Abstract
The rabbit has been recognized as a valuable model in various biomedical and biological research fields because of its intermediate size and phylogenetic proximity to primates. However, the technology for precise genome manipulations in rabbit has been stalled for decades, severely limiting its applications in biomedical research. Novel genome editing technologies, especially CRISPR/Cas9, have remarkably enhanced precise genome manipulation in rabbits, and shown their superiority and promise for generating rabbit models of human genetic diseases. In this review, we summarize the brief history of transgenic rabbit technology and the development of novel genome editing technologies in rabbits.
Keywords: rabbit, transgenic animal, gene targeting, genome editing, base editing
Introduction
Rabbit (Oryctolagus cuniculus) is a classic research animal model and is increasingly becoming a translational model of choice, serving to bridge the gap between rodent models and larger animal models[1–5]. Similar to rodents, the rabbit has several advantages to be a classic research animal model, such as relatively inexpensive housing, facile breeding, short gestation (28–32 days for New Zealand white rabbit, NZW) and large litter size (4–10 kits per litter for NZW). Additionally, rabbits share characteristics with larger animals that are phylogenetically, physiologically, and anatomically closer to humans[6–7]. As early as the nineteenth century, rabbits were used to develop the rabies vaccine by Louis Pasteur[8] and also played a vital role in the discovery of Mycobacterium tuberculosis[9]. The rabbit model has provided tremendous breakthroughs and insights into understanding the molecular and cellular mechanisms of atherosclerosis, including the discoveries of low-density lipoprotein receptor deficiency as a cause for human familial hypercholesterolemia[10] and statin, the most potent lipid-lowering drug[10-11] which is prescribed annually for more than 30 million hyperlipidemic patients worldwide.
Both embryo transfer and in vitro fertilization, two of the basic techniques for transgenic animal technology, were developed first in rabbits[12–13]. A transgenic animal carries an exogenous DNA cassette, and the foreign DNA cassette that could be transmitted through the germ line. Transgenic animals represent unique models that are custom tailored to address specific biological questions. They therefore provide a very powerful tool for dissecting complex biological processes and systems and identifying specific gene functions. Since the first transgenic rabbit produced by Hammer and his colleagues[14], transgenic rabbit models have been generated by different laboratories worldwide for different research purposes: (a) human disease models, like infectious disease[4], cardiovascular disease[3,5], ophthalmic disease[15], respiratory disease[16], and nervous system disease[17]; (b) production of human proteins as mammary gland bioreactors[18]; (c) toxicity tests[17] and (d) antibody production[18]. Most of these models were produced by microinjection of DNA into fertilized eggs which results in random integrations of the exogenous DNA into the rabbit genome. This can lead to multi-copy integration, unpredictable expression of the transgene and inadvertent disruption of neighboring genes or the gene at the insertion site[19]. To solve this problem, gene targeting technology was developed in mice, in which foreign DNA could be targeted and integrated into a specific locus by homologous recombination (HR) in pluripotent stem cells (PSCs) followed by chimera production. Gene targeting allowed precise manipulation of the genome to avoid the problems caused by random insertion. More importantly, it allowed scientists to study human genetic diseases by removing or adding, specific mutations in the animal genome in order to establish the roles of individual genes in healthy and diseased animals. In large animals such as pigs, sheep and cattle, in which the germline competent PSCs are not available, gene targeting could be achieved by HR in cultured somatic cells, followed by somatic cell nuclear transfer (SCNT) (Fig. 1). However, due to lack of germline competent PSC lines and the extreme difficulty of SCNT in rabbits, the gene targeting technology in rabbits lagged behind that in rodents as well as in pigs.
Figure 1.
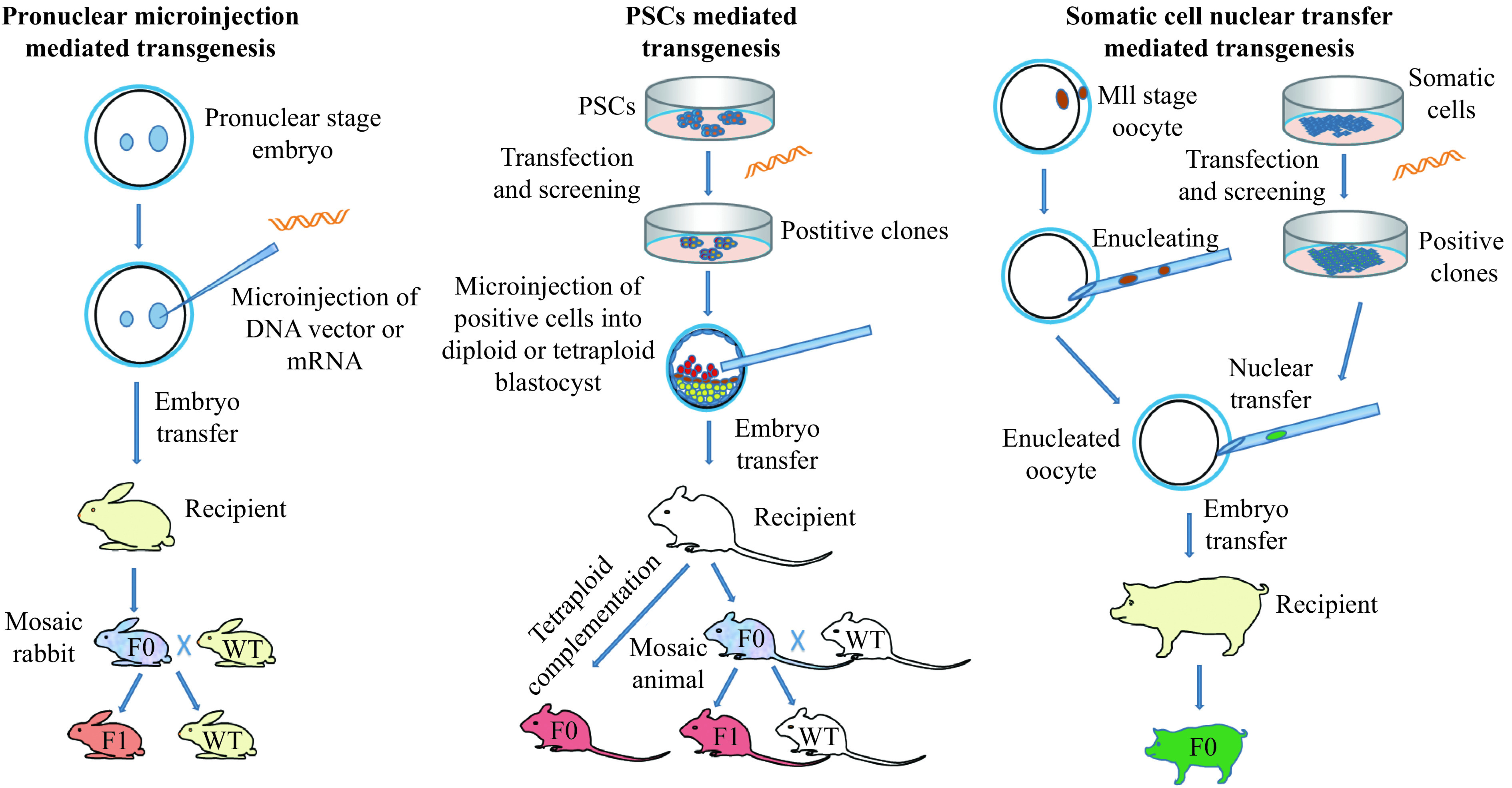
Three major approaches to generate transgenic animal models.
While the first success in rabbit pronuclear microinjection was achieved within 5 years from the first in mouse, it took 24 more years to achieve gene targeting in rabbits after it was first reported in mouse (Fig. 2). Due to the technical lagging, the development and application of genetically engineered rabbit models are much limited. Recently, with the advent of gene editing nucleases, including, zinc finger nuclease (ZFN), transcription activator-like effector nuclease (TALEN), and clustered regularly interspaced short palindromic repeats/CRISPR-associated protein-9 (CRISPR/Cas9), it is now possible to modify the rabbit genome precisely and efficiently. In this review, we summarize the brief history of transgenic rabbit technology and the progress of engineered endonuclease-mediated precise genome editing in rabbits.
Figure 2.
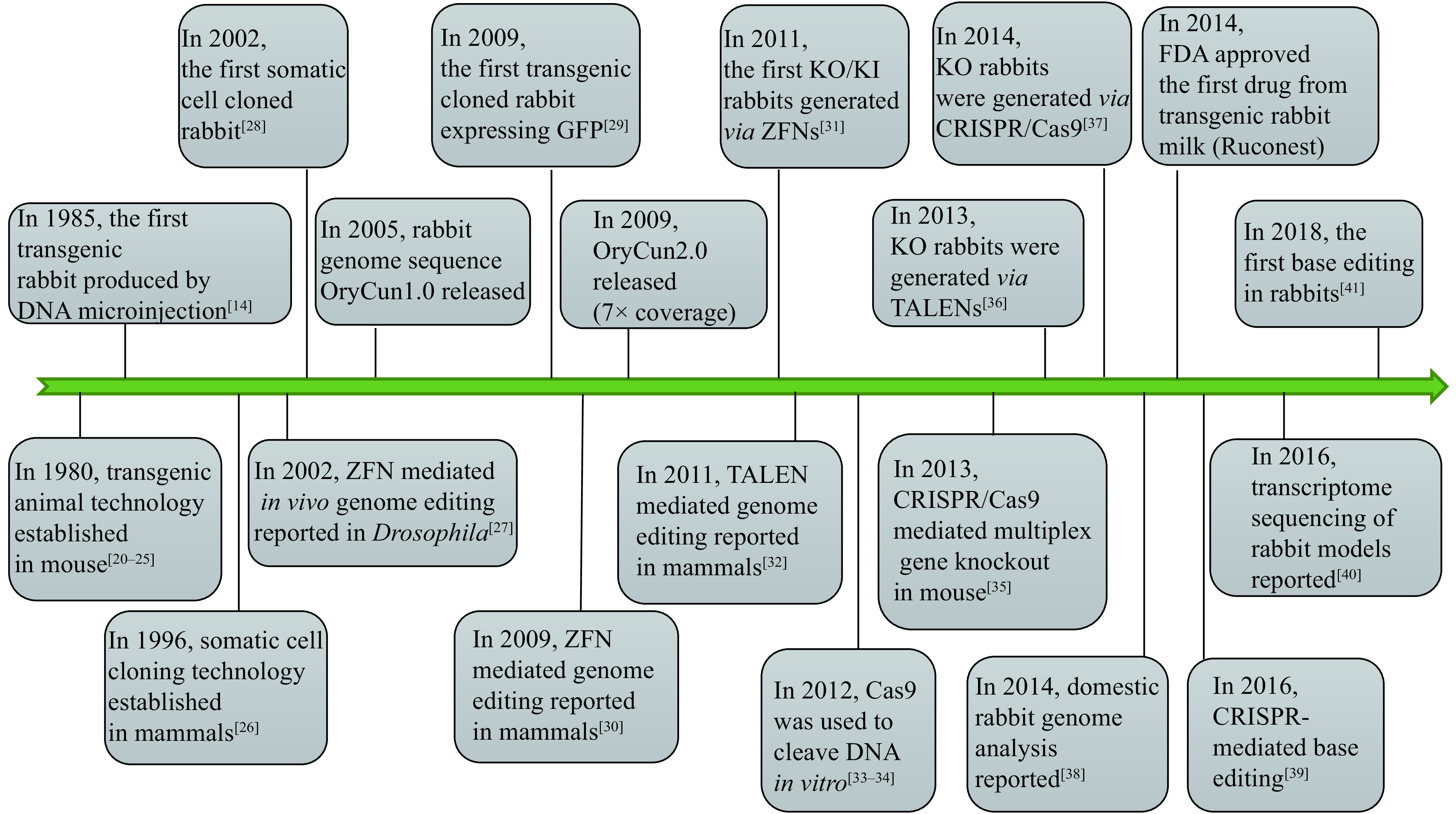
Emerging key technologies for genomic engineering and their first application in rabbits.
ZFNs: zinc finger nucleases; TALENs: transcription activator-like effector nucleases; CRISPR/Cas9: clustered regularly interspaced short palindromic repeats/CRISPR-associated protein-9.
Development of traditional methods of transgenic animal production in rabbits
There are mainly three traditional methods for generation of transgenic animals, including DNA microinjection-mediated transgenesis, PSCs-mediated transgenesis and SCNT-mediated transgenesis (Fig. 1). Other methods of transgenic animal production like sperm-mediated gene transfer[42], and viral-mediated gene transfer[43], have also been reported, but these methods have considerable problems with reproducibility and transgene rearrangement as well as low efficiency[43–45].
DNA microinjection mediated transgenesis in rabbits
In the early 1980s, the technology for generation of transgenic mice was reported by several groups[21–25], which described the direct microinjection of the DNA fragment of interest into the zygotic pronuclei. It was the first effective technique to produce a transgenic mammal, subsequently widely applied to generate transgenic animals in a wide variety of species in the following decades. In 1985, the first transgenic rabbit was produced by Robet E. Hammer and colleagues[14]. They injected the pronuclear stage embryos with a fusion gene consisting of the human growth hormone driven by the mouse metallothionein-I promoter (MT-hGH). The efficiency of MT-hGH integration into the rabbit genome reached 12% and the foreign gene was inherited through germline transmission. Afterwards, due to the advantage of being straightforward and reliable, the DNA microinjection method has been used to establish numerous transgenic rabbit models for research in cardiovascular disease, infectious disease, bioreactors and so on[2,18], and the method remained to be the most reliable and commonest means of producing transgenic rabbits until recently. Despite numerous attempts to improve the efficiency of microinjection-mediated transgenic animal production[46], none of these efforts could solve one of the most prominent disadvantages of the pronuclear injection mediated transgenesis, that is, random integration. Recently, with the development of transposon mediated gene transfer technology, pronuclear microinjection of Sleeping Beauty transposons achieved promising results in rabbits, with single copy, stable and reproducible gene expression, though the integration site is still uncontrollable[47–48].
PSCs-mediated gene transfer
Gene targeting is the process of creating designed genomic modifications at a specific sequence in a genome. Such modifications could be deletion, insertion or replacement of endogenous sequence. PSCs-mediated gene targeting in mice has revolutionized the study of developmental biology and human medicine. The PSCs can be derived from preimplantation embryos (embryonic stem cells, ES cells) or reprogrammed from differentiated cells (induced pluripotent stem cells, iPS cells). The PSCs possess robust proliferative ability and pluripotency which makes the PSCs bear capability to differentiate into various cell types, including germ cells. To precisely modify the endogenous gene or deliver a foreign gene fragment into the target site, the PSCs in culture were transfected to introduce the desired modification via HR. After screening, the PSCs with the desired modifications could be injected into a blastocyst stage embryo, and the gene modified PSCs will differentiate and incorporate into the host germline. Then, following breeding could thus generate the gene targeted animal model (Fig. 1). In 1981, two groups of scientists successfully isolated and cultured ES cells from the mouse[49–50]. These cells were shown to be able to differentiate into multiple cell types including germline and could generate transgenic offspring when the genetically altered ES cells were injected into mouse blastocysts[51–52]. Since HR events in ES cells could be screened and enriched in a petri dish, ES cells have subsequently been widely used as vehicles to produce gene-targeted mouse models[53]. More recently, with the development of iPS technology[54–55], live pups were generated using pluripotent iPS cells through tetraploid complementation in different independent laboratories[56–58], indicating iPS cells could also be used for generating gene targeted animal models. Using PSCs-mediated gene targeting, genetically modified mice have been produced to perform gain- and loss-of-function genetic studies, to follow gene expression or determine cell lineage, and/or to control the effects of mutations spatially and temporally[59]. This versatile technology has rendered the mouse a powerful and indispensable experimental model in fundamental basic and medical research and thus a dominant animal species in the biomedical sciences.
Unfortunately, PSCs technology in rabbits has lagged behind considerably compared to that in mice. Although a number of groups[60–66] have reported the successful establishment of rabbit ES cells and iPS cells, which exhibit the characteristic features of primed pluripotency similar to those of human pluripotent stem cells, and some cell lines can be passaged for at least 50 times or more than 1 year, almost all of the established cell lines have low or no chimeric competency, and no germline transmission has been reported yet. Interested readers can refer to a comprehensive review of rabbit PSCs[67].
SCNT-mediated transgenesis in rabbits
Since the first cloned sheep, Dolly, was reported in 1996[26], the technique of SCNT combined with somatic cell gene modification provided an alternative for gene targeting in mammals, especially in large animal species that do not have germline competent PCS lines available[68–70]. In this approach, the genetic manipulation can be performed in cultured somatic cells, and then single cell clones with the expected modification are screened and transferred into enucleated oocytes. The reconstituted embryos are then transferred into the oviducts of surrogate females for animal production (Fig. 1). All cloned offspring would have the same genotype as the donor cell.
The rabbit is one of the earliest mammals used for nuclear transfer studies. In 1988, the first cloned rabbit generated with embryonic blastomere nuclear transfer was reported[71]. In 2002, the first somatic cell cloned rabbit was successfully produced by Renard's group in France using freshly collected cumulus cells as nuclear donor rather than cultured somatic cells[28]. It turned out that the SCNT in rabbits is extremely difficult with high miscarriage rates and perinatal mortality[72]. In 2006, Li et al produced cloned rabbits from cultured adult fibroblasts[73], which enabled genetic manipulation and screening before nuclear transfer. After that, transgenic rabbits expressing green fluorescent protein were cloned using in vitro transfected adult fibroblasts[29] and mesenchymal stem cells[66,74]. In 2015, the first SCNT-mediated gene knockout (KO) rabbit was finally reported[75] using recombinant adeno-associated virus-mediated HR and SCNT. In the study, 18 cloned kits were born from 38 recipients with 784 embryos transferred, and only 5 cloned kits were alive at weaning. In addition, the authors noted that delivery of the HR vectors to rabbit fibroblasts is the major barrier, and no targeted cellular clones were identified using electroporation- and liposome-mediated DNA delivery. The limited somatic cell proliferation and low frequency of HR in adult fibroblasts and the extremely low efficiency of SCNT in rabbits limited the application of SCNT in rabbit gene targeting.
Engineered nucleases mediated genome editing in rabbits
Recently ZFN, TALEN, and CRISPR/Cas9 have emerged as powerful programmable tools for genome editing[76–77]. These engineered endonucleases are efficient in generating double-strand breaks (DSB) in the specific genomic loci that can be repaired either by error-prone, non-homologous end joining (NHEJ) leading to frameshifts which may result in a functional KO of the target gene or by homology directed repair (HDR) to integrate a mutation or a DNA insert at a specific locus. Thanks to the very high rates of targeted DSB, gene targeted animals can be now readily derived by direct injection of the engineered endonucleases together with or without donor DNA into pronuclear stage embryos, bypassing the need for germline competent PSCs or highly efficient SCNT[37,78].
Zinc-finger nucleases
The prototype enzymes for demonstrating DSB stimulation of gene targeting were meganucleases, I-SceI and HO, both of which recognize long DNA target (18 bp for I-SceI, 24 bp for HO)[79]. While they provided very useful information on the efficiency and mechanisms of DSB repair, they were limited in their utility because it is difficult to alter their recognition sequences, rendering them non-programmable. ZFNs were the first generation of the programmable genome editing nucleases. Kim et al, in 1996[80], first designed and cleaved target DNA in vitro using a ZFN pair that consisted of two different zinc finger proteins (DNA-binding domains) each linked with a Fok I endonuclease domain. Zinc finger domains can be engineered to target specific DNA sequences and this enables ZFNs to target unique sequences within complex genomes. During the process of DNA-protein recognition, zinc finger proteins recognize the 18 bp target sequence through 6 tandem fingers, each of which primarily recognizes 3 bp of DNA. Once both zinc finger proteins bind to target sites that lie in inverted orientation, the two Fok I endonuclease domains form a functional dimer to cleave the target DNA sequence. Then, the DNA repair pathway is subsequently triggered and the mutation is generated.
ZFNs were first applied to in vivo genome editing in the fruit fly[27], and later widely used to modify target genes in other organisms[30,81–84]. In rabbit, ZFNs were first engineered to KO the immunoglobin gene through pronuclear stage embryo microinjection with ZFN mRNA[31]. The phenotypes of immunoglobin locus KO were confirmed by serum IgM and IgG deficiency and lack of IgM+ and IgG+ B lymphocytes. The study also found that targeted sequence replacement (knock-in, KI) occurred in the target site with the presence of a linear DNA donor. Our lab and others also employed ZFNs to generate rabbit models for lipid metabolism[85] and atherosclerosis[86–88] studies. However, the expensive Zinc Finger library, the laborious engineering and assembly process of ZFNs and the complexity of ZF-DNA recognition restrict its practical application in genome editing.
Transcription activator-like effector nucleases
TALENs are considered an upgraded version of programmable nucleases that are similar to ZFNs but much simpler to design and assemble. The DNA binding domain in TALENs was derived from Xanthomonas spp. bacteria[89–90]. While TALENs utilize the same Fok I endonuclease domain as ZFNs, its DNA binding domain contains a repeated highly conserved 33–34 amino acid sequence with Repeat Variable Diresidue (RVD) at 12th and 13th amino acids, which recognizes specific nucleotides, for example, NN for guanine, NI for adenine, HD for cytosine and NG for thymine. This straightforward relationship between amino acid sequence and DNA recognition has allowed for easier engineering of sequence specific binding domains than with ZFNs[91–92].
Since the TALEN domains allow for a more predictable, modular creation of binding domains, they gained immediate popularity after ZFNs. To date, TALENs have already been utilized to introduce various gene modifications in a large number of organisms and cultured cell lines. In rabbits, this technique was first used to target Rag1 and Rag2 genes to generate immunodeficent rabbits by microinjection of TALENs mRNA into single-cell stage embryos[36]. In this study, extremely high efficiency of KO, in both in vitro cultured embryos and in offspring, were achieved in rabbits. The gene targeted rabbits show typical pathological phenotypes, such as absence of the thymus and consequent deficiency of T and B lymphocytes, which are consistent with those of immunodeficient patients. Like ZFNs, TALENs rely on protein-DNA recognition and thus need to be designed and assembled according to each target, which is laborious and time consuming.
CRISPR/Cas9
The CRISPR/Cas9 technology is the most recent version of the programmable genome editing nucleases, and is even more rapid and modular than the TALEN platform. Cas9 is an endonuclease enzyme playing a protective role against foreign nucleic acids in the CRISPR adaptive immune system in bacteria. The feature of bacterial CRISPR immune system is that genetic materials taken up from previous invasive elements are expressed in crRNA, which could direct the Cas9 endonuclease to cut foreign DNA elements containing the same sequence[33–34]. Therefore, to modify the sequence of the eukaryotic genome, the CRISPR/Cas9 system has been remolded from bacterial immune system to genome editing tool, using a designed guide RNA to target Cas9 nuclease to a specific DNA sequence[93]. Binding of Cas9 nuclease on a specific protospacer adjacent motif (PAM) sequence on the genome (NGG for spCas9) will unwind the adjacent sequence, allowing the RNA: DNA pairing, which activates the nuclease domains in Cas9 to cut DNA double strands. While ZFNs and TALENs rely on protein-DNA recognition, which is less predictable for design and more labor and time consuming for assembly because of the difficulties of protein construction, synthesis, purification and validation, the CRISPR/Cas9 system relies on the RNA–DNA recognition, which is much simpler and more predictable.
In 2014, our lab was first to report CRISPR/Cas9 system mediated gene targeting in rabbits[37] (Fig. 3). In this study, we found that the efficiency of KO introduced by CRISPR/Cas9 was up to 100% in rabbit embryos and 84% in rabbit kits, and no off-target effects were detected at bioinformatically predicted off-target sites in the offspring. We refer the interested readers to detailed protocols for this technology in rabbits[94]. In the years since, more than a dozen of genetically modified rabbits were reported through CRISPR/Cas9 system (Table 1), among which, human disease models are dominant, including models for hyperlipidemia[87], Marfan syndrome[96], muscular dystrophy[97–98], immunodeficiency[114–115], congenital cataracts[99–100], Wilson disease[111], tyrosinemia type 1[95] and premature aging syndrome[107]. Using CRISPR/Cas9 technology, most of the complex genetic manipulations, including point mutation, large fragment replacement, large deletion, conditional KO, etc., that were previously only achievable in rodents could now be fulfilled in rabbits. For example, two groups efficiently introduced disease causing point mutations in rabbits using CRISPR/Cas9 system combined with single-strand oligodeoxyribonucleotide (ssODNs) as donor to establish rabbit model of oculocutaneous albinism and Wilson disease, respectively[111–112]. To find a safe harbor locus in rabbit genome for the KI of transgenes and its stable expression, we identified and characterized the rabbit Rosa26 locus (rbRosa26), and achieved highly efficient KI rates and constitutive, ubiquitous foreign gene expression[118]. To improve large fragment KI efficiency, we found that an HDR enhancer, RS-1, is beneficial for TALEN and CRISPR/Cas9 mediated large fragment KI, with approximately 2 –4 fold increase at two rabbit specific loci[119]. To achieve large fragment deletion, dual guide RNAs were employed to target the rabbit genome, and up to 105 kb fragment deletion was achieved in 10% and 17.67% of rabbit embryos and live pups, respectively[113].
Figure 3.
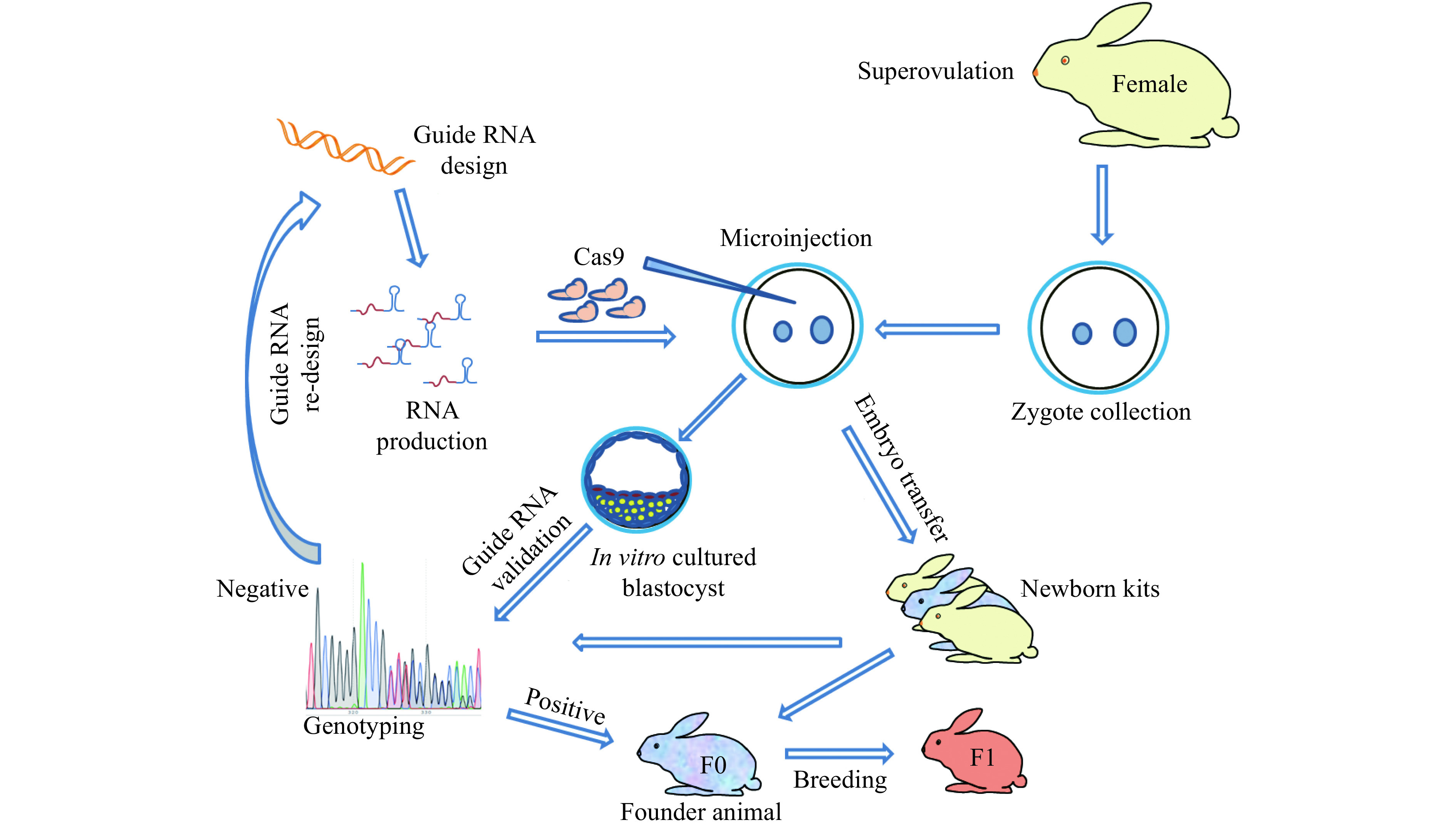
Procedure for rabbit genome editing using CRISPR/Cas9.
Table 1. Summary of genetically modified rabbits using engineered nucleases.
| Nucleases | Genes | Modification | Components | Injected site | Application | Reference |
| KO: knock-out; KI: knock-in. | ||||||
| ZFN | IGM | KO/KI | ZFN mRNA | Cytoplasm | Immunology | |
| APOCIII | KO | ZFN mRNA | Cytoplasm | Lipid metabolism and atherosclerosis | [85] | |
| APOE | KO | ZFN mRNA | Pronuclear | Lipid metabolism and atherosclerosis | [86] | |
| CETP | KO | ZFN mRNA | Cytoplasm | Lipid metabolism and atherosclerosis | [88] | |
| TALENs | RAG1 | KO | TALEN mRNA | Cytoplasm | Immunodeficiency | [36] |
| RAG2 | KO | |||||
| FAH | KO | TALEN mRNA | Cytoplasm | Hereditary tyrosinemia type 1 | [95] | |
| CRISPR/
Cas9 |
FBN1 | KO | Cas9 mRNA, sgRNA | Cytoplasm | Marfanoid-progeroid- lipodystrophy syndrome | [96] |
| DMD | KO | Cas9 mRNA, sgRNA | Cytoplasm | Duchenne muscular dystrophy | [97] | |
| ANO5 | KO | Cas9 mRNA, sgRNA | Cytoplasm | Muscular dystrophy | [98] | |
| aA-Crystallin | KO | Cas9 mRNA, dual sgRNA | Cytoplasm | Congenital cataracts | [99] | |
| GJA8 | KO | Cas9 mRNA, dual sgRNA | Cytoplasm | Congenital cataracts | [100] | |
| APOE | KO | Cas9 mRNA, sgRNA | Cytoplasm | Lipid metabolism and atherosclerosis | [37] | |
| LDLR | KO | |||||
| CD36 | KO | |||||
| RYR2 | KO | Cas9 mRNA, sgRNA | Cytoplasm | Heart arrhythmia. | ||
| LDLR | KO | Cas9 mRNA, sgRNA | Cytoplasm | Lipid metabolism and atherosclerosis | [101] | |
| MSTN | KO | Cas9 mRNA, sgRNA | Cytoplasm | Muscle hypertrophy | [102-103] | |
| SRY | KO | Cas9 mRNA, sgRNA | Cytoplasm | Sex reversal syndromes and hermaphroditism syndromes | [104-105] | |
| PHEX | KO | Cas9 mRNA, dual sgRNA | Cytoplasm | X-linked hypophosphatemia | [106] | |
| LMNA | KO | Cas9 mRNA, sgRNA | Cytoplasm | Premature aging syndrome | [107] | |
| WRN | KO | Cpf1 mRNA, sgRNA | Cytoplasm | Werner syndrome | [108] | |
| TYR | KO | Circular plasmid | Pronuclear | Coat color | [109] | |
| TYR | KO | Cas9 mRNA, dual sgRNAs | Cytoplasm | Coat color | [110] | |
| ATP7B | Precision point
mutation |
Cas9 mRNA, 2sgRNA, ssODN | Cytoplasm | Wilson Disease | [111] | |
| TYR | Precision point
mutation |
Cas9 mRNA, sgRNA, ssODN | Cytoplasm | Coat color | [112] | |
| TYR | KO, large fragment
deletion |
Cas9 mRNA, dual sgRNAs | Cytoplasm | Coat color | [113] | |
| FOXN1 | Multiplex gene KO | Cas9 mRNA, sgRNA | Cytoplasm | Immunodeficiency | [114-115] | |
| PRKDC | ||||||
| RAG1 | Multiplex gene KO | Cas9 mRNA, sgRNA | Cytoplasm | Immunodeficiency | [114-116] | |
| RAG2 | ||||||
| IL2RG | ||||||
| TIKI1 | Multiplex gene KO | Cas9 mRNA, sgRNA | Cytoplasm | Head development | [28] | |
| ALB | Serum albumin | |||||
| FUT1 | Multiplex gene KO using
a single sgRNA |
Cas9 mRNA, sgRNA | Cytoplasm | Fucosyltransferases enzymes activity | [117] | |
| FUT2 | ||||||
| SEC1 | ||||||
| ROSA 26 | KI | Cas9 mRNA, sgRNA, dsDNA | Cytoplasm | Safe harbor gene | [118-119] | |
| APOA1 | Cytoplasm | Lipid metabolism and atherosclerosis | ||||
| Base
editor |
MSTN | Base editing | BE3 mRNA | Cytoplasm | Muscle hypertrophy | [41] |
| LMNA | Hutchinson−Gilford progeria syndrome | |||||
| TYR | Coat color | |||||
| DMD | Base editing | ABE7.10 mRNA | Cytoplasm | Duchenne muscular dystrophy | ||
By attaching functional domains to nuclease inactive Cas9, it is possible to target many functionalities to a specific genomic location. Recently, a series of targeted base editing technologies were developed by fusion of cytidine deaminase or adenine deaminase with nuclease inactive Cas9[39,120–122]. Under the guide and binding of inactive Cas9 or Cas9 nickase, the deaminase converts target C:G-to-T:A or A:T-to-G:C, which holds great promise to treat various genetic diseases caused by point mutations. In rabbits[41], the cytidine base editor (CBE) and adenine base editor (ABE) were reported to highly induce site-specific conversion of C:G-to-T:A or A:T-to-G:C with efficiency rates of 53%–88% or 44%–100%, respectively. The CBE and ABE systems provide a powerful route to efficiently introduce point mutations, with much less off-target effects than the DSB-based HR method, and could be effective methods to create rabbit models of human genetic diseases associated with single nucleotide variants.
Concerns of using engineered nucleases mediated genome editing in rabbits
The development of the programmable nucleases such as ZFNs, TALENs and the powerful CRISPR/Cas9 system has revolutionized the field of genome editing in rabbits. Meanwhile, challenges still remain that need to be overcome. A major concern and potential limitation of the genome editing nuclease system is the off-target effects, that is the mutations induced at sites other than the intended on-target site, which may lead to genetic instability and disrupt normal gene functions[123]. Although the off-target events reported so far are relatively very low in frequency, these studies do nevertheless serve as a timely reminder of the need for caution when the technology is proposed for human therapy. In animal model production, the off-target concerns are not as critical as in human therapy. Most studies in rabbits so far detected no or only minimal off-target mutations in the predicted sites, and the unintended mutations, if any, may be diluted by a serial breeding program. In a recent study, Keith Joung's lab proved that appropriately designed guide RNAs can direct efficient in vivo gene editing in mouse livers with no detectable off-target mutations as examined by deep sequencing[124–125]. Key tools and strategies for optimizing CRISPR-Cas9 genome editing specificity have been summarized in a review[126–128]. New mutation detection methods were employed to predict and analyze the potential off-target sites[128]. With carefully designed guide RNA and new versions of Cas9 protein with higher specificity, combined with a well-designed breeding program, off-target effects could be effectively avoided in rabbit model production.
In addition to the classic off-target events, which are caused by inaccurate recognition of the target sequence, a recent study[129] has found that the use of CRISPR-Cas9 had caused unintended on-target large deletions or rearrangements in as long as several thousand base pairs range, which may affect non-target genes or regulatory sequences around the target gene. These on-target large deletions or rearrangements, though in low frequency, may require carefully designed characterization to determine the exact nature of the mutation in the target gene. In animal models, using lines from different founder animals could minimize the potential effects of the unwanted on-target mutations, though it's a challenge in rabbit models due to cost. If possible, using tools that avoid DSB formation, such as base editors, may avoid these unwanted on-target damages. Engineered nucleases have been widely used to disrupt gene function by creating premature stop codons in target genes. Recently, several studies have reported unexpected exon skipping in gene-edited cells and mice generated with the CRISPR/Cas9 system, which partially preserve the wild-type gene function[130–132]. By analyzing 22 gene-edited rabbit lines generated with CRISPR/Cas9, Tingting Sui et al found evidence of exon skipping at relatively high frequency in rabbits, suggesting that CRISPR-mediated exon skipping depends on exon location[133]. In one of our KO rabbit model, although a specific small deletion of 10 bp on the first exon resulted in a premature stop codon, this small deletion made an upstream out-of-frame ATG to become in frame and produce a transcript that is only slightly different from the wild-type transcript at the 5′ end (unpublished data). These studies suggest the necessity of carefully performed genotyping, including sequencing, and expression validation of the rabbit models produced by nuclease mediated genome targeting.
Conclusions
The precise genome editing in rabbits has boomed within the past 4 years thanks to the newly developed CRISPR/Cas9 technology. The availability of the rabbit whole genome sequence[40,134] combined with the versatility of rabbit genome editing technology has greatly increased their value to biomedicine, motivating efforts to develop novel rabbit models that better replicate human disease to help 'bridge the gap between bench and bedside'. In the future, novel genomic engineered rabbit models will sustain the promise for understanding the pathophysiology of human disease and for development of new therapeutics.
Contributor Information
Y. Eugene Chen, Email: echenum@umich.edu.
Dongshan Yang, Email: doyang@umich.edu.
References
- 1.Esteves PJ, Abrantes J, Baldauf HM, et al The wide utility of rabbits as models of human diseases. Exp Mol Med. 2018;50:1–10. doi: 10.1038/s12276-018-0094-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fan JL, Watanabe T Transgenic rabbits as therapeutic protein bioreactors and human disease models. Pharmacol Ther. 2003;99(3):261–282. doi: 10.1016/S0163-7258(03)00069-X. [DOI] [PubMed] [Google Scholar]
- 3.Peng XW Transgenic rabbit models for studying human cardiovascular diseases. https://pubmed.ncbi.nlm.nih.gov/23561880/ Comp Med. 2012;62(6):472–429. [PMC free article] [PubMed] [Google Scholar]
- 4.Peng XW, Knouse JA, Hernon KM Rabbit models for studying human infectious diseases. https://pubmed.ncbi.nlm.nih.gov/26678367/ Comp Med. 2015;65(6):499–507. [PMC free article] [PubMed] [Google Scholar]
- 5.Fan JL, Kitajima S, Watanabe T, et al Rabbit models for the study of human atherosclerosis: from pathophysiological mechanisms to translational medicine. Pharmacol Ther. 2015;146:104–119. doi: 10.1016/j.pharmthera.2014.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li WH, Gouy M, Sharp PM, et al Molecular phylogeny of Rodentia, Lagomorpha, Primates, Artiodactyla, and Carnivora and molecular clocks. Proc Natl Acad Sci U S A. 1990;87(17):6703–6707. doi: 10.1073/pnas.87.17.6703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Graur D, Duret L, Gouy M Phylogenetic position of the order Lagomorpha (rabbits, hares and allies) Nature. 1996;379(6563):333–335. doi: 10.1038/379333a0. [DOI] [PubMed] [Google Scholar]
- 8.Pearce J Louis Pasteur and rabies: a brief note. J Neurol Neurosurg Psychiatry. 2002;73(1):82. doi: 10.1136/jnnp.73.1.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cambau E, Drancourt M Steps towards the discovery of Mycobacterium tuberculosis by Robert Koch, 1882 . Clin Microbiol Infect. 2014;20(3):196–201. doi: 10.1111/1469-0691.12555. [DOI] [PubMed] [Google Scholar]
- 10.Goldstein JL, Kita T, Brown MS Defective lipoprotein receptors and atherosclerosis—Lessons from an animal counterpart of familial hypercholesterolemia. N Engl J Med. 1983;309(5):288–296. doi: 10.1056/NEJM198308043090507. [DOI] [PubMed] [Google Scholar]
- 11.Endo A Regulation of cholesterol synthesis, as focused on the regulation of HMG-CoA reductase (author's transl) Seikagaku (in Japanese) 1980;52(10):1033–1049. [PubMed] [Google Scholar]
- 12.Biggers JD Walter Heape, FRS: a pioneer in reproductive biology. Centenary of his embryo transfer experiments. J Reprod Fert. 1991;93(1):173–186. doi: 10.1530/jrf.0.0930173. [DOI] [PubMed] [Google Scholar]
- 13.Chang MC Fertilization of rabbit ova in vitro . Nature. 1959;184(4684):466–467. doi: 10.1038/184466a0. [DOI] [PubMed] [Google Scholar]
- 14.Hammer RE, Pursel VG, Rexroad CE Jr, et al Production of transgenic rabbits, sheep and pigs by microinjection. Nature. 1985;315(6021):680–683. doi: 10.1038/315680a0. [DOI] [PubMed] [Google Scholar]
- 15.Zernii EY, Baksheeva VE, Iomdina EN, et al Rabbit models of ocular diseases: new relevance for classical approaches. CNS Neurol Disord Drug Targets. 2016;15(3):267–291. doi: 10.2174/1871527315666151110124957. [DOI] [PubMed] [Google Scholar]
- 16.Kamaruzaman NA, Kardia E, Kamaldin N, et al The rabbit as a model for studying lung disease and stem cell therapy. Biomed Res Int. 2013;2013:691830. doi: 10.1155/2013/691830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Burkholder TH, Linton G, Hoyt RF Jr, et al. The rabbit as an experimental model[M]//Suckow MA, Stevens KA, Wilson RP. The Laboratory Rabbit, Guinea Pig, Hamster, and Other Rodents. Boston: Academic Press, 2012: 529–560.
- 18.Christensen ND, Peng XW. Rabbit genetics and transgenic models[M]//Suckow MA, Stevens KA, Wilson RP. The Laboratory Rabbit, Guinea Pig, Hamster, and Other Rodents. Boston: Academic Press, 2012: 165–193.
- 19.Isola LM, Gordon JW Transgenic animals: a new era in developmental biology and medicine. https://pubmed.ncbi.nlm.nih.gov/2007198/ Biotechnology. 1991;16:3–20. [PubMed] [Google Scholar]
- 20.Gordon JW, Scangos GA, Plotkin DJ, et al Genetic transformation of mouse embryos by microinjection of purified DNA. Proc Natl Acad Sci U S A. 1980;77(12):7380–7384. doi: 10.1073/pnas.77.12.7380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Brinster RL, Chen HY, Trumbauer M, et al Somatic expression of herpes thymidine kinase in mice following injection of a fusion gene into eggs. Cell. 1981;27(1):223–231. doi: 10.1016/0092-8674(81)90376-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Costantini F, Lacy E Introduction of a rabbit β-globin gene into the mouse germ line. Nature. 1981;294(5836):92–94. doi: 10.1038/294092a0. [DOI] [PubMed] [Google Scholar]
- 23.Gordon JW, Ruddle FH Integration and stable germ line transmission of genes injected into mouse pronuclei. Science. 1981;214(4526):1244–1246. doi: 10.1126/science.6272397. [DOI] [PubMed] [Google Scholar]
- 24.Wagner EF, Stewart TA, Mintz B The human beta-globin gene and a functional viral thymidine kinase gene in developing mice. Proc Natl Acad Sci U S A. 1981;78(8):5016–5020. doi: 10.1073/pnas.78.8.5016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wagner TE, Hoppe PC, Jollick JD, et al Microinjection of a rabbit beta-globin gene into zygotes and its subsequent expression in adult mice and their offspring. Proc Natl Acad Sci U S A. 1981;78(10):6376–6380. doi: 10.1073/pnas.78.10.6376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Campbell KHS, McWhir J, Ritchie WA, et al Sheep cloned by nuclear transfer from a cultured cell line. Nature. 1996;380(6569):64–66. doi: 10.1038/380064a0. [DOI] [PubMed] [Google Scholar]
- 27.Bibikova M, Golic M, Golic KG, et al Targeted chromosomal cleavage and mutagenesis in Drosophila using zinc-finger nucleases. https://pubmed.ncbi.nlm.nih.gov/12136019/ Genetics. 2002;161(3):1169–1175. doi: 10.1093/genetics/161.3.1169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chesne P, Adenot PG, Viglietta C, et al Cloned rabbits produced by nuclear transfer from adult somatic cells. Nat Biotechnol. 2002;20(4):366–369. doi: 10.1038/nbt0402-366. [DOI] [PubMed] [Google Scholar]
- 29.Li SG, Guo Y, Shi JJ, et al Transgene expression of enhanced green fluorescent protein in cloned rabbits generated from in vitro-transfected adult fibroblasts. Transgenic Res. 2009;18(2):227–235. doi: 10.1007/s11248-008-9227-y. [DOI] [PubMed] [Google Scholar]
- 30.Geurts AM, Cost GJ, Freyvert Y, et al Knockout rats via embryo microinjection of zinc-finger nucleases . Science. 2009;325(5939):433. doi: 10.1126/science.1172447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Flisikowska T, Thorey IS, Offner S, et al Efficient immunoglobulin gene disruption and targeted replacement in rabbit using zinc finger nucleases. PLoS One. 2011;6(6):e21045. doi: 10.1371/journal.pone.0021045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tesson L, Usal C, Menoret S, et al Knockout rats generated by embryo microinjection of TALENs. Nat Biotechnol. 2011;29(8):695–696. doi: 10.1038/nbt.1940. [DOI] [PubMed] [Google Scholar]
- 33.Gasiunas G, Barrangou R, Horvath P, et al Cas9-crRNA ribonucleoprotein complex mediates specific DNA cleavage for adaptive immunity in bacteria. Proc Natl Acad Sci U S A. 2012;109(39):E2579–E2586. doi: 10.1073/pnas.1208507109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jinek M, Chylinski K, Fonfara I, et al A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 2012;337(6096):816–821. doi: 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang H, Yang H, Shivalila CS, et al One-step generation of mice carrying mutations in multiple genes by CRISPR/Cas-mediated genome engineering. Cell. 2013;153(4):910–918. doi: 10.1016/j.cell.2013.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Song J, Zhong J, Guo XG, et al Generation of RAG 1- and 2-deficient rabbits by embryo microinjection of TALENs. Cell Res. 2013;23(8):1059–1062. doi: 10.1038/cr.2013.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yang D, Xu J, Zhu T, et al Effective gene targeting in rabbits using RNA-guided Cas9 nucleases. J Mol Cell Biol. 2014;6(1):97–99. doi: 10.1093/jmcb/mjt047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Carneiro M, Rubin CJ, Di Palma F, et al Rabbit genome analysis reveals a polygenic basis for phenotypic change during domestication. Science. 2014;345(6200):1074–1079. doi: 10.1126/science.1253714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Komor AC, Kim YB, Packer MS, et al Programmable editing of a target base in genomic DNA without double-stranded DNA cleavage. Nature. 2016;533(7603):420–424. doi: 10.1038/nature17946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang Z, Zhang JF, Li H, et al Hyperlipidemia-associated gene variations and expression patterns revealed by whole-genome and transcriptome sequencing of rabbit models. Sci Rep. 2016;6:26942. doi: 10.1038/srep26942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liu Z, Chen M, Chen S, et al Highly efficient RNA-guided base editing in rabbit. Nat Commun. 2018;9(1):2717. doi: 10.1038/s41467-018-05232-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shen W, Li L, Pan QJ, et al Efficient and simple production of transgenic mice and rabbits using the new DMSO-sperm mediated exogenous DNA transfer method. Mol Reprod Dev. 2006;73(5):589–594. doi: 10.1002/mrd.20401. [DOI] [PubMed] [Google Scholar]
- 43.Hiripi L, Negre D, Cosset FL, et al Transgenic rabbit production with simian immunodeficiency virus-derived lentiviral vector. Transgenic Res. 2010;19(5):799–808. doi: 10.1007/s11248-009-9356-y. [DOI] [PubMed] [Google Scholar]
- 44.Smith KR. Sperm-mediated gene transfer: concepts and controversies[M]. Sharjah, UAE: Bentham Science, 2012.
- 45.Kuznetsov AV, Kuznetsova IV, Schit IY DNA interaction with rabbit sperm cells and its transfer into ova in vitro and in vivo. Mol Reprod Dev. 2000;56(S2):292–297. doi: 10.1002/(SICI)1098-2795(200006)56:2+<292::AID-MRD18>3.0.CO;2-Z. [DOI] [PubMed] [Google Scholar]
- 46.Houdebine LM The methods to generate transgenic animals and to control transgene expression. J Biotechnol. 2002;98(2-3):145–160. doi: 10.1016/S0168-1656(02)00129-3. [DOI] [PubMed] [Google Scholar]
- 47.Katter K, Geurts AM, Hoffmann O, et al Transposon-mediated transgenesis, transgenic rescue, and tissue-specific gene expression in rodents and rabbits. FASEB J. 2013;27(3):930–941. doi: 10.1096/fj.12-205526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ivics Z, Hiripi L, Hoffmann OI, et al Germline transgenesis in rabbits by pronuclear microinjection of Sleeping beauty transposons . Nat Protoc. 2014;9(4):794–809. doi: 10.1038/nprot.2014.009. [DOI] [PubMed] [Google Scholar]
- 49.Evans MJ, Kaufman MH Establishment in culture of pluripotential cells from mouse embryos. Nature. 1981;292(5819):154–156. doi: 10.1038/292154a0. [DOI] [PubMed] [Google Scholar]
- 50.Martin GR Isolation of a pluripotent cell line from early mouse embryos cultured in medium conditioned by teratocarcinoma stem cells. Proc Natl Acad Sci U S A. 1981;78(12):7634–7638. doi: 10.1073/pnas.78.12.7634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gossler A, Doetschman T, Korn R, et al Transgenesis by means of blastocyst-derived embryonic stem cell lines. Proc Natl Acad Sci U S A. 1986;83(23):9065–9069. doi: 10.1073/pnas.83.23.9065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Robertson E, Bradley A, Kuehn M, et al Germ-line transmission of genes introduced into cultured pluripotential cells by retroviral vector. Nature. 1986;323(6087):445–448. doi: 10.1038/323445a0. [DOI] [PubMed] [Google Scholar]
- 53.Kuehn MR, Bradley A, Robertson EJ, et al A potential animal model for Lesch-Nyhan syndrome through introduction of HPRT mutations into mice. Nature. 1987;326(6110):295–298. doi: 10.1038/326295a0. [DOI] [PubMed] [Google Scholar]
- 54.Okita K, Ichisaka T, Yamanaka S Generation of germline-competent induced pluripotent stem cells. Nature. 2007;448(7151):313–317. doi: 10.1038/nature05934. [DOI] [PubMed] [Google Scholar]
- 55.Wernig M, Meissner A, Foreman R, et al In vitro reprogramming of fibroblasts into a pluripotent ES-cell-like state . Nature. 2007;448(7151):318–324. doi: 10.1038/nature05944. [DOI] [PubMed] [Google Scholar]
- 56.Boland MJ, Hazen JL, Nazor KL, et al Adult mice generated from induced pluripotent stem cells. Nature. 2009;461(7260):91–94. doi: 10.1038/nature08310. [DOI] [PubMed] [Google Scholar]
- 57.Kang L, Wang JL, Zhang Y, et al iPS cells can support full-term development of tetraploid blastocyst-complemented embryos. Cell Stem Cell. 2009;5(2):135–138. doi: 10.1016/j.stem.2009.07.001. [DOI] [PubMed] [Google Scholar]
- 58.Zhao XY, Li W, Lv Z, et al iPS cells produce viable mice through tetraploid complementation. Nature. 2009;461(7260):86–90. doi: 10.1038/nature08267. [DOI] [PubMed] [Google Scholar]
- 59.Capecchi MR Gene targeting in mice: functional analysis of the mammalian genome for the twenty-first century. Nat Rev Genet. 2005;6(6):507–512. doi: 10.1038/nrg1619. [DOI] [PubMed] [Google Scholar]
- 60.Fang ZF, Gai H, Huang YZ, et al Rabbit embryonic stem cell lines derived from fertilized, parthenogenetic or somatic cell nuclear transfer embryos. Exp Cell Res. 2006;312(18):3669–3682. doi: 10.1016/j.yexcr.2006.08.013. [DOI] [PubMed] [Google Scholar]
- 61.Wang SF, Tang XH, Niu YY, et al Generation and characterization of rabbit embryonic stem cells. Stem Cells. 2007;25(2):481–489. doi: 10.1634/stemcells.2006-0226. [DOI] [PubMed] [Google Scholar]
- 62.Honda A, Hirose M, Inoue K, et al Stable embryonic stem cell lines in rabbits: potential small animal models for human research. Reprod Biomed Online. 2008;17(5):706–715. doi: 10.1016/S1472-6483(10)60320-3. [DOI] [PubMed] [Google Scholar]
- 63.Osteil P, Tapponnier Y, Markossian S, et al Induced pluripotent stem cells derived from rabbits exhibit some characteristics of naïve pluripotency. Biol Open. 2013;2(6):613–628. doi: 10.1242/bio.20134242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Xue F, Ma YH, Chen YE, et al Recombinant rabbit leukemia inhibitory factor and rabbit embryonic fibroblasts support the derivation and maintenance of rabbit embryonic stem cells. Cell Reprogram. 2012;14(4):364–376. doi: 10.1089/cell.2012.0001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Du FL, Chen CH, Li Y, et al Derivation of rabbit embryonic stem cells from vitrified-thawed embryos. Cell Reprogram. 2015;17(6):453–462. doi: 10.1089/cell.2015.0044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zakhartchenko V, Flisikowska T, Li S, et al Cell-mediated transgenesis in rabbits: chimeric and nuclear transfer animals. Biol Reprod. 2011;84(2):229–237. doi: 10.1095/biolreprod.110.087098. [DOI] [PubMed] [Google Scholar]
- 67.Tancos Z, Nemes C, Polgar Z, et al Generation of rabbit pluripotent stem cell lines. Theriogenology. 2012;78(8):1774–1786. doi: 10.1016/j.theriogenology.2012.06.017. [DOI] [PubMed] [Google Scholar]
- 68.McCreath KJ, Howcroft J, Campbell KHS, et al Production of gene-targeted sheep by nuclear transfer from cultured somatic cells. Nature. 2000;405(6790):1066–1069. doi: 10.1038/35016604. [DOI] [PubMed] [Google Scholar]
- 69.Lai LX, Kolber-Simonds D, Park KW, et al Production of α-1,3-galactosyltransferase knockout pigs by nuclear transfer cloning. Science. 2002;295(5557):1089–1092. doi: 10.1126/science.1068228. [DOI] [PubMed] [Google Scholar]
- 70.Richt JA, Kasinathan P, Hamir AN, et al Production of cattle lacking prion protein. Nat Biotechnol. 2007;25(1):132–138. doi: 10.1038/nbt1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Stice SL, Robl JM Nuclear reprogramming in nuclear transplant rabbit embryos. Biol Reprod. 1988;39(3):657–664. doi: 10.1095/biolreprod39.3.657. [DOI] [PubMed] [Google Scholar]
- 72.Du FL, Xu J, Zhang JF, et al Beneficial effect of young oocytes for rabbit somatic cell nuclear transfer. Cloning Stem Cells. 2009;11(1):131–140. doi: 10.1089/clo.2008.0042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Li SG, Chen XJ, Fang ZF, et al Rabbits generated from fibroblasts through nuclear transfer. Reproduction. 2006;131(6):1085–1090. doi: 10.1530/rep.1.01065. [DOI] [PubMed] [Google Scholar]
- 74.Li SG, Flisikowska T, Kessler B, et al Production of cloned transgenic rabbits from mesenchymal stem cells. Reprod Fertil Dev. 2010;22(1):192–192. doi: 10.1071/RDv22n1Ab67. [DOI] [Google Scholar]
- 75.Yin MR, Jiang WH, Fang ZF, et al Generation of hypoxanthine phosphoribosyltransferase gene knockout rabbits by homologous recombination and gene trapping through somatic cell nuclear transfer. Sci Rep. 2015;5:16023. doi: 10.1038/srep16023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Gaj T, Gersbach CA, Barbas III CF ZFN, TALEN, and CRISPR/Cas-based methods for genome engineering. Trends Biotechnol. 2013;31(7):397–405. doi: 10.1016/j.tibtech.2013.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Conklin BR Sculpting genomes with a hammer and chisel. Nat Methods. 2013;10(9):839–840. doi: 10.1038/nmeth.2608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Cui XX, Ji DA, Fisher DA, et al Targeted integration in rat and mouse embryos with zinc-finger nucleases. Nat Biotechnol. 2011;29(1):64–67. doi: 10.1038/nbt.1731. [DOI] [PubMed] [Google Scholar]
- 79.Paques F, Duchateau P Meganucleases and DNA double-strand break-induced recombination: perspectives for gene therapy. Curr Gene Ther. 2007;7(1):49–66. doi: 10.2174/156652307779940216. [DOI] [PubMed] [Google Scholar]
- 80.Kim YG, Cha J, Chandrasegaran S Hybrid restriction enzymes: zinc finger fusions to Fok I cleavage domain. Proc Natl Acad Sci U S A. 1996;93(3):1156–1160. doi: 10.1073/pnas.93.3.1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Yang DS, Yang HQ, Li W, et al Generation of PPARγ mono-allelic knockout pigs via zinc-finger nucleases and nuclear transfer cloning . Cell Res. 2011;21(6):979–982. doi: 10.1038/cr.2011.70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Yu SL, Luo JJ, Song ZY, et al Highly efficient modification of beta-lactoglobulin (BLG) gene via zinc-finger nucleases in cattle . Cell Res. 2011;21(11):1638–1640. doi: 10.1038/cr.2011.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Perez EE, Wang JB, Miller JC, et al Establishment of HIV-1 resistance in CD4+ T cells by genome editing using zinc-finger nucleases . Nat Biotechnol. 2008;26(7):808–816. doi: 10.1038/nbt1410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Meng XD, Noyes MB, Zhu LJ, et al Targeted gene inactivation in zebrafish using engineered zinc-finger nucleases. Nat Biotechnol. 2008;26(6):695–701. doi: 10.1038/nbt1398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Yang DS, Zhang JF, Xu J, et al Production of apolipoprotein C-III knockout rabbits using zinc finger nucleases. J Vis Exp. 2013;(81):e50957. doi: 10.3791/50957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Ji DA, Zhao GJ, Songstad A, et al Efficient creation of an APOE knockout rabbit. Transgenic Res. 2015;24(2):227–235. doi: 10.1007/s11248-014-9834-8. [DOI] [PubMed] [Google Scholar]
- 87.Niimi M, Yang DS, Kitajima S, et al ApoE knockout rabbits: a novel model for the study of human hyperlipidemia. Atherosclerosis. 2016;245:187–193. doi: 10.1016/j.atherosclerosis.2015.12.002. [DOI] [PubMed] [Google Scholar]
- 88.Zhang JF, Niimi M, Yang DS, et al Deficiency of cholesteryl ester transfer protein protects against atherosclerosis in rabbits. Arterioscler Thromb Vasc Biol. 2017;37(6):1068–1075. doi: 10.1161/ATVBAHA.117.309114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Miller JC, Tan SY, Qiao GJ, et al A TALE nuclease architecture for efficient genome editing. Nat Biotechnol. 2011;29(2):143–148. doi: 10.1038/nbt.1755. [DOI] [PubMed] [Google Scholar]
- 90.Christian M, Cermak T, Doyle EL, et al Targeting DNA double-strand breaks with TAL effector nucleases. Genetics. 2010;186(2):757–761. doi: 10.1534/genetics.110.120717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Boch J, Scholze H, Schornack S, et al Breaking the code of DNA binding specificity of TAL-type III effectors. Science. 2009;326(5959):1509–1512. doi: 10.1126/science.1178811. [DOI] [PubMed] [Google Scholar]
- 92.Moscou MJ, Bogdanove AJ A simple cipher governs DNA recognition by TAL effectors. Science. 2009;326(5959):1501. doi: 10.1126/science.1178817. [DOI] [PubMed] [Google Scholar]
- 93.Hsu PD, Lander ES, Zhang F Development and applications of CRISPR-Cas9 for genome engineering. Cell. 2014;157(6):1262–1278. doi: 10.1016/j.cell.2014.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Yang DS, Xu J, Chen YE. Generation of rabbit models by gene editing nucleases[M]//Liu CY, Du YB. Microinjection. New York: Humana Press, 2019: 327–345.
- 95.Li L, Zhang QJ, Yang HQ, et al Fumarylacetoacetate hydrolase knock-out rabbit model for hereditary tyrosinemia type 1. J Biol Chem. 2017;292(11):4755–4763. doi: 10.1074/jbc.M116.764787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Chen M, Yao B, Yang QB, et al Truncated C-terminus of fibrillin-1 induces Marfanoid-progeroid-lipodystrophy (MPL) syndrome in rabbit. Dis Model Mech. 2018;11(4):dmm031542. doi: 10.1242/dmm.031542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Sui TT, Lau YS, Liu D, et al A novel rabbit model of Duchenne muscular dystrophy generated by CRISPR/Cas9. Dis Model Mech. 2018;11(6):dmm032201. doi: 10.1242/dmm.032201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Sui TT, Xu L, Lau YS, et al Development of muscular dystrophy in a CRISPR-engineered mutant rabbit model with frame-disrupting ANO5 mutations. Cell Death Dis. 2018;9(6):609. doi: 10.1038/s41419-018-0674-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Yuan L, Yao HB, Xu YX, et al CRISPR/cas9-mediated mutation of αA-crystallin gene induces congenital cataracts in rabbits. Invest Ophthalmol Vis Sci. 2017;58(6):BIO34–BIO41. doi: 10.1167/iovs.16-21287. [DOI] [PubMed] [Google Scholar]
- 100.Yuan L, Sui TT, Chen M, et al CRISPR/Cas9-mediated GJA8 knockout in rabbits recapitulates human congenital cataracts. Sci Rep. 2016;6:22024. doi: 10.1038/srep22024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Lu R, Yuan T, Wang Y, et al Spontaneous severe hypercholesterolemia and atherosclerosis lesions in rabbits with deficiency of low-density lipoprotein receptor (LDLR) on exon 7. EBioMedicine. 2018;36:29–38. doi: 10.1016/j.ebiom.2018.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Guo R, Wan Y, Xu D, et al Generation and evaluation of Myostatin knock-out rabbits and goats using CRISPR/Cas9 system. Sci Rep. 2016;6:29855. doi: 10.1038/srep29855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Lv Q, Yuan L, Deng J, et al Efficient Generation of Myostatin Gene Mutated Rabbit by CRISPR/Cas9. Sci Rep. 2016;6:25029. doi: 10.1038/srep25029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Song Y, Liu T, Wang Y, et al Mutation of the Sp1 binding site in the 5' flanking region of SRY causes sex reversal in rabbits. Oncotarget. 2017;8(24):38176–38183. doi: 10.18632/oncotarget.16979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Song Y, Xu Y, Liang M, et al CRISPR/Cas9-mediated mosaic mutation of SRY gene induces hermaphroditism in rabbits. Biosci Rep. 2018;38(2):BSR20171490. doi: 10.1042/BSR20171490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Sui T, Yuan L, Liu H, et al CRISPR/Cas9-mediated mutation of PHEX in rabbit recapitulates human X-linked hypophosphatemia (XLH) Hum Mol Genet. 2016;25(13):2661–2671. doi: 10.1093/hmg/ddw125. [DOI] [PubMed] [Google Scholar]
- 107.Sui T, Liu D, Liu T, et al LMNA-mutated rabbits: a model of premature aging syndrome with muscular dystrophy and dilated cardiomyopathy. Aging Dis. 2019;10(1):102–115. doi: 10.14336/AD.2018.0209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Wu H, Liu Q, Shi H, et al Engineering CRISPR/Cpf1 with tRNA promotes genome editing capability in mammalian systems. Cell Mol Life Sci. 2018;75(19):3593–3607. doi: 10.1007/s00018-018-2810-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Honda A, Hirose M, Sankai T, et al Single-step generation of rabbits carrying a targeted allele of the tyrosinase gene using CRISPR/Cas9. Exp Anim. 2015;64(1):31–37. doi: 10.1538/expanim.14-0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Song Y, Xu Y, Deng J, et al CRISPR/Cas9-mediated mutation of tyrosinase (Tyr) 3' UTR induce graying in rabbit. Sci Rep. 2017;7(1):1569. doi: 10.1038/s41598-017-01727-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Jiang WH, Liu LL, Chang QR, et al Production of Wilson disease model rabbits with homology-directed precision point mutations in the ATP7B gene using the CRISPR/Cas9 system. Sci Rep. 2018;8:1332. doi: 10.1038/s41598-018-19774-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Song YN, Zhang YX, Chen M, et al Functional validation of the albinism-associated tyrosinase T373K SNP by CRISPR/Cas9-mediated homology-directed repair (HDR) in rabbits. EBioMedicine. 2018;36:517–525. doi: 10.1016/j.ebiom.2018.09.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Song YN, Yuan L, Wang Y, et al Efficient dual sgRNA-directed large gene deletion in rabbit with CRISPR/Cas9 system. Cell Mol Life Sci. 2016;73(15):2959–2968. doi: 10.1007/s00018-016-2143-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Song J, Wang GS, Hoenerhoff MJ, et al Bacterial and Pneumocystis infections in the lungs of gene-knockout rabbits with severe combined immunodeficiency . Front Immunol. 2018;9:429. doi: 10.3389/fimmu.2018.00429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Song J, Yang DS, Ruan JX, et al Production of immunodeficient rabbits by multiplex embryo transfer and multiplex gene targeting. Sci Rep. 2017;7(1):12202. doi: 10.1038/s41598-017-12201-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Yan Q, Zhang Q, Yang H, et al Generation of multi-gene knockout rabbits using the Cas9/gRNA system. Cell Regen (Lond) 2014;3(1):12. doi: 10.1186/2045-9769-3-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Liu H, Sui T, Liu D, et al Multiple homologous genes knockout (KO) by CRISPR/Cas9 system in rabbit. Gene. 2018;647:261–267. doi: 10.1016/j.gene.2018.01.044. [DOI] [PubMed] [Google Scholar]
- 118.Yang DS, Song J, Zhang JF, et al Identification and characterization of rabbit ROSA26 for gene knock-in and stable reporter gene expression. Sci Rep. 2016;6:25161. doi: 10.1038/srep25161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Song J, Yang DS, Xu J, et al RS-1 enhances CRISPR/Cas9- and TALEN-mediated knock-in efficiency. Nat Commun. 2016;7:10548. doi: 10.1038/ncomms10548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Gaudelli NM, Komor AC, Rees HA, et al Programmable base editing of A·T to G·C in genomic DNA without DNA cleavage. Nature. 2017;551(7681):464–471. doi: 10.1038/nature24644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Komor AC, Zhao KT, Packer MS, et al Improved base excision repair inhibition and bacteriophage Mu Gam protein yields C: G-to-T: a base editors with higher efficiency and product purity. Sci Adv. 2017;3(8):eaao4774. doi: 10.1126/sciadv.aao4774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Koblan LW, Doman JL, Wilson C, et al Improving cytidine and adenine base editors by expression optimization and ancestral reconstruction. Nat Biotechnol. 2018;36(9):843–846. doi: 10.1038/nbt.4172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Hsu PD, Scott DA, Weinstein JA, et al DNA targeting specificity of RNA-guided Cas9 nucleases. Nat Biotechnol. 2013;31(9):827–832. doi: 10.1038/nbt.2647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Schmid-Burgk JL, Gao LY, Li D, et al Highly parallel profiling of cas9 variant specificity. Mol Cell. 2020 doi: 10.1016/j.molcel.2020.02.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Tycko J, Myer VE, Hsu PD Methods for optimizing crispr-cas9 genome editing specificity. Mol Cell. 2016;63(3):355–370. doi: 10.1016/j.molcel.2016.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Chen SM, Yao YF, Zhang YC, et al CRISPR system: discovery, development and off-target detection. Cell Signal. 2020;70:109577. doi: 10.1016/j.cellsig.2020.109577. [DOI] [PubMed] [Google Scholar]
- 127.Li JJ, Hong SY, Chen WJ, et al Advances in detecting and reducing off-target effects generated by CRISPR-mediated genome editing. J Genet Genomics. 2019;46(11):513–521. doi: 10.1016/j.jgg.2019.11.002. [DOI] [PubMed] [Google Scholar]
- 128.Zischewski J, Fischer R, Bortesi L Detection of on-target and off-target mutations generated by CRISPR/Cas9 and other sequence-specific nucleases. Biotechnol Adv. 2017;35(1):95–104. doi: 10.1016/j.biotechadv.2016.12.003. [DOI] [PubMed] [Google Scholar]
- 129.Kosicki M, Tomberg K, Bradley A Repair of double-strand breaks induced by CRISPR-Cas9 leads to large deletions and complex rearrangements. Nat Biotechnol. 2018;36(8):765–771. doi: 10.1038/nbt.4192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Mou HW, Smith JL, Peng LT, et al CRISPR/Cas9-mediated genome editing induces exon skipping by alternative splicing or exon deletion. Genome Biol. 2017;18(1):108. doi: 10.1186/s13059-017-1237-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Prykhozhij SV, Steele SL, Razaghi B, et al A rapid and effective method for screening, sequencing and reporter verification of engineered frameshift mutations in zebrafish. Dis Model Mech. 2017;10(6):811–822. doi: 10.1242/dmm.026765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Sharpe JJ, Cooper TA Unexpected consequences: exon skipping caused by CRISPR-generated mutations. Genome Biol. 2017;18(1):109. doi: 10.1186/s13059-017-1240-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Sui TT, Song YN, Liu ZQ, et al CRISPR-induced exon skipping is dependent on premature termination codon mutations. Genome Biol. 2018;19(1):164. doi: 10.1186/s13059-018-1532-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Li P, Estrada J, Zhang F, et al Isolation, characterization, and nuclear reprogramming of cell lines derived from porcine adult liver and fat. Cell Reprogram. 2010;12(5):599–607. doi: 10.1089/cell.2010.0006. [DOI] [PubMed] [Google Scholar]


