Abstract
Tillering is a key agronomy trait for wheat (Triticum aestivum L.) production. Previously, we have reported a dwarf-monoculm wheat mutant (dmc) obtained from cultivar Guomai 301 (wild type, WT), and found growth regulating factors (GRFs) playing important roles in regulating wheat tillering. This study is to systematically investigate the roles of all the wheat GRFs (T. aestivum GRFs, TaGRFs) in regulating tillering, and screen out the key regulators. A total of 30 TaGRFs were identified and their physicochemical properties, gene structures, conserved domains, phylogenetic relationships and tissue expression profiles were analyzed. The expression levels of all the TaGRFs were significantly lower in dmc than those in WT at early tillering stage, and the abnormal expressions of TaGRF2-7(A, B, D), TaGRF5-7D, TaGRF10-6(A, B, D) and TaGRF11-2A were major causes constraining the tillering of dmc. The transcriptions of TaGRFs were significantly affected by exogenous indole acetic acid (IAA) and gibberellin acid (GA3) applications, which suggested that TaGRFs as well as IAA, GA signaling were involved in controlling wheat tillering. This study provided valuable clues for functional characterization of GRF genes in wheat.
Keywords: Wheat (Triticum aestivum L.), Tillering, Growth regulating factor, Expression profiles, IAA, GA
Introduction
Transcription factors are the most important regulators in plants. They are involved in various biological processes such as plant growth and development, metabolism, reproduction and differentiation (Chen & Cao, 2015; Kim & Tsukaya, 2015). Up to now, more than 60 transcription factors have been found in plants (Zhao et al., 2019). Growth-regulating factor (GRF) is a kind of plant-specific transcription factor, which regulates plant growth and development, plant cell size, participates in chloroplast proliferation and abiotic stress response (Chen & Rajewsky, 2007; Omidbakhshfard et al., 2015; Ruan et al., 2018). The most prominent feature of the GRF proteins is that there are two conserved domains in the N-terminal region, namely QLQ (Gln, Leu, Gln) and WRC (Trp, Arg, Cys) domains (Kim & Kende, 2004).
The first GRF gene OsGRF1 was discovered in rice (Oryza sativa L.), and it affects stem elongation by regulating gibberellin signaling (Van der Knaap, Kim & Kende, 2000). With the continuous analyses of plant genomic sequences, GRF genes have been identified in various plant species, such as Arabidopsis (Arabidopsis thaliana) (nine) (Kim, Choi & Kende, 2003), rice (12) (Choi, Kim & Kende, 2004), tomato (Solanum lycopersicum) (13) (Yuan et al., 2017), barley (Hordeum vulgare) (12) (Xue et al., 2020), soybean (Glycine max (Linn.) Merr) (22) (Chen et al., 2019), quinoa (Chenopodium quinoa Willd.) (18) (Shi et al., 2019), tea plant (Camellia sinensis) (11) (Wang et al., 2019b), and tobacco (Nicotiana tabacum) (30) (Dong et al., 2020).
In recent years, many studies have demonstrated that GRFs play a variety of important regulatory roles in plant growth and development (Ma et al., 2017). GRFs participate in the growth and development of root (Bao et al., 2014), stem (Kim & Lee, 2006), leaf (Horiguchi, Kim & Tsukaya, 2005; Liu et al., 2012, 2015) and flower (Liang et al., 2014; Lee et al., 2015), and play an important role in signal transduction and stress response of plants (Liu et al., 2008; Hewezi et al., 2012; Casadevall et al., 2013). OsGRF4 controls grain shape, panicle length and seed shattering in rice (Sun et al., 2016). However, there is almost no evidence of the GRFs regulating tiller development in wheat. A recent study indicates that TaGRFs may be involved in regulating wheat (Triticum aestivum L.) tillering (Wang et al., 2019a).
Common wheat is one of the most important food crops worldwide. Previously, we have reported a dwarf-monoculm wheat mutant (dmc) derived from Guomai 301 (He et al., 2018), and found that wheat miR396b (tae-miR396b) and several of its target TaGRFs are involved in regulating wheat tillering in dmc (An et al., 2019). This provides a good opportunity to design experiments exploring the molecular mechanisms of wheat tiller development. Here, we attempt to thoroughly investigate the expression profiles of all TaGRFs in Guomai 301 and mutant dmc under normal growth and development condition, as well as exogenous indole acetic acid (IAA) and gibberellin acid (GA) applications. The results provided a theoretical base for further functional characterization of GRF genes in wheat.
Materials and Methods
Plant materials and growth conditions
The mutant dmc was obtained according to the method described in the previous article (An et al., 2019). All the plant materials were planted in our experimental field (34°25′ N, 115°39′ E, 49 m a.s.l.). The field experiments were carried out in a completely randomized design as described by Li et al. (2019). Fertilizer and weed management were similar to that of wheat breeding (Li et al., 2014).
IAA and GA treatments
For hormone treatments, the wheat seedlings at the early three-leaf stage were sprayed with 10−5 mol L−1 IAA solution, and the wheat seedlings at the three-leaf stage were sprayed with 2 × 10−4 mol L−1 GA solution on the leaves until all the leaves were wet. The tillers of seedlings were sampled at 0 (untreated control), 1 and 2 h after hormone treatments. The RNAs of all the samples were immediately extracted.
Tiller sample preparation
Three bulks of tiller samples were prepared separately at the three-leaf stage (WT1, dmc1; sampling date: 15 November 2018), the over-winter stage (WT2, dmc2; sampling date: 6 January 2019) and the rising to jointing stage (WT3, dmc3; sampling date: 16 February 2019) for RNA extraction. The RNAs were used for qRT-PCR analysis. Wheat tillering had been completed at the rising to jointing stage. All the samples had three biological replicates.
Identification of TaGRFs
The genome sequences and protein sequences of latest wheat genome assembly version IWGSC refseqv1.1 (http://plants.ensembl.org/) was used to identify wheat GRF family members. The longest transcript sequence corresponding to each candidate gene was selected as the final sequence.
The Hidden Markov Model (HMM) of WRC (PF08879) and QLQ (PF08880) domains were obtained from the Pfam website (http://pfam.xfam.org/) and were used to identify the conserved domains of wheat GRF proteins with HMMER software (https://www.ebi.ac.uk/Tools/hmmer/). Besides, the protein sequences of Arabidopsis GRF family members and rice GRF family members were downloaded from PlantTFDB (http://planttfdb.cbi.pku.edu.cn/), and these sequences were used as input sequences to BLAST in the wheat protein database. All output protein sequences with e-value ≤ 1 × 10−10 were collected, removing the redundant sequences. The domains of the candidate GRF family members were confirmed by searching Pfam website and the Conserved Domain Search at NCBI (https://www.ncbi.nlm.nih.gov/Structure/bwrpsb/bwrpsb.cgi). The TaGRF genes were named mainly to refer to the annotation information from the UniProt database (https://www.uniprot.org/) and their chromosomal locations.
The lengths of amino acid sequences, molecular weights, pIs and other characteristics of wheat GRF proteins were analyzed using the online analysis software of EXPASY website (https://www.expasy.org/). All identified TaGRF genes were located on chromosomes, and the gene map was drawn using TBtools software (Chen et al., 2020).
Construction of the phylogenetic tree
Amino acid sequences of the GRF proteins in wheat, Arabidopsis and rice were used to conduct multi-sequence alignment. Based on the results of sequence alignment, the phylogenetic tree was constructed using the Neighbor-Joining method in MEGA software (https://www.megasoftware.net/). The check parameter bootstrap value was 1,000, and the default value of the system was used for other parameters.
Conserved motif and gene structure analysis of TaGRFs
The online website MEME (http://meme-suite.org/) was used to analyze the motifs of TaGRFs protein sequences. Parameters were set as follows: the motif discovery mode was classic mode, the site distribution was Zero or One Occurrence Per Sequence (zoops), the maximum number of motifs to find was 10, and other parameters were default. The DNA and cDNA sequences corresponding to predicted proteins from the wheat genome database were downloaded from the wheat genome database. Then, TBtools software was used to draw motif distribution and the structure distribution map of TaGRFs, and the Logo diagram of amino acid conservation was drawn on the WEBLOGO website.
Analysis of the cis-acting elements in TaGRF promoters
The 2,000 bp upstream sequences before transcription start positions of TaGRFs were extracted from wheat genome sequence, and the cis-acting elements were analyzed using the PlantCARE database (http://bioinformatics.psb.ugent.be/webtools/plantcare/html).
Gene duplication and synteny analysis of TaGRFs
Multiple Collinearity Scan toolkit (MCScanX) program with the default parameters was adopted to analyze the gene duplication events of TaGRFs. To exhibit the synteny relationship of the orthologous GRF genes obtained from wheat and other selected species, the syntenic analysis maps were constructed using the Dual Synteny Plotter software (https://github.com/CJ-Chen/TBtools). According to the results of MCScanX, the nonsynonymous substitution rate (Ka) and synonymous substitution rate (Ks) of duplicated GRF genes were calculated by KaKs Calculator 2.0 software. The Ka/Ks ratios for TaGRF genes were used to assess the selection pressure on duplicated genes and Ka/Ks ratio >1, <1, or =1 indicated positive, negative, or neutral evolution, respectively.
Tissue specific expression analysis of TaGRFs
The raw gene expression data were downloaded from the Wheat Expression Browser (http://www.wheat-expression.com/) (Table S1). A total of eight mRNA-seq data from wheat cultivar Chinese Spring and Azhurnaya were analyzed. The samples were prepared from four tissues, including spike (anthesis stage), grain (milk grain stage), leaves/shoots (flag leaf stage) and roots (flag leaf stage). TBtools software was used to draw the expression heat map of TaGRFs in wheat. The expression profiles were performed based on the transcripts per million (TPM) values of TaGRF genes. The Gene Ontology database (GO, geneontology.org) was used for functional annotation of the GRF genes. GO annotations were mapped according to molecular functions, biological processes and cellular components (Table S2).
RNA-seq and gene expression analysis
The samples for RNA-seq analysis were prepared at the three-leaf stage to four-leaf stage in 2017. Two super bulk samples of the mutant dmc (T01, T02 and T03) and WT (T04, T05 and T06) with three biological replicates were prepared. The transcript abundance of TaGRF genes was calculated as fragments per kilobase of exon model per million mapped reads (FPKM) (He et al., 2018). The expression profiles of all the TaGRF genes in Guomai 301 and mutant dmc were analyzed based on the RNA-seq data in this study (Table S3). The heatmaps were drawn by HemI1.0 based on the transformed data of log2 (FPKM + 1) values (Deng et al., 2014).
qRT-PCR
qRT-PCR was performed as described previously by An et al. (2019). The primers of TaGRFs were designed using Primer-Blast of NCBI website (https://www.ncbi.nlm.nih.gov/tools/primer-blast/). All the primer sequences were listed in Table S4. The β-actin gene was used as an internal control and each reaction was performed with triplicates. The relative expression of TaGRFs was calculated by 2−ΔΔCt methods (Livak & Schmittgen, 2001).
Results
Typical traits of Guomai 301 and mutant dmc
The plant height and tiller number between mutant dmc and Guomai 301 were significantly different at heading stage (Fig. 1A). The average plant height and tiller number of Guomai 301 were 64.9 cm and 20.33, the average plant height of mutant dmc was 48.0 cm and most dmc plants had no tiller. At the three-leaf stage (Fig. 1B), the tiller buds of WT formed two primary tillers (PTs) at the base of the main culm (Fig. 1E). Meanwhile, only one tiny protuberance formed at the main culm base of dmc (Fig. 1H). At the over-winter stage (Fig. 1C), the tiller number of WT had reached 11–14, which mainly consisted of PTs and secondary tillers (Fig. 1F), while there were only two tiny tiller primordia (TPs) at the base of the dmc (Fig. 1I). Between the rising stage and the jointing stage (Fig. 1D), the tiny TPs of dmc were almost unchanged as before (Fig. 1J); but the tiller number of WT had reached its maximum value (Fig. 1G) (An et al., 2019).
Figure 1. The tiller micromorphology of Guomai 301 and mutant dmc.
(A) The plant phenotype of Guomai 301 (left) and mutant dmc (right). (B) Guomai 301 (left) and dmc (right) at the three-leaf stage. (C) Guomai 301 (left) and dmc (right) at the over-winter stage.(D) Guomai 301 (left) and dmc (right) at the rising to jointing stage. (E–G) WT at the three-leaf stage, over-winter stage, and the rising to jointing stage. (H–J) dmc at the three-leaf stage, over-winter stage, and the rising to jointing stage. MC, main culm; TP, tiller primordium; PT, primary tiller; ST, secondary tiller. (A) Scale bar = 10 cm; (B)–(D) Scale bar = 2 cm; (E) and (H)–(J) scale bar = 1 mm; (F) and (G) Scale bar = 1 cm.
Identification of TaGRFs
A total of 30 TaGRF genes (including 13 homoeologous groups) (Table 1) were finally identified from the wheat genome using nine Arabidopsis GRF and 12 rice GRF genes as input sequences. The lengths of TaGRF proteins ranged from 211 to 611 aa and the mean value was 359 aa; the molecular weight ranged from 22.3 to 64.3 kDa and the average value was 38.8 kDa. The TaGRF6-4B was the heaviest and TaGRF10-6B was the lightest. The isoelectric point (pI) was between 4.72 and 9.90, with an average value of 7.9, and the TaGRF10-6D was the highest and TaGRF11-2B was the lowest. Among them, seven TaGRFs with pIs less than seven were slightly acidic, while the remaining 23 TaGRFs with pIs greater than seven were slightly alkaline. These data indicated that most TaGRFs contained more basic amino acids. The fat solubility of TaGRFs was between 45.83 and 70.6, with an average value of 57.056. Their hydrophobic indices ranged from −0.882 to −0.35, indicating that TaGRFs had good hydrophilicity (Grand average of hydropathicity, GRAVY).
Table 1. Some basic characteristics of wheat GRF genes.
| Gene name | Gene ID | Physical position and gene direction | Protein length (aa) | Isoelectric point (pI) | Molecular weight (Da) | Aliphatic index | GRAVY |
|---|---|---|---|---|---|---|---|
| TaGRF1-6A | TraesCS6A02G335900.1 | 568515906–568517951 | 409 | 7.22 | 44,786.45 | 46.65 | −0.833 |
| TaGRF1-6B | TraesCS6B02G366700.1 | 639532839–639534932 | 410 | 7.21 | 44,724.4 | 45.83 | −0.821 |
| TaGRF1-6D | TraesCS6D02G315700.1 | 423814859–423816885 | 414 | 7.24 | 45,263.95 | 46.09 | −0.835 |
| TaGRF2-6A | TraesCS6A02G174800.1 | 188250927–188252018 | 315 | 8.12 | 33,604.85 | 70.6 | −0.35 |
| TaGRF2-7A | TraesCS7A02G165600.1 | 121056090–121057606 | 309 | 8.55 | 34,214.05 | 51.33 | −0.841 |
| TaGRF2-7B | TraesCS7B02G070200.1 | 76903837–76905287 | 316 | 8.55 | 34,886.68 | 49.59 | −0.882 |
| TaGRF2-7D | TraesCS7D02G166400.1 | 117132680–117134270 | 320 | 8.26 | 35,405.24 | 50.5 | −0.882 |
| TaGRF3-2A | TraesCS2A02G435100.1 | 687048698–687052485 | 384 | 6.76 | 42,326.85 | 54.61 | −0.623 |
| TaGRF3-2D | TraesCS2D02G435200.1 | 546208318–546212200 | 391 | 7.04 | 42,780.33 | 53.4 | −0.62 |
| TaGRF4-2B | TraesCS2B02G458400.1 | 653016354–653019980 | 387 | 7.01 | 42,454.97 | 52.4 | −0.621 |
| TaGRF4-6A | TraesCS6A02G269600.1 | 496010434–496017229 | 408 | 7.65 | 43,457.29 | 52.03 | −0.579 |
| TaGRF4-6B | TraesCS6B02G296900.1 | 532899954–532903804 | 406 | 8.46 | 43,435.28 | 53.03 | −0.571 |
| TaGRF4-6D | TraesCS6D02G245300.1 | 347433246–347436961 | 409 | 8.16 | 43,620.48 | 52.4 | −0.559 |
| TaGRF5-4A | TraesCS4A02G434900.1 | 705509912–705513259 | 371 | 8.5 | 39,941.33 | 47.44 | −0.726 |
| TaGRF5-7A | TraesCS7A02G049100.1 | 22937762–22940558 | 370 | 8.78 | 40,161.6 | 46.24 | −0.762 |
| TaGRF5-7D | TraesCS7D02G044200.1 | 22586526–22589173 | 368 | 8.57 | 39,895.24 | 46.77 | −0.75 |
| TaGRF6-4A | TraesCS4A02G255000.1 | 567181384–567185511 | 607 | 6.87 | 63,953.28 | 65.58 | −0.416 |
| TaGRF6-4B | TraesCS4B02G060000.1 | 51679625–51683720 | 611 | 6.72 | 64,277.53 | 65.01 | −0.423 |
| TaGRF6-4D | TraesCS4D02G059600.1 | 35476875–35481038 | 578 | 6.58 | 61,162.1 | 64.43 | −0.45 |
| TaGRF9-4A | TraesCS4A02G291500.1 | 594532829–594535533 | 408 | 9 | 45,327.7 | 55.78 | −0.842 |
| TaGRF9-4D | TraesCS4D02G020300.1 | 8777205–8779937 | 415 | 8.82 | 45,994.94 | 56.75 | −0.794 |
| TaGRF10-6A | TraesCS6A02G257600.1 | 479833651–479834702 | 212 | 9.54 | 22,597.56 | 67.36 | −0.384 |
| TaGRF10-6B | TraesCS6B02G267500.1 | 481089083–481090143 | 211 | 9.64 | 22,347.22 | 67.68 | −0.382 |
| TaGRF10-6D | TraesCS6D02G238900.1 | 339389350–339390413 | 215 | 9.9 | 22,750.77 | 68.74 | −0.36 |
| TaGRF11-2A | TraesCS2A02G238700.1 | 323528641–323530709 | 319 | 4.89 | 34,683.79 | 66.68 | −0.492 |
| TaGRF11-2B | TraesCS2B02G256600.2 | 298324714–298327379 | 263 | 4.72 | 28,160.15 | 61.63 | −0.689 |
| TaGRF11-2D | TraesCS2D02G246600.1 | 288225741–288228187 | 264 | 4.76 | 28,182.2 | 62.16 | −0.692 |
| TaGRF12-2A | TraesCS2A02G398300.2 | 651752444–651753590 | 225 | 9.82 | 23,823.85 | 64.36 | −0.436 |
| TaGRF12-2B | TraesCS2B02G416300.1 | 594982278–594983776 | 227 | 9.82 | 24,077.17 | 62.51 | −0.479 |
| TaGRF12-2D | TraesCS2D02G395900.1 | 506941472–506942539 | 229 | 9.57 | 24,221.3 | 64.1 | −0.443 |
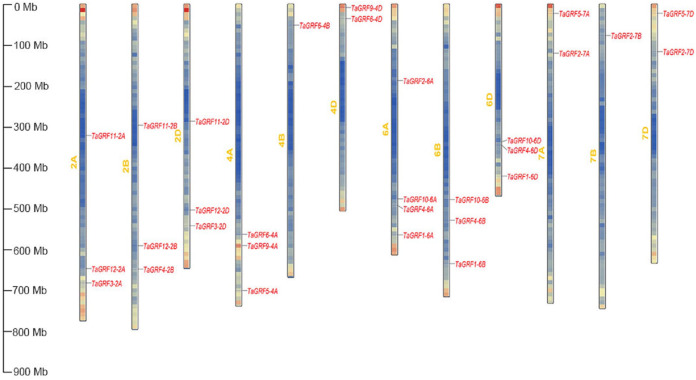
The 30 TaGRFs were located on 12 of the 21 wheat chromosomes. Each of the 12 chromosomes had 1–4 TaGRFs. Among them, chromosome 6A had four TaGRFs, 4B and 7B had one TaGRF and 4D, 7A and 7D had two TaGRFs, respectively (Fig. 2).
Figure 2. The chromosomal distributions of TaGRF genes.
The density of genes on each chromosome is uneven, red and blue denote higher and lower gene density, respectively.
Phylogenetic tree of wheat GRF proteins
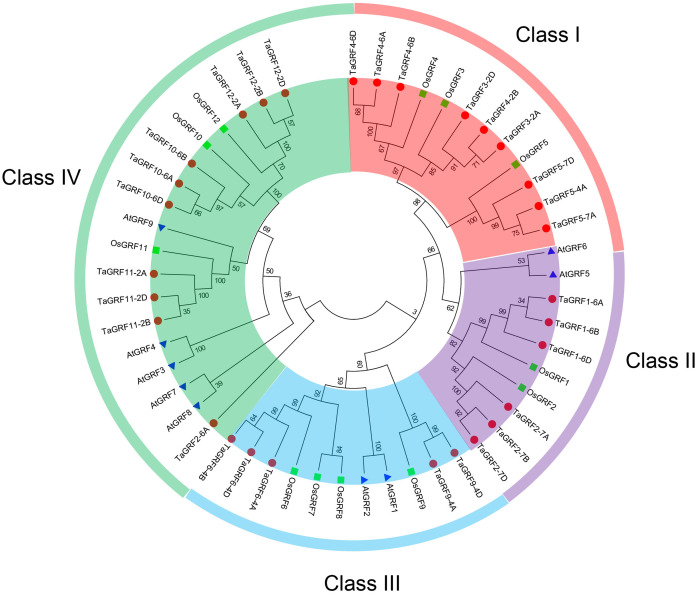
In order to better understand the phylogenetic relationship between TaGRFs and those in other plant species. The phylogenetic tree was constructed (Fig. 3), including 30 TaGRFs, nine AtGRFs and 12 OsGRFs. According to the branches of the phylogenetic tree, the 51 GRFs were clustered into four classes: Class I, II, III and IV. There were nine TaGRFs in Class I, six TaGRFs in Class II, five TaGRFs in Class III and 10 TaGRFs in Class IV. Phylogenetic analysis indicated that wheat GRFs were more closely related to rice GRFs than Arabidopsis GRFs.
Figure 3. The phylogenetic tree of the GRFs in T. aestivum L. (Ta), A. thaliana (At), and O. sativa (Os).
Structures and conserved motifs of wheat GRF genes
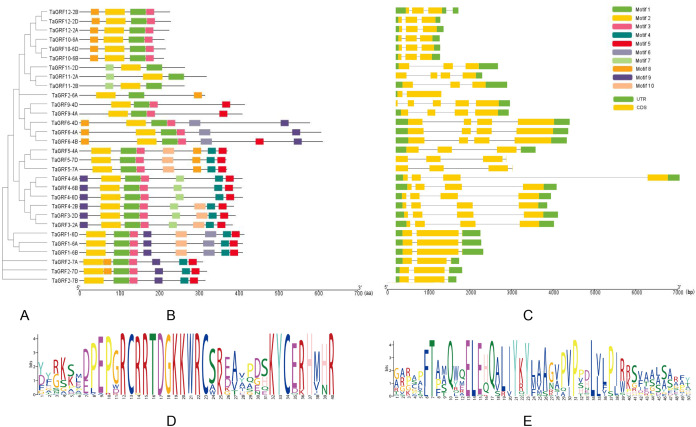
Based on the gene structural information, the phylogenetic tree of TaGRFs was built (Fig. 4A). A total of 10 conservative motifs were predicted in wheat GRF proteins (Fig. 4B). Motif 1 was the WRC domain and motif 2 was the QLQ domain, both were highly conserved and existed in the protein sequences of all wheat GRF proteins (Figs. 4D and 4E). In addition, most TaGRFs also contained motif 3. The exon-intron structure diagram of TaGRFs showed that their genomic DNA sequence lengths were significantly different (Fig. 4C). The longest was TaGRF4-6A, its length was about 7,000 bp, and the shortest was about 1,000 bp. The exon number of TaGRFs was 2–5, and the exon numbers of TaGRFs were closely related to their classes in the phylogenetic tree of TaGRFs (Fig. 3). For example, each of the six TaGRFs in Class II had two exons. Among the 30 TaGRFs, 25 TaGRFs had both 5′-UTR (untranslated region) and 3′-UTR, two TaGRFs only contained 3′-UTR, one TaGRF only contained 5′-UTR, and the remaining two TaGRFs had no UTRs.
Figure 4. Phylogenetic relationships, gene and protein structures of TaGRFs.
(A) The phylogenetic tree was constructed based on the full-length sequences of wheat GRF proteins using MEGA software. (B) The motif compositions of TaGRFs. The motifs, numbers 1–10, are displayed in different colored boxes. (C) Exon-intron structures of TaGRFs. Green boxes indicate untranslated 5′- and 3′-regions; yellow boxes indicate exons; black lines indicate introns. (D and E) The conserved sequences of motif 1 (left) and motif 2 (right) in TaGRFs. The lengths of the proteins (B, aa) and DNAs (C, bp) can be estimated using the scales at the bottoms.
Cis-acting elements in the promoters of TaGRFs and GO annotation
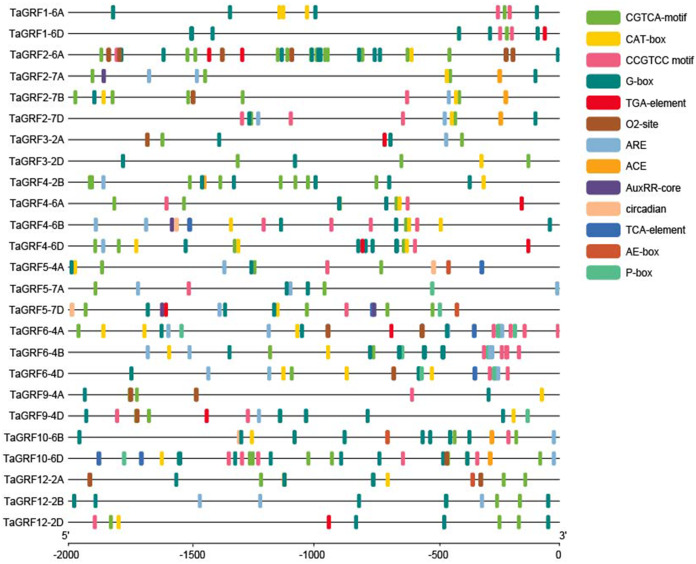
Among the members of the wheat GRF gene family, the promoter sequences of TaGRF1-6B, TaGRF10-6A, TaGRF11-2A, TaGRF11-2B and TaGRF11-2D contained a large number of ‘N’, so they hadn’t been analyzed (Fig. 5). Except for a large number of CAAT-box and TATA-box elements, there were also a large number of cis-acting regulatory elements related to growth and development, hormones and stress responsiveness in the promoter sequences of TaGRFs, including some cis-acting elements involving in auxin (AuxRR-core, TGA-element), gibberellin (P-box), light response (ACE, AEbox and G-box), methyl jasmonate reaction (CGTCA-motif), salicylic acid reaction (TCA-element), circadian rhythm control (circadian), regulation of gliadin metabolism (O2-site), and regulation of meristematic expression (CAT-box and CCGTCC motif). These cis-acting elements implied TaGRFs played various potential roles in regulating wheat growth and development, and response to hormones and stresses.
Figure 5. The Cis-acting elements in the promoters of TaGRFs.
Gene Ontology annotation revealed that the TaGRF genes are mainly involved in regulation of transcription, DNA-templated (GO:0006355) and response to deep water (GO:0030912). Both TaGRF1 and TaGRF2 are involved in response to gibberellin (GO:0009739) (Table S2).
Synteny analysis of TaGRFs
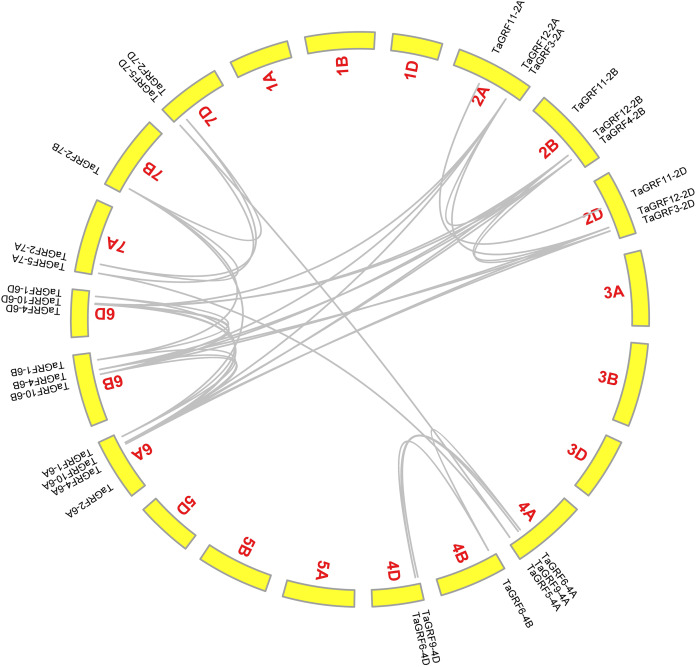
A total of 39 TaGRF gene pairs were probably segmental duplicated genes, including 21 pairs (24 genes) of homoeologous genes, and they distributed on different chromosomes (Fig. 6; Table S5). According to the methodology of Holub (2001), there were no tandem duplication genes in wheat GRF gene family, and most TaGRFs were generated by large-scale repeated events or segmental duplications. It implied that the segmental duplication played an important role in the evolution of the TaGRFs.
Figure 6. Schematic diagram of the chromosome distribution and interchromosome relationships of TaGRFs.
The grey lines indicate probably duplicated TaGRF gene pairs.
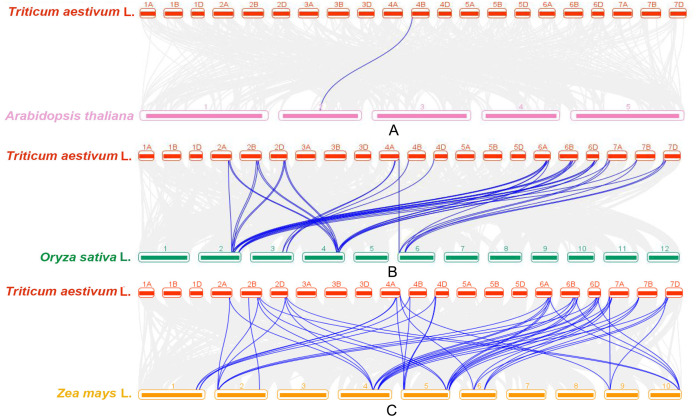
Three comparative syntenic maps of wheat were constructed compared with three representative plant species, including Arabidopsis, rice and maize (Fig. 7). Only one wheat GRF gene, TaGRF6-4B, had a syntenic gene in Arabidopsis (AtGRF1) (Fig. 7A). The numbers of syntenic gene pairs between wheat and rice (Fig. 7B), wheat and maize (Fig. 7C) were 40 and 51 (Table S6). It indicated that the evolutions of GRF genes in wheat, rice and maize were similar.
Figure 7. Synteny analyses of GRFs between wheat and three representative plant species.
(A) The synteny analyses of GRFs between wheat and Arabidopsis. (B) The synteny analyses of GRFs between wheat and rice. (C) The synteny analyses of GRFs between wheat and maize. Gray lines in the background indicate all collinear blocks within wheat and other plant genomes, while the blue lines highlight the syntenic GRF gene pairs.
To better understand the evolutionary constraints acting on TaGRFs, we calculated the Ka/Ks ratios of these syntenic gene pairs. The results showed that the Ka/Ks ratios of most TaGRF syntenic gene pairs were less than 1 (Table S7), suggesting that the TaGRFs might have undergone purifying selection processes.
The expression patterns of TaGRFs in different tissues
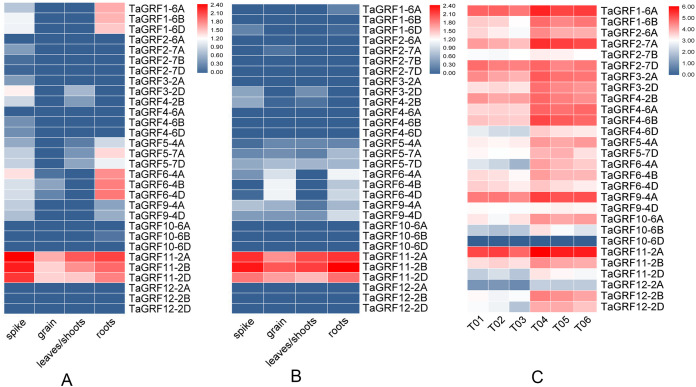
The expression profiles of TaGRFs in spike, grain, leaf/shoot and root showed that about half of 30 TaGRFs had similar expression patterns in Chinese Spring and Azhurnaya, and the others expressed differentially in the four tissues of Chinese Spring (Fig. 8A). The expression patterns of most TaGRFs in Azhurnaya were similar to that in Chinese Spring. However, the expression profiles of TaGRF1, TaGRF6 and TaGRF9 were significantly different between Azhurnaya and Chinese Spring (Fig. 8B). The expression levels of all the three homoeologous genes of TaGRF11-2(A, B, D) in the four tissues were high, while the expressions of TaGRF2, TaGRF4, TaGRF10 and TaGRF12 were not detected or their levels were very low, implying they played no important roles in the development of the four tissues. TaGRF1, TaGRF5, TaGRF6 and TaGRF9 expressed highly in spike and root, implying their roles in regulating spike and root development. Tissue-specific expression profiles of some TaGRFs between Azhurnaya and Chinese Spring were slightly different, which was probably caused by genotype, sample, and experiment variations. These results can be used as a reference for functional studies of TaGRFs.
Figure 8. Expression profiles of the TaGRF genes in different wheat tissues.
(A) Heatmap of expression profiles of TaGRFs in Chinese Spring (CS). (B) Heatmap of expression profiles of TaGRFs in Azhurnaya. (C) Heatmap of TaGRF genes in tiller primordia of WT and dmc based on transcriptome data. Three biological replicates were set up in the mutant dmc (T01, T02 and T03) and WT (T04, T05 and T06), and each sample bulk of tiller primordia included more than 10 independent individuals. Note: Blue: Low expression level; Red: High expression level; The gene expression values present as log2-transformed normalized TPM values.
Expressions of TaGRFs in TPs of the mutant dmc
The expression patterns of all 30 TaGRFs were investigated using available transcriptome data (Fig. 8C). Among the 30 TaGRFs, TaGRF1-6D and TaGRF5-7A had not been detected, TaGRF10-6D was almost not expressed, and TaGRF12-2A expressed very lowly in all detected samples, these four genes probably were not necessary for wheat tiller development (Fig. 8C). TaGRF1-6A (FPKM > 20 in dmc), TaGRF9-4A and TaGRF11-2A (FPKM > 24 in dmc) expressed relatively highly indicated their basic functions in wheat tiller development. All TaGRFs expressed at a reduced level in dmc than in WT. Compared to WT, the expression level of TaGRF6-4A decreased by 66.2%, TaGRF12-2B decreased by 57.8%, and other 15 TaGRFs decreased by more than 40% in dmc. This indicated that the expressions of all the TaGRFs played positive roles at early tillering stage, the constrained tillering of the dmc was associated with the lower expression levels of TaGRFs.
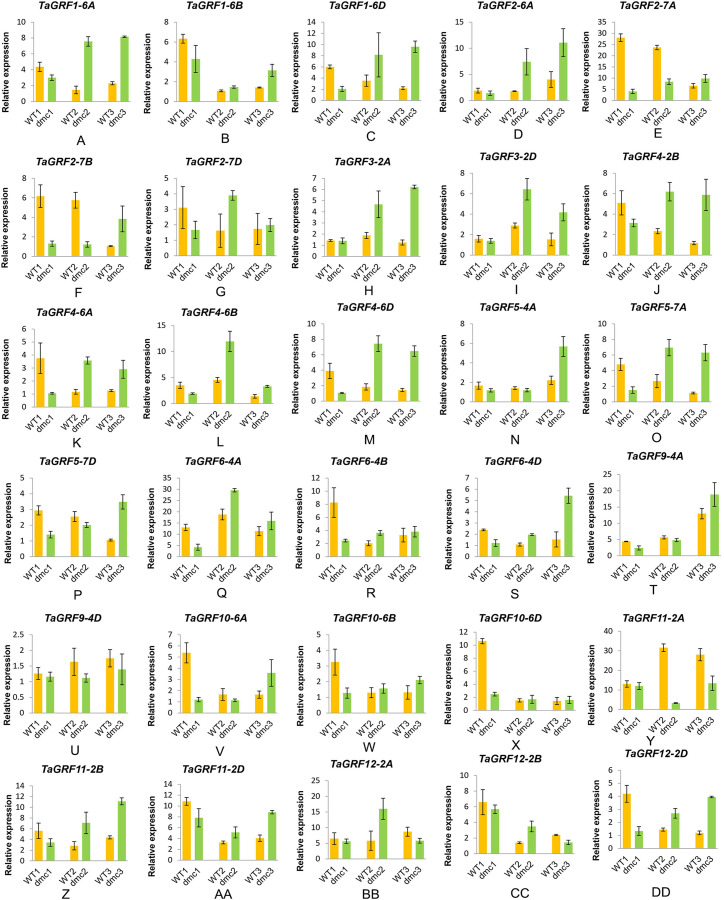
qRT-PCR was performed to analyze the expression patterns of TaGRFs in the TPs of WT and mutant dmc at three tiller developmental stages (Fig. 9). The 30 TaGRF genes had various expression patterns at three tillering stages. All the 30 TaGRFs expressed at a reduced level in dmc at the three-leaf stage, which was consistent with the transcriptome data. A total of 20 TaGRFs expressed highly in dmc at the over-winter stage and the rising to jointing stage, while TaGRF12-2A and TaGRF12-2B expressed lowly in dmc at the rising to jointing stage. Among these genes, TaGRF1-6D, TaGRF2-7D, TaGRF4-6A, TaGRF4-6D, TaGRF5-7A, TaGRF6-4B, TaGRF6-4D and TaGRF12-2D expressed significantly differentially in dmc and WT at wheat tillering stages, implying their important roles in the abnormal tillering in dmc. The expression levels of TaGRF2-7A, TaGRF2-7B, TaGRF5-4A, TaGRF5-7D, TaGRF9-4A, TaGRF9-4D, TaGRF10-6A and TaGRF11-2A were always lower in dmc at two tillering stages, the expression levels of TaGRF2-7A, TaGRF2-7B and TaGRF11-2A in dmc were significantly different from that in WT. TaGRF9-4D and TaGRF11-2A expressed lowly in dmc compared to that in WT at each stage, implying they might affect the tillering of dmc. There were 15 TaGRFs expressed at an upward trend in dmc. TaGRF2-6A, TaGRF9-4A and TaGRF9-4D expressed at an upward trend in WT, implying their roles in regulating tiller development. TaGRF10-6D and TaGRF12-2B expressed at a downward trend in dmc. The expression patterns of TaGRF genes were complex. In summary, the abnormal expressions of TaGRF2-7(A, B, D), TaGRF5-7D, TaGRF10-6(A, B, D) and TaGRF11-2A were major causes for constraining the tillering of dmc.
Figure 9. Expression analysis of 30 TaGRF genes at three tillering stages by qRT-PCR.
(A–DD) The qRT-PCR results of 30 TaGRF genes in the tiller primordia of WT and dmc at three tillering stages. WT1, dmc1: the three-leaf stage; WT2, dmc2: the over-winter stage; WT3, dmc3: the rising to jointing stage. Data were normalized to β-actin gene and vertical bars indicated standard deviation.
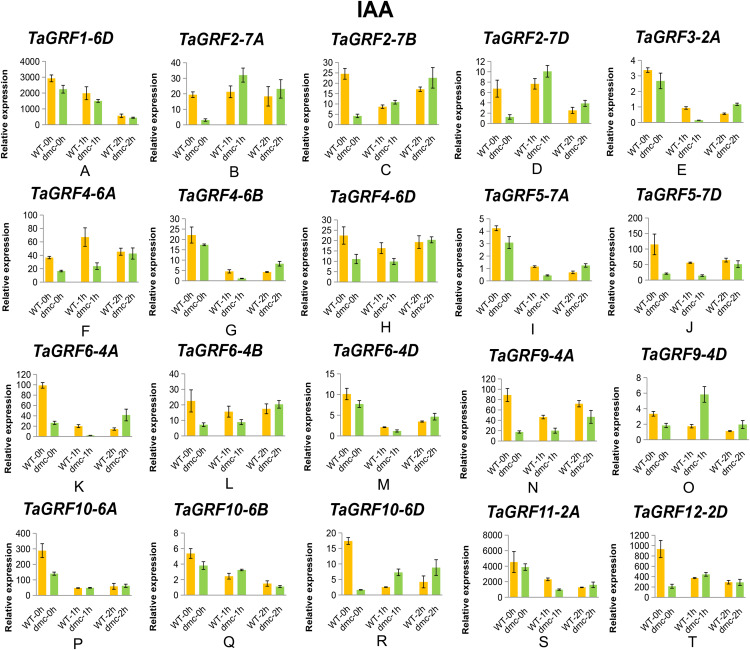
Expression patterns of TaGRFs in response to IAA and GA treatments
A total of 20 representative TaGRFs were selected from the 30 TaGRF genes to investigate whether their expressions were affected by IAA and GA treatments. These TaGRF genes were carefully selected based on the cis-acting elements in their promoters (Fig. 5) and their expression levels in tillers of dmc (Fig. 9). The expressions of 20 TaGRFs were affected by IAA treatment in various degrees (Fig. 10). The expressions of 9 TaGRFs in WT and 2 TaGRFs (TaGRF1-6D and TaGRF10-6B) in dmc were continuously significantly repressed by IAA treatment, their expressions decreased by more than 50%. Among these genes, there was a cis-acting element involving in auxin (AuxRR-core, TGA-element) in the 2,000 bp upstream sequences before transcription start positions of TaGRF1-6D, TaGRF3-2A, TaGRF4-6B, TaGRF6-4A, TaGRF9-4D and TaGRF12-2D. The expression levels of 8 TaGRFs in Guomai 301 and 9 TaGRFs in dmc was down-regulated at 1 hour and up-regulated at 2 h after IAA treatment. There were 2 and 3 cis-acting elements involving in auxin in the 2,000 bp upstream sequences before transcription start positions of TaGRF4-6D and TaGRF5-7D, respectively. Therefore, the expressions of most TaGRFs were repressed by IAA at 1 h after treatment. The expression levels of TaGRF4-6A, TaGRF4-6D, TaGRF6-4B and TaGRF10-6A were almost the same in WT and dmc at 2 h after IAA treatment. The expressions of TaGRF2-7B, TaGRF4-6A, TaGRF6-4B, TaGRF9-4A and TaGRF10-6D in dmc were significantly up-regulated by IAA treatment, which suggested these TaGRFs played key roles in regulating wheat tillering.
Figure 10. Expression profiles of 20 representative TaGRF genes in response to IAA treatment.
(A–T) The qRT-PCR results of 20 TaGRF genes in response to IAA treatment. Data were normalized to β-actin gene and vertical bars indicated standard deviation.
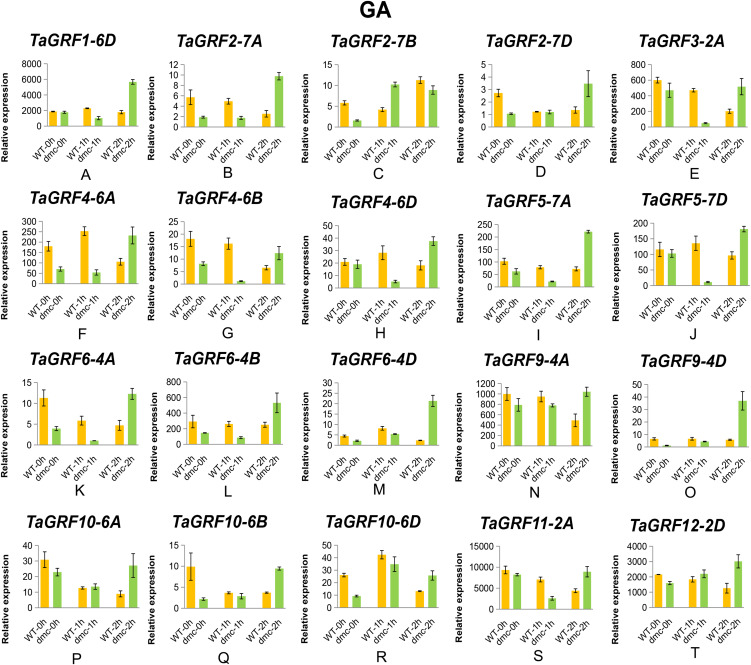
The expressions of the 20 TaGRFs were also affected by GA treatment in various degrees (Fig. 11). It was worth mentioning that most of the homoeologous genes showed similar expression patterns in response to GA treatment, such as TaGRF4, TaGRF5, TaGRF6, TaGRF9 and TaGRF10. The expressions of TaGRF1-6D, TaGRF6-4B and TaGRF9-4D in WT were hardly affected by GA treatment, while they were significantly up-regulated in dmc at 2 h after GA treatment. There were 10 TaGRFs in dmc were significantly repressed at 1 h and significantly up-regulated at 2 h after GA treatment. The expressions of all the detected TaGRFs were significantly up-regulated in dmc after GA treatment. There were 1–3 cis-acting elements (indicated in the parentheses behind the gene symbols) involving in gibberellin (P-box) in the 2,000 bp upstream sequences before transcription start positions of TaGRF5-7A (1), TaGRF6-4A (3), TaGRF6-4B (2), TaGRF6-4D (2), TaGRF9-4D (1) and TaGRF10-6D (1), respectively. The expressions of all TaGRFs were affected by both IAA and GA treatments indicated that phytohormone IAA and GA were involved in regulating wheat tillering.
Figure 11. Expression profiles of 20 representative TaGRF genes in response to GA treatment.
(A–T) The qRT-PCR results of 20 TaGRF genes in response to GA treatment. Data were normalized to β-actin gene and vertical bars indicated standard deviation.
Discussion
Characteristics and evolution of wheat GRF gene family members
Plant GRF proteins typically have two conserved domains, QLQ and WRC, in the N-terminal regions. The QLQ domain interacts with GRF-interacting factors (GIFs) to form transcription activating factor that participates in the biological processes of plant growth and development (Kim & Kende, 2004). The WRC domain interacts with the cis-acting regions of its downstream genes and plays the biological function of GRF. The genome-wide analyses of GRF gene families have been widely carried out in various plant species whose genomes have been sequenced (Cao et al., 2016). In this study, a total of 30 wheat GRF proteins with the two conserved domains, QLQ and WRC, were identified. Phylogenetic and gene structure analyses showed that most of the GRF genes in the same subfamily had similar exon/intron structures, which provided clues to the evolutionary relationships of plant GRFs (Hu & Liu, 2011). These data indicated that the GRF genes with similar structures have similar evolution histories and functions (Babenko et al., 2004; Roy & Penny, 2007). Here, we found wheat homoeologous genes of TaGRF3-2(A, B, D) and TaGRF10-6(A, B, D) were highly homologous to OsGRF3 and OsGRF10. The distributions of the introns and exons in TaGRF genes were similar to those of rice GRF genes (Cao et al., 2016), suggesting that TaGRF genes and OsGRF genes might have similar functions. The gene duplication events are usually derived from the polyploidization or tandem and segmental duplication, segmental duplication occurs most frequently in plants because most plant species are diploidized polyploids (Moore & Purugganan, 2005; Adams & Wendel, 2005). No tandem repeat genes were found in TaGRFs, there were 24 homoeologous genes in 27 segmental duplication GRF genes, and not every GRF had three homoeologous genes on the homoeologous chromosomes A, B and D. It had been reported that some homoeologous genes could be lost during the polyploidization of the genome (Lynch & Force, 2000). Hence, it was highly possible that the segmental duplication played an important role in the expansion of the TaGRFs. A large number of cis-acting elements related to growth and development, hormones and stress regulation were found in the promoter regions of TaGRF genes, which implied their various functions.
Various functions of TaGRFs
Plant GRFs regulate growth and development. AtGRF5 stimulates Arabidopsis chloroplast division, photosynthesis and leaf longevity (Vercruyssen et al., 2015). Overexpression of ZmGRF10 can reduce leaf size and plant height of maize (Wu et al., 2014). OsGRF1 not only regulates growth at the juvenile stage, but also regulates heading in rice (Luo et al., 2005). AtGRF4 and AtGRF5 are involved in the development of leaf size and leaf senescence in Arabidopsis (Horiguchi, Kim & Tsukaya, 2005; Kim & Lee, 2006). As the target genes of miR396, the expressions of GRF genes are post-transcriptionally down-regulated by miR396 (Hewezi & Baum, 2012; Lee & Kim, 2014). There are six GRF genes (AtGRF1-AtGRF3 and AtGRF7-AtGRF9) are regulated by miR396 (miR396a and miR396b) in Arabidopsis (Liu et al., 2009). Down-regulated expressions of AtGRF genes by miR396 overexpression resulted in Arabidopsis with enhanced leaf adaxial-abaxial defects, narrower leaves, abnormal development of the pistil and compromises the shoot meristem, etc (Jones-Rhoades, Bartel & Bartel, 2006; Rodriguez et al., 2010, 2015; Wang et al., 2011). Overexpression of Ptc-miR396 from Populus trichocarpa in tobacco resulted in altered plant growth and flower development (Baucher et al., 2013). miR396 plays an important role in controlling carpel number and pistil development via regulation of the GRF/GIF complex (Liang et al., 2014). OsmiR396d regulates the expressions of OsGRF genes and participates in the regulation of flower organ development in rice (Liu et al., 2014).
The functional diversity of genes can be predicted by their tissue-specific expression patterns. GRF genes not only have different distributions and molecular structures, but also have different tissue-specific expression patterns. Studies have shown that TaGRF genes are highly expressed in active development tissues or organs, and they are relatively lowly expressed in mature tissues or organs (Choi, Kim & Kende, 2004; Wu, Wang & Zhuang, 2017). TaGRFs express highly in young spikes and roots, which is consistent with the putative function based on the meristem-related cis-acting elements in the promoter regions of TaGRFs. AtGRF1 and AtGRF3 are the most highly expressed genes in Arabidopsis roots, which are essential for root development (Szakasits et al., 2009; Hewezi et al., 2012). Overexpression of AtMIR396a decreased the transcript levels of AtGRF genes and resulted in a shorter root phenotype (Bao et al., 2014). TaGRF11 and its homoeologous genes (TaGRF11-2A, 2B, 2D) were highly expressed in various tissues, indicating they played essential roles in wheat growth and development (Fig. 8). TaGRF1-6A, TaGRF6-4A and their homoeologous genes expressed highly in roots suggesting their functions in root development (Fig. 8). The data indicated that TaGRFs functioned differently during wheat development.
The key TaGRFs involved in tiller development
Plant miR396/GRF is a conservative plant growth regulation module, but there is no definite research report on its involvement in plant branching and tillering (Liebsch & Palatnik, 2020). Overexpressions of OsGRF3 and OsGRF10 reduce formations of tillers and internodes in rice (Kuijt et al., 2014). Therefore, we speculated that the homologous genes of TaGRF3 and TaGRF10 might be related to the growth and development of tiller in wheat. The tiller ability of the transgenic wheat overexpression miR396 is significantly decreased (Song, 2016). Our miRNome and transcriptome integrative analysis about the mutant dmc and WT found that the highly expressed tae-miR396b (T. aestivum microRNA396b) down-regulated the expressions of many TaGRFs (TaGRF1, TaGRF2, TaGRF5, TaGRF9, TaGRF10 and TaGRF12) in dmc during tillering (He et al., 2018; An et al., 2019). It demonstrated that the miR396/GRF regulatory module played a key role in wheat tiller development. Compared with the WT, the expression levels of all TaGRFs in dmc were significantly decreased at early tillering stage, which is positively related to the phenotype of dmc (Fig. 9). All the 30 TaGRFs have different expression patterns in WT and dmc, but only those significantly differentially expressed TaGRFs in TPs are the key tiller development regulators. In this case, TaGRF1-6D, TaGRF2, TaGRF4, TaGRF5-7A, TaGRF11-2B and TaGRF12-2D were significantly differentially expressed at early tillering stage, indicating their important roles in regulating tiller numbers in wheat. In summary, TaGRF2-7(A, B, D), TaGRF5-7D, TaGRF10-6(A, B, D) and TaGRF11-2A were key regulators in wheat tiller growth and development, especially in dmc.
Exogenous IAA and GA affect the expressions of TaGRFs
There is considerable evidence that GRFs play significant roles in regulating plant growth and development, and play a variety of regulatory roles in signal transduction and stress response of plants. There are a large number of cis-acting elements related to hormones in TaGRF promoters, including those related to IAA (AuxRR-core, TGA-element) and GA (P-box). IAA regulates plant morphological formation, such as plant tropism growth, root development and tiller formation (Li et al., 2000; Ren, Dai & Liu, 2012). OsmiR167a represses its targets OsARF12 (auxin response factor, ARF), OsARF17 and OsARF25, to control rice tiller angle by fine-tuning auxin asymmetric distribution in shoots (Li et al., 2020). GA participates in regulation of many physiological processes in wheat including the spike development, stem elongation, plant height and stress responses (Zhang et al., 2007; Pavlista, Santra & Baltensperger, 2013). KNOX (KNOTTED1—like homeobox) proteins contribute to the regulation of meristem maintenance by negatively regulating the production of GA (Sakamoto et al., 2001; Bolduc & Hake, 2009). Repression the activities of KNOX genes is a conserved function of GRFs (Kuijt et al., 2014). These results suggested that the GRF genes positively regulated the production of GAs, and GAs in turn up-regulated GRF gene expressions (Wang et al., 2014). OsGRF1 may play a regulatory role in GA-induced stem elongation (Van der Knaap, Kim & Kende, 2000). GA treatment increases the expressions of some OsGRFs (OsGRF1, 2, 3, 7, 8, 10 and 12) in rice, while the expression of OsGRF9 decreases (Bao et al., 2014). Interactions between exogenous cytokinin and nitrogen treatment can regulate tiller bud growth in winter wheat (Yang et al., 2020). There are many evidences of IAA and GA affect tiller developments (Lee, Foster & Morgan, 1998; Frantz et al., 2004; Liu et al., 2011; Assuero et al., 2012; Cai et al., 2013). In previous studies, we found that the contents of IAA were significantly lower, but the contents of GA were significantly higher in the tiller tissues of dmc (An et al., 2019). The results of qRT-PCR also confirmed that the expressions of TaGRFs were significantly affected by IAA and GA applications (Figs. 10 and 11). According to these data, it was considered that TaGRFs as well as IAA and GA signaling were involved in regulating wheat tiller development.
Conclusions
A total of 30 TaGRF transcription factors with both typical QLQ and WRC conserved domains were identified in wheat. TaGRF genes distribute on 12 of the 21 wheat chromosomes, and their promoter regions have a large number of cis-acting elements related to plant growth and development, hormone signal pathway and stress response. The expression levels of all the TaGRF genes are significantly lower in dmc at the early tillering stage (three-leaf stage), and the expressions also are significantly affected by exogenous IAA and GA. TaGRF1-6A, TaGRF2-7D, TaGRF9-4A and TaGRF11-2A play basic roles in wheat tiller development. The abnormal expressions of TaGRF2-7(A, B, D), TaGRF5-7D, TaGRF10-6(A, B, D) and TaGRF11-2A are major causes constraining the tillering of dmc. TaGRFs and IAA, GA signaling are involved in controlling wheat tillering.
Supplemental Information
Acknowledgments
We are grateful for the assistance by Shangqiu Academy of Agricultural and Forestry Sciences. We thank the National Centre of Engineering and Technological Research of Wheat for the technical support for the cultivations.
Funding Statement
This study was supported by the open project fund of National Key Laboratory of Wheat and Maize Crop Science, Henan Agricultural University (2020) and National Key R&D Program of China (2017YFD0301101). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Contributor Information
Lei Li, Email: lilei@henau.edu.cn.
Jishan Niu, Email: jsniu@henau.edu.cn.
Additional Information and Declarations
Competing Interests
The authors declare that they have no competing interests.
Author Contributions
Jing Zhang conceived and designed the experiments, performed the experiments, analyzed the data, prepared figures and/or tables, and approved the final draft.
Junchang Li conceived and designed the experiments, performed the experiments, analyzed the data, prepared figures and/or tables, and approved the final draft.
Yongjing Ni performed the experiments, authored or reviewed drafts of the paper, selected the mutant, and approved the final draft.
Yumei Jiang performed the experiments, authored or reviewed drafts of the paper, contributed to the field experiments and maintenance of the wheat accessions, and approved the final draft.
Zhixin Jiao performed the experiments, prepared figures and/or tables, helped with sowing and sample preparation, and approved the final draft.
Huijuan Li performed the experiments, prepared figures and/or tables, and approved the final draft.
Ting Wang performed the experiments, prepared figures and/or tables, helped with sowing and sample preparation, and approved the final draft.
Peipei Zhang performed the experiments, prepared figures and/or tables, helped with sowing and sample preparation, and approved the final draft.
Mengyao Han performed the experiments, prepared figures and/or tables, helped with sowing and sample preparation, and approved the final draft.
Lei Li conceived and designed the experiments, authored or reviewed drafts of the paper, and approved the final draft.
Hongjie Liu performed the experiments, authored or reviewed drafts of the paper, selected the mutant, and approved the final draft.
Qiaoyun Li performed the experiments, authored or reviewed drafts of the paper, contributed to the field experiments and maintenance of the wheat accessions, and approved the final draft.
Jishan Niu conceived and designed the experiments, authored or reviewed drafts of the paper, and approved the final draft.
DNA Deposition
The following information was supplied regarding the deposition of DNA sequences:
Raw sequencing reads are available at NCBI under BioProject ID PRJNA670838.
Data Availability
The following information was supplied regarding data availability:
The raw measurements for qRT-PCR are available in the Supplemental File.
References
- Adams & Wendel (2005).Adams K, Wendel JF. Polyploidy and genome evolution in plants. Current Opinion in Plant Biology. 2005;8(2):135–141. doi: 10.1016/j.pbi.2005.01.001. [DOI] [PubMed] [Google Scholar]
- An et al. (2019).An J, Niu H, Ni Y, Jiang Y, Zheng Y, He R, Li J, Jiao Z, Zhang J, Li H, Li Q, Niu J. The miRNA–mRNA networks involving abnormal energy and hormone metabolisms restrict tillering in a wheat mutant dmc. International Journal of Molecular Sciences. 2019;20(18):4586. doi: 10.3390/ijms20184586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Assuero et al. (2012).Assuero SG, Lorenzo M, Pérez Ramírez NM, Velázquez LM, Tognetti JA. Tillering promotion by paclobutrazol in wheat and its relationship with plant carbohydrate status. New Zealand Journal of Agricultural Research. 2012;55(4):347–358. doi: 10.1080/00288233.2012.706223. [DOI] [Google Scholar]
- Babenko et al. (2004).Babenko VN, Rogozin IB, Mekhedov SL, Koonin EV. Prevalence of intron gain over intron loss in the evolution of paralogous gene families. Nucleic Acids Research. 2004;32(12):3724–3733. doi: 10.1093/nar/gkh686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao et al. (2014).Bao M, Bian H, Zha Y, Li F, Sun Y, Bai B, Chen Z, Wang J, Zhu M, Han N. miR396a-mediated basic helix–loop–helix transcription factor bHLH74 repression acts as a regulator for root growth in Arabidopsis seedlings. Plant and Cell Physiology. 2014;55(7):1343–1353. doi: 10.1093/pcp/pcu058. [DOI] [PubMed] [Google Scholar]
- Baucher et al. (2013).Baucher M, Moussawi J, Vandeputte OM, Monteyne D, Mol A, Pérez-Morga D, El Jaziri M. A role for the miR396/GRF network in specification of organ type during flower development, as supported by ectopic expression of Populus trichocarpa miR396c in transgenic tobacco. Plant Biology. 2013;15(5):892–898. doi: 10.1111/j.1438-8677.2012.00696.x. [DOI] [PubMed] [Google Scholar]
- Bolduc & Hake (2009).Bolduc N, Hake S. The maize transcription factor KNOTTED1 directly regulates the gibberellin catabolism gene ga2ox1. Plant Cell. 2009;21(6):1647–1658. doi: 10.1105/tpc.109.068221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai et al. (2013).Cai T, Xu HC, Yin YP, Yang WB, Wang ZL. Mechanisms of tiller occurrence affected by exogenous IAA, GA, and ABA in wheat with different spike-types. Acta Agronomica Sinica. 2013;39(10):1835. doi: 10.3724/SP.J.1006.2013.01835. [DOI] [Google Scholar]
- Cao et al. (2016).Cao Y, Han Y, Jin Q, Lin Y, Cai Y. Comparative genomic analysis of the GRF genes in Chinese pear (Pyrus bretschneideri Rehd), poplar (Populous), grape (Vitis vinifera), Arabidopsis and rice (Oryza sativa) Frontiers in Plant Science. 2016;7(577):1750. doi: 10.3389/fpls.2016.01750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casadevall et al. (2013).Casadevall R, Rodriguez RE, Debernardi JM, Palatnik JF, Casati P. Repression of growth regulating factors by the microRNA396 inhibits cell proliferation by UV-B radiation in Arabidopsis leaves. Plant Cell. 2013;25(9):3570–3583. doi: 10.1105/tpc.113.117473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen & Cao (2015).Chen Y, Cao J. Comparative analysis of dof transcription factor family in maize. Plant Molecular Biology Reporter. 2015;33(5):1245–1258. doi: 10.1007/s11105-014-0835-9. [DOI] [Google Scholar]
- Chen et al. (2020).Chen C, Chen H, Zhang Y, Thomas HR, Frank MH, He Y, Xia R. TBtools: an integrative toolkit developed for interactive analyses of big biological data. Molecular Plant. 2020;13(8):1194–1202. doi: 10.1016/j.molp.2020.06.009. [DOI] [PubMed] [Google Scholar]
- Chen & Rajewsky (2007).Chen K, Rajewsky N. The evolution of gene regulation by transcription factors and microRNAs. Nature Reviews Genetics. 2007;8(2):93–103. doi: 10.1038/nrg1990. [DOI] [PubMed] [Google Scholar]
- Chen et al. (2019).Chen F, Yang Y, Luo X, Zhou W, Dai Y, Zheng C, Liu W, Yang W, Shu K. Genome-wide identification of GRF transcription factors in soybean and expression analysis of GmGRF family under shade stress. BMC Plant Biology. 2019;19(1):269. doi: 10.1186/s12870-019-1861-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi, Kim & Kende (2004).Choi D, Kim JH, Kende H. Whole genome analysis of the OsGRF gene family encoding plant-specific putative transcription activators in rice (Oryza sativa L.) Plant and Cell Physiology. 2004;45(7):897–904. doi: 10.1093/pcp/pch098. [DOI] [PubMed] [Google Scholar]
- Deng et al. (2014).Deng W, Wang Y, Liu Z, Cheng H, Xue Y. HemI: a toolkit for illustrating heatmaps. PLOS ONE. 2014;9(11):e111988. doi: 10.1371/journal.pone.0111988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong et al. (2020).Dong YP, Yang SM, Zhao DG, Song L. Bioinformatics analysis of tobacco TIR-NBS gene family. Guihaia. 2020;40(6):891–900. doi: 10.11931/guihaia.gxzw201909045. [DOI] [Google Scholar]
- Frantz et al. (2004).Frantz JM, Pinnock D, Klassen S, Bugbee B. Characterizing the environmental response of a gibberellic acid-deficient rice for use as a model crop. Agronomy Journal. 2004;96(4):1172–1181. doi: 10.2134/agronj2004.1172. [DOI] [Google Scholar]
- He et al. (2018).He R, Ni Y, Li J, Jiao Y, Zhu X, Jiang Y, Li Q, Niu J. Quantitative changes in the transcription of phytohormone-related genes: some transcription factors are major causes of the wheat mutant dmc not tillering. International Journal of Molecular Sciences. 2018;19(5):1324. doi: 10.3390/ijms19051324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hewezi & Baum (2012).Hewezi T, Baum TJ. Complex feedback regulations govern the expression of miRNA396 and its GRF target genes. Plant Signaling & Behavior. 2012;7(7):749–751. doi: 10.4161/psb.20420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hewezi et al. (2012).Hewezi T, Maier TR, Nettleton D, Baum TJ. The Arabidopsis microRNA396-GRF1/GRF3 regulatory module acts as a developmental regulator in the reprogramming of root cells during cyst nematode infection. Plant Physiology. 2012;159(1):321–335. doi: 10.1104/pp.112.193649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holub (2001).Holub EB. The arms race is ancient history in Arabidopsis, the wildflower. Nature Reviews Genetics. 2001;2(7):516–527. doi: 10.1038/35080508. [DOI] [PubMed] [Google Scholar]
- Horiguchi, Kim & Tsukaya (2005).Horiguchi G, Kim GT, Tsukaya H. The transcription factor AtGRF5 and the transcription coactivator AN3 regulate cell proliferation in leaf primordia of Arabidopsis thaliana. Plant Journal. 2005;43(1):68–78. doi: 10.1111/j.1365-313X.2005.02429.x. [DOI] [PubMed] [Google Scholar]
- Hu & Liu (2011).Hu L, Liu S. Genome-wide identification and phylogenetic analysis of the ERF gene family in cucumbers. Genetics and Molecular Biology. 2011;34(4):624–634. doi: 10.1590/S1415-47572011005000054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones-Rhoades, Bartel & Bartel (2006).Jones-Rhoades MW, Bartel DP, Bartel B. MicroRNAs and their regulatory roles in plants. Annual Review of Plant Biology. 2006;57(1):19–53. doi: 10.1146/annurev.arplant.57.032905.105218. [DOI] [PubMed] [Google Scholar]
- Kim, Choi & Kende (2003).Kim JH, Choi D, Kende H. The AtGRF family of putative transcription factors is involved in leaf and cotyledon growth in Arabidopsis. Plant Journal. 2003;36(1):94–104. doi: 10.1046/j.1365-313X.2003.01862.x. [DOI] [PubMed] [Google Scholar]
- Kim & Kende (2004).Kim JH, Kende H. A transcriptional co-activator, AtGIF1, is involved in regulating leaf growth and morphology in Arabidopsis. Proceedings of the National Academy of Sciences of the United States of America. 2004;101(36):13374–13379. doi: 10.1073/pnas.0405450101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim & Lee (2006).Kim JH, Lee BH. Growth-regulating factor4 of Arabidopsis thaliana is required for development of leaves, cotyledons, and shoot apical meristem. Journal of Plant Biology. 2006;49(6):463–468. doi: 10.1007/BF03031127. [DOI] [Google Scholar]
- Kim & Tsukaya (2015).Kim JH, Tsukaya H. Regulation of plant growth and development by the GROWTH-REGULATING FACTOR and GRF-INTERACTING FACTOR duo. Journal of Experimental Botany. 2015;66(20):6093–6107. doi: 10.1093/jxb/erv349. [DOI] [PubMed] [Google Scholar]
- Kuijt et al. (2014).Kuijt SJH, Greco R, Agalou A, Shao J, Hoen CCJ, Övernäs E, Osnato M, Curiale S, Meynard D, Gulik RV, Maraschin SDF, Meijer AH, Ouwerkerk PBF. Interaction between the GROWTH-REGULATING FACTOR and KNOTTED1-LIKE HOMEOBOX families of transcription factors. Plant physiology. 2014;164(4):1952–1966. doi: 10.1104/pp.113.222836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, Foster & Morgan (1998).Lee IJ, Foster KR, Morgan PW. Effect of gibberellin biosynthesis inhibitors on native gibberellin content, growth and floral initiation in Sorghum bicolor. Journal of Plant Growth Regulation. 1998;17(4):185–195. doi: 10.1007/PL00007034. [DOI] [PubMed] [Google Scholar]
- Lee et al. (2015).Lee BH, Jeon JO, Lee MM, Kim JH. Genetic interaction between GROWTH-REGULATING FACTOR and CUP-SHAPED COTYLEDON in organ separation. Plant Signaling & Behavior. 2015;10(2):e988071. doi: 10.4161/15592324.2014.988071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee & Kim (2014).Lee BH, Kim JH. Spatio-temporal distribution patterns of GRF-INTERACTING FACTOR expression and leaf size control. Plant Signaling & Behavior. 2014;9(9):e29697. doi: 10.4161/psb.29697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li et al. (2020).Li Y, Li J, Chen Z, Wei Y, Qi Y, Wu C. OsmiR167a-targeted auxin response factors modulate tiller angle via fine-tuning auxin distribution in rice. Plant Biotechnology Journal. 2020;18(10):2015–2026. doi: 10.1111/pbi.13360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li et al. (2014).Li Q-Y, Qin Z, Jiang Y-M, Shen C-C, Duan Z-B, Niu J-S. Screening wheat genotypes for resistance to black point and the effects of diseased kernels on seed germination. Journal of Plant Diseases and Protection. 2014;121(2):79–88. doi: 10.1007/BF03356495. [DOI] [Google Scholar]
- Li et al. (2019).Li J, Zhang J, Li H, Niu H, Xu Q, Jiao Z, An J, Jiang Y, Li Q, Niu J. The major factors causing the microspore abortion of genic male sterile mutant NWMS1 in wheat (Triticum aestivum L.) International Journal of Molecular Sciences. 2019;20(24):6252. doi: 10.3390/ijms20246252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li et al. (2000).Li CX, Zhao GC, Dai XM, Jiang LN, Shang YL. Research on the relationship between wheat tillering dynamics and endogenous hormone. Acta Agronomica Sinica. 2000;6:963–968. [Google Scholar]
- Liang et al. (2014).Liang G, He H, Li Y, Wang F, Wu D. Molecular mechanism of microRNA396 mediating pistil development in Arabidopsis. Plant Physiology. 2014;164(1):249–258. doi: 10.1104/pp.113.225144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liebsch & Palatnik (2020).Liebsch D, Palatnik J. MicroRNA miR396, GRF transcription factors and GIF co-regulators: a conserved plant growth regulatory module with potential for breeding and biotechnology. Current Opinion in Plant Biology. 2020;53:31–42. doi: 10.1016/j.pbi.2019.09.008. [DOI] [PubMed] [Google Scholar]
- Liu et al. (2011).Liu Y, Gu D, Ding Y, Wang Q, Li G, Wang S. The relationship between nitrogen, auxin and cytokinin in the growth regulation of rice (Oryza sativa L.) tiller buds. Australian Journal of Crop Science. 2011;5(4):1019. doi: 10.1111/j.1439-0523.2010.01842.x. [DOI] [Google Scholar]
- Liu et al. (2014).Liu H, Guo S, Xu Y, Li C, Zhang Z, Zhang D, Xu S, Zhang C, Chong K. OsmiR396d-regulated OsGRFs function in floral organogenesis in rice through binding to their targets OsJMJ706 and OsCR4. Plant Physiology. 2014;165(1):160–174. doi: 10.1104/pp.114.235564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu et al. (2012).Liu J, Hua W, Yang HL, Zhan GM, Li RJ, Deng LB, Wang XF, Liu GH, Wang HZ. The BnGRF2 gene (GRF2-like gene from Brassica napus) enhances seed oil production through regulating cell number and plant photosynthesis. Journal of Experimental Botany. 2012;63(10):3727–3740. doi: 10.1093/jxb/ers066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu et al. (2009).Liu D, Song Y, Chen Z, Yu D. Ectopic expression of miR396 suppresses GRF target gene expression and alters leaf growth in Arabidopsis. Physiologia Plantarum. 2009;136(2):223–236. doi: 10.1111/j.1399-3054.2009.01229.x. [DOI] [PubMed] [Google Scholar]
- Liu et al. (2008).Liu HH, Tian X, Li YJ, Wu CA, Zheng CC. Microarray-based analysis of stress-regulated microRNAs in Arabidopsis thaliana. RNA. 2008;14(5):836–843. doi: 10.1261/rna.895308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu et al. (2015).Liu Q, Wang FD, Zhang YH, Qiu NW, Gao JW. Reaserch progress of plant growth-regulating factor. Plant Physiology Journal. 2015;51:1775–1779. doi: 10.13592/j.cnki.ppj.2015.0288. [DOI] [Google Scholar]
- Livak & Schmittgen (2001).Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods. 2001;25(4):402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Luo et al. (2005).Luo AD, Liu L, Tang ZS, Bai XQ, Cao SY, Chu CC. Down-regulation of OsGRF1 gene in rice rhd1 mutant results in reduced heading date. Journal of Integrative Plant Biology. 2005;47(6):745–752. doi: 10.1111/j.1744-7909.2005.00071.x. [DOI] [Google Scholar]
- Lynch & Force (2000).Lynch M, Force A. The probability of duplicate gene preservation by subfunctionalization. Genetica. 2000;108(3):285–295. doi: 10.1023/A:1004191625603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma et al. (2017).Ma C, Yuan JL, Zhang S, Jia QS, Feng YL. The molecular mechanisms of growth-regulating factors (GRFs) in plant growth, development and stress response. Journal of Nuclear Agricultural Sciences. 2017;31:2145–2153. doi: 10.11869/j.issn.100-8551.2017.11.2145. [DOI] [Google Scholar]
- Moore & Purugganan (2005).Moore RC, Purugganan MD. The evolutionary dynamics of plant duplicate genes. Current Opinion in Plant Biology. 2005;8(2):122–128. doi: 10.1016/j.pbi.2004.12.001. [DOI] [PubMed] [Google Scholar]
- Omidbakhshfard et al. (2015).Omidbakhshfard MA, Proost S, Fujikura U, Mueller-Roeber B. Growth-Regulating Factors (GRFs): a small transcription factor family with important functions in plant biology. Molecular Plant. 2015;8(7):998–1010. doi: 10.1016/j.molp.2015.01.013. [DOI] [PubMed] [Google Scholar]
- Pavlista, Santra & Baltensperger (2013).Pavlista AD, Santra DK, Baltensperger DD. Bioassay of winter wheat for gibberellic acid sensitivity. American Journal of Plant Sciences. 2013;4(10):2015–2022. doi: 10.4236/ajps.2013.410252. [DOI] [Google Scholar]
- Ren, Dai & Liu (2012).Ren Y, Dai S, Liu W. Auxin transport and its roles in signal transduction and plant development. Biotechnology Bulletin. 2012;29:9–16. [Google Scholar]
- Rodriguez et al. (2015).Rodriguez RE, Ercoli MF, Debernardi JM, Breakfield NW, Mecchia MA, Sabatini M, Cools T, Veylder LD, Benfey P, Palatnik JF. MicroRNA miR396 regulates the switch between stem cells and transit-amplifying cells in Arabidopsis roots. Plant Cell. 2015;27(12):3354–3366. doi: 10.1105/tpc.15.00452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez et al. (2010).Rodriguez RE, Mecchia MA, Debernardi JM, Schommer C, Weigel D, Palatnik JF. Control of cell proliferation in Arabidopsis thaliana by microRNA miR396. Development. 2010;137(1):103–112. doi: 10.1242/dev.043067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy & Penny (2007).Roy SW, Penny D. On the incidence of intron loss and gain in paralogous gene families. Molecular Biology and Evolution. 2007;24(8):1579–1581. doi: 10.1093/molbev/msm082. [DOI] [PubMed] [Google Scholar]
- Ruan et al. (2018).Ruan X, Wang J, Liu H, Zhao J. Identification and basic characteristic analysis of GRF gene family in Brassica napus L. Molecular Plant Breeding. 2018;16:2420–2428. [Google Scholar]
- Sakamoto et al. (2001).Sakamoto T, Kamiya N, Ueguchi-Tanaka M, Iwahori S, Matsuoka M. KNOX homeodomain protein directly suppresses the expression of a gibberellin biosynthetic gene in the tobacco shoot apical meristem. Genes & Development. 2001;15(5):581–590. doi: 10.1101/gad.867901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi et al. (2019).Shi P, He B, Fei Y, Wang J, Wang W, Wei F, Lu Y, Gu M. Identification and expression analysis of GRF transcription factor family of Chenopodium quinoa. Acta Agronomica Sinica. 2019;45(5):1841–1850. doi: 10.3724/SP.J.1006.2019.84084. [DOI] [Google Scholar]
- Song (2016).Song L. Transformation of gene MiR396 into wheat varieties by microprojectile bombardment and Agrobacterium tumefaciens infection. 2016. Thesis, Huazhong Agricultural University.
- Sun et al. (2016).Sun P, Zhang W, Wang Y, He Q, Shu F, Liu H, Wang J, Wang J, Yuan L, Deng H. OsGRF4 controls grain shape, panicle length and seed shattering in rice. Journal of Integrative Plant Biology. 2016;58(10):836–847. doi: 10.1111/jipb.12473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szakasits et al. (2009).Szakasits D, Heinen P, Wieczorek K, Hofmann J, Wagner F, Kreil DP, Sykacek P, Grundler FMW, Bohlmann H. The transcriptome of syncytia induced by the cyst nematode Heterodera schachtii in Arabidopsis roots. Plant Journat. 2009;57(5):771–784. doi: 10.1111/j.1365-313X.2008.03727.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van der Knaap, Kim & Kende (2000).Van der Knaap E, Kim JH, Kende KH. A novel gibberellin-induced gene from rice and its potential regulatory role in stem growth. Plant Physiology. 2000;122(3):695–704. doi: 10.1104/pp.122.3.695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vercruyssen et al. (2015).Vercruyssen L, Tognetti VB, Gonzalez N, Dingenen JV, Milde LD, Bielach A, Rycke RD, Breusegem FV, Inzé D. GROWTH REGULATING FACTOR5 stimulates Arabidopsis chloroplast division, photosynthesis, and leaf longevity. Plant Physiology. 2015;167(3):817–832. doi: 10.1104/pp.114.256180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang et al. (2011).Wang L, Gu X, Xu D, Wang W, Wang H, Zeng M, Chang Z, Huang H, Cui X. miR396-targeted AtGRF transcription factors are required for coordination of cell division and differentiation during leaf development in Arabidopsis. Journal of Experimental Botany. 2011;62(2):761–773. doi: 10.1093/jxb/erq307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang et al. (2014).Wang F, Qiu N, Ding Q, Li J, Zhang Y, Li H, Gao J. Genome-wide identification and analysis of the growth-regulating factor family in Chinese cabbage (Brassica rapa L. ssp. pekinensis) BMC genomics. 2014;15(1):807. doi: 10.1186/1471-2164-15-807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang et al. (2019a).Wang Z, Shi H, Yu S, Zhou W, Li J, Liu S, Deng M, Ma J, Wei Y, Zheng Y, Liu Y. Comprehensive transcriptomics, proteomics, and metabolomics analyses of the mechanisms regulating tiller production in low-tillering wheat. Theoretical and Applied Genetics. 2019a;132(8):2181–2193. doi: 10.1007/s00122-019-03345-w. [DOI] [PubMed] [Google Scholar]
- Wang et al. (2019b).Wang P, Zheng Y, Yi L, Zhou Z, Yang J, Ye N. Genome-wide identification and expression analysis of GRF gene family in Camellia sinensis. Acta Botanica Boreali-Occidentalia Sinica. 2019b;39:0413–0421. doi: 10.7606/j.issn.1000-4025.2019.03.0413. [DOI] [Google Scholar]
- Wu, Wang & Zhuang (2017).Wu Z, Wang W, Zhuang J. Developmental processes and responses to hormonal stimuli in tea plant (Camellia sinensis) leaves are controlled by GRF and GIF gene families. Functional & Integrative Genomics. 2017;17(5):503–512. doi: 10.1007/s10142-017-0553-0. [DOI] [PubMed] [Google Scholar]
- Wu et al. (2014).Wu L, Zhang D, Xue M, Qian J, He Y, Wang S. Overexpression of the maize GRF10, an endogenous truncated growth-regulating factor protein, leads to reduction in leaf size and plant height. Journal of Integrative Plant Biology. 2014;56(11):1053–1063. doi: 10.1111/jipb.12220. [DOI] [PubMed] [Google Scholar]
- Xue et al. (2020).Xue Z, Wang S, Yang Y, Gao Z, Feng H. Genome -wide identification and bio-informatic analysis of growth regulating factor (GRF) family in barley. 2020. http://kns.cnki.net/kcms/detail/46.1068.S.20200326.1152.006.html http://kns.cnki.net/kcms/detail/46.1068.S.20200326.1152.006.html
- Yang et al. (2020).Yang D, Luo Y, Kong X, Huang C, Wang Z. Interactions between exogenous cytokinin and nitrogen application regulate tiller bud growth via sucrose and nitrogen allocation in winter wheat. Journal of Plant Growth Regulation. 2020;40(1):329–341. doi: 10.1007/s00344-020-10106-3. [DOI] [Google Scholar]
- Yuan et al. (2017).Yuan Q, Zhang C, Zao T, Xu X. Research advances of GRF transcription factor in plant. Genomics and Applied Biology. 2017;36:3145–3151. doi: 10.13417/j.gab.036.003145. [DOI] [Google Scholar]
- Zhang et al. (2007).Zhang Y, Ni Z, Yao Y, Nie X, Sun Q. Gibberellins and heterosis of plant height in wheat (Triticum aestivum L.) BMC Genetics. 2007;8(1):40. doi: 10.1186/1471-2156-8-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao et al. (2019).Zhao KK, Fan R, Liu Y, Shi Y, Jiang M, Chen Q. Genome wide identificaton and expression analysis of GRF gene family in Gossypium barbadense. Journal of Xinjiang Agicultural University. 2019;2:151–160. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The following information was supplied regarding data availability:
The raw measurements for qRT-PCR are available in the Supplemental File.