Abstract
Serratiopeptidase is a proteolytic enzyme extensively used as an anti-inflammatory and analgesic drug. Present work reports a thermoactive serratiopeptidase from Serratia marcescens AD-W2, a soil isolate from the North-Western Himalayan region of India. The extracellular metalloprotease has been purified by a simple two-step procedure resulting in a specific activity of 20,492 Units/mg protein with 5.28-fold purification. The molecular mass of the metalloprotease, as determined by SDS-PAGE was ~ 51 kDa. The purified serratiopeptidase presented optimum activity at pH 9.0, temperature 50 °C and stability in wide pH and temperature range. Critical temperature of 50 °C confirmed the thermoactivity of the purified serratiopeptidase. The kinetic studies of the purified serratiopeptidase revealed Vmax and Km of 57,256 Units/mL and 1.57 mg/mL, respectively, for casein. The purified serratiopeptidase from S. marcescens AD-W2 was found to be 100% identical to serralysin from Serratia marcescens ATCC 21074/E-15. The catalytic domain comprising of Zn coordinated with three histidine residues (His192, His196, His202), along with glutamate (Glu193) and tyrosine (Tyr232) residues, further confirmed that the purified protein is identical to serralysin.
Supplementary Information
The online version contains supplementary material available at 10.1186/s13568-021-01215-7.
Keywords: Serratia marcescens, Metalloprotease, Serratiopeptidase, Serralysin, Homology modelling
Key points
Thermoactive serratiopeptidase from S. marcescens AD-W2 was purified with simple two step purification method.
Purified serratiopeptidase has specific activity of 20,492 Units/mg protein and critical temperature of 50 °C.
Mass fingerprint and predicted catalytic site confirms that the purified protein is serralysin.
Introduction
Serratiopeptidase (EC.3.4.24.40), also known as serrapeptase or serralysin, is an extracellular metalloprotease enzyme known for its pharmaceutical importance. It is widely used as an anti-inflammatory, analgesic, anti-oedemic, and anti-biofilm formulation agent (Ethiraj and Gopinath 2017). Serratiopeptidase has gained attention as a potent analgesic and anti-inflammatory drug with rapidly increasing market demands in the recent past (Olmstead 2017; Olmstead 2018; Srivastava et al. 2019). This drug also finds applications in chronic inflammatory diseases such as fibrocystic breast disease, sinusitis, carpal tunnel syndrome, bronchitis, arthritis, and atherosclerosis (Pakhale and Bhagwat 2016).
Serratiopeptidase was first isolated from Enterobacterium Serratia E-15, an isolate from the gut of Bombyx mori (Miyata et al. 1970). Serratiopeptidase is reportedly produced from S. marcescens, a Gram-negative small rod belonging to the family Enterobacteriaceae. Although S. marcescens is one of the most common serratiopeptidase producers (Romero et al. 2001; Nam et al. 2013; Ethiraj and Gopinath 2017), other bacteria have also been reported for serratiopeptidase production. These include other Serratia sp., Bacillus licheniformis, Streptomyces hydrogenans (Salarizadeh et al. 2014; Wagdarikar et al. 2015; Nageswara et al. 2019).
Keeping in view, the increasing demand of anti-inflammatory therapeutics in the global market, there is an immediate need to evolve efficient processes for serratiopeptidase production with efficient activity. In this context, our group has been engaged in isolation and characterization of serratiopeptidase producing microorganisms from NW Himalayan region and has recently reported S. marcescens isolated from mulberry phyllosphere for serratiopeptidase production (Koul et al. 2020). The present study relates to the isolation, characterization and purification of serratiopeptidase from S. marcescens AD-W2, a soil isolate from NW Himalayas. The purified serratiopeptidase has been investigated for its characteristics and structural properties vis-a-vis serralysin from S. marcescens ATCC 21074/E-15.
Materials and methods
Isolation and characterization of S. marcescens AD-W2
Serratia marcescens AD-W2, used in the present study was isolated from soil, collected from NW-Himalayas at an altitude of about 1000ft by serial dilution method (James and Natalie 2014), and was maintained on tryptone soya agar (TSA). The culture was identified by 16S rRNA gene sequencing and the sequence was submitted in NCBI GenBank. The phylogenetic tree was made using the minimum evolution method (Rzhetsky and Nei 1992), using MEGA X (Kumar et al. 2018). The culture has been deposited in Sir Col. R. N. Chopra Microbial Resource Centre, Jammu, India, with accession number MRCJ-997.
Serratiopeptidase production from S. marcescens AD-W2
The isolated microorganism was subjected to growth in a 500 mL Erlenmeyer flask containing 100 mL tryptone soya broth in a rotary shaker at 200 RPM at 30 °C. In order to evaluate the production of enzyme, several production media were used for enzyme production followed by evaluation of protease activity up to 72 h. Based on the screening of various production media, soyabean meal and casein based medium (constituents in g/L: Soyabean meal—20.0, Casein—15.0, dihydrogen ammonium phosphate—7.5, soya oil—5.0, NaCl—0.5, KCl—0.1, MgSO4—0.1, ZnSO4—0.1, dextrose anhydrous-20.0, pH 7.5) was selected for protease production for further studies.
Extracellular protease assay and protein estimation
The qualitative protease assay of the culture supernatant was performed on the milk agar plate (James and Natalie 2014), while the quantitative assay was performed according to Cupp-Enyard (2008) with slight modifications. One Unit of Protease (serratiopeptidase) activity was defined as micrograms of tyrosine released per minute on hydrolysis of casein under standard assay conditions (pH 8.0, 37 °C). Protein estimation was performed by the Bradford method (Bradford 1976).
Purification of serratiopeptidase from S. marcescens AD-W2
The culture supernatant having the desired specific activity was partially purified by ammonium sulphate up to 30–80% saturation. The precipitates were dissolved in 0.05 M buffer (pH 8.0), dialyzed against the same buffer for enzyme activity or protein content evaluation, and further subjected to Ion exchange chromatography using the MonoQ5/50 GL column (GE make) installed with Bio-Rad DuoFlow Chromatography system. 40 mg of partially purified protein was loaded onto the column, and elution was performed using NaCl gradient in phosphate buffer (pH 6.0). 0.5 mL fractions were collected and evaluated for protease (serratiopeptidase) activity.
Sodium dodecylsulphate-polyacrylamide gel electrophoresis (SDS-PAGE) and zymography
The crude, partially purified and purified enzymes were subjected to 10% SDS-PAGE according to Laemmli discontinuous system (Laemmli 1970). Zymography was performed as per protocol given by Lantz and Ciborowski (1994) with few modifications.
Characterization of serratiopeptidase from S. marcescens AD-W2
In order to evaluate the optimum activity, the assay was carried out, keeping all the assay conditions the same, except the variables (pH, temperature, substrate concentration, or inhibitors). The relative activities were calculated, considering the initial activity as 100%. The effect of temperature on serratiopeptidase activity was studied at 30–60 °C, while the effect of pH on the serratiopeptidase activity was evaluated at varied pH range, i.e., pH 6.0–7.0 (phosphate buffer), 8.0–9.0 (Tris–HCl buffer), pH 10.0 (Glycine NaOH), and pH 11.0–12.0 (Sodium bicarbonate buffer). The effect of inhibitors was studied with 2 mM inhibitors (Olajuyigbe and Falade 2014). The kinetic studies of purified serratiopeptidase were performed by activity assay with varying concentrations of casein under standard assay conditions to determine Vmax. Km values were determined by the double reciprocal Lineweaver–Burk plot. The critical temperature of the purified serratiopeptidase and activation energies below and above critical temperatures were determined by the Arrhenius plot generated from the temperature-activity dataset.
Identification of purified serratiopeptidase
The purified protein band from the SDS-PAGE gel was trypsin digested by the procedure discribed in Sigma Proteoprep kit, while reduction and alkylation steps were omitted for rapid processing (Shevchenko et al. 2006). The mass fingerprints of the peptides were obtained on Bruker UltrafleXtreme MALDI TOF/TOF using multiple laser shots. 2 µL of the tryptic digest was mixed with α-cyano-4-hydroxycinnamic acid (10 mg/mL CHCA in 50% acetonitrile and 0.1% TFA) in a 1:1 ratio, transferred on the target MALDI plate and was allowed to dry at room temperature. Bovine serum albumin was used as a positive control. The mass spectra were searched in the Swiss-prot database along with the contaminant database on the MASCOT server for identification of the protein (Perkins et al. 1999). The sequence obtained using the MASCOT software after MALDI-TOF analysis was subjected to BLAST in the UNIPROT database. The top six results were used for multiple sequence alignment in SEAVIEW software (Galtier et al. 1996). The three-dimensional structure of serratiopeptidase was built using the SWISS-MODEL server (Schwede et al. 2003) using serralysin protein from Serratia sp. (PDB entry 1srp) as the template.
Results
Isolation and characterization of S. marcescens AD-W2
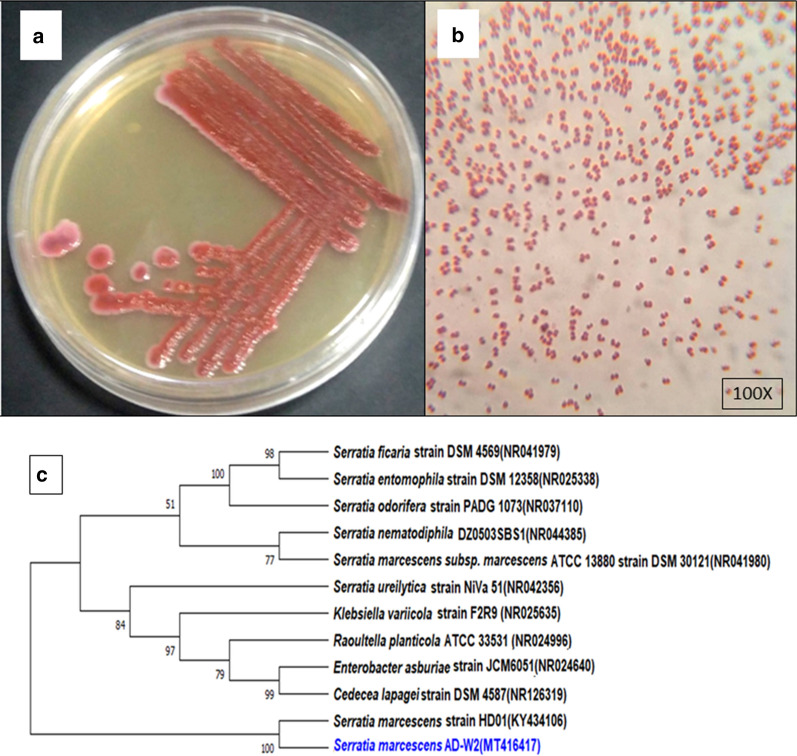
The isolated strain AD-W2 presented the characteristic morphological features of Serratia sp. (Zhang et al. 2009) like red pigmentation, short Gram-negative rod shape are shown in Fig. 1a, b. The 16S rRNA gene sequence revealed that the isolate has a 99.85% similarity with both S. nematodiphila and S. marcescens type strain. The negetive results for lactose utilization, xylose utilization, and arginine dihyrolase, confirmed that the isolate is S. marcescens and not S. nematodiphila. The sequence was submitted to the NCBI GenBank with Accession number MT416417. As per the phylogenetic tree made using the Minimum evolution method in Fig. 1c, the strain had the highest similarity with S. marcescens. The culture has been deposited in Sir R. N. Chopra, Microbial Resource Centre Jammu, India with Accession number MRCJ-997.
Fig. 1.
a Morphology of pure isolate AD-W2 on Nutrient Agar plate. b Microscopic image of pure isolate AD-W2 after Gram staining. c Phylogenetic tree of AD-W2 with closely related 16S rRNA gene sequences of Type strains
Purification of serratiopeptidase from S. marcescens AD-W2
Serratiopeptidase production profile of S. marcescens AD-W2 in the production medium has been demonstrated in Fig. 2. The maximum extracellular protease activity of 3528 Units/mL was obtained in 60 h. Partial purification by ammonium sulphate precipitation at 30–80% saturation led to the specific activity of 7659 Units/mg protein. The compilation of purification of serratiopeptidase from S. marcescens AD-W2 along with specific activities and purification folds, has been shown in Table 1. The purification of around two fold was achieved in ammonium sulphate precipitation. However, Romero et al. (2001) achieved 3.6 fold purification after 50–70% ammonium sulphate saturation. Further purification by MonoQ ion-exchange chromatography (Fig. 3) resulted in the specific activity of 20,492 (Units/mg protein) with a purification fold of 5.28.
Fig. 2.
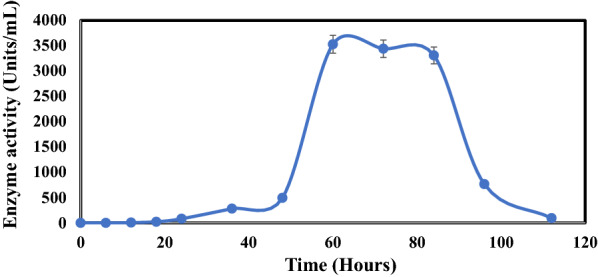
Time course profile of S. marcescens AD-W2 for serratiopeptidase production
Table 1.
The parameters of purification of serratiopeptidase from culture supernatant up to ion-exchange chromatography
| Sample | Total protease unit | Total protein content (mg) | Specific activity (units of enzyme activity/mg protein) | Purification fold |
|---|---|---|---|---|
| Culture supernatant | 613,851.8 | 158.3 | 3877.466 | 1 |
| Ammonium sulphate (40–80%) | 49,156.85 | 6.42 | 7658.901 | 1.97 |
| Ion exchange chromatography (MonoQ column) | 39,533.85 | 1.93 | 20,492.59 | 5.28 |
Fig. 3.
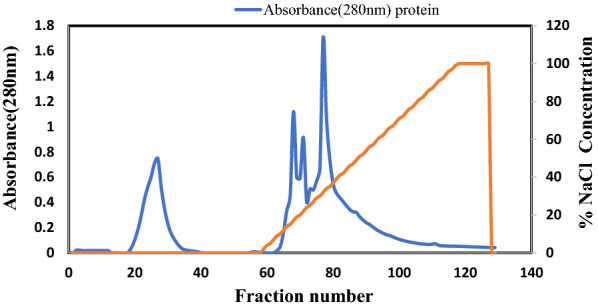
The protein elution profile during Ion exchange chromatography using MonoQ 5/50 GL column
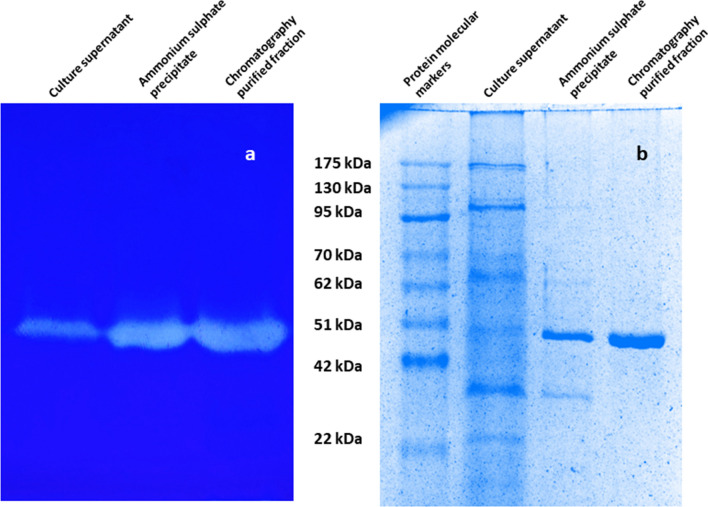
SDS-PAGE and zymography of serratiopeptidase from S. marcescens AD-W2
SDS-PAGE of crude extracellular protease, partially purified and purified serratiopeptidase, has been demonstrated in Fig. 4. Zymogram of the crude culture supernatant, partially purified and purified serratiopeptidase indicates that the purified protein is indeed protease with clear zones appeared due to hydrolysis of the substrate (casein). The molecular weight of purified serratiopeptidase molecule was predicted to be ~ 51 kDa, which is in the range of other serratiopeptidase reported from various sources (Devi et al. 2013; Nam et al. 2013; Gupte and Luthra 2017; Ethiraj and Gopinath 2017).
Fig.4.
a Casein Zymogram and b SDS-PAGE of serratiopeptidase from S. marcescens AD-W2
Characterization of purified serratiopeptidase from S. marcescens AD-W2
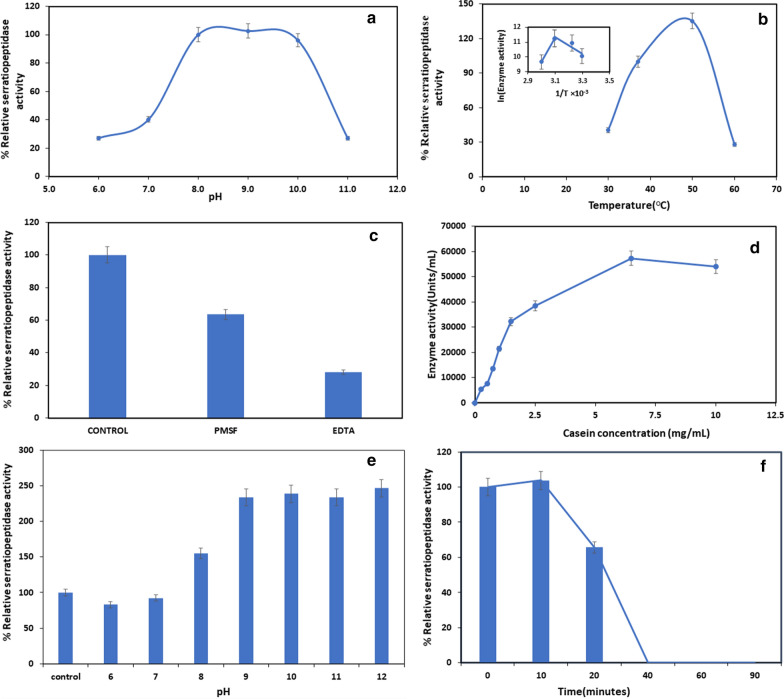
The characteristics of purified serratiopeptidase from S. marcescens AD-W2 have been demonstrated in Fig. 5. The optimum pH (Fig. 5a) and temperature (Fig. 5b) of serratiopeptidase from S. marcescens AD-W2 were observed to be pH 9.0 and 50 °C, respectively. Critical temperature, as determined by the Arrhenius plot (Fig. 5b inset), was 50 °C, above which the enzyme gets inactivated. The activation energy for the reaction to proceed below critical temperature, i.e., 50 °C, was − 45 kJ/mol, while above critical temperature, it was calculated as 141 kJ/mol. The inhibition studies indicate that EDTA inhibits 71.2% activity (Fig. 5c). However, inhibition by PMSF was not significant. As shown in Fig. 5d, The activity increased with substrate concentration until 6.5 mg/mL, thereafter it did not increase. Vmax and Km values of purified serratiopeptidase from S. marcescens AD-W2 are 57,256 Units/mL and 1.57 mg/mL, respectively, for casein. The stability studies indicate that the purified serratiopeptidase is stable under a wide temperature range up to 50 °C and pH range with higher conformational stability at higher pH (Fig. 5e). The stability profile also indicates that the purified serratiopeptidase retains 65% activity after 20 min incubation at 50 °C and 15% at 55 °C activity after 10 min incubation (Fig. 5f).
Fig. 5.
Effect of various reaction conditions and stability of serratiopeptidase from S. marcescens AD-W2: a pH; b temperature (Arrhenius plot shown in inset); c inhibitors d substrate concentration; e pH stability; f temperature stability at 50 °C
Identification of purified serratiopeptidase
The protein mass fingerprint (Additional file 1: Fig. S1) was searched in the MASCOT database and the search report revealed that the purified protein from S. marcescens AD-W2 was similar to serralysin (UNIPROT Accession number P07268) with 65% sequence coverage and a negligible chance of being false. The protein scores and sequence coverage of serratiopeptidase from S. marcescens AD-W2 with the reported serralysin are available in the (Additional file 1: Figs. S2 and S3).
The multiple sequence alignment of serralysin from different sources with the sequence of purified protein from S. marcescens AD-W2 has been shown in Fig. 6. The three-dimensional homology model of serratiopeptidase from S. marcescens AD-W2, as shown in Additional file 1: Fig. S1, was generated using online SWISS-MODEL software (http://swissmodel.expasy.org/) (Schwede et al. 2003) (Fig. 7). The template used by the modeller for building model was the crystal structure of serralysin protein from Serratia sp. (PDB entry 1srp), the first serralysin reported and characterized (Hamada et al. 1996). The biocatalytic qualities of the predicted models were confirmed by the PROCHECK server. Figure 8 demonstrates the Ramachandran plot analysis of the model protein. The stereo view of the catalytic domain (Fig. 9), from the depicted catalytic domain of serratiopeptidase from S. marcescens AD-W2 comprises of Zinc coordinated with three main residues, i.e. His192, His196, His202 along with Glu193 and Tyr232.
Fig. 6.
Multiple amino-acid sequence alignment of the serratiopeptidase from S. marcescens AD-W2, PRZN_SERME serralysin from S. marcescens (E-15) with 100% sequence identity, PRZN_SERMA Serralysin from S. marcescens with 98.6% sequence identity, 0A240AD01_SERFI Serralysin from S. ficaria with 92.6% sequence identity, D4E064_SEROD Serralysin from S. odorifera DSM 4582 with 82.8% sequence identity and E0SC28_DICD3 Secreted protease A from Dickeya dadantii with 64.8% sequence identity. The protein sequence from serratiopeptidase was aligned to see the similarities, differences, and conservation of various amino acid residues using multiple sequence alignment. The alignment was carried out by using Clustal W platform in Seaview software
Fig. 7.
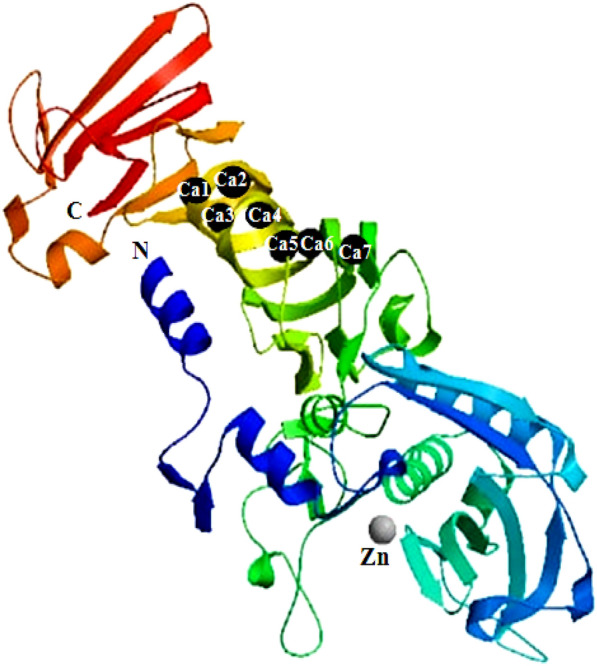
Cartoon view of the overall three-dimensional structure of Serratiopeptidase, a ribbon diagram showing the positions of the secondary structure elements was built using online SWISS-MODEL software (https://swissmodel.expasy.org/) and colored according to secondary-structure element. β-Strands are depicted as arrows, the zinc ion is shown by a grey ball, and calcium ions by black balls
Fig. 8.
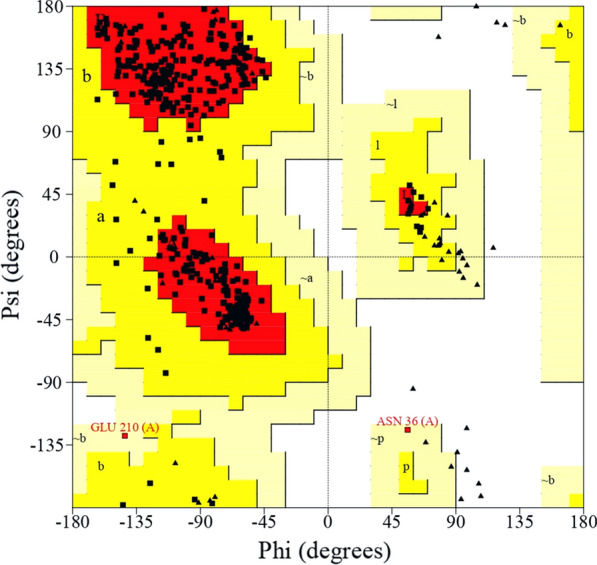
Ramachandran plot of the chimeric protein. The Ramachandran plot analysis (PROCHECK server); Laskowski et al. (1993), revealed that the modelled structure projected 91.0% of amino acid residues in most favoured regions of the Ramachandran plot, 8.5% are in additional allowed regions, 0.5% residues in generously allowed regions and 0.0% residues in disallowed regions
Fig. 9.
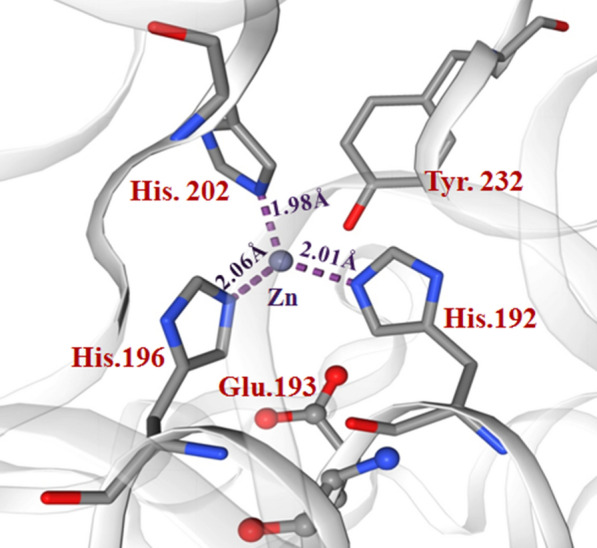
Stereoview of the zinc-binding site in the Serratiopeptidase. The zinc ion is shown as a purple ball. The active site of serratiopeptidase from S. marcescens AD-W2 shows the zinc ion coordinated as Zn.1, His.192, His.196, His.202, Glu.193, and Tyr.232 is forming a distorted trigonal bipyramidal catalytic active center
Discussion
Serratiopeptidase is an industrially important enzyme being used in the pharmaceutical industry as therapeutics (Ethiraj and Gopinath 2017). S. marcescens is known as the largest producer of serratiopeptidase. However, other bacteria are also known for Serratiopeptidase production (Salarizadeh et al. 2014; Wagdarikar et al. 2015; Nageswara et al. 2019). During the present study, S. marcescens AD-W2 has been demonstrated as a source of thermoactive serratiopeptidase. Although, the strain has a similarity with S. marcescens as well as S. nematodiphila, the biochemical tests like lactose utilization, xylose utilization, and arginine dihyrolase test revealed its close relation with S. marcescens (Zhang et al. 2009). The phylogenetic tree generated by the minimum evolution method also confirmed our observation to conclude the identification of the isolate as S. marcescens.
The serratiopeptidase from S. marcescens AD-W2 was purified by a simple two-step procedure presenting the specific activity of 20,492 Units/mg protein with an overall 5.28-fold purification. The molecular weight of purified serratiopeptidase molecule was found to be ~ 51 kDa, which is in the range of other serratiopeptidase reported from various sources (Devi et al. 2013; Nam et al. 2013; Gupte and Luthra 2017; Ethiraj and Gopinath 2017). Further, the zymogram confirmed that the purified protein was indeed the protease enzyme.
The characterization of the purified enzyme was carried out for optimum pH & temperature and the results are in agreement with previously demonstrated optimum pH from 8.5 to 10.0 (Miyata et al. 1970; Salamone and Wodzinski 1997; Nam et al. 2013; Salarizadeh et al. 2014). However, the optimum temperature was found to be higher than the previously reported temperature optima (Salamone and Wodzinski 1997; Nam et al. 2013; Salarizadeh et al. 2014). This is also higher than 40 °C as reported by Miyata et al. (1970) from Serratia marcescens E-15.
An attempt was made to determine the critical temperature of serratiopeptidase from S. marcescens AD-W2, which was calculated by the Arrhenius equation as 50 °C, above which the enzyme gets inactivated. To the best of our knowledge, this was the first attempt to determine the critical temperature of serratiopeptidase. This could be because of the positive activation energy for the reaction to proceed below the critical temperature, i.e., 50 °C. The inhibition studies indicate significant inhibition by EDTA and insignificant inhibition by PMSF, which confirms that the enzyme is a metalloprotease. The results from enzyme kinetics studies corroborate with the previously reported serralysin-like metalloprotease (Nageswara et al. 2019). The stability studies indicate that the purified serratiopeptidase is stable under wide temperature and pH range with higher conformational stability at higher pH. The stability profile has been observed to be better than previous reports showing 15% activity after 20 min of incubation at 50 °C (Nam et al. 2013).
Homology modelling of purified protein was performed using the template from the crystal structure of serralysin protein from Serratia sp. PDB entry 1srp (Hamada et al. 1996). Ramachandran plot indicates that there were 91.0% residues in the most favourable region, 8.5% residues in the additional allowed region, and 0.5% in the generously allowed region. There were no residues in the disallowed region and, the predicted overall three-dimensional structure of serratiopeptidase from S. marcescens AD-W2 shares an identical fold (100% identity) with its homolog, serralysin from Serratia sp. E-15. The 3D structure of serratiopeptidase from S. marcescens AD-W2 is a monomer, identical to the template molecule comprising of two domains and an extended N-terminal tail (Hamada et al. 1996). The serratiopeptidase from S. marcescens AD-W2 was predicted to be an elongated molecule with ligands 7 × CA (Calcium Ion) and 1 × ZN (Zinc Ion). It consists of an extended N-terminal tail linked with the C-terminus and two main domains i.e. N-terminal proteolytic domain and a catalytic domain with a Zinc ion. The stereoview (Fig. 9) of the depicted catalytic domain of serratiopeptidase from S. marcescens AD-W2 comprises of Zinc coordinated with three main residues, i.e. His192, His196, His202 along with Glu193 and Tyr232 residues. The predicted active site also corroborates with that of the crystal structure of serralysin (Wu et al. 2016; Nageswara et al. 2019).
In conclusion, serratiopeptidase from S. marcescens AD-W2 was successfully purified with a specific activity of 20,492 units/mg. The thermoactive serratiopeptidase was found to have stability in wide pH and temperature range with Km of 1.57 mg/mL and Vmax of 57,256 Units/mL, respectively. The predicted active site in the catalytic domain re-confirmed that the purified protein from S. marcescens AD-W2 is a serratiopeptidase. Further, the process for the production of serratiopeptidase may be developed either using the wild-type strain or by developing the recombinant strain.
Supplementary Information
Additional file 1. Fig. S1: MALDI-MS/MS mass fingerprint of purified serratiopeptidase from Serratia marcescens AD-W2. Fig. S2: Protein scores obtained from MASCOT server with MALDI-MS/MS mass fingerprint. Fig. S3: Protein sequence of the purified serratiopeptidase from Serratia marcescens AD-W2. The red colour region shows sequence coverage (65%) in peptide mass fingerprint obtained through peptide mass fingerprint searches in the Mascot server. Fig. S4: gene sequence of serralysin was translated into protein and then aligned with protein sequences having highest similarity in UNIPROT database, we could see that the Zinc binding domain is highly conserved in nature. Even in proteins predicted from the plant genome oryza sativa there is very high similarity with that from microbial sources. This might be an indication of close relation of Serratia marcescens in plants not only limited to mulberry (Koul et al. 2020) and Cucurbita pepa (Selvakumar et al. 2008) and also much greater role of serralysin as natural insecticidal in plants (Kaviyarasi and Suryanarayan 2016).
Acknowledgements
Authors are thankful to the Director CSIR-IIIM for his interest in this work. DC thanks UGC for JRF. The manuscript bears the institutional communication number CSIR-IIIM/IPR/00192.
Accession numbers
The 16S rRNA gene sequence of S. marcescens AD-W2 has been submitted to NCBI with accession number MT416417. The culture has been submitted to Col. Sir R. N. Chopra Microbial Resource Centre, Jammu, India with Accession number MRCJ-997.
Authors’ contributions
DC carried out most of the experiments and wrote the manuscript, JKK carried out molecular modelling work and wrote the bioinformatics part. DK contributed in enzyme production, characterization experiments and formatting of the manuscript, MMH helped in executing protein purification, MJD supervised Ion Exchange Chromatography experiments, AC conceptualized, coordinated, supervised the entire work and corrected the manuscript. All authors read and approved the final manuscript.
Funding
Not applicable.
Availability of data and materials
The datasets generated during and/or analysed during the current study are available from the corresponding author on reasonable request.
Declarations
Ethics approval and consent to participate
This article does not contain any studies with human participants or animals performed by any of the authors.
Consent for publication
All authors have provided consent for this publication.
Competing interests
All the authors declare that they do not have any conflict of interest.
Footnotes
The original online version of this article was revised: The error in the coauthor name has been corrected.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Change history
5/4/2021
A Correction to this paper has been published: 10.1186/s13568-021-01223-7
References
- Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- Cupp-Enyard C. Sigma’s non-specific protease activity assay—casein as a substrate. J Vis Exp. 2008 doi: 10.3791/899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devi CS, Joseph RE, Saravanan H, Naine SJ, Srinivansan VM. Screening and molecular characterization of Serratia marcescens VITSD2: a strain producing optimum serratiopeptidase. Front Biol. 2013;8:632–639. doi: 10.1007/s11515-013-1284-9. [DOI] [Google Scholar]
- Ethiraj S, Gopinath S. Production, purification, characterization, immobilization, and application of serrapeptase: a review. Front Biol. 2017;12:333–348. doi: 10.1007/s11515-017-1461-3. [DOI] [Google Scholar]
- Galtier N, Gouy M, Gautier C. SEAVIEW and PHYLO_WIN: two graphic tools for sequence alignment and molecular phylogeny. Bioinformatics. 1996;12:543–548. doi: 10.1093/bioinformatics/12.6.543. [DOI] [PubMed] [Google Scholar]
- Gupte V, Luthra U. Analytical techniques for serratiopeptidase: a review. J Pharm Anal. 2017;7:203–207. doi: 10.1016/j.jpha.2017.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamada K, Hata Y, Katsuya Y, Hiramatsu H, Fujiwara T, Katsube Y. Crystal structure of serratia protease, a zinc-dependent proteinase from Serratia sp. E-15, containing a β-sheet coil motif at 2.0 AÅ resolution. J Biochem. 1996;119:844–851. doi: 10.1093/oxfordjournals.jbchem.a021320. [DOI] [PubMed] [Google Scholar]
- James C, Natalie S. Microbiology. A laboratory manual. Pearson Education; 2014. [Google Scholar]
- Koul D, Chander D, Manhas RS, Chaubey A. Isolation and characterization of serratiopeptidase producing bacteria from mulberry phyllosphere. Curr Microbiol. 2020 doi: 10.1007/s00284-020-02280-0. [DOI] [PubMed] [Google Scholar]
- Kumar S, Stecher G, Li M, Knyaz C, Tamura K, Battistuzzi FU. MEGA X: molecular evolutionary genetics analysis across computing platforms. Molec Biol Evol. 2018;35(6):1547–1549. doi: 10.1093/molbev/msy096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Lantz MS, Ciborowski P. Methods in enzymology. Elsevier; 1994. [45] Zymographic techniques for detection and characterization of microbial proteases; pp. 563–594. [DOI] [PubMed] [Google Scholar]
- Laskowski RA, MacArthur MW, Moss DS, Thornton JM. PROCHECK: a program to check the stereochemical quality of protein structures. J Appl Crystall. 1993;26:283–291. doi: 10.1107/S0021889892009944. [DOI] [Google Scholar]
- Miyata K, Maejima K, Tomoda K, Isono M. Serratia protease: Part I. Purification and general properties of the enzyme. Agric Biol Chem. 1970;34:310–318. [Google Scholar]
- Nageswara S, Guntuku G, Yakkali BL. Purification, characterization, and structural elucidation of serralysin-like alkaline metalloprotease from a novel source. J Genet Eng Biotechnol. 2019;17:1. doi: 10.1186/s43141-019-0002-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nam MS, Whang KS, Choi SH, Bae HC, Kim YK, Park YW. Purification, characterization, and properties of an alkaline protease produced by Serratia marcescens S3-R1 inhabiting Korean ginseng rhizosphere. J Sci Food Agric. 2013;93:3876–3882. doi: 10.1002/jsfa.6363. [DOI] [PubMed] [Google Scholar]
- Olajuyigbe FM, Falade AM. Purification and partial characterization of serine alkaline metalloprotease from Bacillus brevis MWB-01. Bioresour Bioprocess. 2014;1:8. doi: 10.1186/s40643-014-0008-6. [DOI] [Google Scholar]
- Olmstead SF (2017) Compositions and methods comprising Serratia peptidase for inhibition of bacterial vaginosis, bacterial vaginitis or fungal vaginitis (U.S. Patent No. 9,737,591). U.S. Patent and Trademark Office
- Olmstead SF (2018) Methods of comprising serratia peptidase for inhibition and treatment of biofilms related to certain conditions (U.S. Patent No. 9,931,381). U.S. Patent and Trademark Office
- Pakhale SV, Bhagwat SS. Purification of serratiopeptidase from Serratia marcescens NRRL B 23112 using ultrasound assisted three phase partitioning. UltrasonSono Chem. 2016;31:532–538. doi: 10.1016/j.ultsonch.2016.01.037. [DOI] [PubMed] [Google Scholar]
- Perkins DN, Pappin DJ, Creasy DM, Cottrell JS. Probability-based protein identification by searching sequence databases using mass spectrometry data. Electrophor Int J. 1999;20:3551–3567. doi: 10.1002/(SICI)1522-2683(19991201)20:18<3551::AID-ELPS3551>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- Romero FJ, García LA, Salas JA, Díaz M, Quirós LM. Production, purification and partial characterization of two extracellular proteases from Serratia marcescens grown in whey. Process Biochem. 2001;36:507–515. doi: 10.1016/S0032-9592(00)00221-1. [DOI] [Google Scholar]
- Rzhetsky A, Nei M. A simple method for estimating and testing minimum-evolution trees. Molec Biol Evol. 1992;9:945–967. [Google Scholar]
- Salamone P, Wodzinski R. Production, purification and characterization of a 50-kDa extracellular metalloprotease from Serratia marcescens. Appl Microbiol Biotechnol. 1997;48:317–324. doi: 10.1007/s002530051056. [DOI] [PubMed] [Google Scholar]
- Salarizadeh N, Hasannia S, Akbari Noghabi K, Hassan Sajedi R. Purification and characterization of 50 kda extracellular metalloprotease from Serratia sp. ZF03. Iran J Biotechnol. 2014;12:18–27. doi: 10.15171/ijb.1009. [DOI] [Google Scholar]
- Schwede T, Kopp J, Guex N, Peitsch MC. SWISS-MODEL: an automated protein homology-modeling server. Nucleic Acids Res. 2003;31:3381–3385. doi: 10.1093/nar/gkg520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shevchenko A, Tomas H, Havli J, Olsen JV, Mann M. In-gel digestion for mass spectrometric characterization of proteins and proteomes. Nat Protoc. 2006;1:2856–2860. doi: 10.1038/nprot.2006.468. [DOI] [PubMed] [Google Scholar]
- Srivastava V, Mishra S, Chaudhuri TK. Enhanced production of recombinant serratiopeptidase in Escherichia coli and its characterization as a potential biosimilar to native biotherapeutic counterpart. Microb Cell Factories. 2019;18:1–15. doi: 10.1186/s12934-019-1267-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagdarikar MJ, Joshi AM, Shaikh AA. Media optimization studies for enhanced production of serratiopeptidase from Bacillus licheniformis (NCIM 2042) Asian J Biomed Pharm Sci. 2015;5:19. doi: 10.15272/ajbps.v5i42.677. [DOI] [Google Scholar]
- Wu D, Ran T, Wang W, Xu D. Structure of a thermostable serralysin from Serratia sp. FS14 at 1.1 Å resolution. Acta Crystallogr Sect F Struct Biol Commun. 2016;72:10–15. doi: 10.1107/S2053230X15023092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C-X, Yang S-Y, Xu M-X, Sun J, Liu H, Liu J-R, Liu H, Kan F, Sun J, Lai R. Serratia nematodiphila sp. nov., associated symbiotically with the entomopathogenic nematode Heterorhabditidoides chongmingensis (Rhabditida: Rhabditidae) Int J Syst Evol Microbiol. 2009;59:1603–1608. doi: 10.1099/ijs.0.003871-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1. Fig. S1: MALDI-MS/MS mass fingerprint of purified serratiopeptidase from Serratia marcescens AD-W2. Fig. S2: Protein scores obtained from MASCOT server with MALDI-MS/MS mass fingerprint. Fig. S3: Protein sequence of the purified serratiopeptidase from Serratia marcescens AD-W2. The red colour region shows sequence coverage (65%) in peptide mass fingerprint obtained through peptide mass fingerprint searches in the Mascot server. Fig. S4: gene sequence of serralysin was translated into protein and then aligned with protein sequences having highest similarity in UNIPROT database, we could see that the Zinc binding domain is highly conserved in nature. Even in proteins predicted from the plant genome oryza sativa there is very high similarity with that from microbial sources. This might be an indication of close relation of Serratia marcescens in plants not only limited to mulberry (Koul et al. 2020) and Cucurbita pepa (Selvakumar et al. 2008) and also much greater role of serralysin as natural insecticidal in plants (Kaviyarasi and Suryanarayan 2016).
Data Availability Statement
The datasets generated during and/or analysed during the current study are available from the corresponding author on reasonable request.