Abstract
The multifunctional protein clusterin (CLU) is a secreted glycoprotein ubiquitously expressed throughout the body, including in the eye. Its primary function is to act as an extracellular molecular chaperone, preventing the precipitation and aggregation of misfolded extracellular proteins. Clusterin is commonly identified at fluid-tissue interfaces, and has been identified in most body fluids. It is a component of exfoliation material, and CLU mRNA is reduced in eyes with exfoliation syndrome compared with controls. SNPs located in the CLU genomic region have been associated with Alzheimer disease (AD) at the genome-wide level and several CLU SNPs located in an apparent regulatory region have been nominally associated with XFS/XFG in Caucasians with European ancestry and in south Indians. Interestingly, clusterin associates with altered elastic fibers in human photo-aged skin and prevents UV-induced elastin aggregation in vitro. In light of the known geographic risk factors for XFS/XFG, which could include UV light, investigations of CLU-geographic interactions could be of interest. Future studies investigating rare CLU variation and other complex interactions including gene-gene interactions in XFS/XFG cases and controls may also be fruitful. While CLU has been considered as a therapeutic target in AD, cancer and dry eye, a role for clusterin in XFS/XFG needs to be better defined before therapeutic approaches involving CLU can be entertained.
Keywords: exfoliation syndrome, clusterin, chaperone, elastic fibers
Exfoliation syndrome (XFS) and the associated glaucoma (XFG) are genetically complex traits.1 LOXL1 (lysyl oxidase-like 1) is a major risk factor for XFS/XFG. While LOXL1 genetic variants were initially identified in Iceland2, the association with XFS/XFG has been replicated in populations throughout the world, including the United States.3–7 Although LOXL1 is a major risk factor for XFS/XFG, the association is complex. While the LOXL1 risk alleles are present in the majority of cases worldwide, they are also frequently found in controls, arguing that other genetic and/or environmental factors are necessary for the disease to be fully manifested.8 A recent multi-ethnic genome wide association study (GWAS) identified common variants near CACNA1A (calcium channel, voltage-dependent, P/Q type, alpha 1A subunit) significantly associated with XFS.9 Here we review data suggesting that clusterin (CLU), an extracellular chaperone protein known to reduce protein aggregation, could contribute to XFS/XFG pathogenesis.
Clusterin is a secreted glycoprotein ubiquitously expressed throughout the body, including in the eye. Its primary function is to act as an extracellular molecular chaperone, preventing the precipitation and aggregation of misfolded extracellular proteins.10 Clusterin is commonly identified at fluid-tissue interfaces, and has been found in most body fluids.10 The protein is encoded by a single gene (CLU) that includes a 14-bp element in the promoter that is specifically recognized by the transcription factor HSF1 (heat shock factor 1) allowing for heat-shock-induced transcription.11 CLU mRNA is also up-regulated in response to a broad range of cellular stress signals and conditions: oncogene activity, growth factors and cytokines (including TGFβ), and other stress- or apoptosis-inducing agents including UVA, UVB, proteotoxic stress, heavy metals, oxidants, hypoxia, ionizing radiation, and chemotherapeutic drugs.12–14
The translated clusterin preprotein is cleaved prior to secretion from the cell to produce A and B subunits, and post-translational modifications include glycosylation at 6 different asparagine residues (N-glycosylation (Asn86, Asn103, Asn145, Asn291, Asn354, Asn374).15 Mature clusterin contains a disulfide-linked core region that involves at least 4 and possibly 5 disulfide bonds.16 Under normal conditions clusterin is constitutively secreted from cells and is found in extracellular fluids.17 Mature, post-translationally modified clusterin can be released from the ER/Golgi under conditions of ER stress, when it may also have a role in intracellular processes occurring outside the secretory system.18
After secretion, clusterin’s primary role is to bind to hydrophobic complexes and denatured proteins to promote their removal.19 Clusterin is a highly potent inhibitor of protein precipitation, with more activity than heat-shock proteins.20 Interestingly, clusterin can form self-oligomers depending on the pH of the environment.21–23 In the mildly acidic environment of the anterior chamber, clusterin would be expected to be disassociated (monomers) and have maximal chaperone activity.21 In some instances clusterin has been reported to form high molecular weight complexes with particular ligands such as fibrinogen.22
Clusterin has been implicated in a wide variety of biological and pathological processes (Figure 1). The protein appears to be particularly important in pathological conditions related to cellular stress. CLU knockout mice are indistinguishable from their wild type littermates under normal conditions. With age or stress however, CLU knockout mice develop glomerular neuropathy, potentially due to the accumulation of pathological protein deposits in the absence of clusterin activity.24
Figure 1.
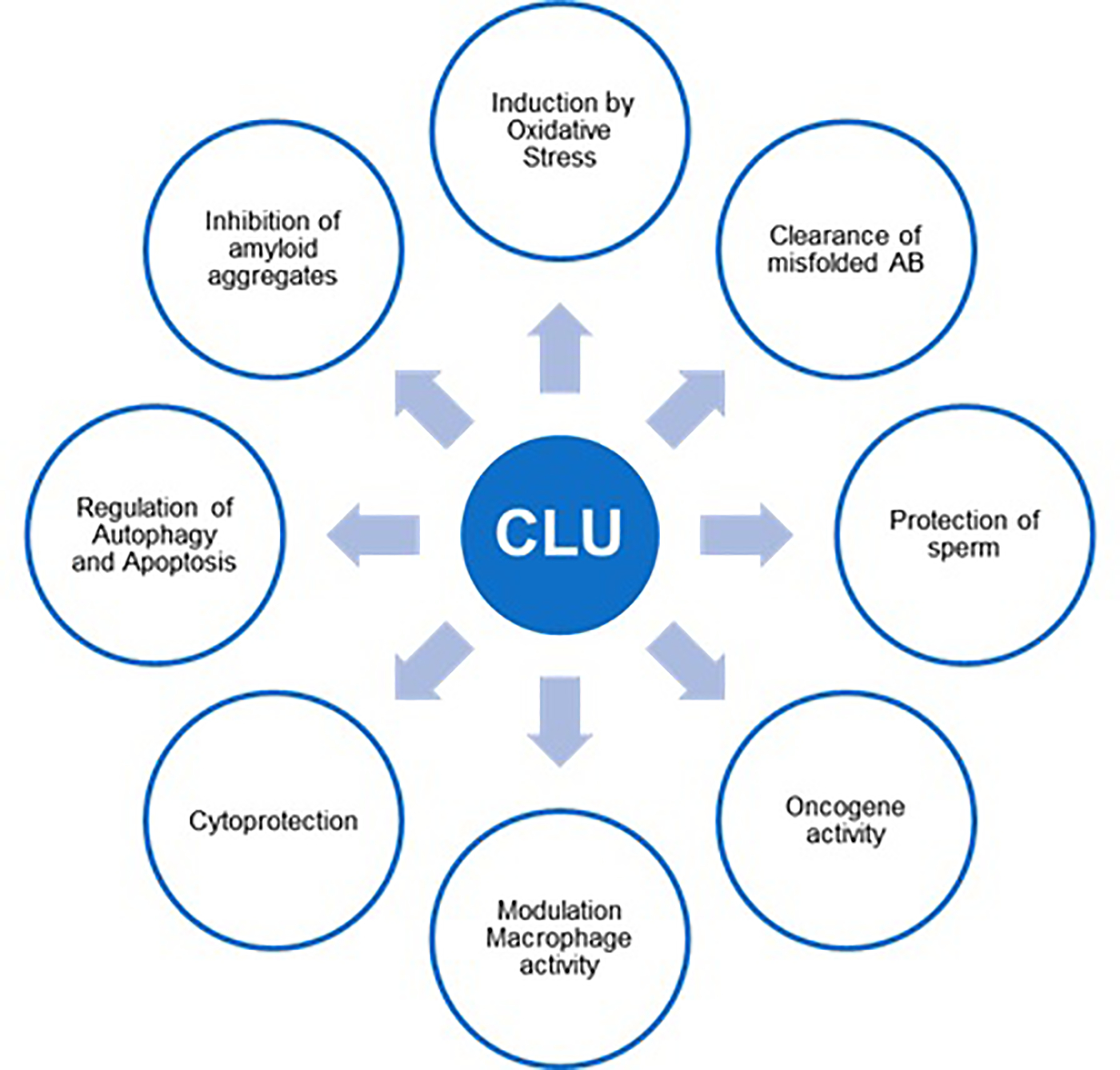
Biological and pathological processes involving clusterin.
Specific pathological or biological processes involving clusterin include induction by oxidative stress,25 clearance of misfolded Alzheimer amyloid beta (Aβ) peptide across the blood brain barrier,26 association with sperm cells to prevent their rejection by the female immune system,27 modulation of macrophage activity,28 induction of PI3K/AKT-signaling and other cytoprotective effects,29 and alteration of autophagy and apoptosis.30,31 Clusterin is known to interact with many proteins and ligands are part of these processes.
In addition to XFS/XFG, clusterin has been implicated in a number of human disorders including Alzheimer disease,32 various types of cancer and cancer therapies,33 dry eye,34,35 AMD,36 and more recently diabetic retinopathy37. The inhibition of formation of amyloid aggregates by clusterin is an important feature of Alzheimer disease. Binding to misfolded proteins is thought to be related to clusterin’s surface hydrophobicity which is enhanced by acidic pH.21
Clusterin in Exfoliation syndrome and Exfoliation glaucoma
Clusterin is known to be expressed in ocular tissues relevant to XFS/EG including the iris and ciliary body.38,39 Clusterin is found in higher amounts in aqueous humor from XFS patients compared with POAG or cataract patients.40 Three different studies identified clusterin as a component of exfoliation material.41–43 The relatively acidic pH of the aqueous humor would be expected to promote clusterin chaperone activity.21
An important study provided evidence that CLU mRNA expression is reduced in the iris, lens, and ciliary processes of XFS patients44 and a follow-up study showed that expression of both CLU mRNA and protein was reduced in eyes of XFS patients with and without glaucoma in comparison to normal and POAG eyes.45 This study also showed that TGFβ reduced CLU expression in ciliary body epithelial cells in vitro, suggesting that TGFβ levels could regulate CLU expression in XFS eyes.
CLU genetic associations
Alzheimer’ Disease (AD)
Clusterin inhibits the aggregation of amyloid beta (Aβ) peptide and participates in the clearance of Aβ via the blood-brain barrier,32,46 processes integrally related to AD risk. There are other interesting similarities when comparing clusterin activities in AD and XFS/XFG. Similar to clusterin’s presence in aqueous humor and in exfoliation material, in the brain, CLU is highly expressed in both the cerebrospinal fluid,47 amyloid plaques48 and neurofibrillary tangles.49 Aβ and clusterin combine together in normal cerebrospinal fluid, suggesting that reduced CLU expression may contribute in Aβ aggregation.49
Genome-wide association studies have identified SNPs located in the CLU genomic region significantly associated with AD at the genome-wide level.50,51 These studies have been replicated in most populations, with exceptions of African-Americans and Hispanics.52 Mutations in the Cys-rich region of the clusterin protein result in reduced secretion of clusterin, providing additional support for a critical role in disease development.53
Exfoliation syndrome and exfoliation glaucoma
While CLU appears to be a promising candidate for an XFS/XFG risk factor, genetic studies have not provided consistent association results. A single CLU SNP (rs3087554) located in the 3’ untranslated region (Figure 2) was nominally associated with XFS in the Blue Mountain Eye Study (86 cases and 2,422 controls) at the genotypic level (P = 0.044), but not at the allelic level or when the age of controls was restricted to those over age 73 (P >0.07).54 Significant association between the rs3087554 variant and XFS was not observed in a German (661 cases and 342 controls; P > 0.08) and Italian (209 cases and 190 controls; P > 0.70) case-control set, although a positive association for another CLU SNP, rs2279590 (also associated with AD) was reported in the German dataset only (P = 0.035).55 A study of 136 cases and 89 controls from India did not find an association of rs3087554 with XFS (P >0.06), but did find a significant association with rs2279590 (P = 0.039).56 However, a recent study of 299 cases and 224 controls from South India did not find significant association of rs3087554 or rs2279590 with XFS (P >0.43).57 Additionally, a recent GWAS in a Japanese dataset of 1,484 cases and 1,188 controls showed nominal association of rs3087554 with XFS (P = 0.029), although the direction of effect for the minor allele ‘G’ is in the opposite direction compared with the effect in Caucasians with European ancestry.9 rs2279590 was not included in the Japanese study.
Figure 2. Features of the Clusterin gene (CLU) genomic region.

Clusterin gene (CLU) genomic region. The genomic location of the CLU exons (larger blue rectangles) and introns (blue lines) as depicted in the UCSC genome browser (http://ucsc.genome.edu) on chromosome 8 are shown. Beneath the exon/intron are the locations of the SNPs associated with Alzheimer disease (AD) (green type) and XFS/XFG (black type). rs2279590 is associated with both AD and XFS/XFG. The layered H3K27Ac histone marks (frequently found in regulatory regions) for 8 cell types studied by ENCODE are shown beneath the SNPs. rs3087554 and rs2279590 fall in a H3K27Ac peak in HUVEC cells (human umbilical vein). Rs3087554 and rs2279590 are also in the vicinity of a CLU miRNA binding site (miR-370) indicated in green below the H3K27Ac peaks.
Our group performed a meta-analysis of data from two US and Israeli datasets (314 cases and 446 controls) with published data from other Caucasian populations.58 We found a statistically significant association of rs2279590 with XFS/XFG overall (summary OR = 1.18, 95% CI: 1.03–1.33, P = 0.01). No significant association was found for rs3087554 in Caucasian populations (summary OR = 0.90, 95% CI: 0.77–1.05, P = 0.17). Other CLU SNPs were not investigated.
Despite these important observations, genome-wide association for common CLU variants and XFS/XFG has not yet been observed. Significant genetic heterogeneity for CLU association has been observed among populations,58 which could confound results in multi-ethnic GWAS. Alternatively, common CLU genetic variation may not underlie any clusterin function that could be involved in XFS/XFG. Rare CLU variation and complex genetic processes such as gene-gene and gene-environment interactions have not yet been fully evaluated in XFS/XFG cases and controls.
Rare CLU genetic variants have been evaluated in AD. In a sequencing study of AD cases an increased frequency of rare non-synonymous mutations in the CLU β-chain domain were identified in AD patients.59 A subsequent report showed that three patient-specific CLU mutations in the B-chain caused abnormal subcellular CLU localization and diminished CLU transport through the secretory pathway.53
Complex genetics and interactions
Complex genetic interactions such as gene-gene and gene-environment interactions are expected to contribute to diseases with complex inheritance such as XFS/XFG. CLU is an excellent candidate genetic risk factor for XFS/XFG, and while individual CLU SNPs have demonstrated only nominal association with disease,58 these results do not exclude the possibility of involvement of CLU in complex interactions.
Interestingly, clusterin associates with altered elastic fibers in human photo-aged skin and prevents UV-induced elastin aggregation in vitro.60 Recent studies suggest that CLU expression and clusterin glycosylation may be markers of cellular redox and oxidative stress which can be induced by UV light, suggesting that clusterin could link environmental factors related to UV light exposure with disease risk. Considering the significant effects of latitude on XFS risk,61–64 which may involve ocular UV light exposures, gene-environment studies investigating interactions for UV light exposure and CLU SNPs could be of interest in a well-powered sample.
In AD, a significant gene-gene interaction has been identified for CLU and MS4A4E, coding for a membrane protein of unknown function65. The odds ratio (OR) for the interaction on disease risk was substantially higher than that for CLU or MS4A4E alone, providing good support that the interaction could contribute to disease pathogenesis.
Clusterin as a therapeutic target
The important molecular chaperone activity of CLU, and its increased expression in response to cellular stress suggests that modulating CLU expression could have therapeutic potential, especially in diseases such as AD32,48 and cancer68 where CLU expression is increased. CLU is also a therapeutic target in dry eye.34,35
CLU expression is increased in several cancers and is also increased after administration of chemotherapy, especially cisplatin, doxorubicin, Herceptin, Hsp90 inhibitors, and HDAC inhibitors.66 In prostrate cancer decreasing CLU expression using an antisense oligonucleotide molecule reduced TGFβ signaling and blocked metastases in mice.69,70 Subsequent studies have shown that reducing CLU expression improved the cytotoxicity of hormone-, radiation-, and chemo-therapies.68,71 A CLU antisense molecule (OGX-011) showed promising safety and efficacy in phase I and II clinical trials for prostate and lung cancers in humans72–74 but failed to demonstrate improved survival in a large phase III study for prostate cancer.75
CLU’s role in protecting cells at fluid-tissue interfaces from the toxicity of protein oligomers has identified CLU as a possible therapeutic target in AD. However, while CLU is clearly involved in mediating the safe clearance and disposal of excess Aβ,47 CLU activity in animal models is complex, making it difficult to design a therapeutic molecule or strategy. For example, in a mouse model of AD, reduction in CLU expression also reduced Aβ deposition and neurotoxicity; however if APOE was also reduced (also expected in AD patients) then reducing CLU resulted in increased Aβ amyloid deposition.78,79 In in vitro studies, CLU can either potently inhibit Aβ amyloid formation or, at low ratios of CLU:Aβ, (e.g. 1:500), actually increase amyloid formation.49 Therefore, currently its not clear if increasing or decreasing CLU expression would be beneficial in AD, a complex situation that could be similar to XFS/XFG (see discussion below).
In the eye, topical CLU has been shown to protect the corneal surface from desiccating stress in a pre-clinical mouse model of dry eye. The protection offered by CLU is likely due to its interaction with LGALS3 (Galectin-3),80 a lectin known to be present on corneal epithelial cells, and MMP9, a metalloproteinase known to be involved in dry eye development.81 CLU is present in tears and perhaps protects the eye from desiccating stress by ‘sealing’ the surface through its interaction with these proteins.34,35
Conclusions
Clusterin is an important extracellular molecular chaperone that protects cells at fluid-tissue interfaces from physical stress and removes potentially toxic protein oligomers. These features suggest that CLU could have a significant role in the development of XFS/XFG, a disease characterized by the accumulation of protein aggregates in fluid-filled spaces. The observations that clusterin is a component of XFS material and that CLU expression is altered in XFS/XFG also support a role for clusterin in disease pathogenesis. Genetically, CLU common variants are associated at the genome-wide level with AD, a disease also involving accumulation of aggregated protein. In XFS/XFG common CLU variants are only associated with disease risk at the nominal level. Future studies investigating rare CLU variation and complex interactions including gene-gene and gene-environment interactions in XFS/XFG cases and controls could be of interest.
Other than genetic studies there are important areas of research that could provide insight into the role of CLU in EXF/XFG. First, it is interesting that CLU expression is decreased in XFS/XFG when in other pathogenic conditions, such as AD and cancer, the expression is increased. Are there secondary factors that decrease CLU expression in XFS/XFG? Does the reduction in CLU expression contribute to increased aggregation of proteins? In this regard, the demonstration that increased TGFβ can decrease CLU expression may be relevant. It could also be of interest to investigate the ocular phenotype of CLU knock-out mice at older ages and in stressed environments.
Similar to AD, is it also possible that CLU contributes to the formation of the XFS aggregated protein? CLU is known to be a component of XFS aggregate material and is also known to have enhanced binding to ligands in mildly acidic environments such as would be expected in aqueous humor.
Finally, is clusterin a therapeutic target for XFS/XFG? It is tempting to speculate that increasing clusterin protein activity could result in reduction and/or removal of aggregated protein complexes in XFS/XFG eyes. However, the experience in AD suggests that the CLU-aggregated protein interaction is complex. Further investigations into the effects of overexpression of CLU, as well as reduction of CLU expression, ideally in relevant animal models, are needed before approaching CLU as a therapeutic target for XFS/XFG.
Acknowledgments
Funding: This work is funded by the NIH grants EY020928 (JLW), EY015473 (LRP), the Harvard Glaucoma Center of Excellence
Footnotes
Competing Interest: None
References
- 1.Sein J, Galor A, Sheth A, Kruh J, Pasquale LR, Karp CL. Exfoliation syndrome: new genetic and pathophysiologic insights. Curr Opin Ophthalmol. 2013. March;24(2):167–74. [DOI] [PubMed] [Google Scholar]
- 2.Thorleifsson G, Magnusson KP, Sulem P, Walters GB, Gudbjartsson DF, Stefansson H, Jonsson T, Jonasdottir A, Jonasdottir A, Stefansdottir G, Masson G, Hardarson GA, Petursson H, Arnarsson A, Motallebipour M, Wallerman O, Wadelius C, Gulcher JR, Thorsteinsdottir U, Kong A, Jonasson F, Stefansson K. Common sequence variants in the LOXL1 gene confer susceptibility to exfoliation glaucoma. Science. 2007;317:1397–1400. [DOI] [PubMed] [Google Scholar]
- 3.Fingert JH, Alward WL, Kwon YH, Wang K, Streb LM, Sheffield VC, Stone EM. LOXL1 mutations are associated with exfoliation syndrome in patients from the midwestern United States. Am. J. Ophthalmol. 2007;144:974–975. [DOI] [PubMed] [Google Scholar]
- 4.Aragon-Martin JA, Ritch R, Liebmann J, O’Brien C, Blaaow K, Mercieca F, Spiteri A, Cobb CJ, Damji KF, Tarkkanen A, Rezaie T, Child AH, Sarfarazi M. Evaluation of LOXL1 gene polymorphisms in exfoliation syndrome and exfoliation glaucoma. Mol. Vis. 2008;14:533–541. [PMC free article] [PubMed] [Google Scholar]
- 5.Challa P, Schmidt S, Liu Y, Qin X, Vann RR, Gonzalez P, Allingham RR, Hauser MA. Analysis of LOXL1 polymorphisms in a United States population with pseudoexfoliation glaucoma. Mol. Vis. 2008;14:146–149. [PMC free article] [PubMed] [Google Scholar]
- 6.Fan BJ, Pasquale L, Grosskreutz CL, Rhee D, Chen T, DeAngelis MM, Kim I, delBono E, Miller JW, Li T, Haines JL, Wiggs JL. DNA sequence variants in the LOXL1 gene are associated with pseudoexfoliation glaucoma in a US clinic-based population with broad ethnic diversity. BMC. Med. Genet. 2008;9:5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yang X, Zabriskie NA, Hau VS, Chen H, Tong Z, Gibbs D, Farhi P, Katz BJ, Luo L, Pearson E, Goldsmith J, Ma X, Kaminoh Y, Chen Y, Yu B, Zeng J, Zhang K, Yang Z. Genetic association of LOXL1 gene variants and exfoliation glaucoma in a Utah cohort. Cell. Cycle. 2008;7:521–524. [DOI] [PubMed] [Google Scholar]
- 8.Fan BJ, Pasquale LR, Rhee D, Li T, Haines JL, Wiggs JL. LOXL1 promoter haplotypes are associated with exfoliation syndrome in a US Caucasian population. Invest. Ophthalmol. Vis. Sci. 2011;52:2372–2378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Aung T, Ozaki M, Mizoguchi T, Allingham RR, Li Z, Haripriya A, Nakano S, Uebe S, Harder JM, Chan AS, Lee MC, Burdon KP, Astakhov YS, Abu-Amero KK, Zenteno JC, Nilgün Y, Zarnowski T, Pakravan M, Safieh LA, Jia L, Wang YX, Williams S, Paoli D, Schlottmann PG, Huang L, Sim KS, Foo JN, Nakano M, Ikeda Y, Kumar RS, Ueno M, Manabe S, Hayashi K, Kazama S, Ideta R, Mori Y, Miyata K, Sugiyama K, Higashide T, Chihara E, Inoue K, Ishiko S, Yoshida A, Yanagi M, Kiuchi Y, Aihara M, Ohashi T, Sakurai T, Sugimoto T, Chuman H, Matsuda F, Yamashiro K, Gotoh N, Miyake M, Astakhov SY, Osman EA, Al-Obeidan SA, Owaidhah O, Al-Jasim L, Al Shahwan S, Fogarty RA, Leo P, Yetkin Y, Oğuz Ç, Kanavi MR, Beni AN, Yazdani S, Akopov EL, Toh KY, Howell GR, Orr AC, Goh Y, Meah WY, Peh SQ, Kosior-Jarecka E, Lukasik U, Krumbiegel M, Vithana EN, Wong TY, Liu Y, Koch AE, Challa P, Rautenbach RM, Mackey DA, Hewitt AW, Mitchell P, Wang JJ, Ziskind A, Carmichael T, Ramakrishnan R, Narendran K, Venkatesh R, Vijayan S, Zhao P, Chen X, Guadarrama-Vallejo D, Cheng CY, Perera SA, Husain R, Ho SL, Welge-Luessen UC, Mardin C, Schloetzer-Schrehardt U, Hillmer AM, Herms S, Moebus S, Nöthen MM, Weisschuh N, Shetty R, Ghosh A, Teo YY, Brown MA, Lischinsky I; Blue Mountains Eye Study GWAS Team.; Wellcome Trust Case Control Consortium 2., Crowston JG, Coote M, Zhao B, Sang J, Zhang N, You Q, Vysochinskaya V, Founti P, Chatzikyriakidou A, Lambropoulos A, Anastasopoulos E, Coleman AL, Wilson MR, Rhee DJ, Kang JH, May-Bolchakova I, Heegaard S, Mori K, Alward WL, Jonas JB, Xu L, Liebmann JM, Chowbay B, Schaeffeler E, Schwab M, Lerner F, Wang N, Yang Z, Frezzotti P, Kinoshita S, Fingert JH, Inatani M, Tashiro K, Reis A, Edward DP, Pasquale LR, Kubota T, Wiggs JL, Pasutto F, Topouzis F, Dubina M, Craig JE, Yoshimura N, Sundaresan P, John SW, Ritch R, Hauser MA, Khor CC. A common variant mapping to CACNA1A is associated with susceptibility to exfoliation syndrome. Nat Genet. 2015. April;47(4):387–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wilson MR, Easterbrook-Smith SB. Clusterin is a secreted mammalian chaperone. Trends Biochem Sci. 2000. March;25(3):95–8. [DOI] [PubMed] [Google Scholar]
- 11.Michel D, Chatelain G, North S, Brun G. Stress-induced transcription of the clusterin/apoJ gene. Biochem J. 1997. November 15;328 ( Pt 1):45–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zoubeidi A, Gleave M. Small heat shock proteins in cancer therapy and prognosis. Int J Biochem Cell Biol. 2012;44:1646–1656. [DOI] [PubMed] [Google Scholar]
- 13.Viard I, Wehrli P, Jornot L, Bullani R, Vechietti JL, Schifferli JA, Tschopp J, French LE. Clusterin gene expression mediates resistance to apoptotic cell death induced by heat shock and oxidative stress. J Invest Dermatol 1999;112: 290–6. [DOI] [PubMed] [Google Scholar]
- 14.Loison F, Debure L, Nizard P, le Goff P, Michel D, le Drean Y. Up-regulation of the clusterin gene after proteotoxic stress: implication of HSF1-HSF2 heterocomplexes. Biochem J 2006;395:223–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rohne P, Prochnow H, Wolf S, Renner B, Koch-Brandt C. The chaperone activity of clusterin is dependent on glycosylation and redox environment. Cell Physiol Biochem. 2014;34(5):1626–39. [DOI] [PubMed] [Google Scholar]
- 16.Choi-Miura NH, Takahashi Y, Nakano Y, Tobe T, Tomita M. Identification of the disulfide bonds in human plasma protein SP-40,40 (apolipoprotein-J). J Biochem. 1992;112(4):557–61. [DOI] [PubMed] [Google Scholar]
- 17.Nizard P, Tetley S, Le Drean Y, et al. Stress-induced retrotranslocation of clusterin/ApoJ into the cytosol. Traffic. 2007;8:554–565. [DOI] [PubMed] [Google Scholar]
- 18.Balantinou E, Trougakos IP, Chondrogianni N, et al. Transcriptional and posttranslational regulation of clusterin by the two main cellular proteolytic pathways. Free Radic Biol Med. 2009;46:1267–1274. [DOI] [PubMed] [Google Scholar]
- 19.Wilson MR, Easterbrook-Smith SB. Clusterin is a secreted mammalian chaperone. Trends in biochemical sciences. 2000;25(3):95–8. [DOI] [PubMed] [Google Scholar]
- 20.Humphreys DT, Carver JA, Easterbrook-Smith SB, et al. Clusterin has chaperone-like activity similar to that of small heat-shock proteins. J Biol Chem. 1999;274:6875–6881. [DOI] [PubMed] [Google Scholar]
- 21.Poon S, Rybchyn MS, Easterbrook-Smith SB, Carver JA, Pankhurst GJ, Wilson MR. Mildly acidic pH activates the extracellular molecular chaperone clusterin. J Biol Chem 2002; 277: 39532–40. [DOI] [PubMed] [Google Scholar]
- 22.Wyatt AR, Yerbury JJ, Wilson MR. Structural characterization of clusterin-chaperone client protein complexes. J Biol Chem 2009; 284: 21920–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hochgrebe T, Pankhurst GJ, Wilce J, Easterbrook-Smith SB. pH-dependent changes in the in vitro ligand-binding properties and structure of human clusterin. Biochemistry 2000; 39: 1411–9. [DOI] [PubMed] [Google Scholar]
- 24.Rosenberg ME, Girton R, Finkel D, et al. Apolipoprotein J/clusterin prevents progressive glomerulopathy of aging. Mol Cell Biol. 2002;22:1893–1902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Trougakos IP. The molecular chaperone apolipoprotein J/clusterin as a sensor of oxidative stress: implications in therapeutic approaches - a mini-review. Gerontology. 2013;59(6):514–23. [DOI] [PubMed] [Google Scholar]
- 26.Miners JS, Clarke P, Love S. Clusterin Levels are Increased in Alzheimer’s Disease and Influence the Regional Distribution of Aβ. Brain Pathol. 2016. June 1. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ronquist G Prostasomes: Their Characterisation: Implications for Human Reproduction: Prostasomes and Human Reproduction. Adv Exp Med Biol. 2015;868:191–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shim YJ, Tae YK, Kang BH, Park JS, Jeon SY, Min BH. Toll-like receptor 4 signaling is required for clusterin-induced tumor necrosis factor-α secretion in macrophage. Biochem Biophys Res Commun. 2017. January 22;482(4):1407–1412. [DOI] [PubMed] [Google Scholar]
- 29.Ma X, Bai Y. IGF-1 activates the P13K/AKT signaling pathway via upregulation of secretory clusterin. Mol Med Rep. 2012. December;6(6):1433–7. [DOI] [PubMed] [Google Scholar]
- 30.Zhang F, Kumano M, Beraldi E, Fazli L, Du C, Moore S, Sorensen P, Zoubeidi A, Gleave ME. Clusterin facilitates stress-induced lipidation of LC3 and autophagosome biogenesis to enhance cancer cell survival. Nat Commun. 2014. December 12;5:5775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Arumugam P, Samson A, Ki J, Song JM. Knockdown of clusterin alters mitochondrial dynamics, facilitates necrosis in camptothecin-induced cancer stem cells. Cell Biol Toxicol. 2017. January 7. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 32.Weinstein G, Beiser AS, Preis SR, Courchesne P, Chouraki V, Levy D, Seshadri S. Plasma clusterin levels and risk of dementia, Alzheimer’s disease, and stroke. Alzheimers Dement (Amst). 2016. July 9;3:103–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tellez T, Garcia-Aranda M, Redondo M. The Role of Clusterin in Carcinogenesis and its Potential Utility as Therapeutic Target. Curr Med Chem. 2016;23(38):4297–4308. [DOI] [PubMed] [Google Scholar]
- 34.Fini ME, Bauskar A, Jeong S, Wilson MR. Clusterin in the eye: An old dog with new tricks at the ocular surface. Exp Eye Res. 2016. June;147:57–71. [DOI] [PubMed] [Google Scholar]
- 35.Bauskar A, Mack WJ, Mauris J, Argüeso P, Heur M, Nagel BA, Kolar GR, Gleave ME, Nakamura T, Kinoshita S, Moradian-Oldak J, Panjwani N, Pflugfelder SC, Wilson MR, Fini ME, Jeong S. Clusterin Seals the Ocular Surface Barrier in Mouse Dry Eye. PLoS One. 2015. September 24;10(9):e0138958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kim JH, Kim JH, Jun HO, Yu YS, Min BH, Park KH, Kim KW. Protective effect of clusterin from oxidative stress-induced apoptosis in human retinal pigment epithelial cells. Invest Ophthalmol Vis Sci. 2010. January;51(1):561–6. [DOI] [PubMed] [Google Scholar]
- 37.Zhang C, Nie J, Feng L, Luo W, Yao J, Wang F, Wang H. The emerging roles of clusterin on reduction of both blood retina barrier breakdown and neural retina damage in diabetic retinopathy. Discov Med. 2016. April;21(116):227–37. [PubMed] [Google Scholar]
- 38.Borrás T The cellular and molecular biology of the iris, an overlooked tissue: the iris and pseudoexfoliation glaucoma. J Glaucoma. 2014. October-November;23(8 Suppl 1):S39–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Carnes MU, Allingham RR, Ashley-Koch A, Hauser MA. Transcriptome analysis of adult and fetal trabecular meshwork, cornea, and ciliary body tissues by RNA sequencing. Exp Eye Res. 2016. November 30. pii: S0014–4835(16)30504–8. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 40.Hardenborg E, Botling-Taube A, Hanrieder J, Andersson M, Alm A, Bergquist J. Protein content in aqueous humor from patients with pseudoexfoliation (PEX) investigated by capillary LC MALDI-TOF/TOF MS. Proteomics Clin Appl. 2009. March;3(3):299–306. [DOI] [PubMed] [Google Scholar]
- 41.Clusterin and complement activation in exfoliation glaucoma. Doudevski I, Rostagno A, Cowman M, Liebmann J, Ritch R, Ghiso J. Invest Ophthalmol Vis Sci. 2014. April 17;55(4):2491–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Proteomic analysis of exfoliation deposits. Ovodenko B, Rostagno A, Neubert TA, Shetty V, Thomas S, Yang A, Liebmann J, Ghiso J, Ritch R. Invest Ophthalmol Vis Sci. 2007. April;48(4):1447–57. [DOI] [PubMed] [Google Scholar]
- 43.Detecting protein aggregates on untreated human tissue samples by atomic force microscopy recognition imaging. Creasey R, Sharma S, Craig JE, Gibson CT, Ebner A, Hinterdorfer P, Voelcker NH. Biophys J. 2010. September 8;99(5):1660–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zenkel M, Pöschl E, von der Mark K, Hofmann-Rummelt C, Naumann GO, Kruse FE, Schlötzer-Schrehardt U. Differential gene expression in pseudoexfoliation syndrome. Invest Ophthalmol Vis Sci. 2005. October;46(10):3742–52. [DOI] [PubMed] [Google Scholar]
- 45.Clusterin deficiency in eyes with pseudoexfoliation syndrome may be implicated in the aggregation and deposition of pseudoexfoliative material. Zenkel M, Kruse FE, Jünemann AG, Naumann GO, Schlötzer-Schrehardt U. Invest Ophthalmol Vis Sci. 2006. May;47(5):1982–90. [DOI] [PubMed] [Google Scholar]
- 46.Wyatt AR, Yerbury JJ, Berghofer P, Greguric I, Katsifis A, Dobson CM, Wilson MR. Clusterin facilitates in vivo clearance of extracellular misfolded proteins. Cell Mol Life Sci 2011; 68: 3919–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Deming Y, Xia J, Cai Y, Lord J, Holmans P, Bertelsen S, Holtzman D, Morris JC, Bales K, Pickering EH, Kauwe J, Goate A, Cruchaga C; Alzheimer’s Disease Neuroimaging Initiative. A potential endophenotype for Alzheimer’s disease: cerebrospinal fluid clusterin. Neurobiol Aging. 2016. January;37:208.e1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Baig S, Palmer LE, Owen MJ, Williams J, Kehoe PG, Love S. Clusterin mRNA and protein in Alzheimer’s disease. J Alzheimers Dis. 2012;28(2):337–44. [DOI] [PubMed] [Google Scholar]
- 49.Yerbury JJ, Poon S, Meehan S, Thompson B, Kumita JR, Dobson CM, Wilson MR. The extracellular chaperone clusterin influences amyloid formation and toxicity by interacting with prefibrillar structures. FASEB J. 2007. August;21(10):2312–22. [DOI] [PubMed] [Google Scholar]
- 50.Genome-wide association study identifies variants at CLU and CR1 associated with Alzheimer’s disease. Lambert JC et al. Nat Genet. 2009. October;41(10):1094–9. [DOI] [PubMed] [Google Scholar]
- 51.Genome-wide association study identifies variants at CLU and PICALM associated with Alzheimer’s disease. Harold D, et al. Nat Genet. 2009. October;41(10):1088–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Du W, Tan J, Xu W, Chen J, Wang L. Association between clusterin gene polymorphism rs11136000 and late-onset Alzheimer’s disease susceptibility: A review and meta-analysis of case-control studies. Exp Ther Med. 2016. November;12(5):2915–2927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bettens K, Vermeulen S, Van Cauwenberghe C, Heeman B, Asselbergh B, Robberecht C, Engelborghs S, Vandenbulcke M, Vandenberghe R, De Deyn PP, Cruts M, Van Broeckhoven C, Sleegers K. Reduced secreted clusterin as a mechanism for Alzheimer-associated CLU mutations. Mol Neurodegener. 2015. July 16;10:30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Burdon KP, Sharma S, Hewitt AW, McMellon AE, Wang JJ, Mackey DA, Mitchell P, Craig JE. Genetic analysis of the clusterin gene in pseudoexfoliation syndrome. Mol. Vis. 2008;14:1727–1736. [PMC free article] [PubMed] [Google Scholar]
- 55.Krumbiegel M, Pasutto F, Mardin CY, Weisschuh N, Paoli D, Gramer E, Zenkel M, Weber BH, Kruse FE, Schlötzer-Schrehardt U, Reis A. Exploring functional candidate genes for genetic association in german patients with pseudoexfoliation syndrome and pseudoexfoliation glaucoma. Invest. Ophthalmol. Vis. Sci. 2009;50:2796–2801. [DOI] [PubMed] [Google Scholar]
- 56.Padhy B, Nanda GG, Chowdhury M, Padhi D, Rao A, Alone DP. Role of an extracellular chaperone, clusterin in the pathogenesis of pseudoexfoliation syndrome and pseudoexfoliation glaucoma. Exp. Eye. Res. 2014;127C:69–76. [DOI] [PubMed] [Google Scholar]
- 57.Dubey SK, Hejtmancik JF, Krishnadas SR, Sharmila R, Haripriya A, Sundaresan P. Lysyl oxidase-like 1 gene in the reversal of promoter risk allele in pseudoexfoliation syndrome. JAMA. Ophthalmol. 2014;132:949–955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Fan BJ, Pasquale LR, Kang JH, Levkovitch-Verbin H, Haines JL, Wiggs JL. Association of clusterin (CLU) variants and exfoliation syndrome: An analysis in two Caucasian studies and a meta-analysis. Exp Eye Res. 2015. October;139:115–22.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Vardarajan BN, Ghani M, Kahn A, Sheikh S, Sato C, Barral S, Lee JH, Cheng R, Reitz C, Lantigua R, Reyes-Dumeyer D, Medrano M, Jimenez-Velazquez IZ, Rogaeva E, St George-Hyslop P, Mayeux R. Rare coding mutations identified by sequencing of Alzheimer disease genome-wide association studies loci. Ann Neurol. 2015. September;78(3):487–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Janig E, Haslbeck M, Aigelsreiter A, Braun N, Unterthor D, Wolf P, Khaskhely NM, Buchner J, Denk H, Zatloukal K. Clusterin associates with altered elastic fibers in human photoaged skin and prevents elastin from ultraviolet-induced aggregation in vitro. Am J Pathol. 2007. November;171(5):1474–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Pasquale LR, Jiwani AZ, Zehavi-Doriphthalmol. Solar exposure and residential geographic history in relation to exfoliation syndrome in the United States and Israel. JAMA Ophthalmol. 2014. December;132(12):1439–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kang JH, Wiggs JL, Pasquale LR. Relation between time spent outdoors and exfoliation glaucoma or exfoliation glaucoma suspect. Am J Ophthalmol. 2014. September;158(3):605–14.e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kang JH, Loomis S, Wiggs JL, Stein JD, Pasquale LR. Demographic and geographic features of exfoliation glaucoma in 2 United States-based prospective cohorts. Ophthalmology. 2012. January;119(1):27–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Stein JD, Pasquale LR, Talwar N, Kim DS, Reed DM, Nan B, Kang JH, Wiggs JL, Richards JE. Geographic and climatic factors associated with exfoliation syndrome. Arch Ophthalmol. 2011. August;129(8):1053–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ebbert MT, Boehme KL, Wadsworth ME, Staley LA; Alzheimer’s Disease Neuroimaging Initiative.; Alzheimer’s Disease Genetics Consortium., Mukherjee S, Crane PK, Ridge PG, Kauwe JS. Interaction between variants in CLU and MS4A4E modulates Alzheimer’s disease risk. Alzheimers Dement. 2016. February;12(2):121–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zoubeidi A, Chi K, Gleave M. Targeting the cytoprotective chaperone, clusterin, for treatment of advanced cancer. Clin Cancer Res. 2010;16:1088–1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Yamamoto Y, Lin PJ, Beraldi E, Zhang F, Kawai Y, Leong J, Katsumi H, Fazli L, Fraser R, Cullis PR, Gleave M. siRNA Lipid nanoparticle potently silences clusterin and delays progression when combined with androgen receptor cotargeting in enzalutamide-resistant prostate cancer. Clin Cancer Res. 2015. November 1;21(21):4845–55. [DOI] [PubMed] [Google Scholar]
- 68.Trougakos IP, Gonos ES. Clusterin/apolipoprotein J in human aging and cancer. Int J Biochem Cell Biol. 2002;34:1430–1448. [DOI] [PubMed] [Google Scholar]
- 69.Shiota M, Zardan A, Takeuchi A, Kumano M, Beraldi E, Naito S, Zoubeidi A, Gleave ME. Clusterin mediates TGF-β-induced epithelial-mesenchymal transition and metastasis via Twist1 in prostate cancer cells. Cancer Res. 2012. October 15;72(20):5261–72. [DOI] [PubMed] [Google Scholar]
- 70.Zellweger T, Chi K, Miyake H, et al. Enhanced radiation sensitivity in prostate cancer by inhibition of the cell survival protein clusterin. Clin Cancer Res. 2002;8:3276–3284. [PubMed] [Google Scholar]
- 71.Zoubeidi A, Chi K, Gleave M. Targeting the cytoprotective chaperone, clusterin, for treatment of advanced cancer. Clin Cancer Res. 2010;16:1088–1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Chi KN, Siu LL, Hirte H, et al. A Phase I Study of OGX-011, a 2ʹ-methoxyethyl phosphorothioate antisense to clusterin, in combination with docetaxel in patients with advanced cancer. Clin Cancer Res. 2008;14:833–839. [DOI] [PubMed] [Google Scholar]
- 73.Laskin J, Hao D, Canil C, et al. A phase I/II study of OGX-011 and a gemcitabine (GEM)/platinum regimen as first-line therapy in 85 patients with advanced non-small cell lung cancer. J Clin Oncol. 2007;25:7596. [Google Scholar]
- 74.Laskin JJ, Nicholas G, Lee C, et al. Phase I/II Trial of Custirsen (OGX-011), an inhibitor of clusterin, in combination with a gemcitabine and platinum regimen in patients with previously untreated advanced non-small cell lung cancer. J Thorac Oncol. 2012;7:579–586. [DOI] [PubMed] [Google Scholar]
- 75.Wilson MR, Zoubeidi A. Clusterin as a therapeutic target. Expert Opin Ther Targets. 2017. Feb;21(2):201–213. [DOI] [PubMed] [Google Scholar]
- 76.DeMattos RB, O’dell MA, Parsadanian M, et al. Clusterin promotes amyloid plaque formation and is critical for neuritic toxicity in a mouse model of Alzheimer’s disease. Proc Natl Acad Sci USA. 2002;99:10843–10848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.DeMattos RB, Cirrito JR, Parsadanian M, et al. ApoE and clusterin co-operatively suppress Aβ levels and deposition: evidence that apoE regulates extracellular Aβ metabolism in vivo. Neuron. 2004;41:193–202. [DOI] [PubMed] [Google Scholar]
- 80.Fujii A, Shearer TR, Azuma M. Galectin-3 enhances extracellular matrix associations and wound healing in monkey corneal epithelium. Exp Eye Res. 2015. August;137:71–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Aragona P, Aguennouz M, Rania L, Postorino E, Sommario MS, Roszkowska AM, De Pasquale MG, Pisani A, Puzzolo D. Matrix metalloproteinase 9 and transglutaminase 2 expression at the ocular surface in patients with different forms of dry eye disease. Ophthalmology. 2015. January;122(1):62–71. [DOI] [PubMed] [Google Scholar]


