Abstract
Clonal haematopoiesis, which is highly prevalent in older individuals, arises from somatic mutations that endow a proliferative advantage to haematopoietic cells. Clonal haematopoiesis increases the risk of myocardial infarction and stroke independently of traditional risk factors1. Among the common genetic variants that give rise to clonal haematopoiesis, the JAK2V617F (JAK2VF) mutation, which increases JAK–STAT signalling, occurs at a younger age and imparts the strongest risk of premature coronary heart disease1,2. Here we show increased proliferation of macrophages and prominent formation of necrotic cores in atherosclerotic lesions in mice that express Jak2VF selectively in macrophages, and in chimeric mice that model clonal haematopoiesis. Deletion of the essential inflammasome components caspase 1 and 11, or of the pyroptosis executioner gasdermin D, reversed these adverse changes. Jak2VF lesions showed increased expression of AIM2, oxidative DNA damage and DNA replication stress, and Aim2 deficiency reduced atherosclerosis. Single-cell RNA sequencing analysis of Jak2VF lesions revealed a landscape that was enriched for inflammatory myeloid cells, which were suppressed by deletion of Gsdmd. Inhibition of the inflammasome product interleukin-1β reduced macrophage proliferation and necrotic formation while increasing the thickness of fibrous caps, indicating that it stabilized plaques. Our findings suggest that increased proliferation and glycolytic metabolism in Jak2VF macrophages lead to DNA replication stress and activation of the AIM2 inflammasome, thereby aggravating atherosclerosis. Precise application of therapies that target interleukin-1β or specific inflammasomes according to clonal haematopoiesis status could substantially reduce cardiovascular risk.
Atherosclerotic cardiovascular disease (ACVD) is the major cause of death and disability in the developed world3. A large burden of residual ACVD risk remains despite current therapies, including intensive lowering of low-density lipoprotein levels3, which highlights the need for new treatments. In the Canakinumab Antiinflammatory Thrombosis Outcomes Study (CANTOS), inhibition of IL-1β reduced cardiovascular events, thereby validating the contribution of inflammation to ACVD4. However, canakinumab therapy was associated with a small risk of infections and has not been approved for cardiovascular conditions. Thus, a more precise way to identify patients who may benefit most from anti-inflammatory therapy is required. Clonal haematopoiesis usually arises from somatic mutations in haematopoietic stem and progenitor cells (HSPCs) in one of four genes (TET2, ASXL1, DNMT3A or JAK2), which lead to clonal expansion of haematopoietic cells. The prevalence of clonal haematopoiesis increases with age, and it affects more than 10% of people who are over 70 years old1. Although clonal haematopoiesis conferred an increased risk of haematological malignancies of 0.5–1% per year, this modest increase was not nearly enough to account for the 40% increase in mortality, which was mainly due to ACVD1. Although studies in mouse models of Tet2−/− clonal haematopoiesis have shown an increase in atherosclerosis and increased macrophage inflammation, insights into the underlying mechanisms remain limited1,5. It is unclear whether different variants of clonal haematopoiesis increase atherosclerosis through a common mechanism, and the potential for using clinically relevant anti-IL-1 therapies to treat clonal haematopoiesis has not been assessed.
JAK2VF is a gain-of-function mutation that is associated with myeloproliferative neoplasms (MPNs) and an increased risk of atherothrombotic disease6. Although JAK2VF-associated MPNs are uncommon, the JAK2VF mutation was found in 3.1% of a general European population, most of whom did not have features of MPNs7. Hyperlipidaemic mice in which all haematopoietic cells contain heterozygousJak2VF mutations, thus modelling MPN, have increased atherosclerosis, abnormalities in multiple cell lineages and inflammasome activation in splenocytes8. However, the mechanisms that link these changes to atherosclerosis, the specific role of the macrophage inflammasome, and the relevance of these findings to the much more prevalent milder form of clonal haematopoiesis and its therapeutic targeting are unclear.
Atherosclerosis in Jak2VF mice
To gain insights into these mechanisms, we used an Mx1-driven Cre recombinase to generate mice expressing Jak2VF (ref. 9); this recombinase induces expression of the transgene in HSPCs, thereby modelling the acquisition of mutations in human clonal haematopoiesis. We performed RNA sequencing (RNA-seq) on CD11b+ splenic myeloid cells from wild-type or Ldlr−/− mice following transplantation of wild-type or Mx1-cre Jak2VF bone marrow. Gene ontology analysis showed that expression of Jak2VF was associated with enrichment for genes involved in cellular proliferation, DNA damage repair, and metabolic pathways (Extended Data Fig. 1a, b). A hallmark of the early stage of atherosclerosis is the accumulation of macrophages derived from blood monocytes. Local proliferation of macrophages may sustain the macrophage population in advanced atherosclerotic lesions10. To determine whether macrophage proliferation was increased in atherosclerotic lesions, we generated transgenic mice in which the expression of Jak2VF was controlled by tamoxifen-inducible, macrophage-specific Cx3cr1-Cre (Cx3cr1-cre Jak2VF). After 15 weeks on a Western-type diet, tamoxifen-treated Ldlr−/− mice transplanted with Cx3cr1-cre Jak2VF bone marrow displayed increased lesion areas, proliferation of macrophages and formation of necrotic cores (Fig. 1a-d, Extended Data Fig. 2a). The Jak2VF allele burden increased in blood monocytes without any change in blood cell counts or plasma cholesterol (Extended Data Fig. 2b, c). By contrast, S100A8-cre Jak2VF mice, in which Jak2VF expression is increased specifically in neutrophils (Extended Data Fig. 2d, e), showed no changes in plasma cholesterol, plaque area or morphology (Extended Data Fig. 2f-k). These findings indicate that expression of Jak2VF specifically in macrophages drives their proliferation and fosters the formation of necrotic cores.
Fig. 1 ∣. Increased atherosclerosis in mice with macrophage Jak2VF expression and Jak2VF clonal haematopoiesis.
a, Immunofluorescence staining of aortic root plaques from mice with Cx3cr1-cre Jak2VF bone marrow. White arrows, Ki67+ nuclei. b–d, Quantification of lesion area (b), percentage of macrophage nuclei that were Ki67+ (c; n = 18 control, n = 21 Jak2VF mice) and necrotic core normalized to lesion area (d; n = 17 control, n = 21 Jak2VF mice). e, Representative haematoxylin and eosin (H&E) images of aortic root lesions from mice with Jak2VF clonal haematopoiesis. Scale bar, 200 μm. f, Lesion area for mice in e (n = 19 mice). g, Immunofluorescence staining of aortic roots. Arrows, Ki67+ nuclei; yellow dashed lines, lesions. Scale bar, 50 μm. h, Increased CD45.2 cells in Jak2VF lesions relative to blood monocytes (n = 18 control, n = 15 Jak2VF mice). i, Percentage Ki67+ CD45.1 or CD45.2 cells in lesions (n = 18 control, n = 14 Jak2VF mice). Mean ± s.e.m.; two-tailed t-test (b–d, f), two-way ANOVA with Bonferroni multiple comparison (h, i).
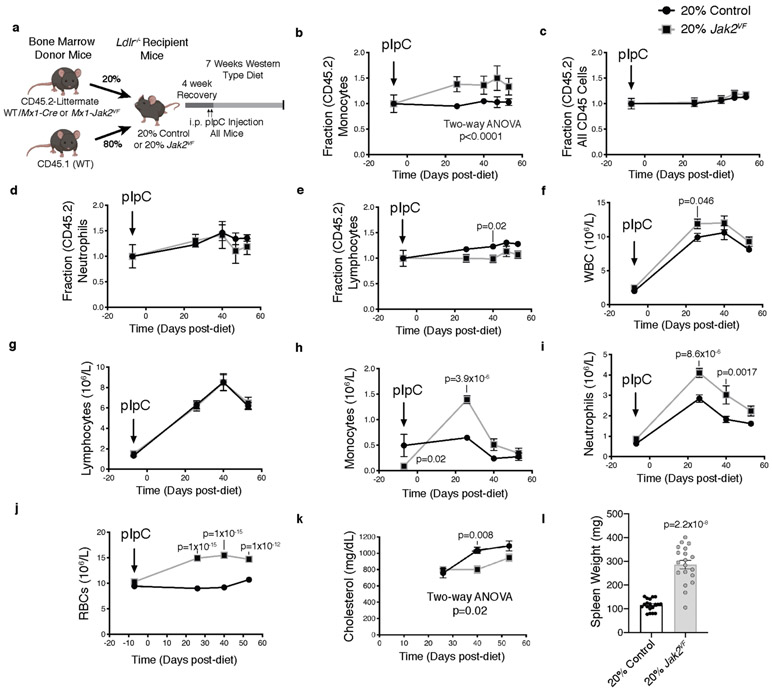
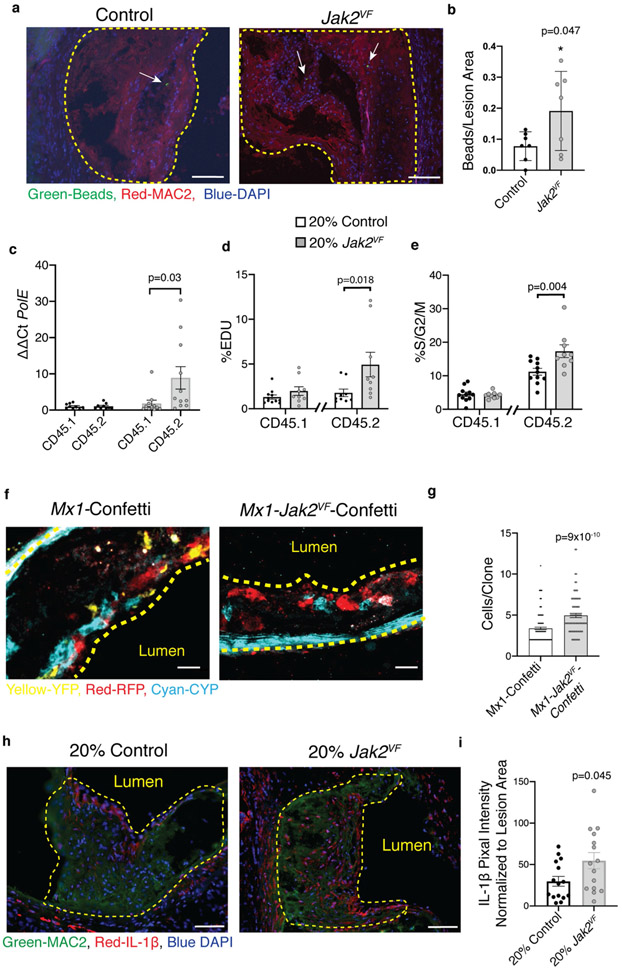
To study the link between CVD risk and Jak2VF-clonal haematopoiesis, we generated chimeric Ldlr−/− mice with a mixture of bone marrow from Mx1-cre Jak2VF mice (CD45.2+, 20%) and wild-type mice (CD45.1, 80%) or from CD45.2 wild-type mice (20%) and CD45.1 wild-type mice (80%) (Extended Data Fig. 3a). The Jak2VF burden in blood cells, blood cell counts, and plasma cholesterol did not change substantially during the study (Extended Data Fig. 3b-l). Jak2VF chimeric mice displayed a twofold increase in lesion area (Fig. 1e, f). Although Jak2VF monocytes made up only about 30% of the circulating population, Jak2VF macrophages comprised about 60% of aortic root macrophages (Fig. 1g, h). In part this may reflect increased expression of integrins in Jak2VF blood leukocytes11 leading to increased binding to endothelium over plaques8; indeed, bead trapping studies demonstrated increased entry of Jak2VF monocytes into lesions (Extended Data Fig. 4a, b). However, Jak2VF (CD45.2+) macrophages exhibited increased markers of proliferation, indicating that macrophage replication in plaques also contributes to the enrichment of Jak2VF macrophages (Fig. 1i, Extended Data Fig. 4c). Notably, CD45.1 macrophages within the same lesions, which encountered a similar milieu to CD45.2+ (Jak2VF) cells, showed no increase in proliferation. Flow cytometry of plaque leucocytes also showed both increased incorporation of 5-ethynyl-2′-deoxyuridine (EdU) and an increased fraction of CD45.2 cells in the G2M phase of the cell cycle (Extended Data Fig. 4d, e). Consistently, mice with Jak2VF Confetti bone marrow12 (in which haematopoietic cells express one of several fluorescent proteins in a stochastic manner) showed increased expansion of haematopoietic cell clones in lesions compared to controls (Extended Data Fig. 4f, g, Supplementary Videos 1, 2). Thus, the increased proliferation of Jak2VF macrophages has a critical cell-intrinsic component that leads to clonal expansion.
Inflammasomes and plaque stability
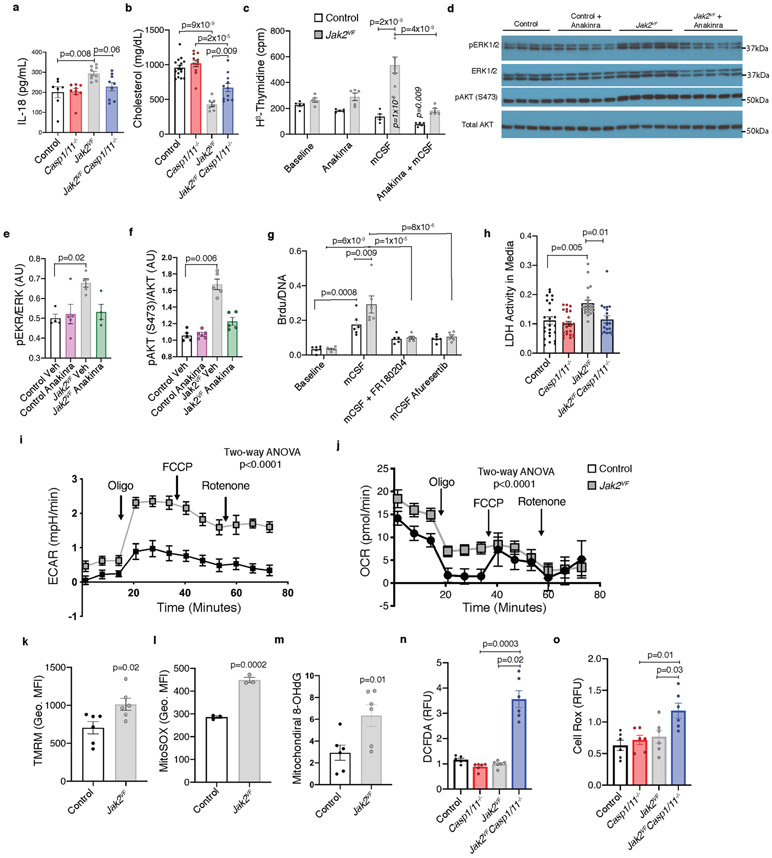
Atherosclerotic lesions in mice with Jak2VF clonal haematopoiesis showed increased macrophage staining for IL-1β (Extended Data Fig. 4h, i), suggesting that inflammasome activation was enhanced. To evaluate the role of inflammasomes in plaque development, we generated Jak2VF mice with concurrent deletion of the downstream inflammasome components caspase 1 and 11 (Jak2VFCasp1/11−/−). Jak2VF mice exhibited an increase in serum levels of the inflammasome product IL-18, and a decrease in cholesterol levels (Extended Data Fig. 5a, b), similar to humans2,13 These changes were partly reversed by Casp1/11 deficiency, without changes in spleen weight or circulating leukocyte counts (Supplementary Table 1). Deletion of Casp1/11 in Jak2VF mice abrogated increased proliferation of macrophages in lesions (Fig. 2a, b). Moreover, features of plaque stability, including macrophage accumulation, cap thickness, and necrotic core area, improved markedly in mice with Jak2VFCasp1/11−/− bone marrow (Fig. 2c-e). Changes in lesion size were not significant (Fig. 2f), possibly because Jak2VF mice show a decrease in cholesterol of more than 50% that is partially reversed by Casp1/11 deficiency. These results show that activation of the inflammasome contributes to increased macrophage proliferation within atheromas and to the promotion of plaque characteristics that are linked to destabilization–a key trigger of the thrombotic complications of atherosclerosis, including myocardial infarction14.
Fig. 2 ∣. Inflammasome activation promotes macrophage proliferation and features of plaque instability in mice with Jak2VF bone marrow.
a, Immunofluorescence images of aortic root plaques. Arrows, Ki67+ nuclei. Scale bar, 50 μm. b, Percentage Ki67+ cells in lesions. c, Macrophage number normalized to total cells in lesions (n = 8 mice (b, c)). d, Necrotic core area (n = 9 control, n = 8 Casp1/11−/−Jak2VF and Jak2VFCasp1/11−/− mice). e, Cap thickness (n = 8 mice). f, Lesion area (n = 9 control, n = 8 Casp1/11−/−Jak2VF and Jak2VFCasp1/11−/− mice). g, Immunofluorescence staining of aortic root lesions from mice with Jak2VF clonal haematopoiesis. Arrows, pγH2AX+ nuclei. Scale bars, 100 μm. h, pγH2AX+MAC2+ cells normalized to lesion area (n = 19 control, n = 20 Jak2VF mice). i, Representative immunofluorescence staining of aortic roots. Yellow dashed lines, lesion. j, 8-oxo-deoxyguanosine (8-OHdG) fluorescence intensity normalized to lesion area (n = 17 mice). k, Representative H&E images of aortic root lesions from mice with Jak2VF or Jak2VFNlrp3−/− bone marrow. Yellow dashed lines, necrotic cores. l, m, Quantification of lesion area (l; n = 15 mice) and percentage necrotic core area (m; n = 19 mice). n, Representative H&E images of aortic root lesions from mice with Jak2VF (n = 24) or Jak2VFAim2−/− (n = 25) bone marrow. Yellow dashed lines, necrotic core. o, p, Lesion area (o) and percentage necrotic core area (p). Mean ± s.e.m.; two-tailed t-test (l, m, o, p); Kruskal–Wallis two-tailed test with Dunn’s comparison (b, c, e); one-way ANOVA with Tukey’s post hoc analysis (d, f); two-way ANOVA (e); two-tailed Mann–Whitney test (h, j).
To understand the mechanisms that link activation of inflammasomes to macrophage proliferation and plaque destabilization, we treated cultured bone marrow-derived macrophages (BMDMs) with M-CSF, a major promoter of macrophage burden in atherosclerotic plaques15,16. Jak2VF BMDMs showed increased M-CSF-mediated proliferation, which was completely dependent on IL-1 as it was abolished by the IL-1 antagonist anakinra (Extended Data Fig. 5c). In parallel, the effects of Jak2VF and IL-1β signalling converged to increase activity in ERK and AKT signalling pathways, which increase macrophage differentiation and proliferation17 (Extended Data Fig. 5d-f). Treatment with ERK and AKT inhibitors reversed the increase in proliferation (Extended Data Fig. 5g). Jak2VF BMDMs also showed increased CASP1/11-dependent release of lactate dehydrogenase (LDH), which suggests that they were undergoing pyroptotic cell death (Extended Data Fig. 5h). These data suggest that inflammasome activation in Jak2VF macrophages increases secretion of IL-1β, which acts onJak2VF macrophages to increase AKT and ERK signalling and thereby leads to both increased proliferation and pyroptotic cell death.
Paralleling their increased proliferative capacity, Jak2VF macrophages displayed increased glycolysis and mitochondrial respiration (Extended Data Fig. 5i-k), as well as increased mitochondrial reactive oxygen species (ROS) and oxidized DNA (Extended Data Fig. 5l, m). Even though increased mitochondrial ROS could be secondary to inflammasome activation, rather than causative, BMDMs expressing Jak2VF and lacking Casp1/11 showed markedly enhanced cellular accumulation of ROS (Extended Data Fig. 5n, o) thereby situating ROS-dependence upstream of inflammasome activation. Increased mitochondrial ROS can lead to oxidation of DNA and activation of NLRP3 inflammasomes18 but can also promote damage to nuclear DNA, formation of double-stranded DNA breaks and activation of AIM2 inflammasomes19. Indeed, immunofluorescence staining of lesions from mice with Jak2VF clonal haematopoiesis showed accumulation of oxidized DNA and pγH2AX-positive macrophages (Fig. 2g-j), which suggest the presence of ROS-mediated oxidative DNA damage and replication stress. Jak2VF BMDMs showed increases in both polydAdT-mediated AIM2 activation and ATP-mediated NLRP3 inflammasome activation (Extended Data Fig. 6a, b). We also found increased AIM2 and NLRP3 inflammasome activation in human JAK2VF macrophages derived from induced pluripotent stem cells (iPS cells) compared to isogenic controls (Extended Data Fig. 6c, d). However, in cultured Jak2VF BMDMs, AIM2 protein was increased, whereas NLRP3 was decreased (Extended Data Fig. 6e-g). As Aim2 is regulated by IFNγ, which is induced by JAK–STAT signalling20,21, we administered IFNγ-neutralizing antibodies to BMDMs. This reversed the increase in Aim2 expression in Jak2VF BMDMs, indicating a direct link between JAK2 gain of function, IFNγ signalling and Aim2 expression (Extended Data Fig. 6h).
Experiments in mice have established the role of the NLRP3 inflammasome in accelerated atherosclerosis22. However, the above observations implicated the AIM2 inflammasome in aggravated atherogenesis resulting from the Jak2VF mutation. To assess the respective roles of the NLRP3 and AIM2 inflammasomes23 in atherosclerosis, we transplanted Jak2VFNlrp3−/− or Jak2VFAim2−/− bone marrow into Ldlr−/− mice and fed them a Western-type diet for 12 weeks. Both interventions moderately reduced cholesterol levels (Extended Data Fig. 6i-k). Whereas Nlrp3 deficiency had no significant effect on the areas of lesions or necrotic cores (Fig. 2k-m), Aim2 deficiency markedly reduced both (Fig. 2n-p). In multivariable analyses adjusted for plasma cholesterol levels, the effect of Aim2 on lesion area remained significant. Consistent with cell culture studies, AIM2 expression was increased in Jak2VF lesions (Extended Data Fig. 6l, m).
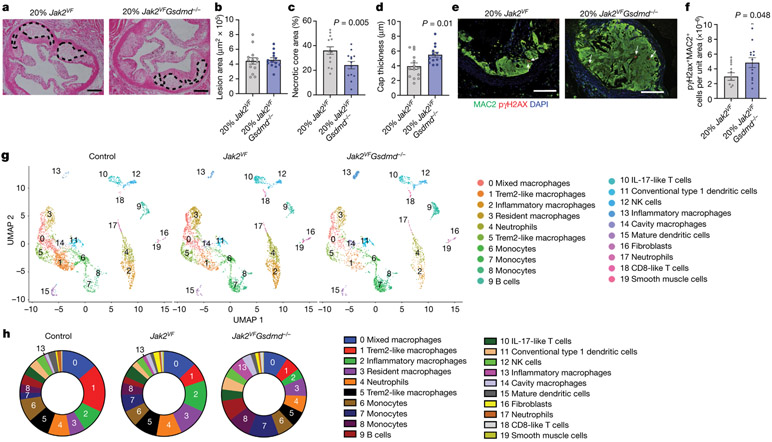
Activation of AIM2 or NLRP3 inflammasomes leads to pyroptotic cell death as a result of CASP1-dependent proteolytic cleavage of gasdermin D (GSMDM). GSDMD N-terminal fragments form pores in the plasma membrane that allow the release of mature IL-1β and LDH24. To assess the role of GSDMD in Jak2VF clonal haematopoiesis, we transplanted bone marrow (either 80% wild-type and 20% Jak2VF or 80% wild-type and 20% Jak2VFGsdmd−/−) into Ldlr−/− mice. Gsdmd deficiency markedly enhanced the Jak2VF allele burden in blood, potentially owing to suppression of pyroptosis (Extended Data Fig. 7a-h), with no alteration in cholesterol (Extended Data Fig. 7i). Although there was no change in overall lesion area, there was a decrease in necrotic core formation and a reciprocal increase in cap thickness in Gsdmd−/− mice (Fig. 3a-d). In a parallel experiment, we transplanted Gsdmd−/− or littermate control bone marrow into Ldlr−/− control mice and found no change in lesion area or necrotic core formation (Extended Data Fig. 7j-l). Mice with haematopoietic expression of Jak2VF demonstrated a further increase in pγH2ax staining as a result of Gsdmd deficiency (Fig. 3e, f), consistent with increased ROS-mediated formation of double-stranded DNA breaks acting upstream of AIM2 inflammasome activation and pyroptosis.
Fig. 3 ∣. Jak2VF promotes pyroptosis and inflammation in lesions.
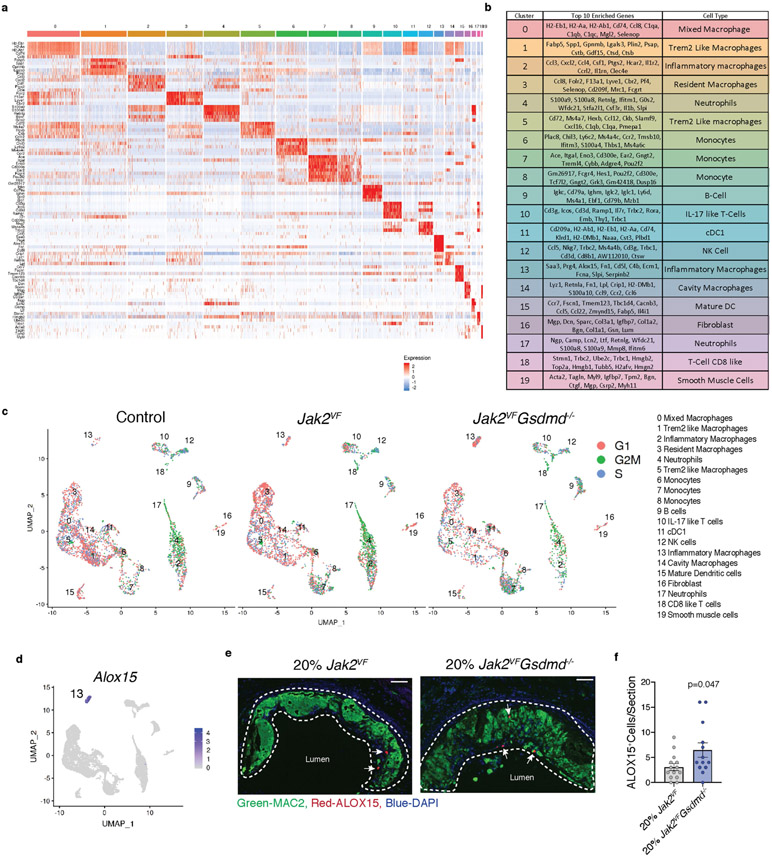
a, Representative H&E images of aortic root lesions from mice with Jak2VF or Jak2VFGsdmd−/− bone marrow. Dashed lines, necrotic core. Scale bars, 200 μm. b–d, Lesion area (b), percentage necrotic core area (c), and cap thickness (d; n = 14 Jak2VF, n = 13 Jak2VFGsdmd−/− mice). e, Aortic root lesions were stained for pγH2AX (white arrows). Scale bars, 100 μm. f, Quantification of pγH2AX+MAC2+ cells (n = 14 Jak2VF, n = 13 Jak2VFGsdmd−/− mice). g, Single-cell RNA-seq of plaque cells. UMAP plot of CD45+ cells from aortas of Ldlr−/− mice with 100% bone marrow of indicated genotypes (control 8 mice pooled; Jak2VF 16 mice pooled; Jak2VFGsdmd−/− 8 mice pooled). h, Percentage cell populations within clusters. Mean ± s.e.m.; two-tailed t-test (b–d, f).
Single-cell RNA-seq
Single-cell RNA-seq studies have uncovered complex changes in immune cell profiles in atherosclerotic lesions in mice, with prominent accumulation of several distinct macrophage populations25. To gain further insights into immune cell populations in atherosclerotic lesions, we carried out single-cell RNA-seq in cells dissociated from the aortae of Ldlr−/− mice with haematopoietic expression of Jak2VF or Jak2VFGsdmd−/−. Clustering visualized on uniform manifold approximation and projection (UMAP) showed 19 distinct cell populations, with a predominance of monocyte and macrophage subsets, but also neutrophils, dendritic cells, T cells and B cells (Fig. 3g, h, Extended Data Fig. 8a, b). Clusters 2 (inflammatory macrophages) and 3 (resident macrophages) were increased in Jak2VF mice, whereas cluster 1 was reduced in both Jak2VF and Jak2VFGsdmd−/− mice (Extended Data Table 1). Cluster 1 had characteristics of Trem2high, foamy non-inflammatory macrophages, consistent with the reduction in LDL cholesterol in Jak2VF mice. A population of inflammatory macrophages (cluster 2) rose in Jak2VF mice and fell with Gsdmd deficiency. These cells appeared to be highly inflammatory, with increased expression of multiple chemokines and Il1r compared to other clusters (Extended Data Fig. 8a, b, Supplementary Table 2). A formal assessment of cell cycle genes by Seurat analysis revealed a significant excess of expression of G2M-associated genes in Jak2VF cells that appeared especially prominent in clusters 2 and 4 (Extended Data Fig. 8c). Gsdmd deficiency reduced all major macrophage populations but increased monocytes in lesional cells (clusters 6–8) (Extended Data Table 1). The clustering analysis also revealed a rare population of macrophages in control mice that were slightly increased in Jak2VF mice and greatly increased in Jak2VFGsdmd−/− mice (cluster 13). These macrophages had high expression of Alox15 and Saa3, as well as pro-thrombotic and inflammasome genes (F5, Serpinb2, and Pycard). This cluster could represent a distinct population of inflammatory macrophages that cannot complete pyroptosis owing to Gsdmd deficiency (‘zombie’ macrophages). We confirmed the existence of a potentially similar intimal macrophage population by ALOX15 staining of Gsdmd−/− lesions (Extended Data Fig. 8d-f). Overall, these studies confirmed an increase in inflammatory, proliferative myeloid populations in Jak2VF lesions that are reduced by Gsdmd deficiency, but also revealed complex changes, with a reduced population of non-inflammatory Trem2High macrophages and a potentially adverse increase in monocytes and ‘zombie’ macrophages in Gsdmd−/− mice.
Potential therapeutic interventions
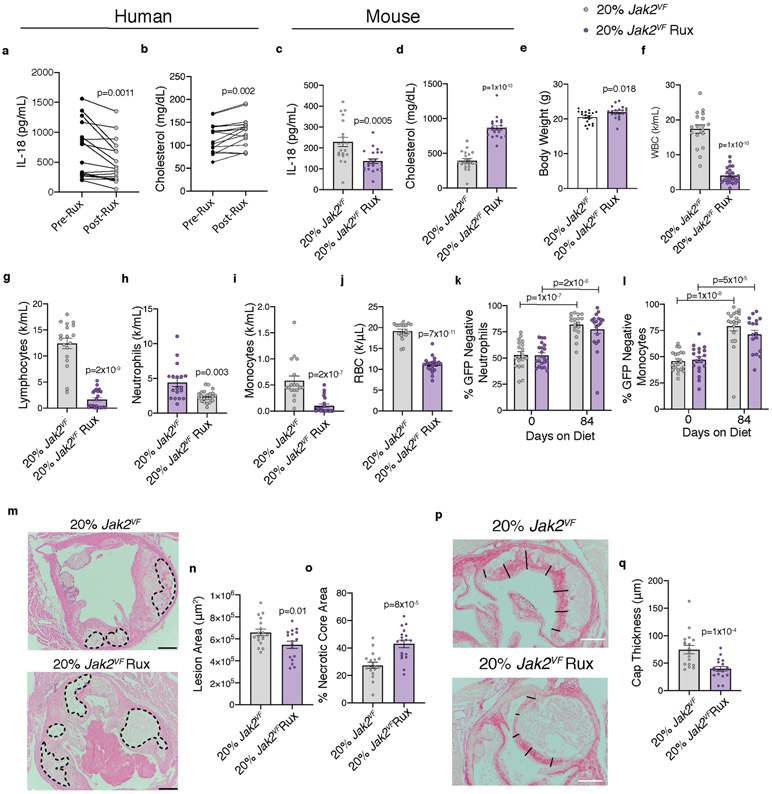
To assess the potential of available therapies that could be repurposed for JAK2VF-driven ACVD, we first evaluated the JAK inhibitor ruxolitinib (Rux), which has been approved for treatment of JAK2VF-associated haematological malignancies. Treatment with Rux lowered IL-18 levels but also increased cholesterol levels in patients with MPNs (Extended Data Fig. 9a, b), consistent with earlier findings2,26. Rux treatment similarly reduced IL-18 and increased cholesterol levels in Ldlr−/− mice with haematopoietic Jak2VF expression (Extended Data Fig. 9c, d). As found in humans and mouse models of MPN, Rux administration increased mouse body weight and markedly suppressed blood cell counts without altering Jak2VF burden (Extended Data Fig. 9e-l). These changes were associated with the development of slightly smaller atherosclerotic lesions with markedly increased necrotic cores and thinner caps (Extended Data Fig. 9m-q), indicating that Rux therapy promoted lesional necrosis. Rux inhibits both JAK1 and JAK2 and thus could have broad effects on signalling pathways that are required for cell survival. Rux also reduces oxidative metabolism of glucose in Jak2VF cells, leading to depletion of NADPH and glutathione with the potential to promote oxidative cell death27.
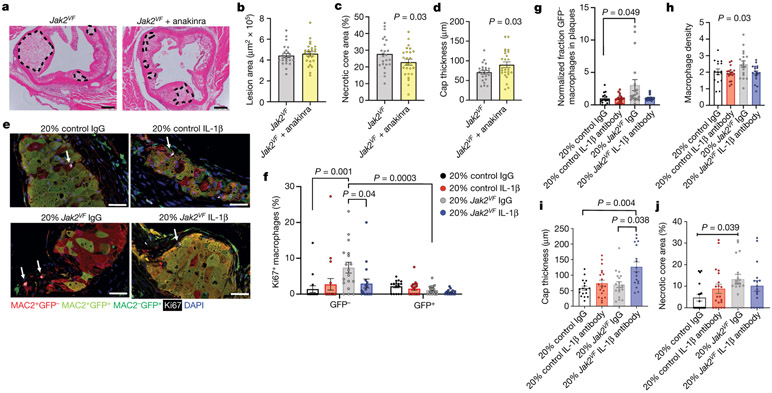
To assess the effects of inhibition of the inflammasome product IL-1, we administered the IL-1 receptor antagonist anakinra to Ldlr−/− mice with haematopoietic expression of Jak2VF. Anakinra had no effect on circulating cell counts or cholesterol, but completely normalized macrophage proliferation and density in early plaques (Extended Data Fig. 10a-f), confirming that IL-1 promotes macrophage proliferation in lesions. Moreover, in mice with Jak2VF bone marrow and more advanced atherosclerosis, anakinra reduced the areas of necrotic cores and increased cap thickness in lesions (Fig. 4a-d, Extended Data Fig. 10g). Thus, while not changing overall lesion size, IL-1 antagonism reduced features that have been implicated in instability of human atherosclerotic plaques.
Fig. 4 ∣. Effects of anakinra or IL-1β antibodies on atherosclerotic plaque development.
a, Representative H&E images of aortic root lesions from mice with 100% Jak2VF bone marrow treated with anakinra. Dashed lines, necrotic core. Scale bar, 200 μm. b–d, Lesion area (b), percentage necrotic core area (c; n = 24 Jak2VF, n = 25 Jak2VF + anakinra mice), and cap thickness (d; n = 23 Jak2VF, n = 25 Jak2VF + anakinra mice). e, Representative immunofluorescence image of aortic roots stained for GFP (Jak2VF-negative cells), MAC2, Ki67 and DAPI. Arrows, Ki67+ nuclei. Scale bars, 25 μm. f, Percentage Ki67+ macrophages (n = 16 control, n = 18 Jak2VF mice). g, Fraction of macrophages in aortic roots normalized to fraction in blood monocytes (n = 16 control, n = 19 Jak2VF mice). h–j, Quantification of aortic root lesion macrophage density (h), cap thickness (i), and necrotic core area (j) (n = 16 control, n = 20 Jak2VF mice). Mean ± s.e.m.; two-tailed t-test (b–d); one-way ANOVA with Tukey’s post hoc analysis (h); two-way ANOVA with Tukey’s post hoc analysis (f); Kruskal–Wallis two-tailed test with Dunn’s comparison (g, i, j).
Anakinra inhibits both IL-1β and IL-1α; to inhibit IL-1β specifically, we administered an antibody that selectively targets mouse IL-1β to Ldlr−/− mice transplanted with 80% wild-type (GFP+) and 20% control (GFP−) or 80% wild-type (GFP+) and 20% Jak2VF (GFP−) bone marrow (Extended Data Fig. 10h). The anti-IL-1β antibodies normalized macrophage proliferation in lesions (white Ki67 staining), specifically in Jak2VF macrophages (GFP−, red staining), and restored Jak2VF macrophage burden to levels similar to controls (Fig. 4e-g). Mice with Jak2VF clonal haematopoiesis showed no change in plasma cholesterol and modestly increased neutrophil and red cell counts that were not affected by IL-1β antibody treatment (Extended Data Fig. 10i, Supplementary Table 1). Treatment with antibodies against IL-1β did not alter Jak2VF blood monocyte proliferation (Extended Data Fig. 10j). Jak2VF chimeric mice treated with antibodies against IL-1β showed enhanced features of plaque stability, including decreased macrophage density, decreased necrotic cores and increased fibrous cap thickness (Fig. 4h-j, Extended Data Fig. 10k, l) These reciprocal changes did not result in an overall change in plaque size. Even though mice do not reliably rupture atherosclerotic plaques, changes in macrophage density, necrotic core size, and fibrous cap thickness reflect features that have been implicated in plaque stability in humans14. These findings parallel the beneficial clinical effects of IL-1β antibody therapy in CANTOS; in an imaging study, these were not associated with changes in lesion volume28.
Discussion
Our studies suggest that the Jak2VF mutation causes cellular metabolic and proliferative changes that lead to DNA oxidative damage, replication stress, AIM2 inflammasome activation and IL-1β production, which further drives cell proliferation. Increased macrophage proliferation depends on both intrinsic JAK2(VF) function and IL-1β, which suggests that inflammasome activation creates a feedforward loop to promote proliferation but also leads to pyroptotic cell death. These events lead to an overall increase in the burden of inflammatory macrophages within atherosclerotic lesions but also to increased necrotic core formation (Extended Data Fig. 10m). This is consistent with emerging evidence that inflammasome-dependent GSDMD-mediated formation of membrane pores initially allows secretion of active IL-1β but eventually leads to pyroptotic cell death29. Even though both NLRP3 and AIM2 inflammasomes could be activated in cultured Jak2VF macrophages, the AIM2 inflammasome predominated in promoting lesion formation in vivo, probably reflecting increased expression of AIM2 downstream of JAK/STAT signalling. In atherosclerosis-prone mice with Tet2 deficiency, increased lesion formation was not associated within macrophage proliferation within plaques and was reversed using an inhibitor of the NLRP3 inflammasome5, indicating that distinct mechanisms promote atherosclerosis in clonal haematopoiesis caused by different mutations. Inhibition of IL-1β or its downstream mediators such as IL-630 could be particularly effective and potentially broadly beneficial in ACVD associated with clonal haematopoiesis, while targeting of upstream inflammasome components such as AIM2 or NLRP3 may need to be tailored to the specific genetic factors responsible for clonal haematopoiesis.
Methods
Mice
All mice used for these studies were female and on a C57BL/6J background. Mice were housed in a specific pathogen-free facility under standard conditions of temperature (about 23 °C) with a 12-h light–dark cycle and food available ad lib (humidity was not noted). Cages and water were changed every 14–21 days. All mouse experiments were conducted in accordance with the Institutional Animal Care and Use Committee of Columbia University guidelines. Mice containing the Jak2V617F conditional knockin allele were generated as previously described9. For transgene expression, Jak2V617F mice were then crossed to either Mx1-Cre (The Jackson Laboratory, B6.Cg-Tg(Mx1-Cre)1Cgn/J (003556)), B6.Cg-Tg(S100A8 Cre-EGFP) (21614), or Cx3cr1-Cre (The Jackson Laboratory, B6.129P2(C) Cx3cr1tm2.1 (Cre/ERT2)Jung/J (020940)) mice to generate Mx1-creJak2VF, Cx3cr1-creJak2VF, and S100A8-cre Jak2VF, respectively. For the generation of Jak2VFCasp1/11−/− mice, Mx1-cre Jak2VF mice were crossed to Casp1/11−/− mice (The Jackson Laboratory, B6N.129S2-Casp1tm1Flv/J (016621)). Jak2VF Confetti mice were generated by crossing Mx-Cre Jak2VF mice with Confetti mice (The Jackson Laboratory, B6.129P2-Gt(ROSA)26Sortm1 (CAG-Brainbow2.1) Cle/J mice (17492)). Jak2VFAim2−/− and Jak2VFNlrp3−/− mice were generated by crossing Mx1-Jak2VF mice to Aim2−/− (The Jackson Laboratory, B6.129P2-Aim2Gt(CSG445)Byg/J) (13144)) or Nlrp3−/− (The Jackson Laboratory, (B6.129S6-Nlrp3tm1Bhk/J) (21302)) mice, respectively. Other animal colonies were purchased from The Jackson Laboratory: wild-type C57BL/6J (000664)), Ldlr−/− (B6.129S7-Ldlrtm1Her/J (002207)), CD45.1 (B6.SJL-Ptprca Pepcb/BoyJ (002014)), and GFP ((C57BL/6-Tg(CAGEGFP)1310sb/LeySopJ (06567)). Gsdmd knockout mice were obtained from Genentech. Littermate control mice contained either an allele with Cre or the Jak2VF transgene, but not both.
Bone marrow transplantation
Bone marrow transplantations (BMT) were conducted as previously described31 except that female recipient mice aged 8–12 weeks (C57BL/6J (wild-type) or Ldlr−/−) were lethally irradiated once with 10.5 Gy from a caesium gamma source. Within 24 h of irradiation, bone marrow was collected from female donor mice aged 6–15 weeks old with the indicated genotypes and the total bone marrow cell number was quantified. Irradiated mice were randomized to treatment groups and then anaesthetized with isoflurane before receiving 4 × 106 total bone marrow cells each through intravenous (i.v.) injection (100 μl final volume). Three weeks after BMT, cohorts involving Mx1-Cre mice were injected intraperitonially (i.p.) with 200 μg/mouse/day polyinosinic:polycytidylic acid (pIpC) twice, with a one-day rest between treatments.
Atherosclerosis studies
For atherosclerosis studies, sample sizes were power based on the assumption of a coefficient of variation of about 25–30% that will enable an 80% chance of detecting a >33% difference in atherosclerosis lesion size, necrotic core formation, and macrophage proliferation with P < 0.05. Four weeks after BMT, Ldlr−/− recipient mice were fed with a Western-type diet (WTD) (ENVIGO, cat. no. TD.88137) for the indicated time. Isoflurane-anaesthetized mice were repeatedly bled though cheek puncture for serum, complete blood counts, and flow cytometric analysis at the indicated time points. Following diet, aortic roots were collected and fixed with 4% paraformaldehyde for 24 h at 4 °C, then embedded in paraffin (except when CD45.1/2 chimeric mice were used, and for bead recruitment studies, in which case aortic roots were frozen in optimal cutting temperature (OCT) compound).
Histological analysis
Lesion area and necrotic core area.
Lesion area was quantified as previously reported31. Researchers blinded to genotype processed paraffinized tissue and sectioned lesions at similar points (mouse to mouse) in the aortic root over a 150-μm span for a total of six sections (spaced 30 μm apart). Slides were then stained with H&E and imaged with a Nikon Labophot 2 and Image Pro Plus software (Media Cybernetics version 7.0.0.591). Lesion area was then quantified in a blinded manner and the average of six slides was used to determine lesion area. Similarly, necrotic core area was quantified in a blinded manner using the same six H&E sections. When presented as percentage necrotic core area, total lesion area was used as the denominator.
Cap thickness.
Aortic root sections (one per mouse) removed from similar regions (mouse to mouse) were stained with picrosirius red. Cap thickness was measured in the largest lesion on each section at even intervals 2 μm apart. Then the average thickness of lesion was reported in length units. All lesion analysis was conducted with FIJI software32.
Immunohistochemistry and immunofluorescence
Slides of aortic roots from similar regions (mouse to mouse) were deparaffinized and suspended in a citric acid-based antigen retrieval solution (Vector Laboratories, H3300) and heated under pressure for 20 min. Slides were then cooled, washed 4 times with PBS containing 0.1% Tween 20 (PBST), blocked with 10% goat serum, and incubated with primary antibodies overnight at 4 °C: Ki67 (KI-67 Vector Laboratories VP-K451 1:1,500; KI-67 Abcam ab15580 1:100), IL-1B (Novus NB600-633 1:250), GFP (Abcam ab13970 1:1,000), MAC2 (Cedarlane CL8942AP 1:10,000), ACTA2 (Millipore Sigma C6198 1:500), CD45.2 (Biolegend 109804 1:100), CD45.1 (Biolegend 110750 1:100), DNA/RNA damage (clone 15A3, Abcam ab62623 1:250), pγH2AX (Cell Signaling 9718S 1:500), AIM2 (Abcam ab119791 1:1,000) and/or ALOX15 (Abcam ab244205 1:1,000). Slides were then washed 4 times with PBST, incubated with secondary antibodies (Invitrogen) for 1 h at room temperature, washed 4 times with PBST, and mounted in Prolong Gold antifade reagent with DAPI (Thermo Fisher Scientific). Slides were imaged on a Leica Fluorescent DMI 6000B wide-field microscope. Image analysis was conducted in a blinded manner in Fiji software. For immunohistochemical analysis, slides were incubated with secondary antibodies conjugated to HRP following overnight primary antibody incubation, exposed (Vector Laboratories, SK-4600), and counterstained with haematoxylin. All immunofluorescence analysis was done in a blinded manner using FIJI software.
Ki67.
To determine the percentage of Ki67+ macrophages in lesions, one slide per mouse was stained and imaged as indicated above. Then Ki67+DAPI+MAC2+ events were recorded as well as all MAC2+DAPI+ events and the percentage of Ki67+ events was calculated. When indicated, CD45.1/2 or GFP were also used in staining to stratify for genotype in the clonal haematopoiesis model. For CD45.1/2 staining, MAC2 staining was not present.
Macrophage density.
Macrophage density was determined from one aortic root slide per mouse (from similar regions mouse to mouse) stained for MAC2, DAPI, and ACTA2. Total DAPI+MAC2+ nuclei were determined per lesion unit area.
Clonal expansions of Confetti cells
To assess clonal expansion of bone marrow-derived macrophages within atherosclerotic lesions, lethally irradiated (2 × 650 rad with an interval of 4 h) Ldlr−/− mice were transplanted with bone marrow cells expressing Mx1-cre Jak2VF Confetti (B6.129P2-Gt(ROSA)26Sortm 1(CAG-Brainbow2.1)Cle/J (17492) or Mx1-cre Confetti (5 × 106 cells per mouse i.v.). Three weeks after transplantation, expression of Cre recombinase was induced by two intraperitoneal injections of pIpC (200 μg/mouse/day) followed by feeding with a high-fat diet (ENVIGO, cat. no. TD.88137) for 16 weeks. Immediately after death, the heart and aorta were perfused with 4% methanol-free PFA for 10 min, washed with PBS and embedded in OCT and frozen at −80 °C for 30 min. Frozen samples were cut with a cryotome (CryoStar NX70, ThermoFisher Scientific) into 50-μm sections at aortic valve level, mounted with DAKO mounting medium and imaged within 24 h.
Imaging was performed using a Leica SP8 WLL DIVE FALCON (fast lifetime contrast) system with one- and two-photon excitation, pulsed two-photon excitation from 700 to 1,300 nm. The objective was a HC IRAPO L 25×/1.00 W motCORR (WD 2.6 mm). Fluorescence lifetime imaging (FLIM) was used to identify fluorescent macrophages from the highly autofluorescent environment (nuclear GFP (the fourth colour in the confetti model) was not unequivocally identified within lesions due to autofluorescence, and therefore not quantified) in atherosclerotic lesions, and images were analysed using Leica Application Suite X (LAS X) and FIJI (ImageJ) software. Clones were defined as at least two cells of the same colour (of four possible colours of the confetti construct) in close proximity within lesions.
Bead recruitment studies
After 11 weeks WTD feeding, 1-μm latex fluoresbrite green fluorescent microspheres (Polysciences) in PBS were administered i.v. to mice. Twenty-four hours later, mice were bled and bead incorporation into monocytes was determined. Four days later, aortic roots were removed into OCT, frozen and sectioned. Bead accumulation was quantified histologically.
Clonal haematopoiesis model
Chimeric mice were generated by mixing bone marrow from Jak2VF or control mice expressing CD45.2 with bone marrow expressing CD45.1 or GFP; 8 × 105 (Jak2VF or control): 3.2 × 106 (CD45.1 or GFP) total cells. Bone marrow mixtures were then transplanted into lethally irradiated Ldlr−/− mice. WTD feeding was commenced four weeks after BMT for a duration of 7.5 weeks. For IL-1β antibody experiments, GFP bone marrow instead of CD45.1 bone marrow was used and IL-1β antibodies or an IgG control (provided by Novartis) were administered subcutaneously weekly as previously described33.
Flow cytometry analysis of blood cells for fraction CD45.1/2 or GFP positive/negative was conducted throughout the course of study. Cell populations were determined through flow cytometry staining with antibodies to total CD45 (clone 30-F11), CD45.1 (clone A20), CD45.2 (clone 104), GR1 (clone RB6-8C5), CD115 (clone AFS98), CD3 (clone 17A), CD4 (clone GK1.5), CD45R (clone RA3-6B2), and/or CD11b (clone M1/72). At termination of the IL-1β antibody study, blood monocytes were fixed, permeabilized, stained with 500 nM propidium iodide (Thermo Fisher Scientific, P3566), and analysed by flow cytometry.
Cx3c1-cre Jak2VF atherosclerosis study
Mice containing the Jak2VF conditional knockin were crossed with mice expressing a tamoxifen-inducible Cx3cr1-Cre recombinase (The Jackson Laboratory). Bone marrow from these mice and littermate controls was transplanted into lethally irradiated Ldlr−/− mice. Four weeks later, all mice were subjected to a high-cholesterol diet equivalent to ENVIGO (TD.88137) with the addition of 250 mg/kg tamoxifen for 15 weeks. At terminal isolation, blood was collected and pooled from four mice per sample for monocyte and neutrophil isolation with CD115 beads (Miltenyi Biotec, 130-096-354) as well as Ly6G beads (Miltenyi Biotec, 130-120-337), and mRNA was isolated for Jak2V617F burden quantification as previously described34. In addition, blood was pooled from two mice and stained for pSTAT5 (Y694) and analysed via flow cytometry. Each spleen was weighed following terminal isolation.
Ruxolitinib atherosclerosis study
Chimeric Ldlr−/− mice with 20% Mx1-Jak2VF and 80% GFP bone marrow were administered WTD ENVIGO (TD.88137) supplemented with ruxolitinib (2 g/kg)35,36 for 12 weeks.
Plasma from patients with MPN treated with ruxolitinib
Plasma was collected from patients with diagnosed with MPN before and at least 3 months after initiation of ruxolitinib therapy at Memorial Sloan Kettering. Samples were obtained with informed consent in accordance with all ethical regulations and were approved by the Institutional Review Board at memorial Sloan Kettering Cancer Center. IL-18 concentrations were determined via ELISA (MBL 7620). Total plasma cholesterol was determined via Cholesterol E assay (Wako, cat. no. 999-02601).
Anakinra atherosclerosis study
Mx1-creJak2VF and littermate control bone marrow was transplanted into Ldlr−/− mice. Four weeks later, WTD was initiated and 10 mg/kg anakinra or saline vehicle was administered intraperitoneally on 6 days per week for 7 weeks (short term) or 12 weeks (long term) to assess features of plaque stability.
CD11b+ splenocyte isolation
Freshly collected spleens were filtered through 60-μm cell filters with pressure in red blood cell lysis buffer (Biolegend). Filtered cells were then centrifuged at 800g for 10 min, suspended in MACs buffer (0.5% BSA, 2 mM EDTA, PBS, pH 7.4), filtered once more and centrifuged again. Cells were incubated with CD11b conjugated beads (Miltenyi Biotec, 130-049-601) for 20 min at 4 °C, washed with MACs buffer (PBS, 0.1% BSA, 2 mM EDTA), and filtered through 60-μm cell filters. Cells were then used for the indicated assays.
Bone marrow derived macrophage cultures
Bone marrow was flushed from hindlimbs with Hanks balanced salt solution (HBSS) and filtered in 60-μm cell filters on ice. Cells were centrifuged at 800g for 10 min at 4°C and suspended in DMEM with 10% FBS and 20% L-cell medium (LCM). Bone marrow cells were then incubated in non-tissue culture treated flasks for 5 days, quantified, and plated into new dishes overnight for the indicated assays.
Bulk cell RNA-seq
CD11b+ splenocytes were isolated from wild-type recipient mice 11 weeks after BMT. For hypercholesteraemia experiments, BMT was conducted in Ldlr−/− mice, four weeks after which a WTD was administered and sustained for seven weeks. Following WTD, mice were killed and CD11b+ splenocytes were collected and isolated into TRIzol reagent (Thermo Fisher Scientific). Aortic roots were also removed. RNA was isolated with RNeasy kits (Qiagen) and RNA-seq experiments were conducted as previously described37 on a NexSeq 500 (illumina). Genes were considered differentially expressed if they were significant at 5% false discovery rate (FDR) by DESeq2 and were at least twofold different in average reads per kilobase of transcript per million mapped reads (RPKM). Gene ontology analysis was conducted using the PANTHER database38. Extended Data Fig. 1 displays RNA-seq data. All data have been deposited into a public repository.
ROS analysis
BMDMs were plated into 96-well plates and stained with 3 μM MitoSOX (Thermo Fisher Scientific, M36008) for 20 min. Cells were then washed and suspended in DMEM with 1 mM sodium pyruvate and analysed using a SpectraMax M2 (Molecular Devices). Data were normalized to cell number.
For quantification of oxidized mitochondrial DNA, BMDMs were collected and mitochondria isolated using a microcentrifuge kit (Thermo Fisher Scientific, 89874). Mitochondrial DNA was then isolated using AllPrep DNA/RNA Mini Kit (QIAGEN). Isolated mitochondrial DNA was normalized based on DNA content and analysed for 8-hydroxy 2-deoxyguanosine through ELISA (Abcam, ab201734).
BMDMs were incubated with 5 μM 2′,7′-dichlorodihydrofluorescein diacetate (H2DCFDA) (Thermo Fisher Scientific, cat. no. D399) for 30 min in HBSS. BMDMs were then washed, resuspended in DMEM, and analysed on a SpectraMax M2; relative fluorescent units (RFU) were calculated and the data were normalized to cell number. BMDMs were incubated with 5 μM CellROX (Thermo Fisher Scientific, C10422) was for 30 min in HBSS. Cells were then washed and suspended in DMEM, and RFU was determined with data normalized to cell number.
Human iPS cell generation and macrophage differentiation
JAK2(V617F) and isogenic JAK2 wild-type iPS cells were generated from peripheral blood mononuclear cells of a patient with primary myelofibrosis, obtained from the Hematological Malignancies Tissue Bank of Mount Sinai with informed consent under a protocol approved by a Mount Sinai Institutional Review Board, using Sendai virus reprogramming, as previously described39. iPS cells were cultured on mitotically inactivated mouse embryonic fibroblasts with human embryonic stem cell medium supplemented with 6 ng/ml FGF2, as previously described39. Haematopoietic lineage specification was performed following a previously described spin-embryoid body-based protocol to generate haematopoietic progenitor cells through a haemogenic endothelium intermediate39. On day 10, the cells were transferred to macrophage differentiation culture consisting of StemPro-34 SFM medium with 1% nonessential amino acids (NEAA), 1 mM L-glutamine and 0.1 mM β-mercaptoethanol (2ME), supplemented with 100 ng/ml macrophage colony-stimulating factor (M-CSF) and 25 ng/ml interleukin 3 (IL-3) for 11 days with medium changes every two days. Following differentiation, macrophages were preincubated with lipopolysaccharide (LPS; 20 ng/ml) for 3 h then incubated with Nigericin at the indicated concentrations, and IL-1β in the medium was measured using ELISA. For pdAdT administration LPS was not added to the culture medium, and pdAdT was administered in lipofectamine 2000 for 4 h.
Inflammasome activation in BMDMs
NLRP3 inflammasome activation was assessed in BMDMs through treatment with 20 ng/ml LPS (Millipore Sigma, L4391) for three hours, followed by 1mM ATP (Millipore Sigma, A7699). One hour later, the medium was removed and analysed for IL-1β protein content with ELISA (R&D Systems, DY401). For AIM2 inflammasome activation, BMDMs were treated with 20 ng/ml LPS + pdAdT with lipofectamine 2000 (Invivogen, cat. no. tlrl-patn) for 6 h. Medium was then collected and IL-1β protein content measured with ELISA. NLRP3 and AIM2 protein expression in BMDMs were assessed in naive BMDMS. For Aim2 mRNA analysis IFNγ-neutralizing antibodies (Biolegend 505848) were incubated with BMDMs for 24 h and harvested into TRIazol. mRNA was isolated with a RNeasy kit (Qiagen) and cDNA was generated using a cDNA synthesis kit (Thermo Fisher Scientific, K1671). qPCR was conducted for Aim2 (GATTCAAAGTGCAGGTGCGG and TCTGAGGCTTAGCTTGAGGAC) normalized to M36b4.
LDH activity
Medium was taken from BMDMs incubated in LCM medium for 7 days and analysed for LDH activity with a CyQUANT LDH Cytotoxicity Assay (Thermo Fisher Scientific, C20301). Data were normalized to cell number at 5 days post incubation.
Blood isolation
Blood from mice was collected at the indicated time points through cheek bleed into EDTA-coated tubes. Blood cell numbers were then quantified with a VetScan HM5 Hematology system (Abaxis). For flow cytometry analysis red blood cells (RBCs) were lysed with RBC lysis buffer, washed in PBS with 0.5% BSA and 2 mM EDTA, then stained with the indicated antibodies.
Serum analysis
Mouse serum was isolated through centrifugation of blood at 12,000g for 10 min at 4 °C. Serum was evaluated for IL-18 through ELISA (MBL International, cat. no. 7625). Total serum cholesterol was determined using a cholesterol E assay (Wako, cat. no. 999-02601).
Immunoblot
Protein lysates from the indicated cell type were suspended in radioimmunoprecipitation assay (RIPA) buffer with protease and phosphatase inhibitors (Thermo Fisher Scientific, 78440). Protein concentrations were quantified with bicinchoninic acid (BCA) analysis (Thermo Fisher Scientific, 23225). Proteins were then loaded into polyacrylamide gels (BioRad) and transferred to nitrocellulose or PDVF membranes, blocked with 5% milk, and then incubated overnight with the indicated primary antibody (Cell Signaling pERK 4370T, ERK 4695T, pAKT 4060S, AKT 4298, AIM2 63660S, or NLRP3 15101) in 5% bovine serum albumin (BSA). Membranes were then washed and incubated with secondary conjugated to HRP, washed again, and then visualized with HRP chemiluminescent substrates (Thermo Fisher Scientific, 34580).
BMDM proliferation
H3-thymidine incorporation assays were conducted on BMDMs incubated with LCM medium for 6 days and plated into 12-well plates with normalized cell counts. After a 6-h adhesion period, cells were washed and suspended in DMEM (no FBS or LCM) with H3-thymidine and the indicated chemicals for 16 h. BMDMs were then washed and collected as previously described17. Scintillation counts were determined using an LS6500 scintillation counter (Beckman Coulter) and normalized to cell number before H3-thymidine addition.
For ERK and AKT inhibitor studies, 5-bromo-2′-deoxyuridine (BrdU) incorporation into BMDMs was monitored. BMDMs were differentiated and incubated in DMDM without FBS in the presence of 5 μm BrdU for 16 h in the presence of 100 ng/ml M-CSF and 500 nM afuresertib or 500nM FR180204 (Cayman Chemical). BrdU incorporation was measured with Roche cell proliferation ELISA and normalized to total cellular DNA (ThermoFisher Quant-iT PicoGreen dsDNA Kit).
BDMD incubation with anakinra
BMDMs were incubated with 75 μg/ml anakinra for four hours and harvested into TRIazol. mRNA was isolated with an RNeasy kit (Qiagen) and cDNA was generated using a cDNA synthesis kit (Thermo Fisher Scientific, K1671). qPCR was conducted for Spi1 normalized to M36b4.
For protein analysis, BMDMs that were incubated in LCM medium for 5 days were washed and suspended in DMEM with anakinra for 12 h. BMDMs were then stimulated with 1 ng/ml recombinant M-CSF (Pepro-Tech, 315-02) for 30 min, and protein was isolated in RIPA buffer with protease and phosphatase inhibitors. Immunoblots were conducted as described above and protein optical density was quantified in Fiji software.
Seahorse
CD11b+ splenocytes were collected as described above and suspended in DMEM containing 25 mM glucose, 1 mM pyruvate, and 2 mM glutamate. Thirty thousand cells per well were plated into 96-well seahorse plates, centrifuged, and analysed with a Seahorse XF96 as previously described40.
Mitochondrial potential
BMDMs were stained with 100 ng/ml tetramethylrhodamine (TMRM) (Thermo Fisher Scientific, cat. no. I34361) for 20 min in DMEM with LCM. Cells were then washed, brought into solution with cell stripper (Corning, 25-056-Cl), and suspended in DMEM with LCM. Geometric mean fluorescence (Geo. MFI) was then obtained using flow cytometry. Following an initial reading, 5 μM carbonyl cyanide 3-chlorophenylhydrazone (CCCP) (Millipore Sigma, C2759) was added to BDMDs and 10 min later a second Geo. MFI was determined. Final Geo. MFI measurements were recorded as Geo. MFIinitial – Geo. MFIFinal.
Aortic single-cell digest
Following termination of WTD, aortas were isolated, digested with 2.5 mg/ml liberase (Millipore Sigma, LIBTM-RO), 120 U/ml hyaluronidase (Millipore Sigma, H3506), and 160 U/ml DNase I (Millipore Sigma, DN25) for 45 min at 37°C as previously described31.
For qPCR analysis, cells were washed and stained with CD11b-APC, BV786-Ly6G, CD45.1-PE and CD45.2-BV421. Cells were sorted on a BD Influx sorter and isolated directly into TRIzol. qPCR analysis was conducted as described above.
Aortic macrophage proliferation
Ldlr−/− mice with 20% Jak2VF or control bone marrow and 80% CD45.1 bone marrow were subjected to a 12-week WTD. Twenty-four hours before cell collection, mice were injected i.p. with 5-ethynyl-2′-deoxyuridine (EdU; 0.1 mg/mouse). Following aortic digest, cells were stained for CD45.1, CD45.2, and CD68 and washed. EdU incorporation was then determined using Click-iT Plus EdU flow cytometry Assay Kit (ThermoFisher C10363). Cells were then stained with propidium iodine (1 μg/ml), treated with ribonuclease A, then analysed using flow cytometry.
Single-cell RNA-seq
For single-cell RNA-seq (scRNA-seq), after 12 weeks WTD, Ldlr−/− mice with control, Mx1-Jak2VF, or Mx1-Jak2VFGsdmd−/− bone marrow (not in a chimeric model) were killed, and aortas were digested as indicated above, except that necrosulfonamide (20 μM, Torcis) was supplemented in all media to prevent ex vivo necroptosis and gasdermin D oligomerization. Cell digests were sorted by flow cytometry for DAPI−CD45+ events, then run on Genomics scRNA-seq.
Analysis of scRNA-seq data
Fastq files were processed using the Cell Ranger v.3.1.0 pipeline. In brief, reads were aligned to mouse reference genome build mm10 and a mouse transcriptome built on Ensembl 93 annotation. A unique molecular identifier (UMI) count matrix was generated for each sample, followed by cell-calling implemented in Cell Ranger. Downstream analysis was performed using Seurat v.3.1.141 in R. The count matrices were filtered to include 1) genes expressed in ≥10 cells; 2) cells with ≥200 genes expressed; and 3) cells with ≤10% reads mapped to mitochondrial genes. Filtering on the maximum number of genes and the maximum number of UMIs per cell was also applied, with slightly different thresholds owing to differences in sequencing depth. A summary of the filtering parameters are provided in Extended Data Table 1. Filtered count matrices were integrated using the SCTransform workflow in Seurat. First, the top 500 variable genes were identified from each sample. Second, the top 1,500 variable genes from the union of variable genes across four samples were selected as integration features. The first 30 dimensions from canonical correlation analysis were used to identify integration anchors. UMAP was conducted using the first 20 principal components. An SNN graph was built on the integrated data with 20 nearest neighbours and 20 dimensions of principal components. Louvain clustering was applied with a resolution parameter of 0.5. To identify differentially expressed genes for each cluster, we first normalized UMI counts by the total number of UMIs in each cell, multiplied the values by 10,000, and then log-transformed the values with a pseudo-count of 1. A model-based analysis of single-cell transcriptomics test was performed on genes that were expressed in ≥25% of cells in the cluster tested or in other cells. Fold change ≥1.5 and Bonferroni corrected P < 0.05 were used to define differentially expressed genes.
Cell-cycle scoring of scRNA-seq data
Cell-cycle scoring analysis was performed in Seurat. A list of cell-cycle markers42 was loaded, including 54 G2/M phase genes and 43 S phase genes. The human gene symbols were converted to mouse gene symbols using the biomaRt43 package in R, which successfully converted 52 G2/M phase genes and 40 S phase genes to mouse gene symbols. Each cell was assigned cell phase classification (G2M/G1/S) based on the expression of the above markers.
Flow cytometric analysis
Samples were analysed on a BD LSRFortessa with Facs Software. For sorting, a BD influx sorter was used with FACs Diva software. Gating strategies are provided in Supplementary Figs. 1-4.
Statistics
Statistical analyses were conducted using Excel (Microsoft Office 2019) and GraphPad prism 8. Data presented were mean ± s.e.m. All measurements were taken from distinct samples unless otherwise noted. When applicable, all statistical test were conducted using two-sided analyses. Data were tested for normality in Prism 8 using a Kolmogorov–Smirnov test. Formal outliers were removed using a Grubb’s test (Prism). RNA-seq statistical analysis was conducted as previously described37.
Reporting summary
Further information on research design is available in the Nature Research Reporting Summary linked to this paper.
Data availability
The datasets generated and/or analysed during the current study are available in the Gene Expression Omnibus with the following accession numbers: mouse CD11b+ cells for bulk RNA-seq analysis (Extended Data Fig. 1) (GSE163689), mouse CD45+ aortic cells for scRNA-seq (GSE163536). All other relevant data are available from the corresponding authors upon reasonable request. Source data are provided with this paper.
Code availability
Gene ontology analysis used PANTHER (http://www.pantherdb.org/).
Extended Data
Extended Data Fig. 1 ∣. Bulk RNA-seq analysis of CD11b+ splenocytes.
a, b, Gene ontology analysis of CD11b+ splenocytes from wild-type (a; n = 6 control, n = 7 Jak2VF mice) and Ldlr−/− recipient mice (b; n = 6 control, n = 4Jak2VF mice) containing bone marrow from control or Mx1-Jak2VF transgenic mice.
Extended Data Fig. 2 ∣. Monocyte/macrophage- and neutrophil-specific expression of Jak2VF.
a, Scheme of experimental procedures for atherosclerosis studies in mice with monocyte/macrophage-specific expression of Jak2VF. b, qPCR analysis of Jak2VF mRNA in blood monocytes (n = 4 samples (3 mice pooled per sample,12-total mice per genotype)) and neutrophils (n = 5 (3 mice pooled per sample, 15 total mice per genotype)). c, Serum cholesterol levels (n = 14, 18, 19 control, n = 13, 18, 19 Jak2VF mice for day 21, 56, 83, respectively). d, Scheme of experimental procedure for studies of mice with neutrophil-specific expression of Jak2VF. e, qPCR analysis of Jak2VF mRNA in blood monocytes and neutrophils. Each point indicates four mice pooled together (n = 3 pooled samples, 12 total mice per genotype). f, Serum cholesterol levels. g–k, Quantification of lesion area (g), percentage necrotic core area (h), percentage macrophage area (i), percentage collagen area (j), and percentage smooth muscle cell (SMC) area (k; n = 12 control, n = 9 Jak2VF mice). Mean ± s.e.m.; two-tailed Mann–Whitney test (g), two-tailed t-test (f, h–k), two-way ANOVA followed by Bonferroni multiple comparison post hoc test (b, c, e).
Extended Data Fig. 3 ∣. Jak2VF allele burden and blood cell counts in mice with Jak2VF clonal haematopoiesis.
a, Experimental design for Jak2VF clonal haematopoiesis mice. b–e, Fraction CD45.2:CD45.1 (Jak2VF/control:CD45.1) in blood monocytes (b), all CD45+ cells (c), neutrophils (d), and lymphocytes (e); black arrow, induction of Mx1-cre (n = 10, 19, 18, 9, 20 control, n = 9, 18, 19, 9, 17 Jak2VF mice, for day −7, 26, 40, 47, 53, respectively). f–, Blood cell counts of white blood cells (f; WBC), lymphocytes (g), monocytes (h), neutrophils (i), and red blood cells (j; RBCs) (n = 17, 19, 10, 21 control, n = 17, 16, 10, 13 Jak2VF mice, for day −7, 26, 40, 53, respectively). k, Serum cholesterol (n = 17, 18, 19 control, n = 17, 12, 18 Jak2VF mice, for day 26, 40, 53, respectively). l, Spleen weight (n = 19 mice). Mean ± s.e.m.; two-tailed Mann–Whitney test (l), two-way ANOVA followed by Bonferroni multiple comparison post hoc test (b–k).
Extended Data Fig. 4 ∣. Jak2VF monocytes/macrophages have increased recruitment and proliferation in lesions.
a, Representative immunofluorescence images of aortic roots stained for latex beads (green), MAC2 (red), and DAPI (blue). White arrows, beads; scale bars, 100 μm. b, Quantification of beads in lesions. c, qPCR analysis of mRNA from CD11b+ aortic cells sorted for CD45.1 or CD45.2; PolE (n = 9 control, n = 11 Jak2VF mice). d, e, CD68+ cells isolated from aortas quantified for percentage EdU incorporation (d) and S/G2/M phase with propidium iodine staining (e; n = 11 control, n = 9 Jak2VF mice). f, Representative images of lesions from mice with Mx1-Confetti or Mx1-Jak2VF-Confetti expression in bone marrow. Cyan, cyan fluorescent protein (membrane-tethered CFP); yellow, membrane yellow fluorescent protein (cytoplasmic YFP); red, red fluorescent protein (cytoplasmic RFP). Yellow dashed lines, lesion boundary; scale bars, 25 μm. g, Quantification of the number of cells per clone in Confetti lesions (n = 149 control, n = 82 Jak2VF clones; four mice per group). h, Representative immunofluorescence images of aortic roots stained for MAC2 (green), IL-1β (red), and DAPI (blue). Scale bars, 50 μm. i, Quantification of total IL-1β fluorescent intensity normalized to area (n = 15 mice). Mean ± s.e.m.; two-tailed t-test (b), two-tailed Mann–Whitney test (d, e, g, i), Kruskal–Wallis two-tailed test with Dunn’s comparison (c).
Extended Data Fig. 5 ∣. IL-1β promotes increased ERK/AKT-driven proliferation of Jak2VF macrophages associated with increased glycolytic metabolism and mitochondrial ROS generation.
a, b, Quantification of serum IL-18 (a; n = 7 control, n = 9 Casp1/11−/−, n = 9 Jak2VF, n = 9 Casp1/11−/−Jak2VF mice) and cholesterol (b; n = 16 control, n = 11 Casp1/11−/−, n = 7 Jak2VF, n = 11 Casp1/11−/−Jak2VF mice) following 12-week WTD. c, Macrophage proliferation marked by 3H-thymidine incorporation into BMDMs during 16-h incubations (n = 4 biological replicates, replicated twice). d, Representative immunoblot analysis of BMDMs co-incubated with anakinra. e, f, Densitometric quantification of pERK1/2 (e; n = 4 control, n = 5 control + anakinra, n = 5 Jak2VF, n = 4 Jak2VF + anakinra; biological replicates from three mice pooled, representative of two experiments) and pAKT (f; n = 5 biological replicates from three mice pooled, representative of two experiments). g, Incorporation of BrdU into BMDMs co-incubated for 16 h with 100 ng ml−1 M-CSF and the indicated inhibitors (n = 6 biological replicates from three mice pooled, n = 5 M-CSF + FR180204). h, LDH release from non-stimulated BMDMs following 7-day culture (n = 24 control, n = 24 Casp1/11−/−, n = 22 Jak2VF, n = 19 Jak2VFCasp1/11−/− biological replicates from three mice repeated four times). i,j, Glycolysis rate indicated by the extracellular acidification rate (ECAR) (i) and mitochondrial respiration marked by oxygen consumption rate (OCR) (j; n = 15 control, n = 18 Jak2VF biological replicates repeated twice). k, Mitochondrial potential in CD11b+ splenocytes measured by tetramethylrhodamine, methyl ester, perchlorate (TMRM) geometric mean fluorescence intensity (geo. MFI) (n = 6 mice). l, MitoSOX geo. MFI in BMDMs (n = 3 mice). m, Quantification of mitochondrial localized 8-OHdG in BMDMs (n = 6 mice). n, o, Total cellular ROS in BMDMs marked by relative fluorescence units (RFU) of DCFDA (n) and Cell ROX (o; n = 6 biological replicates from three mice pooled). Mean ± s.e.m.; one-way ANOVA followed by Tukey’s post hoc test (a, b, e, o), Kruskal–Wallis two-tailed test with Dunn’s comparison (f, h, n), two-way ANOVA followed by Bonferroni’s multiple comparison post hoc test (c, g, i, j), two-tailed t-test (k–m).
Extended Data Fig. 6 ∣. JAK2VF macrophages display increased NLRP3 and AIM2 inflammasome activation.
a, b, IL-1β was quantified in medium from mouse BMDMs treated with pdAdT for 6 h (a) or LPS (20 ng ml−1; b) followed by a 1-h incubation with ATP (n = 6 biological replicates, representative of five experiments). Vertical P values are relative to LPS within the same genotype. c, d, IL-1β release from human iPSC-macrophages stimulated with pdAdT (c; n = 12 baseline, n = 6 treatments biological replicates representative of two experiments) and nigericin (d; n = 6 biological replicates, representative of two experiments). Vertical P values are relative to baseline within the same genotype. e, Immunoblot analysis of NLRP3 and AIM2 in protein lysates from BMDMs. f, g, Densiometric quantification of AIM2 (f) and NLRP3 (g; n = 8 biological replicates from four mice per group pooled together). h, Aim2 mRNA expression in BMDMs incubated in the presence of IFNY-neutralizing antibodies for 24 h (n = 6 biological replicates). i, Experimental scheme of atherosclerosis studies conducted in mice with Jak2VF bone marrow deficient in Nlrp3 or Aim2. j, k, Plasma cholesterol following 12-week WTD in Jak2VFNlrp3−/− (j; n = 15 Jak2VF, n = 19 Jak2VFNlrp3−/− mice) and Jak2VFAim2−/− mice (k; n = 24 Jak2VF, n = 25 Jak2VFAim2−/− mice). l. Representative immunofluorescence image of aortic roots stained for DAPI (blue) and AIM2 (magenta). Yellow dashed lines, lesions; scale bars, 50 μm. m, Quantification of AIM2 mean fluorescence intensity in lesions (n = 16 control, n = 18 Jak2VF mice). Mean ± s.e.m.; two-way ANOVA followed by Tukey’s post hoc test (a–d), one-way ANOVA followed by Tukey’s post hoc test (h), two-tailed t-test (j, k), two-tailed Mann–Whitney test (f, g, m).
Extended Data Fig. 7 ∣. Haematological parameters and atherosclerosis in Ldlr−/− mice transplanted with Gsdmd−/− bone marrow.
a, WBCs; b, lymphocytes (n = 14 Jak2VF, n = 13 Jak2VFGsdmd−/− mice); c, monocytes (n = 14 Jak2VF, n = 12 Jak2VFGsdmd−/− mice); d, neutrophils (n = 14 Jak2VF, n = 13 Jak2VFGsdmd−/− mice); e, RBCs (n = 13 mice). f–h, Jak2VF burden in blood lymphocytes (f), neutrophils (g), and monocytes (h; n = 14 Jak2VF, n = 13 Jak2VFGsdmd−/− mice). i, Serum cholesterol (n = 13 mice). j, Representative H&E images of aortic root lesions from Ldlr−/− mice with wild-type (WT) or Gsdmd−/− bone marrow fed a WTD for 12 weeks. Dashed lines, necrotic core; scale bars 200 μm. k, Quantification of lesion area. l, Percentage necrotic core area (n = 12 wild-type, n = 14 Gsdmd−/− mice). Mean ± s.e.m.; two-tailed t-test (a, b, d, e, i, k, l), two-tailed Mann–Whitney test (c, f–h).
Extended Data Fig. 8 ∣. scRNA-seq reveals increased proliferation and inflammatory macrophages.
a, Heat map of top five differentially expressed genes enriched in each cluster. b, Table showing top ten differentially expressed genes in each cluster with cell annotations (each sample pooled from eight mice). c, Cell cycle classification based on cell cycle gene expression using Seurat analysis; one-tailed X2 test, P = 1.415 × 10−10 indicates G2M cells enriched in Jak2VF lesions. d, UMAP plot of cluster 13 enriched Alox15. Blue indicates relative gene expression. e, Representative immunofluorescence images of aortic root lesions stained for MAC2 (green), ALOX15 (magenta) and DAPI (blue). White dashed lines, plaque; arrows, ALOX15+ cells; scale bars, 50 μm. f, Quantification of ALOX15+ cells per aortic root section (n = 14 Jak2VF, n = 13 Jak2VFGsdmd−/− mice). Mean ± s.e.m.; *P < 0.05 with respect to control genotype, two-tailed Mann–Whitney test (f).
Extended Data Fig. 9 ∣. Ruxolitinib reduces IL-18 and increases plasma cholesterol in individuals with MPN and mice with JAK2VF mutations.
a, b, Plasma from patients with MPN isolated before and after ruxolitinib (Rux) therapy were analysed for IL-18 (a) and total cholesterol (b; n = 17 patients). c, d, Serum from mice with chimeric 20% Jak2VF bone marrow was analysed for IL-18 (c) and total cholesterol (d; n = 20 Jak2VF, n = 22 Jak2VF + Rux mice). e, Body weight following 12 weeks WTD (n = 19 Jak2VF, n = 20 Jak2VF + Rux mice). f–j, Blood counts of WBCs (f), lymphocytes (g), neutrophils (h), monocytes (i) and RBCs (j; n = 18 Jak2VF, n = 22 Jak2VF + Rux mice). k, l, Jak2VF burden in blood neutrophils (k) and monocytes (l; n = 17 Jak2VF, n = 20 Jak2VF + Rux mice). m, Representative H&E images of aortic root lesions from mice with 20% Jak2VF chimeric bone marrow treated with Rux. Dashed lines, necrotic core; scale bars, 200 μm. n, o, Quantification of lesion area (n; n = 17 Jak2VF, n = 18 Jak2VF + Rux mice) and percentage necrotic core area (o; n = 18 mice). p, Representative picrosirius red-stained lesions (black lines indicate cap thickness). Scale bars, 200 μm. q, Quantification of cap thickness (n = 18 mice). Mean ± s.e.m.; Wilcoxon paired two-tailed test (a), two-tailed Student’s paired t-test (b); two-tailed Mann–Whitney test (c, e–j, n–p), two-tailed t-test (d), two-way ANOVA followed by Tukey’s post hoc test (k, l).
Extended Data Fig. 10 ∣. Inhibition of IL-1 reduces macrophage proliferation and density in plaques.
a, Terminal serum cholesterol levels in mice on a WTD and treated with anakinra for 7 weeks (n = 9 control, n = 8 control + anakinra, n = 7 Jak2VF, n = 9 Jak2VF + anakinra mice). b, Representative images of aortic roots following 7-week administration of anakinra or vehicle. MAC2 (green), phalloidin (cyan), Ki67 (red), DAPI (blue); arrows, Ki67+ nuclei; scale bars, 50 μm. c–e, Quantification of percentage Ki67+ cells (c), macrophage density (d) and lesion area (e; n = 8 control, n = 8 control + anakinra, n = 6 Jak2VF, n = 9 Jak2VF + anakinra mice). f, Serum cholesterol following 12 weeks WTD, related to late lesions in Fig. 4 (n = 24 Jak2VF, n = 25 Jak2VF + anakinra mice). g, Representative picrosirius red-stained lesions from mice treated with anakinra for 12 weeks. Black lines indicate cap thickness; scale bars, 200 μm. h, Experimental design for Jak2VF clonal haematopoiesis mice using GFP+ cells. i, Terminal serum cholesterol (n = 18 control IgG, n = 19 control IL-1, n = 19 Jak2VF IgG, n = 17 Jak2VF IL-1 mice). j, G2-M phase monocytes from blood identified with propidium iodine staining (n = 18 control IgG, n = 20 control IL-1, n = 18 Jak2VF IgG, n = 17 Jak2VF IL-1 mice). k, Representative picrosirius red-stained aortic root lesions from mice as in j. Black lines indicate cap thickness; scale bars, 200 μm. l, Representative H&E images of aortic roots. Dashed lines, necrotic core; scale bars, 200 μm. m, Overall summary scheme. Mean ± s.e.m.; one-way ANOVA followed by Tukey’s multiple comparison post hoc test (a, c–e), two-tailed t-test (f); Kruskal–Wallis two-tailed test with Dunn’s comparison (i, j).
Extended Data Table 1 ∣.
Characteristics of scRNA-seq analysis
| a | ||||
|---|---|---|---|---|
| Control vs. Jak2VF |
Jak2VF vs. Jak2VFGsdmd−/− |
Control vs Jak2VFGsdmd−/− |
||
| Cluster 1 Trem2- Like Macrophages | 5.9x10−49 | 6.2x10−53 | 0.04 | |
| Cluster 2 Inflammatory Macrophages | 7.1x10−5 | 1.6x10−16 | 1.6x10−33 | |
| Cluster 3 Resident Macrophages | 3.5x10−15 | 0.47 | 2.7x10−9 | |
| Cluster 4 Neutrophils | 0.17 | 0.01 | 6.5x10−6 | |
| Cluster 5 Trem2 Like Macrophages | 0.96 | 8x10−4 | 8.2x10−6 | |
| Cluster 6 Monocytes | 5.2X10−5 | 1.3x10−3 | 1 | |
| Cluster 7 Monocytes | 1.9x10−15 | 2.7x10−46 | 8.6x10−13 | |
| Cluster 8 Monocytes | 0.01 | 1.8x10−2 | 4.8x10−12 | |
| Cluster 13 Inflammatory Macrophages | 5.9x10−5 | 3.0x10−36 | 4.6x10−23 | |
| Cluster 14 Cavity Macrophages | 0.15 | 4.7x10−9 | 3.8x10−5 | |
| Cluster 17 Neutrophils | 3.5x10−3 | 8.3x10−3 | 1 |
| b | ||||||||
|---|---|---|---|---|---|---|---|---|
| Sample | Genotype | Min. # genes |
Max. # genes |
Max. # UMIs |
Max. % mitochondrial reads |
# genes | # cells | |
| 1 | Control | 200 | 4,000 | 20,000 | 10 | 13,399 | 3,546 | |
| 2 | Jak2VF | 200 | 4,000 | 20,000 | 10 | 13,547 | 2,592 | |
| 3 | Jak2VF | 200 | 5,000 | 30,000 | 10 | 13,678 | 1,481 | |
| 4 | Jak2VFGasd−/− | 200 | 4,000 | 25,000 | 10 | 14,184 | 2,973 |
a, scRNA-seq population distribution statistics. Bonferroni-corrected P values of two-tailed tests of equal proportions between genotypes in a subset of macrophages, monocytes, and neutrophils. Tests of proportions were conducted for cells assigned to macrophages, monocytes, and neutrophils between each pair of genotypes with the null of equal proportions. b, Filtering parameters for Ldlr−/− mouse scRNA-seq data. The minimum number of genes, maximum number of genes, maximum number of UMIs, and maximum percentage of reads mapped to mitochondrial genes applied to each cell are shown for each sample. The number of genes and the number of cells after filtering are also listed.
Supplementary Material
Acknowledgements
Supported by the Leducq Foundation (TNE-18CVD04) and NIH grants HL-118567, HL-155431, HL-148071, HL-137663, CA108671, CA008748, HL-080472, HL-134892. T.P.F. was supported by a Columbia Clinical and Translational Science Award (CTSA) (TL1-TR-001875), and F32HL151051-01. D.G.T. was supported by F30HL137327. E.P.P. is supported by the US National Institutes of Health (NIH) grants R01HL137219 and R01CA225231, by the New York State Stem Cell Board, and by a Scholar Award from the Leukemia and Lymphoma Society (LLS). C.S. and S.M. were supported by the German Research Foundation (DFG), CRC 1123, grants A07 (C.S.) and B06 (S.M.), as well as the DZHK (German Centre for Cardiovascular Research) and the BMBF (German Ministry of Education and Research) (grant 81Z0600204 to C.S.). We thank M. Gonzalez PisfiL, S. Dietzel and the Core Facility Bioimaging at the Biomedical Center, Ludwig Maximilian University, Planegg-Martinsried, Germany, for technical support. M.W. was supported by a Rosalind Franklin Fellowship from the University Medical Center Groningen and by Netherlands Organization of Scientific Research VIDI grant 917.15.350. Research reported in this publication was performed in the CCTI Flow Cytometry Core, supported in part by the Office of the Director, National Institutes of Health under awards S10RR027050 and S10OD020056. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. Images were collected and/or image processing and analysis for this work was performed in the Confocal and Specialized Microscopy Shared Resource of the Herbert Irving Comprehensive Cancer Center at Columbia University, supported by NIH grant P30 CA013696.
Footnotes
Online content
Any methods, additional references, Nature Research reporting summaries, source data, extended data, supplementary information, acknowledgements, peer review information; details of author contributions and competing interests; and statements of data and code availability are available at https://doi.org/10.1038/s41586-021-03341-5.
Competing interests R.L.L. is on the supervisory board of Qiagen and is a scientific advisor to Loxo, Imago, C4 Therapeutics and Isoplexis, which include equity interest. He receives research support from and consulted for Celgene and Roche and has consulted for Lilly, Janssen, Astellas, Morphosys and Novartis. He has received honoraria from Roche, Lilly and Amgen for invited lectures and from Gilead for grant reviews. P.L. is an unpaid consultant to, or involved in clinical trials for Amgen, AstraZeneca, Esperion Therapeutics, Ionis Pharmaceuticals, Kowa Pharmaceuticals, Novartis, Pfizer, Sanofi-Regeneron, and XBiotech, Inc. P.L. is a member of the scientific advisory board for Amgen, Corvidia Therapeutics, DalCor Pharmaceuticals, IFM Therapeutics, Kowa Pharmaceuticals, Olatec Therapeutics, Medimmune, Novartis, and XBiotech, Inc. P.L.’s laboratory has received research funding in the last two years from Novartis. A.R.T. is a scientific advisory board member for Amgen, Staten Biotech and Fortico Biotech and is a consultant for Janssen, CSL and the Medicines Company. B.E. has research funding from Celgene and Deerfield, and consulting fees from Grail. E.P.P. has received honoraria from Celgene and Merck and research support from Incyte for research not related to this study.
Supplementary information The online version contains supplementary material available at https://doi.org/10.1038/s41586-021-03341-5.
Peer review information Nature thanks Clinton Robbins, Eicke Latz and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Jaiswal S et al. Clonal hematopoiesis and risk of atherosclerotic cardiovascular disease. N. Engl. J. Med 377, 111–121 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bick AG et al. Inherited causes of clonal haematopoiesis in 97,691 whole genomes. Nature 586, 763–768 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Benjamin EJ et al. Heart disease and stroke statistics—2019 update: a report from the American Heart Association. Circulation 139, e56–e528 (2019). [DOI] [PubMed] [Google Scholar]
- 4.Ridker PM et al. Antiinflammatory therapy with Canakinumab for atherosclerotic disease. N. Engl. J. Med 377, 1119–1131 (2017). [DOI] [PubMed] [Google Scholar]
- 5.José J et al. Clonal hematopoiesis associated with TET2 deficiency accelerates atherosclerosis development in mice. Science 355, 842–847 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Landolfi R et al. Efficacy and safety of low-dose aspirin in polycythemia vera. N. Engl. J. Med 350, 114–124 (2004). [DOI] [PubMed] [Google Scholar]
- 7.Cordua S et al. Prevalence and phenotypes of JAK2 V617F and calreticulin mutations in a Danish general population. Blood 134, 469–479 (2019). [DOI] [PubMed] [Google Scholar]
- 8.Wang W et al. Macrophage inflammation, erythrophagocytosis, and accelerated atherosclerosis in Jak2V617F mice. Circ. Res 123, e35–e47 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mullally A et al. Physiological Jak2V617F expression causes a lethal myeloproliferative neoplasm with differential effects on hematopoietic stem and progenitor cells. Cancer Cell 17, 584–596 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Robbins CS et al. Local proliferation dominates lesional macrophage accumulation in atherosclerosis. Nat. Med 19, 1166–1172 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Edelmann B et al. JAK2-V617F promotes venous thrombosis through β1/β2 integrin activation. J. Clin. Invest 128, 4359–4371 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Snippert HJ et al. Intestinal crypt homeostasis results from neutral competition between symmetrically dividing Lgr5 stem cells. Cell 143, 134–144 (2010). [DOI] [PubMed] [Google Scholar]
- 13.Liu DJ et al. Exome-wide association study of plasma lipids in >300,000 individuals. Nat. Genet 49, 1758–1766 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hansson GK, Libby P & Tabas I Inflammation and plaque vulnerability. J. Intern. Med 278, 483–493 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Anderson KL et al. PU.1 and the granulocyte- and macrophage colony-stimulating factor receptors play distinct roles in late-stage myeloid cell differentiation. Blood 94, 2310–2318 (1999). [PubMed] [Google Scholar]
- 16.Rajavashisth T et al. Heterozygous osteopetrotic (op) mutation reduces atherosclerosis in LDL receptor-deficient mice. J. Clin. Invest 101, 2702–2710 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Celada A et al. The transcription factor PU.1 is involved in macrophage proliferation. J. Exp. Med 184, 61–69 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shimada K et al. Oxidized mitochondrial DNA activates the NLRP3 inflammasome during apoptosis. Immunity 36, 401–414 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hu B et al. The DNA-sensing AIM2 inflammasome controls radiation-induced cell death and tissue injury. Science 354, 765–768 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schwartz DM et al. JAK inhibition as a therapeutic strategy for immune and inflammatory diseases. Nat. Rev. Drug Discov 16, 843–862 (2017). [DOI] [PubMed] [Google Scholar]
- 21.Katherine L et al. Cloning a novel member of the human interferon-inducible gene family associated with control of tumorigenicity in a model of human melanoma. Oncogene 15, 453–457 (1997). [DOI] [PubMed] [Google Scholar]
- 22.Duewell P et al. NLRP3 inflammasomes are required for atherogenesis and activated by cholesterol crystals. Nature 464, 1357–1361 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Paulin N et al. Double-strand DNA sensing Aim2 inflammasome regulates atherosclerotic plaque vulnerability. Circulation 138, 321–323 (2018). [DOI] [PubMed] [Google Scholar]
- 24.Kayagaki N et al. Caspase-11 cleaves gasdermin D for non-canonical inflammasome signalling. Nature 526, 666–671 (2015). [DOI] [PubMed] [Google Scholar]
- 25.Zernecke A et al. Meta-analysis of leukocyte diversity in atherosclerotic mouse aortas. Circ. Res 127, 402–426 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mesa RA et al. Effects of ruxolitinib treatment on metabolic and nutritional parameters in patients with myelofibrosis from COMFORT-I. Clin. Lymphoma Myeloma Leuk 15, 214–221.e1 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rao TN et al. JAK2-mutant hematopoietic cells display metabolic alterations that can be targeted to treat myeloproliferative neoplasms. Blood 134, 1832–1846 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Choudhury RP et al. Arterial effects of Canakinumab in patients with atherosclerosis and type 2 diabetes or glucose intolerance. J. Am. Coll. Cardiol 68, 1769–1780 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Evavold CL et al. The pore-forming protein gasdermin D regulates interleukin-1 secretion from living macrophages. Immunity 48, 35–44.e6 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bick AG et al. Genetic interleukin 6 signaling deficiency attenuates cardiovascular risk in clonal hematopoiesis. Circulation 141, 124–131 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Westerterp M et al. Cholesterol efflux pathways suppress inflammasome activation, NETosis, and atherogenesis. Circulation 138, 898–912 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schindelin J et al. Fiji: an open-source platform for biological-image analysis. Nat. Methods 9, 676–682 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vromman A et al. Stage-dependent differential effects of interleukin-1 isoforms on experimental atherosclerosis. Eur. Heart J 40, 2482–2491 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mullally A et al. Distinct roles for long-term hematopoietic stem cells and erythroid precursor cells in a murine model of Jak2V617F-mediated polycythemia vera. Blood 120, 166–172 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.lacobucci I et al. Truncating erythropoietin receptor rearrangements in acute lymphoblastic leukemia. Cancer Cell 29, 186–200 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Maude SL et al. Efficacy of JAK/STAT pathway inhibition in murine xenograft models of early T-cell precursor (ETP) acute lymphoblastic leukemia. Blood 125, 1759–1767 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Thomas DG et al. LXR suppresses inflammatory gene expression and neutrophil migration through cis-repression and cholesterol efflux. Cell Rep. 25, 3774–3785.e4 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Thomas PD et al. PANTHER: a library of protein families and subfamilies indexed by function. Genome Res. 13, 2129–2141 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kotini AG et al. Stage-specific human induced pluripotent stem cells map the progression of myeloid transformation to transplantable leukemia. Cell Stem Cell 20, 315–328.e7 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fidler TP et al. Glucose metabolism is required for platelet hyperactivation in a murine model of type 1 diabetes. Diabetes 68, 932–938 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Butler A, Hoffman P, Smibert P, Papalexi E & Satija R Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat. Biotechnol 36, 411–420 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tirosh I et al. Dissecting the multicellular ecosystem of metastatic melanoma by single-cell RNA-seq. Science 352, 189–196 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Durinck S, Spellman PT, Birney E & Huber W Mapping identifiers for the integration of genomic datasets with the R/Bioconductor package biomaRt. Nat. Protocols 4, 1184–1191 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated and/or analysed during the current study are available in the Gene Expression Omnibus with the following accession numbers: mouse CD11b+ cells for bulk RNA-seq analysis (Extended Data Fig. 1) (GSE163689), mouse CD45+ aortic cells for scRNA-seq (GSE163536). All other relevant data are available from the corresponding authors upon reasonable request. Source data are provided with this paper.