Abstract
Background and Aim
Obesity affects the gut microbiome, which in turn increases the risk for colorectal cancer. Several studies have shown the mechanisms by which some bacteria may influence the development of colorectal cancer; however, gut microbiome characteristics in obese patients with colorectal cancer remain unclear. Therefore, this study evaluated their gut microbiome profile and its relationship with metabolic markers.
Methods
The study assessed fecal samples from 36 consecutive patients with colorectal cancer and 38 controls without colorectal cancer. To identify microbiotic variations between patients with colorectal cancer and controls, as well as between nonobese and obese individuals, 16S rRNA gene amplicon sequencing was performed.
Results
Principal coordinate analysis showed significant differences in the overall structure of the microbiome among the study groups. The α‐diversity, assessed by the Chao1 index or Shannon index, was higher in patients with colorectal cancer versus controls. The relative abundance of the genera Enterococcus, Capnocytophaga, and Polaribacter was significantly altered in obese patients with colorectal cancer, whose serum low‐density lipoprotein concentrations were positively correlated with the abundance of the genus Enterococcus; among the most abundant species was Enterococcus faecalis, observed at lower levels in obese versus nonobese patients.
Conclusions
This study demonstrated several compositional alterations of the gut microbiome in patients with colorectal cancer and showed that a reduced presence of E. faecalis may be associated with obesity‐related colorectal cancer development. The gut microbiome may provide novel insights into the potential mechanisms in obesity‐related colorectal carcinogenesis.
Keywords: Capnocytophaga, colorectal neoplasms, enterococcus, gastrointestinal microbiome, obesity
This study demonstrated that a reduced presence of Enterococcus faecalis may be associated with the obesity‐related colorectal cancer development. The gut microbiome may provide novel insights for the potential mechanisms in obesity‐related colorectal carcinogenesis.

Introduction
Colorectal cancer (CRC) is the third most common cancer and the second most common cause of cancer death, estimated at 881 000 deaths globally in 2018, accounting for about 1 in 10 cancer deaths. 1 The average risk of developing colon cancer before 75 years of age is estimated to be 31%. 2 Evidence is sufficient for a causal relationship between the risk of CRC and excess body weight or obesity, which is thought to be one of the attributes of the global burden of cancer. 3 The underlying mechanisms linking obesity to CRC remain a matter of debate; however, metabolic syndrome, insulin resistance, and alterations in adipocytokine concentrations appear to be of great importance. 4 The authors of this study previously demonstrated that visceral fat obesity and metabolic syndrome, insulin resistance following hyperinsulinemia, 5 impaired secretion of adipocytokine 6 , 7 , 8 or incretin, 9 , 10 and systemic low chronic inflammation 11 , 12 contribute to the development of CRC or colorectal adenoma, which is a well‐known precancerous lesion of CRC.
Obesity, physical activity, and dietary patterns may affect the gut microbiome. 13 , 14 Obesity has been associated with phylum‐level changes and reduced the diversity of the gut microbiome. 15 The relative abundance of the phylum Firmicutes and the phylum Bacteroidetes was increased in obese individuals compared with lean individuals; this proportion decreased with weight loss achieved by a fat‐restricted or carbohydrate‐restricted low‐calorie diet. 16 Microbiome transplantation was demonstrated to change the capacity for energy harvest from the diet 17 or insulin resistance. 18 Several studies have shown the association between weight loss and enrichment of Akkermansia muciniphila, the administration of which may prevent the development of obesity and its complications. 19 This evidence implies the significant role of microbial composition in the pathogenesis of obesity. However, another study reported no relationship between body mass index and the ratio of Firmicutes/Bacteroidetes in Japanese individuals. 20 To understand the influence of a microbiome on obesity, studies must investigate the relationships in various environments considering factors such as race; lifestyle, including diet and physical activity; socioeconomic status; and disease.
Emerging evidence suggests that the gut microbiome is also associated with increasing risk of CRC development. 13 , 21 , 22 In patients with CRC, a higher abundance of Fusobacterium nucleatum has been associated with unfavorable outcomes. 23 , 24 Enrichment of F. nucleatum on CRC tissue altered recruitment of tumor‐infiltrating immune cells, resulting in attenuation of the antitumor immune response. 25 , 26 Several specific bacteria, including Streptococcus bovis, Helicobacter pylori, Bacteroides fragilis, E. faecalis, Clostridium septicum, and Escherichia coli, have been identified and are suspected to play a role in CRC development. 27 In addition, recent large‐cohort multiomics data indicated that shifts in the microbiome and metabolome were apparent in cases of colorectal adenomas and intramucosal carcinomas, in addition to more advanced lesions. 28
These findings seem to provide new opportunities to take advantage of the existing knowledge of the gut microbiome for the pathogenesis of obesity‐related CRC development. However, the characteristics of the microbiome in obese patients with CRC remain vastly underexplored. Therefore, this study evaluated the gut microbiome profile and its relationship with metabolic markers in obese patients with CRC.
Methods
Study population
From June 2018 to July 2019, fecal samples were obtained from 36 consecutive patients with CRC at Yamagata University Hospital, Yamagata, Japan, and 38 individuals without CRC (controls) as confirmed on total colonoscopy as part of the health checkup conducted at Tohoku Central Hospital, Yamagata. The control participants in the present study were asymptomatic adults who underwent health checkups; they were not patients who visited the hospital. No obvious gastrointestinal abnormalities were found in the results of the health checkups, including colonoscopy. Exclusion criteria included a history of bowel resection, inflammatory bowel disease, and irritable bowel syndrome for both groups and a history or diagnosis of malignant neoplasm in the controls. None of the participants were taking antibiotics or acid‐suppressing drugs or were undergoing antiobesity treatment.
This study was approved by the Ethics Committee of Yamagata University Faculty of Medicine (#2018‐260) and Tohoku Central Hospital (#807‐6) and was conducted in accordance with the Declaration of Helsinki. Written informed consent was obtained from all participants before beginning the study.
Methods for obtaining clinical data, colonoscopy, and stool collection are described in the Supporting information.
Stool DNA extraction
Fecal DNA was isolated using the KingFisher Duo Prime Purification System with MagMAX CORE Nucleic Acid Purification kit (Thermo Fisher Scientific, Waltham, MA, USA). Approximately 120 μL of the stored fecal sample was mixed with one volume of sterilized phosphate‐buffered salts; the supernatant was collected after centrifugation at 100g for 1 min and then incubated at 55°C for 30 min after mixing with both 10 μL of proteinase K and its buffer. The mixture was centrifuged at 15000g for 2 min; then, the supernatant was collected. The subsequent extraction procedure was performed according to the manufacturer's instructions. The yield and quality of the extracted DNA were evaluated with the Qubit HS assay kit (Thermo Fisher Scientific).
Sequencing of 16S rRNA gene
Multiplex polymerase chain reactions (PCRs) were performed for the purified DNA samples using two primer pools corresponding to the V2, V4, and V8 regions (size of amplicon was 250 base pairs [bp], 288 bp, and 295 bp, respectively) or V3, V6–7, and V9 regions (sizes of amplicon were 215 bp, 260 bp, and 209 bp, respectively) of the 16S rRNA (16S metagenomics kit, Thermo Fisher Scientific). According to the procedure manual of the Ion AmpliSeq library kit (Thermo Fisher Scientific), adapter sequences were added to the PCR products to obtain sequence libraries. To ensure the quality and size of each library, digital electrophoresis using a D1000 ScreenTape on a 2200 Tape Station (Agilent Technologies, Santa Clara, CA, USA) was performed. Amplified 30–140 pM libraries were submitted to emulsion PCR. After purification using biotin‐coated beads, the PCR products were loaded onto an Ion P1 chip v3. Ion semiconductor sequencing was performed using Ion Proton (Thermo Fisher Scientific). To obtain taxonomy assignment, >101 bp read lengths in sequence files were run through the assembly programs constructed with Bowtie2 version 2.3.5 program along with consecutive mapping on Greengenes database (published May 2011, including 16Sr RNA gene sequence information of 406 998 strains). The read depths against the 16S rRNA gene regions were calculated by using the bedtools software version 2.29.0. To optimize mapping, microbiome data were used that could confirm multiple reads in ±3 bp over the size of amplicon against the V1–V9 regions using the In House R script.
Statistical analysis
Continuous variables and categorical variables were analyzed using the two‐tailed Wilcoxon test and the Fisher exact test, respectively. Multiple comparisons were performed using the Kruskal–Wallis test to compare continuous variables following a post‐hoc Steel–Dwass test. A value of P < 0.05 was considered statistically significant. Statistical calculations were conducted using JMP 14 (SAS Institute) and R programming language software, version 3.6.1.
Results
Select characteristics of participants
In the patients with CRC (n = 36), the number of current smokers and those with diabetes or hyperlipidemia; median age; concentrations of fasting plasma glucose (FPG), fasting plasma insulin (FPI), and hemoglobin A1c (HbA1c); and the homeostasis model assessment of insulin resistance (HOMA‐IR) were significantly higher, whereas body mass index (BMI), total cholesterol (TC), and high‐density lipoprotein (HDL) and low‐density lipoprotein (LDL) concentrations were significantly lower versus controls (n = 38, Table S1, Supporting information). No significant differences were observed between patients with CRC and controls for the proportion of participants who were male, obese, or a current drinker or for triglyceride (TG) concentrations. In the controls, the proportion of males, FPI and TG concentrations, and HOMA‐IR were significantly higher, whereas HDL concentration was lower in obese (n = 17) versus nonobese participants (n = 21, Table S2). In patients with CRC, no significant differences were noted in clinical characteristics and CRC stages between obese (n = 9) and nonobese (n = 27) participants. In obese participants, mean age, the proportion of diabetes and hyperlipidemia, and FPG concentrations were higher, whereas TC and LDL concentrations were lower in patients with CRC versus controls.
Altered diversity of the gut microbiome in patients with CRC
Principal coordinates analysis demonstrated a significant difference in the overall microbiota structure between patients with CRC and controls (Fig. 1a) or between obese and nonobese participants in each group (Fig. 1b,c) or between obese patients with CRC and obese controls (Fig. 1d). The median Shannon index, which estimated both operational taxonomic unit (OTU) evenness and richness, tended to be higher in patients with CRC (Fig. 2a, Table S3). No significant intergroup differences in the Shannon index were observed between nonobese and obese controls or patients with CRC (Fig. 2b). The median Chao1 index, which estimated OTU richness, was significantly higher in patients with CRC versus controls (Fig. 2c). In either nonobese or obese participants, the median Chao1 index tended to be higher in patients with CRC versus controls (Fig. 2d).
Figure 1.
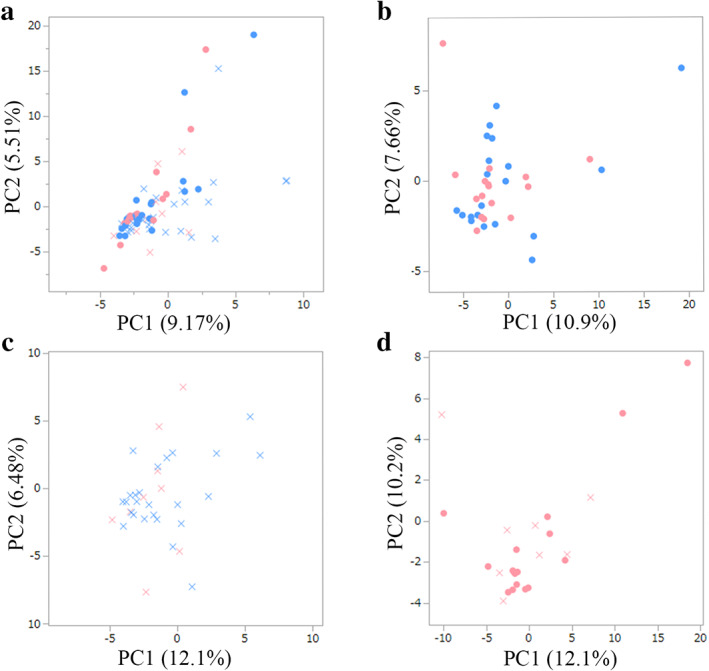
Principal coordinate analysis to assess the beta diversity based on OTU abundance at the genus levels. Unweighted UniFrac distances were calculated between the patients with CRC (cross) and the controls (circle) (a), or between obese (pink) and nonobese participants (blue) in controls (b) or patients with CRC (c), or between obese patients with CRC and obese controls (d). Microbial structural differences were observed in each of the comparisons. Controls, individuals without colorectal cancer; CRC, colorectal cancer; OTU, operational taxonomic unit; PC, principal component.  , Non‐obese control;
, Non‐obese control;  , obese control;
, obese control;  , non‐obese CRC;
, non‐obese CRC;  , obese CRC.
, obese CRC.
Figure 2.
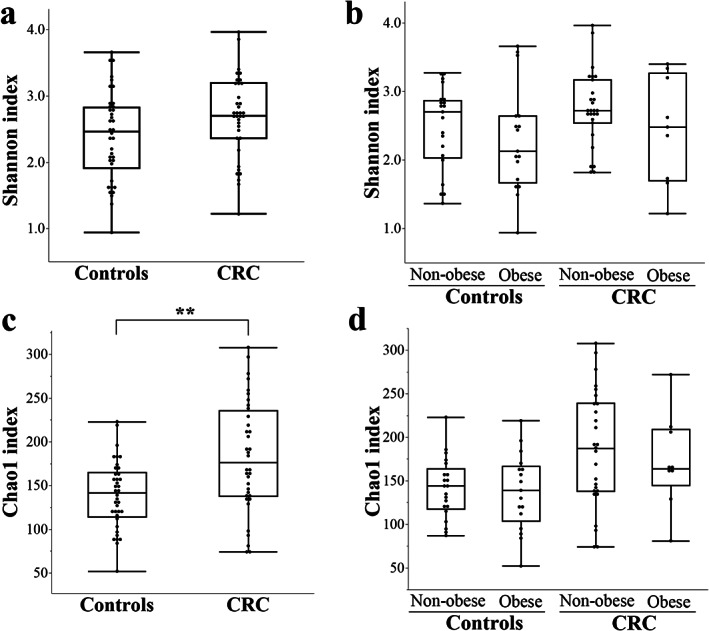
Alpha diversity in the patients with CRC and the controls with or without obesity. The observed Shannon and Chao1 phylogenetic diversity indices were calculated by the “vegan” version 2.5‐6 package of R version 3.6.1. The line inside the box represents the median, while the whiskers represent the lowest and highest values within the 1.5 IQR (Table S3). No significant differences were observed in the median Shannon index among the groups (a, b). The median Chao1 index was significantly higher in patients with CRC versus controls (c) in either case of nonobese or obese participants (d; Kruskal–Wallis test, P = 0.02; Steel–Dwass test showed no significant differences among the groups). No significant intergroup differences were noted in the Choa1 index between nonobese and obese controls or patients with CRC. **P < 0.01. CRC, colorectal cancer; IQR, internal quartile range.
Characteristics of gut microbiome in obese patients with CRC
The relative abundance rates (Tables S4,S5) of the phyla Actinobacteria, Fusobacteria, and Tenericutes were significantly higher in patients with CRC versus controls (Fig. 3a,b). Relative abundance rates of 38 genera were significantly higher, whereas those of 9 genera were significantly lower in patients with CRC versus controls (Fig. 4, Table S6). In patients with CRC, comparison of these genera between obese and nonobese participants revealed that the relative abundance of the genus Enterococcus was significantly lower in obese patients, whereas the relative abundance of the genera Capnocytophaga and Polaribacter were significantly higher in obese patients with CRC (Fig. 5a–c, Table S7). In controls, no significant differences were observed in the relative abundance of Enterococcus and Capnocytophaga between obese and nonobese participants (Fig. 5d,e). The genus Polaribacter was observed in none of the controls nor in seven patients (19.4%) with CRC.
Figure 3.
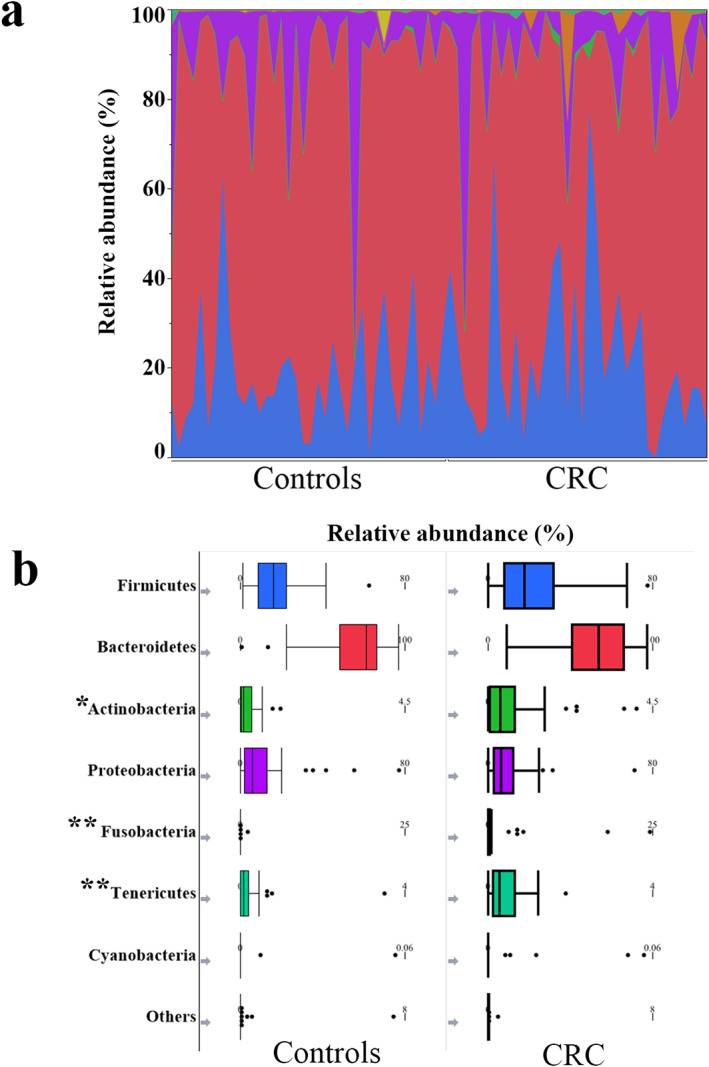
Phylum‐level microbial composition. The phylum‐level taxonomic composition of all participants (a). Comparison between the patients with CRC and the controls; the median relative abundances of the phyla Actinobacteria (P = 0.03), Fusobacteria (P < 0.001), and Tenericutes (P < 0.001) were significantly higher, whereas no significant differences were observed in the median relative abundance of the phyla Firmicutes (P = 0.42), Bacteroidetes (P = 0.16), Proteobacteria (P = 0.77), and Cyanobacteria (P = 0.21). *P < 0.05, **P < 0.01. CRC, colorectal cancer.  , Firmicutes;
, Firmicutes;  , Bacteroidetes;
, Bacteroidetes;  , Actinobacteria;
, Actinobacteria;  , Proteobacteria;
, Proteobacteria;  , Fusobacteria;
, Fusobacteria;  , Tenericutes;
, Tenericutes;  , Cyanobacteria;
, Cyanobacteria;  , others.
, others.
Figure 4.
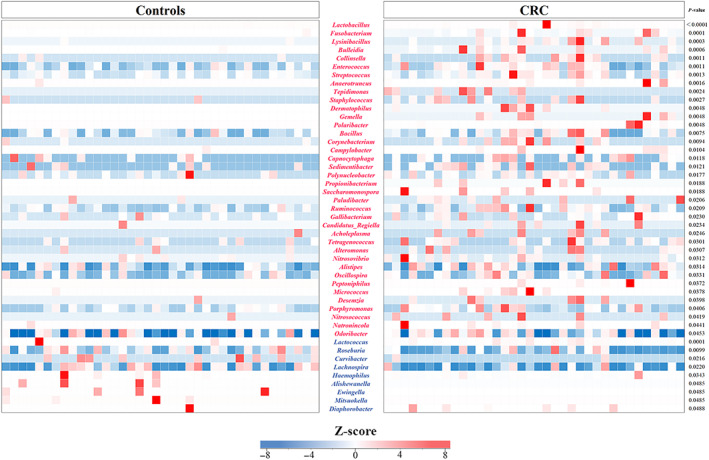
Heatmap visualizing the Z‐score distribution of the genera in patients with colorectal cancer (CRC) and the controls. The Z‐scores were used to assess the observation's deviation of microbiome components from the mean values. The proportion of 38 genera (violet‐red) was significantly higher in the patients with CRC than in the controls: Lactobacillus, Fusobacterium, Lysinibacillus, Bulleidia, Collinsella, Enterococcus, Streptococcus, Anaerotruncus, Tepidimonas, Staphylococcus, Dermatophilus, Gemella, Polaribacter, Bacillus, Corynebacterium, Campylobacter, Capnocytophaga, Sedimentibacter, Polynucleobacter, Propionibacterium, Saccharomonospora, Paludibacter, Ruminococcus, Gallibacterium, Candidatus Regiella, Acholeplasma, Tetragenococcus, Alteromonas, Nitrosovibrio, Alistipes, Oscillospira, Peptoniphilus, Micrococcus, Desemzia, Porphyromonas, Nitrosococcus, Natronincola, and Odoribacter. In contrast, the proportion of nine genera (blue) was significantly lower in patients with CRC: Lactococcus, Roseburia, Curvibacter, Lachnospira, Haemophilus, Alishewanella, Ewingella, Mitsuokella, and Diaphorobacter.
Figure 5.
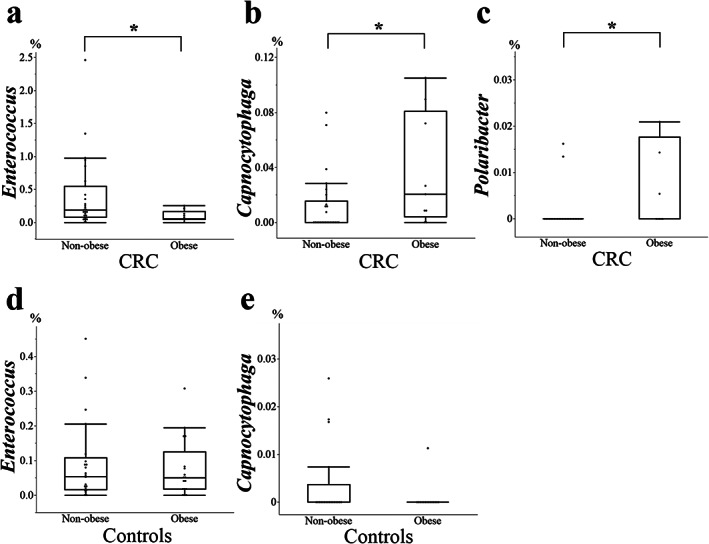
Comparison of the genera between obese and nonobese patients with colorectal cancer (CRC). In the obese patients with CRC, the genus Enterococcus (P = 0.04) was significantly lower, and the genera Polaribacter (P = 0.03) and Capnocytophaga (P = 0.03) were significantly higher than in the nonobese patients with CRC. In the controls, no significant differences were observed in the genera Enterococcus (P = 0.65) and Capnocytophaga (P = 0.74) between obese and nonobese participants. None of the controls has Polaribacter. *P < 0.05.
Relationship between genera and metabolic factors in patients with CRC
For Enterococcus, Capnocytophaga, and Polaribacter, the relative abundance of which was significantly altered in obese patients with CRC, their correlation with metabolic factors was evaluated. The relative abundance of the genus Enterococcus positively correlated with LDL concentrations in patients with CRC (Fig. 6a, Table S8), particularly in obese patients (Fig. 6b).
Figure 6.
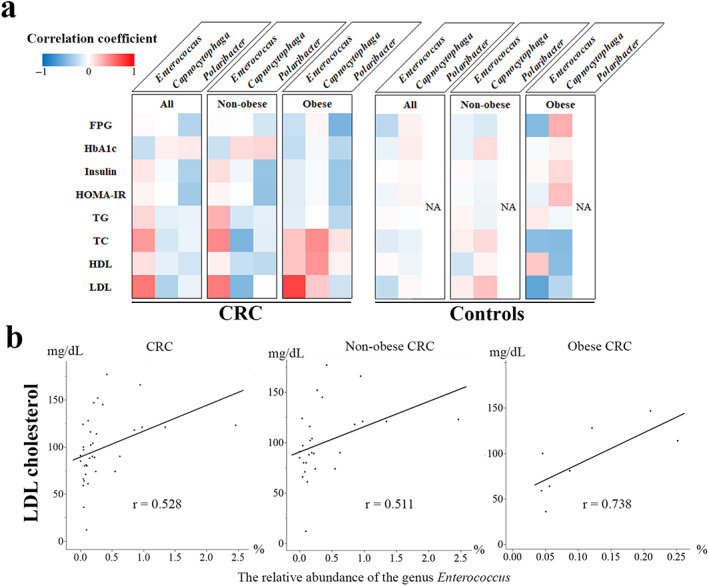
Association with metabolic markers. The Spearman correlation coefficient was used for metabolic markers to assess the correlations (a). The genus Polaribacter was observed in none of the controls. In the patients with CRC, particularly in the obese patients, the LDL concentrations were associated with the relative abundance of the genus Enterococcus (b). No strong correlation between the relative abundance of the genus Capnocytophaga or Polaribacter and metabolic markers, including FPG, HbA1c, FPI, TG, TC, HDL, and LDL concentrations and HOMA‐IR, was observed in patients with CRC. In controls, no significant correlation between the genus Enterococcus or Capnocytophaga and metabolic markers was noted. CRC, colorectal cancer; FPG, fasting plasma glucose; HOMA‐IR, homeostasis model assessment of insulin resistance; HDL, high‐density lipoprotein; LDL, low‐density lipoprotein; NA, not applicable; TC, total cholesterol; TG, triglyceride.
Enterococcus species in obese patients with CRC
The most abundant species in the genus Enterococcus was E. faecalis, which was significantly lower in obese versus nonobese patients with CRC (Fig. 7a). In controls, the relative abundance of E. faecalis was higher in obese versus nonobese participants. The relative abundance of E. faecalis against all microbiome components in the present study was significantly lower in obese versus nonobese patients with CRC (Fig. 7b); in contrast, no significant difference was observed in those components between obese and nonobese controls. Furthermore, no significant differences were found in the relative abundance of E. faecalis among the CRC stages (Fig. S1) and participant ages (Fig. S2), although patients with CRC were significantly older than controls. No significant differences were noted in the relative abundance of the other identified Enterococcus species between obese and nonobese patients with CRC.
Figure 7.
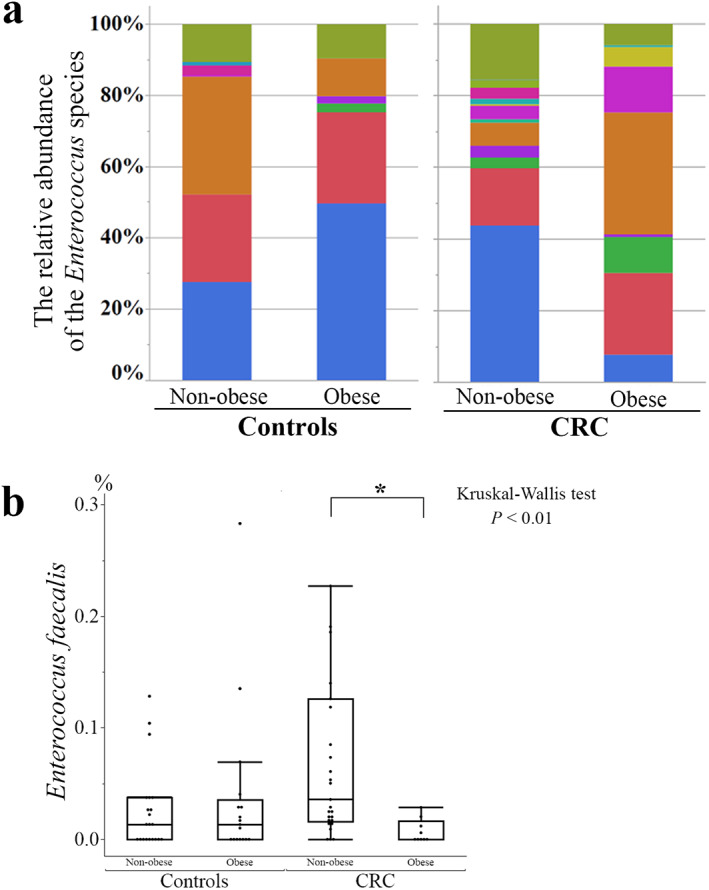
Analysis of Enterococcus species. E. faecalis was the most abundant species of the genus Enterococcus in the participants. In patients with colorectal cancer (CRC), the relative abundance of E. faecalis against all Enterococcus species (P < 0.01, a) or all microbiome components (P = 0.01, b) was significantly lower in obese versus nonobese patients (Kruskal–Wallis test, P = 0.003; Steel–Dwass test, P = 0.01). *P < 0.05.  , E. faecalis;
, E. faecalis;  , E. saccharolyticus;
, E. saccharolyticus;  , E. sulfureus;
, E. sulfureus;  , E. lactis;
, E. lactis;  , E. casseliflavus;
, E. casseliflavus;  , E. hirae;
, E. hirae;  , E. gallinarum;
, E. gallinarum;  , E. avium;
, E. avium;  , E. hermanniensis;
, E. hermanniensis;  , E. columbae;
, E. columbae;  , E. cecorum;
, E. cecorum;  , E. haemoperoxidus;
, E. haemoperoxidus;  , unclassified.
, unclassified.
Discussion
In the present study, the gut microbiome compositions were altered between obese and nonobese participants both in the controls and in patients with CRC. Increased diversity of the gut microbiome was observed in patients with CRC. The relative abundance of the genera Enterococcus, Capnocytophaga, and Polaribacter was significantly altered in obese patients with CRC. Furthermore, these results were the first to demonstrate the correlation between serum LDL concentrations and the relative abundance of the genus Enterococcus and that E. faecalis was significantly reduced in obese patients with CRC. Thus, the present study is the first to describe the association of E. faecalis in obesity‐related CRC development.
A meta‐analysis showed that the gut microbiome in patients with CRC had higher richness than in controls, partly due to expansions of species typically derived from the oral cavity. 28 In the present study, the estimated α‐diversity, assessed by the Chao1 index or Shannon index, was higher in patients with CRC than in controls. The α‐diversity has been shown to be decreased in obese patients. 29 In the present study, lower α‐diversity was observed in obese versus nonobese participants both in the control and CRC groups, although no significant differences were noted. Principal component analysis revealed differences in the gut microbiota composition between controls and patients with CRC for either nonobese or obese cases. The gut microbiome in obese patients with CRC had unique characteristics that may be associated with the development of obesity‐related CRC.
Consistent with the previous reports, 28 , 30 , 31 , 32 , 33 , 34 this study could identify associations between patients with CRC and the phyla Fusobacteria and several genera: Alistipe, Anaerotruncus, Campylobacter, Collinsella, Enterococcus, Fusobacterium, Gemella, Porphyromonas, Roseburia, Ruminococcus, and Streptococcus. In particular, the association between Fusobacterium 31 , 32 , 34 or Enterococcus 30 , 35 and CRC development has been well studied. In addition to these bacteria, an association was found between 36 genera and the patients with CRC; these candidate genera have potential roles to enhance CRC development. The present study included a limited number of participants and focused on the analysis of obese patients with CRC. Additional validation studies are therefore required to establish these associations because they may be influenced by various environmental factors such as dietary habits, drug treatments, and lifestyle, all of which are known to have an impact on microbiota composition. 36
The relative abundance of the genera Polaribacter and Capnocytophaga was significantly higher in obese versus nonobese patients with CRC in the present study. Polaribacter has been detected in the small intestine of mice, 37 but to date, no reports have shown the association between this genus and obesity or CRC. The genus Capnocytophaga is the oral microbiota. The increased abundance of Capnocytophaga is reported to be associated in obese patients who have periodontal disease, 38 in individuals with diabetes, 39 and in hyperglycemic individuals. 39 Diabetic or hyperglycemic individuals are known to have an increased risk of CRC development. These findings imply the important roles of the oral microbiota Capnocytophaga in the development of obesity‐related CRC, similar to Fusobacterium, which is also one of the periodontal pathogens known to be associated with type 2 diabetes or hyperglycemia 40 and CRC development. 24 , 25 , 34
In the present study, the relative abundance of the genus Enterococcus was reduced in obese patients with CRC and was positively correlated with LDL concentrations, particularly in obese patients with CRC. The most abundant species in the genus Enterococcus was E. faecalis. Administration of heat‐treated E. faecalis FK‐23 (FK‐23) inhibits body weight gain in high‐fat diet (HFD)‐induced mice. 41 In general, patients with CRC, especially with advanced disease, tended to reduce their body weight. Reduced E. faecalis in obese patients with CRC may have an inhibitory effect on their body weight loss. Furthermore, FK‐23 supplementation in mice could attenuate HFD‐induced liver steatosis by upregulating gene expression involved in lipid metabolism, such as acetyl‐CoA carboxylase and fatty acid synthase. 41 The E. faecalis strain ATCC19433 may exert cholesterol‐lowering effects on hypercholesterolemic mice by promoting ATP‐binding cassette subfamily G member 5/8‐mediated cholesterol transportation. 42 Aligned with these findings, E. faecalis seems to have a serum cholesterol‐lowing effect. However, abundance of the genus Enterococcus was positively correlated with serum LDL concentrations in patients with CRC, particularly in the obese patients in the present study. The potential effect of E. faecalis on cholesterol metabolism may differ for patients with CRC.
Several studies demonstrated the association between E. faecalis and CRC development. In a human study, E. faecalis was significantly increased in patients with CRC compared with healthy individuals. 35 , 43 The E. faecalis strain OG1RF could produce an extracellular free radical that promotes chromosomal instability associated with CRC development in intestinal epithelial cell lines and a rat model of intestinal colonization. 44 M1 macrophages, which are polarized by the E. faecalis strain OG1RFSS, activate Wnt/β‐catenin signaling in murine primary colon epithelial cells and enhance the expression of transcription factors involved in induced pluripotency and CRC stem cell markers. 45 These findings might promote the role of E. faecalis in the carcinogenesis of CRC. In contrast, it has been reported that the heat‐killed E. faecalis strain EC‐12 suppresses intestinal polyp development in Apc Min/+ mice, partially due to attenuated β‐catenin signaling. 46 Furthermore, the heat‐killed E. faecalis strain KH2 may attenuate NLRP3 inflammasome activation in macrophages and ameliorate the severity of dextran sulfate sodium‐induced colitis and the development of colitis‐associated CRC in mice. 47 These conflicting roles of E. faecalis in CRC development may be attributed to the different isolated and investigated strains. 48 , 49 Given that obesity leads to increased risk for CRC, the authors of the present study speculated that reduced E. faecalis in obese patients with CRC could be a promoting factor for the development of obesity‐related CRC.
No significant differences were noted in the relative abundance of E. faecalis among the CRC stages in the present study. In either stage, the relative abundance of E. faecalis tended to be lower in obese versus nonobese patients with CRC. Reduced E. faecalis was widely observed from the early to the advanced stages of disease. Reduction of E. faecalis thus seems to be associated with CRC promotion rather than the results of CRC progression.
Age may have an impact on the abundance of E. faecalis. In fact, the patients with CRC were significantly older than the controls. However, no significant correlation was noted between the relative abundance of E. faecalis and age for either patients with CRC or controls. In the patients with CRC, the relative abundance of E. faecalis tended to be lower in obese versus nonobese patients despite age. Therefore, age did not appear to be major potential confounding factor affecting the link between the abundance of E. faecalis and obesity‐related CRC development.
In the present study, patients with CRC were older and had a lower proportion of obesity and lower BMI than controls. However, abnormalities in glucose metabolism and high insulin resistance were found. Aging and reduced food intake due to CRC development may have altered the weight of patients with CRC, particularly in the advanced stage. Nevertheless, glucose intolerance and insulin resistance were observed in patients with CRC, which is consistent with previous reports 4 , 5 , 6 , 7 , 8 , 9 , 10 , 11 , 12 that metabolic abnormalities associated with obesity are independent risk factors of CRC development. In addition, TC and LDL levels were lower in patients with CRC than in controls, which may be attributable to the higher proportion of patients with hyperlipidemia in the CRC group. In the present study, we evaluated microbiome features of patients with obesity‐related CRC by comparing obese and nonobese individuals in the control and CRC groups, respectively. Although age and CRC stage had no significant effect on microbiome features, the replication of these alterations must be confirmed in a well‐controlled study using a population with weight changes and treatment details.
This study has several important limitations. First, observations were restricted to the limited number of participants, which may affect the conclusions. Various factors, such as food, exercise habits, race, lifestyle, physical activity, environment, drug, and bowel habits, impact the composition of the gut microbiome. In the present study, we could not perform a comparison by adjusting for all these factors. Future studies with larger populations and adjustments to various environments are required to verify these results. Second, this observational study could not evaluate the causal relationship. Microbiome alterations observed in the present study may have a combined effect on both CRC development and obesity. However, the relative abundance of Enterococcus was significantly lower in obese patients with CRC, although no significant difference in its relative abundance was found between the obese and nonobese participants in the control group. Moreover, the relative abundance of E. faecalis was significantly lower in obese patients with CRC. Therefore, we believe that this microbiome alteration may be a specific finding in patients with obesity‐related CRC rather than in those with CRC or obesity alone. Obesity is a well‐known risk factor of CRC development but does not always lead to CRC, and some specific findings might lead to obesity‐related CRC development. Thus, we believe that E. faecalis may play a role in its pathogenesis. Further investigations must clarify the mechanisms underlying the association between the microbiome alteration, especially E. faecalis, and the development of obesity‐associated CRC. In addition to the analysis of the composition of gut microbiome, examinations of the functional characteristics such as metabolites and meta‐transcriptomes would help in the understanding of the mechanisms.
In conclusion, this study described several compositional alterations, including the genera Enterococcus, Polaribacter, and Capnocytophaga, in obese patients with CRC in whom E. faecalis was reduced. Thus, E. faecalis may be an active participant in the development of obesity‐related CRC. Understanding how the gut microbiome communicates the link between obesity and colorectal carcinogenesis may provide an opportunity to better describe the mechanistic insights into obesity‐related colorectal carcinogenesis and to identify potential significant implications for therapeutic strategies for CRC.
Supporting information
Appendix S1. Supplementary methods.
Figure S1. Comparison of the relative abundance of Enterococcus faecalis among disease stages of patients with CRC.
Figure S2. Correlation between patient age and the relative abundance of Enterococcus faecalis.
Table S1. Selected characteristics of patients with colorectal cancer and controls.
Table S2. Comparison of characteristics between nOb and Ob both in the controls and the patients with CRC.
Table S3. Comparison of alpha diversity.
Table S4. Relative abundance of the phyla in all participants.
Table S5. Relative abundance of the genera in all participants.
Table S6. Z‐scores of the genera in patients with colorectal cancer and controls.
Table S7. Comparison of the relative abundance of the genera between obese and non‐obese patients with colorectal cancer.
Table S8. Spearman correlation coefficients of metabolic markers and the relative abundance of the genus Enterococcus, Capnocytophaga, or Polaribacter.
Acknowledgments
We thank all subjects who participated in this study. This work was supported in part by the Japan Society for the Promotion of Science KAKENHI Grant Numbers JP17K15920 (to Yu Sasaki), JP17K15922 (to Makoto Yagi), JP19K17389 (to Takashi Kon), JP20K16239 (to Makoto Yagi), and JP20K08350 (to Yu Sasaki) and by the Japan Mutual Aid Association of Public‐School Teachers Grant‐in‐Aid for Occupational Research on Healthcare and Medical Treatment (to Shoichi Nishise).
Declaration of conflict of interest: None
Author contribution: Masakuni Shoji, Yu Sasaki, and Yasuhiko Abe contributed to the study concept and design. Masakuni Shoji, Yu Sasaki, Shoichi Nishise, Takao Yaoita, Makoto Yagi, Naoko Mizumoto, Takashi Kon, Yusuke Onozato, Takayuki Sakai, Matsuki Umehara, and Minami Ito collected the clinical samples and information. Masakuni Shoji, Ayumi Koseki, Ryoko Murakami, Yuki Miyano, and Hidenori Sato performed 16S rRNA metagenome experiments. Masakuni Shoji, Yu Sasaki, Ryoko Murakami, Yuki Miyano, and Hidenori Sato. performed bioinformatics analyses. Masakuni Shoji and Yu Sasaki wrote the manuscript. Yasuhiko Abe and Yoshiyuki Ueno supervised the study.
References
- 1. Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2018; 68: 394–424. [DOI] [PubMed] [Google Scholar]
- 2. Ferlay J, Colombet M, Soerjomataram I et al. Estimating the global cancer incidence and mortality in 2018: GLOBOCAN sources and methods. Int. J. Cancer. 2019; 144: 1941–53. [DOI] [PubMed] [Google Scholar]
- 3. Sung H, Siegel RL, Torre LA et al. Global patterns in excess body weight and the associated cancer burden. CA Cancer J. Clin. 2019;69: 88–112. [DOI] [PubMed] [Google Scholar]
- 4. Bardou M, Barkun AN, Martel M. Obesity and colorectal cancer. Gut. 2013; 62: 933–47. [DOI] [PubMed] [Google Scholar]
- 5. Sato T, Takeda H, Sasaki Y, Kawata S. Increased homeostasis model assessment‐insulin resistance is a risk factor for colorectal adenoma in Japanese males. Tohoku J. Exp. Med. 2011; 223: 297–303. [DOI] [PubMed] [Google Scholar]
- 6. Otake S, Takeda H, Suzuki Y et al. Association of visceral fat accumulation and plasma adiponectin with colorectal adenoma: evidence for participation of insulin resistance. Clin. Cancer Res. 2005; 11: 3642–6. [DOI] [PubMed] [Google Scholar]
- 7. Otake S, Takeda H, Fujishima S et al. Decreased levels of plasma adiponectin associated with increased risk of colorectal cancer. World J. Gastroenterol. 2010; 16: 1252–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Yagi M, Sasaki Y, Abe Y et al. Association between high levels of circulating chemerin and colorectal adenoma in men. Digestion. 2020; 101: 571–8. [DOI] [PubMed] [Google Scholar]
- 9. Sasaki Y, Takeda H, Sato T et al. Increased levels of serum glucose‐dependent insulinotropic polypeptide as a novel risk factor for human colorectal adenoma. Metabolism. 2011; 60: 1253–8. [DOI] [PubMed] [Google Scholar]
- 10. Sasaki Y, Abe Y, Takeda H et al. Impaired secretion of glucagon‐like peptide 1 in patient with colorectal adenoma after an oral glucose load. Digestion. 2018; 97: 324–32. [DOI] [PubMed] [Google Scholar]
- 11. Sasaki Y, Takeda H, Sato T et al. Serum interleukin‐6, insulin, and HOMA‐IR in male individuals with colorectal adenoma. Clin. Cancer Res. 2012; 18: 392–9. [DOI] [PubMed] [Google Scholar]
- 12. Yaoita T, Sasaki Y, Yokozawa J et al. Treatment with anti‐interleukin‐6 receptor antibody ameliorates intestinal polyposis in Apc(Min/+) mice under high‐fat diet conditions. Tohoku J. Exp. Med. 2015; 235: 127–34. [DOI] [PubMed] [Google Scholar]
- 13. Song M, Chan AT, Sun J. Influence of the gut microbiome, diet, and environment on risk of colorectal cancer. Gastroenterology. 2020; 158: 322–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kim B, Choi H‐N, Yim J‐E. Effect of diet on the gut microbiota associated with obesity. J. Obes. Metab. Syndr. 2019; 28: 216–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Turnbaugh PJ, Hamady M, Yatsunenko T et al. A core gut microbiome in obese and lean twins. Nature. 2009; 457: 480–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Ley RE, Turnbaugh PJ, Klein S, Gordon JI. Microbial ecology: human gut microbes associated with obesity. Nature. 2006; 444: 1022–3. [DOI] [PubMed] [Google Scholar]
- 17. Turnbaugh PJ, Ley RE, Mahowald MA, Magrini V, Mardis ER, Gordon JI. An obesity‐associated gut microbiome with increased capacity for energy harvest. Nature. 2006; 444: 1027–31. [DOI] [PubMed] [Google Scholar]
- 18. Vrieze A, Van Nood E, Holleman F et al. Transfer of intestinal microbiota from lean donors increases insulin sensitivity in individuals with metabolic syndrome. Gastroenterology. 2012; 143: 913–916.e7. [DOI] [PubMed] [Google Scholar]
- 19. Plovier H, Everard A, Druart C et al. A purified membrane protein from Akkermansia muciniphila or the pasteurized bacterium improves metabolism in obese and diabetic mice. Nat. Med. 2017; 23: 107–13. [DOI] [PubMed] [Google Scholar]
- 20. Takagi T, Naito Y, Inoue R et al. Differences in gut microbiota associated with age, sex, and stool consistency in healthy Japanese subjects. J. Gastroenterol. 2019; 54: 53–63. [DOI] [PubMed] [Google Scholar]
- 21. Renehan AG, Zwahlen M, Egger M. Adiposity and cancer risk: new mechanistic insights from epidemiology. Nat. Rev. Cancer. 2015; 15: 484–98. [DOI] [PubMed] [Google Scholar]
- 22. Dekker E, Tanis PJ, Vleugels JLA, Kasi PM, Wallace MB. Colorectal cancer. Lancet. 2019; 394: 1467–80. [DOI] [PubMed] [Google Scholar]
- 23. Flanagan L, Schmid J, Ebert M et al. Fusobacterium nucleatum associates with stages of colorectal neoplasia development, colorectal cancer and disease outcome. Eur. J. Clin. Microbiol. Infect. Dis. 2014; 33: 1381–90. [DOI] [PubMed] [Google Scholar]
- 24. Mima K, Nishihara R, Qian ZR et al. Fusobacterium nucleatum in colorectal carcinoma tissue and patient prognosis. Gut. 2016; 65: 1973–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kostic AD, Chun E, Robertson L et al. Fusobacterium nucleatum potentiates intestinal tumorigenesis and modulates the tumor‐immune microenvironment. Cell Host Microbe. 2013; 14: 207–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Mima K, Sukawa Y, Nishihara R et al. Fusobacterium nucleatum and T cells in colorectal carcinoma. JAMA Oncol. 2015; 1: 653–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Gagnière J, Raisch J, Veziant J et al. Gut microbiota imbalance and colorectal cancer. World J. Gastroenterol. 2016; 22: 501–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Thomas AM, Manghi P, Asnicar F et al. Metagenomic analysis of colorectal cancer datasets identifies cross‐cohort microbial diagnostic signatures and a link with choline degradation. Nat. Med. 2019; 25: 667–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Bouter KE, van Raalte DH, Groen AK, Nieuwdorp M. Role of the gut microbiome in the pathogenesis of obesity and obesity‐related metabolic dysfunction. Gastroenterology. 2017; 152: 1671–8. [DOI] [PubMed] [Google Scholar]
- 30. Wang T, Cai G, Qiu Y et al. Structural segregation of gut microbiota between colorectal cancer patients and healthy volunteers. ISME J. 2012; 6: 320–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Ahn J, Sinha R, Pei Z et al. Human gut microbiome and risk for colorectal cancer. J. Natl. Cancer Inst. 2013; 105: 1907–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Wu N, Yang X, Zhang R et al. Dysbiosis signature of fecal microbiota in colorectal cancer patients. Microb. Ecol. 2013; 66: 462–70. [DOI] [PubMed] [Google Scholar]
- 33. Chen W, Liu F, Ling Z, Tong X, Xiang C. Human intestinal lumen and mucosa‐associated microbiota in patients with colorectal cancer. PLoS One. 2012; 7: e39743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Feng Q, Liang S, Jia H et al. Gut microbiome development along the colorectal adenoma‐carcinoma sequence. Nat. Commun. 2015; 6: 6528. [DOI] [PubMed] [Google Scholar]
- 35. Balamurugan R, Rajendiran E, George S, Samuel GV, Ramakrishna BS. Real‐time polymerase chain reaction quantification of specific butyrate‐producing bacteria, Desulfovibrio and Enterococcus faecalis in the feces of patients with colorectal cancer. J. Gastroenterol. Hepatol. 2008; 23: 1298–303. [DOI] [PubMed] [Google Scholar]
- 36. Cani PD. Human gut microbiome: hopes, threats and promises. Gut. 2018; 67: 1716–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Onishi JC, Campbell S, Moreau M et al. Bacterial communities in the small intestine respond differently to those in the caecum and colon in mice fed low‐ and high‐fat diets. Microbiology. 2017; 163: 1189–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Silva‐Boghossian CM, Cesário PC, Leão ATT, Colombo APV. Subgingival microbial profile of obese women with periodontal disease. J. Periodontol. 2018; 89: 186–94. [DOI] [PubMed] [Google Scholar]
- 39. Graves DT, Corrêa JD, Silva TA. The oral microbiota is modified by systemic diseases. J. Dent. Res. 2019; 98: 148–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Gurung M, Li Z, You H et al. Role of gut microbiota in type 2 diabetes pathophysiology. EBioMedicine. 2020; 51: 102590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Kondoh M, Shimada T, Fukada K et al. Beneficial effects of heat‐treated Enterococcus faecalis FK‐23 on high‐fat diet‐induced hepatic steatosis in mice. Br. J. Nutr. 2014; 112: 868–75. [DOI] [PubMed] [Google Scholar]
- 42. Zhu Y, Li T, Din AU, Hassan A, Wang Y, Wang G. Beneficial effects of Enterococcus faecalis in hypercholesterolemic mice on cholesterol transportation and gut microbiota. Appl. Microbiol. Biotechnol. 2019; 103: 3181–91. [DOI] [PubMed] [Google Scholar]
- 43. Geravand M, Fallah P, Yaghoobi MH et al. Investigation of Enterococcus faecalis population in patients with polyp and colorectal cancer in comparison of healthy individuals. Arq. Gastroenterol. 2019; 56: 141–5. [DOI] [PubMed] [Google Scholar]
- 44. Huycke MM, Abrams V, Moore DR. Enterococcus faecalis produces extracellular superoxide and hydrogen peroxide that damages colonic epithelial cell DNA. Carcinogenesis. 2002; 23: 529–36. [DOI] [PubMed] [Google Scholar]
- 45. Wang X, Yang Y, Huycke MM. Commensal‐infected macrophages induce dedifferentiation and reprogramming of epithelial cells during colorectal carcinogenesis. Oncotarget. 2017; 8: 102176–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Miyamoto S, Komiya M, Fujii G et al. Preventive effects of heat‐killed Enterococcus faecalis strain EC‐12 on mouse intestinal tumor development. Int. J. Mol. Sci. 2017; 18: 826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Chung I‐C, OuYang C‐N, Yuan S‐N et al. Pretreatment with a heat‐killed probiotic modulates the NLRP3 inflammasome and attenuates colitis‐associated colorectal cancer in mice. Nutrients. 2019; 11: 516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. de Almeida CV, Taddei A, Amedei A. The controversial role of Enterococcus faecalis in colorectal cancer. Therap. Adv. Gastroenterol. 2018; 11: 1756284818783606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. De Almeida C, Lulli M, di Pilato V et al. Differential responses of colorectal cancer cell lines to Enterococcus faecalis' strains isolated from healthy donors and colorectal cancer patients. J. Clin. Med. 2019; 8: 388. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix S1. Supplementary methods.
Figure S1. Comparison of the relative abundance of Enterococcus faecalis among disease stages of patients with CRC.
Figure S2. Correlation between patient age and the relative abundance of Enterococcus faecalis.
Table S1. Selected characteristics of patients with colorectal cancer and controls.
Table S2. Comparison of characteristics between nOb and Ob both in the controls and the patients with CRC.
Table S3. Comparison of alpha diversity.
Table S4. Relative abundance of the phyla in all participants.
Table S5. Relative abundance of the genera in all participants.
Table S6. Z‐scores of the genera in patients with colorectal cancer and controls.
Table S7. Comparison of the relative abundance of the genera between obese and non‐obese patients with colorectal cancer.
Table S8. Spearman correlation coefficients of metabolic markers and the relative abundance of the genus Enterococcus, Capnocytophaga, or Polaribacter.


