Abstract
Background
The black soldier fly (Hermetia illucens) has significant economic potential. The larvae can be used in financially viable waste management systems, as they are voracious feeders able to efficiently convert low-quality waste into valuable biomass. However, most studies on H. illucens in recent decades have focused on optimizing their breeding and bioconversion conditions, while information on their biology is limited.
Methods
About 200 fifth instar well-fed larvae were sacrificed in this work. The liquid chromatography-tandem mass spectrometry and scanning electron microscopy were employed in this study to perform a proteomic and ultrastructural analysis of the peritrophic matrix (PM) of H. illucens larvae.
Results
A total of 565 proteins were identified in the PM samples of H. illucen, of which 177 proteins were predicted to contain signal peptides, bioinformatics analysis and manual curation determined 88 proteins may be associated with the PM, with functions in digestion, immunity, PM modulation, and others. The ultrastructure of the H. illucens larval PM observed by scanning electron microscopy shows a unique diamond-shaped chitin grid texture.
Conclusions
It is the first and most comprehensive proteomics research about the PM of H. illucens larvae to date. All the proteins identified in this work has been discussed in details, except several unnamed or uncharacterized proteins, which should not be ignored and need further study. A comparison of the ultrastructure between H. illucens larval PM and those of other insects as observed by SEM indicates that the PM displays diverse textures on an ultra-micro scale and we suscept a unique diamond-shaped chitin grid texture may help H. illucens larval to hold more food. This work deepens our understanding of the molecular architecture and ultrastructure of the H. illucens larval PM.
Supplementary Information
The online version contains supplementary material available at 10.1186/s12953-021-00175-x.
Keywords: Hermetia illucens, Liquid chromatography-tandem mass spectrometry, Midgut, Peritrophic matrix, Proteomic analysis, Scanning electron microscopy
Background
The peritrophic matrix (PM) is an acellular, semi-permeable structure lining the digestive tracts of certain invertebrates to encase food particles, as first reported in lepidopteran larvae [1, 2]. The original name was “peritrophic membrane” because of its appearance, but this term has been gradually replaced by “peritrophic matrix” in past decades; not only to avoid the misleading term “membrane” reserved in biology for lipid bilayers, but also to emphasize that the PM is actually an extracellular matrix with complex properties [3]. There are two types of PM: Type I PM is a multi-layered sleeve secreted by the epithelial cells along the entire midgut, and primarily observed in larval Lepidoptera and adult, blood-feeding Diptera. Secretion of Type I PM is mostly food-stimulated, as in female mosquitoes, but it can also be formed continuously, as in locusts. The more common, Type II PM is a highly ordered, 1–3 layers, sleeve-like structure secreted by a group of cells at the junction of the foregut and midgut. The physiological significance of these different types of PM is unclear [4, 5]. The PM is thought to be homologous to mucus secretions in the mammalian digestive tract [6], acting as a physical barrier to protect the midgut from rough food particles and digestive enzymes [7]. In addition, the PM can compartmentalize the midgut, promote digestion [8], neutralize toxic compounds [9], and act as an anti-infective barrier [10]. To understand how the PM performs these essential functions, one must study its molecular architecture [11].
The main components of the PM are protein and chitin. Chitin is a polysaccharide polymer of N-acetylglucosamine whose chains form strong, crystal microfibers by hydrogen bonding [12–14]. The PMs of different insects exhibit structural differences adapted to different food sources or physiological and immune challenges. In the typical microfiber arrangement, about 10 chitin microfibers are arranged in parallel to form a microfiber bundle. These bundles intersect to form a 60° or 90° grid structure, or they align in random directions to form a dense network structure. The pores of these grid structures are filled with many different proteins and carbohydrates.
PM-associated proteins can be divided into four classes according to their extractability [2]. The class I PM-associated proteins can be easily eluted from the PM with physiological buffers, such as many digestive enzymes. The class II PM-associated proteins can be removed with a mild detergent such as SDS to disrupt the weak ionic interactions. The class III proteins are tightly bound to the PM and require a strong denaturing agent such as urea to be extracted. The class IV proteins are thought to be covalently bound to the PM and so cannot be extracted by the above three methods. Proteins in the PM can also be divided into structural proteins, or peritrophins, and non-structural proteins. Structural proteins associate with the chitin scaffold and modify the integrity, elasticity, and permeability of the PM [15]. They usually contain more than one chitin-binding domain (CBD). Non-structural proteins are hydrolases and chitin-modifying enzymes, which regulate the structure of the PM, such as changing the pore size and permeability of the PM to adapt to physiological activities [16, 17]. In addition, many proteomic studies have shown that a large number of digestive enzymes are embedded in the PM, suggesting that the PM serves as a scaffold to which digestive enzymes associate [18, 19].
This paper examines the PM of the black solder fly (BSF; Hermetia illucens Linnaeus; Diptera: Stratiomyidae). BSF is thought to originate in the America and is currently widely spread in tropical, subtropical, and temperate regions [20]. Adults do not need to feed, do not bite, and are not disease vectors. The larvae have great potential utility in industry. 1) BSF larvae are a suitable protein source for poultry, swine, and several valuable fish species [21–23]. 2) BSF larvae are a potential source of bioactive substances like antimicrobial peptides, and enzymes such as cellulose-, chitin-, and lignin-degrading enzymes [24]. 3) BSF larvae are ideal for bioconversion, capable of efficiently transforming low-quality biomass like organic waste, kitchen waste, and agricultural by-products into high-quality protein [20, 25]. How they are able to digest so much so well is unknown. Most studies on BSF in recent decades have focused on the optimization of breeding conditions and the rearing of the larvae, but information on their biology is very limited [26–29].
In this work, mass spectrometry and scanning electron microscopy were employed to analyze the proteome and ultrastructure of the PM, to produce a more comprehensive understanding of the molecular architecture and ultrastructure of this essential structure.
Materials and methods
BSF larvae rearing and isolation of larval PM
BSF larvae were kept in an artificial climate chamber at a temperature of 27 °C, relative humidity of 70–80%, and a photoperiod of 12 h: 12 h light:dark. All the larvae were fed with commercially available wheat bran and water. About 200 fifth instar larvae were divided into three groups and sacrificed in this work. The larvae were anesthetized on ice, the larval epidermis was cut open, and the midgut was taken out. The PM was pulled out of the midgut, then washed in sterile, pre-cooled, phosphate buffered saline solution (PBS, 140 mM NaCl, 1 mM KCl, 6 mM phosphate buffer, pH 7.4) to remove the food debris, and then flash-frozen in liquid nitrogen. Several PMs were immersed in paraformaldehyde for scanning electron microscopy observations.
Total protein extraction
The total protein of the BSF larvae was extracted with protein lysis buffer (8 M urea with appropriate protease inhibitor), and treated twice with a high-throughput tissue crusher Wonbio-96c (Shanghai Wanbo Biotechnology Co., Ltd) for 40 s. The mixture was incubated on ice for 30 min, vortexed 5–10 s every 5 min, and then centrifuged at 16000×g at 4 °C for 30 min. Protein supernatants were mixed with four- fold volumes of cold acetone for 12 h to precipitate the protein and improve the final concentration. After centrifugation at 12000×g for 20 min, the pellet was rinsed twice with 90% acetone and dried under vacuum. The acetone dry powder was resuspended in protein lysis buffer (8 M urea with appropriate protease inhibitor), incubated on ice for 30 min, vortexed 5–10 s every 5 min, and then centrifuged at 16000×g at 4 °C for 30 min. Total protein concentration was determined by the bicinchoninic acid (BCA) method with a BCA Protein Assay Kit (Beyotime Biotechnology). Protein quantification was performed according to the kit protocol.
Protein digestion
From the extracted proteins, in each group 80 μg protein were collected, and tetraethylammonium bromide (TEAB) was added to a final concentration of 100 mM. The mixture was reduced with 10 mM Tris (2-carboxyethyl) phosphine (TCEP) at 37 °C for 60 min and alkylated with 40 mM iodoacetamide (IAM) at room temperature for 40 min in darkness. Six- fold volumes of cold acetone were added to precipitate the protein at − 20 °C for 4 h. After centrifugation for 20 min at 10000×g at 4 °C, the pellet was rinsed with 90% acetone. Trypsin was added at a 1:50 trypsin-to-protein mass ratio and incubated at 37 °C overnight, then desalted with HLB and dried under vacuum.
LC-MS/MS analysis
The trypsin-digested peptides were analyzed by online nano-flow liquid chromatography tandem mass spectrometry performed on an EASY-nLC system (Thermo, USA) connected to a Q Exactive HF-X Quadrupole Orbitrap mass spectrometer (Thermo, USA) through a nanoelectrospray ion source. Briefly, the C18-reversed phase column (75 μm × 25 cm, Thermo, USA) was equilibrated with solvent A (2% ACN with 0.1% formic acid) and solvent B (80% ACN with 0.1% formic acid). The peptides were eluted using the following gradient: 0–1 min, linear gradient of solvent B from 0 to 5%; 1–41 min, linear gradient of solvent B from 5 to 23%; 41–51 min, linear gradient of solvent B from 23 to 29%; 51–59 min, 29%–100% B; 59–65 min, gradient of solvent B 100%; 65–90 min, linear gradient of solvent B from 100 to 0%. The tryptic peptides were separated at a flow rate of 300 nL/min. The Q Exactive HF-X instrument was operated in data-dependent acquisition mode (DDA) to automatically switch between full scan MS and MS/MS acquisition. The survey of full scan MS spectra (m/z 350–1300) was acquired in the Orbitrap with 60,000 resolution. The automatic gain control (AGC) target was 3e6 and the maximum fill time was 20 ms. The top 20 most intense precursor ions were selected for fragmentation by higher-energy collision dissociation (HCD) in collision cells. The MS/MS resolution was set at 15000 (at m/z 100), the automatic gain control (AGC) target at 1e5, the maximum fill time at 50 ms, and dynamic exclusion at 18 s.
Protein identification and bioinformatics analysis
The RAW data files were analyzed using ProteomeDiscoverer (Thermo Scientific, Version 2.2) against an in-house H. illucens L transcriptome database. The main MS/MS search parameters were as follows: Mass tolerance of 10 ppm for MS and 0.05 Da for MS/MS tolerance, trypsin as the enzyme with 2 missed cleavage allowed, carbamidomethylation of cysteine as fixed modification, and methionine oxidation as dynamic modifications. High confidence peptides were used for protein identifications by setting a target false discovery rate (FDR) threshold of 1% at the peptide level. Only proteins that had at least two unique peptides were used for protein identification. The preliminarily identified proteins were further analyzed through multiple databases, including the presence of signal peptides (SignalP http://www.cbs.dtu.dk/services/SignalP/), subcellular location (TargetP: http:// www.cbs.dtu.dk/services/TargetP/), motif analysis (https://myhits.sib.swiss/cgi-bin/motif_scan), and annotation based on the MEROPS database, UniProt database, and NCBI BLAST analysis. Gene ontology (GO) analysis and Kyoto Encyclopedia of Genes and Genomes (KEGG) analysis were performed using OmicShare tools, which is a free online platform for data analysis (www.omicshare.com/tools).
Data availability
All raw LC-MS/MS data are available on the Mass Spectrometry Interactive Virtual Environment (MassIVE) data repository at ftp://massive.ucsd.edu/MSV000086950/
Field emission scanning electron microscopy (FESEM)
The PM were immersed in paraformaldehyde for three hours. Then the PM were washed with PBS and dehydrated with a gradient alcohol series (30, 50, 70, 80, 90, 100%), 10 min each, repeated twice. After that, the PM were critical point dried with a FEI CPD-030 dryer and sputter-coated with 3–4 nm platinum. The morphological characteristics of the PM were observed under a Hitachi SU8010 scanning electron microscope (Hitachi, Tokyo, Japan).
Results
Identification of PM proteins
Due to the lack of complete H. illucens proteome data in the existing databases, we constructed an in-house EST contigs through transcriptome sequencing, and the raw data files from the LC-MS/MS were analyzed using ProteomeDiscoverer (Thermo Scientific, Version 2.2) against the derived protein sequences from the contigs.
Proteomic analysis identified a total of 565 proteins from the PM samples of H. illucens (Table S1). Since a few midgut epithelial cells may have been attached during the dissection of the PM, coupled with the high sensitivity of mass spectrometry, the results may have contained proteins from the epithelial cells. Therefore, we artificially screened the proteins to remove these. Since PM is a non-cellular structure, the proteins in the PM are mostly released from the microvilli of the columnar midgut cells into the extracellular space through apocrine secretion. Such secreted proteins usually contain a signal peptide sequence to guide them into the extracellular space, and usually do not contain a transmembrane domain. Some of these proteins did not have homologous proteins in public databases, and were thus suspected to be newly discovered proteins, for which further studies are needed to identify them. Combining the results of protein subcellular localization, a total of 88 proteins (Table 1) were identified as possible PM proteins.
Table 1.
Proteins associated with the PM of H. illucens larvae
| Contig name | Protein descriptiona | pI | MW (kDa) | Main Feature |
|---|---|---|---|---|
| 1. Enzymes | ||||
| 1.1 Protein metabolism | ||||
| cds. TRINITY_DN12248_c0_g1_m.1812 | Trypsin-2-like | 5.07 | 28.1 | Tryp_SPc |
| cds. TRINITY_DN13311_c0_g1_m.3204 | Trypsin-1 | 4.92 | 27.2 | Tryp_SPc |
| cds. TRINITY_DN14779_c0_g1_m.5149 | Trypsin-7 | 5.43 | 28.6 | Tryp_SPc |
| cds. TRINITY_DN13668_c0_g1_m.3716 | Trypsin delta/gamma | 6.04 | 28.6 | Tryp_SPc |
| cds. TRINITY_DN18689_c0_g1_m.11218 | Trypsin-like protease | 9.31 | 24.9 | Tryp_SPc |
| cds. TRINITY_DN15755_c2_g1_m.6527 | Trypsin delta/gamma-like protein | 5.85 | 26.0 | Tryp_SPc |
| cds. TRINITY_DN11388_c0_g1_m.848 | Serine protease | 5.94 | 30.4 | Tryp_SPc super family |
| cds. TRINITY_DN12733_c0_g1_m.2440 | Serine proteases 1/2 | 5.6 | 31.2 | Tryp_SPc |
| cds. TRINITY_DN17609_c0_g2_m.9414 | Serine proteases 1/2 | 8.09 | 30.0 | Tryp_SPc |
| cds. TRINITY_DN12169_c0_g1_m.1711 | serine protease1/2 | 5.66 | 28.3 | Tryp_SPc super family |
| cds. TRINITY_DN16378_c0_g2_m.7436 | Serine protease | 5.68 | 44.5 | Tryp_SPc; CLIP_1 |
| cds. TRINITY_DN15101_c0_g1_m.5599 | Serine protease easter | 6.42 | 41.8 | Tryp_SPc; CLIP_1 |
| cds. TRINITY_DN803_c0_g1_m.17778 | Brachyurin-like | 6.00 | 30.8 | Tryp_SPc |
| cds. TRINITY_DN10692_c0_g1_m.300 | Brachyurin-like | 5.30 | 31.0 | Tryp_SPc |
| cds. TRINITY_DN11863_c0_g1_m.1372 | Chymotrypsin | 5.87 | 27.4 | Tryp_SPc |
| cds. TRINITY_DN17320_c1_g1_m.8906 | Chymotrypsin BI | 6.23 | 31.5 | Tryp_SPc |
| cds. TRINITY_DN10739_c0_g1_m.333 | Chymotrypsin-2 | 7.20 | 27.3 | Tryp_SPc |
| cds. TRINITY_DN13390_c0_g1_m.3313 | Chymotrypsin BI | 5.77 | 32.2 | Tryp_SPc |
| cds. TRINITY_DN10743_c0_g1_m.337 | Chymotrypsin BI | 5.31 | 31.7 | Tryp_SPc |
| cds. TRINITY_DN21565_c0_g1_m.17048 | Chymotrypsin BI | 5.06 | 32.4 | Tryp_SPc |
| cds. TRINITY_DN19986_c0_g1_m.13768 | Carboxypeptidase B | 6.57 | 45.9 | Propep_M14 |
| cds. TRINITY_DN19294_c0_g1_m.12373 | Angiotensin-converting enzyme | 5.20 | 73.6 | Peptidase_M2 |
| 1.2 Lipid metabolism | ||||
| cds. TRINITY_DN13210_c0_g1_m.3048 | Pancreatic triacylglycerol lipase | 5.27 | 37.4 | Pancreat_lipase_like |
| cds. TRINITY_DN13611_c0_g2_m.3637 | Lipase | 8.38 | 35.0 | Pancreat_lipase_like |
| cds. TRINITY_DN16324_c0_g1_m.7354 | Lipase | 5.53 | 37.2 | Pancreat_lipase_like |
| cds. TRINITY_DN13611_c0_g1_m.3635 | Lipase | 6.15 | 35.3 | Pancreat_lipase_like |
| 1.3 Glycosyl hydrolase | ||||
| cds. TRINITY_DN16881_c0_g3_m.8198 | Alpha-amylase | 6.47 | 53.8 | Aamy_C |
| cds. TRINITY_DN18580_c0_g1_m.10995 | Maltases | 4.97 | 69.8 | AmyAc_maltase |
| 1.4 Others | ||||
| cds. TRINITY_DN11418_c0_g1_m.880 | Chitinase-like protein | 7.47 | 48.9 | GH18_IDGF |
| cds. TRINITY_DN20328_c0_g1_m.14492 | Sphingomyelin phosphodiesterase | 5.83 | 70.3 | MPP_ASMase; SapB |
| cds. TRINITY_DN14434_c0_g1_m.4713 | Adenosine deaminase | 5.62 | 62.8 | Adm_rel super family |
| 2. Transporters | ||||
| cds. TRINITY_DN15890_c0_g1_m.6730 | Transferrin | 6.18 | 78.8 | PBP2_transferrin |
| cds. TRINITY_DN16029_c2_g1_m.6931 | Ferritin | 6.38 | 25.5 | Euk_Ferritin |
| 3. Odorant-binding protein | ||||
| cds. TRINITY_DN13988_c0_g1_m.4120 | Odorant-binding protein | 7.33 | 14.3 | PBP_GOBP |
| cds. TRINITY_DN11658_c0_g1_m.1123 | Odorant-binding protein | 7.20 | 15.0 | PBP_GOBP |
| cds. TRINITY_DN11155_c0_g1_m.640 | Odorant-binding protein | 7.65 | 15.8 | PBP_GOBP |
| cds. TRINITY_DN11257_c0_g1_m.728 | Odorant-binding protein | 5.24 | 15.3 | PBP_GOBP |
| cds. TRINITY_DN13076_c0_g1_m.2863 | Odorant-binding protein | 6.76 | 15.6 | PBP_GOBP |
| cds. TRINITY_DN9614_c0_g1_m.18143 | Odorant-binding protein | 7.08 | 16.5 | PBP_GOBP |
| cds. TRINITY_DN16710_c0_g8_m.7928 | Odorant-binding protein | 8.13 | 16.5 | PBP_GOBP |
| cds. TRINITY_DN16710_c0_g3_m.7924 | Odorant-binding protein | 6.95 | 14.2 | PBP_GOBP |
| 4. Immunity/defense | ||||
| cds. TRINITY_DN16169_c3_g1_m.7152 | SVWC domain-containing protein | 5.08 | 15.8 | SVWC |
| cds. TRINITY_DN16192_c2_g1_m.7187 | Lysozyme c | 8.31 | 14.8 | LYZ_C_invert |
| cds. TRINITY_DN13537_c1_g3_m.3530 | MD-2-related lipid-recognition protein | 6.57 | 17.1 | ML super family |
| cds. TRINITY_DN11748_c0_g1_m.1238 | MD-2-related lipid-recognition protein | 7.83 | 17.0 | ML super family |
| cds. TRINITY_DN19333_c0_g1_m.12455 | Hemocytin | 5.69 | 435.7 | 5 × VWD; 5 × TIL; 2 × FA58C |
| cds. TRINITY_DN17913_c0_g1_m.9886 | 3-glucan binding protein | 7.4 | 53.7 | CBM39; GH16_beta_GRP |
| cds. TRINITY_DN18830_c0_g1_m.11490 | Beta-1,3-glucan-binding protein | 5.39 | 42.5 | GH16_CCF |
| 5. Chitin binding | ||||
| cds. TRINITY_DN19733_c1_g5_m.13257 | Peritrophin-48-like | 4.82 | 20.7 | ChtBD2 |
| cds. TRINITY_DN16786_c0_g1_m.8036 | Peritrophin-48-like | 4.68 | 34.8 | ChtBD2 |
| cds. TRINITY_DN18694_c0_g3_m.11232 | Peritrophin-44 | 5.11 | 31.8 | ChtBD2 |
| cds. TRINITY_DN11775_c1_g1_m.1271 | Peritrophin-48-like | 5.53 | 35.1 | 2 × CBM_14 |
| cds. TRINITY_DN17674_c0_g2_m.9518 | Unnamed protein product | 4.55 | 39.6 | ChtBD2 |
| cds. TRINITY_DN14642_c0_g4_m.4965 | Peritrophic matrix protein 14 | 4.89 | 36.2 | 2 × CBM_14 |
| cds. TRINITY_DN18631_c0_g1_m.11102 | Uncharacterized protein | 4.58 | 33.7 | 2 × ChtBD2; 3 × CBM_14 |
| cds. TRINITY_DN19733_c1_g3_m.13254 | Uncharacterized protein | 5.3 | 36.8 | 2 × ChtBD2 |
| cds. TRINITY_DN17744_c0_g2_m.9623 | Peritrophin-44-like | 4.93 | 36.0 | ChtBD2 |
| cds. TRINITY_DN16319_c1_g1_m.7350 | Peritrophin-44 | 4.89 | 36.9 | 2 × ChtBD2 |
| cds. TRINITY_DN12826_c0_g1_m.2569 | Uncharacterized protein | 6.81 | 16.7 | CBM_14 |
| cds. TRINITY_DN12769_c0_g1_m.2499 | Uncharacterized protein | 4.67 | 24.8 | ChtBD2 |
| cds. TRINITY_DN11775_c1_g2_m.1272 | Unnamed protein product | 4.64 | 33.5 | 2 × ChtBD2 |
| cds. TRINITY_DN21192_c8_g1_m.16673 | Uncharacterized protein | 4.30 | 39.2 | CBM_14 |
| cds. TRINITY_DN12094_c0_g1_m.1629 | Uncharacterized protein | 7.69 | 15.0 | CBM_14; ChtBD2 |
| cds. TRINITY_DN13289_c0_g1_m.3171 | Uncharacterized protein | 7.71 | 15.0 | CBM_14; ChtBD2 |
| 6. Other protein | ||||
| cds. TRINITY_DN20571_c0_g2_m.15053 | Hexamerin | 8.31 | 67.1 | Hemocyanin_M; Hemocyanin_N; Hemocyanin_C |
| cds. TRINITY_DN19822_c1_g1_m.13436 | Hexamerin | 6.95 | 124.6 | Hemocyanin_M; Hemocyanin_N; Hemocyanin_C |
| cds. TRINITY_DN19826_c0_g1_m.13447 | Hexamerin | 9.01 | 93.2 | Hemocyanin_M; Hemocyanin_N; Hemocyanin_C |
| cds. TRINITY_DN17686_c0_g2_m.9540 | Hexamerin 2 beta | 6.55 | 84.1 | Hemocyanin_M; Hemocyanin_N; Hemocyanin_C |
| cds. TRINITY_DN16262_c0_g1_m.7277 | Hexamerin | 6.37 | 83.1 | Hemocyanin_M; Hemocyanin_N; Hemocyanin_C |
| cds. TRINITY_DN17535_c2_g2_m.9284 | Laminin | 5.43 | 413.1 | LamG |
| cds. TRINITY_DN14691_c0_g1_m.5031 | Calumenin | 4.67 | 38.5 | EFh_CREC_Calumenin_like |
| cds. TRINITY_DN13489_c0_g1_m.3457 | Nidogen | 4.78 | 147.6 | EGF_3; LY |
| cds. TRINITY_DN18659_c0_g3_m.11159 | Uncharacterized protein | 4.53 | 107.7 | 5 × DUF753 |
| cds. TRINITY_DN12676_c0_g1_m.2371 | Uncharacterized protein | 4.12 | 49.5 | Tryp_SPc super family |
| cds. TRINITY_DN18428_c0_g1_m.10766 | Uncharacterized protein | 4.53 | 74.1 | 4 × DUF753 |
| cds. TRINITY_DN4827_c0_g1_m.17337 | Unnamed protein product | 4.7 | 28.6 | JHBP |
| cds. TRINITY_DN16431_c0_g1_m.7515 | Uncharacterized protein | 4.27 | 50.9 | 2 × DUF753 |
| cds. TRINITY_DN11073_c0_g1_m.579 | Uncharacterized protein | 8.12 | 27.5 | JHBP |
| cds. TRINITY_DN12629_c0_g1_m.2312 | Uncharacterized protein | 4.92 | 33.2 | DUF1397 |
| cds. TRINITY_DN17619_c0_g2_m.9425 | Unnamed protein product | 4.88 | 23.5 | None |
| cds. TRINITY_DN13426_c0_g1_m.3363 | Uncharacterized protein | 4.77 | 24.4 | None |
| cds. TRINITY_DN14613_c0_g1_m.4924 | Uncharacterized protein | 7.47 | 58.3 | None |
| cds. TRINITY_DN18647_c0_g1_m.11141 | Uncharacterized protein | 4.55 | 74.3 | 3 × DUF753 |
| cds. TRINITY_DN12553_c0_g4_m.2207 | Unnamed protein product | 4.65 | 17.4 | None |
| cds. TRINITY_DN14642_c0_g5_m.4966 | Unnamed protein product | 4.86 | 18.8 | None |
| cds. TRINITY_DN12917_c0_g1_m.2687 | Uncharacterized protein | 5.49 | 16.9 | None |
| cds. TRINITY_DN16299_c0_g1_m.7327 | Unnamed protein product | 4.78 | 59.7 | 3 × EB |
| cds. TRINITY_DN15832_c0_g1_m.6643 | Uncharacterized protein | 5.29 | 53.7 | NUC |
Tryp_SPc Trypsin-like serine protease, CLIP_1 Serine protease Clip domain PPAF-2, Propep_M14 Carboxypeptidase activation peptide, ChtBD2 Chitin-binding domain type 2, CBM_14 Chitin binding Peritrophin-A domain, MPP_ASMase acid sphingomyelinase and related proteins, metallophosphatase domain, SapB Saposin (B) Domains, Adm_rel super family adenosine deaminase-related growth factor, LYZ_C_invert C-type invertebrate lysozyme, DUF753 Protein of unknown function
aAnnotation based on MEROPS database, UniProt database and NCBI BLAST analysis
Functional annotation of PM proteins by GO and KEGG analyses
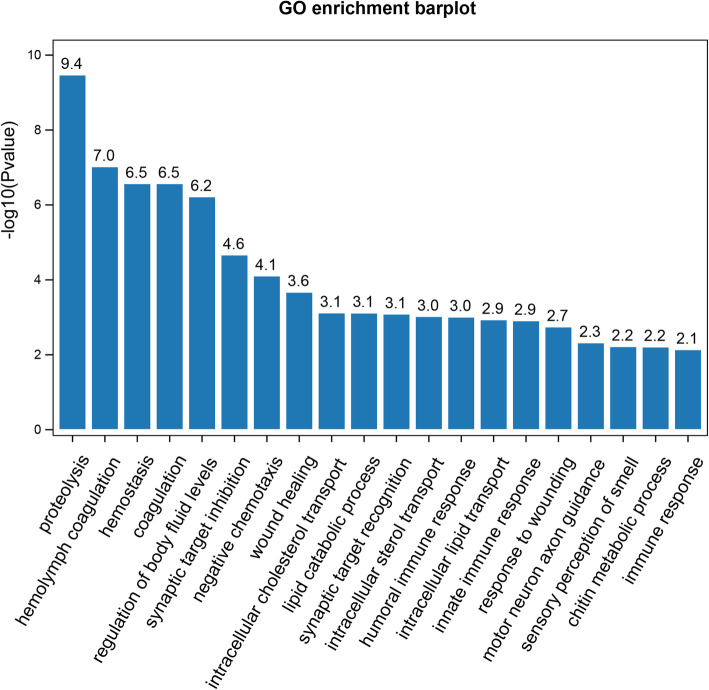
Enrichment analyses were performed to elucidate the biological function of the PM proteins. The results of the GO analysis demonstrated that the enriched GO terms were ‘proteolysis (GO:0006508)’, ‘hemolymph coagulation (GO:0042381)’, ‘regulation of body fluid levels (GO:0050878)’, ‘wound healing (GO:0042060)’, ‘innate immune response (GO:0045087)’, ‘chitin metabolic process (GO:0006030)’ etc. (Fig. 1), demonstrating that most queried proteins were involved in biological processes related to metabolic processes, immunity and so on. KEGG analysis revealed that PM proteins were enriched in metabolism, organismal systems, Environmental information processing, human disease and cellular processes (Fig. 2).
Fig. 1.
GO enrichment analysis of the predicted PM proteins. GO, Gene Ontology
Fig. 2.
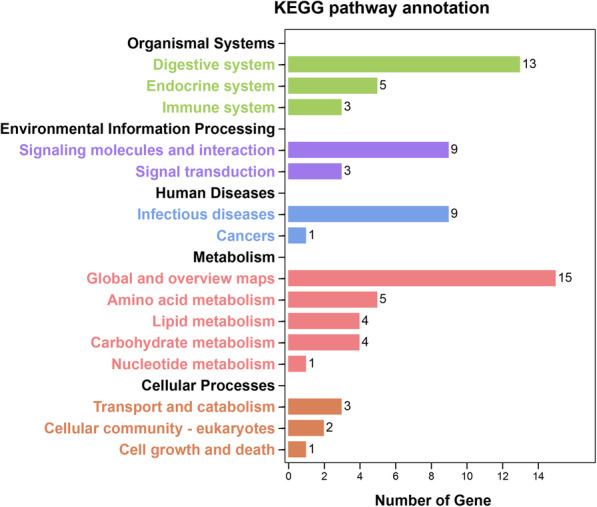
KEGG pathway enrichment of the predicted PM proteins. KEGG, Kyoto Encyclopedia of Genes and Genomes
Ultrastructure of the PM
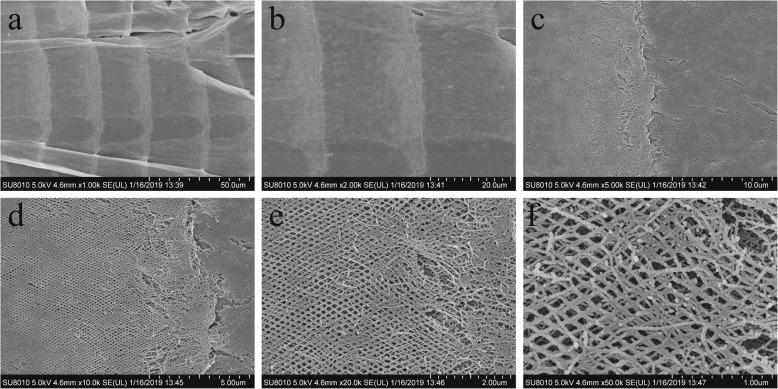
The SEM results showed that the outermost layer of the PM was arranged in continuous segments, and the length of each segment was approximately the same, about 25 μm (Fig. 3a). At the junction of every two sections, chitinous fibers intersect each other in a grid shape. The fibers at the anterior end of the outer PM layer are arranged loosely and appear brighter, while fibers at the posterior end are densely arranged and darker (Fig. 3b, c). Under high magnification, the fibers cross each other into diamond-shaped grids, which are filled with various proteins (Fig. 3d, e). Under higher magnification, one sees that many proteins are embed in the chitin scaffold (Fig. 3f).
Fig. 3.
Morphology of H. illucens larval PM observed by SEM under different magnification. a was observed under ×1000 magnification, the outermost layer of the PM was arranged in continuous segments, and the length of each segment was approximately the same, about 25 μm; b and c was observed under ×2000 and ×5000 magnification respectively, the chitinous fibers at the anterior end of the outer PM layer are arranged loosely, and appears brighter, while chitinous fibers at the posterior end are densely arranged and is darker; d and e was observed under × 10, 000 and × 20, 000 magnification respectively, chitinous fibers cross each other into diamond-shaped grids, and are filled with various proteins; f The chitinous-protein fibers can be clearly observed under × 50, 000 magnification
Discussion
Proteins involved in digestion and metabolism
One of the functions of the PM is to compartmentalize the midgut and divide it into the endoperitrophic, ectoperitrophic, and intraperitrophic spaces. After ingested food enters the midgut, digestive pro-enzymes are secreted from the anterior region of the midgut into the midgut lumen through extracellular secretion or apocrine secretion, then proteolytically activated. Digestion begins in the anterior region of the midgut as digestive enzymes cleave food macromolecules. Through peristaltic contraction of the PM, food and digestive enzymes (nucleases, lipases, proteases, etc.) are moved into the median midgut for further digestion. In the ectoperitrophic space, aminopeptidase, carboxypeptidase and amylase perform the final digestion. By compartmentalizing the midgut, damage to the enzyme catalytic sites and midgut epithelial cells caused by the accumulation of digestive intermediates are avoided, promoting food digestion [30–35].
In the putative PM proteome, we found twenty proteins belonging to the Chymotrypsin(S1) family, including six trypsin-like proteases, six serine proteases, two Brachyurin-like proteins, and six chymotrypsins. Trypsin is a kind of endoprotease that mainly cleaves the peptide chain at the carboxyl part of lysine or arginine. Trypsin is a major protease in the midgut of most insects (including Orthoptera, Hymenoptera, Diptera, Lepidoptera and Coleoptera) with a molecular weight of 20–35 kDa, an isoelectric point of 4–5, and an optimum pH range of 8–11 [36–38]. Brachyurin, also known as collagenase, was first found in the hepatopancreas of the fiddler crabs Uca pugilator. The site for cleavage of collagen is usually located 3/4 away from the amino end of the collagen chain. Brachyurin has a wide range of specificity for peptide bonds, but exhibits low activity for the main substrates of trypsin and chymotrypsin [39]. Chymotrypsin is a major digestive enzyme in the midgut of Diptera. Its cleavage site is usually located at the C-terminal peptide bond of a hydrophobic amino acid, and is more inclined to long-chain substrates. Chymotrypsin can catalyze the decomposition of proteins to produce free amino acids necessary for the growth and development of insects [40, 41]. There are two types of catalytically active chymotrypsin found in the midgut of the fruit fly. One is located in the midgut lumen with high catalytic activity, and the other is located in the cell membrane with weak catalytic activity [42]. Serine protease is the main digestive enzyme in insect midguts, and thus the target of most pest control strategies based on protease inhibitors [43]. In the course of evolution, plants produced some protease inhibitors to prevent insects from feeding, and insects correspondingly secreting excessive proteases or proteases that are not sensitive to these inhibitors. Therefore, in the past decades, a large number of protease-encoding genes were found in the digestive system of insects.
In the putative PM proteome, we found two metallopeptidases, one in the carboxypeptidase (M14) family, and one in the peptidyl-dipeptidase A (M2) family. Digestive carboxypeptidase is an exonuclease, which requires divalent metal cations (such as Zn2+) to catalyze the hydrolysis of the peptide bond at the carboxyl terminal of the polypeptide. The members of the carboxypeptidase M14 family can be divided into carboxypeptidase A and carboxypeptidase B according to the types of amino acids released by their catalytic functions. Carboxypeptidase A is more prone to hydrolyze polypeptides with aromatic or hydrophobic amino acids at the carboxyl terminal. Carboxypeptidase B is more inclined to hydrolyze peptides with lysine and arginine at the carboxyl terminal [44]. Besides digestion, carboxypeptidase can be involved in other physiological activities. For example, carboxypeptidase was found in the molting fluid of Helicoverpa armigera larvae and pre-pupa, and is believed to be related to apolysis [45]. Carboxypeptidase B in the midgut of Anopheles gambiae is related to the sexual development of Plasmodium, and is a potential target for malaria prevention and control [46]. One protein belonging to the peptidyl-dipeptidase A (M2) family was identified as angiotensin-converting enzymes (ACEs). In insects, ACE is involved in the metabolic inactivation of peptide neurotransmitters and processing prohormones into active peptide hormones [47–49]. ACE is expressed in many tissues in Drosophila, such as reproductive tissues, larval and adult midguts, larval tracheae, and adult salivary glands. ACE expression has also been detected in both sexes’ reproductive tissues in other insects, indicating that it plays an important role in reproduction [50–52].
Lipase plays an important role in the acquisition, storage, and metabolism of fat [53, 54]. In this study, a total of four lipases were found, including one pancreatic triacylglycerol lipase (TAG-lipase). TAG-lipases can hydrolyze the outer ester link of TAG and act on the water-lipid interface. Insects’ TAG-lipases need to be activated by calcium ions, and will preferentially hydrolyze unsaturated fatty acids, similar to mammalian pancreas TAG-lipases [55, 56]. Compared to mammals, few studies on insect pancreatic TAG-lipases exist, such as a pancreatic lipase-related protein as found in the brown planthopper, Nilaparvata lugens, that is essential for the hatching of eggs [57, 58]. A functional study using RNAi found that pancreatic TAG-lipases may be related to the virulence of BPH to rice varieties [59].
Carbohydrates are important energy sources in different developmental stages of insects. Three carbohydrases were found in this study: one α-amylases, and one maltase. In insects, the digestion of starch usually depends on these two enzymes. Maltose and glucose generated by amylase through hydrolyzing the α-1,4 glycosidic bond of starch are the energy sources for insect development, reproduction, and flight, with further digestion performed by maltase through hydrolyzing the α-1,4 or α-1,6 glycosidic bonds at the non-reducing ends of these oligosaccharides [60, 61]. Insect amylase relies on calcium and chloride ions to maintain activity and structural integrity, but it has also been reported that amylase in lepidopteran insects does not require chloride ions to activate [61, 62]. Maltase, also known as α-glucosidase, is a typical exo-type hydrolase, divided into family I and family II according to its structure. Insect α-glucosidase is classified as family I. Family I α-glucosidase has four conserved regions, which can also be found in α-amylase, but there is no sequence similarity between them [63]. Carbohydrases are often used as targets for pesticides, such as amylases or glucosidase inhibitors [64, 65].
We found a sphingomyelin phosphodiesterase (SMPD), an adenosine deaminase, and a chitinase-like protein in the PM proteome of H. illucens. SMPD is a hydrolase involved in the metabolism of sphingolipids. It can hydrolyze sphingomyelin into phosphocholine and ceramide, and plays an important role in signal transduction. In the bumble bee (Bombus lantschouensis), SMPD is expressed in various tissues, with higher expression levels in the ovary and midgut [66]. Differential expression of its homologous protein in Anopheles gambiae infected by different bacteria suggests it may also be involved in invertebrate immune response [67, 68]. Chitinase belongs to family 18 of the glycoside hydrolases, and can hydrolyze chitin into small, soluble oligosaccharides. Chitinase is very important for the regulation of the thickness and permeability of the PM, and is also involved in the degradation of the PM during molting. The products of chitin hydrolysis can be recycled to synthesize new chitin [69].
Transporter
A transferrin and a ferritin were found in the PM proteome of H. illucens. Iron participates in a variety of physiological activities, such as oxygen metabolism, amino acid production, and DNA biosynthesis. In some arthropods, it also participates in egg development and offspring production [70, 71]. However, iron can also cause production of reactive oxygen species and highly reactive radicals, which may cause cell death and tissue damage. Therefore, vertebrates and arthropods have evolved a variety of specific proteins, such as transferrin and ferritin, to keep iron in a safe form [72]. Transferrin is a secreted protein with high affinity for iron ions. In insects, it not only participates in iron transport, but also has some immune functions [71, 73, 74]. Ferritin is a sphere complex with 24 subunits, which can hold thousands of iron atoms and maintain a nontoxic state. It is the main protein that stores iron in organisms [75]. For insects, the secretion of iron-laden ferritin through the midgut and Malpighian tubules is the main way for insects to excrete iron [76].
Proteins involved in signaling
We found eight odorant-binding proteins (OBP) in the PM proteome of H. illucens. OBPs are soluble proteins with a small molecular weight, usually between 17 and 22 kDa, first found in the antennae of the male giant moth (Antheraea polyphemus). They can bind, solubilize, and deliver odor molecules or pheromones to their receptors [77–79]. There exists a relatively conserved sequence in insect OBPs, which contains six cysteine residues and can form three disulfide bonds, which also makes the tertiary structure of OBP conserved. The number of OBP genes in different insect genomes varies from a few to hundreds [80–82]. In addition to their abundant expression in insect antennae, some OBP family members are expressed in other tissues, such as the abdomen and wings of Periplaneta americana [83], and the midgut and head of diamondback moths (Plutella xylostella) [84]. Four OBPs in honeybees are similarly highly expressed in regions with low chemosensory receptor expression [85]. These cases indicate that OBP may participate in other life activities besides chemosensation, such as responses to adverse environmental conditions and the scavenging of various, small, hydrophobic molecules [86, 87].
Proteins involved in larval innate immunity
BSF is an omnivorous insect with a wide range of larval diets, so its food may carry various bacteria, viruses, fungi, or parasites that can infect the insect when it eats. In the past few decades, there have been many studies on how insects fight these infections, and the PM is considered to be the first line of defense [3]. In fact, early reports hypothesized that PM production was primarily to protect midgut epithelial cells from infection by pathogens in food [69]. In addition to the aforementioned immune-related serine proteases and other proteins, some other proteins related to larval immunity were also found in this study, including two beta-1,3-glucan-binding protein (BGRP), one protein with an SVWC domain, one C-type lysozyme, two MD-2 related lipid recognition proteins and one hemocytin. The insect innate immune system uses a large number of pattern recognition proteins to recognize the molecular characteristics of pathogenic microorganisms, thereby triggering a series of immune defense mechanisms. BGNP is a main pattern recognition receptors in insects and is more specific to fungi [88]. SVWC is a short-sequence protein in arthropods that can respond to changes in nutritional status or environmental changes such as bacterial and viral infections. In the European bumblebee (Bombus terrestris), SVWC participates in the host’s antiviral immunity and is related to the expression of antimicrobial peptides [89]. Lysozyme is an antibacterial enzyme widely found in bacteriophages, plants, and animals. It can dissolve bacteria by hydrolyzing the cell walls of bacteria. Animal lysozymes are divided into c-type, g-type and i-type, with c-type lysozyme expression affected by bacterial challenge [90]. MD-2 related lipid recognition protein is a type of secretory or luminal protein that binds to lipids and is mainly expressed in the gut. It can also bind to bacterial lipopolysaccharides and interact with cell receptors, which are important for innate immunity [91]. Hemocytin is a protein in insects homologous to mammalian von Willebrand factor. The expression of hemocytin in the silkworm is up-regulated following bacterial infection and before pupation, indicating it may play an important role in immunity and metamorphosis [92].
PM proteins with chitin-binding domains
In addition to the above-mentioned, non-structural proteins, some structural proteins, namely peritrophin, were found in the PM proteome of H. illucens. Peritrophins usually contain more than one chitin-binding domain (CBD), which are called the peritrophin-A domain (PAD), peritrophin-B domain, and peritrophin-C domain with 6, 8, or 10 cysteine residues respectively that can form 3–5 disulfide bonds, of which PAD is the most common (2). PAD is also called type-2 chitin-binding domain (ChtBD2), and its sequence feature is CX13–20CX5–6CX9–19CX10–14CX4–14C, where X represents any amino acid except cysteine [2, 93]. PAD is mainly found in peritrophin, but it is also found in midgut chitinase, as well as some proteins in the Malpighian tube, rectum, and epidermis [94, 95]. Peritrophin-44 is the first peritrophin found in the PM of Lucilia cuprina larvae with several CBDs. These tandem CBDs may facilitate cross-linking chitin fibrils within the PM. Peritrophin-44 and another protein, Peritrophin-48, are the main proteins involved in the construction of PM, and represent approximately 70% of the protein mass of the PM [96–99]. In this study, in addition to Peritrophin-44 and Peritrophin-48, we also found a variety of unknown proteins containing ChtBD2 domains. The functions of these proteins need further study.
Other PM proteins
Among the other proteins in the BSF PM proteome are hexamerins. Hexamerins can store a large amount of amino acids, which play an important role in insect metamorphosis and reproduction, and when suffering adverse environments such as food shortages [100]. Hexamerin is expressed in all insect life stages, reaching a peak in the last instar. Hexamerin is usually expressed in the fat body and stored in the hemolymph, but is also moderately expressed in the midgut of Apriona germari, and in the midgut, epidermis, and Malpighian tubules of Spodoptera exigua [101, 102]. Expression of hexamerin in tussah silkworm (Antheraea pernyi) was significantly increased when they were infected with microorganisms, so it may be related to immune response [103].
A large number of uncharacterized or unnamed proteins were also discovered. These proteins should not be ignored, and more research is needed on them, because they may have important significance to the molecular architecture of the PM or lead to better breeding and utilization of BSF larvae.
We also compared the identified PM proteome of H.illucens with other reported insects, especially the Musca domestica which also belongs to Diptera. In terms of the number of proteins identified, there are 47 species in the 5th instar Bombyx mori [18], 71 secreted proteins in M.domestica [104], and 115 in Aedes aegypti [105]. From the point of view of protein function, the PM proteome of different insects are all involved in the physiological processes of digestion, metabolism and immunity. In terms of digestion, trypsin, amylase, and carboxypeptidase were found in the PM proteome of H.illucens and M.domestica [104]. In addition, some Brachyurin-like proteins which can participate in the metabolism of collagen were also found in H.illucens. Peritrophin is a kind of important protein in the PM proteome. Only 4 peritrophin are found in A.aegypti [105], 8 in M.domestica, and 16 in H.illucens. This may be due to the PM of A.aegypti is usually induced following a blood meal. While the larva of M.domestica and H.illucens constantly feed before pupation, but the instar of H.illucens is much longer than that of M.domestica. From the molecular weight point of view, most of the peritrophin are between 20 and 40 kDa, and a number of peritrophin of about 16 kDa have been found in H.illucens. The function of these peritrophin needs further verification.
Ultrastructure of the PM
The SEM results showed that the outermost layer of the PM was arranged in continuous segments, and the length of each segment was approximately the same, about 25 μm (Fig. 1a). It was observed in well-fed Locusta migratoria, the midgut secretes PM every 15 min. The inner PM will move posteriorly with the food, making the PM appear as a telescope-like structure (3), as we observed in the BSF PM (Fig. 1b, c). A grid-like structure of chitin fibers observed in the BSF PM (Fig. 1d, e) may be found in the PM of other insects, but the currently known arrangements are very different. For example, the PM from larval Trichoplusia ni is organized in a random, felt-like structure of fibers [106], the Ostrinia nubilalis PM in an orthogonal structure of fibers [107], and Anomala cuprea PM in a hexagonal structure of fibers. The significance of the differences in arrangement are not clear yet. We speculate that it may be related to the feeding habits of the insects. BSF larvae are described as voracious feeders in most studies, and a diamond-shaped chitin grid is more flexible when deformed, so it may enable their PM to hold more food.
Conclusions
The main function of the PM is to protect the midgut epithelial cells from coarse food particles, pathogens, and toxins, and it can also compartmentalize the midgut to promote digestion. In this study, we discovered a large number of digestive enzymes bound to the BSF PM, which can digest a variety of proteins, fats, and other ingested food particles. We also discovered a variety of immune-related proteins known to play roles in the identification of and defense against pathogens. The findings of some transporters suggest that the PM maybe involved in ion transport and excretion. The findings of some odorant-binding proteins also indicate that the PM is an important place for insects to interact with their external environment. Scanning electron microscopy revealed clearly the ultrastructure of the BSF PM, and found that it is a scaffold that can hold a variety of proteins. PMs in insects display diverse textures on this ultramicro scale, but the reason for this diversification needs further study. To summarize, our work used liquid chromatography-tandem mass spectrometry and scanning electron microscopy to conduct a more in-depth study of the PM of BSF larvae, and deepen our understanding of its molecular architecture and ultrastructure.
Supplementary Information
Additional file 1: Table S1. all the identified protein from peritrophic matrix sample
Acknowledgements
The authors would like to express their gratitude to EditSprings (https://www.editsprings.com/) for the expert linguistic services provided.
Abbreviations
- BSF
Black soldier fly
- PM
Peritrophic matrix
- SEM
Scanning electron microscopy
- CBD
Chitin-binding domain
- BCA
Bicinchoninic acid
- TCEP
Tris (2-carboxyethyl) phosphine
- IAM
Iodoacetamide
- DDA
Data-dependent acquisition mode
- AGC
Automatic gain control
- HCD
Higher-energy collision dissociation
- FDR
False discovery rate
- FESEM
Field emission scanning electron microscopy
- S1 family
Chymotrypsin family
- M14 family
Carboxypeptidase family
- M2 family
Peptidyl-dipeptidase A family
- ACE
Angiotensin-converting enzymes
- TAG
Pancreatic triacylglycerol
- SMPD
Sphingomyelin phosphodiesterase
- OBP
Odorant-binding proteins
- BGRP
Beta-1,3-glucan-binding protein
- PAD
Peritrophin-A domain
- ChtBD2
Type-2 chitin-binding domain
Authors’ contributions
JX and DH conceived and designed this study; YL and JR performed the experiment and analyzed the data; XW and ZS participated in manuscript preparation; YL wrote the manuscript; All authors have read and agreed to the published version of the manuscript. All authors read and approved the final manuscript.
Funding
This work was financially supported by the National Natural Science Foundation of China (Nos. 31830084, 31970440).
Availability of data and materials
All data generated or analysed during this study are included in this published article and its supplementary information files or from the corresponding author on reasonable request.
Declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Yu-Bo Lin and Jing-Jing Rong contributed equally to this work.
References
- 1.Lyonet P. Traite anatomique de la chenille, qui ronge le bois de saul, augmente d'une explication des planches: Gosse et Pinet; 1762.
- 2.Tellam RL, Wijffels G, Willadsen P. Peritrophic matrix proteins. Insect Biochem Mol Biol. 1999;29(2):87–101. doi: 10.1016/S0965-1748(98)00123-4. [DOI] [PubMed] [Google Scholar]
- 3.Cohen E, Moussian B. Extracellular composite matrices in arthropods: Springer; 2016. 10.1007/978-3-319-40740-1.
- 4.Peters W. Peritrophic membranes: Springer Berlin Heidelberg; 1992. 10.1007/978-3-642-84414-0.
- 5.Ryerse JS, Purcell JP, Sammons RD, Lavrik PB. Peritrophic membrane structure and formation in the larva of a moth, Heliothis. Tissue Cell. 1992;24(5):751–71. 10.1016/0040-8166(92)90047-B. [DOI] [PubMed]
- 6.Hegedus D, Erlandson M, Gillott C, Toprak U. New insights into peritrophic matrix synthesis, architecture, and function. Annu Rev Entomol. 2009;54(1):285–302. doi: 10.1146/annurev.ento.54.110807.090559. [DOI] [PubMed] [Google Scholar]
- 7.Sudha P, Muthu SP. Damage to the midgut epithelium caused by food in the absence of peritrophic membrane. Curr Sci. 1988;57(11):624–625. [Google Scholar]
- 8.Kelkenberg M, Odman-Naresh J, Muthukrishnan S, Merzendorfer H. Chitin is a necessary component to maintain the barrier function of the peritrophic matrix in the insect midgut. Insect Biochem Mol Biol. 2015;56:21–28. doi: 10.1016/j.ibmb.2014.11.005. [DOI] [PubMed] [Google Scholar]
- 9.Pérez-Hedo M, López C, Albajes R, Eizaguirre M. Low susceptibility of non-target Lepidopteran maize pests to the Bt protein Cry1Ab. Bull Entomol Res. 2012;102(6):737–743. doi: 10.1017/S0007485312000351. [DOI] [PubMed] [Google Scholar]
- 10.Weiss BL, Savage AF, Griffith BC, Wu Y, Aksoy S. The peritrophic matrix mediates differential infection outcomes in the tsetse fly gut following challenge with commensal, pathogenic, and parasitic microbes. J Immunol (Baltimore, Md : 1950) 2014;193(2):773–782. doi: 10.4049/jimmunol.1400163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Toprak U, Erlandson M, Baldwin D, Karcz S, Wan L, Coutu C, Gillott C, Hegedus DD. Identification of the Mamestra configurata (Lepidoptera: Noctuidae) peritrophic matrix proteins and enzymes involved in peritrophic matrix chitin metabolism. Insect Sci. 2016;23(5):656–674. doi: 10.1111/1744-7917.12225. [DOI] [PubMed] [Google Scholar]
- 12.Liu X, Zhang H, Li S, Zhu KY, Ma E, Zhang J. Characterization of a midgut-specific chitin synthase gene (LmCHS2) responsible for biosynthesis of chitin of peritrophic matrix in Locusta migratoria. Insect Biochem Mol Biol. 2012;42(12):902–910. doi: 10.1016/j.ibmb.2012.09.002. [DOI] [PubMed] [Google Scholar]
- 13.Kaya M, Baran T, Erdoğan S, Menteş A, Özüsağlam MA, Çakmak YS. Physicochemical comparison of chitin and chitosan obtained from larvae and adult Colorado potato beetle (Leptinotarsa decemlineata) Mater Sci Eng C Mater Biol Appl. 2014;45:72–81. doi: 10.1016/j.msec.2014.09.004. [DOI] [PubMed] [Google Scholar]
- 14.Kaya M, Erdogan S, Mol A, Baran T. Comparison of chitin structures isolated from seven Orthoptera species. Int J Biol Macromol. 2015;72:797–805. doi: 10.1016/j.ijbiomac.2014.09.034. [DOI] [PubMed] [Google Scholar]
- 15.Toprak U, Erlandson M, Hegedus DD. Peritrophic matrix proteins. Trends Entomol. 2010;6:23–51. [Google Scholar]
- 16.Dixit R, Arakane Y, Specht CA, Richard C, Kramer KJ, Beeman RW, Muthukrishnan S. Domain organization and phylogenetic analysis of proteins from the chitin deacetylase gene family of Tribolium castaneum and three other species of insects. Insect Biochem Mol Biol. 2008;38(4):440–451. doi: 10.1016/j.ibmb.2007.12.002. [DOI] [PubMed] [Google Scholar]
- 17.Toprak U, Baldwin D, Erlandson M, Gillott C, Hou X, Coutu C, Hegedus DD. A chitin deacetylase and putative insect intestinal lipases are components of the Mamestra configurata (Lepidoptera: Noctuidae) peritrophic matrix. Insect Mol Biol. 2008;17(5):573–585. doi: 10.1111/j.1365-2583.2008.00829.x. [DOI] [PubMed] [Google Scholar]
- 18.Hu X, Chen L, Xiang X, Yang R, Yu S, Wu X. Proteomic analysis of peritrophic membrane (PM) from the midgut of fifth-instar larvae, Bombyx mori. Mol Biol Rep. 2012;39(4):3427–3434. doi: 10.1007/s11033-011-1114-6. [DOI] [PubMed] [Google Scholar]
- 19.Zhong X, Zhang L, Zou Y, Yi Q, Zhao P, Xia Q, Xiang Z. Shotgun analysis on the peritrophic membrane of the silkworm Bombyx mori. BMB Rep. 2012;45(11):665–670. doi: 10.5483/BMBRep.2012.45.11.261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang YS, Shelomi M. Review of black soldier fly (Hermetia illucens) as animal feed and human food. Foods. 2017;6(10):91. 10.3390/foods6100091. [DOI] [PMC free article] [PubMed]
- 21.Newton G, Booram C, Barker R, Hale O. Dried Hermetia illucens larvae meal as a supplement for swine. J Anim Sci. 1977;44(3):395–400. doi: 10.2527/jas1977.443395x. [DOI] [Google Scholar]
- 22.Hale O. Dried Hermetia illucens larvae (Diptera: Stratiomyidae) as a feed additive for poultry. Ga Entomol Soc J. 1973;8:16–20.
- 23.St-Hilaire S, Sheppard C, Tomberlin JK, Irving S, Newton L, McGuire MA, et al. Fly prepupae as a feedstuff for rainbow trout, Oncorhynchus mykiss. J World Aquacult Soc. 2007;38(1):59–67. doi: 10.1111/j.1749-7345.2006.00073.x. [DOI] [Google Scholar]
- 24.Müller A, Wolf D, Gutzeit HO. The black soldier fly, Hermetia illucens - a promising source for sustainable production of proteins, lipids and bioactive substances. Z Naturforsch C J Biosci. 2017;72(9–10):351–363. doi: 10.1515/znc-2017-0030. [DOI] [PubMed] [Google Scholar]
- 25.van Huis A. Potential of insects as food and feed in assuring food security. Annu Rev Entomol. 2013;58(1):563–583. doi: 10.1146/annurev-ento-120811-153704. [DOI] [PubMed] [Google Scholar]
- 26.Harnden LM, Tomberlin JK. Effects of temperature and diet on black soldier fly, Hermetia illucens (L.) (Diptera: Stratiomyidae), development. Forensic Sci Int. 2016;266:109–116. doi: 10.1016/j.forsciint.2016.05.007. [DOI] [PubMed] [Google Scholar]
- 27.Leong SY, Kutty SR, Malakahmad A, Tan CK. Feasibility study of biodiesel production using lipids of Hermetia illucens larva fed with organic waste. Waste Manag (New York, NY) 2016;47(Pt A):84–90. doi: 10.1016/j.wasman.2015.03.030. [DOI] [PubMed] [Google Scholar]
- 28.Bonelli M, Bruno D, Caccia S, Sgambetterra G, Cappellozza S, Jucker C, Tettamanti G, Casartelli M. Structural and functional characterization of Hermetia illucens larval Midgut. Front Physiol. 2019;10:204. doi: 10.3389/fphys.2019.00204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Barragan-Fonseca KB, Dicke M, van Loon JJ. Nutritional value of the black soldier fly (Hermetia illucens L.) and its suitability as animal feed–a review. J Insects Food Feed. 2017;3(2):105–120. doi: 10.3920/JIFF2016.0055. [DOI] [Google Scholar]
- 30.Santos CD, Terra WR. Distribution and characterization of oligomeric digestive enzymes from Erinnyis ello larvae and inferences concerning secretory mechanisms and the permeability of the peritrophic membrane. J Insect Biochem. 1986;16(4):691–700. doi: 10.1016/0020-1790(86)90013-2. [DOI] [Google Scholar]
- 31.Takesue Y, Yokota K, Miyajima S, Taguchi R, Ikezawa H. Membrane anchors of alkaline phosphatase and trehalase associated with the plasma membrane of larval midgut epithelial cells of the silkworm, Bombyx mori. J Biochem. 1989;105(6):998–1001. doi: 10.1093/oxfordjournals.jbchem.a122794. [DOI] [PubMed] [Google Scholar]
- 32.Ferreira C, Terra WR. Spatial organization of digestion, secretory mechanisms and digestive enzyme properties in Pheropsophus aequinoctialis (Coleoptera: Carabidae) J Insect Biochem. 1989;19(4):383–391. doi: 10.1016/0020-1790(89)90043-7. [DOI] [Google Scholar]
- 33.Jordão BP, Terra WR. Regional distribution and substrate specificity of digestive enzymes involved in terminal digestion in Musca domestica hind-midguts. Arch Insect Biochem Physiol. 1991;17(2–3):157–168. doi: 10.1002/arch.940170209. [DOI] [PubMed] [Google Scholar]
- 34.Jordão BP, Capella AN, Terra WR, Ribeiro AF, Ferreira C. Nature of the anchors of membrane-bound aminopeptidase, amylase, and trypsin and secretory mechanisms in Spodoptera frugiperda (Lepidoptera) midgut cells. J Insect Physiol. 1999;45(1):29–37. doi: 10.1016/S0022-1910(98)00098-5. [DOI] [PubMed] [Google Scholar]
- 35.Bolognesi R, Terra WR, Ferreira C. Peritrophic membrane role in enhancing digestive efficiency. Theoretical and experimental models. J Insect Physiol. 2008;54(10–11):1413–1422. doi: 10.1016/j.jinsphys.2008.08.002. [DOI] [PubMed] [Google Scholar]
- 36.Tamaki FK, Pimentel AC, Dias AB, Cardoso C, Ribeiro AF, Ferreira C, Terra WR. Physiology of digestion and the molecular characterization of the major digestive enzymes from Periplaneta americana. J Insect Physiol. 2014;70:22–35. doi: 10.1016/j.jinsphys.2014.08.007. [DOI] [PubMed] [Google Scholar]
- 37.Hemmati SA, Sajedi RH, Moharramipour S, Taghdir M, Rahmani H, Etezad SM, et al. Biochemical characterization and structural analysis of trypsin from Plodia interpunctella midgut: implication of determinants in extremely alkaline pH activity profile. J Arch Phytopathol. 2017;42(4):307–318. [Google Scholar]
- 38.Hemmati SA, Mehrabadi M. Structural ensemble-based computational analysis of trypsin enzyme genes discovered highly conserved peptide motifs in insects. J Arch Phytopathol Plant Prot. 2020;53(7–8):335–354. doi: 10.1080/03235408.2020.1744978. [DOI] [Google Scholar]
- 39.Tsu CA, Craik CS. Substrate recognition by recombinant serine collagenase 1 from Uca pugilator. J Biol Chem. 1996;271(19):11563–11570. doi: 10.1074/jbc.271.19.11563. [DOI] [PubMed] [Google Scholar]
- 40.Barrett AJ, Rawlings N, Woessner JF. Handbook of Proteolytic Enzymes: Second Edition. 2004. pp. 1–1140. [Google Scholar]
- 41.Spit J, Zels S, Dillen S, Holtof M, Wynant N, Vanden BJ. Effects of different dietary conditions on the expression of trypsin- and chymotrypsin-like protease genes in the digestive system of the migratory locust, Locusta migratoria. Insect Biochem Mol Biol. 2014;48:100–109. doi: 10.1016/j.ibmb.2014.03.002. [DOI] [PubMed] [Google Scholar]
- 42.Tamaki FK, Padilha MH, Pimentel AC, Ribeiro AF, Terra WR. Properties and secretory mechanism of Musca domestica digestive chymotrypsin and its relation with Drosophila melanogaster homologs. Insect Biochem Mol Biol. 2012;42(7):482–490. doi: 10.1016/j.ibmb.2012.03.005. [DOI] [PubMed] [Google Scholar]
- 43.Gatehouse JA. Prospects for using proteinase inhibitors to protect transgenic plants against attack by herbivorous insects. Curr Protein Pept Sci. 2011;12(5):409–416. doi: 10.2174/138920311796391142. [DOI] [PubMed] [Google Scholar]
- 44.Ferreira C, Rebola KG, Cardoso C, Bragatto I, Ribeiro AF, Terra WR. Insect midgut carboxypeptidases with emphasis on S10 hemipteran and M14 lepidopteran carboxypeptidases. Insect Mol Biol. 2015;24(2):222–239. doi: 10.1111/imb.12151. [DOI] [PubMed] [Google Scholar]
- 45.Sui YP, Liu XB, Chai LQ, Wang JX, Zhao XF. Characterization and influences of classical insect hormones on the expression profiles of a molting carboxypeptidase a from the cotton bollworm (Helicoverpa armigera) Insect Mol Biol. 2009;18(3):353–363. doi: 10.1111/j.1365-2583.2009.00879.x. [DOI] [PubMed] [Google Scholar]
- 46.Mongkol W, Arunyawat U, Surat W, Kubera A. Active compounds against Anopheles minimus Carboxypeptidase B for malaria transmission-blocking strategy. J Med Entomol. 2015;52(6):1322–1332. doi: 10.1093/jme/tjv133. [DOI] [PubMed] [Google Scholar]
- 47.Isaac RE, Siviter RJ, Stancombe P, Coates D, Shirras AD. Conserved roles for peptidases in the processing of invertebrate neuropeptides. Biochem Soc Trans. 2000;28(4):460–464. doi: 10.1042/bst0280460. [DOI] [PubMed] [Google Scholar]
- 48.Schoofs L, Veelaert D, De Loof A, Huybrechts R, Isaac E. Immunocytochemical distribution of angiotensin I-converting enzyme-like immunoreactivity in the brain and testis of insects. Brain Res. 1998;785(2):215–227. doi: 10.1016/S0006-8993(97)01398-X. [DOI] [PubMed] [Google Scholar]
- 49.Lamango NS, Nachman RJ, Hayes TK, Strey A, Isaac RE. Hydrolysis of insect neuropeptides by an angiotensin-converting enzyme from the housefly, Musca domestica. Peptides. 1997;18(1):47–52. doi: 10.1016/S0196-9781(96)00232-X. [DOI] [PubMed] [Google Scholar]
- 50.Chintapalli VR, Wang J, Dow JA. Using FlyAtlas to identify better Drosophila melanogaster models of human disease. Nat Genet. 2007;39(6):715–720. doi: 10.1038/ng2049. [DOI] [PubMed] [Google Scholar]
- 51.Rylett CM, Walker MJ, Howell GJ, Shirras AD, Isaac RE. Male accessory glands of Drosophila melanogaster make a secreted angiotensin I-converting enzyme (ANCE), suggesting a role for the peptide-processing enzyme in seminal fluid. J Exp Biol. 2007;210(Pt 20):3601–3606. doi: 10.1242/jeb.009035. [DOI] [PubMed] [Google Scholar]
- 52.Xu J, Baulding J, Palli SR. Proteomics of Tribolium castaneum seminal fluid proteins: identification of an angiotensin-converting enzyme as a key player in regulation of reproduction. J Proteome. 2013;78:83–93. doi: 10.1016/j.jprot.2012.11.011. [DOI] [PubMed] [Google Scholar]
- 53.Santana CC, Barbosa LA, Júnior IDB, Nascimento TGD, Dornelas CB, Grillo LAM. Lipase activity in the larval midgut of Rhynchophorus palmarum: biochemical characterization and the effects of reducing agents. Insects. 2017;8(3):100. 10.3390/insects8030100. [DOI] [PMC free article] [PubMed]
- 54.Javed MA, Coutu C, Theilmann DA, Erlandson MA, Hegedus DD. Proteomics analysis of Trichoplusia ni midgut epithelial cell brush border membrane vesicles. Insect Sci. 2019;26(3):424–440. doi: 10.1111/1744-7917.12547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Grillo LA, Majerowicz D, Gondim KC. Lipid metabolism in Rhodnius prolixus (Hemiptera: Reduviidae): role of a midgut triacylglycerol-lipase. Insect Biochem Mol Biol. 2007;37(6):579–588. doi: 10.1016/j.ibmb.2007.03.002. [DOI] [PubMed] [Google Scholar]
- 56.Hoffman AG. End product specificity of triacylglycerol lipases from intestine, fat body, muscle and haemolymph of the American cockroach, Periplaneta americana L. Lipids. 1979;14(11):893–899. doi: 10.1007/BF02533502. [DOI] [PubMed] [Google Scholar]
- 57.Gilham D, Labonté ED, Rojas JC, Jandacek RJ, Howles PN, Hui DY. Carboxyl ester lipase deficiency exacerbates dietary lipid absorption abnormalities and resistance to diet-induced obesity in pancreatic triglyceride lipase knockout mice. J Biol Chem. 2007;282(34):24642–24649. doi: 10.1074/jbc.M702530200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Xu L, Huang HJ, Zhou X, Liu CW, Bao YY. Pancreatic lipase-related protein 2 is essential for egg hatching in the brown planthopper, Nilaparvata lugens. Insect Mol Biol. 2017;26(3):277–285. doi: 10.1111/imb.12290. [DOI] [PubMed] [Google Scholar]
- 59.Yuan LY, Hao YH, Chen QK, Rui PA, Zhang WQ. Pancreatic triglyceride lipase is involved in the virulence of the brown planthopper to rice plants. J Integr Agric. 2020;19(11):2758–2766. doi: 10.1016/S2095-3119(20)63188-4. [DOI] [Google Scholar]
- 60.Cruz WO, Sinhori GGC, de Lima CAR, Pontes EG. Biochemical properties of α-amylase from Midgut of Alphitobius diaperinus (panzer) (Coleoptera: Tenebrionidae) larvae. Neotrop Entomol. 2018;47(5):698–708. doi: 10.1007/s13744-018-0590-y. [DOI] [PubMed] [Google Scholar]
- 61.Pimentel AC, Barroso IG, Ferreira JMJ, Dias RO, Ferreira C, Terra WR. Molecular machinery of starch digestion and glucose absorption along the midgut of Musca domestica. J Insect Physiol. 2018;109:11–20. doi: 10.1016/j.jinsphys.2018.05.009. [DOI] [PubMed] [Google Scholar]
- 62.Kaur R, Kaur N, Gupta AK. Structural features, substrate specificity, kinetic properties of insect α-amylase and specificity of plant α-amylase inhibitors. Pestic Biochem Physiol. 2014;116:83–93. doi: 10.1016/j.pestbp.2014.09.005. [DOI] [PubMed] [Google Scholar]
- 63.da Costa SG, Bates P, Dillon R, Genta FA. Characterization of α-Glucosidases from Lutzomyia longipalpis reveals independent hydrolysis Systems for Plant or blood sugars. Front Physiol. 2019;10:248. doi: 10.3389/fphys.2019.00248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Mehrabadi M, Bandani AR, Saadati F. Inhibition of Sunn pest, Eurygaster integriceps, α-amylases by α-amylase inhibitors (T-αAI) from Triticale. J Insect Sci. 2010;10:179. doi: 10.1673/031.010.14139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Memarizadeh N, Zamani P, Sajedi R, Ghadamyari M. Purification and characterization of Midgut α-Glucosidase from larvae of the Rice green Caterpillar, Naranga aenescens Moore. J Agric Sci Technol. 2014;16:1229–1240. [Google Scholar]
- 66.Han L, Ding G, Liu Y, Huang J, Wu J. Characterization of sphingomyelin phosphodiesterase expression in bumblebee (Bombus lantschouensis). J Insect Sci. 2018;18(5):20. 10.1093/jisesa/iey106. [DOI] [PMC free article] [PubMed]
- 67.Aguilar R, Jedlicka AE, Mintz M, Mahairaki V, Scott AL, Dimopoulos G. Global gene expression analysis of Anopheles gambiae responses to microbial challenge. Insect Biochem Mol Biol. 2005;35(7):709–719. doi: 10.1016/j.ibmb.2005.02.019. [DOI] [PubMed] [Google Scholar]
- 68.Xu X, Dong Y, Abraham EG, Kocan A, Srinivasan P, Ghosh AK, Sinden RE, Ribeiro JMC, Jacobs-Lorena M, Kafatos FC, Dimopoulos G. Transcriptome analysis of Anopheles stephensi-Plasmodium berghei interactions. Mol Biochem Parasitol. 2005;142(1):76–87. doi: 10.1016/j.molbiopara.2005.02.013. [DOI] [PubMed] [Google Scholar]
- 69.Lehane MJ. Peritrophic matrix structure and function. Annu Rev Entomol. 1997;42(1):525–550. doi: 10.1146/annurev.ento.42.1.525. [DOI] [PubMed] [Google Scholar]
- 70.Andrews NC. Forging a field: the golden age of iron biology. Blood. 2008;112(2):219–230. doi: 10.1182/blood-2007-12-077388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Geiser DL, Winzerling JJ. Insect transferrins: multifunctional proteins. Biochim Biophys Acta. 2012;1820(3):437–451. doi: 10.1016/j.bbagen.2011.07.011. [DOI] [PubMed] [Google Scholar]
- 72.Arosio P, Ingrassia R, Cavadini P. Ferritins: a family of molecules for iron storage, antioxidation and more. Biochim Biophys Acta. 2009;1790(7):589–599. doi: 10.1016/j.bbagen.2008.09.004. [DOI] [PubMed] [Google Scholar]
- 73.Lambert LA. Molecular evolution of the transferrin family and associated receptors. Biochim Biophys Acta. 2012;1820(3):244–255. doi: 10.1016/j.bbagen.2011.06.002. [DOI] [PubMed] [Google Scholar]
- 74.Zhou G, Velasquez LS, Geiser DL, Mayo JJ, Winzerling JJ. Differential regulation of transferrin 1 and 2 in Aedes aegypti. Insect Biochem Mol Biol. 2009;39(3):234–244. doi: 10.1016/j.ibmb.2008.12.004. [DOI] [PubMed] [Google Scholar]
- 75.Andrews NC. Molecular control of iron metabolism. Best Pract Res Clin Haematol. 2005;18(2):159–169. doi: 10.1016/j.beha.2004.10.004. [DOI] [PubMed] [Google Scholar]
- 76.Nichol H, Locke M. The localization of ferritin in insects. Tissue Cell. 1990;22(6):767–777. doi: 10.1016/0040-8166(90)90042-8. [DOI] [Google Scholar]
- 77.Guo D, Hao C, Cui X, Wang Y, Liu Z, Xu B, et al. Molecular and functional characterization of the novel odorant-binding protein gene AccOBP10 from Apis cerana cerana. J Biochem. 2020;169(2):215–25. 10.1093/jb/mvaa103. [DOI] [PubMed]
- 78.Pelosi P, Iovinella I, Zhu J, Wang G, Dani FR. Beyond chemoreception: diverse tasks of soluble olfactory proteins in insects. Biol Rev Camb Philos Soc. 2018;93(1):184–200. doi: 10.1111/brv.12339. [DOI] [PubMed] [Google Scholar]
- 79.Vogt RG, Riddiford LM. Pheromone binding and inactivation by moth antennae. Nature. 1981;293(5828):161–163. doi: 10.1038/293161a0. [DOI] [PubMed] [Google Scholar]
- 80.Zeng Y, Yang YT, Wu QJ, Wang SL, Xie W, Zhang YJ. Genome-wide analysis of odorant-binding proteins and chemosensory proteins in the sweet potato whitefly, Bemisia tabaci. Insect Sci. 2019;26(4):620–634. doi: 10.1111/1744-7917.12576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Gong DP, Zhang HJ, Zhao P, Xia QY, Xiang ZH. The odorant binding protein gene family from the genome of silkworm, Bombyx mori. BMC Genomics. 2009;10(1):332. doi: 10.1186/1471-2164-10-332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Robertson HM, Baits RL, Walden KKO, Wada-Katsumata A, Schal C. Enormous expansion of the chemosensory gene repertoire in the omnivorous German cockroach Blattella germanica. J Exp Zool B Mol Dev Evol. 2018;330(5):265–278. doi: 10.1002/jez.b.22797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Li ZQ, He P, Zhang YN, Dong SL. Molecular and functional characterization of three odorant-binding protein from Periplaneta americana. PLoS One. 2017;12(1):e0170072. doi: 10.1371/journal.pone.0170072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Cai LJ, Zheng LS, Huang YP, Xu W, You MS. Identification and characterization of odorant binding proteins in the diamondback moth, Plutella xylostella. Insect Sci. 2020. 10.1111/1744-7917.12817. [DOI] [PubMed]
- 85.Langeswaran K, Jeyaraman J, Mariadasse R, Soorangkattan S. Insights from the molecular modeling, docking analysis of illicit drugs and bomb compounds with honey bee odorant binding proteins (OBPs) Bioinformation. 2018;14(5):219–231. doi: 10.6026/97320630014219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Grolli S, Merli E, Conti V, Scaltriti E, Ramoni R. Odorant binding protein has the biochemical properties of a scavenger for 4-hydroxy-2-nonenal in mammalian nasal mucosa. FEBS J. 2006;273(22):5131–5142. doi: 10.1111/j.1742-4658.2006.05510.x. [DOI] [PubMed] [Google Scholar]
- 87.Uchida K. 4-Hydroxy-2-nonenal: a product and mediator of oxidative stress. Prog Lipid Res. 2003;42(4):318–343. doi: 10.1016/S0163-7827(03)00014-6. [DOI] [PubMed] [Google Scholar]
- 88.Morozova O, Marra MA. Applications of next-generation sequencing technologies in functional genomics. Genomics. 2008;92(5):255–264. doi: 10.1016/j.ygeno.2008.07.001. [DOI] [PubMed] [Google Scholar]
- 89.Wang H, Smagghe G, Meeus I. The role of a single gene encoding the single von Willebrand factor C-domain protein (SVC) in bumblebee immunity extends beyond antiviral defense. Insect Biochem Mol Biol. 2017;91:10–20. doi: 10.1016/j.ibmb.2017.10.002. [DOI] [PubMed] [Google Scholar]
- 90.Tanaka T, Kawano S, Nakao S, Umemiya-Shirafuji R, Rahman MM, Boldbaatar D, Battur B, Liao M, Fujisaki K. The identification and characterization of lysozyme from the hard tick Haemaphysalis longicornis. Ticks Tick-Borne Dis. 2010;1(4):178–185. doi: 10.1016/j.ttbdis.2010.09.001. [DOI] [PubMed] [Google Scholar]
- 91.Horáčková J, Rudenko N, Golovchenko M, Havlíková S, Grubhoffer L. IrML - a gene encoding a new member of the ML protein family from the hard tick, Ixodes ricinus. J Vector Ecol. 2010;35(2):410–418. doi: 10.1111/j.1948-7134.2010.00100.x. [DOI] [PubMed] [Google Scholar]
- 92.Yamakawa M, Tanaka H. Immune proteins and their gene expression in the silkworm, Bombyx mori. Dev Comp Immunol. 1999;23(4–5):281–289. doi: 10.1016/S0145-305X(99)00011-7. [DOI] [PubMed] [Google Scholar]
- 93.Bolognesi R, Arakane Y, Muthukrishnan S, Kramer KJ, Terra WR, Ferreira C. Sequences of cDNAs and expression of genes encoding chitin synthase and chitinase in the midgut of Spodoptera frugiperda. Insect Biochem Mol Biol. 2005;35(11):1249–1259. doi: 10.1016/j.ibmb.2005.06.006. [DOI] [PubMed] [Google Scholar]
- 94.Gaines PJ, Walmsley SJ, Wisnewski N. Cloning and characterization of five cDNAs encoding peritrophin-a domains from the cat flea, Ctenocephalides felis. Insect Biochem Mol Biol. 2003;33(11):1061–1073. doi: 10.1016/S0965-1748(03)00096-1. [DOI] [PubMed] [Google Scholar]
- 95.He N, Botelho JM, McNall RJ, Belozerov V, Dunn WA, Mize T, et al. Proteomic analysis of cast cuticles from Anopheles gambiae by tandem mass spectrometry. Insect Biochem Mol Biol. 2007;37(2):135–146. doi: 10.1016/j.ibmb.2006.10.011. [DOI] [PubMed] [Google Scholar]
- 96.Jasrapuria S, Arakane Y, Osman G, Kramer KJ, Beeman RW, Muthukrishnan S. Genes encoding proteins with peritrophin A-type chitin-binding domains in Tribolium castaneum are grouped into three distinct families based on phylogeny, expression and function. Insect Biochem Mol Biol. 2010;40(3):214–227. doi: 10.1016/j.ibmb.2010.01.011. [DOI] [PubMed] [Google Scholar]
- 97.Wang P, Li G, Granados RR. Identification of two new peritrophic membrane proteins from larval Trichoplusia ni: structural characteristics and their functions in the protease rich insect gut. Insect Biochem Mol Biol. 2004;34(3):215–227. doi: 10.1016/j.ibmb.2003.10.001. [DOI] [PubMed] [Google Scholar]
- 98.Schorderet S, Pearson RD, Vuocolo T, Eisemann C, Riding GA, Tellam RL. cDNA and deduced amino acid sequences of a peritrophic membrane glycoprotein, 'peritrophin-48′, from the larvae of Lucilia cuprina. Insect Biochem Mol Biol. 1998;28(2):99–111. doi: 10.1016/S0965-1748(97)00103-3. [DOI] [PubMed] [Google Scholar]
- 99.Elvin CM, Vuocolo T, Pearson RD, East IJ, Riding GA, Eisemann CH, Tellam RL. Characterization of a major peritrophic membrane protein, peritrophin-44, from the larvae of Lucilia cuprina. cDNA and deduced amino acid sequences. J Biol Chem. 1996;271(15):8925–8935. doi: 10.1074/jbc.271.15.8925. [DOI] [PubMed] [Google Scholar]
- 100.Zhu YC, Muthukrishnan S, Kramer KJ. cDNA sequences and mRNA levels of two hexamerin storage proteins PinSP1 and PinSP2 from the Indianmeal moth, Plodia interpunctella. Insect Biochem Mol Biol. 2002;32(5):525–536. doi: 10.1016/S0965-1748(01)00131-X. [DOI] [PubMed] [Google Scholar]
- 101.Tang B, Wang S, Zhang F. Two storage hexamerins from the beet armyworm Spodoptera exigua: cloning, characterization and the effect of gene silencing on survival. BMC Mol Biol. 2010;11(1):65. doi: 10.1186/1471-2199-11-65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Kim SR, Yoon HJ, Park NS, Lee SM, Moon JY, Seo SJ, Jin BR, Sohn HD. Molecular cloning, expression, and characterization of a cDNA encoding the arylphorin-like hexameric storage protein from the mulberry longicorn beetle, Apriona germari. Arch Insect Biochem Physiol. 2003;53(2):49–65. doi: 10.1002/arch.10085. [DOI] [PubMed] [Google Scholar]
- 103.Liu C, Zhu J, Ma J, Zhang J, Wang X, Zhang R. A novel hexamerin with an unexpected contribution to the prophenoloxidase activation system of the Chinese oak silkworm, Antheraea pernyi. Arch Insect Biochem Physiol. 2020;103(4):e21648. doi: 10.1002/arch.21648. [DOI] [PubMed] [Google Scholar]
- 104.Wang Y, Xiu JF, Cheng JZ, Man L, Peng Z, Shang XL, et al. Proteomic analysis of the peritrophic matrix from the midgut of third instar larvae, Musca domestica. Biomed Environ Sci. 2016;29(1):56–65. doi: 10.3967/bes2015.006. [DOI] [PubMed] [Google Scholar]
- 105.Whiten SR, Ray WK, Helm RF, Adelman ZN. Characterization of the adult Aedes aegypti early midgut peritrophic matrix proteome using LC-MS. PLoS One. 2018;13(3):e0194734. doi: 10.1371/journal.pone.0194734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Adang MJ, Spence KD. Surface morphology of peritrophic membrane formation in the cabbage looper, Trichoplusia ni. Cell Tissue Res. 1981;218(1):141–147. doi: 10.1007/BF00210100. [DOI] [PubMed] [Google Scholar]
- 107.Hopkins TL, Harper MS. Lepidopteran peritrophic membranes and effects of dietary wheat germ agglutinin on their formation and structure. Arch Insect Biochem Physiol. 2001;47(2):100–109. doi: 10.1002/arch.1040. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Table S1. all the identified protein from peritrophic matrix sample
Data Availability Statement
All raw LC-MS/MS data are available on the Mass Spectrometry Interactive Virtual Environment (MassIVE) data repository at ftp://massive.ucsd.edu/MSV000086950/
All data generated or analysed during this study are included in this published article and its supplementary information files or from the corresponding author on reasonable request.