Abstract
Purpose:
The objective was to study temporal changes in tumor vascular physiological indices in a period of 24 h in a 9L gliosarcoma rat model.
Methods:
Fischer-344 rats (N = 14) were orthotopically implanted with 9L cells. At 2 weeks post-implantation, they were imaged twice in a 24 h interval using dynamic contrast enhanced magnetic resonance imaging (DCE-MRI). Data-driven model-selection-based analysis was used to segment tumor regions with varying vascular permeability characteristics. The region with the maximum number of estimable parameters of vascular kinetics was chosen for comparison across the two time points. It provided estimates of three parameters for an MR contrast agent (MRCA): i) plasma volume (vp), ii) forward volumetric transfer constant (Ktrans) and interstitial volume fraction (ve, ratio of Ktrans to reverse transfer constant, kep). In addition, MRCA extracellular distribution volume (VD) was estimated in the tumor and its borders, along with tumor blood flow (TBF) and peritumoral MRCA flux. Descriptors of parametric distributions were compared between the two times. Tumor extent was examined by hematoxylin and eosin (H & E) staining. Picrosirus red staining of secreted collagen was performed as an additional index for 9L cells.
Results:
Test-retest differences between population summaries for any parameter were not significant (paired t and Wilcoxon signed rank tests). Bland-Altman plots showed no apparent trends between the differences and averages of the test-retest measures for all indices. The intraclass correlation coefficients showed moderate to almost perfect reproducibility for all of the parameters, except vp. H & E staining showed tumor infiltration in parenchyma, perivascular space and white matter tracts. Collagen staining was observed along the outer edges of main tumor mass.
Conclusion:
The data suggest the relative stability of these MR indices of tumor microenvironment over a 24 h duration in this gliosarcoma model.
Keywords: DCE-MRI, Glioblastoma, Ktrans, Model selection, Test-retest
1. Introduction
Gliomas, including glioblastoma multiforme, comprise the most prevalent primary brain tumors. Persons with these tumors have a very short life expectancy following diagnosis even after the most aggressive current therapy, i.e., tumor debulking followed by radiotherapy (RT) and chemotherapy [1]. Although primary gliosarcomas are rarer than gliomas, it is known that recurrent gliomas tend to present gliosarcoma-like features with both glial and mesenchymal differentiations [2]. Except for the absence of minimal epidermal growth factor receptor (EGFR) amplification, the gliosarcoma genotypic profile is similar to that of glioma and the same general treatment plan is recommended for both [3–5]. As with glioma, recurrent gliosarcoma is also highly aggressive and always fatal. Considering their relative inaccessibility for biopsies, minimally invasive analyses of glioma grading and treatment effects are essential in longitudinal evaluations of disease progress and/or therapy efficacy [6].
Dynamic contrast enhanced MRI (DCE-MRI) is being widely used for such evaluations in both human primary brain tumors and their analogs, animal glioma models. Some parameters of interest are plasma volume fraction (vp), forward volume transfer constant (Ktrans), and, provided that reverse transfer constant (kep) can be estimated, interstitial volume fraction (ve) [7–9], along with tumor blood flow (TBF) and apparent diffusion coefficient (ADC) of water. Recently it was shown that the distribution volume (VD), defined as the sum of plasma volume and interstitial volume fraction, could also be estimated from equilibrating DCE-MRI data using a Logan graphical approach [10,11]. Underlining its physiological relevance, VD was also shown to inversely correlate with tumor cellularity [10]. Taken together, these estimates of microvascular function using T1-weighted DCE-MRI can produce measures of tumor physiology that may serve as tumor biomarkers [12,13].
Test-retest stability of these MRI indices and their potential as biomarkers of therapeutic effects in a U251 glioma model have been previously investigated [14]. In the present study, to confirm and extend their utility across tumor models, test-retest measurements of vascular physiology parameters were performed in the widely used, syngeneic, orthotopic, 9L gliosarcoma model. Two identical imaging studies were carried out in a 24 h time interval to examine the relative stability or variability of the DCE-MRI parameters. In addition to previously described indices, several novel aspects of this model such as its tumor-normal tissue boundary characteristics and peritumoral magnetic resonance contrast agent (MRCA) flux, and their relevance to tumor microenvironment were also examined. Hematoxylin and eosin (H & E) histology was performed to confirm tumor boundary demarcation observed on MRI. Since this cell line is known to secrete collagen in vitro [15], presence and distribution of 9L cells was also confirmed by staining for secreted collagen.
2. Methods
2.1. Tumor implantation
All experimental procedures were approved by the Institutional Animal Care and Use Committee according to federal regulations of Policy on Humane Care and Use of Laboratory Animals. Fifteen male Fischer-344 rats (~7 weeks old; Charles River, Kingston, NY) were intracerebrally implanted with 9L cells following published procedures [16,17]. Briefly, animals were anesthetized with isoflurane (4% for induction, 0.75 to 1.5% for maintenance, balance N2O:O2 = 2:1). The scalp was swabbed with Betadine and alcohol, the eyes coated with Lacri-lube (Allergan Inc., Parsippany-Troy Hills, NJ) and the head immobilized in a rat stereotactic device (Kopf, Cayunga, CA). A 1 cm incision was made 2 mm right of the midline and the skull exposed. A burr hole was drilled 3.5 mm to the right of bregma, without penetrating the dura. A #2701 10 μL Hamilton syringe with a 26 gauge needle containing 9L tumor cells freshly harvested from log phase growth (5 × 104 in 10 μL of phosphate buffered saline) was lowered to a depth of 3.0 mm and then raised back to a depth of 2.5 mm. Cells were then injected at a rate of 0.5 μL/10 s until the entire volume was injected. The needle was gently withdrawn, the burr hole sealed with sterile bone wax and the scalp sutured.
2.2. MRI studies
For the MRI study, between 12 and 14 days after tumor induction, each animal was anesthetized with the isoflurane gas mixture and allowed to spontaneously respire. A tail vein was cannulated for MRCA administration. Body temperature was maintained at 37 °C with warm air and monitored via an intra-rectal type T thermocouple. Identical procedures were carried out in the two MRI studies conducted 24 h apart for each animal.
All MRI studies were performed using a 7 Tesla magnet in a Varian/Agilent (Santa Clara, CA) 20 cm bore system with a Direct Drive spectrometer and console. Gradient maximum strengths and rise times were 250 mT/m and 120 μs. All MRI image sets were acquired with a 32 mm × 32 mm FOV. The transmit coil was a RAPID (RAPID MR International, Columbus, OH) Quadrature Birdcage coil, and the receive coil was RAPID two-channel phased-array surface coil for rat brain imaging. Arterial spin labeling (ASL), DCE-MRI, Look-Locker (LL), T1-weighted, and diffusion-weighted imaging (DWI) image sets were acquired and used to derive the tumor physiological estimates employed herein. A pulsed gradient spin-echo DWI sequence was run in three directions (x, y, z) to generate a parametric map of ADC. DWI sequence parameters were as follows: matrix 128 × 64, 13 slices, 0.8 mm thickness, 0.2 mm gap, repetition time (TR) = 1500 ms, echo time (TE) = 40 ms, number of experiments = 1, b-values = 0, 600, 1217 s/mm2, gradient amplitude = 107 mT/m, gradient duration = 10 ms. ASL data was acquired to estimate cerebral blood flow in a single central slice, as previously described [7]. Sets of MRI parameters were obtained with alternating gradients and frequency offsets in combinations of four as follows: matrix = 128 × 64, one 1.0 mm slice, number of averages (NA) = 2, TE/TR = 24/1500. Arterial labeling = 1 s. Total time = 13 min.
Preceding, and immediately succeeding the DCE-MRI sequence two LL sequences were run so that a voxel-by-voxel estimate of longitudinal relaxation time (T1) in the tissue could be made pre- and post-MRCA administration. LL sequence parameters were as follows: FA = 15°, matrix 128 × 64, five 2.0 mm slices, no gap. NE = 24 inversion-recovery echoes on 50 ms intervals, TE/TR = 4.0/2000 ms. The DCE-MRI sequence was a dual-echo gradient-echo (2GE) sequence, the “mgems” sequence in the Agilent VnmrJ library. The 2GE sequence acquired a set of three slices on 2 mm centers (1.8 mm slice, 0.2 mm gap). The slice set was centered on the tumor and 150 image sets at 4.0 s intervals were acquired with the following parameters: flip angle (FA) = 25°, matrix = 128 × 64, NE = 2, NA = 1, TE1/TE2/TR = 2.0/4.0/60 ms. Total run time was about 10 min. At image 15 of the 2GE sequence, a bolus injection of the MRCA gadopentetate dimeglumine (Magnevist, Bayer Healthcare Pharmaceuticals, Wayne, NJ), 0.25 mmol/kg at undiluted concentration, no flush, was performed by hand push.
Prior to the pre-contrast LL sequence, and after the post-contrast LL sequence, two high-resolution T1-weighted spin-echo images were acquired pre- and post-CA, to locate the tumor and its size, with the following parameters: FA = 45°, 180°, matrix 256 × 192, 27 slices, 0.4 mm thickness, 0.1 mm gap, NE = 1, NA = 4, TE/TR = 16/800 ms.
2.3. Tumor vascular parameters
DCE-MRI data was initially analyzed using a model selection paradigm [18,19] based on Patlak, extended Patlak [8,20] and Logan [21] graphical methods. Model selection is a data-driven segmentation technique that applies the 3-parameter standard model, or nested subsets of this model, voxel-by-voxel to DCE-MRI studies. It requires a concentration-time trace of an indicator in both arterial blood and tissue. Common practice uses the dynamic change in longitudinal relaxation rate (ΔR1; R1 = 1/T1) to estimate the dynamic change in MRCA blood and tissue concentration. Once a time trace of R1 is calculated, the pharmacokinetic model that best accounts for the variation of the signal can be selected using an F-test [18].
Following this technique, model selection generated maps of brain regions and labeled them with the number of parameters used to describe the data contained therein. This resulted in virtual segmentation of the tumor as: i) Model 1 region: essentially normal vasculature with no leakage, where the filling of the vasculature with MRCA in the plasma occurs (vp ≠ 0, Ktrans=kep = 0); ii) Model 2 region: tissue regions with leakage without measurable backflux (vp ≠ 0, Ktrans ≠ 0, kep = 0); or iii) Model 3 region: highly leaky vessels with measurable backflux of MRCA from the interstitial space to the microvasculature (vp ≠ 0, Ktrans ≠ 0, kep ≠ 0). In the Model 3 region chosen in this study for further for analyses, the MRI vascular parameters measured were: i) tumor blood flow (TBF) via ASL [22,23]; ii) ADC via DWI; and iii) the parameters vp, Ktrans and ve, via Patlak analysis [14,20] and, iv) VD in the tumor and its leaky rim (VD-inner edge) and in the peritumoral tissue (VD-outer edge) via Logan analysis [10,21]. In addition, exudate fluid flux across the boundary of the tumor was estimated following published methods [19].
Quantitative maps of MRI vascular parameters were generated for test-retest imaging sessions. The MRI slice with the largest tumor cross-section was selected for the analysis. The model selection paradigm was employed to delineate the tumor region of interest (ROI), and the ROI of the tumor periphery, which is mostly normal tissue. In all animals, post-contrast T1-weighted images and H & E-stained sections were used to confirm the tumor boundary defined previously by model selection.
2.4. Histology
After the second MRI study, the animals were continued on isoflurane anesthesia and transcardially perfused with normal saline followed by the fixative, 4% paraformaldehyde. After the brains were carefully removed, they were stored in the fixative overnight. Two millimeter thick coronal slices through the tumor were obtained using a rat-brain matrix (Activational Systems, Inc., Warren, MI). The brain tissue was processed using a VIP Tissue Tek Processing center and embedded in paraffin. Seven micrometer thick sections were cut from the paraffin block corresponding to the selected MR imaging slice and placed on Superfrost Plus (Fisher Scientific) slides. It contained the largest tumor area and was stained with H & E for evaluation of tumor ROIs. Additional sections were stained for visualizing tumor collagen distribution using Picrosirius red staining. Images were collected using a Nikon Eclipse E800 microscope equipped with ACT1C software. An ROI was chosen within the contralateral hemisphere for comparison, usually within the caudate putamen.
2.5. Statistical analyses
A previously published method for MR map analysis and data acquisition was followed [14]. It was observed in that study that collecting voxel-wise information (temporal-spatial analysis or T-S) from any given region of interest (ROI) resulted in more exhaustive and physiologically useful data summary than averaging the information across that ROI. Accordingly, a similar analysis approach was also employed in this study that generated pixel-by-pixel estimates of parameter values for an ROI voxel that encompassed the Model 3 area. Using the complete range of values for a given parameter, summary estimates of median, mean, variance, skewness and kurtosis were compared between the test and retest imaging measures via paired t-tests and/or Wilcoxon signed rank tests to characterize the stability of the summary estimates. Bland-Altman plots [24] were constructed to visually assess any potential relationship or trends between the difference in the test-retest measurement and the average of the two measurements for tumor volume, TBF, ADC and VD. In these plots, the mean values along with the 95% limits of agreement for the difference in test-retest measurements were included. In addition, Kendall-tau tests were used to formally test for trends.
Intraclass correlation coefficients (ICC) were computed to assess the reproducibility of the MRI parameters measured 24 h apart. Landis and Koch proposed the following cutpoints for interpreting the degree of reproducibility: < 0 none, 0–0.2 slight, 0.21–0.4 fair, 0.41–0.6 moderate, 0.61–0.8 substantial and 0.81–1.0 almost perfect [25]. In addition, the means and standard deviations for test and retest, along with the mean, standard deviation and 95% confidence intervals for the differences between the two time points were computed. Depending on the variability in the differences, these intervals could be used to evaluate potential treatment effects in future studies. All of the above analyses were performed on the summary estimates of mean and median from the ROI, because these summary estimates are the ones most likely to be used in studies.
Paired t-tests were done to compare the tumor volume between the two time points. All analyses were done using SAS version 9.4.All testing was done at the alpha = 0.05 level. Since each rat served as its own control, no blinding of investigators was done.
3. Results
3.1. General observations
At 12–14 days after tumor implantation, the typical largest diameter of the tumor on a coronal image was ~5 mm. The extent of tumor observed on MRI was matched by the corresponding H & E-stained sections. An example of T1, CBF and model selection maps for retest imaging with their corresponding H & E stained brain section from one rat are shown in Fig. 1. Higher magnification imaging of these specimens showed tumor cell acini breaking off from the main tumor mass and invading the host tissue. Individual tumor cells and clusters were also seen in perivascular spaces and along white matter tracks (Fig. 2). Picrosirius staining for collagen exhibited strands of pink-stained collagen in the tumor interstitial space (Fig. 3). Such staining was absent in tumor-free regions of the ipsilateral hemisphere and also in the contralateral hemisphere. Collagen staining was visible only around blood vessels in such non-tumor regions (Fig. 3).
Fig. 1.
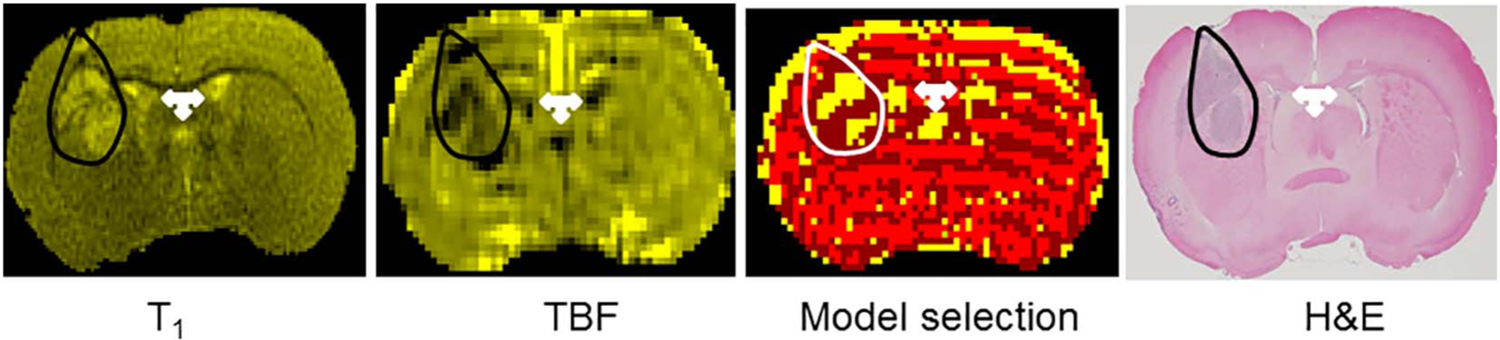
Top panel: From left to right, i) a T1-weighted image; ii) tumor blood flow (TBF) map acquired during retest of a 9L tumor in a Fischer-344 rat; iii) the corresponding model selection map; and iv) H & E-stained brain section. A close agreement in distribution of tumor tissue (hyperintense regions on T1 image, darker regions on CBF map and densely staining tissue on H & E) can be seen between the images (outlined in black). Tumor tissue grew as two nearly separate masses in this rat and all three methods reproduce it distinctly. A small aggregate of tumor cells near the cortical edge can also be seen in both T1 and H & E. The model selection map shows in yellow pixels within the white outline the Model 3 regions selected for a typical analysis. Note that in this example not all tumor tissue is encompassed by yellow pixels or model 3 region. The dark red pixels around the yellow pixels represent Model 2 regions with leakage, but no backflux. The bright red regions are Model 1 regions with no MRCA leakage. The other hyperintense regions on T1, dark areas on CBF and yellow pixels on model selection map are water/proton-rich brain ventricles (white 3-headed arrow) and pial vasculature.
Fig. 2.

High magnification images of H & E-stained tumor tissue. A higher than normal cell density compared to brain tissue typically seen in cerebral tumors was observed (A; arrows, mitotic bodies). Acini of tumor cells moving away into host tissue could be seen at the tumor boundary (B). Migrating tumor cells were also present in the perivascular space (C) and along white matter tracts (D).
Fig. 3.
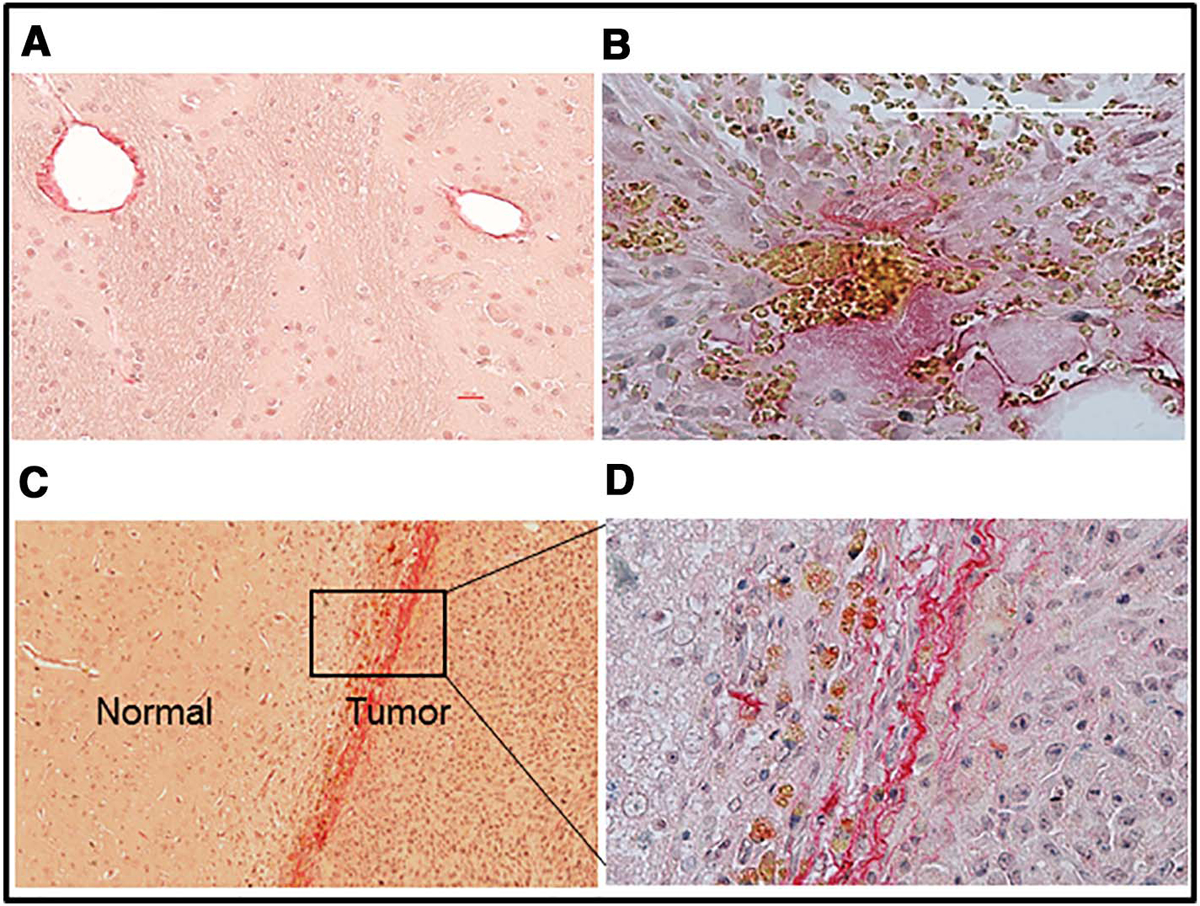
Distribution of collagen visualized by Picrosirius red staining. Positive staining is seen only around blood vessels on the contralateral side (A) whereas dense staining is seen in the tumor interstitial space (B). In some cases, collagen staining was also observed along the boundary between normal and tumor tissue (C and D). These brain sections were counterstained with Meyer’s hematoxylin.
3.2. Image analyses
Contrast-enhancing tumors were visible in all 15 rats following post-contrast high-resolution T1-weighted imaging. However, movement-induced imaging artifacts in one experiment precluded its inclusion in final analyses and, thus, 14 sets of data were used for the final analyses. A set of maps of model parameters was produced in three 2 mm slices centered on the tumor for each of the two examinations performed in a 24 h interval. Fig. 4 depicts, in a single MRI slice, a complete set of parameter maps (vp, Ktrans, ve, model selection) derived from DCE-MRI and used to characterize tumor vascular status in this rat (test, top row; retest, bottom row).
Fig. 4.
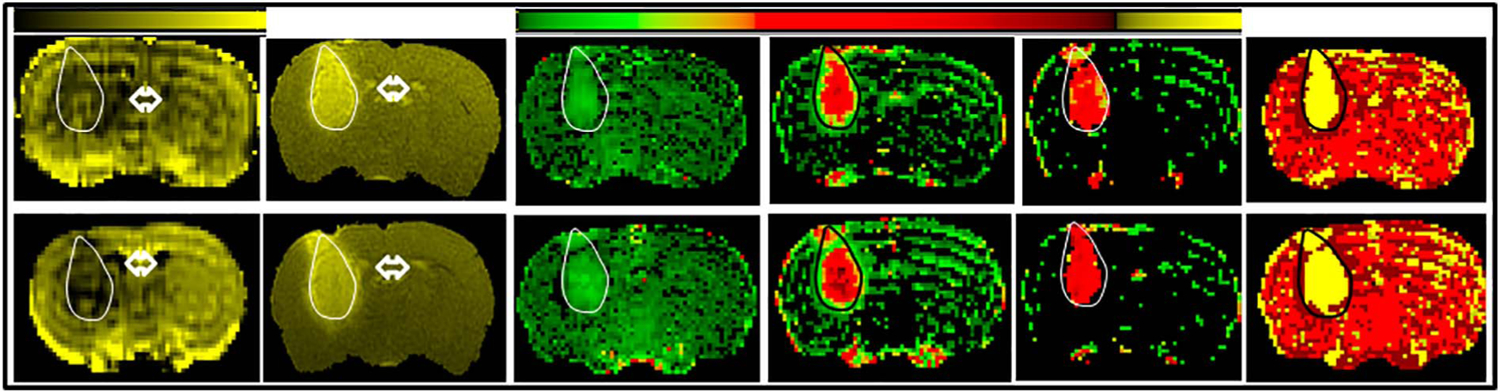
A representative set of maps acquired at the two time points from one rat. From left to right, they are: TBF, T1, vp, Ktrans, ve and Model selection. Top row depicts maps from ‘test’ and the bottom row from ‘retest’. The scale bar on top to the left is for TBF maps with increasing values from black to yellow. Range of values for TBF test is: 0–150 and TBF retest is: 0–180. The scale bar on the right is for vp, Ktrans and ve, with increasing values from green to yellow. Range of values for test are: vp: 0–0.94; Ktrans: 0–0.13; ve: 0–0.35; and for retest: vp: 0–0.40; Ktrans: 0–0.05; ve: 0–0.21. Tumor mass is visible on the left side as an oval shaped mass extending into the striatum. It is outlined in either white or black for identification. Fluid-filled regions such as ventricles that are also show up due to accumulation of the contrast agent are indicted by double headed arrows. A visual examination of these maps suggests that the range of values among the measured parameters, vp, Ktrans and ve, did not appreciably change between the two imaging sessions. Note the relative uniformity of pixels for vp and ve, but the varying pixel colors for Ktrans that suggest the differing permeability of tumor vasculature in the core and periphery. The model selection maps (yellow pixels outlined in black) also followed a similar pattern and did not change between the two imaging times. However, the area covered by the pixels increased slightly from ‘test’ to ‘retest’ in direct relation to the increase in tumor size.
The average tumor volume increased between the two time points (38.1 + 26 vs 47.7 + 37, p = 0.022). Keeping with this increase, the average number of pixels with Model 3 acceptance increased during this interval (168 ± 93 at test and 201 ± 111 at retest; p < 0.01). Tables 1–6 contain the descriptive information for the test and retest for each summary estimate, along with the differences between the test and retest measures. None of the differences between the test and retest measures were significant, and this was true for both paired t-tests and Wilcoxon signed rank tests. Each parameter in the T-S sample exhibited a positive skewness, albeit with the summary measures of skewness and kurtosis having large variability. The ICC’s for the mean and median can also be found in Tables 1–6. For all but vp the reproducibility measures were in the moderate to almost perfect range. However, for vp there appears to be no reproducibility between test and retest. There was also a large amount of variability in the differences between these two measures of vp. The highest levels of reproducibility were observed for Ktrans. Even though the ICC measures for TBF were in the moderate to substantial ranges, it should be noted that there remained a high degree of variability in the differences between the test and retest measures which lead to large confidence intervals.
Table 1.
Summary statistics for plasma volume, vp.
| Statistical parameters | Median (× 10−3) | Mean (× 10−3) | Variance (× 10−4) | Skewness | Kurtosis | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| Mean | SD | Mean | SD | Mean | SD | Mean | SD | Mean | SD | |
| Test values | 6.73 | 2.70 | 9.28 | 8.06 | 4.22 | 8.0 | 0.18 | 3.64 | 13.29 | 30.88 |
| Retest values | 6.27 | 6.56 | 6.47 | 8.21 | 1.85 | 3.43 | 0.39 | 2.33 | 6.88 | 10.71 |
| Mean difference | −0.46 | 7.14 | −2.82 | 13.62 | 2.37 | 5.63 | 0.20 | 3.74 | −6.40 | 34.20 |
| Paired t-test: p-value | 0.81 | 0.45 | 0.14 | 0.84 | 0.49 | |||||
| Wilcoxon sign test: p-value | 0.54 | 0.66 | 0.29 | 0.54 | 0.46 | |||||
| 95% CI for mean difference | (−4.58, 3.67) | (−10.68, 5.05) | ||||||||
| Intra-class correlation coefficient | −0.013 | −0.401 | ||||||||
The differences were not statistically significant for any sampling statistic in the period of 24 h. Standard deviations (the square root of the variances) for the sampling distribution of the statistical parameters across animals are provided as an indication of the characteristic spread of these parameters, with no expectation that these parameters are normally distributed.
Table 6.
Summary statistics for interstitial distribution volume (VD) measured in tumor, tumor inner rim and outer normal edge.
| Statistical parameters | VD tumor (× 10−1) | VD inner edge (× 10−1) | VD outer edge (× 10−1) | Flux (× 10−3) | ||||
|---|---|---|---|---|---|---|---|---|
| Mean | SD | Mean | SD | Mean | SD | Mean | SD | |
| Test values | 1.98 | 0.66 | 1.90 | 0.75 | 1.00 | 0.37 | 1.87 | 1.18 |
| Retest values | 2.24 | 0.70 | 1.99 | 0.80 | 0.99 | 0.43 | 1.89 | 1.83 |
| Mean difference | 0.26 | 0.65 | 0.09 | 0.85 | −0.01 | 0.38 | 0.02 | 1.13 |
| Paired t-test: p-value | 0.14 | 0.48 | 0.86 | 0.72 | ||||
| Wilcoxon sign test: p-value | 0.15 | 0.25 | 0.76 | 0.97 | ||||
| 95% CI for mean difference | (−0.10, 0.62) | (−0.40, 0.58) | (−0.23, 0.20) | (−0.63, 0.67) | ||||
| Intra-class correlation coefficient | 0.54 | 0.41 | 0.56 | 0.73 | ||||
Values from the normal outer edge were lower than from those from tumor inner rim, suggesting a decrease in interstitial space or compression of the peritumoral normal tissue. Flux values indicate the outward flow of the contrast agent, contributing to peritumoral edema often observed in solid tumors. Other details are same as in for Table 1.
Values for VD in the rim decreased progressively from tumor inner edge to outer edge, but their test-retest values for a given region did not change. The MRCA flux values were consistent across the two time points tested (Table 6).
The summary data for each parameter were analyzed for differences. Bland-Altman plots for the summary measures of median, mean and variance for vp, Ktrans, ve, TBF, VD and ADC demonstrated no apparent trends between the differences and averages of the test-retest measures. (Supplemental data: Figs. 1–18). Also, none of the Kendall-tau tests for trend were significant suggesting that stable estimates of most vascular parameters over a 24 h period are available in this model at this stage of tumor progression.
4. Discussion
The purposes of the following analyses were to assess the reproducibility (or repeatability) of taking DCE-MRI measurements 24 h apart in a rat tumor model and to provide confidence intervals around the observed differences between the two measurements. The latter could then be potentially used to evaluate “true” treatment effects in future studies. The data demonstrate that over a period of 24 h at this stage of tumor growth, several vascular parameters in the 9L orthotopic gliosarcoma model remain stable. Since a rapidly worsening tumor status may represent end-stage tumor burden on brain, the tumors investigated this study presumably represent a stage that can respond to treatments. In such a scenario, these parameters can serve as non-invasive, MRI biomarkers of tumor pathophysiology and also of treatment efficacy. Utilizing them can assist in longitudinal assessments of a putative drug on progression-free as well as long-term survival. Supporting this conclusion, estimates of vascular permeability parameters were stably reproducible, thus supporting their use as biomarkers, particularly when used as paired comparisons. Paired comparisons of the 14 median values of Ktrans, a parameter employed in several studies as an index of treatment effects on tumor [18,26–29], produced a mean difference of ~8% of the grouped mean values of that statistic. Since the 9L gliosarcoma is among the most widely used rodent models for brain tumors [30–32], it is likely that these data will be of significant interest to a number of investigators. Additionally, the persistent positive skewness in the voxel-based sampling of vascular parameters suggests that the median may be a better summary statistic for single-point reporting of trends in the sampled parameters, as in previous work in the U251 model of cerebral tumor [14] and also in human glioma evaluations [33]. It is noteworthy that despite the absence of significant differences between the mean values from test and retest imaging sessions, the values did not closely cluster around the mean hinting at the range of pixel-wise variations within a given ROI. This information can be important when therapeutic efficacies are being evaluated to understand whether a putative drug’s effects are limited by the pre-existing magnitude of the change it is expected to target.
Histologically, tumor mass was identifiable by its increased cell density, leading to darker shades of staining. Distinct varied cell types typical of normal brain architecture were mostly absent (Fig. 2-A). Mitotic cells could be seen within the tumor mass. Most tumors did not seem well circumscribed, showing tumor infiltration into normal tissue. These migration patterns followed at least three separate routes. There were spherical- and oblong-shaped clusters of tumor cells moving away from tumor borders into adjacent normal brain (Fig. 2-B), infiltration along perivascular/Virchow-Robin spaces (Fig. 2-C), and/or along white matter pathways (Fig. 2-D). Such presentations of these tumors on H & E staining were similar to those previously reported for this model [6].
Along with typical presentation on H & E, presence of 9L cells was also discernible with Picrosirius red staining. It was observed that distribution of collagen within the tumor mass was in spots usually toward its periphery with some cases of partial tumor encapsulation (Fig. 3-B, C). The collagen subtypes could not be discriminated by this staining, although collagen Type I and IV are known to be expressed in gliomas [15,34]. Different collagen subtypes are reported to demarcate varied levels of differentiation in other cancers such as oral squamous cell carcinoma [35], but such collagen content/subtype-based classifications in gliomas are yet to be done.
Phenotypic presentations of 9L tumors on DCE-MRI were typical of human gliomas [36,37]. They presented a fairly well defined border with a ring of contrast enhancement and peritumoral fluid flux. However, characteristic features of the tumor-normal tissue boundary that contribute to the flux and peritumoral edema have not been reported before for this model. The measured values for VD from tumor, rim and edge in these tumors and the MRCA flux data provided some insight into the possible interactions of tumor periphery and normal tissue in this model. VD values from tumor tissue represent a method to estimate tumor cell density and, in addition, represent a novel measure of the extravascular, extracellular space available for MRCA flow. The MRCA Magnevist neither binds to any transporters nor is taken up by cells, but diffuses through this space after extravasation. The absence of leaky vessels beyond the tumor edge prevents its backflux via vasculature. Thus, an estimate of VD from this region along with flux rates can be of help in estimating its plausible driving forces, e.g., TBF, interstitial fluid pressure and/or peritumoral compression. These physical forces determine tumor perfusion and resistance to drug penetration. Thus, their estimation may assist in evaluating tumor staging and aggressiveness. However, underlining the importance of tumor model-dependent differences, the magnitude of several of these imaging biomarkers varied between the orthotopic 9L and U251 models. In these studies, VD values from the tumor and inner rim were about 20%, in comparison to tumor outer normal edge values of 10%. Tumor and inner edge values from 9L vary from a U251 glioblastoma model that gave VD values of ~7.5% and 15% in the tumor mass and its rim, respectively [14], although comparable VD in the outer normal edge as in the U251 model (10%) suggest peripheral tumor tissue compression to a similar degree in both models. However, owing probably due to the larger VD values intratumorally, i.e., suggestive of more available interstitial space for flow, the MRCA flux values for 9L were far lower than those in U251. This was also reflected by decreased extent of peritumoral ring of contrast enhancement in 9L, in comparison to U251 (unpublished observations).
Utility of these various parameters is dependent on the effect being tested. Some are not amenable to serve as acute imaging biomarkers. For instance, Wilkins et al. reported that tumor volume was not affected by radiotherapy over a period of 7 days despite tumor necrosis observed on histopathology [38]. In the present studies tumor volume increased significantly from test to retest and is unlikely to be affected by a treatment in between. Vascular volume, vp, was observed to be variable and, thus, may not be a reliable biomarker. Of the other parameters generated by model selection analysis of tumor tissue, Ktrans is known to be sensitive in general to vasoactive agents [39–41]. Along with ve, Ktrans has been shown to be of use in grading gliomas [42]. Responses of TBF and Ktrans have been shown to mirror single and combination treatment effects of cediranib and SC6889 in a 4C8 mouse glioma model [43]. Using model selection approach, temporal responses of glioma to experimental, single and combination therapies in other glioma models have been reported [44,45]. We have shown here that Ktrans was the most stable parameter, supporting its utility as a tumor vascular biomarker. However, no other report has explored utilizing graded alterations in Ktrans values spanning the tumor mass to demarcate regions with varying vascular characteristics. Model selection provides us with tools for such an analysis and modulating effects of smaller and greater extents of underlying damage may have on treatment efficacy. The other accompanying imaging indices from model selection analysis presented herein, viz., various VD values and flux provide additional indices of tumor physiological status and their use as biomarkers needs to be established. With the increasing applications of DCE-MRI for clinical tumor evaluations, it has been recommended that tracer kinetic analysis and model selection, with their generation of physiological indices of tumor biology, are superior to such indices as signal intensity, contrast enhancement ratio and area under the curve, which have no clear physiological interpretations [46]. In contrast, Semi-automated, interactive tumor segmentation being currently the preferred choice in clinical imaging [47], model selection can generate several unbiased indices to aid in more effective tumor grading and longitudinal evaluations.
To summarize, this study has showed the relative stability of several MRI biomarkers that reflect the physiological state of the tumor and its surround. Model selection paradigms generated stable and reproducible ROIs for data summary. The 9L model replicates several phenotypic features of human glioma and is being employed in laboratories to test experimental mono- and combination therapies [6,48,49]. DCE-MRI with model selection, a minimally invasive and quantitative method, can be very useful for such evaluations and for their quick translation into clinical applications.
Supplementary Material
Table 2.
Summary statistics for forward volume transfer constant, Ktrans.
| Statistical parameters | Median (× 10−2) | Mean (× 10−2) | Variance (× 10−3) | Skewness | Kurtosis | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| Mean | SD | Mean | SD | Mean | SD | Mean | SD | Mean | SD | |
| Test values | 3.10 | 2.21 | 3.69 | 2.43 | 1.58 | 2.10 | 2.66 | 3.32 | 17.74 | 36.40 |
| Retest values | 2.93 | 1.85 | 3.60 | 2.09 | 2.65 | 7.66 | 1.63 | 2.15 | 9.41 | 15.0 |
| Mean difference | −0.16 | 1.32 | −0.10 | 0.85 | 1.07 | 6.24 | −1.03 | 3.39 | −8.33 | 38.62 |
| Paired t-test: p-value | 0.65 | 0.67 | 0.53 | 0.27 | 0.43 | |||||
| Wilcoxon sign test: p-value | > 0.99 | 0.90 | 0.39 | 0.17 | 0.76 | |||||
| 95% CI for mean difference | (−0.93, 0.6) | (−0.59, 0.39) | ||||||||
| Intra-class correlation coefficient | 0.79 | 0.93 | ||||||||
The differences were not statistically significant for any sampling statistic. Other details are same as in for Table 1.
Table 3.
Summary statistics for interstitial volume fraction, ve.
| Statistical parameters | Median (× 10−1) | Mean (× 10−1) | Variance (× 10−2) | Skewness | Kurtosis | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| Mean | SD | Mean | SD | Mean | SD | Mean | SD | Mean | SD | |
| Test values | 1.88 | 0.68 | 1.90 | 0.68 | 0.96 | 1.14 | 0.99 | 2.93 | 10.24 | 30.69 |
| Retest values | 1.94 | 0.65 | 2.00 | 0.60 | 1.18 | 1.46 | 0.34 | 0.80 | 1.71 | 2.69 |
| Mean difference | 0.06 | 0.72 | 0.11 | 0.53 | 0.23 | 1.22 | −0.65 | 2.95 | −8.53 | 30.90 |
| Paired t-test: p-value | 0.76 | 0.46 | 0.49 | 0.42 | 0.32 | |||||
| Wilcoxon sign test: p-value | 0.50 | 0.50 | 0.80 | 0.90 | 0.26 | |||||
| 95% CI for mean difference | (−0.36, 0.47) | (−0.20, 0.42) | ||||||||
| Intra-class correlation coefficient | 0.42 | 0.65 | ||||||||
The differences were not statistically significant for any sampling statistic. Other details are same as in for Table 1.
Table 4.
Summary statistics for tumor blood flow (TBF).
| Statistical parameters | Median | Mean | Variance | Skewness | Kurtosis | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| Mean | SD | Mean | SD | Mean | SD | Mean | SD | Mean | SD | |
| Test values | 14.91 | 16.40 | 20.58 | 14.24 | 417.1 | 382.2 | 1.48 | 0.98 | 2.60 | 2.61 |
| Retest values | 9.81 | 10.85 | 15.35 | 9.39 | 332.0 | 209.8 | 1.94 | 1.14 | 5.34 | 6.16 |
| Mean difference | −5.01 | 10.31 | −5.07 | 11.84 | −72.1 | 429.1 | 0.48 | 1.04 | 2.78 | 5.55 |
| Paired t-test: p-value | 0.12 | 0.16 | 0.57 | 0.13 | 0.11 | |||||
| Wilcoxon sign test: p-value | 0.08 | 0.42 | 0.91 | 0.09 | 0.12 | |||||
| 95% CI for mean difference | (−11.56, 1.54) | (−12.59, 2.45) | ||||||||
| Intra-class correlation coefficient | 0.73 | 0.53 | ||||||||
The differences were not statistically significant for any sampling statistic. Other details are same as in for Table 1.
Table 5.
Summary statistics for apparent diffusion coefficient (ADC).
| Statistical parameters | Median (× 10−3) | Mean (× 10−3) | Variance (× 10−7) | Skewness | Kurtosis | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| Mean | SD | Mean | SD | Mean | SD | Mean | SD | Mean | SD | |
| Test values | 3.40 | 0.54 | 3.42 | 0.52 | 3.78 | 3.40 | 0.51 | 1.20 | 2.77 | 3.95 |
| Retest values | 3.34 | 0.50 | 3.40 | 0.48 | 2.92 | 2.74 | 1.22 | 1.71 | 6.41 | 14.07 |
| Mean difference | −0.06 | 0.45 | 0.02 | 0.44 | −0.86 | 3.47 | 0.71 | 2.11 | 3.65 | 15.47 |
| Paired t-test: p-value | 0.64 | 0.86 | 0.38 | 0.23 | 0.39 | |||||
| Wilcoxon sign test: p-value | 0.90 | > 0.99 | 0.42 | 0.29 | 0.35 | |||||
| 95% CI for mean difference | (−0.31, 0.20) | (−0.28, 0.23) | ||||||||
| Intra-class correlation coefficient | 0.63 | 0.61 | ||||||||
The differences were not statistically significant for any sampling statistic. Other details are same as in for Table 1.
Acknowledgments
Funding
Research reported in this publication was supported by National Cancer Institute of the National Institutes of Health under award number R01CA135329. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Abbreviations:
- DCE-MRI
dynamic contrast enhanced magnetic resonance imaging
- MRCA
magnetic resonance contrast agent
- vp
plasma volume
- Ktrans
forward volumetric transfer constant
- ve
interstitial volume fraction
- kep
reverse transfer constant
- VD
extracellular distribution volume
- TBF
tumor blood flow
- H & E
hematoxylin and eosin
- RT
radiotherapy
- EGFR
epidermal growth factor receptor
- ADC
apparent diffusion coefficient
- ASL
arterial spin labeling
- DWI
diffusion-weighted imaging
- LL
Look-Locker
- 2GE
dual echo gradient echo
- ROI
region of interest
- TR
repetition time
- TE
echo time
- FA
flip angle
- NA
number of averages
Footnotes
Appendix A. Supplementary data
Supplementary data to this article can be found online at http://dx.doi.org/10.1016/j.mri.2017.09.003.
References
- [1].Stupp R, Mason WP, van den Bent MJ, Weller M, Fisher B, Taphoorn MJB, et al. National Cancer Institute of Canada Clinical Trials Group: radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N Engl J Med 2005;352:987–96. [DOI] [PubMed] [Google Scholar]
- [2].McAleer MF, Brown PD. Therapeutic management of gliosarcoma in the temozolomide era. CNS Oncol 2015;4:171–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Lutterbach J, Guttenberger R, Pagenstecher A. Gliosarcoma: a clinical study. Radiother Oncol 2001;61:57–64. [DOI] [PubMed] [Google Scholar]
- [4].Cachia D, Kamiya-Matsuoka C, Mandel JJ, Olar A, Cykowski MD, Armstrong TS, et al. Primary and secondary gliosarcomas: clinical, molecular and survival characteristics. J Neurooncol 2015:401–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Damodaran O, van Heerden J, Nowak AK, Bynevelt M, McDonald K, Marsh J, et al. Clinical management and survival outcomes of gliosarcomas in the era of multi-modality therapy. J Clin Neurosci 2014;21:478–81. 10.1016/j.jocn.2013.07.042. [DOI] [PubMed] [Google Scholar]
- [6].Bouchet A, Bidart M, Miladi I, Le Clec’h C, Serduc R, Coutton C, et al. Characterization of the 9L gliosarcoma implanted in the Fischer rat: an orthotopic model for a grade IV brain tumor. Tumour Biol 2014;35:6221–33. [DOI] [PubMed] [Google Scholar]
- [7].Ewing JR, Knight RA, Nagaraja TN, Yee JS, Nagesh V, Whitton PA, et al. Patlak plots of Gd-DTPA MRI data yield blood-brain transfer constants concordant with those of 14C-sucrose in areas of blood-brain opening. Magn Reson Med 2003;50:283–92. [DOI] [PubMed] [Google Scholar]
- [8].Patlak CS, Blasberg RG, Fenstermacher JD. Graphical evaluation of blood-to-brain transfer constants from multiple-time uptake data. J Cereb Blood Flow Metab 1983;3:1–7. [DOI] [PubMed] [Google Scholar]
- [9].Tofts PS, Brix G, Buckley DL, Evelhoch JL, Henderson E, Knopp MV, et al. Estimating kinetic parameters from dynamic contrast-enhanced T(1)-weighted MRI of a diffusable tracer: standardized quantities and symbols. J Magn Reson Imaging 1999;10:223–32. [DOI] [PubMed] [Google Scholar]
- [10].Aryal MP, Nagaraja TN, Keenan KA, Bagher-Ebadian H, Panda S, Brown SL, et al. Dynamic contrast enhanced MRI parameters and tumor cellularity in a rat model of cerebral glioma at 7 T. Magn Reson Med 2014;71:2206–14. 10.1002/mrm.24873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Logan J Graphical analysis of PET data applied to reversible and irreversible tracers. Nucl Med Biol 2000;27:661–70. [DOI] [PubMed] [Google Scholar]
- [12].Li X, Huang W, Morris EA, Tudorica LA, Seshan VE, Rooney WD, et al. Dynamic NMR effects in breast cancer dynamic-contrast-enhanced MRI. Proc Natl Acad Sci U S A 2008;105:17937–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Li X, Rooney WD, Varallyay CG, Gahramanov S, Muldoon LL, Goodman JA, et al. Dynamic-contrast-enhanced-MRI with extravasating contrast reagent: rat cerebral glioma blood volume determination. J Magn Reson 2010;206:190–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Aryal MP, Nagaraja TN, Brown SL, Lu M, Bagher-Ebadian H, Ding G, et al. Intratumor distribution and test-retest comparisons of physiological parameters quantified by dynamic contrast-enhanced MRI in rat U251 glioma. NMR Biomed 2014;27:1230–8. 10.1002/nbm.3178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Ghahary A, Bhatnagar R, Price K, Forsyth NL, Shen YJ, Tredget TE, et al. Rat glioma cell lines C6 and 9L synthesize type 1 collagen in vitro. Brain Res Bull 1992;28:47–56. [DOI] [PubMed] [Google Scholar]
- [16].Ewing JR, Brown SL, Lu M, Panda S, Ding G, Knight RA, et al. Model selection in magnetic resonance imaging measurements of vascular permeability: gadomer in a 9L model of rat cerebral tumor. J Cereb Blood Flow Metab 2006;26:310–20. [DOI] [PubMed] [Google Scholar]
- [17].Nagaraja TN, Croxen RL, Panda S, Knight RA, Keenan KA, Brown SL, et al. Application of arsenazo III in the preparation and characterization of an albumin-linked, gadolinium-based macromolecular magnetic resonance contrast agent. J Neuosci Methods 2006;157:238–45. [DOI] [PubMed] [Google Scholar]
- [18].Ewing JR, Bagher-Ebadian H. Model selection in measures of vascular parameters using dynamic contrast-enhanced MRI: experimental and clinical applications. NMR Biomed 2013;26:1028–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Ewing JR, Nagaraja TN, Aryal MP, Keenan KA, Elmghirbi R, Bagher-Ebadian H, et al. Peritumoral tissue compression is predictive of exudate flux in a rat model of cerebral tumor: an MRI study in an embedded tumor. NMR Biomed 2015;28:1557–69. 10.1002/nbm.3418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Patlak C, Blasberg R. Graphical evaluation of blood to brain transfer constants from multiple time up take data. Generalizations. J Cereb Blood Flow Metab 1985;5:584–90. [DOI] [PubMed] [Google Scholar]
- [21].Logan J, Fowler JS, Volkow ND, Wolf AP, Dewey SL, Schlyer DJ, et al. Graphical analysis of reversible radioligand binding from time-activity measurements applied to [N-11C-methyl]-(−)-cocaine PET studies in human subjects. J Cereb Blood Flow Metab 1990;10:740–7. [DOI] [PubMed] [Google Scholar]
- [22].Ewing JR, Wei L, Knight RA, Pawa S, Nagaraja TN, Brusca T, et al. Direct comparison of local cerebral blood flow rates measured by MRI arterial spin-tagging and quantitative autoradiography in a rat model of experimental cerebral ischemia. J Cereb Blood Flow Metab 2003;23(2):198–209. [DOI] [PubMed] [Google Scholar]
- [23].Williams DS, Detre JA, Leigh JS, Koretsky AP. Magnetic resonance imaging of perfusion using spin inversion of arterial water. Proc Natl Acad Sci 1992;89:212–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Bland JM, Altman DG. Measuring agreement in method comparison studies. Stat Methods Med Res 1999;8:135–60. [DOI] [PubMed] [Google Scholar]
- [25].Landis JR, Koch GG. The measurement of observer agreement for categorical data. Biometrics 1977;33(1):159–74. . [PubMed] [Google Scholar]
- [26].Jackson A, Jayson GC, Li KL, Zhu XP, Checkley DR, Tessier JJ, et al. Reproducibility of quantitative dynamic contrast-enhanced MRI in newly presenting glioma. Br J Radiol 2003;76:153–62. [DOI] [PubMed] [Google Scholar]
- [27].Kickingereder P, Sahm F, Wiestler B, Roethke M, Heiland S, Schlemmer H-P, et al. Evaluation of microvascular permeability with dynamic contrast-enhanced MRI for the differentiation of primary CNS lymphoma and glioblastoma: radiologic-pathologic correlation. AJNR Am J Neuroradiol 2014;35:1503–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Nagaraja TN, Aryal MP, Brown SL, Bagher-Ebadian H, Mikkelsen T, Yang JJ, et al. Cilengitide-induced temporal variations in transvascular transfer parameters of tumor vasculature in a rat glioma model: identifying potential MRI biomarkers of acute effects. PLoS One 2013;8:e84493 10.1371/journal.pone.0084493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Padhani AR, Hayes C, Landau S, Leach MO. Reproducibility of quantitative dynamic MRI of normal human tissues. NMR Biomed 2002;15:143–53. [DOI] [PubMed] [Google Scholar]
- [30].Barth RF, Kaur B. Rat brain tumor models in experimental neurooncology: the C6, 9L, T9, RG2, F98, BT4C, RT-2 and CNS-1 gliomas. J Neurooncol 2009;94:299–312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Huszthy PC, Daphu I, Niclou SP, Stieber D, Nigro JM, Sakariassen PO, et al. In vivo models of primary brain tumors: pitfalls and perspectives. Neuro Oncol 2012;14(8):979–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Jacobs VL, Valdes PA, Hickey WF, De Leo JA. Current review of in vivo GBM rodent models: emphasis on the CNS-1 tumour model. ASN Neuro 2011;3(3). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Patankar TF, Haroon HA, Mills SJ, Balériaux D, Buckley DL, GJM Parker, et al. Is volume transfer coefficient (Ktrans) related to histologic grade in human gliomas? AJNR Am J Neuroradiol 2005;26:2455–65. [PMC free article] [PubMed] [Google Scholar]
- [34].Ogawa K, Oguchi M, Nakashima Y, Yamabe H. Distribution of collagen type IV in brain tumors: an immunohistochemical study. J Neurooncol 1989;7:357–66. [DOI] [PubMed] [Google Scholar]
- [35].Patankar SR, Wankhedkar DP, Tripathi NS, Bhatia SN, Sridharan G. Extracellular matrix in oral squamous cell carcinoma: Friend or foe? Indian J Dent Res 2016;27:184–9. 10.4103/0970-9290.183125. [DOI] [PubMed] [Google Scholar]
- [36].Bagher-Ebadian H, Jain R, Nejad-Davarani SP, Mikkelsen T, Lu M, Jiang Q, et al. Model selection for DCE-T1 studies in glioblastoma. Magn Reson Med 2011;68:241–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Jain R, Ellika SK, Scarpace L, Schultz LR, Rock JP, Gutierrez J, et al. Quantitative estimation of permeability surface-area product in astroglial brain tumors using perfusion CT and correlation with histopathologic grade. AJNR Am J Neuroradiol 2008;29:694–700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Wilkins DE, Raaphorst GP, Saunders JK, Sutherland GR, Smith IC. Correlation between Gd-enhanced MR imaging and histopathology in treated and untreated 9L rat brain tumors. Magn Reson Imaging 1995;13(1):89–96. [DOI] [PubMed] [Google Scholar]
- [39].O’Neill AF, Qin L, Wen PY, de Groot JF, Van den Abbeele AD, Yap JT. Demonstration of DCE-MRI as an early pharmacodynamic biomarker of response to VEGF Trap in glioblastoma. J Neurooncol 2016;130(3):495–503. 10.1007/s11060-016-2243-5. [DOI] [PubMed] [Google Scholar]
- [40].Park J, Aryal M, Vykhodtseva N, Zhang YZ, McDannold N. Evaluation of permeability, doxorubicin delivery, and drug retention in a rat brain tumor model after ultrasound-induced blood-tumor barrier disruption. J Control Release 2016. 10.1016/j.jconrel.2016.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Zhang L, Zhao W, Liang C, Yi X, Pei Y, Lin Y, et al. VEGFR-1 targeted DNAzyme via transcatheter arterial delivery influences tumor vasculature assessed through dynamic contrast-enhanced magnetic resonance imaging. Oncol Rep 2016;36(3):1339–44. 10.3892/or.2016.4933. [DOI] [PubMed] [Google Scholar]
- [42].Zhang N, Zhang L, Qiu B, Meng L, Wang X, Hou BL. Correlation of volume transfer coefficient Ktrans with histopathologic grades of gliomas. J Magn Reson Imaging 2012;36:355–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Lobo MR, Kukino A, Tran H, Schabel MC, Springer CS Jr., Gillespie GY, et al. Synergistic antivascular and antitumor efficacy with combined cediranib and SC6889 in intracranial mouse glioma. PLoS One 2015;10(12):e0144488 10.1371/journal.pone.0144488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Brown SL, Nagaraja TN, Aryal MP, Panda S, Cabral G, Keenan KA, et al. MRI-tracked tumor vascular changes in the hours after single-fraction irradiation. Radiat Res 2015;183:713–21. [DOI] [PubMed] [Google Scholar]
- [45].Elmghirbi R, Nagaraja TN, Brown SL, Panda S, Aryal MP, Keenan KA, et al. Acute temporal changes of MRI-tracked tumor vascular parameters after combined anti-angiogenic and radiation treatments in a rat glioma model: identifying signatures of synergism. Radiat Res 2016. 10.1667/rr14358.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Duan C, Kallehauge JF, Bretthorst GL, Tanderup K, Ackerman JJ, Garbow JR. Are complex DCE-MRI models supported by clinical data? Magn Reson Med 2016. 10.1002/mrm.26189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Gordillo N, Montseny E, Sobrevilla P. State of the art survey on MRI brain tumor segmentation. Magn Reson Imaging 2013;31:1426–38. [DOI] [PubMed] [Google Scholar]
- [48].Huynh NT, Morille M, Bejaud J, Legras P, Vessieres A, Jaouen G, et al. Treatment of 9L gliosarcoma in rats by ferrociphenol-loaded lipid nanocapsules based on a passive targeting strategy via the EPR effect. Pharm Res 2011;28:3189–98. 10.1007/s11095-011-0501-y. [DOI] [PubMed] [Google Scholar]
- [49].Zhang R, Saito R, Shibahara I, Sugiyama S, Kanamori M, Sonoda Y, et al. Temozolomide reverses doxorubicin resistance by inhibiting P-glycoprotein in malignant glioma cells. J Neurooncol 2016;126:235–42. 10.1007/s11060-015-1968-x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


