Abstract
Amyotrophic lateral sclerosis (ALS) is a late-onset neurodegenerative disease selectively affecting motor neurons, leading to progressive paralysis. Although most cases are sporadic, ∼10% are familial. Similar proteins are found in aggregates in sporadic and familial ALS, and over the last decade, research has been focused on the underlying nature of this common pathology. Notably, TDP-43 inclusions are found in almost all ALS patients, while FUS inclusions have been reported in some familial ALS patients. Both TDP-43 and FUS possess ‘low-complexity domains’ (LCDs) and are considered as ‘intrinsically disordered proteins’, which form liquid droplets in vitro due to the weak interactions caused by the LCDs. Dysfunctional ‘liquid–liquid phase separation’ (LLPS) emerged as a new mechanism linking ALS-related proteins to pathogenesis. Here, we review the current state of knowledge on ALS-related gene products associated with a proteinopathy and discuss their status as LLPS proteins. In addition, we highlight the therapeutic potential of targeting LLPS for treating ALS.
Keywords: phase separation, stress granule, motor neuron, ALS therapy
Introduction
Background of amyotrophic lateral sclerosis (ALS)
ALS is a fatal late-onset neurodegenerative disease affecting primarily upper and lower motor neurons. The average age of onset is between 51 and 66 years old (Longinetti and Fang, 2019). ALS is a progressive disease and initial muscle weakness progresses into paralysis and death of the patient, most often due to respiratory failure typically 2–5 years after displaying the first symptoms (Brown and Al-Chalabi, 2017).
Genetics of ALS
ALS cases are predominantly sporadic (sALS), with ∼10% being inherited (familial ALS; fALS). Mutations in >25 genes have been linked to this disease (Nguyen et al., 2018). However, the majority of fALS can be explained by alterations in four main genes, namely C9orf72, SOD1, TARDBP, and FUS. Hexanucleotide repeat expansions in the chromosome 9 open reading frame 72 (C9orf72) gene are the most common genetic cause of fALS, accounting for ∼40% of fALS (Majounie et al., 2012). Mutations in the superoxide dismutase 1 (SOD1) gene account for ∼20% of fALS (Rosen et al., 1993), while mutations in the genes encoding DNA/RNA-binding proteins TAR DNA-binding protein 43 (TARDBP, encoding TDP-43) and fused in sarcoma (FUS) are causal to ∼5% and ∼4% of fALS, respectively (Kwiatkowski et al., 2009; Vance et al., 2009; Taylor et al., 2016).
Pathology of ALS
A characteristic pathological feature of ALS is protein aggregation in the cytoplasm of motor neurons and sometimes also in other cell types, such as glial cells (Brown and Al-Chalabi, 2017). In 97% of ALS patients, these inclusions contain TDP-43 (Neumann et al., 2006).
ALS belongs to a disease spectrum also including frontotemporal dementia (FTD), a disease affecting mainly the frontal and temporal lobes associated with dementia. Indeed, both diseases sometimes occur simultaneously (ALS/FTD) and exhibit clinical and pathological overlap, as well as common genetic causes. In post-mortem material of some FTD patients, TDP-43 or FUS aggregates can be found in the absence of pathological mutations in the genes encoding these proteins (Blokhuis et al., 2013).
During the last decades, researchers sought a unifying mechanism responsible for the complex nature of ALS pathology. However, ALS seems to be a polygenic disease in which multiple small hits (risk variants) combined with yet unclear environmental factors could lead to pathological changes in multiple pathways, concluding in motor neuron death. This multistage model is consistent with the observation that the disease is caused by a six-step process (Al-Chalabi et al., 2014). Liquid–liquid phase separation (LLPS) is one process implicated in major pathways in ALS-related pathogenicity and will be the focus of this review.
Potential role of LLPS in ALS
LLPS drives the formation of membraneless organelles
LLPS is the phenomenon in which a homogenous fluid de-mixes into two distinct liquid phases, one being condensed and the other one dilute, resulting in a membraneless boundary that allows for selective passage (Brangwynne, 2013; Hyman et al., 2014). This can be observed in vitro, where phase-separating proteins produce spherical droplets visible under the microscope or even by eye as a cloudy state. This state is reversible and dynamic, but the droplets can later change physical state, going from liquid to gel, and even further to solid. LLPS plays an important role in normal cell physiology and is used to regulate cellular functions such as cellular compartment control, metabolic processing, and signalling using membraneless organelles (Hyman et al., 2014; Lin et al., 2015). Proteins that drive phase separation engage in multivalent interactions, which stem from domain–domain, domain–motif, and motif−motif interactions and direct interactions of intrinsically disordered regions (IDRs). IDRs typically do not encode a particular 3D conformation and are often composed of a limited number of amino acids and/or repetitive sequence elements. This means that they have a low sequence complexity and, therefore, these domains are termed low-complexity domains (LCDs) that are rich in charged amino acids (Kato et al., 2012; Molliex et al., 2015). Many of these LCD-containing proteins can be found in membraneless organelles (Markmiller et al., 2018).
Stress granules (SGs) are an example of membraneless organelles. Assembly of these granules is induced in the cytoplasm in response to adverse conditions, such as immediate stresses, concentration changes, or pH fluctuations. SGs sequester free mRNA and RNA-binding proteins (RBPs) in order to halt translation and to conserve energy for acute cellular needs (Kedersha and Anderson, 2002, 2007). As conditions improve, LLPS is reversed. Advanced microscopy techniques have revealed that SGs consist of a highly concentrated core made up of mRNA and proteins, surrounded by the shell that is more dynamic in nature (Protter and Parker, 2016). Online databases for SG proteomes have been created to understand the nature of proteins collected within these organelles (http://rnagranuledb.lunenfeld.ca/). This type of analysis showed that SGs are enriched in intrinsically disordered proteins (Reijns et al., 2008; Han et al., 2012; Jain et al., 2016), but not limited to LCD proteins (Youn et al., 2019).
ALS-associated proteins aggregate into membraneless organelles
TDP-43 and FUS are associated with pathological ALS aggregates that could arise from improperly disassembled SGs (Dewey et al., 2012). TDP-43 and FUS have been associated with other membraneless organelles, such as DNA repair sites, paraspeckles, and transport granules (Andersson et al., 2008; Dormann et al., 2010). As the assembly and disassembly of LLPS are tightly regulated, it is not surprising that the dysfunction in this process could be implicated in disease aetiology. Despite the SG being the most studied of all membraneless organelles, other noteworthy organelles assist in RNA processing and ribosomal assembly.
Here, we will summarize the information on ALS-related proteins predicted and found to phase separate and the role of this process in pathogenicity. Meanwhile, we will discuss potential therapeutic avenues targeting LLPS.
ALS-LLPS proteins
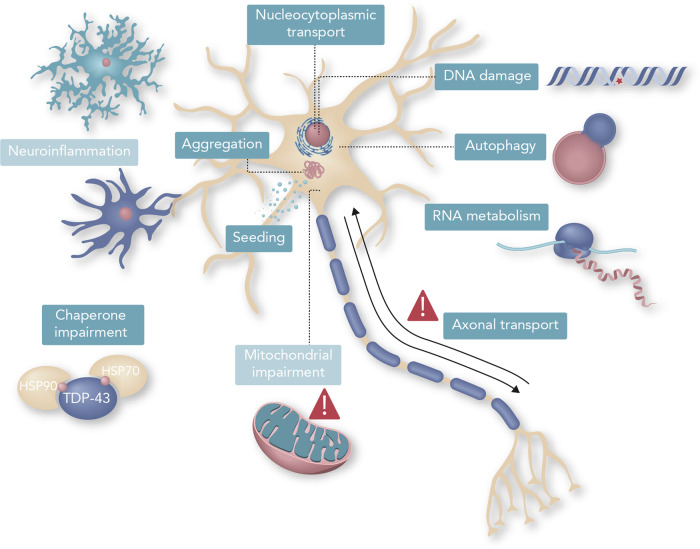
Many pathogenic pathways involved in ALS rely on LLPS. Indeed, evidence implicates phase separation in nucleocytoplasmic transport, RNA metabolism, DNA repair, mitochondrial function, protein aggregation, among others (Figure 1 and Table 1).
Figure 1.
Mechanisms related to ALS associated or not with LLPS. Schematic illustration of the most important pathological processes related to ALS that rely on or are influenced by the functioning of LLPS in motor neurons. Grey boxes are for processes not directly linked to LLPS yet. Detailed evidence for the mechanisms involved in LLPS can be found in Table 1.
Table 1.
Mechanisms in ALS and evidence supporting involvement of phase separation.
| Mechanism | LLPS | General effect | Evidence | References |
|---|---|---|---|---|
| Autophagy | Yes | Autophagy proteins regulating LLPS | p62 is involved in the aggregation of polyubiquitinated misfolded proteins by LLPS and forms droplets in vivo with liquid-like properties | Sun et al. (2018) |
| UBQLN2 disruption | ALS-linked mutations modulate Ubiquilin-2 LLPS | Dao et al. (2018) | ||
| Axonal transport | Yes | DPRs causing transport defects | Altered mRNA transport and local translation; LLPS influences cytoskeletal function and microtubule-based transport by steric hindrance | De Vos and Hafezparast (2017); Burk and Pasterkamp (2019); Guo et al. (2019) |
| DPRs cause perturbation of kinesin-1 and dynein-1 motors along microtubules in vitro and in vivo | Fumagalli et al. (2019) | |||
| Chaperone impairment | Yes | Chaperone binding impaired by LLPS proteins | Kapβ2 inhibits LLPS of FUS | Yoshizawa et al. (2018) |
| Hsp104 mediates disaggregation of TDP-43 and FUS fibrils | Shorter (2016) | |||
| Mutant UBQLN2 lacks recognition by HSP70 causing accumulation of misfolded/aggregated proteins | Hjerpe et al. (2016) | |||
| TNPO1/Kapβ2 are FUS chaperones mediating phase separation and SG association of FUS | Hofweber et al. (2018) | |||
| Hsp27 chaperones FUS to undergo LLPS in response to stress-induced phosphorylation | Liu et al. (2020) | |||
| DNA repair | Yes | DNA repair impeded | 53BP1 determines liquid-like behaviour of DNA repair compartments | Penndorf et al. (2018) |
| FUS involved in DNA repair | Wang et al. (2013) | |||
| Activation of PARP-1 directs FUS to DNA damage sites | Singatulina et al. (2019) | |||
| Inflammation | Maybe | No effect | No direct evidence; TIA1 and G3BP are linked with microglia function in Alzheimer’s disease | Ghosh and Geahlen (2015) |
| Mitochondrial dysfunction | Maybe | Mitochondrial nucleoids formed by LLPS | TFAM spontaneously phase separates | Smith et al. (2019) |
| Nucleocytoplasmic transport | Yes + No | DPRs and nuclear pore interactions influencing LLPS behaviour | (G4C2)58 expression in Drosophila salivary glands leads to abnormal nuclear membranes | Freibaum et al. (2015) |
| PR50 and GR50 interact with nuclear pore proteins in HEK cells | Lee et al. (2016) | |||
| Poly-PR peptides lead to blockage of nuclear pores in U2OS cells treated with PR20 peptides | Shi et al. (2017) | |||
| Drosophila salivary glands expressing (G4C2)30 repeats show impaired import by sequestration of RanGap in nuclear RNA foci | Zhang et al. (2015) | |||
| HEK293 cells transfected with PR50/GR50 show sequestration to SGs of proteins involved in nuclear transport | Zhang et al. (2018) | |||
| Poly-GR and poly-PR expressed in motor neurons and cell lines do not directly interfere with nucleocytoplasmic transport | Vanneste et al. (2019) | |||
| Nucleocytoplasmic transport factors identified as modifiers of DPR pathology in a Drosophila screen | Boeynaems et al. (2016) | |||
| Protein aggregation | Yes | Proteins in droplets evolving to protein aggregates | ALS mutations of FUS and TDP-43 cause an acceleration of aberrant phase transition | Patel et al. (2015); Conicella et al. (2016) |
| RNA metabolism | Yes | Droplets sequestering the machinery of RNA metabolism and translation, causing significant reductions in new protein synthesis | Pathological inclusions are driven by aberrant interactions between LCDs of TDP-43 that can be antagonized with RNA | Mann et al. (2019) |
| Non-functional transcription machinery causes general reduction in protein synthesis | Butti and Patten (2019) | |||
| LLPS of poly(GR) and poly(PR) in vitro enhances the multivalent interactions of the liquid phase of SGs in cells | Boeynaems et al. (2017) | |||
| Seeding of proteins | Yes | Recombinant TDP-43 seeds forming droplets of protein | Exposure to TDP-43 seeds in neuronal-like cells leads to the formation of protein droplets | Gasset-Rosa et al. (2019) |
Pathogenic phenotypes are shown in blue background and non-pathogenic phenotypes are shown in beige background. Yes/no results in the LLPS column refer to the presence/absence of direct involvement of LLPS in a particular ALS-related mechanism.
DPRs, arginine-rich dipeptide repeat proteins; Kapβ2, karyopherin-β2; TFAM, mitochondrial transcription factor A; TNPO1, transportin 1.
The major findings in phase separation related to ALS reveal (i) that pathogenic mutations in ALS-related genes lead to faster maturation of droplets to solid fibrillar aggregates (Johnson et al., 2009; Conicella et al., 2016; Schmidt and Rohatgi, 2016), (ii) that several ALS-related proteins such as TIA1 (Mackenzie et al., 2017), TDP-43 (Colombrita et al., 2009), FUS, Taf15 (Andersson et al., 2008; Abramzon et al., 2012), heterogeneous nuclear ribonucleoprotein A1 (hnRNPA1) and hnRNPA2 (McDonald et al., 2011), and Profilin1 (Wu et al., 2012) are found in SGs, and (iii) that the ALS pathological proteins FUS and TDP-43 have an LCD, making them prone to participate in weak and dynamic interactions (Kato et al., 2012; Molliex et al., 2015). The overexpression of these proteins produced SGs, similar to the ones formed after the addition of arsenite (Winton et al., 2008; Liu-Yesucevitz et al., 2010; Boeynaems et al., 2017). Moreover, liquid granule proteins G3BP1 and TIAR-2 are essential for granule formation implicated in axon regeneration, an important feature for motor neurons.
In Table 2, we list genes encoding proteins associated with ALS. The scores were obtained using PSPer (Orlando et al. 2019), an unsupervised tool, which provides an estimation of the probability that the target proteins undergo LLPS using a molecular mechanism similar to FUS-like proteins. The aggregation propensity for each protein was also evaluated using TANGO (Fernandez-Escamilla et al., 2004) and PASTA2 (Walsh et al., 2014). To estimate the global aggregation propensity of a protein, we used the maximum TANGO score, likely representing the aggregation propensity of the strongest aggregate-prone region.
Table 2.
Genes associated with ALS/FTD with LLPS and aggregation scores.
| Gene | LLPS score | Phase separation confirmed | Aggregation score | Found in aggregates |
|---|---|---|---|---|
| ADAR2 | 0.242 | Not yet | 98.368 | Yes |
| ANXA11 | 0.405 | Yes | 76.584 | Yes |
| ATXN2 | 0.633 | Yes | 93.428 | Yes |
| C9orf72 | 0.137 | No (DPRs only) | 99.952 | DPR yes |
| DNAJC7 | 0.289 | Not yet | 62.24 | Unknown |
| EWS | 0.857 | Yes | 4.521 | Yes |
| FUS | 0.915 | Yes | 4.473 | Yes |
| FIG4 | 0.285 | Not yet | 99.592 | Yes |
| hnRNPA1 | 0.603 | Yes | 92.027 | Yes |
| hnRNPA2B1 | 0.649 | Yes | 55.529 | Yes |
| hnRNPA3 | 0.643 | Yes | 92.132 | Yes |
| KIF5A | 0.236 | Not yet | 60.092 | Unknown |
| Matrin 3 | 0.422 | Not yet | 7.724 | Yes |
| NEK1 | 0.389 | Yes | 94.682 | Unknown |
| OPTN | 0.197 | Not yet | 79.727 | Yes |
| SOD1 | 0.223 | Not yet | 14.877 | Yes |
| SGMR1 | 0.173 | Not yet | 99.951 | Yes |
|
SYNCRIP (hnRNPQ) |
0.571 | Yes | 74.295 | Yes |
| SQSTM | 0.321 | Not yet | 6.455 | Yes |
| TARDBP | 0.376 | Yes | 97.627 | Yes |
| TAF15 | 0.801 | Yes | 4.652 | Yes |
| TIA1 | 0.422 | Yes | 47.614 | Yes |
| TBK1 | 0.194 | Not yet | 96.825 | No |
| UBQLN2 | 0.241 | Yes | 98.684 | Yes |
| VCP | 0.288 | Not yet | 61.728 | Yes |
Scores indicated in red background: highly likely to aggregate/undergo LLPS, LLPS score >0.6, aggregation score >60.
Scores indicated in orange background: likely to aggregate/undergo LLPS, LLPS score >0.3, aggregation score >40.
Scores indicated in beige background: somewhat likely to aggregate/undergo LLPS, LLPS score >0.2, aggregation score >10.
Scores indicated in white background: unlikely to aggregate/undergo LLPS, LLPS score <0.2, aggregation score <10.
TDP-43
TDP-43 is the most studied protein in ALS, as it is the main protein found in pathological end-stage inclusions. This aggregated form of TDP-43 is ubiquitinated, mostly phosphorylated (Arai et al., 2006; Neumann et al., 2009a), and/or acetylated in ALS cases (Cohen et al., 2015). In healthy individuals, TDP-43 resides in the nucleus, binds to DNA and RNA, and is involved in mRNA stability and splicing. The most puzzling observation in ALS-related TDP-43 pathology remains the appearance of TDP-43 aggregates without mutation in the TARDBP gene.
Structurally, TDP-43 contains an LCD in the C-terminal region, with a glycine-rich region, and a strong hydrophobic region rich in glutamine/asparagine (making it ‘prion-like’) (Gitler and Shorter, 2011). This LCD harbours most ALS-linked mutations (Kabashi et al., 2008; Sreedharan et al., 2008).
In the nucleus, TDP-43 is found in dimers, trimers, tetramers, and higher-order oligomers by the interaction of their N-terminal domains (Afroz et al., 2017). Under pathological conditions, TDP-43 leaves the nucleus and forms complexes that are post-translationally modified (Neumann et al., 2006, 2009a; Cohen et al., 2015; Nonaka et al., 2016). Recombinant TDP-43 readily aggregates and forms oligomeric species, which are toxic to cells (Fang et al., 2014). In addition, the C-terminal domain of TDP-43 is also known to aggregate (Johnson et al., 2009). Using solution nuclear magnetic resonance, it was shown that in the droplet state, the TDP-43 LCD forms monomeric states bearing potential for self-aggregation (Conicella et al., 2016). Increasing the concentration of the TDP-43 LCD leads to more gel-like formations. Moreover, expressing proteins with ALS-linked mutations also decreases the liquid properties of the TDP-43 LCD (Conicella et al., 2016). These observations provide direct evidence that ALS mutations disrupt the phase-separating properties of this protein.
TDP-43 was found in the SG proteome (Ayala et al., 2008; Colombrita et al., 2009), and the overexpression of TDP-43 was reported to cause spontaneous SG formation (Boeynaems et al., 2017). However, these findings are debated (Colombrita et al., 2009; Dewey et al., 2011), which could be due to the different types of constructs driving overexpression, as well as the appearance of transfection-related SGs. Recent data support TDP-43 aggregation independent of SGs, where TDP-43 can bypass SGs and produce aggregates irrespective of SG localization (McGurk et al., 2018; Gasset-Rosa et al., 2019; Mann et al., 2019).
Using biochemical assays, cleaved forms of TDP-43 of 35 kDa (C35) and 25 kDa (C25) are found, and this aberrant cleavage enhances aggregation and cellular toxicity (Zhang et al., 2009). However, these cleaved fractions have been shown to bypass SGs, C35 being partially impaired and C25 being almost fully impaired from going into SGs (McGurk et al., 2018).These data indicate that the N-terminal region of TDP-43 is important for the recruitment to SGs and could explain why fragments lacking this region are found in patient material and/or disease models. C-terminal fragments accumulate in the brains, not spinal cords, of ALS and FTD patients and are therefore described as a neuropathological signature of these diseases (Hasegawa et al., 2008; Igaz et al., 2008; Kwong et al., 2014). By generating >50000 mutations in TDP-43, variants that increase aggregation were found to strongly decrease toxicity, while toxic variants promoted the formation of liquid-like condensates (Bolognesi et al., 2019). These results advocate for pro-aggregate species over toxic liquid condensates. However, whether or not aggregates are toxic remains an open question.
FUS
Mutations in FUS can cause fALS (Kwiatkowski et al., 2009; Vance et al., 2009). Although only contributing to 4%–5% of fALS, this gene is heavily linked to FTD-FUS and partially to ALS-FUS pathology. FUS encodes a DNA/RNA-binding protein playing a crucial role in RNA metabolism and DNA repair (Ratti and Buratti, 2016). Protein mislocalization and inclusions have been found in FUS-associated ALS, both in neurons and glia (Kwiatkowski et al., 2009; Deng et al., 2014a). However, FUS inclusions found in FTD cases do not bear disease-related mutations (Neumann et al., 2009,b). FUS, together with EWSR1 and TAF15, is part of the FET proteins family and has an N-terminal LCD, RGG (arginine/glycine)-rich domains, a zinc-finger (ZnF) domain, and an RNA recognition motif (RRM) domain. The nuclear localization signal (NLS) domain keeps FUS predominantly nuclear and harbours most ALS-related mutations (Vance et al., 2009), which were shown to interfere with the FUS nuclear/cytoplasmic balance (Dormann et al., 2010).
Like TDP-43, the FUS LCD has a prion-like motif, with a high tendency to form cross β-amyloid assemblies (Kwon et al., 2013). The phase-separating behaviour of the FUS LCD is impeded upon mutational change of tyrosine residues to serines (Han et al., 2012; Kato et al., 2012). In contrast to TDP-43, FUS aggregation is prevented by phosphorylation (Kwon et al., 2013; Schwartz et al., 2013; Monahan et al., 2017), which in cells occurs during DNA damage response (Deng et al., 2014b). Recombinant FUS was able to form amyloid-like cross β filaments in vitro at high concentrations, but truncation of the LCD abrogates this behaviour (Kato et al., 2012). At physiological concentrations and in the presence of molecular crowding agents, the FUS LCD was also able to phase separate in vitro (Patel et al., 2015). This suggests that LLPS precedes the cross β aggregation of FUS. Full-length FUS was found to form droplets in vitro, and RNA: protein ratios were shown to modulate LLPS. Low RNA concentrations, similar to what is found in the cytoplasm, decreased the liquid-like behaviour of FUS (Burke et al., 2015; Maharana et al., 2018).
FUS methylation of arginine residues was found to disrupt LLPS and decrease its aggregation propensity (Hofweber et al., 2018; Qamar et al., 2018). Methylated FUS plays distinct pathological roles in FTD-FUS and ALS-FUS. Pathological inclusions containing FUS with asymmetrically dimethylated arginines are only found in ALS-FUS cases (Dormann et al., 2012), and inclusions containing unmethylated/monomethylated arginine at the RGG3 region of FUS are only observed in FTD-FUS cases (Suárez-Calvet et al., 2016). In addition, RGG2 arginines promote phase separation of FUS cell free systems and in cells (Bogaert et al., 2018). Inclusions in FTD-TDP and ALS-TDP, but not FTD-FUS proteinopathy, have amyloid properties, as shown by the use of aggregation dyes (Bigio et al., 2013), suggesting that LLPS could potentially account for this variability.
TAF15 and EWS
Despite TAF15, EWS, and FUS being structurally similar, they do not follow the same patterns of accumulation (Neumann et al., 2011). Some variants in EWSR1 have been described in sALS, although follow-up studies are lacking (Couthouis et al., 2012). Furthermore, EWSR1 and TAF15 were localized to cytoplasmic puncta in sALS but were not found in aggregates (Couthouisa et al., 2011), an observation supported by the absence of these proteins in post-mortem studies. As a consequence, the contribution of these proteins to overall ALS-related LLPS seems to be limited.
SOD1
Mutations in SOD1 were the first discovered genetic cause of ALS (Deng et al., 1993; Rosen et al., 1993) and, as such, extensive research has focused on this gene and the SOD1 protein it encodes. Localization of SOD1 is mainly cytoplasmic. However, it was also found in the nucleus, lysosomes, and mitochondria (Tafuri et al., 2015). The main function of SOD1 is to eliminate free superoxide radicals, a major cause of oxidative stress. SOD1 pathology is based on increased aggregation, dimer destabilization, and oligomerization of the protein. Mutant SOD1 causes ALS via a toxic gain-of-function, which could include oxidative stress, excitotoxicity, and mitochondrial dysfunction, amongst others (Wong et al., 1995). SOD1 and TDP-43 inclusions are never reported to co-localize in sALS, indicating that these are produced via different pathways (Farrawell et al., 2015).
Unlike TDP-43 or FUS, SOD1 is not directly involved in phase separation. SOD1 misfolds into amyloid aggregates, and its aggregating behaviour seems directly linked to ALS (Bruijn et al., 1998; Prudencio et al., 2009). Accordingly, the LLPS and aggregation score of SOD1 in our study were low.
C9orf72
Individuals with C9-ALS/FTD have hexanucleotide repeats in the non-coding region of C9orf72 ranging from 66 to >4400 units in contrast to non-disease individuals carrying 2–30 repeats in the intronic region (Gijselinck et al., 2016; Balendra and Isaacs, 2018). These expansions are the most common genetic cause of ALS, although they occur more frequently in individuals with European descent (Ishiura and Tsuji, 2015). C9orf72 is translated into a guanine nucleotide exchange factor and is involved in regulating vesicular trafficking and autophagy (Iyer et al., 2018). Mechanisms proposed for C9-ALS pathology include three non-mutually exclusive hypotheses: loss-of-function through a lower expression of the C9orf72 gene and toxic RNA gain-of-function or toxic protein gain-of-function via the generation of dipeptide repeat proteins (DPRs) due to non-ATG-mediated translation from the repeat transcripts.
The latter leads to the formation of DPRs from sense and antisense transcripts, producing five different DPRs (Ash et al., 2013; Mori et al., 2013). Arginine-rich DPRs undergo LLPS, induce phase separation of some important ALS proteins, and are shown to disrupt mitochondrial function, RNA processing, SG dynamics (Boeynaems et al., 2017), as well as axonal transport (Fumagalli et al., 2019). These DPRs, however, cannot directly affect nucleocytoplasmic transport (Vanneste et al., 2019).
Other proteins
Ataxin-2
Mutations in ATXN2 (encoding Ataxin-2) are considered as risk factors in ALS and can modify TDP-43 toxicity (Elden et al., 2010). Ataxin-2, unlike TDP-43 or FUS, is a cytoplasmic protein. This protein can contain an intermediate polyglutamine repeat expansion in ALS cases, with a repeat length between 24 and 39 (Elden et al., 2010; Lee et al., 2011). The LCD of the ATXN2 yeast orthologue PAB1-binding protein 1 (Pbp1) phase separates, forms droplets (Yang et al., 2019), and behaves similarly to the FUS LCD. However, the LCD of Pbp1 is not enriched in aromatic amino acids but contains unusually high concentrations of methionine residues, unlike FUS or TDP-43 (Kato et al., 2019). In yeast, increased expression of Pbp1 enhances and loss-of-function of Pbp1 suppresses TDP-43 toxicity (Elden et al., 2010). Furthermore, the redox state controls the phase-separating properties of Pbp1 via reversible oxidation of its methionine-rich LCD (Kato et al., 2019). Ataxin-2 is involved in RNA metabolism and was identified as a component of SGs (Kaehler et al., 2012). This could be due to its interaction with the DEAD/H-Box RNA helicase DDX6, a component of SGs and P-bodies (Nonhoff et al., 2007). IDRs in Ataxin-2 mediate LLPS and SG assembly and deletion of the IDRs is sufficient to prevent C9orf72 or FUS-induced neurodegeneration (Bakthavachalu et al., 2018).
In flies, the Ataxin-2 homologue Atx2 has a similar dose-dependent effect (Elden et al., 2010). Reduction of Ataxin-2 extends lifespan and reduces pathology in TDP-43 mice, (Becker et al., 2017) and rescues motor defects in both TDP-43 and spinocerebellar ataxia type 2 mouse models (Becker et al., 2017; Scoles et al., 2017). Ataxin-2 positive aggregates were detected in the spinal cord of ALS patients (Elden et al., 2010; Blokhuis et al., 2013). Therefore, Ataxin-2 aggregation could affect local translation by sequestering proteins and RNA, thereby changing the RNA-to-protein ratio in the cell (Nonhoff et al., 2007).
ADARB1
ADARB1 (also called ADAR2) is the main enzyme responsible for RNA editing in humans. It mediates adenosine-to-inosine (A-to-I) editing at the Q/R position (Higuchi et al., 2000). Inefficient RNA editing was reported in sALS cases (Aizawa et al., 2010) and ADAR2 is mislocalized and forms cytoplasmic accumulations and aggregates in the spinal cord of C9orf72 ALS/FTD patients (Moore et al., 2019). ADAR2 is typically concentrated in the nucleolus and FRAP imaging revealed that ADAR2 can shuttle between the nucleolus (a membraneless organelle) and the nucleoplasm (not a membraneless organelle by definition) (Sansam et al., 2003). In addition, the prediction in Table 2 suggests that ADAR2 could phase separate although this has not yet been confirmed experimentally.
hnRNPA2/B1
hnRNPA2/B1, two isoforms differing from each other by 12 amino acids, are a major subclass of evolutionarily conserved RNPs belonging to the same protein family as FUS and TDP-43. hnRNPs have at least one RNA-binding motif and prion-like domain (Chaudhury et al., 2010). Mutations in the RRM domain are associated with multisystem proteinopathy and ALS characterized by TDP-43-positive cytoplasmic inclusions and increase the aggregation propensity of these proteins (Kim et al., 2013; Paul et al., 2017). In addition, hnRNPA2 LCD and the TDP-43 C-terminal domain co-phase separate and induce co-aggregation (Ryan et al., 2018). Finally, while it was suggested that hnRNPA2 undergoes amyloid aggregation (Xiang et al., 2015; Ryan et al., 2018), the resulting aggregates often appear amorphous and are not a typical amyloid in morphology.
hnRNPA1
An ALS-related variant in hnRNPA1 was identified by linkage analysis and exome sequencing, and the mutant protein was found in aggregates in ALS patient tissue (Kim et al., 2013). Although the estimation of mutation frequencies for this gene is extremely low, hnRNPA1, similar to hnRNPA2/B1, shuttles between the nucleus and the cytoplasm and interacts with TDP-43 (Buratti et al., 2005). hnRNPA1 undergoes LLPS and the LCD is sufficient to mediate this process. However, the RRM domains influence both phase-separating properties and kinetics (Molliex et al., 2015). As shown in Table 2, this protein is predicted to phase separate and potentially aggregate, with a score just below that of hnRNPA2/B1.
Optineurin (OPTN)
OPTN is a membrane-bound protein involved in inflammation, autophagy, Golgi maintenance, and vesicular transport. Mutations in OPTN were found in ALS cases (Maruyama et al., 2010). These variants are mostly autosomal recessive and it is not yet clear whether they are a real cause of ALS (Kamada et al., 2014). Induced pluripotent stem cells-derived motor neurons from SOD1 and ALS patients show accumulations of insoluble OPTN, suggesting that this protein is aggregation-prone (Seminary et al., 2018). It is currently unclear whether OPTN is found in aggregates and/or whether it undergoes LLPS. However, prediction software (Table 2) generates a score close to 0 for LLPS, but a score high for aggregation, suggesting that OPTN could potentially be found in aggregate data sets.
Cell-restricted intracellular antigen-1 (TIA1)
TIA1 has been proposed as a novel ALS-related gene, as exome sequencing in an ALS/FTD family with TDP-43 pathology identified a mutation in the TIA1 LCD (Mackenzie et al., 2017). This protein is commonly found in SGs (Kedersha et al., 2000). While this is compelling in terms of its link to the ALS disease mechanisms, the causality of mutations is disputed (Van der Spek et al., 2018). To date, TIA1 was not detected in ALS aggregates, although TDP-43 pathology is observed in patients bearing TIA1 mutations (Mackenzie et al., 2017). The disordered domain of TIA1 undergoes LLPS in the presence of RNA (Lin et al., 2015), and ALS-related mutations in full-length TIA1 promote LLPS in vivo, altering SG dynamics by inhibiting the disassembly (Mackenzie et al., 2017).
Kinesin heavy chain isoform 5A (KIF5A)
Mutations in KIF5A were recently identified in ALS patients (Brenner et al., 2018; Nicolas et al., 2018) and primarily located at the C-terminal cargo-binding domain. The mutations are rare and no studies have yet shown KIF5A in aggregates. KIF5B and KIF5C, two other homologous proteins also encoding kinesin-2 family members, were found in RNA granules (Trendel et al., 2019; Urdaneta et al., 2019). KIF5A is exclusively neuronal, which could explain why it was not found in these studies using non-neuronal human cell lines.
ANXA11
Another relatively new risk gene for sporadic and familial ALS is ANXA11 (Smith et al., 2017). This gene encodes annexin 11, a phospholipid-binding protein involved in vesicle transport. Annexin 11 has also been shown to tether membraneless RNA granules to actively transport lysosomes via its intrinsic membrane-binding and phase-separating properties. ALS-associated ANXA11 mutations impair this tethering function and hence disrupt RNA transport (Liao et al., 2019). Annexin 11-positive protein aggregates were found in spinal cord motor neurons and hippocampal neuronal axons of an ALS patient carrying a mutation in ANXA11, suggesting that the mutated protein is aggregation-prone (Smith et al., 2017). The predicted LLPS score of ANXA11 (Table 2) is similar to that of TIA1, a protein that was experimentally shown to phase separate.
Ubiquilin-2 (UBQLN2)
UBQLN2 is a member of the ubiquilin family implicated in the degradation of misfolded and redundant proteins through the ubiquitin–proteasome system and macroautophagy. Mutations in this gene were found in ALS/FTD patient cohorts, and UBQLN2 was detected in cytosolic inclusions in degenerating motor neurons of fALS and sALS patients (Renaud et al., 2019). The predicted aggregation score is very high, which is in line with these data. However, the predicted LLPS score of UBQLN2 is low (Table 2), although it was found to exhibit LLPS in vitro under physiological conditions (Dao et al., 2018).
HSP40
Mutations in DNAJC7 were found in ALS cases in 2019 (Farhan et al., 2019). This gene encodes HSP40, a member of the heat-shock protein family, which, together with HSP70, facilitates protein homeostasis. More specifically, it aids in the folding of newly synthesized polypeptides and the clearance of misfolded proteins. Protein misfolding, like phase separation, is a mechanism often proposed to be involved in ALS pathology. HSP40 is depleted in fibroblasts from ALS patients carrying mutations in DNAJC7 and localizes to SGs in HEK cells (Markmiller et al., 2018). However, its presence has not been confirmed in aggregates present in ALS patient tissue.
Matrin 3
Matrin 3 (MATR3) is a nuclear matrix protein, hypothesized to stabilize messenger RNA (Johnson et al., 2014). Matrin 3 has been linked to cellular transport defects in ALS due to its role in the regulation of mRNA nuclear export (Boehringer et al., 2017). This protein could potentially phase separate according to its predicted LLPS score (Table 2). While not found in SG proteomic datasets, it was shown to interact with TDP-43 in cytosolic aggregates in spinal neurons of sALS cases (Tada et al., 2018). Despite being found in aggregates, it is predicted to have a low aggregation score (Table 2).
TANK-binding kinase 1 (TBK1)
Mutations in TBK1 (encoding TBK1) were recently identified as an important genetic cause of FTD and ALS. Loss-of-function variants in this gene are associated with cytoplasmic TDP-43 aggregates (Cirulli et al., 2015; Freischmidt et al., 2015).
Recently, it was found that a loss of TBK1 in motor neurons increases SOD1 aggregation and accelerates disease onset (Gerbino et al., 2020). However, the TBK1 protein has not yet been found in aggregates present in ALS patient tissue. No current studies support LLPS for this protein, and the predicted score was accordingly low. However, the aggregation score was similar to that of TDP-43 (Table 2).
NEK1
Gene-burden analysis identified a significant association between loss-of-function NEK1 variants and fALS risk (Kenna et al., 2016). NEK1 has been linked to cilia formation, DNA-damage response, microtubule stability, neuronal morphology, and axonal polarity (Shalom et al., 2008; Higelin et al., 2018). The predicted score for NEK1 suggests mild LLPS behaviour but a high aggregation propensity based on its sequence (Table 2).
Potential therapeutic implications
Because the underlying mechanisms leading to ALS are still incompletely understood, the elaboration of effective treatment is arduous. Below are the current avenues involving LLPS in ALS therapeutic approaches.
Chaperones
Chaperones such as heat-shock proteins and karyopherins were proposed as a form of ‘disaggregases’ to mitigate the toxic misfolding of proteins such as TDP-43 and FUS. One example is yeast Hsp104 (Afroz et al., 2017). Using the cells’ own protein degradation and chaperone machinery, disaggregases could be generated to remove aggregated proteins. A heat-shock protein, Hsp27, inhibits FUS LLPS via weak interactions; when this heat-shock protein is phosphorylated, it causes an enhanced amyloid inhibition of FUS (Liu et al., 2020). This switchable activity of Hsp27 could serve as a potential target for therapy. Recent research identified karyopherin abnormalities associated with the mislocalization and accumulation of disease-related proteins. In addition to their classical function in nuclear import and export, karyopherins also act as chaperones preventing misfolding, accumulation, and irreversible LLPS, as shown for transportin proteins, which can protect against aberrant phase separation (Guo et al., 2018; Yoshizawa et al., 2018). Transportin has been shown to control FUS phase separation (Qamar et al., 2018) by binding to FUS proline–tyrosine NLS (PY-NLS) (Zhang and Chook, 2012) and the RGG3 domain (Dormann et al., 2012). In addition, it directly interacts with arginine, thus interfering with the weak intermolecular interactions of arginines with other residues and eventually suppressing arginine-driven phase transitions. Downregulation of Exportin 1 (XPO1), a nuclear export receptor, prevents FUS-induced neurotoxicity and reduces the LLPS propensity of FUS (Steyaert et al., 2018). Targeting karyopherins is not a novel approach but already used to treat many cancers (Çağatay and Chook, 2018).
Antisense oligonucleotides (ASOs)
ASOs are an interesting therapeutic avenue for ALS. Phase 1 clinical trial of ASO-mediated therapy showed reduced levels of mutant SOD1 (Miller et al., 2013; https://clinicaltrials.gov/ct2/show/NCT03070119, ClinicalTrials.gov Identifier: NCT03070119), while results of the C9orf72 clinical trial (Jiang et al., 2016; https://clinicaltrials.gov/ct2/show/NCT03626012, ClinicalTrials.gov Identifier: NCT03626012) are pending. The use of ASOs is consequently admissible for non-essential, monogenic gain-of-function conditions (such as SOD1), but it would not be an option for TDP-43 or FUS. A way to overcome this is targeting modifiers of these essential proteins. One example is Ataxin-2 (Becker et al., 2017), which is a modifier of TDP-43 toxicity. Indeed, Ataxin-2 ASOs abolished SGs. Because the majority of ALS patients present with TDP-43 proteinopathy, an Ataxin-2 ASO approach could be very promising, as it could be proposed to most patients. Additional targets for a therapeutic ASO approach are Stathmin-2 ASOs. RNAseq studies revealed that this gene is downregulated in TDP-43-depleted cells and decreased in human motor neurons and spinal cord sections of ALS patients, although it is not yet clear whether it is the only and/or most important target (Klim et al., 2019; Melamed et al., 2019).
Small-molecule strategies
Small molecules to disrupt phase separation are currently explored. Using cell- and protein-based screens, it was shown that lipoamide can reduce SG protein aggregation in vitro, in Drosophila, in Caenorhabditis elegans, and in patient-derived motor neurons (Wheeler et al., 2019). Lipoamide and lipoic acid, two non-toxic drugs, alter FUS aggregation specifically by modulating its phase-separating behaviour. This was linked to changes in the cellular stress response of the cells, improving mitochondrial health and leading to restoration of mislocalized FUS to the nucleus. Unlike what is seen in vitro with 1,6 hexandiol, these drugs do not disrupt other membraneless organelles, a concern for developing specific SG targets, as side effects may arise from the inhibition of an essential process. Small-molecule screens in cells showed a reduction of TDP-43 in SGs, and molecules with planar moieties disrupted accumulation of ALS-associated RBPs in SGs (Fang et al., 2019). In addition, RNA can be used to suppress TDP-43 LLPS, as demonstrated by the use of bait oligonucleotide RNA (Mann et al., 2019).
Post-translational modifications
Interfering with post-translational modifications is another therapeutic approach that may be used to alter aberrant LLPS. Examples include the targeting of known phosphorylation sites of regulatory kinases (Kumar Rai et al., 2018). Similarly, alterations of methylation sites could be used to modify the phase-separating behaviour and toxicity of FUS and/or DPRs (Dormann et al., 2012; Qamar et al., 2018; Gittings et al., 2020), paving the way for new therapies. The methylation of arginine residues maintains the positive charge but diminishes its hydrogen-bonding capacity by removing a hydrogen atom for each methyl group, where the methyl group essentially adds hydrophobicity to the side chain. The use of pharmacological tools to alter arginine demethylase activity would allow the modulation of epigenetic control of transcription (Blanc and Richard, 2017).
Activation of autophagy
Autophagy activators inducing SG removal are a controversial route for potential therapeutics. There is evidence that SGs can be targeted to lysosomes for autophagy and this process can be inhibited by depletion of valosine-containing protein (VCP) encoded by VCP, a gene mutated in ALS and FTD (Buchan et al., 2013). Furthermore, this autophagic process is mediated by SG surveillance chaperone complexes, which could also represent novel therapeutic targets (Ganassi et al., 2016). The enhancement of autophagy has recently been found to reduce cytoplasmic FUS mislocalization and rescue FUS phenotypes in vivo (Marrone et al., 2019). Recently, inhibition of lysosomal mTORC1 signalling using the small molecule EN6 was found to clear TDP-43 aggregates in a lysosome-dependent manner (Chung et al., 2019).
Discussion
In the past 5 years, there has been a radical change in the view on LLPS granules. Gasset-Rosa et al. (2019) and McGurk et al. (2018) suggested that SG-independent TDP-43 foci are particularly prone to a conversion into aggregates. This suggests that SGs could be the initial checkpoints serving a protective function. This role was initially hypothesized in 2017, as chaperones were found to be present within SGs (Ganassi et al., 2016; Jain et al., 2016). Therapeutic targets have been suggested for the aggregates and SGs but lacking for oligomeric forms of these mislocalized proteins. Targeting intermediate soluble oligomers of TDP-43 should be considered at earlier time point to ensure prevention.
Novel methods for studying SGs using optodroplets confirmed that transient SGs can indeed turn into solid aggregates (Zhang et al., 2019). The tight control of SG dynamics is only recently being unravelled by looking at the relationship between G3BP1 and its binding partners (Guillé N-Boixet et al., 2020; Sanders et al., 2020; Yang et al., 2020).
In this review, we summarized known aggregating proteins and discussed their potential LLPS properties based on published evidence and predicted scores. These predictions suggest that we could investigate LLPS behaviour and LLPS-related aggregation by studying non-phase-separating ALS proteins. Indeed, SOD1 and MATR3 have low LLPS scores and are not associated to SGs, but they are found in aggregates of ALS patient tissue, despite having low aggregation score predictions. On another note, while TDP-43 and FUS share similarities in terms of domain organization, their pathologies appear to be mutually exclusive, which could potentially be explained by their differences in terms of LLPS and aggregation, as predicted by their contrasting scores (Table 2).
Neurons are post-mitotic cells and as such are particularly vulnerable to proteostasis stress, as these cells are unable to dilute their cellular content by cell division. The correct balance and function of both LLPS and clearance are therefore crucial in order to prevent the accumulation of proteins and the formation of aggregates. Because the cell body lies far from the synapse, motor neurons rely heavily on axonal transport to maintain neuronal health and to ensure proper communication between the different cellular compartments. There are three points to consider. (i) Axonal transport impairments are observed in cell lines derived from fALS and sALS patients. (ii) Pathological RNP granules in motor neurons might be due to these proteins’ unique function. Indeed, they are involved in the transport of RNA granules, relying on LLPS over long axons, and hence contain a much higher amount of RNA granules and RBPs. (iii) This LLPS occurs intentionally at low sterical hindrance to increase transport efficiency, which unfortunately makes this system sensitive to disruptions (Wolozin, 2014).
Some ambiguities remain in ALS-LLPS research. First, many proteins found in SGs are not present in the aggregates observed in ALS patient tissue and often these proteins do not bear ALS-related pathogenic mutations. Second, SGs are currently thought to serve a protective function. Third, it is not merely the existence of SGs, which is put into question, but rather their disassembly. Perhaps the nature of these SG proteins results in partial colocalization with aggregates and, therefore, there is a bias for finding such proteins in association. In addition, while fibres have been shown to grow out of these droplets in vitro and advanced optodroplet techniques have been applied, these systems fail to show fibres growing out of the same droplets in a cellular context.
Future perspectives
We do not fully understand the link between TDP-43, FUS, SOD1, and DPRs, yet they are the main proteins used to classify post-mortem ALS cases. The mislocalization of FUS and TDP-43 precedes their aggregate formation, and RBPs are unable to mislocalize without interacting with these aggregates. In sALS, FUS mislocalization is a hallmark of ALS (Tyzack et al., 2019) without the need of aggregation, suggesting that the initial mislocalization of these nuclear proteins is more significant in terms of pathogenicity than the formation of aggregates.
Moreover, it is not possible to confirm whether all these aggregates result from aberrant phase separation, as the aggresome proteome of ALS/FTD has not yet been completed. Indeed, we currently only know this for a handful of ALS aggregate-positive proteins, for which different morphologies occur in aggregates. In addition, while the impact of LLPS condensates on insoluble proteins has been studied, research on the effect of these condensates on the soluble protein state remains to be determined. Using novel techniques such as laser-capture mass spectrometry, this aggresome will likely be teased out in the near future, but the tools to study aberrant phase separation in an endogenous cell model remain limited.
Funding
Research of the authors is supported by VIB, KU Leuven (C1 and ‘Opening the Future’ Fund), the ‘Fund for Scientific Research Flanders’ (FWO-Vlaanderen), the Agency for Innovation by Science and Technology in Flanders, the Thierry Latran Foundation, the ‘Association Belge contre les Maladies neuro-Musculaires’ (ABMM), the Muscular Dystrophy Association (MDA), Target ALS, the ALS Liga België (A Cure for ALS), and the ALS Association (ALSA). D.P. is funded by the VIB International Life Sciences PhD Program. V.B. is supported by a postdoctoral fellowship from the FWO-Vlaanderen. G.O. acknowledges funding by the Research Foundation Flanders (FWO)—project nr. G.0328.16N.
Conflict of interest: none declared.
References
- Abramzon Y., Johnson J.O., Scholz S.,. et al. (2012). Valosin-containing protein (VCP) mutations in sporadic amyotrophic lateral sclerosis. Neurobiol. Aging 33, 2231.e1–2231.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Afroz T., Hock E.-M., Ernst P., et al. (2017). Functional and dynamic polymerization of the ALS-linked protein TDP-43 antagonizes its pathologic aggregation. Nat. Commun. 8, 45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aizawa H., Sawada J., Hideyama T., et al. (2010). TDP-43 pathology in sporadic ALS occurs in motor neurons lacking the RNA editing enzyme ADAR2. Acta Neuropathol. 120, 75–84. [DOI] [PubMed] [Google Scholar]
- Al-Chalabi A., Calvo A., Chio A., et al. (2014). Analysis of amyotrophic lateral sclerosis as a multistep process: a population-based modelling study. Lancet Neurol. 13, 1108–1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersson M.K., Ståhlberg A., Arvidsson Y., et al. (2008). The multifunctional FUS, EWS and TAF15 proto-oncoproteins show cell type-specific expression patterns and involvement in cell spreading and stress response. BMC Cell Biol. 9, 37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arai T., Hasegawa M., Akiyama H., et al. (2006). TDP-43 is a component of ubiquitin-positive tau-negative inclusions in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Biochem. Biophys. Res. Commun. 351, 602–611. [DOI] [PubMed] [Google Scholar]
- Ash P.E.A., Bieniek K.F., Gendron T.F., et al. (2013). Unconventional translation of C9ORF72 GGGGCC expansion generates insoluble polypeptides specific to c9FTD/ALS. Neuron 77, 639–646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayala Y.M., Zago P., D’Ambrogio A., et al. (2008). Structural determinants of the cellular localization and shuttling of TDP-43. J. Cell Sci. 121, 3778–3785. [DOI] [PubMed] [Google Scholar]
- Bakthavachalu B., Huelsmeier J., Sudhakaran I.P., et al. (2018). RNP-granule assembly via Ataxin-2 disordered domains is required for long-term memory and neurodegeneration. Neuron 98, 754–766.e4. [DOI] [PubMed] [Google Scholar]
- Balendra R., Isaacs A.M. (2018). C9orf72-mediated ALS and FTD: multiple pathways to disease. Nat. Rev. Neurol. 14, 544–558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker L.A., Huang B., Bieri G., et al. (2017). Therapeutic reduction of ataxin-2 extends lifespan and reduces pathology in TDP-43 mice. Nature 544, 367–371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bigio E.H., Wu J.Y., Deng H.X., et al. (2013). Inclusions in frontotemporal lobar degeneration with TDP-43 proteinopathy (FTLD-TDP) and amyotrophic lateral sclerosis (ALS), but not FTLD with FUS proteinopathy (FTLD-FUS), have properties of amyloid. Acta Neuropathol. 125, 463–465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanc R.S., Richard S. (2017). Arginine methylation: the coming of age. Mol. Cell 65, 8–24. [DOI] [PubMed] [Google Scholar]
- Blokhuis A.M., Groen E.J.N., Koppers M., et al. (2013). Protein aggregation in amyotrophic lateral sclerosis. Acta Neuropathol. 25, 777–794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boehringer A., Garcia-Mansfield K., Singh G., et al. (2017). ALS associated mutations in Matrin 3 alter protein-protein interactions and impede mRNA nuclear export. Sci. Rep. 7, 14529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boeynaems S., Bogaert E., Kovacs D., et al. (2017). Phase separation of C9orf72 dipeptide repeats perturbs stress granule dynamics. Mol. Cell 65, 1044–1055.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boeynaems S., Bogaert E., Michiels E., et al. (2016). Drosophila screen connects nuclear transport genes to DPR pathology in c9ALS/FTD. Sci. Rep. 6, 20877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogaert E., Boeynaems S., Kato M., et al. (2018). Molecular dissection of FUS points at synergistic effect of low-complexity domains in toxicity. Cell Rep. 24, 529–537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolognesi B., Faure A.J., Seuma M., et al. (2019). The mutational landscape of a prion-like domain. Nat. Commun. 10, 4162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brangwynne C.P. (2013). Phase transitions and size scaling of membrane-less organelles. J. Cell Biol. 203, 875–881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenner D., Stem Yilmaz R., Mü Ller K., et al. (2018). Hot-spot KIF5A mutations cause familial ALS. Brain 141, 688–697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown R.H., Al-Chalabi A. (2017). Amyotrophic lateral sclerosis. N. Engl. J. Med. 377, 162–172. [DOI] [PubMed] [Google Scholar]
- Bruijn L.I., Houseweart M.K., Kato S, et al. (1998). Aggregation and motor neuron toxicity of an ALS-linked SOD1 mutant independent from wild-type SOD1. Science 281, 1851–1854. [DOI] [PubMed] [Google Scholar]
- Buchan J.R., Kolaitis R.M., Taylor J.P., et al. (2013). Eukaryotic stress granules are cleared by autophagy and Cdc48/VCP function. Cell 153, 1461–1474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buratti E., Brindisi A., Giombi M., et al. (2005). TDP-43 binds heterogeneous nuclear ribonucleoprotein A/B through its C-terminal tail: An important region for the inhibition of cystic fibrosis transmembrane conductance regulator exon 9 splicing. J. Biol. Chem. 280, 37572–37584. [DOI] [PubMed] [Google Scholar]
- Burk K., Pasterkamp R.J. (2019). Disrupted neuronal trafficking in amyotrophic lateral sclerosis. Acta Neuropathol. 137, 859–877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burke K.A., Janke A.M., Rhine C.L.,. et al. (2015). Residue-by-residue view of in vitro FUS granules that bind the C-terminal domain of RNA polymerase II. Mol. Cell 60, 231–241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butti Z., Patten S.A. (2019). RNA dysregulation in amyotrophic lateral sclerosis. Front. Genet. 9, 712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Çağatay T., Chook Y.M. (2018). Karyopherins in cancer. Curr. Opin. Cell Biol. 52, 30–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaudhury A., Hussey G.S., Ray P.S., et al. (2010). TGF-β-mediated phosphorylation of hnRNP E1 induces EMT via transcript-selective translational induction of Dab2 and ILEI. Nat. Cell Biol. 12, 286–293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung C.Y.S., Shin H.R., Berdan C.A., et al. (2019). Covalent targeting of the vacuolar H+-ATPase activates autophagy via mTORC1 inhibition. Nat. Chem. Biol. 15, 776–785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cirulli E.T., Lasseigne B.N., Petrovski S., et al. (2015). Exome sequencing in amyotrophic lateral sclerosis identifies risk genes and pathways. Science 347, 1436–1441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen T.J., Hwang A.W., Restrepo C.R., et al. (2015). An acetylation switch controls TDP-43 function and aggregation propensity. Nat. Commun. 6, 5845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colombrita C., Zennaro E., Fallini C., et al. (2009). TDP-43 is recruited to stress granules in conditions of oxidative insult. J. Neurochem. 111, 1051–1061. [DOI] [PubMed] [Google Scholar]
- Conicella A.E., Zerze G.H., Mittal J., et al. (2016). ALS mutations disrupt phase separation mediated by α-helical structure in the TDP-43 low-complexity C-terminal domain. Structure 24, 1537–1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Couthouis J., Hart M.P., Erion R., et al. (2012). Evaluating the role of the FUS/TLS-related gene EWSR1 in amyotrophic lateral sclerosis. Hum. Mol. Genet. 21, 2899–2911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Couthouisa J., Harta M.P., Shorter J., et al. (2011). A yeast functional screen predicts new candidate ALS disease genes. Proc. Natl Acad. Sci. USA 108, 20881–20890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dao T.P., Kolaitis R.M., Kim H.J., et al. (2018). Ubiquitin modulates liquid–liquid phase separation of UBQLN2 via disruption of multivalent interactions. Mol. Cell 69, 965–978.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Vos K.J., Hafezparast M. (2017). Neurobiology of axonal transport defects in motor neuron diseases: opportunities for translational research? Neurobiol. Dis. 105, 283–299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng H., Gao K., Jankovic J. (2014a). The role of FUS gene variants in neurodegenerative diseases. Nat. Rev. Neurol. 10, 337–348. [DOI] [PubMed] [Google Scholar]
- Deng H.X., Hentati A., Tainer J.A., et al. (1993). Amyotrophic lateral sclerosis and structural defects in Cu, Zn superoxide dismutase. Science 261, 1047–1051. [DOI] [PubMed] [Google Scholar]
- Deng Q., Holler C.J., Taylor G., et al. (2014b). FUS is phosphorylated by DNA-PK and accumulates in the cytoplasm after DNA damage. J. Neurosci. 34, 7802–7813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dewey C.M., Cenik B., Sephton C.F., et al. (2011). TDP-43 is directed to stress granules by sorbitol, a novel physiological osmotic and oxidative stressor. Mol. Cell. Biol. 31, 1098–1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dewey C.M., Cenik B., Sephton C.F., et al. (2012). TDP-43 aggregation in neurodegeneration: are stress granules the key? Brain Res. 1462, 16–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dormann D., Madl T., Valori C.F., et al. (2012). Arginine methylation next to the PY-NLS modulates Transportin binding and nuclear import of FUS. EMBO J. 31, 4258–4275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dormann D., Rodde R., Edbauer D., et al. (2010). ALS-associated fused in sarcoma (FUS) mutations disrupt transportin-mediated nuclear import. EMBO J. 29, 2841–2857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elden A.C., Kim H.J., Hart M.P., et al. (2010). Ataxin-2 intermediate-length polyglutamine expansions are associated with increased risk for ALS. Nature 466, 1069–1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang M.Y., Markmiller S., Vu A.Q., et al. (2019). Small-molecule modulation of TDP-43 recruitment to stress granules prevents persistent TDP-43 accumulation in ALS/FTD. Neuron 103, 802–819.e11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang Y.S., Tsai K.J., Chang Y.J., et al. (2014). Full-length TDP-43 forms toxic amyloid oligomers that are present in frontotemporal lobar dementia-TDP patients. Nat. Commun. 5, 4824. [DOI] [PubMed] [Google Scholar]
- Farhan S.M.K., Howrigan D.P., Abbott L.E., et al. (2019). Exome sequencing in amyotrophic lateral sclerosis implicates a novel gene, DNAJC7, encoding a heat-shock protein. Nat. Neurosci. 22, 1966–1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farrawell N.E., Lambert-Smith I.A., Warraich S.T., et al. (2015). Distinct partitioning of ALS associated TDP-43, FUS and SOD1 mutants into cellular inclusions. Sci. Rep. 5, 13416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez-Escamilla A.-M., Rousseau F., Schymkowitz J., et al. (2004). Prediction of sequence-dependent and mutational effects on the aggregation of peptides and proteins. Nat. Biotechnol. 22, 1302–1306. [DOI] [PubMed] [Google Scholar]
- Freibaum B.D., Lu Y., Lopez-Gonzalez R., et al. (2015). GGGGCC repeat expansion in C9orf72 compromises nucleocytoplasmic transport. Nature 525, 129–133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freischmidt A., Wieland T., Richter B., et al. (2015). Haploinsufficiency of TBK1 causes familial ALS and fronto-temporal dementia. Nat. Neurosci. 18, 631–636. [DOI] [PubMed] [Google Scholar]
- Fumagalli L., Young F.L., Boeynaems S., et al. (2019). C9orf72-derived arginine-containing dipeptide repeats associate with axonal transport machinery and impede microtubule-based motility. bioRxiv, 10.1101/835082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganassi M., Mateju D., Bigi I., et al. (2016). A surveillance function of the HSPB8–BAG3–HSP70 chaperone complex ensures stress granule integrity and dynamism. Mol. Cell 63, 796–810. [DOI] [PubMed] [Google Scholar]
- Gasset-Rosa F., Lu S., Yu H., et al. (2019). Cytoplasmic TDP-43 de-mixing independent of stress granules drives inhibition of nuclear import, loss of nuclear TDP-43, and cell death. Neuron 102, 339–357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerbino V., Kaunga E., Ye J., et al. (2020). The loss of TBK1 kinase activity in motor neurons or in all cell types differentially impacts ALS disease progression in SOD1 mice. Neuron 106, 789–805.e5. [DOI] [PubMed] [Google Scholar]
- Ghosh S., Geahlen R.L. (2015). Stress granules modulate SYK to cause microglial cell dysfunction in Alzheimer’s disease. EBioMedicine 2, 1785–1798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gijselinck I., Van Mossevelde S., Van Der Zee J., et al. (2016). The C9orf72 repeat size correlates with onset age of disease, DNA methylation and transcriptional downregulation of the promoter. Mol. Psychiatry 21, 1112–1124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gitler A.D., Shorter J. (2011). RNA-binding proteins with prion-like domains in ALS and FTLD-U. Prion 5, 179–187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gittings L.M., Boeynaems S., Lightwood D., et al. (2020). Symmetric dimethylation of poly-GR correlates with disease duration in C9orf72 FTLD and ALS and reduces poly-GR phase separation and toxicity. Acta Neuropathol. 139, 407–410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guillé N-Boixet J., Kopach A., Holehouse A.S., et al. (2020). RNA-induced conformational switching and clustering of G3BP drive stress granule assembly by condensation. Cell 181, 346–361.e17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo L., Kim H.J., Wang H., et al. (2018). Nuclear-import receptors reverse aberrant phase transitions of RNA-binding proteins with prion-like domains. Cell 173, 677–692.e20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo W., Stoklund Dittlau K., Van Den Bosch L. (2019). Axonal transport defects and neurodegeneration: molecular mechanisms and therapeutic implications. Semin. Cell Dev. Biol. 99, 133–150. [DOI] [PubMed] [Google Scholar]
- Han T.W., Kato M., Xie S., et al. (2012). Cell-free formation of RNA granules: bound RNAs identify features and components of cellular assemblies. Cell 149, 768–779. [DOI] [PubMed] [Google Scholar]
- Hasegawa M., Arai T., Nonaka T., et al. (2008). Phosphorylated TDP-43 in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Ann. Neurol. 64, 60–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higelin J., Catanese A., Semelink-Sedlacek L.L., et al. (2018). NEK1 loss-of-function mutation induces DNA damage accumulation in ALS patient-derived motoneurons. Stem Cell Res. 30, 150–162. [DOI] [PubMed] [Google Scholar]
- Higuchi M., Maas S., Single F.N., et al. (2000). Point mutation in an AMPA receptor gene rescues lethality in mice deficient in the RNA-editing enzyme ADAR2. Nature 406, 78–81. [DOI] [PubMed] [Google Scholar]
- Hjerpe R., Bett J.S., Keuss M.J., et al. (2016). UBQLN2 mediates autophagy-independent protein aggregate clearance by the proteasome. Cell 166, 935–949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofweber M., Hutten S., Bourgeois B., et al. (2018). Phase separation of FUS is suppressed by its nuclear import receptor and arginine methylation. Cell 173, 706–719.e13. [DOI] [PubMed] [Google Scholar]
- Hyman A.A., Weber C.A., Jülicher F. (2014). Liquid–liquid phase separation in biology. Ann. Rev. Cell Dev. Biol. 30, 39–58. [DOI] [PubMed] [Google Scholar]
- Igaz L.M., Kwong L.K., Xu Y., et al. (2008). Enrichment of C-terminal fragments in TAR DNA-binding protein-43 cytoplasmic inclusions in brain but not in spinal cord of frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Am. J. Pathol. 173, 182–194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishiura H., Tsuji S. (2015). Epidemiology and molecular mechanism of frontotemporal lobar degeneration/amyotrophic lateral sclerosis with repeat expansion mutation in C9orf72. J. Neurogenet. 29, 85–94. [DOI] [PubMed] [Google Scholar]
- Iyer S., Subramanian V., Acharya K.R. (2018). C9orf72, a protein associated with amyotrophic lateral sclerosis (ALS) is a guanine nucleotide exchange factor. PeerJ 6, e5815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain S., Wheeler J.R., Walters R.W., et al. (2016). ATPase-modulated stress granules contain a diverse proteome and substructure. Cell 164, 487–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang J., Zhu Q., Gendron T.F., et al. (2016). Gain of toxicity from ALS/FTD-linked repeat expansions in C9ORF72 is alleviated by antisense oligonucleotides targeting GGGGCC-containing RNAs. Neuron 90, 535–550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson B.S., Snead D., Lee J.J., et al. (2009). TDP-43 is intrinsically aggregation-prone, and amyotrophic lateral sclerosis-linked mutations accelerate aggregation and increase toxicity. J. Biol. Chem. 284, 20329–20339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson J.O., Pioro E.P., Boehringer A., et al. (2014). Mutations in the Matrin 3 gene cause familial amyotrophic lateral sclerosis. Nat. Neurosci. 17, 664–666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabashi E., Valdmanis P.N., Dion P., et al. (2008). TARDBP mutations in individuals with sporadic and familial amyotrophic lateral sclerosis. Nat. Genet. 40, 572–574. [DOI] [PubMed] [Google Scholar]
- Kaehler C., Isensee J., Nonhoff U., et al. (2012). Ataxin-2-like is a regulator of stress granules and processing bodies. PLoS One 7, e50134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamada M., Izumi Y., Ayaki T., et al. (2014). Clinicopathologic features of autosomal recessive amyotrophic lateral sclerosis associated with optineurin mutation. Neuropathology 34, 64–70. [DOI] [PubMed] [Google Scholar]
- Kato M., Han T.W., Xie S., et al. (2012). Cell-free formation of RNA granules: low complexity sequence domains form dynamic fibers within hydrogels. Cell 149, 753–767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato M., Yang Y.S., Sutter B.M., et al. (2019). Redox state controls phase separation of the yeast ataxin-2 protein via reversible oxidation of its methionine-rich low-complexity domain. Cell 177, 711–721.e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kedersha, N.,, and Anderson, P. ( 2007). Mammalian stress granules and processing bodies. Methods Enzymol. 431, 61–81. [DOI] [PubMed] [Google Scholar]
- Kedersha, N., and, Anderson, P. (2002) Stress granules: sites of mRNA triage that regulate mRNA stability and translatability. Biochem. Soc. Trans. 30, 963–969. [DOI] [PubMed] [Google Scholar]
- Kedersha N., Cho M.R., Li W., et al. (2000). Dynamic shuttling of TIA-1 accompanies the recruitment of mRNA to mammalian stress granules. J. Cell Biol. 151, 1257–1268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenna K.P., Van Doormaal P.T.C., Dekker A.M., et al. (2016). NEK1 variants confer susceptibility to amyotrophic lateral sclerosis. Nat. Genet. 48, 1037–1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H.J., Kim N.C., Wang Y.D., et al. (2013). Mutations in prion-like domains in hnRNPA2B1 and hnRNPA1 cause multisystem proteinopathy and ALS. Nature 495, 467–473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klim J.R., Williams L.A., Limone F., et al. (2019). ALS-implicated protein TDP-43 sustains levels of STMN2, a mediator of motor neuron growth and repair. Nat. Neurosci. 22, 167–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar Rai A., Chen J.-X., Selbach M., et al. (2018). Kinase-controlled phase transition of membraneless organelles in mitosis. Nature 559, 211–216. [DOI] [PubMed] [Google Scholar]
- Kwiatkowski T.J., Bosco D.A., LeClerc A.L., et al. (2009). Mutations in the FUS/TLS gene on chromosome 16 cause familial amyotrophic lateral sclerosis. Science 323, 1205–1208. [DOI] [PubMed] [Google Scholar]
- Kwon I., Kato M., Xiang S., et al. (2013). Phosphorylation-regulated binding of RNA polymerase II to fibrous polymers of low-complexity domains. Cell 155, 1049–1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwong L.K., Irwin D.J., Walker A.K., et al. (2014). Novel monoclonal antibodies to normal and pathologically altered human TDP-43 proteins. Acta Neuropathol. Commun. 2, 33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao, Y.-C., Fernandopulle M.S., Wang G., et al. (2019). RNA granules hitchhike on lysosomes for long-distance transport, using annexin A11 as a molecular tether. Cell 179, 147–164.e20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee K.H., Zhang P., Kim H.J., et al. (2016). C9orf72 dipeptide repeats impair the assembly, dynamics, and function of membrane-less organelles. Cell 167, 774–788.e17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee T., Li Y.R., Ingre C., et al. (2011). Ataxin-2 intermediate-length polyglutamine expansions in European ALS patients. Hum. Mol. Genet. 20, 1697–1700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Y., Protter, et al. (2015). Formation and maturation of phase-separated liquid droplets by RNA-binding proteins. Mol. Cell 60, 208–219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu-Yesucevitz L., Bilgutay A., Zhang Y.-J., et al. (2010). Tar DNA binding protein-43 (TDP-43) associates with stress granules: analysis of cultured cells and pathological brain tissue. PLoS One 5, e13250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z., Zhang S., Gu J., et al. (2020). Hsp27 chaperones FUS phase separation under the modulation of stress-induced phosphorylation. Nat. Struct. Mol. Biol. 27, 363–372. [DOI] [PubMed] [Google Scholar]
- Longinetti E., Fang F. (2019). Epidemiology of amyotrophic lateral sclerosis: an update of recent literature. Curr. Opin. Neurol. 32, 771–776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackenzie I.R., Nicholson A.M., Sarkar M., et al. (2017). TIA1 mutations in amyotrophic lateral sclerosis and frontotemporal dementia promote phase separation and alter stress granule dynamics. Neuron 95, 808–816.e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maharana S., Wang J., Papadopoulos D.K., et al. (2018). RNA buffers the phase separation behavior of prion-like RNA binding proteins. Science 360, 918–921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majounie E., Renton A.E., Mok K., et al. (2012). Frequency of the C9orf72 hexanucleotide repeat expansion in patients with amyotrophic lateral sclerosis and frontotemporal dementia: a cross-sectional study. Lancet Neurol. 11, 323–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mann J.R., Gleixner A.M., Mauna J.C., et al. (2019). RNA binding antagonizes neurotoxic phase transitions of TDP-43. Neuron 102, 321–338.e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markmiller S., Soltanieh S., Server K.L., et al. (2018). Context-dependent and disease-specific diversity in protein interactions within stress granules. Cell 172, 590–604.e13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marrone L., Drexler H.C.A., Wang J., et al. (2019). FUS pathology in ALS is linked to alterations in multiple ALS-associated proteins and rescued by drugs stimulating autophagy. Acta Neuropathol. 138, 67–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maruyama H., Morino H., Ito H., et al. (2010). Mutations of optineurin in amyotrophic lateral sclerosis. Nature 465, 223–226. [DOI] [PubMed] [Google Scholar]
- McDonald K.K., Aulas A., Destroismaisons L., et al. (2011). TAR DNA-binding protein 43 (TDP-43) regulates stress granule dynamics via differential regulation of G3BP and TIA-1. Hum. Mol. Genet. 20, 1400–1410. [DOI] [PubMed] [Google Scholar]
- McGurk L., Gomes E., Guo L., et al. (2018). Poly(ADP-Ribose) prevents pathological phase separation of TDP-43 by promoting liquid demixing and stress granule localization. Mol. Cell 71, 703–717.e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melamed Z., López-Erauskin J., Baughn M.W., et al. (2019). Premature polyadenylation-mediated loss of stathmin-2 is a hallmark of TDP-43-dependent neurodegeneration. Nat. Neurosci. 22, 180–190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller T.M., Pestronk A., David W., et al. (2013). An antisense oligonucleotide against SOD1 delivered intrathecally for patients with SOD1 familial amyotrophic lateral sclerosis: a phase 1, randomised, first-in-man study. Lancet Neurol. 12, 435–442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molliex A., Temirov J., Lee J., et al. (2015). Phase separation by low complexity domains promotes stress granule assembly and drives pathological fibrillization. Cell 163, 123–133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monahan Z., Ryan V.H., Janke A.M., et al. (2017). Phosphorylation of the FUS low-complexity domain disrupts phase separation, aggregation, and toxicity. EMBO J. 36, 2951–2967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore S., Alsop E., Lorenzini I., et al. (2019). ADAR2 mislocalization and widespread RNA editing aberrations in C9orf72-mediated ALS/FTD. Acta Neuropathol. 138, 49–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori K., Weng S.M., Arzberger T., et al. (2013). The C9orf72 GGGGCC repeat is translated into aggregating dipeptide-repeat proteins in FTLD/ALS. Science 339, 1335–1338. [DOI] [PubMed] [Google Scholar]
- Neumann M., Sampathu D.M., Kwong L.K., et al. (2006). Ubiquitinated TDP-43 in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Science 314, 130–133. [DOI] [PubMed] [Google Scholar]
- Neumann M., Bentmann E., Dormann D., et al. (2011). FET proteins TAF15 and EWS are selective markers that distinguish FTLD with FUS pathology from amyotrophic lateral sclerosis with FUS mutations. Brain 134, 2595–2609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neumann M., Kwong L.K., Lee E.B., et al. (2009a). Phosphorylation of S409/410 of TDP-43 is a consistent feature in all sporadic and familial forms of TDP-43 proteinopathies. Acta Neuropathol. 117, 137–149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neumann M., Rademakers R., Roeber S., et al. (2009b). A new subtype of frontotemporal lobar degeneration with FUS pathology. Brain 132, 2922–2931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen H.P., Van Broeckhoven C., van der Zee J. (2018). ALS genes in the genomic era and their implications for FTD. Trends Genet. 34, 404–423. [DOI] [PubMed] [Google Scholar]
- Nicolas A., Kenna K.P., Renton A.E., et al. (2018). Genome-wide analyses identify KIF5A as a novel ALS gene. Neuron 97, 1268–1282.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nonaka T., Suzuki G., Tanaka Y., et al. (2016). Phosphorylation of TAR DNA-binding protein of 43 kDa (TDP-43) by truncated casein kinase 1δ triggers mislocalization and accumulation of TDP-43. J. Biol. Chem. 291, 5473–5483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nonhoff U., Ralser M., Welzel F., et al. (2007). Ataxin-2 interacts with the DEAD/H-box RNA helicase DDX6 and interferes with P-bodies and stress granules. Mol. Biol. Cell 18, 1385–1396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orlando G., Raimondi D., Tabaro F., et al. (2019.). Computational identification of prion-like RNA-binding proteins that form liquid phase-separated condensates. Bioinformatics 35, 4617–4623. [DOI] [PubMed] [Google Scholar]
- Patel A., Lee H.O., Jawerth L., et al. (2015). A liquid-to-solid phase transition of the ALS protein FUS accelerated by disease mutation. Cell 162, 1066–1077. [DOI] [PubMed] [Google Scholar]
- Paul K.R., Molliex A., Cascarina S., et al. (2017). Effects of mutations on the aggregation propensity of the human prion-like protein hnRNPA2B1. Mol. Cell. Biol. 37, e00652-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penndorf D., Witte O.W., Kretz A. (2018). DNA plasticity and damage in amyotrophic lateral sclerosis. Neural Regen. Res. 13, 173–180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Protter, D.S.W., and, Parker R. (2016) . Principles and properties of stress granules. Trends Cell Biol. 26, 668–679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prudencio M., Hart P.J., Borchelt D.R., et al. (2009). Variation in aggregation propensities among ALS-associated variants of SOD1: correlation to human disease. Hum. Mol. Genet. 18, 3217–3226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qamar S., Wang G., Randle S.J., et al. (2018). FUS phase separation is modulated by a molecular chaperone and methylation of arginine cation-π interactions. Cell 173, 720–734.e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ratti A., Buratti E. (2016). Physiological functions and pathobiology of TDP-43 and FUS/TLS proteins. J. Neurochem. 138, 95–111. [DOI] [PubMed] [Google Scholar]
- Reijns, M.A.M.,, Alexander, R.D.,, Spiller, M.P., et al. (2008). A role for Q/N-rich aggregation-prone regions in P-body localization. J. Cell Sci. 121, 2463–2472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renaud L., Picher-Martel V., Codron P., et al. (2019). Key role of UBQLN2 in pathogenesis of amyotrophic lateral sclerosis and frontotemporal dementia. Acta Neuropathol. Commun. 7, 103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosen D.R., Siddique T., Patterson D., et al. (1993). Mutations in Cu/Zn superoxide dismutase gene are associated with familial amyotrophic lateral sclerosis. Nature 362, 59–62. [DOI] [PubMed] [Google Scholar]
- Ryan V.H., Dignon G.L., Burke K.A. (2018). Mechanistic view of hnRNPA2 low-complexity domain structure, interactions, and phase separation altered by mutation and arginine methylation. Mol. Cell 69, 465–479.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders D.W., Kedersha N., Lee D.S.W., et al. (2020). Competing protein−RNA interaction networks control multiphase intracellular organization. Cell 181, 306–324.e28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sansam C.L., Wells K.S., Emeson R.B. (2003). Modulation of RNA editing by functional nucleolar sequestration of ADAR2. Proc. Natl Acad. Sci. USA 100(Suppl. 2), 14018–14023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt H.B., Rohatgi R. (2016). In vivo formation of vacuolated multi-phase compartments lacking membranes. Cell Rep. 16, 1228–1236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz J.C., Wang X., Podell E.R., et al. (2013). RNA seeds higher-order assembly of FUS protein. Cell Rep. 5, 918–925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scoles D.R., Meera P., Schneider M.D., et al. (2017). Antisense oligonucleotide therapy for spinocerebellar ataxia type 2. Nature 544, 362–366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seminary E.R., Sison S.L., Ebert A.D. (2018). Modeling protein aggregation and the heat shock response in ALS iPSC-derived motor neurons. Front. Neurosci. 12, 86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shalom O., Shalva N., Altschuler Y., et al. (2008). The mammalian Nek1 kinase is involved in primary cilium formation. FEBS Lett. 582, 1465–1470. [DOI] [PubMed] [Google Scholar]
- Shi K.Y., Mori E., Nizami Z.F., et al. (2017). Toxic PRn poly-dipeptides encoded by the C9orf72 repeat expansion block nuclear import and export. Proc. Natl Acad. Sci. USA 114, E1111–E1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shorter J. (2016). Engineering therapeutic protein disaggregases. Mol. Biol. Cell 27, 1556–1560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singatulina A.S., Hamon L., Sukhanova M.V., et al. (2019). PARP-1 activation directs FUS to DNA damage sites to form PARG-reversible compartments enriched in damaged DNA. Cell Rep. 27, 1809–1821.e5. [DOI] [PubMed] [Google Scholar]
- Smith, B.N., Topp S.D., Fallini C.,. et al. (2017). Mutations in the vesicular trafficking protein Annexin A11 are associated with amyotrophic lateral sclerosis. Sci. Transl. Med. 9, eaad9157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith E.F., Shaw P.J., De Vos K.J. (2019). The role of mitochondria in amyotrophic lateral sclerosis. Neurosci. Lett. 710, 132933. [DOI] [PubMed] [Google Scholar]
- Sreedharan J., Blair I.P., Tripathi V.B., et al. (2008). TDP-43 mutations in familial and sporadic amyotrophic lateral sclerosis. Science 319, 1668–1672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steyaert J., Scheveneels W., Vanneste J., et al. (2018). FUS-induced neurotoxicity in Drosophila is prevented by downregulating nucleocytoplasmic transport proteins. Hum. Mol. Genet. 27, 4103–4116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suárez-Calvet M., Neumann M., Arzberger T., et al. (2016). Monomethylated and unmethylated FUS exhibit increased binding to Transportin and distinguish FTLD-FUS from ALS-FUS. Acta Neuropathol. 131, 587–604. [DOI] [PubMed] [Google Scholar]
- Sun D., Wu R., Zheng J., et al. (2018). Polyubiquitin chain-induced p62 phase separation drives autophagic cargo segregation. Cell Res. 28, 405–415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tada M., Doi H., Koyano S., et al. (2018). Matrin 3 is a component of neuronal cytoplasmic inclusions of motor neurons in sporadic amyotrophic lateral sclerosis. Am. J. Pathol. 188, 507–514. [DOI] [PubMed] [Google Scholar]
- Tafuri F., Ronchi D., Magri F., et al. (2015). SOD1 misplacing and mitochondrial dysfunction in amyotrophic lateral sclerosis pathogenesis. Front. Cell. Neurosci. 9, 336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor J.P., Brown R.H., Cleveland D.W. (2016). Decoding ALS: From genes to mechanism. Nature 539, 197–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trendel J., Schwarzl T., Horos R., et al. (2019). The human RNA-binding proteome and its dynamics during translational arrest. Cell 176, 391–403.e19. [DOI] [PubMed] [Google Scholar]
- Tyzack G.E., Luisier R., Taha D.M., et al. (2019). Widespread FUS mislocalization is a molecular hallmark of amyotrophic lateral sclerosis. Brain 142, 2572–2580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urdaneta E.C., Vieira-Vieira C.H., Hick T., et al. (2019). Purification of cross-linked RNA-protein complexes by phenol-toluol extraction. Nat. Commun. 10, 990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van der Spek R.A., van Rheenen W., Pulit S.L., et al. (2018). Reconsidering the causality of TIA1 mutations in ALS. Amyotroph. Lateral Scler. Frontotemporal Degener. 19, 1–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vance C., Rogelj B., Hortobágyi T., et al. (2009). Mutations in FUS, an RNA processing protein, cause familial amyotrophic lateral sclerosis type 6. Science 323, 1208–1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanneste J., Vercruysse T., Boeynaems S., et al. (2019). C9orf72-generated poly-GR and poly-PR do not directly interfere with nucleocytoplasmic transport. Sci. Rep. 9, 15728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh I., Seno F., Tosatto S.C.E., et al. (2014). PASTA 2.0: an improved server for protein aggregation prediction. Nucleic Acids Res. 42, 301–307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W.Y., Pan L., Su S.C., et al. (2013). Interaction of FUS and HDAC1 regulates DNA damage response and repair in neurons. Nat. Neurosci. 16, 1383–1391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheeler R.J., Lee H.O., Poser I., et al. (2019). Small molecules for modulating protein driven liquid–liquid phase separation in treating neurodegenerative disease. bioRxiv, 10.1101/721001 [DOI] [Google Scholar]
- Winton M.J., Igaz L.M., Wong M.M., et al. (2008). Disturbance of nuclear and cytoplasmic TAR DNA-binding protein (TDP-43) induces disease-like redistribution, sequestration, and aggregate formation. J. Biol. Chem. 283, 13302–13309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolozin B. (2014). Physiological protein aggregation run amuck: stress granules and the genesis of neurodegenerative disease. Discov. Med. 17, 47–52. [PMC free article] [PubMed] [Google Scholar]
- Wong P.C., Pardo C.A., Borchelt D.R., et al. (1995). An adverse property of a familial ALS-linked SOD1 mutation causes motor neuron disease characterized by vacuolar degeneration of mitochondria. Neuron 14, 1105–1116. [DOI] [PubMed] [Google Scholar]
- Wu C.H., Fallini C., Ticozzi N., et al. (2012). Mutations in the profilin 1 gene cause familial amyotrophic lateral sclerosis. Nature 488, 499–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang S., Kato M., Wu L.C.., et al. (2015). The LC domain of hnRNPA2 adopts similar conformations in hydrogel polymers, liquid-like droplets, and nuclei. Cell 163, 829–839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang P., Mathieu C., Kolaitis R.M., et al. (2020). G3BP1 is a tunable switch that triggers phase separation to assemble stress granules. Cell 181, 325–345.e28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y.S., Kato M., Wu X., et al. (2019). Yeast ataxin-2 forms an intracellular condensate required for the inhibition of TORC1 signaling during respiratory growth. Cell 177, 697–710.e17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshizawa T., Ali R., Jiou J., et al. (2018). Nuclear import receptor inhibits phase separation of FUS through binding to multiple sites. Cell 173, 693–705.e22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Youn, J.-Y., , Dyakov, B.J.A.,, Zhang, J., et al. (2019) . Properties of stress granule and P-body proteomes. Mol. Cell 76, 286–294. [DOI] [PubMed] [Google Scholar]
- Zhang K., Daigle J.G., Cunningham K.M., et al. (2018). Stress granule assembly disrupts nucleocytoplasmic transport. Cell 173, 958–971.e17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang K., Donnelly C.J., Haeusler A.R., et al. (2015). The C9orf72 repeat expansion disrupts nucleocytoplasmic transport. Nature 525, 56–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang P., Fan B., Yang P., et al. (2019). Chronic optogenetic induction of stress granules is cytotoxic and reveals the evolution of ALS-FTD pathology. eLife 8, e39578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y.J., Xu Y.F., Cook C., et al. (2009). Aberrant cleavage of TDP-43 enhances aggregation and cellular toxicity. Proc. Natl Acad. Sci. USA 106, 7607–7612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z.C., Chook Y.M. (2012). Structural and energetic basis of ALS-causing mutations in the atypical proline-tyrosine nuclear localization signal of the Fused in Sarcoma protein (FUS). Proc. Natl Acad. Sci. USA 109, 12017–12021. [DOI] [PMC free article] [PubMed] [Google Scholar]