Abstract
Scaffolds hybridization is a well-known drug design strategy for antitumor agents. Herein, series of novel indolyl-pyrimidine hybrids were synthesized and evaluated in vitro and in vivo for their antitumor activity. The in vitro antiproliferative activity of all compounds was obtained against MCF-7, HepG2, and HCT-116 cancer cell lines, as well as against WI38 normal cells using the resazurin assay. Compounds 1–4 showed broad spectrum cytotoxic activity against all these cancer cell lines compared to normal cells. Compound 4g showed potent antiproliferative activity against these cell lines (IC50 = 5.1, 5.02, and 6.6 μM, respectively) comparable to the standard treatment (5-FU and erlotinib). In addition, the most promising group of compounds was further evaluated for their in vivo antitumor efficacy against EAC tumor bearing mice. Notably, compound 4g showed the most potent in vivo antitumor activity. The most active compounds were evaluated for their EGFR inhibitory (range 53–79%) activity. Compound 4g was found to be the most active compound against EGFR (IC50 = 0.25 µM) showing equipotency as the reference treatment (erlotinib). Molecular modeling study was performed on compound 4g revealed a proper binding of this compound inside the EGFR active site comparable to erlotinib. The data suggest that compound 4g could be used as a potential anticancer agent.
Keywords: cancer, pyrimidine, indole, drug design, EGFR, molecular modeling
1. Introduction
Cancer is an abnormal cell growth. It has the potential of migrating from organ to organ in the human body, this process is called metastasis [1,2,3]. The high mortality rate from cancer is due to its ability to metastasis and spread among multiple organs [4]. Cancer is generally formed by self-cell gene mutations or by manipulating normal cell differentiation [5]. The leading factors that could initiate cancer formation are drugs, viruses, radiation, smoking, or even diet [6]. Cancer is classified by the World Health Organization (WHO) as the second leading cause of death worldwide with 9.6 million deaths in 2018 [7]. The death rate from cancer is projected to continue rising worldwide, with an estimated 13 million deaths in 2030 [8]. In fact, cancer treatment is challenging because of the lack of specificity of the treatment on only malignant cells; normal cells are also affected. The current standard care for cancer patients involves surgery, radiation, immunotherapy, and chemotherapy [9]. Given all of that, there is a high demand for the discovery of new, safer, and efficient anticancer agents.
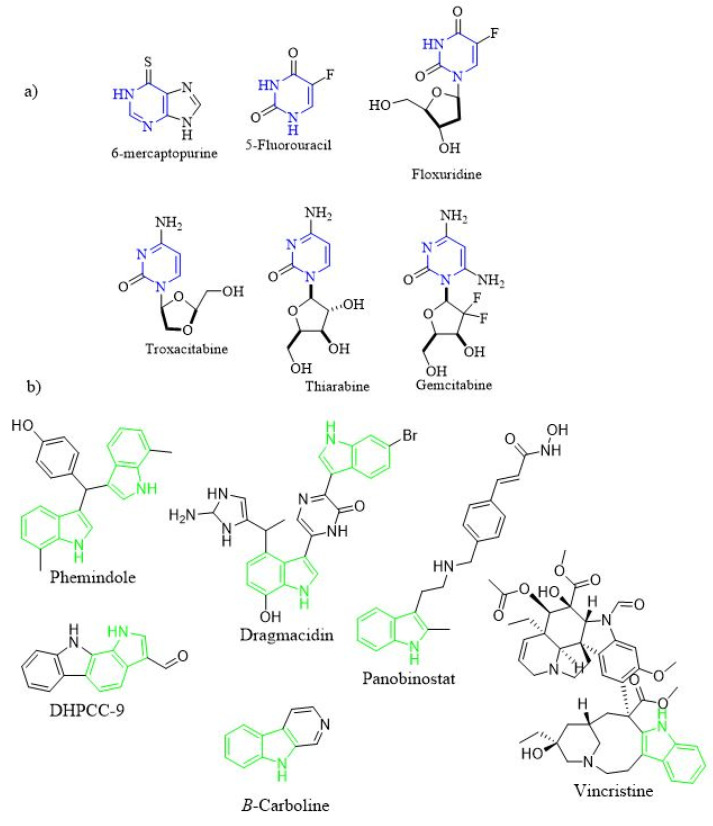
Heterocyclic scaffolds have attracted considerable attention in the design of many biologically active synthetic compounds [10,11,12,13,14]. Pyrimidine, six-membered ring heterocyclic ring with two nitrogen atoms, is widely present in several natural and synthetic drugs [15]. Thus, there are many synthetic pyrimidine derivatives with exhibited several pharmacological actions such as antimicrobial [16,17], anticancer [18,19,20], antioxidant [18], antifungal [21], antithyroid [22], antiviral [23,24,25], anticonvulsant [26], anti-inflammatory [27], antitubercular [28], antileishmanial [29], antimalarial [30], anthelminthic [31], anti-HIV [32], antitubercular [33], and antidepressant activities [34]. In addition, pyrimidine ring prevails in naturally formed structures such alkaloids [35], nucleic bases [36], vitamins [37], and numerous other pharmacophores. Pyrimidine ring is considered the core scaffold of the genetic material and hence it is crucial for anticancer activity [38]. In recent years, several research groups revealed many pyrimidine containing compounds that have anticancer activity (Figure 1a). The FDA approved pyrimidine derivatives with anticancer activity was classified into three primary classes based on their mechanism of action (MOA) [39]: (1) thiopurines: such as 6-mercaptopurine; (2) fluoropyrimidines: such as 5-Fluorouracil (5-FU) and Floxuridine; (3) deoxynucleoside analogues: such as Gemcitabine, Thiarabine and Troxacitabine. Moreover, other pyrimidine compounds with other MOA were discovered such as: (1) glycogen synthase kinase (GSK) inhibitors [40]; (2) cyclin-dependent kinase (CDK) inhibitors [41], and (3) epidermal growth factor receptor (EGFR) inhibitors [42].
Figure 1.
(a) Pyrimidine containing compounds as anticancer agents; (b) indole containing compounds with antitumor activity.
Another important heterocyclic nucleus is the indole ring. It is found in many natural and synthetic compounds including anticancer agents [43] (Figure 1b). Such compounds were found to act as antitumor agents via different MOAs [44] i.e., antimitotic agents (i.e., Vincristine), histone deacetylases (HDACs) (i.e., Panobinostat), PIM kinases (i.e., DHPCC-9), and DNA topoisomerases (i.e., β-carboline).
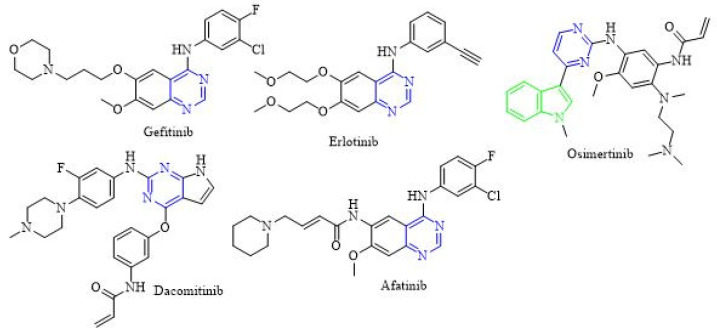
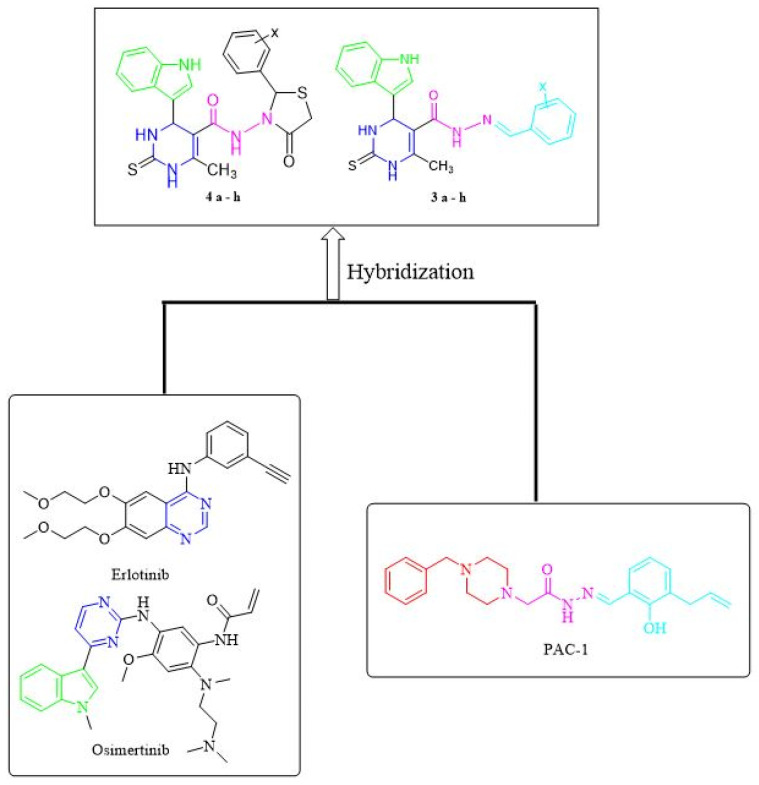
Scaffold hybridization is a drug design approach that is widely used in modern medicinal chemistry. It involves the combination of two or more known pharmacophores/scaffolds into the same structure. In this study, two major classes of anticancer scaffolds are being hybridized to form our novel compounds. The first class of anticancer scaffold is PAC-1 [45]. Dissecting PAC-1 structure, it consists of three regions (A-region, B-region, and C-regions). The B-region usually serves as a linker between A and C-regions. Previous studies on the structure-activity relationships of PAC-1 and its analogues have revealed that the acyl hydrazone moiety (B-region) plays a crucial role for their broad anticancer activity. The second class of anticancer compounds is EGFR inhibitors. Epidermal growth factor receptor (EGFR), also known as HER1 (human epidermal growth factor receptor 1), is an important subfamily of the protein kinases. EGFR is a key mediator that plays a crucial role in cellular functions [46]. The overexpression of EGFR was extensively studied because of its direct role in the development of many human solid tumors such as non-small cell lung cancer (NSCLC) [47] and breast cancer [48]. Therefore, it is expected that inhibition of EGFR protein kinase could be a potential therapeutic treatment for some prominent cancer types. There are two main classes of EGFR inhibitors [49]: (a) Small molecule tyrosine kinase inhibitors (TKIs): This class of compounds is three generations (Figure 2): (1) The first-generation EGFR inhibitors i.e., erlotinib and gefitinib; (2) The second-generation EGFR inhibitors i.e., afatinib and dacomitinib; (3) The third-generation EGFR inhibitors i.e., osimertinib; (b) Monoclonal antibodies (mAbs): There are two approved mAbs Cetuximab and Panitumumab.
Figure 2.
Examples of tyrosine kinase inhibitors that inhibit EGFR.
In this study, the design of our novel compounds was performed using a scaffold hybridization approach. These compounds are designed based on the combination of two pharmacophores (pyrimidine and indole) to form a hybrid scaffold (Figure 3). In addition, the acyl hydrazone moiety was used to bioisosterically replace the NH- of the EGFR inhibitor nucleus. All the synthesized indolyl pyrimidine hybrids were screened for their in vitro antiproliferative activity against three human cancer cell lines i.e., breast (MCF-7), liver (HepG2) and colon (HCT-116) as well as against lung fibroblast (WI38) normal cell line. This was followed-up by an in vivo antitumor evaluation using EAC tumor bearing mice for the most promising compounds. Moreover, these compounds were tested against EGFR enzymes to point out their anticancer mode of action. This complete study of structure–activity relationship (SAR) on these newly discovered compounds is expected to point out the route for more effective heterocyclic compounds as anticancer agents.
Figure 3.
Rational design for the novel indolyl pyrimidine scaffold.
2. Results and Discussion
2.1. Chemistry
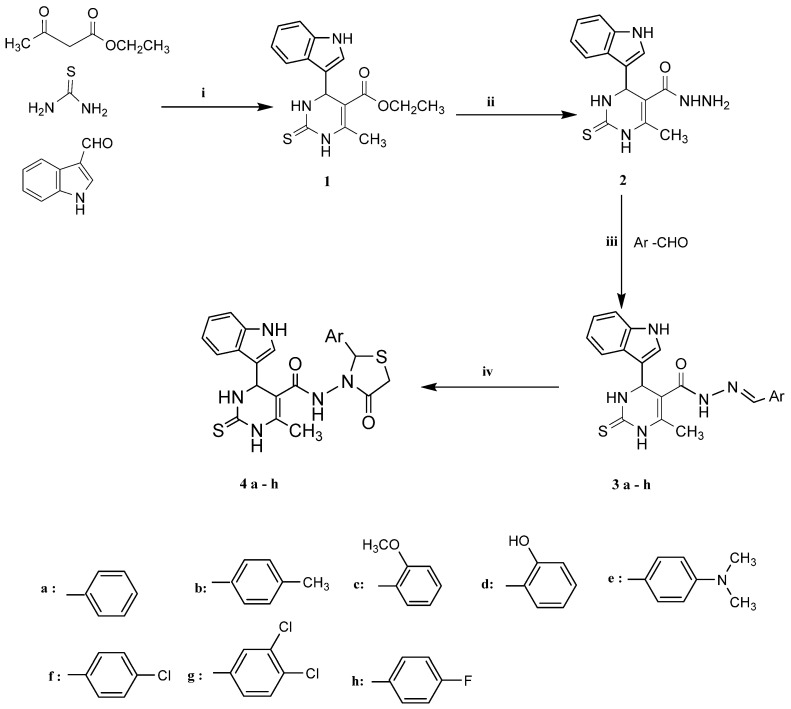
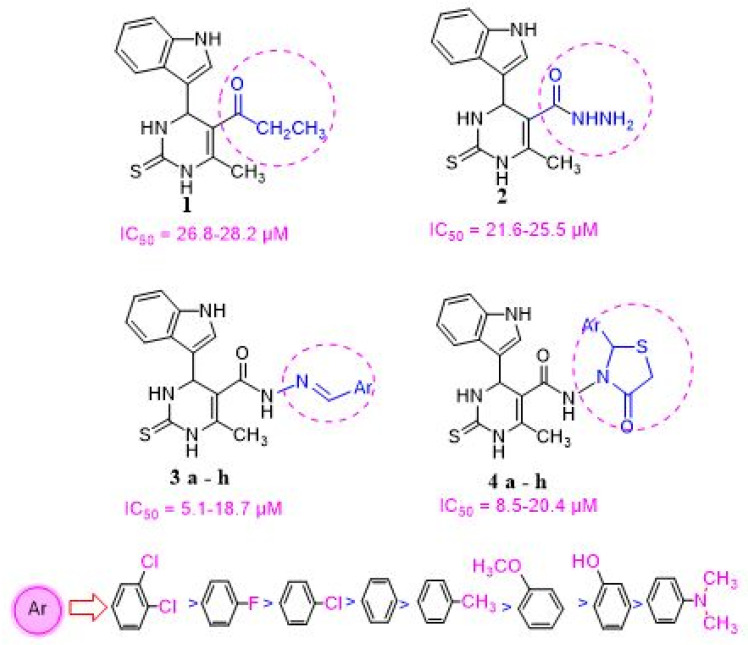
Compound 1 was synthesized through one-pot three component Biginelli reaction [50,51] using an indole-3-carbaldehyde, ethyl acetoacetate, and thiourea. The mixture was stirred under reflux in ethanol containing few drops of 37% HCl. This was followed-up by hydrazinolysis of the ester group of intermediate 1 using hydrazine hydrate in ethanol to form compound 2. Then, hydrazone derivatives 3a–h were prepared from compound 2 via heating with the appropriate aromatic aldehydes either in absolute ethanol only or with acetic acid as a catalyst [52] (Scheme 1).
Scheme 1.
Synthesis of the designed compounds (4a–h). Reagents and conditions: (i) EtOH, HCl; (ii) NH2NH2, EtOH, reflux 10 h; (iii) AcOH, reflux 5–8 h; (iv) thioglycolic acid, AcOH, reflux 5–8 h.
Furthermore, cyclo-condensation of compounds 3a–h with thioglycolic acid in glacial acetic acid was performed to afford compounds 4a–h (Scheme 1). The structure confirmation of these compounds was performed using MS, IR, 1H-NMR, 13C-NMR as well as elemental analysis.
2.2. Molecular Modeling
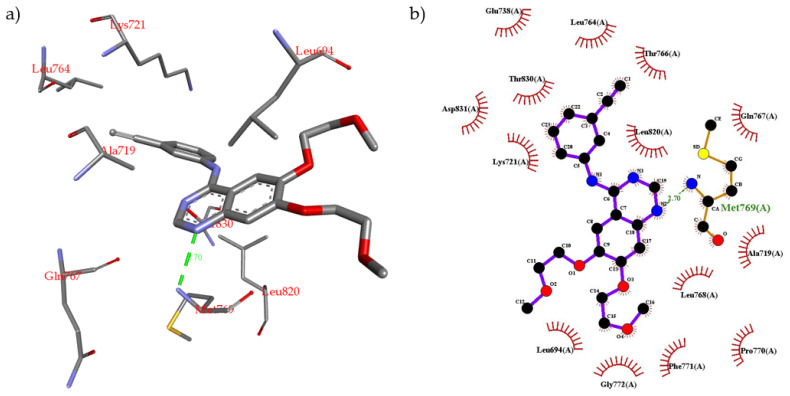
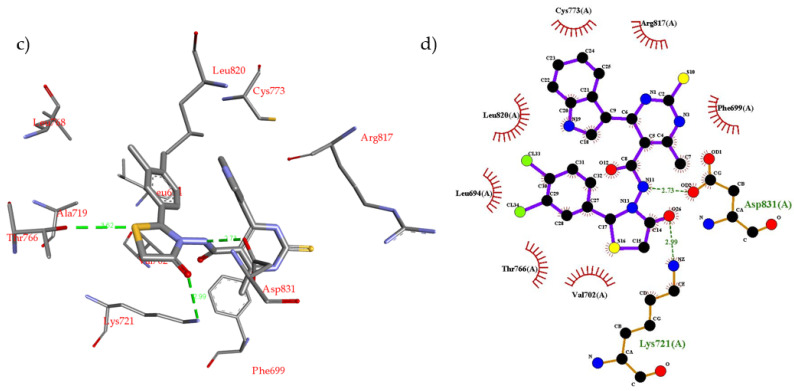
One of the cancer treatment approaches is to block the over activation of receptor tyrosine kinases (TKIs) signaling pathway i.e., inhibition of EGFR. The example for EGFR inhibitors is erlotinib, 1st generation TKIs. To investigate the protein–ligand interactions between EGFR and the novel scaffold, and the possible binding modes were predicted by molecular docking using Autodock Vina version 1.1.2 [53]. The binding mode of the most promising compound 4g was compared to that of erlotinib in complex with the EGFR binding site. The crystal structure of EGFR kinase domain in complex with erlotinib was obtained from the protein data bank (PDB ID: 1M17) [54]. The docking parameters were validated by docking erlotinib into the EGFR binding site, and the binding mode was compared to the X-ray crystal structure. The docking results of erlotinib show nine possible conformers with binding energy ranging from −6.8 kcal/mol to −7.3 kcal/mol. The most stable (lowest energy) conformer was compared to the X-ray crystal structure. This conformer closely reproduced the binding mode observed crystallographically in PDB entry 1M17 forming the same interactions with EGFR active site (see Supplementary Materials). Analysis of X-ray crystal structure of erlotinib inbound to EGFR reveals that erlotinib (N-1 of quinazoline) interacts with the backbone NH of amino acid Met769 via an H-bond (2.70 Å). In addition, erlotinib phenyl rings form several hydrophobic interactions with proximal hydrophobic residues i.e., Val702, Ala719, Lys721, Leu764, Thr766, Leu820, and Thr830 (Figure 4a,b). Compound 4g was docked inside the EGFR binding site using the same parameters used for erlotinib. It was found that compound 4g binds more favorably than erlotinib inside the EGFR active site. The docking results of compound 4g show nine possible conformers with binding energy ranging from −8.8 kcal/mol to −9.6 kcal/mol. Compound 4g fits well inside the EGFR binding site and forms several non-covalent interactions (Figure 4c,d). The most stable conformer was used to interpret these non-covalent interactions. Compound 4g was found to form three H-bonding interactions as follows: (1) The -NH of the linker forms an H-bond (2.73 Å) with Asp831 residue; (2) The -C=O of the thiazolidine ring forms an H-bond (2.99 Å) with Lys721 residue; (3) The S-atom of the thiazolidine ring forms weak H-bond (3.62 Å) with Thr766 residue. In addition, several strong hydrophobic interactions between the aromatic rings of compound 4g and the proximal amino acid hydrophobic residues, i.e., Leu694, Phe699, Val702, Cys737, Thr766, and Leu820, were observed.
Figure 4.
(a) Non-covalent interactions of erlotinib inside the EGFR active site; (b) 2D schematic representation of erlotinib inside the EGFR binding site; (c) non-covalent interactions of docked compound 4g inside the EGFR active site; (d) 2D schematic representation of compound 4g inside the EGFR binding site.
2.3. Biological Screening
2.3.1. In Vitro Antiproliferative Activity
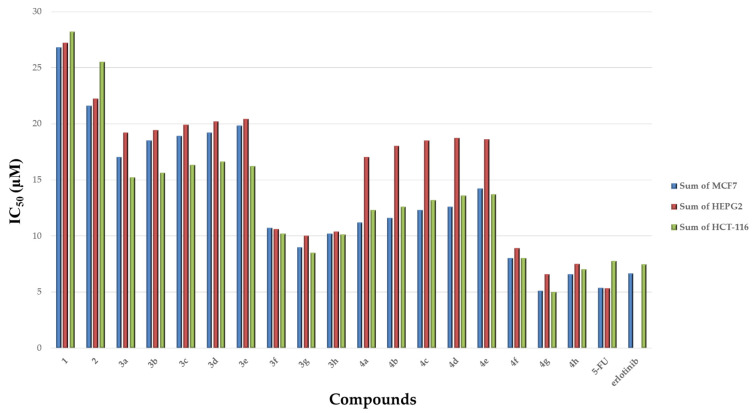
Compounds 1–4 were assessed for their in vitro cytotoxic activity against three human cell lines i.e., breast (MCF-7), liver (HepG2), colon (HCT-116) cancer cell lines as well as in lung fibroblast (WI38) normal cell line using the Resazurin Cell Growth Inhibition Assay [55]. Both erlotinib and 5-Fluorouracil (5-FU) were used as reference drugs. Concentrations of the tested compounds that give about 50% inhibition of cell viability (IC50, mean of triplicate) in µM were obtained. The results revealed that most of the tested compounds showed significant in vitro antiproliferative activity when compared to normal cells. Compounds 1–2 were found to possess moderate antiproliferative activity against the three cell lines when compared to normal cell lines. Compound 4g showed the most potent inhibitory profile against MCF-7, HepG2, and HCT-116 when compared to normal cell lines. In addition, compounds 4f and 4h showed promising inhibitory activity against all cell lines, whereas compounds 3f, 3g, and 3h showed notable effectiveness against these cell lines. The IC50 range of the tested compounds was 8.30 to 20.4 μM. To interpret the in vitro results and correlate it to compounds’ structures (structure activity relation (SAR)), it was found that the presence of -COOEt (compound 1) or -CONHNH2 (compound 2) substituents on C5 of the pyrimidine nucleus resulted in moderate efficacy against MCF-7, HepG2, and HCT-116 cell line. However, if replaced by a hydrazone substituent, a more prominent efficacy toward the three cancer cell lines was observed. Notably, the inhibitory activity of compounds 3a-h is influenced by the type of the hydrazone formed. The most promising in vitro antiproliferative activity was obtained when the hydrazone is bearing electron-withdrawal groups i.e., 4-chlorobenzylidene moiety (compound 3f), 3,4-dichloro benzylidene moiety (compound 3g) and 4-fluorobenzylidene moiety (compound 3h) compared to the standard treatment (Table 1, Figure 5). The inhibitory activity of compounds was reduced when the hydrazone group is bearing electron donating groups i.e., 4-methyl group (compound 3b), 2-methoxy group (compound 3c), 4-hydroxy (compound 3d) and 4-dimethylamino group (compound 3e). The trend of antiproliferative activity of compounds 3a–e against all tested cell lines has the following descending order: (3g > 3h >3f >3a >3b > 3c > 3d > 3e). Furthermore, the addition of the thiazolidinone ring via a condensation reaction showed a more profound effectiveness against the three cancer cell lines compared to the standard treatment. The most potent compounds were found to follow what was revealed before that presence of an electron withdrawing group, i.e., 4-chlorophenyl (compound 4f), 3,4-dichloro (compound 4g) and 4-fluorophenyl (compound 4h), improves the antiproliferative activity. The higher the lipophilic character of the electron withdrawing groups i.e., Cl on the phenyl ring, the more potent is the antiproliferative activity. In addition, installing an electron donating group was found to have no effect on the activity of these compounds (Table 1, Figure 4). All of the tested compounds 4a–h showed in vitro antiproliferative activities with IC50 values ranging from 5.02 to 18.62 μM. The trend of antiproliferative activity of these hybrids 4a–e against all tested cancer cell lines had the following descending order: (4g > 4h > 4f > 4a > 4b > 4c > 4d > 4e). Compound 4g showed the most potent activity against MCF-7, HepG2 and HCT-116 with IC50 = 5.1 ± 1.14, 5.02 ± 1.19 and 6.6 ± 1.40 μM, respectively, compared to the reference treatment (Table 1). This is mainly because this compound has the highest lipophilicity as well as less electron density as it has 2-Cl atoms (more electronegativity) on the indolyl pyrimidine-thiazolidinone.
Table 1.
Cytotoxic activity (IC50) of compounds 1–4 against MCF-7, HCT-116, HepG2 and WI-38 cell lines.
| Compounds | * Cytotoxic Activity IC50
a (μM) |
|||
|---|---|---|---|---|
| MCF-7 | HCT-116 | HepG2 | WI-38 | |
| 1 | 26.8 ± 2.20 | 27.2 ± 1.11 | 28.8 ± 2.25 | 30.0 ± 1.20 |
| 2 | 21.6 ± 1.8 | 22.2 ± 1.5 | 25.5 ± 2.17 | 40.50 ± 2.25 |
| 3a | 17.0 ± 1.80 | 15.2 ± 2.32 | 19.2 ± 1.50 | 38.23 ± 2.10 |
| 3b | 18.50 ± 2.11 | 15.6 ± 3.15 | 19.4 ± 2.19 | 39.13 ± 1.30 |
| 3c | 18.9 ± 2.00 | 16.3 ± 4.16 | 19.9 ± 2.06 | 37.42 ± 1.49 |
| 3d | 19.2± 2.35 | 16.6 ± 1.27 | 20.2 ± 2.33 | 35.26 ± 2.42 |
| 3e | 19.8 ± 2.70 | 16.2 ± 1.15 | 20.4 ± 2.30 | 40.01 ± 2.31 |
| 3f | 10.7 ± 2.30 | 10.20 ± 1.18 | 10.6 ± 2.32 | 29.82 ± 1.51 |
| 3g | 9.0 ± 2.40 | 8.5 ± 2.30 | 10.0 ± 2.46 | 30.01 ± 3.72 |
| 3h | 10.2 ± 2.01 | 10.11 ± 2.45 | 10.4 ± 2.08 | 28.10 ± 4.22 |
| 4a | 11.2 ± 4.31 | 12.3 ± 2.17 | 17.0 ± 4.30 | 38.03 ± 5.51 |
| 4b | 11.6 ± 3.22 | 12.6 ± 1.05 | 18.0 ± 3.20 | 39.8 ± 4.51 |
| 4c | 12.3 ± 4.08 | 13.2 ± 1.03 | 18.5 ± 4.38 | 29.5 ± 4.02 |
| 4d | 12.6 ± 1.70 | 13.6 ± 1.09 | 18.7 ± 1.20 | 29.9 ± 3.63 |
| 4e | 14.2 ± 0.74 | 13.7 ± 0.55 | 18.62 ± 0.94 | 20.24 ± 5.52 |
| 4f | 8.01 ± 1.83 | 8.02 ± 0.26 | 8.9 ± 1.73 | 19.33 ± 5.87 |
| 4g | 5.1 ± 1.14 | 5.02± 1.19 | 6.6 ± 1.40 | 16.32 ± 3.21 |
| 4h | 6.6 ± 1.28 | 7.02 ± 0.46 | 7.5 ± 1.29 | 18.12 ± 2.06 |
| 5-FU | 5.38 ± 0.24 | 7.88 ± 0.2 | 5.34 ± 0.4 | 5.70 ± 1.50 |
| erlotinib | 6.65 ± 0.82 | 7.49 ± 0.65 | nd | 22.50 ± 0.65 |
* Three independent experiments were performed for each concentration. a IC50 values represent mean ± SD of three experiments.
Figure 5.
Cytotoxic activity (IC50) of compounds 1–4, 5-FU and erlotinib in different cell lines.
As shown in Figure 6, there is a consistent relation between the lipophilicity and/or electronic property of the substituent groups on the Ar-group and the antiproliferative activity. It was found that the nature of substitution on the Ar group affected the order of activity of tested compounds against tested cell lines: (3,4-(Cl)2 > F > Cl > H > CH3 > OCH3 > OH > N(CH3)2). Installment of a more electronegative and a lipophilic substituent to phenyl ring on either the pyrimidine and thiazolidinone respectively enhance cytotoxic activity against MCF-7, HepG2, and HCT-116 cancer cells. In addition, the sensitivity of the tested cell lines to compounds 1–4 was found to be in the following descending order: (MCF-7, HCT-116 > HepG2). This is mainly because these cancer cell lines have different expression levels for EGFR.
Figure 6.
Structure activity relationship (SAR) of the pyrimidine derivatives against different cell lines.
2.3.2. In Vivo Antitumor Evaluation
The most promising compounds with the best in vitro antitumor profile i.e., compounds 3g, 4f, 4g and 4h were further evaluated for their in vivo antitumor efficacy against Ehrlich ascites carcinoma (EAC) in mice. The antitumor activity of these compounds was assessed by the change in tumor volume, viable cell count, mean survival time (MST) and % increase in lifespan (% ILS) of the EAC tumor bearing mice and 5-FU (standard treatment) [56]. The mean survival time (MST) and % increase in lifespan (% ILS) of EAC tumor bearing mice, as well as the influence of these compounds on viable tumor cells and the tumor volume, are shown in Table 2. The hematological profile of these mice was also assessed in comparison with the standard treatment with 5-FU, and no obvious effect on RBCs or WBCs was observed. It was found that compound 4g exhibited the highest % ILS of mice, as well as results illustrated that treatment with compound 4g resulted in the lowest tumor volume and viable tumor cell count. Tumor volume and viable tumor cell count were also determined, and results suggest that treatment with compound 4g resulted in the lowest tumor volume and viable tumor cell count.
| MST = (day of first death + day of last death)/2 |
| %ILS = [(MST of treated group/MST of control group) − 1] × 100 |
Table 2.
Effect of treatment by compounds 3g, 4f, 4g, and 4h on MST, % ILS, tumor volume, and viable tumor cell count in EAC tumor bearing mice.
| Group | MST (day) a | % ILS a | Tumor Volume (mL) a | Viable Tumor Cell Count (106/mL) a |
|---|---|---|---|---|
| Normal | nd | nd | nd | Nd |
| EAC only | 16.5 | nd | 8.01 | 80.25 |
| 3g | 30 | 110.0 | 2.97 | 48.00 |
| 4f | 37 | 130.0 | 2.50 | 40.70 |
| 4g | 46 | 230.0 | 0.89 | 21.70 |
| 4h | 40 | 180.0 | 1.18 | 28.50 |
| 5-FU | 53.0 | 265.5 | 0.80 | 19.07 |
a Results are the mean of two experiments. nd, not determined.
2.3.3. In Vitro EGFR Inhibition Assay
The correlation between the cytotoxic activity of compounds and the level of EGFR expression in the cancerous cell lines used was the cornerstone for an in vitro testing of the new hybrid compounds against EGFR. The data revealed that the sensitivity of the tested compounds to cell lines MCF-7, HCT-116 was significantly more than to the HepG2 cell lines. This is mainly because these cancerous cell lines exhibit different EGFR expression levels. The HepG2 cell line was found to have undetectable EGFR expression [57]. On the other hand, both MCF-7 and HCT-116 cell lines were found to have considerable EGFR expression [58,59]. WI38 cell line does not express EGFR. A first-generation EGFR inhibitor, erlotinib, was used as standard treatment. EGFR was treated with 10 µM of the chosen compounds as well as the reference treatment with erlotinib. The in vitro screening of these analogs against EGFR revealed inhibitory activity range 53–79%, and the best inhibitory activity 79% was shown by compound 4g followed by 4f (71%). The % inhibition of EGFR activity was measured for compounds 3g (53%), and for compound 4h (70%) (Table 3). IC50 for these compounds was determined. Most of the tested analogs showed comparable inhibitory activities to erlotinib (0.30 μM) as shown in Table 3. The tested analogs showed EGFR inhibitory activity with IC50 values ranging from 0.25 to 0.50 µM. Compound 4g was found to be the most active one against EGFR comparable to erlotinib. In addition, compounds 4f and 4h exhibited good activity with IC50 value of 0.38 and 0.39 µM, respectively.
Table 3.
EGFR % inhibition at 10 µM concentration and IC50 (µM) of compounds 3g, 4f, 4g, and 4h.
| Compound | % Inhibition of EGFR | IC50 (µM) a |
|---|---|---|
| 3g | 53 | 0.50 ± 0.04 |
| 4f | 71 | 0.38 ± 0.02 |
| 4g | 79 | 0.25 ± 0.01 |
| 4h | 70 | 0.39 ± 0.02 |
| erlotinib | 81 | 0.30 ± 0.01 |
a mean of two independent replicates ± SD.
3. Materials and Methods
3.1. Instruments
All melting points were determined using Electro-thermal IA 9100 apparatus (Shimadzu, Kyoto, Japan) and were uncorrected. FT-IR spectra were recorded using potassium bromide pellets on a PerkinElmer 1650 spectrophotometer (USA). 1H-NMR and 13C-NMR spectra were obtained in DMSO-d6 using a Varian Mercury (300 MHz and 75 MHz respectively) spectrometer (Varian, Crawley, UK), and chemical shifts were given as ppm from TMS as internal reference. Mass spectra were recorded on 70 eV EI Ms-QP 1000 EX. Microanalyses were performed using Vario, Elementar apparatus, Organic Microanalysis Unit, Faculty of Science, Cairo University, Cairo, Egypt, and the results were within the accepted range (0.40) of the calculated values. Column Chromatography was performed on (Merck, Darmstadt, Germany) Silica gel 60 (particle size 0.06–0.20 mm).
3.2. Chemistry
3.2.1. Ethyl 4-(1H-indol-3-yl)-6-methyl-2-thioxo-1,2,3,4-tetrahydropyrimidine-5-Carboxylate (1)
A mixture of thiourea (10 mmol), indole-3-carbaldehyde (10 mmol), and ethyl acetoacetate (10 mmol) in absolute ethanol (25 mL) containing 37% HCl (7 drops) was heated to reflux for 7 h, the reaction mixture was allowed to cool down to room temperature and was neutralized with 25% ammonia (0.5 mL). The precipitate formed was filtered off, washed several times with 50% ethanol, and was finally recrystallized from ethanol to give compound 1, m.p. 245–247 °C as reported [50].
3.2.2. 4-(1H-indol-3-yl)-6-methyl-2-thioxo-1,2,3,4-tetrahydropyrimidine-5-carbohydrazide (2)
A mixture of the ester derivative 1 (10 mmol) and hydrazine hydrate (50 mmol, 99%) in 20 mL ethanol was refluxed for 10 h, then was cooled down to room temperature and poured into ice/water mixture. The produced precipitate was filtered off, dried, and was crystallized from ethanol to give compound 2, m.p. 208–210 °C as reported [50].
3.2.3. General Procedure for the Preparation of Compounds (3a–h)
A mixture of compound 2 (10 mmol) and the appropriate aldehyde (20 mmol) in glacial acetic acid (35 mL) was refluxed for 5–8 h. The solution was cooled down to room temperature and ice was added, the formed precipitate was filtered off, washed with water, and was further crystallized from ethanol to give the desired hydrazone (3a–h). The titled compounds 3a, 3d, 3f, and 3h were synthesized according to the reported method [50].
4-(1H-indol-3-yl)-6-methyl-2-thioxo-1,2,3,4-tetrahydropyrimidine-5-carboxylic acid (4-methyl-benzylidene)-hydrazide (3b)
Yellow crystals, yield 80%, m.p. 240–242 °C. IR (KBr) v max (cm−1): 3340 (NH), 1685 (C=O), 1590 (C=N). 1H-NMR (300 MHz, DMSO-d6) 2.1 (s, 3H, CH3), 2.5 (s, 3H, CH3, pyrim), 5.2 (s, 1H, pyrimidine), 6.6–8.2 (m, 9H, Ar–H), 8.7 (s, 1H, CH=N), 10.4,10.7,10.9, 11.5 (4 s, 4H, 4NH, D2O exchangeable). 13C-NMR (300 MHz, DMSO-d6) 18.0 and 20.0 (2CH3), 55.4 (C-4 pyrimidine), 106.5 (C-5 pyrimidine), 111.2–140.2 (aromatic C), 151.1 (C-6 pyrimidine), 156.0 (C=N), 168.1 (C=O), 178.2 (C=S); MS(EI): m/z 403 [M+] (14%); Anal. Calcd. for C22H21N5OS (403.5): C, 65.49; H, 5.25; N, 17.36; Found: C, 65.40; H, 5.20; N, 17.31.
4-(1H-indol-3-yl)-6-methyl-2-thioxo-1,2,3,4-tetrahydropyrimidine-5-carboxylic acid (2-methoy- benzylidene)-hydrazide (3c)
Yellow crystals, yield 82%, m.p. 220–222 °C. IR (KBr) v max (cm−1): 3366 (NH), 1689 (C=O), 1604 (C=N). 1H-NMR (300 MHz, DMSO-d6) 2.5 (s, 3H, CH3, pyrim), 3.3 (s, 3H, OCH3), 5.0 (s, 1H, pyrimidine), 7.0–8.7 (m, 9H, Ar–H), 8.8 (s, 1H, CH=N), 9.6,10.0,10.4,11.6 (4s, 4H, 4NH, D2Oexchangeable). 13C-NMR (300 MHz, DMSO-d6) 18.3 (CH3), 58.2 (OCH3), 55.3 (C-4 pyrimidine), 106.8 (C-5 pyrimidine), 118.6–149.0 (aromatic C), 150.7 (C-6 pyrimidine), 155.2 (C=N), 165.2 (C=O), 177.6 (C=S). MS (EI): m/z 419 [M+] (11%). Anal. Calcd. for C22H21N5O2S (419.49): C, 62.99; H, 5.05; N, 16.69; Found: C, 62.90; H, 5.00; N, 16.61.
4-(1H-indol-3-yl)-6-methyl-2-thioxo-1,2,3,4-tetrahydropyrimidine-5-carboxylic acid (4-dimethylamino-benzylidene)-hydrazide (3e)
Yellowish brown solid, yield 76%, m.p. 180–182 °C. IR (KBr) v max (cm−1): 3442 (NH),1685 (C=O), 1603 (C=N). 1H-NMR (300 MHz, DMSO-d6) 2.0 (s, 3H, CH3, pyrim), 2.5 (s, 6H, CH3), 5.0 (s, 1H, pyrimidine), 6.7–8.2 (m, 9H, Ar–H), 8.7(s, 1H, CH=N), 9.6,10.0,10.4, 11.0 (4s, 4H, 4NH, D2Oexchangeable). 13C-NMR (300 MHz, DMSO-d6) 18.0 (CH3), 43.6 (N-CH3), 55.0 (C-4 pyrimidine), 111.6-137.0 (aromatic Cs), 108.0 (C-5 pyrimidine), 153.0 (C-6 pyrimidine), 158.0 (C=N), 168.8 (C=O), 178.2 (C=S). MS(EI): m/z 432 [M+] (15 %). Anal. Calcd. for C23H24N6OS (432.17): C, 63.87; H, 5.59; N, 19.43; Found: 63.80; H, 5.51; N, 19.42.
4-(1H-indol-3-yl)-6-methyl-2-thioxo-1,2,3,4-tetrahydropyrimidine-5-carboxylic acid (3,4-dichloro- benzylidene)-hydrazide (3g)
Brown crystals, yield 64%, m.p. 160–162 °C. IR (KBr) v max (cm−1): 3350 (NH), 1685 (C=O), 1686 (C=N). 1H-NMR (300 MHz, DMSO-d6) 2.50 (s, 3H, CH3, pyrim), 5.2 (s, 1H, pyrimidine), 7.2–8.2 (m, 8H, Ar-H), 8.7 (s, 1H, CH=N), 9.9, 10.0,10.1,11.1 (4s, 4H, 4NH, D2O exchangeable). 13C-NMR (300 MHz, DMSO-d6) 18.2 (CH3), 55.3 (C-4 pyrimidine), 106.6 (C-5 pyrimidine), 111.0–140.4 (aromatic Cs), 151.3 (C-6 pyrimidine),154.2 (C=N), 178.4 (C=S), 186.6 (C=O). MS (EI): m/z 457 [M+] (12%), 459 [M+2] (4%). Anal. Calcd. for C21H17Cl2N5OS (458.3636): C, 55.03; H, 3.74; N, 15.28; Found: C, 55.05; H, 3.77; N, 15.20.
3.2.4. General Procedure for the Preparation of Compounds (4a–h)
A mixture of compounds 3a–h (10 mmol) and thioglycolic acid (10 mmol) in glacial acetic acid (30 mL) was refluxed for 5–8 h. The excess solvent was evaporated under vacuum, and the obtained residue was poured on crushed ice. The formed solid product was filtered off, washed with water, and was crystallized from ethanol to obtain the desired products 4a–h, respectively.
4-(1H-indol-3-yl)-6-methyl-N-(4-oxo-2-phenylthiazolidin-3-yl)-2-thioxo-1,2,3,4-tetrahydropyrimidine-5-carboxamide (4a)
Yellow solid, yield 80%, m.p. 270–272 °C. IR (KBr) v max (cm−1): 3390 (NH), 1654,1704 (2 C=O), 3060 (CH-Ar); 1H-NMR (300 MHz, DMSO-d6) 2.5 (s, 3H, CH3), 3.8 (s, 2H, CH2 of thiazolidinone ring), 5.9 (1H, s, CH of thiazolidinone ring), 5.1 (s, 1H, pyrimidine), 7.4–8.7 (m, 10H, Ar-H), 9.8,10.1,10.8, 11.0 (4s, 4H, 4NH, D2O exchangeable). 13C-NMR (300 MHz, DMSO-d6) 18.9 (CH3), 58.2 (CH), 39.3 (CH2), 55.5 (C-4 pyrimidine), 113.9–148.4 (aromatic Cs), 106.0 (C-5 pyrimidine), 155.3 (C-6 pyrimidine), 165.6 and 169.6 (2 C=O), 179.4 (C=S); MS (EI): m/z 463 [M+] (5%). Anal. Calcd. for C23H21N5O2S2 (463.57): C, 59.59; H, 4.57; N, 15.11; Found: C, 59.55; H, 4.55; N, 15.10.
4-(1H-indol-3-yl)-6-methyl-N-(4-oxo-2-(p-tolyl)thiazolidin-3-yl)-2-thioxo-1,2,3,4-tetrahydropyrimidine-5-carboxamide (4b)
Off-white solid, yield 80%, m.p. 220–222 °C. IR (KBr) v max (cm−1): 3260 (NH), 1665 and 1690 (2 C=O), 3050 (CH-Ar), 2910 (CH-sp3), 1H-NMR (300 MHz, DMSO-d6) 2.5–2.6 (s, 6H, 2CH3), 3.90 (2H, s, CH2 of thiazolidinone ring), 5.90 (1H, s, CH of thiazolidinone ring), 5.1 (s, 1H,pyrimidine), 6.7–8.5 (m, 10H, Ar-H), 10.0, 10.2, 10.6, 11.0 (4 s, 4H, 4NH, D2O exchangeable); 13C-NMR (300 MHz, DMSO-d6) 18.1 and 20.1 (2 CH3), 57.0 (CH), 38.0 (CH2), 55.2 (C-4 pyrimidine), 111.0–140.1 (aromatic Cs), 106.1 (C-5 pyrimidine), 152.1 (C-6 pyrimidine), 165.0 and 168.1 (2 C=O), 179.0 (C=S); MS(EI): m/z 477 [M+] (8%). Anal. Calcd. for C24H23N5O2S2 (477.60): C, 60.36; H, 4.85; N, 14.66; Found: C, 60.34; H, 4.82; N, 14.60.
4-(1H-indol-3-yl)-N-(2-(2-methoxyphenyl)-4-oxothiazolidin-3-yl)-6-methyl-2-thioxo-1,2,3,4-tetrahydropyrimidine-5-carboxamide (4c)
Buff solid, yield 80%, m.p. 200–202 °C. IR (KBr) v max (cm−1): 3255 (NH), 1660 and 1693 (2 C=O), 3090 (CH-Ar), 2940 (CH-sp3). 1H-NMR (300 MHz, DMSO-d6) 2.3 (s, 3H, CH3), 3.3 (s, 3H, OCH3), 4.0 (2H, s, CH2 of thiazolidinone ring), 5.9 (1H, s, CH of thiazolidinone ring), 5.1 (s, 1H, pyrimidine), 7.2–7.3 (m, 9H, Ar-H), 9.6, 10.3, 10.8, 11.2 (4s, 4H, 4NH, D2O exchangeable); 13C-NMR (300 MHz, DMSO-d6) 18.5 (CH3), 59.9 (OCH3), 58.5 (CH), 39.3 (CH2), 55.2 (C-4 pyrimidine), 113.5-145.3 (aromatic Cs), 108.1 (C-5 pyrimidine), 155.5 (C-6 pyrimidine), 166.3 and 168.4 (2 C=O), 179.3 (C=S). MS(EI): m/z 493 [M+] (10%). Anal. Calcd. for C24H23N5O3S2 (493.12): C, 58.40; H, 4.70; N, 14.19; Found: C, 58.45; H, 4.77; N, 14.15.
N-(2-(2-hydroxyphenyl)-4-oxothiazolidin-3-yl)-4-(1H-indol-3-yl)-6-methyl-2-thioxo-1,2,3,4-tetrahydropyrimidine-5-carboxamide (4d)
Yellow crystals, yield 75%, m.p.180–182 °C. IR (KBr) v max (cm−1): 3250(NH), 1662 and 1692 (2 C=O), 3070 (CH-Ar), 2930 (CH-sp3); 1H-NMR (300 MHz, DMSO-d6) 2.6 (s, 3H, CH3), 3.92 (2H, s, CH2 of thiazolidinone ring), 5.92 (1H, s, CH of thiazolidinone ring), 5.0 (s, 1H, pyrimidine), 6.5-8.6 (m, 9H, Ar-H), 11.2 (s, 1H, OH, D2Oexchangeable), 10.0, 10.2, 10.8, 11.0 (s, 4H, 4NH, D2O exchangeable); 13C-NMR (300 MHz, DMSO-d6) 18.2 (CH3), 57.0 (CH), 38.0 (CH2), 55.0 (C-4 pyrimidine), 112.0–140.0 (aromatic Cs), 106.2 (C-5 pyrimidine), 152.0 (C-6 pyrimidine), 165.2 and 168.3 (2 C=O), 179.2 (C=S). MS (EI): m/z 479 [M+] (11%). Anal. Calcd. for C23H21N5O3S2 (479.57): C, 57.60; H, 4.41; N, 14.60; Found: C, 57.62; H, 4.44; N, 14.65.
N-(2-(4-(dimethylamino)phenyl)-4-oxothiazolidin-3-yl)-4-(1H-indol-3-yl)-6-methyl-2-thioxo-1,2,3,4-tetrahydropyrimidine-5-carboxamide (4e)
Light yellow solid, yield 55%, m.p.170–172 °C. IR (KBr) v max (cm−1): 3275 (NH), 1665 and 1695 (2 C=O), 3080 (CH-Ar), 2920 (CH-sp3). 1H-NMR (300 MHz, DMSO-d6) 2.3 (s, 3H, CH3), 2.5 (s, 3H, 2CH3), 3.8 (2H, s, CH2 of thiazolidinone ring), 5.9 (1H, s, CH of thiazolidinone ring), 5.1 (s, 1H, pyrimidine), 6.8–8.4 (m, 9H, Ar-H), 9.0, 9.9, 10.2, 10.7 (4 s, 4H, 4NH, D2O exchangeable); 13C-NMR (300 MHz, DMSO-d6) 18.2 (CH3), 43.6 (N-CH3), 58.0 (CH), 37.0 (CH2), 55.2 (C-4 pyrimidine), 113.0-140.0 (aromatic Cs), 106.0 (C-5 pyrimidine), 152.2 (C-6 pyrimidine), 165.0, 168.0 (2 C=O), 179.3 (C=S). MS (EI): m/z 506 [M+] (14%). Anal. Calcd. for C25H26N6O2S2 (506.64): C, 59.27; H, 5.17; N, 16.59; Found: C, 59.28; H, 5.19; N, 16.58.
N-(2-(4-chlorophenyl)-4-oxothiazolidin-3-yl)-4-(1H-indol-3-yl)-6-methyl-2-thioxo-1,2,3,4-tetrahydropyrimidine-5-carboxamide (4f)
Yellow solid, yield 65%, m.p. 140–142 °C. IR (KBr) v max (cm−1): 3265 (NH), 1660 and 1694 (2 C=O), 3055 (CH-Ar), 2925 (CH-sp3). 1H-NMR (300 MHz, DMSO-d6) 2.5 (s, 3H, CH3), 3.9 (2H, s, CH2 of thiazolodinone ring), 5.9 (1H, s, CH of thiazolidinone ring), 5.1(s, 1H, pyrimidine), 7.1–7.5 (m, 9H, Ar-H), 9.6, 9.9, 10.3, 11.2 (s, 4H, 4NH, D2O exchangeable); 13C-NMR (300 MHz, DMSO-d6) 18.2 (CH3), 57.3 (CH), 38.3 (CH2), 55.2 (C-4 pyrimidine), 111.0-142.0 (aromatic Cs), 106.0 (C-5 pyrimidine), 152.4 (C-6 pyrimidine), 165.1 and 168.0 (2 C=O), 179.5 (C=S); MS(EI): m/z 497 [M+](10 %), 499 [M+2] (3%). Anal. Calcd. for C23H20ClN5O2S2 (498.02): C, 55.47; H, 4.05; N, 14.06; Found: C, 55.44; H, 4.02; N, 14.04.
N-(2-(3,4-dichlorophenyl)-4-oxothiazolidin-3-yl)-4-(1H-indol-3-yl)-6-methyl-2-thioxo-1,2,3,4-tetrahydropyrimidine-5-carboxamide (4g)
Yellow crystals, yield 73%, m.p. 150–152 °C. IR (KBr) v max (cm−1): 3270 (NH), 1660 and 1698 (2 C=O), 3065 (CH-Ar), 2945 (CH-sp3); 1H-NMR (300 MHz, DMSO-d6) 2.6 (s, 3H, CH3), 3.95 (s, CH2 of thiazolidinone ring), 5.92 (1H, s, CH of thiazolidinone ring), 5.0 (s, 1H, pyrimidine), 6.78–8.27 (m, 8H, Ar-H), 10.0, 10.4, 10.6, 11.0 (4 s, 4H, 4NH, D2O exchangeable); 13C-NMR (300 MHz, DMSO-d6) 18.3 (CH3), 57.5 (CH), 38.0 (CH2). 55.3 (C-4 pyrimidine), 113.0–140.0 (aromatic Cs), 106.0 (C-5 pyrimidine), 152.5 (C-6pyrimidine), 165.9 and 168.0 (2 C=O), 179.5 (C=S). MS (EI): m/z 531 [M+] (16%), 533 [M+2] (5%). Anal. Calcd. for C23H19Cl2N5O2S2 (532.46): C, 51.88; H, 3.60; N,13.15; Found: C, 51.85; H, 3.63; N, 13.17.
N-(2-(4-fluorophenyl)-4-oxothiazolidin-3-yl)-4-(1H-indol-3-yl)-6-methyl-2-thioxo-1,2,3,4-tetrahydropyrimidine-5-carboxamide (4h)
White solid, yield 75%, m.p.155–158 °C. IR (KBr) v max (cm−1): 3280 (NH), 1665 and 1697 (2 C=O), 3075 (CH-Ar), 2955 (CH-sp3); 1H-NMR (300 MHz, DMSO-d6) 2.6 (s, 3H, CH3), 3.90 (2H, s, CH2 of thiazolidinone ring), 5.90 (1H, s, CH of thiazolidinone ring), 5.1 (s, 1H, pyrimidine), 6.6–8.5 (m, 8H, Ar-H), 10.0, 10.2, 10.6, 11.0 (4 s, 4H, 4NH, D2O exchangeable); 13C-NMR (300 MHz, DMSO-d6) 18.4 (CH3), 57.1 (CH), 38.1 (CH2), 55.4 (C-4 pyrimidine), 112.0-143.0 (aromatic Cs), 106.0 (C-5 pyrimidine), 152.5 (C-6 pyrimidine), 165.5 and168.0 (2 C=O), 179.5 (C=S); MS (EI): m/z 481 [M+] (10%). Anal. Calcd. for C23H20FN5O2S2 (481.56): C, 57.36; H, 4.19; N, 14.54; Found: C, 57.34; H, 4.16; N, 14.52.
3.3. Biological Evaluation
3.3.1. In Vitro Cytotoxicity Assay
In vitro cytotoxicity was measured according to the Resazurin Cell Growth Inhibition Assay [53,55,59,60] Alamar Blue or Resazurin (Promega, Mannheim, Germany) reduction assay was used to assess the cytotoxicity of the studied samples. The assay tests cellular viability and mitochondrial function. Briefly, adherent cells were grown in tissue culture flasks, and then harvested by treating the flasks with 0.025% trypsin and 0.25 mM EDTA for 5 min. Once detached, cells were washed, counted, and an aliquot (5 × 103 cells) were placed in each well of a 96-well cell culture plate in a total volume of 100 mL. Cells were allowed to attach overnight and then treated with samples. The final concentration of samples ranged from 0 to 100 mM. After 48 h, 20 mL Resazurin 0.01% w/v solution was added to each well and the plates were incubated at 37 °C for 1–2 h. Fluorescence was measured on an automated 96-well Infinite M2000 ProTM plate reader (Tecan, Crailsheim, Germany) using an excitation wavelength of 544 nm and an emission wavelength of 590 nm. 5-FU as well as erlotinib were used as standard treatment. Each assay was performed as triplicates. It should be noted that DMSO was used as a solvent control such that the vehicle concentration does not exceed 0.05% in all concentrations used. Cancer cells were treated with gradually increasing concentrations of our compounds from (0.01 μM to 100 μM). The viability was compared based on a comparison with untreated cells. IC50 (on cancer cells) were the concentration of sample required to inhibit 50% of the cell proliferation and were calculated from a calibration curve by a linear regression using Microsoft Excel.
3.3.2. In Vivo Antitumor Assay
In vivo antitumor evaluation was carried out as previously reported [56,60] (see Supplementary Materials).
3.3.3. In Vitro EGFR Inhibitory Assay
The most potent analogs 3g, 4f, 4g, and 4h against MCF-7, HepG2 and HCT-116 cell lines were further tested for EGFR inhibitory activity using Enzy Chrom TM Kinase Assay Kit (EKIN-400) as previously described [60]. In addition, 20 µL reaction mixture containing the EGFR kinase, ATP, and substrate in the provided assay buffer. Set up a blank control that contains ATP and substrate but no enzyme. Incubate at desired temperature for 30 min. Prepare 900 µL 10µM (adenosine diphosphate) ADP premix by mixing 3 µL of 3 mM standard and 897 µL distilled water. Transfer 20 µL standards into separate wells of the plate. Prepare enough working reagent for each well. Add 40 µL working reagent to each assay well. Tap plate to mix. Incubate at room temperature for 10 min. Read fluorescence intensity at λexc = 530nm and λem = 590 nm, and then calculate kinase activity.
3.3.4. Data Analysis
Results are represented as M ± S.E.M of at least three independent experiments. Data analysis has been done using Graph Pad Prism and Sigma Plot 11 software.
3.4. Molecular Modeling Procedure
The crystal structure of the human EGFR co-crystallized with erlotinib (PDB ID: 1M17) was used for the docking analysis. 3D structures of erlotinib and 4g were obtained using the Discovery Studio software (Accelrys Inc., San Diego, CA, USA). Auto Dock Tools (The Scripps Research Institute, La Jolla, CA, USA) [61] was used to prepare the ligands and receptor as pdbqt files after removing water, adding polar hydrogen atoms and Gasteiger charges respectively. The docking grid box size used was 40 × 40 × 40 Å3, encompassing the entire EGFR binding pocket. An exhaustiveness value of 8 was used while keeping the other parameters with their default values. The best docking pose (most stable) was selected for binding mode comparison with that of erlotinib. Visualization of ligand-protein non-covalent interactionswas performed using Discovery Studio software. The schematic 2D representations of enzyme-ligand complexes were generated using LIGPLOT version 2.2.4 (European Molecular Biology Laboratory, Cambridge, UK).
4. Conclusions
This study reveals the discovery of novel indolyl pyrimidine hybrids having hydrazide compound 2, hydrazone compounds 3a–h or condensed thiazolidinone moiety compounds 4a–h with promising antitumor activities. Results of the in vitro cytotoxic evaluation suggest that the hybridization of indole, pyrimidine, and thiazolidinone rings is a promising antitumor scaffold. The most potent antiproliferative compound in this study was compound 4g in vitro against MCF-7, HepG2 and HCT-116 with IC50 = 5.1 ± 1.14, 5.02 ± 1.19 and 6.6 ± 1.40 μM. The presence of electron withdrawal group such as Cl atoms was found to be a potential reason for such activity. Compounds 3g, 4f, 4g, and 4h were the most promising antiproliferative agents against all the tested cell lines. The cytotoxicity of all compounds was more specific to cancer cells compared to WI38 normal cells. Furthermore, compound 4g showed the highest in vivo antitumor efficacy against EAC tumor bearing mice. In addition, compound 4g showed a similar effect in in vitro EGFR inhibition assay comparable to erlotinib. Molecular docking of compound 4g into the binding site of EGFR kinase enzyme was performed to explain the efficacy against EGFR. It was revealed from the docking study that compound 4g tends to bind more favorably in the EGFR active site than erlotinib. To sum up, the new scaffold reported in this study may represent a new scaffold for developing more effective anticancer agents.
Acknowledgments
We would like to thank colleagues at Faculty of Pharmacy, Helwan University for their help and support. All authors would like to thank Mossad Sayed Mohamed for helpful discussions.
Abbreviations
| MCF-7 | Michigan Cancer Foundation-7 cell line |
| HepG2 | Hepatoma G2 cell line |
| HCT116 | human colorectal carcinoma cell line |
| 5-FU | 5-Fluorouracil |
| EGFR | Epidermal Growth Factor Receptor |
| SAR | Structure-Activity Relationship |
| HCl | Hydrochloric acid |
| MS | Mass Spectrometry |
| IR | Infrared Spectroscopy |
| NMR | Nuclear Magnetic Resonance |
| IC50 | Concentrations of tested compounds that give about 50% inhibition of cell viability |
| EAC | Ehrlich ascites carcinoma |
Supplementary Materials
Supplementary data includes molecular modeling, biological evaluation details as well as representative spectral data.
Author Contributions
N.M.A. and A.M.S. synthesized, characterized, and analyzed spectral data of all compounds; M.M.Y. and M.K.S. performed the biological testing for all compounds; A.M.S. performed molecular modeling for chosen compounds; N.M.A. and A.M.S. contributed to early manuscript drafts; A.M.S. contributed to writing and finalizing the manuscript and its supplementary materials in their final versions; N.M.A., M.M.Y., M.K.S., and A.M.S. analyzed and interpreted the data for all compounds and contributed equally to the design of this study. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
The study was conducted according to the guidelines of the Declaration of Helsinki and was approved by the Animal Ethics Committee of the Faculty of Pharmacy, Helwan University (ethical code number: 05A2019; date: October 2019). The study was conducted in accordance with the EC, directive 86/609/EEC.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Conflicts of Interest
The authors declare no conflict of interest.
Sample Availability: Sample availability of the compounds 4a–h is very limited from the authors.
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Said A.M., Parker M.W., Vander Kooi C.W. Design, synthesis, and evaluation of a novel benzamidine-based inhibitor of VEGF-C binding to Neuropilin-2. Bioorg. Chem. 2020;100:103856. doi: 10.1016/j.bioorg.2020.103856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Seyfried T.N., Huysentruyt L.C. On the origin of cancer metastasis. Crit Rev. Oncog. 2013;18:43–73. doi: 10.1615/CritRevOncog.v18.i1-2.40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Guan X. Cancer metastases: Challenges and opportunities. Acta Pharm. Sin. B. 2015;5:402–418. doi: 10.1016/j.apsb.2015.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dillekås H., Rogers M.S., Straume O. Are 90% of deaths from cancer caused by metastases? Cancer Med. 2019;8:5574–5576. doi: 10.1002/cam4.2474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Baskar R., Lee K.A., Yeo R., Yeoh K.W. Cancer and radiation therapy: Current advances and future directions. Int. J. Med. Sci. 2012;9:193–199. doi: 10.7150/ijms.3635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Parsa N. Environmental factors inducing human cancers. Iran. J. Public Health. 2012;41:1–9. [PMC free article] [PubMed] [Google Scholar]
- 7.Siegel R.L., Miller K.D., Jemal A. Cancer statistics, 2018. CA A Cancer J. Clin. 2018;68:7–30. doi: 10.3322/caac.21442. [DOI] [PubMed] [Google Scholar]
- 8.Rahib L., Smith B.D., Aizenberg R., Rosenzweig A.B., Fleshman J.M., Matrisian L.M. Projecting cancer incidence and deaths to 2030: The unexpected burden of thyroid, liver, and pancreas cancers in the United States. Cancer Res. 2014;74:2913–2921. doi: 10.1158/0008-5472.CAN-14-0155. [DOI] [PubMed] [Google Scholar]
- 9.Thakor A.S., Gambhir S.S. Nanooncology: The future of cancer diagnosis and therapy. CA Cancer J. Clin. 2013;63:395–418. doi: 10.3322/caac.21199. [DOI] [PubMed] [Google Scholar]
- 10.Jain K.S., Arya N., Inamdar N.N., Auti P.B., Unawane S.A., Puranik H.H., Sanap M.S., Inamke A.D., Mahale V.J., Prajapati C.S., et al. The Chemistry and Bio-Medicinal Significance of Pyrimidines & Condensed Pyrimidines. Curr. Top. Med. Chem. 2016;16:3133–3174. doi: 10.2174/1568026616666160609100410. [DOI] [PubMed] [Google Scholar]
- 11.Mabkhoot Y.N. Synthesis and chemical characterisation of new bis-thieno [2,3-b]thiophene derivatives. Molecules. 2010;15:3329–3337. doi: 10.3390/molecules15053329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sayed A.R., Gomha S.M., Abdelrazek F.M., Farghaly M.S., Hassan S.A., Metz P. Design, efficient synthesis and molecular docking of some novel thiazolyl-pyrazole derivatives as anticancer agents. BMC Chem. 2019;13:116. doi: 10.1186/s13065-019-0632-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Abu-Melha S., Edrees M.M., Salem H.H., Kheder N.A., Gomha S.M., Abdelaziz M.R. Synthesis and Biological Evaluation of Some Novel Thiazole-Based Heterocycles as Potential Anticancer and Antimicrobial Agents. Molecules. 2019;24:539. doi: 10.3390/molecules24030539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Abdel-aziz H.M., Gomha S.M., El-Sayed A.A., Mabkhot Y.N., Alsayari A., Muhsinah A.B. Facile synthesis and antiproliferative activity of new 3-cyanopyridines. BMC Chem. 2019;13:137. doi: 10.1186/s13065-019-0652-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ali E.M.H., Abdel-Maksoud M.S., Oh C.H. Thieno[2,3-d]pyrimidine as a promising scaffold in medicinal chemistry: Recent advances. Bioorg. Med. Chem. 2019;27:1159–1194. doi: 10.1016/j.bmc.2019.02.044. [DOI] [PubMed] [Google Scholar]
- 16.Abdelhamid A.O., Abdelall E.K.A., Abdel-Riheem N.A., Ahmed S.A. Synthesis and Antimicrobial Activity of Some New 5-Arylazothiazole, Pyrazolo[1,5-a] Pyrimidine, [1,2,4]Triazolo[4,3-a]Pyrimidine, and Pyrimido[1,2-a]Benzimidazole Derivatives Containing the Thiazole Moiety. Phosphorus Sulfur Silicon Relat. Elem. 2010;185:709–718. doi: 10.1080/10426500902922933. [DOI] [Google Scholar]
- 17.Liu H.B., Gao W.W., Tangadanchu V.K.R., Zhou C.H., Geng R.X. Novel aminopyrimidinyl benzimidazoles as potentially antimicrobial agents: Design, synthesis and biological evaluation. Eur. J. Med. Chem. 2018;143:66–84. doi: 10.1016/j.ejmech.2017.11.027. [DOI] [PubMed] [Google Scholar]
- 18.Ahmed N.M., Youns M., Soltan M.K., Said A.M. Design, synthesis, molecular modelling, and biological evaluation of novel substituted pyrimidine derivatives as potential anticancer agents for hepatocellular carcinoma. J. Enzym. Inhib. Med. Chem. 2019;34:1110–1120. doi: 10.1080/14756366.2019.1612889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Xie F., Zhao H., Zhao L., Lou L., Hu Y. Synthesis and biological evaluation of novel 2,4,5-substituted pyrimidine derivatives for anticancer activity. Bioorg. Med. Chem. Lett. 2009;19:275–278. doi: 10.1016/j.bmcl.2008.09.067. [DOI] [PubMed] [Google Scholar]
- 20.Nagarapu L., Vanaparthi S., Bantu R., Ganesh Kumar C. Synthesis of novel benzo[4,5]thiazolo[1,2-a]pyrimidine-3-carboxylate derivatives and biological evaluation as potential anticancer agents. Eur. J. Med. Chem. 2013;69:817–822. doi: 10.1016/j.ejmech.2013.08.024. [DOI] [PubMed] [Google Scholar]
- 21.Sun L., Wu J., Zhang L., Luo M., Sun D. Synthesis and Antifungal Activities of Some Novel Pyrimidine Derivatives. Molecules. 2011;16:5618–5628. doi: 10.3390/molecules16075618. [DOI] [Google Scholar]
- 22.Awad S.M., Zohny Y.M., Ali S.A., Mahgoub S., Said A.M. Design, Synthesis, Molecular Modeling, and Biological Evaluation of Novel Thiouracil Derivatives as Potential Antithyroid Agents. Molecules. 2018:23. doi: 10.3390/molecules23112913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dragún M., Rada B., Novotný L., Beránek J. Antiviral activities of pyrimidine nucleoside analogues: Some structure—Activity relationships. Acta Virol. 1990;34:321–329. [PubMed] [Google Scholar]
- 24.Hoffmann H.-H., Kunz A., Simon V.A., Palese P., Shaw M.L. Broad-spectrum antiviral that interferes with de novo pyrimidine biosynthesis. Proc. Natl. Acad. Sci. USA. 2011;108:5777–5782. doi: 10.1073/pnas.1101143108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Krečmerová M., Dračínský M., Snoeck R., Balzarini J., Pomeisl K., Andrei G. New prodrugs of two pyrimidine acyclic nucleoside phosphonates: Synthesis and antiviral activity. Bioorg. Med. Chem. 2017;25:4637–4648. doi: 10.1016/j.bmc.2017.06.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shaquiquzzaman M., Khan S.A., Amir M., Alam M.M. Synthesis, anticonvulsant and neurotoxicity evaluation of some new pyrimidine-5-carbonitrile derivatives. Saudi Pharm J. 2012;20:149–154. doi: 10.1016/j.jsps.2011.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mohamed M.S., Awad S.M., Sayed A.I. Synthesis of certain pyrimidine derivatives as antimicrobial agents and anti-inflammatory agents. Molecules. 2010;15:1882–1890. doi: 10.3390/molecules15031882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Inoyama D., Paget S.D., Russo R., Kandasamy S., Kumar P., Singleton E., Occi J., Tuckman M., Zimmerman M.D., Ho H.P., et al. Novel Pyrimidines as Antitubercular Agents. Antimicrob. Agents Chemother. 2018:62. doi: 10.1128/AAC.02063-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Suryawanshi S.N., Kumar S., Shivahare R., Pandey S., Tiwari A., Gupta S. Design, synthesis and biological evaluation of aryl pyrimidine derivatives as potential leishmanicidal agents. Bioorg. Med. Chem. Lett. 2013;23:5235–5238. doi: 10.1016/j.bmcl.2013.06.060. [DOI] [PubMed] [Google Scholar]
- 30.Iman M., Davood A., Khamesipour A. Design of antimalarial agents based on pyrimidine derivatives as methionine aminopeptidase 1b inhibitor: Molecular docking, quantitative structure activity relationships, and molecular dynamics simulation studies. J. Chin. Chem. Soc. 2020;67:880–890. doi: 10.1002/jccs.201900165. [DOI] [Google Scholar]
- 31.Partridge F.A., Forman R., Willis N.J., Bataille C.J.R., Murphy E.A., Brown A.E., Heyer-Chauhan N., Marinič B., Sowood D.J.C., Wynne G.M., et al. 2,4-Diaminothieno[3,2-d]pyrimidines, a new class of anthelmintic with activity against adult and egg stages of whipworm. PLoS Negl. Trop. Dis. 2018;12:e0006487. doi: 10.1371/journal.pntd.0006487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Romeo R., Iannazzo D., Veltri L., Gabriele B., Macchi B., Frezza C., Marino-Merlo F., Giofrè S.V. Pyrimidine 2,4-Diones in the Design of New HIV RT Inhibitors. Molecules. 2019;24:1718. doi: 10.3390/molecules24091718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liu P., Yang Y., Tang Y., Yang T., Sang Z., Liu Z., Zhang T., Luo Y. Design and synthesis of novel pyrimidine derivatives as potent antitubercular agents. Eur. J. Med. Chem. 2019;163:169–182. doi: 10.1016/j.ejmech.2018.11.054. [DOI] [PubMed] [Google Scholar]
- 34.Park D.I., Dournes C., Sillaber I., Uhr M., Asara J.M., Gassen N.C., Rein T., Ising M., Webhofer C., Filiou M.D., et al. Purine and pyrimidine metabolism: Convergent evidence on chronic antidepressant treatment response in mice and humans. Sci. Rep. 2016;6:35317. doi: 10.1038/srep35317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Funayama S., Cordell G.A., editors. Alkaloids. Academic Press; Boston, MA, USA: 2015. Chapter 11—Alkaloids Derived from Nucleic Acids and Related Compounds; pp. 193–208. [DOI] [Google Scholar]
- 36.Daly M.M., Allfrey V.G., Mirsky A.E. Purine and pyrimidine contents of some desoxypentose nucleic acids. J. Gen. Physiol. 1950;33:497–510. doi: 10.1085/jgp.33.5.497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cerecedo L.R. Physiology of Pyrimidines. Nature. 1940;146:274. doi: 10.1038/146274a0. [DOI] [Google Scholar]
- 38.Sondhi S.M., Goyal R.N., Lahoti A.M., Singh N., Shukla R., Raghubir R. Synthesis and biological evaluation of 2-thiopyrimidine derivatives. Bioorg. Med. Chem. 2005;13:3185–3195. doi: 10.1016/j.bmc.2005.02.047. [DOI] [PubMed] [Google Scholar]
- 39.Parker W.B. Enzymology of purine and pyrimidine antimetabolites used in the treatment of cancer. Chem. Rev. 2009;109:2880–2893. doi: 10.1021/cr900028p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Miyazaki Y., Maeda Y., Sato H., Nakano M., Mellor G.W. Rational design of 4-amino-5,6-diaryl-furo[2,3-d]pyrimidines as potent glycogen synthase kinase-3 inhibitors. Bioorg. Med. Chem. Lett. 2008;18:1967–1971. doi: 10.1016/j.bmcl.2008.01.113. [DOI] [PubMed] [Google Scholar]
- 41.Kim D.C., Lee Y.R., Yang B.S., Shin K.J., Kim D.J., Chung B.Y., Yoo K.H. Synthesis and biological evaluations of pyrazolo[3,4-d]pyrimidines as cyclin-dependent kinase 2 inhibitors. Eur. J. Med. Chem. 2003;38:525–532. doi: 10.1016/S0223-5234(03)00065-5. [DOI] [PubMed] [Google Scholar]
- 42.Joshi G., Nayyar H., Kalra S., Sharma P., Munshi A., Singh S., Kumar R. Pyrimidine containing epidermal growth factor receptor kinase inhibitors: Synthesis and biological evaluation. Chem. Biol. Drug Des. 2017;90:995–1006. doi: 10.1111/cbdd.13027. [DOI] [PubMed] [Google Scholar]
- 43.Dadashpour S., Emami S. Indole in the target-based design of anticancer agents: A versatile scaffold with diverse mechanisms. Eur. J. Med. Chem. 2018;150:9–29. doi: 10.1016/j.ejmech.2018.02.065. [DOI] [PubMed] [Google Scholar]
- 44.Prakash B., Amuthavalli A., Edison D., Sivaramkumar M.S., Velmurugan R. Novel indole derivatives as potential anticancer agents: Design, synthesis and biological screening. Med. Chem. Res. 2018;27:321–331. doi: 10.1007/s00044-017-2065-9. [DOI] [Google Scholar]
- 45.Putt K.S., Chen G.W., Pearson J.M., Sandhorst J.S., Hoagland M.S., Kwon J.T., Hwang S.K., Jin H., Churchwell M.I., Cho M.H., et al. Small-molecule activation of procaspase-3 to caspase-3 as a personalized anticancer strategy. Nat. Chem. Biol. 2006;2:543–550. doi: 10.1038/nchembio814. [DOI] [PubMed] [Google Scholar]
- 46.Seshacharyulu P., Ponnusamy M.P., Haridas D., Jain M., Ganti A.K., Batra S.K. Targeting the EGFR signaling pathway in cancer therapy. Expert Opin. Targets. 2012;16:15–31. doi: 10.1517/14728222.2011.648617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chen Y., Wu J., Wang A., Qi Z., Jiang T., Chen C., Zou F., Hu C., Wang W., Wu H., et al. Discovery of N-(5-((5-chloro-4-((2-(isopropylsulfonyl)phenyl)amino)pyrimidin-2-yl)amino)-4-methoxy-2-(4-methyl-1,4-diazepan-1-yl)phenyl)acrylamide (CHMFL-ALK/EGFR-050) as a potent ALK/EGFR dual kinase inhibitor capable of overcoming a variety of ALK/EGFR associated drug resistant mutants in NSCLC. Eur. J. Med. Chem. 2017;139:674–697. doi: 10.1016/j.ejmech.2017.08.035. [DOI] [PubMed] [Google Scholar]
- 48.Chang J., Ren H., Zhao M., Chong Y., Zhao W., He Y., Zhao Y., Zhang H., Qi C. Development of a series of novel 4-anlinoquinazoline derivatives possessing quinazoline skeleton: Design, synthesis, EGFR kinase inhibitory efficacy, and evaluation of anticancer activities in vitro. Eur. J. Med. Chem. 2017;138:669–688. doi: 10.1016/j.ejmech.2017.07.005. [DOI] [PubMed] [Google Scholar]
- 49.Cheng H., Nair S.K., Murray B.W. Recent progress on third generation covalent EGFR inhibitors. Bioorg. Med. Chem Lett. 2016;26:1861–1868. doi: 10.1016/j.bmcl.2016.02.067. [DOI] [PubMed] [Google Scholar]
- 50.Desai N.C., Bhatt M.J. Catalytic synthesis and antimicrobial activity of N-(3-chloro-2-oxo-4-phenylazetidin-1-yl)-4-(1H-indol-3-yl)-6-methyl-2-thioxo-1,2,3,4-tetrahydropyrimidine-5-carboxamides. Heterocycl. Commun. 2016;22:131–136. doi: 10.1515/hc-2016-0032. [DOI] [Google Scholar]
- 51.Viveka S., Nagaraja G.K., Shama P., Basavarajaswamy G., Rao K.P., Yanjarappa Sreenivasa M. One pot synthesis of thiazolo[2,3-b]dihydropyrimidinone possessing pyrazole moiety and evaluation of their anti-inflammatory and antimicrobial activities. Med. Chem. Res. 2018;27:171–185. doi: 10.1007/s00044-017-2058-8. [DOI] [Google Scholar]
- 52.AbdElhameid M.K., Labib M.B., Negmeldin A.T., Al-Shorbagy M., Mohammed M.R. Design, synthesis, and screening of ortho-amino thiophene carboxamide derivatives on hepatocellular carcinomaas VEGFR-2Inhibitors. J. Enzym. Inhib Med. Chem. 2018;33:1472–1493. doi: 10.1080/14756366.2018.1503654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Trott O., Olson A.J. AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 2010;31:455–461. doi: 10.1002/jcc.21334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Stamos J., Sliwkowski M.X., Eigenbrot C. Structure of the epidermal growth factor receptor kinase domain alone and in complex with a 4-anilinoquinazoline inhibitor. J. Biol. Chem. 2002;277:46265–46272. doi: 10.1074/jbc.M207135200. [DOI] [PubMed] [Google Scholar]
- 55.Nasr T., Bondock S., Youns M. Anticancer activity of new coumarin substituted hydrazide-hydrazone derivatives. Eur. J. Med. Chem. 2014;76:539–548. doi: 10.1016/j.ejmech.2014.02.026. [DOI] [PubMed] [Google Scholar]
- 56.Ahmed O.M., Ahmed R.R. Anti-proliferative and apoptotic efficacy of diallyl disulfide on Ehrlich ascites carcinoma. Hepatoma Res. 2015;1:67–74. doi: 10.4103/2394-5079.157602. [DOI] [Google Scholar]
- 57.Zhao P., Yang X., Qi S., Liu H., Jiang H., Hoppmann S., Cao Q., Chua M.S., So S.K., Cheng Z. Molecular imaging of hepatocellular carcinoma xenografts with epidermal growth factor receptor targeted affibody probes. Biomed. Res. Int. 2013;2013:759057. doi: 10.1155/2013/759057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Moerkens M., Zhang Y., Wester L., van de Water B., Meerman J.H. Epidermal growth factor receptor signalling in human breast cancer cells operates parallel to estrogen receptor α signalling and results in tamoxifen insensitive proliferation. BMC Cancer. 2014;14:283. doi: 10.1186/1471-2407-14-283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yang J.L., Qu X.J., Russell P.J., Goldstein D. Regulation of epidermal growth factor receptor in human colon cancer cell lines by interferon alpha. Gut. 2004;53:123–129. doi: 10.1136/gut.53.1.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Maher M., Kassab A.E., Zaher A.F., Mahmoud Z. Novel pyrazolo[3,4-d]pyrimidines: Design, synthesis, anticancer activity, dual EGFR/ErbB2 receptor tyrosine kinases inhibitory activity, effects on cell cycle profile and caspase-3-mediated apoptosis. J. Enzym. Inhib Med. Chem. 2019;34:532–546. doi: 10.1080/14756366.2018.1564046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Morris G.M., Huey R., Lindstrom W., Sanner M.F., Belew R.K., Goodsell D.S., Olson A.J. AutoDock4 and AutoDockTools4: Automated docking with selective receptor flexibility. J. Comput. Chem. 2009;30:2785–2791. doi: 10.1002/jcc.21256. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Not applicable.