Abstract
Background
Neutralization tests (NT) are the gold standard for detecting and quantifying anti-SARS-CoV-2 neutralizing antibodies (NAb), but their complexity restricts them to research settings or reference laboratories. Antibodies against S protein receptor binding domain (RBD) have been shown to confer a neutralizing activity against SARS-CoV-2. Assays quantitatively measuring anti-S1-RBD-SARS-CoV-2 antibodies could be of great value for NAb screening of potential donors for convalescent-phase plasma therapy, assessing natural or vaccine-induced immunity, stratifying individuals for vaccine receipt, and documenting vaccine response.
Methods
Elecsys Anti-SARS-CoV-2 S (Elecsys-S), a high-throughput automated electrochemiluminescence double-antigen sandwich immunoassay for quantitative measurement of pan-anti-S1-RBD-SARS-CoV-2 antibodies, was evaluated against NT on 357 patients with PCR-confirmed SARS-CoV-2 infection. NT was performed in a BSL-3 laboratory using a Slovenian SARS-CoV-2 isolate; the NT titer ≥1:20 was considered positive.
Results
Elecsys-S detected pan-anti-S1-RBD-SARS-CoV-2 antibodies in 352/357 (98.6 %) samples. NAb were identified by NT in 257/357 (72 %) samples. The Elecsys-S/NT agreement was moderate (Cohen’s kappa 0.56). High NT titer antibodies (≥1:160) were detected in 106/357 (30 %) samples. Elecsys-S’s pan-anti-S1-RBD-SARS-CoV-2 antibody concentrations correlated with individual NT titer categories (the lowest concentrations were identified in NT-negative samples and the highest in samples with NT titer 1:1,280), and the Elecsys-S cutoff value for reasonable prediction of NAb generated after natural infection was established (133 BAU/mL).
Conclusion
Although NT should remain the gold standard for assessing candidates for convalescent-phase plasma donors, selected commercial anti-SARS-CoV-2 assays with optimized cutoff, like Elecsys-S, could be used for rapid, automated, and large-scale screening of individuals with clinically relevant NAb levels as suitable donors.
Keywords: SARS-CoV-2, Antibody, Electrochemiluminescence, Immunoassay, Neutralization
1. Introduction
Accurate assays for detecting antibodies against severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) are needed to inform diagnostic, therapeutic, and public health decisions [1,2]. Many commercial and laboratory-developed anti-SARS-CoV-2 assays are available to measure neutralizing and non-neutralizing anti-SARS-CoV-2 antibodies against a range of viral proteins/isotopes with various analytical techniques [3,4].
Neutralization assays are the gold standard for detecting and quantifying anti-SARS-CoV-2 neutralizing antibodies (NAb), mainly used in research settings or offered as laboratory-developed tests by reference laboratories [3]. Antibodies against the receptor binding domain (RBD) of the S1 subunit of the spike (S) protein have shown neutralizing activity against SARS-CoV-2 [5]. Assays quantitatively measuring anti-S1-RBD-SARS-CoV-2 antibodies are very promising for screening potential donors for convalescent-phase plasma therapy, assessing natural or vaccine-induced immunity, stratifying individuals for vaccine receipt, and documenting vaccine response, which could inform return-to-work and travel decisions and other public health measures [1,6].
Elecsys Anti-SARS-CoV-2 S (Elecsys-S) (Roche Diagnostics, Mannheim, Germany) is a recently launched rapid (18-minute) high-throughput automated electrochemiluminescence double-antigen sandwich immunoassay for quantitative measurement of pan-anti-S1-RBD-SARS-CoV-2 antibodies. Elecsys-S was launched in Europe in September 2020 and received FDA EUA on November 25, 2020. The assay has been extensively evaluated by the manufacturer, showing 100.00 % (95 % confidence interval (CI), 99.7–100 %) analytical specificity on 1,100 samples, 99.98 % (95 %CI, 99.91–100 %) clinical specificity on 5991 samples, and 98.8 % (95 %CI, 98.1–99.3 %) clinical sensitivity on 1423 samples obtained 14 days or later after SARS-CoV-2 PCR-confirmation. The only published Elecsys-S evaluation shows clinical specificity of 99.8 % (95 %CI, 99.4–99.9 %) on 1159 pre−COVID-19 samples and 97.6 % (95 %CI, 93.2–99.1 %) clinical sensitivity on 125 sera taken 43–144 days after symptom onset [7].
Here we present the first manufacturer-independent performance evaluation of Elecsys-S against a neutralization test assessed on 357 patients with PCR-confirmed SARS-CoV-2 infection.
2. Material and methods
The study population comprised 357 anonymized plasma samples collected during July-November 2020 from 357 Slovenian patients with PCR-confirmed SARS-CoV-2 infection. In all patients, SARS-CoV-2 infection was confirmed by cobas 6800 SARS-CoV-2 (Roche Diagnostics) [8]. Blood samples were collected from 35 days up to 6 months after symptom onset or first PCR positivity.
To determine the concentration of pan-anti-S1-RBD-SARS-CoV-2 antibodies, Elecsys-S was performed on a cobas e411 analyzer, using the manufacturer’s cut-off value for positive result of ≥0.8 U/mL. The manufacturer reports a linear measurement range from 0.4 to 250 U/mL, which can be expanded by dilution. Recently, World Health Organization (WHO) introduced International Standard for anti-SARS-CoV-2 immunoglobulin [9]. The neat sample was assigned to contain 1000 binding antibody units (BAU)/mL. A mathematical transposition of Elecsys-S specific U to BAU follow the equation: Elecsys-S U = 0.972 x BAU. Since transposition results in minimal numeric changes only that are well within the range of typical assay variance, according to the manufacturer’s recommendations Elecsys-S U/mL were considered to be equivalent to the BAU/mL of the WHO standard, and all Elecsys-S results were expressed in BAU/mL.
A neutralization test (NT) was performed in a BSL-3 laboratory using SARS-CoV-2 virus isolated from a Slovenian patient in April 2020 (strain Slovenia/SI-4265/20, D614 G; EVA-GLOBAL-Ref-SKU: 005V-03961; B.1.1 Pango lineage). Pango lineage B.1.1 was the most common SARS-CoV-2 genetic variant present in Slovenia from March to October 2020 thus we consider strain Slovenia/SI-4265/20 as reference strain for our study population. NT was based on the standardized protocol [10]. Briefly, Vero E6 cells were seeded in a concentration of 105/well (96-well plate) 1 day before NT was performed. Serial dilutions of heat-inactivated plasma samples (56 °C, 30 min) were incubated with 100 TCID50 of SARS-CoV-2 for 1 h at 37 °C. Each plasma dilution-virus mixture was added to the cells in triplicate and incubated at 37 °C with 5 % CO2 for 4 days. The neutralization endpoint titer was determined as the endpoint plasma dilution that inhibited the SARS-CoV-2–induced cytopathic effect in at least 2 out of 3 parallels. The NT titer ≥1:20 was considered positive.
Correlation between Elecsys-S and NT was assessed using an ordinal one-way ANOVA followed by post-hoc Bonferroni’s multiple comparison test [11].
Further, we tested Elecsys-S’s ability to differentiate between plasma samples with high NT antibody titer (≥1:160) and low titer (<1:160). The 1:160 cutoff was based on the NAb titer for COVID-19 convalescent-phase plasma therapy recommended by the FDA [12]. The 357 samples were randomized into two groups: 184 were used to develop a univariate logistic regression model with logarithmic Elecsys-S values as a predictive variable and high/low NT titer as dependent variable, and 173 for model validation. A chi-squared test was performed to identify any significant difference between the groups. To establish the optimal cutoff value, a receiver-operating-characteristic (ROC) curve was plotted in the validation group. The Elecsys-S cutoff value was determined by identifying an inflection point on the validation ROC curve, corresponding to a combination of high specificity (>84 %) and sensitivity (>74 %). We chose higher specificity compared to sensitivity to develop an algorithm for a sufficiently high probability of high NT titer with a lower number of samples chosen.
A contingency table used validation data to assess overall agreement with 95 % CIs. Agreement between both tests was assessed using Cohen’s kappa statistics. All statistical analyses used Excel (Microsoft, Redmond, WA) and GraphPad Prism version 8 (GraphPad Software, San Diego, CA).
3. Results
Out of 357 samples, 352 (98.6 %) tested positive for pan-anti-S1-RBD-SARS-CoV-2 antibodies; all five Elecsys-S-negative samples were NT-negative. NAb were detected by NT in 257/357 (72 %) samples (Table 1 ). The agreement between Elecsys-S and NT was moderate (Cohen’s kappa 0.56; 95 %CI, 0.42–0.69). All but one NT-negative sample (99/100) tested positive using Elecsys Anti-SARS-CoV-2 (Elecsys-N), a pan-anti-SARS-CoV-2 assay targeting the SARS-CoV-2 nucleoprotein (N) [13]; the mean Elecsys-N value was 36 (range 0.9–166).
Table 1.
Mean, minimum, and maximum concentrations of pan-anti-S1-RBD-SARS-CoV-2 antibodies (BAU/mL) measured by the Elecsys Anti-SARS-CoV-2 S assay in plasma samples of 357 patients with PCR-confirmed SARS-CoV-2 infection across different neutralization antibody titer (NT titer) categories.
| Anti-SARS-CoV-2 S (BAU/mL) |
||||
|---|---|---|---|---|
| NT (titer) | N | Mean | Minimum | Maximum |
| <1:10 | 100 | 29.5 | 0.40 | 248.3 |
| 1:20 | 49 | 66.5 | 4.77 | 468.8 |
| 1:40 | 50 | 126.5 | 2.55 | 821.5 |
| 1:80 | 52 | 243.3 | 3.18 | 1936 |
| 1:160 | 44 | 470.0 | 1.45 | 2223 |
| 1:320 | 27 | 584.2 | 8.61 | 2445 |
| 1:640 | 23 | 527.8 | 12.84 | 1906 |
| 1:1280 | 12 | 858.9 | 139.40 | 3362 |
TOTAL: 357
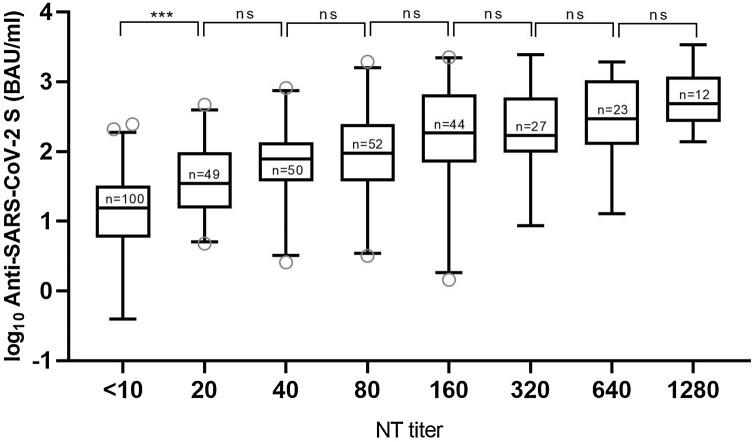
High NT titer antibodies (≥1:160) were detected in 106/357 (30 %) samples (Table 1). Pan-anti-S1-RBD-SARS-CoV-2 concentrations significantly differed regarding corresponding NT titer categories (ordinary one-way ANOVA: F (7, 349) = 34.6; p < 0.0001; Fig. 1 ). The lowest pan-anti-S1-RBD-SARS-CoV-2 concentrations were identified in NT-negative samples (post-hoc Bonferroni’s test, p < 0.001), and the highest in samples with NT titer 1:1,280 (Fig. 1, Table 1).
Fig. 1.
Correlation between neutralization antibody titers (NT titers) measured by neutralization test and logarithmic values (BAU/mL) of pan-anti-S1-RBD-SARS-CoV-2 antibodies measured by Elecsys Anti-SARS-CoV-2 S assay (Elecsys-S) in plasma samples of 357 patients with PCR-confirmed SARS-CoV-2 infection. Whiskers include data between the 2.5th and 97.5th percentiles. Differences in logarithmic Elecsys-S values were statistically significant among NT titers (ordinary one-way ANOVA: F (7, 349) = 34.6; p < 0.0001). *** = p < 0.001, ns = p > 0.05.
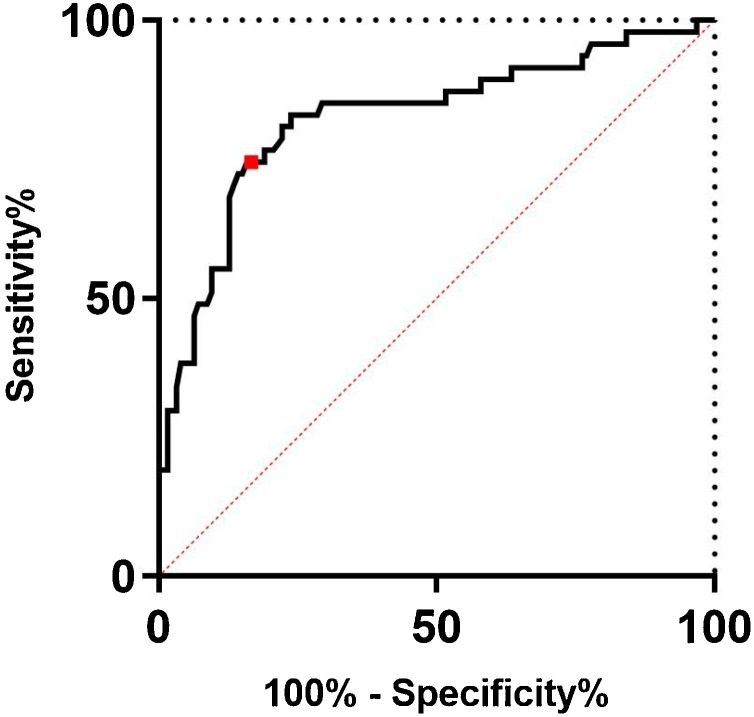
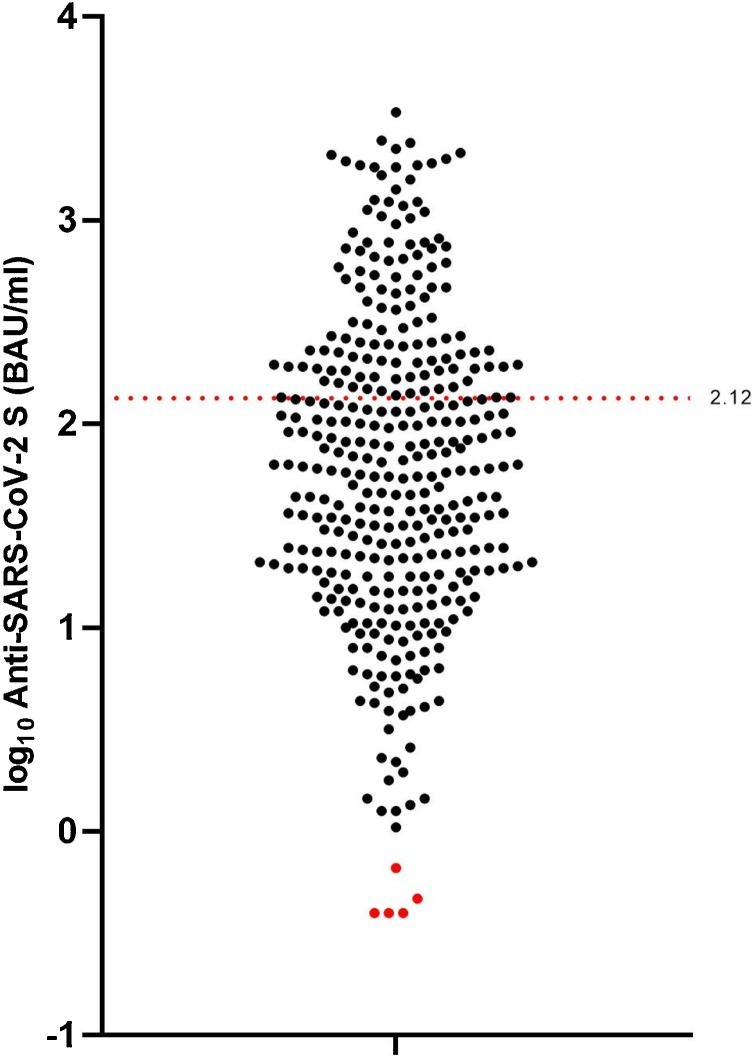
To establish the optimal Elecsys-S cutoff value for prediction of NAb presence, a univariate logistic regression model was developed and validated. As shown in Table 2 , no statistically significant differences in the distribution of low vs. high NT titer samples were found between the model development and model validation groups (p = 0.311). As shown in Fig. 2 , the selected Elecsys-S cutoff of 2.12 log10 BAU/mL or 133 BAU/mL resulted in a specificity of 84 % and sensitivity of 74.5 % for prediction of NAb generated after natural infection. Out of 357 samples tested, 118 (33 %) contained pan-anti-S1-RBD-SARS-CoV-2 antibodies in concentrations above selected Elecsys-S cutoff (Fig. 3 ).
Table 2.
Distribution of plasma samples with high neutralization (NT) antibody titer (≥1:160) and low NT antibody titer (<1:160) in model development (N = 184) and model validation (N = 173) groups of samples used for development and validation of the univariate logistic regression model, respectively.
| Sample group |
|||
|---|---|---|---|
| NT titer | Model development | Model validation | TOTAL |
| ≥1:160 | 59 (32 %) | 47 (27.2 %) | 106 (29.7 %) |
| <1:160 | 125 (68 %) | 126 (72.8 %) | 251 (70.3 %) |
| TOTAL | 184 (100 %) | 173 (100 %) | 357 (100 %) |
Fig. 2.
Receiver-operating-characteristic (ROC) curve representing the discriminating ability of the logistic model on validation data with area under the curve (AUC) 0.82 (95 % CI 0.75–0.90). The red dot represents an inflection point on the ROC curve to gain optimal discriminative measures (i.e., specificity >84 % and sensitivity >74.5 %). (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
Fig. 3.
Distribution of logarithmic values (U/mL) of pan-anti-S1-RBD-SARS-CoV-2 antibodies measured by the Elecsys Anti-SARS-CoV-2 S assay (Elecsys-S) in plasma samples of 357 patients with PCR-confirmed SARS-CoV-2 infection. Black dots represent Elecsys-S positive results (N = 352) and red dots negative results (N = 5) using the Elecsys-S manufacturer’s cutoff (≥0.8 BAU/mL). The red dotted line represents the selected Elecsys-S cutoff of 2.12 log10 BAU/mL or 133 BAU/mL for prediction of the presence of neutralizing antibodies generated after natural infection. Out of 357 samples tested, 118 (33 %) were above the selected Elecsys-S cutoff. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
4. Conclusions
As far as we know, apart from limited manufacturer’s data, for which Elecsys-S showed positive agreement of 92.3 % (95 %CI, 63.97–99.81 %) for 15 samples compared to a vesicular stomatitis virus-based pseudo-neutralization assay [14], there are no data in peer-reviewed literature concerning Elecsys-S performance against neutralization assay(s).
This study showed that, among Elecsys-S positive convalescent-phase individuals, two-thirds contained measurable NAb levels (≥1:20) and one-third in high titers (≥1:160). Pan-anti-S1-RBD-SARS-CoV-2 antibody concentrations correlated with individual NT titer categories, and an Elecsys-S cutoff value for reasonable prediction of NAb presence after natural infection was established (133 BAU/mL). If this Elecsys-S cutoff value for reasonable prediction of NAb presence is also applicable in post-vaccination sera needs further research.
Despite substantial interest, the results of randomized trials or matched treatment–control studies of convalescent-phase plasma therapy remain inconclusive [3,15]. However, in a recent study of 3082 hospitalized patients not requiring mechanical ventilation, a dose-dependent effect relative to donors’ NAb titers was observed, with higher NAb titers associated with a lower risk of death [15]. These again emphasize a need for exact quantification of NAb in donors’ plasma. Our results confirmed a recent suggestion that, although NT should remain the gold standard for assessing convalescent-phase plasma donors, selected commercial anti-SARS-CoV-2 assays with an optimized cutoff, like Elecsys-S, could be used to maximize the positive predictive value (PPV), making it possible to select individuals with clinically relevant NAb levels as suitable donors [3,16,17].
Previous studies showed that concomitant use of two different anti-SARS-CoV-2 assays improves the PPV compared with an individual assay alone for identifying potential convalescent-phase plasma donors while maintaining the negative predictive value [1,[17], [18], [19]]. This study showed that Elecsys-S with an optimized cutoff can provide such information in less than 20 min without the need to be backed with another anti-SARS-CoV-2 assay. In addition, the high throughput of cobas e analyzers allows large-scale screening of potential donors if desirable [20].
Funding
This study was funded by the Institute of Microbiology and Immunology, Faculty of Medicine, University of Ljubljana, by the Slovenian Research Agency (grants P3-0083 and V3-2034), and by the European Virus Archive Global (EVAg) project, which has received funding from the European Union Horizon 2020 research and innovation program under grant agreement no. 653316.
CRediT authorship contribution statement
Katarina Resman Rus: Methodology, Validation, Data curation, Investigation, Writing - original draft, Writing - review & editing. Miša Korva: Methodology, Validation, Writing - review & editing. Nataša Knap: Methodology, Validation, Writing - review & editing. Tatjana Avšič Županc: Methodology, Validation, Writing - review & editing, Supervision. Mario Poljak: Conceptualization, Methodology, Writing - original draft, Writing - review & editing, Supervision.
Declaration of Competing Interest
The authors declare no conflicts of interest.
Acknowledgements
We thank Robert Krošelj and Maja Accetto Kos for their excellent laboratory assistance.
References
- 1.Turbett S.E., Anahtar M., Dighe A.S., Garcia Beltran W., Miller T., Scott H., et al. Evaluation of three commercial SARS-CoV-2 serologic assays and their performance in two-test algorithms. J. Clin. Microbiol. 2020;59:e01892–20. doi: 10.1128/JCM.01892-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Watson J., Richter A., Deeks J. Testing for SARS-CoV-2 antibodies. BMJ. 2020;370:m3325. doi: 10.1136/bmj.m3325. [DOI] [PubMed] [Google Scholar]
- 3.Hanson K.E., Caliendo A.M., Arias C.A., Englund J.A., Hayden M.K., Lee M.J., et al. Infectious Diseases Society of America guidelines on the diagnosis of COVID-19: serologic testing. Clin. Infect. Dis. 2020:ciaa134. doi: 10.1093/cid/ciaa1343. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Theel E.S., Couturier M.R., Filkins L., Palavecino E., Mitchell S., Campbell S., et al. Application, verification, and implementation of SARS-CoV-2 serologic assays with emergency use authorization. J. Clin. Microbiol. 2020;59:e02148. doi: 10.1128/JCM.02148-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ju B., Zhang Q., Ge J., Wang R., Sun J., Ge X., Yu J., et al. Human neutralizing antibodies elicited by SARS-CoV-2 infection. Nat. Cell Biol. 2020;584:115–119. doi: 10.1038/s41586-020-2380-z. [DOI] [PubMed] [Google Scholar]
- 6.Mekonnen D., Mengist H.M., Derbie A., Nibret E., Munshea A., He H., et al. Diagnostic accuracy of serological tests and kinetics of severe acute respiratory syndrome coronavirus 2 antibody: a systematic review and meta-analysis. Rev. Med. Virol. 2020:e2181. doi: 10.1002/rmv.2181. In press. [DOI] [PubMed] [Google Scholar]
- 7.Schaffner A., Risch L., Aeschbacher S., Risch C., Weber M.C., Thiel S.L., et al. Characterization of a pan-immunoglobulin assay quantifying antibodies directed against the receptor binding domain of the SARS-CoV-2 S1-Subunit of the spike protein: a population-based study. J. Clin. Med. 2020;9 doi: 10.3390/jcm9123989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Poljak M., Korva M., Knap Gašper N., Fujs Komloš K., Sagadin M., Uršič T., et al. Clinical evaluation of the cobas SARS-CoV-2 test and a diagnostic platform switch during 48 hours in the midst of the COVID-19 pandemic. J. Clin. Microbiol. 2020;58:e00599–20. doi: 10.1128/JCM.00599-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mattiuzzo G., Bentley E.M., Hassall M., Routley S., Richardson S., Beranasconi V., et al. World Health Organization; Hertfordshire: 2020. Establishment of the WHO International Standard and Reference Panel for anti-SARS-CoV-2 Antibody.https://www.who.int/publications/m/item/WHO-BS-2020.2403 [Google Scholar]
- 10.Rijkers G., Murk J.L., Wintermans B., van Looy B., van den Berge M., Veenemans J., et al. Differences in antibody kinetics and functionality between severe and mild severe acute respiratory syndrome coronavirus 2 infections. J. Infect. Dis. 2020;222:1265–1269. doi: 10.1093/infdis/jiaa463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Armstrong R.A. When to use the Bonferroni correction. Ophthalmic Physiol. Opt. 2014;34:502–508. doi: 10.1111/opo.12131. [DOI] [PubMed] [Google Scholar]
- 12.Lee W.T., Girardin R.C., Dupuis A.P., Kulas K.E., Payne A.F., Wong S.J., et al. Neutralizing antibody responses in COVID-19 convalescent sera. J. Infect. Dis. 2021;223:47–55. doi: 10.1093/infdis/jiaa673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Harley K., Gunsolus I.L. Comparison of the clinical performances of the abbott alinity IgG, abbott architect IgM, and Roche elecsys total SARS-CoV-2 antibody assays. J. Clin. Microbiol. 2020;59:e02104–02120. doi: 10.1128/JCM.02104-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Muench P., Jochum S., Wenderoth V., Ofenloch-Haehnle B., Hombach M., Strobl M., et al. Development and validation of the Elecsys Anti-SARS-CoV-2 immunoassay as a highly specific tool for determining past exposure to SARS-CoV-2. J. Clin. Microbiol. 2020;58 doi: 10.1128/jcm.01694-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Joyner M.J., Carter R.E., Senefeld J.W., Klassen S.A., Mills J.R., Johnson P.W., et al. Convalescent plasma antibody levels and the risk of death from Covid-19. NEJM. 2021;384:1015–1027. doi: 10.1056/NEJMoa2031893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bal A., Trabaud M.A., Fassier J.B., Rabilloud M., Saker K., Langlois-Jacques C., et al. Six-month antibody response to SARS-CoV-2 in healthcare workers assessed by virus neutralisation and commercial assays, in press. Clin. Microbiol. Infect. 2021 doi: 10.1016/j.cmi.2021.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zilla M., Wheeler B.J., Keetch C., Mitchell G., McBreen J., Wells A., et al. Variable performance in 6 commercial SARS-CoV-2 antibody assays may affect convalescent plasma and seroprevalence screening. Am. J. Clin. Pathol. 2020;155:343–353. doi: 10.1093/ajcp/aqaa228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hicks S.M., Pohl K., Neeman T., McNamara H.A., Parsons K.M., He J.S., et al. A dual antigen ELISA allows the assessment of SARS-CoV-2 antibody seroprevalence in a low transmission setting. J. Infect. Dis. 2020;223:10–14. doi: 10.1093/infdis/jiaa623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lisboa Bastos M., Tavaziva G., Abidi S.K., Campbell J.R., Haraoui L.P., Johnston J.C., et al. Diagnostic accuracy of serological tests for covid-19: systematic review and meta-analysis. BMJ. 2020;370:m2516. doi: 10.1136/bmj.m2516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Manthei D.M., Whalen J.F., Schroeder L.F., Sinay A.M., Li S.H., Valdez R., et al. Differences in performance characteristics among four high-throughput assays for the detection of antibodies against SARS-CoV-2 using a common set of patient samples. Am. J. Clin. Pathol. 2020;155:267–279. doi: 10.1093/ajcp/aqaa200. [DOI] [PMC free article] [PubMed] [Google Scholar]