Abstract
Pancreatic ductal adenocarcinoma (PDAC) is one of the most lethal malignancies, which is usually diagnosed at an advanced stage. The late disease diagnosis, the limited availability of effective therapeutic interventions and lack of robust diagnostic biomarkers, are some of the primary reasons for the dismal 5-year survival rates (~8%) in patients with PDAC. The pancreatic cancer develops through accumulation of a series of genomic and epigenomic alterations which lead to the transformation of normal pancreatic epithelium into an invasive carcinoma – a process that can take up to 15-20 years to develop, from the occurrence of first initiating mutational event. These facts highlight a unique window of opportunity for the earlier detection of PDAC, which could allow timely disease interception and improvement in the overall survival outcomes in patients suffering from this fatal malignancy. Non-coding RNAs (ncRNAs) have been recognized to play a central role in PDAC pathogenesis and are emerging as attractive candidates for biomarker development in various cancers, including PDAC. More specifically, the ncRNAs play a pivotal role in PDAC biology as they affect tumor growth, migration, and invasion by regulating cellular processes including cell cycle, apoptosis, and epithelial-mesenchymal transition. In this review, we focus on three types of well-established ncRNAs — microRNAs (miRNAs), long noncoding RNAs (lncRNAs), and circular RNAs (circRNAs) — and discuss their potential as diagnostic, prognostic and predictive biomarkers in PDAC.
Keywords: Pancreatic ductal adenocarcinoma, Non-coding RNAs, Diagnostic biomarkers, Predictive biomarkers, Prognostic biomarkers
1. Introduction
Pancreatic ductal adenocarcinoma (PDAC) ranks as the 11th most commonly diagnosed cancer in the United States, with an estimated 57,600 new cases in 2020 [1]. While the relative incidence of PDAC may not be as frequent, it remains one of the most lethal malignancies and the 3rd leading cause of cancer-related deaths in the United States [2]. The rising mortality rates from this cancer has projected it to become the second leading cause of cancer-related mortality by the year 2030. This is partly because at initial diagnosis most patients with PDAC already are at an advanced stage, with only ~10-20% patients with a resectable disease. Not surprisingly, in spite of all the medical advances made in the last decade, the overall 5-year survival rates for metastatic PDAC remain only ~8% [2].
At present, surgical resection is the only curative treatment option available for patients with PDAC; however, 5-year survival rates following surgical resection alone still remain relatively low [3, 4]. As per the National Comprehensive Cancer Network (NCCN) guidelines, gemcitabine-based or fluorouracil (5-FU)-based adjuvant treatment is the mainstay for most PDAC patients with a resectable disease. However, due to an elevated risk of complications or poor performance status, some patients are not ideal candidates for receiving such adjuvant treatments. To overcome this clinical challenge, data gathered in the past decade has highlighted the potential therapeutic impact of neoadjuvant chemotherapy or chemoradiotherapy for improving patient survival and tumor resectability status, particularly in PDAC patients with a locally advanced disease [5–11]. In addition, while the introduction of newer gemcitabine-based adjuvant chemotherapy regimens referred to as modified FOLFIRINOX (mFOLFIRINOX) that include a combination of fluorouracil, leucovorin, irinotecan, and oxaliplatin, have somewhat improved the prognosis of patients with resectable PDAC, a significant number of patients still experience disease relapse and their prognosis remains relatively poor [4, 12]. Taken together, this highlights the underlying issue that for a complete cure or improved survival, early detection of cancer might have the greatest impact on patient outcomes. This certainly has been observed in several other cancers, including breast, colon, prostate, and cervical – in each instance, the survival outcomes were significantly improved due to earlier detection of the cancer [13–17]. However, this has not yet been possible in PDAC, because currently there is a lack of robust diagnostic modalities available for the early detection of this fatal disease.
While imaging-based methods such as computed tomography (CT) and magnetic resonance imaging (MRI) are routinely used for PDAC diagnosis, their diagnostic sensitivity and specificity remains quite poor for the detection of early-stage lesions [18]. Likewise, in terms of non-invasive markers, currently carbohydrate antigen 19-9 (CA19-9) and carcinoembryonic antigen (CEA) are the only tumor markers currently used in the clinic; however, these suffer from poor diagnostic accuracy [19]. The present challenges with the imaging-based approaches, and the issues with tumor markers as robust biomarkers for the early-detection of PDAC highlight the need for development of molecular biomarkers that are more robust and can be used in the clinic for the identification of patients at risk for developing this fatal disease.
One such molecular substrate that has garnered a lot of attention in the past several years is the development of various non-coding RNAs (ncRNAs) as biomarkers for early diagnosis and determining prognosis of patients with PDAC. The ncRNAs not only function as important epigenetic regulators in PDAC pathogenesis but have also emerged as attractive targets for their potential clinical use as diagnostic, prognostic, and predictive biomarkers in various human cancers including PDAC. In recent years, technological advancements such as genome-wide sequencing have discovered novel ncRNAs, and this list is continuously growing each day. In this review article, we highlight the importance of early detection in PDAC and discuss the accumulating evidence for ncRNAs in terms of their potential as diagnostic, prognostic, and predictive biomarkers in this malignancy.
2. Importance of early detection in PDAC
Late onset of symptoms and a rapid progression to death are hallmarks of PDAC. Due to the asymptomatic nature of the disease, most patients present with non-resectable, locally advanced or metastatic disease at initial diagnosis – at which time surgical resection is not possible; hence, limiting the availability of treatment options in these patients. To muddy the waters further, the currently available treatment options in PDAC are less effective when offered to patients with an advanced disease.
2.1. Pancreatic precursor lesions and progression to invasive carcinoma – a window of opportunity for early detection
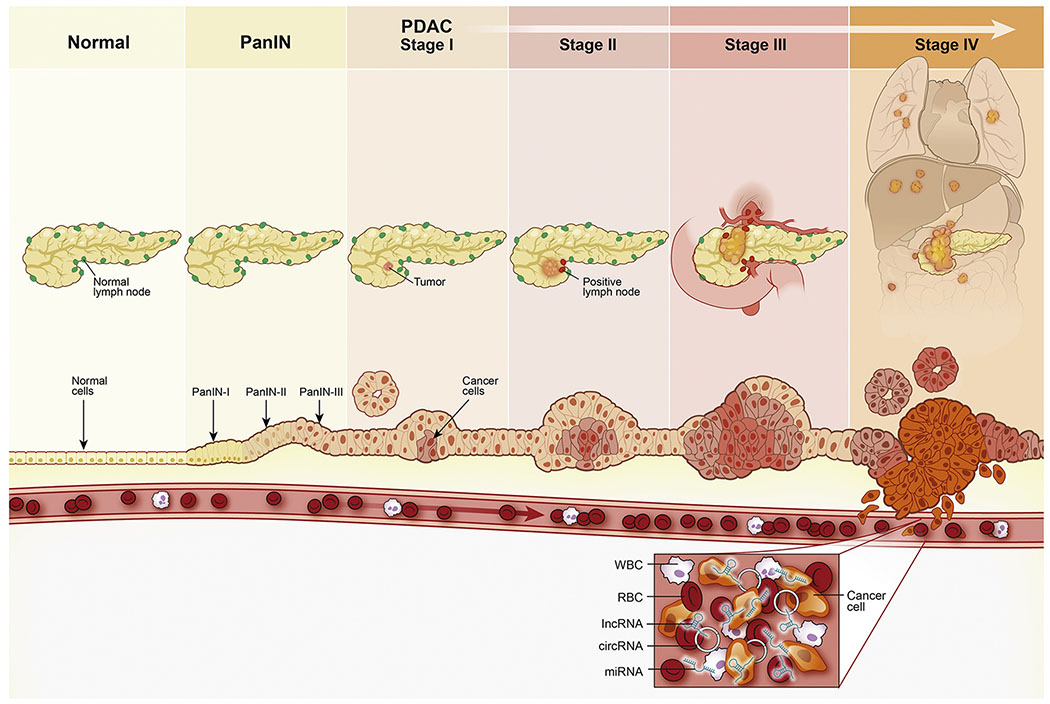
As depicted in Figure 1, the development of PDAC from non-invasive stages is a slow and gradual process [20]. More specifically, invasive PDAC develops from precursor lesions that are classified into three different histological categories: pancreatic intraepithelial neoplasia (PanIN), intraductal pancreatic mucinous neoplasia (IPMN), and mucinous cystic neoplasia (MCN) [21, 22]. Among these, PanINs are the most common precursor lesions of PDAC. PanINs are microscopic lesions (often <5 mm in size) originating from the small pancreatic ducts and are composed of columnar and cuboidal cells with varying amounts of mucin [23, 24]. Based on a number of molecular and histological alterations that transform a normal pancreatic duct into PanINs, these can be classified into low-grade dysplasia (PanIN-IA and -IB) and high-grade dysplasia (PanIN-II and -III) [25]. Similar to PanINs, the IPMNs are mucin-producing epithelial neoplasms that can arise within the main duct of pancreas (MD-IPMN), in one of its side branch-ducts (BD-IPMN), or both (mixed-IPMN). Further classification of IPMN is based on the degree of dysplasia, which ranges from low-grade dysplasia (IPMN, low-grade), intermediate-grade dysplasia (IPMN, low grade) and IPMN with high-grade dysplasia or carcinoma in-situ (IPMN, high grade) [21, 26]. In terms of their neoplastic potential, the MD-IPMNs are associated with a higher risk of developing invasive disease (~63%) compared to BD-IPMNs (~15%) [27]. Lastly, MCNs have a distinct ovarian-type stroma and are more common in women than men [28, 29]. All types of cystic neoplasms of the pancreas carry a risk of malignant transformation, albeit to varying degrees; thus, finding biomarkers that can accurately predict which of the cystic neoplasms will progress to invasive carcinomas may improve surgical management and treatment decisions. From an early-detection standpoint, there is substantive evidence that it can take more than a decade for a normal pancreatic epithelial cell to progress into an invasive, metastatic pancreatic carcinoma from the time point of first genetic event acquired within the tumor-initiating cells [30]. This is particularly attractive from an early-detection viewpoint as this long timeframe provides a window of opportunity for the detection of early precursors and PDAC lesions, at which time therapeutic intervention could be employed to drastically improve the survival rates in patients suffering from this disease [31].
Figure 1:
The evolution and progression of PDAC: The PDAC develops gradually over a period of time in which a series of genomic, epigenomic and morphological alterations are initiated in the normal cells. Initial genetic and epigenomic changes occur slowly during the transformation of normal epithelium through premalignant lesions (PanIN I- III); where the changes are not visible at the organ level making detection of premalignant lesions difficult. The premalignant cells continue to grow leading to tumor formation which gradually invades nearby lymph nodes, ultimately enter the systemic circulation and metastasize to distant organ sites. Tumor cells that reach the blood circulation also release their cellular contents (e.g. DNA, RNA, proteins etc.), which are interrogated for the development of blood-based liquid biopsy assays for the early disease detection. PanIN – pancreatic intraepithelial neoplasia; PDAC – pancreatic ductal adenocarcinoma; lncRNA – long non-coding RNA; circRNA – circular RNA; miRNA – microRNA; RBC – Red blood cell, WBC – White blood cell
2.2. Limitations of current diagnostic approaches in PDAC – the need for development of robust molecular biomarkers for its early detection
Currently, various technical and molecular approaches are used for the diagnosis of PDAC (Table 1); with each of these modalities with their own inherent advantages and disadvantages. Imaging-based approaches that utilize multi-detector CT accompanied by 3D-reconstruction and MRI is frequently used to stage PDAC patients, prior to surgery [32, 33]. However, these imaging tools for diagnosing and staging PDAC patients lack adequate sensitivity, are beset with false negative results, and frequently fail to detect small and potentially curable pancreatic lesions [18]. Endoscopic ultrasound, which is another commonly used screening tool for high-risk PDAC patients, is an expensive and invasive modality [34–36]. In addition, due to an overall lower incidence rate of PDAC and high cost-to-benefit ratio, the existing methods for the screening of average-risk general population for PDAC are unlikely to have a substantial impact on patient outcomes.
Table 1.
A list of clinical and pre-clinical diagnostic approaches in patients with PDAC
| Methods | Evidence | Sensitivity | Specificity | References | |||
|---|---|---|---|---|---|---|---|
| Imaging modalities | Endoscopic Ultrasound | Clinical (stage T1-T2) | 72% | 90% | [167] | ||
| Clinical (stage T3-T4) | 90% | 72% | |||||
| Multi-detector CT | Clinical | 76–92% | 67% | [168–171] | |||
| MRI | Clinical | 78–100% | 72–99% | [172–174] | |||
| Molecular Imaging | Clinical | 87–89% | 70% | [175] | |||
| Tumor marker | CA19-9 | Clinical | 78.2% | 82.8% | [176] | ||
| Mutational markers | TP53, SMAD4, PIK3CA, PTEN, AKT1, MUC3, MUC4 | Clinical | 32–79% 76–89% |
96–100% 96–100% |
[177–187] | ||
| ncRNA markers | miRNAs | miR-21 | Pre-clinical | 67–96% | 61–100% | [64, 188–191] | |
| miR-155 | Pre-clinical | 53–93% | 73–100% | [64, 190, 192] | |||
| miR-196a | Pre-clinical | 43–100% | 84–90% | [64, 87] | |||
| miR-196b | Pre-clinical | 78–100% | 78–100% | [64, 87] | |||
| lncRNAs | HOTAIR | Pre-clinical | 78–80% | 86–90% | [193] | ||
| PVT1 | Pre-clinical | 69–96% | 64–95% | [193] | |||
| MALAT1 | Pre-clinical | 66% | 72% | [139] | |||
In addition to imaging-based approaches, there are only two noninvasive, serological diagnostic biomarkers that are often used for PDAC diagnosis in the clinic – the CA19-9 and CEA; both of which suffer from limited sensitivity and specificity for PDAC. In addition, elevated expression of these biomarkers is usually associated with advanced disease stage but may also indicate the presence of diseases other than PDAC [37, 38]; highlighting their inadequacy for the early detection of PDAC. Moreover, 5-10% of patients who lack fucosyltransferase activity due to germline variants do not produce CA19-9 [39]. Various protein and DNA biomarkers have been reported that show diagnostic and prognostic potential in PDAC patients either alone or in combination with CA19-9 [39–44]. However, there are currently no available reliable liquid biopsy assays with high sensitivity and specificity to detect early pancreatic cancer.
In view of the limitations of existing methods described above, there has been unprecedented interest in developing novel diagnostic biomarkers for the detection of PDAC in its earliest stages. There is no question that detecting the disease early will have a direct impact on the management of PDAC and improving patient survival.
3. ncRNAS and their important role in PDAC
The ncRNAs are important functional components of the human genome that are transcribed from DNA but are not translated into proteins. While there is lack of clear consensus on the various categories of ncRNAs, based on size these can be classified into small ncRNAs (<200 nucleotides in length) and long non-coding RNAs (lncRNAs; size>200 nucleotides) [45]. While the nomenclature and discovery of various ncRNAs continue to evolve, at this time, small ncRNAs primarily encompass microRNAs (miRNAs), small inhibitory RNAs (siRNAs), PlWI-interacting RNAs (piRNAs), and small nucleolar RNAs (snoRNAs). In contrast, lncRNAs mostly consist of large intergenic non-coding RNAs (lincRNAs), transcribed ultraconserved regions (T-UCRs), and circular RNAs (circRNAs) [45]. Even though ncRNAs are not translated into proteins, they have been recognized to be critical regulators for various biological processes such as DNA replication, translation, RNA splicing, and epigenetic regulation (Figure 2). Deregulation of ncRNAs has been reported in many diseases, including cancer [45–47]. Additionally, with the advent of next-generation sequencing technologies, various categories of ncRNAs are continuously being discovered, characterized for their biological roles, and have been developed as potential disease biomarkers through their expression analysis not only in tissues, but as well as in other bodily fluids such as blood (plasma and serum), saliva, pancreatic juice, cerebrospinal fluid and urine – highlighting their promise as potential biomarkers for cancer diagnosis, prognosis and disease monitoring [48–51]. Moreover, some ncRNAs, specially miRNAs and lncRNAs have also shown therapeutic potential and different delivery systems for ncRNA-based therapeutics have also been previously described [52].
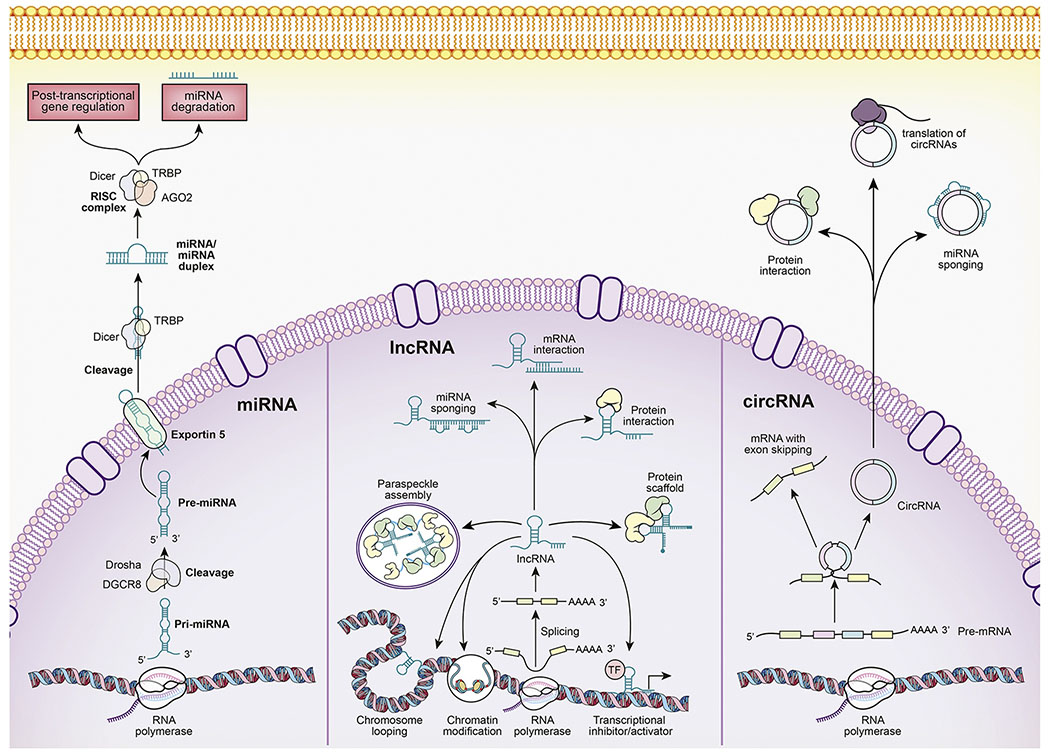
Figure 2:
Overview of the biogenesis and functions of key ncRNAs: The biogenesis of miRNAs (left panel) involves the transcription of primary miRNA (Pri-miRNA) by RNA polymerase (II or III) from the miRNA gene. The Pri-miRNAs are long, RNA stem-loop structures, which are eventually processed by the DROSHA–DiGeorge syndrome critical region 8 (DGCR8) complex resulting in the cleavage of the Pri-miRNA and production of a smaller product called, the precursor miRNA (Pre-miRNA), which is approximately 60 nucleotides in length. The Pre-miRNA is exported to the cytoplasm from the nucleus by Exportin-5 protein. The Pre-miRNA is processed further in the cytoplasm by the ribonuclease DICER protein in conjunction with the RNA-binding protein transactivation response element RNA-binding protein (TRBP). The DICER-TRBP complex cleaves the Pre-miRNA to form a miRNA/miRNA duplex. One of the strands from this duplex is degraded while the other functional strand binds to the Argonaute 2 (AG02) protein and is incorporated into the RNA-induced silencing complex (RISC) involving DICER and TRBP. The miRNA strand guides the RISC complex to target mRNAs causing translational repression or degradation. The lncRNAs (middle panel) are transcribed by the RNA polymerase and are usually adenylated at the 3’ end and capped at 5’ end. Their expression is cell type- and cell state-specific and they can undergo alternative splicing leading to different isoforms. lncRNAs can regulate genes in many ways. For example, they can activate (enhancer RNAs) or inhibit the transcription of nearby genes by either directly interacting with the RNA polymerase or transcription factors. lncRNAs can also interact with DNA by virtue of their sequence complementarity to single stranded DNA mediating chromosomal looping. They can also induce changes in the chromatin structure and interact with nucleolar (paraspeckle) proteins forming paraspeckle assembly. Their other functions include acting as miRNA sponges, regulating mRNA stability, interaction with proteins or acting as a scaffold for proteins. The circRNAs (right panel) are also transcribed by the RNA polymerase similar to mRNAs however unlike mRNAs, circRNAs can be processed through alternative splicing of both, exons and introns of the pre-mRNA. The circular shape of circRNAs is the result of back-splicing which is described by lariat-driven circularization and intron-pairing-driven circularization. circRNAs can act as miRNA sponges due to the presence of multiple miRNA response elements in their sequence. Some circRNAs can also interact with proteins harboring RNA-binding sites. Additionally, they can also encode for and translate into various proteins.
Herein, we discuss the biomarker potential of three types of ncRNAs in PDAC – the miRNAs, lncRNAs, and circRNAs, and provide a succinct yet comprehensive state of literature for their emerging role as diagnostic, prognostic and predictive biomarkers.
4. miRNAs as potential biomarkers in PDAC
Among the ncRNAs, miRNAs are the most widely explored as biomarkers in malignant diseases. miRNAs are 18–25 nucleotides long and regulate gene expression at the post-transcriptional level [53, 54]. Abnormal expression levels of miRNAs have been implicated in the pathogenesis of PDAC [55, 56]. One of the most attractive features of miRNAs is their stability in bodily fluids (blood, urine, saliva), which makes them ideal targets for liquid biopsies.
4.1. Diagnostic significance of miRNAs in pancreatic cancer
The last decade has witnessed an explosion in the number of studies that have focused on the identification of miRNAs that are differentially expressed in clinical specimens from patients with PDAC compared to healthy controls (Table 2).
Table 2.
The potential miRNA biomarkers for the identification of patients with pancreatic precursor lesions and PDAC.
| Pathologic condition | Source | miRNA | Expression | References | |
|---|---|---|---|---|---|
| PDAC | Tissue | miR-21 | Increased | [55, 57, 62, 63, 76, 86, 88, 90, 92, 96, 194] | |
| miR-31 | Increased | [56] | |||
| miR-96 | Decreased | [56, 194, 195] | |||
| miR-143 | Increased | [56] | |||
| miR-146a | Increased | [56, 194] | |||
| miR-148a | Decreased | [56, 89, 196] | |||
| miR-150 | Increased | [56] | |||
| miR-155 | Increased | [56, 57, 62, 63, 88, 89, 92] | |||
| miR-181a,b,d | Increased | [57] | |||
| miR-196a,b | Increased | [56, 66, 92, 194, 197–199] | |||
| miR-210 | Increased | [56, 67, 200] | |||
| miR-212 | Increased | [55, 65] | |||
| miR-216 | Decreased | [20, 55, 56] | |||
| miR-217 | Decreased | [56, 89, 194] | |||
| miR-222 | Increased | [55, 57] | |||
| miR-223 | Increased | [56] | |||
| miR-375 | Decreased | [56] | |||
| miR-483 | Increased | [96] | |||
| miR-494 | Decreased | [56] | |||
| miR-1290 | Increased | [68] | |||
| Blood (plasma or serum) | miR-16 | Increased | [70] | ||
| miR-18a | Increased | [201] | |||
| miR-20a | Increased | [202, 203] | |||
| miR-21 | Increased | [64, 96, 203] | |||
| miR-155 | Increased | [64, 192] | |||
| miR-185 | Increased | [70, 202, 203] | |||
| miR-191 | Increased | [65, 203] | |||
| miR-192 | Increased | [65, 204] | |||
| miR-194 | Increased | [65] | |||
| miR-196a,b | Increased | [64, 70, 198] | |||
| miR-210 | Increased | [205] | |||
| miR-212 | Increased | [65, 206] | |||
| miR-483 | Increased | [96] | |||
| miR-492 | Decreased | [207] | |||
| miR-508 | Decreased | [65] | |||
| miR-513a | Decreased | [65] | |||
| miR-602 | Increased | [65] | |||
| miR-630 | Decreased | [65] | |||
| miR-663a | Decreased | [207] | |||
| miR-801 | Increased | [65] | |||
| miR-887 | Decreased | [65] | |||
| miR-923 | Decreased | [65] | |||
| miR-1246 | Increased | [208] | |||
| miR-3976 | Increased | [208] | |||
| miR-4306 | Increased | [208] | |||
| miR-4644 | Increased | [208] | |||
| Pancreatic Juice | miR-21 | Increased | [62, 92] | ||
| miR-155 | Increased | [62, 92, 192] | |||
| miR-196a,b | Increased | [92, 199] | |||
| miR-210 | Increased | [199] | |||
| miR-1427 | Increased | [199] | |||
| Stool | miR-21 | Increased | [200] | ||
| miR-181a,b,d | Increased | [197] | |||
| miR-210 | Increased | [197, 200] | |||
| miR-216 | Decreased | [190, 200] | |||
| Saliva | miR-940 | Increased | [193] | ||
| miR-3679 | Decreased | [193] | |||
| PanIN | Tissue | miR-21 | Increased | [86, 88, 90] | |
| miR-29b | Increased | [86] | |||
| miR-148a | Decreased | [89] | |||
| miR-155 | Increased | [88–90] | |||
| miR-182 | Increased | [89] | |||
| miR-196a,b | Increased | [66] | |||
| miR-217 | Decreased | [89] | |||
| miR-425 | Increased | [86] | |||
| miR-708 | Increased | [86] | |||
| miR-874 | Increased | [86] | |||
| Blood (plasma or serum) | miR-21 | Increased | [64, 96, 203] | ||
| miR-196a,b | Increased | [70] | |||
| IPMN | Tissue | miR-21 | Increased | [63, 86, 92] | |
| miR-99a,b | Decreased | [98] | |||
| miR-155 | Increased | [63, 92] | |||
| miR-196a,b | Increased | [92] | |||
| Blood (plasma or serum) | miR-21 | Increased | [96] | ||
| miR-212 | Decreased | [206] | |||
| Pancreatic juice | miR-21 | Increased | [92] | ||
| miR-92a | Increased | [206] | |||
| miR-99a,b | Decreased | [206] | |||
| miR-100 | Decreased | [206] | |||
| miR-125b | Increased | [206] | |||
| miR-145 | Increased | [206] | |||
| miR-155 | Increased | [92] | |||
| miR-196a,b | Increased | [92] | |||
4.1.1. Diagnostic potential in PDAC
Due to the presence of thick stromal layer and persistent inflammation in patients with chronic pancreatitis (CP) and PDAC, the diagnostic utility of imaging-based approaches has been clinically challenging. In an effort to overcome this issue for improved discrimination between CP and PDAC, several studies have focused on the interrogation of miRNA expression profiles. In one of the first such studies, Bloomston et al. identified a panel of 25 miRNAs that were significantly deregulated in PDAC compared to adjacent healthy tissues [57]. Of these, miR-196a, miR-196b, miR-203, miR-210, miR-217, miR-222,, and miR-375 were dysregulated only in PDAC, whereas miR-29c, miR-96, miR-143, miR-145, miR-148b, and miR-150 were abnormally expressed in both CP and PDAC. These findings suggested that the latter group of miRNAs were likely responsible for causing a desmoplastic reaction, as opposed to tumorigenesis. More specifically, several studies have noted significant downregulation of miR-92, miR-132, miR-148a, miR-216a, and miR-217 in PDAC [56, 58–61]. In contrast, miR-31, miR-143, miR-145, miR-146a, miR-150, miR-155, miR-194, miR-196a, miR-196b, miR-210, miR-222, and miR-223 were observed to be markedly upregulated in PDAC compared to healthy specimens [55–57, 62–67].
These initial studies for the observed deregulation of miRNA expression became the basis for subsequent studies that began evaluating their biomarker potential. For example, Lee et al. identified upregulation of miR-21, miR-301, and miR-376a in PDAC tissues compared to normal pancreatic tissues, indicating their potential to detect patients with PDAC from healthy individuals [55]. Later studies reported elevated expression of miR-1290 in patients with early-stage PDAC patients, which also exhibited significantly superior diagnostic potential compared to the classic tumor marker, CA19-9, in differentiating early-stage PDAC from controls [68]. Significant upregulation of miR-135b was observed in PDAC vs. non-diseased control specimens, and could also distinguish patients with PDAC from those with CP with relatively high sensitivity and specificity [69].
Following the observation of encouraging results in tissues, researchers started to explore the feasibility of translating these miRNA biomarkers in blood (serum or plasma) specimens. In one such effort, the previously identified miRNAs miR-21, miR-155, and miR-196a were also found to be upregulated in sera from patients with PDAC compared with healthy subjects [70]. While individual miRNAs have shown potential to detect PDAC, a combination of circulating miRNAs often results in an increased diagnostic accuracy. For instance, a combination of miR-196a and miR-217 expression levels discriminated PDAC from healthy controls and CP cases [56]. More intriguingly, diagnostic performance of circulating miRNAs together with tumor markers such as CA19-9 has also been analyzed. In this context, plasma levels of miR-16 and miR-196a combined with CA19-9 offered a significantly superior diagnosis of early stage PDAC [70].
As the enthusiasm for miRNA-based biomarkers in PDAC continues to build, various researchers also began to systematically examine the functional significance of specific miRNAs, their downstream target genes (mRNAs), the signaling pathways and cellular processes controlled by individual miRNAs in PDAC (Figure 3). For example, miR-217, which has been found to be significantly downregulated in PDAC tissues and cell lines, was revealed to target KRAS mRNA [71]. Subsequent studies supported this notion and illustrated that miR-217 acts as a tumor suppressor by inhibiting KRAS, thereby reducing the constitutive phosphorylation of the downstream signal transducer AKT and eventually inhibiting cell proliferation. Kent et al. characterized miRNA expression profiles in multiple experimental model systems in which KRAS was constitutively activated due to the presence of a KRASG12D mutation [72]. These studies revealed that activated KRAS signaling led to the repression of the miR-143/145 cluster, which is required to maintain the tumorigenic potential of pancreatic cancer cells. Using a dual-reporter luciferase assay, it was demonstrated that downregulation of miR-143/145 was achieved by binding of the KRAS-responsive element-binding protein (RREB1) to the miR-143/145 promoter region. Likewise, miR-375, which is upstream of PI3K/AKT signaling, acts as a tumor suppressor by inhibiting the growth of PDAC cells through AKT signaling pathway [73, 74]. Specific attention has been placed on the role of miR-21 in PDAC, as it is implicated in tumorigenesis, tumor cell invasion, and metastasis of pancreatic tumor cells, as well as desmoplastic reaction and has been consistently observed to be overexpressed in PDAC compared to healthy and/or CP samples [75–78]. Phosphatase and tensin homolog (PTEN), a tumor suppressor gene known to suppress PI3K-AKT-mTOR signaling and control various cellular processes, is targeted by miR-21, miR-221, and miR-181a [79–81]. One of the downregulated miRNAs, miR-216a, targets JAK2 mRNA and inhibits its expression, lead to reduction in the tumor volume in an animal model by increasing tumor cell apoptosis and decreasing cell proliferation [82]. Furthermore, transfection with miR-216a inhibited the growth of pancreatic cells and the transcription of the survivin gene and other apoptotic genes located downstream of the JAK/STAT pathway [83]. miR-155 is also associated with the JAK/STAT pathway, and pancreatic cells with its knockdown resulted in increased expression of suppressor of cytokine signaling 1 (SOCS1) – a tumor suppressor protein that negatively regulates the JAK/STAT3 signaling pathway [84]. Collectively, these data provide a strong evidence supporting the importance of miRNAs as potential biomarkers for the diagnosis of PDAC.
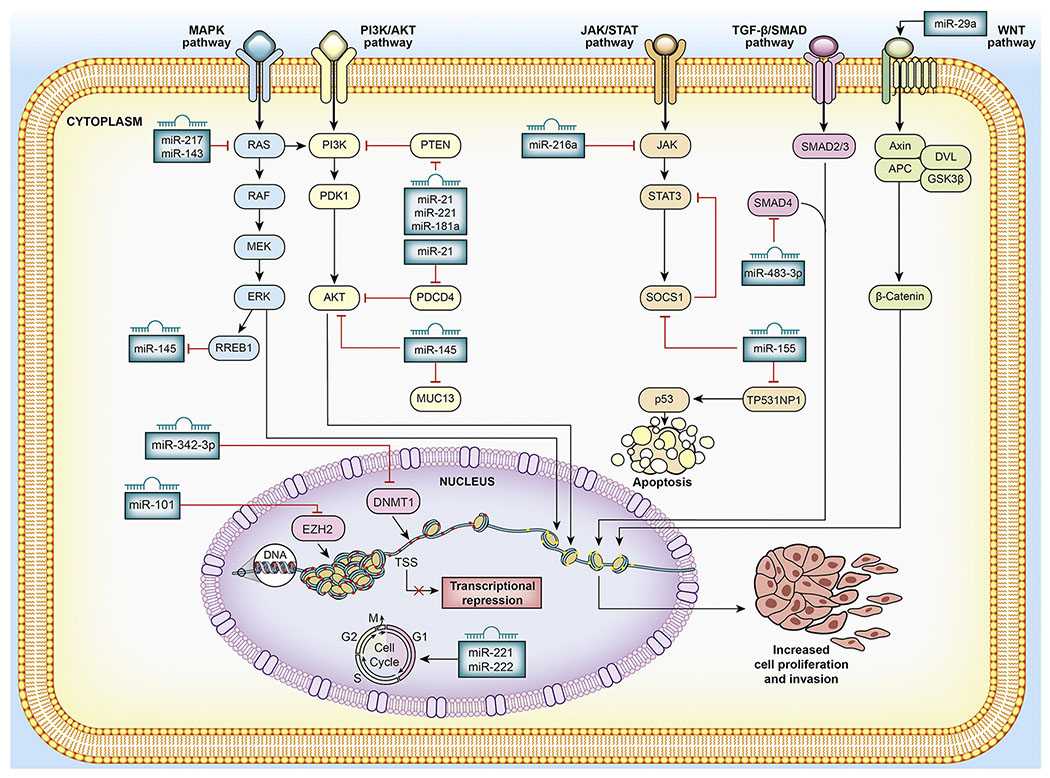
Figure 3:
Mechanistic role of the miRNAs in PDAC: miRNAs can act both as tumor suppressors and oncogenes by regulating different key downstream gene targets that mediate cellular growth signaling pathways. For example, miR-217 which is often found downregulated in PDAC, acts as a tumor suppressor and targets KRAS oncogene which endows proliferation, survival and invasion properties onto cancer cells through the activation of several downstream effector pathways such as the PI3K/AKT and the RAF/ERK pathway. On the other hand, miR-21, which is usually found upregulated in PDAC, targets the tumor suppressor genes PTEN and PDCD4. miR-21 mediated inhibition of PTEN leads into activated downstream signaling of PI3K/AKT pathway. PDCD4, a tumor suppressor gene involved in apoptosis, invasion is inhibited by miR-21 and is potentially involved in PI3K/AKT signaling pathway. miRNAs can also induce transcriptional repression or activation by modulating chromatin structure through the targeting of epigenetic regulatory genes. For example, miR-342-3p can act as a tumor suppressor by inhibiting cancer cell proliferation and invasion through targeting DNMT1.
4.1.2. Diagnostic potential in PanINs
There exist histological and molecular differences between PDAC and its precursor lesions, as well as between different types of precursor lesions – PanINs and IPMNs [85]. Moreover, based on the idea that the transformation of normal pancreatic tissue into an invasive PDAC manifests over several years through the accumulation of a series of genetic and epigenetic changes, miRNA expression profiles have been evaluated in low- and high-grade PanINs to identify biomarkers that can detect the disease in the earliest stages of neoplastic transformation.
In a PanIN progression model based on miRNA expression profiles, 735 human miRNAs were interrogated in PanINs, PDAC, and healthy tissues which led to the identification of specific miRNAs that discriminated PanINs from healthy and PDAC [86]. This report also revealed that the expression profiles of specific miRNAs correlated with different stages of premalignant lesions. Thirteen miRNAs (miR-21, miR-29b, miR-146a, miR-182, miR-193a-3p, miR-193b, miR-200a, miR-200b, miR-200c, miR-425, miR-486-3p, miR-708, and miR-874) were significantly upregulated in PanIN-II & III lesions compared to PanIN-I lesions and normal pancreatic tissues. More specifically, miR-196b was significantly upregulated in PanIN-III compared to normal pancreatic tissues [86]. In a subsequent effort, Slater et al. studied miRNA expression profiles in serum specimens from patients with PDAC and PanINs, as well as healthy controls [66, 87]. Among the five miRNAs evaluated (miR-21, miR-155, miR-196a, miR-196b, and miR-210), only miR-196a and miR-196b showed significantly increased expression in patients with PDAC or PanIN-II & III lesions compared to those with PanIN-I lesions or healthy controls. Of note, a combination of miR-196a and miR-196b expression levels demonstrated a perfect sensitivity and specificity of 100% for discriminating between patients with PanIN-II & III lesions and healthy controls [66, 87]. Ryu et al. compared the relative expression of three candidate miRNAs (miR-21, miR-155, and miR-221, all reported to be overexpressed in PDAC) between PanIN lesions of various histological grades and non-neoplastic pancreatic ductal epithelium [88]. These researcher found that miR-21 was marginally upregulated in PanIN-III, while miR-155 was significantly overexpressed in PanIN-II & III cases compared to PanIN-I or healthy controls; highlighting that miR-155 overexpression is likely an early event in the multi-step progression model of PDAC [88]. Several follow-up studies have since then corroborated the upregulated expression of miR-21 and miR-155 in PanIN lesions [89, 90]. Increased expression of other miRNAs (miR-10b, miR-200, miR-205, miR-221, and miR-222) has also been reported during the progression from PanIN to PDAC [89, 90].
In a detailed functional study, miR-145 expression was shown to decrease with increasing stages of PanINs [91]. While investigating the biological mechanism underlying this phenomenon, it was reported that miR-145 regulated the expression of MUC13 and acted as a tumor suppressor. Ectopic expression of miR-145 in cultured cells and animal models resulted in decreased cellular invasion and reduced tumor growth, respectively, highlighting the role of miR-145 in the development and progression of pancreatic cancer [91]. Likewise, expression of miR-148 was also found to inversely correlate with disease progression, with higher expression in normal tissue compared to PanINs and PDAC [89] – all of which support the role of distinct miRNAs involved in neoplastic disease progression and their biomarker potential.
4.1.3. Diagnostic potential in IPMNs
The first study of miRNA expression profiles in IPMNs was conducted in 2009 [92]. In a small cohort of 15 IPMN tissues, the expression of a panel of 12 miRNAs was measured, which led to the identification that 10 of the 12 miRNAs were significantly overexpressed in IPMNs compared to normal pancreatic tissues. The most promising of these miRNAs were miR-21 and miR-155 – and the elevated expression of these two miRNAs also correlated with greater cellular atypia in the IPMNs [92]. As described earlier, miR-21 has been implicated in repressing the activity of the tumor suppressor genes PTEN and programmed cell death 4 (PDCD4), resulting in the activation of the AKT signaling pathway and increased cellular transformation and metastases, respectively. Similarly, miR-155 was reported to repress a pro-apoptotic gene, tumor protein 53-induced nuclear protein 1 (TP53INP1), and increased tumorigenicity in an animal model [93].
In a larger cohort study comprising of 65 invasive IPMNs, 16 non-invasive IPMNs and 5 normal pancreatic ductal tissues, miR-21 and miR-155 were yet again confirmed to be overexpressed in surgically resected IPMN specimens [63]. Furthermore, the overexpression of these miRNAs was more prominent in invasive vs. non-invasive IPMNs. The same study also reported downregulation of miR-101 in invasive IPMNs [63]. Subsequently, Nakahara et al. confirmed these results and demonstrated that from a mechanistic viewpoint, miR-101 targets oncogenic enhancer of zeste 2 homolog (EZH2). The authors proposed that downregulation of miR-101 leads to the overexpression of EZH2, which could promote the transformation of premalignant IPMNs into PDAC [94].
A subsequent study analyzed pancreatic juice specimens and FFPE tissues for the alterations in expression of miR-196a in specific subtypes (intestinal vs. non-intestinal) of IPMNs. This study reported that miR-196a was specifically upregulated in intestinal-type IPMNs and showed a diagnostic performance, with an area under the curve (AUC) value of 0.85 for the receiver operating characteristic (ROC) curve analysis [95]. The expression of circulating miR-483-3p has also been shown to differentiate IPMNs from PDAC and healthy controls [96]. Combining the expression of miR-483-3p with miR-21 further increased the diagnostic potential for discriminating patients with IPMNs from those with PDAC and healthy controls, and was comparable to the traditional markers CA19-9 and CEA [96]. The miR-483-3p is located within intron 2 of the IGF2 locus and its overexpression has been reported to suppress the expression of the transcription factor DPC4/Smad4 in PDAC tissues [97]. DPC4/Smad4 act as mediators of the TGF-β signaling pathway and are involved in TGF-β-induced epithelial-mesenchymal transition (EMT) [97].
In a genome-wide miRNA expression profiling effort, miR-99a, miR-99b, miR-100, miR-126, miR-130a, and miR-342-3p were found to be under expressed in high-risk vs. low-risk IPMNs [98]. In addition, low levels of miR-99b in cystic fluid from patients with IPMNs were associated with main duct involvement and hence associated with an increased risk for developing into malignant neoplasms. The authors also evaluated expression of the downstream target mRNAs of these downregulated miRNAs and found that some of them, such as IRS-1 (miR-126 target), ATG2B and MEOX2 (miR-130a targets), and DNMT1 (miR-342-3p target), were upregulated in high-risk vs. low-risk IPMNs [98]. More recently, miRNA expression profiles were shown to differentiate pancreatic cystic neoplasms, wherein miR-31-5p, miR-99a-5p, miR-375, and miR-4830-5p were characteristic of serous cyst adenomas (SCAs) and distinguished SCAs from MD- and BD-IPMNs [99].
Taken together, these studies underscore the importance of miRNAs for their clinical significance as diagnostic biomarkers for differentiating patients with IPMNs from healthy controls, as well for discriminating between various subtypes of pancreatic lesions – thereby potentially improving the diagnosis and management of these patients.
4.2. Prognostic significance of miRNAs in PDAC
In addition to early detection, identifying prognostic biomarkers that offer improved prediction of patient survival are also of critical importance. Several studies have found associations between miRNA expression and patient outcomes in PDAC [100]. Not only higher expression of miR-21 was observed in PDAC compared to healthy tissues, it was also a superior predictor of poorer outcomes in patients with PDAC [76, 101]. High expression of miR-21 also led to reduced gemcitabine sensitivity and apoptosis in PDAC cells [77]. Further downstream analysis revealed that overexpression of miR-21 potentially downregulated expression of PTEN and activated the PI3K/AKT signaling pathway.
It was reported that expression of a panel of six miRNAs (miR-30a-3p, miR-105, miR-127, miR-187, miR-452, and miR-518a-2) was significantly associated with improved prognosis in patients with PDAC [57]. In a study of 225 PDAC patients, high expression of miR-212 and miR-675 and low expression of miR-148a, miR-187, and let-7g were independent predictors of poor prognosis in PDAC [102]. Recently, Liang et al. conducted a TCGA database study to evaluate the predictive value of several miRNAs [103]. These researchers reported that a 5-miRNA signature consisting of miR-125a, miR-328, miR-376b, miR-376c, and miR-1301 had the highest prognostic potential, with a corresponding hazard ratio (HR) of 0.139 (95% CI, 0.043–0.443; P<0.001). To better understand the underlying molecular mechanisms of these five miRNAs, the authors performed gene set enrichment and gene ontology analyses. They discovered that the target genes of the candidate miRNAs were involved in a variety of critical biological processes including developmental process, cell differentiation and anatomical structure morphogenesis [103]. Finally, miR-15a miR-155, miR-200c, miR-203, miR-210, miR-222, miR-302, and miR-506 were also shown to correlate with the prognosis in patients with PDAC [104–107]; emphasizing the clinical significance of miRNAs as prognostic biomarkers in PDAC.
5. LncRNAs as Biomarkers in PDAC
The lncRNAs are >200 nucleotides long and are transcribed by RNA polymerase II. The lncRNAs play important roles in an array of diverse biological, developmental, and pathological processes, as they are involved in RNA regulatory mechanisms and control the expression of their downstream target genes. As illustrated in Figure 4, in addition, they mediate a diverse array of cellular processes through chromatin reprogramming, cis or trans regulation at neighboring genes, and post-transcriptional regulation of mRNA processing [108–110]. Dysregulation of lncRNAs has been implicated in several human diseases including cancer [111–113]. As shown in Table 3, increasing evidence supports that aberrant expression of lncRNAs plays an oncogenic or tumor-suppressive role in various cancers including PDAC [114–120]. As a result, there is great degree of interest in the identification of lncRNAs that can be developed for their clinical application as cancer biomarkers. Additionally, the lncRNAs are highly tissue- and disease-specific, which makes them attractive candidates for development as disease biomarkers.
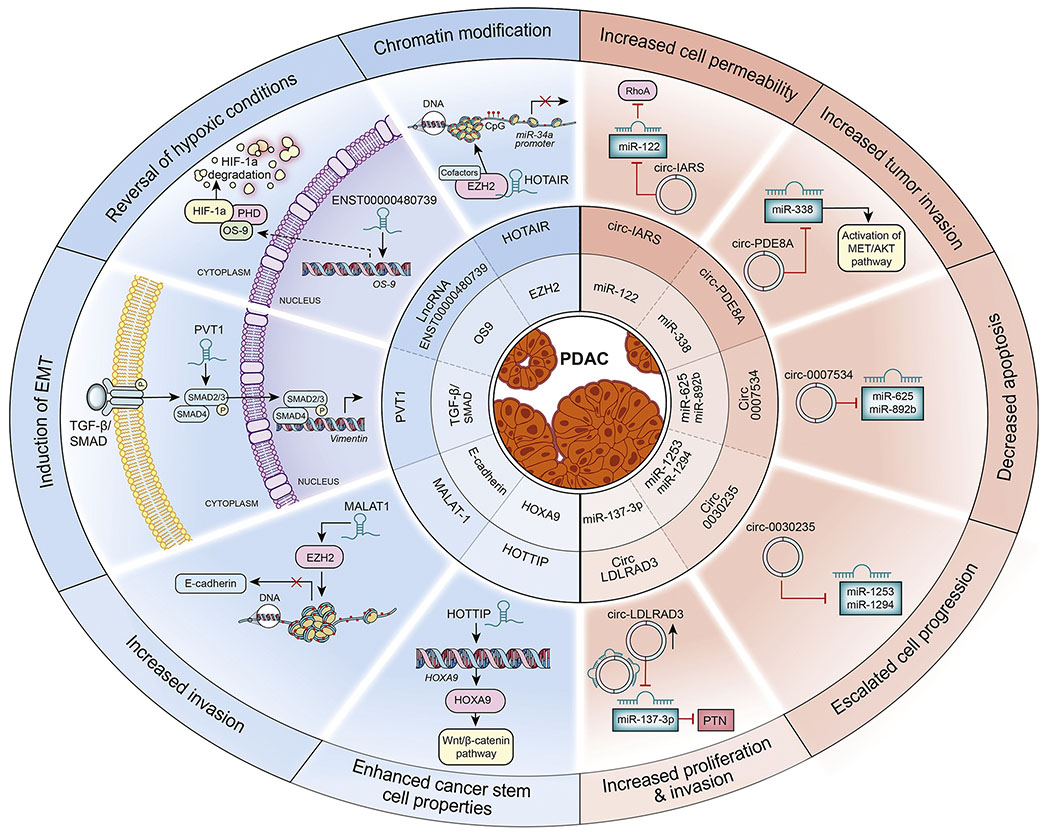
Figure 4:
Functional roles of lncRNAs and circRNAs in PDAC: The lncRNAs and circRNAs implicated in PDAC are shown. The left half of the circle shows the lncRNAs and their downstream targets, cellular phenotype and target genes or pathways affected. For example, HOTAIR interacts with EZH2 and binds to the promoter region of miR-34a. This binding leads to chromatin modification due to hypermethylation of miR-34a promoter repressing its expression. The lncRNA ENST00000480739 acts as an enhancer for OS9, increasing the expression of OS9. OS9 potentially binds to the ubiquitination complex comprised of prolyl hydroxylase (PHD) and other proteins which lead to the degradation of HIF-1α protein changing the hypoxic condition in the cells. The lncRNA PVT1 interacts with the SMAD2/3 proteins of the TGF-β/SMAD pathway, activating the expression of vimentin and inducing EMT changes in pancreatic cancer cells. The lncRNA MALAT1 interacts with EZH2 which in turn leads to the hypermethylation of E-cadherin promoter inhibiting its expression and increasing invasive potential of pancreatic cancer cells. The lncRNA HOTTIP regulate the expression of HOXA9, activating the Wnt-β-catenin pathway and increasing cancer stem cell properties of the pancreatic cancer cells. CircRNAs and their target miRNAs are shown in the right half of the circle. For example, circLDLRAD3 sequester miR-137-3p. In the absence of miR-137-3p, pleiotrophin (PTN) is expressed and leads to increased proliferation and invasion of pancreatic cancer cells. Circ0030235 inhibits the expression of miR-1253 and miR-1294 and leads to increased cell progression. Circ0007534 acts as a sponge for miR-625 and miR-892b and affects apoptosis. Circ-PDE8A targets miR-338 activating the MET/AKT pathway and increase invasive potential of pancreatic cancer cells. Circ-IARS increases the expression of RhoA by inhibiting miR-122 increasing the permeability and invasive potential of pancreatic cancer cells.
Table 3.
A list of candidates lncRNAs and circRNAs implicated in PDAC.
| LncRNA | Expression | Targets | Pathway | Effect on biological process | References |
|---|---|---|---|---|---|
| HOTAIR | Upregulated |
GDF15
NOTCH 3 DR5 miR-34 |
WNT | Increased proliferation Increased invasion Decreased apoptosis |
[126, 127] |
| HOTTIP | Upregulated |
PD-L1
HOXA13 |
WNT/ β-Catenin | Increased H3K27 trimethylation Increased cancer stem cell proliferation Increased cell survival Increased migration |
[140–142] |
| MALAT-1 | Upregulated | E-cadherin promoters NORG-1 CCND, MAPK8 VEGFA |
Hippo-YAP PI3K-AKT NF-κb mTOR MAPK WNT |
Increased growth increased invasion Increased EMT |
[135, 139] |
| ENST00000480739 | Down regulated |
HIF-1α OS-9 |
HIF | Increased invasion | [129] |
| PVT1 | Upregulated | MYC promoters | TGF- β/Smad | Increased cytoprotective autophagy Increased EMT |
[126] |
| CircRNA | Expression | Target miRNA | Pathway | Effect on biological process | References |
| circ-LDLRAD3 | Upregulated | miR-137-3p | PTN | Increased invasion Increased metastasis |
[156, 158] |
| circ_0030235 | Upregulated | miR-1253 miR-1294 |
- | Increased invasion Increased migration |
[159] |
| circ_0007534 | Upregulated | miR-625 and miR-892b | - | Increased proliferation Increased invasion Decreased apoptosis |
[162] |
| circ-PDE8A | Upregulated | miR-338 | MACC/MET/ERK | Increased invasion | [163] |
| circ-IARS | Upregulated | miR-122 | - | Increased metastasis | [164] |
| ciRS-7 | Upregulated | miR-7 | EGFR/STAT3 | Increased proliferation Increased metastasis |
[157] |
5.1. HOTAIR
The HOX antisense transcript intergenic RNA (HOTAIR) is a well-characterized lncRNA whose aberrant expression has been documented in several cancer types. It is transcribed from the homeobox C (HOXC) locus located on chromosome 12 [121, 122]. In PDAC, HOTAIR promotes cellular proliferation and metastasis [123]. HOTAIR interacts with human EZH2, a component of the polycomb repressive complex 2 (PRC2) and binds to the promoter region of miR-34. This binding leads to histone 3 lysine 27 (H3K27) trimethylation and eventually to the transcriptional repression of miR-34, an increase in cellular proliferation, and a decrease in apoptosis [123]. Kim et al. reported that HOTAIR targeted and bound the tumor suppressor gene GDF15, repressing its expression, which led to increased proliferation of pancreatic cancer cells [124]. These researchers also used a publicly-available database to illustrate that HOTAIR was overexpressed in PDAC specimens compared to normal pancreatic tissues, and in more aggressive tumors characterized by higher tumor and lymph node staging [124].
In a large study involving 900 PDAC and equal number of control specimens, a functional single nucleotide polymorphism (SNP) located within the 3’ UTR region of the HOTAIR gene was associated with a significantly higher risk for developing pancreatic cancer and higher HOTAIR expression [125]. Xie and colleagues noted that expression of HOTAIR was significantly higher in pancreatic tumor tissues [126], subsequently they validated their findings in salivary samples and noted that HOTAIR expression was also significantly higher in saliva from patients with PDAC vs. patients with benign tumors [126]. In another study, HOTAIR expression exhibited impressive diagnostic potential which was superior than the conventional CA19-9 marker for discriminating PDAC from benign pancreatic lesions and healthy controls. Increased levels of HOTAIR in serum also correlated with advanced PDAC stages [127]. HOTAIR was also shown to regulate the expression of the death receptor 5 (DR5) gene through epigenetic modulation and to contribute to tumor necrosis factor-related apoptosis-inducing ligand (TRAIL) resistance – suggesting it to serve as a potential target for sensitization of pancreatic cancer cells resistant to TRAIL therapy [128].
5.2. lncRNA ENST00000480739
The lncRNA ENST00000480739 functions as a tumor suppressor. In a study comprising of 35 patients with PDAC, expression of ENST00000480739 was lower in PDAC specimens compared to the adjacent normal tissues [129]. In addition, its expression negatively correlated with tumor stage and lymph node metastasis status. The authors demonstrated that in cultured cells, ENST00000480739 directly increased the expression of the osteosarcoma amplified 9 (OS-9) gene, both at the mRNA and protein level by activating its promoter region. Further knockdown experiments demonstrated that lncRNA ENST00000480739 negatively regulated HIF-1α by upregulating OS-9, thereby leading to a reduction in PDAC cell invasion [129].
5.3. PVT1
The lncRNA plasmacytoma variant translocation 1 (PVT1) is located at the 8q24.21 locus, and has been shown to interact with MYC gene promoter [130]. Several functions of PVT1 have been reported in pancreatic cancer cells, including promotion of EMT via the TGF- β/Smad pathway [131], acting as a miR-448 sponge to promote proliferation and migration [132], and modulating cytoprotective autophagy to promote PDAC development [133]. Xie et al. reported that PVT1 expression was higher in PDAC tissue and serum specimens compared to healthy specimens, and could serve as a diagnostic biomarker in this malignancy [126]. In comparison to the tumor marker CA19-9, PVT1 expression demonstrated higher sensitivity and specificity. In addition, the progression-free survival of patients with locally advanced or advanced PDAC treated with gemcitabine as first-line treatment was significantly higher in patients with low expression of PVT1; highlighting its potential as a prognostic biomarker in patients with PDAC treated with first-line gemcitabine-based treatment [134].
5.4. MALAT-1
The metastasis-associated lung adenocarcinoma transcript-1 (MALAT-1) is another lncRNA that is frequently overexpressed in PDAC. Expression of MALAT-1 also positively correlates with tumor size, clinical stage, lymph node metastasis, distant metastasis, and prognosis [135]. A negative correlation was observed in PDAC patients with higher MALAT-1 expression levels and disease-free survival [136]. Various transcriptional targets of MALAT-1 have been identified, including the promoter of E-cadherin [137], N-myc downregulated gene-1 (NDRG-1) [138], cyclin D1 (CCND1), RAF-mitogen-activated kinase 8 (MAPK8), and vascular endothelial growth factor A (VEGFA) [139]. Through RNA immunoprecipitation assays, MALAT1 was demonstrated to physically bind to EZH2 and negatively correlate with E-cadherin expression. Further investigations revealed that silencing of EZH2 increased E-cadherin expression in pancreatic cancer cells suggesting that MALAT1 potentially promotes pancreatic cancer cell migration and invasion through the repression of tumor suppressor gene E-cadherin [137]. Altogether, the findings suggest that MALAT1 plays an important role in PDAC pathogenesis, warranting further investigation.
5.5. HOTTIP
The HOXA distal transcript antisense RNA (HOTTIP) lncRNA is a HOX-related lncRNA. Similar to HOTAIR, the HOTTIP lncRNA is associated with chromatin-modification complexes and enhances H3K27 trimethylation to repress the expression of multiple HOXA genes [140]. In the context of PDAC, the HOTTIP lncRNA seems to regulate HOX genes i.e. HOXA9, HOXA10 and HOXA11. The HOXA9 gene has been shown to promote cancer stem cell proliferation through the Wnt/β-catenin signaling pathway [141, 142]. Likewise, the HOXA10 and HOXA11 genes regulate expression of matrix metalloproteinase 3 and 2 genes which promote invasion in pancreatic cancer cells [142]. Increased expression of HOTTIP has been documented in various pancreatic cancer cell lines as well as tissue specimens [143]. The same study also demonstrated that downregulation of HOTTIP expression increased the anti-tumor effects of gemcitabine in cultured cells and animal models.
6. CircRNAs as Biomarkers in PDAC
CircRNAs are another class of ncRNAs that are currently a popular research topic. circRNAs are circular in shape and lack a 5’ cap or 3’ Poly-A tail terminal ends. These ncRNAs are more stable than other endogenous mRNAs due to their circular shape, which protects them from traditional RNA-mediated degradation [144]. circRNAs have been proposed to function as miRNA sponges that suppress the ability of miRNAs to bind to their target mRNAs [145,146]. The role of miRNAs in various cellular processes and diseases are well established. Considering that circRNAs interact with miRNAs, their association is also implicated in various diseases, including cancer [147–151]. The role of circRNAs in cancer and the regulation of key cellular processes by circRNAs are reviewed elsewhere [152].
A number of studies have reported aberrant expression of circRNAs in PDAC (Table 3). Li et al. performed one of the very first studies to investigate the expression of circRNAs in PDAC, using a microarray expression approach in six pairs of PDAC samples and matched adjacent normal tissues [153]. They identified a number of up- and downregulated circRNAs between PDAC and adjacent normal tissues, among which they further validated seven circRNAs using quantitative real-time PCR assays [153]. A subsequent study identified 115 upregulated and 141 downregulated circRNAs in PDAC compared to adjacent normal mucosa specimens, through the analysis of publicly-available microarray datasets [154]. These authors also considered potential circRNA:miRNA interactions and performed pathway analysis to discover that the B-Raf proto-oncogene, serine/threonine kinase (BRAF) and Dual specificity mitogen-activated protein kinase kinase 2 (MAP2K2) interacted with the most number of pathways, indicating the involvement of MAPK signaling pathway. Li et al. investigated circRNA expression in extracellular vesicles derived from the plasma of patients with PDAC and uncovered 453 circRNAs that were differentially expressed between patients and healthy controls [155]. As illustrated in Figure 4, several studies have also shown the aberrant expression of circRNAs in PDAC and their association with pancreatic cancer cell proliferation and metastasis [156, 157].
In view of their stable expression, circRNAs represent potentially viable biomarkers for PDAC. Yang et al. found circ-LDLRAD3 was upregulated in PDAC tissues, plasma, and PDAC cell lines. They also reported a positive correlation between the expression of circ-LDLRAD3 and clinicopathological factors, such as venous and lymphatic invasion, in the patient specimens [158]. Moreover, to investigate the suitability of circ-LDLRAD3 as a PDAC diagnostic biomarker, the authors also found that when combined with CA19-9, circ-LDLRAD3 exhibited superior diagnostic sensitivity and specificity vs. CA19-9 [158]. The circ-LDLRAD3 was shown to directly target miR-137-3p and to regulate proliferation, migration, and invasion in pancreatic cancer cells through the miR-137-3p/pleiotrophin (PTN) axis [156].
Likewise, the circ_0030235 is another circRNA overexpressed in PDAC tissues and cell lines [153, 159]. Functional studies revealed that overexpression of circ_0030235 associated with advanced tumor stage and positive lymph node metastasis in patients with PDAC, suggesting that it may function as an oncogene. Additionally, circ_0030235 is thought to act as a molecular sponge for miR-1253 and miR-1294, as the two miRNAs were found to be significantly downregulated in its presence [159]. These two miRNAs have been shown to act as tumor suppressors in several other cancers as well. The miR-1253 inhibits cell growth and invasion by targeting WNT5A [160] while miR-1294 has been shown to inhibit cell proliferation by targeting c-Myc [161]. In a similar fashion, circ_0007534 was found to be upregulated in PDAC tissues and correlated with aggressive phenotypes [162]. Furthermore, circ_0007534 was shown to promote the oncogenic potential of cells by enhancing cell proliferation, migration, and invasion in PDAC cell lines; and miR-625 and miR-892b were identified as the targets of circ_0007534 [162].
The stable nature of circRNAs in serum provides a rationale for investigating circulating circRNAs as cancer biomarkers [163, 164]. In this context, circ-PDE8A was identified in exosomes derived from the plasma of patients with PDAC, and it was shown that its high expression associated with disease progression and poor prognosis. Further mechanistic investigations revealed that circ-PDE8A could activate the MET tyrosine kinase receptor and act as a sponge for miR-338 [163]. The circ-IARS was also found to be highly expressed in the exosomes of patients with PDAC and positively correlated with tumor and lymph node metastasis status [164]. Functional assays revealed that circ-IARS increases the activity of RhoA by absorbing miR-122, resulting in an increased permeability of the cells promoting metastasis [164].
Another circRNA that is implicated in PDAC is ciRS-7. Previously, ciRS-7 was shown to act as a miR-7 sponge in colorectal and hepatocellular cancer [165, 166]. In pancreatic cancer, ciRS-7 was found to be upregulated in PDAC tissues as compared to normal tissues. Using pancreatic cancer cells, ciRS-7 was shown to act as a miR-7 sponge, and ciRS-7 knockdown led to a decrease in EGFR and STAT3 expression suppressing proliferation and reducing invasion of PDAC cells [157].
Although only a limited number of studies have investigated the role of circRNAs in PDAC to date, these studies suggest that aberrantly expressed circRNAs might be involved in the regulation of PDAC and could potentially serve as diagnostic makers.
7. Conclusion and Future Perspectives
The only chance of a complete cure in PDAC is highly dependent on early disease diagnosis, upon which curative treatments can be implemented. Thus, there remains an unmet clinical need for the identification and development of highly sensitive and specific diagnostic biomarkers that can detect PDAC and its precursor lesions at their earliest stage. Given that the effective treatment options in PDAC patients are still limited, availability of improved prognostic and predictive biomarkers are also desirable in PDAC – which can help adequate risk-stratification and selection of patients for specific treatment regimens. Various studies have revealed that ncRNAs are important regulators of a multitude of cancer-associated mechanisms in PDAC, including cellular proliferation, invasion and apoptotic deregulation – making them attractive candidates for diagnostic, predictive, and prognostic biomarkers.
In spite of the substantial enthusiasm for miRNAs as robust biomarkers in PDAC, to date, not a single miRNA biomarker has made it to the clinic for the diagnosis of patients with PDAC. This can be attributed to various limitations of the existing studies, including lack of a systematic biomarker discovery and validation approach, analysis of well-defined clinical cohorts with adequate statistical considerations, and the use of inconsistent analytical and biomarker normalization approaches. In the case of cell-free miRNAs, their heterogenous origin is also a potential limiting factor. With regards to lncRNAs, while the data to date seems promising, further studies are needed to systematically dissect the full potential of lncRNAs as biomarkers in PDAC. Finally, circRNAs are an exciting and emerging concept; however, the research in this regard is still in its infancy. As it stands currently, more sophisticated bioinformatic approaches are needed to identify the entire compendium of circRNAs in PDAC, followed by their functional characterization – which eventually will provide the springboard for their development into diagnostic, prognostic, and predictive biomarkers in PDAC.
Taken together, the expression profiles of various ncRNAs offer an exciting opportunity for development into biomarkers for PDAC. However, before we get to that point, substantial challenges must be overcome, including careful planning of large-scale prospective human studies to validate the clinical significance of most promising PDAC-associated ncRNA biomarkers. In addition, concurrent functional studies to unravel the functional underpinnings for their mechanistic role will lend further credence to their biomarker potential; which collectively will provide the much needed rationale and confidence for their translation into the clinic as we usher into the exciting era of precision oncology.
ACKNOWLEDGEMENTS
We would like to thank Dr. Sarah T. Wilkinson for her help in editing of the manuscript and Tiffany S. DaVanzo for her assistance in developing figure illustrations for this article.
Funding: The present work was supported by the CA72851, CA187956, CA202797 and CA214254 grants from the National Cancer Institute, National Institute of Health and through a pilot research award from the City of Hope Ludwig Cancer Research-Hilton Foundation Partnership award.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of Interest: None of the authors has any potential conflicts to disclose
REFERENCES
- [1].National Cancer Institute. Surveillance, epidemiology, and end results program. Cancer stat facts: pancreas cancer., 2019. http://seer.cancer.gov/statfacts/html/pancreas.html. .
- [2].Siegel RL, Miller KD, Jemal A, Cancer statistics, 2018, CA Cancer J Clin 68(1) (2018) 7–30. [DOI] [PubMed] [Google Scholar]
- [3].Oettle H, Neuhaus P, Hochhaus A, Hartmann JT, Gellert K, Ridwelski K, Niedergethmann M, Zulke C, Fahlke J, Arning MB, Sinn M, Hinke A, Riess H, Adjuvant chemotherapy with gemcitabine and long-term outcomes among patients with resected pancreatic cancer: the CONKO-001 randomized trial, JAMA 310(14) (2013) 1473–81. [DOI] [PubMed] [Google Scholar]
- [4].Neoptolemos JP, Stocken DD, Friess H, Bassi C, Dunn JA, Hickey H, Beger H, Fernandez-Cruz L, Dervenis C, Lacaine F, Falconi M, Pederzoli P, Pap A, Spooner D, Kerr DJ, Buchler MW, C. European Study Group for Pancreatic, A randomized trial of chemoradiotherapy and chemotherapy after resection of pancreatic cancer, N Engl J Med 350(12) (2004) 1200–10. [DOI] [PubMed] [Google Scholar]
- [5].Bockhorn M, Uzunoglu FG, Adham M, Imrie C, Milicevic M, Sandberg AA, Asbun HJ, Bassi C, Buchler M, Charnley RM, Conlon K, Cruz LF, Dervenis C, Fingerhutt A, Friess H, Gouma DJ, Hartwig W, Lillemoe KD, Montorsi M, Neoptolemos JP, Shrikhande SV, Takaori K, Traverso W, Vashist YK, Vollmer C, Yeo CJ, Izbicki JR, S. International Study Group of Pancreatic, Borderline resectable pancreatic cancer: a consensus statement by the International Study Group of Pancreatic Surgery (ISGPS), Surgery 155(6) (2014) 977–88. [DOI] [PubMed] [Google Scholar]
- [6].Gillen S, Schuster T, Meyer Zum Buschenfelde C, Friess H, Kleeff J, Preoperative/neoadjuvant therapy in pancreatic cancer: a systematic review and meta-analysis of response and resection percentages, PLoS Med 7(4) (2010) e1000267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Golcher H, Brunner TB, Witzigmann H, Marti L, Bechstein WO, Bruns C, Jungnickel H, Schreiber S, Grabenbauer GG, Meyer T, Merkel S, Fietkau R, Hohenberger W, Neoadjuvant chemoradiation therapy with gemcitabine/cisplatin and surgery versus immediate surgery in resectable pancreatic cancer: results of the first prospective randomized phase II trial, Strahlenther Onkol 191(1) (2015) 7–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Heinrich S, Pestalozzi B, Lesurtel M, Berrevoet F, Laurent S, Delpero JR, Raoul JL, Bachellier P, Dufour P, Moehler M, Weber A, Lang H, Rogiers X, Clavien PA, Adjuvant gemcitabine versus NEOadjuvant gemcitabine/oxaliplatin plus adjuvant gemcitabine in resectable pancreatic cancer: a randomized multicenter phase III study (NEOPAC study), BMC Cancer 11 (2011) 346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Tachezy M, Gebauer F, Petersen C, Arnold D, Trepel M, Wegscheider K, Schafhausen P, Bockhorn M, Izbicki JR, Yekebas E, Sequential neoadjuvant chemoradiotherapy (CRT) followed by curative surgery vs. primary surgery alone for resectable, non-metastasized pancreatic adenocarcinoma: NEOPA- a randomized multicenter phase III study (NCT01900327, DRKS00003893, ISRCTN82191749), BMC Cancer 14 (2014) 411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Hackert T, Sachsenmaier M, Hinz U, Schneider L, Michalski CW, Springfeld C, Strobel O, Jager D, Ulrich A, Buchler MW, Locally Advanced Pancreatic Cancer: Neoadjuvant Therapy With Folfirinox Results in Resectability in 60% of the Patients, Ann Surg 264(3) (2016) 457–63. [DOI] [PubMed] [Google Scholar]
- [11].Murphy JE, Wo JY, Ryan DP, Jiang W, Yeap BY, Drapek LC, Blaszkowsky LS, Kwak EL, Allen JN, Clark JW, Faris JE, Zhu AX, Goyal L, Lillemoe KD, DeLaney TF, Fernandez-Del Castillo C, Ferrone CR, Hong TS, Total Neoadjuvant Therapy With FOLFIRINOX Followed by Individualized Chemoradiotherapy for Borderline Resectable Pancreatic Adenocarcinoma: A Phase 2 Clinical Trial, JAMA Oncol 4(7) (2018) 963–969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Conroy T, Hammel P, Hebbar M, Ben Abdelghani M, Wei AC, Raoul JL, Chone L, Francois E, Artru P, Biagi JJ, Lecomte T, Assenat E, Faroux R, Ychou M, Volet J, Sauvanet A, Breysacher G, Di Fiore F, Cripps C, Kavan P, Texereau P, Bouhier-Leporrier K, Khemissa-Akouz F, Legoux JL, Juzyna B, Gourgou S, O’Callaghan CJ, Jouffroy-Zeller C, Rat P, Malka D, Castan F, Bachet JB, G. Canadian Cancer Trials, G.I.P.G. the Unicancer, FOLFIRINOX or Gemcitabine as Adjuvant Therapy for Pancreatic Cancer, N Engl J Med 379(25) (2018) 2395–2406. [DOI] [PubMed] [Google Scholar]
- [13].Patz EF Jr., Greco E, Gatsonis C, Pinsky P, Kramer BS, Aberle DR, Lung cancer incidence and mortality in National Lung Screening Trial participants who underwent low-dose CT prevalence screening: a retrospective cohort analysis of a randomised, multicentre, diagnostic screening trial, Lancet Oncol 17(5) (2016) 590–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Berry DA, Cronin KA, Plevritis SK, Fryback DG, Clarke L, Zelen M, Mandelblatt JS, Yakovlev AY, Habbema JD, Feuer EJ, Cancer I, C. Surveillance Modeling Network, Effect of screening and adjuvant therapy on mortality from breast cancer, N Engl J Med 353(17) (2005) 1784–92. [DOI] [PubMed] [Google Scholar]
- [15].Hoffman RM, Clinical practice. Screening for prostate cancer, N Engl J Med 365(21) (2011) 2013–9. [DOI] [PubMed] [Google Scholar]
- [16].Levin TR, Corley DA, Jensen CD, Schottinger JE, Quinn VP, Zauber AG, Lee JK, Zhao WK, Udaltsova N, Ghai NR, Lee AT, Quesenberry CP, Fireman BH, Doubeni CA, Effects of Organized Colorectal Cancer Screening on Cancer Incidence and Mortality in a Large Community-Based Population, Gastroenterology 155(5) (2018) 1383–1391 e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].A.C. Society, Cancer Prevention & Early Detection Facts & Figures 2019-2020, American Cancer Society (2019) 1–59. [Google Scholar]
- [18].Rahib L, Smith BD, Aizenberg R, Rosenzweig AB, Fleshman JM, Matrisian LM, Projecting cancer incidence and deaths to 2030: the unexpected burden of thyroid, liver, and pancreas cancers in the United States, Cancer Res 74(11) (2014) 2913–21. [DOI] [PubMed] [Google Scholar]
- [19].Xing H, Wang J, Wang Y, Tong M, Hu H, Huang C, Li D, Diagnostic Value of CA 19-9 and Carcinoembryonic Antigen for Pancreatic Cancer: A Meta-Analysis, Gastroenterol Res Pract 2018 (2018)8704751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Hruban RH, Goggins M, Parsons J, Kern SE, Progression model for pancreatic cancer, Clin Cancer Res 6(8) (2000) 2969–72. [PubMed] [Google Scholar]
- [21].Hruban RH, Maitra A, Kern SE, Goggins M, Precursors to pancreatic cancer, Gastroenterol Clin North Am 36(4) (2007) 831–49, vi. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Matthaei H, Schulick RD, Hruban RH, Maitra A, Cystic precursors to invasive pancreatic cancer, Nat Rev Gastroenterol Hepatol 8(3) (2011) 141–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Hruban RH, Adsay NV, Albores-Saavedra J, Compton C, Garrett ES, Goodman SN, Kern SE, Klimstra DS, Kloppel G, Longnecker DS, Luttges J, Offerhaus GJ, Pancreatic intraepithelial neoplasia: a new nomenclature and classification system for pancreatic duct lesions, Am J Surg Pathol 25(5) (2001) 579–86. [DOI] [PubMed] [Google Scholar]
- [24].Zamboni G, Hirabayashi K, Castelli P, Lennon AM, Precancerous lesions of the pancreas, Best Pract Res Clin Gastroenterol 27(2) (2013) 299–322. [DOI] [PubMed] [Google Scholar]
- [25].Hruban RH, Wilentz RE, Kern SE, Genetic progression in the pancreatic ducts, Am J Pathol 156(6) (2000) 1821–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Basturk O, Hong SM, Wood LD, Adsay NV, Albores-Saavedra J, Biankin AV, Brosens LA, Fukushima N, Goggins M, Hruban RH, Kato Y, Klimstra DS, Kloppel G, Krasinskas A, Longnecker DS, Matthaei H, Offerhaus GJ, Shimizu M, Takaori K, Terris B, Yachida S, Esposito I, Furukawa T, M. Baltimore Consensus, A Revised Classification System and Recommendations From the Baltimore Consensus Meeting for Neoplastic Precursor Lesions in the Pancreas, Am J Surg Pathol 39(12) (2015) 1730–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Sahani DV, Lin DJ, Venkatesan AM, Sainani N, Mino-Kenudson M, Brugge WR, Fernandez-Del-Castillo C, Multidisciplinary approach to diagnosis and management of intraductal papillary mucinous neoplasms of the pancreas, Clin Gastroenterol Hepatol 7(3) (2009) 259–69. [DOI] [PubMed] [Google Scholar]
- [28].Crippa S, Salvia R, Warshaw AL, Dominguez I, Bassi C, Falconi M, Thayer SP, Zamboni G, Lauwers GY, Mino-Kenudson M, Capelli P, Pederzoli P, Castillo CF, Mucinous cystic neoplasm of the pancreas is not an aggressive entity: lessons from 163 resected patients, Ann Surg 247(4) (2008) 571–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Yamao K, Yanagisawa A, Takahashi K, Kimura W, Doi R, Fukushima N, Ohike N, Shimizu M, Hatori T, Nobukawa B, Hifumi M, Kobayashi Y, Tobita K, Tanno S, Sugiyama M, Miyasaka Y, Nakagohri T, Yamaguchi T, Hanada K, Abe H, Tada M, Fujita N, Tanaka M, Clinicopathological features and prognosis of mucinous cystic neoplasm with ovarian-type stroma: a multi-institutional study of the Japan pancreas society, Pancreas 40(1) (2011) 67–71. [DOI] [PubMed] [Google Scholar]
- [30].Yachida S, Jones S, Bozic I, Antal T, Leary R, Fu B, Kamiyama M, Hruban RH, Eshleman JR, Nowak MA, Velculescu VE, Kinzler KW, Vogelstein B, Iacobuzio-Donahue CA, Distant metastasis occurs late during the genetic evolution of pancreatic cancer, Nature 467(7319) (2010) 1114–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Lennon AM, Wolfgang CL, Canto MI, Klein AP, Herman JM, Goggins M, Fishman EK, Kamel I, Weiss MJ, Diaz LA, Papadopoulos N, Kinzler KW, Vogelstein B, Hruban RH, The early detection of pancreatic cancer: what will it take to diagnose and treat curable pancreatic neoplasia?, Cancer Res 74(13) (2014) 3381–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Kaneko OF, Lee DM, Wong J, Kadell BM, Reber HA, Lu DS, Raman SS, Performance of multidetector computed tomographic angiography in determining surgical resectability of pancreatic head adenocarcinoma, J Comput Assist Tomogr 34(5) (2010) 732–8. [DOI] [PubMed] [Google Scholar]
- [33].Adamek HE, Albert J, Breer H, Weitz M, Schilling D, Riemann JF, Pancreatic cancer detection with magnetic resonance cholangiopancreatography and endoscopic retrograde cholangiopancreatography: a prospective controlled study, Lancet 356(9225) (2000) 190–3. [DOI] [PubMed] [Google Scholar]
- [34].Poruk KE, Firpo MA, Adler DG, Mulvihill SJ, Screening for pancreatic cancer: why, how, and who?, Ann Surg 257(1) (2013) 17–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Canto MI, Harinck F, Hruban RH, Offerhaus GJ, Poley JW, Kamel I, Nio Y, Schulick RS, Bassi C, Kluijt I, Levy MJ, Chak A, Fockens P, Goggins M, Bruno M, International C Cancer of Pancreas Screening, International Cancer of the Pancreas Screening (CAPS) Consortium summit on the management of patients with increased risk for familial pancreatic cancer, Gut 62(3) (2013) 339–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Harinck F, Konings IC, Kluijt I, Poley JW, van Hooft JE, van Dullemen HM, Nio CY, Krak NC, Hermans JJ, Aalfs CM, Wagner A, Sijmons RH, Biermann K, van Eijck CH, Gouma DJ, Dijkgraaf MG, Fockens P, Bruno MJ, i. Dutch research group on pancreatic cancer surveillance in high-risk, A multicentre comparative prospective blinded analysis of EUS and MRI for screening of pancreatic cancer in high-risk individuals, Gut 65(9) (2016) 1505–13. [DOI] [PubMed] [Google Scholar]
- [37].Locker GY, Hamilton S, Harris J, Jessup JM, Kemeny N, Macdonald JS, Somerfield MR, Hayes DF, Bast RC Jr., Asco, ASCO 2006 update of recommendations for the use of tumor markers in gastrointestinal cancer, J Clin Oncol 24(33) (2006) 5313–27. [DOI] [PubMed] [Google Scholar]
- [38].Goggins M, Molecular markers of early pancreatic cancer, J Clin Oncol 23(20) (2005) 4524–31. [DOI] [PubMed] [Google Scholar]
- [39].Capello M, Bantis LE, Scelo G, Zhao Y, Li P, Dhillon DS, Patel NJ, Kundnani DL, Wang H, Abbruzzese JL, Maitra A, Tempero MA, Brand R, Firpo MA, Mulvihill SJ, Katz MH, Brennan P, Feng Z, Taguchi A, Hanash SM, Sequential Validation of Blood-Based Protein Biomarker Candidates for Early-Stage Pancreatic Cancer, J Natl Cancer Inst 109(4) (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Kim J, Bamlet WR, Oberg AL, Chaffee KG, Donahue G, Cao XJ, Chari S, Garcia BA, Petersen GM, Zaret KS, Detection of early pancreatic ductal adenocarcinoma with thrombospondin-2 and CA19-9 blood markers, Sci Transl Med 9(398) (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Kaur S, Smith LM, Patel A, Menning M, Watley DC, Malik SS, Krishn SR, Mallya K, Aithal A, Sasson AR, Johansson SL, Jain M, Singh S, Guha S, Are C, Raimondo M, Hollingsworth MA, Brand RE, Batra SK, A Combination of MUC5AC and CA19-9 Improves the Diagnosis of Pancreatic Cancer: A Multicenter Study, Am J Gastroenterol 112(1) (2017) 172–183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Cohen JD, Javed AA, Thoburn C, Wong F, Tie J, Gibbs P, Schmidt CM, Yip-Schneider MT, Allen PJ, Schattner M, Brand RE, Singhi AD, Petersen GM, Hong SM, Kim SC, Falconi M, Doglioni C, Weiss MJ, Ahuja N, He J, Makary MA, Maitra A, Hanash SM, Dal Molin M, Wang Y, Li L, Ptak J, Dobbyn L, Schaefer J, Silliman N, Popoli M, Goggins MG, Hruban RH, Wolfgang CL, Klein AP, Tomasetti C, Papadopoulos N, Kinzler KW, Vogelstein B, Lennon AM, Combined circulating tumor DNA and protein biomarker-based liquid biopsy for the earlier detection of pancreatic cancers, Proc Natl Acad Sci U S A 114(38) (2017) 10202–10207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Dahlem C, Barghash A, Puchas P, Haybaeck J, Kessler SM, The Insulin-Like Growth Factor 2 mRNA Binding Protein IMP2/IGF2BP2 is Overexpressed and Correlates with Poor Survival in Pancreatic Cancer, Int J Mol Sci 20(13) (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Golob-Schwarzl N, Puchas P, Gogg-Kamerer M, Weichert W, Goppert B, Haybaeck J, New Pancreatic Cancer Biomarkers eIF1, eIF2D, eIF3C and eIF6 Play a Major Role in Translational Control in Ductal Adenocarcinoma, Anticancer Res 40(6) (2020) 3109–3118. [DOI] [PubMed] [Google Scholar]
- [45].Esteller M, Non-coding RNAs in human disease, Nat Rev Genet 12(12) (2011) 861–74. [DOI] [PubMed] [Google Scholar]
- [46].Adams BD, Parsons C, Walker L, Zhang WC, Slack FJ, Targeting noncoding RNAs in disease, J Clin Invest 127(3) (2017) 761–771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Anfossi S, Babayan A, Pantel K, Calin GA, Clinical utility of circulating non-coding RNAs - an update, Nat Rev Clin Oncol 15(9) (2018) 541–563. [DOI] [PubMed] [Google Scholar]
- [48].Ferrero G, Cordero F, Tarallo S, Arigoni M, Riccardo F, Gallo G, Ronco G, Allasia M, Kulkarni N, Matullo G, Vineis P, Calogero RA, Pardini B, Naccarati A, Small non-coding RNA profiling in human biofluids and surrogate tissues from healthy individuals: description of the diverse and most represented species, Oncotarget 9(3) (2018) 3097–3111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Yeri A, Courtright A, Reiman R, Carlson E, Beecroft T, Janss A, Siniard A, Richholt R, Balak C, Rozowsky J, Kitchen R, Hutchins E, Winarta J, McCoy R, Anastasi M, Kim S, Huentelman M, Van Keuren-Jensen K, Total Extracellular Small RNA Profiles from Plasma, Saliva, and Urine of Healthy Subjects, Sci Rep 7 (2017) 44061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Umu SU, Langseth H, Bucher-Johannessen C, Fromm B, Keller A, Meese E, Lauritzen M, Leithaug M, Lyle R, Rounge TB, A comprehensive profile of circulating RNAs in human serum, RNA Biol 15(2) (2018) 242–250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Yuan T, Huang X, Woodcock M, Du M, Dittmar R, Wang Y, Tsai S, Kohli M, Boardman L, Patel T, Wang L, Plasma extracellular RNA profiles in healthy and cancer patients, Sci Rep 6 (2016) 19413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Taucher V, Mangge H, Haybaeck J, Non-coding RNAs in pancreatic cancer: challenges and opportunities for clinical application, Cell Oncol (Dordr) 39(4) (2016) 295–318. [DOI] [PubMed] [Google Scholar]
- [53].Krol J, Loedige I, Filipowicz W, The widespread regulation of microRNA biogenesis, function and decay, Nat Rev Genet 11(9) (2010) 597–610. [DOI] [PubMed] [Google Scholar]
- [54].Thomson JM, Newman M, Parker JS, Morin-Kensicki EM, Wright T, Hammond SM, Extensive post-transcriptional regulation of microRNAs and its implications for cancer, Genes Dev 20(16) (2006) 2202–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Lee EJ, Gusev Y, Jiang J, Nuovo GJ, Lerner MR, Frankel WL, Morgan DL, Postier RG, Brackett DJ, Schmittgen TD, Expression profiling identifies microRNA signature in pancreatic cancer, Int J Cancer 120(5) (2007) 1046–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Szafranska AE, Davison TS, John J, Cannon T, Sipos B, Maghnouj A, Labourier E, Hahn SA, MicroRNA expression alterations are linked to tumorigenesis and non-neoplastic processes in pancreatic ductal adenocarcinoma, Oncogene 26(30) (2007) 4442–52. [DOI] [PubMed] [Google Scholar]
- [57].Bloomston M, Frankel WL, Petrocca F, Volinia S, Alder H, Hagan JP, Liu CG, Bhatt D, Taccioli C, Croce CM, MicroRNA expression patterns to differentiate pancreatic adenocarcinoma from normal pancreas and chronic pancreatitis, JAMA 297(17) (2007) 1901–8. [DOI] [PubMed] [Google Scholar]
- [58].Hanoun N, Delpu Y, Suriawinata AA, Bournet B, Bureau C, Selves J, Tsongalis GJ, Dufresne M, Buscail L, Cordelier P, Torrisani J, The silencing of microRNA 148a production by DNA hypermethylation is an early event in pancreatic carcinogenesis, Clin Chem 56(7) (2010) 1107–18. [DOI] [PubMed] [Google Scholar]
- [59].Zhang S, Hao J, Xie F, Hu X, Liu C, Tong J, Zhou J, Wu J, Shao C, Downregulation of miR-132 by promoter methylation contributes to pancreatic cancer development, Carcinogenesis 32(8) (2011) 1183–9. [DOI] [PubMed] [Google Scholar]
- [60].Vychytilova-Faltejskova P, Kiss I, Klusova S, Hlavsa J, Prochazka V, Kala Z, Mazanec J, Hausnerova J, Kren L, Hermanova M, Lenz J, Karasek P, Vyzula R, Slaby O, MiR-21, miR-34a, miR-198 and miR-217 as diagnostic and prognostic biomarkers for chronic pancreatitis and pancreatic ductal adenocarcinoma, Diagn Pathol 10 (2015) 38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Long MM, Zhan M, Xu SW, Yang RM, Chen W, Zhang SL, Shi YH, He Q, Mohan M, Liu Q, Wang J, miR-92b-3p acts as a tumor suppressor by targeting Gabra3 in pancreatic cancer, Molecular Cancer 16 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Sadakari Y, Ohtsuka T, Ohuchida K, Tsutsumi K, Takahata S, Nakamura M, Mizumoto K, Tanaka M, MicroRNA expression analyses in preoperative pancreatic juice samples of pancreatic ductal adenocarcinoma, JOP 11(6) (2010) 587–92. [PubMed] [Google Scholar]
- [63].Caponi S, Funel N, Frampton AE, Mosca F, Santarpia L, Van der Velde AG, Jiao LR, De Lio N, Falcone A, Kazemier G, Meijer GA, Verheul HM, Vasile E, Peters GJ, Boggi U, Giovannetti E, The good, the bad and the ugly: a tale of miR-101, miR-21 and miR-155 in pancreatic intraductal papillary mucinous neoplasms, Ann Oncol 24(3) (2013) 734–741. [DOI] [PubMed] [Google Scholar]
- [64].Wang J, Chen J, Chang P, LeBlanc A, Li D, Abbruzzesse JL, Frazier ML, Killary AM, Sen S, MicroRNAs in plasma of pancreatic ductal adenocarcinoma patients as novel blood-based biomarkers of disease, Cancer Prev Res (Phila) 2(9) (2009) 807–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Zhang J, Zhao CY, Zhang SH, Yu DH, Chen Y, Liu QH, Shi M, Ni CR, Zhu MH, Upregulation of miR-194 contributes to tumor growth and progression in pancreatic ductal adenocarcinoma, Oncol Rep 31(3) (2014) 1157–64. [DOI] [PubMed] [Google Scholar]
- [66].Slater EP, Strauch K, Rospleszcz S, Ramaswamy A, Esposito I, Kloppel G, Matthai E, Heeger K, Fendrich V, Langer P, Bartsch DK, MicroRNA-196a and-196b as Potential Biomarkers for the Early Detection of Familial Pancreatic Cancer, Translational Oncology 7(4) (2014) 464–471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Schultz NA, Werner J, Willenbrock H, Roslind A, Giese N, Horn T, Wojdemann M, Johansen JS, MicroRNA expression profiles associated with pancreatic adenocarcinoma and ampullary adenocarcinoma, Mod Pathol 25(12) (2012) 1609–22. [DOI] [PubMed] [Google Scholar]
- [68].Li A, Yu J, Kim H, Wolfgang CL, Canto MI, Hruban RH, Goggins M, MicroRNA array analysis finds elevated serum miR-1290 accurately distinguishes patients with low-stage pancreatic cancer from healthy and disease controls, Clin Cancer Res 19(13) (2013) 3600–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Munding JB, Adai AT, Maghnouj A, Urbanik A, Zollner H, Liffers ST, Chromik AM, Uhl W, Szafranska-Schwarzbach AE, Tannapfel A, Hahn SA, Global microRNA expression profiling of microdissected tissues identifies miR-135b as a novel biomarker for pancreatic ductal adenocarcinoma, International Journal of Cancer 131(2) (2012) E86–E95. [DOI] [PubMed] [Google Scholar]
- [70].Liu J, Gao J, Du Y, Li Z, Ren Y, Gu J, Wang X, Gong Y, Wang W, Kong X, Combination of plasma microRNAs with serum CA19-9 for early detection of pancreatic cancer, Int J Cancer 131(3) (2012) 683–91. [DOI] [PubMed] [Google Scholar]
- [71].Zhao WG, Yu SN, Lu ZH, Ma YH, Gu YM, Chen J, The miR-217 microRNA functions as a potential tumor suppressor in pancreatic ductal adenocarcinoma by targeting KRAS, Carcinogenesis 31(10) (2010) 1726–33. [DOI] [PubMed] [Google Scholar]
- [72].Kent OA, Chivukula RR, Mullendore M, Wentzel EA, Feldmann G, Lee KH, Liu S, Leach SD, Maitra A, Mendell JT, Repression of the miR-143/145 cluster by oncogenic Ras initiates a tumor-promoting feed-forward pathway, Genes Dev 24(24) (2010) 2754–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Song SD, Zhou J, Zhou J, Zhao H, Cen JN, Li DC, MicroRNA-375 targets the 3-phosphoinositide-dependent protein kinase-1 gene in pancreatic carcinoma, Oncol Lett 6(4) (2013) 953–959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Zhou J, Song S, He S, Zhu X, Zhang Y, Yi B, Zhang B, Qin G, Li D, MicroRNA-375 targets PDK1 in pancreatic carcinoma and suppresses cell growth through the Akt signaling pathway, Int J Mol Med 33(4) (2014) 950–6. [DOI] [PubMed] [Google Scholar]
- [75].Kadera BE, Li L, Toste PA, Wu N, Adams C, Dawson DW, Donahue TR, MicroRNA-21 in pancreatic ductal adenocarcinoma tumor-associated fibroblasts promotes metastasis, PLoS One 8(8) (2013)e71978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Dillhoff M, Liu J, Frankel W, Croce C, Bloomston M, MicroRNA-21 is overexpressed in pancreatic cancer and a potential predictor of survival, J Gastrointest Surg 12(12) (2008) 2171–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].Giovannetti E, Funel N, Peters GJ, Del Chiaro M, Erozenci LA, Vasile E, Leon LG, Pollina LE, Groen A, Falcone A, Danesi R, Campani D, Verheul HM, Boggi U, MicroRNA-21 in pancreatic cancer: correlation with clinical outcome and pharmacologic aspects underlying its role in the modulation of gemcitabine activity, Cancer Res 70(11) (2010) 4528–38. [DOI] [PubMed] [Google Scholar]
- [78].Hwang JH, Voortman J, Giovannetti E, Steinberg SM, Leon LG, Kim YT, Funel N, Park JK, Kim MA, Kang GH, Kim SW, Del Chiaro M, Peters GJ, Giaccone G, Identification of microRNA-21 as a biomarker for chemoresistance and clinical outcome following adjuvant therapy in resectable pancreatic cancer, PLoS One 5(5) (2010) e10630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [79].Liu J, Xu D, Wang Q, Zheng D, Jiang X, Xu L, LPS induced miR-181a promotes pancreatic cancer cell migration via targeting PTEN and MAP2K4, Dig Dis Sci 59(7) (2014) 1452–60. [DOI] [PubMed] [Google Scholar]
- [80].Sarkar S, Dubaybo H, Ali S, Goncalves P, Kollepara SL, Sethi S, Philip PA, Li Y, Down-regulation of miR-221 inhibits proliferation of pancreatic cancer cells through up-regulation of PTEN, p27(kip1), p57(kip2), and PUMA, Am J Cancer Res 3(5) (2013) 465–77. [PMC free article] [PubMed] [Google Scholar]
- [81].Park JK, Lee EJ, Esau C, Schmittgen TD, Antisense inhibition of microRNA-21 or -221 arrests cell cycle, induces apoptosis, and sensitizes the effects of gemcitabine in pancreatic adenocarcinoma, Pancreas 38(7) (2009) e190–9. [DOI] [PubMed] [Google Scholar]
- [82].Hou BH, Jian ZX, Cui P, Li SJ, Tian RQ, Ou JR, miR-216a may inhibit pancreatic tumor growth by targeting JAK2, FEBS Lett 589(17) (2015) 2224–32. [DOI] [PubMed] [Google Scholar]
- [83].Wang S, Chen X, Tang M, MicroRNA-216a inhibits pancreatic cancer by directly targeting Janus kinase 2, Oncol Rep 32(6) (2014) 2824–30. [DOI] [PubMed] [Google Scholar]
- [84].Huang C, Li H, Wu W, Jiang T, Qiu Z, Regulation of miR-155 affects pancreatic cancer cell invasiveness and migration by modulating the STAT3 signaling pathway through SOCS1, Oncol Rep 30(3) (2013) 1223–30. [DOI] [PubMed] [Google Scholar]
- [85].Yonezawa S, Higashi M, Yamada N, Goto M, Precursor lesions of pancreatic cancer, Gut Liver 2(3) (2008) 137–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [86].Yu J, Li A, Hong SM, Hruban RH, Goggins M, MicroRNA alterations of pancreatic intraepithelial neoplasias, Clin Cancer Res 18(4) (2012) 981–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [87].Slater EP, Strauch K, Rospleszcz S, Ramaswamy A, Esposito I, Kloppel G, Matthai E, Heeger K, Fendrich V, Langer P, Bartsch DK, MicroRNA-196a and -196b as Potential Biomarkers for the Early Detection of Familial Pancreatic Cancer, Transl Oncol 7(4) (2014) 464–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [88].Ryu JK, Hong SM, Karikari CA, Hruban RH, Goggins MG, Maitra A, Aberrant MicroRNA-155 expression is an early event in the multistep progression of pancreatic adenocarcinoma, Pancreatology 10(1) (2010) 66–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [89].Xue Y, Abou Tayoun AN, Abo KM, Pipas JM, Gordon SR, Gardner TB, Barth RJ, Suriawinata AA, Tsongalis GJ, MicroRNAs as diagnostic markers for pancreatic ductal adenocarcinoma and its precursor, pancreatic intraepithelial neoplasm, Cancer Genet-Ny 206(6) (2013) 217–221. [DOI] [PubMed] [Google Scholar]
- [90].du Rieu MC, Torrisani J, Selves J, Al Saati T, Souque A, Dufresne M, Tsongalis GJ, Suriawinata AA, Carrere N, Buscail L, Cordelier P, MicroRNA-21 is induced early in pancreatic ductal adenocarcinoma precursor lesions, Clin Chem 56(4) (2010) 603–12. [DOI] [PubMed] [Google Scholar]
- [91].Khan S, Ebeling MC, Zaman MS, Sikander M, Yallapu MM, Chauhan N, Yacoubian AM, Behrman SW, Zafar N, Kumar D, Thompson PA, Jaggi M, Chauhan SC, MicroRNA-145 targets MUC13 and suppresses growth and invasion of pancreatic cancer, Oncotarget 5(17) (2014) 7599–609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [92].Habbe N, Koorstra JB, Mendell JT, Offerhaus GJ, Ryu JK, Feldmann G, Mullendore ME, Goggins MG, Hong SM, Maitra A, MicroRNA miR-155 is a biomarker of early pancreatic neoplasia, Cancer Biol Ther 8(4) (2009) 340–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [93].Gironella M, Seux M, Xie MJ, Cano C, Tomasini R, Gommeaux J, Garcia S, Nowak J, Yeung ML, Jeang KT, Chaix A, Fazli L, Motoo Y, Wang Q, Rocchi P, Russo A, Gleave M, Dagorn JC, Iovanna JL, Carrier A, Pebusque MJ, Dusetti NJ, Tumor protein 53-induced nuclear protein 1 expression is repressed by miR-155, and its restoration inhibits pancreatic tumor development, Proc Natl Acad Sci U S A 104(41) (2007) 16170–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [94].Nakahara O, Takamori H, Iwatsuki M, Baba Y, Sakamoto Y, Tanaka H, Chikamoto A, Horino K, Beppu T, Kanemitsu K, Honda Y, Iyama K, Baba H, Carcinogenesis of intraductal papillary mucinous neoplasm of the pancreas: loss of microRNA-101 promotes overexpression of histone methyltransferase EZH2, Ann Surg Oncol 19 Suppl 3 (2012) S565–71. [DOI] [PubMed] [Google Scholar]
- [95].Aso T, Ohtsuka T, Tamura K, Ideno N, Kono H, Nagayoshi Y, Ohuchida K, Ueda J, Takahata S, Shindo K, Aishima S, Oda Y, Mizumoto K, Tanaka M, Elevated expression level of microRNA-196a is predictive of intestinal-type intraductal papillary mucinous neoplasm of the pancreas, Pancreas 43(3) (2014) 361–6. [DOI] [PubMed] [Google Scholar]
- [96].Abue M, Yokoyama M, Shibuya R, Tamai K, Yamaguchi K, Sato I, Tanaka N, Hamada S, Shimosegawa T, Sugamura K, Satoh K, Circulating miR-483–3p and miR-21 is highly expressed in plasma of pancreatic cancer, Int J Oncol 46(2) (2015) 539–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [97].Hao J, Zhang S, Zhou Y, Hu X, Shao C, MicroRNA 483-3p suppresses the expression of DPC4/Smad4 in pancreatic cancer, FEBS Lett 585(1) (2011) 207–13. [DOI] [PubMed] [Google Scholar]
- [98].Permuth-Wey J, Chen YA, Fisher K, McCarthy S, Qu X, Lloyd MC, Kasprzak A, Fournier M, Williams VL, Ghia KM, Yoder SJ, Hall L, Georgeades C, Olaoye F, Husain K, Springett GM, Chen DT, Yeatman T, Centeno BA, Klapman J, Coppola D, Malafa M, A genome-wide investigation of microRNA expression identifies biologically-meaningful microRNAs that distinguish between high-risk and low-risk intraductal papillary mucinous neoplasms of the pancreas, PLoS One 10(1) (2015) e0116869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [99].Lee LS, Szafranska-Schwarzbach AE, Wylie D, Doyle LA, Bellizzi AM, Kadiyala V, Suleiman S, Banks PA, Andruss BF, Conwell DL, Investigating MicroRNA Expression Profiles in Pancreatic Cystic Neoplasms, Clin Transl Gastroenterol 5 (2014) e47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [100].Frampton AE, Krell J, Jamieson NB, Gall TM, Giovannetti E, Funel N, Mato Prado M, Krell D, Habib NA, Castellano L, Jiao LR, Stebbing J, microRNAs with prognostic significance in pancreatic ductal adenocarcinoma: A meta-analysis, Eur J Cancer 51(11) (2015) 1389–404. [DOI] [PubMed] [Google Scholar]
- [101].Papaconstantinou IG, Manta A, Gazouli M, Lyberopoulou A, Lykoudis PM, Polymeneas G, Voros D, Expression of microRNAs in patients with pancreatic cancer and its prognostic significance, Pancreas 42(1) (2013) 67–71. [DOI] [PubMed] [Google Scholar]
- [102].Schultz NA, Andersen KK, Roslind A, Willenbrock H, Wojdemann M, Johansen JS, Prognostic microRNAs in cancer tissue from patients operated for pancreatic cancer--five microRNAs in a prognostic index, World J Surg 36(11) (2012) 2699–707. [DOI] [PubMed] [Google Scholar]
- [103].Liang L, Wei DM, Li JJ, Luo DZ, Chen G, Dang YW, Cai XY, Prognostic microRNAs and their potential molecular mechanism in pancreatic cancer: A study based on The Cancer Genome Atlas and bioinformatics investigation, Mol Med Rep 17(1) (2018) 939–951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [104].Ohuchida K, Mizumoto K, Kayashima T, Fujita H, Moriyama T, Ohtsuka T, Ueda J, Nagai E, Hashizume M, Tanaka M, MicroRNA expression as a predictive marker for gemcitabine response after surgical resection of pancreatic cancer, Ann Surg Oncol 18(8) (2011) 2381–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [105].Greither T, Grochola LF, Udelnow A, Lautenschlager C, Wurl P, Taubert H, Elevated expression of microRNAs 155, 203, 210 and 222 in pancreatic tumors is associated with poorer survival, Int J Cancer 126(1) (2010) 73–80. [DOI] [PubMed] [Google Scholar]
- [106].Guo S, Xu X, Tang Y, Zhang C, Li J, Ouyang Y, Ju J, Bie P, Wang H, miR-15a inhibits cell proliferation and epithelial to mesenchymal transition in pancreatic ductal adenocarcinoma by down-regulating Bmi-1 expression, Cancer Lett 344(1) (2014) 40–46. [DOI] [PubMed] [Google Scholar]
- [107].Li J, Wu H, Li W, Yin L, Guo S, Xu X, Ouyang Y, Zhao Z, Liu S, Tian Y, Tian Z, Ju J, Ni B, Wang H, Downregulated miR-506 expression facilitates pancreatic cancer progression and chemoresistance via SPHK1/Akt/NF-kappaB signaling, Oncogene 35(42) (2016) 5501–5514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [108].Rinn JL, Chang HY, Genome regulation by long noncoding RNAs, Annu Rev Biochem 81 (2012) 145–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [109].Iyer MK, Niknafs YS, Malik R, Singhal U, Sahu A, Hosono Y, Barrette TR, Prensner JR, Evans JR, Zhao S, Poliakov A, Cao X, Dhanasekaran SM, Wu YM, Robinson DR, Beer DG, Feng FY, Iyer HK, Chinnaiyan AM, The landscape of long noncoding RNAs in the human transcriptome, Nat Genet 47(3) (2015) 199–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [110].Guttman M, Rinn JL, Modular regulatory principles of large non-coding RNAs, Nature 482(7385) (2012)339–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [111].Zhou Q, Chung AC, Huang XR, Dong Y, Yu X, Lan HY, Identification of novel long noncoding RNAs associated with TGF-beta/Smad3-mediated renal inflammation and fibrosis by RNA sequencing, Am J Pathol 184(2) (2014) 409–17. [DOI] [PubMed] [Google Scholar]
- [112].Martens-Uzunova ES, Bottcher R, Croce CM, Jenster G, Visakorpi T, Calin GA, Long noncoding RNA in prostate, bladder, and kidney cancer, Eur Urol 65(6) (2014) 1140–51. [DOI] [PubMed] [Google Scholar]
- [113].Prensner JR, Chinnaiyan AM, The emergence of lncRNAs in cancer biology, Cancer Discov 1(5) (2011) 391–407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [114].Gibb EA, Brown CJ, Lam WL, The functional role of long non-coding RNA in human carcinomas, Mol Cancer 10 (2011) 38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [115].Li J, Chen Z, Tian L, Zhou C, He MY, Gao Y, Wang S, Zhou F, Shi S, Feng X, Sun N, Liu Z, Skogerboe G, Dong J, Yao R, Zhao Y, Sun J, Zhang B, Yu Y, Shi X, Luo M, Shao K, Li N, Qiu B, Tan F, Chen R, He J, LncRNA profile study reveals a three-lncRNA signature associated with the survival of patients with oesophageal squamous cell carcinoma, Gut 63(11) (2014) 1700–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [116].Qi P, Du X, The long non-coding RNAs, a new cancer diagnostic and therapeutic gold mine, Mod Pathol 26(2) (2013) 155–65. [DOI] [PubMed] [Google Scholar]
- [117].Zeng S, Xiao YF, Tang B, Hu CJ, Xie R, Yang SM, Li BS, Long Noncoding RNA in Digestive Tract Cancers: Function, Mechanism, and Potential Biomarker, Oncologist 20(8) (2015) 898–906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [118].Cui L, Dong Y, Wang X, Zhao X, Kong C, Liu Y, Jiang X, Zhang X, Downregulation of long noncoding RNA SNHG1 inhibits cell proliferation, metastasis, and invasion by suppressing the Notch-1 signaling pathway in pancreatic cancer, J Cell Biochem 120(4) (2019) 6106–6112. [DOI] [PubMed] [Google Scholar]
- [119].Huang X, Zhi X, Gao Y, Ta N, Jiang H, Zheng J, LncRNAs in pancreatic cancer, Oncotarget 7(35) (2016) 57379–57390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [120].Xu B, Gong X, Zi L, Li G, Dong S, Chen X, Li Y, Silencing of DLEU2 suppresses pancreatic cancer cell proliferation and invasion by upregulating microRNA-455, Cancer Sci 110(5) (2019) 1676–1685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [121].Zhao J, Ohsumi TK, Kung JT, Ogawa Y, Grau DJ, Sarma K, Song JJ, Kingston RE, Borowsky M, Lee JT, Genome-wide identification of polycomb-associated RNAs by RIP-seq, Mol Cell 40(6) (2010) 939–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [122].Tsai MC, Manor O, Wan Y, Mosammaparast N, Wang JK, Lan F, Shi Y, Segal E, Chang HY, Long noncoding RNA as modular scaffold of histone modification complexes, Science 329(5992) (2010) 689–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [123].Li CH, Xiao Z, Tong JH, To KF, Fang X, Cheng AS, Chen Y, EZH2 coupled with HOTAIR to silence MicroRNA-34a by the induction of heterochromatin formation in human pancreatic ductal adenocarcinoma, Int J Cancer 140(1) (2017) 120–129. [DOI] [PubMed] [Google Scholar]
- [124].Kim K, Jutooru I, Chadalapaka G, Johnson G, Frank J, Burghardt R, Kim S, Safe S, HOTAIR is a negative prognostic factor and exhibits pro-oncogenic activity in pancreatic cancer, Oncogene 32(13) (2013) 1616–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [125].Jiang D, Xu L, Ni J, Zhang J, Cai M, Shen L, Functional polymorphisms in LncRNA HOTAIR contribute to susceptibility of pancreatic cancer, Cancer Cell Int 19 (2019) 47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [126].Xie Z, Chen X, Li J, Guo Y, Li H, Pan X, Jiang J, Liu H, Wu B, Salivary HOTAIR and PVT1 as novel biomarkers for early pancreatic cancer, Oncotarget 7(18) (2016) 25408–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [127].Ma Y, Hu M, Zhou L, Ling S, Li Y, Kong B, Huang P, Long non-coding RNA HOTAIR promotes cancer cell energy metabolism in pancreatic adenocarcinoma by upregulating hexokinase-2, Oncol Lett 18(3) (2019) 2212–2219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [128].Yang SZ, Xu F, Zhou T, Zhao X, McDonald JM, Chen Y, The long non-coding RNA HOTAIR enhances pancreatic cancer resistance to TNF-related apoptosis-inducing ligand, J Biol Chem 292(25) (2017) 10390–10397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [129].Sun YW, Chen YF, Li J, Huo YM, Liu DJ, Hua R, Zhang JF, Liu W, Yang JY, Fu XL, Yan T, Hong J, Cao H, A novel long non-coding RNA ENST00000480739 suppresses tumour cell invasion by regulating OS-9 and HIF-1alpha in pancreatic ductal adenocarcinoma, Br J Cancer 111(11) (2014) 2131–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [130].Wolpin BM, Rizzato C, Kraft P, Kooperberg C, Petersen GM, Wang Z, Arslan AA, Beane-Freeman L, Bracci PM, Buring J, Canzian F, Duell EJ, Gallinger S, Giles GG, Goodman GE, Goodman PJ, Jacobs EJ, Kamineni A, Klein AP, Kolonel LN, Kulke MH, Li D, Malats N, Olson SH, Risch HA, Sesso HD, Visvanathan K, White E, Zheng W, Abnet CC, Albanes D, Andreotti G, Austin MA, Barfield R, Basso D, Berndt SI, Boutron-Ruault MC, Brotzman M, Buchler MW, Bueno-de-Mesquita HB, Bugert P, Burdette L, Campa D, Caporaso NE, Capurso G, Chung C, Cotterchio M, Costello E, Elena J, Funel N, Gaziano JM, Giese NA, Giovannucci EL, Goggins M, Gorman MJ, Gross M, Haiman CA, Hassan M, Helzlsouer KJ, Henderson BE, Holly EA, Hu N, Hunter DJ, Innocenti F, Jenab M, Kaaks R, Key TJ, Khaw KT, Klein EA, Kogevinas M, Krogh V, Kupcinskas J, Kurtz RC, LaCroix A, Landi MT, Landi S, Le Marchand L, Mambrini A, Mannisto S, Milne RL, Nakamura Y, Oberg AL, Owzar K, Patel AV, Peeters PH, Peters U, Pezzilli R, Piepoli A, Porta M, Real FX, Riboli E, Rothman N, Scarpa A, Shu XO, Silverman DT, Soucek P, Sund M, Talar-Wojnarowska R, Taylor PR, Theodoropoulos GE, Thornquist M, Tjonneland A, Tobias GS, Trichopoulos D, Vodicka P, Wactawski-Wende J, Wentzensen N, Wu C, Yu H, Yu K, Zeleniuch-Jacquotte A, Hoover R, Hartge P, Fuchs C, Chanock SJ, Stolzenberg-Solomon RS, Amundadottir LT, Genome-wide association study identifies multiple susceptibility loci for pancreatic cancer, Nat Genet 46(9) (2014) 994–1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [131].Zhang X, Feng W, Zhang J, Ge L, Zhang Y, Jiang X, Peng W, Wang D, Gong A, Xu M, Long noncoding RNA PVT1 promotes epithelialmesenchymal transition via the TGFbeta/Smad pathway in pancreatic cancer cells, Oncol Rep 40(2) (2018) 1093–1102. [DOI] [PubMed] [Google Scholar]
- [132].Zhao L, Kong H, Sun H, Chen Z, Chen B, Zhou M, LncRNA-PVTl promotes pancreatic cancer cells proliferation and migration through acting as a molecular sponge to regulate miR-448, J Cell Physiol 233(5) (2018) 4044–4055. [DOI] [PubMed] [Google Scholar]
- [133].Huang F, Chen W, Peng J, Li Y, Zhuang Y, Zhu Z, Shao C, Yang W, Yao H, Zhang S, LncRNA PVT1 triggers Cyto-protective autophagy and promotes pancreatic ductal adenocarcinoma development via the miR-20a-5p/ULK1 Axis, Mol Cancer 17(1) (2018) 98. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- [134].Wang CJ, Shi SB, Tian J, Xu J, Niu ZX, lncRNA MALAT1, HOTTIP and PVT1 as predictors for predicting the efficacy of GEM based chemotherapy in first-line treatment of pancreatic cancer patients, Oncotarget 8(56) (2017) 95108–95115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [135].Pang EJ, Yang R, Fu XB, Liu YF, Overexpression of long non-coding RNA MALAT1 is correlated with clinical progression and unfavorable prognosis in pancreatic cancer, Tumour Biol 36(4) (2015) 2403–7. [DOI] [PubMed] [Google Scholar]
- [136].Liu JH, Chen G, Dang YW, Li CJ, Luo DZ, Expression and prognostic significance of lncRNA MALAT1 in pancreatic cancer tissues, Asian Pac J Cancer Prev 15(7) (2014) 2971–7. [DOI] [PubMed] [Google Scholar]
- [137].Han T, Jiao F, Hu H, Yuan C, Wang L, Jin ZL, Song WF, Wang LW, EZH2 promotes cell migration and invasion but not alters cell proliferation by suppressing E-cadherin, partly through association with MALAT-1 in pancreatic cancer, Oncotarget 7(10) (2016) 11194–207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [138].Cheng Y, Imanirad P, Jutooru I, Hedrick E, Jin UH, Rodrigues Hoffman A, Leal de Araujo J, Morpurgo B, Golovko A, Safe S, Role of metastasis-associated lung adenocarcinoma transcript-1 (MALAT-1) in pancreatic cancer, PLoS One 13(2) (2018) e0192264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [139].Xie ZC, Dang YW, Wei DM, Chen P, Tang RX, Huang Q, Liu JH, Luo DZ, Clinical significance and prospective molecular mechanism of MALAT1 in pancreatic cancer exploration: a comprehensive study based on the GeneChip, GEO, Oncomine, and TCGA databases, Onco Targets Ther 10 (2017) 3991–4005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [140].Wang KC, Yang YW, Liu B, Sanyal A, Corces-Zimmerman R, Chen Y, Lajoie BR, Protacio A, Flynn RA, Gupta RA, Wysocka J, Lei M, Dekker J, Helms JA, Chang HY, A long noncoding RNA maintains active chromatin to coordinate homeotic gene expression, Nature 472(7341) (2011) 1204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [141].Fu Z, Chen C, Zhou Q, Wang Y, Zhao Y, Zhao X, Li W, Zheng S, Ye H, Wang L, He Z, Lin Q, Li Z, Chen R, LncRNA HOTTIP modulates cancer stem cell properties in human pancreatic cancer by regulating HOXA9, Cancer Lett 410 (2017) 68–81. [DOI] [PubMed] [Google Scholar]
- [142].Cheng Y, Jutooru I, Chadalapaka G, Corton JC, Safe S, The long non-coding RNA HOTTIP enhances pancreatic cancer cell proliferation, survival and migration, Oncotarget 6(13) (2015) 10840–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [143].Li Z, Zhao X, Zhou Y, Liu Y, Zhou Q, Ye H, Wang Y, Zeng J, Song Y, Gao W, Zheng S, Zhuang B, Chen H, Li W, Li H, Li H, Fu Z, Chen R, The long non-coding RNA HOTTIP promotes progression and gemcitabine resistance by regulating HOXA13 in pancreatic cancer, J Transl Med 13 (2015) 84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [144].Chen LL, Yang L, Regulation of circRNA biogenesis, RNA Biol 12(4) (2015) 381–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [145].Li J, Yang J, Zhou P, Le Y, Zhou C, Wang S, Xu D, Lin HK, Gong Z, Circular RNAs in cancer: novel insights into origins, properties, functions and implications, Am J Cancer Res 5(2) (2015) 47280. [PMC free article] [PubMed] [Google Scholar]
- [146].Qu S, Yang X, Li X, Wang J, Gao Y, Shang R, Sun W, Dou K, Li H, Circular RNA: A new star of noncoding RNAs, Cancer Lett 365(2) (2015) 141–8. [DOI] [PubMed] [Google Scholar]
- [147].Lukiw WJ, Circular RNA (circRNA) in Alzheimer’s disease (AD), Front Genet 4 (2013) 307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [148].Xie H, Ren X, Xin S, Lan X, Lu G, Lin Y, Yang S, Zeng Z, Liao W, Ding YQ, Liang L, Emerging roles of circRNA_001569 targeting miR-145 in the proliferation and invasion of colorectal cancer, Oncotarget 7(18) (2016) 26680–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [149].Nair AA, Niu N, Tang X, Thompson KJ, Wang L, Kocher JP, Subramanian S, Kalari KR, Circular RNAs and their associations with breast cancer subtypes, Oncotarget 7(49) (2016) 80967–80979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [150].Zhong Z, Lv M, Chen J, Screening differential circular RNA expression profiles reveals the regulatory role of circTCF25-miR-103a-3p/miR-107-CDK6 pathway in bladder carcinoma, Sci Rep 6 (2016) 30919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [151].Xia W, Qiu M, Chen R, Wang S, Leng X, Wang J, Xu Y, Hu J, Dong G, Xu PL, Yin R, Circular RNA has_circ_0067934 is upregulated in esophageal squamous cell carcinoma and promoted proliferation, Sci Rep 6 (2016) 35576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [152].Li J, Sun D, Pu W, Wang J, Peng Y, Circular RNAs in Cancer: Biogenesis, Function, and Clinical Significance, Trends Cancer 6(4) (2020) 319–336. [DOI] [PubMed] [Google Scholar]
- [153].Li H, Hao X, Wang H, Liu Z, He Y, Pu M, Zhang H, Yu H, Duan J, Qu S, Circular RNA Expression Profile of Pancreatic Ductal Adenocarcinoma Revealed by Microarray, Cell Physiol Biochem 40(6) (2016) 1334–1344. [DOI] [PubMed] [Google Scholar]
- [154].Zhang Q, Wang JY, Zhou SY, Yang SJ, Zhong SL, Circular RNA expression in pancreatic ductal adenocarcinoma, Oncol Lett 18(3) (2019) 2923–2930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [155].Li Q, Geng S, Yuan H, Li Y, Zhang S, Pu L, Ge J, Niu X, Li Y, Jiang H, Circular RNA expression profiles in extracellular vesicles from the plasma of patients with pancreatic ductal adenocarcinoma, FEBS Open Bio 9(12) (2019) 2052–2062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [156].Yao J, Zhang C, Chen Y, Gao S, Downregulation of circular RNA circ-LDLRAD3 suppresses pancreatic cancer progression through miR-137-3p/PTN axis, Life Sci 239 (2019) 116871. [DOI] [PubMed] [Google Scholar]
- [157].Liu L, Liu FB, Huang M, Xie K, Xie QS, Liu CH, Shen MJ, Huang Q, Circular RNA ciRS-7 promotes the proliferation and metastasis of pancreatic cancer by regulating miR-7-mediated EGFR/STAT3 signaling pathway, Hepatobiliary Pancreat Dis Int 18(6) (2019) 580–586. [DOI] [PubMed] [Google Scholar]
- [158].Yang F, Liu DY, Guo JT, Ge N, Zhu P, Liu X, Wang S, Wang GX, Sun SY, Circular RNA circ-LDLRAD3 as a biomarker in diagnosis of pancreatic cancer, World J Gastroenterol 23(47) (2017) 8345–8354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [159].Xu Y, Yao Y, Gao P, Cui Y, Upregulated circular RNA circ_0030235 predicts unfavorable prognosis in pancreatic ductal adenocarcinoma and facilitates cell progression by sponging miR-1253 and miR-1294, Biochem Biophys Res Commun 509(1) (2019) 138–142. [DOI] [PubMed] [Google Scholar]
- [160].Liu M, Zhang Y, Zhang J, Cai H, Zhang C, Yang Z, Niu Y, Wang H, Wei X, Wang W, Gao P, Li H, Zhang J, Sun G, MicroRNA-1253 suppresses cell proliferation and invasion of non-small-cell lung carcinoma by targeting WNT5A, Cell Death Dis 9(2) (2018) 189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [161].Wang Z, Yan J, Zou T, Gao H, MicroRNA-1294 inhibited oral squamous cell carcinoma growth by targeting c-Myc, Oncol Lett 16(2) (2018) 2243–2250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [162].Hao L, Rong W, Bai L, Cui H, Zhang S, Li Y, Chen D, Meng X, Upregulated circular RNA circ_0007534 indicates an unfavorable prognosis in pancreatic ductal adenocarcinoma and regulates cell proliferation, apoptosis, and invasion by sponging miR-625 and miR-892b, J Cell Biochem 120(3) (2019) 3780–3789. [DOI] [PubMed] [Google Scholar]
- [163].Li Z, Yanfang W, Li J, Jiang P, Peng T, Chen K, Zhao X, Zhang Y, Zhen P, Zhu J, Li X, Tumor-released exosomal circular RNA PDE8A promotes invasive growth via the miR-338/MACC1/MET pathway in pancreatic cancer, Cancer Lett 432 (2018) 237–250. [DOI] [PubMed] [Google Scholar]
- [164].Li J, Li Z, Jiang P, Peng M, Zhang X, Chen K, Liu H, Bi H, Liu X, Li X, Circular RNA IARS (circ-IARS) secreted by pancreatic cancer cells and located within exosomes regulates endothelial monolayer permeability to promote tumor metastasis, J Exp Clin Cancer Res 37(1) (2018) 177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [165].Weng W, Wei Q, Toden S, Yoshida K, Nagasaka T, Fujiwara T, Cai S, Qin H, Ma Y, Goel A, Circular RNA ciRS-7-A Promising Prognostic Biomarker and a Potential Therapeutic Target in Colorectal Cancer, Clin Cancer Res 23(14) (2017) 3918–3928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [166].Xu L, Zhang M, Zheng X, Yi P, Lan C, Xu M, The circular RNA ciRS-7 (Cdrlas) acts as a risk factor of hepatic microvascular invasion in hepatocellular carcinoma, J Cancer Res Clin Oncol 143(1) (2017) 17–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [167].Li JH, He R, Li YM, Cao G, Ma QY, Yang WB, Endoscopic ultrasonography for tumor node staging and vascular invasion in pancreatic cancer: a meta-analysis, Dig Surg 31(4-5) (2014) 297–305. [DOI] [PubMed] [Google Scholar]
- [168].Kauhanen SP, Komar G, Seppanen MP, Dean KI, Minn HR, Kajander SA, Rinta-Kiikka I, Alanen K, Borra RJ, Puolakkainen PA, Nuutila P, Ovaska JT, A prospective diagnostic accuracy study of 18F-fluorodeoxyglucose positron emission tomography/computed tomography, multidetector row computed tomography, and magnetic resonance imaging in primary diagnosis and staging of pancreatic cancer, Ann Surg 250(6) (2009) 957–63. [DOI] [PubMed] [Google Scholar]
- [169].Palazzo L, Roseau G, Gayet B, Vilgrain V, Belghiti J, Fekete F, Paolaggi JA, Endoscopic ultrasonography in the diagnosis and staging of pancreatic adenocarcinoma. Results of a prospective study with comparison to ultrasonography and CT scan, Endoscopy 25(2) (1993) 143–50. [DOI] [PubMed] [Google Scholar]
- [170].Sheridan MB, Ward J, Guthrie JA, Spencer JA, Craven CM, Wilson D, Guillou PJ, Robinson PJ, Dynamic contrast-enhanced MR imaging and dual-phase helical CT in the preoperative assessment of suspected pancreatic cancer: a comparative study with receiver operating characteristic analysis, AJR Am J Roentgenol 173(3) (1999) 583–90. [DOI] [PubMed] [Google Scholar]
- [171].Heinrich S, Goerres GW, Schafer M, Sagmeister M, Bauerfeind P, Pestalozzi BC, Hany TF, von Schulthess GK, Clavien PA, Positron emission tomography/computed tomography influences on the management of resectable pancreatic cancer and its cost-effectiveness, Ann Surg 242(2) (2005) 235–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [172].Motosugi U, Ichikawa T, Morisaka H, Sou H, Muhi A, Kimura K, Sano K, Araki T, Detection of pancreatic carcinoma and liver metastases with gadoxetic acid-enhanced MR imaging: comparison with contrast-enhanced multi-detector row CT, Radiology 260(2) (2011) 446–53. [DOI] [PubMed] [Google Scholar]
- [173].Park MJ, Kim YK, Choi SY, Rhim H, Lee WJ, Choi D, Preoperative detection of small pancreatic carcinoma: value of adding diffusion-weighted imaging to conventional MR imaging for improving confidence level, Radiology 273(2) (2014) 433–43. [DOI] [PubMed] [Google Scholar]
- [174].Rao SX, Zeng MS, Cheng WZ, Yao XZ, Jin DY, Ji Y, Small solid tumors (< or = 2 cm) of the pancreas: relative accuracy and differentiation of CT and MR imaging, Hepatogastroenterology 58(107-108) (2011) 996–1001. [PubMed] [Google Scholar]
- [175].Toft J, Hadden WJ, Laurence JM, Lam V, Yuen L, Janssen A, Pleass H, Imaging modalities in the diagnosis of pancreatic adenocarcinoma: A systematic review and meta-analysis of sensitivity, specificity and diagnostic accuracy, Eur J Radiol 92 (2017) 17–23. [DOI] [PubMed] [Google Scholar]
- [176].Poruk KE, Gay DZ, Brown K, Mulvihill JD, Boucher KM, Scaife CL, Firpo MA, Mulvihill SJ, The clinical utility of CA 19-9 in pancreatic adenocarcinoma: diagnostic and prognostic updates, Curr Mol Med 13(3) (2013) 340–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [177].Yu J, Sadakari Y, Shindo K, Suenaga M, Brant A, Almario JAN, Borges M, Barkley T, Fesharakizadeh S, Ford M, Hruban RH, Shin EJ, Lennon AM, Canto MI, Goggins M, Digital next-generation sequencing identifies low-abundance mutations in pancreatic juice samples collected from the duodenum of patients with pancreatic cancer and intraductal papillary mucinous neoplasms, Gut 66(9) (2017) 1677–1687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [178].Singhi AD, McGrath K, Brand RE, Khalid A, Zeh HJ, Chennat JS, Fasanella KE, Papachristou GI, Slivka A, Bartlett DL, Dasyam AK, Hogg M, Lee KK, Marsh JW, Monaco SE, Ohori NP, Pingpank JF, Tsung A, Zureikat AH, Wald AI, Nikiforova MN, Preoperative next-generation sequencing of pancreatic cyst fluid is highly accurate in cyst classification and detection of advanced neoplasia, Gut 67(12) (2018) 2131–2141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [179].Springer S, Wang Y, Dal Molin M, Masica DL, Jiao Y, Kinde I, Blackford A, Raman SP, Wolfgang CL, Tomita T, Niknafs N, Douville C, Ptak J, Dobbyn L, Allen PJ, Klimstra DS, Schattner MA, Schmidt CM, Yip-Schneider M, Cummings OW, Brand RE, Zeh HJ, Singhi AD, Scarpa A, Salvia R, Malleo G, Zamboni G, Falconi M, Jang JY, Kim SW, Kwon W, Hong SM, Song KB, Kim SC, Swan N, Murphy J, Geoghegan J, Brugge W, Fernandez-Del Castillo C, Mino-Kenudson M, Schulick R, Edil BH, Adsay V, Paulino J, van Hooft J, Yachida S, Nara S, Hiraoka N, Yamao K, Hijioka S, van der Merwe S, Goggins M, Canto MI, Ahuja N, Hirose K, Makary M, Weiss MJ, Cameron J, Pittman M, Eshleman JR, Diaz LA Jr., Papadopoulos N, Kinzler KW, Karchin R, Hruban RH, Vogelstein B, Lennon AM, A combination of molecular markers and clinical features improve the classification of pancreatic cysts, Gastroenterology 149(6) (2015) 1501–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [180].Singhi AD, Nikiforova MN, Fasanella KE, McGrath KM, Pai RK, Ohori NP, Bartholow TL, Brand RE, Chennat JS, Lu X, Papachristou GI, Slivka A, Zeh HJ, Zureikat AH, Lee KK, Tsung A, Mantha GS, Khalid A, Preoperative GNAS and KRAS testing in the diagnosis of pancreatic mucinous cysts, Clin Cancer Res 20(16) (2014) 4381–9. [DOI] [PubMed] [Google Scholar]
- [181].Nikiforova MN, Khalid A, Fasanella KE, McGrath KM, Brand RE, Chennat JS, Slivka A, Zeh HJ, Zureikat AH, Krasinskas AM, Ohori NP, Schoedel KE, Navina S, Mantha GS, Pai RK, Singhi AD, Integration of KRAS testing in the diagnosis of pancreatic cystic lesions: a clinical experience of 618 pancreatic cysts, Mod Pathol 26(11) (2013) 1478–87. [DOI] [PubMed] [Google Scholar]
- [182].Rosenbaum MW, Jones M, Dudley JC, Le LP, Iafrate AJ, Pitman MB, Next-generation sequencing adds value to the preoperative diagnosis of pancreatic cysts, Cancer Cytopathol 125(1) (2017) 41–47. [DOI] [PubMed] [Google Scholar]
- [183].Jones M, Zheng Z, Wang J, Dudley J, Albanese E, Kadayifci A, Dias-Santagata D, Le L, Brugge WR, Fernandez-del Castillo C, Mino-Kenudson M, Iafrate AJ, Pitman MB, Impact of next-generation sequencing on the clinical diagnosis of pancreatic cysts, Gastrointest Endosc 83(1) (2016) 140–8. [DOI] [PubMed] [Google Scholar]
- [184].Garcia-Carracedo D, Chen ZM, Qiu W, Huang AS, Tang SM, Hruban RH, Su GH, PIK3CA mutations in mucinous cystic neoplasms of the pancreas, Pancreas 43(2) (2014) 245–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [185].Garcia-Carracedo D, Turk AT, Fine SA, Akhavan N, Tweel BC, Parsons R, Chabot JA, Allendorf JD, Genkinger JM, Remotti HE, Su GH, Loss of PTEN expression is associated with poor prognosis in patients with intraductal papillary mucinous neoplasms of the pancreas, Clin Cancer Res 19(24) (2013) 6830–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [186].Schonleben F, Qiu W, Ciau NT, Ho DJ, Li X, Allendorf JD, Remotti HE, Su GH, PIK3CA mutations in intraductal papillary mucinous neoplasm/carcinoma of the pancreas, Clin Cancer Res 12(12) (2006) 3851–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [187].Schonleben F, Qiu W, Remotti HE, Hohenberger W, Su GH, PIK3CA, KRAS, and BRAF mutations in intraductal papillary mucinous neoplasm/carcinoma (IPMN/C) of the pancreas, Langenbecks Arch Surg 393(3) (2008) 289–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [188].Alemar B, Izetti P, Gregorio C, Macedo GS, Castro MA, Osvaldt AB, Matte U, Ashton-Prolla P, miRNA-21 and miRNA-34a Are Potential Minimally Invasive Biomarkers for the Diagnosis of Pancreatic Ductal Adenocarcinoma, Pancreas 45(1) (2016) 84–92. [DOI] [PubMed] [Google Scholar]
- [189].Que R, Ding G, Chen J, Cao L, Analysis of serum exosomal microRNAs and clinicopathologic features of patients with pancreatic adenocarcinoma, World J Surg Oncol 11 (2013) 219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [190].Yang JY, Sun YW, Liu DJ, Zhang JF, Li J, Hua R, MicroRNAs in stool samples as potential screening biomarkers for pancreatic ductal adenocarcinoma cancer, Am J Cancer Res 4(6) (2014) 663–73. [PMC free article] [PubMed] [Google Scholar]
- [191].Goto T, Fujiya M, Konishi H, Sasajima J, Fujibayashi S, Hayashi A, Utsumi T, Sato H, Iwama T, Ijiri M, Sakatani A, Tanaka K, Nomura Y, Ueno N, Kashima S, Moriichi K, Mizukami Y, Kohgo Y, Okumura T, An elevated expression of serum exosomal microRNA-191, - 21, -451a of pancreatic neoplasm is considered to be efficient diagnostic marker, BMC Cancer 18(1) (2018) 116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [192].Cote GA, Gore AJ, McElyea SD, Heathers LE, Xu H, Sherman S, Korc M, A pilot study to develop a diagnostic test for pancreatic ductal adenocarcinoma based on differential expression of select miRNA in plasma and bile, Am J Gastroenterol 109(12) (2014) 1942–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [193].Xie Z, Yin X, Gong B, Nie W, Wu B, Zhang X, Huang J, Zhang P, Zhou Z, Li Z, Salivary microRNAs show potential as a noninvasive biomarker for detecting resectable pancreatic cancer, Cancer Prev Res (Phila) 8(2) (2015) 165–73. [DOI] [PubMed] [Google Scholar]
- [194].Hong TH, Park IY, MicroRNA expression profiling of diagnostic needle aspirates from surgical pancreatic cancer specimens, Ann Surg Treat Res 87(6) (2014) 290–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [195].Yu S, Lu Z, Liu C, Meng Y, Ma Y, Zhao W, Liu J, Yu J, Chen J, miRNA-96 suppresses KRAS and functions as a tumor suppressor gene in pancreatic cancer, Cancer Res 70(14) (2010) 6015–25. [DOI] [PubMed] [Google Scholar]
- [196].Liffers ST, Munding JB, Vogt M, Kuhlmann JD, Verdoodt B, Nambiar S, Maghnouj A, Mirmohammadsadegh A, Hahn SA, Tannapfel A, MicroRNA-148a is down-regulated in human pancreatic ductal adenocarcinomas and regulates cell survival by targeting CDC25B, Lab Invest 91(10) (2011) 1472–9. [DOI] [PubMed] [Google Scholar]
- [197].Ren Y, Gao J, Liu JQ, Wang XW, Gu JJ, Huang HJ, Gong YF, Li ZS, Differential signature of fecal microRNAs in patients with pancreatic cancer, Mol Med Rep 6(1) (2012) 201–9. [DOI] [PubMed] [Google Scholar]
- [198].Kong X, Du Y, Wang G, Gao J, Gong Y, Li L, Zhang Z, Zhu J, Jing Q, Qin Y, Li Z, Detection of differentially expressed microRNAs in serum of pancreatic ductal adenocarcinoma patients: miR-196a could be a potential marker for poor prognosis, Dig Dis Sci 56(2) (2011) 602–9. [DOI] [PubMed] [Google Scholar]
- [199].Wang J, Raimondo M, Guha S, Chen J, Diao L, Dong X, Wallace MB, Killary AM, Frazier ML, Woodward TA, Wang J, Sen S, Circulating microRNAs in Pancreatic Juice as Candidate Biomarkers of Pancreatic Cancer, J Cancer 5(8) (2014) 696–705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [200].Link A, Becker V, Goel A, Wex T, Malfertheiner P, Feasibility of fecal microRNAs as novel biomarkers for pancreatic cancer, PLoS One 7(8) (2012) e42933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [201].Morimura R, Komatsu S, Ichikawa D, Takeshita H, Tsujiura M, Nagata H, Konishi H, Shiozaki A, Ikoma H, Okamoto K, Ochiai T, Taniguchi H, Otsuji E, Novel diagnostic value of circulating miR-18a in plasma of patients with pancreatic cancer, Br J Cancer 105(11) (2011) 1733–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [202].Wang WS, Liu LX, Li GP, Chen Y, Li CY, Jin DY, Wang XL, Combined serum CA19-9 and miR-27a-3p in peripheral blood mononuclear cells to diagnose pancreatic cancer, Cancer Prev Res (Phila) 6(4) (2013) 331–8. [DOI] [PubMed] [Google Scholar]
- [203].Liu R, Chen X, Du Y, Yao W, Shen L, Wang C, Hu Z, Zhuang R, Ning G, Zhang C, Yuan Y, Li Z, Zen K, Ba Y, Zhang CY, Serum microRNA expression profile as a biomarker in the diagnosis and prognosis of pancreatic cancer, Clin Chem 58(3) (2012) 610–8. [DOI] [PubMed] [Google Scholar]
- [204].Zhao C, Zhang J, Zhang S, Yu D, Chen Y, Liu Q, Shi M, Ni C, Zhu M, Diagnostic and biological significance of microRNA-192 in pancreatic ductal adenocarcinoma, Oncol Rep 30(1) (2013) 276–84. [DOI] [PubMed] [Google Scholar]
- [205].Ho AS, Huang X, Cao H, Christman-Skieller C, Bennewith K, Le QT, Koong AC, Circulating miR-210 as a Novel Hypoxia Marker in Pancreatic Cancer, Transl Oncol 3(2) (2010) 109–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [206].Henry JC, Bassi C, Giovinazzo F, Bloomston M, MicroRNA from pancreatic duct aspirate differentiates cystic lesions of the pancreas, Ann Surg Oncol 20 Suppl 3 (2013) S661–6. [DOI] [PubMed] [Google Scholar]
- [207].Lin MS, Chen WC, Huang JX, Gao HJ, Sheng HH, Aberrant expression of microRNAs in serum may identify individuals with pancreatic cancer, Int J Clin Exp Med 7(12) (2014) 5226–34. [PMC free article] [PubMed] [Google Scholar]
- [208].Madhavan B, Yue S, Galli U, Rana S, Gross W, Muller M, Giese NA, Kalthoff H, Becker T, Buchler MW, Zoller M, Combined evaluation of a panel of protein and miRNA serum-exosome biomarkers for pancreatic cancer diagnosis increases sensitivity and specificity, Int J Cancer 136(11) (2015) 2616–27. [DOI] [PubMed] [Google Scholar]