Abstract
Fusarium oxysporum is an important soilborne fungal pathogen with many different formae speciales that can colonize the plant vascular system and cause serious crop wilt disease worldwide. We found a glycoside hydrolase family 12 protein FoEG1, secreted by F. oxysporum, that acted as a pathogen‐associated molecular pattern (PAMP) targeting the apoplast of plants to induce cell death. Purified FoEG1 protein triggered cell death in different plants and induced the plant defence response to enhance the disease resistance of plants. The ability of FoEG1 to induce cell death was mediated by leucine‐rich repeat (LRR) receptor‐like kinases BAK1 and SOBIR1, and this ability was independent of its hydrolase activity. The mutants of cysteine residues did not affect the ability of FoEG1 to induce cell death, and an 86 amino acid fragment from amino acid positions 144 to 229 of FoEG1 was sufficient to induce cell death in Nicotiana benthamiana. In addition, the expression of FoEG1 was strongly induced in the early stage of F. oxysporum infection of host plants, and FoEG1 deletion or loss of enzyme activity reduced the virulence of F. oxysporum. Therefore, our results suggest that FoEG1 can contribute to the virulence of F. oxysporum depending on its enzyme activity and can also act as a PAMP to induce plant defence responses.
Keywords: FoEG1, Fusariumoxysporum, PAMP, plant immunity, virulence
The secreted protein FoEG1 from Fusarium oxysporum triggers cell death and induces plant immunity by targeting to the apoplast of plants, and also contributes to the virulence of F. oxysporum.

1. INTRODUCTION
Plants' growth and development are often confronted with a variety of pathogenic microorganisms. With the interaction between plants and pathogens, disease resistance of plants and the pathogenicity of pathogens are in a dynamic balance of coevolution. The defence response of plants to pathogens is mainly expressed in two ways: PAMP‐triggered immunity (PTI) and effector‐triggered immunity (ETI). PTI is a plant defence response triggered by highly conserved pathogen‐associated molecular patterns (PAMPs), and represents the innate immune system and the basic defence response of plants. This process is accomplished by plants recognizing PAMPs using pattern recognition receptors (PRRs) on the cell membrane. The best known or classical PAMPs are flagellin flg22 (Felix et al., 1999), EF‐Tu (Kunze et al., 2004), ethylene‐inducing xylanase (EIX) (Jürg et al., 1999), glucans (Fliegmann et al., 2004), and chitin (Shinya et al., 2015) in bacteria, oomycetes, or fungi. After recognizing PAMPs, plants activate a series of downstream defence‐related responses, including cell death, reactive oxygen species (ROS) bursts, callose deposition, calcium ion (Ca2+) level elevation, and induced expression of defence‐related genes (Boller & Felix, 2009; Zipfel, 2009).
The plant cell wall is the first barrier encountered when pathogens infect plants and it is mainly composed of polysaccharide components such as pectins, hemicelluloses, celluloses, and β‐1,3‐glucans (Lai & Liou, 2018). Pathogens secrete numerous cell‐wall‐degrading enzymes (CWDEs), such as pectinases, cellulases, hemicellulases, and ligninases, to overcome the plant cell wall (Kubicek et al., 2014). Previous studies have shown that many CWDEs are virulence factors and are involved in pathogen infection processes (Quoc & Chau, 2017). For example, the endoxylanase gene xynB has been demonstrated to affect the virulence of Xanthomonas oryzae pv. oryzae on rice (Rajeshwari et al., 2005), and Xyn11, a xylanase of glycoside hydrolase family 11 from Botrytis cinerea, can induce cell death in plant leaves and is required for virulence in B. cinerea (Brito et al., 2006). Endo‐β‐1,4‐xylanase SsXyl1 deletion strains of Sclerotinia sclerotiorum produce aberrant sclerotia that cannot germinate to form apothecia and also lose virulence to the hosts (Yu et al., 2016). The deletion of VmXyl1, an endo‐β‐1,4‐xylanase of glycoside hydrolase family 10, from Valsa mali significantly reduces pycnidia formation and reduces virulence to apple leaves and twigs although it does not affect mycelial growth (Yu et al., 2018). In the plant pathogenic oomycete Phytophthora parasitica, two GH10 xylanases, ppxyn1 and ppxyn2, were found to be essential for virulence towards Nicotiana benthamiana and tomato plants (Lai & Liou, 2018).
Some CWDEs act as PAMPs, in addition to the virulence effect mentioned above, which can induce the immune response of plants. For instance, PsXEG1 is a glycoside hydrolase family 12 (GH12) protein with xyloglucanase activity, that contributes to the virulence of Phytophthora sojae (Ma et al., 2015). However, soybean recognizes PsXEG1 to induce an immune response, which is then inhibited by pathogen RXLR effectors. BcXYG1 is a xyloglucanase of the GH12 family from B. cinerea that can induce strong necrosis and resistance responses in dicot plants (Zhu et al., 2017). VdEG1 and VdEG3, two GH12 proteins with cellulase activity from Verticillium dahliae, were also found to have the function of PAMPs and can trigger cell death and an immune response independent of their enzymatic activity in N. benthamiana (Gui et al., 2017). Moreover, the pectate lyase VdPEL1 and the cutinase VdCUT11 from V. dahliae, and the xylanase BcXyl1 from B. cinerea can also contribute to virulence and simultaneously trigger plant immunity as PAMPs (Gui et al., 2018; Yang, Yang, et al., 2018; Yang, Zhang, et al., 2018). However, the way in which these CWDEs induce the plant immune response may be different. Some CWDEs can be directly recognized as PAMPs by plant receptors (Nurnberger et al., 2004). For example, fungal endopolygalacturonases can be recognized as PAMPs by the Arabidopsis thaliana leucine‐rich repeat receptor‐like protein (LRR‐RLP) AtRLP42 (Zhang et al., 2014). Another hydrolytic enzyme, ethylene‐inducing xylanase (EIX), can be recognized as a PAMP by the tomato LRR‐RLPs LeEIX1 and LeEIX2, although only LeEIX2 could transmit the signal that induced the hypersensitive response (HR) (Ron & Avni, 2004). Recently, a novel EIX‐like protein VdEIX3, identified from V. dahliae, was found to be recognized by N. benthamiana NbEIX2 resulting in immunity induction in N. benthamiana (Yin et al., 2020). Moreover, there are some CWDEs that induce plant immune responses indirectly. These CWDEs are not directly recognized by plants, but plants perceive the cell wall fragments produced by the action of CWDEs and elicit the plant defence response. For example, pectin in the plant cell wall can be degraded to produce oligogalacturonides under the action of polygalacturonase, which can trigger the plant immune response (D’Ovidio et al., 2004; Prade et al., 1999). The plant immunity induced by the extracellular cutinase VdCUT11 from V. dahliae may be mediated by the degradation of plant cell wall polymers that leads to the release of plant cell wall fragments (damage‐associated molecular patterns, DAMPs), which trigger the plant defence response (Gui et al., 2018). Most of these identified CWDEs belong to families of protein with the glycoside hydrolase domain, which indicates that there may be many protein hydrolases in the glycoside hydrolase families that are related to pathogen infection or plant immunity induction.
The genus Fusarium contains many important plant pathogens that can cause severe plant diseases in many important crops worldwide, resulting in major yield and quality losses (Kazan & Gardiner, 2018). For example, Fusarium head blight, Fusarium wilt, Fusarium crown rot, and Fusarium root rot are important plant diseases caused by Fusarium pathogens. Among these Fusarium pathogens, Fusarium oxysporum is a typical soilborne fungus that can colonize the plant vascular system and cause serious crop wilt disease (de Sain & Rep, 2015). The species affects more than 120 plant species, including some important crops such as banana, cotton, tomato, melon, and soybean (de Sain & Rep, 2015; Edel‐Hermann & Lecomte, 2019; Xu et al., 2020). However, strains usually only selectively infect one or a few host species (de Sain & Rep, 2015); these strains are classified as different formae speciales (ff. sp.) based on this host specificity (Edel‐Hermann & Lecomte, 2019). Fusarium wilt caused by F. oxysporum has been reported in many countries worldwide. However, it is very difficult to control this pathogen due to its soilborne nature and ability to advance to the parenchymatous cells (Abro et al., 2019; Vethavalli & Sudha, 2012). Moreover, another reason for the difficulty in controlling F. oxysporum is its persistence as thick‐walled chlamydospores in the soil. This feature enables it to withstand adverse environmental conditions such as high temperature and drying. No effective control method has so far been found in agricultural production to completely control this disease (Abro et al., 2019), which caused large yield losses that could rise up to 45% in severe cases (McGovern, 2015). Therefore, it is necessary to develop new control strategies to control it. The secreted proteins with GH12 domains mentioned above are widely present in oomycetes, fungi, and bacteria (Ma et al., 2015) such as P. sojae, V. dahliae, and B. cinerea, and play important roles in plant immunity induction and pathogenicity of pathogens. Especially in view of its important function in plant immunity induction, GH12 proteins may have the potential to enhance plant disease resistance. There are, however, no reported studies on glycoside hydrolase 12 family proteins of Fusarium spp. and their involvement in the interaction between pathogens and plants.
In this study, we identified a GH12 family protein, FoEG1, secreted by F. oxysporum that can trigger plant cell death and analysed the function of FoEG1 in plant immunity induction, functional domain, and pathogenicity. In addition, our study also found that FoEG1 has some characteristics differing from the homologous proteins of other pathogenic fungi. Overall, the results of this study provide an experimental basis for studies on the interaction between F. oxysporum and plants, and the pathogenic mechanism of the fungus.
2. RESULTS
2.1. FoEG1, a glycoside hydrolase family 12 protein from F. oxysporum, induces cell death and targets the apoplast of plants
A total of five proteins containing the GH12 domain were identified in F. oxysporum f. sp. vasinfectum by searching the genome sequence. These five proteins (FOTG_02663, FOTG_11380, FOTG_10947, FOTG_12529, and FOTG_17138) were categorized as secreted proteins that contain a signal peptide but lack transmembrane domains. To determine the ability of inducing plant cell death, the transient expression of these proteins was carried out in N. benthamiana. The full‐length coding sequences of these proteins were cloned into pGR107, followed by agroinfiltration in N. benthamiana with green fluorescent protein (GFP) and INF1 used as negative and positive controls, respectively. Among these proteins, only FOTG_02663 (named FoEG1) showed strong cell death‐inducing activity in N. benthamiana similar to the positive control INF1, while the negative control GFP and the other four proteins, FOTG_11380, FOTG_10947, FOTG_12529, and FOTG_17138, did not induce plant cell death (Figure 1a,b), indicating that cell death was specifically caused by FoEG1.
FIGURE 1.
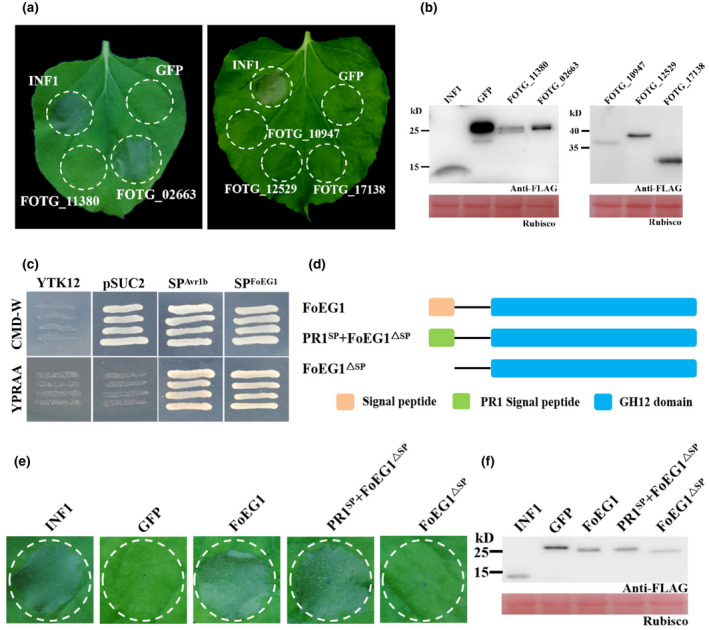
FoEG1 (FOTG_02663) can induce cell death in Nicotiana benthamiana and the signal peptide (SP) is required for FoEG1‐induced cell death. (a) The cell death‐inducing ability of glycoside hydrolase 12 proteins of Fusarium oxysporum was determined in N. benthamiana. The leaves of 4‐week‐old N. benthamiana were infiltrated with Agrobacterium tumefaciens carrying the indicated genes. INF1 and green fluorescent protein (GFP) were used as positive and negative controls, respectively. Photographs were taken 5 days postagroinfiltration (dpa). (b) Immunoblot analysis of transiently expressed glycoside hydrolase 12 proteins fused to the FLAG‐tag in N. benthamiana leaves 48 hr after infiltration. (c) Functional validation of the SP of FoEG1 by yeast signal trap assay. The yeast strain YTK12 could not grow on CMD−W medium (lacking tryptophan). The strain containing the pSUC2 vector can grow due to the function of the Trp operon. Fusion of the functional SP of FoEG1 in‐frame with mature yeast invertase enabled secretion of invertase, resulting in growth on YPRAA medium. The known functional SP of Avr1b was used as a positive control. (d) The determination of the cell death‐inducing ability of FoEG1 with or without SP. (e) Cell death induction was detected in N. benthamiana leaves 5 dpa with the A. tumefaciens carrying the indicated proteins. (f) Immunoblot of proteins from N. benthamiana leaves transiently expressing the indicated proteins
FoEG1 contains 249 amino acids, including three cysteine residues, and is a secreted protein with a predicted N‐terminal signal peptide (amino acids 1–16, SP). The secretory function of FoEG1 was tested based on the yeast signal trap assay system (Jacobs et al., 1997) and the signal peptide of FoEG1 was shown to be sufficient for the secretion of invertase in yeast (Figure 1c). This result demonstrated that FoEG1 was indeed a secreted protein. To determine whether FoEG1 targets the apoplast to induce plant cell death, the N‐terminal signal peptide of FoEG1 was deleted to produce FoEG1ΔSP, and the original signal peptide of FoEG1 was replaced with the signal peptide from plant pathogenesis‐related protein 1 (PR1) to produce PR1SP + FoEG1ΔSP (Figure 1d). Transient expression of the above proteins was carried out in N. benthamiana by the potato virus X (PVX) expression system, and the results showing that FoEG1ΔSP could not induce cell death in N. benthamiana, whereas PR1SP + FoEG1ΔSP could induce cell death like FoEG1 (Figure 1e). Immunoblot analysis confirmed that all proteins were expressed in N. benthamiana following A. tumefaciens‐mediated transient expression (Figure 1f). To determine that the cell death‐inducing ability of FoEG1 was not attributed to its signal peptide, we fused the signal peptide of FoEG1 with GFP and transiently expressed it in N. benthamiana. The results showed that GFP fused with the signal peptide of FoEG1 could not cause cell death in N. benthamiana (Figure S2), indicating that the ability of FoEG1 to induce cell death was not driven by its signal peptide. To further ascertain whether FoEG1 targets the apoplast of a plant to induce cell death, we agroinfiltrated pBinGFP4‐FoEG1, pBinGFP‐FoEG1 Δ SP, and pBinGFP4‐PR1 SP + FoEG1 Δ SP vectors in N. benthamiana. The results of confocal microscopy showed that FoEG1 and PR1SP + FoEG1Δ SP, but not FoEG1Δ SP, can be observed in the apoplast of N. benthamiana cells after plasmolysis (Figure S3a,b). This further confirmed the apoplastic location of FoEG1. These results indicate that the ability of FoEG1 to induce cell death of N. benthamiana depends on its signal peptide and is targeted to the apoplast of plants. Moreover, based on a BLAST search on the genome of F. oxysporum, we found that FoEG1 was highly conserved in various formae speciales (Figure S4).
2.2. FoEG1 induces cell death in different plants
To further verify the ability of FoEG1 to induce plant cell death, FoEG1 was expressed in Escherichia coli (Figure S5a) and the purified protein with varying concentrations ranging from 0.1 to 10 μM was infiltrated into the leaves of N. benthamiana using a syringe without a needle. The results showed that the recombinant protein of FoEG1 can induce cell death in the tested concentration range of 0.3–10 μM (Figure 2a). To test whether FoEG1 could induce cell death in other plants, the purified protein of FoEG1 was injected into the leaves of tobacco (Nicotiana tabacum), tomato (Solanum lycopersicum), cotton (Gossypium hirsutum), soybean (Glycine max), and maize (Zea mays). The results showed that FoEG1 could cause cell death in tobacco, tomato, and cotton, but not in soybean and maize (Figure 2b).
FIGURE 2.
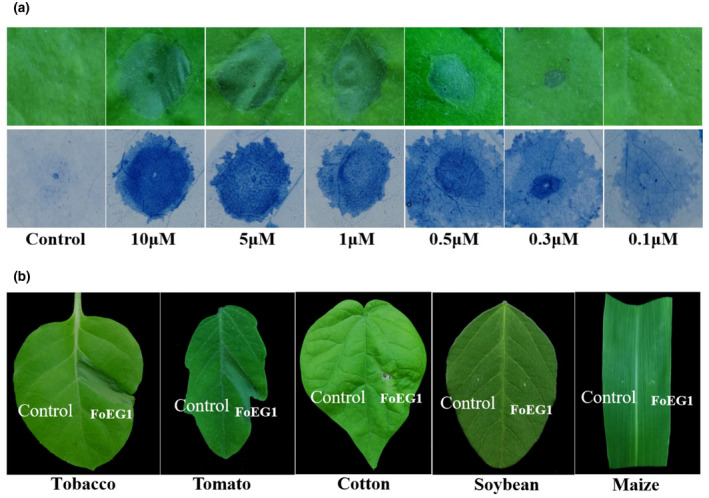
FoEG1 can induce cell death in various plant species. (a) Nicotiana benthamiana leaves were infiltrated with purified FoEG1 protein. Buffer was used as control. Treated N. benthamiana leaves were photographed 48 hr postinfiltration and stained with trypan blue. (b) Treatment of tobacco, tomato, cotton, soybean, and maize leaves with purified FoEG1 and buffer control. Treated leaves of different plants were photographed 48 hr postinfiltration
2.3. The cell death‐inducing activity of FoEG1 is independent of its glycoside hydrolase activity
FoEG1, which is a typical GH12 family protein, showed significant sequence conservation by multiple sequences alignment and phylogenetic analysis (Figure S6). The proteins of the GH12 family have two known conserved enzyme active sites that affect their hydrolase activity (Sandgren et al., 2001); the corresponding catalytic residues in FoEG1 were E143 and E230 (Figure S6a). According to previous studies on GH12 family proteins (Gui et al., 2017; Ma et al., 2015; Zhu et al., 2017), we determined the enzyme activity of FoEG1 and found that it has cellulase activity, while site‐directed mutagenesis of the two conserved catalytic residues in FoEG1 resulted in almost complete loss of cellulase activity (Figure S5c). To determine whether the hydrolase activity of FoEG1 affects its ability to induce plant cell death, site‐directed mutations of the two conserved catalytic residues of FoEG1 were carried out (Figure 3a). The mutant proteins were then expressed in N. benthamiana following Agrobacterium tumefaciens‐mediated transient expression. The results revealed that, irrespective of the simultaneous or individual mutagenesis of the two catalytic residues of FoEG1, the mutant proteins could still strongly trigger cell death (Figure 3b,d,f,h). The presence of all mutant proteins in N. benthamiana was confirmed using immunoblot analysis (Figure 3c,e,g,i). The same results were obtained in the activity test of the recombinant protein of each of the mutants in N. benthamiana (Figure S5b,d). Moreover, to eliminate the effect of the types of amino acids used in site‐directed mutations on the results, we replaced the two conserved catalytic residues with different amino acids (Gly, Ala, Ser, or Asp) during site‐directed mutations, and still obtained the same results (Figure 3b,d,f,h). This was an indication that the ability of FoEG1 to induce cell death was independent of the glycoside hydrolase activity.
FIGURE 3.
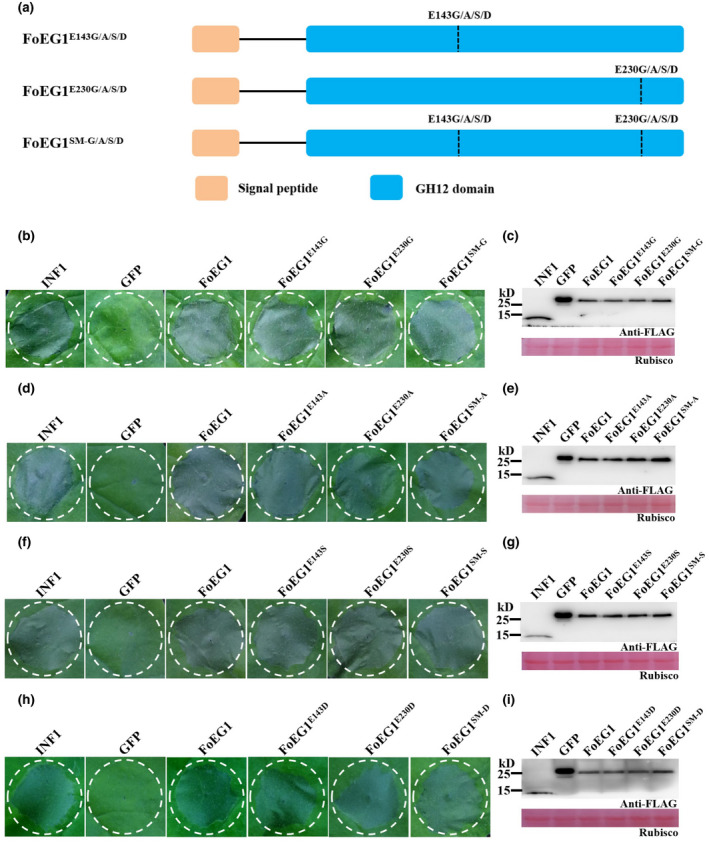
Cell death‐inducing activity of FoEG1 is independent of its hydrolase activity in Nicotiana benthamiana. (a) Schematic presentation of FoEG1 mutants. E143G/A/S/D and E230G/A/S/D represent the replacement of Glu at position 143 or 230 of FoEG1 with Gly, Ala, Ser, or Asp, respectively. SM‐x represents the replacement of Glu at both positions 143 and 230 of FoEG1 with x amino acid, simultaneously. (b), (d), (f), and (h) Cell death‐inducing activity of the indicated mutant proteins was assessed 5 days after agroinfiltration by transient expression in 4‐week‐old N. benthamiana leaves; INF1 and green fluorescent protein (GFP) were used as positive and negative controls, respectively. (c), (e), (g), and (i) Immunoblot analysis of proteins from N. benthamiana leaves transiently expressing the indicated mutant proteins
2.4. FoEG1 induces plant immunity responses
The HR is a type of programmed cell death that is generally considered to be closely related to the plant defence response (Camagna & Takemoto, 2018). To determine whether FoEG1‐induced cell death was associated with plant defence responses, the accumulation of ROS and the deposition of callose in N. benthamiana after infiltration of purified FoEG1 protein were measured. The results showed that the leaves of N. benthamiana produced significant accumulation of ROS and callose deposition 24 hr after infiltration (Figure 4a).
FIGURE 4.
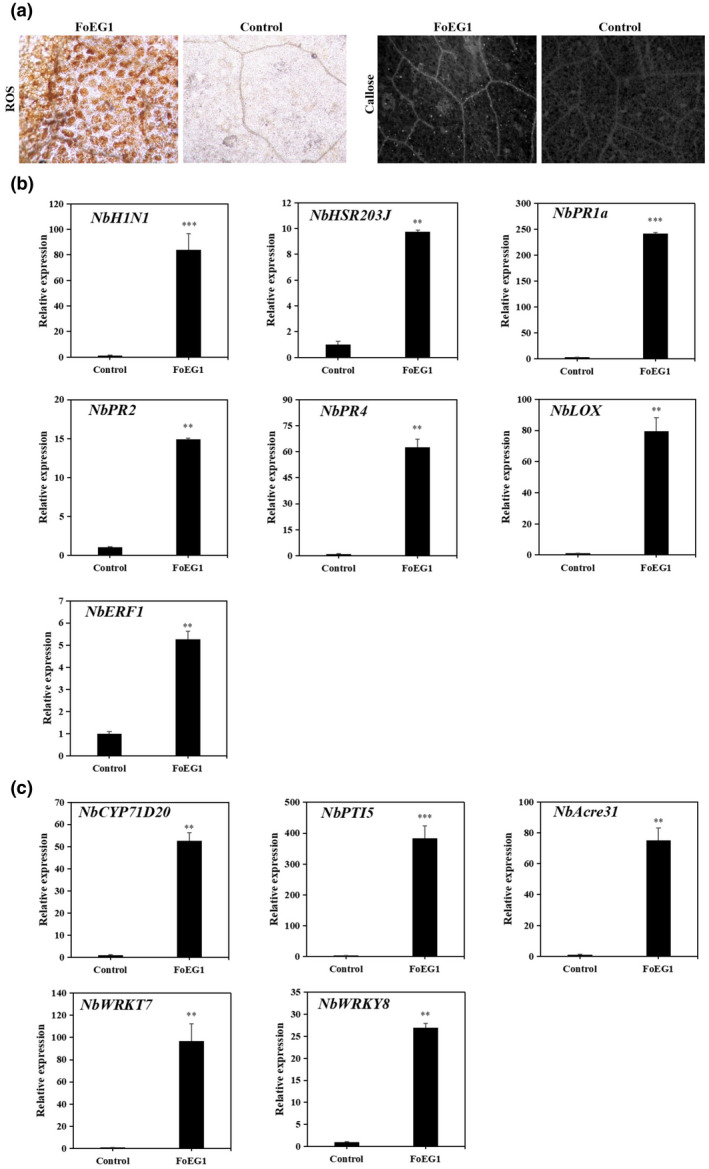
FoEG1 induces plant immunity responses in Nicotiana benthamiana. (a) Accumulation of reactive oxygen species (ROS) and callose deposition in N. benthamiana. The leaves of N. benthamiana were infiltrated with 0.1 μM purified FoEG1 protein or buffer control for 24 hr. (b) Relative expression of hypersensitive response (HR)‐specific marker genes and defence‐related marker genes in N. benthamiana. (c) Relative expression of pathogen‐associated molecular pattern (PAMP)‐triggered immunity (PTI) marker genes induced by FoEG1 in N. benthamiana. The leaves of N. benthamiana were infiltrated with 0.1 μM purified FoEG1 protein or buffer control for 24 hr. Total RNA was extracted and transcript levels were analysed by quantitative reverse transcription PCR. NbActin and NbEF‐1α were used as the internal reference genes. The leaves of N. benthamiana were infiltrated with 0.1 μM purified FoEG1 protein or buffer control for 24 hr. Asterisks at the top of the bars indicate statistical significance (**p < .01; ***p < .001)
The relative expressions of NbHIN1 and NbHSR203J, the HR marker genes, were significantly increased after infiltration by FoEG1 (Figure 4b), which indicates that FoEG1 can trigger HR‐associated immunity of N. benthamiana. Moreover, the relative expressions of some genes related to the defence response or hormone signalling pathways, such as NbPR1a, NbPR2, NbPR4, NbLOX, and NbERF1, were significantly increased compared with the control in N. benthamiana after infiltration by FoEG1 (Figure 4b). NbPR1a, NbPR2, NbPR4, NbLOX, and NbERF1 are the marker genes of salicylic acid (SA)‐, jasmonic acid (JA)‐, or ethylene‐dependent immunity; thus, FoEG1 may induce the plant immune response by activating the SA‐, JA‐ or ethylene‐mediated defence pathways.
Due to the conservation of FoEG1 in fungi and its ability to induce cell death and the plant immune response, we speculate that FoEG1 may function as a PAMP. To ascertain the veracity of FoEG1 acting as a PAMP, the relative expressions of some PTI marker genes were measured. The results showed that the relative expression of the PTI marker genes NbCYP71D20, NbPTI5, NbACRE31, NbWRKY7, and NbWRKY8 increased significantly in N. benthamiana after infiltration by FoEG1 (Figure 4c), suggesting that FoEG1 may function as a PAMP and its involvement in induced immune response in plants may be a PTI process.
2.5. BAK1/SERK3 and SOBIR1 are required for FoEG1‐induced cell death in N. benthamiana
Plants can perceive PAMPs through PRRs such as receptor‐like kinases (RLKs) or receptor‐like proteins (RLPs), and this process then activates the plant immune response (Gui et al., 2017). The receptor‐associated kinases BAK1/SERK3 and SOBIR1 are important components in the regulation of PRRs, which can promote intracellular signalling after perception of pathogens PAMPs (Heese et al., 2007; Liebrand et al., 2013, 2014; Nie et al., 2019). To further test whether FoEG1 functions as a PAMP to participate in the induction of plant cell death, virus‐induced gene silencing (VIGS) constructs targeting NbBAK1 or NbSOBIR1 expression in N. benthamiana were generated using a tobacco rattle virus (TRV) vector. The leaves of N. benthamiana were agroinfiltrated with FoEG1 or INF1 3 weeks after agroinfiltration with the VIGS constructs. The results showed that FoEG1 failed to induce cell death in both BAK1‐ and SOBIR1‐silenced plants similar to the positive control INF1 (Figure 5a,d). FoEG1 and INF1 could, however, still cause cell death in N. benthamiana treated with TRV2:GFP (Figure 5a,d). Immunoblot analysis confirmed that all proteins were successfully expressed in silenced N. benthamiana (Figure 5b,e). The expression of BAK1 and SOBIR1 was confirmed to be significantly reduced in corresponding plants by quantitative reverse transcription PCR (RT‐qPCR) analysis (Figure 5c,f). For further understanding of the signalling components involved in FoEG1‐induced cell death, EDS1‐, NDR1‐, and RXEG1‐silenced N. benthamiana plants were generated, as these may also be involved in PTI responses or the recognition of the GH12 protein XEG1 (Nie et al., 2019; Wang et al., 2018). The leaves of N. benthamiana were agroinfiltrated with FoEG1 or INF1. The results, however, showed that the silencing of EDS1, NDR1, and RXEG1 did not significantly affect FoEG1‐induced cell death (Figure S7), indicating that FoEG1 requires BAK1 and SOBIR1 but not EDS1, NDR1, or RXEG1 for cell death activation in N. benthamiana.
FIGURE 5.
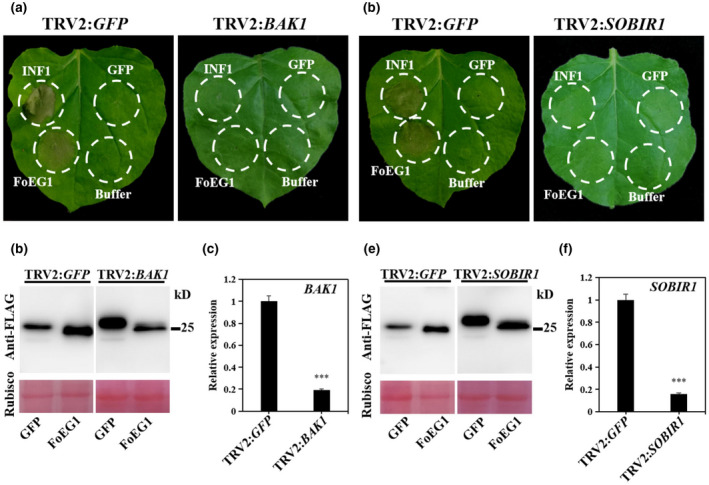
Cell death‐inducing activity of FoEG1 in Nicotiana benthamiana was mediated by the receptor‐like kinases BAK1 and SOBIR1. (a) and (d) BAK1 and SOBIR1 were required for FoEG1‐induced cell death in N. benthamiana. N. benthamiana plants were subjected to virus‐induced gene silencing (VIGS) by inoculation with tobacco rattle virus (TRV) constructs for 3 weeks to obtain BAK1‐ or SOBIR1‐silenced plants. FoEG1 was transiently expressed in the gene‐silenced leaves. Photographs were taken 4 days after agroinfiltration. (b) and (e) Immunoblot analysis of FoEG1 protein fused with FLAG tag transiently expressed in BAK1‐ or SOBIR1‐silenced N. benthamiana leaves. The total protein loading control is shown by Ponceau S‐stained RuBisCO protein. (c) and (f) The silencing efficiencies of BAK1 and SOBIR1 were determined by quantitative reverse transcription PCR. NbActin was used as the internal reference gene. Means and standard errors were calculated from three biological replicates. Asterisks at the top of the bars indicate statistical significance (***p < .001)
2.6. An 86 amino acid fragment of FoEG1 is sufficient for its cell death‐inducing activity
A disulphide bond can be formed between two cysteine residues, which is essential for the maintenance of protein structure stability as spatial structure of a protein has a great influence on its function (Sevier & Kaiser, 2002). In this study, the FoEG1 protein contained 249 amino acids, including three cysteine residues, C33, C62, and C237. The first two (C33 and C62) are conserved in all the FoEG1 homologs (Figure S6a). According to previous studies on the structure of fungal GH12 proteins, all the GH12 proteins have these two conserved cysteine residues that can form a disulphide bond between them (Sandgren et al., 2001, 2005; Zhu et al., 2017). A disulphide bond was also predicted between the two conserved cysteine residues C33 and C62 of FoEG1 by bioinformatics analysis. To determine whether these cysteine residues affect the ability of FoEG1 to induce cell death, site‐directed mutations of all three cysteine residues in FoEG1 were carried out (Figure 6a) and the mutant proteins were then expressed in N. benthamiana by A. tumefaciens‐mediated transient expression. The results showed that no matter whether the three cysteine residues of FoEG1 were mutated individually or simultaneously, the mutant proteins could still strongly induce cell death (Figure 6b). Immunoblot analysis confirmed that all mutant proteins were successfully expressed in N. benthamiana (Figure 6c). The destruction of cysteine residues did not affect the ability of FoEG1 to induce cell death, so we speculated that there may be an important fragment in the FoEG1 protein that was the core component of its ability to regulate the induction of cell death. To test this hypothesis, truncated mutants of the N‐terminal or C‐terminal of the FoEG1 protein were generated and examined for their ability to induce cell death by agroinfiltration in the leaves of N. benthamiana. The results showed that truncated proteins FoEG1‐C1 and FoEG1‐C2 still maintained the ability to induce cell death in N. benthamiana, whereas FoEG1‐C3 did not trigger cell death (Figure 6d). The truncated mutant FoEG1‐N1 was observed to induce cell death while FoEG1‐N2 resulted in the loss of cell death‐inducing activity. Furthermore, the truncated protein FoEG1‐C4 could still induce the same cell death as the full‐length FoEG1 protein (Figure 6d). The protein fragment with 86 residues spanning amino acids 144 to 229 was identified as a fragment of FoEG1 for triggering cell death in N. benthamiana. Moreover, this 86 amino acid fragment could also induce active HR and accumulation of ROS in N. benthamiana like FoEG1. These results indicate that the 86 amino acid fragment is the core part of FoEG1‐induced cell death.
FIGURE 6.
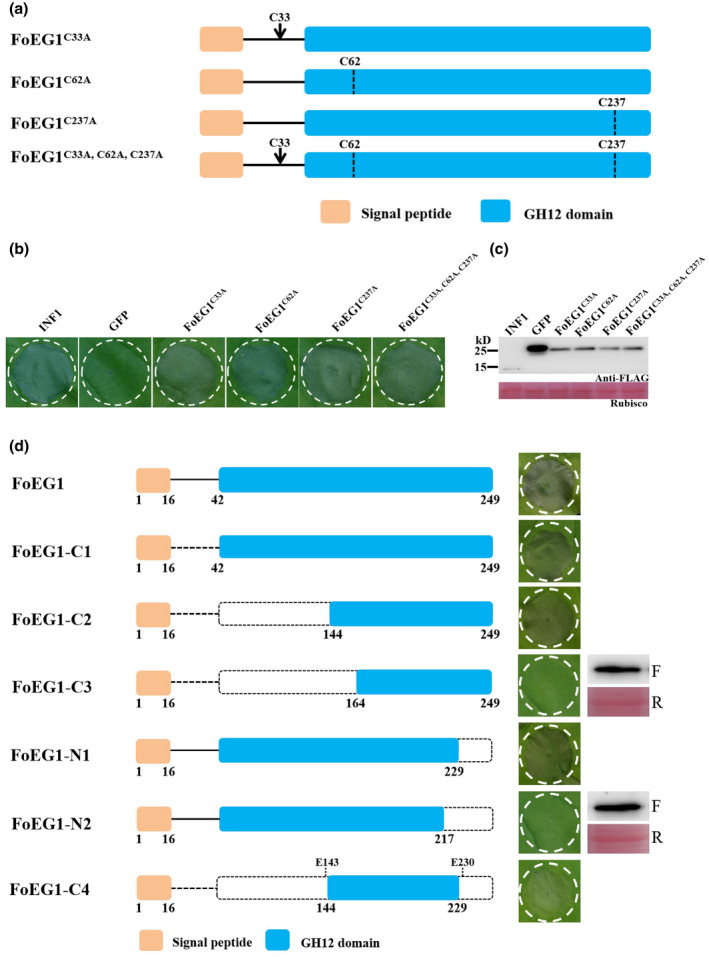
An 86 amino acid fragment of FoEG1 was sufficient to induce cell death in Nicotiana benthamiana. (a) Schematic presentation of FoEG1 mutants (replaced C33, C62, and C237 with Ala). (b) Cell death‐inducing activity of the indicated mutant proteins was determined 5 days after agroinfiltration by transient expression in 4‐week‐old N. benthamiana leaves, and INF1 and green fluorescent protein (GFP) were used as positive and negative controls, respectively. (c) Immunoblot analysis of proteins from N. benthamiana leaves transiently expressing the indicated mutant proteins. (d) Various truncated mutants of FoEG1 were constructed and transiently expressed by agroinfiltration in 4‐week‐old N. benthamiana leaves. Photographs were taken 5 days after agroinfiltration. Transient expression of FoEG1 mutants that could not cause cell death was confirmed by immunoblot analysis, with anti‐FLAG antibody labelled “F” and Ponceau S‐stained RuBisCO protein labelled “R”
2.7. FoEG1 contributes to the pathogenicity of F. oxysporum
To determine the biological function of FoEG1 in the growth, development, and pathogenicity of F. oxysporum, the expression profile of FoEG1 in the infection stage of F. oxysporum to host plants was first analysed by RT‐qPCR. The results showed that the expression of FoEG1 was significantly induced in the early stage of infection from 24 to 48 hr (Figure S8), indicating that FoEG1 may be related to the infection of F. oxysporum to the host. Further investigation into the biological role of FoEG1 was carried out. We generated gene deletion and complementation mutants (Figure S9). It was found that all mutants had no significant influence on colony growth, mycelial and conidial morphology, as well as conidial production and germination ability (Figure S9). To determine the potential role of FoEG1 in the pathogenicity of F. oxysporum, the wild‐type strains and mutant strains were inoculated onto cotton. Disease symptom observations showed that the virulence of deletion mutants to cotton was significantly reduced compared with that of the wild‐type strains (Figure 7). The complementation mutants produced by reintroducing the FoEG1 gene into the deletion mutant recovered their virulence to the host plants in a similar fashion as the strain of the wild type. However, the reintroduction of the catalytic site‐directed mutagenized gene (FoEG1 SM) to the FoEG1 deletion mutants of F. oxysporum could not restore the virulence, which indicates that the enzymatic activity of FoEG1 is required for the full virulence of F. oxysporum in cotton.
FIGURE 7.
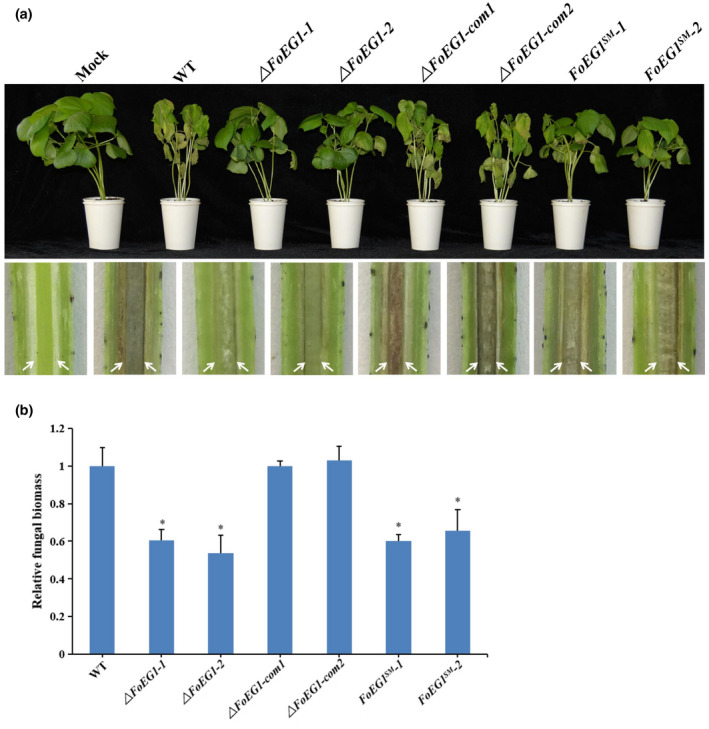
Determination of the virulence function of FoEG1 in pathogenicity of Fusarium oxysporum to cotton. (a) Phenotypes of cotton seedlings inoculated with FoEG1 deletion and complementation mutants (com). The disease symptoms 4 weeks after inoculation are shown at the top, and the discolouration of the inoculation shoot longitudinal sections is shown at the bottom. The order of the bottom pictures is the same as that of the top pictures. The white arrows indicate the location for discolouration in vascular tissues. (b) The relative fungal biomasses of the gene deletion and complementation mutants on cotton were determined by quantitative PCR. The relative fungal biomass of wild‐type (WT) strain was set as 1 as the control (*p < .05)
2.8. FoEG1 enhances plant resistance to fungal pathogens
After detecting the ROS, callose deposition, and the expression of disease resistance‐related genes, we found that FoEG1 can induce the immune response of plants. To investigate FoEG1 ability in modulating plant resistance against fungal pathogens, N. benthamiana leaves were infiltrated with purified FoEG1 protein and then inoculated with B. cinerea 24 hr after infiltration. We found that the development of lesions on the treated leaves was significantly restricted (Figure S10). Different formae speciales of F. oxysporum were used to inoculate cotton and tomato after these host plants were infiltrated with purified FoEG1 protein for 24 hr. Disease symptom observation showed that FoEG1‐treated plants exhibited greatly enhanced resistance to F. oxysporum compared to the control‐treated plants (Figure 8). The treatment with FoEG1 significantly delayed the development of disease symptoms. These results suggest that FoEG1 can increase plant resistance to fungal pathogens to some extent.
FIGURE 8.
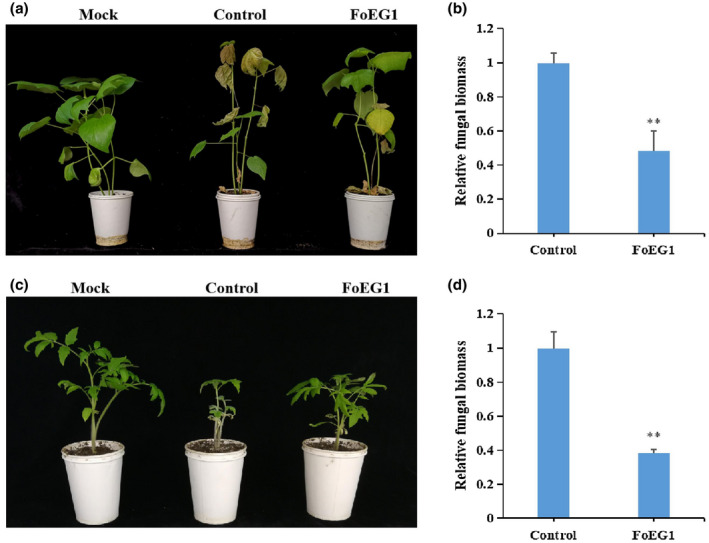
FoEG1 enhances the resistance of cotton and tomato to Fusarium oxysporum infection. (a) and (c) Phenotypes of pretreated cotton or tomato seedlings inoculated with F. oxysporum, respectively. Cotton or tomato plants were pretreated with 0.3 μM purified FoEG1 and then inoculated with the conidial suspension of F. oxysporum at 5 × 106 conidia/ml 24 hr after treatment. Phenotypes were observed and photographed at 4 weeks postinoculation. (b) and (d) The relative fungal biomasses on cotton and tomato, respectively, were determined by quantitative PCR. Asterisks at the top of the bars indicate statistical significance (**p < .01)
3. DISCUSSION
F. oxysporum is a soilborne pathogenic fungus that usually invades from root wounds of the host plant, or directly through the root epidermis and root hairs to gain entrance to the host plant to colonize the xylem vessels of roots and stems (Enkerli et al., 1999; Gui et al., 2017; Ma et al., 2015; Pietro et al., 2003; Poinssot et al., 2003; Zhang et al., 2014; Zhu et al., 2017). According to genomic annotations, the genome of F. oxysporum encodes numerous CWDEs such as glycoside hydrolases (de Sain & Rep, 2015), but many of them have not been verified for their biological functions in the interaction between F. oxysporum and hosts. Previous studies have found that GH12 family proteins are secreted proteins with CWDE activity that play important roles in the interaction between pathogens and host. However, there is no related report of GH12 family proteins in Fusarium pathogens, especially in F. oxysporum. In this study, we explored the functions of the proteins with a glycoside hydrolase family 12 domain secreted by F. oxysporum. We found five secreted proteins containing the GH12 domain by analysing the genome of F. oxysporum f. sp. vasinfectum, but only one protein among the five proteins, designated FoEG1, had the ability to trigger cell death and plant immunity by targeting plant apoplast. This protein can also play a critical role in fungal pathogenicity as a virulence factor during host infection and colonization.
The apoplastic space between plant cells is a complex battlefield where many important interactions characterizing the relationship between plants and pathogens are played out in the intercellular apoplastic space (Doehlemann & Hemetsberger, 2013; Mott et al., 2014). A PAMP VmE02 from V. mali and a cerato‐platanin (CP) protein SsCP1 from S. sclerotiorum can cause cell death with or without signal peptide (Nie et al., 2019; Yang, Tang, et al., 2018). The full‐length FoEG1, which was different from VmE02 and SsCP1, was found to induce strong cell death in this study, but lost this ability after the signal peptide was truncated. In addition, the subcellular localization showed that FoEG1 was localized in the plant apoplast. These results indicate that apoplastic space is very important for the function of FoEG1. This is similar to the protein PAMP RcCDI1 identified from ascomycete fungus Rhynchosporium commune that can also induce cell death in solanaceous plants and requires the presence of a signal peptide (Franco‐Orozco et al., 2017). The action of FoEG1 on the apoplast induced plant defence responses, including ROS, callose deposition, and up‐regulation of a series of resistance genes involved in SA‐ or JA‐signalling pathways. FoEG1 also significantly activated the up‐regulation of PTI‐related marker genes. These results suggest that FoEG1 is an extracellular cell death‐inducing protein that functions in the plant apoplast.
PAMPs can be recognized by PRRs on the surface of the plant cell membrane and plants rely on two kinds of PRRs to detect PAMPs, namely RLKs and RLPs (Liebrand et al., 2014), which can trigger immune signalling to promote plant resistance against pathogens (van der Burgh et al., 2019). There is a large number of receptor proteins in the cell membrane of plants, but due to the lack of intracellular kinase regions, these receptor proteins need to bind to specific receptor kinases in order to transmit signals to the cell to activate a downstream immune response (van der Burgh et al., 2019). Previous studies have found that plant‐derived cell surface receptors such as RLP23, ELR, and RXEG1 can specifically recognize apoplastic cell death‐inducing proteins (Albert et al., 2015; Du et al., 2015; Wang et al., 2018). For example, RXEG1, an LRR‐RLP identified from N. benthamiana through a high‐throughput VIGS screen, can specifically recognize the GH12 family cell death‐inducing protein PsXEG1 (Wang et al., 2018). However, some cell surface receptors, such as RLP23, ELR, and RXEG1, have extracellular LRRs but lack a cytoplasmic signalling domain (Li et al., 2020). The LRR receptor‐like kinases (LRR‐RLKs) BAK1 and SOBIR1 were shown to be essential for RLP23, ELR, or RXEG1‐induced cell death and immune responses that act as coreceptors to transduce signals to downstream components (Li et al., 2020). Moreover, BAK1 was required for the attenuation of EIX‐induced defence responses by the decoy receptor LeEIX1 (Bar et al., 2010), and SOBIR1 can interact with tomato RLPs SlEIX2, which was involved in plant immunity (Liebrand et al., 2013). Fungal endopolygalacturonases can be recognized by Arabidopsis thaliana LRR‐RLP AtRLP42, and Arabidopsis SOBIR1 can interact with AtRLP42 and was essential for AtRLP42‐mediated responsiveness to endopolygalacturonases (Zhang et al., 2014). Therefore, BAK1 and SOBIR1 are the central hubs of defence signal transmission, which can combine with most of the PRRs to form a complex and transmit the PAMP signals perceived by the receptors to the cell interior (Heese et al., 2007; Liebrand et al., 2013). In this study, it was found that BAK1 and SOBIR1 were required for FoEG1 to induce cell death in N. benthamiana; when BAK1 or SOBIR1 was silenced, FoEG1 could not induce cell death. Thus, FoEG1 is an apoplastic cell death‐inducing protein similar to VdEG1 (Gui et al., 2017) and VdCUT11 (Gui et al., 2018) from V. dahliae, RcCDI1 (Franco‐Orozco et al., 2017) from ascomycete fungi R. commune, and BcXYG1 (Zhu et al., 2017) from B. cinerea, which are also mediated by BAK1 and SOBIR1 to induce cell death. We speculate that the putative FoEG1 receptor in plants will form a LRR‐RLPs/SOBIR1/BAK1 complex with BAK1 and SOBIR1 after perceiving FoEG1 to complete the signal transduction in cells and then activate the downstream immune response to trigger cell death. Unlike FoEG1, VdEG3 is only mediated by BAK1 and not SOBIR1 (Gui et al., 2017), indicating that although they all function as PAMP proteins with the GH12 domain, they induce plant immunity in different ways.
Many cell death‐inducing proteins contained nonfull‐length functional regions or peptides, which play a critical role in cell death induction or plant immune response activation. For example, a 26 amino acid peptide of BcXyl1 in B. cinerea or a 63 amino acid small peptide (EG3‐MF) from the GH12 domain in VdEG3 from V. dahliae is sufficient to induce cell death in N. benthamiana (Gui et al., 2017; Yang, Yang, et al., 2018). The plant cell death‐inducing potential of elicitor EIX has been attributed to an essential five amino acid region (Rotblat et al., 2002). In this study, it was found that the 86 amino acid residues region from amino acid positions 144 to 229 produced by truncating the FoEG1 protein was sufficient to induce cell death in N. benthamiana. This characteristic is different from PsXEG1 or VdEG1. The ability of PsXEG1 and VdEG1 to cause cell death requires the integrity of their proteins, and any deletion at the C‐ or N‐terminal would render them incapable of inducing cell death (Gui et al., 2017; Ma et al., 2015). In addition, although fragments of both FoEG1 and VdEG3 can cause cell death, these segments being located between the two enzyme active sites, the sizes of these fragments are different: the small peptide of VdEG3 is shorter than that of FoEG1. It was found that the homologous fragment (FoEG1164−227) of VdEG3 in FoEG1 could not induce cell death in this study. Therefore, we speculate that although these proteins are all GH12 family proteins, their mechanism of inducing cell death may be different or the plants might have different recognition epitopes for these proteins.
Fungal pathogenicity is reliant on many secreted proteins, including CWDEs, during infection and colonization (de Sain & Rep, 2015; Jonkers et al., 2009; Tzima et al., 2011). The genome of F. oxysporum f. sp. lycopersici encodes eight different polygalacturonases, and the single deletion mutants lacking either pg1 or pgx6 show reduced polygalacturonase activity and the Δpg1Δpgx6 double mutant is significantly reduced in virulence (Bravo et al., 2016). In addition, some of the CWDEs mentioned above, such as PsXEG1, VdEG1, VdEG3, and VdPEL1, can act as virulence factors (Gui et al., 2017; Ma et al., 2015; Yang, Zhang, et al., 2018; Zhu et al., 2017). In this study, the absence of FoEG1 reduced the virulence of F. oxysporum to cotton, in a similar fashion to PsXEG1, VdPEL1, VdEG1, and VdEG3, suggesting that FoEG1 contributes to the virulence of F. oxysporum. However, not all fungal CWDEs have been conclusively shown to be involved in pathogenicity and virulence. For example, the deletion mutant strain of a pectate lyase gene PelA from Fusarium graminearum does not show attenuated virulence during the infection of wheat coleoptiles (Blum, 2017). CWDEs secreted by plant‐pathogenic fungi are important for host infection and colonization. Whilst some CWDEs such as FoEG1 appear to be critical, others may be less important or redundant because several genes serve the same function. Moreover, although FoEG1 was highly conserved between different formae speciales of F. oxysporum and other pathogenic fungi, its role in pathogenicity of other formae speciales of F. oxysporum remains to be further investigated.
In conclusion, our results indicate that FoEG1 has the ability to induce plant cell death and can also act as a PAMP to trigger plant immune responses and significantly enhance the disease resistance of plants. Moreover, FoEG1 plays a critical role in the pathogenicity of F. oxysporum f. sp. vasinfectum. Although FoEG1 may be capable of triggering plant immune responses in isolation as a transgene or a recombinant protein, the pathogen has evolved mechanisms to suppress this outcome and allow FoEG1 to function as a critically important effector during host infection and colonization.
4. EXPERIMENTAL PROCEDURES
4.1. Fungal strains and plants growth conditions
F. oxysporum f. sp. vasinfectum and F. oxysporum f. sp. lycopersici wild‐type or mutant strains were cultured and maintained on potato dextrose agar (PDA) at 25 °C in the dark. E. coli DH5α and BL21(DE3) cultured in lysogeny broth (LB) at 37 °C were used for plasmid construction and expression of recombinant protein. A. tumefaciens GV3101, cultured in LB at 28 °C, was used for A. tumefaciens‐mediated transient expression of proteins in plant leaves. N. benthamiana, N. tabacum, G. hirsutum, S. lycopersicum, G. max, and Z. mays seedlings were grown in a greenhouse at 25 °C and a 16 hr photoperiod supplemented with fluorescent lighting and 60% relative humidity.
4.2. Plasmid construction
The tested genes were amplified from F. oxysporum cDNA using the indicated primers (Table S1), including FoEG1, FOTG_11380, FOTG_10947, FOTG_12529, FOTG_17138, FoEG1 gene without signal peptide (FoEG1 ΔSP), site‐directed mutagenesis sequences of catalytic residues of FoEG1 (FoEG1 E143, FoEG1 E230, and FoEG1 E143/E230), truncated FoEG1, SP(FoEG1)‐GFP (sequence encoding the fusion protein of FoEG1 signal peptide and GFP), and PR1 SP + FoEG1 ΔSP (sequence encoding a PR1 signal peptide + FoEG1ΔSP fusion protein). For transient expression in N. benthamiana, all sequences of the above genes or fragments were cloned separately into the PVX vector pGR107. The INF1 and GFP genes were also cloned into pGR107 and used as positive and negative controls, respectively. For functional assays of signal peptides in yeast, the predicted signal peptide‐encoding sequences of FoEG1 were fused in frame to the secretion‐defective invertase gene in the vector pSUC2 (Jacobs et al., 1997) to form the recombinant construct pSUC2‐FoEG1 SP. To study the subcellular localization of FoEG1 in planta, the full‐length FoEG1, FoEG1 ΔSP, and PR1 SP + FoEG1 ΔSP were cloned into pBinGFP4 vector to generate the recombinant expression vectors pBinGFP4‐FoEG1, pBinGFP4‐FoEG1 ΔSP, and pBinGFP4‐PR1 SP + FoEG1 ΔSP. For generation of FoEG1 gene expression construct, the FoEG1 sequence without the signal peptide was cloned into pET‐32a vector. Constructs used for VIGS in N. benthamiana were generated in the pTRV2 vector as described by Liu et al. (2002) using N. benthamiana cDNA as template for gene fragment amplification. To create constructs for gene complementation, the entire coding region of FoEG1 with its native promoter and terminator was cloned into pKOV21 vector. All constructs were validated by sequencing at Sangon (Sangon Biotech).
4.3. Expression and purification of recombinant protein and enzymatic activity assays
For protein expression and purification, the expression vector pET‐32a‐FoEG1 was transformed into E. coli BL21(DE3) cells. Recombinant protein expression was induced by adding IPTG to a final concentration of 0.2 mM for 20 hr at 16 °C. Cells were collected by centrifugation at 5,000 × g for 10 min and resuspended in lysis buffer (50 mM Tris‐HCl, 200 mM NaCl, pH 8.0), followed by sonication and centrifugation at 10,000 × g for 10 min. Recombinant protein was purified by affinity chromatography using Ni‐NTA resin (GE Healthcare Life Sciences) as described in the manufacturer's instructions. Enzyme activities were measured as described previously using carboxymethylcellulose (Sigma) as the substrate (Ma et al., 2015).
4.4. Transient expression in N. benthamiana and protein infiltration assays
All transient expression vectors were transformed into A. tumefaciens GV3101 and these strains were subsequently cultured in LB at 28 °C in a shaking incubator at 200 rpm for 24 hr. The bacteria cells were harvested by centrifugation at 5,000 × g for 5 min, resuspended in MES buffer (10 mM MgCl2, 10 mM MES, 150 μM acetosyringone, pH 5.7), adjusted to a final OD600 of 1.0, and left in darkness for 3 hr at room temperature before infiltration. The A. tumefaciens cell suspension was infiltrated into 5–6‐week‐old N. benthamiana leaves using a 1‐ml syringe without a needle. Symptom development was monitored at 3–5 days after agroinfiltration for N. benthamiana. Transient protein expression in N. benthamiana was verified by western blot using anti‐FLAG antibodies.
To test the ability of recombinant proteins to induce plant cell death, different concentrations of purified protein were infiltrated into the leaves of N. benthamiana. The recombinant protein was also tested to determine its ability to induce cell death in other plants. Purified protein was infiltrated into the leaves of tobacco, cotton, tomato, soybean, and maize. Leaves were monitored and photographed at 1–5 days after infiltration. To further investigate cell death, trypan blue staining was performed as described by Qi et al. (2016). All experiments were repeated at least three times.
4.5. Yeast signal sequence trap system
The functional validation of the predicted signal peptide of FoEG1 was performed by the yeast signal sequence trap system as previously described (Jacobs et al., 1997). The pSUC2‐FoEG1 SP plasmid construct was transformed into the Saccharomyces cerevisiae strain YTK12 and the transformants were grown on CMD−W (lacking tryptophan) medium (0.67% yeast N base without amino acids, 0.075% tryptophan dropout supplement, 2% sucrose, 0.1% glucose, and 2% agar). Clones were identified by PCR using vector‐specific primers (Table S1). The positive clones were incubated on YPRAA medium (1% yeast extract, 2% peptone, 2% raffinose, and 2 μg/ml antimycin A) for invertase secretion. YTK12 transformed with pSUC2‐Avr1b SP and the empty vector pSUC2 were used as positive and negative controls, respectively (Wang et al., 2020).
4.6. Subcellular localization assay
To examine the subcellular localization of FoEG1, the constructs pBinGFP4‐FoEG1, pBinGFP4‐FoEG1 ΔSP, and pBinGFP4‐PR1 SP + FoEG1 ΔSP were transformed into A. tumefaciens GV3101 and agroinfiltrated into 4‐week‐old N. benthamiana leaves. The empty pBinGFP4 vector was used as control. At 2 days postinfiltration, the N. benthamiana leaves were harvested and imaged under a laser scanning confocal microscope (LSM 980; Zeiss) with excitation at 488 nm and emission at 510 nm for GFP. For fluorescence detection after plasmolysis, N. benthamiana leaves were treated with 1 M NaCl to induce plasmolysis before observation.
4.7. ROS activity, callose deposition, and disease resistance induction assays
To detect the ROS activity and callose deposition, the recombinant protein was infiltrated into the leaves of N. benthamiana. The accumulation of ROS in the plant leaves of N. benthamiana was detected using 3,3′‐diaminobenzidine (DAB) solution as described previously (Bindschedler et al., 2006). To detect the deposition of callose, the N. benthamiana leaves were stained with aniline blue 24 hr after infiltration as described previously (Chen et al., 2012).
To test the induction of resistance by recombinant proteins in plants, the whole leaves of 5‐week‐old N. benthamiana were infiltrated with the purified FoEG1 protein. Fungal agar plugs from 5–7‐day‐old plates of B. cinerea were inoculated into the leaves of N. benthamiana 24 hr after infiltration. The inoculated plants were then moved into a greenhouse at 25 °C with 14 hr of supplemental fluorescent light. The lesion development on the N. benthamiana leaves was evaluated by measuring the average lesion area 72 hr after inoculation. Induced systemic resistance responses in plants were also assessed in 2‐week‐old tomato and cotton seedlings. These plants were treated with 0.3 μM purified protein and then inoculated with the conidial suspension of different formae speciales of F. oxysporum at 5 × 106 conidia/ml by the root‐dip method 24 hr after treatment. The resulting disease symptoms were evaluated 4 weeks postinoculation and the discolouration in vascular tissues was observed. All the experiments were repeated three times.
4.8. RNA extraction and RT‐qPCR analysis
The leaf samples of N. benthamiana were collected 24 hr after infiltration with 0.1 μM recombinant protein. Total RNA was then extracted using an EasyPure Plant RNA Kit (TransGen Biotech) according to the manufacturer's instructions. First‐strand cDNA was synthesized from 1 μg of total RNA using the TransScript All‐in‐One First‐Strand cDNA Synthesis SuperMix (TransGen Biotech), followed by qPCR using the RealStar Green Fast Mixture (GenStar). The genes NbActin and NbEF‐1α in N. benthamiana were used as endogenous controls (Gui et al., 2017; Sainsbury & Lomonossoff, 2008). Relative expression levels were determined using the 2−ΔΔ C t method with three independent biological replicates.
4.9. Virus‐induced gene silencing in N. benthamiana
For VIGS assays in N. benthamiana, the plasmid vectors pTRV1 and pTRV2 with target genes were transformed into A. tumefaciens GV3101. The bacterial cells were harvested and resuspended in MES buffer (10 mM MgCl2, 10 mM MES, 200 μM acetosyringone, pH 5.7), adjusted to a final OD600 of 1.0, and left in darkness for 3 hr at room temperature. A. tumefaciens cell suspension carrying TRV2 vectors was mixed with A. tumefaciens cell suspension carrying TRV1 in a 1:1 ratio before infiltration. The mixed cell suspensions were then infiltrated into two or three primary leaves of N. benthamiana seedlings at the four‐leaf stage. The vectors pTRV2:PDS and pTRV2:GFP were used as controls. The silencing efficiency of the gene was validated by RT‐qPCR analysis 3 weeks after agroinfiltration. The experiments were repeated three times using at least six plants at each assay.
4.10. Generation of gene deletion and complementation mutants and virulence tests
For generation of FoEG1 deletion mutants in F. oxysporum, transformants of F. oxysporum were obtained using the polyethylene glycol (PEG)‐mediated protoplast transformation method as described previously (Zhang et al., 2017) (Figure S1). All positive transformants were identified using PCR with the corresponding primers (Table S1). To obtain FoEG1 complementation mutants, the linearized pKOV21 vector with FoEG1 and its native promoter and terminator was introduced into the FoEG1 deletion mutant using the same PEG‐mediated protoplast transformation method. To test the role of FoEG1 in the pathogenicity of F. oxysporum, cotton seedlings were inoculated with strains of the wild‐type, deletion, and complementation mutants by the root‐dip method. The disease symptoms were observed 4 weeks postinoculation and the discolouration in vascular tissues was observed. All the experiments were repeated at least three times.
4.11. Bioinformatics analysis
The identified glycoside hydrolase family 12 proteins containing a signal peptide but lacking transmembrane domains were analysed as secreted proteins by the SignalP‐5.0 Server (http://www.cbs.dtu.dk/services/SignalP/), TMHMM Server 2.0 (http://www.cbs.dtu.dk/services/TMHMM/), and SMART MODE (http://smart.embl‐heidelberg.de/smart/change_mode.pl). Identification of disulphide bonds in FoEG1 was performed using DiANNA (http://clavius.bc.edu/~clotelab/DiANNA/). Homologous sequences of FoEG1 in different formae speciales of F. oxysporum or the known GH12 domain‐containing proteins from other pathogens were obtained by querying the FoEG1 protein sequence against the NCBI database using BLAST search programs (Bailey et al., 2009). The multiple sequence alignment of FoEG1 and its homologues in different formae speciales of F. oxysporum or other pathogens was generated using the ClustalX v. 2.0 program (Larkin et al., 2007). Phylogenetic dendrograms were constructed using MEGA 6.0 software with neighbour‐joining tree method (Tamura et al., 2013).
CONFLICT OF INTEREST
The authors declare no conflict of interest.
Supporting information
FIGURE S1 Targeted gene replacement strategy of the HYG‐resistant cassette
FIGURE S2 GFP fused with the signal peptides of FoEG1 cannot induce cell death in Nicotiana benthamiana
FIGURE S3 Subcellular localization of FoEG1, FoEG1ΔSP, and PR1+FoEG1ΔSP in Nicotiana benthamiana leaves
FIGURE S4 Amino acid sequence alignment of FoEG1 proteins in different formae speciales of Fusarium oxysporum
FIGURE S5 The ability of FoEG1 and its mutants to induce cell death
FIGURE S6 Sequence similarity of FoEG1 (EXM34257.1) from Fusarium oxusporum and other known GH12 proteins in different organisms
FIGURE S7 EDS1, NDR1 and RXEF1 are not required for FoEG1 to induce cell death in Nicotiana benthamiana
FIGURE S8 Expression pattern analysis of FoEG1 during the infection stage in cotton and tomato
FIGURE S9 Growth and development characteristics of gene deletion and complementation mutants of FoEG1
FIGURE S10 The disease resistance induced by pretreatment with recombinant FoEG1 protein
TABLE S1 Primers used in this study
ACKNOWLEDGEMENTS
The authors thank Professor Guiliang Jian at the Institute of Plant Protection, Chinese Academy of Agricultural Sciences for providing the F. oxysporum f. sp. vasinfectum strain and Professor Jieyin Chen at the Institute of Plant Protection, Chinese Academy of Agricultural Sciences for providing the vectors pGR107, pSUC2, and pBinGFP4. This study was supported by the National Key Research and Development Program of China (2017YFD0200900).
Zhang L, Yan J, Fu Z, et al. FoEG1, a secreted glycoside hydrolase family 12 protein from Fusarium oxysporum, triggers cell death and modulates plant immunity. Mol Plant Pathol. 2021;22:522–538. 10.1111/mpp.13041
DATA AVAILABILITY STATEMENT
The data that support the findings of this study are available from the corresponding author upon reasonable request.
REFERENCES
- Abro, M.A. , Sun, X. , Li, X. , Jatoi, G.H. & Guo, L. (2019) Biocontrol potential of fungal endophytes against Fusarium oxysporum f. sp. cucumerinum causing wilt in cucumber. The Plant Pathology Journal, 35, 598–608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albert, I. , Böhm, H. , Albert, M. , Feiler, C.E. , Imkampe, J. , Wallmeroth, N. et al. (2015) An RLP23‐SOBIR1‐BAK1 complex mediates NLP‐triggered immunity. Nature Plants, 1, 15140. [DOI] [PubMed] [Google Scholar]
- Bailey, T.L. , Boden, M. , Buske, F.A. , Frith, M. , Grant, C.E. , Clementi, L. et al. (2009) MEME SUITE: Tools for motif discovery and searching. Nucleic Acids Research, 37, W202–W208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bar, M. , Sharfman, M. , Ron, M. & Avni, A. (2010) BAK1 is required for the attenuation of ethylene‐inducing xylanase (Eix)‐induced defense responses by the decoy receptor LeEix1. The Plant Journal, 63, 791–800. [DOI] [PubMed] [Google Scholar]
- Bindschedler, L.V. , Dewdney, J. , Blee, K.A. , Stone, J.M. , Asai, T. , Plotnikov, J. et al. (2006) Peroxidase‐dependent apoplastic oxidative burst in Arabidopsis required for pathogen resistance. Plant Journal, 47, 851–863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blum, A. (2017) Regulation and Cell Biology of Secondary Metabolite Production in Fusarium graminearum and Fusarium pseudograminearum. PhD thesis: The University of Queensland. [Google Scholar]
- Boller, T. & Felix, G. (2009) A renaissance of elicitors: Perception of microbe‐associated molecular patterns and danger signals by pattern‐recognition receptors. Annual Review of Plant Biology, 60, 379–406. [DOI] [PubMed] [Google Scholar]
- Bravo, R.G. , Di Pietro, A. & Roncero, M.I. (2016) Combined action of the major secreted exo‐ and endopolygalacturonases is required for full virulence of Fusarium oxysporum . Molecular Plant Pathology, 17, 339–353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brito, N. , Espino, J.J. & Gonzalez, C. (2006) The endo‐β‐1,4‐xylanase xyn11A is required for virulence in Botrytis cinerea . Molecular Plant‐Microbe Interactions, 19, 25–32. [DOI] [PubMed] [Google Scholar]
- van der Burgh, A.M. , Postma, J. , Robatzek, S. & Joosten, M. (2019) Kinase activity of SOBIR1 and BAK1 is required for immune signalling. Molecular Plant Pathology, 20, 410–422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camagna, M. & Takemoto, D. (2018) Hypersensitive response in plants. Encyclopedia of Life Sciences, 10.1002/9780470015902.a0020103.pub2 [DOI] [Google Scholar]
- Chen, M. , Zeng, H. , Qiu, D. , Guo, L. , Yang, X. , Shi, H. et al. (2012) Purification and characterization of a novel hypersensitive response‐inducing elicitor from Magnaporthe oryzae that triggers defense response in rice. PLoS One, 7, e37654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Ovidio, R. , Mattei, B. , Roberti, S. & Bellincampi, D. (2004) Polygalacturonases, polygalacturonase‐inhibiting proteins and pectic oligomers in plant–pathogen interactions. Biochimica et Biophysica Acta, 1696, 237–244. [DOI] [PubMed] [Google Scholar]
- Doehlemann, G. & Hemetsberger, C. (2013) Apoplastic immunity and its suppression by filamentous plant pathogens. New Phytologist, 198, 1001–1016. [DOI] [PubMed] [Google Scholar]
- Du, J. , Verzaux, E. , Chaparro‐Garcia, A. , Bijsterbosch, G. , Keizer, L.C.P. , Zhou, J.I. et al. (2015) Elicitin recognition confers enhanced resistance to Phytophthora infestans in potato. Nature Plants, 1, 15034. [DOI] [PubMed] [Google Scholar]
- Edel‐Hermann, V. & Lecomte, C. (2019) Current status of Fusarium oxysporum formae speciales and races. Phytopathology, 109, 512–530. [DOI] [PubMed] [Google Scholar]
- Enkerli, J. , Felix, G. & Boller, T. (1999) The enzymatic activity of fungal xylanase is not necessary for its elicitor activity. Plant Physiology, 121, 391–397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felix, G. , Duran, J.D. , Volko, S. & Boller, T. (1999) Plants have a sensitive perception system for the most conserved domain of bacterial flagellin. The Plant Journal, 18, 265–276. [DOI] [PubMed] [Google Scholar]
- Fliegmann, J. , Mithofer, A. , Wanner, G. & Ebel, J. (2004) An ancient enzyme domain hidden in the putative β‐glucan elicitor receptor of soybean may play an active part in the perception of pathogen‐associated molecular patterns during broad host resistance. Journal of Biological Chemistry, 279, 1132–1140. [DOI] [PubMed] [Google Scholar]
- Franco‐Orozco, B. , Berepiki, A. , Ruiz, O. , Gamble, L. , Griffe, L.L. , Wang, S. et al. (2017) A new proteinaceous pathogen‐associated molecular pattern (PAMP) identified in ascomycete fungi induces cell death in Solanaceae. New Phytologist, 214, 1657–1672. [DOI] [PubMed] [Google Scholar]
- Gui, Y. , Chen, J. , Zhang, D. , Li, N. , Li, T. , Zhang, W. et al. (2017) Verticillium dahliae manipulates plant immunity by glycoside hydrolase 12 proteins in conjuction with carbohydrate‐binding module 1: GH12 proteins and CBM1 manipulate plant immunity. Environmental Microbiology, 19, 1914–1932. [DOI] [PubMed] [Google Scholar]
- Gui, Y.J. , Zhang, W.Q. , Zhang, D.D. , Zhou, L. , Short, D. , Wang, J. et al. (2018) A Verticillium dahliae extracellular cutinase modulates plant immune responses. Molecular Plant‐Microbe Interactions, 31, 260–273. [DOI] [PubMed] [Google Scholar]
- Heese, A. , Hann, D.R. , Gimenez‐Ibanez, S. , Jones, A.M.E. , He, K. , Li, J. et al. (2007) The receptor‐like kinase SERK3/BAK1 is a central regulator of innate immunity in plants. Proceedings of the National Academy of Sciences of the United States of America, 104, 12217–12222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs, K.A. , Collins‐Racie, L.A. , Colbert, M. , Duckett, McKeough , Golden‐Fleet, M. , Kelleher, K. et al. (1997) A genetic selection for isolating cDNAs encoding secreted proteins. Gene, 198, 289–296. [DOI] [PubMed] [Google Scholar]
- Jonkers, W. , Rodrigues, C. & Rep, M. (2009) Impaired colonization and infection of tomato roots by the Δfrp1 mutant of Fusarium oxysporum correlates with reduced CWDE gene expression. Molecular Plant‐Microbe Interactions, 22, 507–518. [DOI] [PubMed] [Google Scholar]
- Jürg, E. , Felix, G. & Boller, T. (1999) The enzymatic activity of fungal xylanase is not necessary for its elicitor activity. Plant Physiology, 121, 391–397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazan, K. & Gardiner, D.M. (2018) Fusarium crown rot caused by Fusarium pseudograminearum in cereal crops: Recent progress and future prospects. Molecular Plant Pathology, 19, 1547–1562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubicek, C.P. , Starr, T.L. & Glass, N.L. (2014) Plant cell wall‐degrading enzymes and their secretion in plant‐pathogenic fungi. Annual Review of Phytopathology, 52, 427–451. [DOI] [PubMed] [Google Scholar]
- Kunze, G. , Zipfel, C. , Robatzek, S. , Niehaus, K. , Boller, T. & Felix, G. (2004) The N terminus of bacterial elongation factor Tu elicits innate immunity in Arabidopsis plants. The Plant Cell, 16, 3496–3507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai, M.W. & Liou, R.F. (2018) Two genes encoding GH10 xylanases are essential for the virulence of the oomycete plant pathogen Phytophthora parasitica . Current Genetics, 64, 931–943. [DOI] [PubMed] [Google Scholar]
- Larkin, M.A. , Blackshields, G. , Brown, N.P. , Chenna, R. , McGettigan, P.A. , McWilliam, H. et al. (2007) Clustal W and Clustal X version 2.0. Bioinformatics, 23, 2947–2948. [DOI] [PubMed] [Google Scholar]
- Li, Y.A. , Han, Y. , Qu, M. , Chen, J. , Chen, X. , Geng, X. et al. (2020) Apoplastic cell death‐inducing proteins of filamentous plant pathogens: Roles in plant–pathogen interactions. Frontiers in Genetics, 11, 661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liebrand, T.W.H. , van den Berg, G.C.M. , Zhang, Z. , Smit, P. , Cordewener, J.H.G. , America, A.H.P. et al. (2013) Receptor‐like kinase SOBIR1/EVR interacts with receptor‐like proteins in plant immunity against fungal infection. Proceedings of the National Academy of Sciences of the United States of America, 110, 10010–10015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liebrand, T.W. , van den Burg, H.A. & Joosten, M.H. (2014) Two for all: Receptor‐associated kinases SOBIR1 and BAK1. Trends in Plant Science, 19, 123–132. [DOI] [PubMed] [Google Scholar]
- Liu, Y. , Schiff, M. , Marathe, R. & Dinesh‐Kumar, S.P. (2002) Tobacco Rar1, EDS1 and NPR1/NIM1 like genes are required for N‐mediated resistance to tobacco mosaic virus. The Plant Journal, 30, 415–429. [DOI] [PubMed] [Google Scholar]
- Ma, Z. , Song, T. , Zhu, L. , Ye, W. , Wang, Y. , Shao, Y. et al. (2015) A Phytophthora sojae glycoside hydrolase 12 protein is a major virulence factor during soybean infection and is recognized as a PAMP. The Plant Cell, 27, 2057–2072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGovern, R. (2015) Management of tomato diseases caused by Fusarium oxysporum . Crop Protection, 73, 78–92. [Google Scholar]
- Mott, G.A. , Middleton, M.A. , Desveaux, D. & Guttman, D.S. (2014) Peptides and small molecules of the plant–pathogen apoplastic arena. Frontiers in Plant Science, 5, 677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nie, J. , Yin, Z. , Li, Z. , Wu, Y. & Huang, L. (2019) A small cysteine‐rich protein from two kingdoms of microbes is recognized as a novel pathogen‐associated molecular pattern. New Phytologist, 222, 995–1011. [DOI] [PubMed] [Google Scholar]
- Nurnberger, T. , Brunner, F. , Kemmerling, B. & Piater, L. (2004) Innate immunity in plants and animals: Striking similarities and obvious differences. Immunological Reviews, 198, 249–266. [DOI] [PubMed] [Google Scholar]
- Pietro, A.D. , Madrid, M.P. , Caracuel, Z. , Delgado‐Jarana, J. & Roncero, M.I. (2003) Fusarium oxysporum: Exploring the molecular arsenal of a vascular wilt fungus. Molecular Plant Pathology, 4, 315–325. [DOI] [PubMed] [Google Scholar]
- Poinssot, B. , Vandelle, E. , Bentéjac, M. , Adrian, M. , Levis, C. , Brygoo, Y. et al. (2003) The Endopolygalacturonase 1 from Botrytis cinerea activates grapevine defense reactions unrelated to its enzymatic activity. Molecular Plant‐Microbe Interactions, 16, 553–564. [DOI] [PubMed] [Google Scholar]
- Prade, R.A. , Zhan, D. , Ayoubi, P. & Mort, A.J. (1999) Pectins, pectinases and plant–microbe interactions. Biotechnology and Genetic Engineering Reviews, 16, 361–391. [DOI] [PubMed] [Google Scholar]
- Qi, M. , Link, T.I. , Müller, M. , Hirschburger, D. , Pudake, R.N. , Pedley, K.F. et al. (2016) A small cysteine‐rich protein from the Asian soybean rust fungus, Phakopsora pachyrhizi, suppresses plant immunity. PLoS Pathogens, 12, e1005827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quoc, N.B. & Chau, N. (2017) The role of cell wall degrading enzymes in pathogenesis of Magnaporthe oryzae . Current Protein & Peptide Science, 18, 1019–1034. [DOI] [PubMed] [Google Scholar]
- Rajeshwari, R. , Jha, G. & Sonti, R.V. (2005) Role of an in planta‐expressed xylanase of Xanthomonas oryzae pv. oryzae in promoting virulence on rice. Molecular Plant‐Microbe Interactions, 18, 830–837. [DOI] [PubMed] [Google Scholar]
- Ron, M. & Avni, A. (2004) The receptor for the fungal elicitor ethylene‐inducing xylanase is a member of a resistance‐like gene family in tomato. The Plant Cell, 16, 1604–1615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rotblat, B. , Enshell‐Seijffers, D. , Gershoni, J.M. , Schuster, S. & Avni, A. (2002) Identification of an essential component of the elicitation active site of the EIX protein elicitor. The Plant Journal, 32, 1049–1055. [DOI] [PubMed] [Google Scholar]
- de Sain, M. & Rep, M. (2015) The role of pathogen‐secreted proteins in fungal vascular wilt diseases. International Journal of Molecular Sciences, 16, 23970–23993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sainsbury, F. & Lomonossoff, G.P. (2008) Extremely high‐level and rapid transient protein production in plants without the use of viral replication. Plant Physiology, 148, 1212–1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandgren, M. , Shaw, A. , Ropp, T.H. , Wu, S. , Bott, R. , Cameron, A.D. et al. (2001) The X‐ray crystal structure of the Trichoderma reesei family 12 endoglucanase 3, Cel12A, at 1.9 A resolution. Journal of Molecular Biology, 308, 295–310. [DOI] [PubMed] [Google Scholar]
- Sandgren, M. , Ståhlberg, J. & Mitchinson, C. (2005) Structural and biochemical studies of GH family 12 cellulases: Improved thermal stability, and ligand complexes. Progress in Biophysics and Molecular Biology, 89, 246–291. [DOI] [PubMed] [Google Scholar]
- Sevier, C.S. & Kaiser, C.A. (2002) Formation and transfer of disulphide bonds in living cells. Nature Reviews Molecular Cell Biology, 3, 836–847. [DOI] [PubMed] [Google Scholar]
- Shinya, T. , Nakagawa, T. , Kaku, H. & Shibuya, N. (2015) Chitin‐mediated plant‐fungal interactions: Catching, hiding and handshaking. Current Opinion in Plant Biology, 26, 64–71. [DOI] [PubMed] [Google Scholar]
- Tamura, K. , Stecher, G. , Peterson, D. , Filipski, A. & Kumar, S. (2013) MEGA6: Molecular evolutionary genetics analysis version 6.0. Molecular Biology and Evolution, 30, 2725–2729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzima, A.K. , Paplomatas, E.J. , Rauyaree, P. , Ospina‐Giraldo, M.D. & Kang, S. (2011) VdSNF1, the sucrose nonfermenting protein kinase gene of Verticillium dahliae, is required for virulence and expression of genes involved in cell‐wall degradation. Molecular Plant‐Microbe Interactions, 24, 129–142. [DOI] [PubMed] [Google Scholar]
- Vethavalli, S. & Sudha, S.S. (2012) In vitro and in silico studies on biocontrol agent of bacterial strains against Fusarium oxysporum f. sp. lycopersici . Research Journal of Biotechnology, 3, 22–31. [Google Scholar]
- Wang, D. , Tian, L. , Zhang, D.D. , Song, J. , Song, S.S. , Yin, C.M. et al. (2020) Functional analyses of small secreted cysteine‐rich proteins identified candidate effectors in Verticillium dahliae . Molecular Plant Pathology, 21, 667–685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, Y. , Xu, Y. , Sun, Y. , Wang, H. , Qi, J. , Wan, B. et al. (2018) Leucine‐rich repeat receptor‐like gene screen reveals that Nicotiana RXEG1 regulates glycoside hydrolase 12 MAMP detection. Nature Communications, 9, 594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, Y. , Zhou, H. , Zhao, G. , Yang, J. , Luo, Y. , Sun, S. et al. (2020) Genetical and O‐glycoproteomic analyses reveal the roles of three protein O‐mannosyltransferases in phytopathogen Fusarium oxysporum f.sp. cucumerinum . Fungal Genetics and Biology, 134, 103285. [DOI] [PubMed] [Google Scholar]
- Yang, G. , Tang, L. , Gong, Y. , Xie, J. , Fu, Y. , Jiang, D. et al. (2018) A cerato‐platanin protein SsCP1 targets plant PR1 and contributes to virulence of Sclerotinia sclerotiorum . New Phytologist, 217, 739–755. [DOI] [PubMed] [Google Scholar]
- Yang, Y. , Yang, X. , Dong, Y. & Qiu, D. (2018) The Botrytis cinerea xylanase BcXyl1 modulates plant immunity. Frontiers in Microbiology, 9, 2535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, Y. , Zhang, Y. , Li, B. , Yang, X. , Dong, Y. & Qiu, D. (2018) A Verticillium dahliae pectate lyase induces plant immune responses and contributes to virulence. Frontiers in Plant Science, 9, 1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin, Z. , Wang, N. , Pi, L. , Li, L. , Duan, W. , Wang, X. et al. (2020) Nicotiana benthamiana LRR‐RLP NbEIX2 mediates the perception of an EIX‐like protein from Verticillium dahliae . Journal of Integrative Plant Biology. 10.1111/jipb.13031 [DOI] [PubMed] [Google Scholar]
- Yu, C. , Li, T. , Shi, X. , Saleem, M. , Li, B. , Liang, W. et al. (2018) Deletion of endo‐β‐1,4‐xylanase VmXyl1 impacts the virulence of Valsa mali in apple tree. Frontiers in Plant Science, 9, 663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, Y. , Xiao, J. , Du, J. , Yang, Y. , Bi, C. & Qing, L. (2016) Disruption of the gene encoding endo‐β‐1,4‐xylanase affects the growth and virulence of Sclerotinia sclerotiorum . Frontiers in Microbiology, 7, 1787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, L. , Kars, I. , Essenstam, B. , Liebrand, T.W.H. , Wagemakers, L. , Elberse, J. et al. (2014) Fungal endopolygalacturonases are recognized as microbe‐associated molecular patterns by the Arabidopsis receptor‐like protein RESPONSIVENESS to BOTRYTIS POLYGALACTURONASES1. Plant Physiology, 164, 352–364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, Y. , Gao, X. , Sun, M. , Liu, H. & Xu, J.R. (2017) The FgSRP1 SR‐protein gene is important for plant infection and pre‐mRNA processing in Fusarium graminearum . Environmental Microbiology, 19, 4065–4079. [DOI] [PubMed] [Google Scholar]
- Zhu, W. , Ronen, M. , Gur, Y. , Minz‐Dub, A. , Masrati, G. , Ben‐Tal, N. et al. (2017) BcXYG1, a secreted xyloglucanase from Botrytis cinerea, triggers both cell death and plant immune responses. Plant Physiology, 175, 438–456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zipfel, C. (2009) Early molecular events in PAMP‐triggered immunity. Current Opinion in Plant Biology, 12, 414–420. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
FIGURE S1 Targeted gene replacement strategy of the HYG‐resistant cassette
FIGURE S2 GFP fused with the signal peptides of FoEG1 cannot induce cell death in Nicotiana benthamiana
FIGURE S3 Subcellular localization of FoEG1, FoEG1ΔSP, and PR1+FoEG1ΔSP in Nicotiana benthamiana leaves
FIGURE S4 Amino acid sequence alignment of FoEG1 proteins in different formae speciales of Fusarium oxysporum
FIGURE S5 The ability of FoEG1 and its mutants to induce cell death
FIGURE S6 Sequence similarity of FoEG1 (EXM34257.1) from Fusarium oxusporum and other known GH12 proteins in different organisms
FIGURE S7 EDS1, NDR1 and RXEF1 are not required for FoEG1 to induce cell death in Nicotiana benthamiana
FIGURE S8 Expression pattern analysis of FoEG1 during the infection stage in cotton and tomato
FIGURE S9 Growth and development characteristics of gene deletion and complementation mutants of FoEG1
FIGURE S10 The disease resistance induced by pretreatment with recombinant FoEG1 protein
TABLE S1 Primers used in this study
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.


