Abstract
Simple Summary
Intra-tumor heterogeneity is inherent to all cancers and makes direct mapping of genotype–phenotype relationships challenging. The advent of single-cell multi-omics techniques has allowed us to begin to comprehensively dissect cellular heterogeneity and access biological information unobtainable from bulk analysis. Applications cover many fields and are increasingly numerous. This review aims to summarize the most important applications of single-cell technologies in hematological tumors, providing a translational view. Data show the power of single-cell multi-omics to resolve the complex biology of heterogeneous populations and to derive information that can be used to improve treatment strategies. We discuss, with a practical example, how to make use of these techniques to study the heterogeneity of a specific type of monoclonal gammopathy called Waldenström’s macroglobulinemia.
Abstract
Single-cell sequencing techniques have become a powerful tool for characterizing intra-tumor heterogeneity, which has been reflected in the increasing number of studies carried out and reported. We have rigorously reviewed and compiled the information about these techniques inasmuch as they are relative to the area of hematology to provide a practical view of their potential applications. Studies show how single-cell multi-omics can overcome the limitations of bulk sequencing and be applied at all stages of tumor development, giving insights into the origin and pathogenesis of the tumors, the clonal architecture and evolution, or the mechanisms of therapy resistance. Information at the single-cell level may help resolve questions related to intra-tumor heterogeneity that have not been previously explained by other techniques. With that in mind, we review the existing knowledge about a heterogeneous lymphoma called Waldenström’s macroglobulinemia and discuss how single-cell studies may help elucidate the underlying causes of this heterogeneity.
Keywords: single-cell sequencing, applications, hematology, Waldenström’s macroglobulinemia
1. Introduction
Despite its monoclonal origin, intra-tumor heterogeneity (ITH) is inherent to the majority of cancers and can be associated with genetic variability, epigenetic modifications, gene and protein expression, metabolism, morphology, and other features of tumors [1]. Moreover, ITH is known to play a significant role in tumor progression, preservation of oncogenic potential, cell survival under changing microenvironmental conditions, and resistance to drug therapy [1,2,3], the latter being even more relevant nowadays due to the use of targeted therapies that may favor the selection and expansion of the clones [4,5]. Accurate evaluation of ITH could be essential for efficient diagnosis and for successful treatment and outcome [6]. Determining the pattern of somatic mutations is no longer sufficient, but it is essential to know their distribution in individual tumor cells and clones, and how they interact to influence tumor fitness [7]. Approaches based on molecular genetics have been developed to study tumor behavior and predict treatment efficiency by considering factors and mechanisms for generating ITH. Despite its widespread use and recognized diagnostic capacity, next-generation sequencing of the bulk tumor reveals only a minority of the genetic aberrations present in an entire tumor and can lead to the mutational burden being underestimated. In addition, it is not possible to distinguish which alterations occur in the same clone(s) or to elucidate the order of appearance and co-dependency of mutations. Single biopsies from geographically localized tumor areas cannot recapitulate the complexity of spatial heterogeneity. Such problems inherent to tumor sampling bias due to ITH may be mitigated by circulating tumor DNA (ctDNA) sampling [8,9]. Sequencing ctDNA can provide a wider vision of the tumor genomic landscape but can never reveal how certain genetic abnormalities change the behavior of the cell subset involved.
Single-cell sequencing techniques represent a step forward in the characterization of the complete tumor architecture, revolutionizing the landscape with respect to ITH. Not only that, single-cell genomics allows us to determine the timing of the alterations (early vs. late mutations) and provide information about mutations that cooperate in the development of the disease. Genomic, transcriptomic, and epigenomic assays at the single-cell level are transforming our understanding of cellular complexity, thereby enabling us to access biological information that cannot be obtained from bulk analysis [10].
Conventional single-cell methods are based on manual-picking micropipettes [11] or cell fluorescence-activated cell sorting (FACS) [12]. By contrast, the new methodologies include microfluidic technologies, such as the C1™ system from Fluidigm® (South San Francisco, CA, USA), which isolates single-cells into individual reaction chambers in an integrated fluidic circuit [13], droplet-based microfluidic technologies, such as the Tapestri System (MissionBio, South San Francisco, CA, USA) [14], the Chromium System (10× Genomics, Pleasanton, CA, USA) [15], and the Cellular Indexing of Transcriptomes and Epitopes by Sequencing (CITE-seq) (Technology Innovation Lab, New York Genome Center, New York, NY, USA) [16], and nanowell-based technologies, such as the Rhapsody™ Single-Cell Analysis System (BD Biosciences, San Jose, CA, USA), which uses planar arrays of microwells for cell capture [17], and the Seq-Well platform (Shalek Lab, Massachusetts Institute of Technology and the Broad Institute, Cambridge, MA, USA) [18], to mention the most important ones.
Single-cell methods involve single-cell isolation, barcoding, and sequencing to analyze multiple types of molecules from individual cells, as well as the integrative analysis of the data to characterize cell types and their functions based on molecular signatures. To analyze multiple types of molecules from the same cell, it is essential to isolate the single cells and then to barcode the molecules. The process will be explained in greater details for two of the most widely used single-cell systems. The Tapestri Platform (Mission Bio) allows DNA and protein analysis of each individual cell and is based on a two-step microfluidic workflow. The first step involves encapsulating the cells into sub-nanoliter droplets and isolating DNA and oligo-conjugate antibodies from each single cell. The second step involves cell barcoding and targeted amplification by polymerase chain reaction (PCR) within the droplets, which are then disrupted, before extracting barcoded DNA for library amplification. Final libraries are purified and sequenced. The Chromium Platform (10× Genomics) enables the analysis of gene expression, chromatin accessibility, cell-surface proteins, immune clonotype, antigen specificity, and clustered regularly interspaced short palindromic repeats (CRISPR) edits. It is based on the generation of thousands of single-cell partitions, each containing an identifying barcode for downstream analysis using advanced microfluidics. Within the instrument, barcoded gel beads, coated with a unique oligonucleotide barcode sequence and functionalized sequences to capture molecules of interest, are mixed with cells or nuclei, enzymes, and partitioning oil to form thousands of single-cell emulsion droplets. Each droplet is the location of an individual reaction in which the beads are dissolved and molecules of interest from each cell are captured and barcoded. After barcoding, all fragments from the same cell or nucleus share a common barcode. Barcoded fragments from thousands of cells are pooled for downstream reactions to create sequencing libraries. After sequencing, bioinformatics tools use the identifying barcodes to map sequencing reads back to their single cell or nucleus of origin.
Although there are already many reviews of this topic, most of them have focused on the technical aspects of the technologies. Here, instead, we have addressed their practical aspects, presenting a translational point of view. This is a powerful, emerging technique, as evinced by the increasing rate at which studies are being published that demonstrate its broad range of applications. This review confines itself to considering the applications in the field of hematology to provide a comprehensive view of the potential value of these technologies. It also discusses how these applications can be translated to the study of Waldenström’s macroglobulinemia, a particular type of lymphoma/monoclonal gammopathy that is a good example of ITH. Finally, the existing challenges and limitations are considered.
2. Applications of Single-Cell Multi-Omics in Hematological Malignancies
Single-cell analysis has the potential to contribute to our understanding of cell complexity, and, specifically, there are many areas of hematology where it could be of great interest (Table 1). We consider the most relevant of these below.
Table 1.
Summary of the main applications of single-cell technologies in different areas of hematology. CAR T, chimeric antigen receptor T-cell; Cas9, CRISPR-associated endonuclease Cas9; CRISPR, clustered regularly interspaced short palindromic repeats.
| Area | Application | Reference |
|---|---|---|
| Immune system | Study stem cell-like CD8+ memory T cells to develop immunotherapies and vaccines | [21] |
| Tumor cells | Combine genotyping and immunophenotyping to fully characterize the disease | [23] |
| Clonal hematopoiesis | Distinguish mutations associated with clonal hematopoiesis vs. true leukemia to accurately measure residual disease | [29] |
| Oncogenesis | Establish the sequence of genetic events that occur in the disease development; characterize mutational combinations that promote clonal expansion to select targeted therapies | [32,37] |
| Clonal evolution | Study the predisposition to develop leukemia in Shwachman-Diamond syndrome to identify patients with high-risk clones | [40] |
| Therapy resistance | Characterize clonal complexity to predict clinical relapse; evaluate concurrence of different resistance mechanisms to search for novel treatment strategies | [35,43,46,47] |
| Microenvironment | Define the supportive role of the immune microenvironment to develop new therapeutic approaches | [51] |
| B-cell biology | Model the germinal center to understand lymphoma pathology | [53] |
| Cancer therapy | Identify transcriptomic features in anti-CD19 CAR T-infusion cell products to determine efficacy and toxicity | [55] |
| CRISPR-Cas9 gene editing | Analyze Cas9-introduced gene edits to quantify the abundance of CRISPR-introduced disease drivers and decipher the effects of multiplex gene editing | [56] |
| Bone marrow transplant | Assess donor/host chimerism to monitor bone marrow engraftment and predict relapse after transplant | [57] |
2.1. Characterizing Immune Cell Populations
Single-cell RNA-sequencing studies, with different objectives, have yielded descriptions of a wide range of immune cells. For example, the characterization of human hematopoietic populations at the earliest stage of hematopoiesis (i.e., CD34+ cells) could provide a basis for generating blood and immune cells for clinical purposes [19], the study of CD4+ T-cells allows the specific cell states that drive disease or treatment response to be identified [20], and the analysis of stem cell-like CD8+ memory T-cells may help in the development of immunotherapies and vaccines [21].
2.2. Defining the Transcriptomic, Proteomic, and Epigenomic Identity of Malignant Cells
Genotyping and single-cell RNA-sequencing can be integrated to determine how somatic mutations corrupt the hematopoietic process. Nam et al. showed that malignant hematopoietic progenitors (CD34+ cells) with mutated CALR have a fitness advantage with respect to myeloid differentiation and upregulation of the nuclear factor kappa B (NF-κB) pathway, highlighting the cell-identity dependency of somatic mutations in human hematopoiesis [22].
Single-Cell DNA and Antibody Sequencing (DAb-seq) is a method that integrates DNA profiling and surface proteins of single cells at high throughput, allowing the proteogenomic dynamics of multiple patients to be tracked over multiple treatments and recurrences. Analyzing leukemia patients using this approach identified extensive genotype–phenotype decoupling, with immunophenotypic heterogeneity among cells with the same pathogenic mutation, as well as genotypically diverse cells with a convergent malignant immunophenotype, suggesting that independent phenotype or genotype measurements do not adequately capture the full extent of proteogenomic heterogeneity [23].
A different single-cell approach, established by Granja et al., combines highly multiplexed protein quantification, transcriptome profiling, and analysis of chromatin accessibility to deconvolve aberrant molecular features within blood from patients with mixed-phenotype acute leukemia, with the aim of identifying the causes of the disease. This integrative analysis made it possible to infer that the transcription factor RUNX1 acts as a potential oncogene, regulating malignant genes associated with poor survival [24].
2.3. Detecting Pre-Leukemic Clonal Hematopoiesis Mutations
Another interesting application is the discrimination of mutations associated with age-related clonal hematopoiesis vs. true leukemia, in this case, acute myeloid leukemia (AML) [25]. Mutations in epigenetic regulators DNMT3A and TET2 are frequently found in older individuals and appear to be associated with an increased death risk [26,27]. These mutations are also common in patients with AML [28], making genomic measurement of residual disease very difficult, since detected variants cannot be confidently assigned to the malignancy (particularly in the post-treatment setting) [29,30]. While targeted DNA sequencing cannot determine whether mutations associated with clonal hematopoiesis are also represented within the malignant clone, detection of fusion genes by single-cell DNA and antibody-oligonucleotide sequencing can resolve the leukemic clonal architecture, thereby enabling the comprehensive assessment of AML.
The presence of clonal hematopoiesis has also been assessed by single-cell DNA sequencing in patients with myelodysplastic syndrome-associated phenotypic abnormalities, which are present in ∼10% of newly diagnosed multiple myeloma (MM) cases [31].
2.4. Establishing the Order of Events That Mediate Cancer Origin and Evolution
A comprehensive understanding of disease development may facilitate the rational design of antitumor drugs and prevention strategies. Single-cell sequencing has made it possible to measure the clonal structures of childhood acute lymphoblastic leukemia (ALL) samples at diagnosis, and to establish the sequence of genetic events that underlie the disease; first, most of the structural variants, followed by single-nucleotide variants (SNVs), with KRAS (Kirsten Rat Sarcoma Viral Oncogene Homolog) mutations occurring late in disease development [32]. The order of mutation acquisition and the progenitor cells in which this process is initiated have also been studied in T-cell ALL (T-ALL). Patients showed an early event in a known oncogene (MED12, STAT5B) that was already detectable in multipotent progenitor cells. Intermediate events included copy-number alterations (such as loss of 9p21) and acquisition of fusion genes, while NOTCH1 mutations were typically late events [33], despite having a key role in the development of T-ALL [34].
Reconstructing the complete clonal architecture of the tumor may give insights into its evolution and inform decisions about treatment [35,36]. Myeloid malignancies, including AML, arise from the expansion of clonal hematopoietic stem and progenitor cells that acquire subsequent somatic mutations. Single-cell sequencing has allowed clonal trajectories to be mapped and synergistic combinations of mutations to be revealed that promote clonal expansion and dominance. The work of Miles et al. demonstrated that clonal complexity increased as the disease progressed (from clonal hematopoiesis to AML), and continued to evolve as AML clones acquired mutations in signaling effectors, which were often sub-clonal and not concurrent. In addition, mutational combinations contributed to clonal dominance in various ways: specific co-occurring disease alleles (e.g., NPM1–FLT3-ITD or DNMT3A–IDH2) were associated with clonal dominance, whereas other mutational combinations (e.g., NPM1–RAS) did not promote clonal expansion. Changes in clonal architecture were due to the expansion of pre-existing minor clones and could be detected using single-cell DNA sequencing, drawing attention to the value of searching for therapies targeting these clones before they achieve clonal dominance. Finally, by combining mutational analysis with protein expression, it is possible to identify significant genotype-driven changes in cell-surface protein expression (e.g., mutations in the mitogen-activated protein kinase/extracellular signal-regulated kinases, MAPK/ERK, pathway that led to increased CD11b expression) [37]. Evidence of molecular complexity including intra-lineage clonal evolution was found in another study of myeloproliferative neoplasms: two unusual patients, each harboring two driver mutations (JAK2/CALR and JAK2/MPL), which represented, in both cases, independent clones. These data highlight the importance of routine assessment of the three canonical driver mutations (i.e., JAK2/CALR/MPL) in order to characterize disease biology accurately [38].
2.5. Describing Mechanisms That Lead to Disease Progression and Resistance to Therapy
Much research has been done to understand the underlying etiology of clonal evolution. This helps to determine whether sequential molecular monitoring is of clinical value for the early detection of malignancies, and to assess the utility of preventive interventions. Single-cell techniques have been used to study clonal evolution of severe aplastic anemia to a secondary myeloid malignancy. DNA sequencing at the single-cell level showed that sequential mutation acquisition over a 7-year clonal period occurred within the same clone derived from an initial ASXL1 mutated clonal population [39]. The same strategy was used to study the predisposition to develop leukemia in Shwachman-Diamond syndrome (SDS). The authors linked the development of leukemia to the acquisition of biallelic TP53 alterations that drove progression of TP53 mutated clones. As SDS patients can develop multiple independent TP53 mutated clones, serial monitoring by bulk sequencing would fail to distinguish clinically significant sub-clonal changes in TP53 allelic status. These findings imply the value of integrating of single-cell DNA sequencing into surveillance strategies in order to identify patients with high-risk clones [40]. In MM, single-cell transcriptome analysis revealed that extramedullary progression is associated with alterations in the transcriptional programs of plasma cells and the microenvironment affecting proliferation and immune evasion [41].
Regarding the development of drug resistance, tumors harbor a variety of cell types that are thought to play an important role in this process [42]. Thus, in patients with AML, a single-cell study observed that greater clonal complexity was associated with reduced elimination of all malignant clones with standard chemotherapy regimens, and therefore with a higher risk of resistant clones persisting and eventually causing clinical relapse [43]. Therefore, an important prognostic application would be one that established a diversity index for the tumor, or parts of it, that could predict whether a patient will respond poorly to therapy or have a strong tendency to relapse. Cell composition could also determine a worse clinical outcome. Single-cell RNA-sequencing identified the CD34+CD117dim proportion as an independent factor of poor prognosis in patients with t(8;21) AML, and this leukemic cluster was able to expand at the post-relapse refractory stage after several cycles of chemotherapy [44]. In addition, it is also possible to identify the molecular determinants of clinically relevant outcomes. In patients with AML who are treated with combinations based on venetoclax (a BCL-2 inhibitor), NPM1 and IDH2 mutations are associated with high response rates and durable remissions, while the activation of the FLT3, RAS, and TP53 signaling pathways seems to be linked to resistance development [45]. Emergence of multiple clones, each with distinct mechanisms of resistance, is a common finding following the secondary failure of single-agent targeted therapies for leukemias. An integrated genomic analysis combining DNA sequencing, RNA-sequencing, and a methylation profiling microarray revealed that selection of co-occurring mutations in hematopoietic transcription factor genes (RUNX1/CEBPA) or RAS-RTK (receptor tyrosine kinase) pathway genes were the main drivers of acquired resistance to IDH inhibitors, suggesting that novel strategies targeting certain high-risk co-occurring mutations might improve the therapeutic efficacy of IDH inhibitors in AML [46]. The complex biology of resistance was also highlighted in AML treated with ivosidenib (an inhibitor of mutated IDH1), whereby the concurrence of different mechanisms, particularly receptor tyrosine kinase pathway mutations and IDH-related mutations, contributed to primary and secondary resistance [47]. Likewise, McMahon and colleagues demonstrated that secondary clinical resistance to the FLT3 inhibitor gilteritinib in relapsed AML was commonly mediated by heterogeneous mutations that activate downstream RAS-MAPK pathways [35]. RNA-sequencing identified a CD19 splice variant as a biomarker of the failure of blinatumomab (anti-CD19/CD3) therapy in B-cell progenitor ALL [48], and a USP7 gene signature was associated with resistance to therapy in AML patients [49]. Lastly, a deeper analysis of the CLL response to ibrutinib (a BTK inhibitor) allowed the dissection of the exact cellular and molecular changes induced by this drug and identified candidate molecular markers of therapy response. Chromatin accessibility analysis with ATAC-Seq on FACS-purified immune cell populations was combined with single-cell RNA-sequencing for a subset of timepoints. The high level of detail and biological insight obtained make it particularly well-suited to applications in personalized medicine, whereby each patient may follow a different disease trajectory, and for early-stage clinical trials of new targeted therapies, in which it is critical to understand the molecular and cellular dynamics in order to establish the appropriate dose and identify response biomarkers [50].
2.6. Studying the Microenvironment
The bone marrow microenvironment may support tumor progression and treatment evasion. Witkowsky et al. used single-cell RNA-sequencing and cellular indexing of transcriptomes and epitopes by sequencing (CITE-seq) to show that B-cell ALL remodels the bone marrow immune microenvironment upon disease initiation, as well as the subsequent recovery during conventional chemotherapy [51]. In addition, immune microenvironment profiling identified extrinsic regulators of survival, which would underpin the new immune-based therapeutic approaches for high-risk B-ALL treatment [51].
2.7. Understanding Human B-Cell Biology and Lymphoma Pathology by Modeling the Germinal Center
Integrative single-cell analysis of gene expression, surface phenotype, and the B-cell receptor sequence can be useful for studying the heterogeneous germinal center (GC) B-cell subset [52]. This approach was used to characterize B-cell transcriptional heterogeneity in follicular lymphoma (FL) and compare it with the various states observed in normal GC B-cells. Results showed a major desynchronization of GC-specific gene-expression programs in FL tumor cells, although with distinct FL-specific cell states coexisting within a single patient. This suggests that lymphoma B-cells are not blocked in a GC B-cell state but might adopt dynamic modes of functional diversity [53].
2.8. Cancer Therapy
Transferring T-cells that express tumor-reactive T-cell receptors (TCRs) can induce regression of tumors in patients with advanced cancer. However, isolation and expression of a tumor antigen-specific TCR is a highly complex process. Advances in single-cell sequencing have begun to streamline this process; in particular, single-cell variable (V), diversity (D) and joining (J) genes (VDJ) sequencing data are used to clone antigen-specific TCRs. The same strategy can be used to expand primary human antigen-specific T-cell clones [54].
A different application is the identification of transcriptomic features in anti-CD19 Chimeric Antigen Receptor T-Cell (CAR T)-infusion cell products. This has been associated with efficacy and toxicity in diffuse large B-cell lymphoma (DLBCL). Results have shown that heterogeneity of the cellular and molecular features contributed to the variation in efficacy and toxicity and that the 7-day molecular response might serve as an early predictor of CAR T-cell efficacy [55].
2.9. Other Applications
CRISPR-Cas9 (CRISPR-associated endonuclease Cas9) gene editing allows the rapid interrogation of the functional impact of somatic mutations in human cancers. Single-cell technologies enable the analysis of Cas9-introduced gene edits in thousands of cells, quantifying mutational co-occurrences and zygosity status. As most cancers arise and propagate due to the complex interaction of multiple drivers, these tools allow the contribution of individual genetic drivers to cellular fitness to be assessed and the mutational co-occurrences, interactions of multiple lesions, and clonal evolutionary mechanisms in cellular and mouse models to be studied [56].
Another interesting application is bone marrow engraftment monitoring by the assessment of donor/host chimerism using the individuals’ unique genotype signatures as genetic proxies. Using a droplet microfluidic approach in AML patients at different times (before/after bone marrow transplant and at AML relapse), it was possible to obtain the molecular profile of single nucleotide variants (SNVs) across thousands of cells. This revealed the existence of genetic chimerism after bone marrow transplantation. In addition, comparison of clone number and size across the three times suggested that AML relapse after transplant may result from the aggressive and exclusive expansion of the oncogenic cells carrying tumor-suppressor and/or oncogene mutation(s), and is associated with loss of donor chimerism [57].
2.10. Future Applications
Assays and methods to detect molecular abnormalities will soon be adapted for use at the single-cell level. Fold et al. demonstrated fusion event detection at single-cell resolution using barcoded single-cell RNA-sequencing data in MM, pointing the way forward for the development of fusion methods [58].
3. Single-Cell Techniques in Waldenström’s Macroglobulinemia: Utility and Applicability
As we have seen, single-cell technologies have proven useful in many fields. Now, we will focus on the specific applications that these techniques may have in a particularly heterogeneous hybrid disease. ITH may be the underlying cause of the wide multilevel heterogeneity present in Waldenström’s macroglobulinemia (WM). WM is a rare indolent B-cell lymphoproliferative disorder characterized by bone marrow infiltration by lymphoplasmacytic lymphoma and the presence of an immunoglobulin M (IgM) monoclonal component [59]. The cellular composition of this lymphoma includes malignant lymphocytes and plasma cells. At the clinical level, the disease is consistently heterogeneous, its variety of behaviors being manifested in entities ranging from indolent forms, such as IgM monoclonal gammopathy of undetermined significance (IgM-MGUS) and asymptomatic WM (AWM), to highly symptomatic disease (symptomatic WM, SWM) [59]. Its evolution is highly variable [60], and transformation into aggressive DLBCL as well as leukemia has also been reported [61,62], although the mechanisms responsible are not known. In recent years, progress has been made towards characterizing the genetic profile of WM tumor cells. Whole-genome sequencing has made it possible to identify a recurrent somatic mutation, the MYD88 L265P, as a unifying event in most patients with WM (95–97%) and IgM-MGUS (90%) [63,64,65,66,67]. However, studies in mice have shown that although this alteration might be indispensable for the WM phenotype, it is insufficient by itself for the full development of lymphoma [68]. Whether it may be the tumor-initiating event that confers a competitive advantage on the clone and predisposes it to further genetic alterations remains to be clarified. A recent study has shown the presence of MYD88 L265P in B-cell precursors from 6/10 patients and in residual normal B-cells from 6/10 patients [69]. It would be interesting to evaluate the different cell populations in Ig-MGUS and WM patients at the single-cell level to identify the event(s) that accompany or co-operate with MYD88 mutation in the malignant transformation.
The second most common alteration in WM affects the C-terminal domain of CXCR4 and is present in 40% of patients, almost all of whom are MYD88-mutated [70,71]. Currently, more than 40 different mutations (either nonsense or frameshift) have been described, the most frequent being the nonsense CXCR4 S338X mutation, which is present in ~25% of patients [72,73]. CXCR4 mutations are mostly sub-clonal, with a variable clonal distribution, which suggests they are probably acquired after the MYD88 mutation during WM oncogenesis [71,74,75]. One single-cell study has already addressed the differences between cells that acquire the CXCR4 mutation and those that remain CXCR4-wild-type within a patient. Results did not show a distinct mutation profile for the two populations, but further studies are needed [76].
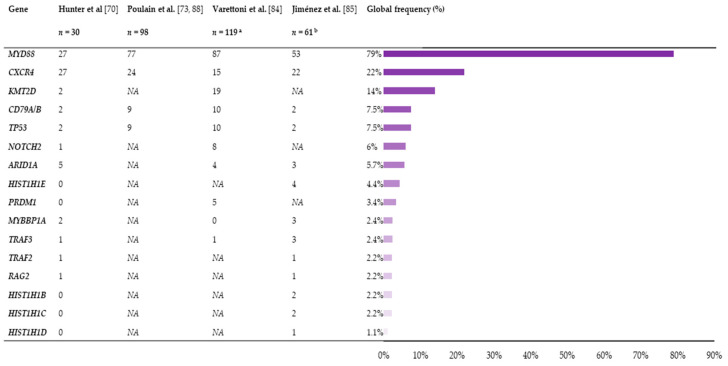
There is increasing evidence about the role of the B-cell receptor pathway in the pathobiology of WM, either alone or in cooperation with the MYD88 signaling axis [77,78,79,80,81,82,83]. Mutations in CD79A and CD79B occur in 8–12% of WM patients, and co-expression of CD79B and MYD88 mutations has been associated with transformation to aggressive lymphoma [84,85,86,87]. Lastly, mutations in TP53 are more rarely observed—they occur in 2–3% of WM—and are associated with poor survival (Figure 1) [77,85,88,89].
Figure 1.
Frequency and distribution of the mutations in the main genes of Waldenström’s macroglobulinemia (WM) according to the different studies. The total number of mutated patients in each study, as well as the global mutation frequency (%) considering the four studies, are displayed. a Corresponding to 57 Immunoglobulin M monoclonal gammopathy of undetermined significance (IgM-MGUS) and 62 WM; b Corresponding to 14 IgM-MGUS, 23 asymptomatic Waldenström’s macroglobulinemia (AWM) and 24 symptomatic Waldenström’s macroglobulinemia (SWM). NA: not applicable.
All this means that, in spite of the monoclonality inherent to all cancer cells, they harbor a substantial degree of variability that could be responsible for the clinical heterogeneity of WM. However, to date, no clear correlation has been found between the mutational profile and the clinical behavior of IgM gammopathies [84,85]. The global frequency of mutations and genomic alterations (copy-number abnormalities, loss of heterozygosity) does increase from IgM-MGUS to SWM, which confirms the association between aggressive clinical behavior and a higher frequency of alterations [84,85,86,90]. Therefore, it appears that these alterations could have a role in the multistep oncogenic process that drives the transition first from IgM-MGUS to AWM, and then to SWM. Considering the heterogeneity of the mutational profile of WM patients (Figure 1), it seems unlikely that the malignant clone has a specific pattern of aberrations. In fact, the evidence suggests that there may be at least two pathways that promote IgM-MGUS progression to symptomatic disease: CXCR4 mutation and 6q21-25 deletion. Loss of chromosome 6q is found in 40–50% of patients with WM [91,92] and appears to be exclusive to CXCR4 in treatment-naïve patients [93]. The increasing frequency of 6q deletions from IgM-MGUS through asymptomatic and symptomatic WM suggests that loss of genes within this region facilitates disease progression [84,91,93,94]. As for CXCR4 mutations, the transcriptomic profile, relative to that of solely MYD88-mutated individuals, shows diminished B-cell differentiation, downregulation of tumor suppressors overexpressed by MYD88 mutation, and alternative activation of the toll-like receptor 7 (TLR7) pathway [95].
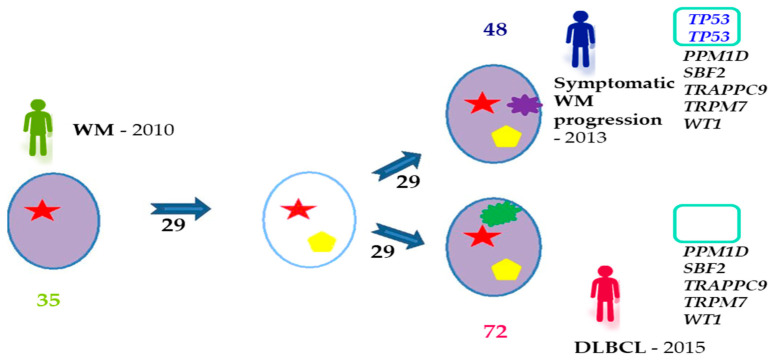
As noted in other tumors, it is not only the type of genetic alteration that matters—the target cell where mutations arise and the order of the events are also of great relevance to the course of the oncogenic process. Therefore, it would be interesting to perform multi-omics analyses in single cells, distinguishing between the three stages of the disease (IgM-MGUS, AWM, and SWM), to resolve the zygosity and co-occurrence/exclusion of mutations, and to examine they extent to which they are correlated with the immunophenotype. With these insights, phylogenetic trees could be reconstructed to determine the order of acquisition of the alterations, the cell of origin, and light could be shed on the putative mechanisms leading to progression. The same strategy could be applied to the transformation from WM to DLBCL or leukemia. It is conceivable that some of the genetic abnormalities driving this process may already be present at diagnosis in certain subclones. Identifying these events would provide a comprehensive view of this transition and would help to develop new diagnostic strategies and targeted therapies. According to previous studies, the transformation process is consistent with a branching model of evolution in which only clones containing driver mutations give rise to more aggressive populations by acquiring new aberrations (Figure 2) [87]. Identical scenarios have been reported in MM [96], FL [97], chronic lymphocytic leukemia [98], AML [99], ALL [100], and even solid tumors [6], although these need to be confirmed in further analyses of single cells. Likewise, it would be interesting to establish whether there is a progenitor tumor cell that is common to both diseases (WM and DLBCL) and that is responsible for their pathogenesis.
Figure 2.
Branching model of tumor evolution observed in WM transformation to DLBCL, adapted from Jiménez, C., Alonso-Álvarez, S., Alcoceba, M. et al. From Waldenström’s macroglobulinemia to aggressive diffuse large B-cell lymphoma: a whole-exome analysis of abnormalities leading to transformation. Blood Cancer J. 7, e591 (2017). https://doi.org/10.1038/bcj.2017.72 [87]. The figure shows an example from a patient who was diagnosed with WM in 2010 and who transformed to DLBCL in 2015, with a symptomatic progression in 2013 before the transformation: 35 mutations were identified at diagnosis, 48 at relapse and 72 at transformation, and 29 of the alterations were conserved throughout the entire process. Mutations in PPM1D, SBF2, TRAPPC9, TRPM7, and WT1 genes were present at progression and transformation. By contrast, two TP53 mutations at relapse that were not observed at transformation. This implies that the transformed final clone did not evolve from the same subclone as was responsible for progression, but from a previous one that would not yet have acquired the TP53 mutations. DLBCL, diffuse large B-cell lymphoma; WM, Waldenström’s macroglobulinemia.
The heterogeneous composition of the bone marrow infiltration in WM, including variable percentages of small lymphocytes, plasmacytoid lymphocytes, and plasma cells [101], is also amenable to study at the single-cell level. The pathophysiology of the plasma cell involvement is poorly understood, and it has been observed that residual plasma cells may be present even in the complete response when the neoplastic B-cell component is absent [102,103]. Moreover, according to a recent study, concordance between the mutational landscape of WM B-cells and plasma cells is not 100%, suggesting that not all WM B-cells differentiate into plasma cells [69]. With more in-depth knowledge of the different populations, it may be possible to determine whether they are part of the neoplastic clone and to identify the origin of the WM cell.
Another interesting outcome of single-cell technologies would be to determine therapy resistance mechanisms throughout the course of treatment. Ibrutinib, a Bruton tyrosine kinase (BTK) inhibitor, is highly active in WM patients [104]. Mutations in BTK at its binding site (Cys481) are common in patients who experience progression on ibrutinib, particularly those with CXCR4 mutations. Multiple BTK mutations can occur within individual patients and coexist with PLCγ2-activating mutations, which are also associated with acquired ibrutinib resistance, both of which lead to downstream ERK1/2 activation [105]. In most cases, BTK mutations are sub-clonal but confer a protective effect against ibrutinib on neighboring BTK wild-type cells through a paracrine mechanism [106]. Single-cell studies may help address the pathways and mechanisms induced in wild-type cells by these alterations, as well as other treatment-emergent mutations, the reason for their co-occurrence, and the particular susceptibility of CXCR4-mutated cells. Such findings would enable us to identify patterns of clonal selection and evolution that mediate clinical resistance, and thereby to use an alternative strategy to overcome resistance in these patients.
It is important not to limit the studies solely to DNA. There may be no direct correspondence between the genetic findings and the final phenotype because important steps must be traversed from one to the other. Transcription and translation, together with all the key regulatory processes involved, play a fundamental role in the pathogenesis of the disease, and everything should be studied and interpreted as a whole. MYD88 and CXCR4 mutations divide WM patients into three major genetic subgroups (MYD88WT, MYD88L265P-CXCR4WHIM, and MYD88L265P-CXCR4WT) that show upregulation and downregulation of different pathways, which is translated into the three final clinical phenotypes with their distinct clinical features [95]. The most clinically relevant consequence of this subclassification of WM is the differential response to certain drugs, such as ibrutinib. Thus, patients with wild-type MYD88 do not achieve an objective response to ibrutinib [107], while those with CXCR4 mutations show lower rates of deep responses, and shorter progression-free survival [108,109]. Therefore, it is crucial that all the information be considered as a whole, and single-cell DNA sequencing studies must be complemented by studies of the phenotypic consequences that a specific genomic abnormality may imply.
4. Limitations of Single-Cell Analysis
As we have shown throughout this review, single-cell technologies have many applications and enormous potential, however, their limitations and challenges must also be recognized. First of all, a large quantity of fresh tumor cells of high viability and purity is essential, but we know that this requirement would be difficult to fulfill for certain diseases. For DNA and protein analysis, it is possible to freeze the samples until they are processed, but for single-cell RNA-sequencing, it is advisable to process the samples on the day of collection to minimize the risk of changes in expression. The workflow is not particularly complicated, but appropriate bioinformatic support and the use of computational methods are absolutely essential. The amount of information generated must be properly processed and analyzed by an integrated pipeline for multi-omics analysis. Allelic dropout, false genotype measurements, sequence-dependent bias, and PCR errors must be considered when analyzing single-cell DNA sequences [110]. For single-cell RNA sequencing, the problems that must be overcome are the poor detection of genes with low-level expression, the 3’ bias, and the noise in the transcriptome data [111]. Different strategies, such as fluorescence-activated cell sorting, Western blotting, metal-tagged antibodies, or oligonucleotide-labeled antibodies, can be used to obtain single-cell proteome profiles, but none of them can detect all the proteins that are expressed [112]. The combination of different single-cell genomic signals is computationally challenging, as these data are intrinsically heterogeneous for experimental, technical, and biological reasons. Multidimensional data obtained from a single cell are integrated and interpreted by computational methods, which provide accurate tools for analyzing multimodal data [113]. Software tools for sequence import and data analysis and visualization are very useful, and some companies already provide them with their products. The great complexity of the data means that results must be interpreted with caution and no clinical decision should be made based on them.
Another limitation of single-cell studies is their expense. As with any emerging technique, up-front costs for consumables, labor, and sequencing are high, creating a barrier to widespread implementation, and excluding these technologies from use in routine testing. Developing high-throughput technologies with a high sample-processing capacity along with easily fabricated and conducted processes could lower the price. The automation of devices would also reduce assay time and minimize human intervention, in addition to limiting user bias and improving reproducibility across laboratories [114].
5. Conclusions
In summary, single-cell analysis can overcome the limitations of bulk sequencing and provide unique insights into tumor heterogeneity, co-dependency of malignant events, and phylogenesis at the cellular level. Furthermore, single-cell multi-omics makes it possible to carry out high-dimensional studies to address complex biological questions, by simultaneously combining the genomic, transcriptomic, and proteomic profiles from the same cell. The integrated information can be used to select the best strategy for the management and treatment of patients.
Acknowledgments
The authors thank the Hematology Department of the University Hospital of Salamanca for its support during the writing of this manuscript.
Author Contributions
Conceptualization, methodology, and investigation, C.J.; writing—original draft preparation, C.J.; writing—review and editing, R.G.-S.; funding acquisition, R.G.-S. Both authors have read and agreed to the published version of the manuscript.
Funding
This study has been funded by Instituto de Salud Carlos III through the project PI18/01866 (Co-funded by European Regional Development Fund/European Social Fund “A way to make Europe”/”Investing in your future”), and Gilead Sciences, Inc., grant number GLD18-00063. C.J. was supported by Instituto de Salud Carlos III (Contrato Sara Borrell CD19/00030).
Conflicts of Interest
The authors declare no conflict of interest.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Michor F., Polyak K. The Origins and Implications of Intratumor Heterogeneity. Cancer Prev. Res. 2010;3:1361–1364. doi: 10.1158/1940-6207.CAPR-10-0234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Andrechek E.R., Nevins J.R. Mouse Models of Cancers: Opportunities to Address Heterogeneity of Human Cancer and Evaluate Therapeutic Strategies. J. Mol. Med. 2010;88:1095–1100. doi: 10.1007/s00109-010-0644-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Somasundaram R., Villanueva J., Herlyn M. Intratumoral Heterogeneity as a Therapy Resistance Mechanism. Role of Melanoma Subpopulations. Adv. Pharmacol. 2012;65:335–359. doi: 10.1016/B978-0-12-397927-8.00011-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gopal P., Sarihan E.I., Chie E.K., Kuzmishin G., Doken S., Pennell N.A., Raymond D.P., Murthy S.C., Ahmad U., Raja S., et al. Clonal Selection Confers Distinct Evolutionary Trajectories in BRAF-Driven Cancers. Nat. Commun. 2019;10:1–14. doi: 10.1038/s41467-019-13161-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Xiao W., Miles L.A., Bowman R.L., Durani V., Tian H.S., DelGaudio N.L., Mishra T., Zhu M., Zhang Y., Stump S.E., et al. A JAK2/IDH1-Mutant MPN Clone Unmasked by Ivosidenib in an AML Patient without Antecedent MPN. Blood Adv. 2020;4:6034–6038. doi: 10.1182/bloodadvances.2020003326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gerlinger M., Rowan A.J., Horswell S., Larkin J., Endesfelder D., Gronroos E., Martinez P., Matthews N., Stewart A., Tarpey P., et al. Intratumor Heterogeneity and Branched Evolution Revealed by Multiregion Sequencing. N. Engl. J. Med. 2012;366:883–892. doi: 10.1056/NEJMoa1113205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vogelstein B., Papadopoulos N., Velculescu V.E., Zhou S., Diaz L.A., Kinzler K.W. Cancer Genome Landscapes. Science. 2013;340:1546–1558. doi: 10.1126/science.1235122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jamal-Hanjani M., Wilson G.A., Horswell S., Mitter R., Sakarya O., Constantin T., Salari R., Kirkizlar E., Sigurjonsson S., Pelham R., et al. Detection of Ubiquitous and Heterogeneous Mutations in Cell-Free DNA from Patients with Early-Stage Non-Small-Cell Lung Cancer. Ann. Oncol. 2016;27:862–867. doi: 10.1093/annonc/mdw037. [DOI] [PubMed] [Google Scholar]
- 9.Russo M., Siravegna G., Blaszkowsky L.S., Corti G., Crisafulli G., Ahronian L.G., Mussolin B., Kwak E.L., Buscarino M., Lazzari L., et al. Tumor Heterogeneity and Lesion-Specific Response to Targeted Therapy in Colorectal Cancer. Cancer Discov. 2016;6:147–153. doi: 10.1158/2159-8290.CD-15-1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhu C., Preissl S., Ren B. Single-Cell Multimodal Omics: The Power of Many. Nat. Methods. 2020;17:11–14. doi: 10.1038/s41592-019-0691-5. [DOI] [PubMed] [Google Scholar]
- 11.Hashimshony T., Wagner F., Sher N., Yanai I. CEL-Seq: Single-Cell RNA-Seq by Multiplexed Linear Amplification. Cell Rep. 2012;2:666–673. doi: 10.1016/j.celrep.2012.08.003. [DOI] [PubMed] [Google Scholar]
- 12.Lohr J.G., Kim S., Gould J., Knoechel B., Drier Y., Cotton M.J., Gray D., Birrer N., Wong B., Ha G., et al. Genetic Interrogation of Circulating Multiple Myeloma Cells at Single-Cell Resolution. Sci. Transl. Med. 2016;8:363ra147. doi: 10.1126/scitranslmed.aac7037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Thompson A.M., Paguirigan A.L., Kreutz J.E., Radich J.P., Chiu D.T. Microfluidics for Single-Cell Genetic Analysis. Lab Chip. 2014;14:3135–3142. doi: 10.1039/C4LC00175C. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pellegrino M., Sciambi A., Treusch S., Durruthy-Durruthy R., Gokhale K., Jacob J., Chen T.X., Geis J.A., Oldham W., Matthews J., et al. High-Throughput Single-Cell DNA Sequencing of Acute Myeloid Leukemia Tumors with Droplet Microfluidics. Genome Res. 2018;28:1345–1352. doi: 10.1101/gr.232272.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hanamsagar R., Reizis T., Chamberlain M., Marcus R., Nestle F.O., de Rinaldis E., Savova V. An Optimized Workflow for Single-Cell Transcriptomics and Repertoire Profiling of Purified Lymphocytes from Clinical Samples. Sci. Rep. 2020;10 doi: 10.1038/s41598-020-58939-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stoeckius M., Hafemeister C., Stephenson W., Houck-Loomis B., Chattopadhyay P.K., Swerdlow H., Satija R., Smibert P. Simultaneous Epitope and Transcriptome Measurement in Single Cells. Nat. Methods. 2017;14:865–868. doi: 10.1038/nmeth.4380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.De Domenico E., Bonaguro L., Schulte-Schrepping J., Becker M., Händler K., Schultze J.L. Optimized Workflow for Single-Cell Transcriptomics on Infectious Diseases Including COVID-19. STAR Protoc. 2020;1:100233. doi: 10.1016/j.xpro.2020.100233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gierahn T.M., Wadsworth M.H., Hughes T.K., Bryson B.D., Butler A., Satija R., Fortune S., Christopher Love J., Shalek A.K. Seq-Well: Portable, Low-Cost Rna Sequencing of Single Cells at High Throughput. Nat. Methods. 2017;14:395–398. doi: 10.1038/nmeth.4179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhu Y., Wang T., Gu J., Huang K., Zhang T., Zhang Z., Liu H., Tang J., Mai Y., Zhang Y., et al. Characterization and Generation of Human Definitive Multipotent Hematopoietic Stem/Progenitor Cells. Cell Discov. 2020;6 doi: 10.1038/s41421-020-00213-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ding J., Smith S.L., Orozco G., Barton A., Eyre S., Martin P. Characterisation of CD4+ T-Cell Subtypes Using Single Cell RNA Sequencing and the Impact of Cell Number and Sequencing Depth. Sci. Rep. 2020;10 doi: 10.1038/s41598-020-76972-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Galletti G., De Simone G., Mazza E.M.C., Puccio S., Mezzanotte C., Bi T.M., Davydov A.N., Metsger M., Scamardella E., Alvisi G., et al. Two Subsets of Stem-like CD8+ Memory T Cell Progenitors with Distinct Fate Commitments in Humans. Nat. Immunol. 2020;21:1552–1562. doi: 10.1038/s41590-020-0791-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nam A.S., Kim K.T., Chaligne R., Izzo F., Ang C., Taylor J., Myers R.M., Abu-Zeinah G., Brand R., Omans N.D., et al. Somatic Mutations and Cell Identity Linked by Genotyping of Transcriptomes. Nature. 2019;571:355–360. doi: 10.1038/s41586-019-1367-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Demaree B., Delley C., Vasudevan H., Peretz C., Ruff D., Smith C., Abate A. Joint Profiling of Proteins and DNA in Single Cells Reveals Extensive Proteogenomic Decoupling in Leukemia. bioRxiv. 2020 doi: 10.1101/2020.02.26.967133. [DOI] [Google Scholar]
- 24.Granja J.M., Klemm S., McGinnis L.M., Kathiria A.S., Mezger A., Corces M.R., Parks B., Gars E., Liedtke M., Zheng G.X.Y., et al. Single-Cell Multiomic Analysis Identifies Regulatory Programs in Mixed-Phenotype Acute Leukemia. Nat. Biotechnol. 2019;37:1458–1465. doi: 10.1038/s41587-019-0332-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dillon L.W., Ghannam J., Nosiri C., Gege G., Goswami M., Calvo K.R., Lindblad K.E., Oetjen K.A., Wilkerson M., Soltis A.R., et al. Personalized Single-Cell Proteogenomics to Distinguish Acute Myeloid Leukemia from Non-Malignant Clonal Hematopoiesis. medRxiv. 2020 doi: 10.1101/2020.10.22.20216028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Young A.L., Challen G.A., Birmann B.M., Druley T.E. Clonal Haematopoiesis Harbouring AML-Associated Mutations Is Ubiquitous in Healthy Adults. Nat. Commun. 2016;7 doi: 10.1038/ncomms12484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jaiswal S., Natarajan P., Silver A.J., Gibson C.J., Bick A.G., Shvartz E., McConkey M., Gupta N., Gabriel S., Ardissino D., et al. Clonal Hematopoiesis and Risk of Atherosclerotic Cardiovascular Disease. N. Engl. J. Med. 2017;377:111–121. doi: 10.1056/NEJMoa1701719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tyner J.W., Tognon C.E., Bottomly D., Wilmot B., Kurtz S.E., Savage S.L., Long N., Schultz A.R., Traer E., Abel M., et al. Functional Genomic Landscape of Acute Myeloid Leukaemia. Nature. 2018;562:526–531. doi: 10.1038/s41586-018-0623-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jongen-Lavrencic M., Grob T., Hanekamp D., Kavelaars F.G., al Hinai A., Zeilemaker A., Erpelinck-Verschueren C.A.J., Gradowska P.L., Meijer R., Cloos J., et al. Molecular Minimal Residual Disease in Acute Myeloid Leukemia. N. Engl. J. Med. 2018;378:1189–1199. doi: 10.1056/NEJMoa1716863. [DOI] [PubMed] [Google Scholar]
- 30.Hourigan C.S., Dillon L.W., Gui G., Logan B.R., Fei M., Ghannam J., Li Y., Licon A., Alyea E.P., Bashey A., et al. Impact of Conditioning Intensity of Allogeneic Transplantation for Acute Myeloid Leukemia with Genomic Evidence of Residual Disease. J. Clin. Oncol. 2020;38:1273–1283. doi: 10.1200/JCO.19.03011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Maia C., Puig N., Cedena M.T., Goicoechea I., Valdes-Mas R., Vazquez I., Chillon M.C., Aguirre P., Sarvide S., Gracia-Aznárez F.J., et al. Biological and Clinical Significance of Dysplastic Hematopoiesis in Patients with Newly Diagnosed Multiple Myeloma. Blood. 2020;135:2375–2387. doi: 10.1182/blood.2019003382. [DOI] [PubMed] [Google Scholar]
- 32.Gawad C., Koh W., Quake S.R. Dissecting the Clonal Origins of Childhood Acute Lymphoblastic Leukemia by Single-Cell Genomics. Proc. Natl. Acad. Sci. USA. 2014;111:17947–17952. doi: 10.1073/pnas.1420822111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.De Bie J., Demeyer S., Alberti-Servera L., Geerdens E., Segers H., Broux M., De Keersmaecker K., Michaux L., Vandenberghe P., Voet T., et al. Single-Cell Sequencing Reveals the Origin and the Order of Mutation Acquisition in T-Cell Acute Lymphoblastic Leukemia. Leukemia. 2018;32:1358–1369. doi: 10.1038/s41375-018-0127-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Albertí-Servera L., Demeyer S., Govaerts I., Swings T., De Bie J., Gielen O., Brociner M., Michaux L.M., Maertens J., Uyttebroeck A., et al. Single-Cell DNA Amplicon Sequencing Reveals Clonal Heterogeneity and Evolution in T-Cell Acute Lymphoblastic Leukemia. Blood. 2020 doi: 10.1182/blood.2020006996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.McMahon C.M., Ferng T., Canaani J., Wang E.S., Morrissette J.J.D., Eastburn D.J., Pellegrino M., Durruthy-Durruthy R., Watt C.D., Asthana S., et al. Clonal Selection with RAS Pathway Activation Mediates Secondary Clinical Resistance to Selective FLT3 Inhibition in Acute Myeloid Leukemia. Cancer Discov. 2019;9:1050–1063. doi: 10.1158/2159-8290.CD-18-1453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Morita K., Wang F., Jahn K., Hu T., Tanaka T., Sasaki Y., Kuipers J., Loghavi S., Wang S.A., Yan Y., et al. Clonal Evolution of Acute Myeloid Leukemia Revealed by High-Throughput Single-Cell Genomics. Nat. Commun. 2020;11 doi: 10.1038/s41467-020-19119-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Miles L.A., Bowman R.L., Merlinsky T.R., Csete I.S., Ooi A.T., Durruthy-Durruthy R., Bowman M., Famulare C., Patel M.A., Mendez P., et al. Single-Cell Mutation Analysis of Clonal Evolution in Myeloid Malignancies. Nature. 2020;587:477–482. doi: 10.1038/s41586-020-2864-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Thompson E.R., Nguyen T., Kankanige Y., Yeh P., Ingbritsen M., McBean M., Semple T., Arnau G.M., Burbury K., Lee N., et al. Clonal Independence of JAK2 and CALR or MPL Mutations in Comutated Myeloproliferative Neoplasms Demonstrated by Single Cell DNA Sequencing. Haematologica. 2021;106:313–315. doi: 10.3324/haematol.2020.260448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Patel B.A., Ghannam J., Groarke E.M., Goswami M., Dillon L., Gutierrez-Rodrigues F., Rios O., Raffo D.Q., Lotter J., Young N.S., et al. Detectable Mutations Precede Late Myeloid Neoplasia in Aplastic Anemia. Haematologica. 2020 doi: 10.3324/haematol.2020.263046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kennedy A.L., Myers K.C., Bowman J., Gibson C.J., Camarda N.D., Furutani E., Muscato G.M., Klein R.H., Ballotti K., Liu S., et al. Distinct Genetic Pathways Define Pre-Leukemic and Compensatory Clonal Hematopoiesis in Shwachman-Diamond Syndrome. bioRxiv. 2020 doi: 10.1101/2020.06.04.134692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ryu D., Kim S.J., Hong Y., Jo A., Kim N., Kim H.J., Lee H.O., Kim K., Park W.Y. Alterations in the Transcriptional Programs of Myeloma Cells and the Microenvironment during Extramedullary Progression Affect Proliferation and Immune Evasion. Clin. Cancer Res. 2020;26:935–944. doi: 10.1158/1078-0432.CCR-19-0694. [DOI] [PubMed] [Google Scholar]
- 42.Pei S., Pollyea D.A., Gustafson A., Stevens B.M., Minhajuddin M., Fu R., Riemondy K.A., Gillen A.E., Sheridan R.M., Kim J., et al. Monocytic Subclones Confer Resistance to Venetoclax-Based Therapy in Patients with Acute Myeloid Leukemia. Cancer Discov. 2020;10:536–551. doi: 10.1158/2159-8290.CD-19-0710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ediriwickrema A., Aleshin A., Reiter J.G., Corces M.R., Kohnke T., Stafford M., Liedtke M., Medeiros B.C., Majeti R. Single-Cell Mutational Profiling Enhances the Clinical Evaluation of AML MRD. Blood Adv. 2020;4:943–952. doi: 10.1182/bloodadvances.2019001181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jiang L., Li X.P., Dai Y.T., Chen B., Weng X.Q., Xiong S.M., Zhang M., Huang J.Y., Chen Z., Chen S.J. Multidimensional Study of the Heterogeneity of Leukemia Cells in t(8;21) Acute Myelogenous Leukemia Identifies the Subtype with Poor Outcome. Proc. Natl. Acad. Sci. USA. 2020;117:20117–20126. doi: 10.1073/pnas.2003900117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.DiNardo C.D., Tiong I.S., Quaglieri A., MacRaild S., Loghavi S., Brown F.C., Thijssen R., Pomilio G., Ivey A., Salmon J.M., et al. Molecular Patterns of Response and Treatment Failure after Frontline Venetoclax Combinations in Older Patients with AML. Blood. 2020;135:791–803. doi: 10.1182/blood.2019003988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang F., Morita K., DiNardo C.D., Furudate K., Tanaka T., Yan Y., Patel K.P., MacBeth K.J., Wu B., Liu G., et al. Leukemia Stemness and Co-Occurring Mutations Drive Resistance to IDH Inhibitors in Acute Myeloid Leukemia Lead Contact. bioRxiv. 2020 doi: 10.1101/2020.10.27.357111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Choe S., Wang H., DiNardo C.D., Stein E.M., de Botton S., Roboz G.J., Altman J.K., Mims A.S., Watts J.M., Pollyea D.A., et al. Molecular Mechanisms Mediating Relapse Following Ivosidenib Monotherapy in IDH1-Mutant Relapsed or Refractory AML. Blood Adv. 2020;4:1894–1905. doi: 10.1182/bloodadvances.2020001503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhao Y., Aldoss I., Qu C., Crawford J.C., Gu Z., Allen E.K., Zamora A.E., Alexander T.B., Wang J., Goto H., et al. Tumor Intrinsic and Extrinsic Determinants of Response to Blinatumomab in Adults with B-ALL. Blood. 2020 doi: 10.1182/blood.2020006287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cartel M., Mouchel P.L., Gotanègre M., David L., Bertoli S., Mas V.M., De Besson A., Sarry J.E., Manenti S., Didier C. Inhibition of Ubiquitin-Specific Protease 7 Sensitizes Acute Myeloid Leukemia to Chemotherapy. Leukemia. 2020 doi: 10.1038/s41375-020-0878-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rendeiro A.F., Krausgruber T., Fortelny N., Zhao F., Penz T., Farlik M., Schuster L.C., Nemc A., Tasnády S., Réti M., et al. Chromatin Mapping and Single-Cell Immune Profiling Define the Temporal Dynamics of Ibrutinib Response in CLL. Nat. Commun. 2020;11 doi: 10.1038/s41467-019-14081-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Witkowski M.T., Dolgalev I., Evensen N.A., Ma C., Chambers T., Roberts K.G., Sreeram S., Dai Y., Tikhonova A.N., Lasry A., et al. Extensive Remodeling of the Immune Microenvironment in B Cell Acute Lymphoblastic Leukemia. Cancer Cell. 2020;37:867–882.e12. doi: 10.1016/j.ccell.2020.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mcheyzer-Williams L.J., Milpied P.J., Okitsu S.L., Mcheyzer-Williams M.G. Class-Switched Memory B Cells Remodel BCRs within Secondary Germinal Centers. Nat. Immunol. 2015;16:296–305. doi: 10.1038/ni.3095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Milpied P., Cervera-Marzal I., Mollichella M.L., Tesson B., Brisou G., Traverse-Glehen A., Salles G., Spinelli L., Nadel B. Human Germinal Center Transcriptional Programs Are De-Synchronized in B Cell Lymphoma. Nat. Immunol. 2018;19:1013–1024. doi: 10.1038/s41590-018-0181-4. [DOI] [PubMed] [Google Scholar]
- 54.Rollins M.R., Spartz E.J., Stromnes I.M. T Cell Receptor Engineered Lymphocytes for Cancer Therapy. Curr. Protoc. Immunol. 2020;129 doi: 10.1002/cpim.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Deng Q., Han G., Puebla-Osorio N., Ma M.C.J., Strati P., Chasen B., Dai E., Dang M., Jain N., Yang H., et al. Characteristics of Anti-CD19 CAR T Cell Infusion Products Associated with Efficacy and Toxicity in Patients with Large B Cell Lymphomas. Nat. Med. 2020;26:1878–1887. doi: 10.1038/s41591-020-1061-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.ten Hacken E., Clement K., Li S., Hernández-Sánchez M., Redd R., Wang S., Ruff D., Gruber M., Baranowski K., Jacob J., et al. High Throughput Single-Cell Detection of Multiplex CRISPR-Edited Gene Modifications. Genome Biol. 2020;21 doi: 10.1186/s13059-020-02174-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Xu L., Durruthy-Durruthy R., Eastburn D.J., Pellegrino M., Shah O., Meyer E., Zehnder J. Clonal Evolution and Changes in Two AML Patients Detected with A Novel Single-Cell DNA Sequencing Platform. Sci. Rep. 2019;9 doi: 10.1038/s41598-019-47297-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Foltz S.M., Gao Q., Yoon C.J., Sun H., Yao L., Li Y., Jayasinghe R.G., Cao S., King J., Kohnen D.R., et al. Evolution and Structure of Clinically Relevant Gene Fusions in Multiple Myeloma. Nat. Commun. 2020;11 doi: 10.1038/s41467-020-16434-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Owen R.G., Treon S.P., Al-Katib A., Fonseca R., Greipp P.R., McMaster M.L., Morra E., Pangalis G.A., San Miguel J.F., Branagan A.R., et al. Clinicopathological Definition of Waldenström’s Macroglobulinemia: Consensus Panel Recommendations from the Second International Workshop on Waldenstrom’s Macroglobulinemia. Semin. Oncol. 2003;30:110–115. doi: 10.1053/sonc.2003.50082. [DOI] [PubMed] [Google Scholar]
- 60.Oza A., Rajkumar S. Waldenström Macroglobulinemia: Prognosis and Management. Blood Cancer J. 2015;5:e296. doi: 10.1038/bcj.2015.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Castillo J.J., Gustine J., Meid K., Dubeau T., Hunter Z.R., Treon S.P. Histological Transformation to Diffuse Large B-Cell Lymphoma in Patients with Waldenström Macroglobulinemia. Am. J. Hematol. 2016;91:1032–1035. doi: 10.1002/ajh.24477. [DOI] [PubMed] [Google Scholar]
- 62.Horsman D.E., Card R.T., Skinnider L.F. Waldenström Macroglobulinemia Terminating in Acute Leukemia: A Report of Three Cases. Am. J. Hematol. 1983;15:97–101. doi: 10.1002/ajh.2830150111. [DOI] [PubMed] [Google Scholar]
- 63.Treon S.P., Xu L., Yang G., Zhou Y., Liu X., Cao Y., Sheehy P., Manning R.J., Patterson C.J., Tripsas C., et al. MYD88 L265P Somatic Mutation in Waldenström’s Macroglobulinemia. N. Engl. J. Med. 2012;367:826–833. doi: 10.1056/NEJMoa1200710. [DOI] [PubMed] [Google Scholar]
- 64.Jiménez C., Sebastián E., Chillón M.C., Giraldo P., Mariano Hernández J., Escalante F., González-López T.J., Aguilera C., García de Coca A., Murillo I., et al. MYD88 L265P Is a Marker Highly Characteristic of, but Not Restricted to, Waldenström’s Macroglobulinemia. Leukemia. 2013;27:1722–1728. doi: 10.1038/leu.2013.62. [DOI] [PubMed] [Google Scholar]
- 65.Xu L., Hunter Z.R., Yang G., Zhou Y., Cao Y., Liu X., Morra E., Trojani A., Greco A., Arcaini L., et al. MYD88 L265P in Waldenstrom’s Macroglobulinemia, IgM Monoclonal Gammopathy, and Other B-Cell Lymphoproliferative Disorders Using Conventional and Quantitative Allele-Specific PCR. Blood. 2013;121:2051–2058. doi: 10.1182/blood-2012-09-454355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Varettoni M., Arcaini L., Zibellini S., Boveri E., Rattotti S., Riboni R., Corso A., Orlandi E., Bonfichi M., Gotti M., et al. Prevalence and Clinical Significance of the MYD88 (L265P) Somatic Mutation in Waldenström’s Macroglobulinemia and Related Lymphoid Neoplasms. Blood. 2013;88:2522–2528. doi: 10.1182/blood-2012-09-457101. [DOI] [PubMed] [Google Scholar]
- 67.Poulain S., Roumier C., Decambron A., Renneville A., Herbaux C., Bertrand E., Tricot S., Daudignon A., Galiègue-Zouitina S., Soenen V., et al. MYD88 L265P Mutation in Waldenstrom Macroglobulinemia. Blood. 2013;121:4504–4511. doi: 10.1182/blood-2012-06-436329. [DOI] [PubMed] [Google Scholar]
- 68.Sewastianik T., Guerrera M.L., Adler K., Dennis P.S., Wright K., Shanmugam V., Huang Y., Tanton H., Jiang M., Kofides A., et al. Human MYD88L265P Is Insufficient by Itself to Drive Neoplastic Transformation in Mature Mouse B Cells. Blood Adv. 2019;3:3360–3374. doi: 10.1182/bloodadvances.2019000588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Rodríguez S., Botta C., Celay J., Goicoechea I., Jiménez C., Garcia-Barchino M.J., Valdés-Mas R., Carrasco Y., Garcés J.J., Larrayoz M., et al. Waldenström’s Macroglobulinemia (WM) Is Preceded By Clonal Lymphopoiesis Including MYD88 L265P in Progenitor B Cells; Proceedings of the 62nd ASH Annual Meeting Exposition; San Diego, CA, USA. 5–8 December 2020; [DOI] [Google Scholar]
- 70.Hunter Z.R., Xu L., Yang G., Zhou Y., Liu X., Cao Y., Manning R.J., Tripsas C., Patterson C.J., Sheehy P., et al. The Genomic Landscape of Waldenström Macroglobulinemia Is Characterized by Highly Recurring MYD88 and WHIM-like CXCR4 Mutations, and Small Somatic Deletions Associated with B-Cell Lymphomagenesis. Blood. 2014;123:1637–1646. doi: 10.1182/blood-2013-09-525808. [DOI] [PubMed] [Google Scholar]
- 71.Xu L., Hunter Z.R., Tsakmaklis N., Cao Y., Yang G., Chen J., Liu X., Kanan S., Castillo J.J., Tai Y.-T., et al. Clonal Architecture of CXCR4 WHIM-like Mutations in Waldenström Macroglobulinaemia. Br. J. Haematol. 2016;172:735–744. doi: 10.1111/bjh.13897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Treon S.P., Cao Y., Xu L., Yang G., Liu X., Hunter Z.R. Somatic Mutations in MYD88 and CXCR4 Are Determinants of Clinical Presentation and Overall Survival in Waldenström Macroglobulinemia. Blood. 2014;123:2791–2796. doi: 10.1182/blood-2014-01-550905. [DOI] [PubMed] [Google Scholar]
- 73.Poulain S., Roumier C., Venet-Caillault A., Figeac M., Herbaux C., Marot G., Doye E., Bertrand E., Geffroy S., Lepretre F., et al. Genomic Landscape of CXCR4 Mutations in Waldenström Macroglobulinemia. Clin. Cancer Res. 2016;22:1480–1488. doi: 10.1158/1078-0432.CCR-15-0646. [DOI] [PubMed] [Google Scholar]
- 74.Roccaro A.M., Sacco A., Jimenez C., Maiso P., Moschetta M., Mishima Y., Aljawai Y., Sahin I., Kuhne M., Cardarelli P., et al. C1013G/CXCR4 Acts as a Driver Mutation of Tumor Progression and Modulator of Drug Resistance in Lymphoplasmacytic Lymphoma. Blood. 2014;123:4120–4131. doi: 10.1182/blood-2014-03-564583. [DOI] [PubMed] [Google Scholar]
- 75.García-Sanz R. WM, MYD88, and CXCR4: Following the Thread. Blood. 2016;128:746–748. doi: 10.1182/blood-2016-06-723924. [DOI] [PubMed] [Google Scholar]
- 76.Jimenez C., Xu L., Tsakmaklis N., Demos M.G., Kofides A., Chan G.G., Guerrera M.L., Chen J.G., Liu X., Munshi M., et al. Comparative Genomics of CXCR4MUT and CXCR4WT Single Cells in Waldenstr¨om’s Macroglobulinemia. Blood Adv. 2020;4:4550–4553. doi: 10.1182/bloodadvances.2020001775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Poulain S., Herbaux C., Bertrand E., Decambron A., Fouquet G., Boyle E., Gay J., Manier S., Duthilleul P., Roumier C., et al. Genomic Studies Have Identified Multiple Mechanisms of Genetic Changes in Waldenström Macroglobulinemia. Clin. Lymphoma Myeloma Leuk. 2013;13:202–204. doi: 10.1016/j.clml.2013.02.008. [DOI] [PubMed] [Google Scholar]
- 78.Ciric B., VanKeulen V., Rodriguez M., Kyle R.A., Gertz M.A., Pease L.R. Clonal Evolution in Waldenstrom Macroglobulinemia Highlights Functional Role of B-Cell Receptor. Blood. 2001;97:321–323. doi: 10.1182/blood.V97.1.321. [DOI] [PubMed] [Google Scholar]
- 79.Varettoni M., Zibellini S., Capello D., Arcaini L., Rossi D., Pascutto C., Rattotti S., Mangiacavalli S., Pochintesta L., Gotti M., et al. Clues to Pathogenesis of Waldenström Macroglobulinemia and Immunoglobulin M Monoclonal Gammopathy of Undetermined Significance Provided by Analysis of Immunoglobulin Heavy Chain Gene Rearrangement and Clustering of B-Cell Receptors. Leuk. Lymphoma. 2013;54:2485–2489. doi: 10.3109/10428194.2013.779689. [DOI] [PubMed] [Google Scholar]
- 80.Young R.M., Staudt L.M. Targeting Pathological B Cell Receptor Signalling in Lymphoid Malignancies. Nat. Rev. Drug Discov. 2013;12:229–243. doi: 10.1038/nrd3937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Leadbetter E.A., Rifkin I.R., Hohlbaum A.M., Beaudette B.C., Shlomchik M.J., Marshak-Rothstein A. Chromatin-IgG Complexes Activate B Cells by Dual Engagement of IgM and Toll-like Receptors. Nature. 2002;416:603–607. doi: 10.1038/416603a. [DOI] [PubMed] [Google Scholar]
- 82.Hou B., Saudan P., Ott G., Wheeler M.L., Ji M., Kuzmich L., Lee L.M., Coffman R.L., Bachmann M.F., DeFranco A.L. Selective Utilization of Toll-like Receptor and MyD88 Signaling in B Cells for Enhancement of the Antiviral Germinal Center Response. Immunity. 2011;34:375–384. doi: 10.1016/j.immuni.2011.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Jefferies C.A., Doyle S., Brunner C., Dunne A., Brint E., Wietek C., Walch E., Wirth T., O’Neill L.A.J. Bruton’s Tyrosine Kinase Is a Toll/Interleukin-1 Receptor Domain-Binding Protein That Participates in Nuclear Factor KappaB Activation by Toll-like Receptor 4. J. Biol. Chem. 2003;278:26258–26264. doi: 10.1074/jbc.M301484200. [DOI] [PubMed] [Google Scholar]
- 84.Varettoni M., Zibellini S., Defrancesco I., Ferretti V.V., Rizzo E., Malcovati L., Gallì A., Giovanni M., Porta D., Boveri E., et al. Pattern of Somatic Mutations in Patients with Waldenström Macroglobulinemia or IgM Monoclonal Gammopathy of Undetermined Significance. Haematologica. 2017;102:2077–2085. doi: 10.3324/haematol.2017.172718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Jiménez C., Prieto-Conde M.I., García-Álvarez M., Alcoceba M., Escalante F., Chillón M.D.C., García de Coca A., Balanzategui A., Cantalapiedra A., Aguilar C., et al. Unraveling the Heterogeneity of IgM Monoclonal Gammopathies: A Gene Mutational and Gene Expression Study. Ann. Hematol. 2018;97:475–484. doi: 10.1007/s00277-017-3207-3. [DOI] [PubMed] [Google Scholar]
- 86.Poulain S., Roumier C., Galiègue-Zouitina S., Daudignon A., Herbaux C., Aiijou R., Lainelle A., Broucqsault N., Bertrand E., Manier S., et al. Genome Wide SNP Array Identified Multiple Mechanisms of Genetic Changes in Waldenstrom Macroglobulinemia. Am. J. Hematol. 2013;88:948–954. doi: 10.1002/ajh.23545. [DOI] [PubMed] [Google Scholar]
- 87.Jiménez C., Alonso-Álvarez S., Alcoceba M., Ordóñez G.R., García-Álvarez M., Prieto-Conde M.I., Chillón M.C., Balanzategui A., Corral R., Marín L.A., et al. From Waldenström’s Macroglobulinemia to Aggressive Diffuse Large B-Cell Lymphoma: A Whole-Exome Analysis of Abnormalities Leading to Transformation. Blood Cancer J. 2017;7 doi: 10.1038/bcj.2017.72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Poulain S., Roumier C., Bertrand E., Renneville A., Caillault-Venet A., Doye E., Geffroy S., Sebda S., Nibourel O., Nudel M., et al. TP53 Mutation and Its Prognostic Significance in Waldenstrom’s Macroglobulinemia. Clin. Cancer Res. 2017;23:6325–6336. doi: 10.1158/1078-0432.CCR-17-0007. [DOI] [PubMed] [Google Scholar]
- 89.Gustine J.N., Tsakmaklis N., Demos M.G., Kofides A., Chen J.G., Liu X., Munshi M., Guerrera M.L., Chan G.G., Patterson C.J., et al. TP53 Mutations Are Associated with Mutated MYD88 and CXCR4, and Confer an Adverse Outcome in Waldenström Macroglobulinaemia. Br. J. Haematol. 2019;184:242–245. doi: 10.1111/bjh.15560. [DOI] [PubMed] [Google Scholar]
- 90.Paiva B., Corchete L.A., Vidriales M.-B., Garcia-Sanz R., Perez J.J., Aires-Mejia I., Sanchez M.-L., Barcena P., Alignani D., Jimenez C., et al. The Cellular Origin and Malignant Transformation of Waldenstrom Macroglobulinemia. Blood. 2015;125:2370–2380. doi: 10.1182/blood-2014-09-602565. [DOI] [PubMed] [Google Scholar]
- 91.Schop R.F.J., Kuehl W.M., Van Wier S.A., Ahmann G.J., Price-Troska T., Bailey R.J., Jalal S.M., Qi Y., Kyle R.A., Greipp P.R., et al. Waldenström Macroglobulinemia Neoplastic Cells Lack Immunoglobulin Heavy Chain Locus Translocations but Have Frequent 6q Deletions. Blood. 2002;100:2996–3001. doi: 10.1182/blood.V100.8.2996. [DOI] [PubMed] [Google Scholar]
- 92.Chang H., Qi X., Xu W., Reader J.C., Ning Y. Analysis of 6q Deletion in Waldenstrom Macroglobulinemia. Eur. J. Haematol. 2007;79:244–247. doi: 10.1111/j.1600-0609.2007.00903.x. [DOI] [PubMed] [Google Scholar]
- 93.Guerrera M.L., Tsakmaklis N., Xu L., Yang G., Demos M., Kofides A., Chan G.G., Manning R.J., Liu X., Chen J.G., et al. MYD88 Mutated and Wild-Type Waldenström’s Macroglobulinemia: Characterization of Chromosome 6q Gene Losses and Their Mutual Exclusivity with Mutations in CXCR4. Haematologica. 2018;103:e408–e411. doi: 10.3324/haematol.2018.190181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.García-Sanz R., Dogliotti I., Zaccaria G.M., Ocio E.M., Rubio A., Murillo I., Escalante F., Aguilera C., García-Mateo A., García de Coca A., et al. 6q Deletion in Waldenström Macroglobulinaemia Negatively Affects Time to Transformation and Survival. Br. J. Haematol. 2020 doi: 10.1111/bjh.17028. [DOI] [PubMed] [Google Scholar]
- 95.Hunter Z.R., Xu L., Yang G., Tsakmaklis N., Vos M., Liu X., Chen J.J.G., Manning R.J., Chen J.J.G., Patterson C.J., et al. Transcriptome Sequencing Reveals a Profile That Corresponds to Genomic Variants in Waldenström Macroglobulinemia. Blood. 2016;128:827–838. doi: 10.1182/blood-2016-03-708263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Bolli N., Avet-Loiseau H., Wedge D.C., Van Loo P., Alexandrov L.B., Martincorena I., Dawson K.J., Iorio F., Nik-Zainal S., Bignell G.R., et al. Heterogeneity of Genomic Evolution and Mutational Profiles in Multiple Myeloma. Nat. Commun. 2014;5:2997. doi: 10.1038/ncomms3997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Okosun J., Bödör C., Wang J., Araf S., Yang C.Y., Pan C., Boller S., Cittaro D., Bozek M., Iqbal S., et al. Integrated Genomic Analysis Identifies Recurrent Mutations and Evolution Patterns Driving the Initiation and Progression of Follicular Lymphoma. Nat. Genet. 2013;46:176–181. doi: 10.1038/ng.2856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Landau D.A., Tausch E., Taylor-weiner A.N., Stewart C., Reiter J.G., Bahlo J., Carter S.L., Cibulskis K., Mertens D., Kluth S., et al. Mutations Driving CLL and Their Evolution in Progression and Relapse. Nature. 2015;526:525–530. doi: 10.1038/nature15395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Ding L., Ley T.J., Larson D.E., Miller C.A., Daniel C., Welch J.S., Ritchey J.K., Young M.A., Lamprecht T., Mclellan M.D., et al. Clonal Evolution in Relapsed Acute Myeloid Leukemia Revealed by Whole Genome Sequencing. Nature. 2012;481:506–510. doi: 10.1038/nature10738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Anderson K., Lutz C., van Delft F.W., Bateman C.M., Guo Y., Colman S.M., Kempski H., Moorman A.V., Titley I., Swansbury J., et al. Genetic Variegation of Clonal Architecture and Propagating Cells in Leukaemia. Nature. 2011;469:356–361. doi: 10.1038/nature09650. [DOI] [PubMed] [Google Scholar]
- 101.Stone M.J., Pascual V. Pathophysiology of Waldenström’s Macroglobulinemia. Haematologica. 2010;95:359–364. doi: 10.3324/haematol.2009.017251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Gajendra S., Sachdev R., Jha B., Goel S., Sahni T. Persistent Monotypic Plasma Cells with Absence of Neoplastic b Cell Component in a Treated Case of Waldenström Macroglobulinemia: A Sign of Residual Disease? J. Clin. Diagnostic Res. 2014;8:FD12–FD13. doi: 10.7860/JCDR/2014/9988.5196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.García-Sanz R., Ocio E., Caballero A., Magalhães R.J.P., Alonso J., López-Anglada L., Villaescusa T., Puig N., Hernández J.M., Fernández-Calvo J., et al. Post-Treatment Bone Marrow Residual Disease >5% by Flow Cytometry Is Highly Predictive of Short Progression-Free and Overall Survival in Patients with Waldenström’s Macroglobulinemia. Clin. Lymphoma. Myeloma Leuk. 2011;11:168–171. doi: 10.3816/CLML.2011.n.040. [DOI] [PubMed] [Google Scholar]
- 104.Treon S.P., Tripsas C.K., Meid K., Warren D., Varma G., Green R., Argyropoulos K.V., Yang G., Cao Y., Xu L., et al. Ibrutinib in Previously Treated Waldenström’s Macroglobulinemia. N. Engl. J. Med. 2015;372:1430–1440. doi: 10.1056/NEJMoa1501548. [DOI] [PubMed] [Google Scholar]
- 105.Xu L., Tsakmaklis N., Yang G., Chen J.G., Liu X., Demos M., Kofides A., Patterson C.J., Meid K., Gustine J., et al. Acquired Mutations Associated with Ibrutinib Resistance in Waldenstrom Macroglobulinemia. Blood. 2017;129:2519–2525. doi: 10.1182/blood-2017-01-761726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Chen J.G., Liu X., Munshi M., Xu L., Tsakmaklis N., Demos M.G., Kofides A., Guerrera M.L., Chan G.G., Patterson C.J., et al. BTKCys481Ser Drives Ibrutinib Resistance via ERK1/2 and Protects BTKwild-Type MYD88-Mutated Cells by a Paracrine Mechanism. Blood. 2018;131:2047–2059. doi: 10.1182/blood-2017-10-811752. [DOI] [PubMed] [Google Scholar]
- 107.Dimopoulos M.A., Trotman J., Tedeschi A., Matous J.V., Macdonald D., Tam C., Tournilhac O., Ma S., Oriol A., Heffner L.T., et al. Ibrutinib for Patients with Rituximab-Refractory Waldenström’s Macroglobulinaemia (INNOVATE): An Open-Label Substudy of an International, Multicentre, Phase 3 Trial. Lancet Oncol. 2017;18:241–250. doi: 10.1016/S1470-2045(16)30632-5. [DOI] [PubMed] [Google Scholar]
- 108.Treon S.P., Gustine J., Meid K., Yang G., Xu L., Liu X., Demos M., Kofides A., Tsakmaklis N., Chen J.G., et al. Ibrutinib Monotherapy in Symptomatic, Treatment-Naïve Patients With Waldenström Macroglobulinemia. J. Clin. Oncol. 2018;36:2755–2761. doi: 10.1200/JCO.2018.78.6426. [DOI] [PubMed] [Google Scholar]
- 109.Castillo J.J., Xu L., Gustine J.N., Keezer A., Meid K., Dubeau T.E., Liu X., Demos M.G., Kofides A., Tsakmaklis N., et al. CXCR4 Mutation Subtypes Impact Response and Survival Outcomes in Patients with Waldenström Macroglobulinaemia Treated with Ibrutinib. Br. J. Haematol. 2019;187:356–363. doi: 10.1111/bjh.16088. [DOI] [PubMed] [Google Scholar]
- 110.Huang L., Ma F., Chapman A., Lu S., Xie X.S. Single-Cell Whole-Genome Amplification and Sequencing: Methodology and Applications. Annu. Rev. Genomics Hum. Genet. 2015;16:79–102. doi: 10.1146/annurev-genom-090413-025352. [DOI] [PubMed] [Google Scholar]
- 111.Amirmahani F., Ebrahimi N., Molaei F., Faghihkhorasani F., Jamshidi Goharrizi K., Mirtaghi S.M., Borjian-Boroujeni M., Hamblin M.R. Approaches for the Integration of Big Data in Translational Medicine: Single-cell and Computational Methods. Ann. N. Y. Acad. Sci. 2021:nyas.14544. doi: 10.1111/nyas.14544. [DOI] [PubMed] [Google Scholar]
- 112.Peterson V.M., Zhang K.X., Kumar N., Wong J., Li L., Wilson D.C., Moore R., Mcclanahan T.K., Sadekova S., Klappenbach J.A. Multiplexed Quantification of Proteins and Transcripts in Single Cells. Nat. Biotechnol. 2017;35:936–939. doi: 10.1038/nbt.3973. [DOI] [PubMed] [Google Scholar]
- 113.Forcato M., Romano O., Bicciato S. Computational Methods for the Integrative Analysis of Single-Cell Data. Brief. Bioinform. 2020 doi: 10.1093/bib/bbaa042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Kong S.L., Li H., Tai J.A., Courtois E.T., Poh H.M., Lau D.P., Haw Y.X., Iyer N.G., Weng Tan D.S., Prabhakar S., et al. Concurrent Single-Cell RNA and Targeted DNA Sequencing on an Automated Platform for Comeasurement of Genomic and Transcriptomic Signatures. Clin. Chem. 2019;65:272–281. doi: 10.1373/clinchem.2018.295717. [DOI] [PubMed] [Google Scholar]