Abstract
Polycyclic aromatic hydrocarbons (PAHs) remediation has received considerable attention due to their significant health concern and environmental pollution. However, PAHs contaminated sites also contain indigenous microbes that can potentially degrade naphthalene. Therefore, this study aimed to isolate, characterise and optimise process parameters for efficient naphthalene degradation. A total of 50 naphthalene degrading bacteria were isolated from Alang-Sosiya ship breaking yard, Bhavnagar, Gujarat and screened for their naphthalene degrading capacity. The selected isolate, Pseudomonas sp. strain SA3 was found to degrade 98.74 ± 0.00% naphthalene at a concentration of 500 ppm after 96 h. Further, optimisation of environmental parameters using one factor at a time approach using different inoculum sizes (v/v), pH, salinity, temperature, carbon and nitrogen source greatly accelerated the degradation process attaining 98.6 ± 0.46% naphthalene degradation after 72 h. The optimised parameters for maximum naphthalene degradation were pH 8, 0.1% peptone as nitrogen source, 8% salinity and 1% (v/v) inoculum size.
Keywords: Naphthalene, Biodegradation, Polycyclic aromatic hydrocarbons (PAHs), Alang-Sosiya ship breaking yard, Pseudomonas sp.
Naphthalene; Biodegradation; Polycyclic aromatic hydrocarbons (PAHs); Alang-Sosiya ship breaking yard; Pseudomonas sp.
1. Introduction
Pollution is one of the leading nuisances in our times, hence particular interest has been generated in the remediation strategies for polycyclic aromatic hydrocarbons (PAHs) to ameliorate their adverse effects. PAHs are an omnipresent set of organic complexes present in air, water, soil and marine sediments containing two or more fused aromatic rings that are recalcitrant remaining in the environment for longer periods (Ofori et al., 2020) The US Environmental Protection Agency (EPA) has marked 16 PAHs as priority pollutants (Ahmadipour et al., 2020). PAHs discharge enters through the water bodies into the oceans thus making the marine ecosystem, the main reservoir of hydrocarbons contaminants causing detrimental effect to marine organisms and human through the food web (Behera et al., 2018). Thus, there is a dire need to remediate these hazardous PAHs by utilising a suitable disposal strategy (Ofori et al., 2020).
Naphthalene is categorised as the first member of PAHs consisting of two fused aromatic rings, which is generally used as a model compound to study PAHs bioremediation (Peters et al., 1999; Hedlund et al., 1999). Jia and Batterman (2010) reported naphthalene exposure to be a cause of hyperplasia and metaplasia in the respiratory and olfactory epithelium respectively. It is found to contaminate the potable water and induces cataractogenic activity in test animals (Samanta et al., 2002). Naphthalene also hinders mitochondrial respiration (Falahatpisheh et al., 2001).
Adeola and Forbes (2020) have discussed technologies like solvent extraction, phytoremediation, chemical oxidation, photocatalytic degradation, electrokinetic, thermal treatment, microbial degradation and integrated remediation technologies. These methods come along with limitations such as high costs, production of toxic by-products and the genesis of a newer pollutant that may affect the environment. Amidst them, the microbial degradation process can be considered as the most effective and eco-friendly means of PAHs degradation where, the PAH molecules are mineralised into carbon dioxide, water, mineral salts and gases with the aid of a single bacterium or microbial consortia (Tirkey et al., 2021). The use of bioremediation technology not only transforms the hazardous molecules into a lesser toxic form but, also re-establish the contaminated area to its natural state.
There are several bacterial species reported for naphthalene degradation including Rabani et al. (2020) have isolated Bacillus licheniformis and Bacillus sonorensis from petroleum contaminated sites of Gwalior, India having the ability to degrade naphthalene. The common genera degrading naphthalene are Pseudomonas, Marinobacter, Mycobacterium, Vibrio and Sphingomonas (Abarian et al., 2018). Abarian et al. (2018) isolated a naphthalene degrading bacteria Sphingobacterium multivorum, from a coal tar mine, Zarand, Iran. Alquati et al. (2005) isolated Rhodococcus, Arthrobacter, Nocardia and Pseudomonas sp. as naphthalene degrading genera from petroleum contaminated soil. Hassanshahian and Boroujeni (2016) isolated and identified naphthalene degrading bacteria as Idiomarina, Halomonas, Marinobacter, Oceanicola, Shewanella, Salegentibacter and Thalassospira from marine sediments and water of the Persian Gulf.
One of the world's largest ship breaking yard is situated at Alang-Sosiya (Latitude: 21.423987°N, Longitude: 72.210799°E) Bhavnagar district of Gujarat, India (Patel et al., 2014). Dismantling of heavy ships round the year generates a massive amount of hazardous petroleum products, PAHs and heavy metals that continuously gets augmented into the ocean and coastal sediments, posing a huge threat to the marine flora and fauna (Barua et al., 2018; Eronat et al., 2019). The contaminants are also found to be harmful to the worker's health who are exposed to this environment (Hossain and Islam, 2006). Coastal areas polluted with such contaminants needs constant clean up. The Alang-Sosiya ship breaking yard (ASSBY) through regular scrapping of ships generates pollutants like petroleum hydrocarbons and heavy metals that contaminate the coastal sediments and water in Alang-Sosiya (Patel et al., 2012b). Therefore ASSBY was chosen to isolate bacteria having the efficiency to degrade naphthalene. The culture conditions of the selected isolate were further optimised using various process parameters like (inoculum size (v/v), pH, salinity, temperature, carbon and nitrogen source) to obtain efficient naphthalene degradation.
2. Materials and methods
2.1. Chemicals and reagents
Naphthalene (99%) was purchased from Sigma-Aldrich, Germany. Bushnell Haas (BH) medium and Zobell marine (ZM) medium were purchased from Hi-Media (Mumbai, India). Acetone and n-hexane were of HPLC grade procured from Sigma-Aldrich and Qualikems respectively. Chemicals such as sodium chloride (NaCl), triphenyl tetrazolium chloride (TTC), yeast extract, glycerol and peptone used in the study were of analytical grade.
2.2. Sampling site
The soil and water samples were collected from ASSBY and their physicochemical parameters like pH, salinity and electrical conductivity were analysed in the laboratory with the aid of a multi-parameter probe (Thermo Fischer, USA).
2.3. Enrichment and isolation of naphthalene consuming bacteria
Soil and water sample were processed through enrichment culture technique to screen potential naphthalene degrading bacteria. In brief, soil (1 g) and water (1 mL) sample enriched with 500 ppm naphthalene was added in a 150 mL Erlenmeyer flasks containing 100 mL BH liquid medium. This was further incubated for 168 h in an incubator shaker (Scigenics Biotech Orbitek) at 120 rpm and 37 °C. According to a study conducted by UNESCO (2004), the mean highest temperature of ASSBY is 34.2 °C. Therefore, 37 °C was selected for the enrichment experiments as mentioned in Patel et al. (2012b). An aliquot of 100 μL enriched sample was then spread on BH agar plates followed by naphthalene spray and incubated at 37 °C for 168 h in dark.
2.4. Bacterial viability using TTC dye
TTC dye was used for screening of potential bacteria that consume naphthalene as a sole carbon source. For the viability test, 500 ppm of naphthalene dissolved in acetone was added to glass test tubes. The acetone was allowed to evaporate completely and 10 mL BH liquid medium was added followed by inoculation of 1% (v/v) inoculum. The tubes without naphthalene served as control. After the addition of TTC dye (0.5% v/v), the tubes were incubated at 120 rpm and 37 °C for 168 h. The selection of the isolates was based on pink or red colour development (Riss et al., 2016; Berridge et al., 2005).
2.5. Identification of the potential isolates
2.5.1. Gram's test and biochemical characteristics of potential isolates
Gram's test and biochemical characteristics of the potential isolates were done using the HiMedia K001 kit and HiMedia KB009 kit respectively, according to the manufacturer's instructions.
2.5.2. Molecular identification
Genomic DNA of potential isolates was extracted according to the protocol mentioned in Wilson (2001). PCR amplification of bacterial 16S rRNA gene was performed using 50 μL reaction mixture of 10x PCR buffer, containing 200 mM MgCl2, 200 μM of dNTPs, 1.25U Taq Polymerase, 0.5 μM of each 27F (5′- AGAGTTTGATCCTGGCTCAG-3′) and 1492R (5′- GGTTACCTTGTTACGACTT-3′) primers and 50 ng of template DNA. The initial temperature in the PCR reaction was at 95 °C for 2 min, followed by 30 cycles of denaturation temperature at 95 °C for 30 s, annealing temperature at 55 °C for 30 s, elongation at 72 °C for 45 s, and followed by a final extension step of 72 °C for 10 min in a PCR machine. PCR amplified products were checked on 1% agarose gel electrophoresis. The amplicons were purified and sequenced bidirectionally. The obtained 16S rRNA sequence were aligned using the BLAST program on the National Center for Biotechnology Information (NCBI) website. The phylogenetic tree was constructed using Neighbor-Joining method sequences based on Kimura 2-parameter model (Kimura, 1980). The evolutionary relationships between identified strains sequences and nearest neighbours were made using software MEGA X.
2.6. Biodegradation experiment
2.6.1. Preparation of inoculum
Cultures grown in ZM broth after achieving OD600 nm -1.0 were centrifuged at 5000 rpm for 10 min. The biomass pellet was washed twice and resuspended in sterilised 0.9% NaCl solution. Further, 1% (v/v) inoculum of this suspension was used in the biodegradation experiment.
2.6.2. Spiking of naphthalene
Naphthalene at a concentration of 500 ppm was dissolved in acetone and 0.5 mL was added in a 150 mL sterile flask. The acetone was allowed to evaporate, leaving the naphthalene inside the flasks. Thereafter, 50 mL of sterile BH liquid medium and 1% (v/v) of inoculum were added to the flask. The un-inoculated flasks containing BH liquid medium and naphthalene served as control. All the flasks were kept in an incubator shaker at 37 °C, 120 rpm for 168 h (Janbandhu and Fulekar, 2011).
2.6.3. Extraction and quantification of residual naphthalene
For extraction of residual naphthalene, 10 mL of n-hexane was added into the flasks and were kept in the incubator shaker for 1 h. The extract was then centrifuged at 8000 rpm for 10 min under room temperature (Patel et al., 2012b). The solvent phase was collected and analysed through Gas Chromatography-Mass Spectrophotometry (GC-MS). The quantification of residual naphthalene concentration was done using a QP-8040 series GC equipped with a Flame Ionization Detector (FID) and SH-Rxi™-5MS column of 30m, i.d. 0.25mm and 0.25μm film thickness. The initial temperature of the column was 40 °C which increased to 300 °C, held for 15 min. Detector and injector temperatures were 250 °C. The concentration of the residual PAH was calculated by area percentage of naphthalene peak in GC at retention time similar to the standard naphthalene. The degradation percentage was calculated using the following formula given by (Li et al., 2008).
| (1) |
where, MT is the final PAHs concentration in each treatment and MI is the initial PAHs concentration in the medium.
2.7. Selection of potential isolate for further biodegradation study
Based on the maximum degradation percentage, the potential isolate was selected for further degradation studies. Samples were collected at every 24 h intervals to analyse the growth of potential isolate through colony forming unit (CFU mL−1) along with naphthalene degradation.
2.8. Effect of environmental parameters on naphthalene degradation
2.8.1. Effect of inoculum size (v/v) on naphthalene degradation
The inoculum size used in the previously mentioned biodegradation experiment with SA3 and SB5 was 1% (v/v). To access the effect of increased inoculum size on naphthalene degradation, three inoculum sizes i.e. 1%, 3% and 5% (v/v) were inoculated in 150 mL flasks containing 50 mL of BH liquid medium amended with 500 ppm of naphthalene at pH 7.0. All the flasks were kept in an incubator shaker at 120 rpm for 96 h at 37 °C. The uninoculated flasks containing naphthalene and BH liquid medium served as the control for all the environmental factors involved in the optimisation experiment. Samples were withdrawn every 24 h for residual naphthalene extraction and quantified using GC-MS.
2.8.2. Effect of different pH on naphthalene degradation
BH liquid having pH value such as 6.0, 7.0, 8.0, 9.0 and 10.0 was adjusted using 0.1 M HCl or NaOH to evaluate the effect of pH on naphthalene degradation. The 150 mL flasks containing 50 mL of BH liquid medium amended with 500 ppm of naphthalene were inoculated with 1% (v/v) inoculum. All the flasks were kept in an incubator shaker at 120 rpm for 96 h at 37 °C.
2.8.3. Effect of salinity on naphthalene degradation
Since the bacteria was isolated from coastal habitat, the effect of saline concentrations such as; 2%, 4%, 6%, and 8% NaCl (w/v) on naphthalene degradation were studied. The flasks containing 50 mL of BH liquid medium adjusted to pH-8.0, amended with 500 ppm of naphthalene were inoculated with 1% (v/v) inoculum. All the flasks were kept in an incubator shaker at 120 rpm for 72 h at 37 °C.
2.8.4. Effect of temperature on naphthalene degradation
Three different temperatures as 25 °C, 30 °C and 37 °C were set to study their impact on naphthalene degradation. The flasks containing 50 mL of BH liquid medium, supplemented with 8% NaCl (w/v), and 500 ppm naphthalene at pH 8.0 was inoculated with 1% (v/v) inoculum. All the flasks were kept in an incubator shaker at 120 rpm for 72 h.
2.8.5. Effect of carbon and nitrogen source on naphthalene degradation
Since naphthalene is a source of only carbon and no other nutrients in it. Therefore 0.1% of glycerol as carbon, peptone and yeast extract as nitrogen source were amended in 50 mL BH liquid medium having 8% (w/v) NaCl. The naphthalene concentration provided was 500 ppm at pH 8.0 and inoculated with 1% (v/v) inoculum. All the flasks were kept in an incubator shaker at 120 rpm for 72 h at 37 °C.
2.9. Statistical analysis
All the experiments were performed in triplicates. The values represented were the average mean ± standard deviation (SD) of the three replicates (n = 3). All statistical significance comparisons between indicated groups were performed using a one-way ANOVA with Fisher's post-test using Infostat software (Version 2016) with p < 0.05.
3. Results and discussion
3.1. Isolation and viability test of naphthalene utilising bacteria
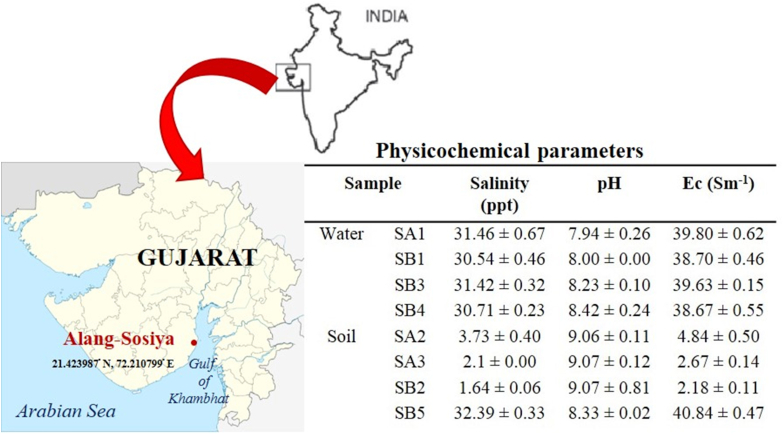
The physicochemical parameters of the collected water and soil sample is presented in Figure 1. The enrichment technique resulted in the isolation and purification of 50 isolates capable of naphthalene utilisation (Table 1). Out of the 50 potential isolate, 15 best isolates were screened through TTC dye which is used as a redox indicator for microbial respiration. Two isolate SA3 100 (3) and SB5 10−2 (2) coded as SA3 and SB5 were able to reduce colourless TTC dye to pink 1,3,5-triphenyl formazan (TPF), were screened for further naphthalene degradation studies.
Figure 1.
Sampling site and physicochemical parameters of soil and water.
Table 1.
Bacterial viability test of 50 isolates.
| S. No. | Bacterial isolates | Colour development |
|---|---|---|
| 1 | SB4 10−2 (3) | - |
| 2 | SB5 10−2 (3) | - |
| 3 | SB3 10−2 (3) | ++ |
| 4 | SB4 10−4 (4) | - |
| 5 | SA3 100 (3) | ++++ |
| 6 | SA3 10−1 (2) | - |
| 7 | SB4 10−2 (6) | - |
| 8 | SB3 10−2 (4) | ++ |
| 9 | SA10−1 (6) | + |
| 10 | SA2 10−2 (1) | - |
| 11 | SA3 10−1(5) | - |
| 12 | SA2 10−1 (2) | - |
| 13 | SA2 10−2(4) | - |
| 14 | SA2 | - |
| 15 | SA1 | - |
| 16 | SB1 100 (1) | +++ |
| 17 | SA2 10−1 (1) | - |
| 18 | SA1 10−3 (3) | + |
| 19 | SB1 10−1 (3) | - |
| 20 | SA2 10−2(3) | - |
| 21 | SB110−1 (1) | - |
| 22 | SB1 10−1(2) | - |
| 23 | SB1 10−2(2) | - |
| 24 | SB110−2 (1) | - |
| 25 | SB3 10−5(2) | + |
| 26 | SA2 10−2 (2) | - |
| 27 | SB1 10−2(4) | +++ |
| 28 | SB4 10−2 (1) | - |
| 29 | SB4 10−3 (2) | - |
| 30 | SA3 10−1(3) | ++ |
| 31 | SB410−5 | - |
| 32 | SA3 10−2(3) | - |
| 33 | SB3 10 −3 (1) | - |
| 34 | SA3 10−2 (2) | - |
| 35 | SB410−4 | - |
| 36 | SA3 10−4 | - |
| 37 | SB5 10−2 (2) | ++++ |
| 38 | SA3 | - |
| 39 | SA3 10−1 (1) | ++ |
| 40 | SA2 10-3(7) | ++ |
| 41 | SA2 10-3 (8) | - |
| 42 | SB3 10-5 4) | - |
| 43 | SA3 10-1(3) | ++ |
| 44 | SA3 10-2(4) | - |
| 45 | SA3 10−1 (2) | +++ |
| 46 | SB5 10−5 (1) | - |
| 47 | SA2 10−3 (4) | - |
| 48 | SB3 10-3 (2) | - |
| 49 | SB5 10-6(2) | - |
| 50 | SA3 10-1(6) | + |
Notes: ++++ denotes high colour intensity, +++ denotes medium colour intensity, ++ denotes low colour intensity, + denotes poor colour intensity and – denotes no colour development.
3.2. Gram's test and biochemical characteristics of potential isolates
Gram's test revealed that both SA3 and SB5 are Gram negative and rod-shaped bacteria. The biochemical characterisation of isolates showed that SA3 was able to utilise inulin, sodium gluconate, glycerol, salicin, rhamose, cellobiose and melezitose whereas isolate SB5 was capable to utilise dextrose, galactose, melibiose, L-arabinose, mannose, D-arabinose, citrate, malonate and sorbose (Table 2).
Table 2.
Biochemical characteristics of potential isolates SA3 and SB5.
| Sugar utilisation | SA3 | SB5 |
|---|---|---|
| Lactose | - | - |
| Xylose | - | ± |
| Maltose | - | - |
| Fructose | - | - |
| Dextrose | - | + |
| Galactose | - | + |
| Raffinose | - | - |
| Trehalose | - | - |
| Melibiose | - | + |
| Sucrose | - | - |
| L-Arabinose | - | + |
| Mannose | - | + |
| Rhamose | + | - |
| Cellobiose | + | - |
| Melezitose | + | - |
| α-M-D-mannoside | - | - |
| Xylitol | - | - |
| ONPG | - | - |
| Esculin | - | ± |
| D-Arabinose | - | + |
| Citrate | - | + |
| Malonate | - | + |
| Sorbose | - | - |
| Inulin | + | - |
| Sodium gluconate | + | - |
| Glycerol | + | - |
| Salicin | + | - |
| Dulcitol | - | - |
| Inositol | - | - |
| Sorbitol | - | - |
| Mannitol | - | - |
| Adonitol | - | - |
| Arabitol | - | - |
| Erythritol | - | - |
| α-M-D-gluconate | - | - |
3.3. Molecular identification of potential isolates
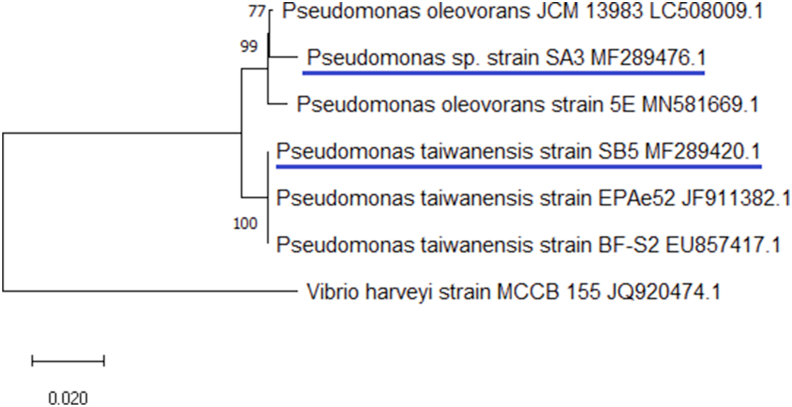
16S rRNA gene sequencing for SA3 and SB5 exhibited 99.22% maximum sequence identity with Pseudomonas oleovorans, and 100% similarity with Pseudomonas taiwanensis respectively. The generated sequences of Pseudomonas sp. strain SA3 and Pseudomonas taiwanensis strain SB5 were deposited in the NCBI Gene Bank with accession numbers MF289476.1 and MF289420.1 respectively and phylogenetic tree was constructed (Figure 2). The evolutionary history was inferred using the Neighbor-Joining method (Saitou and Nei, 1987). The bootstrap consensus tree inferred from 1500 replicates is taken to represent the evolutionary history of the taxa analysed. Branches corresponding to partitions reproduced in less than 50% bootstrap replicates are collapsed. The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test (1500 replicates) are shown next to the branches (Felsenstein, 1985). The evolutionary distances were computed using the Kimura 2-parameter method (Kimura, 1980) and are in the units of the number of base substitutions per site. The results showed that both the strains belonged to the genus Pseudomonas.
Figure 2.
Phylogenetic tree of potential isolates SA3 and SB5. This analysis involved 7 nucleotide sequences. All positions containing gaps and missing data were eliminated (complete deletion option). There were a total of 1281 positions in the final dataset. Evolutionary analyses were conducted in MEGA X (Kumar et al., 2018).
3.4. Biodegradation of naphthalene
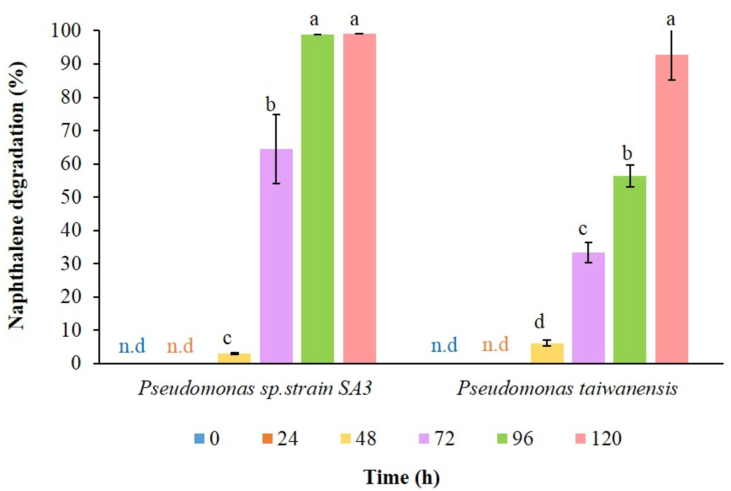
To study the ability to degrade naphthalene by Pseudomonas sp. strain SA3 and P. taiwanensis strain SB5, biodegradation experiments were carried out. The GC-MS results revealed that Pseudomonas sp. strain SA3 and P. taiwanensis strain SB5 degraded 98.74 ± 0.00% and 56.33 ± 3.30% of the 500 ppm naphthalene respectively after 96 h when incubated at 37 °C and 120 rpm (Table 3). Since, a longer incubation time was needed for P. taiwanensis strain SB5 i.e. 120 h to achieve 92.81 ± 7.62% naphthalene degradation and Pseudomonas sp. strain SA3 showed 1.7-fold higher degradation than P. taiwanenesis strain SB5 after 96 h (Figure 3), thus Pseudomonas sp. strain SA3 was selected for further optimisation studies using a one-factor-at-a-time strategy.
Table 3.
Biodegradation of naphthalene by Pseudomonas sp. strain SA3 and Pseudomonas taiwanensis strain SB5.
| Time (h) | Naphthalene degradation (%) |
|
|---|---|---|
| Pseudomonas sp. strain SA3 | Pseudomonas taiwanensis strain SB5 | |
| 0 | 0.00 ± 0.00 | 0.00 ± 0.00 |
| 24 | 0.00 ± 0.00 | 0.00 ± 0.00 |
| 48 | 3.00 ± 0.16 | 6.17 ± 0.84 |
| 72 | 64.44 ± 10.45 | 33.33 ± 3.06 |
| 96 | 98.74 ± 0.00 | 56.33 ± 3.30 |
| 120 | 99.02 ± 0.00 | 92.81 ± 7.62 |
| 144 | 99.56 ± 0.00 | 98.24 ± 0.00 |
Figure 3.
Biodegradation of naphthalene by Pseudomonas sp. strain SA3 and Pseudomonas taiwanensis strain SB5. The values are presented as mean values ±SD, n = 3.Values followed by different alphabets within the treatment are significantly different at p < 0.05 using Fisher's LSD. (n.d: not detectable).
Comparative naphthalene degradation (%) by the selected Pseudomonas sp. strain SA3 with few other reported naphthalene degrading bacteria is presented in Table 4
Table 4.
Comparison of naphthalene degradation (%) by Pseudomonas sp. strain SA3 with few other reported bacterial species.
| Concentration (ppm) | Degradation (%) | Duration (h) | Bacterial species | References |
|---|---|---|---|---|
| 6 | 97.00 | 72 | Pseudomonas mendocina | Barman et al. (2017) |
| 50 | >99.1 | 96 | Bacillus fusiformis (BFN) strain | Lin et al. (2010) |
| 500 | 97.00 | 120 | Halothermophilic bacterial consortium (Ochrobactrum halosaudia strain AJH1, Ochrobactrum halosaudia strain AJH2 and Pseudomonas aeruginosa strain AJH3) | Pugazhendi et al. (2017) |
| 200 | 89.94 | 360 | Shewanella algae strain N1 | Hassanshahian and Boroujeni (2016) |
| 200 | 59.00 | 168 | Sphingobacterium multivorum AHB38N. | Abarian et al. (2018) |
| 100 | 73.00 | 168 | Bacillus licheniformis | Rabani et al. (2020) |
| 200 | 48.00 | 168 | Bacillus licheniformis | Rabani et al. (2020) |
| 100 | 52.00 | 168 | Bacillus sonorensis | Rabani et al. (2020) |
| 200 | 29.00 | 168 | Bacillus sonorensis | Rabani et al. (2020) |
| 500∗ | 98.74 | 96 | Pseudomonas sp. strain SA3 |
The selected strain in this study
3.5. Selection of potential isolate for further biodegradation study
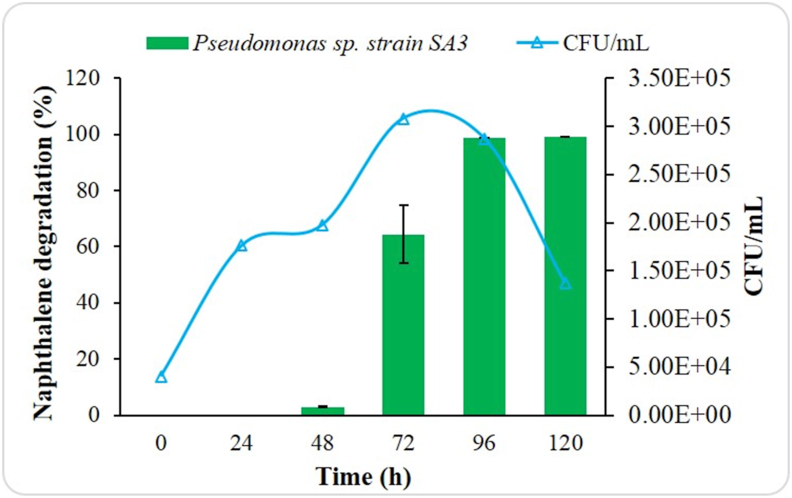
The growth profile (CFU mL−1) of Pseudomonas sp. strain SA3 and simultaneous naphthalene degradation % by the bacteria (initial naphthalene concentration 500 ppm) for 120 h is shown in Figure 4. The result revealed that Pseudomonas sp. strain SA3 was able to achieve 98.74 ± 0.00% naphthalene degradation having 2.8 × 104 CFU mL−1 after 96 h of incubation at 37 °C. Additionally, the degradation % showed a 1.5-fold increment from 72 h to 96 h Abarian et al. (2018) reported the highest growth of Sphingobacterium multivorum strain at 96 h, followed by the stationary phase. The studied strain achieved maximum growth within 72 h, whereas previously reported Sphingobacterium multivorum strain was comparatively slower reaching the maximum growth at 96 h.
Figure 4.
Naphthalene degradation and growth pattern of Pseudomonas sp. strain SA3.
3.6. Effect of environmental parameters on naphthalene degradation
3.6.1. Effect of inoculum size (v/v) on naphthalene degradation
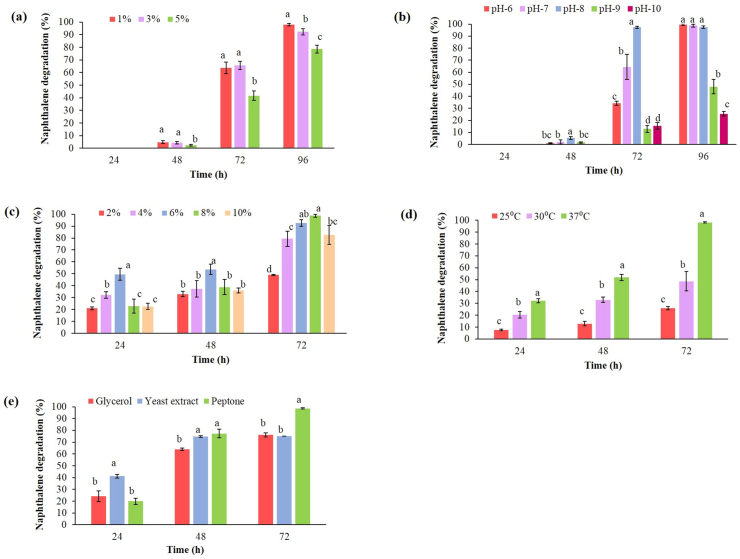
Maximum naphthalene degradation (98 ± 0.00%) was observed at 1% (v/v) inoculum size after 96 h (Figure 5a). Naphthalene degradation using 1% (v/v) inoculum size was significantly higher than 3% (v/v) inoculum size after 96 h and 2-, 1.5- and 1.2-folds higher as compared to 5% (v/v) inoculum size after 48 h, 72 h and 96 h respectively. Moreover, inoculum sizes higher than 1% (v/v) decelerated naphthalene degradation. Therefore, this study suggests that smaller inoculum density (1%v/v) is constructive for maximum xenobiotic degradation which may be owing to its longer lag phase of active bacterial cells capable of rapidly degrading naphthalene which corroborated with Lin et al. (2010). In a similar study, Patel et al. (2012a) reported inoculum of strain DMVP2 exceeding beyond 4% (v/v) led to a sharp decline in phenanthrene degradation. Ghevariya et al. (2011) studied the effect of different inoculum size (102–1010 cells mL−1) and reported more than 60% chrysene degradation with 106 cells mL−1 of inoculum concentration.
Figure 5.
Effect of environmental parameters on naphthalene degradation. (a) Effect of inoculum size (v/v) on naphthalene degradation. (b) Effect of pH on naphthalene degradation. (c) Effect of salinity on naphthalene degradation. (d) Effect of temperature on naphthalene degradation. (e) Effect of carbon and nitrogen source on naphthalene degradation. The values are presented as mean values ± SD, n = 3.Values followed by different alphabets within the treatment are significantly different at p < 0.05 using Fisher's LSD.
3.6.2. Effect of pH on naphthalene degradation
pH is an essential environmental parameter that affects activities like cell membrane transport and catalytic reaction balance. Therefore, it should be taken into consideration for improving process control of the bioremediation system (Al-Hawash et al., 2018). Since the pH of water and soil samples were found to be alkaline (Figure 1), the study range was set from pH 6.0 to pH 10.0. Pseudomonas sp. strain SA3 attained 34.31 ± 1.82%, 64.44 ± 10.45%, 97.38 ± 0.94%, 12.96 ± 2.81% and 15.44 ± 2.66% naphthalene degradation at pH 6.0, pH 7.0, pH 8.0, pH 9.0 and pH 10.0 respectively after 72 h (Figure 5b). Maximum naphthalene degradation (97.38 ± 0.94%) was achieved after 72 h in culture medium having pH 8.0. Therefore, pH 8.0 was selected for further optimisation studies due to its maximum degradation efficiency. According to Dibble and Bartha (1979), marine water is at basic pH, while soil generally has acidic pH. Therefore, often the optimum pH in biodegradation processes are 7.4–7.8. Further, Patel et al. (2012b) reported 100% naphthalene degradation at alkaline pH 8.0–10.0 and Wang et al. (2019) corroborated favourable pH range 6.0–8.0, for the growth of microorganisms for biodegradation. In another study, pH 8.0 was found to be favourable for phenanthrene degradation (Zhao et al., 2009). Al-Hawash et al. (2018) reported many heterotrophic bacteria like to grow in neutral to alkaline conditions.
3.6.3. Effect of salinity on naphthalene degradation
The effect of salinity on naphthalene degradation is depicted in Figure 5c. For the studied strain, up to 48.96 ± 0.52%, 79.31 ± 6.38%, 92.57 ± 2.80%, 98.70 ± 1.20% and 82.61 ± 7.83% naphthalene degradation was attained for 2%, 4%, 6%, 8% and 10% NaCl (w/v) concentration respectively after 72 h. The degradation rate was increased with an increase in salinity to 8% NaCl (w/v) concentration. Barman et al. (2017) conducted a salinity study to check the effect on PAH degradation by Pseudomonas mendocina where they reported a decline in degradation beyond 1.17% salinity. Pugazhendi et al. (2017) have studied the effect of salinity ranging from 4% - 30% on low and high molecular weight PAHs biodegradation where they found a halothermophilic bacterial consortium degraded 97 ± 2.2% of 500 ppm naphthalene after 5 days in 4% NaCl concentration. A halophilic consortium CY-1 at 3% salinity was able to completely degrade phenanthrene after 5 days (Wang et al., 2018). Ashok et al. (1995) reported four bacterial strains including Pseudomonas that could grow under 7.5% (w/v) NaCl condition on 0.1% naphthalene. Somee et al. (2018) also used 7% NaCl (w/v) for the enrichment of diesel degrading consortium whereas in our study, Pseudomonas sp. strain SA3 was not only able to tolerate 8% NaCl (w/v) but also achieved maximum naphthalene degradation (98.70 ± 1.20%) in 72 h, thus making it a significant finding. It was observed that the salinity beyond 8% NaCl (w/v) decelerate the degradation. This shows that high salinity reduces the microbial metabolic rate which could be a reason for a decline in degradation (Shafieiyoun et al., 2020). The degradation rate of naphthalene was greatly influenced by high salinity. Wang et al. (2020) also reported that microbial plasmolysis occurs under high salt concentration which impeded their growth.
3.6.4. Effect of temperature on naphthalene degradation
Temperature affects the physical and chemical nature, composition of microbial community and their metabolism rate during hydrocarbon biodegradation (Al-Hawash et al., 2018). Optimum temperature is an important environmental parameter that support microbial growth during bioaugmentation (Kuppusamy et al., 2016). Naphthalene degradation at 25 °C, 30 °C and 37 °C were found to be 26.05 ± 1.35%, 48.61 ± 8.16% and 98.13 ± 0.55% after 72 h (Figure 5d). In our study, the increase in temperature from 25 °C to 37 °C elicited naphthalene degradation from 26.05 ± 1.35% to 98.13 ± 0.55% after 72 h. This may be due to an increase in PAHs solubility and bioavailability with an increase in temperature (Ghosal et al., 2016). Therefore, approximately 3.8-folds increment in naphthalene degradation was achieved when the incubation temperature increased from 25 °C to 37 °C. Patel et al. (2013) have used temperature 37 °C–40 °C and found maximum phenanthrene degradation (80%) by consortium ASP at 37 °C within 120 h. They also reported a decrease in degradation when the temperature was increased to 45 °C. Leahy and Colwell (1990) has reported temperature in the range of 30 °C–40 °C intensified hydrocarbon metabolism. Our study further agrees with Patel et al. (2012b) where 37 °C was observed to be the optimum temperature for complete naphthalene degradation at a concentration of 1000 ppm by bacterial consortium DV-AL after 36 h.
3.6.5. Effect of carbon and nitrogen source on naphthalene degradation
Nutrients have been considered as an important factor influencing PAH biodegradation. During microbial metabolism of hydrocarbon, existing nutrients are used up, causing a reduction in the population of active PAH degrading microbes (Kuppusamy et al., 2016; Lin et al., 2010). Thus, carbon and nitrogen sources were supplemented to the BH liquid medium to assist Pseudomonas sp. strain SA3 in naphthalene degradation (Figure 5e). The addition of peptone as a nitrogen source (0.1% w/v) revealed maximum naphthalene degradation efficiency (98.58 ± 0.46%) followed by glycerol and yeast extract which were 76.24 ± 1.82% and 75.07 ± 0.08% respectively. Thus, the naphthalene degradation was significantly affected by the supplementation of peptone as a nitrogen source. Patel et al. (2012b) have studied the effect of various co-substrate out of which four co-substrates, peptone, yeast extract, sodium succinate and sodium acetate were found to enhance the naphthalene degradation. They reported 100% naphthalene degradation in the presence of peptone by consortium DV-AL. In this study, addition of 0.1% peptone resulted in 74.72 ± 0.78% naphthalene degradation after 48 h which rose to 98.58 ± 0.46% after 72 h Patel et al. (2013) reported 75% and 88% phenanthrene degradation at a concentration of 300 ppm in the presence of glucose and yeast extract respectively by consortium ASP. Varjani (2017) reviewed that hydrocarbons do not contain nutrients such as nitrogen and phosphorus, and limits the availability of these nutrients and reduces the microbial degradation rate of hydrocarbon. Therefore, supplementation of nutrients such as carbon, nitrogen and phosphorus is essential to promote bioremediation in PAHs polluted environment.
4. Conclusion
Two potential isolates from ASSBY, identified as Pseudomonas sp. strain SA3 and Pseudomonas taiwanensis strain SB5 were able to degrade up to 98.74 ± 0.00% and 92.81 ± 7.62. % naphthalene after 96 h and 120 h respectively. Due to a shorter incubation time and maximum naphthalene degradation Pseudomonas sp. strain SA3 was selected for further optimisation studies. Culture medium supplemented with 0.1% peptone, pH 8.0 and 8% salinity was found to be optimum for maximum naphthalene degradation (98.58 ± 0.46%) by Pseudomonas sp. strain SA3 after 72 h. Thus, the strain can further be utilised for other PAH degradation as a potential candidate for PAH field trials bioremediation.
Declarations
Author contribution statement
Sushma Rani Tirkey: Conceived and designed the experiments; Performed the experiments; Analyzed and interpreted the data; Wrote the paper.
Shristi Ram: Conceived and designed the experiments; Analyzed and interpreted the data.
Sandhya Mishra: Contributed reagents, materials, analysis tools or data.
Funding statement
Sushma Rani Tirkey was supported by University Grants Commission - National Fellowship for Higher Education [F1-17.1/2016-17/NFST-2015-17-ST-JHA-2164]. Shristi Ram was supported by Council of Scientific and Industrial Research-HRDG [31/28(223)2K-17/EMR-I]
Data availability statement
Data will be made available on request.
Declaration of interests statement
The authors declare no conflict of interest.
Additional information
No additional information is available for this paper.
Acknowledgements
All the authors are grateful to the Director, Dr. S. Kannan, CSIR – CSMCRI. Further, SRT and SR would like to acknowledge Academy of Scientific and Innovative Research (AcSIR) for Ph.D. enrolment. All the authors wish to acknowledge Dr. Madhusree Mitra for kind suggestions.
References
- Abarian M., Hassanshahian M., Badoei-Dalfard A. Isolation, screening, and characterization of naphthalene-degrading bacteria from Zarand Mine, Iran. Polycycl. Aromat. Comp. 2018;38(5):410–419. [Google Scholar]
- Adeola A.O., Forbes P.B. Advances in water treatment technologies for removal of polycyclic aromatic hydrocarbons: existing concepts, emerging trends, and future prospects. Water Environ. Res. 2020 doi: 10.1002/wer.1420. [DOI] [PubMed] [Google Scholar]
- Ahmadipour F., Sari A.E., Bahramifar N. Characterization and health risk assessment of particulate-bound polycyclic aromatic hydrocarbons in Tehran, Iran. Air Qual Atmos Health. 2020:1–8. [Google Scholar]
- Al-Hawash A.B., Dragh M.A., Li S., Alhujaily A., Abbood H.A., Zhang X., Ma F. Principles of microbial degradation of petroleum hydrocarbons in the environment. Egypt. J. Aquat. Res. 2018;44(2):71–76. [Google Scholar]
- Alquati C., Papacchini M., Riccardi C., Spicaglia S., Bestetti G. Diversity of naphthalene-degrading bacteria from a petroleum contaminated soil. Ann. Microbiol. 2005;55(4):237. [Google Scholar]
- Ashok B., Saxena S., Musarrat J. Isolation and characterization of four polycyclic aromatic degrading bacteria from soil near an oil refinery. Lett. Appl. Microbiol. 1995;21:246–248. doi: 10.1111/j.1472-765x.1995.tb01052.x. [DOI] [PubMed] [Google Scholar]
- Barman S.R., Banerjee P., Mukhopadhayay A., Das P. Biodegradation of acenapthene and naphthalene by Pseudomonas mendocina: process optimization, and toxicity evaluation. J. Environ. Chem. Eng. 2017;5:4803–4812. [Google Scholar]
- Barua S., Rahman I.M., Hossain M.M., Begum Z.A., Alam I., Sawai H., Maki T., Hasegawa H. Environmental hazards associated with open-beach breaking of end-of-life ships: a review. Environ. Sci. Pollut. Res. 2018;25(31):30880–30893. doi: 10.1007/s11356-018-3159-8. [DOI] [PubMed] [Google Scholar]
- Behera B.K., Das A., Sarkar D.J., Weerathunge P., Parida P.K., Das B.K., Thavamani P., Ramanathan R., Bansal V. Polycyclic Aromatic Hydrocarbons (PAHs) in inland aquatic ecosystems: perils and remedies through biosensors and bioremediation. Environ. Pollut. 2018;241:212–233. doi: 10.1016/j.envpol.2018.05.016. [DOI] [PubMed] [Google Scholar]
- Berridge M.V., Herst P.M., Tan A.S. Tetrazolium dyes as tools in cell biology: new insights into their cellular reduction. Biotechnol. Annu. Rev. 2005;11:127–152. doi: 10.1016/S1387-2656(05)11004-7. [DOI] [PubMed] [Google Scholar]
- Dibble J., Bartha R. Effect of environmental parameters on the biodegradation of oil sludge. Appl. Environ. Microbiol. 1979;37:729–739. doi: 10.1128/aem.37.4.729-739.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eronat A.H., Bengil F., Neşer G. Shipping and ship recycling related oil pollution detection in Çandarlı Bay (Turkey) using satellite monitoring. Ocean Eng. 2019;187:106157. [Google Scholar]
- Falahatpisheh M., Donnelly K., Ramos K. Antagonistic interactions among nephrotoxic polycyclic aromatic hydrocarbons. J. Toxicol. Environ. Health Part A. 2001;62:543–560. doi: 10.1080/152873901300007833. [DOI] [PubMed] [Google Scholar]
- Felsenstein J. Confidence limits on phylogenies: an approach using the bootstrap. Evolution. 1985;39:783–791. doi: 10.1111/j.1558-5646.1985.tb00420.x. [DOI] [PubMed] [Google Scholar]
- Ghevariya C.M., Bhatt J.K., Dave B.P. Enhanced chrysene degradation by halotolerant Achromobacter xylosoxidans using response surface methodology. Bioresour. Technol. 2011;102(20):9668–9674. doi: 10.1016/j.biortech.2011.07.069. [DOI] [PubMed] [Google Scholar]
- Ghosal D., Ghosh S., Dutta T.K., Ahn Y. Current state of knowledge in microbial degradation of polycyclic aromatic hydrocarbons (PAHs): a review. Front. Microbiol. 2016;7:1369. doi: 10.3389/fmicb.2016.01369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hassanshahian M., Boroujeni N.A. Enrichment and identification of naphthalene-degrading bacteria from the Persian Gulf. Mar. Pollut. Bull. 2016;107(1):59–65. doi: 10.1016/j.marpolbul.2016.04.020. [DOI] [PubMed] [Google Scholar]
- Hedlund B.P., Geiselbrecht A.D., Bair T.J., Staley J.T. Polycyclic aromatic hydrocarbon degradation by a new marine bacterium, Neptunomonas naphthovorans gen. nov., sp. nov. Appl. Environ. Microbiol. 1999;65(1):251–259. doi: 10.1128/aem.65.1.251-259.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hossain M.M.M., Islam M.M. Advocacy & Publication Unit, Young Power in Social Action (YPSA).; Chittagong, Bangladesh: 2006. Ship Breaking Activities and its Impact on the Coastal Zone of Chittagong, Bangladesh: towards Sustainable Management. [Google Scholar]
- Janbandhu A., Fulekar M.H. Biodegradation of phenanthrene using adapted microbial consortium isolated from petrochemical contaminated environment. J. Hazard Mater. 2011;187:333–340. doi: 10.1016/j.jhazmat.2011.01.034. [DOI] [PubMed] [Google Scholar]
- Jia C., Batterman S. A critical review of naphthalene sources and exposures relevant to indoor and outdoor air. Int. J. Environ. Res. Publ. Health. 2010;7(7):2903–2939. doi: 10.3390/ijerph7072903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura M. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J. Mol. Evol. 1980;16:111–120. doi: 10.1007/BF01731581. [DOI] [PubMed] [Google Scholar]
- Kumar S., Stecher G., Li M., Knyaz C., Tamura K. Mega X: molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018;35(6):1547–1549. doi: 10.1093/molbev/msy096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuppusamy S., Thavamani P., Megharaj M., Naidu R. Biodegradation of polycyclic aromatic hydrocarbons (PAHs) by novel bacterial consortia tolerant to diverse physical settings–assessments in liquid-and slurry-phase systems. Int. Biodeterior. Biodegrad. 2016;108:149–157. [Google Scholar]
- Leahy J.G., Colwell R.R. Microbial degradation of hydrocarbons in the environment. Microbiol. Mol. Biol. Rev. 1990;54(3):305–315. doi: 10.1128/mr.54.3.305-315.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X., Li P., Lin X., Zhang C., Li Q., Gong Z. Biodegradation of aged polycyclic aromatic hydrocarbons (PAHs) by microbial consortia in soil and slurry phases. J. Hazard Mater. 2008;150:21–26. doi: 10.1016/j.jhazmat.2007.04.040. [DOI] [PubMed] [Google Scholar]
- Lin C., Gan L., Chen Z.L. Biodegradation of naphthalene by strain Bacillus fusiformis (BFN) J. Hazard Mater. 2010;182(1-3):771–777. doi: 10.1016/j.jhazmat.2010.06.101. [DOI] [PubMed] [Google Scholar]
- Ofori S.A., Cobbina S.J., Doke D.A. The occurrence and levels of polycyclic aromatic hydrocarbons (PAHs) in African environments—a systematic review. Environ. Sci. Pollut. Res. 2020;27(26):32389–32431. doi: 10.1007/s11356-020-09428-2. [DOI] [PubMed] [Google Scholar]
- Patel V., Cheturvedula S., Madamwar D. Phenanthrene degradation by Pseudoxanthomonas sp. DMVP2 isolated from hydrocarbon contaminated sediment of Amlakhadi canal, Gujarat, India. J. Hazard Mater. 2012;201:43–51. doi: 10.1016/j.jhazmat.2011.11.002. [DOI] [PubMed] [Google Scholar]
- Patel V., Jain S., Madamwar D. Naphthalene degradation by bacterial consortium (DV-AL) developed from Alang-Sosiya ship breaking yard, Gujarat, India. Bioresour. Technol. 2012;107:122–130. doi: 10.1016/j.biortech.2011.12.056. [DOI] [PubMed] [Google Scholar]
- Patel V., Munot H., Shouche Y.S., Madamwar D. Response of bacterial community structure to seasonal fluctuation and anthropogenic pollution on coastal water of Alang–Sosiya ship breaking yard, Bhavnagar, India. Bioresour. Technol. 2014;161:362–370. doi: 10.1016/j.biortech.2014.03.033. [DOI] [PubMed] [Google Scholar]
- Patel V., Patel J., Madamwar D. Biodegradation of phenanthrene in bioaugmented microcosm by consortium ASP developed from coastal sediment of Alang-Sosiya ship breaking yard. Mar. Pollut. Bull. 2013;74(1):199–207. doi: 10.1016/j.marpolbul.2013.07.001. [DOI] [PubMed] [Google Scholar]
- Peters C.A., Knightes C.D., Brown D.G. Long-term composition dynamics of PAH-containing NAPLs and implications for risk assessment. Environ. Sci. Technol. 1999;33(24):4499–4507. [Google Scholar]
- Pugazhendi A., Qari H., Basahi J.M.A.-B., Godon J.J., Dhavamani J. Role of a halothermophilic bacterial consortium for the biodegradation of PAHs and the treatment of petroleum wastewater at extreme conditions. Int. Biodeterior. Biodegrad. 2017;121:44–54. [Google Scholar]
- Rabani M.S., Sharma R., Singh R., Gupta M.K. Characterization and Identification of naphthalene degrading bacteria isolated from petroleum contaminated Sites and their possible use in bioremediation. Polycycl. Aromat. Comp. 2020:1–12. [Google Scholar]
- Riss T.L., Moravec R.A., Niles A.L., Duellman S., Benink H.A., Worzella T.J., Minor L. Assay Guidance Manual. Eli Lilly & Company and the National Center for Advancing Translational Sciences; 2016. Cell viability assays. [Google Scholar]
- Saitou N., Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987;4(4):406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- Samanta S.K., Singh O.V., Jain R.K. Polycyclic aromatic hydrocarbons: environmental pollution and bioremediation. Trends Biotechnol. 2002;20:243–248. doi: 10.1016/s0167-7799(02)01943-1. [DOI] [PubMed] [Google Scholar]
- Shafieiyoun S., Al-Raoush R.I., Ngueleu S.K., Rezanezhad F., Van Cappellen P. Enhancement of naphthalene degradation by a sequential sulfate injection scenario in a (Semi)-Arid coastal soil: a flow-through reactor experiment. Water Air Soil Pollut. 2020;231(8):1–16. [Google Scholar]
- Somee M.R., Shavandi M., Dastgheib S.M.M., Amoozegar M.A. Bioremediation of oil-based drill cuttings by a halophilic consortium isolated from oil-contaminated saline soil. 3 Biotech. 2018;8(5):229. doi: 10.1007/s13205-018-1261-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tirkey S.R., Ram S., Mishra S. Advanced Oxidation Processes for Effluent Treatment Plants. Elsevier; 2021. Bacteria-mediated remediation: restoring polycyclic aromatic hydrocarbon (PAHs) contaminated marine ecosystems; pp. 119–136. [Google Scholar]
- UNESCO . UNESCO; Paris: 2004. Impacts and Challenges of a Large Coastal industry.Alang-Sosiya Ship-Breaking Yard, Gujarat, India. Coastal Region and Small Island Papers; p. 65. 17. [Google Scholar]
- Varjani S.J. Microbial degradation of petroleum hydrocarbons. Bioresour. Technol. 2017;223:277–286. doi: 10.1016/j.biortech.2016.10.037. [DOI] [PubMed] [Google Scholar]
- Wang C., Huang Y., Zhang Z., Wang H. Salinity effect on the metabolic pathway and microbial function in phenanthrene degradation by a halophilic consortium. Amb. Express. 2018;8:1–13. doi: 10.1186/s13568-018-0594-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H., Zhang H., Zhang K., Qian Y., Yuan X., Ji B., Han W. Membrane fouling mitigation in different biofilm membrane bioreactors with pre-anoxic tanks for treating mariculture wastewater. Sci. Total Environ. 2020;724:138311. doi: 10.1016/j.scitotenv.2020.138311. [DOI] [PubMed] [Google Scholar]
- Wang Z., Wang W., Li Y., Yang Q. Co-metabolic degradation of naphthalene and pyrene by acclimated strain and competitive inhibition kinetics. J. Environ. Sci. Health B. 2019;54(6):505–513. doi: 10.1080/03601234.2019.1586033. [DOI] [PubMed] [Google Scholar]
- Wilson K. Preparation of genomic DNA from bacteria. Curr. Protoc. Mol. Biol. 2001;56:2. doi: 10.1002/0471142727.mb0204s56. 4. 1-2.4. 5. [DOI] [PubMed] [Google Scholar]
- Zhao H.-P., Wu Q.-S., Wang L., Zhao X.-T., Gao H.-W. Degradation of phenanthrene by bacterial strain isolated from soil in oil refinery fields in Shanghai China. J. Hazard Mater. 2009;164:863–869. doi: 10.1016/j.jhazmat.2008.08.098. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data will be made available on request.