Abstract
Simple Summary
Little information is currently available on the epidemiology of parasitic and commensal protist species in captive non-human primates (NHP) and their zoonotic potential. This study investigates the occurrence, molecular diversity, and potential transmission dynamics of parasitic and commensal protist species in a zoological garden in southern Spain. The prevalence and genotypes of the main enteric protist species were investigated in faecal samples from NHP, zookeepers and free-living rats by molecular (PCR and sequencing) methods. A high prevalence of the diarrhoea-causing protists Giardia duodenalis and Blastocystis sp. (but not Cryptosporidium spp.) was observed in captive NHP at the Córdoba Zoo Conservation Centre. NHP can harbour zoonotic genotypes of G. duodenalis, Blastocystis sp., and Enterocytozoon bieneusi. Indeed, strong evidence of the occurrence of Blastocystis zoonotic transmission between NHP and their handlers was provided, despite the use of personal protective equipment and the implementation of strict health and safety protocols. Free-living sympatric rats are infected by host-specific species/genotypes of the investigated protists and seem to play a limited role as source of infections to NHP or humans in this setting. The extent of these findings should be confirmed in similar epidemiological surveys targeting other captive NHP populations.
Abstract
Little information is currently available on the epidemiology of parasitic and commensal protist species in captive non-human primates (NHP) and their zoonotic potential. This study investigates the occurrence, molecular diversity, and potential transmission dynamics of parasitic and commensal protist species in a zoological garden in southern Spain. The prevalence and genotypes of the main enteric protist species were investigated in faecal samples from NHP (n = 51), zookeepers (n = 19) and free-living rats (n = 64) by molecular (PCR and sequencing) methods between 2018 and 2019. The presence of Leishmania spp. was also investigated in tissues from sympatric rats using PCR. Blastocystis sp. (45.1%), Entamoeba dispar (27.5%), Giardia duodenalis (21.6%), Balantioides coli (3.9%), and Enterocytozoon bieneusi (2.0%) (but not Troglodytella spp.) were detected in NHP. Giardia duodenalis (10.5%) and Blastocystis sp. (10.5%) were identified in zookeepers, while Cryptosporidium spp. (45.3%), G. duodenalis (14.1%), and Blastocystis sp. (6.25%) (but not Leishmania spp.) were detected in rats. Blastocystis ST1, ST3, and ST8 and G. duodenalis sub-assemblage AII were identified in NHP, and Blastocystis ST1 in zookeepers. Giardia duodenalis isolates failed to be genotyped in human samples. In rats, four Cryptosporidium (C. muris, C. ratti, and rat genotypes IV and V), one G. duodenalis (assemblage G), and three Blastocystis (ST4) genetic variants were detected. Our results indicate high exposure of NHP to zoonotic protist species. Zoonotic transmission of Blastocysts ST1 was highly suspected between captive NHP and zookeepers.
Keywords: Cryptosporidium, Giardia, Blastocystis, Enterocytozoon bieneusi, Balantioides coli, Troglodytella, non-human primates, rats, zoological garden
1. Introduction
Cryptosporidium spp., Giardia duodenalis, and Entamoeba histolytica are the most frequently identified protozoan parasites causing diarrhoeal disease in humans globally [1]. Clinical manifestations by these infections vary from self-limiting acute diarrhoea in immunocompetent individuals, to fatal chronic diarrhoea in immunocompromised patients [2]. In addition to these well-known enteric pathogens, other potential diarrhoea-causing protist species, including the Stramenopile Blastocystis sp. and the Microsporidia Enterocytozoon bieneusi, have gained wide clinical and scientific interest in recent years [3,4]. These parasites are transmitted via the faecal-oral route either directly (i.e., person-to-person) or indirectly (i.e., waterborne or foodborne). Remarkably, most of the species/genotypes of the above-mentioned protists can be zoonotically transmitted [5,6,7,8]. For this reason, assessing the occurrence and genetic diversity of enteric protists in domestic, captive, and free-living animal hosts is essential to ascertaining their transmission dynamics, including the occurrence and directionality of zoonotic events.
Cryptosporidium spp., G. duodenalis, Blastocystis sp., and E. bieneusi exhibit extensive intra-species genetic diversity leading to the identification of several genotypes/subtypes with marked differences in host and geographical range. At least 40 valid Cryptosporidium species and a similar number of genotypes of unknown species status are currently recognized, with C. hominis and C. parvum causing most of the infections documented in humans and non-human primate (NHP) species [6,9]. Giardia duodenalis is currently regarded as a multi-species complex comprising eight (A to H) distinct assemblages, of which assemblages A and B are frequently reported in humans and NHP [5]. At least 28 subtypes (ST) have been proposed within Blastocystis sp. with apparent loose host specificity. Of them, ST1–9 and ST12 have been documented in humans and/or NHP, among other vertebrates [8,10,11]. A recent evaluation of ST1–ST26 subtypes concluded that only 22 of those subtypes (ST1–ST17, ST21, ST23–ST26) should be acknowledged as legitimate subtypes [10], with the remaining six pending confirmation in future investigations. Finally, nearly 500 E. bieneusi genotypes have been reported and distributed in 11 genetic groups, of which Group 1 (e.g., Type IV, D, and EbpC) and Group 2 (e.g., BEB4, BEB6, I, and J) include most of the potentially zoonotic genotypes [12].
Little is known about the epidemiology of gastrointestinal protist parasites in captive non-human primates (NHP). In Spain, most of the few studies published to date were based on conventional (microscopy) methods and conducted mainly at the zoological gardens of Almuñecar (Granada) and Barcelona [13,14,15,16,17]. Only a single study attempted to characterize the genetic diversity of G. duodenalis in NHP at the Madrid and Valencia zoological gardens [18]. Besides Cryptosporidium spp., G. duodenalis, Blastocystis sp., and E. bieneusi, ciliated protists in NHP have been even poorly studied. This is the case of Balantioides coli, a zoonotic parasite that primarily infect domestic and wild swine, but has also been reported in NHP including gorillas, chimpanzees, bonobos, hamadryas baboons, and Rhesus macaques [19]. Moreover, the commensal ciliate Troglodytella abrassarti has been demonstrated a common finding in the faeces of captive and free-living great apes including eastern and western gorillas, chimpanzees, bonobos, and orangutans [20], but little information is available on its occurrence in captive and free-living lesser apes and monkeys.
This molecular-based epidemiological study aims primarily at assessing the frequency and genetic diversity of generalist and host-adapted enteric protist species in captive NHP and their caretakers at the Córdoba Zoo Conservation Centre (CZCC) in southern Spain, with a special focus on the investigation of potential zoonotic transmission events and their directionality. Secondarily, the same survey was conducted in a free-living sympatric rat population in the same enclosure in order to (i) investigate the role of rodents as transmitters of protist infections to NHP and humans and (ii) assess the suitability of rats as natural reservoirs of Leishmania spp.
2. Materials and Methods
2.1. Study Area
The CZCC extends over 4.5 hectares and include 437 specimens belonging to 102 mammalian, reptilian, and avian species. The CZCC has a small but diverse population of NHP species belonging to 10 genera including Cebuella (n = 2), Cercocebus (n = 4), Cercopithecus (n = 3), Eulemur (n = 2), Hylobates (n = 3), Lemur (n = 5), Macaca (n = 8), Mandrillus (n = 4), Saimiri (n = 3), and Varecia (n = 2). Individuals of the same species are kept in specific enclosures without contact with other NHP, except members of the Lemuridae family (genera Eulemur, Lemur, and Varecia) that share the same enclosure. All NHP are housed in facilities littered with natural materials such as ground bark or earth. The CZCC has strict health and safety protocols in place to ensure that animals, employees, and visitors have a reduced exposure to risk of infection or injury. Employees routinely use appropriate Personal Protective Equipment when in contact with animals or their faecal material.
2.2. Sampling
This cross-sectional study included two sampling periods carried out between December 2018 and January 2019, and between November and December 2019. Fresh faecal samples from NHP were directly collected from the ground at the time of routine cleaning and sanitation of enclosures. Information regarding sex, age, and enclosure sharing with other NHP species was recorded at the time of sampling. In parallel, fresh stool samples were also collected from zookeepers and veterinarians in close contact with NHP that volunteered to participate in the study. Human and NHP stool samples were stored at −20 °C without preservatives at the CZCC Veterinary Laboratory until the end of each sampling campaign, when they were shipped to the Spanish National Centre for Microbiology for downstream molecular analyses.
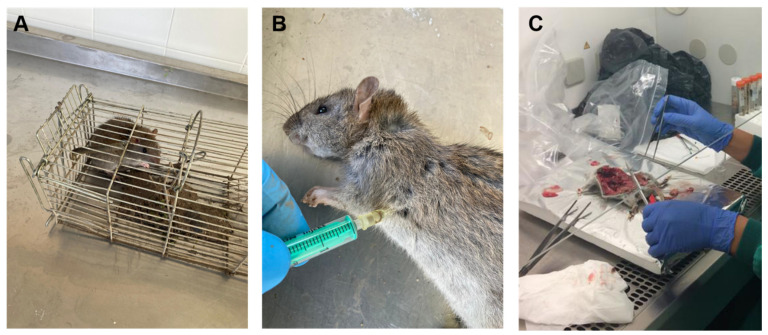
Taking advantage of an ongoing rodent control campaign carried out at the same time as the present study within the CZCC premises undertaken by the local authorities following European guidelines [21], free-living sympatric rats (Rattus spp.) were captured using Tomahawk live traps with bait (chicken, dry dog food, fruit, or peanut butter) (Figure 1A). Most captured rats were identified as brown rats (Rattus norvegicus), but differential detection with black rats (Rattus rattus) was not possible for younger individuals. Traps were placed in the evening and checked for captures the next morning. Rats were anaesthetized with medetomidine (1 mg/kg) and ketamine (50 mg/kg) and then humanely euthanized by an intracardiac injection of sodium pentobarbitone (Dolethal®, Vetoquinol Laboratories, Lure, France) at a dose >150 mg/kg (Figure 1B) [22]. Carcases were frozen at −20 °C and shipped to the Spanish National Centre for Microbiology for necropsy (Figure 1C). After thawing and dissection, the small and large intestine was removed, and the intestinal content extracted for further investigation of enteric protists by molecular methods. Additionally, liver, spleen, and ear skin samples were taken to assess the presence of amastigote forms of Leishmania spp.
Figure 1.
Sampling of rodent specimens within the premises of the Córdoba Zoo Conservation Centre. (A): Capture using live traps; (B): Humanely killing by intracardiac injection of sodium pentobarbitone; (C): Dissection of rat carcasses and organ removal.
2.3. Epidemiological Questionnaire
A standardized questionnaire (Table S1) and an informed consent was provided as part of the sampling kit to be completed by the CZCC personnel that volunteered to participate in the survey. Questions included: (i) demographic characteristics, e.g., age and sex, (ii) behavioural habits, e.g., hand and fruit/vegetable washing and whether there have been any occurrence of diarrhoea in the participant, their family members, and/or pets, (iii) work-related potential risk factors, e.g., contact with faecal material from NHP and/or other animal species, being a food handler, and (iv) additional questions on other risk factors, e.g., types of drinking water, use of recreational waters in the two weeks prior to sample collection, had any contact with pets and any recent travel abroad.
2.4. DNA Extraction and Purification
Genomic DNA was isolated from about 200 mg of each faecal specimen of human, NHP, or rodent origin by using the QIAamp DNA Stool Mini Kit (Qiagen, Hilden, Germany) according to the manufacturer’s instructions, except that samples mixed with InhibitEX buffer were incubated for 10 min at 95 °C. Additionally, genomic DNA from murine tissues (liver, spleen, and ear skin) was isolated using the Speed Tools DNA Extraction Kit (Biotools, Madrid, Spain). To do so, 10–15 mg of each tissue was homogenized in 100 µl of NET-10 buffer (10 mM NaCl, 10 mM EDTA, 10 mM Tris-HCl pH 8) and digested overnight at 56 °C with 100 µL of BT1 buffer (Biotools) and 20 µL Proteinase K (20 mg/mL). After digestion, genomic DNA was extracted according to the manufacturer’s instructions. In all cases, extracted and purified DNA samples were eluted in 200 μL of PCR-grade water and kept at 4 °C until further molecular analysis. A water extraction control was included in each sample batch processed.
2.5. Molecular Detection and Characterization of Giardia Duodenalis
Detection of G. duodenalis DNA was achieved using a real-time PCR (qPCR) method targeting a 62-bp region of the gene codifying the small subunit ribosomal RNA (ssu rRNA) of the parasite [23]. Amplification reactions (25 μL) consisted of 3 μL of template DNA, 0.5 µM of each primer Gd-80F and Gd-127R, 0.4 µM of probe (Table S2), and 12.5 μL TaqMan® Gene Expression Master Mix (Applied Biosystems, CA, USA). Detection of parasitic DNA was performed on a Corbett Rotor GeneTM 6000 real-time PCR system (QIAGEN) using an amplification protocol consisting of an initial hold step of 2 min at 55 °C and 15 min at 95 °C followed by 45 cycles of 15 s at 95 °C and 1 min at 60 °C. Water (no-template) and genomic DNA (positive) controls were included in each PCR run.
Giardia duodenalis isolates that tested positive by qPCR were subsequently assessed by sequence-based multi-locus genotyping of the genes encoding for the glutamate dehydrogenase (gdh), β-giardin (bg), and triose phosphate (tpi) proteins of the parasite. A semi-nested PCR was used to amplify a 432-bp fragment of the gdh gene [24]. PCR reaction mixtures (25 μL) included 5 μL of template DNA and 0.5 μM of the primer pairs GDHeF/GDHiR in the primary reaction and GDHiF/GDHiR in the secondary reaction (Table S2). Both amplification protocols consisted of an initial denaturation step at 95 °C for 3 min, followed by 35 cycles of 95 °C for 30 s, 55 °C for 30 s, and 72 °C for 1 min, with a final extension of 72 °C for 7 min.
A nested PCR was used to amplify a 511 bp-fragment of the bg gene [25]. PCR reaction mixtures (25 μL) consisted of 3 μL of template DNA and 0.4 μM of the primers sets G7_F/G759_R in the primary reaction and G99_F/G609_R in the secondary reaction (Table S2). The primary PCR reaction was carried out with the following amplification conditions: one step of 95 °C for 7 min, followed by 35 cycles of 95 °C for 30 s, 65 °C for 30 s, and 72 °C for 1 min, with a final extension of 72 °C for 7 min. The conditions for the secondary PCR were identical to the primary PCR except that the annealing temperature was 55 °C.
A nested PCR was used to amplify a 530 bp-fragment of the tpi gene [26]. PCR reaction mixtures (50 μL) included 2‒2.5 μL of template DNA and 0.2 μM of the primer pairs AL3543/AL3546 in the primary reaction and AL3544/AL3545 in the secondary reaction (Table S2). Both amplification protocols consisted of an initial denaturation step at 94 °C for 5 min, followed by 35 cycles of 94 °C for 45 s, 50 °C for 45 s, and 72 °C for 1 min, with a final extension of 72 °C for 10 min.
2.6. Molecular Detection and Characterization of Cryptosporidium spp.
The presence of Cryptosporidium spp. was assessed using a nested-PCR protocol to amplify a 587 bp fragment of the ssu rRNA gene of the parasite [27]. Amplification reactions (50 μL) included 3 μL of DNA sample and 0.3 μM of the primer pairs CR-P1/CR-P2 in the primary reaction and CR-P3/CPB-DIAGR in the secondary reaction (Table S2). Both PCR reactions were carried out as follows: one step of 94 °C for 3 min, followed by 35 cycles of 94 °C for 40 s, 50 °C for 40 s and 72 °C for 1 min, concluding with a final extension of 72 °C for 10 min.
2.7. Molecular Detection and Characterization of Blastocystis sp.
Identification of Blastocystis sp. was achieved by a direct PCR protocol targeting the ssu rRNA gene of the parasite [28]. The assay uses the pan-Blastocystis, barcode primer pair RD5/BhRDr to amplify a PCR product of ~600 bp. Amplification reactions (25 μL) included 5 μL of template DNA and 0.5 μM of each primer (Table S2). Amplification conditions consisted of one step of 95 °C for 3 min, followed by 30 cycles of 1 min each at 94, 59 and 72 °C, with an additional 2 min final extension at 72 °C.
In Blastocystis-positive samples from free-living sympatric rats for which Sanger sequencing data were of suboptimal quality, a next-generation amplicon sequencing strategy was used to identify Blastocystis sp. subtypes as previously described [29]. In brief, primers ILMN_Blast505_532F and ILMN_Blast998_1017R were used to generate amplicons. These primers amplify a fragment of the ssu rRNA gene of ~500 bp and are identical to Blast505_532F/Blast998_1017R [30] with the exception of containing the Illumina overhang adapter sequences on the 5′ end. Amplicons from two rats were used to prepare sequencing libraries, and final libraries were quantified by Qubit fluorometric quantitation (Invitrogen, Carlsbad, CA, USA) prior to normalization. A final pooled library concentration of 8 pM with 20% PhiX control was sequenced using Illumina MiSeq 600 cycle v3 chemistry (Illumina, San Diego, CA, USA). Paired end reads were processed and analyzed with an in-house pipeline that uses the BBTools package v38.82 [31], VSEARCH v2.15.1 [32], and BLAST + 2.10.1. Briefly, read pairs were merged, filtered for quality and length, denoised, and checked for chimeric sequences. Clustering and the assignment of centroid sequences to operational taxonomic units (OTUs) was performed within each sample at a 98% identity threshold. Only those OTUs with a minimum of 100 sequences were retained and then checked for chimeras once more. OTUs were then blasted against Blastocystis references from the National Center for Biotechnology Information (NCBI). Hits below an alignment length of 400 bp were removed.
2.8. Molecular Detection and Characterization of Enterocytozoon bieneusi
Detection of E. bieneusi was conducted by a nested PCR protocol to amplify the internal transcribed spacer (ITS) region as well as portions of the flanking large and small subunit of the ribosomal RNA gene as previously described [33]. The outer EBITS3/EBTIS4 and inner EBITS1/EBITS2.4 primer sets (Table S2) were used to generate a PCR product of 390 bp, respectively. Cycling conditions for the primary PCR consisted of one step of 94 °C for 3 min, followed by 35 cycles of amplification (denaturation at 94 °C for 30 s, annealing at 57 °C for 30 s, and elongation at 72 °C for 40 s), with a final extension at 72 °C for 10 min. Conditions for the secondary PCR were identical to the primary PCR except only 30 cycles were carried out with an annealing temperature of 55 °C.
2.9. Molecular Differential Detection of Entamoeba histolytica and Entamoeba dispar
Detection and differential diagnosis between pathogenic E. histolytica and non-pathogenic E. dispar were carried out by a qPCR method targeting a 172-bp fragment of the gene codifying the ssu rRNA gene of the E. histolytica/E. dispar complex [34,35]. Amplification reactions (25 μL) consisted of 3 μL template DNA, 12.5 pmol of the primer set Ehd-239F/Ehd-88R, 5 pmol of each TaqMan® probe (Table S2), and TaqMan® Gene Expression Master Mix (Applied Biosystems). Cycling conditions and data analysis were as described above for the detection of G. duodenalis.
2.10. Molecular Detection of Balantioides coli
Detection of B. coli was attempted by a direct PCR assay to amplify the complete ITS1–5.8s-rRNA–ITS2 region and the last 117 bp (3’ end) of the ssu-rRNA sequence of this ciliate using the primer set B5D/B5RC [36]. PCR reactions (25 μL) consisted of 2 μL of template DNA and 0.4 μM of each primer (Table S2). PCR conditions were as follows: 94 °C for 10 min; 30 cycles of 94 °C for 1 min, 60 °C for 1 min, 72 °C for 1 min, and a final extension for 5 min at 72 °C.
2.11. Molecular Detection of Troglodytella spp.
Detection of Troglodytella spp. was only attempted in captive NHP. Identification of this ciliate mutualist was carried out by a direct PCR method targeting a 401 bp fragment of the ITS region of the rDNA (ITS1-5.8S rDNA-ITS2) of the protist [37]. PCR reactions (25 µl) contained 2 µL of template DNA and 0.8 µM of each primer SSU-end/LSU-start (Table S2). Conditions of PCR for ITS amplification were initial denaturation for 2 min at 94 °C, 35 cycles of 45 s at 94 °C, 45 s at 50 °C, and 90 s at 72 °C, and terminal elongation for 5 min at 72 °C.
2.12. Molecular Detection of Leishmania spp.
Detection of Leishmania spp. was solely attempted in rodents, the only mammalian host for which tissue samples were available. Identification of this kinetoplastida parasite was carried out by a nested PCR protocol to amplify a partial fragment (358 bp) of the ssu rRNA gene of the parasite [38]. The primary PCR reaction (50 µL) contained 10 µL of template DNA and 15 pmol of the primer pair R221/R332 (Table S2). Conditions of PCR for ssu rRNA amplification were initial denaturation for 5 min at 94 °C, 35 cycles of 30 s at 94 °C, 30 s at 60 °C, and 30 s at 72 °C, and terminal elongation for 10 min at 72 °C. In the secondary PCR reaction (25 µL), 10 µL of a 1:40 dilution of the primary PCR product was re-amplified using 7.5 pmol of the primer pair R223/R333 (Table S2). Cycling conditions were as described above except that the annealing temperature was set at 65 °C.
All the direct, semi-nested, and nested PCR protocols described above were conducted on a 2720 Thermal Cycler (Applied Biosystems). Reaction mixes always included 2.5 units of MyTAQTM DNA polymerase (Bioline GmbH, Luckenwalde, Germany), and 5× MyTAQTM Reaction Buffer containing 5 mM dNTPs and 15 mM MgCl2, except for the amplification of Leishmania spp., for which 0.7–1.4 units of Tth DNA polymerase (Biotools B&M Laboratories, S.A., Madrid, Spain) were used. Laboratory-confirmed positive and negative DNA samples of human and animal origin for each parasitic species investigated were routinely used as controls and included in each round of PCR. PCR amplicons were visualized on 1.5–2% D5 agarose gels (Conda, Madrid, Spain) stained with Pronasafe (Conda) or Gel Red (Biotium, Fremont, CA, USA) nucleic acid staining solutions. A 100 bp DNA ladder (Boehringer Mannheim GmbH, Baden-Wurttemberg, Germany) was used for the sizing of obtained amplicons. Positive-PCR products were directly sequenced in both directions using appropriate internal primer sets (Table S2). DNA sequencing was conducted by capillary electrophoresis using the BigDye® Terminator chemistry (Applied Biosystems) on an on ABI PRISM 3130 automated DNA sequencer.
The sequences obtained in this study have been deposited in GenBank under accession numbers MW417420–MW417422 (G. duodenalis), MW414634–MW414644 and MW581486 (Blastocystis sp.), MW406908–MW406921 (Cryptosporidium spp.) and MW414645 (E. bieneusi).
2.13. Statistical Analysis
Prevalence and 95% confidence intervals (95% CI) of any enteric protist infection/carriage, alone or in combination, in the study populations were calculated. Statistically significant differences between the prevalence of enteric protist species in NHP and sampling period were analyzed using the Pearson’s chi-square or Fisher’s exact test with crude odds ratios (OR) and 95% CI, as appropriate. A p value < 0.05 was considered evidence of statistical significance. Data were analyzed using R open-source software. Because of the relatively small sample size, limited number of positives obtained from human stool samples, and associated low statistical power, no attempts were conducted to investigate potential correlations between the occurrence of the detected protist species and the risk factors covered in the epidemiological questionnaire provided to volunteer zookeepers.
3. Results
3.1. Prevalence and Molecular Characterization of Enteroparasites in Captive Non-Human Primates
A total of 51 faecal samples from 10 different species of NHP hosted at the CZCC were collected during the period of study, 28 in the first sampling period and 23 in the second sampling period (Table 1). Members of all 10 NHP species were represented in the two sampling periods. All collected samples could be assigned to individual NHP, except those from the Lemuridae family sharing the same enclosure. Five protist species were detected, including Blastocystis sp. (45.1%, 23/51; 95% CI: 31.1–59.7), E. dispar (27.5%, 14/51; 95% CI: 15.9–41.7), G. duodenalis (21.6%, 11/51; 95% CI: 11.3–35.3), B. coli (3.9%, 2/51; 95% CI: 0.5–13.5), and E. bieneusi (2.0%, 1/51; 95% CI: 0.05–10.5). In contrast, Cryptosporidium spp., E. histolytica, and Troglodytella spp. were not detected in any of the NHP faecal samples analyzed (Table 1). Blastocystis sp. (39.1–50.0%), E. dispar (14.3–43.5%), G. duodenalis (17.9–26.1%), and B. coli (3.6–4.3%) were detected in both sampling campaigns, whereas the only sample that tested positive for E. bieneusi was obtained in the second sampling campaign. Entamoeba dispar was significantly more prevalent in the second sampling campaign than in the first sampling campaign (χ2 = 5.4034, p = 0.0201).
Table 1.
Frequency of enteric protists detected at each sampling campaign in faecal samples from captive non-human primates in the Córdoba Zoo Conservation Centre (Spain).
| First Sampling Campaign | Second Sampling Campaign | All | ||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Frequency Positive Results (%) | Frequency Positive Results (%) | Frequency Positive Results (%) | ||||||||||||||||
| Species | No. | Bl | Ed | Gd | Bc | Eb | No. | Bl | Ed | Gd | Bc | Eb | No. | Bl | Ed | Gd | Bc | Eb |
| Cebuella pygmaea | 1 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 2 | 0.0 | 0.0 | 50.0 | 0.0 | 0.0 | 3 | 0.0 | 0.0 | 33.3 | 0.0 | 0.0 |
| Cercocebus lunulatus | 3 | 100 | 0.0 | 100 | 0.0 | 0.0 | 3 | 33.3 | 100 | 66.7 | 0.0 | 0.0 | 6 | 66.7 | 50.0 | 83.3 | 0.0 | 0.0 |
| Cercopithecus neglectus | 2 | 50.0 | 0.0 | 50.0 | 0.0 | 0.0 | 3 | 66.7 | 66.7 | 0.0 | 0.0 | 0.0 | 5 | 60.0 | 40.0 | 20.0 | 0.0 | 0.0 |
| Eulemur fulvus | 2 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 2 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| Hylobates leucogenys Ogilby | 4 | 75.0 | 100 | 25.0 | 0.0 | 0.0 | 3 | 33.3 | 33.3 | 0.0 | 0.0 | 33.3 | 7 | 57.1 | 71.4 | 14.3 | 0.0 | 14.3 |
| Lemur catta | 2 | 100 | 0.0 | 0.0 | 0.0 | 0.0 | 0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 2 | 100 | 0.0 | 0.0 | 0.0 | 0.0 |
| Macaca sylvanus | 5 | 40.0 | 0.0 | 0.0 | 0.0 | 0.0 | 3 | 33.3 | 100 | 100 | 0.0 | 0.0 | 8 | 37.5 | 37.5 | 37.5 | 0.0 | 0.0 |
| Mandrillus leucophaeus | 5 | 40.0 | 0.0 | 0.0 | 20.0 | 0.0 | 3 | 33.3 | 33.3 | 0.0 | 33.3 | 0.0 | 8 | 37.5 | 12.5 | 0.0 | 25.0 | 0.0 |
| Saimiri sciureus | 2 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 3 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 5 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| Varecia variegata variegata | 2 | 50.0 | 0.0 | 0.0 | 0.0 | 0.0 | 3 | 100 | 0.0 | 0.0 | 0.0 | 0.0 | 5 | 80.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| Total | 28 | 50.0 | 14.3 | 17.9 | 3.6 | 0.0 | 23 | 39.1 | 43.5 | 26.1 | 4.3 | 4.3 | 51 | 45.1 | 27.5 | 21.6 | 3.9 | 2.0 |
Bc: Balantioides coli; Bl: Blastocystis sp., Eb: Enterocytozoon bieneusi; Ed: Entamoeba dispar; Gd: Giardia duodenalis.
Table 2 summarizes the occurrence of the enteric protist species detected in the present survey as single or multiple infections (n = 32). Multiple infections with two protist species were found in 14 samples (43.8%, 14/32) of which five (15.6%, 5/32) were co-infected with Blastocystis sp. and E. dispar, four (12.5%, 4/32) with Blastocystis sp. and G. duodenalis and three (9.4%, 3/32) with G. duodenalis and E. dispar. Three samples (9.4%, 3/32) were co-infected with G. duodenalis, Blastocystis sp., and E. dispar. No associations were demonstrated between Blastocystis sp. and E. dispar (p = 0.630; OR = 1.3, 95% CI: 0.41–4.37) or between Blastocystis sp. and G. duodenalis (p = 0.190; OR = 2.6, 95% CI: 0.54–14.1).
Table 2.
Single and multiple enteric protist infections detected in faecal samples from captive non-human primates in the Córdoba Zoo Conservation Centre (Spain).
| Species Combination | No. of Faecal Samples |
|---|---|
| Blastocystis sp. Only | 10 |
| E. dispar only | 4 |
| G. duodenalis only | 1 |
| Blastocystis sp. + E. dispar | 5 |
| Blastocystis sp. + G. duodenalis | 4 |
| G. duodenalis + E. dispar | 3 |
| Blastocystis sp. + B. coli | 1 |
| E. dispar + E. bieneusi | 1 |
| G. duodenalis + Blastocystis sp. + E. dispar | 3 |
| Total | 32 |
Giardia duodenalis-positive results by qPCR (n = 11) generated cycle threshold (Ct) values ranging from 30.0 to 37.4 (median: 32.4; standard deviation: 2.4). Only a single sample could be genotyped at the bg locus, being identified as sub-assemblage AII (Table 3). Sequence alignment analysis revealed that this sequence was identical to its corresponding reference sequence (GenBank accession number: L40510). Out of the 23 Blastocystis-positive samples at the ssu rDNA (barcode region), the gene of the parasite confirmed by Sanger sequencing revealed the presence of three Blastocystis subtypes (STs), including zoonotic ST1 (39.1%, 9/23), ST3 (34.8%, 8/23), and ST8 (26.1%, 6/23) (Table 3). Additionally, 11 samples yielded amplicons of the expected size but in the form of faint bands on gel electrophoresis. Because their associated Sanger sequences were of poor quality (unreadable), these samples were conservatively considered as negative for Blastocystis sp. Neither mixed infection involving different STs of the parasite nor infections caused by animal-specific (ST10–ST17, ST21, ST23–ST28) subtypes were identified. A moderate genetic diversity was observed within ST1 (alleles 1 and 2, alone or in combination), and ST3 (alleles 34, 32 + 34), but not within ST8, where all isolates were assigned to allele 21. Balantioides coli was unmistakably identified in two isolates, but sequence data of insufficient quality precluded the possibility of determining the genotype of this parasite species. Finally, sequence analysis of the only sample positive to E. bieneusi revealed the presence of genotype D with 100% identity with reference sequence AF101200 (Table 3).
Table 3.
Diversity, frequency, and molecular features of Giardia duodenalis, Blastocystis sp., Balantioides coli, and Enterocytozoon bieneusi in faecal samples from captive non-human primates in the Córdoba Zoo Conservation Centre (Spain). GenBank accession numbers are provided.
| Species | Genotype | Sub-Genotype | Host Species | No. of Isolates | Locus | Reference Sequence | Stretch | Single Nucleotide Polymorphisms | GenBank ID |
|---|---|---|---|---|---|---|---|---|---|
| Giardia duodenalis | A | AII | C.t. | 1 | bg | AY072723 | 205–539 | None | MW417420 |
| Blastocystis sp. | ST1 | Allele 1 | M.l., H.l., C.t. | 4 | ssu rRNA | MK357786 | 4–602 | None | MW414634 |
| Allele 2 | C.t. | 1 | ssu rRNA | MT094302 | 36–539 | None | MW414635 | ||
| Allele 2 | C.n. | 1 | ssu rRNA | MT094302 | 32–539 | C57A, 65InsG, A112G, C128A, C237T, C272T, A458C | MW414636 | ||
| Alleles 1 + 2 | C.t., M.l. | 3 | ssu rRNA | MK357786 | 1–603 | G128R, A474W | MW414637 | ||
| ST3 | Allele 34 | H.l., M.c., C.n. | 7 | ssu rRNA | MK801359 | 1–581 | G114A, A115T, A116G, A159G, T160A, A161T | MW414638 | |
| Alleles 32 + 34 | C.n. | 1 | ssu rRNA | MK801359 | 1–586 | G114A, A115T, A116G, A159K, T160R, A161K, A162R | MW414639 | ||
| ST8 | Allele 21 | V.v.v., L. c. | 6 | ssu rRNA | MT509451 | 1–525 | None | MW414640 | |
| Balantioides coli | Unknown | - | M.l. | 2 | ITS | - | - | - | - |
| Enterocytozoon bieneusi | D | - | H.l. | 1 | ITS | AF101200 | 31–419 | None | MW414645 |
bg: β-giardin; C.n.: Cercopithecus neglectus; C.t.: Cercocebus torquatus; H.l.: Hylobates leucogenys; ITS: Internal transcribed spacer; L.c.: Lemur catta; M.c.: Macaca sylvanus; M.l.: Mandrillus leucophaeus; ssu rRNA: Small subunit ribosomal RNA; V.v.v.: Varecia variegata variegata.
3.2. Prevalence and Molecular Characterization of Enteroparasites in Humans
A total of 19 members of the CZCC personnel, including zookeepers and veterinarians, participated in the study, 15 of them in the first sampling campaign and 11 in the second sampling campaign. Seven zookeepers participated in both sampling campaigns. The male/female ratio was 3.8, and the age range was 21 to 58 years (median: 49 years). Three individuals tested positive for at least one enteroparasite. Two enteric protist species were identified including G. duodenalis (10.5%, 2/19; 95% CI: 1.3–33.1) and Blastocystis sp. (10.5%, 2/19; 95% CI: 1.3–33.1). Giardia duodenalis was detected by qPCR (Ct values: 30.8 and 31.0) in a 58-year-old male and a 49-year-old female, respectively, participating in the second sampling campaign. Both samples failed to be amplified at the gdh, bg, and tpi loci, so the assemblages/sub-assemblages causing the infections were unknown. Blastocystis ST3 allele 34 (GenBank accession number: MW414642) was detected in a 56-year-old male participating in the first sampling campaign, whereas ST1 (GenBank accession number: MW414641) was identified in the same 49-year-old female co-infected with G. duodenalis. Sequence analysis of the later isolate revealed two clear double peaks (R and W) at positions 128 and 264, respectively, of reference sequence MK357786, compatible with mixed infections involving alleles 1, 2, 5 and/or 141. The variables potentially associated with G. duodenalis infections or Blastocystis sp. carriage are summarized in Table 4. The three individuals harbouring G. duodenalis and/or Blastocystis sp. declared no gastrointestinal symptoms at the moment of sampling. All three were food handlers and were regularly in contact with faecal material from NHP and other captive animal species at the CZCC. Other enteric protist species, including Cryptosporidium spp., E. histolytica, E. dispar, E. bieneusi, and B. coli, were apparently absent in the surveyed human population.
Table 4.
Variables potentially associated to G. duodenalis infection and Blastocystis sp. carriage in staff at the Córdoba Zoo Conservation Centre (Spain).
| Variable | Subject 38 | Subject 79 | Subject 86 |
|---|---|---|---|
| Sociodemographic factors | |||
| Sex | Male | Male | Female |
| Age (years) | 56 | 58 | 49 |
| Protist infection/carriage | |||
| Giardia duodenalis | Negative | Positive | Positive |
| Blastocystis sp. | Positive | Negative | Positive |
| Clinical factors | |||
| Diarrhoea in the last 7 days | No | No | No |
| Contact with children <5-years | No | No | No |
| Diarrhoea in family members/relatives | Yes | No | No |
| Work-related factors | |||
| Activity | Veterinarian | Zookeeper | Zookeeper |
| Exposure to faeces from NHP | Yes | Yes | Yes |
| Exposure to faeces from animals other than NHP | Yes | Yes | Yes |
| Any of these animal species with diarrhoea | Yes | Yes | Yes |
| Food handler | Yes | Yes | Yes |
| Behavioural factors | |||
| Recent travel | Yes | No | No |
| Contact with pet dogs | Yes | Yes | Yes |
| Contact with pet cats | Yes | No | Yes |
| Main drinking source—tap | Yes | Yes | Yes |
| Main drinking source—bottled | No | No | No |
| Swimming | No | No | No |
| Handwashing | Frequently | Always | Always |
| Vegetable washing | Always | Always | Always |
3.3. Prevalence and Molecular Characterization of Enteroparasites and Leishmania spp. in Rats
A total of 64 faecal samples of free-living sympatric rats captured within the premises of the zoological garden were available for this study. Three enteric protist species were detected—Cryptosporidium spp. (45.3%, 29/64; 95% CI: 32.8–58.2), G. duodenalis (14.1%, 9/64; 95% CI: 6.6–25.0), and Blastocystis sp. (6.25%, 4/64; 95% CI: 0.4–10.8). None of the samples tested positive for E. bieneusi or B. coli. All the spleen, liver, and skin samples analyzed tested negative for Leishmania spp.
Sequence analyses of the murine Cryptosporidium-positive samples by ssu-PCR revealed the presence of C. muris (10.3%, 3/29), C. ratti (17.2%, 5/29,), rat genotype IV (69.0%, 20/29), and rat genotype V (3.5%, 1/29) (Table 5). All C. muris and C. ratti showed 100% identity with reference sequences AB089284 and MT504541, respectively. Conversely, a high genetic diversity was found within sequences belonging to rat genotype IV, with only four of them being identical to reference sequence JN172970. The remaining 16 sequences varied from JN172970 by 1–5 single nucleotide polymorphisms (SNPs), including a variety of mutations, insertions, deletions, and ambiguous (double peak) positions, the combination 448DelT + G493A being the most frequently detected (Table 5). The only sequence identified as rat genotype V varied from reference sequence MT504543 by a single (A667G) SNP.
Table 5.
Diversity, frequency, and molecular features of Cryptosporidium spp. sequences at the ssu rRNA locus obtained in faecal samples from the rat population under study in Córdoba Zoo Conservation Centre (Spain). GenBank accession numbers are provided.
| Species/Genotype | No. of Isolates | Reference Sequence | Stretch | Single Nucleotide Polymorphisms | GenBank ID |
|---|---|---|---|---|---|
| C. muris | 3 | AB089284 | 504–1012 | None | MW406908 |
| C. ratti a | 6 | MT504541 | 293–751 | None | MW406909 |
| Rat genotype IV | 3 | JN172970 | 377–775 | None | MW406910 |
| 1 | JN172970 | 332–798 | C342T, C410T, C423T, 448DelT, G493A | MW406911 | |
| 1 | JN172970 | 332–752 | C342T, C410T, C423T, 490_491DelTT, G493A | MW406912 | |
| 1 | JN172970 | 328–814 | A428G, 448DelT, G493A | MW406913 | |
| 1 | JN172970 | 328–767 | A445G, 448DelT, G493A, T541A, T542A | MW406914 | |
| 1 | JN172970 | 348–776 | A445G, 448DelT, G493A, T541W, T542W | MW406915 | |
| 5 | JN172970 | 375–814 | 448DelT, G493A | MW406916 | |
| 1 | JN172970 | 454–678 | A459T, A475T, G493A | MW406917 | |
| 1 | JN172970 | 332–788 | A472G, 492InsT, G493A | MW406918 | |
| 3 | JN172970 | 341–814 | 490_491DelTT, G493A | MW406919 | |
| 1 | JN172970 | 481–814 | 490_491DelTT, G493A, G635A | MW406920 | |
| Rat genotype V | 1 | MT504543 | 306–699 | A667G | MW406921 |
a Formerly known as Rat genotype I. See reference [39].
Rodent G. duodenalis-positive samples by qPCR generated Ct values ranging from 24.1 to 36.3 (median: 31.8). Of these, 55.6% (5/9) produced Ct values higher than 30. Two G. duodenalis-positive samples (Ct values: 24.1 and 28.2, respectively) were genotyped as assemblage G at the gdh locus. Both sequences were identical between them and showed an SNP (C262T) compared to reference sequence MF671912. Additionally, one of the sequences was confirmed as assemblage G at the bg locus and showed 100% identity with reference sequence MF671912. None of the two isolates could be amplified at the tpi locus.
The two Blastocystis-positive samples by ssu-PCR and Sanger sequencing were identified as ST4 alleles 92 and 94 (GenBank accession numbers: MW414643 and MW414644), and their sequences were identical with reference sequence MF186667 and MN526920, respectively. As in the case of NHP, three additional samples yielded amplicons of the expected size using barcoding primers but with faint bands on gel electrophoresis that did not produce readable sequences. Those three samples were subjected to a PCR to amplify a different region of the ssu rRNA gene, and two were found positive and subjected to next-generation amplicon sequencing. Those two samples were identified as ST4 and showed 100% identity with the reference sequence U26177. Because of lack of confirmation by Sanger sequencing or failing to amplify with an additional primer set, one sample was conservatively considered as negative for Blastocystis sp.
3.4. Molecular-Based Evidence of Zoonotic Transmission
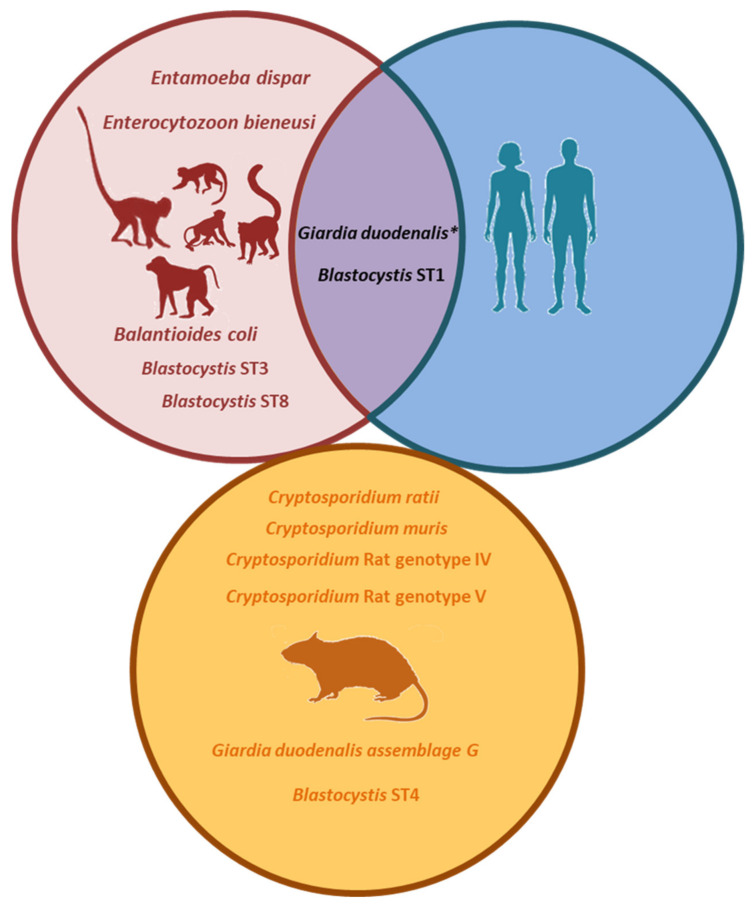
Within NHP, a mangabey (C. lunulatus) investigated during the first sampling campaign was found infected with zoonotic G. duodenalis assemblage AII. Two zookeepers participating in the second sampling campaign were also positive for G. duodenalis, but lack of genotyping data and different sampling intervals precluded the unambiguous demonstration of zoonotic transmission of G. duodenalis infection between NHP and their zookeepers (Figure 2).
Figure 2.
Molecular-based evidence of zoonotic transmission. Enteric protists detected at each species group (non-human primates (NHP), zookeepers and rats) in the Córdoba Zoo Conservation Centre (Spain). * Zoonotic Giardia duodenalis assemblage AII was detected in NHP; in addition, two zookeepers were positive for G. duodenalis of unknown assemblage, potentially making the assessment of zoonotic transmission for this protozoan parasite difficult.
Blastocystis ST1 alleles 1 and 2 (alone or in combination) were consistently detected in mangabeys (C. lunulatus), drills (M. leucophaeus), and gibbons (H. leucogenys Ogilby) along the whole study period, and in a De Brazza´s monkey (C. neglectus) during the second sampling campaign. All these NHP species were housed in close proximity to each other within the CZCC premises. These data strongly suggest that these Blastocystis genetic variants were well established in the NHP population hosted at the CZCC. Interestingly, a zookeeper participating in the second sampling of the survey carried a genetic variant of Blastocystis ST1 compatible with a mixed infection by alleles 1 + 2 (Figure 2). This zookeeper was a food handler and declared regular contact with the faecal material of all NHP in the CZCC. This finding suggests that NHP were acting as a source of Blastocystis infection to the zookeepers responsible for their wellbeing.
Finally, the surveyed rat population was exclusively infected by rodent-specific species/genotypes of Cryptosporidium, G. duodenalis, and Blastocystis (Figure 2), indicating that this host species has a limited role as a source of potential infections for NHP and humans. Additionally, all tested rats were negative to zoonotic Leishmania spp.
4. Discussion
The epidemiology of pathogenic and commensal enteric protists in captive NHP is poorly understood. To fill this gap of knowledge, this survey provides new molecular-based data on the occurrence, transmission, genetic diversity, and zoonotic potential of the protist species that are most relevant from the public health point of view in NHP, their zookeepers/veterinarians, and free-living rats at the CZCC. In addition, the potential role of rodents as a natural reservoir of Leishmania spp. has been investigated.
Cryptosporidium spp. (particularly C. hominis and C. parvum) is, together with rotavirus, Shigella, and enterotoxigenic Escherichia coli, the major contributors to the global burden of diarrhoeal disease [40]. Cryptosporidium spp. is also a common diarrhoea-causing agent in livestock, companion species, and wildlife [6]. Interestingly, Cryptosporidium spp. was absent in the NHP population surveyed here and in the zookeepers/veterinarians that worked in their well care. This agrees with the previous findings observed in NHP (n = 18) from the Almuñecar zoological garden in southern Spain [17] but is in sharp contrast with those from the Barcelona zoological garden, where Cryptosporidium infections were consistently reported in 28–44% of the NHP investigated during a 10-year period [13,15,16]. In those studies, infected NHP were asymptomatic adults with intermittent (up to 10 months) shedding of oocysts irrespectively of the group or lone condition of the animals. This fact suggested that Cryptosporidium reinfection rather than continuous infection was taking part in that setting [17]. In contrast, Cryptosporidium spp. was found at a high prevalence rate (45%) in free-living rats captured within the CZCC enclosure. In the only two previous studies published in Spain, Cryptosporidium spp. infections have been reported in black rats from Catalonia (1/1) and the Canary Islands (14/101) [41,42]. Our sequence analyses revealed the presence of four distinct Cryptosporidium species/genotypes including rat genotype IV (69%), C. ratti (17%), C. muris (10%), and rat genotype V (4%). It should be noted that C. ratti, formerly known as rat genotype I, has been recently proposed as a valid Cryptosporidium species by Martin Kváč’s laboratory [39]. Of these, C. muris and C. ratti (in addition to C. meleagridis and rat genotype II/III, not identified in the present survey) have been previously described in black rats from the Canary Islands [42]. Overall, our data indicate that rats captured at the CZCC were infected by murine-adapted Cryptosporidium species/genotypes and played a limited role as a source of cryptosporidiosis to NHP. Of interest, C. muris and rat genotype III have been sporadically reported in humans and/or companion animals, including dogs and cats [43,44,45,46].
In the present study, G. duodenalis infections were identified in 22% of NHP and 11% of zookeepers. Interestingly, all Giardia-positive cases by qPCR yielded Ct values >30, indicative of moderate-to-low parasite burdens. This agrees with the fact that all positive cases were asymptomatic and produced formed stools, also explaining the low genotyping success rate obtained (7.7%, 1/13). It should be noted that gdh, bg, and tpi are all single-copy genes with limited sensitivity compared with the multiple-copy ssu rRNA gene used in qPCR for detection purposes. An early epidemiological study detected the presence of G. duodenalis in 19.1% of NHP in the Barcelona zoological garden [14], but this parasite was absent in the NHP analyzed at the Almuñecar zoological garden [17]. Our sequence analyses identified the zoonotic sub-assemblage AII in a mangabey (C. lunulatus). In Spain, the G. duodenalis sub-assemblage AII has been found in 15–44% of documented clinical cases [47,48], and in 17–33% of children of paediatric age [49,50]. Although two of the CZCC zookeepers tested positive to this protozoan parasite, we were unable genotype these isolates, so their assemblage/sub-assemblage remained unknown and precluded us to propose a potential source of infection. Of interest, zoonotic AI and BIV have been previously identified in members of the Lemuridae family in the Valencia and Madrid zoological gardens [18].
Blastocystis infection/carriage was demonstrated in 45.1% of the NHP surveyed, a frequency rate considerably lower than those (67–95%) previously reported by conventional microscopy at the Barcelona and Almuñecar zoological gardens, respectively [14,17]. Sequence analyses of Blastocystis isolates revealed interesting data. ST1 was the most prevalent (39%) subtype circulating among captive NHP, being present in two genetic variants, allele 1 and allele 2, either alone or in combination. We have recently reported ST1 as the most common (82%) Blastocystis subtype in wild western chimpanzees (Pan troglodytes verus) in Senegal, although in that survey all the isolates characterized belonged to alleles 7 and 8 [51]. Moreover, we have also demonstrated that human cases of blastocystosis by ST1 in Spain are mainly due to allele 4 (and, to a much lesser extent, allele 77) both in asymptomatic [50,52] and clinical [53] individuals. The fact that one of the CZCC primate handlers carried a genetic variant of Blastocystis ST1 compatible with a mixed infection involving alleles 1, 2, 5 and/or 141 seems to indicate that this ST1 infection is most likely of primate origin and represents a zoonotic transmission event. Similarly, the vast majority of the ST3 isolates detected in NHP at the CZCC belonged to allele 32, a genetic variant not yet described in Spanish human populations [50,52,53]. This fact may indicate that ST3 allele 32 may be better adapted to infect NHP than humans. Finally, Blastocystis ST8 carriage was also a common finding (26%) in NHP at the CZCC. This result was highly expected as this Blastocystis subtype is well-known both in captive [54] and free-living [55] NHP globally. Although rarely reported in humans, the zoonotic potential of ST8 has been demonstrated in a zoological garden in the UK, where this subtype was responsible from one in four Blastocystis infections both in captive NHP and their handlers [56]. Also relevant was the finding of only identifying Blastocystis ST4 in rats. This finding supports that rodents appear to constitute the main animal reservoir of ST4 [56,57]. Additionally, the marked geographical distribution of ST4 in humans (commonly found in Europe but rarely or less frequently present in other geographical areas), together with its clonal structure strongly suggest that ST4 represents a lineage with a recent entry into the human population [58]. In Spain, all human cases carrying Blastocystis ST4 have been assigned to the allele 42 of the protist. As in the case of ST3 allele 32, this fact may indicate that ST4 alleles 92 and 94 (identified in rats in this study) may be particularly adapted to infect/colonize rodent species rather than humans.
Enterocytozoon bieneusi genotype D was detected in a single gibbon (H. leucogenys Ogilby). Genotype D has broad host and geographic ranges and belongs to Group 1 that includes zoonotic E. bieneusi genotypes most frequently found in humans, domestic and wild (including NHP) animal species worldwide [59,60]. In Spain, E. bieneusi genotype D has been described in renal transplant recipients [61], domestic rabbits [62] and cats [63], wild red foxes [62], and environmental (water) samples [64]. This result clearly indicates that NHP may act as suitable reservoirs for human microsporidiosis by E. bieneusi.
Regarding ciliate species, zoonotic B. coli was identified in a low (3.9%) proportion of the NHP investigated, but not in their handlers. At first sight, this result is much lower than those previously documented by microscopy examination in NHP at the Barcelona (38.1%) and Almuñecar (16.6%) zoological gardens [14,17]. However, when considering primate groups, B. coli has been found only in Catarrhini (Mandrillus, present results; Cercocebus, Gorilla, Pan, Papio, and Pongo) [14]. The data from Pérez-Cordón et al. [17] are aggregated and it is not possible to identify the positive primate genera. Our negative results in Strepsirrhini (Eulemur, Lemur, and Varecia) and Plathyrrhini (Saimiri) primates are in accordance with previous data [19] and suggest that these primates are uncommon or not valid hosts for this ciliate. It should be noted that, because B. coli cysts are morphologically indistinguishable from other ciliate species (i.e., Buxtonella spp.), it is possible that some of the microscopy-based prevalence rates described above do indeed represent an overestimation of the true occurrence of the parasite. While B. coli-like cysts are easily identifiable by microscopy, differential diagnosis based on molecular (PCR and Sanger sequencing) methods should be used for the correct identification of B. coli.
Finally, the absence of T. abrassarti in the investigated species is in accordance with previous surveys [65]. This ciliate is commonly reported in wild great apes [20] but there is no conclusive evidence in lesser apes and monkeys (e.g., red colobus, red-tailed monkeys, vervet monkeys, and yellow baboons). In captive chimpanzees, prevalence and infection intensities are influenced by the dietary starch concentration, suggesting a symbiotic function and participation in nourishment degradation [66].
Vector-borne Leishmania infantum, the causative agent of visceral and cutaneous leishmaniasis in Spain, is one of the most important neglected zoonosis in the Mediterranean region. In Spain, and in addition to domestic dogs, leporids such as rabbits and hares have been demonstrated as competent reservoirs of the infection [67]. Micromammals (e.g., mice, shrews) seem to play a limited role in the epidemiology of the parasite [68], although an unanticipated high prevalence rate (33%) of the parasite has been recently described in rats captured in sewers in the city of Barcelona [69]. To confirm the accuracy and extent of these previous findings, we investigated by PCR the occurrence of Leishmania spp. in rat tissues, including liver, spleen, and ear skin. In all cases, we failed to detect the presence of the parasite. It is possible that the limited number of rodent samples (n = 64) analyzed in the present study may have biased the obtained results. Further studies are warranted to assess the role of rodent on Leishmania spp. transmission in these epidemiological scenarios.
5. Conclusions
A high prevalence of the diarrhoea-causing protists G. duodenalis and Blastocystis sp. (but not Cryptosporidium spp.) was observed in captive NHP at the CZCC. NHP can harbour zoonotic genotypes of G. duodenalis, Blastocystis sp., and E. bieneusi. Indeed, strong evidence of the occurrence of Blastocystis zoonotic transmission between NHP and their handlers was provided, despite the use of personal protective equipment and the implementation of strict health and safety protocols. Free-living sympatric rats are infected by host-specific species/genotypes of the investigated protists and seem to play a limited role as a source of infections to NHP or humans in this setting. The extent of these findings should be confirmed in similar epidemiological surveys targeting other captive NHP populations.
Acknowledgments
David González-Barrio was recipient of a ‘Sara Borrell’ postdoctoral fellowship (CD19CIII/00011) funded by the Spanish Ministry of Science, Innovation and Universities.
Supplementary Materials
The following are available online at https://www.mdpi.com/2076-2615/11/3/700/s1, Table S1: English version of the standardized epidemiological questionnaire used in this study, Table S2: Oligonucleotides used for the molecular identification and/or characterization of the intestinal parasitic and commensal protist species investigated in the present study.
Author Contributions
Conceptualization, P.C.K., M.S., C.C., F.J.N., I.G.-B., R.G., D.G.-B., F.P.-G., R.C.-B. and D.C.; methodology, P.C.K., J.G.M., M.S., R.G., I.G.-B., D.C.-T., F.P.-G., R.C.-B. and D.C.; software, P.C.K.; A.D.; J.G.M., D.C.; validation, P.C.K., M.S., F.P.-G., R.C.-B., D.G.-B. and D.C.; formal analysis, P.C.K., A.D., B.B., A.S.M., J.G.M., M.S.; C.C., S.M., D.C.-T., and R.G.; investigation, P.C.K., F.P.-G., R.C.-B. and D.C.; resources, M.S., D.C. and I.G.-B.; data curation, P.C.K., A.D., M.S., C.C. and D.C.; writing—original draft preparation, P.C.K., J.G.M., M.S., F.P.-G., R.C.-B. and D.C.; writing—review and editing, P.C.K., J.G.M., M.S., F.P.-G., R.C.-B., D.G.-B. and D.C.; supervision, F.P.-G., R.C.-B., D.G.-B., and D.C.; project administration, D.C.; funding acquisition, I.G.-B. and D.C. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the Health Institute Carlos III (ISCIII), Ministry of Economy and Competitiveness (Spain), under project PI16CIII/00024. Additional funding was obtained from the University of Córdoba (Spain), under the project UCO-FEDER-1264967.
Institutional Review Board Statement
This study was carried out in accordance with Spanish legislation guidelines (RD 8/2003) and with the International Guiding Principles for Biomedical Research Involving Animals issued by the Council for International Organization of Medical Sciences and the International Council for Laboratory Animal Science (RD 53/2013). This study has been approved by Ethics Committee of the Health Institute Carlos III on 17 December 2018 under the reference number CEI PI 90_2018-v2. Written informed consent was obtained from zookeepers that volunteered to participate in the survey.
Data Availability Statement
All relevant data are within the article and its additional files. The sequences data were submitted to the GenBank database under the accession numbers MW417420–MW417422 (G. duodenalis), MW414634–MW414644 (Blastocystis sp.), MW406908–MW406921 (Cryptosporidium spp.) and MW414645 (E. bieneusi).
Conflicts of Interest
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.DuPont H.L. Persistent diarrhea: A clinical review. JAMA. 2016;315:2712–2723. doi: 10.1001/jama.2016.7833. [DOI] [PubMed] [Google Scholar]
- 2.Hemphill A., Müller N., Müller J. Comparative pathobiology of the intestinal protozoan parasites Giardia lamblia, En-tamoeba histolytica, and Cryptosporidium parvum. Pathogens. 2019;8:116. doi: 10.3390/pathogens8030116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ajjampur S.S., Tan K.S. Pathogenic mechanisms in Blastocystis spp.—Interpreting results from in vitro and in vivo studies. Parasitol. Int. 2016;65:772–779. doi: 10.1016/j.parint.2016.05.007. [DOI] [PubMed] [Google Scholar]
- 4.Li W., Feng Y., Santin M. Host Specificity of Enterocytozoon bieneusi and Public Health Implications. Trends Parasitol. 2019;35:436–451. doi: 10.1016/j.pt.2019.04.004. [DOI] [PubMed] [Google Scholar]
- 5.Ryan U., Cacciò S.M. Zoonotic potential of Giardia. Int. J. Parasitol. 2013;43:943–956. doi: 10.1016/j.ijpara.2013.06.001. [DOI] [PubMed] [Google Scholar]
- 6.Ryan U., Fayer R., Xiao L. Cryptosporidium species in humans and animals: Current understanding and research needs. Parasitology. 2014;141:1667–1685. doi: 10.1017/S0031182014001085. [DOI] [PubMed] [Google Scholar]
- 7.Li W., Feng Y., Zhang L., Xiao L. Potential impacts of host specificity on zoonotic or interspecies transmission of Enterocytozoon bieneusi. Infect. Genet. Evol. 2019;75:104033. doi: 10.1016/j.meegid.2019.104033. [DOI] [PubMed] [Google Scholar]
- 8.Hublin J.S.Y., Maloney J.G., Santin M. Blastocystis in domesticated and wild mammals and birds. Res. Vet. Sci. 2020 doi: 10.1016/j.rvsc.2020.09.031. [DOI] [PubMed] [Google Scholar]
- 9.Widmer G., Köster P.C., Carmena D. Cryptosporidium hominis infections in non-human animal species: Revisiting the concept of host specificity. Int. J. Parasitol. 2020;50:253–262. doi: 10.1016/j.ijpara.2020.01.005. [DOI] [PubMed] [Google Scholar]
- 10.Stensvold C.R., Clark C.G. Pre-empting Pandora’s Box: Blastocystis Subtypes Revisited. Trends Parasitol. 2020;36:229–232. doi: 10.1016/j.pt.2019.12.009. [DOI] [PubMed] [Google Scholar]
- 11.Maloney J.G., Molokin A., Da Cunha M.J.R., Cury M.C., Santin M. Blastocystis subtype distribution in domestic and captive wild bird species from Brazil using next generation amplicon sequencing. Parasite Epidemiol. Control. 2020;9:e00138. doi: 10.1016/j.parepi.2020.e00138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ou Y., Jiang W., Roellig D.M., Wan Z., Li N., Guo Y., Feng Y., Xiao L. Characterizations of Enterocytozoon bieneusi at new genetic loci reveal a lack of strict host specificity among common genotypes and the existence of a canine-adapted Enterocytozoon species. Int. J. Parasitol. 2021;51:215–223. doi: 10.1016/j.ijpara.2020.09.008. [DOI] [PubMed] [Google Scholar]
- 13.Gómez M.S., Gracenea M., Gosalbez P., Feliu C., Enseñat C., Hidalgo R. Detection of oocysts of Cryptosporidium in several species of monkeys and in one prosimian species at the Barcelona zoo. Parasitol. Res. 1992;78:619–620. doi: 10.1007/BF00936462. [DOI] [PubMed] [Google Scholar]
- 14.Soledad Gómez M., Gracenea M., Montoliu I., Feliu C., Monleon A., Fernandez J., Enseñat C. Intestinal parasitism—protozoa and helminths—in primates at the Barcelona Zoo. J. Med. Primatol. 1996;25:419–423. doi: 10.1111/j.1600-0684.1996.tb00038.x. [DOI] [PubMed] [Google Scholar]
- 15.Gómez M.S., Torres J., Gracenea M., Fernandez-Morán J., Gonzalez-Moreno O. Further report on Cryptosporidium in Barcelona zoo mammals. Parasitol. Res. 2000;86:318–323. doi: 10.1007/s004360050049. [DOI] [PubMed] [Google Scholar]
- 16.Gracenea M., Gómez M.S., Torres J., Carné E., Fernández-Morán J. Transmission dynamics of Cryptosporidium in primates and herbivores at the Barcelona zoo: A long-term study. Veter. Parasitol. 2002;104:19–26. doi: 10.1016/S0304-4017(01)00611-2. [DOI] [PubMed] [Google Scholar]
- 17.Pérez Cordón G., Hitos Prados A., Romero D., Sánchez Moreno M., Pontes A., Osuna A., Rosales M.J. Intestinal parasitism in the animals of the zoological garden “Peña Escrita” (Almuñecar, Spain) Vet. Parasitol. 2008;156:302–309. doi: 10.1016/j.vetpar.2008.05.023. [DOI] [PubMed] [Google Scholar]
- 18.Martínez-Díaz R.A., Sansano-Maestre J., Martínez-Herrero M.D.C., Ponce-Gordo F., Gómez-Muñoz M.T. Occurrence and genetic characterization of Giardia duodenalis from captive nonhuman primates by multi-locus sequence analysis. Parasitol. Res. 2011;109:539–544. doi: 10.1007/s00436-011-2281-z. [DOI] [PubMed] [Google Scholar]
- 19.Ponce-Gordo F., García-Rodríguez J.J. Balantioides coli. Res. Veter. Sci. 2020;S0034–5288:31066–31073. doi: 10.1016/j.rvsc.2020.10.028. [DOI] [PubMed] [Google Scholar]
- 20.Pomajbíková K., Petrželková K.J., Profousová I., Petrášová J., Kišidayová S., Váradyová Z., Modrý D. A survey of entodiniomorphid ciliates in chimpanzees and bonobos. Am. J. Phys. Anthr. 2009;142:42–48. doi: 10.1002/ajpa.21191. [DOI] [PubMed] [Google Scholar]
- 21.UNE 16636:2015. [(accessed on 6 February 2021)]; Available online: https://www.aenor.com/normas-y-libros/buscador-de-normas/une/?Tipo=N&c=N0055762.
- 22.Mayer J., Mans C. Rodents. In: Carpenter J.W., Marion C.J., editors. Exotic Animal Formulary. 5th ed. Elsevier; St. Louis, MO, USA: 2018. pp. 673–675. [Google Scholar]
- 23.Verweij J.J., Schinkel J., Laeijendecker D., van Rooyen M.A., van Lieshout L., Polderman A.M. Real-time PCR for the detection of Giardia lamblia. Mol. Cell. Probes. 2003;17:223–225. doi: 10.1016/S0890-8508(03)00057-4. [DOI] [PubMed] [Google Scholar]
- 24.Read C.M., Monis P.T., Thompson R.C. Discrimination of all genotypes of Giardia duodenalis at the glutamate dehydro-genase locus using PCR-RFLP. Infect. Genet. Evol. 2004;4:125–130. doi: 10.1016/j.meegid.2004.02.001. [DOI] [PubMed] [Google Scholar]
- 25.Lalle M., Pozio E., Capelli G., Bruschi F., Crotti D., Cacciò S.M. Genetic heterogeneity at the beta-giardin locus among human and animal isolates of Giardia duodenalis and identification of potentially zoonotic subgenotypes. Int. J. Parasitol. 2005;35:207–213. doi: 10.1016/j.ijpara.2004.10.022. [DOI] [PubMed] [Google Scholar]
- 26.Sulaiman I.M., Fayer R., Bern C., Gilman R.H., Trout J.M., Schantz P.M., Das P., Lal A.A., Xiao L. Triosephosphate isomerase gene characterization and potential zoonotic transmission of Giardia duodenalis. Emerg. Infect. Dis. 2003;9:1444–1452. doi: 10.3201/eid0911.030084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tiangtip R., Jongwutiwes S. Molecular analysis of Cryptosporidium species isolated from HIV-infected patients in Thai-land. Trop. Med. Int. Health. 2002;7:357–364. doi: 10.1046/j.1365-3156.2002.00855.x. [DOI] [PubMed] [Google Scholar]
- 28.Scicluna S.M., Tawari B., Clark C.G. DNA Barcoding of Blastocystis. Protist. 2006;157:77–85. doi: 10.1016/j.protis.2005.12.001. [DOI] [PubMed] [Google Scholar]
- 29.Maloney J.G., Molokin A., Santin M. Next generation amplicon sequencing improves detection of Blastocystis mixed subtype infections. Infect. Genet. Evol. 2019;73:119–125. doi: 10.1016/j.meegid.2019.04.013. [DOI] [PubMed] [Google Scholar]
- 30.Santín M., Gómez-Muñoz M.T., Solano-Aguilar G., Fayer R. Development of a new PCR protocol to detect and subtype Blastocystis spp. from humans and animals. Parasitol. Res. 2011;109:205–212. doi: 10.1007/s00436-010-2244-9. [DOI] [PubMed] [Google Scholar]
- 31.Bushnell B. BBMap Download. [(accessed on 6 February 2021)];2014 Available online: SourceForge.net.
- 32.Rognes T., Flouri T., Nichols B., Quince C., Mahé F. VSEARCH: A versatile open source tool for metagenomics. PeerJ. 2016;4:e2584. doi: 10.7717/peerj.2584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Buckholt M.A., Lee J.H., Tzipori S. Prevalence of Enterocytozoon bieneusi in swine: An 18-month survey at a slaughter-house in Massachusetts. Appl. Environ. Microbiol. 2002;68:2595–2599. doi: 10.1128/AEM.68.5.2595-2599.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Verweij J.J., Oostvogel F., Brienen E.A., Nang-Beifubah A., Ziem J., Polderman A.M. Prevalence of Entamoeba histolytica and Entamoeba dispar in northern Ghana. Trop. Med. Int. Health. 2003;8:1153–1156. doi: 10.1046/j.1360-2276.2003.01145.x. [DOI] [PubMed] [Google Scholar]
- 35.Gutiérrez-Cisneros M.J., Cogollos R., López-Vélez R., Martín-Rabadán P., Martínez-Ruiz R., Subirats M., Merino F.J., Fuentes I. Application of real-time PCR for the differentiation of Entamoeba histolytica and E. dispar in cyst-positive faecal samples from 130 immigrants living in Spain. Ann. Trop. Med. Parasitol. 2010;104:145–149. doi: 10.1179/136485910X12607012373759. [DOI] [PubMed] [Google Scholar]
- 36.Ponce-Gordo F., Fonseca-Salamanca F., Martínez-Díaz R.A. Genetic heterogeneity in internal transcribed spacer genes of Balantidium coli (Litostomatea, Ciliophora) Protist. 2011;162:774–794. doi: 10.1016/j.protis.2011.06.008. [DOI] [PubMed] [Google Scholar]
- 37.Vallo P., Petrželková K.J., Profousová I., Petrášová J., Pomajbíková K., Leendertz F., Hashimoto C., Simmons N., Babweteera F., Machanda Z., et al. Molecular diversity of entodiniomorphid ciliate Troglodytella abrassarti and its coevolution with chimpanzees. Am. J. Phys. Anthropol. 2012;148:525–533. doi: 10.1002/ajpa.22067. [DOI] [PubMed] [Google Scholar]
- 38.Cruz I., Chicharro C., Nieto J., Bailo B., Canavate C., Figueras M.A.-C., Alvar J. Comparison of new diagnostic tools for management of pediatric Mediterranean visceral leishmaniasis. J. Clin. Microbiol. 2006;44:2343–2347. doi: 10.1128/JCM.02297-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ježková J., Prediger J., Holubová N., Sak B., Konečný R., Feng Y., Xiao L., Rost M., McEvoy J., Kváč M. Cryptosporidium ratti n. sp. (Apicomplexa: Cryptosporidiidae) and genetic diversity of Cryptosporidium spp. in brown rats (Rattus norvegicus) in the Czech Republic. Parasitology. 2021;148:84–97. doi: 10.1017/S0031182020001833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kotloff K.L. The burden and etiology of diarrheal illness in developing countries. Pediatr. Clin. North Am. 2017;64:799–814. doi: 10.1016/j.pcl.2017.03.006. [DOI] [PubMed] [Google Scholar]
- 41.Torres J., Gracenea M., Gómez M.S., Arrizabalaga A., González-Moreno O. The occurrence of Cryptosporidium parvum and C. muris in wild rodents and insectivores in Spain. Vet. Parasitol. 2000;92:253–260. doi: 10.1016/S0304-4017(00)00331-9. [DOI] [PubMed] [Google Scholar]
- 42.García-Livia K., Martín-Alonso A., Foronda P. Diversity of Cryptosporidium spp. in wild rodents from the Canary Islands, Spain. Parasites Vectors. 2020;13:445. doi: 10.1186/s13071-020-04330-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Guy R.A., Yanta C.A., Muchaal P.K., Rankin M.A., Thivierge K., Lau R., Boggild A.K. Molecular characterization of Cryptosporidium isolates from humans in Ontario, Canada. Parasites Vectors. 2021;14:1–14. doi: 10.1186/s13071-020-04546-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ayinmode A.B., Obebe O.O., Falohun O.O. Molecular detection of Cryptosporidium species in street-sampled dog faeces in Ibadan, Nigeria. Vet. Parasitol. Reg. Stud. Rep. 2018;14:54–58. doi: 10.1016/j.vprsr.2018.08.005. [DOI] [PubMed] [Google Scholar]
- 45.Yang R., Ying J.L.J., Monis P., Ryan U. Molecular characterisation of Cryptosporidium and Giardia in cats (Felis catus) in Western Australia. Exp. Parasitol. 2015;155:13–18. doi: 10.1016/j.exppara.2015.05.001. [DOI] [PubMed] [Google Scholar]
- 46.Pavlásek I., Ryan U. The first finding of a natural infection of Cryptosporidium muris in a cat. Vet. Parasitol. 2007;144:349–352. doi: 10.1016/j.vetpar.2006.10.005. [DOI] [PubMed] [Google Scholar]
- 47.De Lucio A., Martínez-Ruiz R., Merino F.J., Bailo B., Aguilera M., Fuentes I., Carmena D. Molecular genotyping of Giardia duodenalis isolates from symptomatic individuals attending two major public hospitals in Madrid, Spain. PLoS ONE. 2015;10:e0143981. doi: 10.1371/journal.pone.0143981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Azcona-Gutiérrez J.M., De Lucio A., Hernández-De-Mingo M., García-García C., Soria-Blanco L.M., Morales L., Aguilera M., Fuentes I., Carmena D. Molecular diversity and frequency of the diarrheagenic enteric protozoan Giardia duodenalis and Cryptosporidium spp. in a hospital setting in Northern Spain. PLoS ONE. 2017;12:e0178575. doi: 10.1371/journal.pone.0178575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cardona G.A., Carabin H., Goñi P., Arriola L., Robinson G., Fernández-Crespo J.C., Clavel A., Chalmers R.M., Carmena D. Identification and molecular characterization of Cryptosporidium and Giardia in children and cattle populations from the province of Álava, North of Spain. Sci. Total Environ. 2011;412/413:101–108. doi: 10.1016/j.scitotenv.2011.09.076. [DOI] [PubMed] [Google Scholar]
- 50.Muadica A.S., Köster P.C., Dashti A., Bailo B., Hernández-de-Mingo M., Reh L., Balasegaram S., Verlander N.Q., Ruiz Chércoles E., Carmena D. Molecular diversity of Giardia duodenalis, Cryptosporidium spp. and Blastocystis sp. in asymptomatic school children in Leganés, Madrid (Spain) Microorganisms. 2020;8:466. doi: 10.3390/microorganisms8040466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Renelies-Hamilton J., Noguera-Julian M., Parera M., Paredes R., Pacheco L., Dacal E., Saugar J.M., Rubio J.M., Poulsen M., Köster P.C., et al. Exploring interactions between Blastocystis sp., Strongyloides spp. and the gut microbiomes of wild chimpanzees in Senegal. Infect. Genet. Evol. 2019;74:104010. doi: 10.1016/j.meegid.2019.104010. [DOI] [PubMed] [Google Scholar]
- 52.Paulos S., Köster P.C., De Lucio A., Hernández-De-Mingo M., Cardona G.A., Fernández-Crespo J.C., Stensvold C.R., Carmena D. Occurrence and subtype distribution of Blastocystis sp. in humans, dogs and cats sharing household in northern Spain and assessment of zoonotic transmission risk. Zoonoses Public Health. 2018;65:993–1002. doi: 10.1111/zph.12522. [DOI] [PubMed] [Google Scholar]
- 53.Köster P.C., Molina A., García M., Cifre S., Trelis M., Pérez de Ayala A., Azcona J.M., García C., Paulos S., Hernández-de-Mingo M., et al. Molecular diversity and frequency of Blastocystis sp. subtypes in Spanish clinical patients: A pilot multicentre study; Proceedings of the 2nd International Blastocystis Conference; Bogotá, Colombia. 9–12 October 2018. [Google Scholar]
- 54.Oliveira-Arbex A.P., David É.B., Tenório M.D.S., Cicchi P.J.P., Patti M., Coradi S.T., Lucheis S.B., Jim J., Guimarães S. Diversity of Blastocystis subtypes in wild mammals from a zoo and two conservation units in southeastern Brazil. Infect. Genet. Evol. 2020;78:104053. doi: 10.1016/j.meegid.2019.104053. [DOI] [PubMed] [Google Scholar]
- 55.Helenbrook W.D., Shields W.M., Whipps C.M. Characterization of Blastocystis species infection in humans and mantled howler monkeys, Alouatta palliata aequatorialis, living in close proximity to one another. Parasitol. Res. 2015;114:2517–2525. doi: 10.1007/s00436-015-4451-x. [DOI] [PubMed] [Google Scholar]
- 56.Stensvold C.R., Alfellani M.A., Nørskov-Lauritsen S., Prip K., Victory E.L., Maddox-Hyttel C., Nielsen H.V., Clark C.G. Subtype distribution of Blastocystis isolates from synanthropic and zoo animals and identification of a new subtype. Int. J. Parasitol. 2009;39:473–479. doi: 10.1016/j.ijpara.2008.07.006. [DOI] [PubMed] [Google Scholar]
- 57.Mohammadpour I., Bozorg-Ghalati F., Gazzonis A.L., Manfredi M.T., Motazedian M.H., Mohammadpour N. First molecular subtyping and phylogeny of Blastocystis sp. isolated from domestic and synanthropic animals (dogs, cats and brown rats) in southern Iran. Parasites Vectors. 2020;13:1–11. doi: 10.1186/s13071-020-04225-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Stensvold C.R., Alfellani M., Clark C.G. Levels of genetic diversity vary dramatically between Blastocystis subtypes. Infect. Genet. Evol. 2012;12:263–273. doi: 10.1016/j.meegid.2011.11.002. [DOI] [PubMed] [Google Scholar]
- 59.Li W., Zhong Z., Song Y., Gong C., Deng L., Cao Y., Zhou Z., Cao X., Tian Y., Li H., et al. Human-pathogenic Enterocytozoon bieneusi in captive giant pandas (Ailuropoda melanoleuca) in China. Sci. Rep. 2018;8:6590. doi: 10.1038/s41598-018-25096-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Chen L., Zhao J., Li N., Guo Y., Feng Y., Feng Y., Xiao L. Genotypes and public health potential of Enterocytozoon bie-neusi and Giardia duodenalis in crab-eating macaques. Parasites Vectors. 2019;12:254. doi: 10.1186/s13071-019-3511-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Galván A.L., Sánchez A.M., Valentín M.A., Henriques-Gil N., Izquierdo F., Fenoy S., del Aguila C. First cases of mi-crosporidiosis in transplant recipients in Spain and review of the literature. J. Clin. Microbiol. 2011;49:1301–1306. doi: 10.1128/JCM.01833-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Galván-Díaz A.L., Magnet A., Fenoy S., Henriques-Gil N., Haro M., Gordo F.P., Millán J., Miró G., Del Águila C., Izquierdo F. Microsporidia detection and genotyping study of human pathogenic E. bieneusi in animals from Spain. PLoS ONE. 2014;9:e92289. doi: 10.1371/journal.pone.0092289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Dashti A., Santín M., Cano L., de Lucio A., Bailo B., de Mingo M.H., Köster P.C., Fernández-Basterra J.A., Aramburu-Aguirre J., López-Molina N., et al. Occurrence and genetic diversity of Enterocytozoon bieneusi (Microsporidia) in owned and sheltered dogs and cats in Northern Spain. Parasitol. Res. 2019;118:2979–2987. doi: 10.1007/s00436-019-06428-1. [DOI] [PubMed] [Google Scholar]
- 64.Galván A.L., Magnet A., Izquierdo F., Fenoy S., Rueda C., Fernández Vadillo C., Henriques-Gil N., Del Aguila C. Molecular characterization of human-pathogenic Microsporidia and Cyclospora cayetanensis isolated from various water sources in Spain: A year-long longitudinal study. Appl. Environ. Microbiol. 2013;79:449–459. doi: 10.1128/AEM.02737-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kooriyama T., Hasegawa H., Shimozuru M., Tsubota T., Nishida T., Iwaki T. Parasitology of five primates in Mahale Mountains National Park, Tanzania. Primates. 2012;53:365–375. doi: 10.1007/s10329-012-0311-9. [DOI] [PubMed] [Google Scholar]
- 66.Petrželková K.J., Schovancová K., Profousová I., Kišidayová S., Váradyová Z., Pekár S., Kamler J., Modrý D. The effect of low- and high-fiber diets on the population of entodiniomorphid ciliates Troglodytella abrassarti in captive chimpanzees (Pan troglodytes) Am. J. Primatol. 2012;74:669–675. doi: 10.1002/ajp.22021. [DOI] [PubMed] [Google Scholar]
- 67.Ortega-García M.V., Salguero F.J., Rodríguez-Bertos A., Moreno I., García N., García-Seco T., Luz Torre G., Domínguez L., Domínguez M. A pathological study of Leishmania infantum natural infection in European rabbits (Oryctolagus cuniculus) and Iberian hares (Lepus granatensis) Transbound. Emerg. Dis. 2019;66:2474–2481. doi: 10.1111/tbed.13305. [DOI] [PubMed] [Google Scholar]
- 68.Millán J. Molecular investigation of vector-borne parasites in wild micromammals, Barcelona (Spain) Parasitol. Res. 2018;117:3015–3018. doi: 10.1007/s00436-018-5971-y. [DOI] [PubMed] [Google Scholar]
- 69.Galán-Puchades M.T., Gómez-Samblás M., Suárez-Morán J.M., Osuna A., Sanxis-Furió J., Pascual J., Bueno-Marí R., Franco S., Peracho V., Montalvo T., et al. Leishmaniasis in Norway Rats in Sewers, Barcelona, Spain. Emerg. Infect. Dis. 2019;25:1222–1224. doi: 10.3201/eid2506.181027. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All relevant data are within the article and its additional files. The sequences data were submitted to the GenBank database under the accession numbers MW417420–MW417422 (G. duodenalis), MW414634–MW414644 (Blastocystis sp.), MW406908–MW406921 (Cryptosporidium spp.) and MW414645 (E. bieneusi).