Abstract
Simple Summary
Although the prognosis of acute lymphoblastic leukemia (ALL) has improved significantly during the past decades, ALL remains a major cause of pediatric cancer mortality, and more accurate risk-stratification is required. We investigated IGF2BP3, which has previously been associated with aggressive cancers, and found high and subtype-specific expression of IGF2BP3 in B-cell ALL, that was associated with good outcome in high-risk patients. Results suggest that IGF2BP3 could be useful to improve stratification and prognosis of B-ALL.
Abstract
The oncofetal protein insulin-like growth factor 2 mRNA-binding protein 3 (IGF2BP3) belongs to a family of RNA-binding proteins involved in localization, stability, and translational regulation of target RNAs. IGF2BP3 is used as a diagnostic and prognostic marker in several malignancies. Although the prognosis of pediatric B-cell acute lymphoblastic leukemia (B-ALL) has improved, a subgroup of patients exhibits high-risk features and suffer from disease recurrence. We sought to identify additional biomarkers to improve diagnostics, and we assessed expression of IGF2BP3 in a population-based pediatric cohort of B-ALL using a tissue microarray platform. The majority of pediatric B-ALL cases were positive for IGF2BP3 immunohistochemistry and were associated with an increased proliferative phenotype and activated STAT5 signaling pathway. Two large gene expression data sets were probed for the expression of IGF2BP3—the highest levels were seen among the B-cell lymphomas of a germinal center origin and well-established (KMT2A-rearranged and ETV6-RUNX1) and novel subtypes of B-ALL (e.g., NUTM1 and ETV6-RUNX1-like). A high mRNA for IGF2BP3 was associated with a proliferative “metagene” signature and a high expression of CDK6 in B-ALL. A low expression portended inferior survival in a high-risk cohort of pediatric B-ALL. Overall, our results show that IGF2BP3 shows subtype-specificity in expression and provides prognostic utility in high-risk B-ALL.
Keywords: insulin-like growth factor 2 mRNA-binding protein 3 (IGF2BP3), mRNA, pediatric B-cell acute lymphoblastic leukemia, prognosis, proliferation, protein
1. Introduction
Pediatric B-cell acute lymphoblastic leukemia (B-ALL) is the most common malignancy in childhood. Despite the significantly improved prognosis, a subgroup of patients with either a poor therapy response or high-risk features still often experience a relapse. Better diagnostic tools are needed to enhance treatment stratification and prognosis, and to avoid overtreatment and adverse long-term side-effects [1,2,3].
Insulin-like growth factor II mRNA-binding protein 3 (IGF2BP3), also known as the IGF2BP3 protein, is a 69 kDa protein that localizes mostly to the cytoplasm [4,5]. This oncofetal RNA-binding protein is a member of the IGF2BP-family, which also includes IGF2BP1 and IGF2BP2 proteins, and shares 59–73% similarity with the amino acid sequence with IGF2BP3 [6,7]. IGF2BP3 binds RNA molecules and acts as a regulator of mRNA localization and stability [7,8]. It is expressed only at a low level in most adult tissues, whereas in multiple human malignancies, it is overexpressed [7,8].
Mutations of IGF2BP3 are rare, but the expression is dysregulated at epigenetic, transcriptional, and post-transcriptional levels. At a cellular level, IGF2BP3 drives miRNA biogenesis; intercepts the cytoplasmic export of mRNA; and regulates mRNA stability, degradation, and transportation [7]. In gastrointestinal and urogenital malignancies, IGF2BP3 is highly expressed, and is associated with cell adhesion, tumor invasion, metastasis, and inferior outcomes [7,8,9]. IGF2BP3 exhibits a strong expression in lymphoid malignancies such as B cell lymphomas of a germinal center origin [10,11]. It is expressed in Reed–Sternberg cells and can be used as a supplementary diagnostic marker in Hodgkin’s lymphoma [12,13,14]. Increased expression of IGF2BP3 is associated with proliferative features in many solid tumors, mantle cell lymphoma, and chronic myeloid leukemia blast crisis [7,15,16], and promotes cell survival during ionizing radiation in B-cells [17].
Stoskus et al. [18] explored the expression of IGF2BP family members in hematopoietic tissues and ALL by using isoform-specific RT-qPCR. In healthy stem or mature hematopoietic cells, the expression of IGF2BP3 was either weak or absent in contrast to IGF2BP2. The analysis of different mature cell populations demonstrated that only CD19+ B-cells expressed detectable levels of IGF2BP3, in line with previous literature [10,18,19]. Among B-ALL, the strongest expression was evident in ETV6-RUNX1 and KMT2A-rearranged subtypes. Liao et al. [16] and Palanichamy et al. [20] showed that siRNA or CRISPR-Cas9-mediated the knockdown of IGF2BP3 reduced proliferation and increased apoptosis in several cell lines (K562, RS4;11, and NALM6).
While a growing body of data supports biological significance and prognostic utility of IGF2BP3 in different epithelial and soft tissue tumors, to date, there are only two studies that have explored its expression in lymphoid leukemias [18,20], and no studies that have assessed expression at the protein level. Hence, we investigated the expression of IGF2BP3 across hematological malignancies and in a trephine biopsy sample cohort of pediatric B-ALL and correlated its expression with cell proliferative features and patient survival.
2. Materials and Methods
2.1. Patient Cohort for Tissue Microarray and Immunohistochemistry
The formalin-fixed and paraffin-embedded bone marrow trephine biopsy samples of the pediatric B-ALL patients were collected into a tissue microarray (TMA) with 1.5 mm punches (see also [21]), and 4-micrometer TMA sections were used for immunohistochemistry. An appendix was used as a control material for the IGF2BP3 and CD19/Ki-67 immunostainings. Immunohistochemistry was performed using the Ventana Benchmark Ultra instrument. BCL6 and pSTAT5 (Y694) immunohistochemistry was performed on whole tissue sections using Ventana Benchmark Classic [22]. The following antibodies were used: IGF2BP3 (lot: 11085707, clone: 69.1, manufacturer: Dako, Santa Clara, CA, USA, id: M3626, dilution: 1:100, species: mouse monoclonal, Ig class: IgG2a, kappa), CD19 (lot: 000085227, clone: EP169, manufacturer: Cell Marque, Rocklin, CA, USA, id: 119R-18, dilution: ready-to-use, species: rabbit monoclonal, Ig class: IgG), Ki-67 (lot: F30644, clone: 30-9, manufacturer: Ventana, Tucson, AZ, USA, id: 790-4286, dilution: ready-to-use, species: rabbit monoclonal, Ig class: IgG), BCL6 (lot: 48794, clone: LN22, id: PA0204, species: mouse monoclonal, manufacturer: Leica Biosystems, Newcastle, UK, dilution: 1:50), and pSTAT5 (Y694) (lot: GR208043, clone: E208, id: ab32364, manufacturer: Abcam, Cambridge, UK, dilution: 1:50). For the IGF2BP3 and Ki-67 stainings, we used the OptiView DAB detection kit; for the CD19 stainings, the UltraView Universal Alkaline Phosphatase Red detection kit; and for BCL6 and pSTAT5 (Y694), the Ultraview Universal DAB detection kit. All of the slides were counterstained using hematoxylin. The expression of BCL6 and pSTAT5 was semiquantitatively graded as negative when antigen was expressed in under 20% of leukemic blasts, and positive when expressed in over 20%. Clinical data and the flow cytometry data (e.g., CD34 expression) were retrieved from patient hospital records gathered as described previously [21]. The flow cytometry results were graded as either negative or positive.
2.2. Image Analysis
Slides were scanned with Hamamatsu Nanozoomer XR using 40× magnification. QuPath software (version 0.2.3) [23] was used to detect cytoplasmic IGF2BP3 positivity in TMA-sections from annotated areas with leukemic cells. A pathologist manually set detection parameters and thresholds using the cytoplasmic staining of IGF2BP3 in germinal center cells as a reference, and the nuclear staining in germinal centers and proliferating epithelium as a reference for Ki-67 staining. The stain vectors and intensity thresholds for the cell and antibody detection were adjusted according to the instructions of the QuPath software in visual control. Inadequate samples were removed from the analysis. Areas with artifacts caused by compression or folding of the tissue were disregarded by setting the proper threshold values for background intensity. With the IGF2BP3 and CD19/Ki-67- double-stained slides, stain vectors were adjusted for hematoxylin, 3,3′-diaminobenzidine (DAB), and alkaline phosphatase (AP) staining using a representative region of interest. Hematoxylin-stained cells were detected using the cell detection function in the QuPath, while the nuclear DAB of Ki-67-positive cells were recognized from the CD19-positive (AP) areas. Single intensity thresholds for IGF2BP3, CD19, and Ki-67 were used to assess the proportion of positive cells.
2.3. Microarray and RNA-Sequencing Data Sets
Hemap is a microarray gene expression data set that includes 6832 cancer samples and 1304 B-ALL samples (662 pediatric and 642 adult cases) [24,25]. The RNA-sequencing data set from the PanALL study cohort includes 1988 B-ALL cases (1234 pediatric and 754 adult cases) [26]. For the survival analyses, the TARGET data set, which includes 155 cases of pediatric high-risk B-ALL patients, was retrieved along with the following clinical information: events (relapse, induction failure, death, and second malignancy), survival, age, leukocyte count, minimal residual disease (MRD) at the end of induction (EOI), and the cytogenetic subtype [27,28].
2.4. Statistical Analysis
The statistical analysis was conducted using IBM SPSS Statistics (version 26) and RStudio (version 3.6.1). The Mann–Whitney U test, Kruskal–Wallis U test, chi-squared test, Fisher’s exact, and log-rank test were used to test the significance of the differences between groups. All tests were two-sided, and p-values under 0.05 were considered statistically significant. The ComplexHeatmap package in R was used to create heatmaps [29]. Cox proportional hazards models were fitted for survival data in order to estimate the hazard of individual risk factors.
3. Results
3.1. IGF2BP3 Protein Is Widely Expressed in Pediatric B-ALL
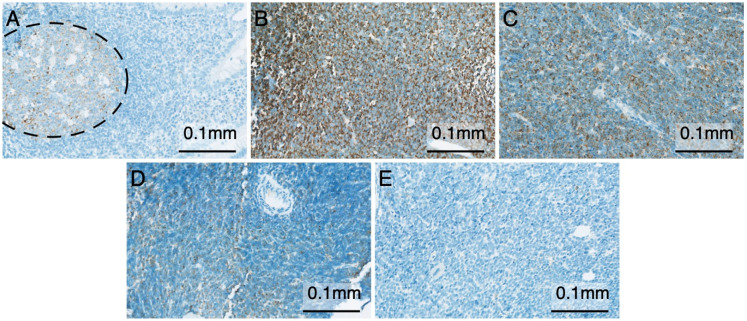
The IGF2BP3 protein has shown diagnostic and prognostic utility in different malignancies [7]. To assess the expression of the IGF2BP3 protein in B-ALL, we employed a population-based pediatric cohort of 83 B-ALL cases, and immunostained the diagnostic bone marrow trephine biopsies embedded in a tissue microarray (TMA) with an antibody against IGF2BP3. The case summary for the TMA samples is shown in Table 1. The appendix was used as a positive control, and it was stained positively in the germinal centers of the lymphoid follicles, as expected (Figure 1A) [10]. Positivity (>1%) to IGF2BP3 was detected in 74 out of 83 patients (89%; Figure 1B–D), while the proportion of positively stained leukemia cells ranged from 1 to 100% (median 34%). IGF2BP3 exhibited a granular staining pattern and was localized mostly to the cytoplasm. Negative IGF2BP3 staining was found in 9 out of 83 B-ALL cases (Figure 1E). No expression of IGF2BP3 was found in the remission bone marrow specimens.
Table 1.
Case summary of the tissue microarray (TMA) cohort.
| Clinical Parameter | Median (IQR) |
|---|---|
| Age (years) | 4.3 (2.7–9.7) |
| WBC (x 10E9/l) | 6.3 (2.7–29.2) |
| MRD (%), EOI | 0.01 (0.00–0.14) |
| n (%) | |
| CNS disease | 5 (6.0) |
| Total | 83 |
| WHO Subtype | |
| Other | 32 (38.6) |
| BCR-ABL1 | 1 (1.2) |
| KMT2A-re | 4 (4.8) |
| ETV6-RUNX1 | 20 (24.1) |
| Hyperdiploid | 22 (26.5) |
| Hypodiploid | 1 (1.2) |
| TCF3-PBX1 | 3 (3.6) |
EOI—end of induction; IQR—interquartile range; KMT2A-re—KMT2A-rearranged; MRD—minimal residual disease; WBC—white blood cell count; WHO—World Health Organization.
Figure 1.
Immunohistochemistry of insulin-like growth factor II mRNA-binding protein 3 (IGF2BP3). (A) Appendix showing positivity (brown color) to IGF2BP3 in the germinal center (dashed circle; 200× magnification). (B) Strongly IGF2BP3-positive bone marrow trephine biopsy of a B-cell acute lymphoblastic leukemia (B-ALL) patient (200× magnification). (C) Pediatric B-ALL case with a heterogeneous pattern of IGF2BP3 expression (200× magnification). (D) Weakly IGF2BP3-positive B-ALL case with only singular positive cells visible (200× magnification). (E) IGF2BP3-negative B-ALL case (200× magnification).
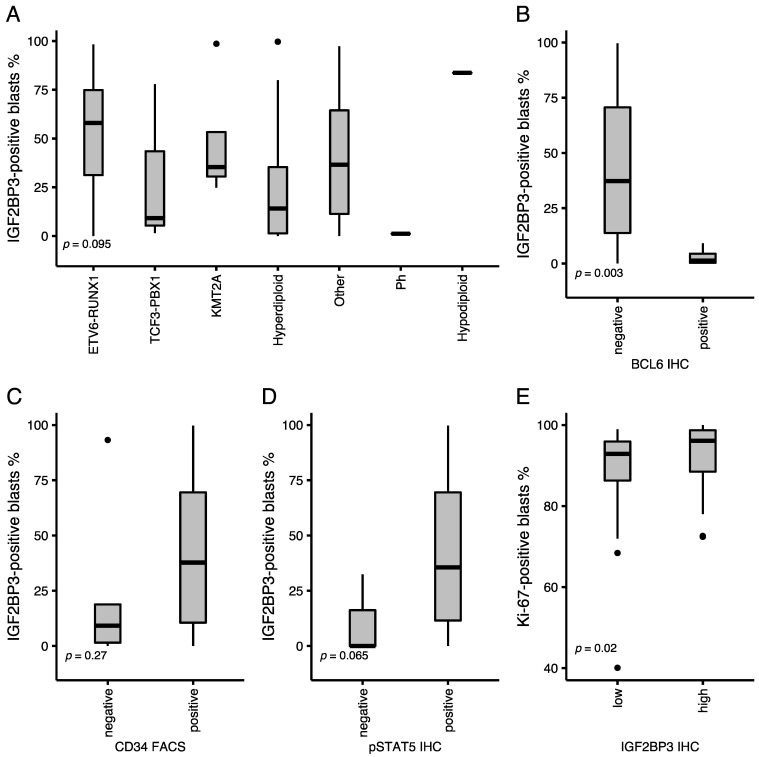
We classified the cases into distinct subtypes according to the WHO 2017 Classification of B-ALL [30]. Expression of IGF2BP3 protein was highest in the ETV6-RUNX1, “Other”, KMT2A-rearranged, and hypodiploid subtypes (Figure 2A). The difference was statistically significant between ETV6-RUNX1 and other subtypes (Mann Whitney U Test p-value = 0.04). The expression of IGF2BP3 protein did not correlate with white blood cell count (WBC), MRD at the end of induction (EOI), CNS disease, or expression of specific cell surface markers.
Figure 2.
Immunophenotype of a B-cell acute lymphoblastic leukemia (B-ALL) tissue microarray cohort. (A) Expression of IGF2BP3 according to the WHO classification of B-ALL. (B) Positivity to IGF2BP3 among cases with either a negative or positive expression of the BCL6 protein. (C) Positivity to IGF2BP3 in cases with negative or positive CD34. (D) Positivity to IGF2BP3 in cases with a negative or positive pSTAT5 (Y694). (E) Expression of Ki-67 among cases with either a low or high IGF2BP3 (median as a cut-off). Dots depict outliers. p-values of (B–E) Mann–Whitney U test and (A) Kruskal–Wallis test are shown.
IGF2BP3 is normally expressed in germinal centers, where the BCL6 protein is active and associated with germinal center-type B-cell lymphomas [10]. We recently showed that the BCL6 protein is also expressed in a fraction of precursor B-ALL [22]. BCL6-positivity, CD34-negativity, and pSTAT5-negativity have been associated with a novel pre-B-cell receptor signaling subtype of B-ALL [31]. Hence, we tested the association between IGF2BP3 and BCL6 proteins and discovered that the IGF2BP3 protein was significantly lower among the BCL6-positive cases (Mann–Whitney U test; p-value = 0.003). Likewise, the mRNA expression of the pre-BCR “metagene” (see below), which is associated with BCL6-positivity [31], exhibited a significantly lower expression among the highest 10th percentile of the IGF2BP3 expressing patients in the PanALL and Hemap data sets (Mann–Whitney U; p-value < 0.001). On the contrary, cases that exhibited phosphorylated STAT5 (pY694) protein or showed a high expression of the stem cell marker CD34 evidenced a higher-than-median level of the IGF2BP3 protein (Figure 2B–D).
3.2. Expression of Ki-67 Is Associated with High IGF2BP3 Protein Expression
A high expression of IGF2BP3 has been associated with proliferative phenotype in malignancies such as mantle cell lymphoma [7,15]. We assessed whether it is associated with cell proliferation in B-ALL by co-staining the trephine biopsy specimens with CD19, a marker of blast cells, and Ki-67, a well-established marker of cell proliferation [32,33]. Overall, the expression of Ki-67 was strong in proliferating cells of germinal centers and the epithelium of appendix (Figure S1A), and in CD19-positive cells of B-ALL samples (Figure S1B,C; proportion of positive cells, median 95%, interquartile range (IQR) 87–98%). A higher-than-median level of IGF2BP3 was significantly associated with the expression of Ki-67 (Mann–Whitney U test; p-value = 0.02; Figure 2E).
3.3. Expression of IGF2BP3 in Hematological Malignancies and B-ALL
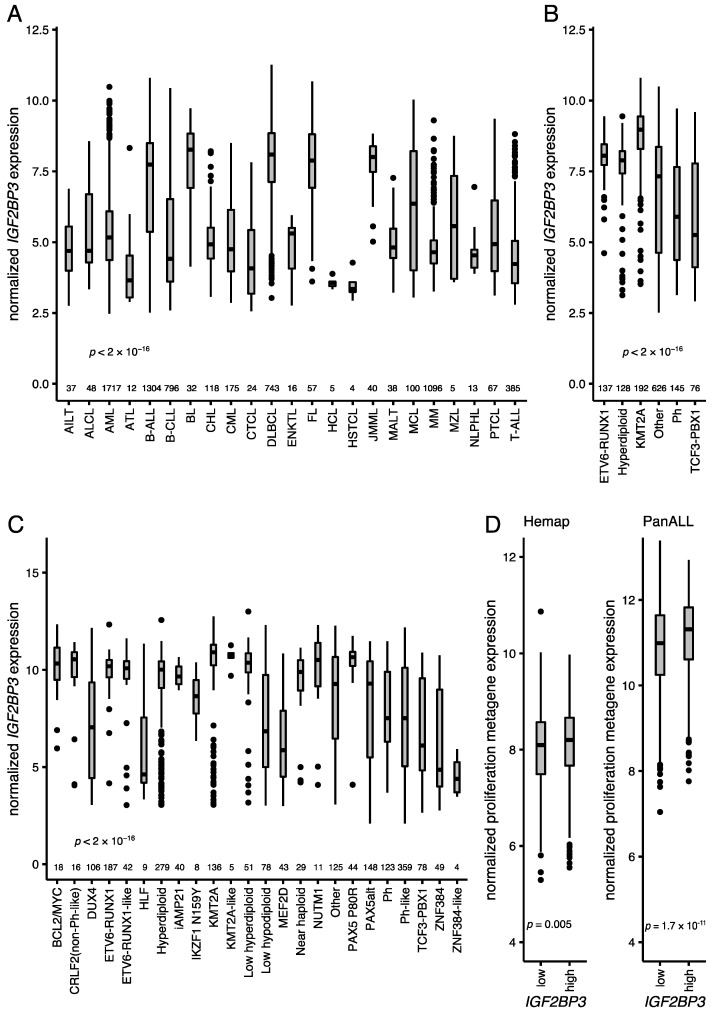
IGF2BP3 is associated with various malignancies of a B-cell origin, and particularly with germinal center lymphomas [10,11]. To get a comprehensive picture across hematological tumors, we assessed IGF2BP3 mRNA levels in 6832 hematological cancers that included 24 different disease entities [24,25]. The median expression of the IGF2BP3 mRNA was the highest in B-ALL, Burkitt lymphoma, diffuse large B-cell lymphoma, follicular lymphoma, mantle cell lymphoma, and juvenile myelomonocytic leukemia, while the lowest median expressions were observed in hairy cell leukemia, hepatosplenic T-cell lymphoma, and adult T-cell leukemia (Figure 3A). The IGFBP3 mRNA was present in all subtypes of B-ALL, with the highest expression in the KMT2A-rearranged and ETV6-RUNX1 subtypes and the lowest in the TCF3-PBX1 and BCR-ABL1 subtypes (Figure 3B). Analysis of the PanALL data set [26], which comprises 1988 B-ALL cases, validated the findings, and also revealed a strong expression in novel subtypes such as NUTM1-rearranged, PAX5-altered, ETV6-RUNX1-like, BCL2/MYC, and CRLF2 (Figure 3C).
Figure 3.
Expression of IGF2BP3 across different hematological malignancies and subtypes of B-ALL. (A) IGF2BP3 expression in the Hemap data set in different hematological malignancies (n = 6832) [24,25]. (B) IGF2BP3 expression in different cytogenetic subtypes of B-ALL (n = 1304) in the Hemap data set [24,25]. (C) IGF2BP3 expression in different B-ALL subtypes of B-ALL in the PanALL study cohort (n = 1988) [26]. (D) Proliferation-associated “metagene” [33] expression in B-ALL in the Hemap and PanALL data sets (median as a cut-off for the IGF2BP3 expression groups). AILT—angioimmunoblastic T-cell lymphoma; ALCL—anaplastic large cell lymphoma; AML—acute myeloid leukemia; ATL—adult T-cell leukemia; B-ALL—B-cell lineage acute lymphoblastic leukemia; B-CLL—B-cell chronic lymphocytic leukemia; BCL2/MYC—BCL2/MYC-rearranged; BL—Burkitt lymphoma; CHL—classic Hodgkin lymphoma; CML—chronic myeloid leukemia; CRLF2—CRLF2 (non-Ph-like); CTCL—cutaneous T-cell lymphoma; DLBCL—diffuse large B-cell lymphoma; DUX4—DUX4-rearranged; ENKTL—extranodal NK/T-cell lymphoma; FL—follicular lymphoma; HCL—hairy cell leukemia; HLF—TCF3/TCF4-HLF; HSTCL—hepatosplenic T-cell lymphoma; iAMP21—intrachromosomal amplification of chromosome 21; IKZF1 N159Y—IKZF1 missense alteration encoding p.Asn159Tyr; JMML—juvenile myelomonocytic leukemia; KMT2A—KMT2A-rearranged; MALT—extranodal marginal zone lymphoma of mucosa-associated lymphoid tissue; MCL—mantle cell lymphoma; MEF2D—MEF2D-rearranged; MM—multiple myeloma; MZL—marginal zone lymphoma; n—number of cases; NLPHL—nodular lymphocyte predominant Hodgkin lymphoma; NUTM1—NUTM1-rearranged; PAX5alt—PAX5 alterations; PAX5 P80R—PAX5 p.Pro80Arg (P80R) alteration; Ph—Philadelphia chromosome (BCR-ABL1); PTCL—peripheral T-cell lymphoma, not otherwise specified; T-ALL—T-cell lineage acute lymphoblastic leukemia; ZNF384—ZNF384-rearranged. Dots depict outliers. p-values of (D) Mann–Whitney U test and (A–C) Kruskal–Wallis test are shown.
3.4. Proliferative “Metagene” Signature in B-ALL
Recently, Giuliano et al. (2018) [33] described a proliferative “metagene” (MKI67, PCNA, CCNB1, MCM2, and TOP2A) that is correlated with cell proliferation. Supporting our earlier observations, the IGF2BP3 mRNA was significantly associated with the proliferative “metagene” when assessed across all hematological malignancies (Mann–Whitney U test; p-value < 0.001). When the analysis was restricted to the B-ALL cases, the “metagene” signature and the MKI67 mRNA showed elevated levels and were significantly associated with a higher-than-median expression of IGF2BP3 (Mann–Whitney U test; p-value = 0.005 and p-value = 0.04, respectively; Figure 3D). A similar analysis in the PanALL data set replicated the findings: the IGF2BP3 mRNA was significantly associated with the high MKI67 mRNA (Mann–Whitney U test, p-value < 0.001) and the proliferation-associated “metagene” with a discretized expression of IGF2BP3 (Mann–Whitney U test, p-value < 0.001; Figure 3D; see also heatmap in Figure S2A–C).
CDK6 and MYC oncoproteins have been reported as targets of the IGF2BP3 protein [20]. In the PanALL data set, CDK6 was higher and MYC was lower among cases with a higher-than-median IGF2BP3 mRNA (Mann–Whitney U test p-value < 0.001, Figure S3A,B). In the Hemap data set, CDK6 was strongly expressed among cases with a higher-than-median IGF2BP3 mRNA, whereas the expression of MYC did not differ (Figure S3C,D).
3.5. High IGF2BP3 mRNA Associates with Favorable Survival in High-Risk B-ALL
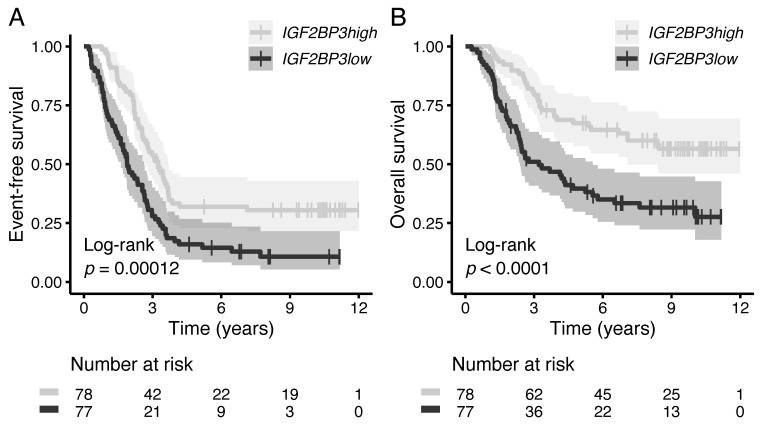
The prognostic value of IGF2BP3 mRNA was evaluated in the TARGET data set that included high-risk pediatric B-ALL cases [27,28]. Higher-than-median IGF2BP3 mRNA showed a statistically significant association with favorable event-free (EFS) and overall survival (OS; Figure 4A,B). In a multivariate analysis that included age, white blood cell count (WBC), and minimal residual disease (MRD) at the EOI as covariates, higher-than-median IGF2BP3 mRNA exhibited a decreased hazard ratio for events (HR 0.46, 95% CI 0.31–0.68) and death (HR 0.50, 95% CI 0.31–0.81; Table 2).
Figure 4.
Association of IGF2BP3 expression at an mRNA level on patient survival. Kaplan–Meier survival analysis for (A) event-free survival and (B) overall survival in the high-risk B-ALL TARGET cohort (n = 155) [27,28]. Statistical significance was tested using the log-rank test, while the median expression of IGF2BP3 was used as a cut-off for the two different patient groups.
Table 2.
| Event-Free Survival | |||||||
|---|---|---|---|---|---|---|---|
| Univariate | Multivariate | ||||||
| HR | 95% CI | p | HR | 95% CI | p | ||
| IGF2BP3 mRNA | ≤median | 1 | 1 | ||||
| >median | 0.49 | 0.34–0.71 | <0.001 | 0.46 | 0.31–0.68 | <0.001 | |
| Age | 1.01 | 0.97–1.05 | 0.69 | 0.97 | 0.93–1.01 | 0.19 | |
| MRD at the EOI | 1.02 | 0.98–1.07 | 0.39 | 1.01 | 0.97–1.06 | 0.58 | |
| Overall survival | |||||||
| Univariate | Multivariate | ||||||
| HR | 95% CI | p | HR | 95% CI | p | ||
| IGF2BP3 mRNA | ≤median | 1 | 1 | ||||
| >median | 0.44 | 0.28–0.68 | <0.001 | 0.5 | 0.31–0.81 | 0.006 | |
| Age | 1.06 | 1.01–1.11 | 0.01 | 1.03 | 0.98–1.08 | 0.24 | |
| WBC | 1 | 1.00–1.00 | 0.58 | 1 | 0.99–1.00 | 0.68 | |
| MRD at the EOI | 1.02 | 0.97–1.08 | 0.47 | 0.99 | 0.94–1.05 | 0.79 |
CI—confidence interval; EOI—end of induction therapy; HR—hazards ratio; MRD—minimal residual disease; WBC—white blood cell count at diagnosis.
In the population-based TMA cohort, positivity to IGF2BP3 protein did not associate with patient survival (data not shown).
4. Discussion
IGF2BP3 is an oncofetal protein that is normally expressed in fetal/embryonic tissues, but is often aberrantly re-expressed in malignant tumors. In solid tumors, its expression is associated with increased proliferation and inferior outcomes. We report here that IGF2BP3 is widely expressed in pediatric B-ALL, and shows a granular staining pattern fitting to the cytoplasmic ribonucleoprotein (RNP) complexes. Its expression is associated with proliferative features and a high level of CDK6, while a low expression confers inferior outcomes in a cohort of high-risk B-ALL patients.
Earlier gene expression studies have associated IGF2BP3 with the ETV6-RUNX1 and KMT2A-rearranged subtypes of B-ALL [18,20]. This aligns well with our results, which were based on large gene expression microarray (Hemap) and RNA-sequencing data sets (PanALL). As novel findings, we report here a strong expression of IGF2BP3 in new subtypes of B-ALL, such as ETV6-RUNX1-like, KMT2A-like, and NUTM1. Moreover, we extended analyses across the whole spectrum of hematological malignancies covering 24 diseases: B cell lymphomas were the strongest expressors, followed by juvenile myelomonocytic leukemia and mantle cell lymphoma, while most diseases of T-cell origin showed a lower expression.
At the protein level, our data are unique. We developed a population-based TMA platform that included 83 pediatric B-ALL cases. Almost 90% of the cases showed moderate or strong positivity to IGF2BP3, often in conjunction with a marker of activated JAK-STAT signaling (pSTAT5A-Y694). Fittingly, the expression of the BCL6 protein, which is not exhibited simultaneously with the activated JAK-STAT5 pathway [34], was absent among IGF2BP3-positive cases. The intracellular staining pattern of IGF2BP3 was granular, possibly referring to the localization of IGF2BP3 to cytoplasmic RNP complexes, where it exerts its function on the target mRNAs. One limitation of the immunohistochemistry results is the possibility of the cross-reactivity of the used IGF2BP3 antibody (and other commercial IGF2BP3 antibodies) with the paralogs of IGF2BP family [7,35,36].
By using the TMA platform, we performed co-stainings with a proliferative marker for Ki-67 and CD19, and showed that the IGF2BP3 protein is associated with active cellular proliferation. This is not a surprise, as B-ALL is an aggressive malignancy with a high proliferative capacity. At the mRNA level, the proliferation-associated “metagene” signature was higher in patients with a strong expression of IGF2BP3. Although the absolute differences were small, the results fit well with earlier data in mantle cell lymphoma and solid tumors, where the expression of IGF2BP3 was similarly associated with proliferation [7,15]. Likewise, a study by Palanichamy et al. [20] showed that the exogenous expression of IGF2BP3 increased the proliferation of bone marrow progenitor cells and provided them with a competitive survival advantage, and that IGF2BP3 was essential for the survival of several B-ALL cell lines. Overall, IGF2BP3 seems to play an active role in the proliferative capacity of B cell blasts, a feature that could possibly be utilized for diagnostic or therapeutic purposes.
We correlated the expression of IGF2BP3 with the patient outcome in a high-risk pediatric B-ALL cohort, in which a higher-than-median IGF2BP3 mRNA level was associated with improved survival. However, no association with outcome was evident for the IGF2BP3 protein in our population-based biopsy cohort, possibly because of its relatively small size or low number of events. A recent single cell analysis of early therapy response in B-ALL showed that patients with high proliferative features are more sensitive to induction chemotherapy, and that therapy-resistant clones are more likely among the quiescent cell populations [37]. Hence, IGF2BP3 expression is associated with cell populations that are actively dividing and more are likely to be killed by chemotherapy, which is reflected in the overall outcome of patients. We note that the prognostic effect was evident only in the high-risk cohort, and therefore in the future, outcome data need further exploration in larger population-based data sets.
5. Conclusions
In conclusion, we found that a high expression of IGF2BP3 is associated with a proliferative phenotype in pediatric B-ALL at mRNA and protein levels, and portends a favorable survival high-risk B-ALL. Our results show that the subtype-specific expression of IGF2BP3 provides diagnostic and prognostic utility in B-ALL.
Acknowledgments
We would like to thank Eini Eskola for aiding in immunohistochemistry.
Supplementary Materials
The following are available online at https://www.mdpi.com/2072-6694/13/7/1505/s1, Figure S1: Immunohistochemical co-staining of CD19 and Ki-67 in B-ALL, Figure S2: Heatmap illustration of expression of proliferation-associated genes (MKI67, PCNA, CCNB1, MCM2, and TOP2A) in cases with either a high or low expression of IGF2BP3, Figure S3: Boxplots showing expression of CDK6 and MYC in cases with either a low or high expression of IGF2BP3 in B-ALL.
Author Contributions
Conceptualization, O.L. and A.M.; data curation, A.M., A.N., J.M. and M.H.; formal analysis, A.M., A.N. and O.L.; funding acquisition, A.M., M.H. and O.L.; investigation, A.M.; methodology, A.M., T.H., T.P. and O.L.; project administration, O.L.; resources, O.L.; software, A.N.; supervision, M.V., M.H., T.P. and O.L.; validation, A.N.; visualization, A.M., L.O. and A.N.; writing—original draft, A.M. and O.L.; writing—review and editing, A.M. and O.L. All authors have read and agreed to the published version of the manuscript.
Funding
The work was supported by the Competitive State Research Financing of the Expert Responsibility at Tampere University Hospital (9X027) and by grants from the Academy of Finland (No. 277816 and No. 310106, O.L.), Sigrid Juselius Foundation (M.H. and O.L.), the Cancer Society of Finland (M.H. and O.L.), the Jane and Aatos Erkko Foundation (O.L. and M.H.), and the Väre Foundation for Paediatric Cancer Research (A.M.).
Institutional Review Board Statement
The study was approved by the Regional Ethics Committee of the Expert Responsibility area of Tampere University Hospital (R16054, R13109, and R19060B) and the National Supervisory Authority for Welfare and Health (Valvira, Dnro: 4243/06.01.03.01/2016 and V/3994112019).
Informed Consent Statement
Not applicable.
Data Availability Statement
The results published here are in whole or part based upon data generated by the Therapeutically Applicable Research to Generate Effective Treatments (https://ocg.cancer.gov/programs/target) initiative, phs000463. The data used for this analysis are available at https://portal.gdc.cancer.gov/projects (accessed on 27 August 2020).
Conflicts of Interest
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Inaba H., Greaves M., Mullighan C.G. Acute lymphoblastic leukaemia. Lancet. 2013;381:1943–1955. doi: 10.1016/S0140-6736(12)62187-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hunger S.P., Mullighan C.G. Acute Lymphoblastic Leukemia in Children. N. Engl. J. Med. 2015;373:1541–1552. doi: 10.1056/NEJMra1400972. [DOI] [PubMed] [Google Scholar]
- 3.Malard F., Mohty M. Acute lymphoblastic leukaemia. Lancet. 2020;395:1146–1162. doi: 10.1016/S0140-6736(19)33018-1. [DOI] [PubMed] [Google Scholar]
- 4.Mueller-Pillasch F., Pohl B., Wilda M., Lacher U., Beil M., Wallrapp C., Hameister H., Knöchel W., Adler G., Gress T.M. Expression of the highly conserved RNA binding protein KOC in embryogenesis. Mech. Dev. 1999;88:95–99. doi: 10.1016/S0925-4773(99)00160-4. [DOI] [PubMed] [Google Scholar]
- 5.Monk D., Bentley L., Beechey C., Hitchins M., Peters J., Preece M.A., Stanier P., Moore G.E. Characterisation of the growth regulating gene IMP3, a candidate for Silver-Russell syndrome. J. Med. Genet. 2002;39:575–581. doi: 10.1136/jmg.39.8.575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nielsen J., Christiansen J., Lykke-Andersen J., Johnsen A.H., Wewer U.M., Nielsen F.C. A Family of Insulin-Like Growth Factor II mRNA-Binding Proteins Represses Translation in Late Development. Mol. Cell. Biol. 1999;19:1262–1270. doi: 10.1128/MCB.19.2.1262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mancarella C., Scotlandi K. IGF2BP3 From Physiology to Cancer: Novel Discoveries, Unsolved Issues, and Future Perspectives. Front. Cell Dev. Biol. 2020;7:1–17. doi: 10.3389/fcell.2019.00363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Burdelski C., Jakani-Karimi N., Jacobsen F., Möller-Koop C., Minner S., Simon R., Sauter G., Steurer S., Clauditz T.S., Wilczak W. IMP3 overexpression occurs in various important cancer types and is linked to aggressive tumor features: A tissue microarray study on 8,877 human cancers and normal tissues. Oncol. Rep. 2018;39:3–12. doi: 10.3892/or.2017.6072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen L., Xie Y., Li X., Gu L., Gao Y., Tang L., Chen J., Zhang X. Prognostic value of high IMP3 expression in solid tumors: A meta-analysis. Onco. Targets. Ther. 2017;10:2849–2863. doi: 10.2147/OTT.S128810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.King R.L., Pasha T., Roullet M.R., Zhang P.J., Bagg A. IMP-3 is differentially expressed in normal and neoplastic lymphoid tissue. Hum. Pathol. 2009;40:1699–1705. doi: 10.1016/j.humpath.2009.05.003. [DOI] [PubMed] [Google Scholar]
- 11.Findeis-Hosey J.J., Xu H. The use of insulin like-growth factor II messenger RNA binding protein-3 in diagnostic pathology. Hum. Pathol. 2011;42:303–314. doi: 10.1016/j.humpath.2010.06.003. [DOI] [PubMed] [Google Scholar]
- 12.Tang H., Wei Q., Ge J., Jian W., Liu J., Zhong L., Fu B., Zhao T. IMP3 as a supplemental diagnostic marker for Hodgkin lymphoma. Hum. Pathol. 2013;44:2167–2172. doi: 10.1016/j.humpath.2013.04.011. [DOI] [PubMed] [Google Scholar]
- 13.Sennekamp J., Seelig H.P. Anti-cytoplasmic autoantibodies in Hodgkin’s lymphoma. Clin. Lab. 2016;62:1579–1584. doi: 10.7754/Clin.Lab.2016.160116. [DOI] [PubMed] [Google Scholar]
- 14.Masoud R., Ibrahiem A., Tantawy D., Eldosoky I. The complementary role of insulin-like growth factor II mRNA-binding protein 3 (IMP3) in diagnosis of Hodgkin’s lymphoma. Ann. Diagn. Pathol. 2019;42:64–68. doi: 10.1016/j.anndiagpath.2019.06.006. [DOI] [PubMed] [Google Scholar]
- 15.Hartmann E.M., Be S., Navarro A., Trapp V., Campo E., Ott G., Rosenwald A. Increased tumor cell proliferation in mantle cell lymphoma is associated with elevated insulin-like growth factor 2 mRNA-binding protein 3 expression. Mod. Pathol. 2012;25:1227–1235. doi: 10.1038/modpathol.2012.84. [DOI] [PubMed] [Google Scholar]
- 16.Liao B., Hu Y., Herrick D.J., Brewer G. The RNA-binding protein IMP-3 is a translational activator of insulin-like growth factor II leader-3 mRNA during proliferation of human K562 leukemia cells. J. Biol. Chem. 2005;280:18517–18524. doi: 10.1074/jbc.M500270200. [DOI] [PubMed] [Google Scholar]
- 17.Liao B., Hu Y., Brewer G. RNA-binding protein insulin-like growth factor mRNA-binding protein 3 (IMP-3) promotes cell survival via insulin-like growth factor II signaling after ionizing radiation. J. Biol. Chem. 2011;286:31145–31152. doi: 10.1074/jbc.M111.263913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Stoskus M., Gineikiene E., Valceckiene V., Valatkaite B., Pileckyte R., Griskevicius L. Identification of characteristic IGF2BP expression patterns in distinct B-ALL entities. Blood Cells Mol. Dis. 2011;46:321–326. doi: 10.1016/j.bcmd.2011.02.005. [DOI] [PubMed] [Google Scholar]
- 19.Natkunam Y., Vainer G., Chen J., Zhao S., Marinelli R.J., Hammer A.S., Hamilton-Dutoit S., Pikarsky E., Amir G., Levy R., et al. Expression of the RNA-binding protein VICKZ in normal hematopoietic tissues and neoplasms. Haematologica. 2007;92:176–183. doi: 10.3324/haematol.10724. [DOI] [PubMed] [Google Scholar]
- 20.Palanichamy J.K., Tran T.M., Howard J.M., Contreras J.R., Fernando T.R., Sterne-Weiler T., Katzman S., Toloue M., Yan W., Basso G., et al. RNA-binding protein IGF2BP3 targeting of oncogenic transcripts promotes hematopoietic progenitor proliferation. J. Clin. Investig. 2016;126:1495–1511. doi: 10.1172/JCI80046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Grönroos T., Mäkinen A., Laukkanen S., Mehtonen J., Nikkilä A., Oksa L., Rounioja S., Marincevic-Zuniga Y., Nordlund J., Pohjolainen V., et al. Clinicopathological features and prognostic value of SOX11 in childhood acute lymphoblastic leukemia. Sci. Rep. 2020;10:2043. doi: 10.1038/s41598-020-58970-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mäkinen A., Nikkilä A., Mehtonen J., Teppo S., Oksa L., Nordlund J., Rounioja S., Pohjolainen V., Laukkanen S., Heinäniemi M., et al. Expression of BCL6 in paediatric B-cell acute lymphoblastic leukaemia and association with prognosis. Pathology. 2021 doi: 10.1016/j.pathol.2021.02.013. in press. [DOI] [PubMed] [Google Scholar]
- 23.Bankhead P., Loughrey M.B., Fernández J.A., Dombrowski Y., McArt D.G., Dunne P.D., McQuaid S., Gray R.T., Murray L.J., Coleman H.G., et al. QuPath: Open source software for digital pathology image analysis. Sci. Rep. 2017;7:1–7. doi: 10.1038/s41598-017-17204-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pölönen P., Mehtonen J., Lin J., Liuksiala T., Häyrynen S., Teppo S., Mäkinen A., Kumar A., Malani D., Pohjolainen V., et al. HEMap: An interactive online resource for characterizing molecular phenotypes across hematologic malignancies. Cancer Res. 2019;79:2466–2479. doi: 10.1158/0008-5472.CAN-18-2970. [DOI] [PubMed] [Google Scholar]
- 25.Mehtonen J., Pölönen P., Häyrynen S., Dufva O., Lin J., Liuksiala T., Granberg K., Lohi O., Hautamäki V., Nykter M., et al. Data-driven characterization of molecular phenotypes across heterogeneous sample collections. Nucleic Acids Res. 2019;47:76. doi: 10.1093/nar/gkz281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gu Z., Churchman M.L., Roberts K.G., Moore I., Zhou X., Nakitandwe J., Hagiwara K., Pelletier S., Gingras S., Berns H., et al. PAX5-driven subtypes of B-progenitor acute lymphoblastic leukemia. Nat. Genet. 2019;51:296–307. doi: 10.1038/s41588-018-0315-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu Y., Easton J., Shao Y., Maciaszek J., Wang Z., Wilkinson M.R., McCastlain K., Edmonson M., Pounds S.B., Shi L., et al. The genomic landscape of pediatric and young adult T-lineage acute lymphoblastic leukemia. Nat. Genet. 2017;49:1211–1218. doi: 10.1038/ng.3909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Roberts K.G., Li Y., Payne-Turner D., Harvey R.C., Yang Y.-L., Pei D., McCastlain K., Ding L., Lu C., Song G., et al. Targetable Kinase-Activating Lesions in Ph-like Acute Lymphoblastic Leukemia. N. Engl. J. Med. 2014;371:1005–1015. doi: 10.1056/NEJMoa1403088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gu Z., Eils R., Schlesner M. Complex heatmaps reveal patterns and correlations in multidimensional genomic data. Bioinformatics. 2016;32:2847–2849. doi: 10.1093/bioinformatics/btw313. [DOI] [PubMed] [Google Scholar]
- 30.Swerdlow S.H., Campo E., Harris N.L., Jaffe E.S., Pileri S.A., Stein H., Thiele J. WHO Classification of Tumours of Haematopoietic and Lymphoid Tissues. 4th ed. International Agency for Research on Cancer (IARC); Lyon, France: 2017. [Google Scholar]
- 31.Geng H., Hurtz C., Lenz K.B., Chen Z., Baumjohann D., Thompson S., Goloviznina N.A., Chen W.Y., Huan J., LaTocha D., et al. Self-Enforcing Feedback Activation between BCL6 and Pre-B Cell Receptor Signaling Defines a Distinct Subtype of Acute Lymphoblastic Leukemia. Cancer Cell. 2015;27:409–425. doi: 10.1016/j.ccell.2015.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Whitfield M.L., George L.K., Grant G.D., Perou C.M. Common markers of proliferation. Nat. Rev. Cancer. 2006;6:99–106. doi: 10.1038/nrc1802. [DOI] [PubMed] [Google Scholar]
- 33.Giuliano C.J., Lin A., Smith J.C., Palladino A.C., Sheltzer J.M. MELK expression correlates with tumor mitotic activity but is not required for cancer growth. eLife. 2018;7 doi: 10.7554/eLife.32838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chan L.N., Murakami M.A., Robinson M.E., Caeser R., Sadras T., Lee J., Cosgun K.N., Kume K., Khairnar V., Xiao G., et al. Signalling input from divergent pathways subverts B cell transformation. Nature. 2020;583:845–851. doi: 10.1038/s41586-020-2513-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lederer M., Bley N., Schleifer C., Hüttelmaier S. The role of the oncofetal IGF2 mRNA-binding protein 3 (IGF2BP3) in cancer. Semin. Cancer Biol. 2014;29:3–12. doi: 10.1016/j.semcancer.2014.07.006. [DOI] [PubMed] [Google Scholar]
- 36.Tschirdewahn S., Panic A., Püllen L., Harke N.N., Hadaschik B., Riesz P., Horváth A., Szalontai J., Nyirády P., Baba H.A., et al. Circulating and tissue IMP3 levels are correlated with poor survival in renal cell carcinoma. Int. J. Cancer. 2019;145:531–539. doi: 10.1002/ijc.32124. [DOI] [PubMed] [Google Scholar]
- 37.Mehtonen J., Teppo S., Lahnalampi M., Kokko A., Kaukonen R., Oksa L., Bouvy-Liivrand M., Malyukova A., Mäkinen A., Laukkanen S., et al. Single cell characterization of B-lymphoid differentiation and leukemic cell states during chemotherapy in ETV6-RUNX1-positive pediatric leukemia identifies drug-targetable transcription factor activities. Genome Med. 2020;12:99. doi: 10.1186/s13073-020-00799-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The results published here are in whole or part based upon data generated by the Therapeutically Applicable Research to Generate Effective Treatments (https://ocg.cancer.gov/programs/target) initiative, phs000463. The data used for this analysis are available at https://portal.gdc.cancer.gov/projects (accessed on 27 August 2020).