Abstract
Simple Summary
Identifying which men at the time of prostate cancer diagnosis have, or will progress to, an aggressive fatal disease will allow clinicians to assist men in making better informed treatment decisions. This will not only be important for those men whose disease is likely to remain indolent and who are currently undergoing unnecessary treatment procedures, but also for those who may need to be targeted with immediate and potentially life-saving therapy. Our case-control study confirms that men who carry BRCA1, BRCA2 and ATM germline pathogenic variants are at increased risk of aggressive disease and provides risk estimates that will be used by clinicians to improve counselling.
Abstract
While gene panel sequencing is becoming widely used for cancer risk prediction, its clinical utility with respect to predicting aggressive prostate cancer (PrCa) is limited by our current understanding of the genetic risk factors associated with predisposition to this potentially lethal disease phenotype. This study included 837 men diagnosed with aggressive PrCa and 7261 controls (unaffected men and men who did not meet criteria for aggressive PrCa). Rare germline pathogenic variants (including likely pathogenic variants) were identified by targeted sequencing of 26 known or putative cancer predisposition genes. We found that 85 (10%) men with aggressive PrCa and 265 (4%) controls carried a pathogenic variant (p < 0.0001). Aggressive PrCa odds ratios (ORs) were estimated using unconditional logistic regression. Increased risk of aggressive PrCa (OR (95% confidence interval)) was identified for pathogenic variants in BRCA2 (5.8 (2.7–12.4)), BRCA1 (5.5 (1.8–16.6)), and ATM (3.8 (1.6–9.1)). Our study provides further evidence that rare germline pathogenic variants in these genes are associated with increased risk of this aggressive, clinically relevant subset of PrCa. These rare genetic variants could be incorporated into risk prediction models to improve their precision to identify men at highest risk of aggressive prostate cancer and be used to identify men with newly diagnosed prostate cancer who require urgent treatment.
Keywords: aggressive prostate cancer, gene panel testing, predisposition, genetic risk factors
1. Introduction
While age, family history, and ethnicity are well-established risk factors for prostate cancer, genetic factors also play an important role and are estimated to account for 57% of its heritability [1]. Genome-wide association studies have thus far identified 269 common variants accounting for about 34% of the familial relative risk [2]. Linkage studies have identified a missense substitution in HOXB13 associated with increased risk of early-onset prostate cancer [3,4]. Rare variants in several genes that were initially implicated in risk for breast or ovarian cancer predisposition (e.g., BRCA1 [5], BRCA2 [6,7], CHEK2 [8], BRIP1 [5], and ATM [9]), as well as the mismatch repair (MMR) genes [10], have also been reported to increase the risk of prostate cancer. Candidate gene approaches, including those using whole-exome sequencing, have contributed to our understanding of genetic risk factors for prostate cancer. For instance, Schaid et al. performed whole-exome sequencing on highly selected prostate cancer cases (n = 491) and controls (n = 429) followed by targeted sequencing of candidate susceptibility genes in an independent dataset comprising 2917 cases and 1899 controls. Eleven genes previously associated with increased risk of prostate cancer (including ATM, BRCA2, and HOXB13) were identified along with ten new candidate genes [11].
The prevalence of germline pathogenic variants in men diagnosed with prostate cancer has been predominantly investigated for DNA repair genes and reported to range between 7.5% and 19% [12,13,14,15,16]. However, these findings have limited clinical utility if they cannot distinguish men whose prostate cancer is likely to remain indolent from those who may need to be targeted with immediate and potentially life-saving therapy. A growing number of studies have therefore used a case-case design to address this issue and aimed to identify germline variants that can distinguish those prostate cancers that will develop into aggressive, clinically relevant disease.
Pritchard et al. reported that germline pathogenic variants in DNA-repair genes were more frequent in men with metastatic prostate cancer than in men with localized disease (82/692, 11.8% and 23/499, 4.6%, respectively; p < 0.001), with the highest number of pathogenic variants being identified in BRCA2 (5.3%), ATM (1.6%), and CHEK2 (1.9%) in men with metastatic disease [13]. Mijuskovic et al. applied the BROCA panel to screen 139 metastatic cases of prostate cancer and 141 indolent cases. They reported a higher frequency of germline protein-truncating variants in men with metastatic disease (n = 17/139, 12.2%) compared with men with indolent disease (n = 4/141, 2.8%) (p = 0.004). The genes containing the highest number of protein-truncating variants in men with metastatic prostate cancer were BRCA2, ATM, and NBN [14].
We recently reported a case-case study of prostate cancer in which we compared the prevalence of germline pathogenic variants in 787 men with aggressive disease and 769 with non-aggressive disease [17]. We found that the proportion of men with aggressive prostate cancer who carried a BRCA2 pathogenic variant exceeded that observed in men with non-aggressive prostate cancer (18/787 carriers, 2.3% and 4/769 carriers, 0.5%, respectively; p = 0.004). We observed a higher proportion of men with aggressive prostate cancer carrying pathogenic variants in ATM than that in men with non-aggressive prostate cancer (14/787 carriers, 0.02% and 5/769 carriers, 0.01%, respectively), although the difference did not reach statistical significance (p = 0.06) [17]. In another case-case study, Darst et al. assessed 2770 men with aggressive and 2775 men with non-aggressive prostate cancer cases from 12 international studies and found that risk for aggressive prostate cancer was associated with rare pathogenic variants in BRCA2 (odds ratio (OR), 95% confidence interval: OR = 3.19, 1.94–5.25, p < 0.001), PALB2 (OR = 6.31, 1.83–21.68, p < 0.001), and ATM (OR = 1.88, 1.10–3.22, p = 0.02) [18].
The possible association between CHEK2 germline pathogenic variants and risk of prostate cancer still requires clarification due to the few and conflicting reports to date. Our case-case study identified 10 (1.3%) men with CHEK2 pathogenic variants and aggressive disease and 5 (0.7%) with the same variants but non-aggressive disease (p = 0.30) [18]. Wu et al. reported a higher proportion of CHEK2 c.1100delC carriers in men with lethal prostate cancer (1.28%) compared with those with low-risk disease (0.16%) [19], but Leongamornlert et al. observed that only “non-1100delC” protein-truncating variants contributed to the aggressive form of the disease [12].
Many commercial laboratories offer clinical genetic testing for hereditary cancer syndromes using panels that range from small cancer syndrome-specific gene panels, based on guidelines, to comprehensive, pan-cancer panels. Some clinical laboratories, such as Ambry Genetics, Invitae, and GeneDx, offer prostate cancer-specific panels that include the following genes: ATM, BRCA1, BRCA2, CHEK2, HOXB13, MLH1, MSH2, MSH6, NBN, PALB2, PMS2, and TP53. Evidence is emerging that genetic information can guide treatment modalities, hence the need to better understand genetic risk factors associated with the risk of aggressive prostate cancer.
In this study, we performed targeted sequencing of 26 genes commonly included on panel tests for cancer predisposition. We defined aggressive prostate cancer as fatal prostate cancer or prostate cancer that met criteria described by Hurwitz et al. [20], i.e., diagnosis of prostate cancer of category T4, N1, or M1 or a Gleason score of 8 or greater. Controls in our study were men who had not been diagnosed with aggressive prostate cancer, i.e., men unaffected with prostate cancer and men with prostate cancer that was not aggressive by the above definition (effectively treating aggressive and non-aggressive prostate cancer as separate diseases).
2. Results
2.1. Prevalence of Germline Pathogenic Variants in Men with Aggressive Prostate Cancer
There were a total of 89 germline pathogenic variants in 85/837 (10%) men with aggressive prostate cancer (individual variant details are provided in Supplementary Table S1).
Of these 89 pathogenic variants, 28 were nonsense, 31 frameshits, 4 inframe deletions, 8 splicing, and 18 missense variants. Pathogenic variants were identified in BRCA2 (number of carriers; prevalence: 20; 2.4%), ATM (16; 1.8%), HOXB13 (12; 1.4%), and CHEK2 (9, 1.1%). There were five carriers (0.6%) of germline pathogenic variants in each of BRCA1, FANCM, and RNASEL. There were four or less carriers of a pathogenic variant in PALB2, MSH6, RECQL, PMS2, BARD1, BRIP1, MSH2, NBN, and RAD51D. All the pathogenic MUTYH variants were monoallelic. No pathogenic variants were identified in CDH1, MLH1, MRE11A, NF1, PTEN, RAD50, RAD51C, TP53, and STK11.
Four men were found to carry more than one germline pathogenic variant. One carried two pathogenic BRCA2 variants known to be in cis and was previously identified by us [17]. One carrier of BRCA2 c.6486_6489del; p.Lys2162Asnfs*5 was found to carry RECQL c.1859C > G; p.Ser620*. One carrier of CHEK2 c.1100delC; p.Thr367Metfs*15 also carried FANCM c.5101C > T; p.Gln1701*. One carrier of ATM c.709dup; p.Thr237Asnfs*17 also carried the nonsense variant HOXB13 c.327C > G; p.Tyr109* (also reported in [17]).
2.2. Associations between Germline Pathogenic Variants and Aggressive Prostate Cancer
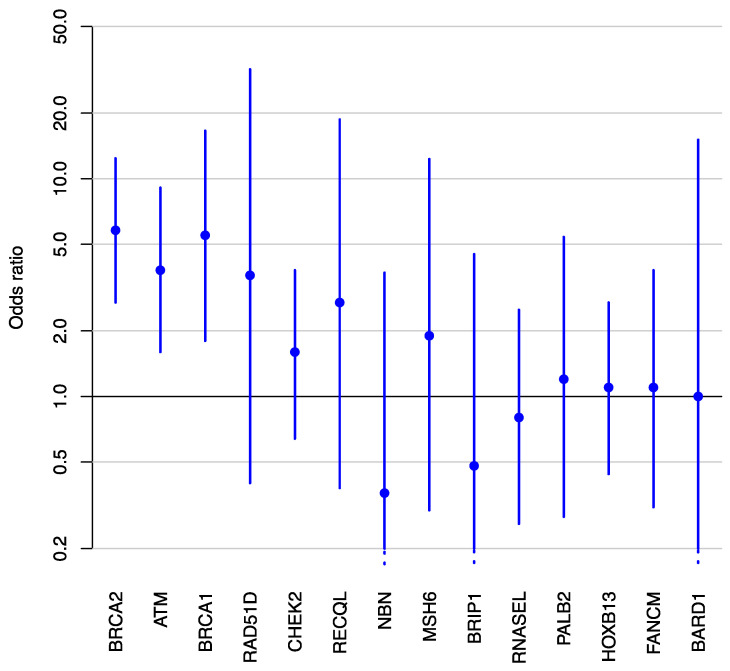
Gene-panel testing of the controls (i.e., men who did not meet criteria for aggressive prostate cancer) identified 273 germline pathogenic variants in 265/7261 (4%) men (Supplementary Table S2), which represents a lower prevalence of pathogenic variants than in aggressive prostate cancer cases (p < 0.0001). After excluding a small number of men with pathogenic variants in more than one gene, a total of 833 cases and 7255 controls were eligible for a case-control analysis. Table 1 presents the clinical characteristics of the men diagnosed with aggressive prostate cancer (cases) included in the statistical analysis. The estimated age-adjusted ORs are presented in Table 2 and Figure 1. We found evidence of an association between aggressive prostate cancer and three genes, with estimated ORs of 5.8 (95% confidence interval (CI): 2.7–12.4) for BRCA2, 5.5 (1.8–16.6) for BRCA1, and 3.8 (1.6–9.1) for ATM. The same three genes were associated with aggressiveness of prostate cancer in a case-case analysis (Supplementary Table S3) but BRCA1 and ATM were not in an analysis where men with non-aggressive prostate cancer were excluded (Supplementary Table S4). Sensitivity analyses showed that the main results were almost unchanged by the inclusion of the small number of men with pathogenic variants in multiple genes (Supplementary Table S5).
Table 1.
Characteristics of the men diagnosed with aggressive prostate cancer who were included in this study.
| Variables and Values | Aggressive PrCa Cases, Number (%) |
|---|---|
| Study | non-missing = 833 |
| ASPREE a | 0 (0%) |
| APC a | 322 (39%) |
| EOPCFS a | 185 (22%) |
| MCCS a | 140 (17%) |
| RFPCS a | 186 (22%) |
| Age at diagnosis in years | non-missing = 833 |
| <60 | 258 (31%) |
| 60–64 | 147 (18%) |
| 65–69 | 262 (31%) |
| ≥70 | 166 (20%) |
| Gleason score | non-missing = 628 |
| 2 | 1 (0%) |
| 3 | 0 (0%) |
| 4 | 5 (1%) |
| 5 | 16 (3%) |
| 6 | 41 (7%) |
| 7 | 105 (17%) |
| 8 | 189 (30%) |
| 9 | 251 (40%) |
| 10 | 20 (3%) |
| Died from prostate cancer | non-missing = 799 |
| No | 324 (41%) |
| Yes | 475 (59%) |
a ASPREE, ASPirin in Reducing Events in the Elderly study; APC, Aggressive Prostate Cancer study; EOPCFS, Early-Onset Prostate Cancer Family Study; MCCS, the Melbourne Collaborative Cohort Study; RFPCFS, Risk Factors for Prostate Cancer Study.
Table 2.
Odds ratios (OR) and 95% confidence intervals (95%CI) for germline pathogenic a variants identified by panel testing of 26 genes in 833 men with aggressive prostate cancer (cases) and in 7255 men without aggressive prostate cancer (controls).
| Gene | Cases (n = 833) | Controls b (n = 7255) | Adjusted OR (95% CI) | p-Value | ||
|---|---|---|---|---|---|---|
| Number of Carriers | % | Number of Carriers | % | |||
| ATM | 14 | 1.7% | 25 | 0.3% | 3.8 (1.6–9.1) | 0.0021 |
| BARD1 | 1 | 0.1% | 8 | 0.1% | 1.0 (0.07–15.1) | 0.97 |
| BRCA1 | 5 | 0.6% | 12 | 0.2% | 5.5 (1.8–16.6) | 0.0023 |
| BRCA2 | 19 | 2.3% | 24 | 0.3% | 5.8 (2.7–12.4) | <0.0001 |
| BRIP1 | 1 | 0.1% | 14 | 0.2% | 0.48 (0.05–4.5) | 0.53 |
| CDH1 | 0 | 0% | 0 | 0% | - | - |
| CHEK2 | 8 | 1% | 41 | 0.6% | 1.6 (0.64–3.8) | 0.32 |
| FANCM | 4 | 0.5% | 23 | 0.3% | 1.1 (0.31–3.8) | 0.89 |
| HOXB13 | 11 | 1.3% | 18 | 0.2% | 1.1 (0.44–2.7) | 0.84 |
| MLH1 | 0 | 0% | 1 | 0% | - | - |
| MRE11A | 0 | 0% | 6 | 0.1% | - | - |
| MSH2 | 1 | 0.1% | 0 | 0% | - | - |
| MSH6 | 3 | 0.4% | 6 | 0.1% | 1.9 (0.3–12.3) | 0.49 |
| MUTYH | 0 | 0% | 0 | 0% | - | - |
| NBN | 1 | 0.1% | 10 | 0.1% | 0.36 (0.03–3.7) | 0.39 |
| NF1 | 0 | 0% | 2 | 0% | - | - |
| PALB2 | 4 | 0.5% | 13 | 0.2% | 1.2 (0.28–5.4) | 0.79 |
| PMS2 | 2 | 0.2% | 0 | 0% | - | - |
| PTEN | 0 | 0% | 0 | 0% | - | - |
| RAD50 | 0 | 0% | 14 | 0.2% | - | - |
| RAD51C | 0 | 0% | 6 | 0.1% | - | - |
| RAD51D | 1 | 0.1% | 6 | 0.1% | 3.6 (0.4–31.8) | 0.25 |
| RECQL | 2 | 0.2% | 5 | 0.1% | 2.7 (0.38–18.7) | 0.33 |
| RNASEL | 5 | 0.6% | 23 | 0.3% | 0.8 (0.26–2.5) | 0.7 |
| STK11 | 0 | 0% | 0 | 0% | - | - |
| TP53 | 0 | 0% | 2 | 0% | - | - |
| Total | 82 | 9.8% | 259 | 3.6% | ||
a Pathogenic (including likely pathogenic) as defined by ClinVar and protein-truncating variants that are absent from ClinVar (accessed November 2020). Excludes protein-truncating variants located in the last coding exon and mono-allelic MUTYH pathogenic variants. For PMS2, panel design avoided regions of homology with the pseudo-gene PMS2CL (as described previously [17]). b Men without aggressive prostate cancer.
Figure 1.
Adjusted odds ratios (large dots) and corresponding 95% confidence intervals (vertical lines) for the association between aggressive prostate cancer and germline pathogenic variants in various genes, sorted by p-value.
3. Discussion
Although the recognition of an important subset of prostate cancer that is aggressive and clinically relevant is not new, there has not been, until very recently, a common evidence-based definition of aggressive prostate cancer. The literature, including reports from epidemiological studies, have used various combinations of clinical parameters, making it difficult to compare and combine studies. In this study, we used the definition recently published by Hurwitz et al., and the hard end point of prostate cancer death to facilitate further elucidation of prostate cancer etiology, including its genetic risk factors, and advance the prevention strategies specifically targeting aggressive prostate cancer [20].
We observed that 10% (85/837) of men with aggressive prostate cancer carried a germline pathogenic variant in a gene commonly included on panel tests for cancer predispostion, more than the 4% (265/7261) of men without aggressive prostate cancer (p < 0.0001). Consistent with the literature, we found that pathogenic variants in BRCA2 and BRCA1 are associated with increased risk of aggressive disease with ORs of 5.8 (2.7–12.4) and 5.5 (1.8–16.6), respectively. The identification of men carrying pathogenic variants in these genes has therapeutic relevance. For instance, PARP inhibitors have been shown to induce substantial responses in patients with metastatic prostate cancer expressing homologous recombination DNA-repair defects [21]. These tumors also appear to be responsive to platinum-based chemotherapy [22], consistent with what has been shown for breast and ovarian cancer diagnosed in women who carry a germline pathogenic variant in BRCA1 and BRCA2. The identification of a germline pathogenic variant also provides information that is highly relevant to relatives, both men and women, as cascade testing can inform risk management strategies for family members.
By combining data from 13 research studies, representing 5560 men with prostate cancer and 3353 unaffected controls, the PRACTICAL consortium conducted the largest gene sequencing study of ATM and estimated an OR for overall prostate cancer risk of 4.4 (2.0–9.5) for pathogenic ATM variant carriers [9]. The PRACTICAL study included 1313 men with prostate cancer whose sequencing data are also included in the present report. Although their definition of “aggressiveness” was similar, the PRACTICAL report also defined sub-groups of aggressiveness: “non-aggressive” (stage T1–T2 and Gleason score 6 disease and, if deceased, death was not due to prostate cancer) and “intermediate aggressive” (those who did not fulfill the criteria for aggressive or non-aggressive disease). When comparing men with aggressive prostate cancer and unaffected controls and men with aggressive and non-aggressive disease, PRACTICAL estimated an OR of 5.4 (2.4–12.5, p < 0.001) and 1.6 (0.9–3.0, p = 0.135), respectively. In our study, the OR associated with risk of aggressive prostate cancer was 3.8 (1.6–9.1) when using men who did not have prostate cancer or did not meet our criteria for aggressive prostate cancer as controls.
One limitation of our study was the fact that the cases and controls were not age-matched. All ORs were adjusted for age, minimizing the impact of this age difference, but we cannot rule out residual confounding or other subtle biases due to this age difference. Another limitation was that our treatment of missing data in the definition of aggressive prostate cancer was conservative, and could cause a small downward bias in our OR estimates. In addition, our approach to targeted sequencing was limited to the coding regions and proximal intron-exon junctions of the genes included on the panel and could only detect single nucleotide variants and small insertions and deletions. Additionally, our analyses focused on germline variants identified as pathogenic in ClinVar. Our findings may thus underestimate the true prevalence of pathogenic missense variants, especially in genes that are less extensively studied than BRCA1 and BRCA2, for which a database of reclassified variants has been established [23].
The strengths of our study include the use of a control dataset recruited from the same country as the cases, for which individual- and variant-level information was available. This is in contrast to other studies including Pritchard et al., who compared the pathogenic variant carrier prevalence of their studies with the ExAC public database. From a bioinformatics processing point of view, although the ideal case-control study design would involve generating sequencing data from the same sequencing platform and applying a single common bioinformatics pipeline [24,25], this is in reality very difficult to achieve, especially in studies requiring large sample sizes to have sufficient power to detect any effect. In our study, although different sequencing platforms were used to generate the raw sequencing data, we reduced potential artefactual variant calls by utilizing the processing pipeline that was the most appropriate for the sequencing technology used to produce the raw sequencing data for the case and the control subjects, then harmonizing the variant calls by (i) restricting calls to regions that are equally able to be called across the three targeted regions and (ii) applying the same filtering and annotation pipelines.
Despite a growing recognition of the role of rare missense variants in cancer predisposition, especially in breast cancer and for genes such as CHEK2 [26] and ATM [27], missense variants individually are currently most commonly classified clinically as variants of uncertain significance. Functional assays have the potential to contribute to the evidence required for interpreting this category of variants, as recently demonstrated with PALB2 [28,29,30,31]. The emerging functional evidence is being incorporated into international efforts aimed at defining the magnitude of cancer risk associated with specific missense variants in cancer predisposition genes and by ClinGen expert panels to support clinical classification of missense variants.
4. Materials and Methods
4.1. Study Subjects
Participants to this study came from (i) the Melbourne Collaborative Cohort Study (MCCS), (ii) the Aggressive Prostate Cancer (APC) study, (iii) the Risk Factors for Prostate Cancer Study (RFPCS), (iv) the Early-Onset Prostate Cancer Family Study (EOPCFS), and (v) the ASPirin in Reducing Events in the Elderly (ASPREE) study. The MCCS, APC, RFPCS, and EOPCFS are Australian research studies of prostate cancer that have been described previously [32,33,34]. The ASPREE study is a randomized, placebo-controlled trial for daily low-dose aspirin. Participants were Australians aged 70 years or older who had no previous diagnosis or current symptoms of atherothrombotic cardiovascular disease, physical disability, or dementia, and no current diagnosis of life-threatening cancer at enrolment. Study design [35], recruitment [36], baseline characteristics [37], and outcomes [38] have been previously described. Our statistical analysis only used ASPREE data that were collected at baseline.
Cases were men diagnosed with aggressive prostate cancer, which was defined to be fatal prostate cancer or prostate cancer that met the criteria described by Hurwitz et al. [20]: cancers that are clinical or pathologic category T4, N1, or M1 or Gleason score greater than or equal to 8. In this definition, missing criteria were assumed to be equal to the lower risk category, e.g., a missing Gleason score was taken to be 7 or lower. A total of 837 aggressive prostate cancer cases were identified from the MCCS, APC, RFPCS, and EOPCFS.
Controls were men who had not been diagnosed with aggressive prostate cancer, i.e., men unaffected by prostate cancer or whose prostate cancer did not meet the above criteria. There were 1238 men identified from MCCS, APC, RFPCS, and EOPCFS who did not have aggressive prostate cancer. Pathology data and precise ages at diagnosis were unavailable for ASPREE participants at baseline, so we excluded ASPREE men with a personal history of aggressive prostate cancer (defined, for the purpose of this exclusion criterion, to be men who died from prostate cancer or had metastatic prostate cancer), leaving a total of 6023 ASPREE controls.
4.2. Gene-Panel Testing
We analyzed rare genetic variants identified in the germline DNA of 837 cases (diagnosed with aggressive prostate cancer) and 7261 controls (not diagnosed with aggressive prostate cancer). Our analysis was restricted to the coding region and proximal intron-exon junctions of 26 genes (ATM: NM_000051, BARD1: NM_000465.2, BRCA1: NM_007294.3, BRCA2: NM_000059.3, BRIP1: NM_032043.2, CDH1: NM_004360.3, CHEK2: NM_007194.3, FANCM: NM_020937.2, HOXB13: NM_006361.5, MLH1: NM_000249.3, MRE11A: NM_005591.3, MSH2: NM_000251.2, MSH6: NM_000179.2, MUTYH: NM_001128425.1, NBN: NM_002485.4, NF1: NM_000267.3, PALB2: NM_024675.3, PMS2: NM_000535.5, PTEN: NM_000314.4, RAD50: NM_005732.3, RAD51C: NM_058216.2, RAD51D: NM_002878.3, RNASEL: NM_021133.3, RECQL: NM_002907.3, STK11: NM_000455.4, TP53: NM_000546.5).
Gene-panel testing had been previously performed for the ASPREE participants [39]. The ASPREE subjects were sequenced using an AmpliSeq panel on the Ion Torrent S5TM XL (Thermo Fisher Scientific, Waltham, MA, USA) and aligned sequencing files (BAMs) were provided for variant calling in this study. Of the 2075 participants from the MCCS, APC, RFPCFS, and EOPCFS, 1553 had been previously tested and reported in a case-case study of prostate cancer aggressiveness and a case-control study focused on ATM [9,18]. Additional gene-panel testing was performed for an additional 522 participants, using methods described previously [18]. Briefly, the germline DNA of these participants was sequenced in-house using a Hi-Plex panel on the NextSeq550 (Illumina, San Diego, CA, USA) [40].
4.3. Variant Calling and Filtering
Reads were mapped to the reference genome (GRch37). Variant calling was performed using VarDict 1.7 [41] and restricted to the overlap of the regions targeted by the two panels. For ASPREE controls sequenced on the Ion Torrent platform, variant calling had also been performed using the Torrent Variant Calling Suite v1.5 as previously described [39] and the intersection with the variant calls from VarDict was used in downstream analyses. Subsequent genetic analyses were restricted to variants with a minimum read depth 50× and variant allele frequency of 0.2. Additionally, for the ASPREE samples, we determined a conservative but high-confidence call set by filtering out variants present in more than 0.05% of all ASPREE participants (n = 65) and variants that had passed our quality filters described above in less than 95% of the calls at a given genomic location.
Variant annotation was performed using VarSeq VSClinical v2.2 (Golden Helix Inc., Bozeman, MT, USA) and included ClinVar annotations from November 2020. This study focused on rare, predicted, protein-truncating variants (PTVs) and pathogenic variants (including likely pathogenic variants). Rare variants were defined as those identified in ExAC v.0.3 with a minor allele frequency ≤0.01 in the non-cancer, non-Finnish European population (NFE non-TCGA). Genetic variants were considered pathogenic if they were annotated as “Pathogenic” or “Likely Pathogenic” in ClinVar. Mono-allelic pathogenic MUTYH variant carriers are reported in Supplementary Table S1 but were not included in our analysis. Predicted PTVs that were classified as “Conflicting” in ClinVar with annotations tending towards pathogenicity (e.g., CHEK2:c.1100delC) were included in this analysis. Also included were PTVs that were absent (unreported) from ClinVar, except if they were located in the last coding exon.
4.4. Statistical Analyses
For each of the genes considered, germline pathogenic variants were combined and an overall OR for their association with aggressive prostate cancer was estimated using unconditional multivariate logistic regression. These analyses were all adjusted for age, the only known prostate cancer risk factor that was available in all our datasets, where the age used was at baseline for ASPREE men and at prostate cancer diagnosis for men in the other studies. Men with prostate cancer that was not aggressive by the above definition were treated as controls for the main analyses since excluding men with non-aggressive disease would mean we could only make inferences about the population of men without non-aggressive prostate cancer.
Excluded from all analyses were women, and male participants in ASPREE with a personal history of aggressive prostate cancer (see Section 4.1) or with no genetic data. A small number of men with germline pathogenic variants in more than one gene were excluded from the main analyses. The effect on our results of this and other analytical choices was investigated with sensitivity analyses. Wald confidence intervals were calculated for each OR, and the likelihood ratio test was used to generate p-values for comparing nested models. All p-values were two-sided and a p-value threshold of 0.05 was used to define statistical significance. Fisher’s exact test was used to test for an overall difference in the prevalence of germline pathogenic variants in men with and without aggressive prostate cancer. All analyses were performed using R version 3.6.1 [42].
5. Conclusions
The cancer risks associated with many of the genes included in prostate cancer susceptibility gene panels are currently not well characterized. Further studies are required to generate the evidence base required for the clinical translation of gene-panel testing. Our study applied a new recommended definition of aggressive prostate cancer and provides further evidence that rare germline pathogenic variants in ATM, BRCA1, and BRCA2 are associated with increased risk of this aggressive, clinically relevant subset of prostate cancer. These rare genetic variants could be incorporated into risk prediction models to improve their precision in identifying men at the highest risk of aggressive prostate cancer and identifying which men, at the time of diagnosis, require urgent treatment, while sparing patients at low risk from the morbidity associated with unnecessary treatment.
Acknowledgments
We would like to acknowledge Daniel J. Park, Bernard J. Pope, and Khalid Mahmood for their contribution to the Hi-Plex technology development.
Supplementary Materials
The following are available online at https://www.mdpi.com/2072-6694/13/7/1495/s1, Table S1: Germline pathogenic variants identified by gene panel testing in men with aggressive prostate cancer; Table S2: Germline pathogenic variants identified by gene panel testing in men without aggressive prostate cancer; Table S3: A case-only analysis where the outcome is aggressive prostate cancer versus all other prostate cancer; Table S4: A sensitivity analysis where men with non-aggressive prostate cancer were excluded rather than treated as controls; Table S5: A sensitivity analysis where a small number of men with germline pathogenic variants in multiple genes were included.
Author Contributions
Conceptualization, T.N.-D. and M.C.S.; methodology, T.N.-D., J.G.D., R.J.M., and M.C.S.; software, T.N.-D. and J.A.S.; validation, T.N.-D., J.A.S., F.H., D.T., and M.M.; formal analysis, T.N-D. and J.G.D.; resources, G.S., D.B., G.G.G., M.C.S., R.L.M., R.J.M., M.R., P.L., R.S., J.M., and E.S.; data curation, T.N.-D. and R.J.M.; writing—original draft preparation, T.N.-D., J.G.D., and M.C.S.; writing—review and editing, T.N.-D., J.G.D., R.J.M., J.A.S., P.-A.D., A.-L.R., F.H., M.M., H.T., M.R., P.L., R.S., J.M., E.S., G.G.G., R.L.M., and M.C.S.; project administration, M.C.S.; funding acquisition, G.G.G., R.L.M., and M.C.S. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by a National Health and Medical Research Council (NMHRC, Australia) Program grant (APP1074383) and Monash University. TN-D is a National Breast Cancer Foundation (Australia) Career Development Fellow (ECF-17-001). M.C.S. is a NMHRC Senior Research Fellow (APP1155163).
Institutional Review Board Statement
The study was conducted according to the guidelines of the Declaration of Helsinki, and approved by the Human Research Committee of the Cancer Council Victoria (0910; 2016) and Monash University (20523; 2019).
Informed Consent Statement
Informed consent was obtained from all subjects involved in the study.
Data Availability Statement
All germline pathogenic variants identified are reported in Supplementary Tables S1 and S2. Sequencing data can be provided upon request to the corresponding author.
Conflicts of Interest
The authors declare no conflict of interest.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Mucci L.A., Hjelmborg J.B., Harris J.R., Czene K., Havelick D.J., Scheike T., Graff R.E., Holst K., Moller S., Unger R.H., et al. Familial Risk and Heritability of Cancer Among Twins in Nordic Countries. JAMA. 2016;315:68–76. doi: 10.1001/jama.2015.17703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Conti D.V., Darst B.F., Moss L.C., Saunders E.J., Sheng X., Chou A., Schumacher F.R., Olama A.A.A., Benlloch S., Dadaev T., et al. Trans-ancestry genome-wide association meta-analysis of prostate cancer identifies new susceptibility loci and informs genetic risk prediction. Nat. Genet. 2021;53:65–75. doi: 10.1038/s41588-020-00748-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ewing C.M., Ray A.M., Lange E.M., Zuhlke K.A., Robbins C.M., Tembe W.D., Wiley K.E., Isaacs S.D., Johng D., Wang Y., et al. Germline mutations in HOXB13 and prostate-cancer risk. N. Engl. J. Med. 2012;366:141–149. doi: 10.1056/NEJMoa1110000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.MacInnis R.J., Severi G., Baglietto L., Dowty J.G., Jenkins M.A., Southey M.C., Hopper J.L., Giles G.G. Population-based estimate of prostate cancer risk for carriers of the HOXB13 missense mutation G84E. PLoS ONE. 2013;8:e54727. doi: 10.1371/journal.pone.0054727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Leongamornlert D., Mahmud N., Tymrakiewicz M., Saunders E., Dadaev T., Castro E., Goh C., Govindasami K., Guy M., O’Brien L., et al. Germline BRCA1 mutations increase prostate cancer risk. Br. J. Cancer. 2012;106:1697–1701. doi: 10.1038/bjc.2012.146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Agalliu I., Karlins E., Kwon E.M., Iwasaki L.M., Diamond A., Ostrander E.A., Stanford J.L. Rare germline mutations in the BRCA2 gene are associated with early-onset prostate cancer. Br. J. Cancer. 2007;97:826–831. doi: 10.1038/sj.bjc.6603929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kote-Jarai Z., Leongamornlert D., Saunders E., Tymrakiewicz M., Castro E., Mahmud N., Guy M., Edwards S., O’Brien L., Sawyer E., et al. BRCA2 is a moderate penetrance gene contributing to young-onset prostate cancer: Implications for genetic testing in prostate cancer patients. Br. J. Cancer. 2011;105:1230–1234. doi: 10.1038/bjc.2011.383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dong X., Wang L., Taniguchi K., Wang X., Cunningham J.M., McDonnell S.K., Qian C., Marks A.F., Slager S.L., Peterson B.J., et al. Mutations in CHEK2 associated with prostate cancer risk. Am. J. Hum. Genet. 2003;72:270–280. doi: 10.1086/346094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Karlsson Q., Brook M.N., Dadaev T., Wakerell S., Saunders E.J., Muir K., Neal D.E., Giles G.G., MacInnis R.J., Thibodeau S.N., et al. Rare Germline Variants in ATM Predispose to Prostate Cancer: A PRACTICAL Consortium Study. Eur. Urol. Oncol. 2021 doi: 10.1016/j.euo.2020.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen Y., Wang J., Fraig M.M., Metcalf J., Turner W.R., Bissada N.K., Watson D.K., Schweinfest C.W. Defects of DNA mismatch repair in human prostate cancer. Cancer Res. 2001;61:4112–4121. [PubMed] [Google Scholar]
- 11.Schaid D.J., McDonnell S.K., FitzGerald L.M., DeRycke L., Fogarty Z., Giles G.G., MacInnis R.J., Southey M.C., Nguyen-Dumont T., Cancel-Tassin G., et al. Two-stage Study of Familial Prostate Cancer by Whole-exome Sequencing and Custom Capture Identifies 10 Novel Genes Associated with the Risk of Prostate Cancer. Eur. Urol. 2020 doi: 10.1016/j.eururo.2020.07.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Leongamornlert D.A., Saunders E.J., Wakerell S., Whitmore I., Dadaev T., Cieza-Borrella C., Benafif S., Brook M.N., Donovan J.L., Hamdy F.C., et al. Germline DNA Repair Gene Mutations in Young-onset Prostate Cancer Cases in the UK: Evidence for a More Extensive Genetic Panel. Eur. Urol. 2019;76:329–337. doi: 10.1016/j.eururo.2019.01.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pritchard C.C., Mateo J., Walsh M.F., De Sarkar N., Abida W., Beltran H., Garofalo A., Gulati R., Carreira S., Eeles R., et al. Inherited DNA-Repair Gene Mutations in Men with Metastatic Prostate Cancer. N. Engl. J. Med. 2016;375:443–453. doi: 10.1056/NEJMoa1603144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mijuskovic M., Saunders E.J., Leongamornlert D.A., Wakerell S., Whitmore I., Dadaev T., Cieza-Borrella C., Govindasami K., Brook M.N., Haiman C.A., et al. Rare germline variants in DNA repair genes and the angiogenesis pathway predispose prostate cancer patients to develop metastatic disease. Br. J. Cancer. 2018;119:96–104. doi: 10.1038/s41416-018-0141-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nicolosi P., Ledet E., Yang S., Michalski S., Freschi B., O’Leary E., Esplin E.D., Nussbaum R.L., Sartor O. Prevalence of Germline Variants in Prostate Cancer and Implications for Current Genetic Testing Guidelines. JAMA Oncol. 2019;5:523–528. doi: 10.1001/jamaoncol.2018.6760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Giri V.N., Hegarty S.E., Hyatt C., O’Leary E., Garcia J., Knudsen K.E., Kelly W.K., Gomella L.G. Germline genetic testing for inherited prostate cancer in practice: Implications for genetic testing, precision therapy, and cascade testing. Prostate. 2019;79:333–339. doi: 10.1002/pros.23739. [DOI] [PubMed] [Google Scholar]
- 17.Nguyen-Dumont T., MacInnis R.J., Steen J.A., Theys D., Tsimiklis H., Hammet F., Mahmoodi M., Pope B.J., Park D.J., Mahmood K., et al. Rare germline genetic variants and risk of aggressive prostate cancer. Int. J. Cancer. 2020;147:2142–2149. doi: 10.1002/ijc.33024. [DOI] [PubMed] [Google Scholar]
- 18.Darst B.F., Dadaev T., Saunders E., Sheng X., Wan P., Pooler L., Xia L.Y., Chanock S., Berndt S.I., Gapstur S.M., et al. Germline sequencing DNA repair genes in 5,545 men with aggressive and non-aggressive prostate cancer. J. Natl. Cancer Inst. 2020 doi: 10.1093/jnci/djaa132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wu Y.S., Yu H.J., Zheng S.L., Na R., Mamawala M., Landis T., Wiley K., Petkewicz J., Shah S., Shi Z.Q., et al. A comprehensive evaluation of CHEK2 germline mutations in men with prostate cancer. Prostate. 2018;78:607–615. doi: 10.1002/pros.23505. [DOI] [PubMed] [Google Scholar]
- 20.Hurwitz L.M., Agalliu I., Albanes D., Barry K.H., Berndt S.I., Cai Q., Chen C., Cheng I., Genkinger J.M., Giles G.G., et al. Recommended definitions of aggressive prostate cancer for etiologic epidemiologic research. J. Natl. Cancer Inst. 2020 doi: 10.1093/jnci/djaa154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mateo J., Carreira S., Sandhu S., Miranda S., Mossop H., Perez-Lopez R., Nava Rodrigues D., Robinson D., Omlin A., Tunariu N., et al. DNA-Repair Defects and Olaparib in Metastatic Prostate Cancer. N. Engl. J. Med. 2015;373:1697–1708. doi: 10.1056/NEJMoa1506859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cheng H.H., Pritchard C.C., Boyd T., Nelson P.S., Montgomery B. Biallelic Inactivation of BRCA2 in Platinum-sensitive Metastatic Castration-resistant Prostate Cancer. Eur. Urol. 2016;69:992–995. doi: 10.1016/j.eururo.2015.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vallee M.P., Francy T.C., Judkins M.K., Babikyan D., Lesueur F., Gammon A., Goldgar D.E., Couch F.J., Tavtigian S.V. Classification of missense substitutions in the BRCA genes: A database dedicated to Ex-UVs. Hum. Mutat. 2012;33:22–28. doi: 10.1002/humu.21629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Guo M.H., Plummer L., Chan Y.-M., Hirschhorn J.N., Lippincott M.F. Burden Testing of Rare Variants Identified through Exome Sequencing via Publicly Available Control Data. Am. J. Hum. Genet. 2018;103:522–534. doi: 10.1016/j.ajhg.2018.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Povysil G., Petrovski S., Hostyk J., Aggarwal V., Allen A.S., Goldstein D.B. Rare-variant collapsing analyses for complex traits: Guidelines and applications. Nat. Rev. Genet. 2019;20:747–759. doi: 10.1038/s41576-019-0177-4. [DOI] [PubMed] [Google Scholar]
- 26.Le Calvez-Kelm F., Lesueur F., Damiola F., Vallée M., Voegele C., Babikyan D., Durand G., Forey N., McKay-Chopin S., Robinot N. Rare, evolutionarily unlikely missense substitutions in CHEK2 contribute to breast cancer susceptibility: Results from a breast cancer family registry case-control mutation-screening study. Breast Cancer Res. 2011;13:R6. doi: 10.1186/bcr2810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tavtigian S.V., Oefner P.J., Babikyan D., Hartmann A., Healey S., Le Calvez-Kelm F., Lesueur F., Byrnes G.B., Chuang S.-C., Forey N., et al. Rare, evolutionarily unlikely missense substitutions in ATM confer increased risk of breast cancer. Am. J. Hum. Genet. 2009;85:427–446. doi: 10.1016/j.ajhg.2009.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wiltshire T., Ducy M., Foo T.K., Hu C., Lee K.Y., Belur Nagaraj A., Rodrigue A., Gomes T.T., Simard J., Monteiro A.N.A., et al. Functional characterization of 84 PALB2 variants of uncertain significance. Genet. Med. 2020;22:622–632. doi: 10.1038/s41436-019-0682-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Boonen R., Rodrigue A., Stoepker C., Wiegant W.W., Vroling B., Sharma M., Rother M.B., Celosse N., Vreeswijk M.P.G., Couch F., et al. Functional analysis of genetic variants in the high-risk breast cancer susceptibility gene PALB2. Nat Commun. 2019;10:5296. doi: 10.1038/s41467-019-13194-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rodrigue A., Margaillan G., Torres Gomes T., Coulombe Y., Montalban G., da Costa E.S.C.S., Milano L., Ducy M., De-Gregoriis G., Dellaire G., et al. A global functional analysis of missense mutations reveals two major hotspots in the PALB2 tumor suppressor. Nucleic. Acids Res. 2019;47:10662–10677. doi: 10.1093/nar/gkz780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Southey M.C., Rewse A., Nguyen-Dumont T. PALB2 Genetic Variants: Can Functional Assays Assist Translation? Trends Cancer. 2020;6:263–265. doi: 10.1016/j.trecan.2020.01.017. [DOI] [PubMed] [Google Scholar]
- 32.Milne R.L., Fletcher A.S., MacInnis R.J., Hodge A.M., Hopkins A.H., Bassett J.K., Bruinsma F.J., Lynch B.M., Dugue P.A., Jayasekara H., et al. Cohort Profile: The Melbourne Collaborative Cohort Study (Health 2020) Int. J. Epidemiol. 2017;46:1757–1757i. doi: 10.1093/ije/dyx085. [DOI] [PubMed] [Google Scholar]
- 33.Papa N.P., MacInnis R.J., English D.R., Bolton D., Davis I.D., Lawrentschuk N., Millar J.L., Pedersen J., Severi G., Southey M.C., et al. Ejaculatory frequency and the risk of aggressive prostate cancer: Findings from a case-control study. Urol. Oncol. 2017;35:530.e7–530.e13. doi: 10.1016/j.urolonc.2017.03.007. [DOI] [PubMed] [Google Scholar]
- 34.Giles G.G., Severi G., Sinclair R., English D.R., McCredie M.R., Johnson W., Boyle P., Hopper J.L. Androgenetic alopecia and prostate cancer: Findings from an Australian case-control study. Cancer Epidemiol. Biomarkers Prev. 2002;11:549–553. [PubMed] [Google Scholar]
- 35.Aspree Investigator Group Study design of ASPirin in Reducing Events in the Elderly (ASPREE): A randomized, controlled trial. Contemp. Clin. Trials. 2013;36:555–564. doi: 10.1016/j.cct.2013.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lockery J.E., Collyer T.A., Abhayaratna W.P., Fitzgerald S.M., McNeil J.J., Nelson M.R., Orchard S.G., Reid C., Stocks N.P., Trevaks R.E., et al. Recruiting general practice patients for large clinical trials: Lessons from the Aspirin in Reducing Events in the Elderly (ASPREE) study. Med. J. Aust. 2019;210:168–173. doi: 10.5694/mja2.12060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.McNeil J.J., Woods R.L., Nelson M.R., Murray A.M., Reid C.M., Kirpach B., Storey E., Shah R.C., Wolfe R.S., Tonkin A.M., et al. Baseline Characteristics of Participants in the ASPREE (ASPirin in Reducing Events in the Elderly) Study. J. Gerontol. A Biol. Sci. Med. Sci. 2017;72:1586–1593. doi: 10.1093/gerona/glw342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.McNeil J.J., Woods R.L., Nelson M.R., Reid C.M., Kirpach B., Wolfe R., Storey E., Shah R.C., Lockery J.E., Tonkin A.M., et al. Effect of Aspirin on Disability-free Survival in the Healthy Elderly. N. Engl. J. Med. 2018;379:1499–1508. doi: 10.1056/NEJMoa1800722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lacaze P., Sebra R., Riaz M., Tiller J., Revote J., Phung J., Parker E.J., Orchard S.G., Lockery J.E., Wolfe R., et al. Medically actionable pathogenic variants in a population of 13,131 healthy elderly individuals. Genet. Med. 2020;22:1883–1886. doi: 10.1038/s41436-020-0881-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hammet F., Mahmood K., Green T.R., Nguyen-Dumont T., Southey M.C., Buchanan D.D., Lonie A., Nathanson K.L., Couch F.J., Pope B.J., et al. Hi-Plex2: A simple and robust approach to targeted sequencing-based genetic screening. Biotechniques. 2019 doi: 10.2144/btn-2019-0026. [DOI] [PubMed] [Google Scholar]
- 41.Lai Z., Markovets A., Ahdesmaki M., Chapman B., Hofmann O., McEwen R., Johnson J., Dougherty B., Barrett J.C., Dry J.R. VarDict: A novel and versatile variant caller for next-generation sequencing in cancer research. Nucleic. Acids Res. 2016;44:e108. doi: 10.1093/nar/gkw227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.R Core Team R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing. [(accessed on 23 March 2021)]; Available online: https://www.r-project.org/
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All germline pathogenic variants identified are reported in Supplementary Tables S1 and S2. Sequencing data can be provided upon request to the corresponding author.