Abstract
Deep convolutional neural network (DCNN) has shown great success in various medical image segmentation tasks, including organ-at-risk (OAR) segmentation from computed tomography (CT) images. However, most studies use the dataset from the same source(s) for training and testing so that the ability of a trained DCNN to generalize to a different dataset is not well studied, as well as the strategy to address the issue of performance drop on a different dataset. In this study we investigated the performance of a well-trained DCNN model from a public dataset for thoracic OAR segmentation on a local dataset and explored the systematic differences between the datasets. We observed that a subtle shift of organs inside patient body due to the abdominal compression technique during image acquisition caused significantly worse performance on the local dataset. Furthermore, we developed an optimal strategy via incorporating different numbers of new cases from the local institution and using transfer learning to improve the accuracy and robustness of the trained DCNN model. We found that by adding as few as 10 cases from the local institution, the performance can reach the same level as in the original dataset. With transfer learning, the training time can be significantly shortened with slightly worse performance for heart segmentation.
INTRODUCTION
Approximately 60 percent of cancer patients are treated with radiation at some time during their course of treatment [1]. As early detection and better treatment has increased the cancer patient survival rate, the importance of protecting normal organs to reduce long term toxicity has gained more attention. Defining organs-at-risk (OARs) by correctly segmenting them from simulation computed tomography (CT) scans is a critical task for radiation oncologists when aiming to optimize the benefit of radiation therapy, with delivery of the maximum dose to the tumor while sparing healthy tissues. Automatic segmentation has the potential to significantly improve the accuracy and reliability while reduce the human efforts for this task [2–3]. In recent years, deep learning (DL) methods using deep convolution neural networks (DCNN) have demonstrated superior performance in multiple OAR segmentation tasks. DCNNs were used to classify voxels in extracted patches to segment OARs [4] and clinical target volumes [5] in head and neck CT images. A dense V-network, which based on the popular U-Net structure [6] but with dense blocks [7], was developed for automatic multi-organ segmentation on abdominal CT [8]. A fully convolutional network with boundary sensitive representation was developed to increase the performance at boundaries for male pelvic organ segmentation [9]. A shape representation model constrained DCNN was developed to utilize the shape information for multi-organ segmentation from head and neck CT [10]. Dilated DCNNs with dilated convolutions were used for clinical target volume and OARs in the planning CT for rectal cancer [11]. A few other studies were also using modified network structure and/or combination with traditional image processing algorithms to improve the segmentation performance [12–16]. Furthermore, to obtain a fair comparison among different algorithms, challenges were often organized in which the same training and testing data sets were provided to all participants. In 2017, the American Association of Physicists in Medicine (AAPM) organized a thoracic auto-segmentation challenge and showed that all top 3 methods were using DCNNs and yielded statistically better results than the rest, including atlas based and other deep learning methods [17]. In addition, the performance achieved by the top 3 methods was compared with human expert contours and found to be “within human contour variation”. As one participating team for this challenge, we developed an optimized DCNN method by using the cropped images and achieved the 2nd place in the live phase and 1st place in the ongoing phase [18].
Despite the superior performance of DCNNs as reported in the literature, one issue that is often overlooked is the ability of the trained networks to generalize to completely unseen datasets, especially if there are systemic differences due to imaging modalities, protocols and practices [19–20]. In the 2017 AAPM challenge, all cases were from the same 3 institutions with roughly equal distribution between the training and testing cases. However, when we directly deployed the trained model to randomly selected clinical cases in our institution (University of Kentucky (UK)), we observed that the segmentation accuracy was much worse compared with the testing cases from the challenge. In particular, heart segmentation completely failed as it extended to the abdomen in many cases. One possible reason for the discrepancy in performance is the subtle differences in CT appearance compared between the local and the challenge dataset. Although the most effective solution for this issue is to add more cases to the training set, the impact of the number of cases and the selection criteria is not well studied. Furthermore, transfer learning [21–22], in which the part or even the whole DCNN can be initialized from another network to reduce the training effort and/or improve the performance, has not been studied in such a scenario. In this study, we first analysed the differences among the datasets and tested the solution of adding cases with different numbers and selection criteria, such as using the cases with the worst performance from the previous network. We also implemented a transfer learning strategy and compared its performance against the traditional method of training from scratch. The segmentation performance was evaluated using contour comparison metrics as well as the time taken by physician to review and modify.
MATERIALS AND METHODS
Segmentation Framework
3D U-Net structure [23] was used for building the thoracic OAR segmentation network. However, the size of clinical CT volumes is often too big to fit into the graphic processing unit (GPU) memory in typical hardware and it’s also inefficient to use the entire volume to segment one organ. To overcome this issue, we utilized a multi-stage process. First, the original CT was down-sampled to fit into GPU memory. A 3D U-Net (localizer) was trained to segment organs in the down-sampled CT. Bounding box for each organ was then generated on the original CT volume. Then, for each organ, a separate 3D U-Net network (segmenter) was trained to provide accurate contouring on high resolution CT. Augmentation and ensembling were used to improve the performance. A detailed description of the segmentation method can be found in our previous paper [18].
Datasets
Two datasets were used in this study: 2017 AAPM Thoracic Auto-segmentation Challenge (challenge dataset) and a private dataset from UK (UK dataset). The challenge dataset contains 60 thoracic CT scans from three clinical sites (MD Anderson Cancer Center (MDACC), Memorial Sloan-Kettering Cancer Center (MSKCC) and MAASTRO clinic), with 20 cases from each institution. The dataset contains CT volumes acquired with different motion management protocols, including average intensity projection of the 4DCT (MDACC), exhale phase of 4DCT (MAASTRO), and free-breathing (MSKCC) contrast-enhanced CT. The datasets were divided into three groups with 36 for training, 12 for offline testing, and 12 for online testing. Each group has 1:1:1 ratio from the three clinical sites.
The UK dataset contains 45 randomly selected thoracic CT scans. The institutional review board (IRB) approval was obtained for retrospective clinical data usage for research. Abdominal compression technique was used for motion management on most patients. CTs were acquired with a GE Lightspeed-16 scanner at 120 kVp. Clinically accepted contours were quality checked to ensure the adherence to the Radiation Therapy Oncology Group (RTOG) 1106 contouring guidelines, as followed by the challenge dataset. The UK dataset was further divided into 30 cases for enhancing the network performance with re-training and 15 for final evaluation.
Dataset Comparison Metrics
Since the model trained with challenge dataset performed much worse in UK dataset, it is likely that the UK dataset contained features that were not found in challenge dataset. The difference may come from the CT scanner setting (kernel, post processing, kVp) that affected image contrast or come from the motion management technique (abdominal compression) that affected the anatomic relationship. Since our CT is routinely calibrated to ensure that water has 0 Hounsfield unit (HU), and our mis-segmentation happens primarily on heart, we computed the HU histogram in heart and used the histogram peak position and full-width-half-maximum of the peak as the comparison. The bin size used for histogram calculation was 10 HU.
Additionally, we used the heart offset, defined as the distance between the top of the right lung and the bottom of the heart in superior-inferior (SI) direction, and the heart SI size, defined as the distance between the superior and inferior end of the heart contour, to evaluate the anatomic changes. Fig. 1 shows the heart offset and heart SI size as the anatomical markers.
Figure 1.

Anatomic information of the heart. Heart offset and heart SI size are defined as illustrated to compare the anatomical changes between datasets.
Contour Evaluation Metrics
We used Dice similarity coefficient (DSC) [26, 27], mean surface distance (MSD) and 95% Hausdorff distance (HD95) to evaluate the agreement between the auto-segmented contours and the ground-truth contours. The DSC is calculated as:
| (1) |
where X and Y are the ground truth and the algorithm segmented contours, respectively. The directed average Hausdorff measure is the average distance of a point in X to its closest point in Y, given as
| (2) |
The mean surface distance (MSD) is then defined as the average of the two directed average Hausdorff measures:
| (3) |
The 95% directed percent Hausdorff measure is the 95th percentile distance over all distances from points in X to their closest point in Y. Denoting the 95th percentile as Rank95, this is given as:
| (4) |
The undirected 95% Hausdorff distance (HD95) is then defined as the average of the two directed distances:
| (5) |
Spinal cord and esophagus are long, tubular structures that span across many slices. However, in our clinical practice, they were often only contoured in slices that were relevant to the treatment. Therefore, when evaluating auto-segmented contours against the clinical accepted contours for these two organs, we only consider those slices that have been contoured in clinical plan.
These metrics provide an objective way to evaluate the relative performance for different models and training schemes. However, it is difficult to assess the quality of the segmentation in the context of clinical practice based on the metric scores alone. Human contour variability can vary with organs, as well as the impact to clinical practice of these evaluation metrics. In addition to the usage of inter-rater variability collected by challenge organizers as the reference, we also recorded the time taken by expert physicians to review and edit the auto-segmented contour as a metric to evaluate whether the model is ready for clinical use.
Model Training and Testing with UK Dataset
To update the model with new cases, we followed the same procedure as in our previous study. The model was first re-trained from scratch with a learning rate of 0.0005 and epochs of 200 for both the localizer and segmenter networks. Alternatively, for the transfer learning experiment, we first loaded the network weights from the original model and fine-tuned the network using the new dataset. As all layers may contribute to the segmentation accuracy, no layer was “frozen” during the fine-tuning process; instead, the same loss was back-propagated to each layer. As the Adam optimizer used in the training process can automatically adapt the learning rate [24], we used the same learning rate of 0.0005; however, the number of epochs were reduced to 100 as the network was initialized with meaning parameters. To reduce overfitting, random 3D translation, rotation and scaling were applied to the input images and the corresponding ground-truth label maps as data augmentation. First, all 30 UK training cases were mixed with the challenge data to train a model from scratch and with transfer learning from the original model, respectively. We then deployed the original model on the 30 UK training cases and ranked them based on the overall segmentation accuracy using the same scoring method as in the challenge. The best performing 10 cases, worst performing 10 cases, and the worst performing 20 cases were mixed with the challenge data respectively to train the corresponding models in order to evaluate the impact of number of added cases on the model performance as well as whether neural network learns more effectively from the cases it did poorly previously. To obtain a fair comparison, all the three models were trained from scratch.
RESULTS
Dataset Comparison
Fig. 2a and 2b show the comparison of two datasets in terms of image contrast and anatomic positions of heart. The challenge data showed greater variations in HU peak position and width as the data came from 3 different institutions, with one of the institution’s data contained a few contrast-enhanced CT. The image contrast variations in UK data were much smaller and were well within the range of the challenge data used for training. For anatomic positions, the UK data showed statistically significant difference with the challenge data in both the heart position (p=0.0036) and the heart size (p=0.0056) in S-I direction. The heart in UK data is smaller in S-I direction and more superior in position relative to the top of the lung. As indicated in the median position in Fig. 2b, more than 50% of the UK data are located below the lower quartile mark of the challenge data. This is likely the reason why the model trained using only the challenge data did not perform well on UK data.
Figure 2.

Comparisons of image contrast and anatomical position for heart between challenge and UK datasets. HU Peak Pos: HU value corresponding to the peak on HU histogram for heart; HU Peak Width: the full-width-half-maximum of the histogram peak; Max Lung_Heart SI: distance between the top of the right lung and the bottom of the heart in superior-inferior (SI) direction; Heart SI: the span of heart contour in SI direction.
To further investigate the reason for systematic differences in heart location and size, we compared the sagittal images from the two datasets. Fig.3 shows the example cases and indicates that the differences originate from the different motion management techniques. In UK, most of our patients underwent abdominal compression so that the heart was pushed higher in thoracic cavity and slightly compressed in the S-I direction.
Figure 3.
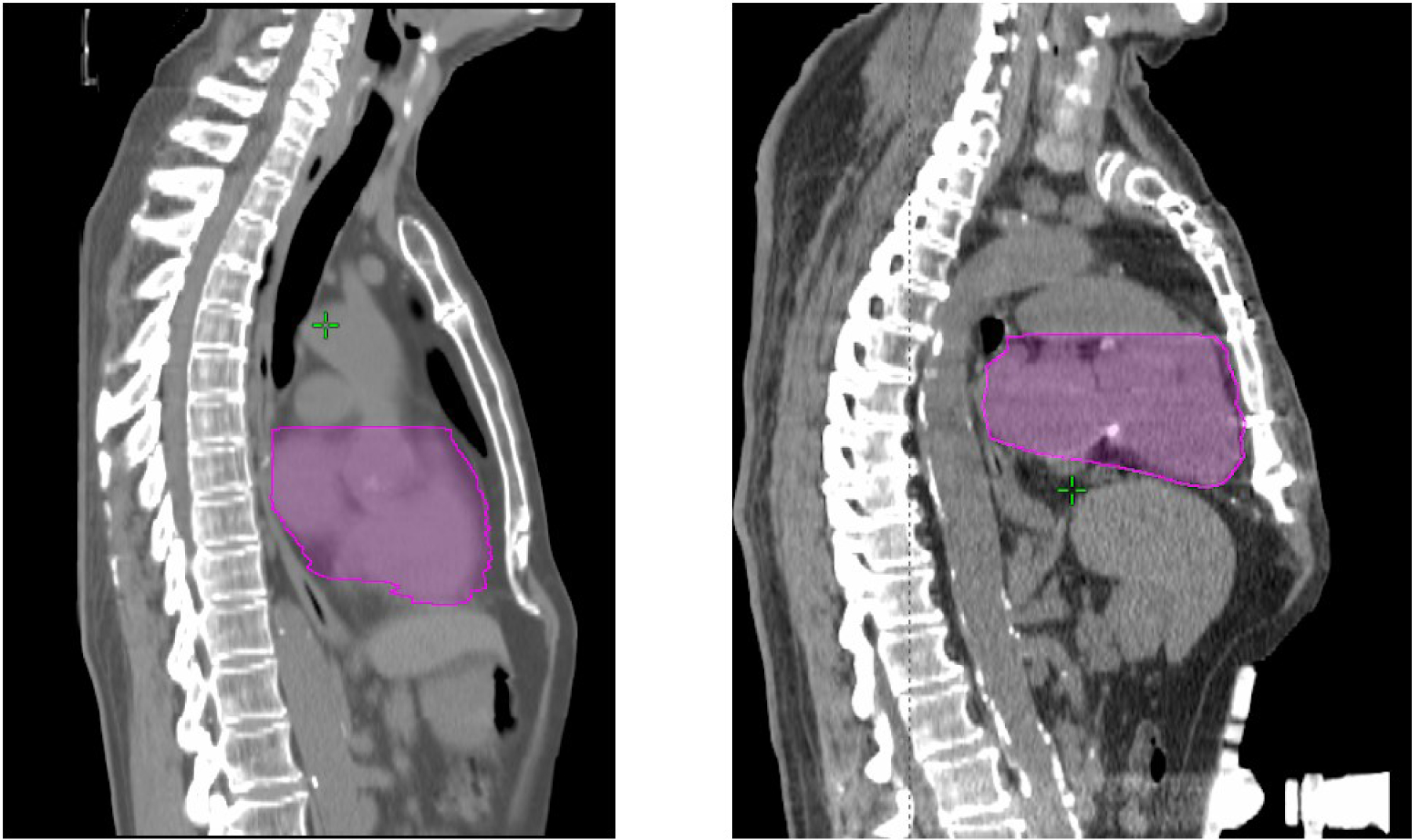
Example cases from the challenge and UK dataset. Left: Challenge case, Right: UK case. The belly compression device is visible in the UK case.
Model Training and Testing with UK Dataset
Fig. 4 shows the DSC, MSD and HD95 achieved on the 15 UK test dataset using the original model (UKorig) and the model trained from scratch with 30 UK cases added to the original challenge training dataset (UKadd30). For a better comparison against the performances achieved in the challenge testing dataset and the inter-observer variability from the experiment conducted by the challenge organizers, the mean and standard deviation of DSC, MSD and HD95 were listed in Table 1, 2 and 3, respectively. The metrics that are better than human expert’s contouring variability are highlighted in bold. When the original model was directly applied to UK data, a large drop in contouring accuracy was observed in heart. As shown in Fig. 4a, while the median DSC is still greater than 0.9, the distribution is heavily tilted to lower scores, with a few cases showing almost completely failed segmentation (DSC < 0.5). The top quantile for HD95 and MSD for heart were much worse as well. Spinal cord contouring accuracy also decreased slightly. The esophagus showed a slightly better performance compared with the challenge, which is likely due to that the esophagus in the UK data is slightly easier to segment. After the model trained with the inclusion of UK data, the performance for all ROIs were improved. The performances in heart and spinal cord increased to similar levels achieved in challenge. Esophagus performance also increased. Overall, 3 of the 5 organs segmented have performances significantly better than human expert’s contouring variability and the remaining 2 organs achieved performances comparable with expert’s contour variability.
Figure 4.
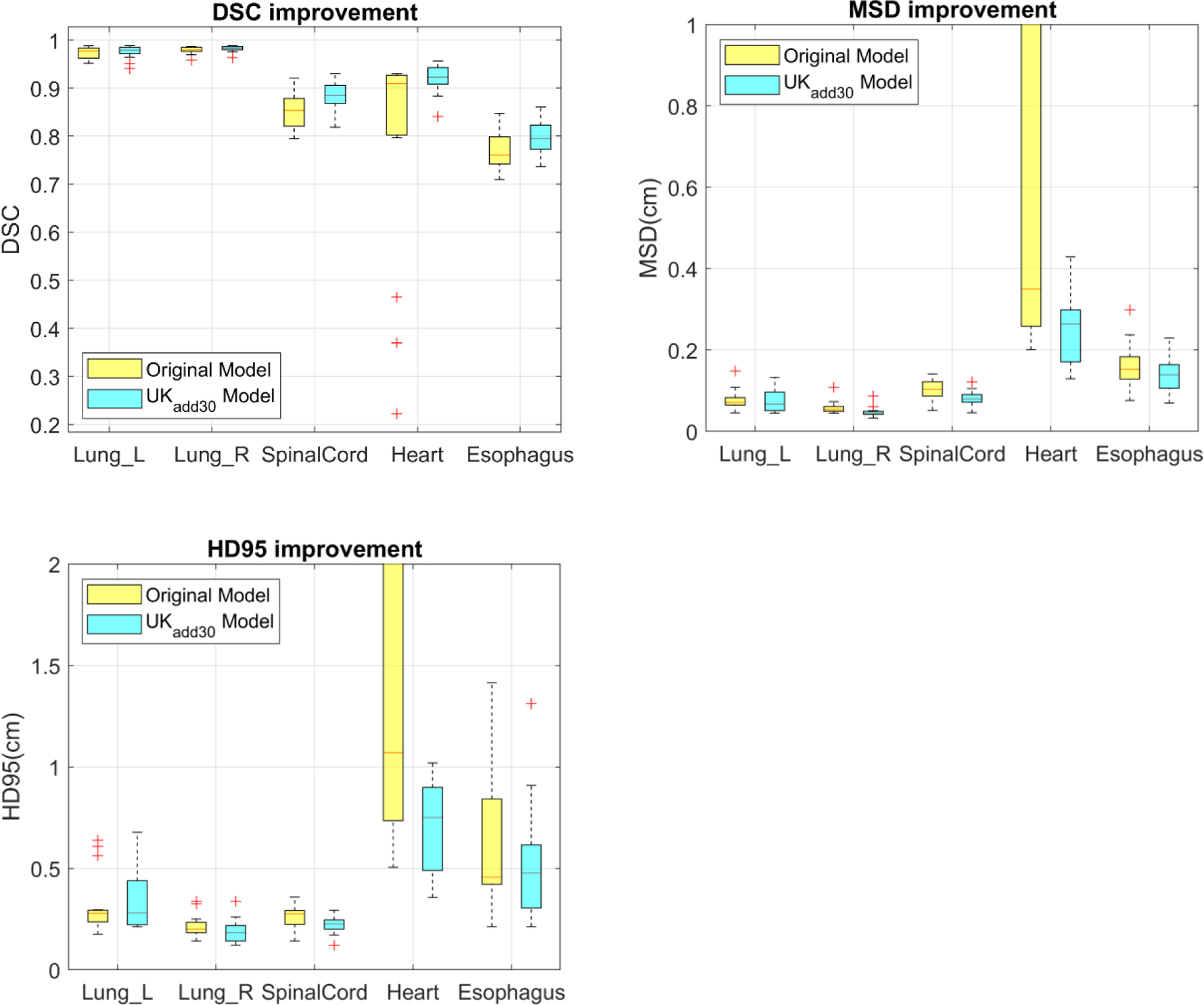
Improvements of DSC, MSD and HD95 after 30 UK cases were added to training. This shows the original model and the model trained from scratch.
Table 1.
DSC achieved by the original model (UKorig) and improved model by adding 30 UK cases and trained from scratch and using transfer learning. The inter-observer value and evaluated on the challenge test cases were also shown as a comparison. Values in Bold indicates better than expert contouring variability.
| Inter observer | Challenge | UKorig | UKadd30 (scratch) | UKadd30 (transfer) | |
|---|---|---|---|---|---|
| SpinalCord | 0.862 | 0.89±0.04 | 0.85±0.04 | 0.88±0.03 | 0.89±0.02 |
| Lung_R | 0.955 | 0.97±0.02 | 0.97±0.01 | 0.98±0.01 | 0.98±0.01 |
| Lung_L | 0.956 | 0.98±0.01 | 0.98±0.01 | 0.98±0.01 | 0.98±0.01 |
| Heart | 0.931 | 0.93±0.02 | 0.79±0.23 | 0.92±0.03 | 0.91±0.09 |
| Esophagus | 0.818 | 0.73±0.09 | 0.77±0.04 | 0.80±0.04 | 0.78±0.04 |
Table 2.
MSD in mm achieved by the original model (UKorig) and improved model by adding 30 UK cases and trained from scratch and using transfer learning. The inter-observer value and evaluated on the challenge test cases were also shown as a comparison. Values in Bold indicates better than expert contouring variability.
| Inter observer | Challenge | UKorig | UKadd30 (scratch) | UKadd30 (transfer) | |
|---|---|---|---|---|---|
| SpinalCord | 0.88 | 0.7±0.3 | 1.0±0.3 | 0.8±0.2 | 0.7±0.1 |
| Lung_R | 1.87 | 0.9±0.6 | 0.8±0.3 | 0.7±0.3 | 0.7±0.2 |
| Lung_L | 1.51 | 0.6±0.3 | 0.6±0.2 | 0.5±0.1 | 0.5±0.1 |
| Heart | 2.21 | 2.3±0.5 | 9.5±11.8 | 2.5±0.8 | 3.0±1.7 |
| Esophagus | 1.07 | 2.3±2.4 | 1.6±0.5 | 1.4±0.4 | 1.5±0.4 |
Table 3.
HD95 in mm achieved by the original model (UKorig) and improved model by adding 30 UK cases and trained from scratch and using transfer learning. The inter-observer value and evaluated on the challenge test cases were also shown as a comparison. Values in Bold indicates better than expert contouring variability.
| Inter observer | Challenge | UKorig | UKadd30 (scratch) | UKadd30 (transfer) | |
|---|---|---|---|---|---|
| SpinalCord | 2.38 | 1.9±0.6 | 2.5±0.6 | 2.2±0.4 | 2.1±0.3 |
| Lung_R | 6.71 | 4.0±2.9 | 3.2±1.5 | 3.5±1.6 | 2.9±1.3 |
| Lung_L | 5.17 | 2.1±0.9 | 2.2±0.5 | 1.8±0.6 | 1.9±0.6 |
| Heart | 6.42 | 6.6±1.5 | 26.7±28.8 | 7.1±2.2 | 10.0±8.1 |
| Esophagus | 3.33 | 8.7±11 | 6.2±3.4 | 5.2±3.0 | 5.3±2.4 |
The tables also show the comparison between training using transfer learning and from scratch. The overall performance is similar; however, for heart segmentation, although the mean DSC values are comparable, there are two cases with suboptimal performances, causing a substantial increase in its standard deviation and the two distance measures. This indicates that although transfer learning can significantly reduce the training time, it is not as effective as training from scratch in terms of “forgetting” the previous model as the failure mode in these two cases with transfer learning is similar as the previous model.
Fig. 5 shows examples of heart and spinal cord segmentation before and after the inclusion of local data in training. The re-training was performed from scratch. As in Fig. 5a, the main issue associated with heart was gross mis-segmentation that included part of liver with the original model. This problem was fixed after inclusion of local data in training (Fig. 5b). For spinal cord, the segmentation with original model was clinically acceptable as shown in Fig. 5c. However, due to the different contouring criteria used in the local ground truth, contouring evaluation metrics showed a slightly poorer score. At our institution, the dosimetrist prefers to contour spinal cord using a circular paintbrush sized appropriately to fit inside the spinal canal. Expansions from the circle are used conservatively. After the inclusion of local data in training, the model learnt this practice and produced segmentation that agrees with local ground truth better as illustrated in Fig. 5d.
Figure 5.
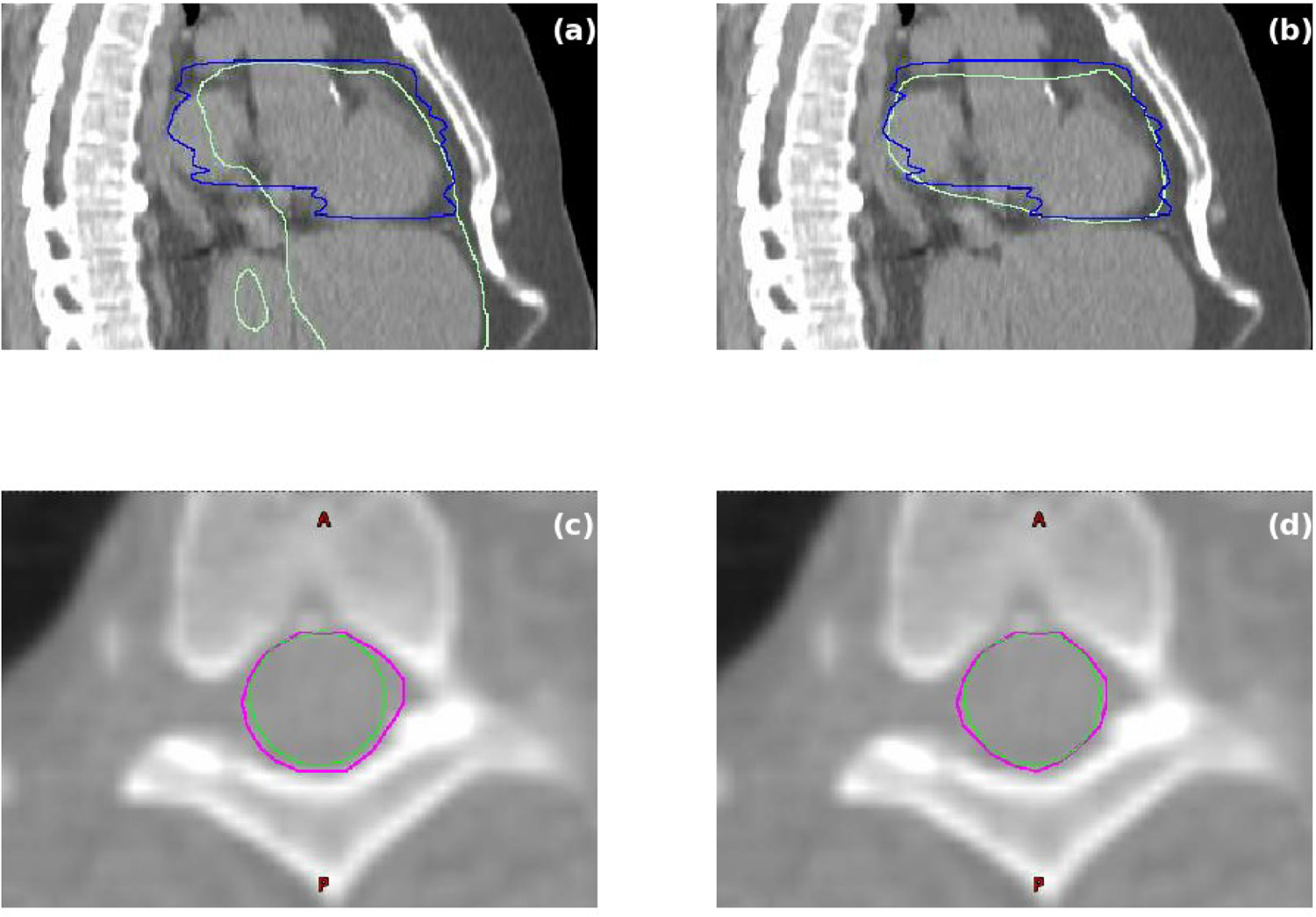
Examples of heart and spinal cord segmentation before and after the inclusion of local data in training. (a) Heart segmentation before the inclusion of local data. (b) Heart segmentation after the inclusion of local data. Blue curves are ground truth heart segmentation. Light green curves are auto-segmentation results. (c) Spinal cord segmentation before the inclusion of local data. (d) Spinal cord segmentation after the inclusion of local data. Green curves are ground truth segmentation. Magenta curves are auto-segmentation results.
The auto segmented contours were then evaluated by an expert physician. The review and editing time taken to produce clinical accepted contours was recorded. The contours from the original model required 7.5±3.8 mins for each case. After adding 30 local cases for re-training from scratch, the review and edit time for each case was reduced to 2.7±1.0 mins.
Fig. 6 shows the improvement of DSC and HD95 metrics in heart, esophagus and spinal cord as the amount and quality of the local data added to the training varied. All models were re-trained from scratch. All organs showed improvements in contouring accuracy and robustness with only 10 worst performing local cases of the original model added to the existing 36-case training dataset (addL10). Further improvements with more local cases (addL20 and add30) were limited. As a comparison, adding 10 best performing cases to the training dataset (addH10) is slightly inferior to the approach of adding 10 worst performing cases (addL10) for esophagus and spinal cord. However, for heart where the most improvement was observed, no such discrepancy in performance could be observed, indicating that the model can adapt well to the systematic differences of the data from multiple institutions with only a few cases.
Figure 6.

Contouring accuracy improvements as the amount and quality of local data used in training varied.
DISCUSSION
We studied the generalizability of DCNN model for thoracic OAR segmentation. We discovered that the drop-off in performance of the original model when applied to the data from our institution is due to the belly compression technique we adopt in our patient simulation, causing systematic differences in the datasets. This performance drop-off highlights the vulnerability of DCNN as it heavily relies on the training data. While it is easy for human to handle this subtle change in anatomy, DCNN often have problems dealing with even simple transformations [23]. DCNN usually has multi-million parameters and those parameters are determined (learned) from the limited examples (training data) provided. Even though data augmentation is performed to prevent overfitting, the variation of image appearance as well as anatomy is still very limited. When presented with a case that is much different from training, it is highly likely that the output from the DCNN is unexpected, as evident from the outlier cases on heart segmentation observed.
While implement data augmentations that simulate all variations in image appearance and anatomy is a possible solution, it is challenging to develop such algorithms to accurately simulate real patient CTs without introducing physically unrealistic distortion artefacts, especially in thoracic regions as different techniques are used to handle respiratory motion. In addition, augmentation only works if we can identify the subtle difference between datasets, which is often difficult to do. An easier solution is to incorporate the local cases into the training of the DCNN and let DCNN learn to handle the subtle differences in datasets. In addition, although consensus guidelines have reduced the contouring variability among physicians and clinics, variations and personal preferences still exists. Adding local cases into the training of the DCNN can also help DCNN learn to adapt to these local contouring preferences, as demonstrated by the improvement in cord contouring accuracy in our study. Our results showed that adding local cases into DCNN training can dramatically improve the accuracy and robustness of the DCNN applied on local data. We also demonstrated that as few as 10 local cases were sufficient in enabling the DCNN to learn those subtle features, despite that the majority of the training data is from other institutions and thus lack those features.
Human usually learns effectively from their mistakes, by reviewing the cases they did poorly against the ground truth. However, our preliminary test showed that, providing DCNN cases that it performed poorly as training data does not have significant advantage over providing it cases that it performed relatively well. This indicates that the DCNN may learn things differently from human being. It also suggests that we don’t have to pay special attention to pick particular cases for training as long as they all contain the desired image or anatomical features.
When comparing the transfer learning and the learning from scratch strategy, our study showed that although transfer learning can significantly reduce the training time, as it trains from the existing model, the performance is slightly reduced, especially for the organ that the previous model failed the worst, even with the same dataset. This is likely due to that the transfer learning is more prone to local minima that is roughly determined by the previous model and therefore may inherit more errors from it. As a comparison, learning from scratch eliminates the previous model and can lead to more robust results.
There are several limitations of this study. First, although we aim to study the systematic differences from multi-institutional environment, we only validated the performance in our local institute with a relatively small dataset (45 cases) and therefore limited the scope of this study. However, we also demonstrated a successful solution and evaluated the quantitative impact of the number of cases as well as the utilization of transfer learning. The encouraging results will lead to continued studies on multiple institutions. The second limitation is we did not fully explore different options of transfer learning, such as freezing multiple layers. However, empirically we hypothesize that it will not change the conclusion, or can make the case even worse as freezing layers further limit the parameter updates.
CONCLUSIONS
We have demonstrated that a DCNN segmentation model trained on public dataset did not perform well on our institution’s data due to the difference in clinical practice. Re-training the model with local cases added to the training data successfully resolved the problem, in which re-training from scratch is slightly more effectiveness than transfer learning in terms of performance enhancement. We have also found that it didn’t take too many local cases for the DCNN to learn about the difference in the underlying patient anatomy. In addition, we did not observe any advantage of collecting cases that DCNN performed poorly as training data. Our study showed that the DCNN based segmentation can overcome the problem of data heterogeneity across different clinical practice with fairly small effort.
ACKNOWLEDGMENTS
The authors would like to thank Nvidia for donating a GPU card that was used in this study. This work was partially supported by NIH Grant R43EB027523.
References
- 1.Miller KD, Siegel RL, Lin CC, Mariotto AB, Kramer JL, Rowland JH, et al. Cancer treatment and survivorship statistics, 2016. CA Cancer J Clin. 2016;66(4):271–89. [DOI] [PubMed] [Google Scholar]
- 2.Sharp G, Fritscher KD, Pekar V, Peroni M, Shusharina N, Veeraraghavan H, et al. Vision 20/20: perspectives on automated image segmentation for radiotherapy. Med Phys. 2014;41(5):050902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cardenas CE, Yang J, Anderson BM, Court LE, Brock KB. Advances in Auto-Segmentation. Semin Radiat Oncol. 2019;29(3):185–197. [DOI] [PubMed] [Google Scholar]
- 4.Ibragimov B, Xing L. Segmentation of organs-at-risks in head and neck CT images using convolutional neural networks. Med Phys. 2017;44(2):547–557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cardenas CE, Anderson BM, Aristophanous M, Yang J, Rhee DJ, McCarroll RE, et al. Auto-delineation of oropharyngeal clinical target volumes using 3D convolutional neural networks. Phys Med Biol. 2018;63(21):215026. [DOI] [PubMed] [Google Scholar]
- 6.Ronneberger O, Fischer P, Brox T. U-Net: Convolutional networks for biomedical image segmentation. arXiv preprint. arXiv:1505.04597. 2015. [Google Scholar]
- 7.Huang G, Liu Z, van der Maaten L, Weinberger KQ. Densely connected convolutional networks. arXiv preprint. arXiv:1608.06993. 2016. [Google Scholar]
- 8.Gibson E, Giganti F, Hu Y, Bonmati E, Bandula S, Gurusamy K, et al. Automatic Multi-Organ Segmentation on Abdominal CT With Dense V-Networks. IEEE Trans Med Imaging. 2018;37(8):1822–1834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang S, He K, Nie D, Zhou S, Gao Y, Shen D. CT male pelvic organ segmentation using fully convolutional networks with boundary sensitive representation. Med Image Anal. 2019;54:168–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tong N, Gou S, Yang S, Ruan D, Sheng K. Fully automatic multi-organ segmentation for head and neck cancer radiotherapy using shape representation model constrained fully convolutional neural networks. Med Phys. 2018;45(10):4558–4567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Men K, Dai J, Li Y. Automatic segmentation of the clinical target volume and organs at risk in the planning CT for rectal cancer using deep dilated convolutional neural networks. Med Phys. 2017;44(12):6377–6389. [DOI] [PubMed] [Google Scholar]
- 12.Zhu W, Huang Y, Zeng L, Chen X, Liu Y, Qian Z, et al. AnatomyNet: Deep learning for fast and fully automated whole-volume segmentation of head and neck anatomy. Med Phys. 2019;46(2):576–589. [DOI] [PubMed] [Google Scholar]
- 13.Dong X, Lei Y, Wang T, Thomas M, Tang L, Curran WJ, et al. Automatic Multi-Organ Segmentation in Thorax CT Images Using U-Net-GAN. Med Phys. 2019;46(5):2157–2168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Seo H, Huang C, Bassenne M, Xiao R, Xing L. Modified U-Net (mU-Net) with Incorporation of Object-Dependent High Level Features for Improved Liver and Liver-Tumor Segmentation in CT Images. IEEE Trans Med Imaging. 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Trullo R, Petitjean C, Dubray B, Ruan S. Multiorgan segmentation using distance-aware adversarial networks. J Med Imaging (Bellingham). 2019;6(1):014001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu X, Guo S, Yang B, Ma S, Zhang H, Li J, et al. Automatic Organ Segmentation for CT Scans Based on Super-Pixel and Convolutional Neural Networks. J Digit Imaging. 2018;31(5):748–760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yang J, Veeraraghavan H, Armato SG 3rd, Farahani K, Kirby JS, Kalpathy-Kramer J, et al. Autosegmentation for thoracic radiation treatment planning: A grand challenge at AAPM 2017. Med Phys. 2018;45(10):4568–4581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Feng X, Qing K, Tustison NJ, Meyer CH, Chen Q. Deep convolutional neural network for segmentation of thoracic organs-at-risk using cropped 3D images. Med Phys. 2019;46(5):2169–2180. [DOI] [PubMed] [Google Scholar]
- 19.Azulay A, Weiss Y. Why do deep convolutional networks generalize so poorly to small image transformations? arXiv preprint. arXiv:1805.12177. 2018. [Google Scholar]
- 20.Chen C, Bai W, Davies RH, Bhuva AN, Manisty C, Moon JC, et al. Improving the generalizability of convolutional neural network-based segmentation on CMR images. arXiv preprint. arXiv: 1907.01268. 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hesamian MH, Jia W, He X, Kennedy P. Deep Learning Techniques for Medical Image Segmentation: Achievements and Challenges. J Digit Imaging. 2019;32(4):582–596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kim J, Lee HS, Song IS, Jung KH. DeNTNet: Deep Neural Transfer Network for the detection of periodontal bone loss using panoramic dental radiographs. Sci Rep. 2019;9(1):17615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cicek O, Abdulkadir A, Lienkamp SS, Brox T, Ronneberger O. 3D U-Net: Learning Dense Volumetric Segmentation from Sparse Annotation. arXiv preprint. arXiv: 1606.06650. 2016. [Google Scholar]
- 24.Kingma DP, Ba J. Adam: A method for stochastic optimization. arXiv preprint. arXiv: 1412.6980. 2014. [Google Scholar]
- 25.Engstrom L, Tran B, Tsipras D, Schmidt L, Madry A. Exploring the Landscape of Spatial Robustness. arXiv preprint. arXiv: 1712.02779. [Google Scholar]
- 26.Dice LR, Measures of the amount of ecologic association between species. Ecology, 1945. 26(3): p. 297–302 [Google Scholar]
- 27.Sorensen TA, A method of establishing groups of equal amplitude in plant sociology based on similarity of species content and its application to analyses of the vegetation on Danish commons. Biol. Skar, 1948. 5: p. 1–34. [Google Scholar]


