Targeted double-strand breaks (DSBs) in genomes can be introduced efficiently by endonucleases (Urnov et al., 2010; Jinek et al., 2012; Joung and Sander, 2013), including zinc-finger nucleases, transcription activator-like effector nucleases, and clustered regularly interspaced palindromic repeats (CRISPR)/Cas9. After DSBs, DNA repair is mainly via homology-directed repair (HDR) and/or non-homologous end joining (NHEJ) (Hustedt and Durocher, 2016). It was reported that genomic DNA replacement can be achieved via HDR at the site of DSBs in multiple organisms (Dickinson et al., 2013; Yang et al., 2013; Zu et al., 2013), but the efficiency is still not enough for general application, in particular for replacing long DNA fragment that is more than hundreds of base pairs (bps). As NHEJ is 10-fold more active than HDR at DSB sites (Mao et al., 2008), we speculated that NHEJ can be utilized to implement long genomic DNA replacement with high efficiency.
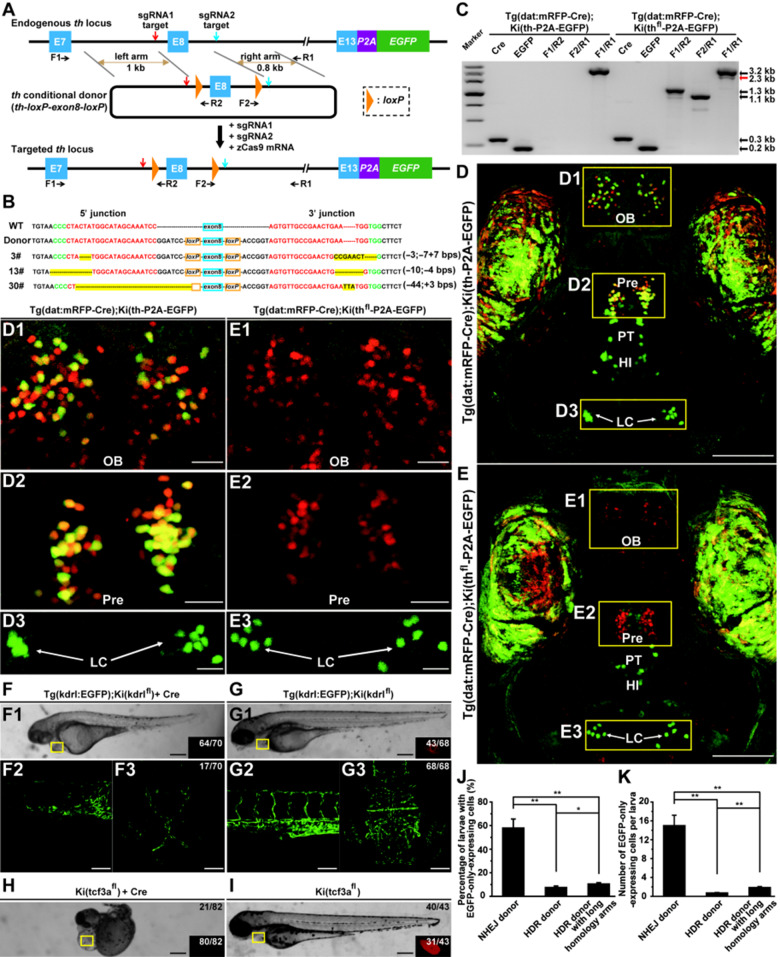
To develop NHEJ-mediated DNA replacement method, we first tried to make a conditional allele of the zebrafish tyrosine hydroxylase (th) gene. Two single guide RNAs (sgRNAs) were designed to specifically target two different non-coding sites in the 7th (for sgRNA1) and 8th (for sgRNA2) introns of the th gene with a cleavage efficiency of 59% and 68%, respectively (Figure 1A;Supplementary Figure S1A). We constructed a th conditional donor th-loxP-exon8-loxP (Figure 1A, middle), and then co-injected it with the zebrafish optimized Cas9 mRNA (zCas9 mRNA) and the two sgRNAs into one-cell-stage embryos of the knockin zebrafish line Ki(th-P2A-EGFP), in which th-expressing cells are labeled by EGFP (Li et al., 2015). Successful DNA replacement in progenies of F0 was verified by polymerase chain reaction (PCR) using th locus-specific (F1 and R1) and donor-specific (F2 and R2) primers (Supplementary Figure S1B), and sequencing of PCR products amplified across the entire edited region with th locus-specific primers (Supplementary Figure S1C). In a total of 41 F0 screened, we identified three replacement founders [Ki(thfl-P2A-EGFP)] with an 18% ± 9% F1 progeny carrying the replacement event (Supplementary Figure S2 and Tables S1 and S2). The indels at both 5ʹ and 3ʹ junctions indicate that the DNA replacement occurred through NHEJ (Figure 1B;Supplementary Figure S1C).
Figure 1.
NHEJ-mediated replacement approach for efficient generation of conditional alleles in zebrafish. (A) Schematic of the strategy for NHEJ-mediated DNA replacement at the th locus of Ki(th-P2A-EGFP) zebrafish. The sgRNA1 and sgRNA2 targets are indicated by red and blue arrows, respectively. E7, E8, and E13 represent the 7th, 8th, and 13th exons, respectively. The primers (F1, R1, F2, R2) for verification are indicated by short black arrows. (B) 5′ and 3′ junction sequences of F1 progenies of three founders carrying DNA replacement in comparison with the sequences of wild-type (WT) zebrafish and the donor vector. The indel mutations are highlighted in yellow, and the protospacer adjacent motif (PAM) and sgRNA target sequences are shown in green and red, respectively. The indels at the 5′ and 3′ junctions are shown in the brackets at right. (C) PCR verification of conditional knockout, showing an additional product at ∼2.3 kb (red arrow), in principle excised from the original 3.2-kb fragment by Cre. (D and E) Projected in vivo confocal images (dorsal view) of [Tg(dat:mRFP-Cre);Ki(th-P2A-EGFP)] (D) and [Tg(dat:mRFP-Cre);Ki(thfl-P2A-EGFP)] (E) larvae at 5 days post-fertilization (dpf), showing that in the conditional knockout embryo, mRFP-Cre-expressing cells (in red) do not express EGFP. The enlarged views of the areas outlined by the rectangles in D and E are shown in D1‒D3 and E1‒E3, respectively. HI, intermediate hypothalamus; LC, locus coeruleus; OB, olfactory bulb; Pre, pretectum; PT, posterior tubercular. (F and G) Generation of the kdrl conditional allele. In vivo bright-filed (top) and projected confocal (bottom) images of 3-dpf [Tg(kdrl:EGFP);Ki(kdrlfl)] intercross larvae with (F) or without (G) Cre mRNA injection at one-cell stage, showing that Cre mRNA injection causes the loss of DsRed expression in the heart (inset in F1) and the defects of blood vessels in the trunk (F2) and brain (F3). (H and I) Generation of the tcf3a conditional allele. Bright-filed image of 2.5-dpf Ki(tcf3afl) intercross larvae with Cre mRNA injection showing the defects of gross morphology (H). (J and K) Comparison of the efficiencies of NHEJ- and HDR-mediated DNA replacement at the gfap locus. (J) Percentage of larvae with EGFP-only-expressing glial cells. (K) Average number of EGFP-only-expressing glial cells per larva. n = 170, 277, and 245. *P < 0.05; **P < 0.01. Data shown are mean ± SEM. Scale bar, 20 µm (D1‒D3, E1‒E3), 100 µm (D, E, F2, F3, G2, G3), 250 µm (F1, G1, H, I).
To examine the excision by Cre, we performed PCR in larvae generated by crossing conditional allele Ki(thfl-P2A-EGFP) with Tg(dat:mRFP-Cre), in which Cre and the monomer red fluorescent protein (mRFP) are specifically expressed in dopaminergic neurons. We found that there was a band with ∼0.9 kb smaller than control (Figure 1C, red arrow), suggesting correct excision by Cre. Furthermore, in embryos of Ki(thfl-P2A-EGFP) zebrafish crossed with Tg(dat:mRFP-Cre), EGFP expression was not observed in mRFP-positive cells in the Pre and OB (Figure 1E, E1 and E2), which was different from the control (Figure 1D, D1 and D2), confirming the occurrence of the frame-shift mutation induced by excising the 8th exon (136 bps in length) between the two loxP sites in the th locus. As a control, EGFP expression was not affected in noradrenergic neurons in the locus coeruleus (LC) where mRFP-Cre did not express (Figure 1D, D3 and E, E3).
Similarly, we also successfully generated two other zebrafish conditional alleles, kdrl (Figure 1F and G; Supplementary Figures S3‒S5) and tcf3a (Figure 1H and I; Supplementary Figures S6 and S7), and two EGFP reporter lines, th (Supplementary Figures S8 and S9) and gfap (Supplementary Figures S10 and S11).
To compare the efficiency of NHEJ- and HDR-mediated DNA replacement approaches, we modified the gfap reporter donor by adding a negative selective marker (SM; Supplementary Figure S12A). The negative SM consisted of an internal ribozyme entry site (IRES), tdTomato (tdT) coding sequence, and SV40 polyA signal (IRES-tdT), which was linked to the 3ʹ side of the right arm (gfap-P2A-EGFP-IRES-tdT). We validated the donor vector by co-injecting with sgRNA1 and zCas9, and found that most of EGFP-expressing cells (197/219 from 16 larvae) also expressed tdT, indicating the insertion of full-length donor vector at the sgRNA1 target site (Supplementary Figure S12B; see also Li et al., 2015). We also constructed two HDR-mediated donors, in which both sgRNA1 and sgRNA2 targets were mutated at protospacer adjacent motifs (Supplementary Figure S12Dand E, left). One HDR-mediated donor had the same homology arms with the NHEJ-mediated donor (Supplementary Figure S12Cand D), while the other had both longer left and right homology arms (Supplementary Figure S12Cand E). By comparing the ratio of the larvae containing EGFP-only-expressing glial cells and the average number of EGFP-only-expressing glial cells in each embryo, we found that both values in embryos injected with the NHEJ donor were ∼5.5 and 8.0 times as large as those with HDR donors with long homology arms, respectively (Figure 1J and K). As multiple-fragment integration rarely occurs (Auer et al., 2014), EGFP-only expression in NHEJ donor-injected embryos can also be due to NHEJ-mediated insertion of the P2A-EGFP fragment at the sgRNA1 target site. Considering the fact that we identified 10 founders carrying DNA replacement and 5 founders carrying insertion in all of our experiments (Supplementary Table S2), we roughly estimated that the DNA replacement efficiency of NHEJ-mediated approach in the gfap locus is ∼3.7‒5.3 times as much as that of HDR-mediated approach.
In this study, we developed an efficient long DNA fragment replacement method by introducing two DSB sites at non-coding genomic regions via the CRISPR/Cas9 system. Interestingly, we found that homology arms extended out of both target sites are important for increasing NHEJ-mediated knockin efficiency (Supplementary Table S3), suggesting that the homology arms are helpful to increase the probability of correct integration when concurrent cleavage happens in both the donor and genome. Taken together, the high efficiency and simplicity make our strategy an applicable DNA replacement method for zebrafish and even other organisms.
[Supplementary material is available at Journal of Molecular Cell Biology online. This work was supported by the Star Program of Shanghai Jiao Tong University (YG2019ZDA20), Key Research Program of Frontier Sciences (QYZDY-SSW-SMC028), and Shanghai Municipal Science and Technology Major Project (18JC1410100 and 2018SHZDZX05).]
Supplementary Material
References
- Auer T.O., Duroure K., De Cian A., et al. (2014). Highly efficient CRISPR/Cas9-mediated knock-in in zebrafish by homology-independent DNA repair. Genome Res. 24, 142–153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickinson D.J., Ward J.D., Reiner D.J., et al. (2013). Engineering the Caenorhabditis elegans genome using Cas9-triggered homologous recombination. Nat. Methods 10, 1028–1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hustedt N., Durocher D. (2016). The control of DNA repair by the cell cycle. Nat. Cell Biol. 19, 1–9. [DOI] [PubMed] [Google Scholar]
- Jinek M., Chylinski K., Fonfara I., et al. (2012). A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 337, 816–821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joung J.K., Sander J.D. (2013). TALENs: a widely applicable technology for targeted genome editing. Nat. Rev. Mol. Cell Biol. 14, 49–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., Zhang B.B., Ren Y.G., et al. (2015). Intron targeting-mediated and endogenous gene integrity-maintaining knockin in zebrafish using the CRISPR/Cas9 system. Cell Res. 25, 634–637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao Z., Bozzella M., Seluanov A., et al. (2008). Comparison of nonhomologous end joining and homologous recombination in human cells. DNA Repair 7, 1765–1771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urnov F.D., Rebar E.J., Holmes M.C., et al. (2010). Genome editing with engineered zinc finger nucleases. Nat. Rev. Genet. 11, 636–646. [DOI] [PubMed] [Google Scholar]
- Yang H., Wang H., Shivalila C.S., et al. (2013). One-step generation of mice carrying reporter and conditional alleles by CRISPR/Cas-mediated genome engineering. Cell 154, 1370–1379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zu Y., Tong X., Wang Z., et al. (2013). TALEN-mediated precise genome modification by homologous recombination in zebrafish. Nat. Methods 10, 329–331. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.