Abstract
Background
Leaf color is an important trait in breeding of leafy vegetables. Y-05, a pakchoi (Brassica rapa ssp. chinensis) cultivar, displays yellow inner (YIN) and green outer leaves (GOU) after cold acclimation. However, the mechanism of this special phenotype remains elusive.
Results
We assumed that the yellow leaf phenotype of Y-05 maybe caused by low chlorophyll content. Pigments measurements and transmission electron microscopy (TEM) analysis showed that the yellow phenotype is closely related with decreased chlorophyll content and undeveloped thylakoids in chloroplast. Transcriptomes and metabolomes sequencing were next performed on YIN and GOU. The transcriptomes data showed that 4887 differentially expressed genes (DEGs) between the YIN and GOU leaves were mostly enriched in the chloroplast- and chlorophyll-related categories, indicating that the chlorophyll biosynthesis is mainly affected during cold acclimation. Together with metabolomes data, the inhibition of chlorophyll biosynthesis is contributed by blocked 5-aminolevulinic acid (ALA) synthesis in yellow inner leaves, which is further verified by complementary and inhibitory experiments of ALA. Furthermore, we found that the blocked ALA is closely associated with increased BrFLU expression, which is indirectly altered by cold acclimation. In BrFLU-silenced pakchoi Y-05, cold-acclimated leaves still showed green phenotype and higher chlorophyll content compared with control, meaning silencing of BrFLU can rescue the leaf yellowing induced by cold acclimation.
Conclusions
Our findings suggested that cold acclimation can indirectly promote the expression of BrFLU in inner leaves of Y-05 to block ALA synthesis, resulting in decreased chlorophyll content and leaf yellowing. This study revealed the underlying mechanisms of leaves color change in cold-acclimated Y-05.
Supplementary Information
The online version contains supplementary material available at 10.1186/s12870-021-02954-2.
Keywords: 5-aminolevulinic acid (ALA), BrFLU, Chlorophyll biosynthesis, Cold acclimation, Leaf color conversion, Pakchoi
Background
Leaf color is determined by pigments, which mainly includes chlorophyll, carotenoids, and anthocyanins [1, 2]. Plant leaf color change are always caused by a variety of factors and can be achieved in many ways [3], such as, chlorophyll biosynthesis and breakdown [4], chloroplast assembly [5], and signal transduction in plant disease resistance [6]. For many plant species, the leaf color is green, which is given by chlorophyll. Chlorophyll, the essential cofactors for photosynthesis [2], has been studied for many years [7, 8]. Chlorophyll enables the plants to use sunlight via photosynthesis. Maintaining chlorophyll normal state and level is essential for photosynthetic efficiency and carbon fixation [9], then directly influence plant growth and development [10, 11].
Numerous Chl-deficient species, also called leaf color mutations, which exhibited significant changes in chlorophyll synthesis or degradation mechanism and various phenotypes in different species, were regarded as suitable materials for exploring the mechanism of chlorophyll biosynthesis [12–15]. For instance, in rice, there were some missense mutations occurred in conserved amino acid of ChlD and ChlI in chl1 and chl9 mutants, respectively. chl1 and chl9 mutants showed poorly stacked grana and underdevelopment chloroplasts [16]. The other color mutant F03–06 in Arabidopsis, controlled by recessive mononuclear gene At5g54810, the gene-silenced plants exhibited similar phenotype with F03–06, yellow leaves with mottled veins, the plants are stunted and growth slowly [17]. A stably inherited Brassica plant etiolated mutation (pem) with DNA sequence variation in the promoter of Bra024218, showed retarded chloroplast development, decreased chlorophyll content and reduced photosynthetic capacity [18]. The chlorophyll (Chl)-deficient mutant pylm of pakchoi had yellow leaves with reduced total Chl content, loose grana lamellae structure and few thylakoid stacks, lower photosynthetic activity and photochemical conversion efficiency, which are caused by the block of Chl a production and down-regulation of genes related with Chl biosynthesis [19]. Mutants with loss-of-function REDUCED CHLOROPLAST COVERAGE (REC) protein, which is homologous with tetratricopeptide repeat (TPR) protein, showed lower chlorophyll contents and smaller chloroplast compartment size compared to wild type in Arabidopsis [20]. Pentatricopeptide repeat (PPR) protein, which motif is assumed to have evolved from a TPR proteins, is associated with various functions including temperature-sensitive chlorosis [21, 22]. Among these leaf color mutations, which exhibits normal or near-normal leaf color at room temperature, but exhibits significant change in leaf color at low temperature, are identified as low temperature-sensitive type [23]. Diverse analytical methods based on multiple omics databases were selected to comprehensively explore chlorophyll biosynthesis involved color conversion in low temperature-sensitive type, including tea (Camellia sinensis L.) [24], rice (Oryza sativa L.) [21], wucai (Brassica campestris L.) [9] at molecular or protein level. However, the mechanism of leaf color change response to low temperature is not totally understood.
Chlorophyll is synthesized in the chloroplast and distributed on the thylakoid membranes in chloroplast [25]. The general process of chlorophyll biosynthesis is composed of three main steps: (1) formation of 5-aminolevulinic acid (ALA) from glutamate, (2) formation of protoporphyrin IX from ALA, and (3) formation of chlorophyll from protoporphyrin IX [26]. ALA has been attracted attention as a key precursor that participated in biosynthesis of chlorophyll and plants greening [27, 28]. Exogenous application of ALA has been shown to increase the chlorophyll content of plants [29–31]. Previous reports indicated that ALA is formed from Glu in the C5 pathway consisting of two steps [32]. First, under the effects of glutamyl-tRNA reductase (GluTR), the Glu-tRNA converts to Glu-1-semialdehyde (GSA). Second, GSA converts to ALA by the effects of GSA-2,1-aminomutase (GSA-AM) [33]. GluTR and GSA-AM are encoded by the nuclear HEMA and GSA genes, respectively. Among the enzymes of ALA synthesis, GluTR is regarded as the main rate-limiting step [34]. Fluorescent in blue light (FLU), a tetratricopeptide repeat (TPR) protein, is the best-known negative regulator of ALA synthesis. Previous researches suggested that FLU protein can interact with the C-terminal segment of GluTR to inactivate ALA synthesis, resulting in reduced chlorophyll content and etiolated seedling [35, 36]. In FLU-overexpression (FLUOE) Arabidopsis, FLUOE lines show pale leaves under medium light and yellow-green leaves under low light owning to decreased chlorophyll contents [37]. In addition, another factor regulating GluTR is GluTR-binding protein (GBP). GBP has been shown to interact with the N-terminal region of GluTR to protect GluTR from degradation, maintaining adequate ALA synthesis [34].
Pakchoi (Brassica rapa ssp. chinensis), a subspecies of Chinese cabbage, is a widely consumed vegetable in Asia, especially in China. Since the main edible organ of pakchoi is leaf, leaf color is an important trait in breeding. Y-05 is a special pakchoi cultivar which displays green leaves grown under normal condition but displays yellow inner (YIN) and green outer (GOU) leaves after cold acclimation. Due to its special phenotype, Y-05 is considered as an interesting and valuable material for cold-acclimated leaf color change research. To date, there has been little research on the leaf color change of Y-05 response to cold acclimation, and its color conversion mechanism remains elusive. Here, we aimed to characterize the changed leaf color of pakchoi Y-05 at the physiological, cellular and molecular levels. Our findings contribute to the understanding of cold acclimated Y-05 displaying yellow inner and green outer leaves. These findings also enrich our understanding of the mechanism of leaf color change.
Results
The inner leaves of cold-acclimated Y-05 exhibit decreased chlorophyll content and undeveloped thylakoids
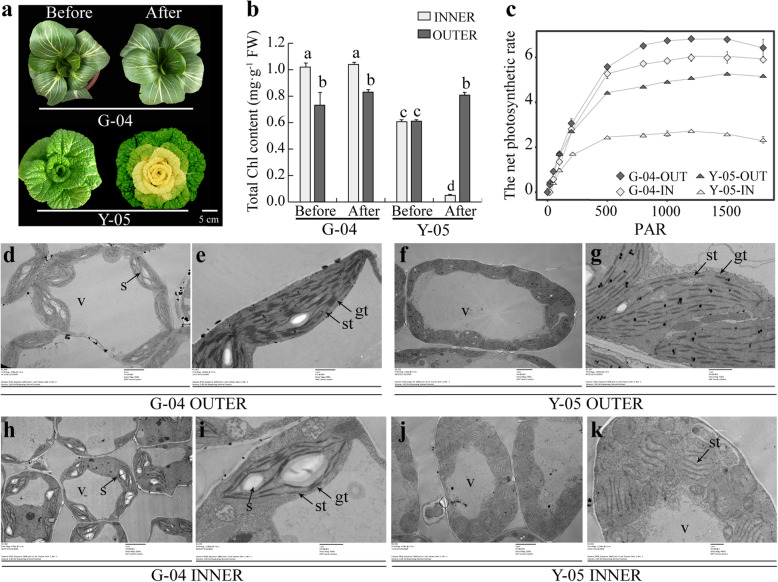
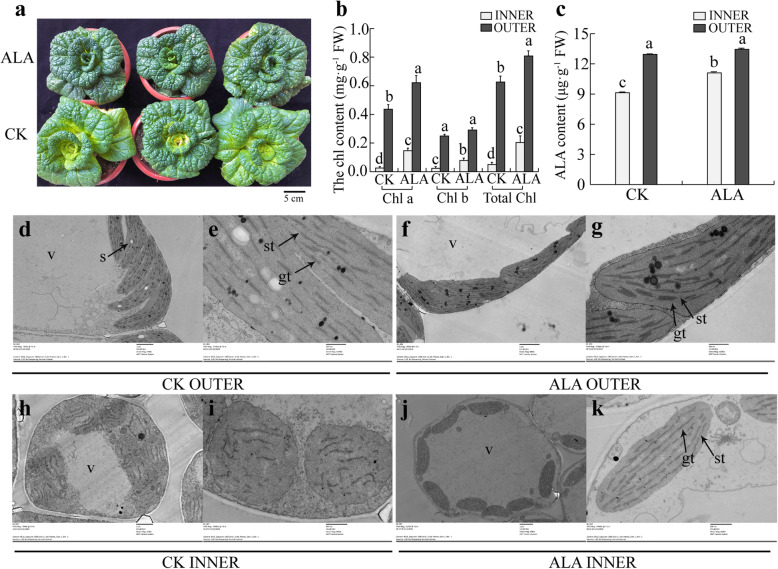
Compared with green leaves of pakchoi cultivar G-04, Y-05 displays yellow inner and green outer leaves after cold acclimation (Fig. 1a). Hence, we took the pigments measurement between G-04 and Y-05 at different growth conditions and periods. Interestingly, all of them show decreased contents in yellow inner leaves compared with leaves of G-04 and green outer leaves of Y-05 (Fig. 1b, Fig. S1). Generally, the content of carotenoids, xanthophylls and anthocyanin increased rather than decreased, which could cause yellow leaf phenotype [1, 2]. So, we proposed that the yellow leaf phenotype of Y-05 maybe caused by low chlorophyll content.
Fig. 1.
The inner leaves of cold-acclimated Y-05 exhibit decreased chlorophyll content and undeveloped thylakoids. a The phenotype of G-04 and Y-05 before and after cold acclimation. For cold acclimation, two-month old Y-05 plants were grown 3 weeks at 4 °C, and then return to 23 °C for continues grown. Before, before cold acclimation. After, after cold acclimation. Bar = 5 cm. b The total chlorophyll content of outer and inner leaves from Y-05 and G-04 before and after cold acclimation. c The net photosynthetic rate (Pn) of outer and inner leaves from Y-05 and G-04 after cold acclimation. PAR, photosynthetic active radiation. Three individual plants of each cultivar were quantified, and the total chlorophyll content and Pn were measured three times. Error bars represent SE (±SE, n = 3). Different letters indicated statistically significant differences at the level of p < 0.05. d-k Chloroplast ultrastructure of outer and inner leaves from G-04 and Y-05 after cold acclimation. v = vacuole, s = starch grains, gt = granum thylakoids, st = stroma thylakoids. In the Fig. 1d, f, h, j, Bar = 4 μm. In the Fig. 1e, g, i, k, Bar = 1 μm
Chlorophyll is the main pigment for photosynthesis in plants and located in the thylakoid membrane of chloroplast [38]. To verify our hypothesis, we checked the photosynthetic capacity (Pn) of outer and inner leaves between cold-acclimated Y-05 and G-04. As our expected, both outer and inner leaves of cold-acclimated Y-05 possess significantly decreased net photosynthetic rate compared with the outer and inner leaves of cold-acclimated G-04 respectively (Fig. 1c).
Next, we further studied the chloroplast ultrastructure of inner and outer leaves from cold-acclimated G-04 and Y-05 by transmission electron microscopy (TEM). Observation of the cold-acclimated G-04 showed lots of mature chloroplasts and thick granum-thylakoids in outer leaves (Fig. 1d, e), and developing chloroplasts and thin granum-thylakoids in inner leaves (Fig. 1h, i). In cold-acclimated Y-05, the green outer leaves show mature chloroplasts but thinner grana stacks compared with outer leaves of G-04 (Fig. 1f, g). However, the yellow inner leaves of Y-05 display undeveloped chloroplasts and almost disappeared granum-thylakoids (Fig. 1j, k). Together, we suggested that the yellow inner leaves of Y-05 are caused by decreased chlorophyll content and undeveloped thylakoids which maybe induced by cold acclimation.
Transcriptomes and metabolomes of cold-acclimated Y-05
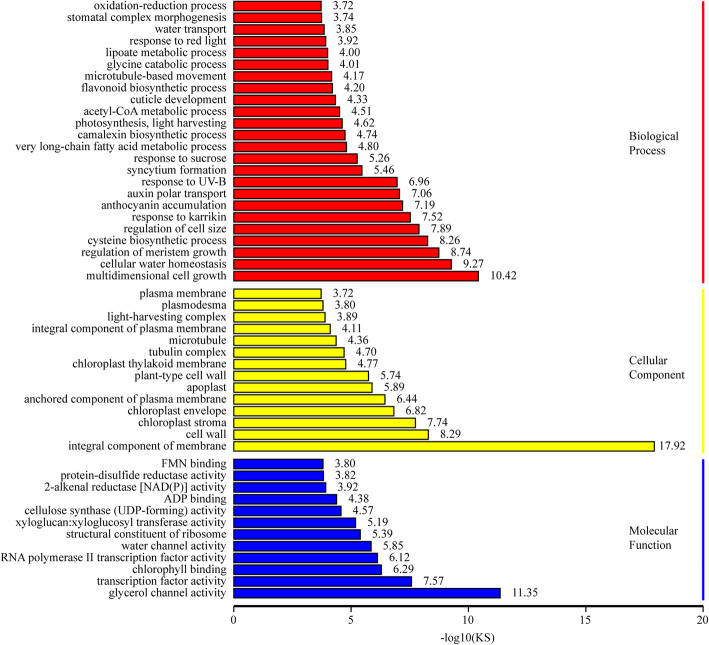
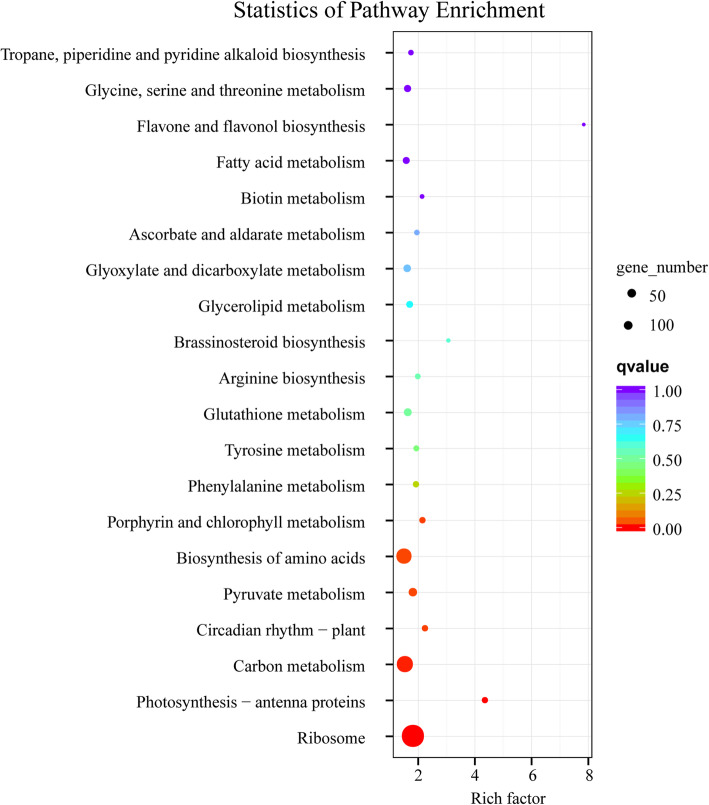
To explore the molecular mechanisms of leaf color change in Y-05 induced by cold acclimation, the transcriptomes of green outer leaves (TOU) and yellow inner leaves (TIN) were conducted (Table S1–4). For both the TOU and TIN samples, three independent biological replicates were set up. The high correlation coefficient indicates a strong linear relationship between biological duplications (Fig. S2a). Totally, 4887 differentially expressed genes (DEGs) were identified between the YIN and GOU, 2239 genes were down-regulated and 2648 genes were up-regulated in the YIN compared with GOU (Fig. S2b, Table S2). Gene Ontology (GO) analysis revealed that the integral component of membrance and chloroplast-related categories were overrepresented in cellular component (Fig. 2, Table S3). Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis revealed significant changes in the ‘Photosynthesis-antenna proteins’(ko00196), ‘circadian rhythm-plant’(ko04712) and ‘Porphyrin and chlorophyll metabolism’(ko00860) pathway with rich factor > 2 (Fig. 3, Table S4).
Fig. 2.
The enriched GO pathway of the differentially expressed genes (DEGs) between TOU and TIN of pak choi Y-05. -log10 (KS) represents the statistical significance of GO term. The bigger of -log10 (KS), the more significant enriched
Fig. 3.
The top 20 enriched KEGG pathway of the differentially expressed genes (DEGs) between TOU and TIN of pak choi Y-05. Each point represents a KEGG pathway, ordinate represents pathway name, and abscissa represents the enrichment factor. The larger the enrichment factors, the more significant the enrichment level of DEGs are showed. The color of the circle represents q-value, lower q-value means the more reliable results. And the size of the circle represents the number of genes enriched in the pathway
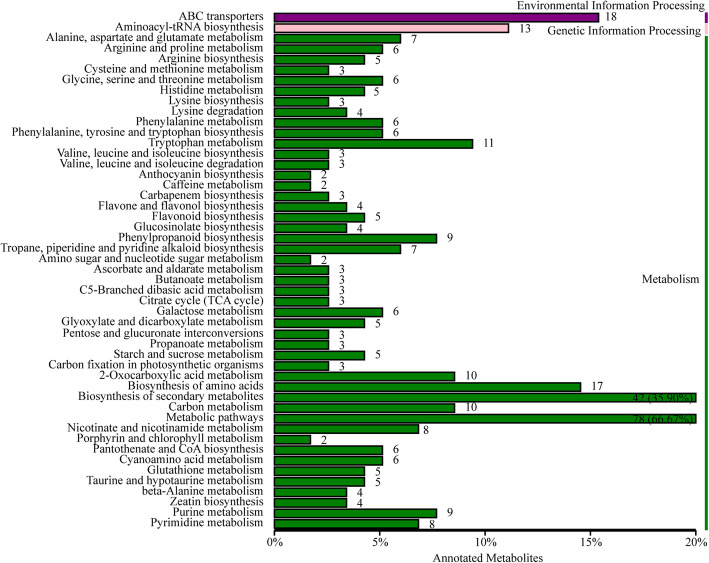
For exploring the differences in the composition of metabolites in inner-yellow (MIN) and outer-green (MOU) leaves of Y-05, the non-targeted metabolomes analysis was performed. Repeatability and correlation analysis between MOU and MIN was assessed to prove data reliability (Fig. S3a). In total, 372 differentially expressed metabolites (DEMs) were identified. One hundred sixty-two metabolites were up-accumulated, and 210 metabolites were down-accumulated in MIN (Fig. S3b, Table S5). Next, we analyzed the DEMs between MIN and MOU. KEGG pathway analysis revealed that most metabolites are mainly enriched in the ‘metabolic pathways’ and ‘biosynthesis of secondary metabolites’ pathway (Fig. 4). Interestingly, the ‘Porphyrin and chlorophyll metabolism’ pathway was also found (Fig. 4), which consistent with our previous results that chlorophyll content changes in Y-05. From the metabolomes data, we found two metabolites involved in chlorophyll biosynthesis, 5-Aminolevulinate (ALA, meta_22) and L-Glutamic acid (meta_277), were down-regulated and up-regulated in MIN compared with MOU respectively (Table S6), meaning that the conversion from L-Glutamic acid to ALA is blocked in inner leaves. Together, we proposed that the decreased chlorophyll content in inner leaves of Y-05 is associated with the inhibition of ALA synthesis.
Fig. 4.
The enriched KEGG pathway of the differentially expressed metabolites (DEMs) between inner-yellow (MIN) and outer-green (MOU) leaves of pakchoi Y-05. Short time-series expression miner (STEM) was used to analyze the metabolites expression pattern. The number of DEMs in each profile was labeled above the frame. The bar represents the proportion metabolites in each profile of the total annotated metabolites
ALA content is critical for the leaf color conversion of pakchoi Y-05
Based on the metabolomes data (Table S5 and S6), the inhibition of ALA synthesis in inner leaves of cold-acclimated Y-05 may result in decreased chlorophyll content and leaf yellowing. To verify our hypothesis, 1 mM ALA was immediately sprayed on leaves of Y-05 after cold acclimation when both outer and inner leaves are still green. The cold-acclimated Y-05 plants sprayed by water were used as control. As our expected, the leaf yellowing is inhibited in ALA-treated inner leaves, while control plants display yellow inner leaves (Fig. 5a). Consistent with the phenotype, the chlorophyll and ALA content showed significantly increased in ALA-treated Y-05 (Fig. 5b, c). Further, we observed the ultrastructure of chloroplasts in ALA-treated and control leaves. The chloroplasts in outer leaves of ALA-treated plants possess thicker granum-thylakoids compared with control plants (Fig. 5d-g). Meanwhile, compared with the undeveloped chloroplasts in inner leaves of control, the inner leaves of ALA-treated plants possess mature chloroplasts and granum-thylakoids (Fig. 5h-k). Together, these results suggested that exogenous application of ALA can rescue the yellow phenotype of Y-05 induced by cold acclimation.
Fig. 5.
Exogenous application of ALA can rescue the yellow leaves phenotype in cold-acclimated Y-05. a The leaf phenotype of ALA-treated plants compared with CK plants. ALA, cold-acclimated Y-05 sprayed by 1 mM ALA. CK, cold-acclimated Y-05 sprayed by water as control. Bar = 5 cm. b The chlorophyll content of ALA-treated plants was higher than CK plants. Chlorophyll a, Chlorophyll b and total chlorophyll abbreviated as Chl a, Chl b, Total Chl, respectively. c The ALA content of ALA-treatment plants was higher than CK plants. Three individual plants of each cultivar were quantified, and the chlorophyll and ALA content were measured three times. Error bars represent SE (±SE, n = 3). Different letters indicated statistically significant differences at the level of p < 0.05. d-k Observation of chloroplast ultrastructure showed that ALA-treated plants possessed mature chloroplasts and granum thylakoids. v = vacuole, gt = granum thylakoids, st = stroma thylakoids. In the Fig. 4d, f, h, j, bar = 2 μm; in the Fig. 4e, g, bar = 500 nm; in the Fig. 4i, k, bar = 800 nm
Next, to further investigate the role of ALA in leaf color conversion of Y-05, gabaculine (3-amino 2,3-dihydrobenzoic acid), an inhibitor of ALA biosynthesis by inactivating GSA-AT activity [39, 40], was used to irrigate one-month-old Y-05 seedlings. The Y-05 seedlings watered by water were used as control. After treatment, the inner leaves of gabaculine-treated Y-05 seedlings display yellow phenotype without cold acclimation, while the control plants still show green inner leaves (Fig. 6a). Consistent with the phenotypes, the chlorophyll and ALA content in inner leaves decreased in gabaculine-treated plants compared with control plants (Fig. 6b, c). These findings further verified that the inhibition of ALA biosynthesis is the key in leaf yellowing of Y-05. Together with the above results, we suggested that the content of ALA is critical for the leaf color conversion of Y-05.
Fig. 6.
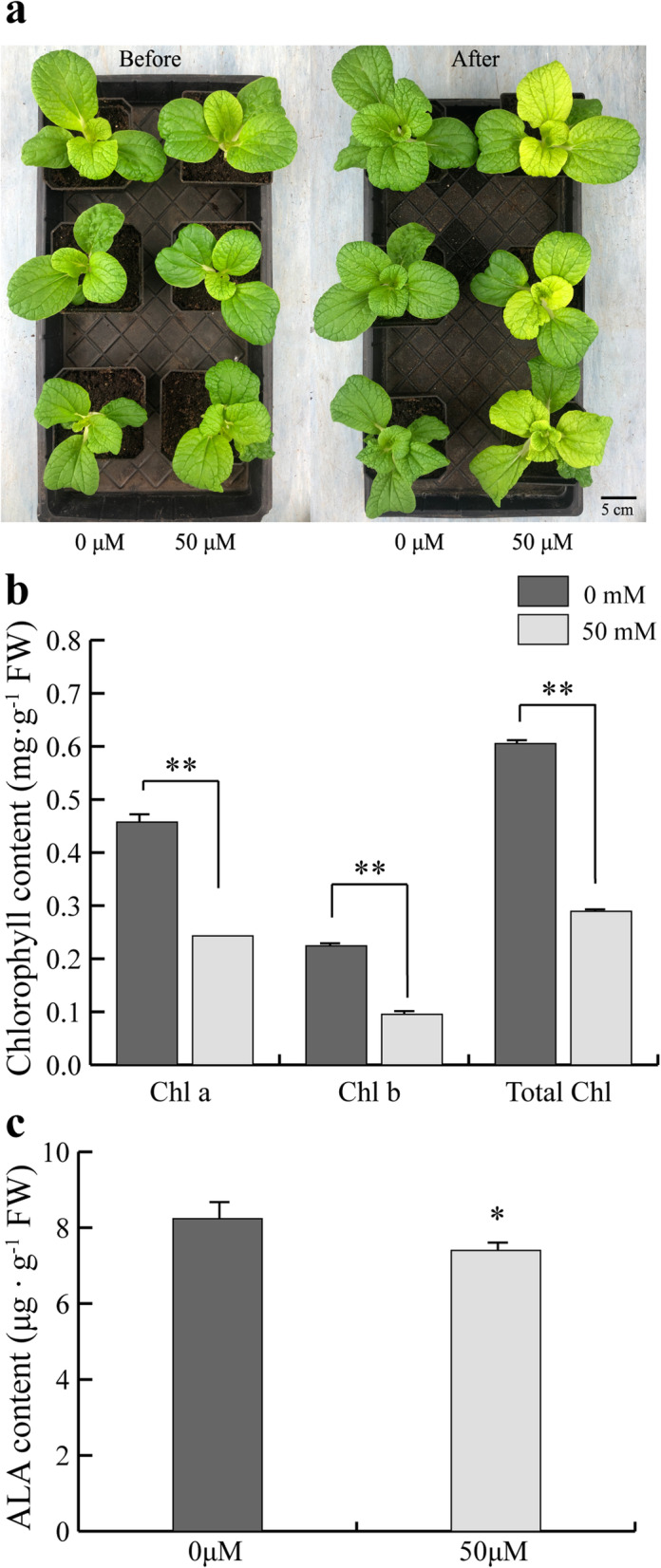
Gabaculine-treated Y-05 seedlings show yellow leaves phenotype without cold acclimation. a The gabaculine-treated plants showed yellow phenotype without cold-acclimation. Before, plants before treated with gabaculine. After, plants after treated with gabaculine. 0 μM and 50 μM represented CK plants and gabaculine-treated plants, respectively. Bar = 5 cm. b The inner leaves of gabaculine-treated plants showed low chlorophyll content compared with CK plants. c The inner leaves of gabaculine-treated plants showed low ALA content compared with CK plants. Three individual plants of each cultivar were quantified, and the chlorophyll and ALA content were measured three times. Error bars represent SE (±SE, n = 3). * represents p < 0.05, ** represents p < 0.01
The upregulation of BrFLU is critical for ALA inhibition in cold-acclimated Y-05
Above data suggested that ALA biosynthesis is blocked in yellow inner leaves of cold-acclimated Y-05. During chlorophyll synthesis process, the conversion of L-Glutamic acid to ALA is the rate-limiting step which is controlled by three positive regulators, GBP, HEMA and GSA, and one negative regulator FLU [34, 41]. To deeply study the molecular mechanism of ALA inhibition in YIN of cold-acclimated Y-05, we firstly studied the homologous genes expressions of GBP, HEMA and GSA between YIN and GOU leaves. The transcription data showed that the expression of BrHEMA1 and BrGSA1 are significantly increased in YIN compared with GOU leaves (Fig. S4, Table S7). Since HEMA and GSA are positive regulators of ALA biosynthesis, the ALA inhibition in YIN leaves should be not associated with increased BrHEMA1 and BrGSA1 expression. As for BrGBP, which showed no significant changes in expression level between YIN and GOU, also should not be the reason for blocked ALA (Fig. S4, Table S7). Previous studies indicated that FLU, a negative regulator of ALA biosynthesis, interacts with the C-terminal of GluTR to inactivate ALA synthesis [37, 42]. Interestingly, BraA05003715 (BrFLU), the homologous gene of Arabidopsis FLU in pakchoi, shows 7.4-fold high expression in YIN compared with GOU (Fig. S4, Table S7), which is consistent with decreased ALA content. Hence, BrFLU was selected as candidate gene of ALA inhibition in yellow inner leaves for next research.
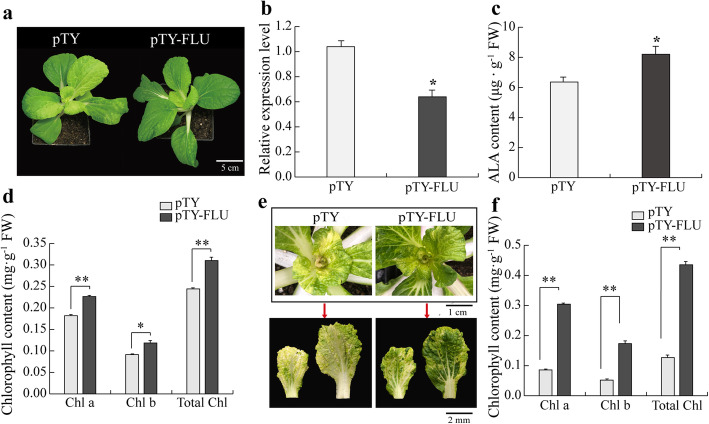
To study the role of BrFLU in leaf color conversion of Y-05, the BrFLU-silenced line (pTY-FLU) of Y-05 was conducted (Fig. 7a, b). The Y-05 plants injected with pTY empty vector were used as control (pTY). Compared with control plants, the silenced plants showed more chlorophyll and ALA content (Fig. 7c, d), suggesting that low BrFLU level makes positive contribution to ALA and chlorophyll biosynthesis in Y-05. Further, if the BrFLU upregulation induced by cold acclimation is closely related with ALA inhibition and decreased chlorophyll content in Y-05 inner leaves, silencing of BrFLU in cold-acclimated Y-05 should increase chlorophyll content and rescue the yellow leaves phenotype, at least in part. To verify our hypothesis, pTY and pTY-FLU lines were treated with 0 °C for 3 weeks. Excitingly, pTY-FLU plants showed green phenotype and higher chlorophyll content compared with control plants (Fig. 7e, f), suggesting that the BrFLU upregulation is closely related with leaf color conversion of Y-05. Together, these data suggested that the upregulation of BrFLU induced by cold acclimation is critical for ALA inhibition, finally resulting in decreased chlorophyll content and yellow inner leaves of Y-05.
Fig. 7.
Silencing of BrFLU in Y-05 is critical for ALA biosynthesis. a The phenotype of pTY plants (CK) and pTY-FLU (BrFLU-silenced) plants. Bar = 5 cm. b The relative expression level of BrFLU significantly decreased in pTY-FLU plants. The inner leaves were selected to confirm the BrFLU expression. c The pTY-FLU plants showed higher ALA content than pTY plants. d The pTY-FLU plants showed higher chlorophyll content than pTY plants. e The phenotype of pTY and pTY-FLU plants after cold acclimation. Bar = 1 cm or 2 mm. f The pTY-FLU plants showed higher chlorophyll content than pTY plants after cold acclimation. Chlorophyll a, Chlorophyll b and total chlorophyll abbreviated as Chl a, Chl b, Total Chl, respectively. Three individual plants of each cultivar were quantified, and the chlorophyll content was measured three times. Error bars represent SE (±SE, n = 3). * represents p < 0.05, ** represents p < 0.01
Furthermore, to explore why BrFLU is specially upregulated in cold-acclimated Y-05, we first compared the BrFLU open reading frame (ORF) sequences between Y-05 and other stay-green pakchoi varieties (G-04, WTC, 2Q, LY, MET, SZQ). Although many single nucleotide polymorphisms in BrFLU were detected between Y-05 and other pakchoi varieties (Fig. S5), the BrFLU amino acid sequences were found no significant different between Y-05 and other varieties (Fig. S6). Since motif appearing or missing in promoter will affect gene expression level, we then analyzed the promoters of BrFLU in Y-05 and other pakchoi varieties. Unfortunately, we did not found any motif specific appearing or missing in promoter of BrFLU from Y-05 (Fig. S7). Taken together, we proposed that the specific upregulation of BrFLU in Y-05 is induced by its upstream regulator which responds to cold acclimation.
Discussion
Leaf color is an important trait in vegetable breeding and marketing. However, previous studies on the mechanism of leaf color conversion were mainly focus on trees and ornamental plants [43, 44]. Although some researches on leaf color conversion were carried out in crops such as rice, maize and wheat [21, 24], there are few studies on vegetables. Here, we studied the mechanism of leaf color conversion in pakchoi. To explore the color conversion mechanism of special phenotype of Y-05 (Fig. 1a), we first measured the pigment contents between yellow and green leaves, including chlorophyll, carotenoids, xanthophyll and anthocyanin. Results showed all pigments decreased in the yellow inner leaves of Y-05 (Fig. S1). Previously, many studies found that leaf color conversion is usually accompanied by senescence [45]. For instance, the chlorophyll was continuously degraded whereas the carotenoids was partial retained during the process of leaf senescence, was thought to be the reason of leaf color change in Ginkgo biloba [46]. However, the leaf color conversion in pakchoi Y-05 only happens in young inner leaves (Fig. 1a), meaning that the conversion is not related with leaf senescence, but response to cold acclimation. Similarly, researchers believed that leaf color mutation is usually accompanied with blocked growth, then causing the economic losses of plants [47]. In our study, the chlorophyll content and Pn value in Y-05 leaves is lower than that in G-04 leaves (Fig. 1b, c), indicating that the development of chloroplasts in the yellow inner leaves of Y-05 is suppressed by cold acclimation. However, Y-05 showed well-developed phenotype (Fig. 1a). Therefore, we suggested that the outer green leaves of Y-05 may supply enough photosynthate for plants to grown. The above findings revealed that the change of inner leaf color is caused by cold acclimation, and wouldn’t affect plant development. However, why the yellow phenotype only happens in young inner leaves needs to be further investigated. One possible is that cold acclimation only plays a role in the early stage of thylakoids development in Y-05. The outer leaves, which have mature chloroplast and developed thylakoids, does not respond to cold acclimation.
Generally, low chlorophyll content is caused by inhibition of chlorophyll synthesis or chlorophyll degradation [4]. Chlorophyll synthesis includes 19 steps from the GluTR to Chl b, a total of 16 enzymes encoded by more than 26 genes are working in this process [48, 49]. For example, ‘White Dove’ is a leaf color mutant in kale (Brassica oleracea). Low temperature induced the low expression of chlorophyll biosynthesis gene POR, lead to chlorophyll content dramatically reduced in ‘White Dove’ [50]. PORA, encoding protochlorophyllide oxidoreductase, play an important role in chlorophyll biosynthesis. Arabidopsis porA-1 seedlings suffer from a drastically reduced chlorophyll content and dwarf phenotype [51]. OsCAO1 mainly controls the synthesis of chl b, its mutation will cause decreased chl b content, resulting in yellow-green leaf color [52]. Meanwhile, there is a dynamic balance between the biosynthesis and catabolism of chlorophyll in plants. The high expression of CHL2 and RCCR genes accelerate the chlorophyll degradation, resulting in leaves yellowing in Cymbidium sinense [53]. Mutations in the degradation pathway of chlorophyll often lead to the phenomenon of green stagnation in plant leaves [54, 55]. Further, chlorophyll degradation in many plants is usually accompanied by senescence or injury [47]. For example, many plant show colorful leaves in autumn, which is related to chlorophyll degradation [56]. Moreover, some stresses including biotic and abiotic stress will trigger cell death and chlorophyll degradation [57]. In our study, the yellow inner leaves of Y-05 are caused by decreased chlorophyll content and undeveloped thylakoids (Fig. 1b, j, k). More important, the yellow leaves phenotype only happened in inner leaves of Y-05 but not in outer leaves. If the yellow leaves phenotype is caused by chlorophyll degradation, both outer and inner leaves should show yellow phenotype after cold acclimation. Hence, we suggested that the low chlorophyll content in yellow inner leaves of Y-05 is caused by the inhibition of chlorophyll synthesis, but not chlorophyll degradation. The idea was also supported by the impaired thylakoids membrane (Fig. 1j, k), since thylakoids membrane is the site of chlorophyll synthesis [58].
To further investigate the mechanism of leaf color conversion in pakchoi Y-05, the transcriptomes of green outer leaves and yellow inner leaves were performed (Figs. 2, 3 and 4, Table S1–5). Both GO and KEGG analyses further confirm the low chlorophyll content (Fig. 1b), weak photosynthetic capacity (Fig. 1c) and impaired chloroplast structure (Fig. 1j, k) in inner leaves of Y-05. Metabolites serve as a bridge between genotype and phenotype. Because of metabolites are closest to phenotypes, and their changes more directly reveal gene functions [59]. Based metabolome data, we found that the low chlorophyll content in yellow inner leaves is closely associated with the block of ALA synthesis (Table S6). Complementary and inhibitory experiments of ALA (Figs. 5 and 6) further support the finding. Moreover, the transcriptomes data revealed that the high expression of BrFLU plays a key role in the block of ALA synthesis (Fig. S4 and Table S7). FLU protein has conserved TPR motifs at its C-terminus, which could interact with GluTR [35, 60], thus influences the rate of synthesis of ALA [41]. In previous studies on Arabidopsis, FLU-overexpressing lines show decreased ALA synthesis and reduced chlorophyll content in the light [60]. In our study, silencing of BrFLU in Y-05 pak choi further confirmed the contribution of BrFLU to the impaired ALA synthesis and decreased chlorophyll content (Fig. 7). Taken together, our results suggested that the impaired ALA synthesis is closely related with enhanced BrFLU expression, resulting in decreased chlorophyll content and leaf yellowing in Y-05. However, we did not found the significant different in the ORF and promoter sequences of BrFLU from Y-05 compared with other varieties (Fig. S5–6). Therefore, the reason of the specific upregulation of BrFLU in cold-acclimated Y-05 is not clear. In other words, the upregulation of BrFLU in Y-05 is regulated by an unknown regulator which responds to cold acclimation (Fig. 8). For the unknown regulator, we suggested three possibilities. One is a BrFLU positive transcription factor which can be induced by cold acclimation, activating BrFLU expression. Second is a BrFLU negative transcription factor which can be depressed by cold acclimation, reducing the inhibition on BrFLU expression. The last possibility is epigenetic regulation such as cold acclimation which may decrease BrFLU methylation level, resulting in enhanced BrFLU expression.
Fig. 8.
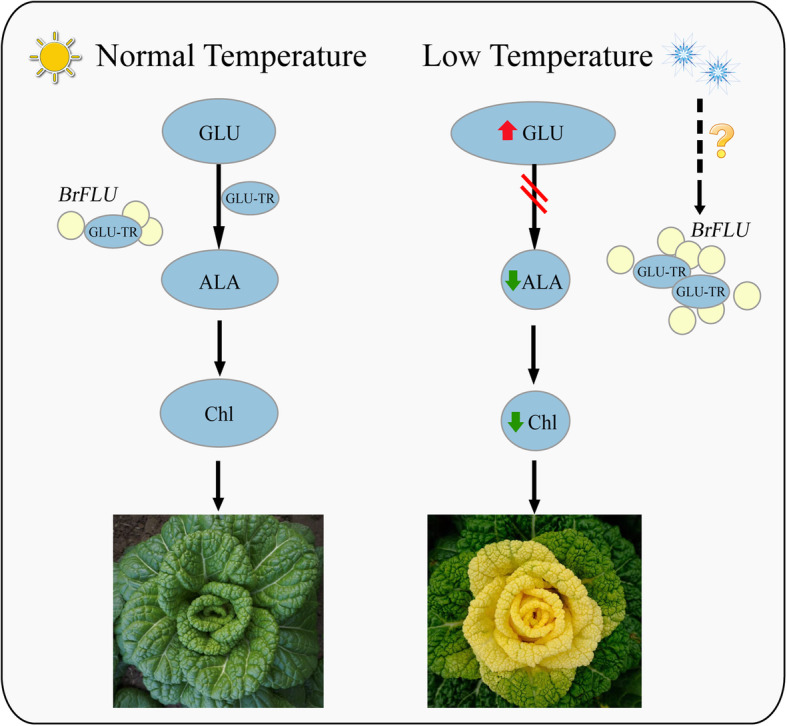
The proposed model of chlorophyll biosynthesis in Y-05 under normal or low temperature. At normal temperature, the expression of BrFLU remains stable, and GLU converts to ALA to maintain normal biosynthesis of chlorophyll. Under low temperature, cold acclimation can trigger an unknown regulator, inducing BrFLU upregulation and the interaction between BrFLU and GLU-TR to block ALA synthesis, resulting in decreased chlorophyll content and leaf yellowing in Y-05. L-Glutamic acid, glutamyl-tRNA reductase, 5-Amino-levulinate and total chlorophyll were abbreviated as Glu, GluTR, ALA and Chl, respectively. Red and green arrow represents up-regulated and down-regulated of compounds, respectively
Conclusions
Y-05 is a special pakchoi cultivar which shows green leaves grown under room temperature but displays yellow inner and green outer leaves after cold acclimation. In cold-acclimated Y-05 pak choi, compared with the green outer leaves, the yellow inner leaves exhibited low chlorophyll content and weak photosynthetic capacity, undeveloped chloroplasts and thylakoids. Through comprehensive analysis of transcriptome and metabonomic sequencing and functional verification, we found that cold acclimation can trigger an unknown regulator, inducing BrFLU upregulation to block ALA synthesis, resulting in decreased chlorophyll content and leaf yellowing (Fig. 8). However, the unknown regulator needs to be further explored. Finally, our findings provide insight into the mechanisms underlying the leaf color change response to cold acclimation.
Methods
Plant materials and growth conditions
All pakchoi inbred lines (Y-05, G-04, WTC, 2Q, LY, MET, SZQ) were grown in pots containing a soil: sand mixture (3: 1) in a controlled artificial climatic chamber with long-day condition (16 h light / 8 h dark) at 23 °C, 70% humidity and 250 μmol·m− 2·s− 1 light. For cold acclimation, two-month-old plants were grown 3 weeks at 4 °C, and then return to 23 °C for continues grown. After 2 weeks grown at 23 °C, cold acclimated Y-05 will exhibit green-outer leaves (GOU) and yellow-inner leaves (YIN). The GOU and YIN leaves were used for next studies. Under the same growing conditions, G-04, which showed stay-green phenotype before and after cold acclimation, was used as the control group for pigments determination and morphological observation experiments. All of the plant materials used in reseach were from State Key Laboratory of Crop Genetics and Germplasm Enhancement of Nanjing Agricultural University.
Measurement of pigments and ALA content
The plant pigments, including chlorophyll, carotenoids contents were measured as described in previous studies [9, 61, 62] with simple modifications. In brief, 0.1 g fresh leaf was soaked in 15 ml extracting solution and then shook with 50 rpm/min for 24 h under dark condition. The absorbance was measured by an UV − vis spectrophotometer (CYTATION3, BioTek, USA) at wavelengths of 665, 649, 642, 485 and 470 nm.
The anthocyanin was measured by method of Huo [63]. In brief, 0.1 g fresh leaf was soaked in 1 ml acidified ethanol (80% ethanol with 0.1% hydrochloric acid) and stood for 24 h under 4 °C dark condition. The absorbance was measured at wavelengths of 536 nm. All the concentrations of pigments were calculated as (mg ∙ g− 1). The ALA concentrations (μg ∙ g− 1) were measured using an enzyme-linked immunosorbent assay (ELISA) kit (Cat No: KT7958-B, Jiangsu Kete Biotechnology Co., Ltd., China).
Transmission electron microscopy (TEM) analysis
Manufacturing methods of ultra-thin slice refer method of Maekawa [64]. In brief, Use a double-sided blade to cut 2 × 2 mm leaves from pakchoi plants and fixed them in 1% (w/v) glutaraldehyde, after washed several times with phosphate buffer, the samples were further fixed in 0.5% (w/v) osmium tetroxide. After infiltrated with resin and cut using an ultramicrotome (EM UC6, Leica Microsystems), the ultra-thin sections were obtained. For transmission electron microscopy, the ultra-thin slice was examined and photographed using a Hitachi (Tokyo, Japan) H-7650 TEM, as previously described [65].
Transcriptome analysis
The third fully expanded leaves from the yellow part and green part from center were sampled 2 weeks after cold acclimation, the same part of three independent plants as biological replicates. After cleaned and cut, the leaf tissues were frozen in liquid nitrogen immediately, then stored in − 80 °C for further research. Total RNA was extracted from YIN and GOU of Y-05 using a TRIzaol reagent (Thermo Fisher Scientific Inc.). The library construction and RNA-seq were performed by Biomarker Technology Co. (Beijing, China). Illumina HiSeq 2500 platform (NEB, USA) was used for library preparations sequencing. After filtering (low Q-value <= 20%), cleaned reads were then assembled and mapped to the Brassica rapa genome (V2.5) (http://brassicadb.org/brad/index.php) using software HISAT2 (version 2.1.0) [66]. Each differentially expressed gene (DEG) function was annotated to these public databases: Nr (NCBI non-redundant protein sequences); KOG/COG (Clusters of Orthologous Groups of proteins); Pfam (Protein family), SwissProt, KEGG (Kyoto Encyclopedia of Genes and Genomes) [67] and GO (Gene Ontology). Differential expression analysis was performed using software DESeq2 [68] based on the expression levels of the genes in different samples. An absolute value of log2 (fold change) ≥2, P-value < 0.05 [69], as the criteria for identifying significantly differential expression.
Untargeted metabolome analysis
The samples for untargeted metabolome analysis are same as transcriptome analysis. But to distinguish them from transcriptomes in data analysis, the metabolites in yellow-inner and green-outer leaves were named MIN and MOU respectively. Metabolite identification and quantification were performed by Biomarker Technology Co. (Beijing, China). In brief, metabolite identification was annotated against following public database: HMDB (http://www.hmdb.ca/), KEGG (Kyoto Encyclopedia of Genes and Genomes), following the standard metabolic operating procedures. Multiple-reaction monitoring (MRM) was used for metabolite quantification. Targeted UPLC-ESI-QTOF/MS profiling and multivariate data analysis (PCA and Opls-da) [70] were used to obtain more reliable information about metabolites. For all differential metabolites which has three biologically repeats, an absolute value of log2 (fold change) ≥ 1 and variable importance in projection (VIP) ≥ 1, P-value < 0.05 [69] were the criteria for identifying significantly differential metabolites.
Treatment of ALA and gabaculine
For ALA treatment, once the cold acclimated Y-05 plants were transferred to 23 °C chamber, the leaves were immediately sprayed by 1 mM ALA (5-Aminolevulinic acid; CAS:5451-09-2; J&K SCIENTIFIC LTD.) solution once every 3 days for 2 weeks. The Y-05 plants sprayed by water were used as control.
For gabaculine treatment, one-month-old Y-05 seedlings without cold acclimation were watered using 50 μM gabaculine aqueous solution (3-amino-2,3-dihydrobenzoic acid; CAS: 59556–17-1, J&K SCIENTIFIC LTD, China.) solution [71] once every three days for two weeks. The Y-05 seedlings watered by water were used as control.
Virus-induced gene silencing (VIGS) analysis
VIGS was used to generate silenced pakchoi as our previously described [72]. A specific 40-bp fragment (5′-CTGAGGAATCAAGAGCCAGAGAAGGCTTTTGAAGAGTTCATGAACTCTTCAA AAGCCTTCTCTGGCTCTTGATTCCTCAG-3′) of BrFLU coding region (underlined) and its antisense sequence was synthesized and inserted into the pTY vector [72]. Then, the construct was introduced into cells of one-month-old Y-05 seedlings using gene gun (Biolistic PDS-1000/He, Bio-rad, USA). The seedlings introduced by empty pTY vector were used as control. After 1 week grown under normal condition, both BrFLU-silenced and control plants were moved in 4 °C chamber for 3 weeks cold acclimation, and then return to 23 °C for continues grown.
RNA isolation and gene expression analysis
Total RNA was extracted from leaves by using TRIzaol reagent (Thermo Fisher Scientific Inc.) [73], and cDNA was synthesized by HiScript II Q RT SuperMix for qPCR (Cat No. r223–01, Vazyme, Nanjing, China). Then analyzed by qPCR using SYBR Green Premix Pro Taq HS qPCR Kit II (Rox Plus) (Cat No. AG11719, Accurate Biotechnology (Hunan) Co.,Ltd., China) on StepOnePlus system (Applied Biosystems, USA). The relative expression of genes was analyzed by the 2−ΔΔCT method [74] and were normalized to the internal control gene BrPP2A for pakchoi. Each reaction was performed in three technical replicates and three independent biological replicates (biological replicates: leaves of the same part of three independent plants; technical replicates: repeat detection and analysis of the same sample). Primers for qPCR analysis were designed by Primer Software Version 5.0 (Premier Biosoft International, CA, USA) and shown in Table S8.
Data analysis
PCA of all samples and Volcano map were generated by BMK Cloud platform (www.biocloud.net). Heatmap and cis-acting analysis of promoter were conducted on TBtools v1.05 [75]. The full-length open reading frame (ORF) of BrFLU was obtained from the NCBI database using BLASTN and sequences amplified from the cultivar including Y-05, G-04, WTC, 2Q, LY, MET and SZQ. The multiple sequence alignment was used online software MultAlin [76]. The amino acid sequence alignment was used software DNAMAN. The 1.2 kb promoter sequence of BrFLU was identified using BLASTN to query the B. rapa genome. The primers were shown in Table S8. Promoter sequence analyzed using the Plant-CARE databases (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/). All methods and materials in our manuscript complied with relevant institutional, national, and international guidelines and legislation.
Supplementary Information
Additional file 1: Table S1. Description of read data, quality control and GC content of TIN and TOU. Table S2. Differential expression analysis and functional annotation of DEGs between TIN and TOU. Table S3. Functional categorization of DEGs between TIN and TOU by Gene ontology (GO) analysis. Table S4 Functional categorization of DEGs between TIN and TOU DEGs enriched in KEGG pathways. Table S5. Differential expression analysis and functional annotation of DEMs between MIN and MOU. Table S6. The DEMs involved in Porphyrin and chlorophyll metabolism (ko00860). Table S7. The expression level of BrHEMA1, BrGSA1, BrGBP, BrFLU between TIN and TOU. Table S8. The primers used in experiments.
Additional file 2: Fig. S1. The pigments content in G-04 and Y-05 leaves. Fig. S2. The correlation between replicates and volcano map of DEGs between TIN and TOU. Fig. S3. The correlation between replicates and volcano map of DEMs between MIN and MOU. Fig. S4. The expression profiles of BrHEMA1, BrGSA1, BrGBP and BrFLU between TIN and TOU. Fig. S5. Alignment of BrFLU nucleotide sequences in seven pakchoi varieties. Fig. S6. Alignment of BrFLU amino acid sequences in seven pakchoi varieties. Fig. S7. The promoter motif analysis of BrFLU in different pakchoi varieties.
Acknowledgements
The authors are thankful to all the lab mates who supported us.
Abbreviations
- YIN
Yellow inner leaves
- GOU
Green outer leaves
- TEM
Transmission electron microscopy
- DEGs
Differentially expressed genes
- ALA
5-aminolevulinic acid
- pem
Plant etiolated mutation (pem)
- REC
REDUCED CHLOROPLAST COVERAGE protein
- TPR
Tetratricopeptide repeat protein
- PPR
Pentatricopeptide repeat protein
- Pn
Net photosynthetic rate
- TOU
transcripts in green outer leaves
- TIN
Transcripts in yellow inner leaves
- PCC
Pearson’s Correlation Coefficient
- GO
Gene Ontology
- KEGG
Kyoto Encyclopedia of Genes and Genomes
- MIN
Metabolites in yellow inner leaves
- MOU
Metabolites in green outer leaves
- DEMs
Differentially expressed metabolites
- Gabaculine
3-amino 2,3-dihydrobenzoic acid
- ELISA
Enzyme-linked immunosorbent assay
- KOG/COG
Clusters of Orthologous Groups of proteins
- Pfam
Protein family
- VIGS
Virus-induced gene silencing
- PCA
Principal component analysis
- GluTR
Glutamyl-tRNA reductase
- GSA
Glu-1-semialdehyd
- GSA-AM
GSA-2,1-aminomutase
- FLU
Fluorescent in blue light
- FLUOE
FLU-overexpressor
- GBP
GluTR-binding protein
Authors’ contributions
TL designed the study. TL, HW, LY and HZ conducted the experiments, analyzed the data and wrote the manuscript. TL and HW revised the manuscript. ZL and XH helped prepared the samples. All authors read and approved the final manuscript.
Funding
This work was supported by the Agricultural Science and Technology Independent Innovation Fund of Jiangsu Province (CX (19)3122), Postdoctoral Science Foundation of Anhui Province (2020B439) and Key research and Development Fund of Anhui Province (202004a06020062).
Availability of data and materials
The datasets used and/or analysed during the current study are available from the corresponding author on reasonable request.
Declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Huiyu Wang, Email: 2019204023@njau.edu.cn.
Zhubo Li, Email: 2019104060@njau.edu.cn.
Lingyun Yuan, Email: ylyun99@163.com.
Hefang Zhou, Email: zhf003200@163.com.
Xilin Hou, Email: hxl@njau.edu.cn.
Tongkun Liu, Email: liutk@njau.edu.cn.
References
- 1.Tanaka Y, Sasaki N, Ohmiya A. Biosynthesis of plant pigments: anthocyanins, betalains and carotenoids. Plant J. 2008;54(4):733–749. doi: 10.1111/j.1365-313X.2008.03447.x. [DOI] [PubMed] [Google Scholar]
- 2.Luo F, Cheng SC, Cai JH, Wei BD, Zhou X, Zhou Q, Zhao YB, Ji SJ. Chlorophyll degradation and carotenoid biosynthetic pathways: gene expression and pigment content in broccoli during yellowing. Food Chem. 2019;297:124964. doi: 10.1016/j.foodchem.2019.124964. [DOI] [PubMed] [Google Scholar]
- 3.Clark ST, Verwoerd WS. A systems approach to identifying correlated gene targets for the loss of colour pigmentation in plants. BMC Bioinformatics. 2011;12(1):343. doi: 10.1186/1471-2105-12-343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Von Wettstein D, Gough S, Kannangara CG. Chlorophyll biosynthesis. Plant Cell. 1995;7(7):1039–1057. doi: 10.2307/3870056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sundberg E. ALBIN03, an Arabidopsis nuclear gene essential for chloroplast differentiation, encodes a chloroplast protein that shows homology to proteins present in bacterial membranes and yeast mitochondria. Plant Cell. 1997;9(5):717–730. doi: 10.1105/tpc.9.5.717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ishikawa A, Okamoto H, Iwasaki Y, Asahi T. A deficiency of coproporphyrinogen III oxidase causes lesion formation in Arabidopsis. Plant J. 2001;27(2):89–99. doi: 10.1046/j.1365-313x.2001.01058.x. [DOI] [PubMed] [Google Scholar]
- 7.Lohscheider JN, Rojas-Stutz MC, Rothbart M, Andersson U, Funck D, Mendgen K, Grimm B, Adamska I. Altered levels of LIL3 isoforms in Arabidopsis lead to disturbed pigment-protein assembly and chlorophyll synthesis, chlorotic phenotype and impaired photosynthetic performance. Plant Cell Environ. 2015;38(10):2115–2127. doi: 10.1111/pce.12540. [DOI] [PubMed] [Google Scholar]
- 8.Xie J, Yao S, Ming J, Deng L, Zeng K. Variations in chlorophyll and carotenoid contents and expression of genes involved in pigment metabolism response to oleocellosis in citrus fruits. Food Chem. 2019;272:49–57. doi: 10.1016/j.foodchem.2018.08.020. [DOI] [PubMed] [Google Scholar]
- 9.Xie S, Nie L, Zheng Y, Wang J, Zhao M. Comparative proteomic analysis reveals that chlorophyll metabolism contributes to leaf color changes in Wucai ( Brassica campestris L.) responding to cold acclimation. J Proteome Res. 2019;18(6):2478–2492. doi: 10.1021/acs.jproteome.9b00016. [DOI] [PubMed] [Google Scholar]
- 10.Reinbothe C, El Bakkouri M, Buhr F, Muraki N, Nomata J, Kurisu G, Fujita Y, Reinbothe S. Chlorophyll biosynthesis: spotlight on protochlorophyllide reduction. Trends Plant Sci. 2010;15(11):614–624. doi: 10.1016/j.tplants.2010.07.002. [DOI] [PubMed] [Google Scholar]
- 11.Wang P, Grimm B. Organization of chlorophyll biosynthesis and insertion of chlorophyll into the chlorophyll-binding proteins in chloroplasts. Photosynth Res. 2015;126(2–3):189–202. doi: 10.1007/s11120-015-0154-5. [DOI] [PubMed] [Google Scholar]
- 12.Zhu L, Zeng X, Chen Y, Yang Z, Qi L, Pu Y, Yi B, Wen J, Ma C, Shen J, Tu J, Fu T. Genetic characterisation and fine mapping of a chlorophyll-deficient mutant (BnaC.ygl) in Brassica napus. Mol Breed. 2014;34(2):603–614. doi: 10.1007/s11032-014-0060-0. [DOI] [Google Scholar]
- 13.Zhang L, Liu C, An X, Wu H, Feng Y, Wang H, Sun D. Identification and genetic mapping of a novel incompletely dominant yellow leaf color gene, Y1718, on chromosome 2BS in wheat. Euphytica. 2017;213(7):141. doi: 10.1007/s10681-017-1894-4. [DOI] [Google Scholar]
- 14.Mueller A, Dockter C, Gough S, Lundqvist U, Wettstein D, Hansson M. Characterization of mutations in barley fch2 encoding Chlorophyllide a Oxygenase. Plant Cell Physiol. 2012;53(7):1232–1246. doi: 10.1093/pcp/pcs062. [DOI] [PubMed] [Google Scholar]
- 15.Kim D-Y, Bovet L, Kushnir S, Noh EW, Martinoia E, Lee Y. AtATM3 is involved in heavy metal resistance in Arabidopsis. Plant Physiol. 2006;140(3):922–932. doi: 10.1104/pp.105.074146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang H, Li J, Yoo J-H, Yoo S-C, Cho S-H, Koh H-J, Seo HS, Paek N-C. Rice Chlorina-1 and Chlorina-9 encode ChlD and ChlI subunits of mg-chelatase, a key enzyme for chlorophyll synthesis and chloroplast development. Plant Mol Biol. 2006;62(3):325–337. doi: 10.1007/s11103-006-9024-z. [DOI] [PubMed] [Google Scholar]
- 17.Bu-Yun P, Jun Z, Li-Jun AN. Genetic screening and isolation of a leaf color mutant F03-06 in Arabidopsis. Hub Agricultural Ences. 2016;55(21):5664–5667. [Google Scholar]
- 18.Li X, Huang S, Liu Z, Hou L, Feng H. Mutation in EMB1923 gene promoter is associated with chlorophyll deficiency in Chinese cabbage (Brassica campestris ssp. pekinensis) Physiol Plant. 2019;166(4):909–920. doi: 10.1111/ppl.12979. [DOI] [PubMed] [Google Scholar]
- 19.Zhang K, Liu Z, Shan X, Li C, Tang X, Chi M, Feng H. Physiological properties and chlorophyll biosynthesis in a Pak-choi (Brassica rapa L. ssp. chinensis) yellow leaf mutant, pylm. Acta Physiologiae Plantarum. 2017;39(1):22. doi: 10.1007/s11738-016-2321-5. [DOI] [Google Scholar]
- 20.Larkin RM, Stefano G, Ruckle ME, Stavoe AK, Sinkler CA, Brandizzi F, Malmstrom CM, Osteryoung KW. REDUCED CHLOROPLAST COVERAGE genes from Arabidopsis thaliana help to establish the size of the chloroplast compartment. PNAS. 2016;113(8):E1116–E1125. doi: 10.1073/pnas.1515741113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lan T, Wang B, Ling Q, Xu C, Tong Z, Liang K, Duan Y, Jin J, Wu W. Fine mapping of cisc(t), a gene for cold-induced seedling chlorosis, and identification of its candidate in rice. Chin Sci Bull. 2010;55(27):3149–3153. doi: 10.1007/s11434-010-4041-3. [DOI] [Google Scholar]
- 22.Barkan A, Small I. Pentatricopeptide repeat proteins in plants. Annu Rev Plant Biol. 2014;65(1):415–442. doi: 10.1146/annurev-arplant-050213-040159. [DOI] [PubMed] [Google Scholar]
- 23.Kusumi K, Iba K. Establishment of the chloroplast genetic system in rice during early leaf development and at low temperatures. Front Plant Sci. 2014;5:386. doi: 10.3389/fpls.2014.00386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Du YY, Chen H, Zhong WL, Wu LY, Ye JH. Effect of temperature on accumulation of chlorophylls and leaf ultrastructure of low temperature induced albino tea plant. Afr J Biotechnol. 2008;7(12):1881–1885. doi: 10.5897/AJB2008.000-5036. [DOI] [Google Scholar]
- 25.Park S, Fischer AL, Steen CJ, Iwai M, Morris JM, Walla PJ, Niyogi KK, Fleming GR. Chlorophyll-carotenoid excitation energy transfer in high-light-exposed thylakoid membranes investigated by snapshot transient absorption spectroscopy. J Am Chem Soc. 2018;140(38):11965–11973. doi: 10.1021/jacs.8b04844. [DOI] [PubMed] [Google Scholar]
- 26.Brzezowski P, Ksas B, Havaux M, Grimm B, Chazaux M, Peltier G, Johnson X, Alric J. The function of PROTOPORPHYRINOGEN IX OXIDASE in chlorophyll biosynthesis requires oxidised plastoquinone in Chlamydomonas reinhardtii. Commun Biol. 2019;2(1):159. doi: 10.1038/s42003-019-0395-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wu Y, Jin X, Liao W, Hu L, Dawuda MM, Zhao X, Tang Z, Gong T, Yu J. 5-Aminolevulinic acid (ALA) alleviated salinity stress in cucumber seedlings by enhancing chlorophyll synthesis pathway. Front Plant Sci. 2018;9:635. doi: 10.3389/fpls.2018.00635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rassadina VV, Averina NG, Koval SF. Disturbance of chlorophyll formation at the level of 5-aminolevulinic acid and Mg-containing porphyrin synthesis in isogenic lines of spring wheat (Triticum aestivum L.) marked with genes cn-A1 and cn-D1. Dokl Biol Sci. 2005;405:472–473. doi: 10.1007/s10630-005-0169-8. [DOI] [PubMed] [Google Scholar]
- 29.Liu D, Kong DD, Fu XK, Ali B, Xu L, Zhou WJ. Influence of exogenous 5-aminolevulinic acid on chlorophyll synthesis and related gene expression in oilseed rape de-etiolated cotyledons under water-deficit stress. Photosynthetica Int J Photosynth Res. 2016;54(3):468–474. [Google Scholar]
- 30.Guo XQ, Li YS, Yu XC. Promotive effects of 5-aminolevulinic acid on photosynthesis and chlorophyll fluorescence of tomato seedlings under suboptimal low temperature and suboptimal photon flux density stress - short communication. Hortic Sci. 2012;39(2):97–99. doi: 10.17221/122/2011-HORTSCI. [DOI] [Google Scholar]
- 31.Li DM, Zhang J, Sun WJ, Li Q, Dai AH, Bai JG. 5-Aminolevulinic acid pretreatment mitigates drought stress of cucumber leaves through altering antioxidant enzyme activity. Sci Hortic. 2011;130(4):820–828. doi: 10.1016/j.scienta.2011.09.010. [DOI] [Google Scholar]
- 32.Hotta Y, Tanaka T, Takaoka H, Takeuchi Y, Konnai M. New physiological effects of 5-Aminolevulinic acid in plants: the increase of photosynthesis, chlorophyll content, and plant growth. Biosci Biotechnol Biochem. 1997;61(12):2025–2028. doi: 10.1271/bbb.61.2025. [DOI] [PubMed] [Google Scholar]
- 33.Kumar AM, Söll D. Antisense HEMA1 RNA expression inhibits Heme and chlorophyll biosynthesis in Arabidopsis. Plant Physiol. 2000;122(1):49–56. doi: 10.1104/pp.122.1.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Apitz J, Nishimura K, Schmied J, Wolf A, Hedtke B, van Wijk KJ, Grimm B. Posttranslational control of ALA synthesis includes GluTR degradation by Clp protease and stabilization by GluTR-binding protein. Plant Physiol. 2016;170(4):2040–2051. doi: 10.1104/pp.15.01945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Goslings D, Meskauskiene R, Kim C, Lee KP, Nater M, Apel K. Concurrent interactions of heme and FLU with Glu tRNA reductase (HEMA1), the target of metabolic feedback inhibition of tetrapyrrole biosynthesis, in dark- and light-grown Arabidopsis plants. Plant J. 2004;40(6):957–967. doi: 10.1111/j.1365-313X.2004.02262.x. [DOI] [PubMed] [Google Scholar]
- 36.Fang Y, Zhao S, Zhang F, Zhao A, Zhang W, Zhang M, Liu L. The Arabidopsis glutamyl-tRNA reductase (GluTR) forms a ternary complex with FLU and GluTR-binding protein. Sci Rep. 2016;6(1):19756. doi: 10.1038/srep19756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hou Z, Yang Y, Hedtke B, Grimm B. FLUORESCENCE IN BLUE LIGHT (FLU) is involved in inactivation and localization of glutamyl-tRNA reductase during light exposure. Plant J 2018. 97(3):517–29. 10.1111/tpj.14138. [DOI] [PubMed]
- 38.Liu J, Wang J, Yao X, Zhang Y, Li J, Wang X, Xu Z, Chen W. Characterization and fine mapping of thermo-sensitive chlorophyll deficit mutant1 in rice (Oryza sativa L.) Breed Sci. 2015;65(2):161–169. doi: 10.1270/jsbbs.65.161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Anderson CM, Gray JC. Effect of Gabaculine on the synthesis of Heme and cytochrome f in etiolated wheat seedlings. Plant Physiol. 1991;96(2):584–587. doi: 10.1104/pp.96.2.584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Drazic G, Bogdanovic M. Gabaculine does not inhibit cytokinin-stimulated biosynthesis of chlorophyll in Pinus nigra seedlings in the dark. Plant Sci. 2000;154(1):23–29. doi: 10.1016/S0168-9452(99)00236-8. [DOI] [PubMed] [Google Scholar]
- 41.Hou Z. Fluorescence in blue light (FLU): functional analysis of its structural domains for light and dark-dependent control of ALA synthesis. 2020. [Google Scholar]
- 42.von Gromoff ED, Alawady A, Meinecke L, Grimm B, Beck CF. Heme, a plastid-derived regulator of nuclear gene expression in Chlamydomonas. Plant Cell. 2008;20(3):552–567. doi: 10.1105/tpc.107.054650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Primka EJ, Smith WK. Synchrony in fall leaf drop: chlorophyll degradation, color change, and abscission layer formation in three temperate deciduous tree species. Am J Bot. 2019;106(3):377–388. doi: 10.1002/ajb2.1247. [DOI] [PubMed] [Google Scholar]
- 44.Shen J, Zou Z, Zhang X, Zhou L, Wang Y, Fang W, Zhu X. Metabolic analyses reveal different mechanisms of leaf color change in two purple-leaf tea plant (Camellia sinensis L.) cultivars. Hortic Res. 2018;5:7. doi: 10.1038/s41438-017-0010-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Teppabut Y, Oyama KI, Kondo T, Yoshida K. Change of Petals' color and chemical components in Oenothera flowers during senescence. Molecules. 2018;23(7):1698. doi: 10.3390/molecules23071698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Matile P. Biochemistry of Indian summer: physiology of autumnal leaf coloration. Exp Gerontol. 2000;35(2):145–158. doi: 10.1016/S0531-5565(00)00081-4. [DOI] [PubMed] [Google Scholar]
- 47.Zhao MH, Li X, Zhang XX, Zhang H, Zhao XY. Mutation mechanism of leaf color in plants: a review. Forests. 2020;11(8):851. doi: 10.3390/f11080851. [DOI] [Google Scholar]
- 48.Beale SI. Green genes gleaned. Trends Plant Sci. 2005;10(7):309–312. doi: 10.1016/j.tplants.2005.05.005. [DOI] [PubMed] [Google Scholar]
- 49.Eckhardt U, Grimm B, Hortensteiner S. Recent advances in chlorophyll biosynthesis and breakdown in higher plants. Plant Mol Biol. 2004;56(1):1–14. doi: 10.1007/s11103-004-2331-3. [DOI] [PubMed] [Google Scholar]
- 50.Zhou S, Hu Z, Zhu M, Zhang B, Deng L, Pan Y, Chen G. Biochemical and molecular analysis of a temperature-sensitive albino mutant in kale named "white dove". Plant Growth Regul. 2013;71(3):281–294. doi: 10.1007/s10725-013-9829-0. [DOI] [Google Scholar]
- 51.Kim A. Arabidopsis light-dependent NADPH: protochlorophyllide oxidoreductase a (PORA) is essential for normal plant growth and development: an addendum. Plant Mol Biol. 2012;80(2):237–240. doi: 10.1007/s11103-012-9944-8. [DOI] [PubMed] [Google Scholar]
- 52.Li Y, Pu X, Li C, Zhong P, Sun C, Li X, Deng X, Wang P. Genetic identification and candidate gene analysis of yellow-green leaf mutant 507ys in rice. Acta Agron Sin. 2014;47:221–229. [Google Scholar]
- 53.Zhu G, Yang F, Shi S, Li D, Wang Z. Transcriptome Characterization of Cymbidium sinense 'Dharma' Using 454 Pyrosequencing and Its Application in the Identification of Genes Associated with Leaf Color Variation. PloS one. 2015;10(6):e0128592. doi: 10.1371/journal.pone.0128592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hu ZL, Deng L, Yan B, Pan Y, Luo M, Chen XQ, Hu TZ, Chen GP. Silencing of the LeSGR1 gene in tomato inhibits chlorophyll degradation and exhibits a stay-green phenotype. Biol Plant. 2011;55(1):27–34. doi: 10.1007/s10535-011-0004-z. [DOI] [Google Scholar]
- 55.Sakuraba Y, Park SY, Paek NC. The divergent roles of STAYGREEN (SGR) homologs in chlorophyll degradation. Mol Cells. 2015;38(5):390–395. doi: 10.14348/molcells.2015.0039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Jiang A, Liu J, Jiang J. Relationships of leaf color and pigment and nutrient elements in senescing leaves of Sassafras tsumu. For Res. 2016;29(03):362–368. [Google Scholar]
- 57.Piao W, Han SH, Sakuraba Y, Paek NC. Rice 7-Hydroxymethyl chlorophyll a reductase is involved in the promotion of chlorophyll degradation and modulates cell death signaling. Mol Cells. 2017;40(10):773–786. doi: 10.14348/molcells.2017.0127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhang MP, Zhang CJ, Yu GH, Jiang YZ, Strasser RJ, Yuan ZY, Yang XS, Chen GX. Changes in chloroplast ultrastructure, fatty acid components of thylakoid membrane and chlorophyll a fluorescence transient in flag leaves of a super-high-yield hybrid rice and its parents during the reproductive stage. J Plant Physiol. 2010;167(4):277–285. doi: 10.1016/j.jplph.2009.09.017. [DOI] [PubMed] [Google Scholar]
- 59.Xu G, Cao J, Wang X, Chen Q, Tian F. Evolutionary metabolomics identifies substantial metabolic divergence between maize and its wild ancestor. Teosinte Plant Cell. 2019;31(9):1990–1909. doi: 10.1105/tpc.19.00111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhang M, Zhang F, Fang Y, Chen X, Chen Y, Zhang W, Dai HE, Lin R, Liu L. The non-canonical Tetratricopeptide repeat (TPR) domain of fluorescent (FLU) mediates complex formation with Glutamyl-tRNA reductase. J Biol Chem. 2015;290(28):17559–17565. doi: 10.1074/jbc.M115.662981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Nouairi I, Jalali K, Zribi F, Barhoumi F, Zribi K, Mhadhbi H. Seed priming with calcium chloride improves the photosynthesis performance of faba bean plants subjected to cadmium stress. Photosynthetica. 2019;57(2):438–445. doi: 10.32615/ps.2019.055. [DOI] [Google Scholar]
- 62.Xu YMWY, Zeng JY. Comparative analysis on the content of carotenoid in the leaves of the three general plants. J Guangxi Normal Univ Nationalities. 2013;30(3):18–19. [Google Scholar]
- 63.Huo L-L, Su P, Lv Y-H. Measurement of Total Anthocyanins in Mulberry by UV-Visible Spectroscopy. Liquor-making. 2005; 4.
- 64.Maekawa S, Takabayashi A, Huarancca Reyes T, Yamamoto H, Tanaka A, Sato T, Yamaguchi J. Pale-green phenotype of atl31 atl6 double mutant leaves is caused by disruption of 5-Aminolevulinic acid biosynthesis in Arabidopsis thaliana. PLoS One. 2015;10(2):e0117662. doi: 10.1371/journal.pone.0117662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Labate MT, Ko K, Ko ZW, Pinto LS, Real MJ, Romano MR, Barja PR, Granell A, Friso G, van Wijk KJ, et al. Constitutive expression of pea Lhcb 1-2 in tobacco affects plant development, morphology and photosynthetic capacity. Plant Mol Biol. 2004;55(5):701–714. doi: 10.1007/s11103-004-1963-7. [DOI] [PubMed] [Google Scholar]
- 66.Kim D, Langmead B, Salzberg SL. HISAT: a fast spliced aligner with low memory requirements. Nat Methods. 2015;12(4):357–360. doi: 10.1038/nmeth.3317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kanehisa M, Furumichi M, Sato Y, Ishiguro-Watanabe M, Tanabe M. KEGG: integrating viruses and cellular organisms. Nucleic Acids Res. 2021;49(D1):D545–DD51. doi: 10.1093/nar/gkaa970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wang L, Feng Z, Wang X, Wang X, Zhang X. DEGseq: an R package for identifying differentially expressed genes from RNA-seq data. Bioinformatics. 2010;26(1):136–138. doi: 10.1093/bioinformatics/btp612. [DOI] [PubMed] [Google Scholar]
- 69.Benjamini Y, Yekutieli D. The control of the false discovery rate in multiple testing under dependency. Ann Stat. 2001;29(4):1165–1188. doi: 10.1214/aos/1013699998. [DOI] [Google Scholar]
- 70.Barri T, Dragsted LO. UPLC-ESI-QTOF/MS and multivariate data analysis for blood plasma and serum metabolomics: effect of experimental artefacts and anticoagulant. Anal Chim Acta. 2013;768:118–128. doi: 10.1016/j.aca.2013.01.015. [DOI] [PubMed] [Google Scholar]
- 71.Tchuinmogne SJ, Huault C, Aoues A, Balange AP. Inhibitory effect of gabaculine on 5-aminolevulinate dehydratase activity in radish seedlings. Plant Physiol. 1989;90(4):1293–1297. doi: 10.1104/pp.90.4.1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Yu J, Yang XD, Wang Q, Gao LW, Yang Y, Xiao D, Liu TK, Li Y, Hou XL, Zhang CW. Efficient virus-induced gene silencing in Brassica rapa using a turnip yellow mosaic virus vector. Biol Plant. 2018;62(4):826–834. doi: 10.1007/s10535-018-0803-6. [DOI] [Google Scholar]
- 73.She H, Qian W, Zhang H, Liu Z, Wang X, Wu J, Feng C, Correll JC, Xu Z. Fine mapping and candidate gene screening of the downy mildew resistance gene RPF1 in spinach. Theor Appl Genet. 2018;131(12):2529–2541. doi: 10.1007/s00122-018-3169-4. [DOI] [PubMed] [Google Scholar]
- 74.Schmittgen TD, Livak KJ. Analyzing real-time PCR data by the comparative CT method. Nat Protoc. 2008;3(6):1101–1108. doi: 10.1038/nprot.2008.73. [DOI] [PubMed] [Google Scholar]
- 75.Chen C, Chen H, Zhang Y, Thomas HR, Frank MH, He Y, Xia R. TBtools: An integrative toolkit developed for interactive analyses of big biological data. Mol Plant. 2020;13(8):1194–1202. doi: 10.1016/j.molp.2020.06.009. [DOI] [PubMed] [Google Scholar]
- 76.Corpet F. Multiple sequence alignment with hierarchical clustering. Nucleic Acids Res. 1988;16(22):10881–10890. doi: 10.1093/nar/16.22.10881. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Table S1. Description of read data, quality control and GC content of TIN and TOU. Table S2. Differential expression analysis and functional annotation of DEGs between TIN and TOU. Table S3. Functional categorization of DEGs between TIN and TOU by Gene ontology (GO) analysis. Table S4 Functional categorization of DEGs between TIN and TOU DEGs enriched in KEGG pathways. Table S5. Differential expression analysis and functional annotation of DEMs between MIN and MOU. Table S6. The DEMs involved in Porphyrin and chlorophyll metabolism (ko00860). Table S7. The expression level of BrHEMA1, BrGSA1, BrGBP, BrFLU between TIN and TOU. Table S8. The primers used in experiments.
Additional file 2: Fig. S1. The pigments content in G-04 and Y-05 leaves. Fig. S2. The correlation between replicates and volcano map of DEGs between TIN and TOU. Fig. S3. The correlation between replicates and volcano map of DEMs between MIN and MOU. Fig. S4. The expression profiles of BrHEMA1, BrGSA1, BrGBP and BrFLU between TIN and TOU. Fig. S5. Alignment of BrFLU nucleotide sequences in seven pakchoi varieties. Fig. S6. Alignment of BrFLU amino acid sequences in seven pakchoi varieties. Fig. S7. The promoter motif analysis of BrFLU in different pakchoi varieties.
Data Availability Statement
The datasets used and/or analysed during the current study are available from the corresponding author on reasonable request.