Abstract
Simple Summary
Bone metastases cause substantial morbidity and implicate worse clinical outcomes for clear-cell renal cell carcinoma patients. MicroRNAs are small RNA molecules that modulate gene translation and are involved in the development of cancer and metastasis. We identified six microRNAs that are potentially specifically involved in metastasis to bone, of which two seem protective and four implicate a higher risk. This aids further understanding of the process of metastasizing to bone. Furthermore, these microRNA hold potential for biomarkers or therapeutic targets.
Abstract
Bone metastasis in clear-cell renal cell carcinoma (ccRCC) leads to substantial morbidity through skeletal related adverse events and implicates worse clinical outcomes. MicroRNAs (miRNA) are small non-protein coding RNA molecules with important regulatory functions in cancer development and metastasis. In this retrospective analysis we present dysregulated miRNA in ccRCC, which are associated with bone metastasis. In particular, miR-23a-3p, miR-27a-3p, miR-20a-5p, and miR-335-3p specifically correlated with the earlier appearance of bone metastasis, compared to metastasis in other organs. In contrast, miR-30b-3p and miR-139-3p were correlated with less occurrence of bone metastasis. These miRNAs are potential biomarkers and attractive targets for miRNA inhibitors or mimics, which could lead to novel therapeutic possibilities for bone targeted treatment in metastatic ccRCC.
Keywords: renal cell carcinoma, bone metastasis, microRNA, miR-23a-3p, miR-20a-5p, miR-27a-3p, miR-335-3p, miR-30b-3p, miR-139-3p, SATB2
1. Introduction
Bone metastases (BM) occur in about 22% to 35% of all metastatic clear-cell renal cell carcinoma (ccRCC) patients [1,2]. They can give rise to skeletal related events (SREs) such as pain, hypercalcemia, pathologic fractures, and myelum compression, causing substantial morbidity. Moreover, BM are associated with poor prognosis in ccRCC when compared to metastases in other organs, and their presence also predicts an inferior outcome in treatment with angiogenesis inhibitors [3,4,5].
Preventing the occurrence of BM in ccRCC patients would provide substantial clinical benefit. Bone resorption inhibitors such as bisphosphonates or denosumab can reduce the number of SRE, though their impact on outcome is not precisely defined [6,7,8]. In order to provide additional therapeutic options to limit the occurrence of BM in ccRCC, better understanding of the underlying molecular mechanisms is required. Previously, we demonstrated that higher receptor activator of nuclear factor κB (RANK) and lower osteoprotegerin (OPG) expression in the primary kidney tumor are correlated with the occurrence of BM [9], findings that were similar to those of Mikami et al. [10].
MicroRNAs (miRNAs) are a class of small non-protein-coding RNA molecules that are principally involved in posttranscriptional regulation by interfering with translation or stability of target transcripts. Through imperfect base pairing, a single miRNA can regulate a large variety of gene transcripts. Dysregulation of miRNAs is frequently associated with cell proliferation, metastasis and apoptosis in various cancers. Several miRNA are currently under development as targets for cancer therapies [11,12,13,14].
In breast cancer (BC) and prostate cancer (PC), diseases with mainly osteoblastic BM, the involvement of miRNAs in the development of BM is well documented. There is less evidence in ccRCC, a malignancy with mainly osteolytic BM. Therefore, our aim was to study the impact of miRNAs on the development of BM in ccRCC.
2. Materials and Methods
2.1. Patient Selection
We used clinical and molecular data available in the University Hospitals Leuven database of ccRCC patients treated with systemic therapies. All patients underwent nephrectomy as first therapeutic intervention.
The study was approved by the Ethics Committee Research UZ/KULeuven (approval number S53479/S63833). Signed consent was obtained from all patients. In some cases, we used biological material from patients who were deceased and for whom general positive advice for the utilization of remaining tissue was foreseen by the institutional board.
2.2. Study Objectives and Endpoints
The principal objective was to investigate miRNA involvement in the development of BM in ccRCCs. Therefore, the primary endpoint of the study was time-to-bone-metastasis (TTBM), defined as the time between initial diagnosis and development of BM. In order to evaluate if the impact on metastasis was specific for BM, we calculated time-to-metastasis-other-than-bone (TTMOTB), defined as the time between initial diagnosis and the development of metastasis in all organs other than BM (MOTB). Furthermore, we included an explorative analysis of the impact of miRNAs on mRNA levels. As BM have a negative prognostic impact in ccRCC, our third objective was to study the impact of the miRNAs involved in the development of BM on clinical outcome, including overall survival (OS), since diagnosis and outcome on first-line anti-vascular endothelial growth factor receptor tyrosine kinase inhibitor (VEGFR-TKI) as measured by response rate (RR), progression free survival (PFS) and OS since start of VEGFR-TKIs.
2.3. MiRNA Extraction
MiRNA analysis was performed at the Centro Nacional de Investigaciones Oncológicas in Madrid as described previously [15]. For miRNA isolation, H&E-stained sections of the tumor samples were examined by a pathologist to confirm the diagnosis and to estimate tumor content. Total RNA was isolated with the Recover All Total Nucleic Acid Isolation kit (Ambion) using 4–6 whole 10-μm sections of the tumor samples. RNA quantity and quality were assessed by NanoDrop Spectrophotometer (Nanodrop Technologies, Wilmington, DE, USA). Quantitative RT-PCR analysis was performed using the Universal miRCURY LNATM microRNA PCR System (Exiqon, Vedbaek, Denmark), following the manufacturer’s instructions. In brief, 10 ng of total RNA was reversely transcribed, using the miRCURY LNATM Universal RT microRNA PCR kit (Exiqon). The resulting cDNA was diluted and PCR reactions were carried out using ExiLENT SYBR® Green Master Mix kit (Exiqon) and LNA™-enhanced microRNA PCR primer sets (Exiqon). MiRNA expression was quantified using the QuantStudio™ 6–7 Flex Real-Time PCR System (Applied Biosystems, Foster City, CA, USA). Stably expressed endogenous control 5s RNA was used for data normalization. Three samples with a low expression of the control 5s RNA were excluded from the analysis. Expression of each miRNA was calculated using the comparative Ct method using the Ct value of the endogenous control to normalize the data. Negative controls were present in all PCR series and assays were carried out in quadruplicates. The expression level of 454 miRNAs was defined in each tumor sample.
2.4. mRNA Extraction
Archived formal-fixed paraffin-embedded (FFPE) samples were retrieved. Tissue blocks with the highest tumoral content were selected for further processing through review of H&E slides. Blank slides with 5 μm thickness were made and underwent macrodissection to exclude non-tumoral tissues, resulting in a total tumoral surface area of 50–1800 mm2. The Maxwell RSC RNA FFPE kit (Promega) was used to perform RNA extraction according to the manufacturer’s instructions. The Forward QuantSeq 3′ mRNA-Seq Library Prep Kit for Illumina (Lexogen) was used to prepare cDNA libraries according to the manufacturer’s instructions with 5 µL of RNA as input and 16 PCR cycles. cDNA concentrations and fragment length were measured with the QubitTM dsDNA HS assay (Thermofisher, Waltham, MA, USA) and Bioanalyzer HS DNA electrophoresis (Agilent, Santa Clara, USA). Clonal clusters were generated with Illumina cBOT. The HiSeq 4000 kit (Illumina) was used for RNAseq according to the manufacturer’s instructions. Raw sequencing reads were trimmed of adaptors and optical duplicates and were subsequently aligned to the human reference genome hg19 with HiSat2 (v2.1.0) and quantified using featureCounts (v1.6.4). Counts were processed using DESeq2 (v1.26.0) and normalized using the VarianceStabilizingTransformation function.
2.5. Selection of miRNAs and Genes
We selected 78 miRNA with previously described involvement in either bone metabolism, oncogenesis, or metastasis (Table 1). For the estimation of TTBM, we selected the subset of patients (n = 106) who developed BM after the initial diagnosis of the ccRCC (metachronous BM), as TTBM was uncertain in patients with BM at diagnosis. For the estimation of TTMOTB, we selected the subset of patients who developed metastasis in organs other than bone after the initial diagnosis (n = 72). Patients with BM at initial diagnosis, but no metastases at other sites, were also included for TTMOTB estimations. We selected 29 genes with known involvement in either bone metabolism, oncogenesis or metastasis (Table 1) for a correlation analysis between miRNA and mRNA expression, for which Spearman correlation was used. For this analysis, we used mRNA and miRNA expression data from all the tumor samples, independently of the timing of development of metastases. As we performed this study from a validation perspective, results with p-value < 0.05 were considered significant and no correction for multiple testing was performed. Correlated miRNA-gene pairs were subsequently searched in DIANA TarBase v8, a database of experimentally validated miRNA-target pairs [16], and miRDB, a database for prediction of functional miRNA targets [17].
Table 1.
Selected miRNAs and genes.
| Selected miRNAs | ||
|---|---|---|
| let-7a-3p [18,19], let-7a-5p [19,20], let-7g-3p [19], let-7g-5p [19], miR-10b-3p [21,22], miR-10b-5p [21,22], miR-15b-3p [23], miR-15b-5p [23], miR-16-1-3p [24], miR-16-2-3p [24], miR-16-5p [24], miR-17-3p [25], miR-17-5p [26], miR-19a-3p [27], miR-20a-5p [28,29], miR-21-3p [30,31], miR-21-5p [26,30], miR-23a-3p [32,33,34], miR-27a-3p [26,34], miR-28-3p [35], miR-30a-3p [36], miR-30a-5p [36], miR-30b-3p [36], miR-30c-5p [36], miR-30d-3p [36], miR-30d-5p [36], miR-30e-3p [36], miR-30e-5p [36], miR-31-5p [26], miR-33a-5p [37], miR-34a-5p [38], miR-34c-5p [39], miR-125a-3p [26], miR-125a-5p [26], miR-126-5p [26,40], miR-133a-3p [41], miR-135a-5p [42], miR-138-5p [43], miR-139-3p [44,45], miR-139-5p [46], miR-141-3p [47], miR-142-3p [48,49], miR-142-5p [48], miR-143-3p [50,51], miR-143-5p [50], miR-145-3p [50,52], miR-145-5p [50,52], miR-148a-3p [27,53], miR-155-5p [54,55], miR-182-5p [56,57], miR-190a-5p [24], miR-190b [24], miR-200a-3p [58], miR-200a-5p [58], miR-200b-3p [58], miR-200b-5p [58], miR-200c-3p [58], miR-203a-3p [42], miR-204-5p [59], miR-210-3p [55,60], miR-211-5p [59], miR-214-3p [61], miR-214-5p [61], miR-218-5p [62], miR-223-3p [47], miR-223-5p [47], miR-296-5p [27], miR-326 [63], miR-335-3p [18], miR-335-5p [64], miR-378a-3p [24], miR-379-5p [65], miR-409-3p [65], miR-409-5p [65], miR-503-5p [24], miR-542-3p [66], miR-542-5p [67], miR-543 [68] | ||
| Selected genes | ||
| Protective for BM | ||
| BMP7 | Bone Morphogenetic Protein 7 | Binds BMPR2 receptor on tumor cells and enhances tumor cell dormancy through ERK/MAPK signaling [69]. |
| BMPR2 | Bone Morphogenetic Protein Receptor Type 2 | Promotes tumor cell dormancy in bone [40]. Inversely correlated with prostate BM occurrence [70]. |
| FOS | Fos Proto-Oncogene | Transcription factor for osteoclastogenesis [71]. |
| NOS3 | Nitric Oxide Synthase 3 | Decreases invasiveness [68]. |
| NOTCH1 | Notch Receptor 1 | Involved in oncogenesis [40]. |
| PDCD4 | Programmed Cell Death 4 | Represses osteoclastogenesis [71]. Downregulation in NSCLC potentially enhances BM [30]. |
| SATB2 | Special AT-Rich Sequence-Binding Protein 2 | Promotes osteoblastogenesis and bone regeneration [72]. Downregulation is associated with metastasis in ccRCC [72]. |
| SRCIN1 | SRC Kinase Signaling Inhibitor 1 | Inhibits osteoclast differentiation [28]. |
| SMAD2 | SMAD Family Member 2 | Decreases invasiveness. Component of the TGFβ pathway. In BC cells, loss of SMAD2 increases BM potential [73]. |
| SMAD4 | SMAD Family Member 4 | Decreases invasiveness [40,74]. |
| OPG | Osteoprotegerin | Decoy receptor preventing RANKL-RANK signaling [9]. |
| Associated with BM | ||
| CD44 | Cluster of differentiator 44 | Promotes invasiveness [40,75]. |
| CDH11 | Cadherin 11 | Promotes osteomimicry [36]. |
| CTGF | Connective Tissue Growth Factor | Stimulates osteoclasts [40]. |
| CXCL8 | C-X-C Motif Chemokine Ligand 8 | Promotes osteoclastogenesis [76]. |
| CXCR4 | C-X-C Motif Chemokine Receptor 4 | Promotes metastasis to osteogenic niches [40,77]. |
| DKK1 | Dickkopf WNT Signaling Pathway Inhibitor 1 | Osteoclast inhibitor [40]. |
| ELK1 | ETS Transcription Factor ELK1 | Promotes osteoblast differentiation [44]. Inducer of c-FOS proto-oncogene and part of ERK/MSK1/Elk-1/Snail signaling pathway (enhances cancer proliferation) [78]. |
| IL11 | Interleukin 11 | Stimulates osteoclasts [40]. |
| ITGA3 | Integrin Subunit Alpha 3 | Promotes invasiveness [40,75]. |
| ITGA5 | Integrin Subunit Alpha 5 | Involved in invasiveness [36], anchors cancer cells to bone [79]. |
| ITGB3 | Integrin Subunit Beta 3 | Integrin promoting osteomimicry and osteolytic metastases [80]. |
| RUNX2 | Runt-Related Transcription Factor 2 | TF essential in osteoblastogenesis and osteomimicry in PC and BC [42]. |
| SMAD1 | SMAD Family Member 1 | Osteoblast differentiation [81]. |
| TCF7 | Transcription Factor 7 | Involved in BM, part of the Wnt/beta-catenin signaling pathway [38]. |
| TGIF2 | TGF-Beta-induced transcription factor 2 | Pro-osteoclastic factor [82]. |
| TGFB1 | Transforming Growth Factor Beta 1 | Stimulates pro-osteoclastic factor production in cancer cells [40]. |
| RANKL | Receptor Activator Of Nuclear Factor Kappa B Ligand | Promotes osteoclastogenesis [10]. |
| RANK | Receptor Activator Of Nuclear Factor Kappa B | Osteoclast activation [83]. |
Abbreviations: miRNA = microRNA, miR = microRNA, BM = bone metastasis, NSCLC = non-small cell lung cancer, ccRCC = clear-cell renal cell carcinoma, BC = breast cancer, TF = transcription factor, PC = prostate cancer.
2.6. Clinical Data
After nephrectomy, patients underwent follow-up medical imagery (with either abdominal ultrasonography and chest X-ray or computed tomography of the chest and abdomen) every three to six months. In patients treated with VEGFR-TKIs, follow-up computed tomography (chest and abdomen) was done every two to three months during treatment. The decision whether or not to perform bone scintigraphy, a modality with low sensitivity to detect osteolytic BM, was made on clinical grounds. Pathology slides were reviewed by expert genitourinary pathologists. The International Metastatic RCC Database Consortium (IMDC) prognostic score was retrospectively calculated for each patient [84].
2.7. Statistical Analysis
Descriptives for time-to-event variables are based on Kaplan–Meier estimates. Cox proportional hazards regression models are used for the analysis of the association between expression data and time-to-event outcomes. Analyses were performed using SAS software (version 9.4) (SAS Institute Inc., Cary, NC, USA) and R (version 4.0.03) (R Core Team, Vienna, Austria).
3. Results
3.1. Included Patients
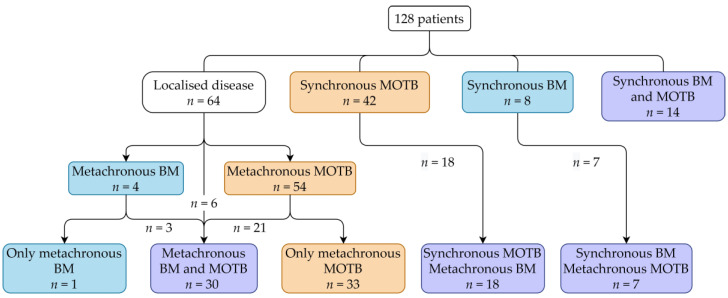
In total, the data of 128 patients were available. Patient characteristics are described in Table 2. All patients reached the metastatic stage and were treated with systemic therapies, among them 109 with VEGFR-TKIs in first-line. Figure 1 shows the occurrence of BM and MOTB over time in the patient cohort. Patients with synchronous BM or MOTB were excluded for analysis of TTBM or TTMOTB, respectively, resulting in patient groups of 106 and 72, respectively. For the OS analysis, all patients were included.
Table 2.
Patient characteristics.
| All Patients | n = 128 | |
|---|---|---|
| Gender: male | 86 | 67% |
| Median age at diagnosis (years) | 62 | IQR: 55–69 |
| Median OS after diagnosis (months) | 49 | IQR: 21–103.25 |
| Median OS after stage IV (months) | 34 | IQR: 16.5–62.25 |
| Bone metastasis | ||
| BM at time of nephrectomy (n) | 22 | 17.2% |
| Metachronous BM (n) | 106 | 82.8% |
| Median time to metachronous BM (months) | 34 | IQR: 24.25–100.5 |
| IMDC risk group at start of first-line therapy | ||
| Favorable (n) | 14 | 11% |
| Intermediate (n) | 81 | 63% |
| Poor (n) | 33 | 26% |
| First-line targeted therapy | ||
| Sunitinib (n) | 68 | 53% |
| Pazopanib (n) | 31 | 24% |
| Sorafenib (n) | 11 | 9% |
| Temsirolimus (n) | 9 | 7% |
| Nivolumab-Ipilimumab (n) | 6 | 5% |
| Other (n) | 3 | 2% |
Abbreviations: IQR = interquartile range, OS = overall survival (months), BM = bone metastasis, IMDC = International Metastatic RCC Database Consortium.
Figure 1.
Clinical stage of included patients. Abbreviations: MOTB = metastasis other than bone, BM = bone metastasis.
3.2. MiRNAs Associated with Longer or Shorter Time-to-Bone-Metastasis
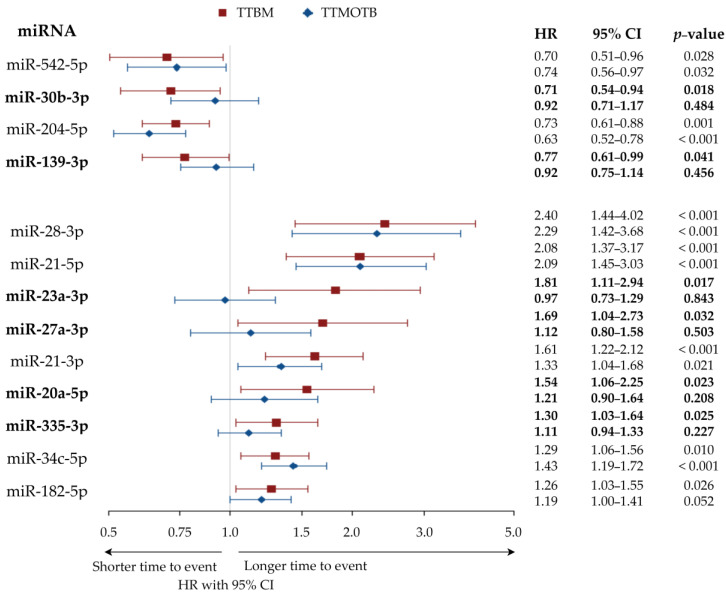
A total of 4 out of 78 miRNA were significantly associated with longer TTBM: miR-204-5p, miR-30b-3p, miR-542-5p, and miR-139-3p. miR-204-5p and miR-542-5p were also correlated to TTMOTB, thus their effect might not be specific for BM. miR-30b-3p and miR-139-3p were not correlated with TTMOTB, thus their effect might be specific for BM.
Nine miRNAs were found to be significantly associated with shorter TTBM: miR-21-5p, miR-21-3p, miR-28-3p, miR-34c-5p, miR-23a-3p, miR-20a-5p, miR-335-3p, miR-182-5p, and miR-27a-3p. However, miR-21-5p, miR-34c-5p, miR-28-3p, and miR-21-3p were also correlated to TTMOTB, thus their effect might not be specific for BM. miR-182-5p showed a trend towards correlation with TTMOTB. miR-20a-5p, miR-335-3p, miR-27a-3p, and miR-23a-3p were clearly not correlated to TTMOTB, thus their effect might be specific for the development of BM.
The 13 miRNAs are displayed with hazard ratio (HR) and confidence interval (CI) in Figure 2. The correlation results for all miRNA are presented in Tables S1 and S2.
Figure 2.
Forest plot displaying HR with 95% CI of TTBM and TTMOTB for 13 miRNA significantly associated with TTBM (univariate Cox proportional hazards model). Note: miRNA correlated with TTBM but not with TTMOTB are highlighted in bold. Abbreviations: miRNA = microRNA, TTBM = time to bone metastasis, TTMOTB = time to metastasis other than bone, miR = microRNA, HR = hazard ratio, CI = confidence interval.
3.3. Correlation of miRNA with Gene Expression
Using the Spearman correlation, we correlated the expression of 13 miRNAs that are correlated to TTBM, with the expression of 29 mRNAs. mRNA expression data were available for 89 of the 128 patients. A non-systematic literature search was able to validate 13 previously described correlations between miRNAs and mRNAs.
Among the miRNAs associated with longer TTBM, miR-204-5p was inversely correlated with RUNX2 and TGFB1. miR-139-3p was correlated with the mRNA expression levels of NOTCH1. miR-30b-3p was inversely correlated with the mRNA expression levels of CXCL8. miR-542-5p was correlated with the mRNA expression levels of SMAD2.
Among the miRNAs associated with shorter TTBM, miR-21-3p was associated with reduced FOS, BMPR2, and SMAD4 expression and increased TFGB1 expression. miR-34c-5p was correlated with reduced SATB2 levels. miR-20-5p was associated with upregulated RUNX2 levels. Finally, miR-182-5p was correlated with lower SATB2 levels.
We studied other possible correlations between miRNAs and mRNAs in an exploratory approach. The significant findings are represented in Table 3. All correlation results are available in Table S3.
Table 3.
Correlations of miRNA with gene expression (Spearman correlation test, p-value < 0.05).
| miRNAs Associated with Longer TTBM | |||
|---|---|---|---|
| miR-30b-3p | BM promoting genes: inverse correlation | ||
| RANKL | rho = −0.33; p = 0.002 | miR-30 family inhibits BM in BC by targeting CXCL8 [36] | |
| ITGA3 | rho = −0.31; p = 0.003 | ||
| TCF7 | rho = −0.25; p = 0.018 | ||
| CD44 | rho = −0.24; p = 0.021 | ||
| CXCL8 | rho = −0.21; p = 0.045 | ||
| miR-139-3p | BM protective genes: positive correlation | ||
| NOTCH1 | rho = 0.29; p = 0.005 | ||
| BMPR2 | rho = 0.27; p = 0.011 | ||
| BM promoting genes: inverse correlation | |||
| TCF7 | rho = −0.23; p = 0.027 | ||
| ITGA3 | rho = −0.22; p = 0.038 | ||
| miR-204-5p | BM protective genes: positive correlation | ||
| FOS | rho = 0.28; p = 0.007 | ||
| BMPR2 | rho = 0.32; p = 0.003 | ||
| SATB2 | rho = 0.34; p = 0.001 | ||
| SMAD4 | rho = 0.35; p = 0.001 | ||
| BM promoting genes: inverse correlation | |||
| CD44 | rho = −0.55; p < 0.001 | miR-204 regulates RUNX2 expression and MSC differentiation [85]. | |
| RANKL | rho = −0.42; p < 0.001 | Sponging of miR-204-5p by lncRNA SNHG4 upregulates RUNX2 and promotes tumor progression in RCC [86]. | |
| ITGA3 | rho = −0.42; p < 0.001 | miR-204-5p inhibits TGF-β-induced IL11 production in BM cells of BC [59]. | |
| RUNX2 ° | rho = −0.41; p < 0.001 | miR-204 inhibits growth and motility of CRC cells by CXCL8 downregulation [87]. | |
| ITGA5 | rho = −0.36; p = 0.001 | ||
| CDH11 ° | rho = −0.39; p < 0.001 | ||
| TGFB1 | rho = −0.33; p = 0.002 | ||
| CXCL8 | rho = −0.3; p = 0.005 | ||
| TCF7 | rho = −0.36; p < 0.001 | ||
| CTGF | rho = −0.26; p = 0.014 | ||
| miR-542-5p | BM protective genes: positive correlation | ||
| SATB2 | rho = 0.34; p = 0.001 | miR-542-5p increases SMAD2 expression levels in ICU weakness [88]. | |
| SMAD2 | rho = 0.29; p = 0.006 | ||
| BM promoting genes: inverse correlation | |||
| TCF7 | rho = −0.37; p < 0.001 | ||
| RUNX2 | rho = −0.31; p = 0.003 | ||
| CD44 | rho = −0.32; p = 0.002 | ||
| CXCR4 | rho = −0.21; p = 0.044 | ||
| miRNAs Associated With Shorter TTBM | |||
| miR-21-5p | BM protective genes: inverse correlation | ||
| SATB2 | rho = −0.22; p = 0.04 | ||
| SMAD4 | rho = −0.24; p = 0.026 | ||
| BM promoting genes: positive correlation | |||
| TCF7 | rho = 0.27; p = 0.011 | ||
| CD44 * | rho = 0.25; p = 0.018 | ||
| ITGA3 | rho = 0.29; p = 0.005 | ||
| miR-21-3p | BM protective genes: inverse correlation | ||
| FOS | rho = −0.45; p < 0.001 | miR-21 deficiency results in cFOS upregulation in periodontal tissues [89]. | |
| BMPR2 | rho = −0.42; p < 0.001 | BMPR2 is directly targeted by miR-21 in PC cells [90]. | |
| SATB2 | rho = −0.31; p = 0.003 | miR-21-3p is involved in proliferation and invasion through SMAD4/Erk signaling in CRC [31]. | |
| SMAD4 | rho = 0.22; p = 0.043 | ||
| BM promoting genes: positive correlation | |||
| ITGA3 | rho = 0.42; p < 0.001 | TGFB1 signaling increases miR-21 expression in renal fibrosis [91]. | |
| RANKL | rho = 0.38; p < 0.001 | ||
| CD44 * | rho = 0.37; p < 0.001 | ||
| TGFB1 | rho = 0.25; p = 0.018 | ||
| CXCL8 * | rho = 0.22; p = 0.043 | ||
| miR-28-3p | BM protective genes: inverse correlation | ||
| SATB2 | rho = −0.24; p = 0.022 | ||
| SRCIN1 | rho = −0.22; p = 0.038 | ||
| BM promoting genes: positive correlation | |||
| CXCR4 | rho = 0.24; p = 0.025 | ||
| miR-34c-5p | BM protective genes: inverse correlation | ||
| SATB2 ° | rho = −0.23; p = 0.033 | miR-34s inhibit osteoblast proliferation and differentiation in mouse by targeting SATB2 [92]. | |
| FOS | rho = −0.26; p = 0.015 | SATB2 targeted by miR-34c-5p suppresses proliferation and metastasis attenuating EMT in CRC [39]. | |
| BM promoting genes: positive correlation | |||
| CD44 | rho = 0.3; p = 0.004 | ||
| ITGA3 | rho = 0.38; p < 0.001 | ||
| RANK | rho = 0.21; p = 0.047 | ||
| TGFB1 | rho = 0.26; p = 0.015 | ||
| miR-23a-3p | BM protective genes: positive correlation | ||
| BMPR2 * | rho = 0.23; p = 0.027 | ||
| BM promoting genes: inverse correlation | |||
| CXCL8 * | rho = −0.25; p = 0.017 | ||
| ELK1 | rho = −0.24; p = 0.021 | ||
| miR-20a-5p | BM protective genes: inverse correlation | ||
| OPG * | rho = −0.21; p = 0.044 | ||
| BM protective genes: positive correlation | |||
| PDCD4 ° | rho = 0.28; p = 0.009 | ||
| BM promoting genes: inverse correlation | |||
| ITGB3 | rho = −0.23; p = 0.03 | ||
| BM promoting genes: positive correlation | |||
| IL11 | rho = 0.27; p = 0.011 | RUNX2 is positively correlated with miR-20a-5p in adipose SC during osteogenic differentiation [93]. | |
| TCF7 | rho = 0.26; p = 0.012 | ||
| RUNX2 | rho = 0.22; p = 0.035 | ||
| miR-335-3p | BM protective genes: inverse correlation | ||
| OPG | rho = −0.23; p = 0.03 | ||
| BM promoting genes: positive correlation | |||
| CD44 | rho = 0.31; p = 0.003 | ||
| ITGA5 | rho = 0.29; p = 0.007 | ||
| RANKL | rho = 0.22; p = 0.04 | ||
| CDH11 | rho = 0.36; p = 0.001 | ||
| ITGA3 | rho = 0.27; p = 0.011 | ||
| TGFB1 | rho = 0.29; p = 0.006 | ||
| TCF7 | rho = 0.35; p = 0.001 | ||
| RUNX2 | rho = 0.3; p = 0.004 | ||
| miR-182-5p | BM protective genes: inverse correlation | ||
| SATB2 *,° | rho = −0.29; p = 0.006 | microRNA-182 targets SATB2 to promote CRC proliferation and metastasis [94]. | |
| BMPR2 * | rho = −0.28; p = 0.009 | ||
| OPG | rho = −0.28; p = 0.007 | ||
| BM promoting genes: positive correlation | |||
| CD44 | rho = 0.37; p < 0.001 | ||
| CDH11 * | rho = 0.25; p = 0.02 | ||
| ITGA3 | rho = 0.37; p < 0.001 | ||
| IL11 * | rho = 0.21; p = 0.046 | ||
| CXCL8 | rho = 0.32; p = 0.002 | ||
| TCF7 | rho = 0.34; p = 0.001 | ||
| RUNX2 | rho = 0.27; p = 0.011 | ||
| miR-27a-3p | BM protective genes: inverse correlation | ||
| BMP7 * | rho = −0.29; p = 0.007 | ||
| BM protective genes: positive correlation | |||
| PDCD4 | rho = 0.23; p = 0.031 | ||
| BM promoting genes: inverse correlation | |||
| CXCL8 * | rho = −0.26; p = 0.013 | ||
BM protective genes: SATB2, BMP7, SMAD4, PDCD4, BMPR2, OPG, NOS3, SMAD2, NOTCH1, FOS, SRCIN1. BM promoting genes: TCF7, DKK1, CD44, ITGA3, CTGF, IL11, CXCL8, RANKL, RUNX2, CDH11, TGFB1, CXCR4, SMAD1, TGIF2, RANK, ELK1, ITGB3, ITGA5. * = listed in DIANA-TarBase v.8 of validated miRNA-target pairs [16]. ° = predicted miRNA target through miRDB.org [17]. Note: miRNA correlated to TTBM and not to TTMOTB are highlighted in bold. Correlations with similar literature findings are highlighted in bold. Abbreviations: miRNA = microRNA, TTBM = time to bone metastasis, miR = microRNA, BM = bone metastasis, BC = breast cancer, MSC = mesenchymal stem cells, lncRNA = long non-coding RNA, SNHG4 = small nucleolar RNA host gene 4, RCC = renal cell carcinoma, CRC = colorectal cancer, PC = prostate cancer, EMT = epithelial-mesenchymal transition, RCC = renal cell carcinoma, SC = stem cells.
3.4. Correlation of miRNA with Clinical Outcome
Subsequently, we checked the impact of the 13 miRNA correlated with TTBM, on outcome. Several of these miRNAs were also correlated with OS (Table 4). miR-204-5p, associated with longer TTBM, is significantly correlated with longer OS. miR-21-5p, miR-21-3p, miR-34c-5p, miR-335-3p, and miR-182-5p, associated with shorter TTBM, were significantly correlated with shorter OS. The full correlation results are available in Table S4.
Table 4.
Correlations of 13 miRNA involved in bone metastasis with OS since diagnosis and PFS and OS on 1st line VEGFR-TKI therapy (univariate Cox regression model).
| miRNA | OS Since Diagnosis | PFS on VEGFR-TKIs | OS on VEGFR-TKIs | |||
| HR (95% CI) | p-value | HR (95% CI) | p-value | HR (95% CI) | p-value | |
| Longer time to bone metastasis | ||||||
| miR-204-5p | 0.73 (0.65–0.82) | <0.001 | 0.84 (0.74–0.94) | 0.003 | 0.81 (0.72–0.92) | 0.001 |
| miR-30b-3p | 0.93 (0.78–1.10) | 0.386 | 1.07 (0.88–1.31) | 0.496 | 0.99 (0.81–1.21) | 0.921 |
| miR-542-5p | 0.85 (0.69–1.05) | 0.133 | 0.96 (0.77–1.20) | 0.728 | 0.89 (0.71–1.11) | 0.296 |
| miR-139-3p | 0.86 (0.73–1.00) | 0.057 | 0.91 (0.76–1.09) | 0.321 | 0.87 (0.73–1.03) | 0.105 |
| Shorter time to bone metastasis | ||||||
| miR-21-5p | 1.69 (1.31–2.18) | <0.001 | 1.14 (0.88–1.47) | 0.327 | 1.29 (1.01–1.65) | 0.043 |
| miR-21-3p | 1.50 (1.31–2.18) | <0.001 | 1.24 (1.04–1.48) | 0.016 | 1.35 (1.13–1.61) | 0.001 |
| miR-28-3p | 1.38 (0.98–1.93) | 0.064 | 1.01 (0.70–1.47) | 0.959 | 1.04 (0.72–1.50) | 0.823 |
| miR-34c-5p | 1.31 (1.16–1.48) | <0.001 | 1.19 (1.05–1.35) | 0.007 | 1.27 (1.11–1.44) | <0.001 |
| miR-23a-3p | 1.05 (0.83–1.33) | 0.697 | 0.98 (0.75–1.30) | 0.903 | 1.01 (0.76–1.34) | 0.953 |
| miR-20a-5p | 1.12 (0.91–1.39) | 0.279 | 0.95 (0.77–1.19) | 0.680 | 0.94 (0.75–1.18) | 0.598 |
| miR-335-3p | 1.22 (1.06–1.41) | 0.005 | 1.10 (0.95–1.26) | 0.193 | 1.11 (0.96–1.28) | 0.155 |
| miR-182-5p | 1.25 (1.09–1.43) | 0.002 | 1.11 (0.96–1.29) | 0.145 | 1.16 (1.01–1.34) | 0.040 |
| miR-27a-3p | 1.13 (0.87–1.47) | 0.351 | 0.87 (0.65–1.17) | 0.365 | 1.03 (0.77–1.39) | 0.831 |
Abbreviations: miRNA = microRNA; BM = bone metastasis; PFS = progression free survival; OS = overall survival; VEGFR-TKI = vascular endothelial growth factor receptor tyrosine kinase inhibitor; HR = hazard ratio; CI = confidence interval; miR = microRNA. Significant correlations (p-value < 0.05) are highlighted in bold.
As the majority of the patients (n = 109) were treated in first-line with VEGFR-TKIs, we compared the expression levels of these 13 miRNA with PFS and OS on VEGFR-TKIs (Tables S5 and S6). miR-204-5p, correlated with longer TTBM, was also correlated with longer PFS and OS on VEGFR-TKIs. miR-21-3p and miR-34c-5p, correlated with shorter TTBM, were also correlated with shorter PFS and OS on VEGFR-TKIs. Finally, miR-21-5p and miR-182-5p, correlated with shorter TTBM, were correlated with shorter OS. miR-204-5p was associated with increased RR (p = 0.03) and miR-34c-5p with decreased RR (p = 0.003).
Additionally, SATB2 was associated with longer PFS (HR = 0.595; p = 0.045) and longer OS on VEGFR-TKI (HR = 0.595; p = 0.039).
3.5. Correlation of mRNA Expression with TTBM and OS
For only 71 of the 106 patients eligible for TTBM analysis, mRNA expression data were available. On this reduced patient series, two significant correlations with TTBM were found: lower SATB2 (p = 0.03) and higher IL11 (p = 0.048) mRNA expression were correlated with shorter TTBM.
There were 89 patients available for the analysis of the correlation between mRNA expression and OS. Higher SATB2 (p < 0.0001), FOS (p = 0.02) and SMAD4 (p = 0.02) mRNA expression were associated with longer OS. Higher CD44 (p < 0.001), ITGA5 (p = 0.004), TGFB1 (p = 0.008), TGIF2 (p = 0.01), SRCIN1 (p = 0.02), RUNX2 (p = 0.02), CHD11 (p = 0.04), and ITGA3 (p = 0.04) were correlated with worse OS. Full results are presented in Table S7.
4. Discussion
Bone metastases in metastatic ccRCC patients occur in up to 35% of patients [1,2]. Several studies have reported a negative impact of BM on clinical outcomes [3,95]. Within ccRCC patients with BM, those with synchronous BM are generally poor risk patients, whereas patients with metachronous BM and longer TTBM were associated with longer OS [96]. To provide new therapeutic options for BM in ccRCC, further research into their pathogenesis is critical. miRNA have attracted much attention in the past years and therapeutic strategies that target them are already used in phase I clinical trials [11,12,13].
Most evidence about the involvement of miRNA in BM comes from BC and PC, malignancies with mostly osteoblastic metastasis. Of osteolytic metastasis, most evidence is in lung and colorectal cancer (CRC). In ccRCC, the amount of evidence is more limited.
Therefore, we studied the impact of miRNAs expressed in primary ccRCC tumors on the development of BM. We report 13 miRNAs that are significantly associated with TTBM. Of these, six miRNA did not have a significant correlation with TTMOTB and might therefore be specific for osteotropic metastasis. We also report correlations between the intratumoral expression of miRNAs and mRNA of genes involved in invasion and metastasis and more specifically in the development of BM. As miRNAs influence gene expression through translation repression or increased degradation of target mRNA, inverse correlations could result from either direct mRNA targeting or from a target upstream in its regulatory pathway. We also reported positive correlations as these could indirectly result from targeting suppressive components of regulatory pathways of the correlated gene. Finally, several miRNAs that are correlated to the development of BM were also associated with outcome.
4.1. miRNA Associated with Longer TTBM
In this study, we identified four miRNA associated with longer TTBM (miR-30b-3p, miR-139-3p, miR-204-5p and miR-542-5p). miR-30b-3p and miR-139-3p were not significantly associated with TTMOTB and might thus be specific for the development of BM. All four miRNAs have demonstrated tumor suppressive properties over a variety of cancer types.
In our series, miR-30b-3p was associated with longer TTBM and inversely correlated to mRNA expression of several genes involved in BM (CD44, ITGA3, TCF7, RANKL and CXLC8). The miR-30 family, to which miR-30b-3p belongs, suppresses BM in BC and PC. These miRNAs inhibit osteoblast differentiation and osteomimicry by targeting RUNX2 [54,97,98,99]. In BC cells, miR-30 inhibition enhances tumorigenesis and metastasis by targeting UBC9 and ITGB3 and osteomimicry of cancer cells by targeting RUNX2, ITGA5 and CDH11 [100]. In BC, miR-30b-3p suppresses BM by targeting CXCL8, IL11, DKK1, RUNX2, CDH11, CTGF, ITGA5, and ITGB3 [36]. Finally, in multiple myeloma, an osteolytic disease, miR-30b-3p has a tumor suppressor role targeting the Wnt/b-Catenin/BCL9 Pathway [101].
We have found that miR-542-5p was associated with longer TTBM and TTMOTB, inversely correlated with the mRNA expression of genes involved in BM (CD44, CXCR4, RUNX2 and TCF7) and positively correlated with the mRNA expression of genes protective against BM (SATB2 and SMAD2). miR-542-5p acts as a tumor suppressor in non-small cell lung cancer (NSCLC) [67], neuroblastoma [102], osteosarcoma [103], CRC [104], BC [105], and hepatocellular carcinoma (HCC) [106], though contradictory evidence exists [107]. A role in bone metabolism has been described for the 3p strand of the miR-542 duplex, as a suppressor of osteoblast cell proliferation and differentiation by targeting BMP7-signalling [66]. miR-542-5p has been shown to increase expression levels of SMAD2 in other diseases [88].
In our series, miR-139-3p was associated with longer TTBM, inversely correlated with the mRNA expression of genes involved in BM (TCF7 and ITGA3), and positively correlated with the mRNA expression of genes protective for BM (BMPR2 and NOTCH1). Additionally, it is also correlated with longer OS. miR-139-3p and its guide strand miR-139-5p are involved in ccRCC pathogenesis and expression levels of both miRNA and several of their target genes are correlated with clinical outcomes [108]. A tumor suppressor role has also been described in bladder cancer [109], CRC [110], glioblastoma multiforme [111], BC [45], and HCC [112]. Additionally, miR-139-3p is downregulated in serum in patients with esophagal squamous cell cancer (SCC), highlighting its potential as a serum biomarker [113]. miR-139-5p is a regulator of osteoblast differentiation and apoptosis by targeting ELK1 and ODSM [44]. In NSCLC, miR-139-5p serum levels are reduced in the presence of lytic BM when compared to metastasis at other sites. Increased expression of miR-139-5p enhances osteogenic differentiation of mesenchymal stem cells (MSC) through NOTCH1 signaling. After exposure to NSCLC cells, this pathway was downregulated [46]. Conditioned media from NSCLC cells have been shown to promote osteoclastogenesis through inhibition of miR-139 and activation of the STAT3/c-FOS/NFATc1 pathway [114]. Activation of the Notch signaling pathway is involved in oncogenesis in a variety of malignancies including ccRCC [115].
Finally, miR-204-5p was associated with longer TTBM and TTMOTB, inversely correlated with the expression of genes involved in BM (CD44, RANKL, ITGA3, RUNX2, CDH11, TCF7, ITGA5, TGFB1, CXCL8 and CTGF), correlated with the expression of genes protective for BM (FOS, BMPR2, SATB2 and SMAD4) and correlated with longer OS. miR-204-5p is usually downregulated in ccRCC tumor tissues [116,117,118] and seems protective for the development of BM. Moreover, miR-204-5p is lower in the more aggressive ccrcc1+4 molecular ccRCC subtypes compared to the less aggressive ccrcc2+3 subtypes [119]. In bone metastatic BC cells, miR-204-5p inhibits TGF-β-induced IL11 production [59]. In laryngeal SCC, miR-204-5p inhibits cell proliferation, invasion, and metastasis [120]. In BC cells, miR-204-5p inhibits viability, proliferation and migration [121]. In oral SCC, it inhibits cell proliferation and metastasis by targeting CXCR4 [122]. Finally, miR-204-5p was shown to inhibit osteogenic differentiation of MSC [123].
4.2. miRNA Associated with Shorter TTBM
In our series, nine miRNA were associated with shorter TTBM. Of these, most were also associated with shorter TTMOTB, however miR-23a-3p, miR-20a-5p, miR-335-3p, and miR-27a-3p were not and might therefore be specifically involved in the development of BM.
We show that miR-23a-3p expression was correlated with TTBM. The oncogenic properties of miR-23a-3p have been previously demonstrated. In ccRCC tissues and cell lines, where it is overexpressed, it blocks apoptosis and enhances proliferation and mobility. Its upregulation in ccRCC is associated with worse OS [33]. Moreover, several studies in gastro-intestinal malignancies [124,125] and BC [126] have highlighted its potential as a serum biomarker. It is part of the miRNA cluster 23a/27a/24-2, which is involved in oncogenesis and inhibits osteoblast differentiation by targeting RUNX2 and SATB2 [34]. Forced expression of this miRNA cluster enhances cell migration and metastasis in BC cells [127]. Inhibition of miR-23a-3p promotes osteoblast proliferation and differentiation [32]. miR-23a-3p also inhibits osteogenic differentiation of human MSC [128]. Surprisingly, in our tumor series, miR-23a-3p expression was inversely correlated with ELK1 and CXCL8 expression and positively correlated with BMPR2 expression. This seems contradictory, as BMPR2 expression is inversely correlated with metastasis occurrence in prostate BM [70].
In our tumor series, miR-20a-5p expression was correlated to TTBM, inversely correlated to the expression of the BM protective gene OPG and correlated to the expression of several genes involved in BM (RUNX2, TCF7 and IL11). In ccRCC, miR-20 has a higher expression in BM compared to primary tumor and to normal renal tissues [27]. Additionally, increased serum expression levels in gastric cancer and CRC show its potential as a biomarker [129,130]. The positive correlation between RUNX2 and miR-20a-5p was previously described in adipose stem cells during osteogenic differentiation. MiR-20a promotes the osteogenesis of MSC through targeting of PPARγ, BAMBI, and CRIM1, which are negative regulators of BMP signaling [93]. Exosomal miR-20a-5p derived from BC cells promotes proliferation and differentiation of osteoclasts [28]. The inverse correlation with OPG, a protein which acts as a decoy receptor for RANK and neutralizes its role in osteoclastogenesis [131], seems biologically consistent with the association of miR-20a-5p with shorter TTBM. Previous reports have however described a positive correlation with miR-20a during osteogenic differentiation [93]. Surprisingly, ITGB3 expression was inversely and PDCD4 expression positively correlated with miR-20a-5p. In bladder and CRC cells, PDCD4 is downregulated by miR-20a [132,133].
We observed that miR-27a-3p was associated with TTBM and inversely correlated with expression of the bone protective gene BMP7. In ccRCC, miR-27a-3p promotes proliferation and metastasis [134,135]. In osteosarcoma, CRC, pancreatic, and gastric cancer, a pro-oncogenic role is also seen [136,137,138,139,140], although in NSCLC [141] and HCC [142] a tumor suppressive function has been proposed. Serum levels of miR-27a-3p are elevated in pancreatic cancer, CRC, and PC, demonstrating its potential as a non-invasive biomarker [143,144,145]. miR-27a-3p is involved in bone metabolism and negatively regulates osteogenic differentiation [146]. It is part of the aforementioned 23a/27a/24-2 miRNA cluster, which is involved in both oncogenesis and osteoblast differentiation by targeting RUNX2 and SATB2 [34]. Surprisingly, miR-27a-3p was also inversely correlated with CXCL8, promotor of osteoclastogenesis, [76] and positively correlated with PDCD4 expression.
In our tumor series, miR-21-3p and miR-21-5p were correlated with TTBM and TTMOTB. miR-21-3p was inversely correlated with the expression of several BM protective genes (FOS, BMPR2, SMAD4 and SATB2) and correlated with the expression of several genes involved in BM (CXCL8, TGFB1, CD44, RANKL and ITGA3). miR-21-5p expression was inversely correlated with the expression of two genes protective for BM (SMAD4 and SATB2) and correlated with the expression of genes involved in BM (TCF7, CD44, and ITGA3). miR-21-5p and miR-21-3p were significantly correlated with shorter OS. Both the 3p and 5p strand of miR-21 are known pro-oncogenic miRNAs across tumor types such as NSCLC, gastric cancer, CRC, and BC [147,148,149]. In ccRCC tissues, miR-21 is upregulated [118,150] and higher expression is also correlated with worse clinical outcomes [151]. In our previous study, miR-21-3p and miR-21-5p were upregulated in the more aggressive ccRCC molecular ccrcc1+4 subtypes [119]. miR-21 plays a pivotal role in RANKL-induced osteoclastogenesis through regulation of PDCD4 and OPG, which is critical for the development of lytic BM [30,71,152]. Exosomal miR-21 from NSCLC cells facilitates osteoclastogenesis by targeting PDCD4 [30] and inhibition of miR-21 impairs osteoclast activity in multiple myeloma by targeting OPG and PIAS3 [152].
In our samples, miR-34c-5p was associated with shorter TTBM and TTMOTB, correlated with the expression of genes involved in BM (RANK, TGFB1, CD44 and ITGA3), inversely correlated with the expression of genes protective for BM (FOS and SATB2) and significantly correlated with shorter OS. miR-34 inhibits osteoblast proliferation and differentiation in the mouse by targeting SATB2 [92]. In bladder cancer, miR-34c-5p enhances proliferation and migration of bladder cancer cells through targeting of NOTCH1 [153]. However, in other malignancies such as NSCLC and laryngeal SCC, a tumor suppressive role is observed [154,155] and a phase I clinical trial has been conducted with a miR-34a mimic in advanced solid tumors [11]. In our previous study, miR-34c-5p was correlated with the more aggressive ccrcc1+4 molecular subtypes [119].
We found that miR-335-3p was correlated with shorter TTBM, inversely associated with OPG expression, positively associated with the expression of genes involved in BM (RANKL, ITGA3, ITGA5, TGFB1, RUNX2, CD44, TCF7 and CDH11), and associated with shorter OS. Unlike this impressive association with the expression of several genes involved in metastasis and BM, the role of miR-335-3p remains unclear. In our previous study, miR-335-3p was correlated with the more aggressive ccrcc1+4 molecular subtypes, [119] but evidence on its prognostic effects remains conflicting [156,157,158]. A pro-oncogenic role was described in astrocytoma [159], but a tumor suppressor role described in BC [160]. In small cell lung carcinoma xenograft models and in PC, miR-335-3p overexpression was protective for BM, however, BM in these diseases are osteoblastic [68,161].
In our tumor series, miR-182-5p expression was correlated with shorter TTBM and TTMOTB, inversely correlated with the expression of genes protective for BM (SATB2, OPG and BMPR2), correlated with the expression of several genes involved in BM (IL11, CDH11, RUNX2, CXCL8, TCF7, CD44, and ITGA3), and correlated with shorter OS. In the literature, conflicting evidence exists for miR-182-5p, as previous studies suggest a tumor suppressive role for this miRNA in ccRCC [162,163,164]. In our previous study, miR-182-5p was correlated with the more aggressive ccrcc1+4 molecular subtypes [119]. However, miR-182 plays a key role in pathological osteoclastogenesis and negative regulation of osteoblast differentiation, and its inhibition could provide new strategies for bone protection [57,165].
Finally, miR-28-3p expression was inversely correlated with the expression of two BM protective genes (SATB2 and SRCIN1), positively correlated with CXCR4 expression, and correlated with shorter OS.
4.3. miRNA-mRNA Associations
We could validate some previously described correlations between miRNAs and mRNA expression of genes involved in the development of BM, although we could not validate other known associations. This can be partly due to the fact that ccRCC is a malignancy with osteolytic BM, whereas most studies have been performed in diseases with osteoblastic BM such as BC and PC. Moreover, only a limited number of samples were included in our study. Nevertheless, our findings on SATB2 in particular are coherent throughout the different parts of this study. This is a DNA binding protein involved in cell differentiation and reprogramming of expression profiles. Its tumor suppressive role has been previously described [72,166], and it holds a critical role in osteoblastogenesis as well [29]. A total of five miRNA associated with shorter TTBM (miR-21-3p, miR-21-5p, miR-28-3p, miR-34c-5p and miR-128-5p) were associated with lower SATB2 levels. A total of two miRNA associated with longer TTBM (miR-204-5p, miR-542-5p) were associated with higher SATB2 expression and could therefore potentially target suppressors of SATB2. SATB2 mRNA levels were correlated with longer TTBM and were also associated with longer OS, since diagnosis and longer PFS and OS since the start of first-line VEGFR-TKI. Several of these associations have previously been described [39,92,94]. Downregulation of SATB2 in ccRCC has previously been associated with metastasis and worse outcome [72].
4.4. Limitations and Strengths of Our Study
Our study has several limitations. All patients included eventually developed metastasis, so this is a selection of ccRCC patients. Intratumoral heterogeneity might also be an important issue as miRNA and mRNA extraction was done on different samples of the tumor. As BM were diagnosed through computed tomography scans and not via magnetic resonance imaging, this may result in underestimation. Finally, with miRNA and mRNA expression from tumor tissues, it is not possible to directly demonstrate the impact of miRNA on mRNA expression, only associations.
However, this study has also its strengths. It is an in-depth analysis of the possible impact of miRNAs on the development of BM in ccRCC, whereas most studies have been done in BC and PC. BC and PC are diseases with mainly osteoblastic BM, with different underlying pathophysiology. Not all of our results are specifically for the development of BM. Many of these miRNAs also seem involved in the global process of metastasis, also towards other organs. However, the development of a new therapy directed against BM and debilitating SREs, even if not specifically targeting BM, will still be of benefit for the patient.
5. Conclusions
In this study, we present a series of 13 miRNA that might be involved in the pathogenesis of BM in ccRCC and describe similar findings in the literature. These miRNAs could be potential biomarkers or attractive targets for miRNA inhibitors or mimics, which could lead to novel therapeutic possibilities for bone targeted treatment in metastatic malignancies.
Supplementary Materials
The following are available online at https://www.mdpi.com/article/10.3390/cancers13071554/s1, Table S1: Correlation of miRNA expression with TTBM, Table S2: Correlation of miRNA expression with TTMOTB, Table S3: Correlations of miRNA with mRNA expression (all), Table S4: Correlation of miRNA expression with OS since diagnosis, Table S5: Correlation of miRNA expression with PFS on VEGFR-TKIs, Table S6: Correlation of miRNA expression with OS on VEGFR-TKIs, Table S7: Correlation of mRNA expression with TTBM and OS.
Author Contributions
Conceptualization, L.K., E.R. and B.B. (Benoit Beuselinck); Data curation, L.K., E.R., B.B. (Bram Boeckx), C.R.-A., O.G.-C., L.I.-P., A.V., J.Z.-R., G.C. and S.C.; Formal analysis, L.K., E.R. and A.L.; Funding acquisition, B.B. (Benoit Beuselinck); Methodology, E.R., A.L. and B.B. (Bram Boeckx); Project administration, B.B. (Benoit Beuselinck); Resources, E.R., D.L., B.B. (Bram Boeckx), C.R.-A., O.G.-C., L.I.-P., A.V., J.Z.-R., G.C., S.C., M.B. and B.B. (Benoit Beuselinck); Supervision, M.A. and B.B. (Benoit Beuselinck); Visualization, L.K.; Writing—original draft, L.K. and B.B. (Benoit Beuselinck); Writing—review and editing, L.K., E.R., D.L., B.B. (Bram Boeckx), L.V., M.A., C.R.-A., O.G.-C., L.I.-P., A.V., J.Z.-R., G.C., S.C., A.L., M.B. and B.B. (Benoit Beuselinck). All authors have read and agreed to the published version of the manuscript.
Funding
L. Kinget has received a grant from “Kom op tegen Kanker” (Stand up to Cancer), the Flemish cancer society. E. Roussel received an unrestricted research grant from Ipsen. A. Verbiest has received a Fonds voor Wetenschappelijk Onderzoek Vlaanderen (Belgium) grant. B. Beuselinck received an unrestricted research grant from Bristol-Myers-Squibb and honorarium from Merck, Pfizer, Bristol-Myers-Squibb, Ipsen and Astra-Zeneca. B. Beuselinck is senior clinical investigator of the Fonds voor Wetenschappelijk Onderzoek Vlaanderen (Belgium). All other authors have nothing to disclose.
Institutional Review Board Statement
The study was conducted according to the guidelines of the Declaration of Helsinki, and approved by the Institutional Review Board/Ethics Committee of UZ/KU Leuven (protocol code S53479/S63833).
Informed Consent Statement
Informed consent was obtained from all subjects involved in the study.
Data Availability Statement
The data presented in this study are available on request from the corresponding author. The data are not publicly available due to privacy concerns.
Conflicts of Interest
The authors declare no conflict of interest.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Chandrasekar T., Klaassen Z., Goldberg H., Kulkarni G.S., Hamilton R.J., Fleshner N.E. Metastatic renal cell carcinoma: Patterns and predictors of metastases—A contemporary population-based series. Urol. Oncol. Semin. Orig. Investig. 2017;35:661.e7–661.e14. doi: 10.1016/j.urolonc.2017.06.060. [DOI] [PubMed] [Google Scholar]
- 2.Santini D., Procopio G., Porta C., Ibrahim T., Barni S., Mazzara C., Fontana A., Berruti A., Berardi R., Vincenzi B., et al. Natural history of malignant bone disease in renal cancer: Final results of an italian bone metastasis survey. PLoS ONE. 2013;8 doi: 10.1371/journal.pone.0083026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Beuselinck B., Oudard S., Rixe O., Wolter P., Blesius A., Ayllon J., Elaidi R., Schöffski P., Barrascout E., Morel A., et al. Negative impact of bone metastasis on outcome in clear-cell renal cell carcinoma treated with sunitinib. Ann. Oncol. 2011;22:794–800. doi: 10.1093/annonc/mdq554. [DOI] [PubMed] [Google Scholar]
- 4.McKay R.R., Kroeger N., Xie W., Lee J.L., Knox J.J., Bjarnason G.A., MacKenzie M.J., Wood L., Srinivas S., Vaishampayan U.N., et al. Impact of bone and liver metastases on patients with renal cell carcinoma treated with targeted therapy. Eur. Urol. 2014;65:577–584. doi: 10.1016/j.eururo.2013.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Motzer R.J., Escudier B., Bukowski R., Rini B.I., Hutson T.E., Barrios C.H., Lin X., Fly K., Matczak E., Gore M.E. Prognostic factors for survival in 1059 patients treated with sunitinib for metastatic renal cell carcinoma. Br. J. Cancer. 2013;108:2470–2477. doi: 10.1038/bjc.2013.236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Beuselinck B., Wolter P., Karadimou A., Elaidi R., Dumez H., Rogiers A., Van Cann T., Willems L., Body J.J., Berkers J., et al. Concomitant oral tyrosine kinase inhibitors and bisphosphonates in advanced renal cell carcinoma with bone metastases. Br. J. Cancer. 2012;107:1665–1671. doi: 10.1038/bjc.2012.385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vrdoljak E., Gore M., Leyman S., Szczylik C., Kharkevich G., Schöffski P., Torday L., Mardiak J., Zhang K., Sajben P., et al. Bisphosphonates in patients with renal cell carcinoma and bone metastases: A sunitinib global expanded-access trial subanalysis. Futur. Oncol. 2015;11:2831–2840. doi: 10.2217/fon.15.140. [DOI] [PubMed] [Google Scholar]
- 8.Keizman D., Ish-Shalom M., Pili R., Hammers H., Eisenberger M.A., Sinibaldi V., Boursi B., Maimon N., Gottfried M., Hayat H., et al. Bisphosphonates combined with sunitinib may improve the response rate, progression free survival and overall survival of patients with bone metastases from renal cell carcinoma. Eur. J. Cancer. 2012;48:1031–1037. doi: 10.1016/j.ejca.2012.02.050. [DOI] [PubMed] [Google Scholar]
- 9.Beuselinck B., Jean-Baptiste J., Couchy G., Job S., De Reynies A., Wolter P., Théodore C., Gravis G., Rousseau B., Albiges L., et al. RANK/OPG ratio of expression in primary clear-cell renal cell carcinoma is associated with bone metastasis and prognosis in patients treated with anti-VEGFR-TKIs. Br. J. Cancer. 2015;113:1313–1322. doi: 10.1038/bjc.2015.352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mikami S., Katsube K.I., Oya M., Ishida M., Kosaka T., Mizuno R., Mochizuki S., Ikeda T., Mukai M., Okada Y. Increased RANKL expression is related to tumour migration and metastasis of renal cell carcinomas. J. Pathol. 2009;218:530–539. doi: 10.1002/path.2567. [DOI] [PubMed] [Google Scholar]
- 11.Hong D.S., Kang Y.K., Borad M., Sachdev J., Ejadi S., Lim H.Y., Brenner A.J., Park K., Lee J.L., Kim T.Y., et al. Phase 1 study of MRX34, a liposomal miR-34a mimic, in patients with advanced solid tumours. Br. J. Cancer. 2020;122:1630–1637. doi: 10.1038/s41416-020-0802-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Van Zandwijk N., Pavlakis N., Kao S.C., Linton A., Boyer M.J., Clarke S., Huynh Y., Chrzanowska A., Fulham M.J., Bailey D.L., et al. Safety and activity of microRNA-loaded minicells in patients with recurrent malignant pleural mesothelioma: A first-in-man, phase 1, open-label, dose-escalation study. Lancet Oncol. 2017;18:1386–1396. doi: 10.1016/S1470-2045(17)30621-6. [DOI] [PubMed] [Google Scholar]
- 13.Reid G., Pel M.E., Kirschner M.B., Cheng Y.Y., Mugridge N., Weiss J., Williams M., Wright C., Edelman J.J.B., Vallely M.P., et al. Restoring expression of miR-16: A novel approach to therapy for malignant pleural mesothelioma. Ann. Oncol. 2013;24:3128–3135. doi: 10.1093/annonc/mdt412. [DOI] [PubMed] [Google Scholar]
- 14.Saliminejad K., Khorram Khorshid H.R., Soleymani Fard S., Ghaffari S.H. An overview of microRNAs: Biology, functions, therapeutics, and analysis methods. J. Cell. Physiol. 2019;234:5451–5465. doi: 10.1002/jcp.27486. [DOI] [PubMed] [Google Scholar]
- 15.García-Donas J., Beuselinck B., Inglada-Pérez L., Graña O., Schöffski P., Wozniak A., Bechter O., Apellániz-Ruiz M., Leandro-García L.J., Esteban E., et al. Deep sequencing reveals microRNAs predictive of antiangiogenic drug response. JCI Insight. 2019;1 doi: 10.1172/jci.insight.86051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Karagkouni D., Paraskevopoulou M.D., Chatzopoulos S., Vlachos I.S., Tastsoglou S., Kanellos I., Papadimitriou D., Kavakiotis I., Maniou S., Skoufos G., et al. DIANA-TarBase v8: A decade-long collection of experimentally supported miRNA-gene interactions. Nucleic Acids Res. 2018;46:D239–D245. doi: 10.1093/nar/gkx1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen Y., Wang X. MiRDB: An online database for prediction of functional microRNA targets. Nucleic Acids Res. 2020;48:D127–D131. doi: 10.1093/nar/gkz757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Avendanõ-Félix M., Fuentes-Mera L., Ramos-Payan R., Aguilar-Medina M., Pérez-Silos V., Moncada-Saucedo N., Marchat L.A., González-Barrios J.A., Ruiz-Garciá E., Astudillo-De La Vega H., et al. A novel osteomirs expression signature for osteoblast differentiation of human amniotic membrane-derived mesenchymal stem cells. Biomed Res. Int. 2019;2019 doi: 10.1155/2019/8987268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dangi-Garimella S., Yun J., Eves E.M., Newman M., Erkeland S.J., Hammond S.M., Minn A.J., Rosner M.R. Raf kinase inhibitory protein suppresses a metastasis signalling cascade involving LIN28 and let-7. EMBO J. 2009;28:347–358. doi: 10.1038/emboj.2008.294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Okuda H., Xing F., Pandey P.R., Sharma S., Watabe M., Pai S.K., Mo Y.Y., Iiizumi-Gairani M., Hirota S., Liu Y., et al. MiR-7 suppresses brain metastasis of breast cancer stem-like cells by modulating KLF4. Cancer Res. 2013;73:1434–1444. doi: 10.1158/0008-5472.CAN-12-2037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhaoa F.L., Hua G.D., Wang X.F., Zhang X.H., Zhang Y.K., Yu Z.S. Serum overexpression of microRNA-10b in patients with bone metastatic primary breast cancer. J. Int. Med. Res. 2012;40:859–866. doi: 10.1177/147323001204000304. [DOI] [PubMed] [Google Scholar]
- 22.Croset M., Goehrig D., Frackowiak A., Bonnelye E., Ansieau S., Puisieux A., Clézardin P. TWIST1 expression in breast cancer cells facilitates bone metastasis formation. J. Bone Miner. Res. 2014;29:1886–1899. doi: 10.1002/jbmr.2215. [DOI] [PubMed] [Google Scholar]
- 23.Vimalraj S., Partridge N.C., Selvamurugan N. A positive role of microRNA-15b on regulation of osteoblast differentiation. J. Cell. Physiol. 2014;229:1236–1244. doi: 10.1002/jcp.24557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ell B., Mercatali L., Ibrahim T., Campbell N., Schwarzenbach H., Pantel K., Amadori D., Kang Y. Tumor-Induced Osteoclast miRNA Changes as Regulators and Biomarkers of Osteolytic Bone Metastasis. Cancer Cell. 2013;24:542–556. doi: 10.1016/j.ccr.2013.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen N., Wu D., Li H., Liu Y., Yang H. MiR-17-3p inhibits osteoblast differentiation by downregulating Sox6 expression. FEBS Open Bio. 2020;10:2499–2506. doi: 10.1002/2211-5463.12979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vimalraj S., Miranda P.J., Ramyakrishna B., Selvamurugan N. Regulation of breast cancer and bone metastasis by MicroRNAs. Dis. Markers. 2013;35:369–387. doi: 10.1155/2013/451248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wotschofsky Z., Liep J., Meyer H.A., Jung M., Wagner I., Disch A.C., Schaser K.D., Melcher I., Kilic E., Busch J., et al. Identification of metastamirs as metastasis-associated microRNAs in clear cell renal cell carcinomas. Int. J. Biol. Sci. 2012;8:1363–1374. doi: 10.7150/ijbs.5106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Guo L., Zhu Y., Li L., Zhou S., Yin G., Yu G., Cui H. Breast cancer cell-derived exosomal miR-20a-5p promotes the proliferation and differentiation of osteoclasts by targeting SRCIN1. Cancer Med. 2019;8:5687–5701. doi: 10.1002/cam4.2454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gong Y., Xu F., Zhang L., Qian Y., Chen J., Huang H., Yu Y. MicroRNA expression signature for Satb2-induced osteogenic differentiation in bone marrow stromal cells. Mol. Cell. Biochem. 2014;387:227–239. doi: 10.1007/s11010-013-1888-z. [DOI] [PubMed] [Google Scholar]
- 30.Xu Z., Liu X., Wang H., Li J., Dai L., Li J., Dong C. Lung adenocarcinoma cell-derived exosomal miR-21 facilitates osteoclastogenesis. Gene. 2018;666:116–122. doi: 10.1016/j.gene.2018.05.008. [DOI] [PubMed] [Google Scholar]
- 31.Hou N., Guo Z., Zhao G., Jia G., Luo B., Shen X., Bai Y. Inhibition of microRNA-21-3p suppresses proliferation as well as invasion and induces apoptosis by targeting RNA-binding protein with multiple splicing through Smad4/extra cellular signal-regulated protein kinase signalling pathway in human colorectal can. Clin. Exp. Pharmacol. Physiol. 2018;45:729–741. doi: 10.1111/1440-1681.12931. [DOI] [PubMed] [Google Scholar]
- 32.Dai Y., Zheng C., Li H. Inhibition of miR-23a-3p promotes osteoblast proliferation and differentiation. J. Cell. Biochem. 2019 doi: 10.1002/jcb.29497. [DOI] [PubMed] [Google Scholar]
- 33.Quan J., Pan X., Li Y., Hu Y., Tao L., Li Z., Zhao L., Wang J., Li H., Lai Y., et al. MiR-23a-3p acts as an oncogene and potential prognostic biomarker by targeting PNRC2 in RCC. Biomed. Pharmacother. 2019;110:656–666. doi: 10.1016/j.biopha.2018.11.065. [DOI] [PubMed] [Google Scholar]
- 34.Hassan M.Q., Gordon J.A.R., Beloti M.M., Croce C.M., Van Wijnen A.J., Stein J.L., Stein G.S., Lian J.B. A network connecting Runx2, SATB2, and the miR-23a∼27a∼24-2 cluster regulates the osteoblast differentiation program. Proc. Natl. Acad. Sci. USA. 2010;107:19879–19884. doi: 10.1073/pnas.1007698107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhu Z., Wen Y., Xuan C., Chen Q., Xiang Q., Wang J., Liu Y., Luo L., Zhao S., Deng Y., et al. Identifying the key genes and microRNAs in prostate cancer bone metastasis by bioinformatics analysis. FEBS Open Bio. 2020;10:674–688. doi: 10.1002/2211-5463.12805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Croset M., Pantano F., Kan C.W.S., Bonnelye E., Descotes F., Alix-Panabieres C., Lecellier C.H., Bachelier R., Allioli N., Hong S.S., et al. miRNA-30 family members inhibit breast cancer invasion, osteomimicry, and bone destruction by directly targeting multiple bone metastasis–associated genes. Cancer Res. 2018;78:5259–5273. doi: 10.1158/0008-5472.CAN-17-3058. [DOI] [PubMed] [Google Scholar]
- 37.Kuo P.L., Liao S.H., Hung J.Y., Huang M.S., Hsu Y.L. MicroRNA-33a functions as a bone metastasis suppressor in lung cancer by targeting parathyroid hormone related protein. Biochim. Biophys. Acta Gen. Subj. 2013;1830:3756–3766. doi: 10.1016/j.bbagen.2013.02.022. [DOI] [PubMed] [Google Scholar]
- 38.Chen W.Y., Liu S.Y., Chang Y.S., Yin J.J., Yeh H.L., Mouhieddine T.H., Hadadeh O., Abou-Kheir W., Liu Y.N. MicroRNA-34a regulates WNT/TCF7 signaling and inhibits bone metastasis in Ras-activated prostate cancer. Oncotarget. 2015;6:441–457. doi: 10.18632/oncotarget.2690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gu J., Wang G., Liu H., Xiong C. SATB2 targeted by methylated miR-34c-5p suppresses proliferation and metastasis attenuating the epithelial-mesenchymal transition in colorectal cancer. Cell Prolif. 2018;51:e12455. doi: 10.1111/cpr.12455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Clézardin P. Pathophysiology of bone metastases from solid malignancies. Jt. Bone Spine. 2017;84:677–684. doi: 10.1016/j.jbspin.2017.05.006. [DOI] [PubMed] [Google Scholar]
- 41.Tang Y., Pan J., Huang S., Peng X., Zou X., Luo Y., Ren D., Zhang X., Li R., He P., et al. Downregulation of miR-133a-3p promotes prostate cancer bone metastasis via activating PI3K/AKT signaling. J. Exp. Clin. Cancer Res. 2018;37:160. doi: 10.1186/s13046-018-0813-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Taipaleenmäki H., Browne G., Akech J., Zustin J., Van Wijnen A.J., Stein J.L., Hesse E., Stein G.S., Lian J.B. Targeting of Runx2 by miR-135 and miR-203 impairs progression of breast cancer and metastatic bone disease. Cancer Res. 2015;75:1433–1444. doi: 10.1158/0008-5472.CAN-14-1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Eskildsen T., Taipaleenmäki H., Stenvang J., Abdallah B.M., Ditzel N., Nossent A.Y., Bak M., Kauppinen S., Kassem M. MicroRNA-138 regulates osteogenic differentiation of human stromal (mesenchymal) stem cells in vivo. Proc. Natl. Acad. Sci. USA. 2011;108:6139–6144. doi: 10.1073/pnas.1016758108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang Y., Wang K., Hu Z., Zhou H., Zhang L., Wang H., Li G., Zhang S., Cao X., Shi F. MicroRNA-139-3p regulates osteoblast differentiation and apoptosis by targeting ELK1 and interacting with long noncoding RNA ODSM. Cell Death Dis. 2018;9:1–16. doi: 10.1038/s41419-018-1153-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhang W., Xu J., Wang K., Tang X., He J. MIR-139-3p suppresses the invasion and migration properties of breast cancer cells by targeting RAB1A. Oncol. Rep. 2019;42:1699–1708. doi: 10.1007/s12094-019-02103-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Xu S., Yang F., Liu R., Li X., Fan H., Liu J., Wei S., Chen G., Chen J., Da Y. Serum microRNA-139-5p is downregulated in lung cancer patients with lytic bone metastasis. Oncol. Rep. 2018;39:2376–2384. doi: 10.3892/or.2018.6316. [DOI] [PubMed] [Google Scholar]
- 47.Ell B., Kang Y. MicroRNAs as regulators of bone homeostasis and bone metastasis. Bonekey Rep. 2014;3:549. doi: 10.1038/bonekey.2014.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shabani P., Izadpanah S., Aghebati-Maleki A., Baghbani E., Baghbanzadeh A., Fotouhi A., Bakhshinejad B., Aghebati-Maleki L., Baradaran B. Role of miR-142 in the pathogenesis of osteosarcoma and its potential as therapeutic approach. J. Cell. Biochem. 2019;120:4783–4793. doi: 10.1002/jcb.27857. [DOI] [PubMed] [Google Scholar]
- 49.Fordham J.B., Guilfoyle K., Naqvi A.R., Nares S. MiR-142-3p is a RANKL-dependent inducer of cell death in osteoclasts. Sci. Rep. 2016;6:1–9. doi: 10.1038/srep24980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Peng X., Guo W., Liu T., Wang X., Tu X., Xiong D., Chen S., Lai Y., Du H., Chen G., et al. Identification of miRs-143 and -145 that Is Associated with Bone Metastasis of Prostate Cancer and Involved in the Regulation of EMT. PLoS ONE. 2011;6:e20341. doi: 10.1371/journal.pone.0020341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Fan X., Chen X., Deng W., Zhong G., Cai Q., Lin T. Up-regulated microRNA-143 in cancer stem cells differentiation promotes prostate cancer cells metastasis by modulating FNDC3B expression. BMC Cancer. 2013;13:1–11. doi: 10.1186/1471-2407-13-61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Guo W., Ren D., Chen X., Tu X., Huang S., Wang M., Song L., Zou X., Peng X. HEF1 promotes epithelial mesenchymal transition and bone invasion in prostate cancer under the regulation of microRNA-145. J. Cell. Biochem. 2013;114:1606–1615. doi: 10.1002/jcb.24502. [DOI] [PubMed] [Google Scholar]
- 53.Banerjee S., Kalyani Yabalooru S.R., Karunagaran D. Identification of mRNA and non-coding RNA hubs using network analysis in organ tropism regulated triple negative breast cancer metastasis. Comput. Biol. Med. 2020;127:104076. doi: 10.1016/j.compbiomed.2020.104076. [DOI] [PubMed] [Google Scholar]
- 54.Eguchi T., Watanabe K., Hara E.S., Ono M., Kuboki T., Calderwood S.K. OstemiR: A Novel Panel of MicroRNA Biomarkers in Osteoblastic and Osteocytic Differentiation from Mesencymal Stem Cells. PLoS ONE. 2013;8 doi: 10.1371/journal.pone.0058796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhang J., Ye Y., Chang D.W., Lin S.H., Huang M., Tannir N.M., Matin S., Karam J.A., Wood C.G., Chen Z.N., et al. Global and Targeted miRNA Expression Profiling in Clear Cell Renal Cell Carcinoma Tissues Potentially Links miR-155-5p and miR-210-3p to both Tumorigenesis and Recurrence. Am. J. Pathol. 2018;188:2487–2496. doi: 10.1016/j.ajpath.2018.07.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chen Q.Y., Des Marais T., Costa M. Deregulation of SATB2 in carcinogenesis with emphasis on miRNA-mediated control. Carcinogenesis. 2019;40:393–402. doi: 10.1093/carcin/bgz020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kim K.M., Park S.J., Jung S.H., Kim E.J., Jogeswar G., Ajita J., Rhee Y., Kim C.H., Lim S.K. MiR-182 is a negative regulator of osteoblast proliferation, differentiation, and skeletogenesis through targeting FoxO1. J. Bone Miner. Res. 2012;27:1669–1679. doi: 10.1002/jbmr.1604. [DOI] [PubMed] [Google Scholar]
- 58.Mock K., Preca B.T., Brummer T., Brabletz S., Stemmler M.P., Brabletz T. The EMT-activator ZEB1 induces bone metastasis associated genes including BMP-inhibitors. Oncotarget. 2015;6:14399–14412. doi: 10.18632/oncotarget.3882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Pollari S., Leivonen S.K., Perälä M., Fey V., Käkönen S.M., Kallioniemi O. Identification of microRNAs inhibiting TGF-β-induced IL-11 production in bone metastatic breast cancer cells. PLoS ONE. 2012;7:e37361. doi: 10.1371/journal.pone.0037361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ren D., Yang Q., Dai Y., Guo W., Du H., Song L., Peng X. Oncogenic miR-210-3p promotes prostate cancer cell EMT and bone metastasis via NF-ΚB signaling pathway. Mol. Cancer. 2017;16:1–16. doi: 10.1186/s12943-017-0688-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Liu J., Li D., Dang L., Liang C., Guo B., Lu C., He X., Cheung H.Y.S., He B., Liu B., et al. Osteoclastic miR-214 targets TRAF3 to contribute to osteolytic bone metastasis of breast cancer. Sci. Rep. 2017;7:1–13. doi: 10.1038/srep40487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hassan M.Q., Maeda Y., Taipaleenmaki H., Zhang W., Jafferji M., Gordon J.A.R., Li Z., Croce C.M., Van Wijnen A.J., Stein J.L., et al. miR-218 directs a Wnt signaling circuit to promote differentiation of osteoblasts and osteomimicry of metastatic cancer cells. J. Biol. Chem. 2012;287:42084–42092. doi: 10.1074/jbc.M112.377515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Valencia K., Martín-Fernández M., Zandueta C., Ormazábal C., Martínez-Canarias S., Bandrés E., de la Piedra C., Lecanda F. MiR-326 associates with biochemical markers of bone turnover in lung cancer bone metastasis. Bone. 2013;52:532–539. doi: 10.1016/j.bone.2012.10.033. [DOI] [PubMed] [Google Scholar]
- 64.Zhang L., Tang Y., Zhu X., Tu T., Sui L., Han Q., Yu L., Meng S., Zheng L., Valverde P., et al. Overexpression of MiR-335-5p Promotes Bone Formation and Regeneration in Mice. J. Bone Miner. Res. 2017;32:2466–2475. doi: 10.1002/jbmr.3230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Gururajan M., Josson S., Chu G.C.Y., Lu C.L., Lu Y.T., Haga C.L., Zhau H.E., Liu C., Lichterman J., Duan P., et al. MiR-154∗ and miR-379 in the DLK1-DIO3 MicroRNA mega-cluster regulate epithelial to mesenchymal transition and bone metastasis of prostate cancer. Clin. Cancer Res. 2014;20:6559–6569. doi: 10.1158/1078-0432.CCR-14-1784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kureel J., Dixit M., Tyagi A.M., Mansoori M.N., Srivastava K., Raghuvanshi A., Maurya R., Trivedi R., Goel A., Singh D. MiR-542-3p suppresses osteoblast cell proliferation and differentiation, targets BMP-7 signaling and inhibits bone formation. Cell Death Dis. 2014;5:e1050. doi: 10.1038/cddis.2014.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.He R.Q., Li X.J., Liang L., Xie Y., Luo D.Z., Ma J., Peng Z.G., Hu X.H., Chen G. The suppressive role of miR-542-5p in NSCLC: The evidence from clinical data and in vivo validation using a chick chorioallantoic membrane model. BMC Cancer. 2017;17:1–15. doi: 10.1186/s12885-017-3646-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Fu Q., Liu X., Liu Y., Yang J., Lv G., Dong S. MicroRNA-335 and -543 suppress bone metastasis in prostate cancer via targeting endothelial nitric oxide synthase. Int. J. Mol. Med. 2015;36:1417–1425. doi: 10.3892/ijmm.2015.2355. [DOI] [PubMed] [Google Scholar]
- 69.Sosa M.S., Bragado P., Aguirre-Ghiso J.A. Mechanisms of disseminated cancer cell dormancy: An awakening field. Nat. Rev. Cancer. 2014;14:611–622. doi: 10.1038/nrc3793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kobayashi A., Okuda H., Xing F., Pandey P.R., Watabe M., Hirota S., Pai S.K., Liu W., Fukuda K., Chambers C., et al. Bone morphogenetic protein 7 in dormancy and metastasis of prostate cancer stem-like cells in bone. J. Exp. Med. 2011;208:2641–2655. doi: 10.1084/jem.20110840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Sugatani T., Vacher J., Hruska K.A. A microRNA expression signature of osteoclastogenesis. Blood. 2011;117:3648–3657. doi: 10.1182/blood-2010-10-311415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Guo C., Xiong D., Yao X., Gu W., Zhang H., Yang B., Peng B., Liu M., Zheng J. Decreased SATB2 expression is associated with metastasis and poor prognosis in human clear cell renal cell carcinoma. Int. J. Clin. Exp. Pathol. 2015;8:3710–3718. [PMC free article] [PubMed] [Google Scholar]
- 73.Petersen M., Pardali E., Van Der Horst G., Cheung H., Van Den Hoogen C., Van Der Pluijm G., Ten Dijke P. Smad2 and Smad3 have opposing roles in breast cancer bone metastasis by differentially affecting tumor angiogenesis. Oncogene. 2010;29:1351–1361. doi: 10.1038/onc.2009.426. [DOI] [PubMed] [Google Scholar]
- 74.Deckers M., Van Dinther M., Buijs J., Que I., Löwik C., Van Der Pluijm G., Ten Dijke P. The tumor suppressor Smad4 is required for transforming growth factor β-induced epithelial to mesenchymal transition and bone metastasis of breast cancer cells. Cancer Res. 2006;66:2202–2209. doi: 10.1158/0008-5472.CAN-05-3560. [DOI] [PubMed] [Google Scholar]
- 75.Pang X., Gong K., Zhang X., Wu S., Cui Y., Qian B.Z. Osteopontin as a multifaceted driver of bone metastasis and drug resistance. Pharmacol. Res. 2019;144:235–244. doi: 10.1016/j.phrs.2019.04.030. [DOI] [PubMed] [Google Scholar]
- 76.Morein D., Erlichman N., Ben-Baruch A. Beyond Cell Motility: The Expanding Roles of Chemokines and Their Receptors in Malignancy. Front. Immunol. 2020;11:952. doi: 10.3389/fimmu.2020.00952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Zlotnik A., Burkhardt A.M., Homey B. Homeostatic chemokine receptors and organ-specific metastasis. Nat. Rev. Immunol. 2011;11:597–606. doi: 10.1038/nri3049. [DOI] [PubMed] [Google Scholar]
- 78.Hsu Y.L., Hou M.F., Kuo P.L., Huang Y.F., Tsai E.M. Breast tumor-associated osteoblast-derived CXCL5 increases cancer progression by ERK/MSK1/Elk-1/Snail signaling pathway. Oncogene. 2013;32:4436–4447. doi: 10.1038/onc.2012.444. [DOI] [PubMed] [Google Scholar]
- 79.Li X.Q., Lu J.T., Tan C.C., Wang Q.S., Feng Y.M. RUNX2 promotes breast cancer bone metastasis by increasing integrin α5-mediated colonization. Cancer Lett. 2016;380:78–86. doi: 10.1016/j.canlet.2016.06.007. [DOI] [PubMed] [Google Scholar]
- 80.Pécheur I., Peyruchaud O., Serre C.M., Guglielmi J., Voland C., Bourre F., Margue C., Cohen-Solal M., Buffet A., Kieffer N., et al. Integrin alpha(v)beta3 expression confers on tumor cells a greater propensity to metastasize to bone. FASEB J. 2002;16:1266–1268. doi: 10.1096/fj.01-0911fje. [DOI] [PubMed] [Google Scholar]
- 81.Katsuno Y., Hanyu A., Kanda H., Ishikawa Y., Akiyama F., Iwase T., Ogata E., Ehata S., Miyazono K., Imamura T. Bone morphogenetic protein signaling enhances invasion and bone metastasis of breast cancer cells through Smad pathway. Oncogene. 2008;27:6322–6333. doi: 10.1038/onc.2008.232. [DOI] [PubMed] [Google Scholar]
- 82.Croset M., Kan C., Clézardin P. Tumour-derived miRNAs and bone metastasis. Bonekey Rep. 2015;4 doi: 10.1038/bonekey.2015.56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Deligiorgi M.V., Panayiotidis M.I., Griniatsos J., Trafalis D.T. Harnessing the versatile role of OPG in bone oncology: Counterbalancing RANKL and TRAIL signaling and beyond. Clin. Exp. Metastasis. 2020;37:13–30. doi: 10.1007/s10585-019-09997-8. [DOI] [PubMed] [Google Scholar]
- 84.Heng D.Y.C., Xie W., Regan M.M., Warren M.A., Golshayan A.R., Sahi C., Eigl B.J., Ruether J.D., Cheng T., North S., et al. Prognostic factors for overall survival in patients with metastatic renal cell carcinoma treated with vascular endothelial growth factor-targeted agents: Results from a large, multicenter study. J. Clin. Oncol. 2009;27:5794–5799. doi: 10.1200/JCO.2008.21.4809. [DOI] [PubMed] [Google Scholar]
- 85.Huang J., Zhao L., Xing L., Chen D. MicroRNA-204 regulates Runx2 protein expression and mesenchymal progenitor cell differentiation. Stem Cells. 2010;28:357–364. doi: 10.1002/stem.288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Wu J., Liu T., Sun L., Zhang S., Dong G. Long noncoding RNA SNHG4 promotes renal cell carcinoma tumorigenesis and invasion by acting as ceRNA to sponge miR-204-5p and upregulate RUNX2. Cancer Cell Int. 2020;20 doi: 10.1186/s12935-020-01606-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Shuai F., Wang B., Dong S. MicroRNA-204 inhibits the growth and motility of colorectal cancer cells by downregulation of CXCL8. Oncol. Res. 2018;26:1295–1305. doi: 10.3727/096504018X15172747209020. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 88.Garros R.F., Paul R., Connolly M., Lewis A., Garfield B.E., Natanek S.A., Bloch S., Mouly V., Griffiths M.J., Polkey M.I., et al. MicroRNA-542 promotes mitochondrial dysfunction and SMAD activity and is elevated in intensive care unit–acquired weakness. Am. J. Respir. Crit. Care Med. 2017;196:1422–1433. doi: 10.1164/rccm.201701-0101OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Chen N., Sui B.D., Hu C.H., Cao J., Zheng C.X., Hou R., Yang Z.K., Zhao P., Chen Q., Yang Q.J., et al. MicroRNA-21 contributes to orthodontic tooth movement. J. Dent. Res. 2016;95:1425–1433. doi: 10.1177/0022034516657043. [DOI] [PubMed] [Google Scholar]
- 90.Qin W., Zhao B., Shi Y., Yao C., Jin L., Jin Y. BMPRII is a direct target of miR-21. Acta Biochim. Biophys. Sin. 2009;41:618–623. doi: 10.1093/abbs/gmp049. [DOI] [PubMed] [Google Scholar]
- 91.Loboda A., Sobczak M., Jozkowicz A., Dulak J. TGF-β 1/Smads and miR-21 in Renal Fibrosis and Inflammation. Mediators Inflamm. 2016;2016 doi: 10.1155/2016/8319283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Wei J., Shi Y., Zheng L., Zhou B., Inose H., Wang J., Guo X.E., Grosschedl R., Karsenty G. miR-34s inhibit osteoblast proliferation and differentiation in the mouse by targeting SATB2. J. Cell Biol. 2012;197:509–521. doi: 10.1083/jcb.201201057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Zhang J.F., Fu W.M., He M.L., Xie W.D., Lv Q., Wan G., Li G., Wang H., Lu G., Hu X., et al. MiRNA-20a promotes osteogenic differentiation of human mesenchymal stem cells by co-regulating BMP signaling. RNA Biol. 2011;8 doi: 10.4161/rna.8.5.16043. [DOI] [PubMed] [Google Scholar]
- 94.Yang M.H., Yu J., Jiang D.M., Li W.L., Wang S., Ding Y.Q. MicroRNA-182 targets special AT-rich sequence-binding protein 2 to promote colorectal cancer proliferation and metastasis. J. Transl. Med. 2014;12:1–11. doi: 10.1186/1479-5876-12-109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Choueiri T.K., Garcia J.A., Elson P., Khasawneh M., Usman S., Golshayan A.R., Baz R.C., Wood L., Rini B.I., Bukowski R.M. Clinical factors associated with outcome in patients with metastatic clear-cell renal cell carcinoma treated with vascular endothelial growth factor-targeted therapy. Cancer. 2007;110:543–550. doi: 10.1002/cncr.22827. [DOI] [PubMed] [Google Scholar]
- 96.Santoni M., Conti A., Procopio G., Porta C., Ibrahim T., Barni S., Guida F.M., Fontana A., Berruti A., Berardi R., et al. Bone metastases in patients with metastatic renal cell carcinoma: Are they always associated with poor prognosis? J. Exp. Clin. Cancer Res. 2015;34:10. doi: 10.1186/s13046-015-0122-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Wu T., Zhou H., Hong Y., Li J., Jiang X., Huang H. miR-30 family members negatively regulate osteoblast differentiation. J. Biol. Chem. 2012;287:7503–7511. doi: 10.1074/jbc.M111.292722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Zhang L., Li G., Wang K., Wang Y., Dong J., Wang H., Xu L., Shi F., Cao X., Hu Z., et al. MiR-30 family members inhibit osteoblast differentiation by suppressing Runx2 under unloading conditions in MC3T3-E1 cells. Biochem. Biophys. Res. Commun. 2020;522:164–170. doi: 10.1016/j.bbrc.2019.11.057. [DOI] [PubMed] [Google Scholar]
- 99.Yi J., Liu D., Xiao J. LncRNA MALAT1 sponges miR-30 to promote osteoblast differentiation of adipose-derived mesenchymal stem cells by promotion of Runx2 expression. Cell Tissue Res. 2019;376:113–121. doi: 10.1007/s00441-018-2963-2. [DOI] [PubMed] [Google Scholar]
- 100.Yu F., Deng H., Yao H., Liu Q., Su F., Song E. Mir-30 reduction maintains self-renewal and inhibits apoptosis in breast tumor-initiating cells. Oncogene. 2010;29:4194–4204. doi: 10.1038/onc.2010.167. [DOI] [PubMed] [Google Scholar]
- 101.Zhao J.J., Lin J., Zhu D., Wang X., Brooks D., Chen M., Chu Z.B., Takada K., Ciccarelli B., Admin S., et al. MiR-30-5p functions as a tumor suppressor and novel therapeutic tool by targeting the oncogenic Wnt/ b-Catenin/BCL9 pathway. Cancer Res. 2014;74:1801–1813. doi: 10.1158/0008-5472.CAN-13-3311-T. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Bray I., Tivnan A., Bryan K., Foley N.H., Watters K.M., Tracey L., Davidoff A.M., Stallings R.L. MicroRNA-542-5p as a Novel Tumor Suppressor in Neuroblastoma. Cancer Lett. 2011;303:56–64. doi: 10.1016/j.canlet.2011.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Li Q., Song S., Ni G., Li Y., Wang X. Serum miR-542-3p as a prognostic biomarker in osteosarcoma. Cancer Biomark. 2018;21:521–526. doi: 10.3233/CBM-170255. [DOI] [PubMed] [Google Scholar]
- 104.Yuan L., Yuan P., Yuan H., Wang Z., Run Z., Chen G., Zhao P., Xu B. miR-542-3p inhibits colorectal cancer cell proliferation, migration and invasion by targeting OTUB1. Am. J. Cancer Res. 2017;7:159–172. [PMC free article] [PubMed] [Google Scholar]
- 105.Wu H.X., Wang G.M., Lu X., Zhang L. miR-542-3p targets sphingosine-1-phosphate receptor 1 and regulates cell proliferation and invasion of breast cancer cells. Eur. Rev. Med. Pharmacol. Sci. 2017;21:108–114. [PubMed] [Google Scholar]
- 106.Tao J., Liu Z., Wang Y., Wang L., Yao B., Li Q., Wang C., Tu K., Liu Q. MiR-542-3p inhibits metastasis and epithelial-mesenchymal transition of hepatocellular carcinoma by targeting UBE3C. Biomed. Pharmacother. 2017;93:420–428. doi: 10.1016/j.biopha.2017.06.070. [DOI] [PubMed] [Google Scholar]
- 107.Cheng D.D., Yu T., Hu T., Yao M., Fan C.Y., Yang Q.C. MiR-542-5p is a negative prognostic factor and promotes osteosarcoma tumorigenesis by targeting HUWE1. Oncotarget. 2015;6:42761–42772. doi: 10.18632/oncotarget.6199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Okada R., Goto Y., Yamada Y., Kato M., Asai S., Moriya S., Ichikawa T., Seki N. Regulation of oncogenic targets by the tumor-suppressive mir-139 duplex (Mir-139-5p and mir-139-3p) in renal cell carcinoma. Biomedicines. 2020;8:599. doi: 10.3390/biomedicines8120599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Yonemori M., Seki N., Yoshino H., Matsushita R., Miyamoto K., Nakagawa M., Enokida H. Dual tumor-suppressors miR-139-5p and miR-139-3p targeting matrix metalloprotease 11 in bladder cancer. Cancer Sci. 2016;107:1233–1242. doi: 10.1111/cas.13002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Zhang L., Dong Y., Zhu N., Tsoi H., Zhao Z., Wu C.W., Wang K., Zheng S., Ng S.S.M., Chan F.K.L., et al. MicroRNA-139-5p exerts tumor suppressor function by targeting NOTCH1 in colorectal cancer. Mol. Cancer. 2014;13 doi: 10.1186/1476-4598-13-124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Dai S., Wang X., Li X., Cao Y. MicroRNA-139-5p acts as a tumor suppressor by targeting ELTD1 and regulating cell cycle in glioblastoma multiforme. Biochem. Biophys. Res. Commun. 2015;467:204–210. doi: 10.1016/j.bbrc.2015.10.006. [DOI] [PubMed] [Google Scholar]
- 112.Zou Z.C., Dai M., Huang Z.Y., Lu Y., Xie H.P., Li Y.F., Li Y., Tan Y., Wang F.L. MicroRNA-139-3p suppresses tumor growth and metastasis in hepatocellular carcinoma by repressing ANXA2R. Oncol. Res. 2018;26:1391–1399. doi: 10.3727/096504018X15178798885361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Wang Y., Fang Q., Tian L., Yuan Z., Tian L., Zhou Z. Expression and Regulatory Network Analysis of MiR-139-3p, a New Potential Serum Biomarker for Esophageal Squamous Cell Carcinoma Based on Bioinformatics Analysis. Technol. Cancer Res. Treat. 2020;19 doi: 10.1177/1533033820920967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Zhang L., Liu M., Liu J., Li X., Yang M., Su B., Lin Y. 27-Hydroxycholesterol enhanced osteoclastogenesis in lung adenocarcinoma microenvironment. J. Cell. Physiol. 2019;234:12692–12700. doi: 10.1002/jcp.27883. [DOI] [PubMed] [Google Scholar]
- 115.Jedroszka D., Orzechowska M., Bednarek A.K. Predictive values of Notch signalling in renal carcinoma. Arch. Med. Sci. 2017;13:1249–1254. doi: 10.5114/aoms.2017.65649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Shu X., Hildebrandt M.A., Gu J., Tannir N.M., Matin S.F., Karam J.A., Wood C.G., Wu X. MicroRNA profiling in clear cell renal cell carcinoma tissues potentially links tumorigenesis and recurrence with obesity. Br. J. Cancer. 2017;116:77–84. doi: 10.1038/bjc.2016.392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Osanto S., Qin Y., Buermans H.P., Berkers J., Lerut E., Goeman J.J., van Poppel H. Genome-wide microRNA expression analysis of clear cell renal cell carcinoma by next generation deep sequencing. PLoS ONE. 2012;7:38298. doi: 10.1371/journal.pone.0038298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Jung M., Mollenkopf H.J., Grimm C., Wagner I., Albrecht M., Waller T., Pilarsky C., Johannsen M., Stephan C., Lehrach H., et al. MicroRNA profiling of clear cell renal cell cancer identifies a robust signature to define renal malignancy. J. Cell. Mol. Med. 2009;13:3918–3928. doi: 10.1111/j.1582-4934.2009.00705.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Verbiest A., van Hoef V., Rodriguez-Antona C., García-Donas J., Graña-Castro O., Albersen M., Baldewijns M., Laenen A., Roussel E., Schöffski P., et al. MicroRNA expression profiles in molecular subtypes of clear-cell renal cell carcinoma are associated with clinical outcome and repression of specific mRNA targets. PLoS ONE. 2020;15 doi: 10.1371/journal.pone.0238809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Gao W., Wu Y., He X., Zhang C., Zhu M., Chen B., Liu Q., Qu X., Li W., Wen S., et al. MicroRNA-204-5p inhibits invasion and metastasis of laryngeal squamous cell carcinoma by suppressing forkhead box C1. J. Cancer. 2017;8:2356–2368. doi: 10.7150/jca.19470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Hong B.S., Ryu H.S., Kim N., Kim J., Lee E., Moon H., Kim K.H., Jin M.S., Kwon N.H., Kim S., et al. Tumor suppressor miRNA-204-5p regulates growth, metastasis, and immune microenvironment remodeling in breast cancer. Cancer Res. 2019;79:1520–1534. doi: 10.1158/0008-5472.CAN-18-0891. [DOI] [PubMed] [Google Scholar]
- 122.Wang X., Li F., Zhou X. miR-204-5p regulates cell proliferation and metastasis through inhibiting CXCR4 expression in OSCC. Biomed. Pharmacother. 2016;82:202–207. doi: 10.1016/j.biopha.2016.04.060. [DOI] [PubMed] [Google Scholar]
- 123.Jiang X., Zhang Z., Peng T., Wang G., Xu Q., Li G. MiR-204 inhibits the osteogenic differentiation of mesenchymal stem cells by targeting bone morphogenetic protein 2. Mol. Med. Rep. 2020;21:43–50. doi: 10.3892/mmr.2019.10791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Yin L., Xu G., Zhu Y., Wang Y. Expression of miR-23a and miR-135 and tumor markers in gastric cancer patients and the significance in diagnosis. Oncol. Lett. 2019;18:5853–5858. doi: 10.3892/ol.2019.10943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Karimi N., Ali Hosseinpour Feizi M., Safaralizadeh R., Hashemzadeh S., Baradaran B., Shokouhi B., Teimourian S. Serum overexpression of miR-301a and miR-23a in patients with colorectal cancer. J. Chinese Med. Assoc. 2019;82:215–220. doi: 10.1097/JCMA.0000000000000031. [DOI] [PubMed] [Google Scholar]
- 126.Wu Q., Lu Z., Li H., Lu J., Guo L., Ge Q. Next-generation sequencing of microRNAs for breast cancer detection. J. Biomed. Biotechnol. 2011;2011 doi: 10.1155/2011/597145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Li X., Liu X., Xu W., Zhou P., Gao P., Jiang S., Lobie P.E., Zhu T. c-MYC-regulated miR-23a/24-2/27a Cluster Promotes Mammary Carcinoma Cell Invasion and Hepatic Metastasis by Targeting Sprouty2 * and the. J. Biol. Chem. 2013 doi: 10.1074/jbc.M113.478560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Li T., Li H., Wang Y., Li T., Fan J., Xiao K., Zhao R.C., Weng X. MicroRNA-23a inhibits osteogenic differentiation of human bone marrow-derived mesenchymal stem cells by targeting LRP5. Int. J. Biochem. Cell Biol. 2016;72:55–62. doi: 10.1016/j.biocel.2016.01.004. [DOI] [PubMed] [Google Scholar]
- 129.Moody L., Dvoretskiy S., An R., Mantha S., Pan Y.-X. The Efficacy of miR-20a as a Diagnostic and Prognostic Biomarker for Colorectal Cancer: A Systematic Review and Meta-Analysis. Cancers. 2019;11:1111. doi: 10.3390/cancers11081111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Yang R., Fu Y., Zeng Y., Xiang M., Yin Y., Li L., Xu H., Zhong J., Zeng X. Serum miR-20a is a promising biomarker for gastric cancer. Biomed. Rep. 2017;6:429–434. doi: 10.3892/br.2017.862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Jung K., Lein M., Stephan C., Von Hösslin K., Semjonow A., Sinha P., Loening S.A., Schnorr D. Comparison of 10 serum bone turnover markers in prostate carcinoma patients with bone metastatic spread: Diagnostic and prognostic implications. Int. J. Cancer. 2004;111:783–791. doi: 10.1002/ijc.20314. [DOI] [PubMed] [Google Scholar]
- 132.Zhong X.L., Yan X., Yang X.K., Xiu H., Zhao M., Wang X.N., Liu J.X. MiR-20a acted as a ceRNA of lncRNA PTENPL and promoted bladder cancer cell proliferation and migration by regulating PDCD4. Eur. Rev. Med. Pharmacol. Sci. 2020;24:2955–2964. doi: 10.26355/eurrev_202003_20660. [DOI] [PubMed] [Google Scholar]
- 133.Jiang Z., Li L., Hou Z., Liu W., Wang H., Zhou T., Li Y., Chen S. LncRNA HAND2-AS1 inhibits 5-fluorouracil resistance by modulating miR-20a/PDCD4 axis in colorectal cancer. Cell. Signal. 2020;66:109483. doi: 10.1016/j.cellsig.2019.109483. [DOI] [PubMed] [Google Scholar]
- 134.Chen Q., Liu T., Bao Y., Zhao T., Wang J., Wang H., Wang A., Gan X., Wu Z., Wang L. CircRNA cRAPGEF5 inhibits the growth and metastasis of renal cell carcinoma via the miR-27a-3p/TXNIP pathway. Cancer Lett. 2020;469:68–77. doi: 10.1016/j.canlet.2019.10.017. [DOI] [PubMed] [Google Scholar]
- 135.Song E.L., Xing L., Wang L., Song W.T., Li D.B., Wang Y., Gu Y.W., Liu M.M., Ni W.J., Zhang P., et al. LncRNA ADAMTS9-AS2 inhibits cell proliferation and decreases chemoresistance in clear cell renal cell carcinoma via the miR-27a-3p/FOXO1 axis. Aging. 2019;11:5705–5725. doi: 10.18632/aging.102154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Liu J., Li M., Liu X., Liu F., Zhu J. MiR-27a-3p promotes the malignant phenotypes of osteosarcoma by targeting ten-eleven translocation 1. Int. J. Oncol. 2018;52:1295–1304. doi: 10.3892/ijo.2018.4275. [DOI] [PubMed] [Google Scholar]
- 137.Liang J., Tang J., Shi H., Li H., Zhen T., Duan J., Kang L., Zhang F., Dong Y., Han A. miR-27a-3p targeting RXRa promotes colorectal cancer progression by activating Wnt/β-catenin pathway. Oncotarget. 2017;8:82991–83008. doi: 10.18632/oncotarget.19635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Su C., Huang D.P., Liu J.W., Liu W.Y., Cao Y.O. miR-27a-3p regulates proliferation and apoptosis of colon cancer cells by potentially targeting BTG1. Oncol. Lett. 2019;18:2825–2834. doi: 10.3892/ol.2019.10629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Rao X., Wan L., Jie Z., Zhu X., Yin J., Cao H. Upregulated miR-27a-3p indicates a poor prognosis in pancreatic carcinoma patients and promotes the angiogenesis and migration by epigenetic silencing of GATA6 and activating VEGFA/VEGFR2 signaling pathway. Onco. Targets. Ther. 2019;12:11241–11254. doi: 10.2147/OTT.S220621. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 140.Zhou L., Liang X., Zhang L., Yang L., Nagao N., Wu H., Liu C., Lin S., Cai G., Liu J. MiR-27a-3p functions as an oncogene in gastric cancer by targeting BTG2. Oncotarget. 2016;7:51943–51954. doi: 10.18632/oncotarget.10460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Yan X., Yu H., Liu Y., Hou J., Yang Q., Zhao Y. miR-27a-3p Functions as a Tumor Suppressor and Regulates Non-Small Cell Lung Cancer Cell Proliferation via Targeting HOXB8. Technol. Cancer Res. Treat. 2019;18 doi: 10.1177/1533033819861971. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 142.Zhao N., Sun H., Sun B., Zhu D., Zhao X., Wang Y., Gu Q., Dong X., Liu F., Zhang Y., et al. miR-27a-3p suppresses tumor metastasis and VM by down-regulating VE-cadherin expression and inhibiting EMT: An essential role for Twist-1 in HCC. Sci. Rep. 2016;6 doi: 10.1038/srep23091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Liu X., Pan B., Sun L., Chen X., Zeng K., Hu X., Xu T., Xu M., Wang S. Circulating exosomal miR-27a and miR-130a act as novel diagnostic and prognostic biomarkers of colorectal cancer. Cancer Epidemiol. Biomarkers Prev. 2018;27:746–754. doi: 10.1158/1055-9965.EPI-18-0067. [DOI] [PubMed] [Google Scholar]
- 144.Wang W.S., Liu L.X., Li G.P., Chen Y., Li C.Y., Jin D.Y., Wang X.L. Combined serum CA19-9 and miR-27a-3p in peripheral blood mononuclear cells to diagnose pancreatic cancer. Cancer Prev. Res. 2013;6:331–338. doi: 10.1158/1940-6207.CAPR-12-0307. [DOI] [PubMed] [Google Scholar]
- 145.Gao W., Hong Z., Huang H., Zhu A., Lin S., Cheng C., Zhang X., Zou G., Shi Z. miR-27a in serum acts as biomarker for prostate cancer detection and promotes cell proliferation by targeting Sprouty2. Oncol. Lett. 2018;16:5291–5298. doi: 10.3892/ol.2018.9274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Xu Y., Li D., Zhu Z., Li L., Jin Y., Ma C., Zhang W. miR-27a-3p negatively regulates osteogenic differentiation of MC3T3-E1 preosteoblasts by targeting osterix. Mol. Med. Rep. 2020;22:1717–1726. doi: 10.3892/mmr.2020.11246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Li X., Wu X. MiR-21-5p promotes the progression of non-small-cell lung cancer by regulating the expression of SMAD7. Onco. Targets. Ther. 2018;11:8445–8454. doi: 10.2147/OTT.S172393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Li Q., Li B., Li Q., Wei S., He Z., Huang X., Wang L., Xia Y., Xu Z., Li Z., et al. Exosomal miR-21-5p derived from gastric cancer promotes peritoneal metastasis via mesothelial-to-mesenchymal transition. Cell Death Dis. 2018;9 doi: 10.1038/s41419-018-0928-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Chen H., Liu H., Zou H., Chen R., Dou Y., Sheng S., Dai S., Ai J., Melson J., Kittles R.A., et al. Evaluation of plasma miR-21 and miR-152 as diagnostic biomarkers for common types of human cancers. J. Cancer. 2016;7:490–499. doi: 10.7150/jca.12351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Wang X., Wang T., Chen C., Wu Z., Bai P., Li S., Chen B., Liu R., Zhang K., Li W., et al. Serum exosomal miR-210 as a potential biomarker for clear cell renal cell carcinoma. J. Cell. Biochem. 2019;120:1492–1502. doi: 10.1002/jcb.27347. [DOI] [PubMed] [Google Scholar]
- 151.Faragalla H., Youssef Y.M., Scorilas A., Khalil B., White N.M.A., Mejia-Guerrero S., Khella H., Jewett M.A.S., Evans A., Lichner Z., et al. The clinical utility of miR-21 as a diagnostic and prognostic marker for renal cell carcinoma. J. Mol. Diagn. 2012;14:385–392. doi: 10.1016/j.jmoldx.2012.02.003. [DOI] [PubMed] [Google Scholar]
- 152.Pitari M.R., Rossi M., Amodio N., Botta C., Morelli E., Federico C., Gullà A., Caracciolo D., Di Martino M.T., Arbitrio M., et al. Inhibition of miR-21 restores RANKL/OPG ratio in multiple myeloma-derived bone marrow stromal cells and impairs the resorbing activity of mature osteoclasts. Oncotarget. 2015;6:27343–27358. doi: 10.18632/oncotarget.4398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Xu Z., Huang B., Zhang Q., He X., Wei H., Zhang D. NOTCH1 regulates the proliferation and migration of bladder cancer cells by cooperating with long non-coding RNA HCG18 and microRNA-34c-5p. J. Cell. Biochem. 2019;120:6596–6604. doi: 10.1002/jcb.27954. [DOI] [PubMed] [Google Scholar]
- 154.Daugaard I., Knudsen A., Kjeldsen T.E., Hager H., Hansen L.L. The association between miR-34 dysregulation and distant metastases formation in lung adenocarcinoma. Exp. Mol. Pathol. 2017;102:484–491. doi: 10.1016/j.yexmp.2017.05.012. [DOI] [PubMed] [Google Scholar]
- 155.Re M., Magliulo G., Gioacchini F.M., Bajraktari A., Bertini A., Çeka A., Rubini C., Ferrante L., Procopio A.D., Olivieri F. Expression Levels and Clinical Significance of miR-21-5p, miR-let-7a, and miR-34c-5p in Laryngeal Squamous Cell Carcinoma. Biomed Res. Int. 2017;2017 doi: 10.1155/2017/3921258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.White N.M.A., Bao T.T., Grigull J., Youssef Y.M., Girgis A., Diamandis M., Fatoohi E., Metias M., Honey R.J., Stewart R., et al. miRNA Profiling for Clear Cell Renal Cell Carcinoma: Biomarker Discovery and Identification of Potential Controls and Consequences of miRNA Dysregulation. J. Urol. 2011;186:1077–1083. doi: 10.1016/j.juro.2011.04.110. [DOI] [PubMed] [Google Scholar]
- 157.Wang K., Chen X., Zhan Y., Jiang W., Liu X., Wang X., Wu B. miR-335 inhibits the proliferation and invasion of clear cell renal cell carcinoma cells through direct suppression of BCL-W. Tumor Biol. 2015;36:6875–6882. doi: 10.1007/s13277-015-3382-6. [DOI] [PubMed] [Google Scholar]
- 158.Qin S., Shi X., Wang C., Jin P., Ma F. Transcription factor and miRNA interplays can manifest the survival of ccRCC patients. Cancers. 2019;11 doi: 10.3390/cancers11111668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Shu M., Zheng X., Wu S., Lu H., Leng T., Zhu W., Zhou Y., Ou Y., Lin X., Lin Y., et al. Targeting oncogenic miR-335 inhibits growth and invasion of malignant astrocytoma cells. Mol. Cancer. 2011;10:59. doi: 10.1186/1476-4598-10-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Tavazoie S.F., Alarcón C., Oskarsson T., Padua D., Wang Q., Bos P.D., Gerald W.L., Massagué J. Endogenous human microRNAs that suppress breast cancer metastasis. Nature. 2008;451:147–152. doi: 10.1038/nature06487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Gong M., Ma J., Guillemette R., Zhou M., Yang Y., Yang Y., Hock J.M., Yu X. MiR-335 inhibits small cell lung cancer bone metastases via IGF-IR and RANKL pathways. Mol. Cancer Res. 2014;12:101–110. doi: 10.1158/1541-7786.MCR-13-0136. [DOI] [PubMed] [Google Scholar]
- 162.Xu X., Wu J., Li S., Hu Z., Xu X., Zhu Y., Liang Z., Wang X., Lin Y., Mao Y., et al. Downregulation of microRNA-182-5p contributes to renal cell carcinoma proliferation via activating the AKT/FOXO3a signaling pathway. Mol. Cancer. 2014;13:109. doi: 10.1186/1476-4598-13-109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Wang X., Li H., Cui L., Feng J., Fan Q. MicroRNA-182 suppresses clear cell renal cell carcinoma migration and invasion by targeting IGF1R. Neoplasma. 2016;63:717–725. doi: 10.4149/neo_2016_508. [DOI] [PubMed] [Google Scholar]
- 164.Fu J.H., Yang S., Nan C.J., Zhou C.C., Lu D.Q., Li S., Mu H.Q. miR-182 affects renal cancer cell proliferation, apoptosis, and invasion by regulating PI3K/ AKT/mTOR signaling pathway. Eur. Rev. Med. Pharmacol. Sci. 2018;22:351–357. doi: 10.26355/eurrev_201801_14179. [DOI] [PubMed] [Google Scholar]
- 165.Inoue K., Deng Z., Chen Y., Giannopoulou E., Xu R., Gong S., Greenblatt M.B., Mangala L.S., Lopez-Berestein G., Kirsch D.G., et al. Bone protection by inhibition of microRNA-182. Nat. Commun. 2018;9 doi: 10.1038/s41467-018-06446-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Sliwinska-Jewsiewicka A., Kowalczyk A.E., Krazinski B.E., Godlewski J., Kwiatkowski P., Kiewisz J., Grzegrzolka J., Dziegiel P., Kmiec Z. Decreased expression of SATB2 associates with tumor growth and predicts worse outcome in patients with clear cell renal cell carcinoma. Anticancer Res. 2018;38:839–846. doi: 10.21873/anticanres.12292. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data presented in this study are available on request from the corresponding author. The data are not publicly available due to privacy concerns.