Abstract
Diesel exhaust (DE) is a major contributor to ambient air pollution around the world. It is a known human carcinogen that targets the respiratory system and increases risk for many diseases, but there is limited research on the effects of DE exposure on the epigenome of human bronchial epithelial cells. Understanding the epigenetic impact of this environmental pollutant can elucidate biological mechanisms involved in the pathogenesis of harmful DE-related health effects. To estimate the causal effect of short-term DE exposure on the bronchial epithelial epigenome, we conducted a controlled single-blinded randomized crossover human experiment of exposure to DE and used bronchoscopy and Illumina 450K arrays for data collection and analysis, respectively. Of the 13 participants, 11 (85%) were male and 2 (15%) were female, and 12 (92%) were White and one (8%) was Hispanic; the mean age was 26 years (SD = 3.8 years). Eighty CpGs were differentially methylated, achieving the minimum possible exact P-value of P = 2.44 × 10−4 (i.e. 2/213). In regional analyses, we found two differentially methylated regions (DMRs) annotated to the chromosome 5 open reading frame 63 genes (C5orf63; 7-CpGs) and unc-45 myosin chaperone A gene (UNC45A; 5-CpGs). Both DMRs showed increased DNA methylation after DE exposure. The average causal effects for the DMRs ranged from 1.5% to 6.0% increases in DNA methylation at individual CpGs. In conclusion, we found that short-term DE alters DNA methylation of genes in target bronchial epithelial cells, demonstrating epigenetic level effects of exposure that could be implicated in pulmonary pathologies.
Keywords: diesel exhaust, respiratory health, epigenetics, DNA methylation, EWAS
Introduction
Outdoor air pollution is a leading cause of morbidity and mortality globally, contributing to 4.2 million premature deaths worldwide in 2016 [1]. A major contributor to air pollution is diesel exhaust (DE) produced from the combustion of diesel fuel, which represents a complex mixture of chemicals. More than 40% of on-road vehicles around the world use diesel for fuel, and the global consumption of diesel has increased by eightfold since 1970 [2]. Exposure to DE is ubiquitous, as it is produced from engines in on-road and off-road vehicles, heavy equipment, and power generators. In the European Union, on-road diesel vehicles contributed to approximately 60% of transportation-attributable emissions of particulate matter <2.5 microns in diameter (PM2.5) in 2015 [3]. On a global scale, the population-weighted transportation-attributable fraction for annual average emissions of PM2.5 and ozone (O3) were 12% and 11%, respectively [3]. The widespread and growing use of diesel as fuel, especially in urban settings, poses a serious threat to public health due to its contribution to air pollution.
DE is a mixture of solid particulate matter, condensed components, and gases that are known to have negative health effects. The solid fraction of DE is primarily composed of elemental carbon (EC) and metals, and the gaseous components include nontoxic inorganic gases and organic molecules like polycyclic aromatic hydrocarbons [4]. Humans exposed to these compounds through inhalation of DE can experience adverse health effects both in localized areas of immediate exposure (i.e. the respiratory tract) and in peripheral organ systems. Exposure to DE can induce oxidative stress, DNA damage, direct cytotoxic effects, and pro-inflammatory signaling that impair the lungs [5–7]. Other organs can become affected due to the transepithelial transport of particles and gases and spread of inflammatory cytokines and biomolecules generated in the lung tissue through blood [4, 8, 9]. On a population-level, several epidemiology studies have found significant associations between occupational exposure to DE and the development of lung and bladder cancers [10–12]. Consequently, the International Agency for Research on Cancer (IARC) classified DE as a known human carcinogen (Group 1) based on sufficient evidence that exposure is associated with an increased risk for lung cancer [13].
The epigenome is at the intersection between genetics, environment, and cellular responsiveness. Epigenetics is defined as DNA modifications, other than to the DNA sequence itself, which are maintained in cell division with stable consequences to gene expression [14]. The epigenome has been shown to be sensitive to environmental toxicant exposure with the potential for both persistence and malleability of changes [15, 16]. The most widely studied epigenetic modification is the addition of a methyl (CH3) group to cytosines within CpG dinucleotides. Epigenome-Wide Association Studies (EWAS) have been widely conducted in the last decade to study diseases and the effects of environmental exposures [17]. However, the majority of human EWAS performed to date face two major challenges. First, observational and cross-sectional epidemiological studies limit the interpretation of findings due to the lack of randomization. Second, most studies analyze DNA from easily accessible tissues and fluids, such blood samples and buccal cell DNA. To overcome these two limitations, we conducted a controlled single-blinded randomized crossover experiment with human participants exposed to DE and clean air and collected target bronchial epithelial cells at each visit. Prior studies have shown that exposure to different air pollutants affects DNA methylation in both animal models and humans [18]. Therefore, we hypothesized that short-term DE would lead to changes in DNA methylation that are present within 24-h following exposure.
Materials and Methods
Study Design
The overall study design, sample, and methods have been described previously [19, 20]. Briefly, healthy participants were recruited under a contract with Westat Corporation (Rockville, MD, USA), Clinical Trial NCT01492517. All study participants were free of cardiopulmonary diseases and allergies, as determined by a detailed medical history, questionnaire, and physical examination. Participants were excluded if they were smokers or pregnant or had a forced vital capacity (FVC) or forced expiratory volume in the first second of expiration (FEV1) <80% predicted for their height and age. Participants were informed of the procedures and potential risks and provided written signed informed consent. Controlled exposures were conducted at the US Environmental Protection Agency (EPA) Human Studies Facility on the campus of the University of North Carolina (UNC), Chapel Hill. The protocol and consent forms were approved by the UNC School of Medicine Committee on the Protection of the Rights of Human Subjects and the US EPA’s Institutional Review Board.
A total of 13 healthy volunteers were randomized to three experimental conditions: DE, O3 (0.300 ppm), and clean air (CA). Results from the O3 exposure experiments have been previously published [20, 21]. The 13 healthy volunteers had a mean age (SD) of 26 years (3.8 years); 11 were male (84.6%), 2 were female (15.4%), 12 were White (92.3%), and one (3.9%) was Hispanic. Relevant to this analysis, six volunteers were exposed to DE at the first study visit, four at the second visit, and the remaining three during a third and final visit. Controlled CA exposure occurred at the first, second, and third study visits for 4, 3, and 6 of the participants, respectively (Table 1). Each study participant was exposed on separate occasions to clean air and up to 300 µg/m3 of DE for 2 h with intermittent periods of exercise in a controlled environment chamber. A minimum washout period of 13 days between exposures was implemented. The exposures were conducted at the same time of the day and on the same day of the week. The exposure chambers were maintained at 40% relative humidity for all exposures. During the 2-h exposure period, participants alternated between 15 min of rest and 15 min of exercise on a cycle ergometer. The cycle ergometer workload was adjusted so that subjects breathed at a ventilatory rate, normalized for body surface area, of 25 l min−1 m−2. DE was obtained from a diesel power generator and introduced into the exposure chamber after dilutions with clean high-efficiency particulate air, charcoal-filtered air, and humidified air to obtain a chamber concentration of up to 300 µg/m3.
Table 1:
Characteristics among participants (n = 13)
| n (%) or mean (SD) | |
|---|---|
| Sex | |
| Male | 11 (84.6%) |
| Female | 2 (15.4%) |
| Race | |
| White | 12 (92.3%) |
| Hispanic | 1 (7.7%) |
| Age (year) | 25.7 (3.8) |
| Visit order for diesel exposure | |
| First | 6 (46.2%) |
| Second | 4 (30.8%) |
| Third | 3 (23.1%) |
| Visit order for clean air exposure | |
| First | 4 (30.8%) |
| Second | 3 (23.1%) |
| Third | 6 (46.1%) |
DNA Methylation Measurements
Within 24-h post-exposure to either clean air or DE, study participants underwent a research bronchoscopy with brush biopsy to obtain bronchial epithelial cells as previously described and following a standard protocol [22, 23]. The cytology brushes containing epithelial cells tips were placed in a 1.5 ml tube with 200 μl Trizol Buffer (ThermoFisher Scientific). DNA was extracted using the Gentra Puregene Buccal Cell Kit (Qiagen, Inc.), and samples were stored frozen at −80°C before analysis. DNA extracted from the bronchial epithelial cells was sent to a commercial laboratory (Expression Analysis, Durham, NC, USA) for DNA methylation assessment using the Illumina HumanMethylation 450K BeadChip array. The extracted DNA samples were placed on four chips. We performed background correction using noob and dye bias correction as well as corrected for probe design bias arising from Type I and Type II probes with the Beta-mixture quantile normalization (BMIQ) method [24, 25]. The DNA methylation β-value, or the proportion of cytosine methylated, at 484 531 CpGs was used for analyses.
Statistical Analyses
The statistical analysis approach has been described previously [21]. Briefly, to estimate the average causal effect (ACE) of DE on the bronchial epithelial epigenome, we assumed no carry-over and no time effects and defined an estimate for the ACE: the mean of the observed participant methylation difference between diesel exposure and clean air for each CpG site (484 531 CpGs). For individual i at study visit j randomized to DE (wi,j = 1) and clean air (wi,j = 0), the observed DNA methylation difference at CpG k (di,k) is defined by the two measured outcomes:
di,k = Yi,j = 2, k(wi, j = 2 = 1) – Yi,j = 1,k (wi,j = 1 = 0) if the participant was exposed to clean air first, and
di,k = Yi,j = 1,k(wi,j = 1 = 1) – Yi,j = 2,k(wi,j = 2 = 0) if the participant was exposed to DE first.
We assume a Bernouilli assignment mechanism and calculate the univariate Fisher-exact P values based on the standard paired t-test statistic and the 213/2 possible randomizations:
With this assignment mechanism and test statistic, the minimum possible observable P-value is 2/213 or P = 2.44 × 10−4. It has been recommended to analyze randomized trials using the exact null randomization distribution, which, in small samples, can substantially differ from its approximating Student’s t distribution [26]. To test for differentially methylated regions (DMRs), we implemented Comb-p, a bioinformatics tool that groups individual CpGs by genomic location and proximity [27]. We used the Fisher-exact P values as input along with the genomic annotation. For the argument, we used a minimum distance of 500 base-pairs and required a P-value of 10−3 to start a region. For each CpG site found within a DMR, we report the ACE and the univariate Fisher-exact paired P-values (and asymptotic P values for comparison). All analyses were performed using the R statistical software.
Results
Demographic characteristics and exposure randomization order among the 13 healthy volunteers are shown in Table 1.
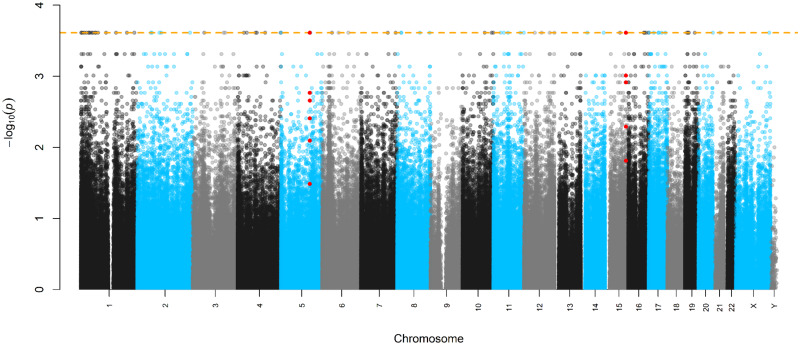
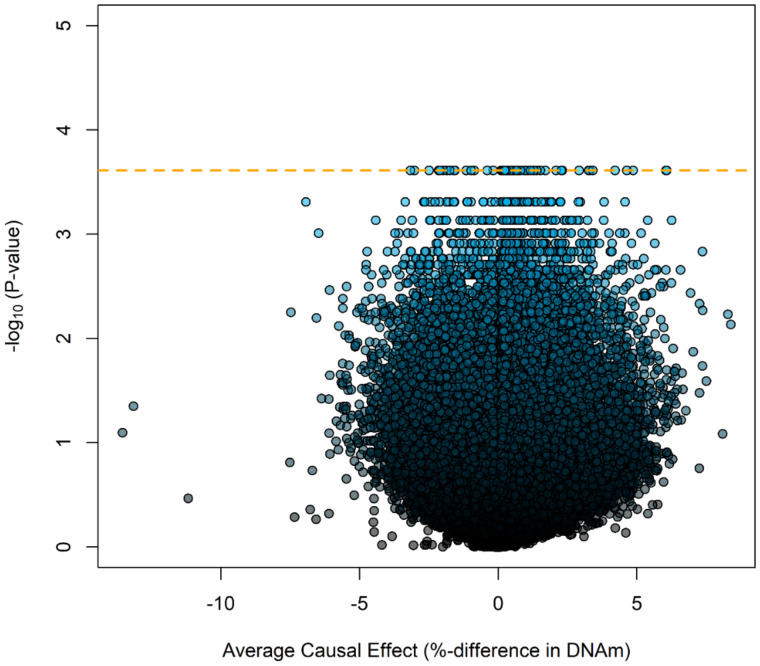
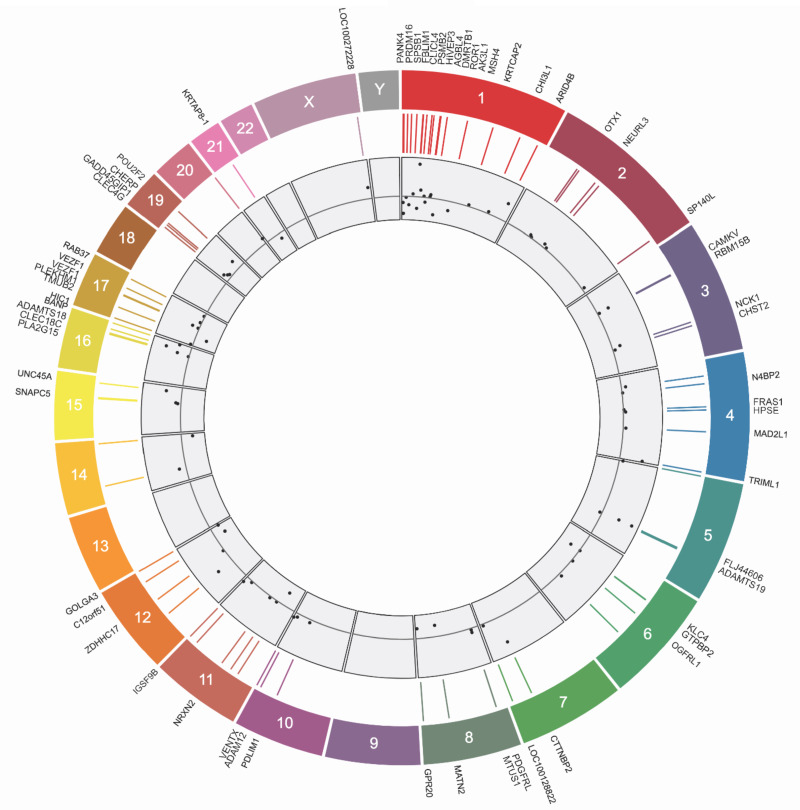
In epigenome-wide analyses, 80 CpGs achieved a minimum possible observable P-value (2/213 or P = 2.44 × 10−4) calculated from the experimental design and shown in Supplementary Table S1. A Manhattan plot of Fisher’s exact P values on the logarithmic scale (y-axis) mapped to each chromosome (x-axis) is displayed in Fig. 1. The volcano plot, displaying the Fisher’s exact P values on the logarithmic scale (y-axis) against the average causal effect (ACE) for each CpG (x-axis), is shown in Fig. 2. Mapping of the individual sites, genes, and effect sizes is displayed in a circular genomic map in Fig. 3. Of the 80 CpGs with the minimum observable P-value, 62 CpGs (77.5%) show increased DNA methylation with DE exposure relative to CA exposure. Among sites achieving the minimum Fisher-exact P-value [P-valuemin or P < (2/213)], the largest ACE was observed at cg12413579 (ACE = 6.07%), which annotated to the Proteasome Subunit Beta 2 (PSMB2) gene. When comparing Fisher-exact P values to asymptotic methods, we observed that 28 CpGs succeed the P-valuemin. Annotated results for the 80 differentially methylated sites based on Fisher-exact tests are available in the Supplementary Table S1. Manhattan, volcano, and quantile–quantile plots of asymptotic paired t-test P values are shown in Supplementary Figs S1–S3, respectively.
Figure 1:
Manhattan plot of exact P values and chromosome position. The dashed orange line is the minimum possible P-value. CpGs annotated to the DMRs of chr5; C5orf63 and chr15: UNC45A are colored in red.
Figure 2:
Volcano plot showing change in DNA methylation in bronchial cells against −log10 of exact P-values. The dashed orange line is the minimum possible P-value from an exact test.
Figure 3:
Circular map of genomic distribution among differentially methylated CpGs. From the outmost ring inward, the map shows gene names, chromosome number, position within the chromosome, and effect sizes among Average Causal Effects (below the gray line is a negative effect size and above the line is a positive effect size).
In regional DNA methylation analyses implemented with Comb-p [27] and based on the Fisher-exact P values, we observed two differentially methylated regions (DMRs). One of the regions was annotated to chromosome 5 (chr5:126 409 062–126 409 311) and covers seven CpGs that mapped to a CpG island on the chromosome 5 open reading frame 63 (C5orf63; clone name: FLJ44606) gene. Two of these seven sites achieved P-valuemin (cg10812634 and cg15405432). All sites within this region showed increased DNA methylation after DE exposure, with ACEs ranging from 2.7% (cg21113900) to 6.0% (cg10812634), and five of the seven sites were annotated to the transcription start site. The second region was found in chromosome 15 (chr15: 91 473 292–91 473 570) and covers five CpGs, with four sites annotated to a CpG island and one to the North Shore region of the unc-45 myosin chaperone A (UNC45A) gene. All five CpGs within this DMR showed increased DNA methylation, with ACEs ranging from 1.5% (cg08267442) to 4.0% (cg08551047) and two that were annotated to the transcription start site. Individual CpGs of DMRs with their ACEs, genomic annotation, and Fisher-exact P values are shown in Table 2.
Table 2:
Regional DNA methylation analyses of controlled human exposures to diesel exhaust utilizing the exact P values
| Genomic coordinates | Gene | Software P-value | CpG sites | Position | Gene group | Relationship to CpG Island | Mean %-DNAm diesel exhaust (DE) | Mean %-DNAm clean air (CA) | Mean causal effect (ACE) | Exact P-value |
|---|---|---|---|---|---|---|---|---|---|---|
| chr5: 126 409 062–126 409 311 | C5orf63 (Clone name; FLJ44606) | 1.44 × 10−4 | cg18710053 | 126409061 | 5'UTR | Island | 24.49% | 19.21% | 5.28% | 3.91 × 10−3 |
| cg27097438 | 126409153 | First Exon | Island | 11.27% | 7.62% | 3.65% | 8.06 × 10−3 | |||
| cg22905511 | 126409198 | TSS200 | Island | 14.89% | 9.64% | 5.25% | 1.71 × 10−3 | |||
| cg10812634 | 126409211 | TSS200 | Island | 16.25% | 10.21% | 6.04% | 2.44 × 10−4 | |||
| cg02304092 | 126409227 | TSS200 | Island | 15.67% | 10.03% | 5.64% | 2.20 × 10−3 | |||
| cg21113900 | 126409308 | TSS200 | Island | 24.84% | 22.10% | 2.74% | 3.27 × 10−2 | |||
| cg15405432 | 126409310 | TSS200 | Island | 10.74% | 7.34% | 3.41% | 2.44 × 10−4 | |||
| chr15: 91 473 292–91 473 570 | UNC45A | 1.71 × 10−7 | cg03291024 | 91473291 | TSS200 | North Shore | 6.45% | 4.39% | 2.06% | 1.22 × 10−3 |
| cg08267442 | 91473386 | TSS200 | Island | 6.99% | 5.54% | 1.45% | 1.54 × 10−2 | |||
| cg01351822 | 91473475 | 5'UTR | Island | 6.17% | 3.15% | 3.03% | 9.77 × 10−4 | |||
| cg16414568 | 91473511 | 5'UTR | Island | 7.46% | 4.55% | 2.91% | 2.44 × 10−4 | |||
| cg08551047 | 91473569 | 5'UTR | Island | 18.60% | 14.57% | 4.04% | 5.13 × 10−3 |
Minimum P values are given in bold.
We performed a gene functional enrichment analysis using the online tool DAVID 6.8 to investigate biological relationships among the genes mapped to the 80 CpGs shown to achieve P-valuemin [28]. The pathways that have an unadjusted P-value < 0.05 are displayed in the Supplementary Table S2. Although none of the pathways achieved statistical significance after multiple testing corrections, the two pathways with the highest enrichment scores include those for epidermal growth factor (EGF) and proteases. Finally, Supplementary Table S3 compares the effect sizes and P values of marginally significant CpGs (P < 0.05) that were found to be differentially methylated in two other experimental human exposure studies [29, 30] with the effect sizes and P values observed in this study.
Discussion
DE emissions from engines around the world contribute to outdoor air pollution that, when inhaled, increases risk for the development and exacerbation of pulmonary and cardiovascular diseases [31, 32]. While epidemiology studies demonstrate significant associations between DE exposure and disease incidence and severity, there is a lack of understanding of pollution-related epigenetic changes which may be involved in mediating or attenuating pathological development. In an epigenome-wide experiment of target bronchial epithelial cells in human subjects exposed to DE, we observed differential DNA methylation at 80 individual CpGs and increased DNA methylation of two regions of the FLJ44606 and UNC45A genes. These results contribute to the growing literature aiming to elucidate the molecular underpinnings of how environmental pollution exposure can affect the epigenome of human bronchial epithelial cells [33, 34].
The FLJ44606 gene encodes for a glutaredoxin-like protein YDR286C homolog. Glutaredoxins are thioltransferases that function as electron carriers in the glutathione-mediated synthesis of DNA nucleotides by ribonucleotide reductase [35]. An EWAS of Korean adult smokers and nonsmokers who had chronic obstructive pulmonary disease found a DMR of FLJ44606 (chr5: 126 408 756–126 409 553) [36], which is in line with our data, showing that airway contaminants might target the FLJ44606 gene.
The UNC45A (Unc-45 Myosin Chaperone A) gene encodes for a regulating component of the heat shock 90 (HSP90) chaperone that is believed to play a role in cell proliferation, the accumulation of myosin during muscle cell development, and the assembly of the progesterone receptor [37]. This protein contributes to tumorigenesis by regulating cancer cell proliferation [38]. In addition, biallelic loss-of-function mutations in this gene cause the development of osteo-oto-hepato-enteric syndrome, which includes the clinical features of cholestasis, congenital diarrhea, impaired hearing, and bone fragility [39]. Therefore, the protein encoded by UNC45A may have important physiological functions in several organ systems. An epigenetic study [40] found differential methylation of the UNC45A gene in patients diagnosed with adrenoleukodystrophy, which is a neurodegenerative disorder characterized by dysregulated long-chain fatty acid metabolism—a pathologic process associated with exposure to traffic-related air pollutants [41]. We found increased DNA methylation of the UNC45A gene associated with DE exposure but in bronchial cells. In addition, DNA methylation of this gene in whole blood samples has been associated with FEV1/FVC, a marker of lung function, among never smokers [42].
The gene functional enrichment analysis we performed for the DMRs showed the highest enrichment scores for EGF and proteases. Both are implicated in several acute and chronic respiratory diseases. EGF stimulates EGF receptors (EGFR) expressed on alveolar and airway epithelial cells to regulate epithelial barrier function in the context of repair and injury [43]. Overexpression or continuous activation of EGFR due to mutations or environmental insults can lead to increased fibrotic processes, hyperproliferation of epithelial cells, increased epithelial permeability, and excessive mucin production. The downstream effects include higher risks for the development of acute lung injury, idiopathic pulmonary fibrosis, chronic obstructive pulmonary disease, non-small cell lung cancer, and other lung conditions [44–48]. Proteolytic enzymes, such as metalloproteases produced by neutrophils and alveolar macrophages, have important physiological roles in tissue remodeling and repair in the lungs and airways [49]. However, dysregulation of proteases mediates the development of acute lung injury and chronic inflammatory lung conditions [50]. Our findings suggest that the biological mechanisms explaining increased risk for lung disease associated with exposure to DE could be linked to methylation of genes that regulate EGF and proteases, but further research is needed.
None of the differential methylation, we found in 80 CpG sites in our participants matched CpG sites in studies examining changes in DNA methylation of blood mononuclear cells in nonsmoking patients with asthma exposed to DE [29] and in bronchial epithelial cells in humans exposed to DE and allergens [30]. However, some of the significant CpG sites that were differentially methylated in both these studies achieved a Fisher-exact P-value < 0.05 in our study. The direction of the effects—decreased methylation—for the CpGs in our study is the same as in the Clifford et al. study, which also examined bronchial cells. Regarding the epigenetic alterations in peripheral blood mononuclear cells in the DE study of Jiang et al., we found overlap in four CpG sites, three of which show the opposite direction of associations. Therefore, differential methylation of certain sites following DE exposure likely varies across tissues, but we were able to replicate findings from six previously reported sites in target bronchial tissue.
The level of DE exposure used in this trial, 300 μg/m3 for 2 h, could be classified in the moderate range. For example, levels can range between 154–1600 μg/m3 for miners, 121 μg/m3 for construction workers, and 10–35 μg/m3 for taxi drivers [51]. However, the composition of DE might play a significant role in the pathogenesis of its effects.
Our study has certain limitations. There is a small sample size with little variation in demographics. This limits the generalizability of our results and the power with which we can statistically detect effects; future studies should include more participants from diverse backgrounds. However, this was a randomized controlled exposure experiment, considered the gold standard to infer causality. The third exposure to O3, not included in this investigation, could have influenced the effects of DE on the epigenome. However, the washout period of 13 days together with the randomization of the order of experimental condition should have minimized this possibility. In fact, none of the signals found for O3 were observed in the findings for DE. Although the statistical power was limited given the number of tests across the epigenome, we found evidence that many sites reached the minimum Fisher-exact P-value. Additionally, the discovery of two robust DMRs with multiple CpG sites that have consistent direction of effects in epigenetically relevant CpG islands further strengthens the evidence for a causal effect of short-term DE exposure on the epigenome. We also did not have access to genetic information from study participants, so we did not exclude sites that might be associated with SNPs; therefore, the possibility of our findings to reflect genetic influence on methylation cannot be excluded. The collection of target bronchial cells is a major strength, as all observational studies have examined leukocyte DNA methylation, and our design provided a unique opportunity to study target tissue.
Conclusions
DE is a significant component of outdoor air pollution, and short-term exposure to DE alters bronchial epithelial cell DNA methylation in a randomized, controlled exposure study of healthy adults. Findings of differential methylation associated with the FLJ44606 and UNC45A genes contribute to our knowledge of the epigenetic alterations that may precipitate the known adverse pulmonary and systemic effects of DE exposure. Functional studies on the target genes and sites may provide further evidence on the molecular impact of DE exposure on human health.
Supplementary data
Supplementary data are available at EnvEpig online.
Data Availability
The datasets used and/or analyzed during the current study are available from the study team on reasonable request.
Funding
This work was supported by the US National Institutes of Health grant R01 ES031259. Research reported in this publication was also supported by the John Harvard Distinguished Science Fellow Program within the FAS Division of Science of Harvard University, and by the Office of the Director, National Institutes of Health under Award Number DP5OD021412. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Conflict of interest statement. None declared.
Supplementary Material
Clinical Trial Registration: ClinicalTrials.gov (U.S. National Library of Medicine), NCT01492517. Registered 15 December 2011—Retrospectively registered, https://clinicaltrials.gov/ct2/show/NCT01492517.
References
- 1. World Health Organization.Ambient Air Pollution: A Global Assessment of Exposure and Burden of Disease. WHO, Geneva, Switzerland; 2016. http://www.who.int/phe/publications/air-pollution-global-assessment/en/ (21 August 2020, date last accessed). [Google Scholar]
- 2. Miller J, Jin L.. Global Progress toward Soot-Free Diesel Vehicles in 2019. The International Council on Clean Transportation (ICCT), San Francisco, CA; 2019. https://theicct.org/publications/global-progress-toward-soot-free-diesel-vehicles-2019 (21 August 2020, date last accessed). [Google Scholar]
- 3. Anenberg S, Miller J, Henze D, Minjares R.. A Global Snapshot of the Air Pollution-Related Health Impacts of Transportation Sector Emissions in 2010 and 2015. The International Council on Clean Transportation (ICCT), San Francisco, CA; 2019. https://theicct.org/publications/health-impacts-transport-emissions-2010-2015 (21 August 2020, date last accessed). [Google Scholar]
- 4. Steiner S, Bisig C, Petri-Fink A, Rothen-Rutishauser B.. Diesel exhaust: current knowledge of adverse effects and underlying cellular mechanisms. Arch Toxicol 2016;90:1541–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Mastrofrancesco A, Alfè M, Rosato E, Gargiulo V, Beatrice C, Di Blasio G, Zhang B, Su DS, Picardo M, Fiorito S.. Proinflammatory effects of diesel exhaust nanoparticles on scleroderma skin cells. J Immunol Res 2014;2014:138751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Benbrahim-Tallaa L, Baan RA, Grosse Y, Lauby-Secretan B, El Ghissassi F, Bouvard V, Guha N, Loomis D, Straif K, International Agency for Research on Cancer Monograph Working Group. Carcinogenicity of diesel-engine and gasoline-engine exhausts and some nitroarenes. Lancet Oncol 2012;13:663–4. [DOI] [PubMed] [Google Scholar]
- 7. Möller W, Felten K, Sommerer K, Scheuch G, Meyer G, Meyer P, Häussinger K, Kreyling WG.. Deposition, retention, and translocation of ultrafine particles from the central airways and lung periphery. Am J Respir Crit Care Med 2008;177:426–32. [DOI] [PubMed] [Google Scholar]
- 8. Topinka J, Milcova A, Schmuczerova J, Mazac M, Pechout M, Vojtisek-Lom M.. Genotoxic potential of organic extracts from particle emissions of diesel and rapeseed oil powered engines. Toxicol Lett 2012;212:11–7. [DOI] [PubMed] [Google Scholar]
- 9. Hesterberg TW, Long CM, Bunn WB, Sax SN, Lapin CA, Valberg PA.. Non-cancer health effects of diesel exhaust: a critical assessment of recent human and animal toxicological literature. Crit Rev Toxicol 2009;39:195–227. [DOI] [PubMed] [Google Scholar]
- 10. Attfield MD, Schleiff PL, Lubin JH, Blair A, Stewart PA, Vermeulen R, Coble JB, Silverman DT.. The diesel exhaust in miners study: a cohort mortality study with emphasis on lung cancer. J Natl Cancer Inst 2012;104:869–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Latifovic L, Villeneuve PJ, Parent M, Johnson KC, Kachuri L, Harris SA, Canadian Cancer Registries Epidemiology Group. Bladder cancer and occupational exposure to diesel and gasoline engine emissions among Canadian men. Cancer Med 2015;4:1948–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Vermeulen R, Silverman DT, Garshick E, Vlaanderen J, Portengen L, Steenland K.. Exposure-response estimates for diesel engine exhaust and lung cancer mortality based on data from three occupational cohorts. Environ Health Perspect 2014;122:172–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. IARC. Diesel and Gasoline Engine Exhausts and Some Nitroarenes. International Agency for Research on Cancer, Lyon, France. 2013. https://publications.iarc.fr/Book-And-Report-Series/Iarc-Monographs-On-The-Identification-Of-Carcinogenic-Hazards-To-Humans/Diesel-And-Gasoline-Engine-Exhausts-And-Some-Nitroarenes-2013 (21 August 2020, date last accessed). [Google Scholar]
- 14. Feinberg AP. The key role of epigenetics in human disease. N Engl J Med 2018;379:400–1. [DOI] [PubMed] [Google Scholar]
- 15. Cardenas A, Rifas-Shiman SL, Agha G, Hivert M-F, Litonjua AA, DeMeo DL, Lin X, Amarasiriwardena CJ, Oken E, Gillman MW, Baccarelli AA.. Persistent DNA methylation changes associated with prenatal mercury exposure and cognitive performance during childhood. Sci Rep 2017;7:1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Cardenas A, Rifas-Shiman SL, Godderis L, Duca R-C, Navas-Acien A, Litonjua AA, DeMeo DL, Brennan KJ, Amarasiriwardena CJ, Hivert M-F, Gillman MW, Oken E, Baccarelli AA.. Prenatal exposure to mercury: associations with global DNA methylation and hydroxymethylation in cord blood and in childhood. Environ Health Perspect 2017;125:087022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Chadwick LH, Sawa A, Yang IV, Baccarelli A, Breakefield XO, Deng H-W, Dolinoy DC, Fallin MD, Holland NT, Houseman EA, Lomvardas S, Rao M, Satterlee JS, Tyson FL, Vijayanand P, Greally JM.. New insights and updated guidelines for epigenome-wide association studies. Neuroepigenetics 2015;1:14–9. [Google Scholar]
- 18. Rider CF, Carlsten C.. Air pollution and DNA methylation: effects of exposure in humans. Clin Epigenet 2019;11:131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Devlin RB, Duncan KE, Jardim M, Schmitt MT, Rappold AG, Diaz-Sanchez D.. Controlled exposure of healthy young volunteers to ozone causes cardiovascular effects. Circulation 2012;126:104–11. [DOI] [PubMed] [Google Scholar]
- 20. Madden MC, Stevens T, Case M, Schmitt M, Diaz-Sanchez D, Bassett M, Montilla TS, Berntsen J, Devlin RB.. Diesel exhaust modulates ozone-induced lung function decrements in healthy human volunteers. Part Fibre Toxicol 2014;11:37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Bind M-AC, Rubin DB, Cardenas A, Dhingra R, Ward-Caviness C, Liu Z, Mirowsky J, Schwartz JD, Diaz-Sanchez D, Devlin RB.. Heterogenous ozone effects on the DNA methylome of bronchial cells observed in a crossover study. Sci Rep 2020;10:15739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Ghio AJ, Kim C, Devlin RB.. Concentrated ambient air particles induce mild pulmonary inflammation in healthy human volunteers. Am J Respir Crit Care Med 2000;162:981–8. [DOI] [PubMed] [Google Scholar]
- 23. Ghio AJ, Bassett M, Chall AN, Levin DG, Bromberg PA.. Bronchoscopy in healthy volunteers. J Bronchol Interv Pulmonol 1998;5:185–94. [Google Scholar]
- 24. Triche TJ, Weisenberger DJ, Van Den Berg D, Laird PW, Siegmund KD.. Low-level processing of Illumina Infinium DNA methylation BeadArrays. Nucleic Acids Res 2013;41:e90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Teschendorff AE, Marabita F, Lechner M, Bartlett T, Tegner J, Gomez-Cabrero D, Beck S.. A beta-mixture quantile normalization method for correcting probe design bias in Illumina Infinium 450 k DNA methylation data. Bioinformatics 2013;29:189–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Bind M-AC, Rubin DB.. When possible, report a Fisher-exact P value and display its underlying null randomization distribution. Proc Natl Acad Sci USA 2020;117:19151–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Pedersen BS, Schwartz DA, Yang IV, Kechris KJ.. Comb-p: software for combining, analyzing, grouping and correcting spatially correlated P-values. Bioinformatics 2012;28:2986–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. DAVID Functional Annotation Bioinformatics Microarray Analysis. 2020. Available from: https://david.ncifcrf.gov/home.jsp (21 August 2020, date last accessed).
- 29. Jiang R, Jones MJ, Sava F, Kobor MS, Carlsten C.. Short-term diesel exhaust inhalation in a controlled human crossover study is associated with changes in DNA methylation of circulating mononuclear cells in asthmatics. Part Fibre Toxicol 2014;11:71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Clifford RL, Jones MJ, MacIsaac JL, McEwen LM, Goodman SJ, Mostafavi S, Kobor MS, Carlsten C.. Inhalation of diesel exhaust and allergen alters human bronchial epithelium DNA methylation. J Allergy Clin Immunol 2017;139:112–21. [DOI] [PubMed] [Google Scholar]
- 31. Kenyon N, Liu F-T.. Pulmonary effects of diesel exhaust. Am J Pathol 2011;179:2678–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Mills NL, Törnqvist H, Gonzalez MC, Vink E, Robinson SD, Söderberg S, Boon NA, Donaldson K, Sandström T, Blomberg A, Newby DE.. Ischemic and thrombotic effects of dilute diesel-exhaust inhalation in men with coronary heart disease. N Engl J Med 2007;357:1075–82. [DOI] [PubMed] [Google Scholar]
- 33. Grilli A, Bengalli R, Longhin E, Capasso L, Proverbio MC, Forcato M, Bicciato S, Gualtieri M, Battaglia C, Camatini M.. Transcriptional profiling of human bronchial epithelial cell BEAS-2B exposed to diesel and biomass ultrafine particles. BMC Genomics 2018;19:302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Glass K, Thibault D, Guo F, Mitchel JA, Pham B, Qiu W, Li Y, Jiang Z, Castaldi PJ, Silverman EK, Raby B, Park J-A, Yuan G-C, Zhou X.. Integrative epigenomic analysis in differentiated human primary bronchial epithelial cells exposed to cigarette smoke. Sci Rep 2018;8:12750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. NCBI. AceView: Gene:FLJ44606, a comprehensive annotation of human, mouse and worm genes with mRNAs or ESTsAceView. Bethesda, MD: National Center for Biotechnology Information, 2020. https://www.ncbi.nlm.nih.gov/IEB/Research/Acembly/av.cgi?db=human&q=FLJ44606 (21 August 2020, date last accessed). [Google Scholar]
- 36. Lee MK, Hong Y, Kim S-Y, London SJ, Kim WJ.. DNA methylation and smoking in Korean adults: epigenome-wide association study. Clin Epigenet 2016;8:103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. NCBI. UNC45A Unc-45 Myosin Chaperone A [Homo sapiens (Human)] - Gene. National Center for Biotechnology Information, Bethesda, MD, 2020. https://www.ncbi.nlm.nih.gov/gene/55898 (21 August 2020, date last accessed). [Google Scholar]
- 38. Eisa NH, Jilani Y, Kainth K, Redd P, Lu S, Bougrine O, Abdul Sater H, Patwardhan CA, Shull A, Shi H, Liu K, Elsherbiny NM, Eissa LA, El-Shishtawy MM, Horuzsko A, Bollag R, Maihle N, Roig J, Korkaya H, Cowell JK, Chadli A.. The co-chaperone UNC45A is essential for the expression of mitotic kinase NEK7 and tumorigenesis. J Biol Chem 2019;294:5246–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Esteve C, Francescatto L, Tan PL, Bourchany A, De Leusse C, Marinier E, Blanchard A, Bourgeois P, Brochier-Armanet C, Bruel A-L, Delarue A, Duffourd Y, Ecochard-Dugelay E, Hery G, Huet F, Gauchez P, Gonzales E, Guettier-Bouttier C, Komuta M, Lacoste C, Maudinas R, Mazodier K, Rimet Y, Rivière J-B, Roquelaure B, Sigaudy S, Stephenne X, Thauvin-Robinet C, Thevenon J, Sarles J, Levy N, Badens C, Goulet O, Hugot J-P, Katsanis N, Faivre L, Fabre A.. Loss-of-function mutations in unc45a cause a syndrome associating cholestasis, diarrhea, impaired hearing, and bone fragility. Am J Hum Genet 2018;102:364–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Schlüter A, Sandoval J, Fourcade S, Díaz-Lagares A, Ruiz M, Casaccia P, Esteller M, Pujol A.. Epigenomic signature of adrenoleukodystrophy predicts compromised oligodendrocyte differentiation. Brain Pathol 2018;28:902–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Ward-Caviness CK, Breitner S, Wolf K, Cyrys J, Kastenmüller G, Wang-Sattler R, Schneider A, Peters A.. Short-term NO2 exposure is associated with long-chain fatty acids in prospective cohorts from Augsburg, Germany: results from an analysis of 138 metabolites and three exposures. Int J Epidemiol 2016;45:1528–38. [DOI] [PubMed] [Google Scholar]
- 42. de Vries M, Nedeljkovic I, van der Plaat DA, Zhernakova A, Lahousse L, Brusselle GG, Amin N, van Duijn CM, Vonk JM, Boezen HM, BIOS Consortium. DNA methylation is associated with lung function in never smokers. Respir Res 2019;20:268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Finigan JH, Downey GP, Kern JA.. Human epidermal growth factor receptor signaling in acute lung injury. Am J Respir Cell Mol Biol 2012;47:395–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Sharma SV, Bell DW, Settleman J, Haber DA.. Epidermal growth factor receptor mutations in lung cancer. Nat Rev Cancer 2007;7:169–81. [DOI] [PubMed] [Google Scholar]
- 45. Vallath S, Hynds RE, Succony L, Janes SM, Giangreco A.. Targeting EGFR signalling in chronic lung disease: therapeutic challenges and opportunities. Eur Respir J 2014;44:513–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Li C, Wei R, Jones-Hall YL, Vittal R, Zhang M, Liu W.. Epidermal growth factor receptor (EGFR) pathway genes and interstitial lung disease: an association study. Sci Rep 2014;4:4893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Shitrit D, Shochet GE, Brook E, Eyal O, Edelstein E.. Epidermal growth factor receptor promotes survival of idiopathic pulmonary fibrosis fibroblasts. Chest 2018;154:725A. [DOI] [PubMed] [Google Scholar]
- 48. Burgel P-R, Nadel JA.. Roles of epidermal growth factor receptor activation in epithelial cell repair and mucin production in airway epithelium. Thorax 2004;59:992–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Pandey KC, De S, Mishra PK.. Role of proteases in chronic obstructive pulmonary disease. Front Pharmacol 2017;8:512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Chakraborti S, Sarkar J, Pramanik PK, Chakraborti T.. Role of proteases in lung disease: a brief overview. In: Chakraborti S, Chakraborti T, Dhalla NS, editors. Proteases in Human Diseases. Singapore: Springer; 2017. p. 333–74. [Google Scholar]
- 51. Pronk A, Coble J, Stewart PA.. Occupational exposure to diesel engine exhaust: a literature review. J Expo Sci Environ Epidemiol 2009;19:443–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the study team on reasonable request.