(3):e1001324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Innes H, Buch S, Hutchinson S, et al. Genome-Wide Association Study for Alcohol-Related Cirrhosis Identifies Risk Loci in MARC1 and HNRNPUL1. Gastroenterology. June 2020. doi: 10.1053/j.gastro.2020.06.014 [DOI] [PubMed] [Google Scholar]
- 42.Khera AV, Won H-H, Peloso GM, et al. Diagnostic Yield of Sequencing Familial Hypercholesterolemia Genes in Patients with Severe Hypercholesterolemia. J Am Coll Cardiol. March 2016. http://linkinghub.elsevier.com/retrieve/pii/S0735109716323993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Emdin C, Khera AV, Aragam K, et al. DNA Sequence Variation in ACVR1C Encoding the Activin Receptor-Like Kinase 7 Influences Body Fat Distribution and Protects Against Type 2 Diabetes. Diabetes. 2019;68(1):226–234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.GTEx Consortium, Laboratory, Data Analysis &Coordinating Center (LDACC)—Analysis Working Group, Statistical Methods groups—Analysis Working Group, et al. Genetic effects on gene expression across human tissues. Nature. 2017;550(7675):204–213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jansen IE, Savage JE, Watanabe K, et al. Genome-wide meta-analysis identifies new loci and functional pathways influencing Alzheimer’s disease risk. Nat Genet. 2019;51(3):404–413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.CARDIoGRAMplusC4D Consortium. A comprehensive 1000 Genomes-based genome-wide association meta-analysis of coronary artery disease. Nat Genet. 2015;47(10):1121–1130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Iida H, Honda M, Kawai HF, et al. Ephrin-A1 expression contributes to the malignant characteristics of {alpha}-fetoprotein producing hepatocellular carcinoma. Gut. 2005;54(6):843–851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Liu DJ, Peloso GM, Yu H, et al. Exome-wide association study of plasma lipids in >300,000 individuals. Nat Genet. 2017;49(12):1758–1766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Vespasiani-Gentilucci U, Dell’Unto C, De Vincentis A, et al. Combining Genetic Variants to Improve Risk Prediction for NAFLD and Its Progression to Cirrhosis: A Proof of Concept Study. Can J Gastroenterol Hepatol. 2018;2018:7564835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Di Costanzo A, Belardinilli F, Bailetti D, et al. Evaluation of Polygenic Determinants of Non-Alcoholic Fatty Liver Disease (NAFLD) By a Candidate Genes Resequencing Strategy. Sci Rep. 2018;8(1):3702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Caussy C, Soni M, Cui J, et al. Nonalcoholic fatty liver disease with cirrhosis increases familial risk for advanced fibrosis. J Clin Invest. 2017;127(7):2697–2704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mathurin P, Bataller R. Trends in the management and burden of alcoholic liver disease. J Hepatol. 2015;62(1 Suppl):S38–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Dulai PS, Singh S, Patel J, et al. Increased risk of mortality by fibrosis stage in nonalcoholic fatty liver disease: Systematic review and meta-analysis. Hepatology. 2017;65(5):15571565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ishigaki K, Akiyama M, Kanai M, et al. Large scale genome-wide association study in a Japanese population identified 45 novel susceptibility loci for 22 diseases. bioRxiv. October 2019:795948. doi: 10.1101/795948 [DOI] [Google Scholar]
- 55.Kiiskinen T, Mars NJ, Palviainen T, et al. Genomic prediction of alcohol-related morbidity and mortality. Transl Psychiatry. 2020;10(1):23. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.