Abstract
Most protein-coding genes in eukaryotes possess at least two poly(A) sites, and alternative polyadenylation is considered a contributing factor to transcriptomic and proteomic diversity. Following transcription, a nascent RNA usually undergoes capping, splicing, cleavage, and polyadenylation, resulting in a mature messenger RNA (mRNA); however, increasing evidence suggests that transcription and RNA processing are coupled. Plants, which must produce rapid responses to environmental changes because of their limited mobility, exhibit such coupling. In this review, we summarize recent advances in our understanding of the coupling of transcription with RNA processing in plants, and we describe the possible spatial environment and important proteins involved. Moreover, we describe how liquid–liquid phase separation, mediated by the C-terminal domain of RNA polymerase II and RNA processing factors with intrinsically disordered regions, enables efficient co-transcriptional mRNA processing in plants.
Keywords: polyadenylation, RNA processing, transcription, coupling regulation, liquid–liquid phase separation (LLPS), gene expression, plant
1. Introduction
Following their transcription from genomic DNA, most nascent RNAs in eukaryotes are processed via multiple steps, including capping, splicing, polyadenylation, and chemical modification, resulting in mature functional messenger RNAs (mRNAs) [1]. Many studies have focused on the mechanisms of splicing and chemical modification, but relatively few studies have considered polyadenylation. Polyadenylation is an important step in gene expression; it affects transcript localization [2], mRNA stability [3,4], translation efficiency [5,6], the nuclear export of mRNAs [7,8], and other essential biochemical processes [9]. The process of polyadenylation includes cleavage at a poly(A) site and the addition of a poly(A) tract [4]. Polyadenylation at different sites of the same gene is called alternative polyadenylation (APA), and it contributes greatly to the complexity of gene expression [1]. APA is widespread among eukaryotes; more than 60% of plant protein-coding genes express alternatively polyadenylated isoforms, and more than 70% of Arabidopsis genes have at least two poly(A) sites [10,11].
In this review, we briefly introduce polyadenylation in plants, and we describe recent advances in our understanding of the regulatory events that couple polyadenylation with transcription, splicing, and the methylation of adenosine at the N6 position (m6A). We then propose a possible spatial environment for this coupling and describe important proteins that are part of the coupling machinery. Finally, we outline a possible pathway whereby such coupling helps regulate gene expression in plants.
2. A Brief Overview of Polyadenylation in Plants
As sessile organisms, plants have evolved specific strategies for responding to environmental changes, and the polyadenylation of pre-mRNAs, which is an important part of gene regulation, plays a key role in plant development and stress responses.
2.1. The Core Polyadenylation Apparatus in Plants
The polyadenylation of most eukaryotic mRNAs involves two steps: cleavage at the appropriate site at the 3′ end of the molecule and the addition of a poly(A) tail. The machinery includes about 85 proteins [12] and multiple sequence elements in the nascent RNA [1]. According to studies of mammalian and yeast cells, the core 3′ cleavage and polyadenylation apparatus includes four complexes: cleavage and polyadenylation specificity factor (CPSF), cleavage stimulation factor (CstF), cleavage factor I (CFI), and cleavage factor II (CFII). These complexes cooperate with poly(A) polymerase (PAP) and various RNA-binding proteins (RBPs) to cleave and polyadenylate nascent RNAs [12,13]. These cleavage and polyadenylation factors work in close collaboration with each other via the recognition of specific sequence elements along the nascent RNA, such as the highly conserved hexamer AAUAAA and its 11 single-nucleotide variants; in humans, up to 82.5% of pre-mRNAs possess this canonical polyadenylation signal (PAS) [13,14]. Nearly all of these core cleavage and polyadenylation factors have homologues in plants (Table 1); however, the hexamer AAUAAA and its 11 single-nucleotide variants are seen in only 8–12% of the transcripts in Arabidopsis thaliana (L.) Heynh and rice [14]. How plants are able to cleave and polyadenylate pre-mRNAs precisely without the use of highly conserved PASs is an interesting question.
Table 1.
Homologues of core cleavage and polyadenylation factors in plants.
| Complex | Subunits | Arabidopsis | References | Recognize Sequence | References |
|---|---|---|---|---|---|
| Cleavage and polyadenylation specificity factor (CPSF) | CPSF160 (CPSF1) | AtCPSF160 | [15] | AAUAAA | [16,17] |
| CPSF100 (CPSF2) | AtCPSF100 | [18] | |||
| CPSF73 (CPSF3) | AtCPSF73-II | [15] | |||
| CPSF30 (CPSF4) | AtCPSF30 | [19] | |||
| WDR33 | FY | [20] | |||
| Fip1 | AtFIP1 | [21] | |||
| Cleavage stimulation factor (CstF) | CstF50 (CstF1) | \ | \ | U/G-rich region | [22] |
| CstF64 (CstF2) | AtCstF64 | [23] | |||
| CstF77 (CstF3) | AtCstF77 | [23] | |||
| Cleavage factor I (CFI) | CFIm25 | \ | \ | UGUA | [24] |
| CFIm68 | \ | \ | |||
| (CFIm59) | \ | \ | |||
| Cleavage factor II (CFII) | CLP1 | Clp1-similar protein 3 (CLPS3) | [25] | G-rich region | [12,26] |
| PCF11 | Pcf11p-similar protein (PCFS4) | [27] |
2.2. Polyadenylation Is Crucial for Plant Development and Stress Responses
A variety of growth defects have been reported in several polyadenylation factor mutants. For example, cstf77-2, a T-DNA insertion mutant of cleavage stimulation factor 77, exhibits curled cotyledons, short roots, a long hypocotyl, and a reduced cell number in the root meristem at the seedling stage, and late flowering and dwarfism in adult plants [28]. Meanwhile, a decreased number of emerged lateral roots, disorganized quiescent center, and reduced stem cell niche were reported in fip1-2 (affecting factor interacting with poly[A] polymerase 1 (Fip1), a core component of the CPSF complex) [29]. Other defects have been detected, like reduced female gametophyte transmission in a mutant of CLPS3 (an ortholog of the human polyadenylation factor CLP1) [25] and altered flowering time in a poly(A) polymerase mutant [30]. Further, homozygous mutations affecting polyadenylation factors have been shown to be lethal [31]. Together, these observations show the indispensable role of polyadenylation in plant development.
Moreover, genome-wide induction of the 3′-untranslated region (UTR) extensions has been observed under conditions of dehydration [32], and increased usage of proximal poly(A) sites (within the 5′-UTR, introns, and protein-coding regions) was observed in response to hypoxia in plants [33]. Global changes in APA have also been detected under conditions of biotic stress (e.g., bacterial blight, rice blast, and rice stripe virus exposure) [34]. Additionally, some polyadenylation factors have been shown to be of great importance in the response of plants to environment changes, including Fip1 and CPSF30 (a core subunit of the CPSF complex). The extent of root inhibition by cadmium was significantly increased in the fip1-2 compared to the wild-type Arabidopsis [29], and AtCPSF30 was shown to play a critical role in modifying the sensitivity of plants to oxidative stress [35] and in resistance to Pseudomonas syringae [36]. These findings suggest that polyadenylation promotes the adaptation of plants to their environment and that correct polyadenylation is crucial for plant development and stress responses.
3. Gene Expression Regulation in Plants via Coupled Transcription and RNA Processing
Nascent RNAs undergo several processing steps, including 5′ capping, splicing, 3′ polyadenylation, and chemical modification, to become mature mRNAs. Emerging evidence has shown interconnections among these processes, which appear to occur in series but may happen co-transcriptionally and even be coupled or interact with each other [37,38,39].
To date, two models have been proposed to explain these coupling processes in yeast and mammalian cells. In the first model, called the recruitment model, transcription is central to the coupling of different RNA processing events, mainly through RNA polymerase II (RNAPII). RNAPII serves as a platform for the recruitment of various processing factors to a nascent RNA, including capping factors, splicing factors, and polyadenylation factors [27]. For example, RNAPII that has been phosphorylated at serine 5 (Ser5P) in its carboxy-terminal domain (CTD) associates, specifically with the spliceosome during co-transcriptional splicing [40]. Besides RNAPII, chromatin itself can act as a platform for different processing factors; for instance, histone H3 trimethylated at lysine 36 (H3K36me3) can recruit RNA-binding proteins [41], and H3K4me3 can recruit the U2 small nuclear ribonucleoprotein (snRNP) to indirectly promote splicing [42].
In the second model, known as the kinetic model or kinetic competition model [43], the relative rates of transcription elongation and splicing or poly(A) site cleavage can affect the output of transcript isoforms and the relative content of different isoforms. The relatively slow elongation rate of RNAPII assists RNA processing factors by allowing more time for spliceosome assembly, the binding of processing factors, and recognition and cleavage at certain poly(A) sites [44]. More recently, it has been reported that the transcription elongation rate can also control RNA processing via changes in nascent RNA folding [45].
Current evidence suggests that these two models function in an interdependent fashion; the transcription elongation rate could influence the recruitment of different processing factors, and, in turn, the transcription elongation rate could be influenced by the differential recruitment of factors that control the elongation rate [37,46]. Studies in which the RNAPII elongation rate was altered suggest that a slower elongation rate allows more time for the recruitment of processing factors to a nascent RNA; however, this is not compatible with both the recruitment coupling and kinetic coupling models [47,48]. High-resolution analyses of transcription elongation rates and the nascent RNA structure are needed to understand the link between these two models. Moreover, it is unknown how these processes are coupled in plants and whether such coupling occurs similarly to that in mammals and yeasts.
3.1. Coupling of the Regulatory Events That Control Gene Expression in Plants
In addition to the coupling of transcription and RNA processing in mammals and yeasts, similar phenomena have been found in plants, particularly in studies of the model plant Arabidopsis thaliana (L.) Heynh. Here, we outline the current progress in understanding such coupling in plants, with a focus on polyadenylation.
3.1.1. Coupling of Polyadenylation and Transcription in Delay of Germination 1 (DOG1) Expression
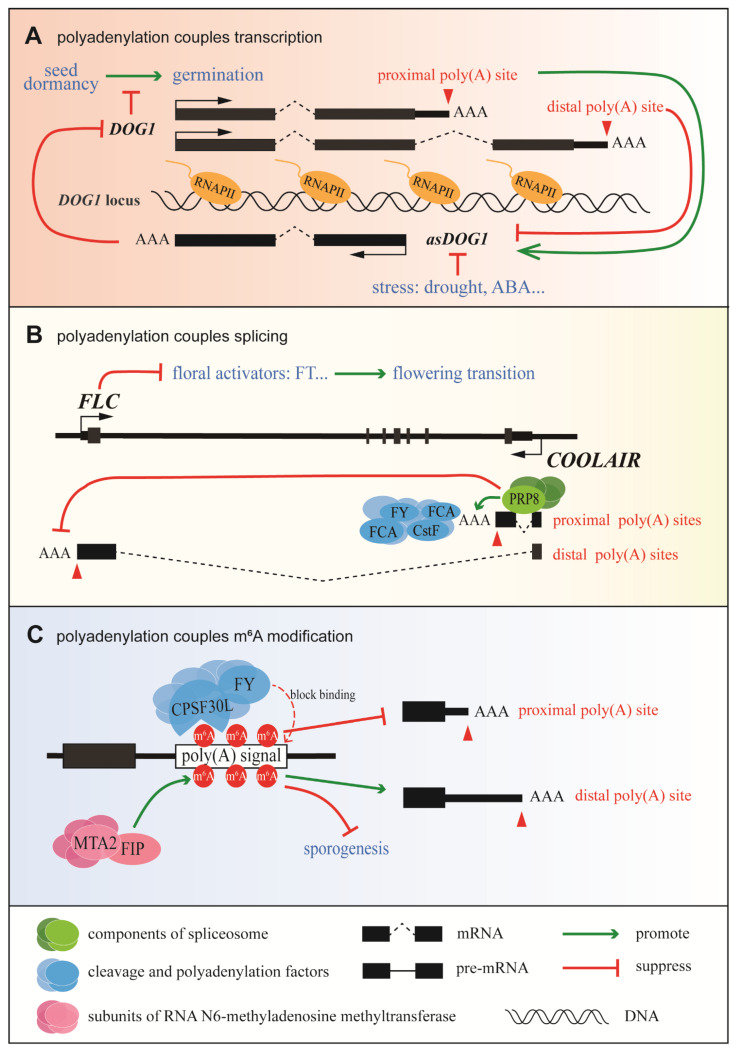
The coupling of polyadenylation and transcription in the control of DOG1 expression is summarized in Figure 1A. Seed dormancy is important for plant survival; it is vital that seeds germinate at the right time [49]. DOG1, which was first identified as a major quantitative trait locus for seed dormancy in natural variations of different Arabidopsis accessions [50], is a key regulator of seed dormancy in Arabidopsis thaliana (L.) Heynh and other plants [51]. DOG1 has two alternative poly(A) sites: polyadenylation of DOG1 at the proximal site results in functional DOG1 (known as shDOG1), while polyadenylation at the less commonly used distal site results in the production of lgDOG1 [52]. 5′-RACE sequence data indicate that the 5′-capped antisense transcript known as asDOG1 (or 1GOD) [53] originates from the end of exon 2, which is close to the proximal poly(A) site of DOG1, and its promoter is located in the region of intron 2 and exon 3 [53]. DOG1 is highly and specifically expressed in seeds, while it is nearly undetectable in seedlings [50,52]. However, asDOG1 is mainly expressed in seedlings and has been shown to suppress DOG1 expression in a cis manner during seed maturation [53]. Under normal conditions, asDOG1 is expressed at a high level in seedlings and the expression of DOG1 is suppressed to promote germination; however, when a stressor is applied, asDOG1 is down-regulated and DOG1 accumulates to promote seed dormancy [54]. Thus, APA of sense transcripts controls the antisense transcription of DOG1 in Arabidopsis thaliana (L.) Heynh [55], and DOG1 affects the expression of asDOG1 in Arabidopsis thaliana (L.) Heynh through APA, polyadenylation, and transcriptional co-regulation of seed dormancy (Figure 1A).
Figure 1.
Schematic diagram of the coupling of transcription and RNA processing in plants. (A) Coupling between polyadenylation and the transcription of delay of germination 1 (DOG1) in Arabidopsis. Increasing usage of the DOG1 proximal poly(A) site promotes the expression of asDOG1, which subsequently suppresses DOG1 expression [55]. (B) Coupling between the polyadenylation and splicing of flowering locus C (FLC) transcripts in Arabidopsis. PRP8 facilitates the efficient splicing of the proximal intron in COOLAIR and promotes usage of the proximal poly(A) site in COOLAIR [31]. (C) Coupling between polyadenylation and m6A modification in plants. The m6A modification blocks binding of the polyadenylation factors CPSF30 and FY to the poly(A) signal, resulting in reduced usage of the proximal poly(A) site [56].
3.1.2. Coupling between Polyadenylation and Splicing in Flowering Locus C (FLC) Expression
The most well-known model for the coupling of polyadenylation and splicing is the regulation of Arabidopsis FLC antisense transcripts (Figure 1B). FLC, which encodes a MADS-box transcription factor, plays a critical role in plant development as the primary inhibitor of flowering. COOLAIR, a collection of long antisense noncoding transcripts produced at the FLC locus, was first detected in 2007 [57]. Based on alternative splicing (AS) and APA, these transcripts can be separated into two classes: class I uses the proximal polyadenylation site located inside intron 6 of FLC, while class II uses the distal polyadenylation site within the FLC promoter. In addition to the use of APA sites, splice variants are produced that are closely related to different FLC expression states. For example, efficient splicing of the intron in class I transcripts promotes the usage of the proximal poly(A) site. This triggers the FLD-dependent demethylation of H3K4me2 in the FLC gene body, downstream of COOLAIR’s proximal poly(A) site, leading to the suppression of FLC expression and the promotion of flowering [31]. Initiation and elongation during FLC transcription are tightly coordinated, and both steps are affected by the status of the chromatin [58], which is thought to be controlled via a kinetic coupling mechanism. When the distal poly(A) site is used, the FLC expression level increases, resulting in late flowering. Adaptive studies of Arabidopsis accessions have shown that single, natural noncoding polymorphisms can significantly change the splicing pattern of class II transcripts [59], resulting in an altered secondary structure of COOLAIR [60] and the up-regulation of FLC expression. Taken together, these results suggest that a precise regulatory network exists during FLC transcription to couple polyadenylation and splicing.
3.1.3. Coupling between Polyadenylation and m6A Modification in Plants
Recent data suggest that m6A functions as a stabilizing factor by suppressing local ribonucleolytic cleavage [61]. Other data show that m6A can suppress the use of proximal poly(A) sites and that adenosines within the AAUAAA signal may be methylated [57] (Figure 1C). CPSF30 (the 30 kDa subunit of CPSF) and WDR33 in mammals are responsible for the recognition of AAUAAA via direct sequence binding [16]. Furthermore, CPSF30L, a homologous isoform of CPSF30, has a YTH domain, enabling it to bind RNAs specifically at the m6A position [17], while flowering locus Y (FLY), a homologue of human WDR33 [16], is associated with the recognition of poly(A) signals in plants [62]. The m6A modification may either directly block the binding of CPSF30 and FLY to poly(A) signals or the binding of m6A by YTH domain-containing proteins, thereby inhibiting the recognition of poly(A) signals by CPSF30 and FLY [50]. In addition, FKBP12-interacting protein (OsFIP) and mRNA adenosine methylase 2 (OsMTA2), two subunits of the RNA m6A methyltransferase complex, which function in sporogenesis, can bind to mRNAs-encoding threonine proteases and NTPases at the early microspore stage, mediating their m6A modification and affecting their expression and/or splicing [63] (Figure 1C). Meanwhile, a transcriptome-wide analysis revealed that m6A modification is highly coordinated with APA site usage [64]. Overall, despite the fact that little is known about m6A modification in plants, evidence suggests that it plays a crucial regulatory role in the coupling of transcription with RNA processing.
In summary, though the regulation of polyadenylation in plants is poorly understood, the above findings confirm that coupling between polyadenylation and transcription, splicing, or m6A modification exists in plants.
3.2. Critical Proteins Involved in the Coupling of Polyadenylation and Other RNA Processing Events
The above-mentioned studies showing the existence of coupling between polyadenylation and other RNA processes in plants add several regulatory layers to gene expression that may help fine-tune the response to stressful conditions or provide a benefit during development. Still, important questions remain. For example, how are these seemingly distinct processes integrated via the collaboration of different processing factors? When and where does this regulation occur? In the next section, we provide a summary of proteins that may participate in the coupling of regulatory processes during gene expression in plants.
3.2.1. RNAPII
RNAPII is the best-known regulatory protein involved in this type of coupling; it has been studied in plants, mammals, and fungi. In eukaryotes, RNAPII plays a fundamental role in gene expression. RNAPII and various phosphorylated CTD isoforms are involved in different stages of the transcription cycle [65,66]. RNAPII interacts with many cleavage and polyadenylation factors, including polyadenylation factor protein 1 of cleavage factor 1 (Pcf11), a component of the CFII cleavage and polyadenylation core complex [67], and Yhh1p/Cft1p, a yeast homologue of human CPSF160, which binds specifically to the phosphorylated RNAPII CTD [68].
A genome-wide analyses of Arabidopsis demonstrated that RNAPII with an unphosphorylated CTD mainly gathers downstream of the transcription start site (TSS), while RNAPII with a Ser5P CTD is required for co-transcriptional splicing; 5′ SS cleavage was achieved through an interaction between the spliceosome complex and Ser5P CTD during the elongation phase of transcription. In addition, RNAPII with a Ser2P CTD paused immediately downstream of the polyadenylation site [69]. Pcf11p-similar protein (PCFS4), a homologue of yeast Pcf11p in Arabidopsis thaliana (L.) Heynh, possesses a C-terminal interaction domain (CID), which is responsible for its interaction with the RNAPII CTD, while its C-terminal region mediates interactions with the polyadenylation factor Clp1-similar protein 3 (CLPS3) [27,67]. RNAPII may thus act in plants as a platform to recruit polyadenylation factors like PCFS4 to promote the cleavage and polyadenylation of a nascent RNA as soon as it is synthetized.
3.2.2. U1 snRNP
Among the proteins that are critical in coupling RNA transcription and processing is U1 snRNP, the most abundant snRNP in most eukaryotes [70]. U1 snRNP is known for its vital role in the initial recognition of 5′ splice sites (SSs) [71]. Purified human U1 snRNP consists of U1 snRNA, Sm proteins, and three U1-specific proteins: U1A, U1C, and U1-70K [72]. Among these subunits, U1A and U1-70K contain a polyA polymerase (PAP) inhibition motif, which interacts with PAP to inhibit polyadenylation [73]. Early studies indicated that U1 snRNP inhibits cleavage and polyadenylation in different ways according to the position of its binding site relative to the poly(A) site. When U1 snRNP binds to the 5′ SS downstream of the proximal poly(A) site, it inhibits cleavage but does not affect the recruitment of other cleavage and polyadenylation factors. In contrast, when U1 snRNP binds to the terminal exon upstream of the PAS, it inhibits polyadenylation [74]. Subsequent genome-wide studies found that the role of U1 snRNP is not carried out in a gene-specific manner [70]. U1 snRNP usually binds to nascent RNAs through base pairing with the U1 snRNA’s 5′ sequence to suppress actionable PASs located in introns, and the binding is associated with RNAPII [66]. U1 snRNP participates in a complex with cleavage and polyadenylation factors (U1-CPAFs) that is distinct from U1–spliceosome complexes; it regulates 3′-end processing and the elongation and termination steps of transcription [75]. In addition, the impact of U1 snRNP is dose dependent [76].
Most protein components of yeast U1 snRNP are conserved in plants [77,78] (Table 2), suggesting a similar function for U1 snRNP in plants. Recently, Arabidopsis U1 (AtU1)A was found to be closely associated with AtU1-70K and AtU1C, confirming its essential role in pre-mRNA splicing in Arabidopsis thaliana (L.) Heynh [79].
Table 2.
Homologues of critical proteins involved in the coupling regulation in plants.
3.2.3. U2 snRNP
Previous studies also found that the interaction between CPSF and U2 snRNP contributes to the coupling of splicing and 3′-end formation in mammals and fungi [83,84]. U2 snRNP binds the intron’s branch site near the 3′ SS through base pairing of the U2 snRNA with the branch site [85]. U2 snRNP auxiliary factor 65 (U2AF65), a splicing factor that promotes pre-spliceosome assembly [86], interacts directly with the 59 kDa subunit of CFIm (CFIm59), and CFIm59/25 heterodimers promote cleavage and polyadenylation [84]. U2 snRNP also interacts with subunits of CPSF directly through its component SF3b [83]. The physical and genetic interactions between the spliceosomal RNA helicase Prp5p and Spt8p/Spt3p, components of the Spt–Ada–Gcn5 acetyltransferase complex, balance transcription initiation/elongation and pre-spliceosome assembly/proofreading in yeast [87].
There are four U2AF65 homologues predicted in the Arabidopsis genome, but only two of them have a U2AF homology motif [80] (Table 2). U2AF65 interacts with Arabidopsis splicing factor 1. Intriguingly, plant SF1 homologues have an additional RNA recognition motif (RRM), which is not present in their yeast and mammalian counterparts [88] and which may be critical in the recognition of nascent RNAs.
3.2.4. Suppressor of Ty 5 (Spt5)
Another critical protein in the control of gene expression via coupled RNA production/processing is the transcription elongation factor Spt5, which is conserved in all domains of life [89]. The C-terminal repeat region (CTR) of Spt5 resembles the RNAPII CTD, which is an intrinsically unstructured extension [90]. The Spt5 CTR sequence varies across species; however, they all contain residues that can be phosphorylated [91]. Dynamic phosphorylation of the Spt5 CTR acts as a switch to promote or suppress transcription elongation [92]. In addition, DRB sensitivity-inducing factor, formed from Spt5 and Spt4, regulates the processivity of RNAPII during elongation through direct interaction with the polymerase [93] and is involved in efficient termination of transcription [94]. When the transcription complex passes over the PAS, Spt5 is dephosphorylated and the Ser2P form of the RNAPII CTD accumulates [95], causing RNAPII to decelerate and become a viable target for the nuclear exonuclease Xrn2 [92]. This is in line with the binding peak of Spt5 discovered downstream of poly(A) sites [94]. Additionally, a physical interaction was detected in yeast between Spt5 and pre-mRNA processing protein 40 (Prp40) [96], which associates with U1 snRNP in the early steps of spliceosome assembly [97]. Further, interactions have been observed between Spt5 and all five spliceosomal snRNAs (U1, U2, U4, U5, and U6). Spts affect the recruitment of U5 snRNP to intron-containing genes [98]. Remarkably, the flexible nature of its CTR enables Spt5 to act as a landing platform for different protein factors, including transcription elongation factors [99], splicing factors, and 3′ cleavage and polyadenylation factors [100]. The first study of Spt5 in plants was published in 2014; thus, research into its function in plants is ongoing [101]. Nevertheless, studies have shown that the Spt4/Spt5 heterodimer is conserved in plants, and Spt5 both co-localizes with RNAPII during transcription [101] and interacts with vernalization independence 5 (VIP5), a core component of the Arabidopsis RNA polymerase II-associated factor 1 complex, through its phosphorylated CTR [81] (Table 2). Therefore, the conserved CTR in Arabidopsis Spt5 may have similar functions to that in yeast and mammals and may be essential for the coupling of multidimensional processes.
3.2.5. The THO/TREX complex
In human and yeast cells, mRNA synthesis and processing are coupled to nuclear mRNA export, during which the TRanscription-EXport (TREX) complex plays an extraordinary role. The TREX complex is composed of the THO complex and a group of additional proteins that are conserved across species, including Saccharomyces cerevisiae, Arabidopsis thaliana (L.) Heynh, Drosophila, and humans [102]. In yeast, the TREX complex is recruited to RNAPII co-transcriptionally through the direct interaction of its subcomplex (THO) with the Ser2-phosphorylated CTD of RNAPII [103]. Intriguingly, the pre-mRNA processing factor 19 splicing complex, also called the NineTeen complex, was found to facilitate the recruitment of the TREX complex to transcribed genes as well [104]. Moreover, the RNA export adaptor yeast RNA annealing protein 1 (Yra1), a subunit of the TREX complex, competes with cleavage factor polyribonucleotide kinase subunit 1 (Clp1) to interact with the 3′-end processing factor Pcf11, and the dynamic balance between Pcf11-Yra1 and Pcf11-Clp1 complexes affects the final selection among different poly(A) sites [105]. Meanwhile, Pcf11 plays a central role in coupling 3′-end processing with transcription via a direct interaction between its CID and the CTD of RNAPII [67]. Thus, the TREX complex, together with these various factors, links transcription, RNA processing, and nuclear export.
Notably, the Arabidopsis THO core complex consists of hyper recombination 1 (HPR1), TEX1 (THO3), THO2, THO5A/B, THO6, and THO7A/B [82] (Table 2). HPR1 and TEX1 are the best studied of these components. Several reports have shown abnormal splicing patterns in hpr1, tex1, and tho2 plants [106,107]. In addition, several splicing factors have been co-purified with TEX1, and TEX1 was found to co-localize with the splicing factor SEINE-ARGININE-RICH (SR) protein family members RSZ22 and RSZ33 [108]. HPR1 was also found to co-localize with the SR protein family member SR33 in the nucleus [106], indicating co-localization between the Arabidopsis THO/TREX complex and splicing factors, consistent with observations in human and yeast cells [104]. More recently, aberrant 3′-UTR extensions were discovered in tex1 and hpr1, the majority of which were shared, suggesting their roles in polyadenylation [109]. PCFS4, the Arabidopsis ortholog of yeast Pcf11, possesses a CID as well, which can interact with the RNAPII CTD [67]. CLPS3, an ortholog of yeast Clp1, is also conserved in Arabidopsis thaliana (L.) Heynh [27]. However, little is known about their roles associated with the TREX complex; instead, plant studies of the THO/TREX complex have focused on the biogenesis of small RNAs, including microRNAs, small interfering RNAs (siRNAs), and trans-acting siRNAs [107,110,111].
3.3. LLPS Enables Efficient Coupling of Transcription and RNA Processing in Plants
In addition to the proteins mentioned above, specific cytosol condensates may provide appropriate spatial environment for the coupling of transcription and RNA processing.
3.3.1. Phase-Separated Membraneless Organelles
Membraneless organelles formed through liquid–liquid phase separation (LLPS) may play a general role in the machinery that is responsible for co-transcriptional mRNA processing; they could provide spatial possibilities for diverse biochemical reactions to take place efficiently at the same time without disturbing each other or perturbing the intracellular environment. Over the past decade, phase separation has become an emerging field, with studies aimed at determining how cells organize diverse functions while maintaining cellular homeostasis [112].
LLPS is a subtype of phase separation that is known to be sensitive to environmental changes; it can respond rapidly to intracellular fluctuations because of its reversible and dynamic nature [113]. Phase separation is driven by interactions among molecules with multivalent domains or intrinsically disordered regions (IDRs) [114]. The proteins involved in LLPS exhibit low sequence complexity and no defined tertiary structure and can alternate between multiple conformations rapidly [115]. The highly flexible structure of intrinsically disordered proteins (IDPs) is of great importance in the establishment and maintenance of nuclear compartments [116]. IDPs are essential for optimal enzyme activity [117], function as hubs in signaling networks [118], act in metabolic regulation [119], and enable stress responses [120,121] through interactions with various partners in membraneless compartments [122].
Various functions in plants rely on proteins with IDRs, including DELLA, CRY, BKI1, BAK1, and ELF3 [121,122,123]. A specific nuclear LLPS condensate of the polyadenylation complex has been observed in Arabidopsis thaliana (L.) Heynh. The RNA-binding protein flowering control locus A (FCA) associates with a coiled-coil protein, FLX-like 2 (FLL2), to promote the formation of liquid-like bodies, which concentrate near 3′-end processing factors at specific poly(A) sites. Both FCA and FLL2 are IDPs with disordered domains [124]. IDPs are widespread in eukaryotes; a genome-wide analyses indicated that the Arabidopsis proteome consists of approximately 30% IDPs [125]. These IDPs can lead to spontaneous nucleation, form phase-separated condensates that resemble non-membrane-bound organelles, and separate from the surrounding phase [126]. A concentrated reaction zone can thus be created, allowing for more efficient biochemical interactions among IDPs and their partners. As a result, different types of regulation (including the coupling of transcription with RNA processing) can take place in distinct spaces in the crowded intracellular environment.
3.3.2. The Underlying Coupling Regulation Model
It is worth stressing that the membraneless compartments and important proteins mentioned above are not isolated from each other, but interrelated. For instance, Spt5 interacts physically with Prp40, a core protein in the U1 snRNP [96]. Additional evidence has been provided by proteomic analyses; numerous interactions among the transcription elongation complex, splicing factors, and cleavage and polyadenylation factors were found in Arabidopsis thaliana (L.) Heynh through co-purification with RNAPII and transcription elongation factors [127]. The RNAPII CTD, which is conserved across eukaryotes, contains tandem heptapeptide repeats that form a mobile extension from the catalytic core of RNAPII [128]. This IDR of the RNAPII CTD confers physicochemical properties beyond those predicted by its sequence that can mediate multivalent interactions during LLPS [129].
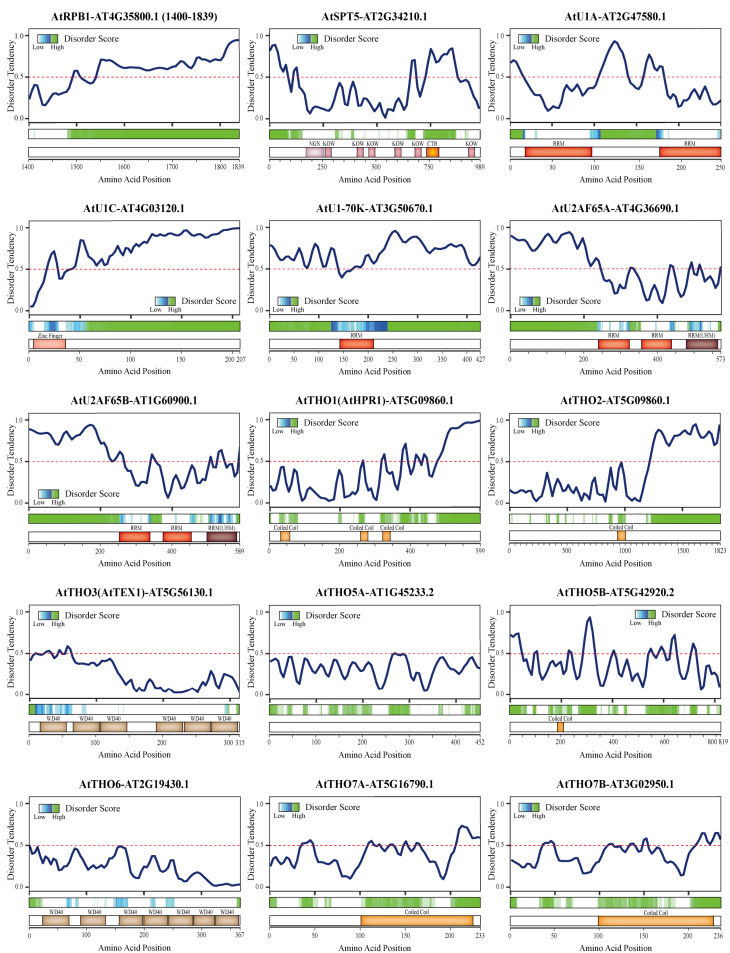
We used a variety of tools to predict the disorder tendency of the other proteins mentioned above, with a focus on proteins from Arabidopsis thaliana (L.) Heynh (Figure 2). Interestingly, all of the proteins and some protein complex subunits were predicted to have highly disordered regions, suggesting that they have the ability to form or promote LLPS. Consistent with our predictions, U1-70K was reported to undergo LLPS via its low-complexity regions [130]. In addition, a few reports have suggested specific roles for the chemical modifications involved in phase separation. For example, H3K9me3 governs heterochromatin formation via phase separation [131], while the number and distribution of m6A modifications affect the phase separation potential of mRNAs [132]. Additionally, phosphorylation of the RNAPII CTD oversees the partition of RNAPII into phase-separated condensates, thereby ensuring that RNAPII functions normally in transcription and RNA processing [133]. Strikingly, the CTR of Spt5, which is a low-complexity region, can be phosphorylated by cyclin-dependent kinase 9, which also controls the phosphorylation state of the CTD of RNAPII [95]. Apart from these reports of proteins (e.g., the RNAPII CTD and U1-70K) and chemical modifications that can undergo or promote LLPS, additional proteins and protein complexes have the potential to participate in LLPS based on our disorder prediction results (Figure 2). Some of them, like the RNAPII CTD, may act as a scaffold or driver of the condensate, while others may participate as client proteins that are mobile depending on the detailed functional requirements, and still others may play different roles in distinct condensates. These membraneless biomolecular condensations can be quite beneficial for achieving efficient regulation in a crowded environment.
Figure 2.
Disorder tendency predictions for proteins involved in the coupling of transcription and RNA processing in plants. All protein sequences were downloaded from TAIR (https://www.arabidopsis.org/ (accessed on 6 November 2020)). (For each figure, top panel) Protein disorder tendency curve. The disorder tendency score of each amino acid was predicted using IUPred2A [134] and subsequently fit to a smooth curve using the R ggplot2 package. The predicted scores were between 0 and 1, with a score above 0.5 (dashed line) indicating disorder. (For each figure, middle panel) Prediction of disordered regions using D2P2 [135]. (For each figure, bottom panel) A domain map of the proteins in Arabidopsis based on previous studies [81,88] and the protein databases UniPro [136], Pfam [137], and SMART [138]. The disorder predictions for full-length proteins, except for AtRPB1 (C-terminal domain), are displayed. Abbreviations: WD40, tryptophan-aspartic acid motif repeats; RRM, RNA recognition motif; UHM, U2AF homology motif; NGN, NusG N-terminal domain; KOW, Kyrpides–Ouzounis–Woese motif; CTR, C-terminal repeat region.
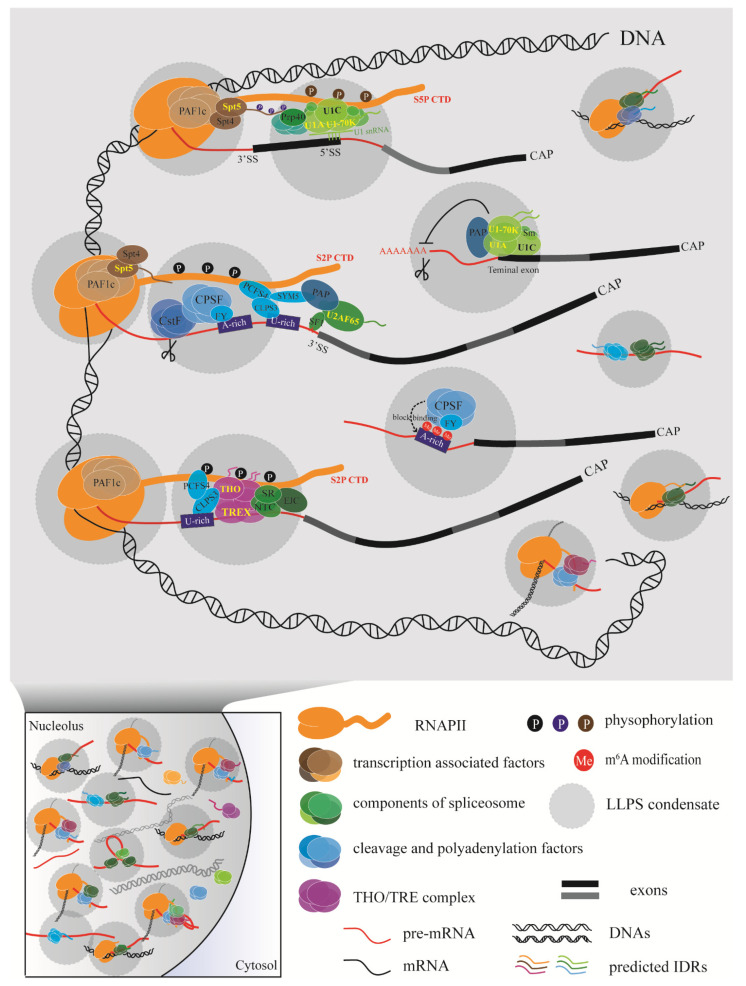
A model of the events involved in coupling transcription with RNA processing has emerged (Figure 3). In this model, diverse factors interact with each other at the right time according to the requirements for transcription and RNA processing. A relatively slow RNAPII elongation rate leaves much more time for polyadenylation factors to assemble properly. Conversely, a fast RNAPII elongation rate may lead to improper polyadenylation, affecting the recognition of a weak poly(A) site or modification site and resulting in chaotic consequences for the cell. With so many processing factors around RNAPII and the nascent RNA, how do the diverse biochemical reactions proceed in order without disturbing each other? Multivalent regulation could allow reactions to occur efficiently inside phase-separated condensates formed by the RNAPII CTD or RNA processing factors with IDRs.
Figure 3.
Schematic diagram of the coupling machinery generated through liquid–liquid phase separation (LLPS) in plants using different processing factors. The various regulatory processes are carried out efficiently inside the phase-separated condensate, which may be driven by the carboxy-terminal domain (CTD) of RNAPII or RNA processing factors with intrinsically disordered regions (IDRs).
4. Conclusions and Perspectives
Once a primary RNA transcript has been synthesized, it must be processed before becoming a fully functional mRNA. Processing includes 5′ capping, splicing within the transcript body, 3′ polyadenylation, and (sometimes) chemical modification. These complex events cannot be easily separated from each other in time and space; however, partitioned membraneless organelles formed by LLPS may greatly improve the accuracy and efficiency of this process.
One interesting question is how plants achieve cleavage and polyadenylation at a precise position using conserved core factors when they lack the conserved PAS sequences found in animals and yeast. One possibility is that plant-specific proteins, such as plant-specific RBPs, are involved in the regulation of polyadenylation in plants. RBPs are of great importance in RNA–protein interactions, which are characterized by the recognition of a specific sequence or structural element in the target RNA by an RNA-binding domain (RBD) such as an RRM or K homology domain [139]. In addition, RBPs appear frequently in LLPS [140]; the sequence-binding specificity of different RBDs may determine the processing of a certain subset of nascent RNAs in a particular droplet. High-throughput studies have shown that the RNA-binding proteomes of plants can be divided into three categories: RBPs containing classical RBDs (22%), RBPs containing non-classical RBDs (39%), and RBPs containing unknown RBDs (39%) [141]. RBPs lacking known RBDs are suspected to be plant specific, and several plant-specific RBPs have been reported to function in plant-specific biological processes such as flowering and photosynthesis (reviewed in [142]). Together, these results provide direction for further study of polyadenylation in plants, with an emphasis on the importance of plant-specific RBPs.
Acknowledgments
We thank Jessica Habashi for critical reading of the manuscript.
Author Contributions
Conceptualization, J.Y., Y.C. and L.M.; writing—original draft preparation, J.Y.; writing—review and editing, J.Y., Y.C. and L.M.; visualization, J.Y.; supervision, L.M.; funding acquisition, Y.C. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by National Natural Science Foundation of China, grant number 31972857.
Conflicts of Interest
The authors declare no conflict of interest.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Desterro J., Bak-Gordon P., Carmo-Fonseca M. Targeting mRNA processing as an anticancer strategy. Nat. Rev. Drug Discov. 2020;19:112–129. doi: 10.1038/s41573-019-0042-3. [DOI] [PubMed] [Google Scholar]
- 2.Andreassi C., Riccio A. To localize or not to localize: mRNA fate is in 3′UTR ends. Trends Cell Biol. 2009;19:465–474. doi: 10.1016/j.tcb.2009.06.001. [DOI] [PubMed] [Google Scholar]
- 3.Hoffman Y., Bublik D.R., Ugalde A.P., Elkon R., Biniashvili T., Agami R., Oren M., Pilpel Y. 3′UTR Shortening Potentiates MicroRNA-Based Repression of Pro-differentiation Genes in Proliferating Human Cells. PLoS Genet. 2016;12:e1005879. doi: 10.1371/journal.pgen.1005879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yu S., Kim V.N. A tale of non-canonical tails: Gene regulation by post-transcriptional RNA tailing. Nat. Rev. Mol. Cell Biol. 2020;21:542–556. doi: 10.1038/s41580-020-0246-8. [DOI] [PubMed] [Google Scholar]
- 5.Bava F.A., Eliscovich C., Ferreira P.G., Miñana B., Ben-Dov C., Guigó R., Valcárcel J., Méndez R. CPEB1 coordinates alternative 3′-UTR formation with translational regulation. Nature. 2013;495:121–125. doi: 10.1038/nature11901. [DOI] [PubMed] [Google Scholar]
- 6.Zhao T., Huan Q., Sun J., Liu C., Hou X., Yu X., Silverman I.M., Zhang Y., Gregory B.D., Liu C.M., et al. Impact of poly(A)-tail G-content on Arabidopsis PAB binding and their role in enhancing translational efficiency. Genome Biol. 2019;20:189. doi: 10.1186/s13059-019-1799-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schmid M., Olszewski P., Pelechano V., Gupta I., Steinmetz L.M., Jensen T.H. The Nuclear PolyA-Binding Protein Nab2p Is Essential for mRNA Production. Cell Rep. 2015;12:128–139. doi: 10.1016/j.celrep.2015.06.008. [DOI] [PubMed] [Google Scholar]
- 8.Stewart M. Polyadenylation and nuclear export of mRNAs. J. Biol. Chem. 2019;294:2977–2987. doi: 10.1074/jbc.REV118.005594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Millevoi S., Vagner S. Molecular mechanisms of eukaryotic pre-mRNA 3′ end processing regulation. Nucleic Acids Res. 2010;38:2757–2774. doi: 10.1093/nar/gkp1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wu X., Liu M., Downie B., Liang C., Ji G., Li Q.Q., Hunt A.G. Genome-wide landscape of polyadenylation in Arabidopsis provides evidence for extensive alternative polyadenylation. Proc. Natl. Acad. Sci. USA. 2011;108:12533–12538. doi: 10.1073/pnas.1019732108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wu X., Zhang Y., Li Q.Q. PlantAPA: A portal for visualization and analysis of alternative polyadenylation in plants. Front. Plant Sci. 2016;7:889. doi: 10.3389/fpls.2016.00889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shi Y., Di Giammartino D.C., Taylor D., Sarkeshik A., Rice W.J., Yates J.R., 3rd, Frank J., Manley J.L. Molecular architecture of the human pre-mRNA 3′ processing complex. Mol. Cell. 2009;33:365–376. doi: 10.1016/j.molcel.2008.12.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tian B., Manley J.L. Alternative polyadenylation of mRNA precursors. Nat. Rev. Mol. Cell Biol. 2017;18:18–30. doi: 10.1038/nrm.2016.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhao Z., Wu X., Ji G., Liang C., Li Q.Q. Genome-wide comparative analyses of polyadenylation signals in eukaryotes suggest a possible origin of the AAUAAA signal. Int. J. Mol. Sci. 2019;20:958. doi: 10.3390/ijms20040958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Xu R., Ye X., Li Q.Q. AtCPSF73-II gene encoding an Arabidopsis homolog of CPSF 73 kDa subunit is critical for early embryo development. Gene. 2004;324:35–45. doi: 10.1016/j.gene.2003.09.025. [DOI] [PubMed] [Google Scholar]
- 16.Chan S.L., Huppertz I., Yao C., Weng L., Moresco J.J., Yates J.R., 3rd, Ule J., Manley J.L., Shi Y. CPSF30 and Wdr33 directly bind to AAUAAA in mammalian mRNA 3′ processing. Genes Dev. 2014;28:2370–2380. doi: 10.1101/gad.250993.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sun Y., Zhang Y., Hamilton K., Manley J.L., Shi Y., Walz T., Tong L. Molecular basis for the recognition of the human AAUAAA polyadenylation signal. Proc. Natl. Acad. Sci. USA. 2018;115:E1419–E1428. doi: 10.1073/pnas.1718723115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Elliott B.J., Dattaroy T., Meeks-Midkiff L.R., Forbes K.P., Hunt A.G. An interaction between an Arabidopsis poly(A) polymerase and a homologue of the 100 kDa subunit of CPSF. Plant Mol. Biol. 2003;51:373–384. doi: 10.1023/A:1022035219500. [DOI] [PubMed] [Google Scholar]
- 19.Delaney K.J., Xu R., Zhang J., Li Q.Q., Yun K.Y., Falcone D.L., Hunt A.G. Calmodulin interacts with and regulates the RNA-binding activity of an Arabidopsis polyadenylation factor subunit. Plant Physiol. 2006;140:1507–1521. doi: 10.1104/pp.105.070672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Simpson G.G., Dijkwel P.P., Quesada V., Henderson I., Dean C. FY is an RNA 3′ end-processing factor that interacts with FCA to control the Arabidopsis floral transition. Cell. 2003;113:777–787. doi: 10.1016/S0092-8674(03)00425-2. [DOI] [PubMed] [Google Scholar]
- 21.Forbes K.P., Addepalli B., Hunt A.G. An Arabidopsis Fip1 homolog interacts with RNA and provides conceptual links with a number of other polyadenylation factor subunits. J. Biol. Chem. 2006;281:176–186. doi: 10.1074/jbc.M510964200. [DOI] [PubMed] [Google Scholar]
- 22.Yang W., Hsu P.L., Yang F., Song J.E., Varani G. Reconstitution of the CstF complex unveils a regulatory role for CstF-50 in recognition of 3′-end processing signals. Nucleic Acids Res. 2018;46:493–503. doi: 10.1093/nar/gkx1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yao Y., Song L., Katz Y., Galili G. Cloning and characterization of Arabidopsis homologues of the animal CstF complex that regulates 3′ mRNA cleavage and polyadenylation. J. Exp. Bot. 2002;53:2277–2278. doi: 10.1093/jxb/erf073. [DOI] [PubMed] [Google Scholar]
- 24.Yang Q., Gilmartin G.M., Doublié S. Structural basis of UGUA recognition by the Nudix protein CFI(m)25 and implications for a regulatory role in mRNA 3′ processing. Proc. Natl. Acad. Sci. USA. 2010;107:10062–10067. doi: 10.1073/pnas.1000848107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xing D., Zhao H., Li Q.Q. Arabidopsis CLP1-SIMILAR PROTEIN3, an ortholog of human polyadenylation factor CLP1, functions in gametophyte, embryo, and postembryonic development. Plant Physiol. 2008;148:2059–2069. doi: 10.1104/pp.108.129817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schäfer P., Tüting C., Schönemann L., Kühn U., Treiber T., Treiber N., Ihling C., Graber A., Keller W., Meister G., et al. Reconstitution of mammalian cleavage factor II involved in 3′ processing of mRNA precursors. RNA. 2018;24:1721–1737. doi: 10.1261/rna.068056.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Xing D., Zhao H., Xu R., Li Q.Q. Arabidopsis PCFS4, a homologue of yeast polyadenylation factor Pcf11p, regulates FCA alternative processing and promotes flowering time. Plant J. 2008;54:899–910. doi: 10.1111/j.1365-313X.2008.03455.x. [DOI] [PubMed] [Google Scholar]
- 28.Zeng W., Dai X., Sun J., Hou Y., Ma X., Cao X., Zhao Y., Cheng Y. Modulation of auxin signaling and development by polyadenylation machinery. Plant Physiol. 2019;179:686–699. doi: 10.1104/pp.18.00782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Téllez-Robledo B., Manzano C., Saez A., Navarro-Neila S., Silva-Navas J., de Lorenzo L., González-García M.P., Toribio R., Hunt A.G., Baigorri R., et al. The polyadenylation factor FIP1 is important for plant development and root responses to abiotic stresses. Plant J. 2019;99:1203–1219. doi: 10.1111/tpj.14416. [DOI] [PubMed] [Google Scholar]
- 30.Kappel C., Trost G., Czesnick H., Ramming A., Kolbe B., Vi S.L., Bispo C., Becker J.D., de Moor C., Lenhard M. Genome-wide analysis of PAPS1-dependent polyadenylation identifies novel roles for functionally specialized poly(A) polymerases in Arabidopsis thaliana. PLoS Genet. 2015;11:e1005474. doi: 10.1371/journal.pgen.1005474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liu F., Marquardt S., Lister C., Swiezewski S., Dean C. Targeted 3′ processing of antisense transcripts triggers Arabidopsis FLC chromatin silencing. Science. 2010;327:94–97. doi: 10.1126/science.1180278. [DOI] [PubMed] [Google Scholar]
- 32.Sun H.X., Li Y., Niu Q.W., Chua N.H. Dehydration stress extends mRNA 3′ untranslated regions with noncoding RNA functions in Arabidopsis. Genome Res. 2017;27:1427–1436. doi: 10.1101/gr.218669.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.De Lorenzo L., Sorenson R., Bailey-Serres J., Hunt A.G. Noncanonical alternative polyadenylation contributes to gene regulation in response to hypoxia. Plant Cell. 2017;29:1262–1277. doi: 10.1105/tpc.16.00746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ye C., Zhou Q., Wu X., Ji G., Li Q.Q. Genome-wide alternative polyadenylation dynamics in response to biotic and abiotic stresses in rice. Ecotoxicol. Environ. Saf. 2019;183:109485. doi: 10.1016/j.ecoenv.2019.109485. [DOI] [PubMed] [Google Scholar]
- 35.Thomas P.E., Wu X., Liu M., Gaffney B., Ji G., Li Q.Q., Hunt A.G. Genome-wide control of polyadenylation site choice by CPSF30 in Arabidopsis. Plant Cell. 2012;24:4376–4388. doi: 10.1105/tpc.112.096107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bruggeman Q., Garmier M., de Bont L., Soubigou-Taconnat L., Mazubert C., Benhamed M., Raynaud C., Bergounioux C., Delarue M. The polyadenylation factor subunit CLEAVAGE AND POLYADENYLATION SPECIFICITY FACTOR30: A key factor of programmed cell death and a regulator of immunity in Arabidopsis. Plant Physiol. 2014;165:732–746. doi: 10.1104/pp.114.236083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bentley D.L. Coupling mRNA processing with transcription in time and space. Nat. Rev. Genet. 2014;15:163–175. doi: 10.1038/nrg3662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mittleman B.E., Pott S., Warland S., Zeng T., Mu Z., Kaur M., Gilad Y., Li Y. Alternative polyadenylation mediates genetic regulation of gene expression. eLife. 2020;9:e57492. doi: 10.7554/eLife.57492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tellier M., Maudlin I., Murphy S. Transcription and splicing: A two-way street. Wiley Interdiscip. Rev. RNA. 2020;11:e1593. doi: 10.1002/wrna.1593. [DOI] [PubMed] [Google Scholar]
- 40.Nojima T., Rebelo K., Gomes T., Grosso A.R., Proudfoot N.J., Carmo-Fonseca M. RNA Polymerase II phosphorylated on CTD Serine 5 interacts with the spliceosome during co-transcriptional splicing. Mol. Cell. 2018;72:369–379.e4. doi: 10.1016/j.molcel.2018.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Luco R.F., Pan Q., Tominaga K., Blencowe B.J., Pereira-Smith O.M., Misteli T. Regulation of alternative splicing by histone modifications. Science. 2010;327:996–1000. doi: 10.1126/science.1184208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sims R.J., 3rd, Millhouse S., Chen C.F., Lewis B.A., Erdjument-Bromage H., Tempst P., Manley J.L., Reinberg D. Recognition of trimethylated histone H3 lysine 4 facilitates the recruitment of transcription postinitiation factors and pre-mRNA splicing. Mol. Cell. 2007;28:665–676. doi: 10.1016/j.molcel.2007.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.De la Mata M., Lafaille C., Kornblihtt A.R. First come, first served revisited: Factors affecting the same alternative splicing event have different effects on the relative rates of intron removal. RNA. 2010;16:904–912. doi: 10.1261/rna.1993510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dujardin G., Lafaille C., de la Mata M., Marasco L.E., Muñoz M.J., Le Jossic-Corcos C., Corcos L., Kornblihtt A.R. How slow RNA polymerase II elongation favors alternative exon skipping. Mol. Cell. 2014;54:683–690. doi: 10.1016/j.molcel.2014.03.044. [DOI] [PubMed] [Google Scholar]
- 45.Saldi T., Fong N., Bentley D.L. Transcription elongation rate affects nascent histone pre-mRNA folding and 3′ end processing. Genes Dev. 2018;32:297–308. doi: 10.1101/gad.310896.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Saldi T., Cortazar M.A., Sheridan R.M., Bentley D.L. Coupling of RNA Polymerase II transcription elongation with pre-mRNA splicing. J. Mol. Biol. 2016;428:2623–2635. doi: 10.1016/j.jmb.2016.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Aslanzadeh V., Huang Y., Sanguinetti G., Beggs J.D. Transcription rate strongly affects splicing fidelity and cotranscriptionality in budding yeast. Genome Res. 2018;28:203–213. doi: 10.1101/gr.225615.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Aslanzadeh V., Beggs J.D. Revisiting the window of opportunity for cotranscriptional splicing in budding yeast. RNA. 2020;26:1081–1085. doi: 10.1261/rna.075895.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Finkelstein R., Reeves W., Ariizumi T., Steber C. Molecular aspects of seed dormancy. Annu. Rev. Plant Biol. 2008;59:387–415. doi: 10.1146/annurev.arplant.59.032607.092740. [DOI] [PubMed] [Google Scholar]
- 50.Bentsink L., Jowett J., Hanhart C.J., Koornneef M. Cloning of DOG1, a quantitative trait locus controlling seed dormancy in Arabidopsis. Proc. Natl. Acad. Sci. USA. 2006;103:17042–17047. doi: 10.1073/pnas.0607877103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Carrillo-Barral N., Rodríguez-Gacio M.D.C., Matilla A.J. Delay of Germination-1 (DOG1): A key to understanding seed dormancy. Plants. 2020;9:480. doi: 10.3390/plants9040480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cyrek M., Fedak H., Ciesielski A., Guo Y., Sliwa A., Brzezniak L., Krzyczmonik K., Pietras Z., Kaczanowski S., Liu F., et al. Seed dormancy in Arabidopsis is controlled by alternative polyadenylation of DOG1. Plant Physiol. 2016;170:947–955. doi: 10.1104/pp.15.01483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Fedak H., Palusinska M., Krzyczmonik K., Brzezniak L., Yatusevich R., Pietras Z., Kaczanowski S., Swiezewski S. Control of seed dormancy in Arabidopsis by a cis-acting noncoding antisense transcript. Proc. Natl. Acad. Sci. USA. 2016;113:E7846–E7855. doi: 10.1073/pnas.1608827113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yatusevich R., Fedak H., Ciesielski A., Krzyczmonik K., Kulik A., Dobrowolska G., Swiezewski S. Antisense transcription represses Arabidopsis seed dormancy QTL DOG1 to regulate drought tolerance. EMBO Rep. 2017;18:2186–2196. doi: 10.15252/embr.201744862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kowalczyk J., Palusinska M., Wroblewska-Swiniarska A., Pietras Z., Szewc L., Dolata J., Jarmolowski A., Swiezewski S. Alternative polyadenylation of the sense transcript controls antisense transcription of DELAY OF GERMINATION 1 in Arabidopsis. Mol. Plant. 2017;10:1349–1352. doi: 10.1016/j.molp.2017.07.011. [DOI] [PubMed] [Google Scholar]
- 56.Parker M.T., Knop K., Sherwood A.V., Schurch N.J., Mackinnon K., Gould P.D., Hall A.J., Barton G.J., Simpson G.G. Nanopore direct RNA sequencing maps the complexity of Arabidopsis mRNA processing and m6A modification. eLife. 2020;9:e49658. doi: 10.7554/eLife.49658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Liu F., Quesada V., Crevillén P., Bäurle I., Swiezewski S., Dean C. The Arabidopsis RNA-binding protein FCA requires a lysine-specific demethylase 1 homolog to downregulate FLC. Mol. Cell. 2007;28:398–407. doi: 10.1016/j.molcel.2007.10.018. [DOI] [PubMed] [Google Scholar]
- 58.Wu Z., Ietswaart R., Liu F., Yang H., Howard M., Dean C. Quantitative regulation of FLC via coordinated transcriptional initiation and elongation. Proc. Natl. Acad. Sci. USA. 2016;113:218–223. doi: 10.1073/pnas.1518369112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Li P., Tao Z., Dean C. Phenotypic evolution through variation in splicing of the noncoding RNA COOLAIR. Genes Dev. 2015;29:696–701. doi: 10.1101/gad.258814.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hawkes E.J., Hennelly S.P., Novikova I.V., Irwin J.A., Dean C., Sanbonmatsu K.Y. COOLAIR antisense RNAs form evolutionarily conserved elaborate secondary structures. Cell Rep. 2016;16:3087–3096. doi: 10.1016/j.celrep.2016.08.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Anderson S.J., Kramer M.C., Gosai S.J., Yu X., Vandivier L.E., Nelson A.D.L., Anderson Z.D., Beilstein M.A., Fray R.G., Lyons E., et al. N6-methyladenosine inhibits local ribonucleolytic cleavage to stabilize mRNAs in Arabidopsis. Cell Rep. 2018;25:1146–1157.e3. doi: 10.1016/j.celrep.2018.10.020. [DOI] [PubMed] [Google Scholar]
- 62.Yu Z., Lin J., Li Q.Q. Transcriptome analyses of FY mutants reveal its role in mRNA alternative polyadenylation. Plant Cell. 2019;31:2332–2352. doi: 10.1105/tpc.18.00545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zhang F., Zhang Y.C., Liao J.Y., Yu Y., Zhou Y.F., Feng Y.Z., Yang Y.W., Lei M.Q., Bai M., Wu H., et al. The subunit of RNA N6-methyladenosine methyltransferase OsFIP regulates early degeneration of microspores in rice. PLoS Genet. 2019;15:e1008120. doi: 10.1371/journal.pgen.1008120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Luo J.H., Wang Y., Wang M., Zhang L.Y., Peng H.R., Zhou Y.Y., Jia G.F., He Y. Natural Variation in RNA m6A Methylation and Its Relationship with Translational Status. Plant Physiol. 2020;182:332–344. doi: 10.1104/pp.19.00987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Haberle V., Stark A. Eukaryotic core promoters and the functional basis of transcription initiation. Nat. Rev. Mol. Cell Biol. 2018;19:621–637. doi: 10.1038/s41580-018-0028-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Chen F.X., Smith E.R., Shilatifard A. Born to run: Control of transcription elongation by RNA polymerase II. Nat. Rev. Mol. Cell Biol. 2018;19:464–478. doi: 10.1038/s41580-018-0010-5. [DOI] [PubMed] [Google Scholar]
- 67.Sadowski M., Dichtl B., Hübner W., Keller W. Independent functions of yeast Pcf11p in pre-mRNA 3′ end processing and in transcription termination. EMBO J. 2003;22:2167–2177. doi: 10.1093/emboj/cdg200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Dichtl B., Blank D., Sadowski M., Hübner W., Weiser S., Keller W. Yhh1p/Cft1p directly links poly(A) site recognition and RNA polymerase II transcription termination. EMBO J. 2002;21:4125–4135. doi: 10.1093/emboj/cdf390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Antosz W., Deforges J., Begcy K., Bruckmann A., Poirier Y., Dresselhaus T., Grasser K.D. Critical Role of Transcript Cleavage in Arabidopsis RNA Polymerase II Transcriptional Elongation. Plant Cell. 2020;32:1449–1463. doi: 10.1105/tpc.19.00891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kaida D. The reciprocal regulation between splicing and 3′-end processing. Wiley Interdiscip. Rev. RNA. 2016;7:499–511. doi: 10.1002/wrna.1348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wahl M.C., Will C.L., Lührmann R. The spliceosome: Design principles of a dynamic RNP machine. Cell. 2009;136:701–718. doi: 10.1016/j.cell.2009.02.009. [DOI] [PubMed] [Google Scholar]
- 72.Pomeranz Krummel D.A., Oubridge C., Leung A.K., Li J., Nagai K. Crystal structure of human spliceosomal U1 snRNP at 5.5 A resolution. Nature. 2009;458:475–480. doi: 10.1038/nature07851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Vagner S., Rüegsegger U., Gunderson S.I., Keller W., Mattaj I.W. Position-dependent inhibition of the cleavage step of pre-mRNA 3′-end processing by U1 snRNP. RNA. 2000;6:178–188. doi: 10.1017/S1355838200991854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Guan F., Caratozzolo R.M., Goraczniak R., Ho E.S., Gunderson S.I. A bipartite U1 site represses U1A expression by synergizing with PIE to inhibit nuclear polyadenylation. RNA. 2007;13:2129–2140. doi: 10.1261/rna.756707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.So B.R., Di C., Cai Z., Venters C.C., Guo J., Oh J.M., Arai C., Dreyfuss G. A complex of U1 snRNP with cleavage and polyadenylation factors controls telescripting, regulating mRNA transcription in human cells. Mol. Cell. 2019;76:590–599. doi: 10.1016/j.molcel.2019.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Berg M.G., Singh L.N., Younis I., Liu Q., Pinto A.M., Kaida D., Zhang Z., Cho S., Sherrill-Mix S., Wan L., et al. U1 snRNP determines mRNA length and regulates isoform expression. Cell. 2012;150:53–64. doi: 10.1016/j.cell.2012.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Koncz C., Dejong F., Villacorta N., Szakonyi D., Koncz Z. The spliceosome-activating complex: Molecular mechanisms underlying the function of a pleiotropic regulator. Front. Plant Sci. 2012;3:9. doi: 10.3389/fpls.2012.00009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Kanno T., Lin W.D., Fu J.L., Chang C.L., Matzke A.J.M., Matzke M. A genetic screen for pre-mRNA splicing mutants of Arabidopsis thaliana identifies putative U1 snRNP components RBM25 and PRP39a. Genetics. 2017;207:1347–1359. doi: 10.1534/genetics.117.300149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Gu J., Xia Z., Luo Y., Jiang X., Qian B., Xie H., Zhu J.K., Xiong L., Zhu J., Wang Z.Y. Spliceosomal protein U1A is involved in alternative splicing and salt stress tolerance in Arabidopsis thaliana. Nucleic Acids Res. 2018;46:1777–1792. doi: 10.1093/nar/gkx1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Park H.Y., Lee H.T., Lee J.H., Kim J.K. Arabidopsis U2AF65 regulates flowering time and the growth of pollen tubes. Front. Plant Sci. 2019;10:569. doi: 10.3389/fpls.2019.00569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Lu C., Tian Y., Wang S., Su Y., Mao T., Huang T., Chen Q., Xu Z., Ding Y. Phosphorylation of SPT5 by CDKD;2 is required for VIP5 recruitment and normal flowering in Arabidopsis thaliana. Plant Cell. 2017;29:277–291. doi: 10.1105/tpc.16.00568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Yelina N.E., Smith L.M., Jones A.M., Patel K., Kelly K.A., Baulcombe D.C. Putative Arabidopsis THO/TREX mRNA export complex is involved in transgene and endogenous siRNA biosynthesis. Proc. Natl. Acad. Sci. USA. 2010;107:13948–13953. doi: 10.1073/pnas.0911341107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Kyburz A., Friedlein A., Langen H., Keller W. Direct interactions between subunits of CPSF and the U2 snRNP contribute to the coupling of pre-mRNA 3′ end processing and splicing. Mol. Cell. 2006;23:195–205. doi: 10.1016/j.molcel.2006.05.037. [DOI] [PubMed] [Google Scholar]
- 84.Millevoi S., Loulergue C., Dettwiler S., Karaa S.Z., Keller W., Antoniou M., Vagner S. An interaction between U2AF 65 and CF I(m) links the splicing and 3′ end processing machineries. EMBO J. 2006;25:4854–4864. doi: 10.1038/sj.emboj.7601331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Lewis J.D., Gunderson S.I., Mattaj I.W. The influence of 5′ and 3′ end structures on pre-mRNA metabolism. J. Cell Sci. Suppl. 1995;19:13–19. doi: 10.1242/jcs.1995.Supplement_19.2. [DOI] [PubMed] [Google Scholar]
- 86.Yu Y., Zhen Z., Qi H., Yuan X., Gao X., Zhang M. U2AF65 enhances milk synthesis and growth of bovine mammary epithelial cells by positively regulating the mTOR-SREBP-1c signalling pathway. Cell Biochem. Funct. 2019;37:93–101. doi: 10.1002/cbf.3378. [DOI] [PubMed] [Google Scholar]
- 87.Shao W., Ding Z., Zheng Z.Z., Shen J.J., Shen Y.X., Pu J., Fan Y.J., Query C.C., Xu Y.Z. Prp5-Spt8/Spt3 interaction mediates a reciprocal coupling between splicing and transcription. Nucleic Acids Res. 2020;48:5799–5813. doi: 10.1093/nar/gkaa311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Jang Y.H., Park H.Y., Lee K.C., Thu M.P., Kim S.K., Suh M.C., Kang H., Kim J.K. A homolog of splicing factor SF1 is essential for development and is involved in the alternative splicing of pre-mRNA in Arabidopsis thaliana. Plant J. 2014;78:591–603. doi: 10.1111/tpj.12491. [DOI] [PubMed] [Google Scholar]
- 89.Decker T.M. Mechanisms of transcription elongation factor DSIF (Spt4-Spt5) J. Mol. Biol. 2020 doi: 10.1016/j.jmb.2020.09.016. [DOI] [PubMed] [Google Scholar]
- 90.Ehara H., Yokoyama T., Shigematsu H., Yokoyama S., Shirouzu M., Sekine S.I. Structure of the complete elongation complex of RNA polymerase II with basal factors. Science. 2017;357:921–924. doi: 10.1126/science.aan8552. [DOI] [PubMed] [Google Scholar]
- 91.Hartzog G.A., Fu J. The Spt4-Spt5 complex: A multi-faceted regulator of transcription elongation. Biochim. Biophys. Acta. 2013;1829:105–115. doi: 10.1016/j.bbagrm.2012.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Cortazar M.A., Sheridan R.M., Erickson B., Fong N., Glover-Cutter K., Brannan K., Bentley D.L. Control of RNA pol II speed by PNUTS-PP1 and Spt5 dephosphorylation facilitates termination by a “sitting duck torpedo” mechanism. Mol. Cell. 2019;76:896–908.e4. doi: 10.1016/j.molcel.2019.09.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Martinez-Rucobo F.W., Sainsbury S., Cheung A.C., Cramer P. Architecture of the RNA polymerase-Spt4/5 complex and basis of universal transcription processivity. EMBO J. 2011;30:1302–1310. doi: 10.1038/emboj.2011.64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Baejen C., Andreani J., Torkler P., Battaglia S., Schwalb B., Lidschreiber M., Maier K.C., Boltendahl A., Rus P., Esslinger S., et al. Genome-wide analysis of RNA polymerase II termination at protein-coding genes. Mol. Cell. 2017;66:38–49.e6. doi: 10.1016/j.molcel.2017.02.009. [DOI] [PubMed] [Google Scholar]
- 95.Parua P.K., Booth G.T., Sansó M., Benjamin B., Tanny J.C., Lis J.T., Fisher R.P. A Cdk9-PP1 switch regulates the elongation-termination transition of RNA polymerase II. Nature. 2018;558:460–464. doi: 10.1038/s41586-018-0214-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Moore M.J., Schwartzfarb E.M., Silver P.A., Yu M.C. Differential recruitment of the splicing machinery during transcription predicts genome-wide patterns of mRNA splicing. Mol. Cell. 2006;24:903–915. doi: 10.1016/j.molcel.2006.12.006. [DOI] [PubMed] [Google Scholar]
- 97.Becerra S., Andrés-León E., Prieto-Sánchez S., Hernández-Munain C., Suñé C. Prp40 and early events in splice site definition. Wiley Interdiscip Rev RNA. 2016;7:17–32. doi: 10.1002/wrna.1312. [DOI] [PubMed] [Google Scholar]
- 98.Maudlin I.E., Beggs J.D. Spt5 modulates cotranscriptional spliceosome assembly in Saccharomyces cerevisiae. RNA. 2019;25:1298–1310. doi: 10.1261/rna.070425.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Liu Y., Warfield L., Zhang C., Luo J., Allen J., Lang W.H., Ranish J., Shokat K.M., Hahn S. Phosphorylation of the transcription elongation factor Spt5 by yeast Bur1 kinase stimulates recruitment of the PAF complex. Mol. Cell Biol. 2009;29:4852–4863. doi: 10.1128/MCB.00609-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Mayer A., Schreieck A., Lidschreiber M., Leike K., Martin D.E., Cramer P. The spt5 C-terminal region recruits yeast 3′ RNA cleavage factor I. Mol. Cell Biol. 2012;32:1321–1331. doi: 10.1128/MCB.06310-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Dürr J., Lolas I.B., Sørensen B.B., Schubert V., Houben A., Melzer M., Deutzmann R., Grasser M., Grasser K.D. The transcript elongation factor SPT4/SPT5 is involved in auxin-related gene expression in Arabidopsis. Nucleic Acids Res. 2014;42:4332–4347. doi: 10.1093/nar/gku096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Heath C.G., Viphakone N., Wilson S.A. The role of TREX in gene expression and disease. Biochem. J. 2016;473:2911–2935. doi: 10.1042/BCJ20160010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Meinel D.M., Burkert-Kautzsch C., Kieser A., O’Duibhir E., Siebert M., Mayer A., Cramer P., Söding J., Holstege F.C., Sträßer K. Recruitment of TREX to the transcription machinery by its direct binding to the phospho-CTD of RNA polymerase II. PLoS Genet. 2013;9:e1003914. doi: 10.1371/journal.pgen.1003914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Chanarat S., Seizl M., Strässer K. The Prp19 complex is a novel transcription elongation factor required for TREX occupancy at transcribed genes. Genes Dev. 2011;25:1147–1158. doi: 10.1101/gad.623411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Johnson S.A., Kim H., Erickson B., Bentley D.L. The export factor Yra1 modulates mRNA 3′ end processing. Nat. Struct. Mol. Biol. 2011;18:1164–1171. doi: 10.1038/nsmb.2126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Xu C., Zhou X., Wen C.K. HYPER RECOMBINATION1 of the THO/TREX complex plays a role in controlling transcription of the REVERSION-TO-ETHYLENE SENSITIVITY1 gene in Arabidopsis. PLoS Genet. 2015;11:e1004956. doi: 10.1371/journal.pgen.1004956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Francisco-Mangilet A.G., Karlsson P., Kim M.H., Eo H.J., Oh S.A., Kim J.H., Kulcheski F.R., Park S.K., Manavella P.A. THO2, a core member of the THO/TREX complex, is required for microRNA production in Arabidopsis. Plant J. 2015;82:1018–1029. doi: 10.1111/tpj.12874. [DOI] [PubMed] [Google Scholar]
- 108.Sørensen B.B., Ehrnsberger H.F., Esposito S., Pfab A., Bruckmann A., Hauptmann J., Meister G., Merkl R., Schubert T., Längst G., et al. The Arabidopsis THO/TREX component TEX1 functionally interacts with MOS11 and modulates mRNA export and alternative splicing events. Plant Mol. Biol. 2017;93:283–298. doi: 10.1007/s11103-016-0561-9. [DOI] [PubMed] [Google Scholar]
- 109.Khan G.A., Deforges J., Reis R.S., Hsieh Y.F., Montpetit J., Antosz W., Santuari L., Hardtke C.S., Grasser K.D., Poirier Y. The transcription and export complex THO/TREX contributes to transcription termination in plants. PLoS Genet. 2020;16:e1008732. doi: 10.1371/journal.pgen.1008732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Jauvion V., Elmayan T., Vaucheret H. The conserved RNA trafficking proteins HPR1 and TEX1 are involved in the production of endogenous and exogenous small interfering RNA in Arabidopsis. Plant Cell. 2010;22:2697–2709. doi: 10.1105/tpc.110.076638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Tao S., Zhang Y., Wang X., Xu L., Fang X., Lu Z.J., Liu D. The THO/TREX complex active in miRNA biogenesis negatively regulates root-associated acid phosphatase activity induced by phosphate starvation. Plant Physiol. 2016;171:2841–2853. doi: 10.1104/pp.16.00680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Sabari B.R. Biomolecular condensates and gene activation in development and disease. Dev. Cell. 2020;55:84–96. doi: 10.1016/j.devcel.2020.09.005. [DOI] [PubMed] [Google Scholar]
- 113.Zhao Y.G., Zhang H. Phase Separation in Membrane Biology: The Interplay between Membrane-Bound Organelles and Membraneless Condensates. Dev. Cell. 2020;55:30–44. doi: 10.1016/j.devcel.2020.06.033. [DOI] [PubMed] [Google Scholar]
- 114.Shin Y., Brangwynne C.P. Liquid phase condensation in cell physiology and disease. Science. 2017;357:eaaf4382. doi: 10.1126/science.aaf4382. [DOI] [PubMed] [Google Scholar]
- 115.Owen I., Shewmaker F. The role of post-translational modifications in the phase transitions of intrinsically disordered proteins. Int. J. Mol. Sci. 2019;20:5501. doi: 10.3390/ijms20215501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Pontvianne F., Liu C. Chromatin domains in space and their functional implications. Curr. Opin. Plant Biol. 2020;54:1–10. doi: 10.1016/j.pbi.2019.11.005. [DOI] [PubMed] [Google Scholar]
- 117.DeForte S., Uversky V.N. Not an exception to the rule: The functional significance of intrinsically disordered protein regions in enzymes. Mol. Biosyst. 2017;13:463–469. doi: 10.1039/C6MB00741D. [DOI] [PubMed] [Google Scholar]
- 118.Wright P.E., Dyson H.J. Intrinsically disordered proteins in cellular signalling and regulation. Nat. Rev. Mol. Cell Biol. 2015;16:18–29. doi: 10.1038/nrm3920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Noree C., Begovich K., Samilo D., Broyer R., Monfort E., Wilhelm J.E. A quantitative screen for metabolic enzyme structures reveals patterns of assembly across the yeast metabolic network. Mol. Biol. Cell. 2019;30:2721–2736. doi: 10.1091/mbc.E19-04-0224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Riback J.A., Katanski C.D., Kear-Scott J.L., Pilipenko E.V., Rojek A.E., Sosnick T.R., Drummond D.A. Stress-triggered phase separation is an adaptive, evolutionarily tuned response. Cell. 2017;168:1028–1040.e19. doi: 10.1016/j.cell.2017.02.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Jung J.H., Barbosa A.D., Hutin S., Kumita J.R., Gao M., Derwort D., Silva C.S., Lai X., Pierre E., Geng F., et al. A prion-like domain in ELF3 functions as a thermosensor in Arabidopsis. Nature. 2020;585:256–260. doi: 10.1038/s41586-020-2644-7. [DOI] [PubMed] [Google Scholar]
- 122.Covarrubias A.A., Romero-Pérez P.S., Cuevas-Velazquez C.L., Rendón-Luna D.F. The functional diversity of structural disorder in plant proteins. Arch Biochem. Biophys. 2020;680:108229. doi: 10.1016/j.abb.2019.108229. [DOI] [PubMed] [Google Scholar]
- 123.Emenecker R.J., Holehouse A.S., Strader L.C. Emerging Roles for Phase Separation in Plants. Dev. Cell. 2020;55:69–83. doi: 10.1016/j.devcel.2020.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Fang X., Wang L., Ishikawa R., Li Y., Fiedler M., Liu F., Calder G., Rowan B., Weigel D., Li P., et al. Arabidopsis FLL2 promotes liquid-liquid phase separation of polyadenylation complexes. Nature. 2019;569:265–269. doi: 10.1016/j.abb.2019.108229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Pietrosemoli N., García-Martín J.A., Solano R., Pazos F. Genome-wide analysis of protein disorder in Arabidopsis thaliana: Implications for plant environmental adaptation. PLoS ONE. 2013;8:e55524. doi: 10.1371/journal.pone.0055524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Wang N., Liu C. Implications of liquid-liquid phase separation in plant chromatin organization and transcriptional control. Curr. Opin. Genet. Dev. 2019;55:59–65. doi: 10.1016/j.gde.2019.06.003. [DOI] [PubMed] [Google Scholar]
- 127.Antosz W., Pfab A., Ehrnsberger H.F., Holzinger P., Köllen K., Mortensen S.A., Bruckmann A., Schubert T., Längst G., Griesenbeck J., et al. The composition of the Arabidopsis RNA polymerase II transcript elongation complex reveals the interplay between elongation and mRNA processing factors. Plant Cell. 2017;29:854–870. doi: 10.1105/tpc.16.00735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Meinhart A., Kamenski T., Hoeppner S., Baumli S., Cramer P. A structural perspective of CTD function. Genes Dev. 2005;19:1401–1415. doi: 10.1101/gad.1318105. [DOI] [PubMed] [Google Scholar]
- 129.Cho W.K., Spille J.H., Hecht M., Lee C., Li C., Grube V., Cisse I.I. Mediator and RNA polymerase II clusters associate in transcription-dependent condensates. Science. 2018;361:412–415. doi: 10.1126/science.aar4199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Xue S., Gong R., He F., Li Y., Wang Y., Tan T., Luo S.Z. Low-complexity domain of U1-70K modulates phase separation and aggregation through distinctive basic-acidic motifs. Sci. Adv. 2019;5:eaax5349. doi: 10.1126/sciadv.aax5349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Wang L., Gao Y., Zheng X., Liu C., Dong S., Li R., Zhang G., Wei Y., Qu H., Li Y., et al. Histone modifications regulate chromatin compartmentalization by contributing to a phase separation mechanism. Mol. Cell. 2019;76:646–659.e6. doi: 10.1016/j.molcel.2019.08.019. [DOI] [PubMed] [Google Scholar]
- 132.Ries R.J., Zaccara S., Klein P., Olarerin-George A., Namkoong S., Pickering B.F., Patil D.P., Kwak H., Lee J.H., Jaffrey S.R. m6A enhances the phase separation potential of mRNA. Nature. 2019;571:424–428. doi: 10.1038/s41586-019-1374-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Guo Y.E., Manteiga J.C., Henninger J.E., Sabari B.R., Dall’Agnese A., Hannett N.M., Spille J.H., Afeyan L.K., Zamudio A.V., Shrinivas K., et al. Pol II phosphorylation regulates a switch between transcriptional and splicing condensates. Nature. 2019;572:543–548. doi: 10.1038/s41586-019-1464-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Mészáros B., Erdos G., Dosztányi Z. IUPred2A: Context-dependent prediction of protein disorder as a function of redox state and protein binding. Nucleic Acids Res. 2018;46:W329–W337. doi: 10.1093/nar/gky384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Oates M.E., Romero P., Ishida T., Ghalwash M., Mizianty M.J., Xue B., Dosztányi Z., Uversky V.N., Obradovic Z., Kurgan L., et al. D²P²: Database of disordered protein predictions. Nucleic Acids Res. 2013;41:D508–D516. doi: 10.1093/nar/gks1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.UniProt Consortium UniProt: A worldwide hub of protein knowledge. Nucleic Acids Res. 2019;47:D506–D515. doi: 10.1093/nar/gky1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Mistry J., Chuguransky S., Williams L., Qureshi M., Salazar G.A., Sonnhammer E.L.L., Tosatto S.C.E., Paladin L., Raj S., Richardson L.J., et al. Pfam: The protein families database in 2021. Nucleic Acids Res. 2020:gkaa913. doi: 10.1093/nar/gkaa913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Letunic I., Khedkar S., Bork P. SMART: Recent updates, new developments and status in 2020. Nucleic Acids Res. 2020:gkaa937. doi: 10.1093/nar/gkaa937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Müller-McNicoll M., Neugebauer K.M. How cells get the message: Dynamic assembly and function of mRNA-protein complexes. Nat. Rev. Genet. 2013;14:275–287. doi: 10.1038/nrg3434. [DOI] [PubMed] [Google Scholar]
- 140.Tian S., Curnutte H.A., Trcek T. RNA granules: A view from the RNA perspective. Molecules. 2020;25:3130. doi: 10.3390/molecules25143130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Bach-Pages M., Homma F., Kourelis J., Kaschani F., Mohammed S., Kaiser M., van der Hoorn R.A.L., Castello A., Preston G.M. Discovering the RNA-binding proteome of plant leaves with an improved RNA interactome capture method. Biomolecules. 2020;10:661. doi: 10.3390/biom10040661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Marondedze C. The increasing diversity and complexity of the RNA-binding protein repertoire in plants. Proc. Biol. Sci. 2020;287:20201397. doi: 10.1098/rspb.2020.1397. [DOI] [PMC free article] [PubMed] [Google Scholar]