Abstract
The discovery and utilization of RNA-guided surveillance complexes, such as CRISPR-Cas9, for sequence-specific DNA or RNA cleavage, has revolutionised the process of gene modification or knockdown. To optimise the use of this technology, an exploratory race has ensued to discover or develop new RNA-guided endonucleases with the most flexible sequence targeting requirements, coupled with high cleavage efficacy and specificity. Here we review the constraints of existing gene editing and assess the merits of exploiting the diversity of CRISPR-Cas effectors as a methodology for surmounting these limitations.
Keywords: CRISPR-Cas systems, gene editing, biological evolution, DNA repair, classification, DNA transposable elements
Introduction
Gene editing is a cornerstone technology for the production of genetically modified organisms with diverse applications spanning the research, medical and pharmaceutical fields. Current approaches to site-specifically edit genomes rely on hijacking the host cell DNA repair pathways responsible for fixing double strand breaks (DSBs) and modify or completely inhibit the function of a target gene[1–2]. In concert with a delivery strategy, such as transfection or electroporation[3–6], a programmable endonuclease is introduced into the host cell, and generates a single or a double-stranded break (DSB) in the DNA[1,7]. A revolution in gene editing in recent years has precipitated as a result of the discovery that a ribonucleoprotein complex called CRISPR-Cas9 (clustered regularly interspaced short palindromic repeats-CRISPR associated protein) can be programmed to specifically target loci in the genome of an organism[1].
To further improve upon the properties of the CRISPR-Cas9 gene editing system, and understand the ecological role of the technology, a significant focus has been exploring the evolutionary diversity of CRISPR-Cas systems. The CRISPR-Cas9 editing complexes were originally discovered from ubiquitous immune systems in prokaryotes, which endow bacteria with a unique memory against past bacteriophage infections that they utilise to mount an acquired immune response[8–9]. As a consequence of this selective pressure, an enormous diversity of different CRISPR-Cas systems exists[10]. Harnessing different interference proteins from this plethora of systems has expanded editing capabilities by providing a set of complementary proteins with different editing efficiencies across diverse organisms as well as different intrinsic specificities and requirements for targeting[11].
Employing big-data style computational pipelines to mine terabyte-scale genome sequencing data for new CRISPR-Cas systems has procured many new such systems including effector proteins used to facilitate interference. These effectors may possess novel or improvement-of-function RNA-guided catalytic activities, and form the basis of improved or alternative gene editing strategies[12–15]. This may overcome inherent issues with current gene editing tools, such as the editing efficiency at different target sites across the genome, or the introduction of large insertions and deletions as a by-product of host-cell DNA repair[8–9,16]. This review will focus on CRISPR-Cas system diversity to create improved or novel gene editing approaches and extend the gene editing toolbox.
The basis of CRISPR-Cas interference and cellular repair
CRISPR-Cas systems are defined as a set of genes, known as Cas associated sequences (Cas), which co-occur with a set of tandem repeat sequences, known as CRISPR[8–9]. During phage invasion in bacteria, a small fragment of DNA (protospacer), produced from phage replication is integrated into CRISPR arrays (Fig. 1)[9,17]. These arrays are structured as alternating segments of fixed-length DNA fragments, mostly acquired from previous infections (spacers) from the phage, and constant pseudo-palindromic repeat sequences (direct repeats)[9,17]. Subsequent transcription of the CRISPR array produces a series of concatenated crRNAs (pre-crRNA), which are cleaved into individual CRISPR RNA (crRNA) fragments[18]. For interference, each crRNA consists of a pseudo-palindromic sequence, which folds into a hairpin structure, used for recognition by the effector protein, and a guide RNA (gRNA) sequence complementary to the target nucleic acid sequence of interest (Fig. 1)[18]. If a phage containing sequences present in a CRISPR array, reinvades the host bacterium, CRISPR-Cas-crRNA complexes use the gRNA segment of the crRNA, in conjunction with a 2 to 5 bp sequence of DNA called a protospacer adjacent motif (PAM) encoded directly adjacent to the target site, to sequence-specifically bind and cleave phage DNA or RNA (Fig. 1)[9,18–20]. In some instances, the gRNA and hairpin domains are split into two separate RNAs. The gRNA contains a crRNA and a hairpin domain with a trans-activating CRISPR RNA (tracrRNA). These two molecules form a duplex at one end of both molecules and function as a single crRNA as observed in other systems. Complementary binding then activates the cleavage activities of the effector, degrading the phage DNA and conferring immunity. This mechanism has been successfully harnessed to use the CRISPR system as a programmable nuclease to edit eukaryotic cells. CRISPR-based editing strategies rely on coercing the host-cell DNA repair pathways to integrate or substitute new genetic material[21]. Unlike other forms of DNA damage, DSBs are unique in that most organisms across the three kingdoms of life possess a homology-directed repair (HDR) pathway, which utilises a segment of DNA homologous to the severed region of the damaged DNA molecule as a template for repair[22–24]. The pathway consists of four basic steps: resection, strand invasion, re-synthesis and Holliday junction resolution[25–26] (Fig. 2).
Figure 1.
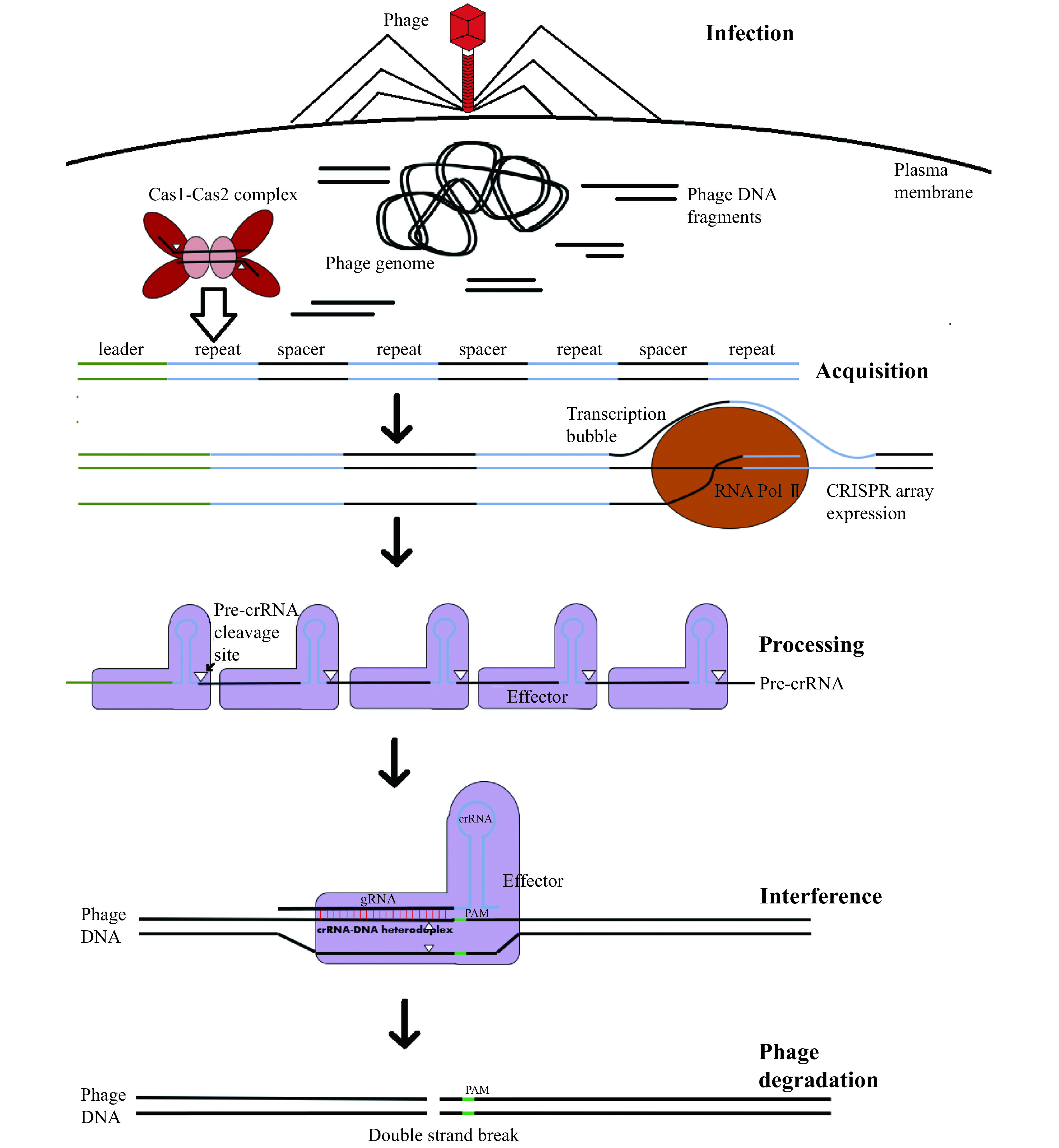
Steps in CRISPR-Cas mediated acquired immunity.
Although there exist countless variations in the exact proteins involved between different prokaryotic organisms, this process involves 3 distinct phases. In the adaptation phase, phage DNA fragments (protospacers) are uptaken and integrated by Cas1-Cas2 complexes into CRISPR loci. Each CRISPR is an alternating series of repeats and fragments of DNA derived from past phage infections (spacers). In response to phage reinfection, these CRISPR-arrays are then expressed and the corresponding long RNA transcripts (pre-crRNA) cleaved into individual CRISPR RNAs (crRNAs). These then complex with interference complexes, which degrade the phage genome in a sequence specific manner dictated by the spacer-derived guide RNA (gRNA) segment of the crRNA.
Figure 2.
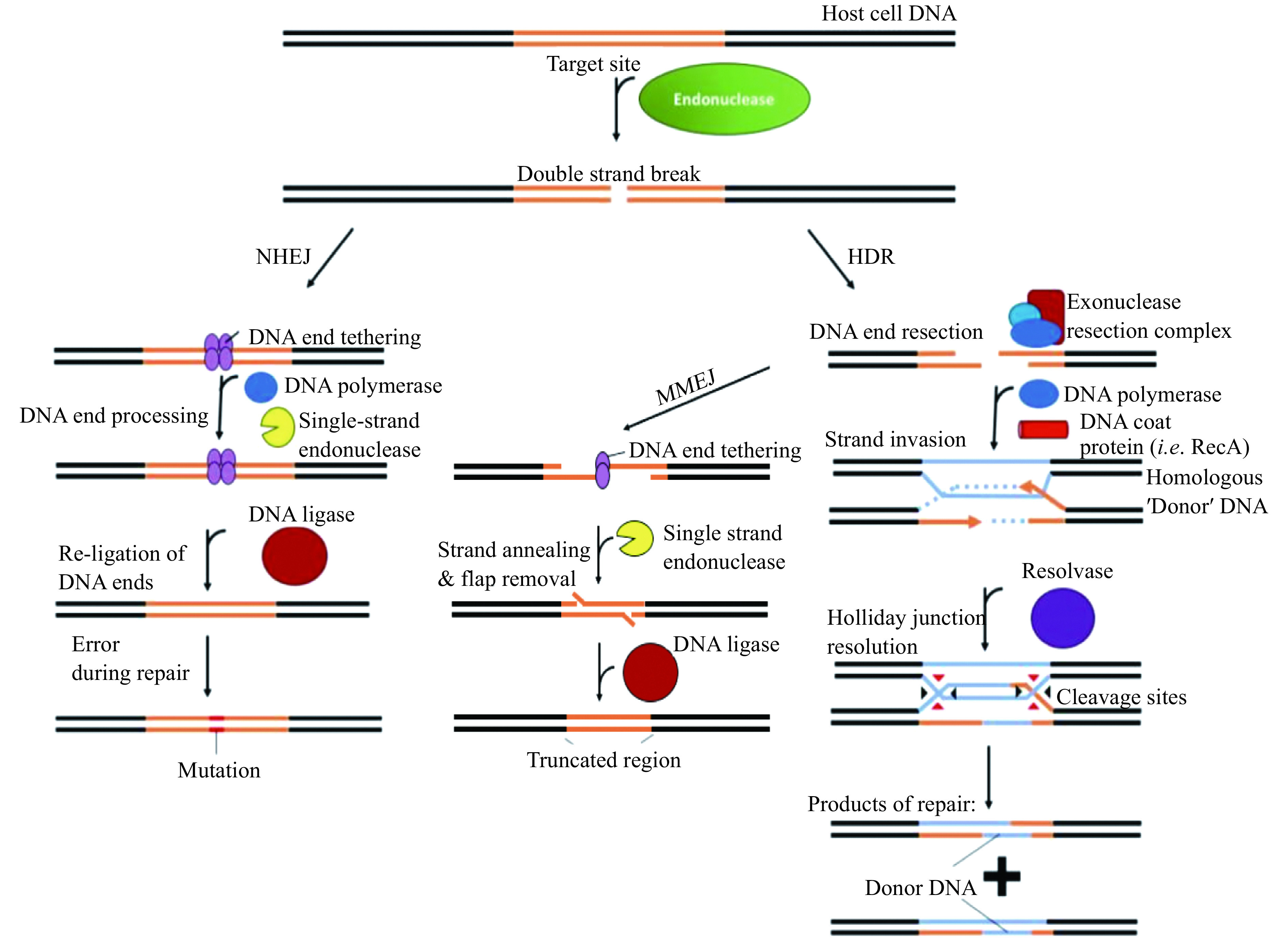
Overview of the main double-stranded break repair mechanisms.
There are 3 main conserved pathways available in most organisms. The NHEJ repair pathway directly tethers and re-joins the severed ends. HDR, in contrast, relies on a piece of homologous DNA (blue), which is utilised as a template for pair, and results in the incorporation of the donor DNA sequence at the break site. MMEJ occurs after the resection step of HDR, and relies on the annealing of large overhangs to re-tether the break site strands. Excision of the overhanging single-stranded DNA reseals the break but results in the deletion of DNA around the break site. NHEJ: non-homologous end joining; HDR: homology-directed repair; MMEJ: microhomology-mediated end joining.
Most modern gene editing platforms leverage this pathway to modify DNA by transfecting the cells of interest with exogenous DNA homologous to the base sequence of the target site. This DNA also encodes the desired mutation to incorporate[21,27–28]. Competing with this pathway, however, is usually a second pathway known as non-homologous end joining (NHEJ), which involves the direct re-joining of the severed DNA ends without the use of a template[29–30]. The accuracy of NHEJ strongly depends on the integrity of the severed DNA ends[31]. If the ends are intact, then repair is usually accurate, however if one of the ends is damaged, or the ends are incompatible, then repair usually results in the incorporation of small insertions or deletions (INDELs) at the position where the strands were severed[31]. This pathway is often used to produce loss-of-function mutations for target genes. Additionally, it is possible for a mutagenic secondary NHEJ pathway, known as microhomology-mediated end joining (MMEJ) to be activated during the initial resection step of HDR[32]. This pathway attempts to re-anneal and re-join large resected regions of single stranded DNA, which always results in DNA deletions as an artefact of the repair process[32]. Which repair pathway facilitates DSB repair for a given host cell thus determines not only the type of modification induced at the target site, also the incidence of artefacts produced from the editing process that can be mutagenic to the host cell (see Burgio et al[33] for more details). In concert with technical issues related to the RNA-guided problems themselves, such as the induction of off-target breaks in other regions of the host genome, as well as limits on the efficacy and target site programmability of various effectors, this has driven a search for editing systems with novel activities that could potentially bypass the dependency of the editing platforms on DSB induction. One of the most likely sources of the building-blocks for such platforms lies in a deeper exploration of CRISPR-Cas system diversity, and unravelling the functions of as yet undiscovered or known or not yet experimentally characterised systems.
Diversity and evolution of CRISPR-Cas systems
All CRISPR-Cas systems are differentiated based on the structure and function of the main effector proteins or complexes, and the presence or absence of specific CRISPR-associated proteins. Class 1 CRISPR-Cas systems (Fig. 3A) utilise large multi-subunit surveillance complexes, for RNA-guided DNA binding. These systems then recruit additional proteins to mediate site-specific DNA degradation[10]. In contrast, class 2 CRISPR-Cas systems all employ a single RNA-guided monomeric effector protein, which effects site-specific strand cleavage of the target nucleic acid strand (Fig. 3B)[10].
Figure 3.
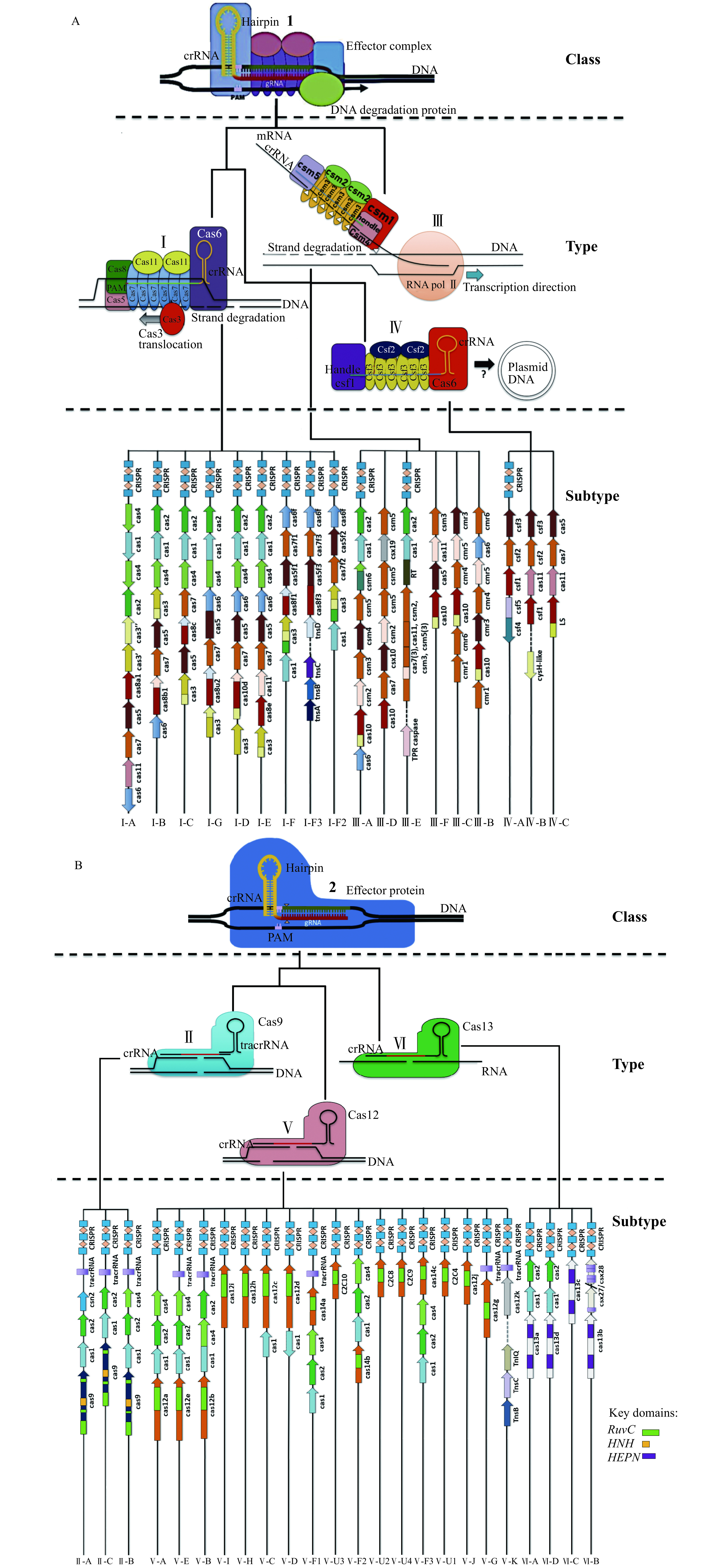
Outline of the current parsimonious classification schemes for CRISPR-Cas systems.
At the highest level, all systems are categorised into either class 1 (A) or 2 (B) based on the presence of either monomeric or multisubunit interference complexes. Each system is then typed based on the mechanism of degradation facilitated by the effector proteins, which corresponds to the catalytic mechanism used by the effector proteins for degradation. Each type is then assigned a variant number based on the lineage of the effector protein and further subtyped based on the co-occurrence of CRISPR-associated accessory proteins unique to each system.
Each of the respective CRISPR-Cas system classes are further classified based on the evolutionary lineage of the effector protein and sub-classified based on the presence or absence of genetically linked accessory proteins (Fig. 3)[10,12]. All CRISPR-Cas systems of the same type possess monomeric effector proteins, or effector protein complexes which descend from the same common ancestral variant[12]. For instance, all Cas9 proteins, which are the defining effector in type Ⅱ CRISPR-Cas systems, are believed to descend from a single ancestral variant. There are three subtypes associated with the type Ⅱ systems all distinct to each other by either the presence of accessory proteins such as Cas4 or Csn2 important for spacer acquisition or interference. Type Ⅱ-A systems include the most commonly used protein for CRISPR gene editing, Streptococcus Pyogenes Cas9[12].
Different types are also assigned a numeric designation based on common catalytic activities, and a potentially related (although not divergent) evolutionary origin followed by a letter designating a unique evolutionary lineage (Fig. 3)[12]. In the case of Class 1 systems with multiple subunits, a new effector type is defined if one of the subunits is novel or unrelated to the others (Fig. 3A).
Type Ⅰ-E systems, such as that found in the K12 strain of Escherichia Coli, contain a surveillance complex called Cascade, which is comprised of five different proteins: Cas8e, Cas11, Cas7, Cas5 and Cas6[12]. By contrast, type Ⅰ-A systems comprise a surveillance complex only composed of four separate subunits, omitting Cas6 from the complex, but including an additional Cas3 protein ortholog used to mediate interference[12].
The diversity of class 2 effectors has been unravelled via big-data mining of assembled bacterial and archaeal genomes and metagenomes
With the exception of Cas12a, which was discovered in Francisella novicia via direct examination of proteins encoded proximal to the CRISPR-arrays, all other Class 2 CRISPR-Cas effector proteins constituting novel systems have been discovered via the utilisation of computational pipelines which employ big-data filtration techniques to predict and isolate effector proteins based on their co-occurrence with conserved motifs in CRISPR-Cas systems, most notably CRISPR arrays (Fig. 4)[10,14,34–38]. The precursor step toward performing this process entails compiling a multi-terabyte sized block of assembled prokaryotic sequencing data from metagenomic and reference sequence databases. The genome data block is then scanned to identify a "seed" or "bait" motif of interest[15]. This can be the tandem repeat pattern which is conserved in CRISPR arrays, or conserved CRISPR-associated proteins such as Cas1/Cas2[14,35–36,34,39]. To avoid missing the detection of systems which employ a minimal structure, such as a single CRISPR-array in the absence of any co-occurring proteins, the union of multiple seeds can be taken, to link potential CRISPR-associated proteins to potentially any other CRISPR-associated proteins[14–15,40].
Figure 4.
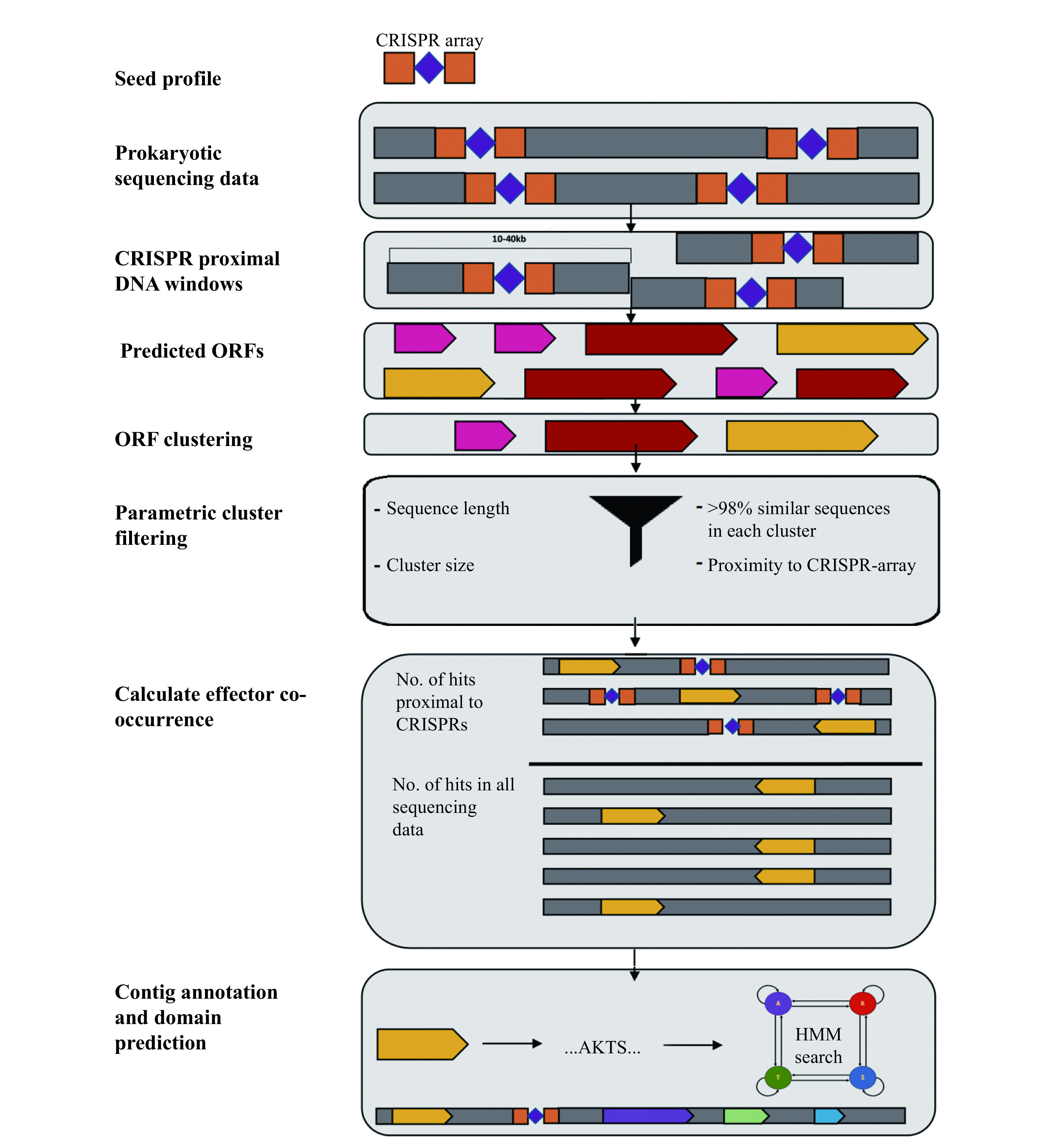
Overview of the computational pipeline used to uncover CRISPR associated proteins.
Proteins were recovered from sequence assemblies by extracting the sequences upstream and downstream of CRISPR arrays and determining whether open reading frames predicted from these sequences co-occurred with CRISPR arrays. A co-occurrence score was calculated as the number of instances of a particular protein homolog encoded within a pre-defined number of bases from a CRISPR-array divided by the total number of instances of the same homolog in all sequencing data used as the starting input to the pipeline.
After seed identification, a 5–20 kb frame of DNA sequence upstream and downstream must be extracted and subject to open reading frame prediction to identify possible candidate CRISPR-associated proteins[15]. These proteins then need to be subject to filtering and clustering to produce putative families of CRISPR-associated proteins[15]. Finally, a "CRISPR-icity" score is then calculated by finding the number of occurrences of each putative CRISPR-associated protein proximal to CRISPR-arrays and dividing this by the total number of occurrences throughout the entire block of genome sequencing data[15]. This proves that the putative CRISPR-associated proteins are linked to their proximally encoded CRISPR arrays. Subsequent analysis of the domains, family sequence diversity and properties of the individual co-occurring CRISPR-associated protein sequences has enabled the deduction of many of these CRISPR-associated proteins to be undiscovered effector proteins, which were then verified experimentally using in vitro cleavage and PAM determination assays[15].
Limitations of CRISPR-Cas system discovery via computational pipelines
There are two main types of limitations, intrinsic and extrinsic with the computational pipelines that have been used to predict new CRISPR-Cas systems to date. The intrinsic limitations of computational pipelines are defined in this instance as the inherent flaws with the pipeline methodology itself, while the extrinsic limitations are those defined as outside the scope of the goals of previously constructed pipelines used for CRISPR-Cas system discovery.
Foremost among the intrinsic limitations is a trade-off between the sensitivity, speed and computational resources required to compute a CRISPR-"icity" score, or equivalent, to validate that putative CRISPR-associated proteins are encoded next to CRISPR-arrays[15].
Using CRISPRs or conserved CRISPR-Cas proteins as seeds comes with the logical corollary that proteins which play important roles in CRISPR-immunity, but are not genetically co-encoded with CRISPR-arrays or conserved proteins, are not detectable[41–43]. Within the members of each cluster of predicted CRISPR-associated proteins, it is assumed that similar homology between related family members implies a conserved function between all members[14,35–38,39,44]. Fewer efforts to date have been made to discriminate the functionality of CRISPR-associated proteins encoded within the same families as opposed to novel families of CRISPR-Cas proteins. Prior studies of the CRISPR-Cas9 effector proteins have already unveiled immense functional diversity from within a single lineage of effector proteins, which implies that similar diversity within other CRISPR-Cas systems exists, and is not characterised by the pipelines, which identify these proteins from within genome sequencing data[45–49].
The pipeline itself also possesses several intrinsic trade-offs between the allowed sensitivity for clustering and co-occurrence score calculation, the false positive rate at which novel putative CRISPR-associated proteins are detected, and the computational requirements to perform the clustering and co-occurrence calculation steps. Because most CRISPR-associated proteins are larger than the maximum size of the short tagged reads which serve as the raw output from metagenome sequencing, only assembled metagenome data can usually be used to detect CRISPR-associated proteins using a bait-based approach[14,35–36,38,44]. Within the pipeline itself, a trade-off exists between the false-discovery rate for CRISPR-associated proteins and the window size[15]. A trade-off also exists between clustering or sequence search algorithm sensitivity and the amount of computational resources required to perform these calculations[50]. For both classes of algorithms, the underlying reason is that a more sensitive search almost always requires performing a larger number of comparisons between the decomposed query sequence (words) and the sequence database[50–52]. As a result, most pipelines have employed algorithms, such as mmseqs2, which optimise this speed/sensitivity trade-off[15,50,53].
Utilisation of the type Ⅰ systems for genome engineering
There have been several attempts to perform both gene knockout and HDR based gene editing using type Ⅰ CRISPR-Cas effectors[56]. This is possible because the single-stranded DNA degradation activity of Cas3, when applied to host-cell genomes, results in a single-stranded resected product anyway from 1 to > 50 kb in size, but most commonly in the range of around 5 to 10 kb [56], which has the potential to trigger the HDR repair pathways to incorporate foreign DNA into the host cell genome within the resected region (Fig. 5A). The key dilemma is that the single stranded progressive degradation catalytic activity of Cas3 when utilised for gene knockout results in lower efficacy in the range of 5% to 60% compared with Cas9 (10%–80%) when the DNA target site and mutation sites overlap[56–57]. Notably, however, Cas3 showed slightly improved HDR knock-in efficacy (0.8%) compared with Cas9 (0.45%) for one target site when the mutation site was significantly downstream of the target site[56].
Figure 5.

Gene editing strategies developed using standalone type I systems.
A: Use of the Cascade Cas3 complex to resect a large region of DNA downstream of the target enables the introduction of large deletions or gene knock-in, via either end-joining or homology-directed repair (HDR) pathways. B: A Tn7 – transposon forms a complex with a Type I-F Cascade complex to integrate DNA containing a gene of interest into the target loci.
An alternate, novel gene editing approach may come from the discovery of type Ⅰ-F systems, which naturally facilitate the programmable integration of Tn7 transposon DNA into a specific locus within a host cell (Fig. 5B). It has recently been discovered that a novel subclass of type Ⅰ and Ⅴ effectors are capable of site-specifically directing the integration of Tn7 transposons[58] with an efficiency in the range of 15% to 60% in bacteria depending on the exact target site specified[59–60]. Unfortunately, there have been no attempts to date to determine whether the use of such system is feasible in mammalian cells. Nevertheless, this represents an important advance a potentially viable alternative approach for integrating DNA into the chromosomes of bacteria.
Type Ⅲ and Ⅳ CRISPR-Cas effectors: Underutilised types with novel editing properties?
Both Type Ⅲ and Ⅳ systems employ a surveillance complex with significant structural and mechanistic similarities to type Ⅰ. In type Ⅲ systems, rather than binding directly to DNA, the complexes natively bind single stranded RNA in a sequence specific manner (Fig. 3A). Csm3 subunits are recruited to the complex upon the formation of a dsRNA duplex and introduce multiple breaks in a target RNA strand[61]. In some systems, the Cas10 subunit of the surveillance complex is able to concurrently degrade the complementary DNA strand. There have been several investigations where Type Ⅲ systems naturally present in bacteria were co-opted for gene editing via the transformation of the target organisms with plasmid encoded CRISPR-arrays containing spacers complementary to a read-out gene of interest. However, to date, no Type Ⅲ has been expressed from vectors in the same manner as type Ⅰ systems for editing purposes, which makes it difficult to assess the potential of these systems as transferable gene editing platforms in other organisms[62–63].
Another potential difficulty associated with the utilisation of type Ⅲ systems is that compared with the CRISPR-Cas systems of other types, the activity of the surveillance complex appears to be more integrated with the host cell defence response[64–65]. Upon target recognition, the surveillance complex synthesises cyclic oligoadenylate molecules whilst simultaneously facilitating processive single stranded DNA degradation of the transcribed gene[64,66–72]. These molecules then act as signalling substrates to stimulate the binding of interference proteins to facilitate RNA strand degradation[64,70]. However, they also activate non-specific nucleases such as NucC, which is found to be co-encoded with certain type Ⅲ systems[65,73]. Upon being activated, this protein non-specifically degrades the host genome[65]. This is a form of abortive infection to prevent the phage infection spreading to other cells in the same colony[65]. This may be an issue when utilising these proteins for gene editing, if the transformation of type Ⅲ CRISPR-Cas systems into a bacterium of interest induces cell death as a side effect. This means that, although potential editing applications exist for type Ⅲ systems, a significant amount of further research would be required to employ these type Ⅲ effectors in bacterial and eukaryotic cells as general purpose editing tools.
Like type Ⅰ systems, Type Ⅳ systems consist of an RNA-guided multi-subunit surveillance complex which sequence specifically binds DNA[74]. However, unlike type Ⅰ and Ⅲ, type Ⅳ systems lack Cas1-Cas2 acquisition proteins and are mainly encoded on plasmids[75]. The spacers from their corresponding CRISPR arrays also appear to map to other plasmids, rather than host cell, or phage genomes wherein they have been observed to mediate interference[74–75]. Intriguingly, this appears to be via a different mechanism to single-stranded DNA cleavage, instead requiring DinG, a protein with distant homology to helicase proteins used in recombinational repair[74]. The key potential advantage of utilising type Ⅳ systems for editing, is that the interference modules consist of fewer subunits than Types Ⅰ/Ⅲ which makes them easier to compact into delivery constructs[10,74–75]. However, given that the exact mechanism interference in these has not yet been characterised[74–75], it remains unclear what, if any applications for gene editing these systems possess.
Class 2 effectors: monomeric endonucleases, which form the cornerstone of all modern gene editing toolboxes
It is widely accepted that the lion's share of gene editing advances enabled by CRISPR based gene editing have come from the discovery and utilisation of class 2 CRISPR-Cas effectors. Diverse lineages of class 2 effector proteins possess several generalisable properties, which grant them a comparative advantage in gene editing and knockdown applications compared with class 1 systems. Class 2 effector proteins are monomeric and sequence specifically degrade DNA at a single loci within the target genome or transcriptome upon binding, usually introducing either a single or double strand break in the target nucleic acid strand[10]. This avoids the possible side effects arising from degrading a large DNA region as is the case in type Ⅰ CRISPR-Cas effector mediated gene editing[56–57]. As a consequence of these properties, the design and operation of computational pipelines, to extract and characterise the full extent of class 2 effectors diversity has been a major priority in the last 5 years. This has unveiled 3 basic types of class 2 effectors (Fig. 3B), subclassified into a plenitude of subtypes with the potential to complement or surpass the traditional first generation SpCas9-CRISPR mediated gene editing platform.
Cas9, the heart of CRISPR-Cas gene editing revolution
It is indisputable that the breakthrough generated from the utilisation of Streptococcus pyogenes Cas9 (SpCas9) to induce programmable RNA-guided Double Stranded cleavage has since become the epicentre of the CRISPR-Cas gene-editing world. Cas9 distinguishes itself from other effector types by its high abundance (present in approximately 10% of bacteria[12]), distinct mechanism of double stranded cleavage (Fig. 6A), high efficacy in diverse organisms and relatively low restrictions on programmability due to small PAM requirements[76–77]. While other type Ⅴ effectors utilise a single RuvC domain to cleave both DNA strands, Cas9 uses its RuvC and HNH domains to cleave the complementary and non target strands almost simultaneously[78]. This cleavage is specific to the target site, with no indiscriminate single stranded DNA (ssDNA) cleavage occurring as a side reaction, which is often observed with the effectors of Type Ⅴ systems such as Cas12a[79].
Figure 6.
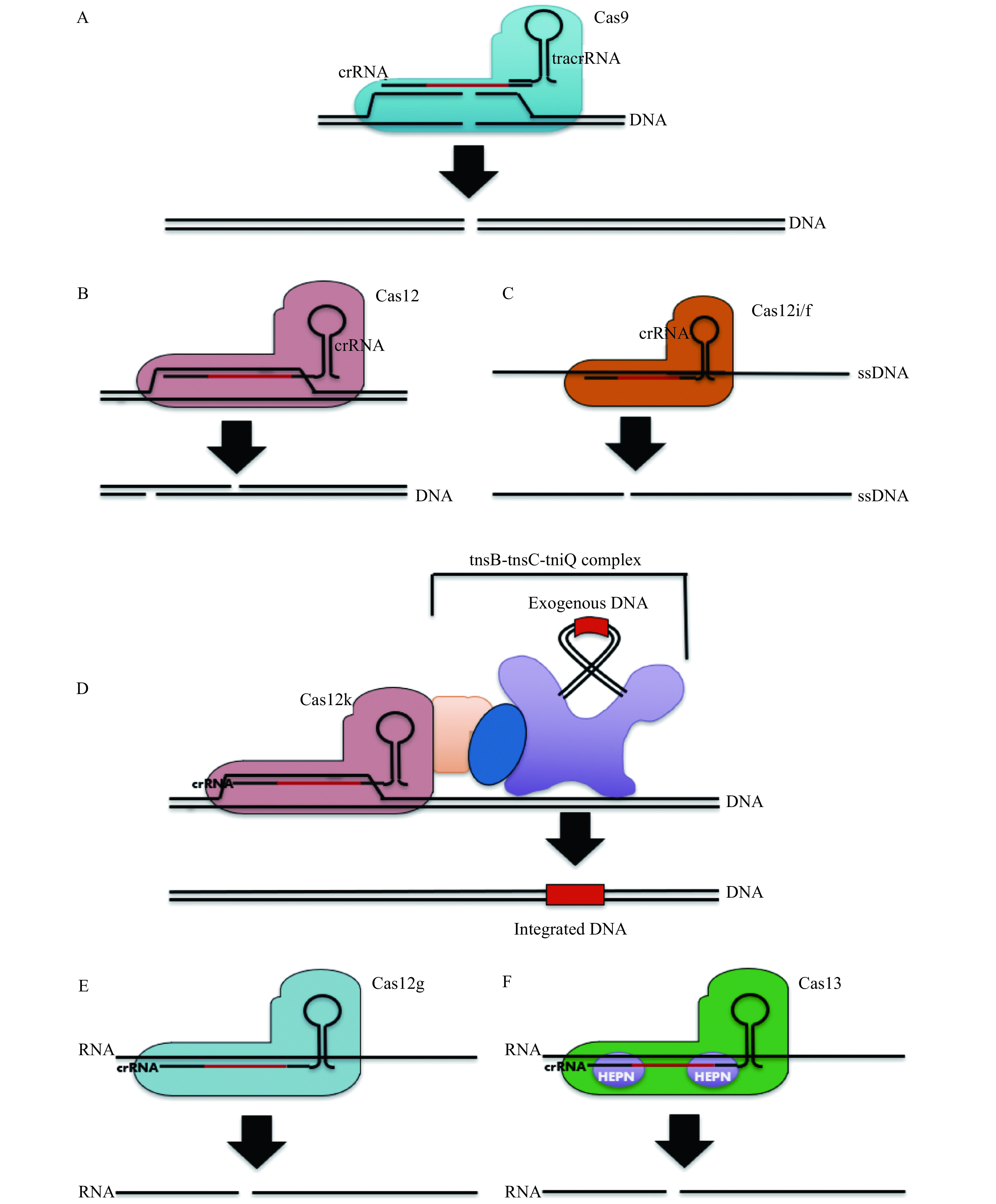
Activities of Class 2 CRISPR-Cas effectors utilized in gene editing systems.
A: The mostly commonly used CRISPR-Cas effector: Cas9, elicits blunt double strand cleavage via two independently functioning RuvC and HNH domains. B: This deviates from most Cas12 effector variants which induce a single staggered DSB at the target site. C: A minority of Cas12 effector types, such as Cas12i and Cas12f, most efficiently induce single stranded DNA breaks, although some representative Cas12f effectors may induce double strand breaks with limited efficacy at the target site as well. D: A special class of Cas12 effectors, designated Cas12k, are recruited by Tn7 transposons to direct the site of transposon integration in a programmable RNA-guided manner. E and F: Both Ca12g (E) and all known Cas13 effectors (F) elicit single strand cleavage of RNA, using an RuvC and HEPN domain respectively.
Due to being the first ortholog discovered, there has been a much greater exploration of Cas9 ortholog diversity and re-engineering of successful orthologs into higher activity variants than for effectors from other types[80] (Table 1)[27,46–47,81,101–103]. The driving motivation for using different Cas9 orthologs for editing lies in their different PAM requirements, protein size to fit into a delivery vector and editing efficacy at different target sites in different organisms. Both Staphylococcus aureus Cas9 (SaCas9, 1053 residues) and Campylobacter jejuni Cas9 (CjeCas9, 984 residues) are smaller than SpCas9, the standard effector used for most editing applications[46,81]. This results in a smaller construct size when genes encoding either of these effector proteins are cloned onto an insertion vector for gene delivery Certain orthologs may also provide an efficacy and specificity improvement when used at certain target sites, due fewer possible off-target sites due to the effector's PAM being more specific to the target site of interest[82–83]. Although overall, there has been relatively little success in finding a naturally occurring Cas9 ortholog, which surpasses SpCas9 in terms of functionality for general purpose use, when used as a collective toolbox for a specific target site, the utilisation of these alternative Cas9 orthologs can significantly increase the specificity and efficacy of the editing reaction, as well as the number of possible sites to induce cleavage in a gene of interest[84].
Table 1. Properties of most commonly used Cas9 orthologs in gene editing.
| Abbreviation | Species | Size (aa) | PAM | gRNA size (bp) | crRNA (5′→3′) | tracrRNA (5′→3′) | Reference |
| spCas9 | Streptococcus Pyogenes | 1368 | NGG | 20 | GUUUUAGAGCUAUGCUGUUUUG | GGAACCAUUCAAAACAGCAUAGCAAGUUAAAAUAAGGCUAGUCCGUUAUCAACUUGAAAAAGUGGCACCGAGUCGGUGCUUUUUUU | Jinek et al, (2012)[101] |
| FnCas9 | Francisella novicida | 1629 | NGG | 20 | CUAACAGUAGUUUACCAAAUAAUUCAGCAACUGAAAC | GUUGUUAGAUUAUUUGGUAUGUACUUGUGUUAGUUUAAAGUAGCUAGAAAAUUCACUUUUAGACCUACUUAUUUUU | Sampson et al, (2013)[47,101] |
| SaCas9 | Staphylococcus aureus | 1053 | NNGRRT | 21 | GUUUUAGUA CUCUGUAAU UUUAGGUAU GAGGUAGAC | AUUGUACUUAUACCUAAAAUUACAGAAUCUACUAAAACAAGGCAAAAUGCCGUGUUUAUCUCGUCAACUUGUUGGCGAGAUUUUU | Ran et al, (2015)[46] |
| NmCas9 |
Neisseria meningitidis
(strain 8013) |
1082 | NNNNGATT | 24 | NGUUGUAGCUCCCUUUCUCAUUUCG | AAAUGAGAACCGUUGCUACAAUAAGGCCGUCUGAAAAGAUGUGCCGCAACGCUCUGCCCCUUAAAGCUUCUGCUUUAAGGGGCAUCGUUUA | Hou et al, (2013)[101-102] |
| St1Cas9 |
Streptococcus thermophilus
(strain LMD-9) |
1121 | NNAGAAW | 20 | GUUAUUGUACUCUCAAGAUUUAUUUUU | GUUACUUAAAUCUUGCAGAAGCUACAAAGAUAAGGCUUCAUGCCGAAAUCAACACCCUGUCAUUUUAUGGCAGGGUGUUUUCGUUAUUUAA | Gasiunas et al, (2012); Cong et al, (2013)[27,101,103] |
| St3Cas9 |
Streptococcus thermophilus
(strain DGC7710) |
1409 | NGGNG | 20 | GUUCGUACUUAGUUUUAGAGCUGUGUUGUUUCG | GUUACUUAAAUCUUGCAGAAGCUACAAAGAUAAGGCUUCAUGCCGAAAUCAACACCCYGUCAUUUUAUGGCAGGGUGUUUUCGUUAUUUAA | Gasiunas et al, (2012); Cong et al, (2013)[27,101,103] |
| CjCas9 | Campylobacter jejuni | 984 | NNNNACAC | 22 | GUUUUAGUCCCUUUUUAAAUUUCUUUAUGGUAAAU | AAGAAAUUUAAAAAGGGACUAAAAUAAAGAGUUUGCGGGACUCUGCGGGGUUACAAUCCCCUAAAACCGCUUUU | Kim et al, (2017)[81,101] |
Cas12 effectors facilitate staggered double strand cleavage in a manner distinct from Cas9
Although all Cas12 effectors possess and utilise a single RuvC domain for nucleic acid strand cleavage, the substrate requirements and mechanism of cleavage differ substantially between different effector types(Fig. 6B-F). To date there are 11 different known Cas12 effector types, alphabetised A to K (Fig. 3B). The mechanism underlying this cleavage has predominantly been studied in Cas12a orthologs, but provides some transferable insight into the process for other Cas12 effectors as well[85–88]. For Cas12a, RNA-DNA heteroduplex formation induces conformational change in the NUC lobe, making the RuvC catalytic residues accessible[88]. Cleavage of the non-target strand must precede cleavage of the target strand[89]. This results in a staggered cleavage pattern with approximately 5 nt 5′ overhangs (Fig. 6B)[89]. This releases the PAM-distal target DNA fragment. However, the ribonucleoprotein complex is still catalytically competent while bound to the PAM-proximal DNA[89–90]. This often results in the activation of a secondary activity wherein indiscriminate cleavage, or 'trans' cleavage of ssDNA (and in some orthologs single stranded RNA (ssRNA), and nicking of dsDNA) by the effector protein occurs[79,89].
Miniature Cas12 effectors are capable for facilitating DNA strand cleavage
One recent advance has been the discovery and characterisation of Cas12j (phi) effectors. These proteins, encoded exclusively on the genomes of large phages and more compact (700 to 800 aa in size) than other Cas12 effectors[92]. While these effectors possess dsDNA cleavage activity, there was a significant difference in efficacy between Cas12j mediated cleavage of the target strand, and Cas12a mediated cleavage of the target strand[38,39,91–92]. This shortcoming means that while the characterisation of Cas12j represents an important step towards more compact, high efficacy editing proteins, it is however unlikely to supersede existing editors such as Cas12a or Cas9.
One of the most exciting potential advances arising from the exploration of CRISPR-Cas effector diversity has been the discovery of tiny, 400 to 700 amino acids long effector proteins. These proteins are small enough to be delivered in a recombinant adenoviral vector (rAAV)[39]. Unfortunately, all Cas14 effectors discovered to date cleave ssDNA with relatively high efficacy, but are unable to cleave dsDNA with comparable efficacy which limits their potential application without protein engineering optimization (Fig. 6E)[39,91]. Nevertheless, there is considerable optimism that this limitation can be surmounted either via direct protein engineering of Cas14 to produce a gain of function variant with higher cleavage activity, or via further exploration and characterization of Cas14 orthologs.
A small subset of Cas12k effectors mediate site-specific integration
A subclade of Cas12 effectors exist that lack functional RuvC catalytic residues and occur in the same operon as Tn7-like transposases. These function as RNA-guided DNA binding proteins and form a complex with Tn7 transposases to direct the site of transposon integration (Fig. 6D). Compared with type Ⅰ-Tn7 transposon integration systems, Cas12k guided systems offer two important distinct advantages in the form of higher insertion efficacy, depending on the loci chosen for targeting, and simpler and smaller construct size, due to the monomeric nature of Cas12k effectors compared with type Ⅰ-F and Ⅰ-B Cascade surveillance complexes[93]. The efficacy of integration by Cas12k proteins (in the range of 15% to 65% in E. Coli[60]) is superior to the measured efficacy in yeast and eukaryotes of prime editing, an alternate means of integrating DNA in host genomes[94]. This efficacy is also competitive with the editing efficacy of Cas9 or Cas12a without the side-effects associated with the utilisation of DSB repair pathways for editing, although further research is needed to demonstrate feasibility in eukaryotic cell lines[94–96].
Utilising RNA-guided gene knockdown as an alternate to gene knockout when producing null mutant strains
A different avenue for bypassing the limitations of DSB based gene knockout protocols is to induce a gene knockdown at the target site. This involves utilizing RNA-guided site-specific riboendonucleases to target and cleave mRNA transcribed by the gene of interest[14,37,38,97–98]. This silences the expression of the target gene. Several Cas12 orthologs have been discovered which possess RNA-guided RNAse activity (Fig. 6C and E), and an entire clade of CRISPR-Cas effectors, designated Cas13, have been discovered which exclusively target and cleave single strand RNA in a manner analogous to RNA-guided DNA targeting CRISPR-Cas effectors[14,37–98–99].
Concluding remarks
Exploring the diversity of CRISPR-Cas systems has unveiled a plethora of possible candidate RNA-guided ribonucleoprotein modules. However, despite the hundreds of thousands of CRISPR-Cas systems, which have been detected in metagenome sequencing data, there has been a much smaller diversity of catalytic activities that could be utilised for alternate gene editing strategies. Some of these systems, employed as emerging technologies have clear potential to become competitive with CRISPR-Cas9 in terms of editing efficacy and target site specificity, while simultaneously lacking the in-built constraints imposed by a reliance on double strand breaks to facilitate genetic recombination. Overall, when added to the existing CRISPR-toolbox, these new RNA-guided interference or integrase systems represent a significant advance when used in specialised cases where the limitations CRISPR-Cas9 are a clear technical obstacle. In the near future we will be able to customise and personalise the gene editing approach by choosing an ideal nuclease for a specific application. In conjunction with other strategies such as either rational, directed evolution or the use of base-editing or prime-editing technology, the exploitation of CRISPR-Cas system diversity remains a promising avenue for developing a general purpose gene editing platform equivalent or superior to the current CRISPR-Cas9 paradigm.
Acknowledgments
We would like to thank Anthony Newman and Jovita De Silva for their efforts in proofreading this work and suggesting improvements. We would like to thank the two anonymous reviewers for their suggestions and comments that have significantly improved the manuscript. Gaetan Burgio is supported by the National Collaborative Research Infrastructure (NCRIS) via Phenomics Australia, the National Health and Medical Research Council of Australia (Grant No. APP1143008), the Australian Research Council (Grant No. DP180101494) and the National Natural Science Foundation of China (Grant No. 81772214).
References
- 1.Jinek M, Chylinski K, Fonfara I, et al A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 2012;337(6096):816–821. doi: 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wang T, Wei JJ, Sabatini DM, et al Genetic screens in human cells using the CRISPR-Cas9 system. Science. 2014;343(6166):80–84. doi: 10.1126/science.1246981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chen SD, Sanjana N, Zheng KJ, et al Genome-wide CRISPR screen in a mouse model of tumor growth and metastasis. Cell. 2015;160(6):1246–1260. doi: 10.1016/j.cell.2015.02.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang HX, Song ZY, Lao YH, et al Nonviral gene editing via CRISPR/Cas9 delivery by membrane-disruptive and endosomolytic helical polypeptide . Proc Natl Acad Sci U S A. 2018;115(19):4903–4908. doi: 10.1073/pnas.1712963115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Teixeira M, Py BF, Bosc C, et al Electroporation of mice zygotes with dual guide RNA/Cas9 complexes for simple and efficient cloning-free genome editing. Sci Rep. 2018;8(1):474. doi: 10.1038/s41598-017-18826-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Del'Guidice T, Lepetit-Stoffaes JP, Bordeleau LJ, et al Membrane permeabilizing amphiphilic peptide delivers recombinant transcription factor and CRISPR-Cas9/Cpf1 ribonucleoproteins in hard-to-modify cells. PLoS One. 2018;13(4):e0195558. doi: 10.1371/journal.pone.0195558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Takeuchi R, Choi M, Stoddard BL Redesign of extensive protein-DNA interfaces of meganucleases using iterative cycles of in vitro compartmentalization . Proc Natl Acad Sci U S A. 2014;111(11):4061–4066. doi: 10.1073/pnas.1321030111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Makarova KS, Grishin NV, Shabalina SA, et al A putative RNA-interference-based immune system in prokaryotes: computational analysis of the predicted enzymatic machinery, functional analogies with eukaryotic RNAi, and hypothetical mechanisms of action. Biol Direct. 2006;1(1):7. doi: 10.1186/1745-6150-1-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Barrangou R, Fremaux C, Deveau H, et al CRISPR provides acquired resistance against viruses in prokaryotes. Science. 2007;315(5819):1709–1712. doi: 10.1126/science.1138140. [DOI] [PubMed] [Google Scholar]
- 10.Makarova KS, Wolf YI, Iranzo J, et al Evolutionary classification of CRISPR–Cas systems: a burst of class 2 and derived variants. Nat Rev Microbiol. 2020;18(2):67–83. doi: 10.1038/s41579-019-0299-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hajizadeh Dastjerdi A, Newman A, Burgio G The expanding class 2 CRISPR toolbox: diversity, applicability, and targeting drawbacks. BioDrugs. 2019;33(5):503–513. doi: 10.1007/s40259-019-00369-y. [DOI] [PubMed] [Google Scholar]
- 12.Makarova KS, Wolf YI, Alkhnbashi OS, et al An updated evolutionary classification of CRISPR-cas systems. Nat Rev Microbiol. 2015;13(11):722–736. doi: 10.1038/nrmicro3569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shmakov S, Smargon A, Scott D, et al Diversity and evolution of class 2 CRISPR-Cas systems. Nat Rev Microbiol. 2017;15(3):169–182. doi: 10.1038/nrmicro.2016.184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yan WX, Chong SR, Zhang HB, et al Cas13d is a compact RNA-targeting Type VI CRISPR effector positively modulated by a WYL-domain-containing accessory protein. Mol Cell. 2018;70(2):327–339.e5. doi: 10.1016/j.molcel.2018.02.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shmakov SA, Faure G, Makarova KS, et al Systematic prediction of functionally linked genes in bacterial and archaeal genomes. Nat Protoc. 2019;14(10):3013–3031. doi: 10.1038/s41596-019-0211-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Horvath P, Romero DA, Coûté-Monvoisin A, et al Diversity, activity, and evolution of CRISPR loci in Streptococcus thermophilus . J Bacteriol. 2008;190(4):1401–1412. doi: 10.1128/JB.01415-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pourcel C, Salvignol G, Vergnaud G CRISPR elements in Yersinia pestis acquire new repeats by preferential uptake of bacteriophage DNA, and provide additional tools for evolutionary studies . Microbiology. 2005;151(3):653–663. doi: 10.1099/mic.0.27437-0. [DOI] [PubMed] [Google Scholar]
- 18.Brouns SJJ, Jore MM, Lundgren M, et al Small CRISPR RNAs guide antiviral defense in prokaryotes. Science. 2008;321(5891):960–964. doi: 10.1126/science.1159689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu L, Li XY, Ma J, et al The molecular architecture for RNA-Guided RNA cleavage by Cas13a. Cell. 2017;170(4):714–726.e10. doi: 10.1016/j.cell.2017.06.050. [DOI] [PubMed] [Google Scholar]
- 20.Marraffini LA, Sontheimer EJ CRISPR interference limits horizontal gene transfer in staphylococci by targeting DNA. Science. 2008;322(5909):1843–1845. doi: 10.1126/science.1165771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Newman A, Starrs L, Burgio G Cas9 cuts and consequences; detecting, predicting, and mitigating CRISPR/Cas9 on‐ and off‐target damage. BioEssays. 2020;42(9):2000047. doi: 10.1002/bies.202000047. [DOI] [PubMed] [Google Scholar]
- 22.Liang F, Han MG, Romanienko PJ, et al Homology-directed repair is a major double-strand break repair pathway in mammalian cells. Proc Natl Acad Sci U S A. 1998;95(9):5172–5177. doi: 10.1073/pnas.95.9.5172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.White MF, Allers T DNA repair in the archaea—an emerging picture. FEMS Microbiol Rev. 2018;42(4):514–526. doi: 10.1093/femsre/fuy020. [DOI] [PubMed] [Google Scholar]
- 24.Ayora S, Carrasco B, Cárdenas PP, et al Double-strand break repair in bacteria: a view from Bacillus subtilis . FEMS Microbiol Rev. 2011;35(6):1055–1081. doi: 10.1111/j.1574-6976.2011.00272.x. [DOI] [PubMed] [Google Scholar]
- 25.Wiktor J, van der Does M, Büller L, et al Direct observation of end resection by RecBCD during double-stranded DNA break repair in vivo . Nucleic Acids Res. 2018;46(4):1821–1833. doi: 10.1093/nar/gkx1290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.van der Heijden T, Modesti M, Hage S, et al Homologous recombination in real time: DNA strand exchange by RecA. Mol Cell. 2008;30(4):530–538. doi: 10.1016/j.molcel.2008.03.010. [DOI] [PubMed] [Google Scholar]
- 27.Cong L, Ran FA, Cox D, et al Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339(6121):819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mali P, Yang L, Esvelt KM, et al RNA-guided human genome engineering via Cas9 . Science. 2013;339(6121):823–826. doi: 10.1126/science.1232033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Davis AJ, Chen DJ DNA double strand break repair via non-homologous end-joining . Transl Cancer Res. 2013;2(3):130–143. doi: 10.3978/j.issn.2218-676X.2013.04.02. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shuman S, Glickman MS Bacterial DNA repair by non-homologous end joining. Nat Rev Microbiol. 2007;5(11):852–861. doi: 10.1038/nrmicro1768. [DOI] [PubMed] [Google Scholar]
- 31.Chang HHY, Watanabe G, Gerodimos CA, et al Different DNA end configurations dictate which NHEJ components are most important for joining efficiency. J Biol Chem. 2016;291(47):24377–24389. doi: 10.1074/jbc.M116.752329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Truong LN, Li YJ, Shi LZ, et al Microhomology-mediated End Joining and Homologous Recombination share the initial end resection step to repair DNA double-strand breaks in mammalian cells. Proc Natl Acad Sci U S A. 2013;110(19):7720–7725. doi: 10.1073/pnas.1213431110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Burgio G, Teboul L Anticipating and identifying collateral damage in genome editing. Trends Genet. 2020;36(12):905–914. doi: 10.1016/j.tig.2020.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shmakov S, Abudayyeh OO, Makarova KS, et al Discovery and functional characterization of diverse class 2 CRISPR-cas systems. Mol Cell. 2015;60(3):385–397. doi: 10.1016/j.molcel.2015.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Burstein D, Harrington LB, Strutt SC, et al New CRISPR–Cas systems from uncultivated microbes. Nature. 2016;542(7640):237–241. doi: 10.1038/nature21059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yan WX, Hunnewell P, Alfonse L, et al Functionally diverse type V CRISPR-Cas systems. Science. 2019;363(6422):88–91. doi: 10.1126/science.aav7271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Smargon AA, Cox DBT, Pyzocha NK, et al Cas13b is a type VI-B CRISPR-associated RNA-guided RNase differentially regulated by accessory proteins Csx27 and Csx28. Mol Cell. 2017;65(4):618–630.e7. doi: 10.1016/j.molcel.2016.12.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Konermann S, Lotfy P, Brideau NJ, et al Transcriptome engineering with RNA-targeting type VI-D CRISPR effectors. Cell. 2018;173(3):665–676.e14. doi: 10.1016/j.cell.2018.02.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Harrington LB, Burstein D, Chen JS, et al Programmed DNA destruction by miniature CRISPR-Cas14 enzymes. Science. 2018;362(6416):839–842. doi: 10.1126/science.aav4294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shmakov SA, Makarova KS, Wolf YI, et al Systematic prediction of genes functionally linked to CRISPR-Cas systems by gene neighborhood analysis. Proc Natl Acad Sci U S A. 2018;115(23):E5307–E5316. doi: 10.1073/pnas.1803440115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Levy A, Goren MG, Yosef I, et al CRISPR adaptation biases explain preference for acquisition of foreign DNA. Nature. 2015;520(7548):505–510. doi: 10.1038/nature14302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Radovčić M, Killelea T, Savitskaya E, et al CRISPR–Cas adaptation in Escherichia coli requires RecBCD helicase but not nuclease activity, is independent of homologous recombination, and is antagonized by 5′ ssDNA exonucleases . Nucleic Acids Res. 2018;46(19):10173–10183. doi: 10.1093/nar/gky799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Deltcheva E, Chylinski K, Sharma CM, et al CRISPR RNA maturation by trans-encoded small RNA and host factor RNase III . Nature. 2011;471(7340):602–607. doi: 10.1038/nature09886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Al-Shayeb B, Sachdeva R, Chen LX, et al Clades of huge phages from across Earth's ecosystems. Nature. 2020;578(7795):425–431. doi: 10.1038/s41586-020-2007-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chatterjee P, Jakimo N, Jacobson JM Minimal PAM specificity of a highly similar SpCas9 ortholog. Sci Adv. 2018;4(10):eaau0766. doi: 10.1126/sciadv.aau0766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ran FA, Cong L, Yan WX, et al In vivo genome editing using Staphylococcus aureus Cas9 . Nature. 2015;520(7546):186–191. doi: 10.1038/nature14299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sampson TR, Saroj SD, Llewellyn AC, et al A CRISPR/Cas system mediates bacterial innate immune evasion and virulence. Nature. 2013;497(7448):254–257. doi: 10.1038/nature12048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dugar G, Leenay RT, Eisenbart SK, et al CRISPR RNA-dependent binding and cleavage of endogenous RNAs by the Campylobacter jejuni Cas9 . Mol Cell. 2018;69(5):893–905.e7. doi: 10.1016/j.molcel.2018.01.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yamada M, Watanabe Y, Gootenberg JS, et al Crystal structure of the minimal Cas9 from Campylobacter jejuni reveals the molecular diversity in the CRISPR-Cas9 systems . Mol Cell. 2017;65(6):1109–1121.e3. doi: 10.1016/j.molcel.2017.02.007. [DOI] [PubMed] [Google Scholar]
- 50.Steinegger M, Söding J MMseqs2 enables sensitive protein sequence searching for the analysis of massive data sets. Nat Biotechnol. 2017;35(11):1026–1028. doi: 10.1038/nbt.3988. [DOI] [PubMed] [Google Scholar]
- 51.Altschul SF, Gish W, Miller W, et al Basic local alignment search tool. J Mol Biol. 1990;215(3):403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 52.Buchfink B, Xie C, Huson DH Fast and sensitive protein alignment using DIAMOND. Nat Methods. 2014;12(1):59–60. doi: 10.15496/publikation-1176. [DOI] [PubMed] [Google Scholar]
- 53.Zhang B, Ye YM, Ye WW, et al Two HEPN domains dictate CRISPR RNA maturation and target cleavage in Cas13d. Nat Commun. 2019;10(1):2544. doi: 10.1038/s41467-019-10507-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Jore MM, Lundgren M, van Duijn E, et al Structural basis for CRISPR RNA-guided DNA recognition by Cascade. Nat Struct Mol Biol. 2011;18(5):529–536. doi: 10.1038/nsmb.2019. [DOI] [PubMed] [Google Scholar]
- 55.Sinkunas T, Gasiunas G, Fremaux C, et al Cas3 is a single-stranded DNA nuclease and ATP-dependent helicase in the CRISPR/Cas immune system. EMBO J. 2011;30(7):1335–1342. doi: 10.1038/emboj.2011.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Morisaka H, Yoshimi K, Okuzaki Y, et al CRISPR-Cas3 induces broad and unidirectional genome editing in human cells. Nat Commun. 2019;10(1):5302. doi: 10.1038/s41467-019-13226-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Dolan AE, Hou ZG, Xiao Yb, et al Introducing a spectrum of long-range genomic deletions in human embryonic stem cells using type I CRISPR-cas. Mol Cell. 2019;74(5):936–950.e5. doi: 10.1016/j.molcel.2019.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Peters JE, Makarova KS, Shmakov S, et al Recruitment of CRISPR-Cas systems by Tn7-like transposons. Proc Natl Acad Sci U S A. 2017;114(35):E7358–E7366. doi: 10.1073/pnas.1709035114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Klompe SE, Vo PLH, Halpin-Healy TS, et al Transposon-encoded CRISPR–Cas systems direct RNA-guided DNA integration. Nature. 2019;571(7764):219–225. doi: 10.1038/s41586-019-1323-z. [DOI] [PubMed] [Google Scholar]
- 60.Strecker J, Ladha A, Gardner Z, et al RNA-guided DNA insertion with CRISPR-associated transposases. Science. 2019;365(6448):48–53. doi: 10.1126/science.aax9181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Mogila I, Kazlauskiene M, Valinskyte S, et al Genetic dissection of the type III-A CRISPR-Cas system csm complex reveals roles of individual subunits. Cell Rep. 2019;26(10):2753–2765.e4. doi: 10.1016/j.celrep.2019.02.029. [DOI] [PubMed] [Google Scholar]
- 62.Li YJ, Pan SF, Zhang Y, et al Harnessing Type I and Type III CRISPR-Cas systems for genome editing. Nucleic Acids Res. 2016;44(4):e34. doi: 10.1093/nar/gkv1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Rahman K, Jamal M, Chen X, et al. Reprogramming the endogenous type III-A CRISPR-Cas system for genome editing, RNA interference and CRISPRi screening in Mycobacterium tuberculosis[EB/OL]. [2020-03-09]. https://www.biorxiv.org/content/10.1101/2020.03.09.983494v1.full.pdf+html.
- 64.Niewoehner O, Garcia-Doval C, Rostøl JT, et al Type III CRISPR–Cas systems produce cyclic oligoadenylate second messengers. Nature. 2017;548(7669):543–548. doi: 10.1038/nature23467. [DOI] [PubMed] [Google Scholar]
- 65.Lau RK, Ye QZ, Birkholz EA, et al Structure and mechanism of a cyclic trinucleotide-activated bacterial endonuclease mediating bacteriophage immunity. Mol Cell. 2020;77(4):723–733.e6. doi: 10.1016/j.molcel.2019.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kazlauskiene M, Kostiuk G, Venclovas Č, et al A cyclic oligonucleotide signaling pathway in type III CRISPR-Cas systems. Science. 2017;357(6351):605–609. doi: 10.1126/science.aao0100. [DOI] [PubMed] [Google Scholar]
- 67.Kazlauskiene M, Tamulaitis G, Kostiuk G, et al Spatiotemporal control of Type III-A CRISPR-Cas immunity: coupling DNA degradation with the target RNA recognition. Mol Cell. 2016;62(2):295–306. doi: 10.1016/j.molcel.2016.03.024. [DOI] [PubMed] [Google Scholar]
- 68.Han WY, Li YJ, Deng L, et al A type III-B CRISPR-Cas effector complex mediating massive target DNA destruction. Nucleic Acids Res. 2017;45(4):1983–1993. doi: 10.1093/nar/gkw1274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Elmore JR, Sheppard NF, Ramia N, et al Bipartite recognition of target RNAs activates DNA cleavage by the Type III-B CRISPR–Cas system. Genes Dev. 2016;30(4):447–459. doi: 10.1101/gad.272153.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Smalakyte D, Kazlauskiene M, Havelund JF, et al Type III-A CRISPR-associated protein Csm6 degrades cyclic hexa-adenylate activator using both CARF and HEPN domains. Nucleic Acids Res. 2020;48(16):9204–9217. doi: 10.1093/nar/gkaa634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Estrella MA, Kuo FT, Bailey S RNA-activated DNA cleavage by the Type III-B CRISPR–Cas effector complex. Genes Dev. 2016;30(4):460–470. doi: 10.1101/gad.273722.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Liu TY, Liu JJ, Aditham AJ, et al Target preference of Type III-A CRISPR-Cas complexes at the transcription bubble. Nat Commun. 2019;10(1):3001. doi: 10.1038/s41467-019-10780-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Chou-Zheng L, Hatoum-Aslan A A type III-A CRISPR-Cas system employs degradosome nucleases to ensure robust immunity. eLife. 2019;8:e45393. doi: 10.7554/eLife.45393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Crowley VM, Catching A, Taylor HN, et al A type IV-A CRISPR-Cas system in pseudomonas aeruginosa mediates RNA-guided plasmid interference in vivo . CRISPR J. 2019;2(6):434–440. doi: 10.1089/crispr.2019.0048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Pinilla-Redondo R, Mayo-Muñoz D, Russel J, Garrett RA, et al Type IV CRISPR–Cas systems are highly diverse and involved in competition between plasmids. Nucleic Acids Res. 2020;48(4):2000–2012. doi: 10.1093/nar/gkz1197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Jiang WZ, Zhou HB, Bi HH, et al Demonstration of CRISPR/Cas9/sgRNA-mediated targeted gene modification in Arabidopsis, tobacco, sorghum and rice. Nucleic Acids Res. 2013;41(20):e188. doi: 10.1093/nar/gkt780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Ma XL, Zhu QL, Chen YL, et al CRISPR/Cas9 Platforms for genome editing in plants: developments and applications. Mol Plant. 2016;9(7):961–974. doi: 10.1016/j.molp.2016.04.009. [DOI] [PubMed] [Google Scholar]
- 78.Raper AT, Stephenson AA, Suo ZC Functional insights revealed by the kinetic mechanism of CRISPR/Cas9. J Am Chem Soc. 2018;140(8):2971–2984. doi: 10.1021/jacs.7b13047. [DOI] [PubMed] [Google Scholar]
- 79.Chen JS, Ma EB, Harrington LB, et al CRISPR-Cas12a target binding unleashes indiscriminate single-stranded DNase activity. Science. 2018;360(6387):436–439. doi: 10.1126/science.aar6245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Gasiunas G, Young JK, Karvelis T, et al A catalogue of biochemically diverse CRISPR-Cas9 orthologs. Nat Commun. 2020;11(1):5512. doi: 10.1038/s41467-020-19344-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Kim E, Koo T, Park SW, et al In vivo genome editing with a small Cas9 orthologue derived from Campylobacter jejuni . Nat Commun. 2017;8(1):14500. doi: 10.1038/ncomms14500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Adli M The CRISPR tool kit for genome editing and beyond. Nat Commun. 2018;9(1):1911. doi: 10.1038/s41467-018-04252-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Hou ZG, Zhang Y, Propson NE, et al Efficient genome engineering in human pluripotent stem cells using Cas9 from Neisseria meningitidis . Proc Natl Acad Sci U S A. 2013;110(39):15644–15649. doi: 10.1073/pnas.1313587110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Gasiunas G, Barrangou R, Horvath P, et al Cas9-crRNA ribonucleoprotein complex mediates specific DNA cleavage for adaptive immunity in bacteria. Proc Natl Acad Sci U S A. 2012;109(39):E2579–E2586. doi: 10.1073/pnas.1208507109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Globyte V, Lee SH, Bae T, et al CRISPR/Cas9 searches for a protospacer adjacent motif by lateral diffusion. EMBO J. 2019;38(4):e99466. doi: 10.15252/embj.201899466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Amrani N, Gao XD, Liu PP, et al NmeCas9 is an intrinsically high-fidelity genome-editing platform. Genome Biol. 2018;19(1):214. doi: 10.1186/s13059-018-1591-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Tang YY, Fu Y Class 2 CRISPR/Cas: an expanding biotechnology toolbox for and beyond genome editing. Cell Biosci. 2018;8(1):59. doi: 10.1186/s13578-018-0255-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Li SY, Cheng QX, Liu JK, et al CRISPR-Cas12a has both cis- and trans-cleavage activities on single-stranded DNA . Cell Res. 2018;28(4):491–493. doi: 10.1038/s41422-018-0022-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Zhang LJ, Sun RR, Yang MY, et al Conformational dynamics and cleavage sites of Cas12a are modulated by complementarity between crRNA and DNA. iScience. 2019;19:492–503. doi: 10.1016/j.isci.2019.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Swarts DC, Jinek M Mechanistic insights into the cis- and trans-acting DNase activities of cas12a . Mol Cell. 2019;73(3):589–600.e4. doi: 10.1016/j.molcel.2018.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Swarts DC, van der Oost J, Jinek M Structural basis for guide RNA processing and seed-dependent DNA Targeting by CRISPR-Cas12a. Mol Cell. 2017;66(2):221–233.e4. doi: 10.1016/j.molcel.2017.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Cofsky JC, Karandur D, Huang CJ, et al CRISPR-Cas12a exploits R-loop asymmetry to form double-strand breaks. eLife. 2020;9:e55143. doi: 10.7554/eLife.55143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Karvelis T, Bigelyte G, Young JK, et al PAM recognition by miniature CRISPR–Cas12f nucleases triggers programmable double-stranded DNA target cleavage. Nucleic Acids Res. 2020;48(9):5016–5023. doi: 10.1093/nar/gkaa208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Pausch P, Al-Shayeb B, Bisom-Rapp E, et al CRISPR-CasΦ from huge phages is a hypercompact genome editor. Science. 2020;369(6501):333–337. doi: 10.1126/science.abb1400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Zetsche B, Gootenberg JS, Abudayyeh OO, et al Cpf1 is a single RNA-guided endonuclease of a class 2 CRISPR-cas system. Cell. 2015;163(3):759–771. doi: 10.1016/j.cell.2015.09.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Rubin BE, Diamond S, Alexander BFC, et al. Targeted genome editing of bacteria within microbial communities[EB/OL]. [2020-07-21]. https://www.biorxiv.org/content/10.1101/2020.07.17.209189v2.
- 97.Anzalone AV, Randolph PB, Davis JR, et al Search-and-replace genome editing without double-strand breaks or donor DNA. Nature. 2019;576(7785):149–157. doi: 10.1038/s41586-019-1711-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Ran FA, Hsu PD, Wright J, et al Genome engineering using the CRISPR-Cas9 system. Nat Protoc. 2013;8(11):2281–2308. doi: 10.1038/nprot.2013.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Teng F, Li J, Cui TT, et al Enhanced mammalian genome editing by new Cas12a orthologs with optimized crRNA scaffolds. Genome Biol. 2019;20(1):15. doi: 10.1186/s13059-019-1620-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Kappel S, Matthess Y, Kaufmann M, Strebhardt K Silencing of mammalian genes by tetracycline-inducible shRNA expression. Nat Protoc. 2007;2(12):3257–3269. doi: 10.1038/nprot.2007.458. [DOI] [PubMed] [Google Scholar]
- 101.Abudayyeh OO, Gootenberg JS, Konermann S, et al C2c2 is a single-component programmable RNA-guided RNA-targeting CRISPR effector. Science. 2016;353(6299):aaf5573. doi: 10.1126/science.aaf5573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Koonin EV, Makarova KS, Zhang F Diversity, classification and evolution of CRISPR-Cas systems. Curr Opin Microbiol. 2017;37:67–78. doi: 10.1016/j.mib.2017.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Abudayyeh OO, Gootenberg JS, Essletzbichler P, et al RNA targeting with CRISPR–Cas13. Nature. 2017;550(7675):280–284. doi: 10.1038/nature24049. [DOI] [PMC free article] [PubMed] [Google Scholar]


