Abstract
Background:
The role of the HLA-DRB1 and HLA-DQB1 genes in the antibody response to hepatitis B (HB) vaccine has been well established; however, the involvement of the HLA-DPB1 allele in the HB vaccine immune response remained to be clarified by a systematic review.
Methods:
A meta-analysis was performed in which databases were searched for relevant studies published in English or Chinese up until June 1, 2020. Six studies were identified and a total of 10 alleles were processed into statistical processing in this meta-analysis.
Results:
Three thousand one hundred forty four subjects (including 2477 responders and 667 non-responders) were included in this research. Alleles HLA-DPB1∗02:02, DPB1∗03:01, DPB1∗04:01, DPB1∗04:02, and DPB1∗14:01 were found to be associated with a significant increase in the antibody response to HB vaccine, and their pooled odds ratios (ORs) were 4.53, 1.57, 3.33, 4.20, and 1.79, respectively; whereas DPB1∗05:01 (OR = 0.73) showed the opposite correlation.
Conclusions:
These findings suggested that specific HLA-DPB1 alleles are associated with the antibody response to HB vaccine.
Keywords: alleles, hepatitis B virus, human leukocyte antigens, vaccine response
1. Introduction
Hepatitis B virus (HBV) infection is a major global health problem. The prevalence of HBV infection in China was approximately 5.49% of the population in 2015,[28] with around 93 million people infected with HBV and approximately 30 million people suffering from HBV-related diseases,[6] making this virus a significant public health burden in China.[31] Hepatitis B (HB) vaccination is an effective measure to reduce the risk of HBV infection. In China, a HBV vaccination program was introduced in 1992 that required all newborns to be vaccinated against HB; however, the cost of vaccination was met by parents and this may have prevented some children from being vaccinated. The China GAVI Project was initiated in 2002 to provide HB vaccine to infants free of charge in the areas of China worst affected by the virus. On March 24, 2005, the State Council promulgated the no. 434 order of “vaccine circulation and prevention inoculation management regulations” to the whole country. Consequently, all newborns were inoculated with HB vaccine free of charge, after which, a significant decline in the HBsAg-positive rate to 0.94% was recorded in China.[10] However, individuals who failed to respond to HB vaccination during infancy may not be fully protected against HBV infection in adolescence. Furthermore, breakthrough and chronic HBV infections were repeatedly reported in individuals who had received complete doses of HB vaccine. It is therefore important to research the mechanism of non-response to HB vaccine to better protect this portion of the population from HB infection.
It had been estimated that the heritability of post-vaccination antibody titers is higher than 70%.[8] A genome wide association study has been proven to be an efficient technique to evaluate the effect of host genetic factors on common traits. Genome wide association studies[5,22,34] revealed that some of the DRB1 and DQB1 genes and related single nucleotide polymorphisms (SNPs) play key roles in the HB vaccine response in different populations. It was reported[22] that 5 HLA-DRB1-DQB1 haplotypes, 2 HLA-DPB1 alleles and BTNL2 genes were associated with the immune response to HB vaccine in a large cohort Japanese population. Another systematic review revealed[17] that 3 HLA-DRB1 and 2 HLA-DQB1 variants were associated with a significant increase in the antibody response to HB vaccine, while 4 DRB1 alleles and 1 DQB1 allele showed the opposite trend. With the exception of the HLA-II alleles, some HLA-II-related SNPs have been found to be related to the HB vaccination response, such as rs9277535,[5] rs3077,[13] rs7770370[34] and rs9271768.[20] Recently, variants of the HLA-DPB1 locus were found to have strong genome-wide associations with the outcomes of HBV infection in several Asian populations.[1,12,15,16] HLA-DPB1, a HLA class II molecule expressed as a cell-surface glycoprotein, not only binds to exogenous antigens and some DPB1 alleles but also binds to endogenous antigens and presents them to CD4+ T cells. Research[5] found that DPB1∗04:02 had a significant association with both the vaccine response and a high-titer response, whereas DPB1∗05:01 was significantly correlated with non-responders. Other research demonstrated[35] that the HLA-DPB1 05:01 and 09:01 alleles significantly correlated with HB low responders, and the 02:01, 02:02, 03:01, 04:01 and 14:01 alleles strongly correlated with HB high responders. To confirm these previous findings and to explore the association of HLA-DPB1 with the response to booster HB vaccination, we performed a meta-analysis to clarify the relationship between HLA-DPB1 polymorphism and the degree of HB vaccination response.
2. Materials and methods
2.1. Selection of studies
We searched the PubMed, EMBASE, Cochrane library, Chinese CNKI, and Wanfang databases using the MeSH terms “Hepatitis B Vaccines”, “HLA Antigens” or “Histocompatibility Antigens”, and the individual corresponding free terms. Furthermore, we performed manual searches by scanning the reference lists of the included articles to locate additional papers related to the topic. The following inclusive criteria were set and reviewed by 2 independent investigators:
-
1.
recombinant vaccine;
-
2.
study's method based on a cohort study design;
-
3.
studies showing interest in the association between HLA alleles and the response to HB (the vaccines were classified into responders (anti-HB ≥10 IU/L) and non-responders (anti-HB < 10 IU/L) after the whole vaccine schedule); and
-
4.
study followed the standard vaccination protocol using the currently licensed HB recombinant vaccines (immunized at 0, 1 and 6 months, with or without 1 additional dose).
The exclusion criteria were as follows:
-
1.
not published as a full text;
-
2.
cohort studies without defined groups of non-responders and responders, or the responder anti-HB threshold was no higher than 10 IU/L;
-
3.
HLA-DPB1 typing had not been obtained for the different groups, or the HLA-DPB1 distribution was not in accordance with Hardy–Weinberg equilibrium.
If data were duplicated between studies, only the study most recently published with the larger number of participants was included.
2.2. Data extraction and synthesis
Two investigators (Guojin Ou and Xiaojuan Liu) carefully examined and extracted the published data independently based on our inclusion and exclusion criteria. Disagreements were resolved by discussion and consensus; with a third reviewer (YongMei Jiang) making the ultimate decision if a consensus was not achieved. We extracted the following information from each study: first author, year of publication, ethnicity, the number of cases and controls for each group, the HLA-DPB1 typing method and the references. The quality of each study was assessed independently by 2 authors using the Newcastle–Ottawa Scale system,[29] which includes 3 aspects: selection (0–4 points), comparability (0–2 points), and exposure (0–3 points). The Newcastle–Ottawa Scale scores ranged from 0 (low quality) to 9 (high quality). Studies scoring more than 7 were considered to be of high quality. This study was approved by the ethics committee of the West China Second University Hospital.
2.3. Statistical analysis
The Hardy–Weinberg equilibrium of the genotype distributions of the HLA-DPB1 alleles were examined using Arlequin 3.5.1.2 software. Review Manager 5.2 software was used to perform meta-analysis. Odd ratios (ORs) with 95% confidence intervals (CIs) were used to assess the strength of associations between HLA-DPB1 polymorphisms and the HBV immune response. The I2 test was performed to assess heterogeneity between studies. If the heterogeneity was not significant (P > 1, I2 < 50.0%), the fixed-effects model was used to pool the ORs; otherwise, the random-effects model was used. Two-sided P values <.05 were considered statistically significant.
3. Results
3.1. Flow diagrams detailing the selection process of eligible studies
The selection process employed to identify eligible studies is displayed in Figure 1. A total of 206 studies were acquired from the PubMed, EMBASE, and China National Knowledge Infrastructure (CNKI) databases. After reviewing the titles, abstracts and full text, we excluded 199 irrelevant studies. Finally, a total of 6 articles were included in the meta-analysis.[5,7,19,22,27,32,35] The main characteristics of all of the eligible studies are shown in Tables 1 and 2. Furthermore, all these studies assessed the association between HLA-DPB1 and the immune response to HB vaccine. The 6 articles included 3144 HB vaccine responders and 667 HB vaccine non-responders used as controls to assess the risk of HLA-DPB1.
Figure 1.
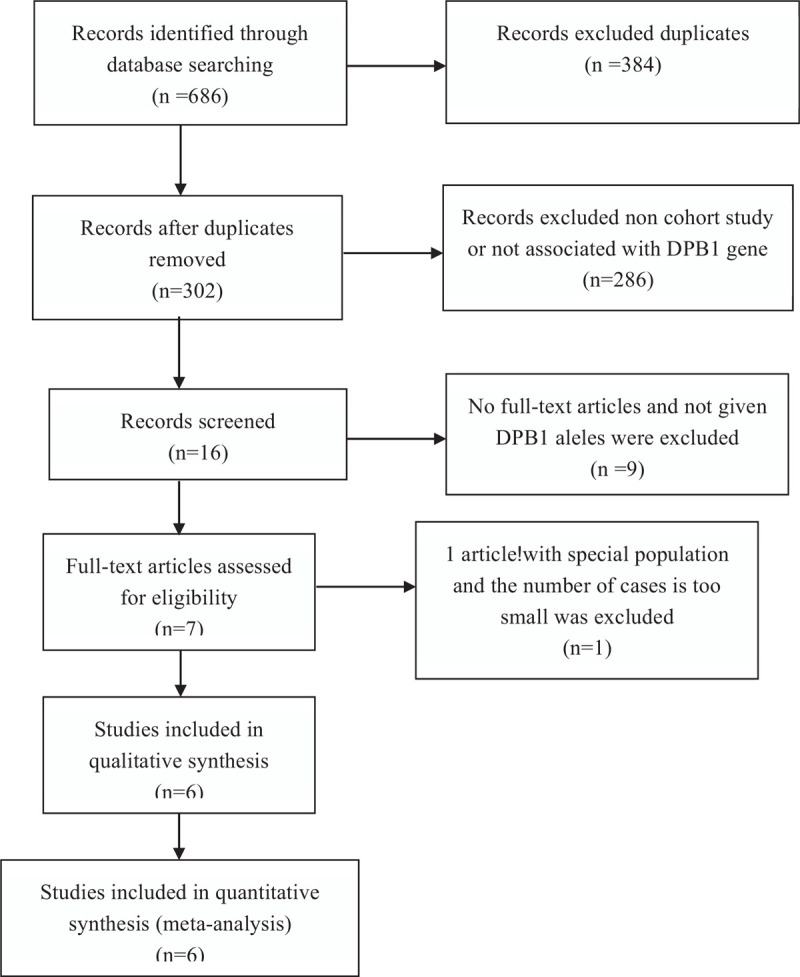
Flow chart of articles election.
Table 1.
Characteristics of the studies regarding hepatitis B vaccine response in different populations.
| Total | |||||||
| First author | Year | Ethnicity | R | NR | Genotype method | NOS | Refs |
| Desombere I | 1998 | Caucasians | 100 | 46 | PCR-SSO | 8 | [7] |
| Wu TW | 2013 | Taiwanese | 510 | 171 | PCR-SBT | 9 | [35] |
| Sakai A | 2016 | Japanese | 418 | 156 | PCR-SSO | 9 | [27] |
| Nishida N | 2018 | Japanese | 1009 | 94 | Array assay | 9 | [22] |
| Wang LY | 2019 | Taiwanese | 207 | 152 | PCR-SBT | 9 | [32] |
| Chung S | 2019 | Korean | 233 | 48 | PCR-SBT | 8 | [5] |
Table 2.
Distribution of HLA-DPB1 alleles in different studies.
| HLA-DPB1 alleles | |||||||||||
| First author (year) | 02:01 | 02:02 | 03:01 | 04:01 | 04:02 | 05:01 | 09:01 | 13:01 | 14:01 | 17:01 | Refs |
| Desombere I (1998) | [7] | ||||||||||
| R, n | 22 | – | 20 | 67 | 21 | 5 | 0 | 2 | 2 | 4 | |
| NR, n | 20 | – | 7 | 12 | 4 | 0 | 4 | 6 | 1 | 1 | |
| Wu TW (2013) | [35] | ||||||||||
| R, n | 142 | 44 | 60 | 52 | 13 | 523 | 7 | 66 | 37 | – | |
| NR, n | 34 | 2 | 6 | 5 | 1 | 231 | 9 | 25 | 5 | – | |
| Sakai A (2016) | [27] | ||||||||||
| R, n | 195 | 26 | 41 | 44 | 100 | 295 | 93 | 16 | 13 | – | |
| NR, n | 62 | 6 | 15 | 5 | 14 | 157 | 35 | 3 | 5 | 1 | |
| Nishida N (2018) | [22] | ||||||||||
| R, n | 469 | – | 107 | – | 194 | 833 | 204 | −25 | – | – | |
| NR, n | 43 | – | 7 | – | 2 | 108 | 20 | – | – | – | |
| Wang LY (2019) | [32] | ||||||||||
| R, n | 49 | 26 | 15 | 32 | 7 | 216 | 6 | 18 | 14 | 2 | |
| NR, n | 28 | 2 | 6 | 4 | 1 | 208 | 8 | 21 | 5 | 2 | |
| Chung S (2019) | [5] | ||||||||||
| R, n | 123 | 25 | 13 | 45 | 51 | 145 | 13 | 25 | 5 | 9 | |
| NR, n | 27 | 1 | 3 | 6 | 1 | 47 | 1 | 7 | – | 7 | |
3.2. Association between HLA-DPB1 and the HB vaccine response
The results of the meta-analysis are presented in Table 3 and Figures 2–7. A total of 10 alleles were subjected to statistical processing. The pooled risk estimates indicated that DPB1∗02:02 (OR = 4.53, 95% CI: 1.64–12.55), DPB1∗03:01 (OR = 1.57, 95% CI: 1.13–2.20), DPB1∗04:01 (OR = 3.33, 95% CI: 2.26–4.90), DPB1∗04:02 (OR = 4.20, 95% CI: 2.70–6.52) and DPB1∗14:01 (OR = 1.79, 95% CI: 1.03–3.12) were associated with a significant increase in the antibody response to HB vaccine. Whereas DPB1∗05:01 (OR = 0.73, 95% CI: 0.69–0.78) showed the opposite trend. These findings suggested that inheriting certain HLA-DPB1 alleles could either strengthen or diminish the HB vaccine response.
Table 3.
Overall results regarding HLA-DPB1 polymorphisms in hepatitis B response patients.
| Heterogeneity | Overall relationship | |||||
| HLA-DPB1∗ | N | I2,% | P | Model | OR (95% CI) | P value |
| 02:01 | 5 | 55 | .05 | R | 1.05 (0.85–1.30) | .06 |
| 02:02 | 4 | 54 | .09 | R | 4.53 (1.64–12.55) | .004 |
| 03:01 | 6 | 22 | .27 | F | 1.57 (1.13–2.20) | .008 |
| 04:01 | 5 | 2 | .40 | F | 3.33 (2.26–4.90) | <.00001 |
| 04:02 | 6 | 0 | .43 | F | 4.20 (2.70–6.52) | <.00001 |
| 05:01 | 5 | 0 | .61 | F | 0.73 (0.69–0.78) | <.00001 |
| 09:01 | 5 | 51 | .08 | R | 0.76 (0.47–1.25) | .28 |
| 13:01 | 5 | 44 | .13 | F | 0.77 (0.56–1.06) | .11 |
| 14:01 | 4 | 0 | .52 | F | 1.79 (1.03–3.12) | .04 |
| 17:01 | 3 | 37 | .21 | F | 0.47 (0.21–1.05) | .07 |
Figure 2.
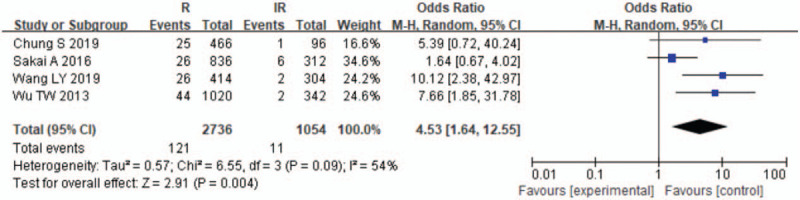
Meta-analysis of correlation of the HLA-DPB1∗02:02 allele polymorphism in HB vaccine response.
Figure 7.
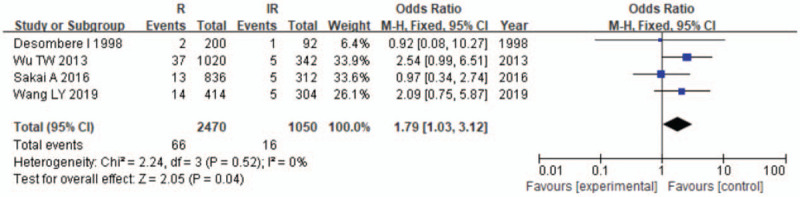
Meta-analysis of correlation of the HLA-DPB1∗14:01 allele polymorphism in HB vaccine response.
Figure 3.
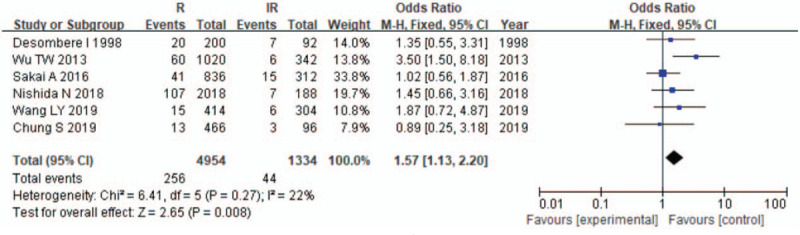
Meta-analysis of correlation of the HLA-DPB1∗03:01 allele polymorphism in HB vaccine response.
Figure 4.
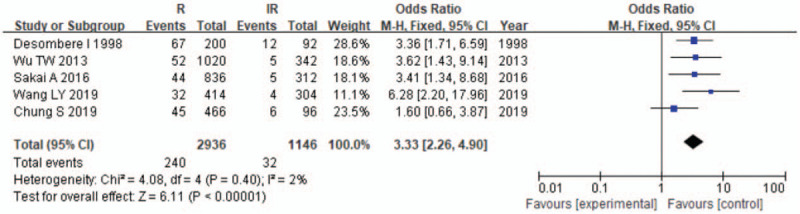
Meta-analysis of correlation of the HLA-DPB1∗04:01 allele polymorphism in HB vaccine response.
Figure 5.
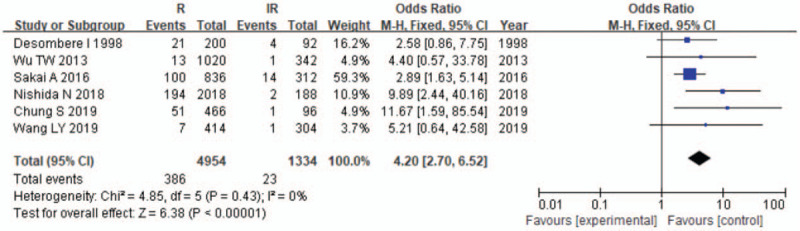
Meta-analysis of correlation of the HLA-DPB1∗04:02 allele polymorphism in HB vaccine response.
Figure 6.
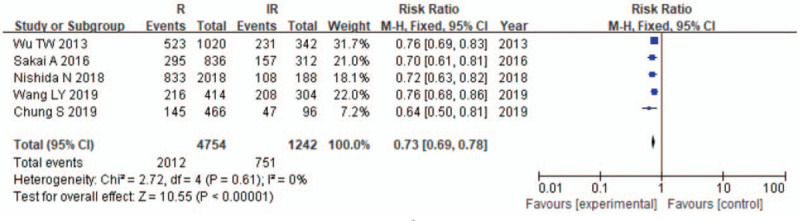
Meta-analysis of correlation of the HLA-DPB1∗05:01 allele polymorphism in HB vaccine response.
3.3. Publication bias
Publication bias of the included articles was assessed using Begg funnel plot. The shape of the funnel plot appeared symmetrical and showed no obvious publication bias for HLA-DPB1 metamorphism (Fig. 8).
Figure 8.
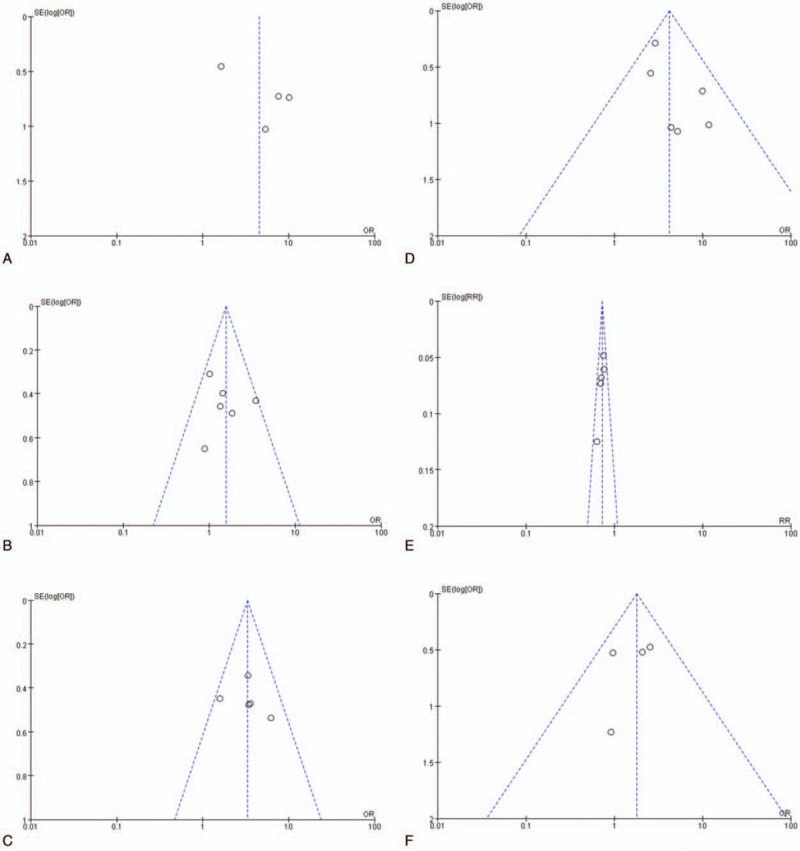
Egger funnel plot for the assessment of HLA-DPB1∗02:02, DPB1∗03:01, DPB1∗04:01, DPB1∗04:02, DPB1∗14:01, and DPB1∗05:01.
4. Discussion
The World Health Organization recommends[4] universal vaccination against HB to ultimately eliminate HBV. This recommendation had been progressively implemented across 168 countries of the world via a universal program by the end of 2006. The China GAVI Project was initiated in 2002 to provide HB vaccine to infants to prevent the consequences of HBV infection in specifically-selected areas. By 2009, coverage of the three-dose HB vaccine regimen had been increased in these areas and inoculation at birth had been increased to almost 90%. This initiative reduced HBV prevalence to <1% among children in these areas, prevented 24 million chronic HBV infections and 4.3 million future deaths due to cirrhosis, hepatocellular carcinoma and acute hepatitis.[3] Active and passive immunizations (HBIG and HB vaccines) are the most cost-effective means of preventing chronic HB infection in infants and have the potential to have a sustained impact on global HBV incidence.[9] However, it has been reported[33] that 58.2% of adolescents who received a primary series of HB vaccinations as infants with GMC had anti-HB levels of <10 mIU/ml. Another study[11] found that 60.0% of such adolescents had anti-HB concentrations <10 mIU/ml on baseline testing. A higher booster vaccine dose should be sufficient to achieve adequate protection by raising the anti-HB level. Even with the additional dose of HB vaccine, 5% to 10% of individuals still failed to respond to the vaccine,[11,38] and the precise mechanisms determining responsiveness to the HB vaccine are poorly understood. Some factors associated with vaccine nonresponsiveness include obesity,[18] age at first vaccination,[36] sex,[30] immune status[14,26] and genetic factors.[17]
A number of cohort studies have assessed the associations between HLA-II gene polymorphisms and the progression of HBV infection, including HBV infection susceptibility, HBV spontaneous clearance, HBV-related HCC development and the HB vaccination response. It has been suggested that an anti-HB titer of 10 mIU/ml is necessary for the prevention of HBV infection. HLA-II genes, including the HLA-DR, HLA-DQ, HLA-DP and DR-DQ-DP haplotypes, were reported to be associated with the immunological response to HB vaccine in healthy people. A meta-analysis[17] of 2308 subjects (including 1215 responders, 873 non-responders and 220 controls) found that DRB1∗01, DRB1∗1301, DRB1∗15, DQB1∗05 (DQB1∗0501), DQB1∗06 and DQB1∗0602 were associated with a significant increase in the antibody response to HB vaccine, whereas DRB1∗03 (DRB1∗0301), DRB1∗04, DRB1∗07, DRB1∗13:02, and DQB1∗02 showed the opposite trend. The DRB1∗01:01-DQB1∗05:01 and DRB1∗08:03-DQB1∗06:01 were associated with a significant increase in the antibody response to HB vaccine, While DRB1∗04:05-DQB1∗04:01, DRB1∗14:06-DQB1∗03:01, and DRB1∗15:01-DQB1∗06:02 showed the opposite trend.[22] The association between HLA-DPB1 polymorphisms and the immunological response to HB vaccine have been investigated in different populations,[5,7,22,27,32,34] but these findings remain controversial or inconclusive. Martinetti and colleagues found[19] that a statistically significant increase in DPBl∗02:01 and DPB1∗03:01 alleles related to a non- or hypo-immune response to HB vaccine, whereas DPB1∗04:01 was related to a positive response to HB vaccine. DPB1∗04:01 was also more abundant in good vaccine responders in the Caucasian population[7] and the Japanese population.[27] DPB1∗04:02 was also related to a high HB vaccine response, whereas DPB1∗05:01 was related to a lower HB vaccine response in the Asian population.[5,22,32] With the exception of DPB1 alleles, DPA1- and DP-related SNPs were also reported to correlate with the HB vaccine response, with DPA1∗01:03,[32] rs3077T[24,25] and rs9277535A[24,25] being significantly more prevalent in HB vaccine responders, and DPA1∗02:02, rs3077C and rs9277535G being significantly less prevalent in HB vaccine responders.
A previous study found that low expression levels of DP mRNA correlated with non-susceptibility to HBV.[23] Whether DP expression is associated with the HB vaccine response remains to be determined. The amino acids at position 84 to 87[39] of the second exon of the antigen presentation sequence of DPB1, including GGPA (with linkage to DPB1∗04:01, 04:02 and 02:02) and DENA (with linkage to DPB1∗05:01, 1401 and 03:01), located at the groove contacting the peptide residues of pocket-1, may be responsible for the different antigen-presenting functions that may lead to the different levels of immune response to HB vaccine. The latest research found that HLA-DPB1 molecules encoding DPGly84[37] (including DPB1∗04:01, DPB1∗04:02 DPB1∗02:01 and DPB1∗02:02) do not bind to the invariant chain (Ii) via the class II-associated invariant chain peptide region to constitutively present endogenous peptides. This means that DPGly84-related alleles can use both class I and II antigen processing pathways to present peptides derived from intracellular and extracellular sources; however, DP84Asp (including DPB1∗05:01) does not possess such endogenous antigen presentation function. Therefore,[2] these polymorphisms have different functions in autoimmune, antiviral and antitumor mechanisms through the different functions of antigen presentation in the CD4+ Tc cell immune response. The DP84Gly genotype not only plays a unique antiviral function in adaptive immunity, but also acts as a ligand of NKp44[21] to activate natural killer (NK) T cells to play an antiviral role in the immune response. Further studies should reveal whether the DP84Gly genotype can promote the immune response to HB vaccine by binding to NKp44 and activating NK cells.
In this meta-analysis, we found that DPB1∗02:02, DPB1∗03:01, DPB1∗04:01, DPB1∗04:02 and DPB1∗14:01 were associated with a significant increase in the antibody response to HB vaccine, with pooled OR values of 4.53, 1.57, 3.33, 4.20, and 1.79, respectively. From the ORs, we found that alleles DPB1∗02:02, DPB1∗04:01 and DPB1∗04:02 were associated with a hyper HB vaccine response, whereas DPB1∗05:01 (OR = 0.73) was related to a hypo HB vaccine response; these results were in accordance with previous reports. However, the correlation between DPB1∗03:01 and DPB1∗14:01 carriers and an increased response to HB vaccine is controversial. In an Italian study involved a small group of infants, DPB1∗03:01 was demonstrated to be related to a non- or hypo-immune response to HB vaccine,[18] whereas the opposite results were found in a group of Taiwanese adolescents,[35] and other studies failed to find an association between DPB1∗03:01 and DPB1∗14:01 and the HB vaccine response.[5,22] Similar to DPB1∗05:01, DPB1∗03:01 and DPB1∗14:01 were shown to display linkage with DP84Asp and were associated with a lower response to HBV antigens, resulting in susceptibility to HBV, a higher likelihood of chronic HBV infections or susceptibility to hepatocellular carcinoma. As the ORs of DPB1∗03:01 and DPB1∗14:01 in the HB vaccine response were low (1.57 and 1.79), whether these alleles are linked to a lower response to HB vaccine should be the subject of future research in larger and more diverse study populations.
To the best of our knowledge, this is the first meta-analysis providing comprehensive insights into the effects of DPB1 on the HB vaccine response. This meta-analysis had several limitations. More precise analysis was not able to be performed because we were unable to obtain the original data for all of the studies, and the subgroup analysis, for example, for age, sex, body mass index and lifestyle, could not be adjusted. Furthermore, our analysis did not consider the possible effects of gene–environment interactions. Therefore, more studies are needed to confirm our findings.
In conclusion, our meta-analysis revealed that HLA-DPB1∗02:02, DPB1∗03:01, DPB1∗04:01, DPB1∗04:02, and DPB1∗14:01 are related to a strengthened HB vaccination response, and HLA-DPB1∗05:01 serves as a low HB vaccination response marker. However, more large-scale studies are warranted to support our findings.
Acknowledgments
We thank Kate Fox, DPhil, from Liwen Bianji, Edanz Group China (www.liwenbianji.cn/ac), for editing the English text of a draft of this manuscript.
Author contributions
Conceptualization: Yongmei Jiang.
Data curation: Guojin Ou, Xiaojuan Liu.
Formal analysis: Guojin Ou, Xiaojuan Liu.
Investigation: Guojin Ou.
Methodology: Xiaojuan Liu.
Project administration: Yongmei Jiang.
Software: Guojin Ou.
Supervision: Yongmei Jiang.
Validation: Yongmei Jiang.
Writing – original draft: Guojin Ou.
Writing – review & editing: Yongmei Jiang.
Footnotes
Abbreviations: CI = confidence interval, HB = hepatitis B, HBV = hepatitis B virus, HLA = human leukocyte antigen, OR = odds ratio, SNP = single nucleotide polymorphism.
How to cite this article: Ou G, Liu X, Jiang Y. HLA-DPB1 alleles in hepatitis B vaccine response: a meta-analysis. Medicine. 2021;100:14(e24904).
This work was supported by the Funding of Sichuan Science and Technology under Contract 2018SZ0395.
The authors have no conflicts of interests to disclose.
All data generated or analyzed during this study are included in this published article [and its supplementary information files].
References
- [1].Al-Qahtani A, Khalak HG, Alkuraya FS, et al. Genome-wide association study of chronic hepatitis B virus infection reveals a novel candidate risk allele on 11q22.3. J Med Genet 2013;50:725–32. [DOI] [PubMed] [Google Scholar]
- [2].Anczurowski M, Hirano N. Mechanisms of HLA-DP antigen processing and presentation revisited. Trends Immunol 2018;39:960–4. [DOI] [PubMed] [Google Scholar]
- [3].Centers for Disease C, Prevention. Progress in hepatitis B prevention through universal infant vaccination China, 1997–2006. MMWR Morb Mortal Wkly Rep 2007;56:441–5. [PubMed] [Google Scholar]
- [4].Centers for Disease C, Prevention. Implementation of newborn hepatitis B vaccination worldwide, 2006. MMWR Morb Mortal Wkly Rep 2008;57:1249–52. [PubMed] [Google Scholar]
- [5].Chung S, Roh EY, Park B, et al. GWAS identifying HLA-DPB1 gene variants associated with responsiveness to hepatitis B virus vaccination in Koreans: Independent association of HLA-DPB1∗04:02 possessing rs1042169 G - rs9277355 C - rs9277356 A. J Viral Hepat 2019;26:1318–29. [DOI] [PubMed] [Google Scholar]
- [6].Cui Y, Jia J. Update on epidemiology of hepatitis B and C in China. J Gastroenterol Hepatol 2013;28: (Suppl 1): 07–10. [DOI] [PubMed] [Google Scholar]
- [7].Desombere I, Willems A, Leroux-Roels G. Response to hepatitis B vaccine: multiple HLA genes are involved. Tissue Antigens 1998;51:593–604. [DOI] [PubMed] [Google Scholar]
- [8].Doi H, Yoshio S, Yoneyama K, et al. Immune determinants in the acquisition and maintenance of antibody to hepatitis B surface antigen in adults after first-time hepatitis B vaccination. Hepatol Commun 2019;3:812–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Guo Y, Zhang W, Zhang Y, et al. Cost-effectiveness analysis of preventing mother-to-child transmission of hepatitis B by injecting hepatitis B immune globulin. Eur J Gastroenterol Hepatol 2012;24:1363–9. [DOI] [PubMed] [Google Scholar]
- [10].Hadler SC, Fuqiang C, Averhoff F, et al. The impact of hepatitis B vaccine in China and in the China GAVI project. Vaccine 2013;31: (Suppl 9): J66–72. [DOI] [PubMed] [Google Scholar]
- [11].Han K, Shao X, Zheng H, et al. Revaccination of non- and low- responders after a standard three dose hepatitis B vaccine schedule. Hum Vaccin Immunother 2012;8:1845–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Huang YH, Liao SF, Khor SS, et al. Large-scale genome-wide association study identifies HLA class II variants associated with chronic HBV infection: a study from Taiwan Biobank. Alimentary Pharmacol Therapeutics 2020;52:682–91. [DOI] [PubMed] [Google Scholar]
- [13].Lau KC, Lam CW, Law CY, et al. Non-invasive screening of HLA-DPA1 and HLA-DPB1 alleles for persistent hepatitis B virus infection: susceptibility for vertical transmission and toward a personalized approach for vaccination and treatment. Clin Chim Acta 2011;412:952–7. [DOI] [PubMed] [Google Scholar]
- [14].Li X, Xu Y, Dong Y, et al. Monitoring the efficacy of infant hepatitis B vaccination and revaccination in 0- to 8-year-old children: protective anti-HBs levels and cellular immune responses. Vaccine 2018;36:2442–9. [DOI] [PubMed] [Google Scholar]
- [15].Li Y, Si L, Zhai Y, et al. Genome-wide association study identifies 8p21.3 associated with persistent hepatitis B virus infection among Chinese. Nat Commun 2016;7:11664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Li Y, Zhai Y, Song Q, et al. Genome-Wide Association Study Identifies a New Locus at 7q21.13 Associated with Hepatitis B Virus-Related Hepatocellular Carcinoma. Clin Cancer Res 2018;24:906–15. [DOI] [PubMed] [Google Scholar]
- [17].Li ZK, Nie JJ, Li J, et al. The effect of HLA on immunological response to hepatitis B vaccine in healthy people: a meta-analysis. Vaccine 2013;31:4355–61. [DOI] [PubMed] [Google Scholar]
- [18].Liu F, Guo Z, Dong C. Influences of obesity on the immunogenicity of hepatitis B vaccine. Hum Vaccin Immunother 2017;13:1014–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Martinetti M, Cuccia M, Daielli C, et al. Anti-HBV neonatal immunization with recombinant vaccine. Part II. Molecular basis of the impaired alloreactivity. Vaccine 1995;13:555–60. [DOI] [PubMed] [Google Scholar]
- [20].McMahon G, Ring SM, Davey-Smith G, et al. Genome-wide association study identifies SNPs in the MHC class II loci that are associated with self-reported history of whooping cough. Human Molecular Genetics 2015;24:5930–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Niehrs A, Garcia-Beltran WF, Norman PJ, et al. A subset of HLA-DP molecules serve as ligands for the natural cytotoxicity receptor NKp44. Nat Immunol 2019;20:1129–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Nishida N, Sugiyama M, Sawai H, et al. Key HLA-DRB1-DQB1 haplotypes and role of the BTNL2 gene for response to a hepatitis B vaccine. Hepatology (Baltimore, Md) 2018;68:848–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].O’Brien TR, Kohaar I, Pfeiffer RM, et al. Risk alleles for chronic hepatitis B are associated with decreased mRNA expression of HLA-DPA1 and HLA-DPB1 in normal human liver. Genes Immun 2011;12:428–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Okada Y, Uno N, Sato S, et al. Strong influence of human leukocyte antigen-DP variants on response to hepatitis B vaccine in a Japanese population. Vaccine 2017;35:5662–5. [DOI] [PubMed] [Google Scholar]
- [25].Roh EY, Yoon JH, In JW, et al. Association of HLA-DP variants with the responsiveness to Hepatitis B virus vaccination in Korean Infants. Vaccine 2016;34:2602–7. [DOI] [PubMed] [Google Scholar]
- [26].Sabry R, Mohamed ZAZ, Abdallah AM. Relationship between Th1 and Th2 cytokine serum levels and immune response to Hepatitis B vaccination among Egyptian health care workers. J Immunoassay Immunochem 2018;39:496–508. [DOI] [PubMed] [Google Scholar]
- [27].Sakai A, Noguchi E, Fukushima T, et al. Identification of amino acids in antigen-binding site of class II HLA proteins independently associated with hepatitis B vaccine response. Vaccine 2017;35:703–10. [DOI] [PubMed] [Google Scholar]
- [28].Schweitzer A, Horn J, Mikolajczyk RT, et al. Estimations of worldwide prevalence of chronic hepatitis B virus infection: a systematic review of data published between 1965 and 2013. Lancet 2015;386:1546–55. [DOI] [PubMed] [Google Scholar]
- [29].Stang A. Critical evaluation of the Newcastle-Ottawa scale for the assessment of the quality of nonrandomized studies in meta-analyses. Eur J Epidemiol 2010;25:603–5. [DOI] [PubMed] [Google Scholar]
- [30].Trevisan A, Frasson C, De Nuzzo D, et al. Significance of anti-HB levels below 10 IU/L after vaccination against hepatitis B in infancy or adolescence: an update in relation to sex. Hum Vaccin Immunother 2020;16:460–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Wang FS, Fan JG, Zhang Z, et al. The global burden of liver disease: the major impact of China. Hepatology 2014;60:2099–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Wang LY, Chen CF, Wu TW, et al. Response to hepatitis B vaccination is co-determined by HLA-DPA1 and -DPB1. Vaccine 2019;37:6435–40. [DOI] [PubMed] [Google Scholar]
- [33].Wang ZZ, Gao YH, Lu W, et al. Long-term persistence in protection and response to a hepatitis B vaccine booster among adolescents immunized in infancy in the western region of China. Hum Vaccin Immunother 2017;13:909–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Wu TW, Chen CF, Lai SK, et al. SNP rs7770370 in HLA-DPB1 loci as a major genetic determinant of response to booster hepatitis B vaccination: results of a genome-wide association study. J Gastroenterol Hepatol 2015;30:891–9. [DOI] [PubMed] [Google Scholar]
- [35].Wu TW, Chu CC, Ho TY, et al. Responses to booster hepatitis B vaccination are significantly correlated with genotypes of human leukocyte antigen (HLA)-DPB1 in neonatally vaccinated adolescents. Human Genetics 2013;132:1131–9. [DOI] [PubMed] [Google Scholar]
- [36].Wu W, Lv J, Liu J, et al. Persistence of immune memory among adults with normal and high antibody response to primary hepatitis B vaccination: results from a five-year follow-up study in China. Hum Vaccin Immunother 2018;14:2485–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Yamashita Y, Anczurowski M, Nakatsugawa M, et al. HLA-DP(84Gly) constitutively presents endogenous peptides generated by the class I antigen processing pathway. Nat Commun 2017;8:15244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Yoshioka N, Deguchi M, Hagiya H, et al. Durability of immunity by hepatitis B vaccine in Japanese health care workers depends on primary response titers and durations. PloS One 2017;12:e0187661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Zhu M, Dai J, Wang C, et al. Fine mapping the MHC region identified four independent variants modifying susceptibility to chronic hepatitis B in Han Chinese. Human Molecular Genetics 2016;25:1225–32. [DOI] [PubMed] [Google Scholar]


