Abstract
The COVID-19, an acute respiratory syndrome caused by SARS-CoV2 is a major catastrophic event of the twenty first century. Relentless efforts for the development of effective pharmaco-therapeutics are in progress but the respite is the development of effective vaccines. However, monotherapy might not always exhibit complete efficacy and may culminate in the rapid evolution of drug-resistant viral strains. Hence, simultaneous modulation of multiple druggable targets not only enhances therapeutic efficacy but also quell the prospects of mutant viruses. Currently, milk peptides have bloomed beyond just being a quintessential part of nutrition to prominent therapeutic implications in human health and diseases. Hence, we have focused on colostrum/milk peptides as they have already been acknowledged for their high potency, target specificity with significantly low or no side effects and bio-toleration. The results presented provide a conceptual strategy for the rational designing of prospective multitargeted peptide inhibitors for SARS-CoV2.
Supplementary Information
The online version contains supplementary material available at 10.1007/s10930-021-09983-8.
Keywords: Milk peptides, SARS-CoV2, Targets, Molecular docking, Molecular dynamics
Introduction
To date, severe acute respiratory syndrome coronavirus 2 (SARS-CoV2) has infected over 38.3 million people and more than 1.09 million people worldwide have succumbed to this deadly virus. Individuals with a chronic immunological deficiency from various disease conditions are more prone to contracting COVID-19 [1]. Along with the chronic illness and death associated with the disease, the person suffering must be socially distanced and hospitalized to check further spreading which overwhelms the medical system and drains the overall economy of the country. This situation exacerbate since many infected individuals are from underprivileged background and hence, it has a pernicious impact on physical, economical and psychological state. The causal organism of COVID-19, SARS-CoV2, is the latest addition of the Betacoronavirus genera and Coronaviridae family. These viruses are inherently different from human coronaviruses (HCoVs) which are considered insignificant pathogens, causing “common cold” in otherwise normal individuals. With pre-existent SARS-CoV and Middle East Respiratory Syndrome (MERS-CoV), novel coronaviruses on other hand are zoonotic, by crossing the species barrier has caused one of the grievous global pandemic of the twenty first century with alarming morbidity and mortality [2]. COVID-19 outbreak has inadvertently highlighted our incapacity in understanding the pathogenic potentials of viruses or perhaps our negligence due to their mild symptoms in the past. Further, it has shown our inabilities in developing safe, effective and quality medicines and vaccines at a rapid rate.
The coronaviruses are master chameleons and modify their genetic makeup frequently. This evolutionary weapon is advantageous to these viruses making them potentially impervious to classical “target specific” drugs making the treatment regime often difficult. Single-target drugs may have lower toxicity owing to their target specificity, but this strategy is ineffective in tackling the pathogens with manifold features such as coronavirus. To tackle the menace of coronavirus efficiently, we need to look beyond the single-target strategy and adopt a multipronged attack. Recently, the multi-target or “magic shotgun” paradigm has gained popularity because of their ability to target a multitude of pathomechanisms of the disease and their superior therapeutic efficiency [3]. Herein, we are introducing milk bioactive peptides as “target-specific shotguns” to commandeer specific inhibitors to both host and SARS-CoV2 targets. Milk peptides are acquiring serious attention for their myriad of positive therapeutic properties such as antihypertensive, antiviral, antimicrobial, antioxidant, immunomodulatory, analgesic, antithrombotic and other bioactivities. The major benefits of employing milk peptides multitargeted drugs are: (i) their production is incredibly cost-effective thus reducing the loss of capital to pharmaceutical companies in case of failure, (ii) they have significantly higher target specificity with low or no side effects and better bioavailability and pharmacokinetics, and (iii) they are easily metabolized thus have no bioaccumulation and hepato-renal toxicities [4]. Recently, peptides of β-lactoglobulin were evaluated by in silico analysis as potential candidates to be used in the treatment of SARS-CoV2 and similarly, lactoferrin peptides were projected as immune boosters [5, 6]. In the present study, we have made an attempt to comprehensively assess the potentials of all the milk proteins in addition to the milk fat globule membrane proteins. Further, we have designed a synergistic computational approach which includes molecular docking and simulation-based screening to explore the plethora of targets including both viral as well as human protein targets in identifying multitargeted peptides against SARS-CoV2.
Materials and Methods
Materials
The peptides carved out from whey proteins and fat globule membrane proteins of Buffalo colostrum and milk [7–11] were used for the present study. More than 300 peptides were selected and culminated in 170 peptides in the range of 5 to 13 amino acids.
Methods
Protein Preparation and Binding Site Prediction
The 3D structures of the protein-targets were fetched from the RCSB database (https://www.rcsb.org/) or built by adopting a homology modeling technique depending on their availability. The 3D-structures were prepared by employing a protein preparation wizard and all redundant non-protein molecules (water and ligands) were removed from the proteins. Further, missing H-atoms were added including the protons essential for the generation of tautomers and appropriate ionization states of charged amino acids viz., Asp, Ser, Glu, Arg and His. Further, it was energy minimized employing the OPLS-2005 force field to alleviate steric clashes.
In order to explore the binding domains of proteins with no prior information, all the potential binding sites on the surface of the binding domain were probed and the suitable sites were selected based on careful observation. Binding sites were identified utilizing the SiteMap tool from Schrodinger [12]. SiteMap predicts energetically potential binding sites through coordination of topological features computed at each site point employing van der Waals probes. SiteMap was lined up to predict the five top-ranked potential receptor sites using the default settings. The site score, druggability score and size were employed to ascertain the most favorable binding pocket.
Docking Based Screening
The peptide libraries employed in this work were prepared using integrated LigPrep and the conformations were generated using the Confgen module with the force field OPLS-3. Molecular docking studies were conducted using Glide software. The grid area for docking was created with standard parameters optimized for peptide docking. The grid box coordinates were centered at the suitable active/binding domain and the size of the computed grid was based on the size of the peptides. Standard precision (SP) docking module integrated into the peptide-docking tool was used in the docking studies. To lower the energy potential of nonpolar entities of peptides, the scaling factor of the van der Waals radii was set to 1.0 with a partial atomic charge of 0.25. In the docking protocol, flexible peptide sampling was used to create different peptide conformations at pH of 7.0 ± 2.0. The OPLS-3 force field was enlisted to construct the state of peptides, to amend the structure, to minimize the conformations and build protonation/tautomerization states of the peptides [13]. Later, the conformations with the lowest minima were administered to a Monte Carlo (MC) protocol that samples the closer torsional minima [14]. No constraints were utilized in the entire docking protocol. After docking, peptides with appropriate hydrogen bond, hydrophobic and π interactions of the best peptide conformations were visualized and inferred using maestro visualizer. Finally, the top-ranked peptide poses with the least energy or docking score were enlisted for further studies.
Molecular Dynamics (MD) Simulations
The top-ranked peptides of ACE2 and Spike proteins were subjected to MD simulations employing the GROMACS (Version-2020) [15], to scrutinize their stability and compatibility in the binding site. Before MD simulation the protein-peptide complexes were scrutinized for any unsuitable deformities. The protein–peptide complexes selected based on the docking studies were represented using the OPLS force field [16]. The complex was solvated by using a single point charge (SPC) water model within a cubical measuring 1 Å on each side. The ionic charge of the system was counterbalanced by appending reverse ions and an overall salt concentration of 0.15 M was maintained. The linear constraint (LINCS) algorithm was employed to fix all hydrogen-related bond lengths, facilitating the use of a 2-fs time step. Electrostatic interactions (long-range) between atoms were handled by using Particle Mesh Ewald (PME), a cut-off of 1.0 nm was implemented to both the PME direct-space constituent of electrostatic features and van der Waals interactions. The 3-stage system-relaxation protocol was engaged before the production simulation. This protocol comprised a sequence minimization and equilibration runs to relax the system gradually without diverging from the primary conformation. The relaxation protocols employed were: (1) 50 ps energy minimization immediately after adding salt to the system, (2) energy equilibration in NVT (constant number of particles, volume and temperature) ensemble at 300 K and (3) 100 ps MD simulation in NPT (constant number of particles, pressure and temperature) ensemble at 300 K.
Finally, production simulations were executed in the NPT ensemble for 50 ns at 300 K temperature and 1.0 bar pressure. For this, the V-rescale (modified Berendsen thermostat) [17] was enforced with a time constant of 0.1 ps and the isotropic Parrinello–Rahman Barostat was used with a time constant of 2 ps. A cut-off of 1.2 Å was employed for evaluating short-range van der Waals interactions. MD trajectories were backed-up by taking frames every 10 ps. After production simulations, root mean square deviations (RMSD), root mean square fluctuations (RMSF), and protein–peptides interactions of the protein-systems were deciphered.
Results
Targeting Entry Points of Coronavirus (ACE2, Spike Protein, TMPRSS, Cathepsin-L, and Furin)
Milk peptides have been extensively exploited for potential health benefits and in addressing several diseases/disorders. In the present study, an attempt has been made to evaluate short peptide (4–13 residues) interactions from the peptide libraries of whey proteins and fat globule membrane proteins characterized from Buffalo colostrum and milk spanning both virus and host-based targets for possible therapeutic interventions. The in silico analysis unraveling possible interactions of single/several peptides for multiple targets are presented.
Angiotensin‐Converting Enzyme 2 (ACE2)
ACE2 receptors are the entry airway for various coronavirus strains viz., SARS-CoV, NL63 and SARS-CoV2. The major functions of the ACE2 are likely to bring homeostasis to the status of the renin–angiotensin system (RAS) by the degradation of angiotensin II to angiotensin. RAS is involved in vasodilation, antiproliferative, anti-hypertrophy, myocardial-protection and renoprotective functions. ACE2 is a universal receptor and present almost in all cells, but greatly expressed in lung, cardiomyocytes, cardiovascular epithelial, intestinal, renal, testicular and neuronal cells. The entry of viral particles via membrane fusion process destroys these receptors leading to partial seize of the catalytic properties thereby inducing a cascade of inflammation and apoptosis [18].
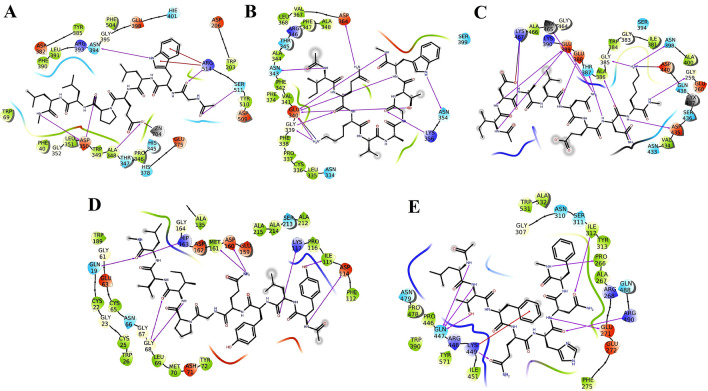
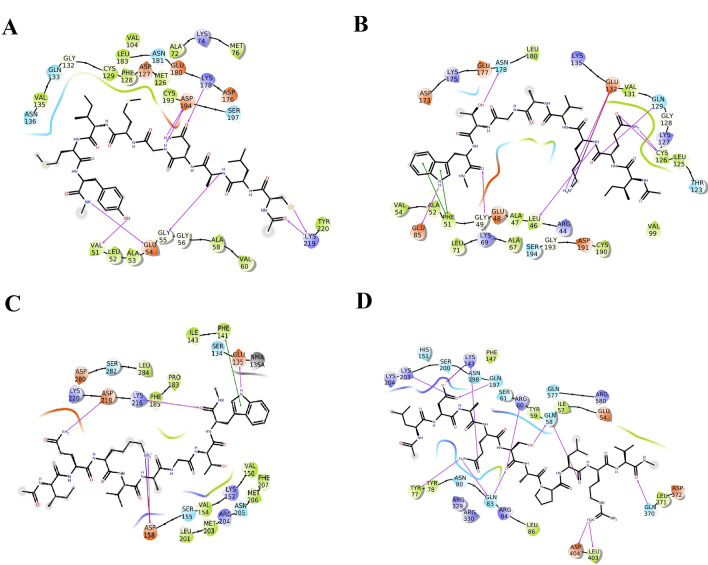
The substrate binding domain of the ACE2 has been used in the development of potent and specific inhibitor, MLN-4760 which is used in numerous studies of ACE2 action in vivo and in vitro. In order to develop similar peptide therapeutics, we have targeted the substrate binding domain (S2 subsite) [19]. Several milk peptides were found to interact with the ACE2 receptor and the best-interacted peptides were listed (Supplementary File 1). The sequence GWLEPLL interacted strongly (− 11.356) with ACE2 by forming nine hydrogen interactions where in Gly1 and Trp2 showed maximum of three hydrogen interactions with Arg514, Ser511 and Asn394 of which Arg514 have been identified as key residue essential for the ligand binding. The α-keto moiety of Gly1 formed a strong interaction with amino moiety of guanidine group of 514. In addition, guanidine group of Arg514 showed two π-cation interactions with both the aromatic rings of Trp2 (P). The pyridine amide group of the Trp2 exhibited strong H-bond interactions with the α-keto moiety of Asp350. The α-keto moiety of Pro5 exhibited H-bond with the α-amino group of Asp350. Similar interactions in vice versa manner i.e. the α-amino group of binding site residues Leu7 exhibited strong H-bond interactions with the Asp350. Other residues that exhibited hydrophobic interactions with peptide were Phe40, Pro346, Ala348, Leu351 and Gly352 (Fig. 1a) (Supplementary 15A).
Fig. 1.
Interactions established after docking, a GWELPL and ACE-2, b IQKVAGTW-Spike, c IDALNENK-TMPRSS2, d YLYQGPIVL-Cathepsin-L and e LTFQHNF-FURIN2. Peptides are in green, while residues of binding sites are represented in three coded format (various color) and thick colored lines encasing peptides represent the active protein site pockets. Solid pink lines depict H-bonds, while hydrophobic interactions are represented in green. Additionally, salt bridges and π-cation stacking, are represented by gradient color (pink and blue lines) and a red line respectively (Color figure online)
Spike Protein
The spike protein of SARS-CoV2 is a multifactorial protein that is crucial for virus entry into host cells. Initially, it associates itself with ACE2 of the host cell via its S1 subunit and then coalesces viral and host cell membranes via its S2 subunit. The receptor-binding S1 subunit of SARS-CoV2 spike protein consists of two distinctive units, S1-N-terminal domain (NTD) and S1-C-terminal domain (CTD), both of which have the ability to be functional receptor-binding domains, RBDs [20].
The binding pocket of spike proteins of both SARS-CoV2 and SARS-CoV are highly conserved. An in silico study to repurpose FDA-approved drugs as SARS-CoV2 spike protein inhibitors via virtual screening revealed that the potent compounds bind at the RBD-ACE2 interface involving diverse amino acids (319–530) of SARS-CoV2 spike protein. A detailed analysis of interface residues revealed that a stretch of the SARS-CoV2 spike protein (339–506) to be interacting majorly with the ACE2 (340–468) and hence, the same site was chosen for docking based screening of milk peptides [21].
The most potential peptide (IQKVAGTW; − 11.03 glide score) was predicted to be positioned on the central shallow pit of the RBD of the Spike-ACE2 complex with strong interactions (Supplementary File 2). The peptide was found to interact with the amino acid residues like Gly339, Glu340, Asp343, Asp354, Lys356 and Asp364 of protein by forming nine hydrogen bonds (Fig. 1b) (Supplementary 15B). The α-keto moiety of Ile1 (P) exhibited H-bond with the α-amino group of Asp343. Similar interactions are seen between Gln2-Gly339 and Trp8-Glu340. The side-chain amide group of Gln2 showed tight interaction with α-keto moiety of Asp364. Similarly, butyl ammonium side chain of Lys3 showed H-bond interactions with α-keto and carboxyl moieties of Gly339 and Glu340 respectively. The α-keto moiety of Gly6 (P) exhibited H-bond with the butyl ammonium side chain of Lys356.
TMPRSS2
As mentioned earlier, one of the key strategies of the SARS-CoV2 is to commandeer cellular proteolytic machinery to undertake the required modification of its spike glycoprotein. The type II transmembrane serine proteases (TMPRSS2), is another example which can be utilized by SARS-CoV2 apparently by cleavage of S-protein at or close to the cell surface. The triggering of Spike by TMPRSS2 leads to the Cathepsin L-independent viral entry into the vulnerable cells. TMPRSS2 is majorly expressed in ACE2-positive human lung cells and hence might play a substantial role in the rapid spread of SARS-CoV2 in the lungs and respiratory tract. Incidentally, TMPRSS2 has been the target of influenza viruses; the hemagglutinin protein of these viruses contains a monobasic cleavage site for TMPRSS2. It is also shown to modify and trigger the F protein of human metapneumovirus, thus numerous respiratory viral pathogens pirate TMPRSS2 and other proteins to promote their virulence [22].
Typically, the TMPRSS2 structure comprises of six key amino acids in the active site make-up of which three residues (His296, Asp345, Ser441) are found at the catalytic site (catalytic triad) and the remaining residues (Asp435, Ser460, Gly462) are at the substrate binding site. The catalytic triad of His296, Asp345 and Ser441 spans the active site with Ser441 on one side and Asp345 and His296 on the other side of the S1 pocket. Asp435 sits at the base of the S1 pocket and Ser436 and Asp440 form the right wall. In this context the entire active site along with the catalytic triad was considered for docking simulations.
The molecular docking results of the milk peptides for TMPRSS2 are summarized in Supplementary File 3. Of the peptides examined in this study, IDALNENK with docking energy − 142.071 kJ/mol and docking score of − 10.74 fits well into the binding site of the TMPRSS2. Though there was low hydrophobic interaction for the peptide, it was coped by the strong hydrophilic interactions and array of H-bond interactions. Generally, in the case of peptide inhibitor binding, we could see many H-bonds between α-keto moieties of peptide residues with α-amino group of receptors and four such H-bonds were seen with Glu389, Thr387 and Asp435. Two salt bridges were observed viz. Asp2-Lys467 and Lys8-Asp440 in both the cases the cationic nitrogen of the butyl ammonium (Lys) formed a salt bridge with carboxyl moiety of aspartate. Ammonium moiety of Lys8 showed three H-bond with ketonic moieties of Gly385, Asn398 (Amido) and Asp440 (Carboxylic). Amide groups of Asn5 and 7 showed interactions with Ala386, Glu389 and Cys465 (Fig. 1c) (Supplementary 15C). Based on the docking simulation results it can be deduced that the milk peptides exhibited strong H-bond interactions with crucial S1 site residues viz. Asp435 and Asp440. The same trend was also observed in Camostat mesylate reported previously explicitly showing numerous polar and non-polar contacts within the binding pocket of TMPRSS2.
Cathepsin-L
Cathepsin-L is a ubiquitously expressed lysosomal endopeptidase that is primarily involved in the terminal degradation of intracellular and endocytosed proteins. It catalyzes the cleavage of the S1 subunit of the SARS-CoV2 spike glycoprotein. This proteolytic activation is essential for coronavirus entry into human host cells which subsequently release viral RNA for replication [23].
Based on our docking experiments, we selected YLYQGPIVL as a potential inhibitor of cathepsin with a docking score of − 12.43 with the corresponding E-model value of − 213.63 (Supplementary File 4). Molecular docking analysis of four potential peptides showed that they shared common intermolecular interactions with the protein. Examination of the binding modes of the peptide revealed that the peptide adopted the classic binding mode of the C-end peptide, in which Ile7 and Leu9 at the C-terminal formed H-bond with three key residues viz. Gly68, Glu19 and His163. Similar to other members of the papain family, the active Cathepsin L comprises of two globular domains, the R-domain (right) and L-domain (left). Active site of Cathepsin-L is subdivided into seven distinguishable regions viz. S1−S4 and ❛S1−S3’. Like all cysteine proteases, the active site of cathepsin-L is composed of a reactive cysteine (Cys25), and a histidine (His163). The peptide spanned the entire active site and showed interaction with the catalytic dyad residue His163 and another crucial residue Glu19 in the S1 region. It is noteworthy that in the S3 pocket, there were two hydrogen bonding contacts between phenyl group of the peptide and the Gly68 of cathepsin L. Gln4 exhibited 2 hydrogen bond interactions with Asp160 and Met161 while Tyr1 formed two H-bond with Asp114 and Ile115. H-bonds between α-keto moieties and α-amino group were seen amongst Tyr1-Asp114 and Ile7-Gly68. The amide group of Gln4 showed two H-bond interactions with the carboxyl of Asp160 and the α-keto moiety of Met161. The α-keto moiety of Leu9 formed strong interactions with the amine group of Gln19 and His163 (Fig. 1d) (Supplementary 15D).
Furin
Furin belongs to a conserved family of proprotein and its major function is endoprotease-convertase activity; it proteolytically actuates numerous precursor-proteins in diverse branches of secretory pathways. Similar to most viral glycoproteins (Furin sites), spike of SARS-CoV2 is proteolytically activated before it can facilitate viral entry into host cells. Because of the ability of furin to cleave important cell surface proteins and SARS-CoV2 having “Furin” mimicing sites increases the potential of the virus to successfully infiltrate into the host. This crucial feature can be seen in other pathogenic viruses like influenza; astonishingly, the other close members (SARS-CoV) of β-coronaviridae family do not contain Furin cleavage site [24].
The binding modes of the lead peptide LTFQHNF in the binding site of human furin were identified using intermolecular flexible docking (Supplementary File 5). Figure 1e depicts the binding conformations of the LTFQHNF in the binding pocket of the furin (Supplementary 15E). The peptide was found to interact with the amino acid residues like Pro266, Ile312, Lys449, Gln447 and Arg490 by forming nine hydrogen bonds. The major portion of the peptide was found encased in the hydrophobic crevice formed by the residues like Trp531, Ala532, Ile312, Tyr313, Pro266, Ala267 and Phe275 contributing strong hydrophobic interactions with the peptide. The α-amino groups of Leu1 and Thr2 formed H-bonds with the α-keto moiety of Gln447. The side chain hydroxyl group of Thr2 interacted with the keto moiety of Gln447. The Phe3 formed a strong π-cationic interaction with Lys449. The amide groups of Gln4 and Asn6 showed H-bond interactions with ammonium and α-amino moieties of Lys449 and Ile312 respectively. The α-keto moiety of His5 formed a strong interaction with amino moiety of guanidine group of Arg490.
The shape and size of Furin binding pockets are known to be large having canyon-like crevice. The active site is made of key amino acid residues including Met189, Asp191, Asn192, Arg193, Glu229, Val231, Asp233, Asp259, Lys261, Arg298, Trp328, and Gln346. Many small molecule inhibitors to the active site have been explored but their ability to completely block the furin activity is yet to be established. Interestingly milk peptides were also found to be well-positioned to interact with the spike protein binding site. The peptide occupied the space to the right of the catalytic triad and extended more to the adjacent pocket and covered the surface area for Furin substrate binding site. The interactions were similar to previously identified peptide like inhibitors m-guanidinomethyl-Phac-RVR-Amba, H-Lys-Arg-Arg-Tle-Lys-4-Amba, c[glutaryl-BVK-Lys-Arg-Arg-Tle-Lys]-4-Amba, and p-guanidinomethyl-Phac-R-Tle-R-Amba [25].
Molecular Dynamics
We have used the all-atom MD technique to explore the affinity profile of the most potent peptide inhibitors of ACE2 and Spike proteins. After the simulation MD trajectories were employed to investigate various criteria such as relative mean square deviation (RMSD), H-bonds interaction profile, potential energy, gyration, and RMS fluctuation analysis.
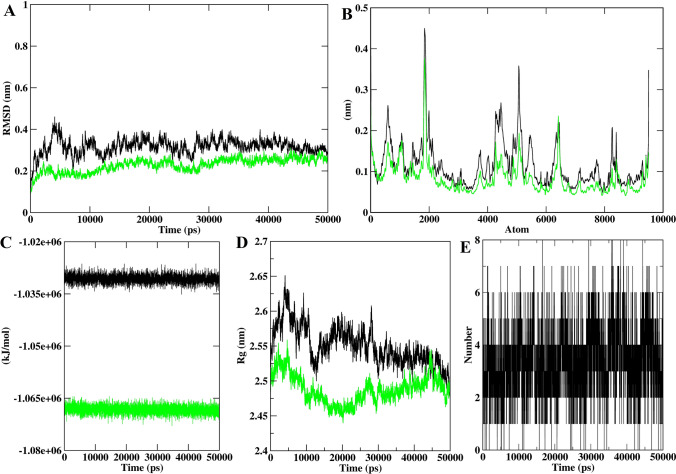
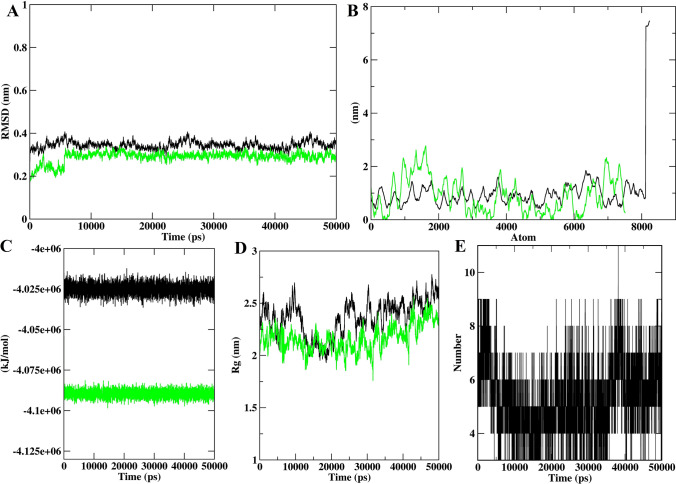
The overall stability of the system during the simulation was evaluated by computing RMSD of the backbone for both protein and protein–peptide complex. The RMSDs till the entire simulation were less than 0.5 Å indicating the stability of ACE2 and Spike protein–peptide systems. The average RMSD of the protein backbone for the ACE2 and ACE2-GWLEPLL complex was found to be 0.32 and 0.23 nm respectively (Fig. 2a). It was also observed that the ACE2-GWLEPLL complex was more stable than the unbound protein, as it showed greater structural perturbations. Similar results were also observed for of Spike-IQKVAGTW complex as well as the average RMSD for protein and complex was 0.345 and 0.289 nm respectively (Fig. 3a). However, both the Spike and Spike-IQKVAGTW complex remained smooth and undisturbed throughout the simulation. To assess the flexibility of the proteins, we computed the RMSF value for both protein and protein-peptide during the simulation. The RMSF range of the ACE2 structure was observed to be between 0.12 and 0.084 nm (Fig. 2b). The average RMSF values for Spike protein and Spike-IQKVAGTW complex were 1.01 and 0.9 nm respectively (Fig. 3b). These observations explicitly reveal that the predicted inhibitor has a close interaction with the binding site of ACE2, thus lowering the fluctuation of the ACE2 and Spike proteins.
Fig. 2.
The results of molecular dynamics simulation, a the backbone RMSD of ACE2 and ACE2-GWELPL for 50 ns MD simulation, b the backbone RMS fluctuation of ACE2, which clearly indicates unbound protein, fluctuates more, c graph depicting the perturbation in potential energy of ACE2 and ACE2-GWELPL, d evaluation of radius of gyration for ACE2 and ACE2-GWELPL and e enumeration of stable H-bond interactions during 50 ns run
Fig. 3.
The results of molecular dynamics simulation, a the backbone RMSD of Spike and IQKVAGTW-Spike for 50 ns MD simulation, b the backbone RMS fluctuation of Spike, which clearly indicates unbound protein, fluctuates more, c graph depicting the perturbation in potential energy of Spike and Spike-IQKVAGTW, d evaluation of radius of gyration for Spike and Spike-IQKVAGTW and e enumeration of stable H-bond interactions during 50 ns run
The alteration in the potential energies brought by the interacting peptides with protein residues during MD simulation was calculated by employing the gmx_energy tool of the Gromacs. The average potential energies of ACE2 and ACE2-GWLEPLL complex were − 1.03e+06 and − 1.1e+06 kJ/mol respectively (Fig. 2c). Similarly, Spike and Spike-IQKVAGTW showed an average of − 4.03e+06 and − 4.1e+06 kJ/mol (Fig. 3c). The total energy values of all the systems were stable around a constant value, which indicated that all the bound systems evolved with constant energies. The number of H-bonds between each peptide and proteins were analyzed using 50 ns simulation trajectories as shown in Figs. 2e and 3e. The ACE2-GWLEPLL complex exhibited an average of five H-bonds throughout the simulation. A similar trend was also observed between IQKVAGTW and the Spike protein. The average H-bond interactions observed during MD studies were between 5 and 8. By analyzing the H-bond profile between the peptides and protein we can deduce that the H-bond interactions were crucial for the efficient binding of ligands to the receptors and stability of the protein complex.
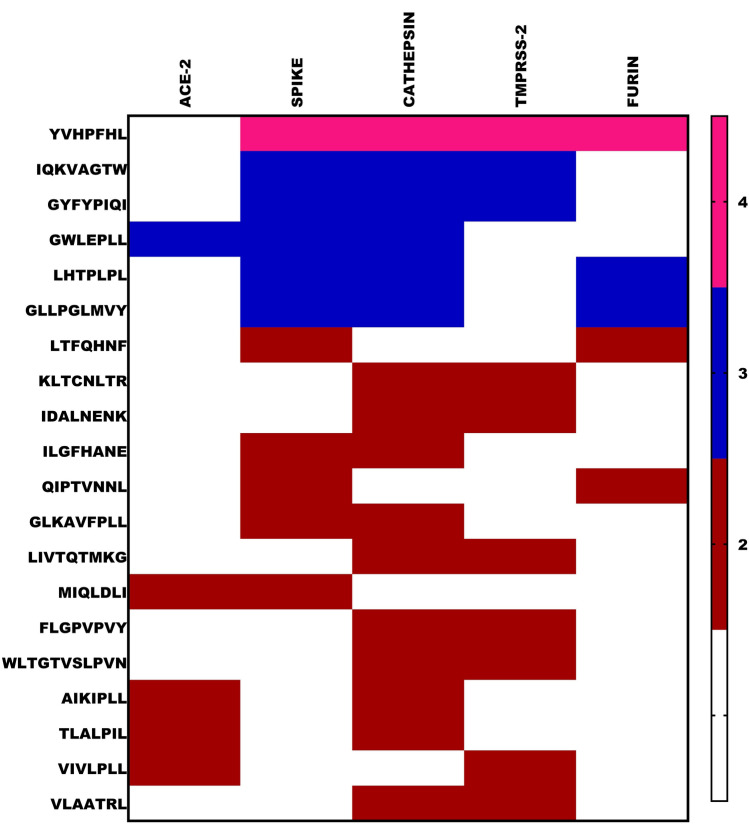
To explore the prospects of milk peptides interacting with multiple targets at the entry point; a heat map analysis was executed (Fig. 4). We observed GWELPLL and MIQLDLI be effective candidates for both ACE2 and the Spike protein. Among all the peptides, YVHPFHL was observed to be a key inhibitory molecule targeting Spike and all the proteases. Further, the peptides IQKVAGTW, GYFYPIQI, LHTPLPL, GLLPGLMVY and GWELPLL were able to interact with at least two proteases as well with the Spike protein, while, 14 different peptide sequences found to harbor interaction to at least two targets.
Fig. 4.
Heat map analysis of Milk peptides displaying greater affinity against ACE-2, Spike, TMPRSS2, Cathepsin-L and Furin. In the gradient ruler, light green color indicated peptide bound to five targets, pink color indicates four, blue color indicates three and brown indicated two targets (Color figure online)
Targeting Endosomal Maturation (AAK1, GAK, PIKfyve and TPC2)
AAK1
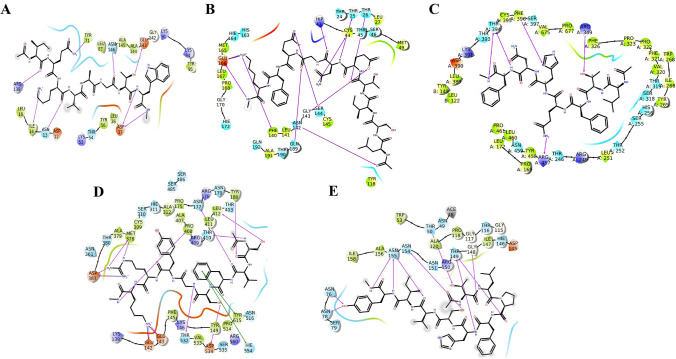
The adaptor associated kinase 1 (AAK1) is the first protein involved in endosome formation thereby it regulates intracellular viral trafficking during entry, assembly and release. AAK1 plays a key role in receptor-mediated endocytosis by specific phosphorylation of adaptor protein 2, which stimulates the binding to cargo proteins [26]. Hence, inhibiting this serine-threonine kinase can disrupt the intracellular viral trafficking and processing. Among the milk peptides screened for the target, CLANGMIMY showed a maximum affinity with a − 12.6 dockscore (Fig. 5a; Supplementary File 6). CLANGMIMY was able to construct a network of strong H-bonds (8 H-bonds) with several amino acid residues inside the enzyme’s active site (e.g. Glu54, Gly55, Lys178, and Asp194) which are found to be crucial for AAK1 activity (Supplementary 16A) [27]. Additionally, Lys219 was able to form two H-bonds with Cys1 of the peptide at dual positions (Carboxyl and Sulfhydryl groups) making it more strong interaction. The lead peptide inhibited the active site by H-bond interactions via Tyr9/Glu54, Ala3/Gly55, and Tyr9/Val51. Among them Asn4 was prominent with triple network of H hydrogen bond targeting the catalytic dyad Asp194 and Lys178. In addition Tyr9 and Cys1 formed H-bonds with Lys219 and Val51 covering the binding pocket from both the terminals.
Fig. 5.
Interactions established after docking, a CLANGMIMY-AAK1, b IQKVAGTW-GAK, c IQKVAGTW-PIKfyve and d LDAQSAPLRV-TPC2. Peptides are in green, while residues of binding sites are represented in three coded format (various color) and thick colored lines encasing peptides represent the active protein site pockets. Solid pink lines depict H-bonds, while hydrophobic interactions are represented in green. Additionally, salt bridges and π-cation stacking, are represented by gradient color (pink and blue lines) and a red line respectively (Color figure online)
GAK
Cyclin G-associated kinase (GAK) mediates the binding of clathrin to the plasma membrane and the trans-Golgi network. It is one of the most important events during endosome formation and maturation for the establishment of virus entry into the cell [28]. Among the docked peptides, IQKVAGTW showed maximum interaction (docking score − 10.5) forming eight hydrogen bonds, one salt bridge and two π–π interactions with Phe residues of the target (Supplementary File 7). In the peptide Trp8 was the strongest moiety forming quadruple interactions—two π stacking with Phe51 and two hydrogen bonds with Glu85 and Lys69. The Gln2 being neutral amino acid interacted in a dual fashion. It formed four hydrogen bonds with residues in the active site viz., Cys126, Gln129 and Leu46 (Fig. 5b) (Supplementary 16B). Secondly, it formed a hydrophobic mesh along with Ile1 and trapped the hydrophobic core of the kinase domain. In addition, Lys3 electrostatically interacted with Glu132 and strongly inhibited the kinase domain through an additional hydrogen bond.
Phosphatidylinositol 3 Phosphate 5 Kinase (PIKfyve)
During endocytosis, one of the key molecules that regulate the dynamic process of endosome maturation is PIKfyve [29]. In addition, it plays a role in several trafficking events associated with the endocytic pathway. In PIKfyve active site resides in Zinc finger domain [30, 31]. Twelve peptides showed maximum interactions and among them, IQKVAGTW showed the most intimacy with a docking score of − 10.13 (Fig. 5c; Supplementary File 8).The zinc finger (158–218) was trapped with the peptide by Gln2 and Lys3 residues via salt bridge formation while, Trp8 formed additional triple non-covalent interactions with the domain ensuring complete inhibition of the target. Among the observed interactions, four were hydrogen bonds, one π stacking and one salt bridge. The peptide residues that were involved in the interactions were Gln2, Lys3, and Try8 displaying binding affinity with the target residues Glu135, Phe141, Asp158 and Asp218 (Supplementary 16C).
Two Pore Channel (TPC2)
Two Pore Channels (TPCs) are intracellular calcium/cation channels located in the membranes of host endolysosomal compartments. Most viruses including SARS-CoV2 use them for compartmentalisation and replication. The two human isoforms of the channel, TPC1 and TPC2, are distinctly present within the endolysosomal system with TPC2 preferentially localizing to late endosomes and lysosomes [32]. Thus TPC2 emerges as a druggable host target for SARS-CoV2 infection. Several peptides showed maximum interaction with the target active pocket. The most common residues interacting with the protein were Gln, Ser, Leu, Arg and Val. The peptide which showed maximum interaction was LDAQSAPLRV with a docking score of -12 (Supplementary File 9). The peptide selectively inhibited domains 2 and 3 of the protein [33]. Ser5 and Asp2 were the strongest interacting residues forming six H-bonds with Gln58/Arg60/Gln83 and Lys203/Lys143/Gln197 respectively. In addition,Val10, Leu8 and Gln4 were found to further strengthen the interaction by forming H-bonds with Gln370, Gln58 and Tyr77 residues of domains 2 and 3 (Fig. 5d) (Supplementary 16D). The residues found in the binding core were Gln58, Arg60, Tyr77, Gln83, Lys143, Lys203 and Asp404. Potentials of milk peptides as multi-directed candidates with target specificity with diverse targets of endosomal membrane trafficking was examined employing a heat map study. Based on the heat map (Fig. 6) it can be deduced that LTFQHNF, IQKVAGTW, HMWPGDIK and KLTCNLTR exhibited strong interactions with all the kinases and TPC2. Further, the analysis showed that the peptides reflected interactions in a diverse pattern with two or more targets.
Fig. 6.
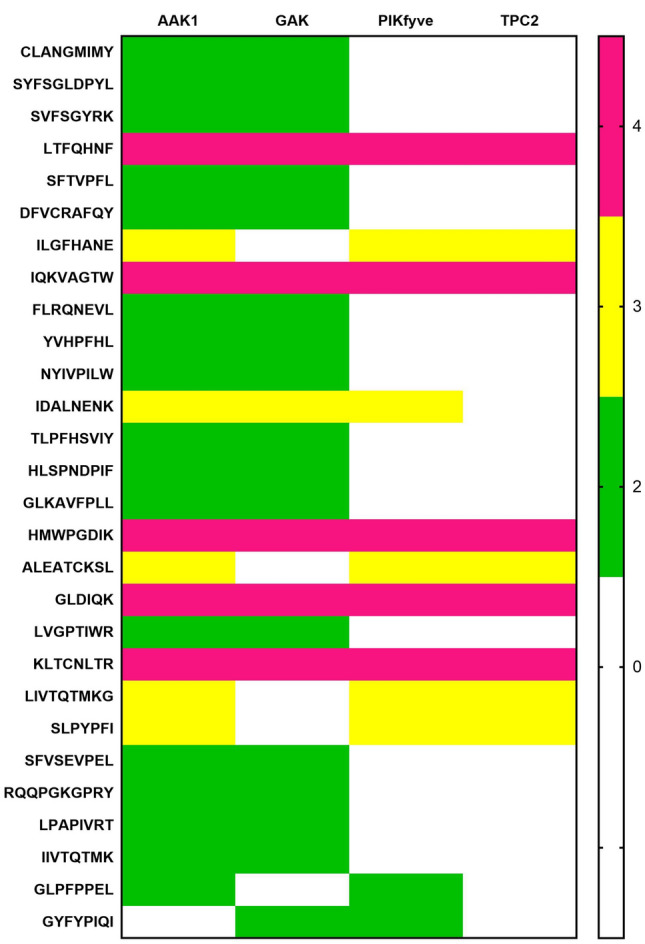
Heat map analysis of Milk peptides displaying greater affinity against AAK, GAK, PIKFyee and TPC2. In the gradient ruler, pink color indicates four, yellow color indicates three targets and green indicates two targets (Color figure online)
Targeting Replication Transcription Complex (PLpro, clpro, Nsp12 and Nsp13) and Virion Assembly (N Protein)
Papain Like Protease (PLpro)
Papain like protease participates in the cleavage of polyproteins produced by translation of viral ORF to produce proteins involved in Replication transcription Complex, RTC [34]. Thus it is considered as one of the key proteins that regulate viral propagation. Six milk peptides were found have maximum interaction, among them, IQKVAGTW had maximum interaction with a docking score of − 9.26 (Supplementary File 10). It was bound to the target through nine hydrogen bonds and the interacting residues at the binding site. The catalytic domain was seized at Asp37 and Glu143 via hydrogen bonds from Trp8 and Thr7. Gly6 and Ile1 act as hydrogen acceptors and interacted with binding site at Asn146 and Arg138 respectively. The interaction was further strengthened by hydrogen bonds viz. Lys3/Asn13, Lys3/ Tyr71 and Ile4/Arg138 (Fig. 7a) (Supplementary 17A).
Fig. 7.
Interactions established after docking, a IQKVAGTW-PLpro, b LSITENGEFK-3CLpro, c LTFQHNF-NSP12, d SVFSGYRK-NSP13 and e TLPFHSVIY-N-Protein. Peptides are in green, while residues of binding sites are represented in three coded format (various color) and thick colored lines encasing peptides represent the active protein site pockets. Solid pink lines depict H-bonds, while hydrophobic interactions are represented in green. Additionally, salt bridges and π-cation stacking, are represented by gradient color (pink and blue lines) and a red line respectively (Color figure online)
3 C-Like Protease (CLpro)
SARS-CoV2 3CLpro is a cysteine protease that possesses two N-terminal domains comprising of two β-barrel chymotrypsin-like folds. The 3CLpro active site spans a fork within the two domains and is categorized by a Cys-His dyad. The protease activity of 3CLpro is crucial for the replication of SARS-CoV2 and it is situated at the 3′ end which exhibits excessive variability [35]. The structure of 3CLpro protein comprises of two catalytic domains, I (R8-101) and II (R102-184) and an III domain (R201-303) essential for protein’s dimerization into an active protease. The catalytic site of 3CLpro is unique in the chymotrypsin-like protein family as it has Cys145 and His41 dyad instead of a canonical Ser(Cys)-His-Asp(Glu) triad. The key residues that lines the active site in addition to Cys145···His41dyad are His163/His172/Glu166. This substrate binding site can fit four substrate residues in positions P1’ through P4, and it is flanked by residues from both domains I and II.
Among the peptides screened, LSITENGEFK showed the lowest binding energy (− 12.247 glide score) and energy potential (− 180.117) for the 3CLpro protein (Supplementary File 11). The peptide was found to interact with the amino acid residues like Thr26, Thr25, His41, Cys44, Phe140, Gly143, Ser144, Glu166 and Gln189 of the protein by forming 10 hydrogen bonds. The α-amino group moiety of Thr4 (P) exhibited H-bond with α-Keto group of Thr26. Similar interactions are seen between Asn6-Cys44. Further, α-amino group of Asn6 demonstrated strong interaction with β-hydroxyl of Thr25 and α-Keto group of the Asn6 formed an H-bond with the imidazole moiety of His41. The carboxyl group of Glu8 showed a better binding affinity towards β-hydroxyl of Ser144. The butyl ammonium side chain of Lys10 showed H-bond interactions with α-Keto of Phe140. Further, the butyl ammonium side chain of Lys10 displayed an H-bond and a salt-bridge interaction with the carboxyl group of Glu166 (Fig. 7b) (Supplementary 17B). In addition, the residues of catalytic dyad were found to interact with the peptide (hydrogen bond with His41 and hydrophobic interaction with Cys145).
Nsp12 (RNA Polymerase)
The replication of ∼ 30-kb SARS-CoV RNA genome is steered by RNA-dependent RNA polymerase (RdRp) domain present in the non-structural protein 12 (Nsp12), one of the key components of viral replicase. The Nsp12 is generally synthesized as a precursor protein, its proteolytic cleavage and conjugation of various NSPs (especially Nsp7 and Nsp8) make polymerase complex. Both Nsp7 and Nsp8 subunits are essential for the proper functioning of the Nsp12 thus Nsp12-Nsp7-Nsp8 subcomplex is regarded as the crucial core unit for mediating coronavirus replication [36].
Architecture of polymerase domain is conserved in viral polymerase family and it contains three subdomains; a finger (Leu366-Ala581 and Lys621-Gly679), a palm (Tyr582-Pro620 and Tyr680-Gln815) and a thumb sub-domain (His816-Glu920). The active site is established in the palm domain by conserved polymerase motifs (A-G). Though the identified peptides missed the active site of the RDRP, we observed that they interacted with a unique allosteric site within the palm domain. This allosteric site was previously shown to be a binding site for many potential inhibitors of RdRp [37, 38]. The docking results showed that LTFQHNF with pose 1 was the most active conformational position and predicts the best energy values (− 134.01) and docking score of − 10.7 (Supplementary File 12) as compared to other docking complexes. The peptide was completely encased in the crevice of the binding site confirming the best-fit pose of the peptide. Further analysis of structural interactions showed that LTFQHNF forms four hydrogen bonds at specific residues (Arg349, Arg457, Phe396 and Thr394) of the target protein. The hydroxyl group of Thr2 interacts with guanidine amine moiety of Arg349. The nitrogen of the amide group in the Gln4 interacted strongly with the α-keto group of the Arg457. The Nitrogen in the imidazole moiety of His5 formed H-bond with an amino group of Phe396. Finally, the α-keto moiety of Phe7 residue interacted with the α-amino group of Thr394 (Fig. 7c) (Supplementary 17C).
Nsp13 (Helicase)
Helicase (nsp13) NTPase/helicase (nsp13) is a critical protein in the replication transcription complex of the virus that catalyzes the separation of duplex oligonucleotides. Due to the fact that this protein is essential for viral RNA synthesis and one of the most conserved proteins among Nidovirales, it is considered as an interesting target for drug development and several chemical inhibitors have been reported [39]. Several peptides interacted maximally and SVFSGYRK was found to be the most prospective candidate molecule (− 12.972; Supplementary File 12). 17 interactions were prevalent with Nsp13 and among them, 10 were H-bonds, three salt bridges and 2 π stacking interactions making the peptide interact intimately with the target (Fig. 7d) (Supplementary 17D). DNA2 and UvrD c of the enzyme domain were the most constrained targets. Lys8 was the strongest interacting residue forming two salt bridges (Glu142, Glu143) and three hydrogen bonds (Pro408, Glu143 and Glu142). Arg7 formed a salt bridge (Asp383) and three hydrogen bonds (two with Met378 and one with Asp383). Ser1 donated a hydrogen (Tyr180) and accepted two hydrogens (Leu412 and Thr410) further strengthened the interaction. In addition, Phe3 and Gly5 formed a hydrophobic mesh and trapped Cys309, Met378 and Ala379 residues of the target.
N Protein
The N protein is the only structural protein that directly links up with the replicase transcriptase complexes, RTCs [40, 41]. This protein binds to the viral RNA genome, which is essential for incorporating the genetic material into viral particles [42, 43]. Its function involves entering the host cell, binding to the viral RNA genome and forming the ribonucleoprotein core [44]. The major feature of this protein is RNA binding domain. More than fifteen peptides bound to the RBD with high affinity (Supplementary File 14). TLPFHSVIY showed a maximum affinity for the target protein with a docking score of − 11.48 accounting for ten hydrogen bonds. The peptide constrains the entire functional domain of the protein. Tyr9 being the prime residue forms three hydrogen bonds with the target (Asn76, Asn78 and Asn155). Thr1 further strengthened the interaction forming two hydrogen bonds (Thr50, Gly117). Other members of the inhibitory team formed three hydrogen bonds Ser6/Asn154, Pro3/Thr149 and His5/Thr149. In addition, Leu2 of the peptide formed a hydrophobic hinge with the Ile147.
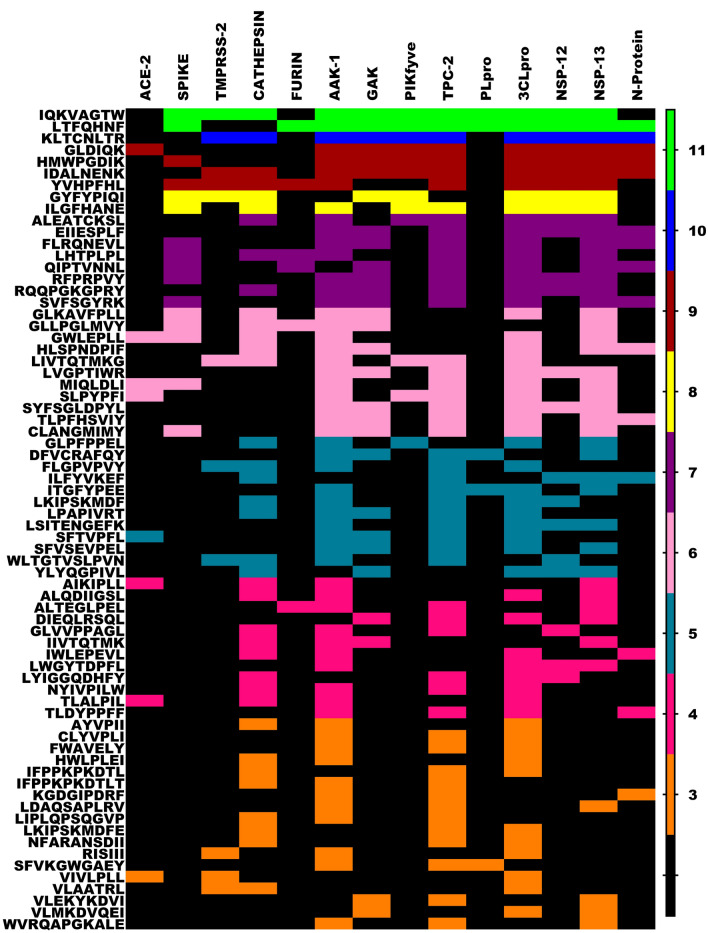
Meticulous assessment of the interaction patterns between the milk peptides and the proteins involved in the replication was evaluated. Several peptides inhibited either PLpro/CLpro or both viral proteases and RTC complex proteins. Incidentally, the novel sequence LTFQHNF displayed effective binding towards all target proteins analyzed in this complex (Fig. 8). The overall heat map analysis of all the targets and their corresponding peptides is given in Fig. 9. As we can observe IQKVAGTW, LTFQHNF and KLTCNLTR were shown to inhibit most of the target proteins considered in this study spanning the entire SARS-CoV2 lifecycle. Similarly, four peptides displayed interactive affinity towards more than nine targets, of which three of the peptides were found to have better interaction for targets involved in endocytic trafficking and replication processes and the majority of entry point targets of Coronavirus. Further, 2, 7, 11, 12, 11 and 18 peptides displayed affirmative interactions with 8, 7, 6, 5, 4 and 3 targets respectively.
Fig. 8.
Heat map analysis of Milk peptides displaying affinity for PLpro, 3CLpro, NSP-12, NSP-13 and N-Protein. In the gradient ruler, light green color indicated peptide bound to five targets, pink color indicates four, blue color indicates three targets and brown indicated two targets (Color figure online)
Fig. 9.
Overall heat map analysis of Milk peptides. The gradient ruler indicates the number of targets interacted by individual peptides: green 11, blue 10, brown 9, yellow 8, violet 7, pink 6, light blue 5, dark pink 4, orange 3 (Color figure online)
Discussion
Targeting Entry Points of Coronavirus
SARS-CoV2 entry into host cells is mediated by its transmembrane Spike (S) glycoprotein that forms homotrimers protruding from the viral surface. Viral entry relies on the specific interaction between the RBD of the viral Spike protein and hosts protein ACE2 on the cell membrane. SARS-CoV2 cannot infect the cells without ACE2, confirming that ACE2 is the cell receptor for SARS-CoV2. The mechanism of binding involves clinging of the extended RBM in the Spike-RBD to the bottom side of the small lobe of ACE2 at the N-terminal domain. A concave crevice is formed on the surface of the RBM that cradles the N-terminal helix of the ACE2. Thus in SARS-CoV2 entry, ACE2 largely functions as recognition and anchoring unit rather than a catalytic enzyme. Once bound, the Spike glycoprotein undergoes a cascade of conformational alterations leading to the fusion of the SARS-CoV2 and the host cell membrane, internalization and subsequent release of viral RNA into the cytoplasm, establishing infection [45]. The whole of ACE2 is internalized together with SARS-CoV2 and it is important to note that protease activity of ACE2 remains intact and doesn’t have any prominent role in the process of viral entry. Hence, targeting the ACE2 binding domain is a pharmacologically better strategy instead of the peptidase domain of the ACE2 [46]. We hypothesize that two-way blocking of the binding mechanism of ACE2 and Spike could be a realistic strategy to prevent the spreading of infection. Herein we targeted binding interphase of both the ACE2 and spike proteins as our multitargeted strategy. Further, we analyzed the binding mechanisms of GWELPLL and IQKVAGTW peptides with the best docking potentials for ACE2 and Spike by employing MD simulation probing. As observed by the MD results we can deduce that both the peptides displayed good interactions and stabilizing properties for both ACE2 and Spike targets. A recent report also discovers a long peptide CP-1 directed to the spike protein blocking the virus-cell fusion process [26].
Another part of the potential therapeutic strategy is targeting the host proteolytic machinery responsible for the activation, priming and substitutions of residues at the S1/S2 or S2 cleavage sites of SARS-CoV2. After the binding of Spike to the ACE2, S1 subunit of Spike protein is cleaved by two important plasma membrane-bound serine proteases viz. TMPRSS2 and Cathepsin-L acting in synergy encouraging large-scale conformational changes in S protein thereby affirming the SARS-CoV2 entry. TMPRSS2 is probably the first protease that acts upon viral proteins as it is a membrane protein and represents a major therapeutic target of host protease machinery. However, its protease activity is principally required until viral endocytosis occurs as it loses its activity after viral internalization with the change in pH [47]. As SARS-CoV2 reaches the intracellular cytosome, the Cathepsin-L is the major protease that brings about the activation of the viral S1 subunit as this cysteinyl protease is active under acidic pH. Further, the SARS-CoV2 requires an acidic pH for its complete activation thus Cathepsin-L represents a key target of pharmacological importance. In addition, Cathepsin-L fusogenetically induces the S protein thereby initiating the fusion of viral and endosomal membranes. Thus serine-protease TMPRSS2 functions as a primary inducer at the host cell membrane and perhaps during endocytosis of the virus and the Cathepsin-L continues S1 subunit degradation in the acidic endosome and in the lysosomal compartments [23]. Another protein that is gaining importance as a crucial host protease is Furin, which mediates cleavage of the Spike at the S1/S2 cleavage site. The S1/S2 junction has a polybasic line of an RRAR motif, matching a substrate sequence for Furin. Further, the activation of Spike protein by Furin has been accepted as a crucial event for the viral fusion and pathogenesis in various pathogenic viruses including SARS-CoV2. Thus Furin represents an appealing therapeutic target for several viral diseases including COVID-19 [48].
Hence, we hypothesize combinatorial mitigation by targeting initial entry points like ACE2, Spike, TMPRSS2, Cathepsin-L and Furin by peptide inhibitors which can significantly cut short the coronavirus infection (Supplementary 15). In the quest of identifying multitargeted peptides, we observed GWELPLL and MIQLDLI to be effective candidates for both ACE2 and the Spike protein. Similarly, several milk peptides were found to interact with different protease targets as well with the Spike protein. Among them, YVHPFHL was found to be a key inhibitory molecule targeting Spike and all the proteases and many such multi-target interactive peptides were found prevalent in the present study. Incidentally, YVHPFHL was also shown to possess antioxidant activity during our previous study [49]. Interestingly, IQKVAGTW, identified as a promising inhibitor of ACE1 was found to possess inhibitory effect to majority of the proteases and the Spike protein demands a detailed investigation to screen whether ACE1 inhibitors can be exploited for targeting host proteases and Spike protein during SARS-CoV2 infection [7]. The data thus supports the role of food-based small molecular weight peptides as potential therapeutic candidates in multi-directed ligand approach to block the entry of SARS-CoV2.
Targeting Endosomal Maturation
Endocytic membrane traffic is an extremely dynamic system and responsible for the intracellular transportation of macromolecules. The cargo molecules designated for transportation to the specific sites are amassed into endocytic vesicles and carried to their target sites by clathrin-mediated trafficking. This mechanism is orchestrated by the AP (adaptor protein) complexes which bind to sorting motifs of the cargo molecule, enlist clathrin and other mediatory proteins, and then focus the load into vesicular transporters, which carry from the donor membrane to the target organelle membrane. This endocytic membrane traffic system is maneuvered by two host kinases namely AAK1 and GAK which control both clathrin-mediated and trans-Golgi network (TGN) transport [50]. Precisely, AAK1 and GAK catalyze phosphate-priming of APs at micro subunits thus increasing their binding affinity for sorting motifs within the cargo. Recently SARS-CoV2 has been shown to seizure this mechanism thereby ensuring safe transport of its genetic material into the host biosynthetic machinery [50].
Another important protein that is involved in inducing endocytic membrane traffic thereby increasing SARS-CoV2 pathogenicity and viral replication is TPC2. The TPCs amplify the membrane polarization (cationic) of endolysosomes, hence play a commanding role in membrane fusion mechanisms. Mounting in vitro and in vivo evidences indicate that TPC2 inhibitors can extenuate the endocytic trafficking and replication of MERS-CoV. Thus, inhibition of the TPC2 channel could be effective in the mitigation of viral replication in the case of SARS-CoV2 as well [51].
The PIKfvye is the only known enzyme that converts phosphatidyl inositol-3-P to phosphatidylinositol 3,5-bisphosphate that plays a crucial role in the maturation of primary into late phagosomes or lysosomes. The replication of SARS-CoV2 also depends on the functioning of PIKfvye for the transfer of viral cargo to the site of replication [52]. Hence, inhibition of PIKfyve leads to the decline in the cellular phosphatidylinositol 3,5-bisphosphate blocking the phagosomal maturation and unloading of virion into the cytosol. Interestingly, several peptides were found to be strong contenders interacting with both the kinases and TPC2, suggesting these peptides as suitable candidates to prevent endosomal trafficking. Although there are quite a few drugs available in the market for these targets individually, we presume that the development of milk peptides with multitargeted potentials can be an alternative therapeutic strategy.
Targeting Replication Transcription Complex and Virion Assembly
After the SARS-CoV2 genome (ssRNA) is held mainly by two non-structural proteins ORF1a and ORF1ab followed by structural proteins. The ORF1a results in the synthesis of two viral proteinases 3CLpro and PLpro while, an elongated polyprotein is encoded by ORF1ab which comprises of several non-structural proteins (Nsps). The major function of PLpro is to cleave at three sites helping in the separation of the nascent polyprotein into its distinct proteins. But, the key protease activity is carried by 3CLpro or Main-protease (Mpro) as it cleaves at eleven sites processing the polyprotein (both 1a and 1b) into 16 Nsps. In addition, proteases assist coronavirus in dodging host innate immune machinery by dismantling ubiquitin and interferon-stimulated gene 15 cofactors from nascent host proteins [53]. Hence, these proteases are appealing targets as they are indispensable not only for viral replication but also to evade the host immune system.
Coronavirus RNA replication is accomplished by RTC, comprising various essential proteins including RdRp, Nsp12 and helicase. It also consists of Nsps with diverse roles such as RNA modification (Nsp14, Nsp16), RNA proofreading (Nsp14), and endoribonuclease activity (Nsp15). In this study, we mainly focused on Nsp12 and 13, as they are the key functional units of RTC and have been principal targets of interest in diverse viral infections viz. hepatitis C, Zika and MERS. They represent highly conserved non-structural proteins amongst coronaviruses having no homology to human proteins with correspondingly minimal cytotoxicity [54]. Another protein involved in the replication process which is gaining major attention is nucleocapsid (N) protein. N-proteins have two forms phosphorylated and non-phosphorylated which are crucial for its functions. The non-phosphorylated form creates nucleocapsid compaction of the RNA genome in the virion. The N protein gets immediately phosphorylated after synthesis and quickly coalesces with RTC (might represent a functional unit of RTC) and modulate subgenomic transcription in the infected cells. Incidentally, the present study unravels LTFQHNF as the most suitable therapeutic candidate addressing all the targets. Although several inhibitors have been identified for many of these proteins, a single molecule offering selective inhibition to all these targets is conspicuously lacking in the current literature. The study thus explores milk peptides as prospective small molecular inhibitory agents against a multitude of targets. Further, our earlier studies indicate IQKVAGTW to possess ACE1 inhibitory [7] and LTFQHNF with thrombolytic activities (unpublished data).
Mounting health-cognizant consumers across the globe coupled with the growing awareness regarding the health benefits of milk proteins and peptides having numerous physiological functionalities have projected these bioactive molecules to be used in various commercial preparations. Milk or early milk bioactive peptides in addition to their nutritional value are also bestowed with several biological properties and have therapeutic effects in health and diseases. Currently, there are more than 400 peptide drugs under global clinical trials, while over 60 already approved for clinical use in the United States, Europe and Japan and as predicted by Coherent Market Insights, the global bioactive protein and peptides market is expected to reach US$ 88.3 Bn by the end of 2027. However, the biggest impediment is to avoid its degradation by digestive enzymes if taken orally and to this end, recently researchers have succeeded in developing target-specific peptide action which can resist gastrointestinal degradation, a step closer to develop oral peptide drugs [55]. Hence, the present study certainly sets a platform to promote virus-based and host-based targeted peptide-therapeutics with novel sequences of milk proteins with a more selective and effective antiviral profile in addition to the recent reports on two milk peptides for possible in vivo and in vitro validations.
Conclusions
In summary, the concurrent blockade of diverse pathways by peptides in the infectious cycle of SARS-CoV2 represents a pharmacologically promising strategy to mitigate COVID-19 menace. Hence, in the current study, we have focused on all the three stages of SARS-CoV2 infection: (i) a combination of multiple selective peptides directed towards targets involved at entry point of Coronavirus viz. ACE2, Spike and proteases, (ii) concomitant obstruction of diverse Kinases/proteins associated in the endosomal maturation, and (iii) a multidirectional inhibition of proteins forming RTC and Virion assembly. Thus, by coalescing the molecular docking, molecular simulation, heat mapping and manual interpretation, we have developed a new strategy to identify plethora of peptides that can block the spread of Coronavirus. Further, we have delineated the molecular interactions essential for binding and selectivity of peptides to the binding sites of the targets effectively. The current study is a promising effort to investigate the potentials of milk peptides as multitargeted drug candidates. However, the multitargeted nature of milk peptides are yet to be explored completely to realize their comprehensive benefit which might be accomplished by employing the advanced high-throughput in vitro binding and activity assays in future.
Supplementary Information
Below is the link to the electronic supplementary material.
Acknowledgements
Dr. Pradeep conveys sincere gratitude to the University Grants Commission (UGC) New Delhi, India for the financial support (UGC/DSKPDF Grant Order No. F.4-2/2006 (BSR)/BL/17-18/0563.
Abbreviation
- 3CLpro
3 C-like protease
- AAK1
Adaptor associated kinase 1
- ACE2
Angiotensin-converting enzyme 2
- GAK
Cyclin G-associated kinase
- HCoVs
Human coronaviruses
- Nsp12
RNA polymerase
- Nsp13
Helicase
- N-Protein
Nucleocapsid (N) protein
- PLpro
Papain like protease
- PIKfyve
Phosphatidylinositol 3phosphate 5 kinase
- RBD
Receptor-binding domains
- RMSD
Relative mean square deviation
- RASÉ
Renin–angiotensin system
- RTC
Replication Transcription Complex
- S1-CTD
S1-C-terminal domain
- S1-NTD
S1-N-terminal domain
- SARS-CoV2
Severe acute respiratory syndrome coronavirus 2
- TGN
Trans-Golgi network
- TPC2
Two pore channel
- TMPRSS2
Type II transmembrane serine proteases
Data Availability
The manuscript has no associated data to disclose.
Declarations
Conflict of interest
The authors declare no conflict of interest, financial or otherwise.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Biesalski HK. Vitamin D deficiency and co-morbidities in COVID-19 patients—a fatal relationship? NFS J. 2020;20:10–21. doi: 10.1016/j.nfs.2020.06.001. [DOI] [Google Scholar]
- 2.Peiris JSM, Lai ST, Poon LLM, et al. Coronavirus as a possible cause of severe acute respiratory syndrome. Lancet. 2003;361:1319–1325. doi: 10.1016/S0140-6736(03)13077-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Liu J, Wang Z. Diverse array-designed modes of combination therapies in Fangjiomics. Acta Pharmacol Sin. 2015;36:680–688. doi: 10.1038/aps.2014.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nongonierma AB, FitzGerald RJ. Enhancing bioactive peptide release and identification using targeted enzymatic hydrolysis of milk proteins. Anal Bioanal Chem. 2018;410:3407–3423. doi: 10.1007/s00216-017-0793-9. [DOI] [PubMed] [Google Scholar]
- 5.Çakır B, Okuyan B, Şener G, Tunali-Akbay T. Investigation of beta-lactoglobulin derived bioactive peptides against SARS-CoV-2 (COVID-19): in silico analysis. Eur J Pharmacol. 2021 doi: 10.1016/j.ejphar.2020.173781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Elnagdy S, Alkhazindar M. The potential of antimicrobial peptides as an antiviral therapy against COVID-19. ACS Pharmacol Transl Sci. 2020;3(4):780–782. doi: 10.1021/acsptsci.0c00059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ashok NR, Aparna HS. Empirical and bioinformatic characterization of buffalo (Bubalus bubalis) colostrum whey peptides and their angiotensin I-converting enzyme inhibition. Food Chem. 2017;228:582–594. doi: 10.1016/j.foodchem.2017.02.032. [DOI] [PubMed] [Google Scholar]
- 8.Rohit AC, Sathisha K, Aparna HS. A variant peptide of buffalo colostrum β-lactoglobulin inhibits angiotensin I-converting enzyme activity. Eur J Med Chem. 2012;53:211–219. doi: 10.1016/j.ejmech.2012.03.057. [DOI] [PubMed] [Google Scholar]
- 9.Mamatha MB, Nishimura SI. Inhibitory potential of Buffalo (Bubalus bubalis) colostrum immunoglobulin G on Klebsiella pneumoniae. Int J Biol Macromol. 2016;88:138–145. doi: 10.1016/j.ijbiomac.2016.03.049. [DOI] [PubMed] [Google Scholar]
- 10.Brijesha N, Aparna HS. Comprehensive characterization of bioactive peptides from Buffalo (Bubalus bubalis) colostrum and milk fat globule membrane proteins. Food Res Int. 2017;97:95–103. doi: 10.1016/j.foodres.2017.03.037. [DOI] [PubMed] [Google Scholar]
- 11.Ashok A, Brijesha N, Aparna HS. Discovery, synthesis, and in vitro evaluation of a novel bioactive peptide for ACE and DPP-IV inhibitory activity. Eur J Med Chem. 2019;180:99–110. doi: 10.1016/j.ejmech.2019.07.009. [DOI] [PubMed] [Google Scholar]
- 12.Halgren TA. Identifying and characterizing binding sites and assessing druggability. J Chem Inf Model. 2009;49:377–389. doi: 10.1021/ci800324m. [DOI] [PubMed] [Google Scholar]
- 13.Harder E, Damm W, Maple J, et al. OPLS3: a force field providing broad coverage of drug-like small molecules and proteins. J Chem Theory Comput. 2016;12:281–296. doi: 10.1021/acs.jctc.5b00864. [DOI] [PubMed] [Google Scholar]
- 14.Alogheli H, Olanders G, Schaal W, et al. Docking of macrocycles: comparing rigid and flexible docking in Glide. J Chem Inf Model. 2017;57:190–202. doi: 10.1021/acs.jcim.6b00443. [DOI] [PubMed] [Google Scholar]
- 15.Van Der Spoel D, Lindahl E, Hess B, et al. GROMACS: fast, flexible, and free. J Comput Chem. 2005;26:1701–1718. doi: 10.1002/jcc.20291. [DOI] [PubMed] [Google Scholar]
- 16.Robertson MJ, Tirado-Rives J, Jorgensen WL. Improved peptide and protein torsional energetics with the OPLS-AA force field. J Chem Theory Comput. 2015;11:3499–3509. doi: 10.1021/acs.jctc.5b00356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bussi G, Donadio D, Parrinello M. Canonical sampling through velocity rescaling. J Chem Phys. 2007 doi: 10.1063/1.2408420. [DOI] [PubMed] [Google Scholar]
- 18.Wu J, Deng W, Li S, Yang X. Advances in research on ACE2 as a receptor for 2019-nCoV. Cell Mol Life Sci. 2020;78(2):531–544. doi: 10.1007/s00018-020-03611-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Clarke NE, Turner AJ. Angiotensin-converting enzyme 2: the first decade. Int J Hypertens. 2012;2012:307315. doi: 10.1155/2012/307315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Huang Y, Yang C, Xu X-F, et al. Structural and functional properties of SARS-CoV-2 spike protein: potential antivirus drug development for COVID-19. Acta Pharmacol Sin. 2020;41:1141–1149. doi: 10.1038/s41401-020-0485-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shekhar N, Sarma P, Prajapat M, et al. In silico structure-based repositioning of approved drugs for Spike glycoprotein S2 domain fusion peptide of SARS-CoV-2: rationale from molecular dynamics and binding free energy calculations. mSystems. 2020 doi: 10.1128/msystems.00382-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zipeto D, da Palmeira JF, Argañaraz GA, Argañaraz ER. ACE2/ADAM17/TMPRSS2 interplay may be the main risk factor for COVID-19. Front Immunol. 2020 doi: 10.3389/fimmu.2020.576745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu T, Luo S, Libby P, Shi GP. Cathepsin L-selective inhibitors: a potentially promising treatment for COVID-19 patients. Pharmacol Ther. 2020;213:107587. doi: 10.1016/j.pharmthera.2020.107587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Singal CMS, Jaiswal P, Seth P. SARS-CoV-2, more than a respiratory virus: its potential role in neuropathogenesis. ACS Chem Neurosci. 2020;11:1887–1899. doi: 10.1021/acschemneuro.0c00251. [DOI] [PubMed] [Google Scholar]
- 25.Vankadari N. Structure of furin protease binding to SARS-CoV-2 spike glycoprotein and implications for potential targets and virulence. J Phys Chem Lett. 2020 doi: 10.1021/acs.jpclett.0c01698. [DOI] [PubMed] [Google Scholar]
- 26.Gil C, Ginex T, Maestro I, et al. COVID-19: drug targets and potential treatments. J Med Chem. 2020 doi: 10.1021/acs.jmedchem.0c00606. [DOI] [PubMed] [Google Scholar]
- 27.Sayed AM, Khalaf AM, Abdelrahim MEA, Elgendy MO. Repurposing of some anti-infective drugs for COVID-19 treatment: a surveillance study supported by an in silico investigation. Int J Clin Pract. 2020 doi: 10.1111/ijcp.13877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chaikuad A, Keates T, Vincke C, et al. Structure of cyclin G-associated kinase (GAK) trapped in different conformations using nanobodies. Biochem J. 2014;459:59–69. doi: 10.1042/BJ20131399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.He K, Marsland R, Upadhyayula S, et al. Dynamics of phosphoinositide conversion in clathrin-mediated endocytic traffic. Nature. 2017;552:410–414. doi: 10.1038/nature25146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shisheva A, Sbrissa D, Ikonomov O. Cloning, characterization, and expression of a novel Zn2+-binding FYVE finger-containing phosphoinositide kinase in insulin-sensitive cells. Mol Cell Biol. 1999;19:623–634. doi: 10.1128/mcb.19.1.623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lees JA, Li PQ, Kumar N, et al. Insights into lysosomal PI(3, 5)P2 homeostasis from a structural-biochemical analysis of the PIKfyve lipid kinase complex. Mol Cell. 2020;80:736–743.e4. doi: 10.1016/j.molcel.2020.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Heister PM, Poston RN. Pharmacological hypothesis: TPC2 antagonist tetrandrine as a potential therapeutic agent for COVID-19. Pharmacol Res Perspect. 2020 doi: 10.1002/prp2.653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.She J, Zeng W, Guo J, et al. Structural mechanisms of phospholipid activation of the human TPC2 channel. eLife. 2019 doi: 10.7554/eLife.45222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mielech AM, Kilianski A, Baez-Santos YM, et al. MERS-CoV papain-like protease has deISGylating and deubiquitinating activities. Virology. 2014;450–451:64–70. doi: 10.1016/j.virol.2013.11.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chou CY, Chang HC, Hsu WC, et al. Quaternary structure of the severe acute respiratory syndrome (SARS) coronavirus main protease. Biochemistry. 2004;43:14958–14970. doi: 10.1021/bi0490237. [DOI] [PubMed] [Google Scholar]
- 36.Peng Q, Peng R, Yuan B, et al. Structural and biochemical characterization of the nsp12-nsp7-nsp8 core polymerase complex from SARS-CoV-2. Cell Rep. 2020 doi: 10.1016/j.celrep.2020.1077742020.107774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pokhrel R, Chapagain P, Siltberg-Liberles J. Potential RNA-dependent RNA polymerase inhibitors as prospective therapeutics against SARS-CoV-2. J Med Microbiol. 2020 doi: 10.1099/jmm.0.001203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Aftab SO, Ghouri MZ, Masood MU, et al. Analysis of SARS-CoV-2 RNA-dependent RNA polymerase as a potential therapeutic drug target using a computational approach. J Transl Med. 2020 doi: 10.1186/s12967-020-02439-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lehmann KC, Snijder EJ, Posthuma CC, Gorbalenya AE. What we know but do not understand about nidovirus helicases. Virus Res. 2015;202:12–32. doi: 10.1016/j.virusres.2014.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zaki AM, van Boheemen S, Bestebroer TM, et al. Isolation of a novel coronavirus from a man with pneumonia in Saudi Arabia. N Engl J Med. 2012;367:1814–1820. doi: 10.1056/nejmoa1211721. [DOI] [PubMed] [Google Scholar]
- 41.Mille JK, Whittaker GR. Host cell entry of Middle East respiratory syndrome coronavirus after two-step, furin-mediated activation of the spike protein. Proc Natl Acad Sci USA. 2014;111:15214–15219. doi: 10.1073/pnas.1407087111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bertram S, Dijkman R, Habjan M, et al. TMPRSS2 activates the human coronavirus 229E for cathepsin-independent host cell entry and is expressed in viral target cells in the respiratory epithelium. J Virol. 2013;87:6150–6160. doi: 10.1128/jvi.03372-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bertram S, Glowacka I, Muller MA, et al. Cleavage and activation of the severe acute respiratory syndrome coronavirus spike protein by human airway trypsin-like protease. J Virol. 2011;85:13363–13372. doi: 10.1128/jvi.05300-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Surjit M, Lal SK. The SARS-CoV nucleocapsid protein: a protein with multifarious activities. Infect Genet Evol. 2008;8:397–405. doi: 10.1016/j.meegid.2007.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ni W, Yang X, Yang D, et al. Role of angiotensin-converting enzyme 2 (ACE2) in COVID-19. Crit Care. 2020;24:422. doi: 10.1186/s13054-020-03120-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Liu M, Wang T, Zhou Y, et al. Potential role of ACE2 in coronavirus disease 2019 (COVID-19) prevention and management. J Transl Intern Med. 2020;8:9–19. doi: 10.2478/jtim-2020-0003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Seth S, Batra J, Srinivasan S. COVID-19: targeting proteases in viral invasion and host immune response. Front Mol Biosci. 2020 doi: 10.3389/fmolb.2020.002152020.00215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wu C, Zheng M, Yang Y, et al. Furin: a potential therapeutic target for COVID-19. iScience. 2020 doi: 10.1016/j.isci.2020.1016422020.101642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ashok NR, Vivek KH, Aparna HS. Antioxidative role of Buffalo (Bubalus bubalis) colostrum whey derived peptides during oxidative damage. Int J Pept Res Ther. 2019;25:1501–1508. doi: 10.1007/s10989-018-9795-5. [DOI] [Google Scholar]
- 50.Neveu G, Ziv-Av A, Barouch-Bentov R, et al. AP-2-associated protein kinase 1 and cyclin G-associated kinase regulate hepatitis C virus entry and are potential drug targets. J Virol. 2015;89:4387–4404. doi: 10.1128/jvi.02705-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Filippini A, D’Amore A, Palombi F, Carpaneto A. Could the inhibition of endo-lysosomal two-pore channels (TPCs) by the natural flavonoid naringenin represent an option to fight SARS-CoV-2 infection? Front Microbiol. 2020 doi: 10.3389/fmicb.2020.009702020.00970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kang YL, Chou YY, Rothlauf PW, et al. Inhibition of PIKfyve kinase prevents infection by Zaire ebolavirus and SARS-CoV-2. Proc Natl Acad Sci USA. 2020;117:20803–20813. doi: 10.1073/pnas.2007837117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rathnayake AD, Zheng J, Kim Y, et al. 3C-like protease inhibitors block coronavirus replication in vitro and improve survival in MERS-CoV–infected mice. Sci Transl Med. 2020 doi: 10.1126/SCITRANSLMED.ABC5332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Elfiky AA. Anti-HCV, nucleotide inhibitors, repurposing against COVID-19. Life Sci. 2020 doi: 10.1016/j.lfs.2020.117477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kong XD, Moriya J, Carle V, et al. De novo development of proteolytically resistant therapeutic peptides for oral administration. Nat Biomed Eng. 2020;4:560–571. doi: 10.1038/s41551-020-0556-3. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The manuscript has no associated data to disclose.