Abstract
Pathogens deploy a wide range of pathogenicity factors, including a plethora of proteases, to modify host tissue or manipulate host defences. Metalloproteases (MPs) have been implicated in virulence in several animal and plant pathogens. Here we investigated the repertoire of MPs in 46 stramenopile species including 37 oomycetes, 5 diatoms, and 4 brown algae. Screening their complete proteomes using hidden Markov models (HMMs) trained for MP detection resulted in over 4,000 MPs, with most species having between 65 and 100 putative MPs. Classification in clans and families according to the MEROPS database showed a highly diverse MP repertoire in each species. Analyses of domain composition, orthologous groups, distribution, and abundance within the stramenopile lineage revealed a few oomycete‐specific MPs and MPs potentially related to lifestyle. In‐depth analyses of MPs in the plant pathogen Phytophthora infestans revealed 91 MPs, divided over 21 protein families, including 25 MPs with a predicted signal peptide or signal anchor. Expression profiling showed different patterns of MP gene expression during pre‐infection and infection stages. When expressed in leaves of Nicotiana benthamiana, 12 MPs changed the sizes of lesions caused by inoculation with P. infestans; with 9 MPs the lesions were larger, suggesting a positive effect on the virulence of P. infestans, while 3 MPs had a negative effect, resulting in smaller lesions. To the best of our knowledge, this is the first systematic inventory of MPs in oomycetes and the first study pinpointing MPs as potential pathogenicity factors in Phytophthora.
Keywords: MEROPS, oomycete, peptidase, plant pathogen, secreted metalloproteases, stramenopile, virulence
Proteome mining and comparative analyses uncovered a large variety of metalloproteases (MPs) in oomycetes. Functional in planta assays pinpointed 12 MPs as potential pathogenicity factors in Phytophthora infestans.

1. INTRODUCTION
Proteases are enzymes that catalyse the breakdown of proteins into smaller polypeptides or single amino acids and play important roles in numerous biochemical processes. They are crucial components of the molecular and cellular machinery in all organisms, not only within the organism itself but also in symbiotic interactions with other organisms. Pathogenic microbes, for example, often produce a variety of proteases to degrade host tissue or to disrupt or modify host defence to create suitable conditions for successful colonization (Figaj et al., 2019; Marshall et al., 2017).
Proteases, also referred to as peptidases, are divided in seven major groups according to their catalytic types: aspartic, cysteine, glutamic, serine, and threonine proteases, metalloproteases (MPs), and asparagine lyases. MEROPS is a peptidase database currently containing over 4,000 entries that groups proteases into clans and families based on amino acid sequence similarity (Rawlings et al., 2018). As such, the MEROPS classification is an excellent tool for categorizing proteases deduced from genome annotations and predicting functions of validated proteases.
MPs function by virtue of a divalent metal cation positioned at their catalytic site. The majority is zinc‐dependent, which is accommodated by two zinc‐binding histidine residues in the HEXXH motif. In animals, MPs are implicated to play a role in receptor modification via a process known as ectodomain shedding. These so‐called sheddases belong to the A Disintegrin and Metalloprotease (ADAM) and Matrix Metalloprotease (MMP) families (Lichtenthaler et al., 2018). They shed the extracellular part of membrane proteins, leading to loss of function of the truncated receptor, alteration of downstream signalling, or loss or gain of function of the released extracellular part (Sanderson et al., 2006).
Microbes also produce MPs and can exploit these as pathogenicity factors (Deu, 2017; Miyoshi & Shinoda, 2000). For example, leishmanolysin GP63, a zinc‐binding MP in protozoan Leishmania species, cleaves protein‐tyrosine phosphatases in macrophages to enhance migration of the pathogen through the extracellular matrix (Gomez et al., 2009). Deletion of GP63 in Leishmania major results in reduced lesion formation in mice (Joshi et al., 2002). In the mammalian pathogen Vibrio cholerae, the extracellular zinc‐dependent MP hemagglutinin, also known as vibriolysin, is involved in modification of toxins, degradation of mucus barriers, and disruption of host intestinal junctions (Benitez & Silva, 2016). In a nematicidal Bacillus thuringiensis strain the MP ColB has a role in the colonization of nematodes (Peng et al., 2015). The fungal pathogen Aspergillus fumigatus uses its MP Mep1p to cleave major complement components, thereby facilitating early immune evasion by disarming defence in the human host (Shende et al., 2018).
In plant pathogens, MPs have been similarly implicated in pathogenicity (Franceschetti et al., 2017). For example, the rice blast fungus Pyricularia oryzae (Magnaporthe oryzae) has an MP‐like protein that is the avirulence factor AVR‐Pita1 and is as such recognized by the rice resistance protein Pi‐ta (Jia et al., 2016). Moreover, comparative genome analyses revealed a novel Pyricularia‐specific MP family that is expanded in several Pyricularia species, among which are potential effector proteins (Gómez Luciano et al., 2019). The fungus Fusarium oxysporum f. sp. lycopersici produces the MP Mep1, which acts in concert with the serine protease Sep1 to cleave chitinases produced by the host as part of its defence machinery. Both FoMep1 and FoSep1 are required for full virulence of F. oxysporum on tomato (Jashni et al., 2015). In the fire blight bacterium Pectobacterium amylovora the lack of PrtA, a secreted extracellular zinc‐binding MP, as well as inhibition of PrtA activity with EDTA, results in reduced host‐plant colonization (Zhang et al., 1999). Similarly, PrtA mutants of Burkholderia glumae, a bacterial pathogen on rice, show significantly reduced virulence (Lelis et al., 2019).
Here we focus on MPs in oomycetes, a diverse class of eukaryotic microbes that belong to the stramenopiles, a lineage also containing diatoms and brown algae. Most oomycetes described so far are plant pathogens, including Phytophthora and Pythium species and the downy mildews (Kamoun et al., 2015). The latter are obligate biotrophs while Phytophthora and Pythium species have a (hemi)biotrophic or necrotrophic lifestyle and can be cultured in vitro. Some Pythium species are pathogens of animals or parasitize on other microbes (mycoparasites), while Saprolegnia species are exclusively animal pathogens (Jiang et al., 2013).
Phytophthora infestans, the causal agent of late blight in potato and tomato, is a hemibiotrophic pathogen that depends on living host tissue for successful infection. Consequently, avoidance of immune recognition is crucial for subsequent host colonization. P. infestans has over 500 genes encoding cytoplasmic effectors and around 200 encoding apoplastic effectors (Haas et al., 2009), several of which contribute to virulence by suppressing host defence (Whisson et al., 2016). Apart from these effector genes, P. infestans has many other putative pathogenicity genes, including genes encoding hydrolases, lipases, proteases, and protease inhibitors (Haas et al., 2009). Examples are the carbohydrate‐active enzymes (CAZymes) (Ospina‐Giraldo et al., 2010), phospholipase Ds (Meijer & Govers, 2006; Meijer et al., 2011, 2019), and aspartic proteases (APs) (Kay et al., 2011; Schoina et al., 2019). One of the APs, PiAP5, has a combination of two consecutive protein domains, referred to as bigram, that is only found in oomycetes. The bigram in PiAP5 consists of an AP domain and a C‐terminal G‐protein‐coupled receptor domain and is preceded by an N‐terminal signal peptide (SP) (Kay et al., 2011). Such a peculiar combination of two consecutive protein domains is a typical example of a Phytophthora‐ or oomycete‐specific bigram (Seidl et al. 2011; Van den Hoogen et al., 2018; Van den Hoogen & Govers, 2018). Phytophthora species possess a relatively high proportion of multidomain proteins with unique bigrams, a feature that presumably provides novel functionality to proteins, and many of these are predicted to be secreted (Seidl et al., 2011). Proteomic analyses of the P. infestans secretome detected several proteases, including MPs, in the extracellular medium (Meijer et al., 2014), collectively suggesting that the pathogen secretes a suite of enzymes that probably play a role in the host–pathogen interface.
While MPs have been shown to play various roles in cellular processes and in pathogenicity, they have hardly been studied in oomycetes. The aims of this study were (a) to make an inventory of the full repertoire of MPs in stramenopiles, and (b) to investigate the potential role of MPs in the virulence of P. infestans. We identified 4,140 putative MPs and divided these into clans and families according to the MEROPS classification. Subsequently, we clustered the MPs in orthologous groups (OGs) and studied the MP distribution and abundance within the stramenopile and oomycete lineages with the aim to identify features potentially related to pathogenicity or lifestyle. We then focused on the MP repertoire of P. infestans and consulted transcriptome data to determine MP expression profiles during asexual development and in planta growth. Finally, we selected P. infestans MPs predicted to be secreted and tested these for their ability to promote or inhibit virulence. This study reveals that oomycetes have a diverse repertoire of MPs and highlights a few MPs that seem to play a role in pathogenicity of the late blight pathogen P. infestans.
2. RESULTS
2.1. Oomycetes have a large diversity of MPs
To gain insight into the MP repertoire and diversity in oomycetes, we identified putative MPs using a bioinformatics procedure with the MEROPS database as the starting point (Figure 1). MEROPS (release 12.1) recognizes 76 MP families (coded as M followed by a number) and 16 clans (a two‐letter code starting with M) (Rawlings et al., 2018). We used all peptidase sequences retrieved from MEROPS to train profile hidden Markov models (HMMs) per MEROPS (sub)family. The HMMs were used to scan the complete proteomes of 46 stramenopiles, including 37 oomycetes, 5 diatoms, and 4 algae (Table S1). Matching protein domains were realigned and used to train stramenopile‐specific MP HMMs. In addition, Pfam metallopeptidase domain HMMs were retrieved (Finn et al., 2016) and all proteins matching these Pfam and/or MEROPS HMMs were considered putative MPs. In total, 4,140 MPs were identified, ranging from 50 to 265 MPs per species (average, 90 MPs), with 70% of the species having between 65 and 100 putative MPs (Figure 2; Table S1). The 4,140 MPs account for 0.60% of the sum of total proteins in the 46 species. At the species level this percentage varies although it is not necessarily proportional to the variation in absolute number of MPs in each species (Table S1). Even though we attempted to design the most reliable computational strategy, we can neither guarantee that we were able to extract all MPs encoded in the 46 genomes, nor exclude false positives.
FIGURE 1.
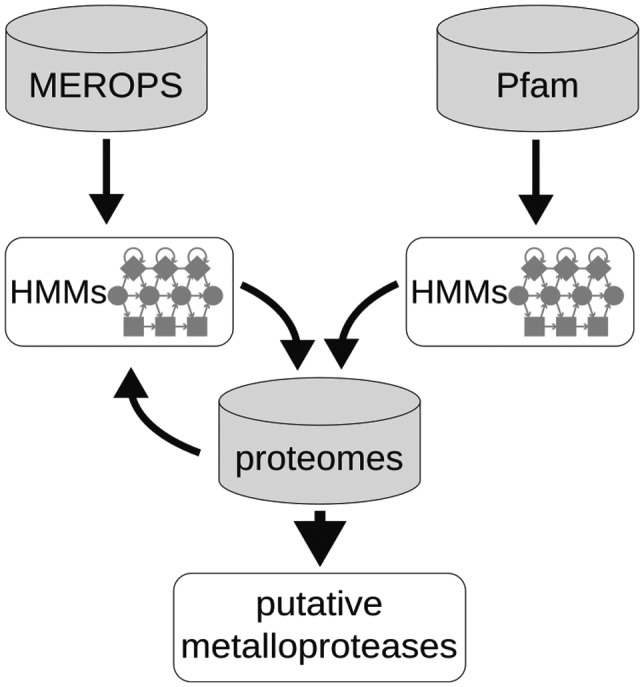
Pipeline for the identification of putative metalloproteases in stramenopiles
FIGURE 2.
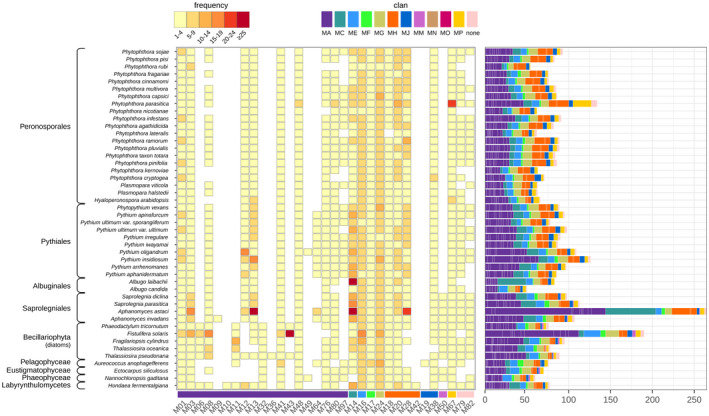
Metalloproteases (MPs) in stramenopiles. MP families (lower bar) were identified in 46 stramenopiles, including 37 oomycetes, and divided over various taxonomic lineages (left). Approximate numbers of MP family members in each species are shown by frequency (centre) and in bar charts (right). MP families are categorized in colour‐coded clans (top right)
The 4,140 stramenopile MPs can be assigned to 9 clans and 33 families (Figure 2), of which 19 (58%) belong to clan MA with the characteristic catalytic HEXXH motif. The other eight clans are represented by up to four families, while families M79 and M82 are not assigned to a clan. Within the oomycetes, the overall presence/absence pattern of MP families is similar for all species, with a few remarkable exceptions. For instance, the M64 family seems to be specific for Pythiaceae and the two Aphanomyces species, whereas M97 is present in nearly all Pythiaceae and most Peronosporaceae but absent in the Albuginales and Saprolegniales. In contrast, M50 is restricted to the Saprolegniales. Albugo laibachii and Aphanomyces astaci both have an exceptionally high number of MPs categorized as M14, while Phytophthora parasitica has a relatively large repertoire of M48, M80, and M67 family members. Initial screenings of the proteomes revealed substantial variation in assembly quality, and in some species even traces of bacterial contamination. This was most prevalent in Phytophthora rubi. Sequences with high identity scores to nonstramenopile sequences were not included in the analyses.
When focusing on nonoomycete species, we observed a similar diversity in MPs as in oomycetes. However, three families are restricted to the diatoms and algae included in this study (i.e., M06, M11, and M32) and one is even diatom‐specific (i.e., M43). Moreover, the M50 family is clearly not limited to the Saprolegniales but is more widely distributed within the stramenopiles. The few families that were only found in a single species (M09, M10, M19, M36, and M42) or limited to two species (M49) were subjected to a closer inspection. Unlike the situation in P. rubi, there were no indications for contamination in the genome assemblies. However, to what extent these MPs are truly species‐specific is questionable given the limited number of diatoms, algae, and Aphanomyces species included in this study.
2.2. Clustering of stramenopile MPs in 85 orthologous groups
To investigate the sequence similarity between MPs, we used OrthoFinder to group all stramenopile MP sequences into OGs, with the exception of those in five MP families that were unique to a single species. OrthoFinder clustered MPs in 85 OGs, with sizes ranging from 2 to 285 MPs per OG (average, 49) (Figure 3; Table S1). Nine MP families each cluster in a single OG, suggesting limited sequence diversity within these MP families, while 19 MP families are each divided over two or more OGs, pointing to increased sequence divergence within these families. Families M16 (MPP β‐subunit) and M24 (aminopeptidases) seem to be the most diverse, each spanning eight OGs.
FIGURE 3.
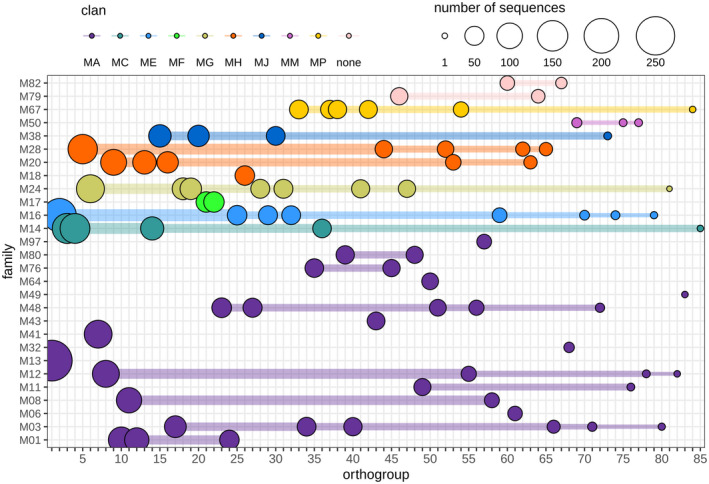
Clustering of stramenopile metalloproteases (MPs) in orthogroups. MP family members (y axis) are clustered in one (one dot) or more (dots connected by horizontal lines) orthogroups. The sizes of the coloured dots correlate with the number of MP sequences per orthogroup. The first orthogroup is the largest with 285 members. For gene IDs clustered per orthogroup see Table S1
We then analysed the distribution of MPs of the same OG over the different species, which showed that two OGs are species‐specific, containing MPs from just one species, while other OGs comprise MPs from either oomycete species or nonoomycete species (Figure S1). In contrast, 14 OGs contain MPs from all (i.e., 46) or nearly all (45) species, with one or more MPs occurring in each of the analysed species. Anticipating that a poorly assembled or misannotated genome can readily lead to missing MPs we define these 14 OGs as the core MP repertoire of stramenopiles comprising MPs from 10 distinct MEROPS families. The largest MP family present in the data set is M13 with 285 members from 42 species that all cluster in the largest OG (Figure 3; Figure S1). Family M13 mainly consists of single‐domain proteins homologous to the human peptidase neprilysin, an MP that cleaves peptides and is known to inactivate several peptide hormones (Rawlings & Salvesen, 2013). M13 is present in all 37 oomycetes and most species encode up to five copies. Strikingly, this MP is more abundant in animal pathogens, including A. astaci (91 copies), Pythium insidiosum (20), and Saprolegnia parasitica (11).
2.3. Stramenopile MPs have diverse domain architectures and N‐terminal targeting sequences
To gain further insight into the functional diversity of stramenopile MPs, we analysed their domain composition and more specifically searched for the occurrence of MPs with distinct domain combinations. This showed that 37% of the MPs in our data set contain at least one accessory domain next to the protease domain. In those multidomain MPs, we detected 204 different Pfam domains in addition to the MP domains (Figure S2; Table S1), and more than 250 bigrams, of which a vast majority occurs in only one or a few species (Figure S3; Table S1). A small proportion of bigrams is more common (around 10%) and present in over 80% of the analysed stramenopiles. The three most ubiquitous bigrams, occurring in nearly all species, consist of (a) the “CAAX‐prenyl protease N‐terminal, five transmembrane helices” domain (Pfam ID PF16491) and an M48 peptidase domain, (b) the “ATPase associated with various cellular activities (AAA)” domain (PF00004) and an M41 peptidase, and (c) the “mitochondrial processing peptidase (MPP) β‐subunit” domain of family M16B combined with a “Middle or third domain of peptidase_M16” (PF16187).
The number of MPs with distinct domain combinations is on average 34 per species and ranges from 21 in Hyaloperonospora arabidopsidis to 58 in Phytophthora lateralis (Figure S4). All Phytophthora and Pythium species have a relatively large number of M28 aminopeptidases with two flanking domains, an N‐terminal “PA (protein‐associated) domain” (PF02225) and a C‐terminal “transferrin receptor‐like dimerization domain” (PF04253). The human homologue is the transferrin receptor (TfR), a cell surface receptor that assists in iron uptake into cells through a cycle of exo‐ and endocytosis of the iron transport protein transferrin (Lawrence, 1999). Ten bigrams were found exclusively in Phytophthora species. The most frequent one, present in seven species, is the “mitochondrial processing peptidase (MPP) β‐subunit” domain of family M16B combined with an N‐terminal “glycosyl hydrolase 10” domain (PF00331). An additional Phytophthora‐specific bigram is a “peptidase dimerization” domain (PF07687) combined with either a “Peptidase family M20/M25/M40” domain or an M20F domain. In Pythium species we found three lineage‐specific bigrams, including an M67C peptidase flanked by a PhoD‐like phosphatase domain (PF09423) and 13 to 17 “membrane occupation and recognition nexus (MORN) repeats” (PF02493), 13 in Pythium aphanidermatum and 17 in Pythium arrhenomanes.
MPs can play a role in different subcellular compartments but they can also be embedded in membranes or secreted to the extracellular environment (Majsec et al., 2017). We assessed all MPs for N‐terminal targeting sequences such as an SP or signal anchor (SA). We used HECTAR, a method developed for predicting the subcellular localization of proteins in stramenopiles (Gschloessl et al., 2008). Notably, in A. astaci and the diatom Fistulifera solaris—the two species with the largest MP repertoire—a strikingly large proportion of the MPs was predicted to have an SP (52% and 53%, respectively), while on average this percentage is much lower (19%) (Figure S5). The SP‐containing proteins were almost exclusively single‐domain MPs (Figure S6). Conceivably, diatoms and algae possess MPs predicted to be targeted to plastids. Remarkably, some of the oomycetes also have MPs with predicted plastid targeting signals, possibly remnants indicative of photosynthetic endosymbionts that were lost during evolution (Wang et al., 2017).
Collectively, these analyses revealed remarkable diversity in stramenopile MPs not only with respect to the subcellular targeting but also in terms of domain composition. Unique bigrams might have specific functions related to the lifestyle of a particular species. Testing this hypothesis warrants a more in‐depth study of the MP repertoire at the single species level.
2.4. The MP repertoire in P. infestans
For further analyses at the single species level we chose to focus on P. infestans. Our computational MP mining approach (Figure 1) resulted in the identification of 91 P. infestans genes encoding putative MPs (Table S2). By carefully examining the predicted gene models and by integration of the alignments with expressed sequence tags (ESTs) and RNA sequencing (RNA‐Seq) data, the majority was found to be correct and a few others were manually corrected. The 91 MPs were categorized in 21 MP families and eight clans. More than one‐third belongs to clan MA (36 MPs divided over 10 families) and 15 group in clan MH (divided over three families; M18, M20, and M28). Three MPs from two families (M78 and M82) are not assigned to a clan (U: unassigned) and the remaining 37 are divided over six clans (MC, ME, MF, MG, MJ, and MP), with family sizes ranging from two to nine (Figure 2; Table S2).
As described above for stramenopile MPs, P. infestans MPs are similarly diverse with respect to their protease domains and domain architecture. Fifty‐four of the 91 P. infestans MPs are single‐domain proteins comprising just the protease domain, as exemplified by all M13 and M14 family members. The other 37 (41%) contain at least one accessory domain next to the protease domain. This can be another MP domain, as for example in five out of eight M16 MPs with one or two additional M16‐like peptidase domains, or accessory domains not directly implicated in MP activity. A clear example of diversity in domain composition among related MPs is M24, a family comprising eight members with five different domain architectures and divided over seven OGs, one of which is an oomycete‐specific OG (Figure 4; Table S2). PITG_12211 and PITG_12220 are paralogues in P. infestans and in most other Phytophthora species, suggesting an ancestral gene duplication that preceded speciation in the Phytophthora genus.
FIGURE 4.
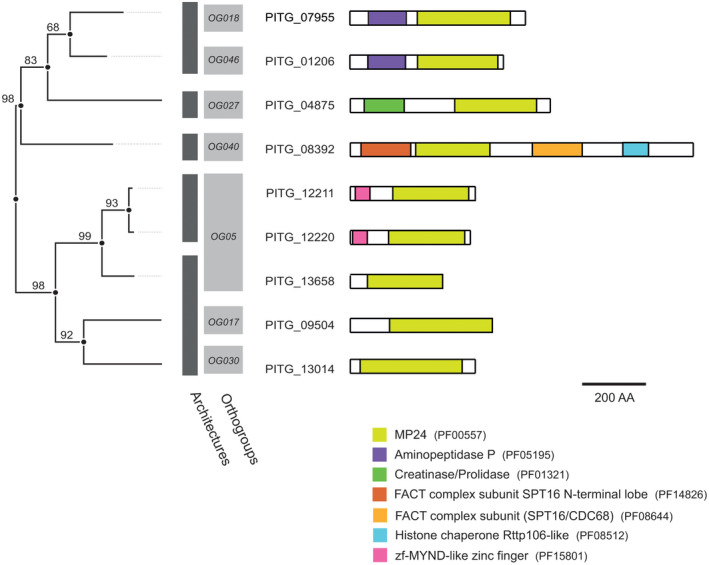
Phylogenetic tree and domain composition of M24 family members in Phytophthora infestans. Phylogram of metalloprotease subdomain sequences with bootstrap values indicated at nodes (left). Clustering based on orthogroup classification (light grey) and domain composition (dark grey) of full‐length P. infestans M24 members (right). Individual domains of M24 members are indicated
In the set of 91, we found three multidomain MPs with a seemingly unique domain architecture. PITG_00577 and PITG_06978, both belonging to the M20 family, have an MP domain preceded by an “Amidohydro_1” domain (PF01979) or a “CENP‐B N‐terminal DNA‐binding domain” (PF04218), respectively. Proteins with such domain architectures are so far only found in species of Pythiaceae and Peronosporaceae. The third one is an M67 MP (PITG_16722) that has an N‐terminal “PWWP” domain (PF00855). MPs with this domain architecture are also found in Albugo and Saprolegnia species but not in nonoomycete lineages.
2.5. Expression profiling of P. infestans MP genes
To determine the expression profiles of the MP genes in P. infestans, we exploited transcriptome data from four in vitro life stages, that is, zoospores, germinating cysts, mycelium, and sporangia, and three in planta infection stages, that is, early, mid, and late infection. The mean transcripts‐per‐million mapped reads (TPM) values were highly variable; eight genes stand out with a mean TPM value above 500, pointing to a relatively high expression in one or more stages (Figure 5). Genes were clustered into eight distinct groups based on their expression profiles (Table S2). Half of the MP genes show the highest transcript levels in early or late infection stages (clusters 2 and 3, respectively). Cluster 2 comprises the three genes with the highest average TPM values, pointing to relatively high transcript levels. MP genes in the other clusters show a peak in one or more in vitro stages, suggesting a relatively high expression in pre‐infection stages. The cluster analysis did not reveal any apparent correlation between expression pattern and specific features of the MPs. Each cluster contains MPs from various clans and families, and also MP genes that are predicted to encode potentially secreted MPs (designated sMPs) are randomly distributed over the clusters (Figure 5).
FIGURE 5.
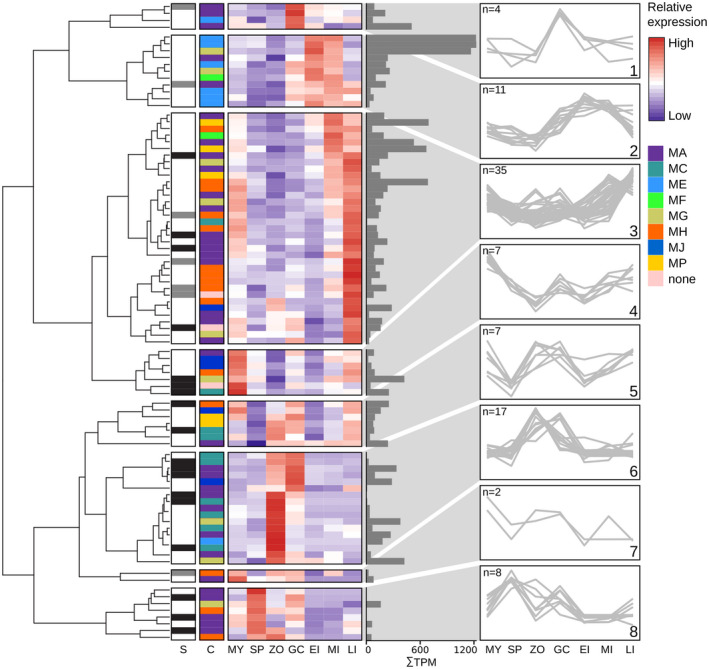
Clustering of Phytophthora infestans metalloprotease (MP) genes based on expression patterns during asexual development and in planta growth. The dendrogram on the left shows the hierarchical clustering of expression patterns. Column S marks MPs with a predicted signal peptide (dark grey) or signal anchor (light grey). Colour‐coded bars in column C indicate MEROPS clans. The heatmap depicts relative expression per gene based on stage‐wise, Z‐score transformed expression values in mycelium (MY), sporangia (SP), zoospores (ZP), germinating cysts (GC), and early, mid, and late infection stages (EI, MI, and LI, respectively). The adjacent bar plot shows mean transcripts‐per‐million mapped reads (TPM) values. Line plots on the right display the expression profiles of the eight clusters and number (n) of genes per cluster. Further details can be found in Table S2
2.6. MPs affecting virulence of P. infestans
To investigate a potential role for P. infestans MPs in virulence, we selected a subset of MPs with an N‐terminal SP or SA and tested their capacity to modulate the virulence of P. infestans. A preliminary selection based on SignalP resulted in a subset of 27 sMPs. A subsequent and more thorough analysis using HECTAR confirmed 22 of the initial 27 as potential sMPs and identified three additional ones (Figure 6). In planta activity was tested by monitoring lesion expansion caused by P. infestans upon inoculation of Nicotiana benthamiana leaves transiently expressing full‐length cDNAs of MP genes. Measurements of lesion sizes at 5 days postinoculation showed significantly larger lesions in nine cases, pointing to a positive effect of these MPs on P. infestans virulence, while in three cases we consistently recorded smaller lesions, suggesting an inhibitory effect (Figure 6). The 12 MPs belong to only six families, out of 21 families identified in P. infestans, and eight out of 12 belong to clan MA. In two families, that is, M12 and M79, all members (three and two, respectively), are sMPs and single‐domain proteins, and they all promote virulence. From family M08, four members were tested, but only the multidomain protein having an EGF‐like domain (PF07974) C‐terminal of the protease domain showed a growth‐promoting effect (PITG_08874). However, it should be noted that in our preliminary selection for sMPs this MP was predicted to be an sMP but HECTAR did not confirm this. This is the same for one of the two M48 MPs that showed a growth‐promoting effect, namely PITG_11607. The other one (PITG_12517) does have an SA but unlike PITG_11607 it is a multidomain protein. It has one of the three most ubiquitous bigrams, occurring in nearly all stramenopile species: an M48 peptidase domain followed by the “CAAX‐prenyl protease N terminal, five transmembrane helices” domain (PF16491). From family M03 all three members were tested in planta, two of which showed an inhibitory effect on virulence. They are both single‐domain proteins but for one, PITG_02711, the initially predicted SP was not confirmed by HECTAR.
FIGURE 6.
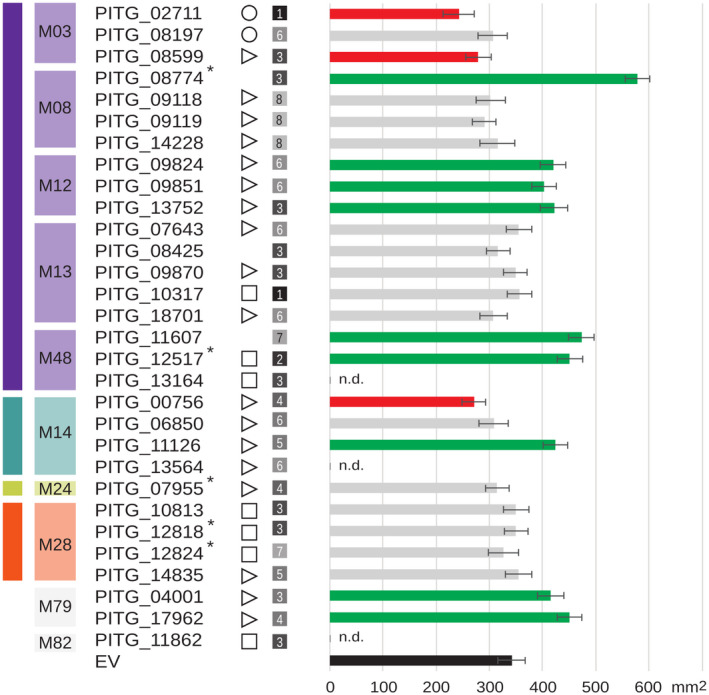
Phytophthora infestans genes encoding putative secreted metalloproteases (MPs) and their capacity to promote or inhibit virulence. MPs are shown with a predicted signal peptide (⊳), signal anchor (◻), or mitochondrial transit peptide (❍). Numbers in grey squares refer to expression clusters (Figure 5). MPs marked with an asterisk (*) have accessory domains. Bars on the right show lesion sizes on Nicotiana benthamiana leaves transiently expressing MP genes at 5 days postinoculation. Inoculation with zoospores of P. infestans strain 14‐3‐GFP was performed one day after agroinfiltration. Bar colours indicate significantly smaller (red) or larger (green) lesions or no effect (grey) when compared to the empty vector control (EV, black; analysis of variance, p < .05). Three MPs (n.d.) were not tested in planta. Bars represent average lesion sizes from three biological repeats (n = 20). Error bars indicate SD
In the M14 family, opposing effects were found with one M14 MP (PITG_00756) hampering and the other one (PITG_11126) promoting virulence. Both are single‐domain proteins, but the latter is larger in size and has signatures of transmembrane regions. Moreover, they are assigned to different subfamilies, M14B and M14C, respectively, and belong to different OGs (Table S2). P. infestans has eight M14 MPs, four of which are predicted to be secreted, including three tested in planta. The third tested M14 MP (M14B; PITG_06850) showed no significant effect.
We did not observe a significant association between expression profile and in planta activity. The 12 MPs that show in planta activity are divided over seven of the eight expression clusters. The ones belonging to one family are mostly assigned to different clusters and their expression profiles often show complementarity. For example, one M03 MP peaks in germinating cysts (cluster 1) and the other in mid and late infection stages (cluster 3). Of the three M12 MPs showing in planta activity, one is in cluster 3 and two in the complementary cluster 6 with relatively high expression in zoospores and germinating cysts. Taken together, these results suggest a role in virulence for a selected set of P. infestans MPs. As yet, there are no data confirming their proteolytic activity and also the question whether proteolytic activity as such is required for virulence remains to be answered.
3. DISCUSSION
Although several studies highlight a direct or indirect role of MPs in microbial pathogenicity (Joshi et al., 2002; Sanz‐Martín et al., 2016), the roles of MPs in pathogenic oomycetes have not yet been explored. Here we investigated the MP repertoire in stramenopiles, with a specific focus on oomycetes, and in particular on P. infestans.
Mining of stramenopile proteomes showed a diverse catalogue of MPs in each species. Of the 33 MP families found in this study, 20 are present in 80% of the analysed stramenopiles, while some families showed striking species‐specific expansions. This is, for example, the case in A. astaci, a pathogen on crustaceans that has several large MP families, mostly comprising single‐domain MPs with an SP. These observations are coherent with recent reports showing that the secretome of A. astaci is enriched for small secreted proteins with peptidase activities, many of which are indeed MPs (Gaulin et al., 2018). One of the expanded families in A. astaci as well as in other animal oomycete pathogens is M13, a family comprising peptidases known as neprilysins, zinc‐metalloenzymes that play a regulatory role in peptide signalling in mammals (Bland et al., 2008). In bacterial pathogens, neprilysins were shown to have a role in virulence, for example PtrA in P. amylovora (Zhang et al., 1999) and PepO in the human pathogen Streptococcus pyogenes (Brouwer et al., 2018). However, in our in planta assays we did not find indications for a virulence function for any of the five P. infestans M13 MPs, suggesting that M13 in oomycetes is probably relevant for animal pathogens but not for plant pathogens.
While the majority (63%) of the 4,140 stramenopile MPs are single‐domain proteins, a substantial number are multidomain proteins with accessory domains potentially required for, for example, dimerization, substrate binding, or targeting to membranes or subcellular compartments. The remarkable versatility in accessory domains implies a broad functionality of MPs. In our analyses we specifically focused on the domain composition of these multidomain MPs and searched for unique bigrams. Most oomycete proteins with unique bigrams seem to be involved in signal transduction (Van den Hoogen & Govers, 2018), but there are also examples of structural proteins, for example, myosin (Seidl et al., 2011). In general, the domain composition of multidomain stramenopile MPs conforms with that of MPs in other organisms. Still, we identified three MPs in P. infestans that have oomycete‐specific bigrams. One is an M67 MP with an N‐terminal “PWWP” domain, named after a conserved motif that binds to methylated histone H4 proteins. The other two are M20 MPs that are even more specific as they are limited to two sublineages within the oomycetes, namely Pythiaceae and Peronosporaceae. One of the M20 MPs has a “CENP‐B N‐terminal DNA‐binding domain” that, similar to the “PWWP” domain, points to a function in the nucleus. There is ample evidence that Phytophthora species exploit epigenetic modifications for tailoring their virulence (Wang et al., 2020) and it would be worth investigating whether or not the oomycete‐specific MPs play a role in that process.
In the set of stramenopile MPs we identified a few other intriguing bigrams. One is present in an M67 MP that is limited to just two Pythium species. In this multidomain protein the MP domain is followed by a PhoD‐like phosphatase and a high number of MORN repeats that function as protein–protein interaction modules (Li et al., 2019). MORN1, a protein in the parasite Toxoplasma gondii with 14 MORN repeats, appears to play a role in cell division and to act as a linker protein between membranes and the cytoskeleton (Gubbels, 2006).
One of the three most widespread bigrams in the entire set is an M48 MP combined with a second M48 domain known as CAAX‐prenyl‐M48 peptidase (PITG_12517). When this MP is expressed in leaves of N. benthamiana its presence has a positive effect on the capacity of P. infestans to form lesions; the lesion sizes increase, suggesting that this MP has a role in virulence of P. infestans. Whether or not both M48 domains in the bigram are essential for its virulence function is questionable, especially as a second single‐domain M48 MP (PITG_11607) also showed a growth‐promoting effect. Furthermore, it is unclear where M48 MPs localize. Although an SA can facilitate secretion, it is unlikely that the CAAX‐prenyl‐M48 part with seven transmembrane spanning regions is secreted. Conceivably, the first M48 domain could be cleaved off and secreted, while the CAAX‐prenyl‐M48 part remains in the endoplasmic reticulum to exert other functions. CAAX‐prenyl‐M48 MPs are widespread in eukaryotes and play a role in posttranslational modification of proteins, in particular in cleavage of the CAAX moiety of farnesylated proteins. A well‐studied CAAX‐prenyl‐M48 MP is Ste24, from Saccharomyces cerevisiae, which has a dual role in mating pheromone maturation, namely CAAX cleavage and removal of the N‐terminal extension (Pryor et al., 2013). Knock‐out of an orthologue of Ste24 in the parasite Leishmania donovani resulted in reduced infectivity and growth (Bhardwaj et al., 2017).
The second P. infestans multidomain MP causing a growth‐promoting effect in planta is PITG_08774, an M08 MP with an accessory EGF‐like domain. Because none of the four single‐domain M08 MPs showed a growth‐promoting effect, it is conceivable that the EGF‐like domain is relevant for virulence. In mammals, the extracellular epidermal growth factor (EGF) binds to the EGF receptor, thereby stimulating growth and development. One could speculate that the EGF‐like domain in PITG_08774 acts as a ligand of a host membrane receptor to bring the MP domain in close proximity to its substrate. Proteolysis of the substrate could then result in suppression of defence. Another example of an M08 MP acting in virulence is leishmanolysin GP63, from Leishmania parasites, but unlike PITG_08774 this is a single‐domain MP (Hallé et al., 2009).
The other P. infestans MPs that showed growth‐promoting or ‐inhibiting effects in in planta assays are all single‐domain MPs divided over four families, M03, M12, M14, and M79. Two of the three P. infestans M03 MPs showed an inhibitory effect on virulence. All three have an SP while in the entire set of stramenopile MPs only 22% (43/193) of the M03 MPs are predicted to be secreted. This inhibitory effect is remarkable, especially because expression seems to be relatively high in germinating cysts and/or mid and late infection stages. Presumably there is a mechanism in place that inhibits the activity of these MPs. One could imagine that the host induces expression of M03‐encoding genes as part of its defence machinery, but that P. infestans in turn produces an effector to suppress M03 activity. To the best of our knowledge this is the first report of M03 MPs with putative roles in a host–pathogen interaction. The same holds for M79, a family with relatively few known members. Similar to M48 MPs, M79 MPs are endopeptidases that release the CAAX moiety from farnesylated proteins (Rawlings & Salvesen, 2013) and both M48 and M79 MPs show the capacity to promote virulence in our in planta assays.
M12 MPs, also known as astacins, have been intensively studied due to their implications in Alzheimer's disease and breast cancer (Hartmann et al., 2013). They function as sheddases and as such modify the activity of membrane‐spanning receptors. Ectodomain shedding has also been observed in pattern recognition receptors acting in plant immunity (Petutschnig et al., 2014). As yet, the proteases involved have not been identified but it is conceivable that such proteases are part of the pathogen's weaponry when attacking plants. P. infestans has three astacin‐like MPs that might have such a role. All three have an SP and showed a consistent growth‐promoting effect in planta. Over 80% of the stramenopile M12 MPs are predicted to be secreted and the M12 MP PITG_09851 was indeed detected in the extracellular proteome (Meijer et al., 2014). The increased expression of PITG_09851 in zoospores and germinated cysts suggests that astacin is present in the pre‐infection stage when encountering the plant surface. The last MP family to consider in relation to virulence is M14, with two out of three tested members affecting lesion size, albeit in opposite manners. The one inhibiting growth is an M14C type MP, and the one promoting growth is an M14B type MP. M14 MPs are carboxypeptidases that hydrolyse single C‐terminal amino acids from polypeptide chains, with each subtype having a specificity for certain amino acids. Possibly the released amino acids act as signal molecules triggering certain responses, either in the host or in the pathogen. Alternatively, modification of the substrate by removal of one amino acid at the C‐terminus could also trigger a cascade of new events affecting the host–pathogen interaction. As yet, literature searches did not reveal examples in which M14 MPs have been implicated in microbial pathogenicity.
This study presents an overview of the MP repertoire in stramenopiles and provides new insights into the immense diversity of MPs in individual species as well the dynamics of genes and gene families related to lifestyle or taxonomic lineage. The lack of certain MPs or MP families could be due to gene losses, with the acquisition of novel unique MPs through gene gains as a counterbalance. Previous studies showed that the evolutionary history of oomycetes is shaped by massive gene gains, duplications, and losses and probably this has been a drive for speciation (Seidl et al., 2012). The more in‐depth analyses were limited to just one species, P. infestans, and were preceded by manual curation of the predicted gene models. During the analyses, we encountered the pitfalls of poorly assembled and annotated genomes, and of traces of contamination in the genome assemblies. For comparative analyses of gene families, one should strive for high‐quality genome assembly and annotation, and manual curation of predicted gene models is paramount for an accurate orthologue inference and family classification of genes and proteins (Vaattovaara et al., 2019).
We pinpointed 12 MPs that potentially affect virulence of P. infestans. Consequently, our finding calls for more in‐depth functional studies on these MPs in Phytophthora and related pathogenic oomycetes. With the broad knowledge gained on MPs in a wide range of organisms, including microbial pathogens, it should be feasible to unravel the mechanisms underlying the growth‐promoting or ‐inhibiting activity of these MPs and to identify compounds that interfere with MP activity. Exploitation of this type of compounds is currently pursued for controlling alveolate parasites such as Plasmodium and Toxoplasma species (Deu, 2017; Escotte‐Binet et al., 2018), and might also be applicable for controlling oomycete diseases in the future.
4. EXPERIMENTAL PROCEDURES
4.1. Annotation of putative metalloproteases
Complete proteomes of 37 oomycete species were retrieved from https://github.com/oomycetes/oomycetes.github.io (McGowan & Fitzpatrick, 2017) (Table S1). Proteomes of four algae and five diatoms were downloaded from Uniprot (Table S1). Because not all proteomes were annotated with equal detail, we used only the longest isoform per gene. Annotation was performed using the pipeline depicted in Figure 1, performing two parallel searches based on either MEROPS (release 12.1) (Rawlings et al., 2018) or Pfam (release 31.0). Prealigned peptidase sequences per (sub)family were downloaded from MEROPS, and were used to train profile HMMs with hmmbuild (HMMER package v3.1b2; http://hmmer.org), generating one HMM per MEROPS (sub)family. The complete proteomes were screened with hmmsearch using these HMMs, and matching sequences at an empirical E value threshold of 1e−10 were kept. Matching subsequences (putative MP domains) were extracted and aligned using MAFFT, v. 7.310 (Katoh & Standley, 2013), with default settings, to train stramenopile‐specific HMMs. These were again matched against all complete proteomes using a lower E value threshold of 1e−25. Matching protein sequences were assigned to their respective best matching MEROPS family. In parallel, we extracted domain HMMs from the Pfam database. HMMs cross‐referenced with MEROPS MP families were extracted, and HMM searches were conducted against all complete proteomes (E value < 1e−20). Proteins matching either a MEROPS MP HMM or a Pfam MP HMM were considered putative MPs and were classified according to MEROPS. In case of annotation discrepancies, that is, both methods annotating a different MP family, MEROPS annotations were maintained. Thus, Pfam‐based candidates were used only to complement the MEROPS‐based candidates. Subsequently, for each candidate MP, BLAST searches were conducted versus the NCBI nonredundant protein database, excluding stramenopile sequences. Candidates matching any nonstramenopile sequence with over 95% sequence identity were considered contamination artifacts and were removed from further analyses.
4.2. Sequence analysis
Putative MP protein sequences were subjected to feature annotation using InterProScan v. 5.30–69.0, which includes SignalP v. 4.1 and Phobius v. 1.01 for SP predictions, and to domain annotation using Pfam release 31.0. HECTAR (https://webtools.sb‐roscoff.fr/) was used to assign MPs to different categories of subcellular targeting. To annotate protein domains, we exploited the hmm search results of Pfam and MEROPS. As HMMER only reports significant local alignments, we included consecutive local alignments to derive full peptidase domains. All nonintersecting Pfam domains reported by InterProScan were considered accessory domains. To assess sequence similarity between MPs, we constructed clusters of orthologous sequences using OrthoFinder v. 2.2.6, with default settings (i.e., using BLASTp and MCL clustering) (Emms & Kelly, 2015).
Gene models of putative P. infestans MPs were manually curated using the P. infestans T30‐4 genome sequence (Haas et al., 2009) in WebApollo (Lee et al., 2013). RNA‐Seq reads of P. infestans were aligned using HiSat2 v. 2.1.0 (Kim et al., 2015) and alignments were used to infer intron–exon boundaries. P. infestans MPs were searched against the NCBI nonredundant database using BLAST to check for homologues outside the stramenopile lineage. Significant hits (E value < 1e−5 and 50% identity) were manually analysed for domain composition using InterPro and Pfam.
4.3. Gene expression analyses
P. infestans strain T20‐2 was grown on rye sucrose medium at 18 °C. Mycelia, sporangia, zoospores, and germinating cysts were isolated as previously described (Meijer et al., 2019). RNA was extracted using a NucleoSpin RNA II extraction kit (Macherey‐Nagel) according to the manufacturer's instructions. RNA‐Seq was performed using an Illumina HiSeq 2000, producing 90‐bp paired‐end reads. In planta expression data from tubers during early, mid, and late infection were downloaded from NCBI BioProject PRJNA361417 (Ah‐Fong et al., 2017). RNA abundance was quantified in TPM values using Kallisto v. 0.43.1 (Bray et al., 2016). Z‐score transformed values were hierarchically clustered using Pearson correlation distance and average linkage. The optimal number of clusters was determined using the gap statistic implemented in NbClust (Charrad et al., 2014) (Table S2).
4.4. Transient expression in N. benthamiana and infection assays
Full‐length coding sequences of MP genes were amplified from cDNA of P. infestans strain 88069 using primers listed in Table S2. Amplified fragments were inserted into plasmid pENTR/D‐TOPO and subsequently in the binary vector pGWB5 by LR recombination. Resulting constructs were transformed into Agrobacterium tumefaciens Agl1. Agroinfiltration in N. benthamiana and infection assays with P. infestans 14‐3‐GFP were performed as described previously (Meijer et al., 2019).
AUTHORS’ CONTRIBUTIONS
C. S., K. B., and F. G. designed the research. C. S., S. R., M. F. S., L. T. L., H. J. G. M., and K. B. performed experiments and analyses. S. R., C. S., K. B., and F. G. integrated data. S. R., C. S., K. B., and F. G. wrote the manuscript. All authors read and approved the manuscript.
COMPETING INTERESTS
The authors declare that they have no competing interests.
Supporting information
FIGURE S1 Clustering of stramenopile MPs in orthogroups. Orthogroup distribution per species (left) and the approximate number of MPs per orthogroup in each species depicted by the sizes of the blue dots. Orthogroup numbering (x axis) starts with ‘1’ for the largest group (OG0000000 with 285 MPs) and ends with ‘85’ (OG0000084). For gene IDs clustered per orthogroup see Table S1
FIGURE S2 Diversity of accessory domains in stramenopile MPs and their frequency of occurrence in MPs in the different species
FIGURE S3 Diversity of bigrams in stramenopile MPs and their frequency of occurrence in MPs in the different species
FIGURE S4 Number of bigrams in MPs in each stramenopile species
FIGURE S5 Percentage of potentially secreted MPs per species based on SP prediction. For each species the ratio secreted MPs over the total number of MPs is shown
FIGURE S6 Percentage of potentially secreted MPs per MP family based on SP prediction. For each MP family the ratio secreted MPs over the total number of stramenopile MPs in that family is shown
TABLE S1 Metalloproteases in stramenopiles. (a) Stramenopile species analysed in this study, including their taxonomic lineages, proteome sizes, total number of predicted MPs and references to data resources. (b) Overview of MPs identified in 47 stramenopiles, including GeneIDs, protein length, MEROPS classification, predicted localization, domain composition, number of accessory domains, orthogroup, and the five best BLAST hits. (c) Clustering of stramenopile MPs in orthogroups. (d) Overview of accessory domains in stramenopile MPs, including GeneID, start and stop position of the domain, Pfam ID and Pfam description. (e) The diversity of accessory domains in stramenopile MPs and their frequency of occurrence in MPs in the different species (depicted in Figure S3)
TABLE S2 Metalloproteases in Phytophthora infestans. (a) Overview of the 91 MPs identified in Phytophthora infestans, including GeneIDs, protein length, MEROPS classification, predicted localization, domain composition, number of accessory domains, orthogroup, and the five best BLAST hits. (b) Overview of accessory domains in P. infestans MPs, including GeneID, start and stop position of the domain, Pfam ID and Pfam description of the domain. (c) Clustering of P. infestans MP genes based on expression profiles. Composition of the eight clusters depicted in Figure 5. (d) TPM expression values for P. infestans MPs. (e) Primers used in this study for cloning the P. infestans MP genes in appropriate vectors for agroinfiltration
ACKNOWLEDGEMENTS
The authors thank Natalie Verbeek‐de Kruif for technical support with cloning and in planta assays. This work was financially supported by the Food‐for‐Thought campaign from the University Fund Wageningen, The Netherlands Organization for Scientific Research (NWO) in the framework of VIDI (H. J. G. M.) and VENI grants (K. B. and M. F. S.) and a fellowship from the Anne van den Ban Fund (L. T. L.).
Schoina C, Rodenburg SYA, Meijer HJG, et al. Mining oomycete proteomes for metalloproteases leads to identification of candidate virulence factors in Phytophthora infestans . Mol Plant Pathol. 2021;22:551–563. 10.1111/mpp.13043
Charikleia Schoina, Sander Y. A. Rodenburg, Klaas Bouwmeester, and Francine Govers contributed equally.
DATA AVAILABILITY STATEMENT
Corrected MP gene models and RNA‐Seq reads analysed in this study are available upon request. All other data generated and analysed in this study are included in the main text and supplementary files.
REFERENCES
- Ah‐Fong, A.M. , Shrivastava, J. & Judelson, H.S. (2017) Lifestyle, gene gain and loss, and transcriptional remodeling cause divergence in the transcriptomes of Phytophthora infestans and Pythium ultimum during potato tuber colonization. BMC Genomics, 18, 764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benitez, J.A. & Silva, A.J. (2016) Vibrio cholerae hemagglutinin(HA)/protease: An extracellular metalloprotease with multiple pathogenic activities. Toxicon, 115, 55–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhardwaj, R. , Das, M. , Singh, S. , Chiranjivi, A.K. , Prabhu, S.V. , Singh, S.K. et al. (2017) Evaluation of CAAX prenyl protease II of Leishmania donovani as potential drug target: Infectivity and growth of the parasite is significantly lowered after the gene knockout. European Journal of Pharmaceutical Sciences, 102, 156–160. [DOI] [PubMed] [Google Scholar]
- Bland, N.D. , Pinney, J.W. , Thomas, J.E. , Turner, A.J. & Isaac, R.E. (2008) Bioinformatic analysis of the neprilysin (M13) family of peptidases reveals complex evolutionary and functional relationships. BMC Evolutionary Biology, 8, 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bray, N.L. , Pimentel, H. , Melsted, P. & Pachter, L. (2016) Near‐optimal probabilistic RNA‐seq quantification. Nature Biotechnology, 34, 525–527. [DOI] [PubMed] [Google Scholar]
- Brouwer, S. , Cork, A.J. , Ong, C.‐L.‐Y. , Barnett, T.C. , West, N.P. , McIver, K.S. et al. (2018) Endopeptidase PepO regulates the SpeB cysteine protease and is essential for the virulence of invasive M1T1 Streptococcus pyogenes . Journal of Bacteriology, 200, e00654‐17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charrad, M. , Ghazzali, N. , Boiteau, V. & Niknafs, A. (2014) NbClust: An R package for determining the relevant number of clusters in a data set. Journal of Statistical Software, 61, 1–36. [Google Scholar]
- Deu, E. (2017) Proteases as antimalarial targets: Strategies for genetic, chemical, and therapeutic validation. FEBS Journal, 284, 2604–2628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emms, D.M. & Kelly, S. (2015) OrthoFinder: Solving fundamental biases in whole genome comparisons dramatically improves orthogroup inference accuracy. Genome Biology, 16, 157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Escotte‐Binet, S. , Huguenin, A. , Aubert, D. , Martin, A.‐P. , Kaltenbach, M. , Florent, I. et al. (2018) Metallopeptidases of Toxoplasma gondii: In silico identification and gene expression. Parasite, 25, 26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Figaj, D. , Ambroziak, P. , Przepiora, T. & Skorko‐Glonek, J. (2019) The role of proteases in the virulence of plant pathogenic bacteria. International Journal of Molecular Sciences, 20, 672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finn, R.D. , Coggill, P. , Eberhardt, R.Y. , Eddy, S.R. , Mistry, J. , Mitchell, A.L. et al. (2016) The Pfam protein families database: Towards a more sustainable future. Nucleic Acids Research, 44, D279–D285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franceschetti, M. , Maqbool, A. , Jiménez‐Dalmaroni, M.J. , Pennington, H.G. , Kamoun, S. & Banfield, M.J. (2017) Effectors of filamentous plant pathogens: Commonalities amid diversity. Microbiology and Molecular Biology Reviews, 81, e00066–e116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaulin, E. , Pel, M.J.C. , Camborde, L. , San‐Clemente, H. , Courbier, S. , Dupouy, M.‐A. et al. (2018) Genomics analysis of Aphanomyces spp. identifies a new class of oomycete effector associated with host adaptation. BMC Biology, 16, 43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gómez Luciano, L.B. , Tsai, I.J. , Chuma, I. , Tosa, Y. , Chen, Y.H. , Li, J.Y. et al. (2019) Blast fungal genomes show frequent chromosomal changes, gene gains and losses, and effector gene turnover. Molecular Biology and Evolution, 36, 1148–1161. [DOI] [PubMed] [Google Scholar]
- Gomez, M.A. , Contreras, I. , Halle, M. , Tremblay, M.L. , McMaster, R.W. & Olivier, M. (2009) Leishmania GP63 alters host signaling through cleavage‐activated protein tyrosine phosphatases. Science Signalling, 2, ra58. [DOI] [PubMed] [Google Scholar]
- Gschloessl, B. , Guermeur, Y. & Cock, J.M. (2008) HECTAR: A method to predict subcellular targeting in heterokonts. BMC Bioinformatics, 9, 393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gubbels, M.‐J. (2006) A MORN‐repeat protein is a dynamic component of the Toxoplasma gondii cell division apparatus. Journal of Cell Science, 119, 2236–2245. [DOI] [PubMed] [Google Scholar]
- Haas, B.J. , Kamoun, S. , Zody, M.C. , Jiang, R.H.Y. , Handsaker, R.E. , Cano, L.M. et al. (2009) Genome sequence and analysis of the Irish potato famine pathogen Phytophthora infestans . Nature, 461, 393–398. [DOI] [PubMed] [Google Scholar]
- Hallé, M. , Gomez, M.A. , Stuible, M. , Shimizu, H. , McMaster, W.R. , Olivier, M. et al. (2009) The Leishmania surface protease GP63 cleaves multiple intracellular proteins and actively participates in p38 mitogen‐activated protein kinase inactivation. Journal of Biological Chemistry, 284, 6893–6908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartmann, M. , Herrlich, A. & Herrlich, P. (2013) Who decides when to cleave an ectodomain? Trends in Biochemical Sciences, 38, 111–120. [DOI] [PubMed] [Google Scholar]
- Jashni, M.K. , Dols, I.H.M. , Iida, Y. , Boeren, S. , Beenen, H.G. , Mehrabi, R. et al. (2015) Synergistic action of a metalloprotease and a serine protease from Fusarium oxysporum f. sp. lycopersici cleaves chitin‐binding tomato chitinases, reduces their antifungal activity, and enhances fungal virulence. Molecular Plant‐Microbe Interactions, 28, 996–1008. [DOI] [PubMed] [Google Scholar]
- Jia, Y. , Zhou, E. , Lee, S. & Bianco, T. (2016) Coevolutionary dynamics of rice blast resistance gene Pi‐ta and Magnaporthe oryzae avirulence gene AVR‐Pita1 . Phytopathology, 106, 676–683. [DOI] [PubMed] [Google Scholar]
- Jiang, R.H.Y. , de Bruijn, I. , Haas, B.J. , Belmonte, R. , Löbach, L. , Christie, J. et al. (2013) Distinctive expansion of potential virulence genes in the genome of the oomycete fish pathogen Saprolegnia parasitica . PLoS Genetics, 9, e1003272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joshi, P.B. , Kelly, B.L. , Kamhawi, S. , Sacks, D.L. & McMaster, W.R. (2002) Targeted gene deletion in Leishmania major identifies leishmanolysin (GP63) as a virulence factor. Molecular and Biochemical Parasitology, 120, 33–40. [DOI] [PubMed] [Google Scholar]
- Kamoun, S. , Furzer, O. , Jones, J.D.G. , Judelson, H.S. , Ali, G.S. , Dalio, R.J. et al. (2015) The top 10 oomycete pathogens in molecular plant pathology. Molecular Plant Pathology, 16, 413–434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh, K. & Standley, D.M. (2013) MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Molecular Biology and Evolution, 30, 772–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kay, J. , Meijer, H.J.G. , Ten Have, A. & Van Kan, J.A.L. (2011) The aspartic proteinase family of three Phytophthora species. BMC Genomics, 12, 254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, D. , Langmead, B. & Salzberg, S.L. (2015) HISAT: A fast spliced aligner with low memory requirements. Nature Methods, 12, 357–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence, C.M. (1999) Crystal structure of the ectodomain of human transferrin receptor. Science, 286, 779–782. [DOI] [PubMed] [Google Scholar]
- Lee, E. , Helt, G.A. , Reese, J.T. , Munoz‐Torres, M.C. , Childers, C.P. , Buels, R.M. et al. (2013) Web Apollo: A web‐based genomic annotation editing platform. Genome Biology, 14, R93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lelis, T. , Peng, J. , Barphagha, I. , Chen, R. & Ham, J.H. (2019) The virulence function and regulation of the metalloprotease gene prtA in the plant‐pathogenic bacterium Burkholderia glumae . Molecular Plant‐Microbe Interactions, 32, 841–852. [DOI] [PubMed] [Google Scholar]
- Li, Y. , Orlando, B.J. & Liao, M. (2019) Structural basis of lipopolysaccharide extraction by the LptB(2)FGC complex. Nature, 567, 486–490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lichtenthaler, S.F. , Lemberg, M.K. & Fluhrer, R. (2018) Proteolytic ectodomain shedding of membrane proteins in mammals–hardware, concepts, and recent developments. EMBO Journal, 37, e99456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majsec, K. , Bhuiyan, N.H. , Sun, Q. , Kumari, S. , Kumar, V. , Ware, D. et al. (2017) The plastid and mitochondrial peptidase network in Arabidopsis thaliana: A foundation for testing genetic interactions and functions in organellar proteostasis. The Plant Cell, 29, 2687–2710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall, N.C. , Finlay, B.B. & Overall, C.M. (2017) Host defenses during infection: Proteases cut to the chase. Molecular & Cellular Proteomics, 16, S161–S171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGowan, J. & Fitzpatrick, D.A. (2017) Genomic, network, and phylogenetic analysis of the oomycete effector arsenal. mSphere, 2, e00408‐17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meijer, H.J.G. & Govers, F. (2006) Genome‐wide analysis of phospholipid signaling genes in Phytophthora spp.: Novelties and a missing link. Molecular Plant‐Microbe Interactions, 19, 1337–1347. [DOI] [PubMed] [Google Scholar]
- Meijer, H.J.G. , Hassen, H.H. & Govers, F. (2011) Phytophthora infestans has a plethora of phospholipase D enzymes including a subclass that has extracellular activity. PLoS One, 6, e17767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meijer, H.J.G. , Mancuso, F.M. , Espadas, G. , Seidl, M.F. , Chiva, C. , Govers, F. et al. (2014) Profiling the secretome and extracellular proteome of the potato late blight pathogen Phytophthora infestans . Molecular & Cellular Proteomics, 13, 2101–2113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meijer, H.J.G. , Schoina, C. , Wang, S. , Bouwmeester, K. , Hua, C. & Govers, F. (2019) Phytophthora infestans small phospholipase D‐like proteins elicit plant cell death and promote virulence. Molecular Plant Pathology, 20, 180–193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyoshi, S. & Shinoda, S. (2000) Microbial metalloproteases and pathogenesis. Microbes and Infection, 2, 91–98. [DOI] [PubMed] [Google Scholar]
- Ospina‐Giraldo, M.D. , Griffith, J.G. , Laird, E.W. & Mingora, C. (2010) The CAZyome of Phytophthora spp.: A comprehensive analysis of the gene complement coding for carbohydrate‐active enzymes in species of the genus Phytophthora . BMC Genomics, 11, 525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng, D. , Lin, J. , Huang, Q. , Zheng, W. , Liu, G. , Zheng, J. et al. (2015) A novel metalloproteinase virulence factor is involved in Bacillus thuringiensis pathogenesis in nematodes and insects. Environmental Microbiology, 18, 846–862. [DOI] [PubMed] [Google Scholar]
- Petutschnig, E.K. , Stolze, M. , Lipka, U. , Kopischke, M. , Horlacher, J. , Valerius, O. et al. (2014) A novel Arabidopsis CHITIN ELICITOR RECEPTOR KINASE 1 (CERK1) mutant with enhanced pathogen‐induced cell death and altered receptor processing. New Phytologist, 204, 955–967. [DOI] [PubMed] [Google Scholar]
- Pryor, E.E. , Horanyi, P.S. , Clark, K.M. , Fedoriw, N. , Connelly, S.M. , Koszelak‐Rosenblum, M. et al. (2013) Structure of the integral membrane protein CAAX protease Ste24p. Science, 340, 1600–1604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rawlings, N.D. , Barrett, A.J. , Thomas, P.D. , Huang, X. , Bateman, A. & Finn, R.D. (2018) The MEROPS database of proteolytic enzymes, their substrates and inhibitors in 2017 and a comparison with peptidases in the PANTHER database. Nucleic Acids Research, 46, D624–D632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rawlings, N.D. & Salvesen, G. (2013) Handbook of Proteolytic Enzymes. Cambridge, USA: Academic Press. [Google Scholar]
- Sanderson, M.P. , Dempsey, P.J. & Dunbar, A.J. (2006) Control of ErbB signaling through metalloprotease mediated ectodomain shedding of EGF‐like factors. Growth Factors, 24, 121–136. [DOI] [PubMed] [Google Scholar]
- Sanz‐Martín, J.M. , Pacheco‐Arjona, J.R. , Bello‐Rico, V. , Vargas, W.A. , Monod, M. , Díaz‐Mínguez, J.M. et al. (2016) A highly conserved metalloprotease effector enhances virulence in the maize anthracnose fungus Colletotrichum graminicola . Molecular Plant Pathology, 17, 1048–1062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoina, C. , Verbeek‐de Kruif, N. , Govers, F. & Bouwmeester, K. (2019) Clade 5 aspartic proteases of Phytophthora infestans are virulence factors implied in RXLR effector cleavage. European Journal of Plant Pathology, 154, 17–29. [Google Scholar]
- Seidl, M.F. , Van den Ackerveken, G. , Govers, F. & Snel, B. (2011) A domain‐centric analysis of oomycete plant pathogen genomes reveals unique protein organization. Plant Physiology, 155, 628–644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seidl, M.F. , Van den Ackerveken, G. , Govers, F. & Snel, B. (2012) Reconstruction of oomycete genome evolution identifies differences in evolutionary trajectories leading to present‐day large gene families. Genome Biology and Evolution, 4, 199–211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shende, R. , Wong, S.S.W. , Rapole, S. , Beau, R. , Ibrahim‐Granet, O. , Monod, M. et al. (2018) Aspergillus fumigatus conidial metalloprotease Mep1p cleaves host complement proteins. Journal of Biological Chemistry, 293, 15538–15555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaattovaara, A. , Leppälä, J. , Salojärvi, J. & Wrzaczek, M. (2019) High‐throughput sequencing data and the impact of plant gene annotation quality. Journal of Experimental Botany, 70, 1069–1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van den Hoogen, D.J. , van den Meijer, H.J.G. , Seidl, M.F. & Govers, F. (2018) The ancient link between G‐protein‐coupled receptors and C‐terminal phospholipid kinase domains. mBio, 9, e02119‐17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van den Hoogen, J. & van den Govers, F. (2018) GPCR‐bigrams: Enigmatic signaling components in oomycetes. PLoS Pathogens, 14, e1007064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, L. , Chen, H. , Li, J. , Shu, H. , Zhang, X. , Wang, Y. et al. (2020) Effector gene silencing mediated by histone methylation underpins host adaptation in an oomycete plant pathogen. Nucleic Acids Research, 48, 1790–1799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, Q. , Sun, H. & Huang, J. (2017) Re‐analyses of “algal” genes suggest a complex evolutionary history of oomycetes. Frontiers in Plant Science, 8, 1540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whisson, S.C. , Boevink, P.C. , Wang, S. & Birch, P.R. (2016) The cell biology of late blight disease. Current Opinion in Microbiology, 34, 127–135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, Y. , Bak, D.D. , Heid, H. & Geider, K. (1999) Molecular characterization of a protease secreted by Erwinia amylovora . Journal of Molecular Biology, 289, 1239–1251. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
FIGURE S1 Clustering of stramenopile MPs in orthogroups. Orthogroup distribution per species (left) and the approximate number of MPs per orthogroup in each species depicted by the sizes of the blue dots. Orthogroup numbering (x axis) starts with ‘1’ for the largest group (OG0000000 with 285 MPs) and ends with ‘85’ (OG0000084). For gene IDs clustered per orthogroup see Table S1
FIGURE S2 Diversity of accessory domains in stramenopile MPs and their frequency of occurrence in MPs in the different species
FIGURE S3 Diversity of bigrams in stramenopile MPs and their frequency of occurrence in MPs in the different species
FIGURE S4 Number of bigrams in MPs in each stramenopile species
FIGURE S5 Percentage of potentially secreted MPs per species based on SP prediction. For each species the ratio secreted MPs over the total number of MPs is shown
FIGURE S6 Percentage of potentially secreted MPs per MP family based on SP prediction. For each MP family the ratio secreted MPs over the total number of stramenopile MPs in that family is shown
TABLE S1 Metalloproteases in stramenopiles. (a) Stramenopile species analysed in this study, including their taxonomic lineages, proteome sizes, total number of predicted MPs and references to data resources. (b) Overview of MPs identified in 47 stramenopiles, including GeneIDs, protein length, MEROPS classification, predicted localization, domain composition, number of accessory domains, orthogroup, and the five best BLAST hits. (c) Clustering of stramenopile MPs in orthogroups. (d) Overview of accessory domains in stramenopile MPs, including GeneID, start and stop position of the domain, Pfam ID and Pfam description. (e) The diversity of accessory domains in stramenopile MPs and their frequency of occurrence in MPs in the different species (depicted in Figure S3)
TABLE S2 Metalloproteases in Phytophthora infestans. (a) Overview of the 91 MPs identified in Phytophthora infestans, including GeneIDs, protein length, MEROPS classification, predicted localization, domain composition, number of accessory domains, orthogroup, and the five best BLAST hits. (b) Overview of accessory domains in P. infestans MPs, including GeneID, start and stop position of the domain, Pfam ID and Pfam description of the domain. (c) Clustering of P. infestans MP genes based on expression profiles. Composition of the eight clusters depicted in Figure 5. (d) TPM expression values for P. infestans MPs. (e) Primers used in this study for cloning the P. infestans MP genes in appropriate vectors for agroinfiltration
Data Availability Statement
Corrected MP gene models and RNA‐Seq reads analysed in this study are available upon request. All other data generated and analysed in this study are included in the main text and supplementary files.


