Abstract
In vitro cultured plant cells, in particular the tobacco BY-2 cell, have demonstrated their potential to provide a promising bioproduction platform for therapeutic proteins by integrating the merits of whole-plant cultivation systems with those of microbial and mammalian cell cultures. Over the past three decades, substantial progress has been made in improving the plant cell culture system, resulting in a few commercial success cases, such as taliglucerase alfa (Elelyso®), the first FDA-approved recombinant pharmaceutical protein derived from plant cells. However, compared to the major expression hosts (bacteria, yeast, and mammalian cells), plant cells are still largely underutilized, mainly due to low productivity and non-human glycosylation. Modern molecular biology tools, in particular RNAi and the latest genome editing technology CRISPR/Cas9, have been used to modulate the genome of plant cells to create new cell lines that exhibit desired “traits” for producing therapeutic proteins. This review highlights the recent advances in cellular engineering of plant cells towards improved recombinant protein production, including creating cell lines with deficient protease levels or humanized glycosylation, and considers potential development in the future.
Keywords: Plant cell culture, Recombinant proteins, Cellular engineering, RNA interference, Genome editing, Glycoengineering
Introduction
Recombinant pharmaceutical proteins obtained through genetic engineering are increasingly being used in the treatment of many diseases such as hepatitis, anemia, diabetes, cancer, and even infectious diseases such as COVID-19 (by vaccines and neutralizing antibodies). The global therapeutic protein market was valued at about $93.14 billion in 2018 and is expected to grow to $172.87 billion in 2022, with an annual growth rate (CAGR) of 16.7%. To date, more than 130 recombinant pharmaceuticals have been approved by the US Food and Drug Administration (FDA) for clinical use, more than 200 are on the market worldwide, and many more are in the clinical development pipeline (Santos et al. 2016; Pham 2018).
Prokaryotic and eukaryotic expression host systems, including bacteria, yeast, insect and mammalian cells, are widely used for the production of recombinant proteins. However, there are limitations associated with these systems in terms of cost, scalability, safety, and the quality/authenticity of proteins. This makes it difficult to utilize them as a comprehensive expression system for the expression of a wide variety of proteins. Plants as hosts for a eukaryotic expression system have emerged as a promising alternative production platform for therapeutic proteins. “Molecular farming” in plants proposes a superior method as compared to other eukaryotic systems in terms of safety, scalability, and cost (Xu et al. 2016). A plant-based production system has also proven effective in mediating the post-translational processing required for many complex proteins (Gecchele et al. 2015; Schillberg et al. 2019). During the SARS-CoV-2 pandemic, plant molecular farming could potentially provide a rapid and scalable supply of protein antigens (as vaccine candidates), antibodies and other therapeutic proteins (Lico et al. 2020). Unlike other bioproduction systems, plant systems have diverse platforms, ranging from whole plants to in vitro cell and tissue cultures, and using either transient expression via viral or nonviral vectors or stable transformation with transgenes targeted to the nuclear or chloroplast genome (Xu et al. 2016). Each platform has its own strengths and weaknesses, which has been widely discussed in the past (Xu et al. 2012, 2016; Schillberg and Finnern 2021), thus are not elaborated in this review.
While field-cultivated plants share many of these features, plant cell suspension culture provides the additional advantage of growth supported by a low-cost medium and controlled environment. These extra benefits help reduce costs and avoid issues associated with pathogen, pesticide, and herbicide contamination. Thus, cGMP (current Good Manufacturing Practice) can be readily implemented throughout the production pipeline. A seminal breakthrough for the commercialization of the plant cell culture platform was achieved in 2012, when the plant cell-produced therapeutic enzyme, taliglucerase alfa (Elelyso®), was approved by the US FDA as an orphan drug for the treatment of Gaucher’s disease (Tekoah et al. 2015). Despite this breakthrough, there exist technical challenges which limit the widespread commercial applications of this platform. These include low productivity, non-human glycosylation, cell clumping, and complex cell morphology, etc. (Fischer et al. 2013; Fischer and Buyel 2020). Therefore, technical innovations that drive the commercial scalability and sustainability of plant cell culture are necessary for the broader adoption of this platform.
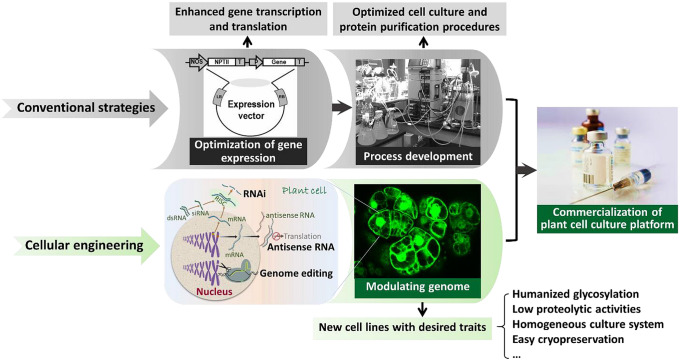
In the past three decades, substantial progress has been made to increase the productivity of plant cell culture systems (Fig. 1). This includes upstream strategies, such as optimizing gene expression constructs and selecting highly productive monoclonal cultures, and downstream strategies, such as optimizing culture conditions (growth medium, bioreactor design and operation) and developing more efficient methods of protein extraction and purification (Xu and Zhang 2014; Santos et al. 2016; Schillberg et al. 2019). On the other hand, cellular engineering by which an intact plant cell genome is modulated to create new cell lines that exhibit desired “traits” has received increasing attention. This has been further promoted by the newly developed CRISPR (Clustered Regularly Interspaced Short Palindromic Repeats)/Cas9 genome editing tool. Like the improvements in bacterial and mammalian cell strains in recent years to increase host’s productivity, cellular engineering of plant cells has the potential to break the current limits of cell line bioproduction to become a commercially competitive bioproduction platform. This review highlights recent advances in cellular engineering of plant cells and considers possible future developments for the improvement of therapeutic protein production.
Fig. 1.
Schematic illustration of strategies used to enhance plant cell culture productivity for bioproduction and commercialization. In addition to the conventional strategies (optimization of gene expression and process development), cellular engineering represents an alternative approach that could potentially revolutionize the use of plant cell culture as a bioproduction platform
Plant cell culture as a bioproduction platform—advantages and challenges
Advantages
Plant cell culture provides a promising alternative bioproduction platform for therapeutic proteins, since it integrates the merits of whole-plant cultivation systems with those of microbial and mammalian cell cultures (Xu et al. 2011). Because plant cells are grown in sterile and controlled environments, cell growth conditions can be precisely controlled, and the culture system is readily adapted to existing bioreactors. This allows for batch-to-batch product consistency and a production process aligned with cGMP (Shih and Doran 2009; Wilson and Roberts 2012). Furthermore, recombinant proteins can be designed to secrete into culture media, making downstream protein purification much more affordable as compared to using whole plants. As such, the decreased regulatory and environmental concerns of plant cell culture bioproduction system make it more appealing to current biopharmaceutical producers than whole plant system (Santos et al. 2016).
The most widely used plant cell line for recombinant protein production is one which was derived from tobacco (Nicotiana tabacum cv. Bright Yellow 2), termed BY-2 cell (Kato et al. 1972). Isolated by Japanese scientists Kato and coworkers in 1972, the BY-2 cell line has many appealing characteristics: fast-growing (doubling time as short as 11 h), robust, and readily able to undergo Agrobacterium-mediated transformation and cell cycle synchronization (Hellwig et al. 2004; Xu et al. 2011). Due to its central role in bioproduction and fundamental research, the BY-2 cell line has been referred to as the “Chinese hamster ovary (CHO)-cell in molecular farming” and the “HeLa cell in the biology of higher plants”. A wide array of functional proteins including antibodies, vaccines, enzymes, growth factors, and cytokines have been successfully expressed in BY-2 cells (Table 1). So far, most cellular engineering studies that have been conducted with the BY-2 cell line aimed to reduce the intracellular proteolytic activities and eliminate the non-human glycosylation (to be discussed in detail below). Besides the BY-2 cell line, other plant cell lines used for bioproduction are those derived from edible crop species, such as rice (Oriza sativa) (Huang et al. 2015), alfalfa (Medicago sativa) (Khoudi et al. 1999), and carrot (Daucus carota) (Park et al. 2020). Particularly, when cultured plant cells are used for oral delivery of biologics, they may be more readily accepted by the general public. Notably, carrot cells have been used by the Israeli company Protalix Biotherapeutics (http://www.protalix.com) to produce taliglucerase alfa (Elelyso®), the first plant cell-made biopharmaceutical approved for the market.
Table 1.
Recombinant proteins produced in tobacco BY-2 cell cultures
| Protein | Expression type | Protein targeting | Protein yield | Reference |
|---|---|---|---|---|
| Hepatitis B surface antigen | Stable | ER | 226 ng/mg TSP | Sojikul et al. (2003) |
| Human monoclonal antibody against hepatitis B virus | Stable | Apoplast | 0.6% TSP | Yano et al. (2004) |
| Human interferon α2b | Stable | Apoplast | 35 mg/L | Xu et al. (2007) |
| Human α -l-iduronidase | Stable | Apoplast | 10 mg/L | Fu et al. (2009) |
| Human monoclonal anti-HIV antibody 2G12 | Stable | Apoplast | 8 mg/L | Holland et al. (2010) |
| Human growth hormone | Stable | Apoplast | 28 mg/L | Xu et al. (2010) |
| Human serum albumin | Stable | Cytosol, Vacuole | 11.88 mg/L | Sun et al. (2011) |
| Human erythropoietin | Stable | ER, Apoplast | N/A | Pires et al. (2012) |
| Human monoclonal anti- vitronectin antibody M12 | Stable | Apoplast | 20–107 mg/L | Kirchhoff et al. (2012), Vasilev et al. (2013), Raven et al. (2015) |
| Human interleukin-10-ELP (elastin‐like polypeptide) | Stable | ER | 3.057% TSP | Kaldis et al. (2013) |
| Green fluorescent protein (GFP) | Stable | Apoplast | 125 mg/L | Zhang et al. (2016a, b) |
| TNFα receptor (TNFR)-Fc fusion | Stable | ER | N/A | Almon et al. 2017), Ilan et al. (2017) |
| DNase I | Stable | Apoplast | N/A | Hanania et al. (2017) |
| GFP-hydrofobin (HFBI) | Stable | Apoplast | 300–1100 mg/L | Reuter et al. (2014, 2016), Hakkinen et al. (2018) |
| Human α1-antitrypsin | Stable | Apoplast | 34.7 mg/L | Zhang et al. (2019) |
| ORF8 from SARS-CoV-2 | Stable (inducible) | Apoplast | 8.8 mg/L | Imamura et al. (2021) |
| GFP | Stable (inducible) | Apoplast | N/A | Sadoch et al. (2021) |
| Human stem cell factor | Stable | Apoplast | 2.5 mg/L | Wang et al. (2021) |
| Viscumin | Transient | Apoplast | 5.0 mg/kg cells | Gengenbach et al. (2019) |
| DsRed (Discosoma sp. red fluorescent protein) | Transient | Apoplast | 70 mg/kg cells | Rademacher et al. (2019) |
| Monoclonal antibody 2G12 | 47 mg/kg cells | |||
| Monoclonal antibody M12 | 175 mg/kg cells | |||
| GFP and DsRed | Transient | Apoplast | ~ 700 mg/kg cells | Poborilova et al. (2020) |
N/A not available, TSP total soluble proteins
Challenges to be overcome by cellular engineering
Despite all these advantages, there are major bottlenecks that must be overcome before the plant cell culture platform can compete with conventional bacterial and mammalian cell systems. These include low production yields, non-human glycosylation, proteolytic degradation, cell aggregation, and culture heterogeneity, as well as difficulty with cryopreservation. Some of them, non-human glycosylation for example, have already been addressed by the cellular engineering strategy.
Low production yields
A major limiting factor for broader applications of the plant cell culture platform is the low yields of recombinant proteins, typically ranging from 0.01 to 10 mg/L (Xu and Zhang 2014). This is partially due to the low copy number of a gene of interest being integrated into the plant cell genome. This is because plant cells lack the “gene amplification” mechanism found in CHO cells which enables the host cells to carry thousands of copies of a gene of interest (Cacciatore et al. 2010). However, recent advances in plant molecular biology have greatly improved the bioproduction capacity of plant cells. An extremely high yield of protein, up to 1.1 g/L hydrofobin (HFBI)-fused green fluorescent protein (GFP), was recently achieved in BY-2 cell culture. This represents the highest titer of recombinant protein ever reported for plant cell culture and a production yield comparable to those generally achieved by yeast and mammalian cell systems (Hakkinen et al. 2018). The mechanism leading to this high rate of protein accumulation remains unknown. It may result from the integration of the GFP-HFBI encoding genes in a transcriptionally active spot of chromosomes (Hakkinen et al. 2018). Nevertheless, this has demonstrated the bioproduction potential of plant cell culture. In fact, plant cells are now moving toward parity with mammalian cells. The productivity of some therapeutic proteins in BY-2 cells, such as the monoclonal antibody M12 whose maximum yield exceeds 100 mg/L, has reached ~ 8 pg/cell/day (Schillberg et al. 2019), which approached the typical rates of 20–40 pg/cell/day for CHO cells (Havenith et al. 2014). This is promising, indicating that it is possible to overcome the low protein productivity by modulating genetic and epigenetic factors in plant cells. So far, there are no reports aiming at increasing protein biosynthesis of plant cells through genome editing technology.
Non-human glycosylation: glycan structure has a major role in the proper functioning of therapeutic proteins, but there is a major difference in the plant and mammalian glycan structure. Glycoproteins produced by plant cells contain N‐linked glycans carrying two plant‐specific residues which are absent from mammalian cells‐derived proteins: β-1,2‐xylose and core α-1,3‐fucose. In contrast, mammalian glycoproteins contain β-1,4-galactose and terminal sialic acid residues that are absent in plants (Gomord et al. 2010; Kallolimath et al. 2016; Strasser 2016) (Fig. 2). The immunogenicity and allergenicity of plant-specific N-glycans have been a key concern in human therapy because human serum contains active antibodies against plant‐specific residues (Gomord et al. 2010). However, recent clinical studies with plant cell-produced proteins, such as taliglucerase alfa, did not indicate that plant sugar residues cause adverse effects in humans (Tekoah et al. 2015; Shaaltiel and Tekoah 2016; Hanania et al. 2017). Nevertheless, extensive research efforts have been made in recent years to modulate the plant-specific glycosylation machinery to humanize glycosylation. These include retaining the recombinant proteins in the ER to produce generic high-mannose glycans, adding inhibitors of enzymes involved in the glycosylation pathway (e.g., kifunesine) directly to the culture media, and engineering the plant cell genome through knocking down or knocking out the enzymes responsible for adding the plant-specific sugar residues (Yin et al. 2011; Bosch et al. 2013; Hanania et al. 2017; Sukenik et al. 2018; Macharoen et al. 2020). On the other hand, exogenous genes encoding β-1,4-galactosyltransferase or polysialyltransferases were introduced into the plant genome to produce more human-like glycans (Paccalet et al. 2007; Castilho and Steinkellner 2012; Kallolimath et al. 2016). In fact, most of the cellular engineering studies reported to date aimed at eliminating plant-specific glycosylation.
Fig. 2.
Schematic illustration of the glycoengineering technologies used on plant cells to humanize the plant N-glycan structures. The complex type of plant glycans is represented here. The knocked-out or knocked-down cell lines can be further engineered (knocked in) with genes encoding the human sialylation pathway to generate plant-derived glycans that carry sialic acids
Cell aggregation and culture heterogeneity
Unlike bacteria and mammalian cells, plant cells tend to grow as aggregates instead of single cells. Aggregation is promoted by extracellular polysaccharides, such as pectin, and occurs when daughter cells fail to separate after cell division (Santos et al. 2016). In addition, the presence of plasmodesmata is beneficial for plant cells growing in aggregates. Due to cell aggregation, generating monoclonal elite transgenic cell lines becomes very challenging. Generally, cell suspension cultures are heterogeneous with varying sizes of cell aggregates and assorted cell morphologies. Cell aggregation and culture heterogeneity could also pose challenges to mixing and mass transfer of oxygen and nutrients when scaling up the cultures in bioreactors (Huang and McDonald 2009). Cellular engineering may provide an essential tool to solve this problem (cell aggregation).
Proteolytic degradation
Post-translational degradation of heterologous proteins by endogenous proteases, either along the secretion pathway or in the culture media, has been regarded as one of the major factors leading to the low protein production in plant cell culture (Pillay et al. 2014; Lallemand et al. 2015; Mandal et al. 2016). Many plant cell expressed proteins, such as plasminogen activators (Schiermeyer et al. 2005), cytokines (Kwon et al. 2003), and monoclonal antibodies (Magy et al. 2014; Hehle et al. 2015) have been shown to undergo proteolytic processing to varying degrees. Several strategies, for example, targeting heterologous proteins to a sub-cellular compartment (e.g., ER), co-expressing proteins with protease inhibitors, or simply altering the pH of the culture were recently applied to reduce proteolytic degradation in plant cell culture (Huang et al. 2009; Jutras et al. 2019). Alternatively, down-regulating the gene expression of certain proteases with RNA interference (RNAi) and antisense RNA technologies could generate plant cell lines with reduced endogenous proteolytic activities, which would increase the accumulation of recombinant proteins (Mandal et al. 2014; Duwadi et al. 2015). While strategies leveraging the genome editing technology to knock out the genes encoding key proteases may be more effective at reducing post-translational protein degradation in plant cells, no research has been reported so far demonstrating its effectiveness.
Cryopreservation and cell banking
Cryopreserving plant cell lines for cell banking is incredibly important for maintaining a consistent supply of well-defined, elite cell lines for bioproduction. Cryopreservation is also necessary to meet cGMP compliance (Schumacher et al. 2015). However, cryopreserving plant cells, usually with liquid nitrogen (− 196 °C) without damaging cells, is much more difficult than cryopreserving animal cells. This is because plant cells contain a large vacuole, housing a substantial amount of cellular water (Schillberg et al. 2019). Submitting these water-filled cells to the dramatically low temperature of liquid nitrogen makes the plant cells prone to injury from expansion. Although cryopreservation protocols have been established to treat the freeze/thawing effects on various plant cell lines (Ogawa et al. 2008; Hakkinen et al. 2018), they are complex and often compromise cell viability. Cellular engineering technologies may provide a solution to this problem by developing a new type of plant cell line containing a more flexible cell wall structure, such as cellulose-free cell wall, which will lead to a great reduction in vacuole and the plant cells size.
In addition to the key challenges discussed above, other important challenges encountered in plant cell culture include somaclonal variation, gene silencing, elite cell line selection, and bioreactor scale-up, etc. These will not be discussed in detail in this review.
Molecular biology tools for cellular engineering
Modern advances in contemporary molecular biology have provided efficient tools for cellular engineering which can potentially help plant cell culture overcome some of the obstacles it currently faces. These include technologies that utilize RNAi or antisense RNA-mediated gene silencing to down regulate target gene expressions and genome editing technologies, particularly CRISPR)/Cas9, to knock out target genes within the plant cell genome.
RNAi and antisense RNA technologies
RNAi is a powerful post-transcriptional gene silencing process which is initiated by a specialized RNase III enzyme, Dicer, that cleaves a long double-stranded RNA (dsRNA) or hairpin-structured RNA (hpRNA) into double-stranded, small (∼20 to 25 bp nucleotides), interfering RNAs (siRNAs) (Vaucheret 2015). The antisense strands of the siRNA duplex are incorporated into an Argonaute (AGO) protein, forming an RNA-induced silencing complex (RISC). The antisense strands then guide the RISC complex to target messenger RNAs (mRNA) in a sequence-specific manner and cleave the mRNA through the action of induced AGO protein, thereby inhibiting protein synthesis. With precise selection of conserved target sequences, the RNAi technology can effectively silence a specific gene or multiple members of a gene family. So far, this RNAi technology has been widely used in plant biotechnology to manage pests and diseases caused by bacteria, fungi, and viruses (Li et al. 2015; Yu et al. 2016) to improve crop yields (Younis et al. 2014) and to generate plants with novel traits (Small 2007). However, this approach suffers from the fact that gene silencing is rarely complete and might not be stable over a long period of time (Mercx et al. 2016).
Compared to RNAi, antisense RNA is a relatively old technology used to regulate gene expression (Oberemok et al. 2018). This technology utilizes a single-stranded RNA to directly bind the coding region of sense RNA, resulting in direct inhibition of translation or mRNA destabilization. With the recombinant DNA method, synthetic genes, encoding antisense RNA molecules, can be introduced into organisms to inhibit the target gene expression.
Genome editing technologies
Recent advances in genome editing technologies mediated by sequence-specific nucleases led to a new era of genome engineering, enabling effective, precise, and rapid modulation of the plant genomes (Wada et al. 2020). In all of these technologies, programmable endonucleases are utilized to induce double strand breaks (DSB) in the target genomic DNA. Mutations in the genome are then generated by the cellular repair system. This is done through either error-prone non-homologous end joining (NHEJ) or homology-directed repair (HDR), which creates insertions, deletions, and substitutions in the genome (El-Mounadi et al. 2020). Three well-established genome editing platforms [zinc finger nucleases (ZFNs), transcription activator-like effector nucleases (TALENs), and CRISPR/Cas9] are currently applied to various organisms, including plants (Bortesi and Fischer 2015). Of which, the CRISPR/Cas9 technology represents the most recent and exciting addition to the genome editing toolbox due to its simplicity, efficiency, specificity, and cost-effectiveness (Wada et al. 2020). This technology is based on the integration of a single guide RNA (sgRNA) that aids the Cas9 endonuclease in creating DSBs in the target genomic DNA, thus generating a null allele (gene disruption). Because of the small size (~ 20 nucleotides) of CRISPR RNA (crRNA) required for precise genome targeting, multiple sgRNAs can be co-constructed with Cas9 on the same vector to achieve multiplex genome editing. However, the application of CRSIPR/Cas9 is associated with off-targeting that introduces unintended mutations at off-target sites within the genome. This off-targeting issue has been demonstrated in a number of reports in plants (Modrzejewski et al. 2020). In order to avoid off-target mutations in plants, strategies include using high-precision CRISPR/Cas nucleases, using CRISPR/Cas nuclease in the form of ribonucleoproteins (RNPs), and carefully selecting target sequences, etc. (Hahn and Nekrasov 2019).
CRISPR/Cas9 has been applied to a growing number of plant species to improve yields and nutritional value, or enhance stress tolerance (El-Mounadi et al. 2020). Nevertheless, only a handful of studies have been reported so far on the applications of this tool to plant suspension cells (Table 2). The successful inactivation of a gene in BY-2 cells was first demonstrated in 2016 by knocking out an exogenous gene encoding the red fluorescent protein (mCherry) that was previously introduced into the BY-2 genome (Mercx et al. 2016). CRISPR/Cas9-mediated targeted-mutagenesis was also achieved in Arabidopsis thaliana and carrot suspension cells by inactivating a reporter protein GFP gene and the flavanone-3-hydroxylase gene, respectively (Klimek-Chodacka et al. 2018; Permyakova et al. 2019).
Table 2.
List of examples in which cellular engineering was performed on plant cells to improve recombinant protein production
| Plant cells | Approach | Target genes | Outcomes | Reference |
|---|---|---|---|---|
| BY-2 | RNAi | Integrated luciferase | Optimization of RNAi protocol for BY-2 cells | Akashi et al. (2004) |
| BY-2 | ZFN | GFP | Targeted integration of GFP gene into BY-2 cells | Cai et al. (2009) |
| BY-2 | RNAi | Cellulose synthase NtCESA1 | Inhibition of cellulose biosynthesis in protoplast | Silva et al. (2010) |
| Rice | RNAi | XylT and FucT | Core α-1,3-fucosylated and/or β-1,2-xylosylated N-glycans on recombinant hGM-CSF significantly reduced | Shin et al. (2011) |
| BY-2 | RNAi | XylT and FucT | Core α-1,3-fucosylated and/or β-1,2-xylosylated N-glycans on soluble proteins significantly reduced | Yin et al. (2011) |
| Rice | RNAi | α-Amylase and Cysteine protease (CysP) | Reduced endogenous α-amylase (94.8%) and CysP (95%); 2.4-fold improvement of recombinant hGM-CSF production | Kim et al. (2013) |
| A. thaliana (Protoplast) | CRISPR/Cas9 | Phytoene Desaturase (PDS3) | A. thaliana PDS3 (Phytoene desaturase) mutation | Li et al. (2013) |
| BY-2 | Anti-sense RNA | Four proteases genes: NtAP, NtCP, NtMMP1, and NtSP | Reduced levels of endogenous protease expression; fourfold increased accumulation of intact antibody 2F5 | Mandal et al. (2014) |
| BY-2 | CRISPR/Cas9 | Integrated red fluorescent protein (RFP) | Site-specific mutation of the RFP gene | Mercx et al. (2016) |
| BY-2 | CRISPR/Cas9 | XylT and FucT | Production of N-linked glycans lacking β-1,2-xylose and/or α-1,3-fucose; recombinant DNase I totally free from any xylose and/or fucose residue | Hanania et al. (2017) |
| Carrot | CRISPR/Cas9 | Flavanone-3-hydroxylase (F3H) | Blockage of the anthocyanin biosynthesis in a purple-colored callus | Klimek-Chodacka et al. (2018) |
| A. thaliana | CRISPR/cas9 | Integrated GFP | Site-specific mutations of the GFP gene and a decrease in GFP transcription | Permyakova et al. (2019) |
| BY-2 | CRISPR/Cas9 | Integrated orange fluorescence protein (pporRFP) | Development of a lipofection-mediated transfection approach for the use of DNA-free Cas9/gRNA RNP for gene editing in plant cells | Liu et al. (2020) |
| BY-2 (protoplasts) | CRISPR/Cas9 | XylT and FucT, T-DNA boundaries | Production of N-linked glycans lacking β-1,2-xylose and/or α-1,3-fucose; heterologous DNA and selectable marker subsequently removed | Sheva et al. (2020) |
| Rice | CRISPR/Cas9 | XylT and FucT | Production of N-linked glycans lacking β-1,2-xylose and α-1,3-fucose; other plant-specific residues of β-1,3-galactose and α-1,4-fucose were still present | Jung et al. (2021) |
Cellular engineering of plant cells
Modulating the plant cell genome towards improved therapeutic protein production is an emerging research field in which many exciting achievements have already been made. Recent utilizations of these modern molecular biology tools for cellular engineering of plant suspension cells are summarized in Table 2. Currently, two groups of genes, including those encoding proteases and glycosyltransferases, are the targets for cellular engineering. Strategically inactivating some of these genes could create new plant cell lines with reduced proteolytic activities and humanized glycosylation.
Creation of plant cell lines with reduced proteolytic activities
In addressing the proteolytic degradation hurdle in plant cell culture, some protease genes have been targeted for inactivation. Plant genomes encode several hundred proteases, which can be distinguished as serine, cysteine, aspartic and metallo-types (four classes) based on the active site residues for catalysis (Pillay et al. 2014; Rawlings et al. 2014, 2016). Given the large number of protease genes and that different products are susceptible to different protease classes, inactivation of certain protease genes in the plant genome for the prevention of product degradation turns out to be quite challenging (Mandal et al. 2016). However, the number of proteases that are active in certain cell types, and specific compartments like the apoplast, is significantly lower (Delannoy et al. 2008). This makes it feasible to inactivate only a small number of protease genes to prevent the degradation of target proteins accumulated in a specific compartment, such as the extracellular space (culture media). This was demonstrated in a recent study, in which four endogenous protease genes (NtAP, NtCP, NtMMP1 and NtSP) encoding for proteases from four catalytic classes (aspartic, cysteine, metallo and serine proteases) of BY-2 cell were simultaneously silenced by the antisense RNA technology. The established new BY-2 cell lines showed substantially reduced proteolytic activity in the culture media (Mandal et al. 2014). Expression of a full-length IgG1(κ) antibody 2F5 with one of the cell lines resulted in a fourfold higher accumulation of the intact antibody heavy chain as compared to the expression of the same antibody in wild-type BY-2 cells (Mandal et al. 2014).
Protease-deficient cell lines were also created for rice cells through the RNAi technology. Expression of recombinant proteins in rice cells driven by the rice α-amylase 3D promoter, which is activated under sucrose-starved conditions, could generate high secreted protein yields up to 247 mg/L of α1-antitrypsin (McDonald et al. 2005). However, this expression system suffers from the accumulation of large amounts of α-amylase (43% of TSP) and proteases in culture media following the induction via sugar depletion. While α-amylase complicates protein purification, secreted proteases, with cysteine proteinase (CysP) being a major class, degrade recombinant proteins. By using the RNAi technology to silence both the α-amylase and CysP genes, a protease-deficient rice cell line was created that showed reduced expression of α-amylase and CysP by 94.8% and 95%, respectively. Expression of a pharmaceutical protein, human granulocyte–macrophage colony-stimulating factor (hGM-CSF) in this cell line reached 288 mg/L, which is 2.45-fold higher than that of transgenic line expressing hGM-CSF only (115 mg/L) (Kim et al. 2013). However, the utility of the antisense RNA and RNAi technologies is limited by the incompleteness of target protein depletion and potential off-target effects (Liang et al. 2016). The CRISPR/Cas9 genome editing tool has been successfully used in plant suspension cells (Mercx et al. 2016; Ren et al. 2016; Hanania et al. 2017). However, the use of this tool for protease gene inactivation in plant cells has yet to be conducted and reported on.
Creation of plant cell lines with humanized glycosylation
The plant-specific residues, β-1,2-xylose and core α-1,3-fucose, are transferred to the developing glycan structure by two resident Golgi enzymes: α-1,3-fucosyltransferase (α-1,3-FucT) and β-1,2-xylosyltransferase (β-1,2-XylT). In order to humanize the plant cell glycosylation, glyco-engineered plant cell lines lacking the machinery to transfer the plant-specific residues could be created by either utilizing RNAi or the CRISPR/Cas9 genome editing technology (Fig. 2).
Glyco-engineering for humanizing glycosylation was first achieved in whole plants. Strategies, including RNAi and random mutagenesis (exposing to ethylmethanesulfonate), were utilized to interfere with the expression of the α-1,3-FucT and β-1,2-XylT genes in different plant species (Cox et al. 2006; Castilho et al. 2011). Glycoengineering of plant suspension cells was then demonstrated in both BY-2 and rice cells by using the RNAi strategy to downregulate the expression of the α-1,3-FucT and β-1,2-XylT genes (Shin et al. 2011; Yin et al. 2011). In the engineered BY-2 cells, the xylosylated and core fucosylated N-glycans isolated from total soluble proteins were significantly reduced, though not completely. In addition, the engineered cell lines were stable, viable, and did not exhibit any obvious phenotypic changes (Yin et al. 2011). Similarly, glyco-engineered rice cells showed normal properties of cell division and proliferation. The recombinant hGM‐CSF that was secreted by established rice cells contained significantly reduced core α-1,3-fucosylated and/or β-1,2-xylosylated glycan structures (Shin et al. 2011).
Alternatively, targeted genome editing strategies were applied to inactivate the α-1,3-FucT and β-1,2-XylT genes in plants. For example, TALEN‐mediated genome editing was used to knock out these glycotransferase genes in N. benthamiana. This generated glycoengineered lines whose N‐glycans lacked β-1,2‐xylose, but exhibited only a 40% reduction in the core α-1,3‐fucose levels as compared to wild‐type plants (Li et al. 2016). Complete knockout of the α-1,3-FucT and β-1,2-XylT genes was achieved in BY-2 cells a year later through utilization of the CRISPR/Cas9 technology (Hanania et al. 2017). The BY-2 cell genome is known to contain two β-1,2-XylT genes and five α-1,3-FucT genes. Thus, a total of 14 loci (seven genes and two alleles per gene) were simultaneously mutated to eliminate the plant-specific glycosylation in BY-2 cells. This also demonstrated the great power of the CRISPR/Cas9 technology for multiplex gene editing in BY-2 cells. The knocked‐out BY-2 cell lines, while exhibiting a typical BY-2 growing rate, did not add the β(1,2)-xylose and/or α(1,3)-fucose to N-linked glycans. Furthermore, recombinant DNaseI expressed in the knocked‐out cell line was totally free from any xylose and fucose residues (Hanania et al. 2017). Thus, these glycoengineered BY-2 cells provide a valuable bioproduction platform for therapeutic proteins with humanized glycosylation. In addition, new glycoengineered rice cells and N. benthamiana plants whose β-1,2-XylT and α-1,3-FucT genes were completely knocked out were recently created through the use of the CRISPR/Cas9 technology (Jansing et al. 2019; Jung et al. 2021).
In addition to eliminating plant-specific sugar moieties through inactivation of certain Xyl and Fuc glycosyltransferase genes, the careful introduction of engineered multigene vectors is also possible. This was demonstrated through the introduction of an engineered multigene vector carrying the human sialylation pathway to enable the controlled generation of protein sialylation in N. benthamiana (Kallolimath et al. 2016). However, there are no reports to date that describe cellular engineering of plant suspension cells for the synthesis of sialic acid structures. Nevertheless, all the aforementioned ellular engineering studies are demonstrative of the exceptional flexibility and capability of the plant cell-based expression platform to engineer and produce complex posttranslational protein modifications.
Future developments
Progress has been made to modulate the plant cell genome for improved therapeutic protein production, particularly the creation of plant cell lines with humanized glycosylation. However, the key technical challenge of the plant cell culture system (low productivity) has yet to be fully addressed. Although many molecular biology approaches, like the optimization of expression vectors (promoters and regulatory regions), have been utilized to achieve considerable increases in expression yields, a tenfold increase in productivity is still required to attain a reasonable profit margin and compete with the yeast and mammalian cell platforms. This leaves much room for yield improvement—through cellular engineering of current plant cell lines. However, clear gene targets for modulation (that could lead to increased productivity) are yet to be discovered in the plant genome. Nevertheless, some other potential aspects of research could be considered seriously. These include down regulating or knocking out genes that are considered nonessential for the propagation of plant cells in vitro, as well as engineering exogenous machinery that will result in higher efficiency transcription and translation of specifically designed gene cassettes.
In contrast to whole plants growing in natural environments which must perform photosynthesis and resist various abiotic and biotic stresses to survive, in vitro cultured plant cells (like BY-2 cells) are supplied with optimized and sterile culture conditions (medium composition, pH, temperature, oxygenation, etc.) to support their rapid heterotrophic propagation. It is not necessary for the plant cells to synthesize some biomolecules in large quantities; for example, the photosynthetic enzyme ribulose-1,5-bisphosphate carboxylase/oxygenase (Rubisco). Inactivation of the genes responsible for the biosynthesis of these biomolecules (through RNAi or genome editing tools) may enable the cultured cells to devote more resources and energy to the synthesis of target proteins (Schillberg et al. 2019). To make this “top-down” engineering strategy work as expected, the genes to be modulated should not be essential to the viability and propagation of the plant cells. Even so, the knocked-out cell lines may still undergo some morphological changes; thus, some adjustments of the culture conditions, particularly the medium composition, may need be made accordingly. On the other hand, engineering exogenous biosynthesis machinery into the plant cell genome may be a more readily achievable strategy to increase protein productivity. Research has been conducted to try and introduce the bacterial T7 expression system into plant systems, including the T7 RNA polymerase gene (in the plant genome) and the T7 promoter (in a gene cassette), but this was done to little avail (McBride et al. 1994; Nguyen et al. 2004).
Alternatively, reducing posttranslational degradation would also contribute to the improved accumulation of target proteins in plant cells. Following the success of creating the protease-deficient BY-2 cell line through the antisense RNA technology, the efficient CRISPR/Cas9 genome editing tool can be applied to completely inactivate some of these protease genes. Because plant cells produce several hundred proteases, knocking out all of these protease genes is certainly impossible. Therefore, it is more practical to create protease-deficient mutants by targeting a specific protein that is accumulated in a specific compartment (the apoplast, for example). This strategy relies on prior knowledge of the specific classes of proteases that are likely lead to the degradation of the target protein. This can be worked out by using protease inhibitors for different classes of proteases to probe the target protein (Mandal et al. 2014) and then merging the experimental results with in silico analyses of the genomic information of plant cells.
In addition to low productivity, other intrinsic properties that halt the use of plant cell culture as a vital bioproduction platform include cell aggregation, presence of large vacuoles, and difficulty in cryopreservation, all of which may be overcome through cellular engineering. These problems can largely be attributed to the distinctive cell wall structure of plant cells, which is absent from animal cells. The cell wall is essential to the survival of the plants growing in their natural habitats, as it provides structure, support, and protection for the cells. However, for undifferentiated plant cells (e.g., BY-2 cells) grown in vitro and in a contained environment, an intact cell wall structure may not be a necessity. Creating cell wall-deficient plant cell lines (e.g., cellulose-free cell lines) through utilization of genome editing tools may prevent cell aggregation and reduce vacuole size, thereby largely overcoming those problems. However, due to gene redundancy and pleiotropic effects in higher plants, creating loss-of-function mutants by simply knocking out target genes may not produce a discernable phenotype. Therefore, concerted efforts in both genome editing experiments and bioinformatics analyses are required to achieve this goal.
Conclusions
In the past thirty years, great progress has been made in plant cell culture technology, and some commercial success have been achieved. However, there are still technical challenges to be overcome before this new bioproduction platform is widely used in commercial applications. The engineering of plant cells through RNAi or genome editing technology is still in early stage of development, however, it shows great potential in dealing with some challenges. Most notably, new glycoengineered cell lines were successfully created to enable the production of glycoproteins devoid of plant-specific glycoepitopes. The next phase of development will be leveraging bioinformatics tools and genome editing technologies to identify the key genes responsible for the traits of interest, and to create next-generation of cell lines that show high productivity while exhibiting desired traits, such as small vacuole, homogeneous cell culture system and easy cryopreservation. These traits are beneficial for bioprocess development and industrial-scale production.
Acknowledgements
This work was supported by the Arkansas IDeA Network of Biomedical Research Excellence—Research Development Grant [Grant No. P20GM103429]; the National Science Foundation [Grant No. 1605564]; and the Arkansas Biosciences Institute, the major research component of the Arkansas Tobacco Settlement Proceeds Act of 2000.
Author contribution statement
UK: conceptualization, writing—original draft preparation. HF: writing—original draft preparation. WG: writing—original draft preparation. CU: writing—reviewing and editing. JX: supervision, conceptualization, writing—reviewing and editing.
Declartions
Conflict of interest
None.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- Akashi H, Miyagishi M, Taira K. RNAi expression vectors in plant cells. Methods Mol Biol. 2004;252:533–543. doi: 10.1385/1-59259-746-7:533. [DOI] [PubMed] [Google Scholar]
- Almon E, Khoury T, Drori A, Gingis-Velitski S, Alon S, Chertkoff R, Mushkat M, Shaaltiel Y, Ilan Y. An oral administration of a recombinant anti-TNF fusion protein is biologically active in the gut promoting regulatory T cells: results of a phase I clinical trial using a novel oral anti-TNFα—based therapy. J Immunol Methods. 2017;446:21–29. doi: 10.1016/j.jim.2017.03.023. [DOI] [PubMed] [Google Scholar]
- Bortesi L, Fischer R. The CRISPR/Cas9 system for plant genome editing and beyond. Biotechnol Adv. 2015;33:41–52. doi: 10.1016/j.biotechadv.2014.12.006. [DOI] [PubMed] [Google Scholar]
- Bosch D, Castilho A, Loos A, Schots A, Steinkellner H. N-glycosylation of plant-produced recombinant proteins. Curr Pharm Des. 2013;19:5503–5512. doi: 10.2174/1381612811319310006. [DOI] [PubMed] [Google Scholar]
- Cacciatore JJ, Chasin LA, Leonard EF. Gene amplification and vector engineering to achieve rapid and high-level therapeutic protein production using the DHFR-based CHO cell selection system. Biotechnol Adv. 2010;28:673–681. doi: 10.1016/j.biotechadv.2010.04.003. [DOI] [PubMed] [Google Scholar]
- Cai CQ, Doyon Y, Ainley WM, Miller JC, Dekelver RC, Moehle EA, Rock JM, Lee YL, Garrison R, Schulenberg L, Blue R, Worden A, Baker L, Faraji F, Zhang L, Holmes MC, Rebar EJ, Collingwood TN, Rubin-Wilson B, Gregory PD, Urnov FD, Petolino JF. Targeted transgene integration in plant cells using designed zinc finger nucleases. Plant Mol Biol. 2009;69:699–709. doi: 10.1007/s11103-008-9449-7. [DOI] [PubMed] [Google Scholar]
- Castilho A, Steinkellner H. Glyco-engineering in plants to produce human-like N-glycan structures. Biotechnol J. 2012;7:1088–1098. doi: 10.1002/biot.201200032. [DOI] [PubMed] [Google Scholar]
- Castilho A, Gattinger P, Grass J, Jez J, Pabst M, Altmann F, Gorfer M, Strasser R, Steinkellner H. N-glycosylation engineering of plants for the biosynthesis of glycoproteins with bisected and branched complex N-glycans. Glycobiology. 2011;21:813–823. doi: 10.1093/glycob/cwr009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox KM, Sterling JD, Regan JT, Gasdaska JR, Frantz KK, Peele CG, Black A, Passmore D, Moldovan-Loomis C, Srinivasan M, Cuison S, Cardarelli PM, Dickey LF. Glycan optimization of a human monoclonal antibody in the aquatic plant Lemna minor. Nat Biotechnol. 2006;24:1591–1597. doi: 10.1038/nbt1260. [DOI] [PubMed] [Google Scholar]
- Delannoy M, Alves G, Vertommen D, Ma J, Boutry M, Navarre C. Identification of peptidases in Nicotiana tabacum leaf intercellular fluid. Proteomics. 2008;8:2285–2298. doi: 10.1002/pmic.200700507. [DOI] [PubMed] [Google Scholar]
- Duwadi K, Chen L, Menassa R, Dhaubhadel S. Identification, characterization and down-regulation of cysteine protease genes in tobacco for use in recombinant protein production. PLoS ONE. 2015;10:e0130556. doi: 10.1371/journal.pone.0130556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Mounadi K, Morales-Floriano ML, Garcia-Ruiz H. Principles, applications, and biosafety of plant genome editing using CRISPR-Cas9. Front Plant Sci. 2020;11:56. doi: 10.3389/fpls.2020.00056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer R, Buyel JF. Molecular farming—the slope of enlightenment. Biotechnol Adv. 2020;40:107519. doi: 10.1016/j.biotechadv.2020.107519. [DOI] [PubMed] [Google Scholar]
- Fischer R, Schillberg S, Buyel JF, Twyman RM. Commercial aspects of pharmaceutical protein production in plants. Curr Pharm Des. 2013;19:5471–5477. doi: 10.2174/1381612811319310002. [DOI] [PubMed] [Google Scholar]
- Fu LH, Miao YS, Lo SW, Seto TC, Sun SSM, Xu ZF, Clemens S, Clarke LA, Kermode AR, Jang L. Production and characterization of soluble human lysosomal enzyme α-iduronidase with high activity from culture media of transgenic tobacco BY-2 cells. Plant Sci. 2009;177:668–675. [Google Scholar]
- Gecchele E, Merlin M, Brozzetti A, Falorni A, Pezzotti M, Avesani L. A comparative analysis of recombinant protein expression in different biofactories: bacteria, insect cells and plant systems. J Vis Exp. 2015;97:52459. doi: 10.3791/52459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gengenbach BB, Keil LL, Opdensteinen P, Muschen CR, Melmer G, Lentzen H, Buhrmann J, Buyel JF. Comparison of microbial and transient expression (tobacco plants and plant-cell packs) for the production and purification of the anticancer mistletoe lectin viscumin. Biotechnol Bioeng. 2019;116:2236–2249. doi: 10.1002/bit.27076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomord V, Fitchette AC, Menu-Bouaouiche L, Saint-Jore-Dupas C, Plasson C, Michaud D, Faye L. Plant-specific glycosylation patterns in the context of therapeutic protein production. Plant Biotechnol J. 2010;8:564–587. doi: 10.1111/j.1467-7652.2009.00497.x. [DOI] [PubMed] [Google Scholar]
- Hahn F, Nekrasov V. CRISPR/Cas precision: do we need to worry about off-targeting in plants? Plant Cell Rep. 2019;38:437–441. doi: 10.1007/s00299-018-2355-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hakkinen ST, Reuter L, Nuorti N, Joensuu JJ, Rischer H, Ritala A. Tobacco BY-2 media component optimization for a cost-efficient recombinant protein production. Front Plant Sci. 2018;9:45. doi: 10.3389/fpls.2018.00045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanania U, Ariel T, Tekoah Y, Fux L, Sheva M, Gubbay Y, Weiss M, Oz D, Azulay Y, Turbovski A, Forster Y, Shaaltiel Y. Establishment of a tobacco BY2 cell line devoid of plant-specific xylose and fucose as a platform for the production of biotherapeutic proteins. Plant Biotechnol J. 2017;15:1120–1129. doi: 10.1111/pbi.12702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Havenith H, Raven N, Di Fiore S, Fischer R, Schillberg S. Image-based analysis of cell-specific productivity for plant cell suspension cultures. Plant Cell Tiss Org Cult. 2014;117:393–399. [Google Scholar]
- Hehle VK, Lombardi R, van Dolleweerd CJ, Paul MJ, Di Micco P, Morea V, Benvenuto E, Donini M, Ma JK. Site-specific proteolytic degradation of IgG monoclonal antibodies expressed in tobacco plants. Plant Biotechnol J. 2015;13:235–245. doi: 10.1111/pbi.12266. [DOI] [PubMed] [Google Scholar]
- Hellwig S, Drossard J, Twyman RM, Fischer R. Plant cell cultures for the production of recombinant proteins. Nat Biotechnol. 2004;22:1415–1422. doi: 10.1038/nbt1027. [DOI] [PubMed] [Google Scholar]
- Holland T, Sack M, Rademacher T, Schmale K, Altmann F, Stadlmann J, Fischer R, Hellwig S. Optimal nitrogen supply as a key to increased and sustained production of a monoclonal full-size antibody in BY-2 suspension culture. Biotechnol Bioeng. 2010;107:278–289. doi: 10.1002/bit.22800. [DOI] [PubMed] [Google Scholar]
- Huang TK, McDonald KA. Bioreactor engineering for recombinant protein production in plant cell suspension cultures. Biochem Eng J. 2009;45:168–184. [Google Scholar]
- Huang TK, Plesha MA, Falk BW, Dandekar AM, McDonald KA. Bioreactor strategies for improving production yield and functionality of a recombinant human protein in transgenic tobacco cell cultures. Biotechnol Bioeng. 2009;102:508–520. doi: 10.1002/bit.22061. [DOI] [PubMed] [Google Scholar]
- Huang LF, Tan CC, Yeh JF, Liu HY, Liu YK, Ho SL, Lu CA. Efficient secretion of recombinant proteins from rice suspension-cultured cells modulated by the choice of signal peptide. PLoS ONE. 2015;10:e0140812. doi: 10.1371/journal.pone.0140812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ilan Y, Gingis-Velitski S, Ben Ya'aco A, Shabbat Y, Zolotarov L, Almon E, Shaaltiel Y. A plant cell-expressed recombinant anti-TNF fusion protein is biologically active in the gut and alleviates immune-mediated hepatitis and colitis. Immunobiology. 2017;222:544–551. doi: 10.1016/j.imbio.2016.11.001. [DOI] [PubMed] [Google Scholar]
- Imamura T, Isozumi N, Higashimura Y, Ohki S, Mori M. Production of ORF8 protein from SARS-CoV-2 using an inducible virus-mediated expression system in suspension-cultured tobacco BY-2 cells. Plant Cell Rep. 2021 doi: 10.1007/s00299-020-02654-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansing J, Sack M, Augustine SM, Fischer R, Bortesi L. CRISPR/Cas9-mediated knockout of six glycosyltransferase genes in Nicotiana benthamiana for the production of recombinant proteins lacking β-1, 2-xylose and core α-1, 3-fucose. Plant Biotechnol J. 2019;17:350–361. doi: 10.1111/pbi.12981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung JW, Shin JH, Lee WK, Begum H, Min CH, Jang MH, Oh HB, Yang MS, Kim SR. Inactivation of the beta (1, 2)-xylosyltransferase and the alpha (1, 3)-fucosyltransferase gene in rice (Oryza sativa) by multiplex CRISPR/Cas9 strategy. Plant Cell Rep. 2021 doi: 10.1007/s00299-021-02667-8. [DOI] [PubMed] [Google Scholar]
- Jutras PV, Grosse-Holz F, Kaschani F, Kaiser M, Michaud D, van der Hoorn RAL. Activity-based proteomics reveals nine target proteases for the recombinant protein-stabilizing inhibitor SlCYS8 in Nicotiana benthamiana. Plant Biotechnol J. 2019;17:1670–1678. doi: 10.1111/pbi.13092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaldis A, Ahmad A, Reid A, McGarvey B, Brandle J, Ma S, Jevnikar A, Kohalmi SE, Menassa R. High-level production of human interleukin-10 fusions in tobacco cell suspension cultures. Plant Biotechnol J. 2013;11:535–545. doi: 10.1111/pbi.12041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kallolimath S, Castilho A, Strasser R, Grunwald-Gruber C, Altmann F, Strubl S, Galuska CE, Zlatina K, Galuska SP, Werner S, Thiesler H, Werneburg S, Hildebrandt H, Gerardy-Schahn R, Steinkellner H. Engineering of complex protein sialylation in plants. Proc Natl Acad Sci U S A. 2016;113:9498–9503. doi: 10.1073/pnas.1604371113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato K, Matsumoto T, Koiwai A, Mizusaki S, Nishida K, Noguchi M, Tamaki E (1972) Liquid suspension culture of tobaco cells. Fermentation technology today: Proceedings of the IVth International Fermentation Symposium, Kyoto, Japan, pp 689–695
- Khoudi H, Laberge S, Ferullo JM, Bazin R, Darveau A, Castonguay Y, Allard G, Lemieux R, Vezina LP. Production of a diagnostic monoclonal antibody in perennial alfalfa plants. Biotechnol Bioeng. 1999;64:135–143. doi: 10.1002/(sici)1097-0290(19990720)64:2<135::aid-bit2>3.0.co;2-q. [DOI] [PubMed] [Google Scholar]
- Kim NS, Jang SH, Yu HY, Chung ND, Kwon TH, Yang MS, Kim TG. Amylase and cysteine proteinase gene knockdown in rice cells using RNA interference for enhancing production of recombinant proteins. Plant Cell Tiss Org Cult. 2013;114:97–107. [Google Scholar]
- Kirchhoff J, Raven N, Boes A, Roberts JL, Russell S, Treffenfeldt W, Fischer R, Schinkel H, Schiermeyer A, Schillberg S. Monoclonal tobacco cell lines with enhanced recombinant protein yields can be generated from heterogeneous cell suspension cultures by flow sorting. Plant Biotechnol J. 2012;10:936–944. doi: 10.1111/j.1467-7652.2012.00722.x. [DOI] [PubMed] [Google Scholar]
- Klimek-Chodacka M, Oleszkiewicz T, Lowder LG, Qi Y, Baranski R. Efficient CRISPR/Cas9-based genome editing in carrot cells. Plant Cell Rep. 2018;37:575–586. doi: 10.1007/s00299-018-2252-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon TH, Seo JE, Kim J, Lee JH, Jang YS, Yang MS. Expression and secretion of the heterodimeric protein interleukin-12 in plant cell suspension culture. Biotechnol Bioeng. 2003;81:870–875. doi: 10.1002/bit.10528. [DOI] [PubMed] [Google Scholar]
- Lallemand J, Bouche F, Desiron C, Stautemas J, de Lemos EF, Perilleux C, Tocquin P. Extracellular peptidase hunting for improvement of protein production in plant cells and roots. Front Plant Sci. 2015;6:37. doi: 10.3389/fpls.2015.00037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li JF, Norville JE, Aach J, McCormack M, Zhang D, Bush J, Church GM, Sheen J. Multiplex and homologous recombination-mediated genome editing in Arabidopsis and Nicotiana benthamiana using guide RNA and Cas9. Nat Biotechnol. 2013;31:688–691. doi: 10.1038/nbt.2654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Guan R, Guo H, Miao X. New insights into an RNAi approach for plant defence against piercing-sucking and stem-borer insect pests. Plant Cell Environ. 2015;38:2277–2285. doi: 10.1111/pce.12546. [DOI] [PubMed] [Google Scholar]
- Li J, Stoddard TJ, Demorest ZL, Lavoie PO, Luo S, Clasen BM, Cedrone F, Ray EE, Coffman AP, Daulhac A, Yabandith A, Retterath AJ, Mathis L, Voytas DF, D'Aoust MA, Zhang F. Multiplexed, targeted gene editing in Nicotiana benthamiana for glyco-engineering and monoclonal antibody production. Plant Biotechnol J. 2016;14:533–542. doi: 10.1111/pbi.12403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang G, Zhang H, Lou D, Yu D. Selection of highly efficient sgRNAs for CRISPR/Cas9-based plant genome editing. Sci Rep. 2016;6:21451. doi: 10.1038/srep21451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lico C, Santi L, Baschieri S, Noris E, Marusic C, Donini M, Pedrazzini E, Maga G, Franconi R, Di Bonito P, Avesani L. Plant molecular farming as a strategy against COVID-19—the Italian perspective. Front Plant Sci. 2020;11:609910. doi: 10.3389/fpls.2020.609910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu W, Rudis MR, Cheplick MH, Millwood RJ, Yang JP, Ondzighi-Assoume CA, Montgomery GA, Burris KP, Mazarei M, Chesnut JD, Stewart CN., Jr Lipofection-mediated genome editing using DNA-free delivery of the Cas9/gRNA ribonucleoprotein into plant cells. Plant Cell Rep. 2020;39:245–257. doi: 10.1007/s00299-019-02488-w. [DOI] [PubMed] [Google Scholar]
- Macharoen K, Li Q, Marquez-Escobar VA, Corbin JM, Lebrilla CB, Nandi S, McDonald KA. Effects of kifunensine on production and N-glycosylation modification of butyrylcholinesterase in a transgenic rice cell culture bioreactor. Int J Mol Sci. 2020;21:6896. doi: 10.3390/ijms21186896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magy B, Tollet J, Laterre R, Boutry M, Navarre C. Accumulation of secreted antibodies in plant cell cultures varies according to the isotype, host species and culture conditions. Plant Biotechnol J. 2014;12:457–467. doi: 10.1111/pbi.12152. [DOI] [PubMed] [Google Scholar]
- Mandal MK, Fischer R, Schillberg S, Schiermeyer A. Inhibition of protease activity by antisense RNA improves recombinant protein production in Nicotiana tabacum cv. Bright yellow 2 (BY-2) suspension cells. Biotechnol J. 2014;9:1065–1073. doi: 10.1002/biot.201300424. [DOI] [PubMed] [Google Scholar]
- Mandal MK, Ahvari H, Schillberg S, Schiermeyer A. Tackling unwanted proteolysis in plant production hosts used for molecular farming. Front Plant Sci. 2016;7:267. doi: 10.3389/fpls.2016.00267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McBride KE, Schaaf DJ, Daley M, Stalker DM. Controlled expression of plastid transgenes in plants based on a nuclear DNA-encoded and plastid-targeted T7 RNA polymerase. Proc Natl Acad Sci USA. 1994;91:7301–7305. doi: 10.1073/pnas.91.15.7301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald KA, Hong LM, Trombly DM, Xie Q, Jackman AP. Production of human α-1-antitrypsin from transgenic rice cell culture in a membrane bioreactor. Biotechnol Prog. 2005;21:728–734. doi: 10.1021/bp0496676. [DOI] [PubMed] [Google Scholar]
- Mercx S, Tollet J, Magy B, Navarre C, Boutry M. Gene inactivation by CRISPR-Cas9 in Nicotiana tabacum BY-2 suspension cells. Front Plant Sci. 2016;7:40. doi: 10.3389/fpls.2016.00040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Modrzejewski D, Hartung F, Lehnert H, Sprink T, Kohl C, Keilwagen J, Wilhelm R. Which factors affect the occurrence of off-target effects caused by the use of CRISPR/Cas: a systematic review in plants. Front Plant Sci. 2020;11:574959. doi: 10.3389/fpls.2020.574959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen HT, Leelavathi S, Reddy VS. Bacteriophage T7 RNA polymerase-directed, inducible and tissue-specific over-expression of foreign genes in transgenic plants. Plant Biotechnol J. 2004;2:301–310. doi: 10.1111/j.1467-7652.2004.00071.x. [DOI] [PubMed] [Google Scholar]
- Oberemok VV, Laikova KV, Repetskaya AI, Kenyo IM, Gorlov MV, Kasich IN, Krasnodubets AM, Gal'chinsky NV, Fomochkina II, Zaitsev AS, Bekirova VV, Seidosmanova EE, Dydik KI, Meshcheryakova AO, Nazarov SA, Smagliy NN, Chelengerova EL, Kulanova AA, Deri K, Subbotkin MV, Useinov RZ, Shumskykh MN, Kubyshkin AV. A half-century history of applications of antisense oligonucleotides in medicine, agriculture and forestry: we should continue the journey. Molecules. 2018;23(6):1302. doi: 10.3390/molecules23061302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogawa Y, Suzuki H, Sakurai N, Aoki K, Saito K, Shibata D. Cryopreservation and metabolic profiling analysis of Arabidopsis T87 suspension-cultured cells. Cryo Lett. 2008;29:427–436. [PubMed] [Google Scholar]
- Paccalet T, Bardor M, Rihouey C, Delmas F, Chevalier C, D'Aoust MA, Faye L, Vezina L, Gomord V, Lerouge P. Engineering of a sialic acid synthesis pathway in transgenic plants by expression of bacterial Neu5Ac-synthesizing enzymes. Plant Biotechnol J. 2007;5:16–25. doi: 10.1111/j.1467-7652.2006.00211.x. [DOI] [PubMed] [Google Scholar]
- Park YJ, Han JE, Lee H, Jung YJ, Murthy HN, Park SY. Large-scale production of recombinant miraculin protein in transgenic carrot callus suspension cultures using air-lift bioreactors. AMB Express. 2020;10:140. doi: 10.1186/s13568-020-01079-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Permyakova NV, Sidorchuk YV, Marenkova TV, Khozeeva SA, Kuznetsov VV, Zagorskaya AA, Rozov SM, Deineko EV. CRISPR/Cas9-mediated GFP gene inactivation in Arabidopsis suspension cells. Mol Biol Rep. 2019;46:5735–5743. doi: 10.1007/s11033-019-05007-y. [DOI] [PubMed] [Google Scholar]
- Pham PV. Medical biotechnology: techniques and applications. In: Azevedo V, Barh D, editors. Omics technologies and bio-engineering: towards improving quqlity of life. Elsevier Science; 2018. pp. 449–469. [Google Scholar]
- Pillay P, Schluter U, van Wyk S, Kunert KJ, Vorster BJ. Proteolysis of recombinant proteins in bioengineered plant cells. Bio Eng. 2014;5:15–20. doi: 10.4161/bioe.25158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pires AS, Rosa S, Castanheira S, Fevereiro P, Abranches R. Expression of a recombinant human erythropoietin in suspension cell cultures of Arabidopsis, tobacco and medicago. Plant Cell Tiss Org Cult. 2012;110:171–181. [Google Scholar]
- Poborilova Z, Plchova H, Cerovska N, Gunter CJ, Hitzeroth II, Rybicki EP, Moravec T. Transient protein expression in tobacco BY-2 plant cell packs using single and multi-cassette replicating vectors. Plant Cell Rep. 2020;39:1115–1127. doi: 10.1007/s00299-020-02544-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rademacher T, Sack M, Blessing D, Fischer R, Holland T, Buyel J. Plant cell packs: a scalable platform for recombinant protein production and metabolic engineering. Plant Biotechnol J. 2019;17:1560–1566. doi: 10.1111/pbi.13081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raven N, Rasche S, Kuehn C, Anderlei T, Klockner W, Schuster F, Henquet M, Bosch D, Buchs J, Fischer R, Schillberg S. Scaled-up manufacturing of recombinant antibodies produced by plant cells in a 200-L orbitally-shaken disposable bioreactor. Biotechnol Bioeng. 2015;112:308–321. doi: 10.1002/bit.25352. [DOI] [PubMed] [Google Scholar]
- Rawlings ND, Waller M, Barrett AJ, Bateman A. MEROPS: the database of proteolytic enzymes, their substrates and inhibitors. Nucleic Acids Res. 2014;42:D503–509. doi: 10.1093/nar/gkt953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rawlings ND, Barrett AJ, Finn R. Twenty years of the MEROPS database of proteolytic enzymes, their substrates and inhibitors. Nucleic Acids Res. 2016;44:D343–350. doi: 10.1093/nar/gkv1118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren C, Liu X, Zhang Z, Wang Y, Duan W, Li S, Liang Z. CRISPR/Cas9-mediated efficient targeted mutagenesis in Chardonnay (Vitis vinifera L.) Sci Rep. 2016;6(1):32289. doi: 10.1038/srep32289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reuter LJ, Bailey MJ, Joensuu JJ, Ritala A. Scale-up of hydrophobin-assisted recombinant protein production in tobacco BY-2 suspension cells. Plant Biotechnol J. 2014;12:402–410. doi: 10.1111/pbi.12147. [DOI] [PubMed] [Google Scholar]
- Reuter LJ, Conley AJ, Joensuu JJ. Continuous flow separation of hydrophobin fusion proteins from plant cell culture extract. Methods Mol Biol. 2016;1385:189–197. doi: 10.1007/978-1-4939-3289-4_14. [DOI] [PubMed] [Google Scholar]
- Sadoch J, Pyc M, Urbanowicz A, Iglewski A, Pilarski R. High-throughput evolutionary optimization of the induction medium towards recombinant protein production in BY-2 tobacco. Biotechnol Bioeng. 2021;118:676–689. doi: 10.1002/bit.27594. [DOI] [PubMed] [Google Scholar]
- Santos RB, Abranches R, Fischer R, Sack M, Holland T. Putting the spotlight back on plant suspension cultures. Front Plant Sci. 2016;7:297. doi: 10.3389/fpls.2016.00297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiermeyer A, Schinkel H, Apel S, Fischer R, Schillberg S. Production of Desmodus rotundus salivary plasminogen activator α1 (DSPAα1) in tobacco is hampered by proteolysis. Biotechnol Bioeng. 2005;89:848–858. doi: 10.1002/bit.20410. [DOI] [PubMed] [Google Scholar]
- Schillberg S, Finnern R. Plant molecular farming for the production of valuable proteins—critical evaluation of achievements and future challenges. J Plant Physiol. 2021;258–259:153359. doi: 10.1016/j.jplph.2020.153359. [DOI] [PubMed] [Google Scholar]
- Schillberg S, Raven N, Spiegel H, Rasche S, Buntru M. Critical analysis of the commercial potential of plants for the production of recombinant proteins. Front Plant Sci. 2019;10:720. doi: 10.3389/fpls.2019.00720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schumacher HM, Westphal M, Heine-Dobbernack E. Cryopreservation of plant cell lines. Methods Mol Biol. 2015;1257:423–429. doi: 10.1007/978-1-4939-2193-5_21. [DOI] [PubMed] [Google Scholar]
- Shaaltiel Y, Tekoah Y. Plant specific N-glycan do not have proven adverse effects in humans. Nat Biotechnol. 2016;34:17–18. doi: 10.1038/nbt.3556. [DOI] [PubMed] [Google Scholar]
- Sheva M, Hanania U, Ariel T, Turbovski A, Rathod VKR, Oz D, Tekoah Y, Shaaltiel Y. Sequential genome editing and induced excision of the transgene in N. tabacum BY2 cells. Front Plant Sci. 2020 doi: 10.3389/fpls.2020.60714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shih SMH, Doran PM. Foreign protein production using plant cell and organ cultures: advantages and limitations. Biotechnol Adv. 2009;27:1036–1042. doi: 10.1016/j.biotechadv.2009.05.009. [DOI] [PubMed] [Google Scholar]
- Shin YJ, Chong YJ, Yang MS, Kwon TH. Production of recombinant human granulocyte macrophage-colony stimulating factor in rice cell suspension culture with a human-like N-glycan structure. Plant Biotechnol J. 2011;9:1109–1119. doi: 10.1111/j.1467-7652.2011.00636.x. [DOI] [PubMed] [Google Scholar]
- Silva AT, Nguyen A, Ye C, Verchot J, Moon JH. Conjugated polymer nanoparticles for effective siRNA delivery to tobacco BY-2 protoplasts. BMC Plant Biol. 2010;10:291. doi: 10.1186/1471-2229-10-291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Small I. RNAi for revealing and engineering plant gene functions. Curr Opin Biotechnol. 2007;18:148–153. doi: 10.1016/j.copbio.2007.01.012. [DOI] [PubMed] [Google Scholar]
- Sojikul P, Buehner N, Mason HS. A plant signal peptide–hepatitis B surface antigen fusion protein with enhanced stability and immunogenicity expressed in plant cells. Proc Natl Acad Sci USA. 2003;100:2209–2214. doi: 10.1073/pnas.0438037100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strasser R. Plant protein glycosylation. Glycobiology. 2016;26:926–939. doi: 10.1093/glycob/cww023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sukenik SC, Karuppanan K, Li Q, Lebrilla CB, Nandi S, McDonald KA. Transient recombinant protein production in glycoengineered Nicotiana benthamiana cell suspension culture. Int J Mol Sci. 2018;19:120. doi: 10.3390/ijms19041205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Q-Y, Ding L-W, Lomonossoff GP, Sun Y-B, Luo M, Li C-Q, Jiang L, Xu Z-F. Improved expression and purification of recombinant human serum albumin from transgenic tobacco suspension culture. J Biotechnol. 2011;155:164–172. doi: 10.1016/j.jbiotec.2011.06.033. [DOI] [PubMed] [Google Scholar]
- Tekoah Y, Shulman A, Kizhner T, Ruderfer I, Fux L, Nataf Y, Bartfeld D, Ariel T, Gingis-Velitski S, Hanania U, Shaaltiel Y. Large-scale production of pharmaceutical proteins in plant cell culture-the protalix experience. Plant Biotechnol J. 2015;13:1199–1208. doi: 10.1111/pbi.12428. [DOI] [PubMed] [Google Scholar]
- Vasilev N, Grömping U, Lipperts A, Raven N, Fischer R, Schillberg S. Optimization of BY-2 cell suspension culture medium for the production of a human antibody using a combination of fractional factorial designs and the response surface method. Plant Biotechnol J. 2013;11:867–874. doi: 10.1111/pbi.12079. [DOI] [PubMed] [Google Scholar]
- Vaucheret H. Gene silencing: mode of miRNA biogenesis matters. Nat Plants. 2015;1:15019. doi: 10.1038/nplants.2015.19. [DOI] [PubMed] [Google Scholar]
- Wada N, Ueta R, Osakabe Y, Osakabe K. Precision genome editing in plants: state-of-the-art in CRISPR/Cas9-based genome engineering. BMC Plant Biol. 2020;20:234. doi: 10.1186/s12870-020-02385-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Karki U, Abeygunaratne H, UnnoldCofre C, Xu J. Plant cell-secreted stem cell factor stimulates expansion and differentiation of hematopoietic stem cells. Process Biochem. 2021;100:39–48. doi: 10.1016/j.procbio.2020.09.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson SA, Roberts SC. Recent advances towards development and commercialization of plant cell culture processes for the synthesis of biomolecules. Plant Biotechnol J. 2012;10:249–268. doi: 10.1111/j.1467-7652.2011.00664.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J, Zhang N. On the way to commercializing plant cell culture platform for biopharmaceuticals: present status and prospect. Pharma Bioprocess. 2014;2:499–518. doi: 10.4155/pbp.14.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J, Tan L, Goodrum KJ, Kieliszewski MJ. High-yields and extended serum half-life of human interferon α2b expressed in tobacco cells as arabinogalactan-protein fusions. Biotechnol Bioeng. 2007;97:997–1008. doi: 10.1002/bit.21407. [DOI] [PubMed] [Google Scholar]
- Xu J, Okada S, Tan L, Goodrum KJ, Kopchick JJ, Kieliszewski MJ. Human growth hormone expressed in tobacco cells as an arabinogalactan-protein fusion glycoprotein has a prolonged serum life. Transgenic Res. 2010;19:849–867. doi: 10.1007/s11248-010-9367-8. [DOI] [PubMed] [Google Scholar]
- Xu J, Ge X, Dolan MC. Towards high-yield production of pharmaceutical proteins with plant cell suspension cultures. Biotechnol Adv. 2011;29:278–299. doi: 10.1016/j.biotechadv.2011.01.002. [DOI] [PubMed] [Google Scholar]
- Xu J, Dolan MC, Medrano G, Cramer CL, Weathers PJ. Green factory: plants as bioproduction platforms for recombinant proteins. Biotechnol Adv. 2012;30:1171–1184. doi: 10.1016/j.biotechadv.2011.08.020. [DOI] [PubMed] [Google Scholar]
- Xu J, Towler M, Weathers PJ. Platforms for plant-based protein production. In: Pavlov A, Bley T, editors. Bioprocessing of plant in vitro systems reference series in phytochemistry. Springer International Publishing AG; 2016. pp. 1–40. [Google Scholar]
- Yano A, Maeda F, Takekoshi M. Transgenic tobacco cells producing the human monoclonal antibody to hepatitis B virus surface antigen. J Med Virol. 2004;73:208–215. doi: 10.1002/jmv.20077. [DOI] [PubMed] [Google Scholar]
- Yin BJ, Gao T, Zheng NY, Li Y, Tang SY, Liang LM, Xie Q. Generation of glyco-engineered BY2 cell lines with decreased expression of plant-specific glycoepitopes. Protein Cell. 2011;2:41–47. doi: 10.1007/s13238-011-1007-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Younis A, Siddique MI, Kim CK, Lim KB. RNA interference (RNAi) induced gene silencing: a promising approach of hi-tech plant breeding. Int J Biol Sci. 2014;10:1150–1158. doi: 10.7150/ijbs.10452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu XD, Liu ZC, Huang SL, Chen ZQ, Sun YW, Duan PF, Ma YZ, Xia LQ. RNAi-mediated plant protection against aphids. Pest Manag Sci. 2016;72:1090–1098. doi: 10.1002/ps.4258. [DOI] [PubMed] [Google Scholar]
- Zhang N, Dolan M, Wu D, Phillips GC, Xu J. Dramatic secretion of recombinant protein expressed in tobacco cells with a designer glycopeptide tag is highly impacted by medium composition. Plant Cell Rep. 2016;35:2513–2522. doi: 10.1007/s00299-016-2051-6. [DOI] [PubMed] [Google Scholar]
- Zhang N, Gonzalez M, Savary B, Xu J. High-yield secretion of recombinant proteins expressed in tobacco cell culture with a designer glycopeptide tag: process development. Biotechnol J. 2016;11:497–506. doi: 10.1002/biot.201500377. [DOI] [PubMed] [Google Scholar]
- Zhang N, Wright T, Caraway P, Xu J. Enhanced secretion of human α1-antitrypsin expressed with a novel glycosylation module in tobacco BY-2 cell culture. Bio Eng. 2019;10:87–97. doi: 10.1080/21655979.2019.1604037. [DOI] [PMC free article] [PubMed] [Google Scholar]