Abstract
Purpose of review
To highlight standard PhenX (consensus measures for Phenotypes and eXposures) measures for nutrition, dietary supplements, and cardiovascular disease research and to demonstrate how these and other PhenX measures can be used to further interdisciplinary genetics research.
Recent findings
PhenX addresses the need for standard measures in large-scale genomic research studies by providing investigators with high-priority, well established, low-burden measurement protocols in a web-based toolkit (https://www.phenxtoolkit.org). Cardiovascular and Nutrition and Dietary Supplements are just 2 of 21 research domains and accompanying measures included in the PhenX Toolkit.
Summary
Genome-wide association studies (GWAS) provide promise for the identification of genomic markers associated with different disease phenotypes, but require replication to validate results. Cross-study comparisons typically increase statistical power and are required to understand the roles of comorbid conditions and environmental factors in the progression of disease. However, the lack of comparable phenotypic, environmental, and risk factor data forces investigators to infer and to compare metadata rather than directly combining data from different studies. PhenX measures provide a common currency for collecting data, thereby greatly facilitating cross-study analysis and increasing statistical power for identification of associations between genotypes, phenotypes, and exposures.
Keywords: cardiovascular disease, genome-wide association studies, metabolic syndrome, nutrition, PhenX (consensus measures for Phenotypes and eXposures)
Introduction
Gene–environment interactions underpin virtually all human diseases, including variations in their severity and clinical presentation. Fundamental understanding of cause of complex disease offers the opportunity to identify and classify genetic risk alleles and to prescribe genetically informed pharmaceutical therapies, as well as to alter environmental exposures to prevent and manage disease initiation and progression. An individual’s genetics influences the capacity to adapt to changing environments and elicits physiological responses in both health and disease. In turn, environmental agents can affect chromatin structure and stability, and gene expression levels. The introduction of high-throughput genotyping to population-based studies has led to the emergence of genome-wide association studies (GWAS) and a revolution in the way that scientists think about the interplay of genetics and common, complex diseases [1].
GWAS provide the potential to inspire new hypotheses and lead to the elucidation of the biochemical and physiological basis of pathogenesis [2]. However, the lack of inclusion of standard measures in GWAS has made it difficult to compare and/or combine these studies despite the fact that many risk factors and phenotypes are common across multiple conditions (e.g. obesity, diet, and smoking) and is a recognized limitation.
PhenX (consensus measures for Phenotypes and eXposures) addresses the need for standard measures in large-scale genomic research studies by providing investigators with high-priority, well established, low-burden measures [3•]. The measures and protocols in the PhenX Toolkit are chosen by content experts and are reviewed by the PhenX Steering Committee. Table 1 shows the 21 high-priority research domains that were selected by the PhenX Steering Committee for inclusion in the toolkit.
Table 1.
PhenX research domains
| Alcohol, tobacco, and other substances | Ocular |
| Anthropometrics | Oral health |
| Cancer | Physical activity and physical fitness |
| Cardiovascular | Psychiatric |
| Demographics | Psychosocial |
| Diabetes | Reproductive health |
| Environmental exposures | Respiratory |
| Gastrointestinal | Skin, bone, and muscle and joint |
| Infectious diseases and immunity | Social environment |
| Neurology | Speech and hearing |
| Nutrition and dietary supplements |
PhenX domains
Each PhenX domain is defined as a field of research with a unifying theme that includes easily enumerated quantitative and qualitative measures. A Working Group of expert scientists is assembled for each domain. Through a consensus process and with input from the broader scientific community, the Working Group selects up to 15 measures to be included in the PhenX Toolkit.
The article focuses on measures from two PhenX domains, Nutrition and Dietary Supplements and Cardiovascular, and the potential application of measures within those domains to assess metabolic syndrome.
Nutrition and dietary supplements
Gene–nutrient interactions influence risk for the initiation and/or progression of common chronic diseases, including diabetes (type 2), metabolic syndrome, cardiovascular and neurological disease, osteoporosis, and cancers [4••,5,6,7••]. Whereas it is established that human genetic variation can confer food and even individual nutrient tolerances/intolerances (e.g. lactose, iron) [8], there is increasing evidence that genetic variation influences individual nutrient requirements required to maintain health [9••] as well as nutrient utilization and metabolism in chronic disease [7••]. GWAS permit the comprehensive identification of genetic variation that interacts with dietary components to confer risk and/or protective phenotypes, and thereby enable the derivation of genetically informed dietary approaches to promote health and prevent and/or manage chronic disease in genetically diverse populations [10].
The scope of the PhenX Nutrition and Dietary Supplements domain is complex because of the number of essential dietary components. There are more than 40 nutrients (minerals, vitamins, fatty acids, amino acids) that are essential for human health. The Working Group was challenged by the structure of the PhenX framework and initially struggled to balance the competing priorities of burden, utility, scope of the domain, and scientific rigor. The Working Group first considered measures and methodologies related to diet, which are best described as dietary exposures or indicators of nutritional status. Dietary exposure levels can be obtained from questionnaires and nutrient composition databases. For some, but not all nutrients, dietary exposure predicts nutritional status, but this relationship is modifiable by genetics and environment. For selenium and vitamin D, diet does not predict nutritional status and therefore biomarker assays are required to quantify nutritional status. This includes physiological measurements of dietary components in human blood or tissue. These nutritional status measurements require analytical chemistry/biochemistry-based methodologies that quantify biomarkers. The Working Group was able to identify both dietary exposure and nutrient status measures that encompassed all essential nutrients to be included in the PhenX Toolkit. The rationale of the Working Group was that all essential nutrients are required for health and therefore should not be ranked or prioritized for GWAS unless the study is driven by a specific and narrow hypothesis.
The Nutrition and Dietary Supplements Working Group chose 13 measures (Table 2) that, taken together, assess all essential nutrients as dietary exposures and/or nutrient status measurements. The Working Group identified ‘gold standard’ methodologies that will enable data to be compared across ethnically diverse populations with acceptable burden to investigators and study participants. The Working Group recommended the multiple pass 24-h recall protocol for measuring total dietary intake [11]. This protocol enables virtually all aspects of diet to be quantified, including dietary intake of most essential nutrients. Because vitamin D and selenium cannot be quantified from a questionnaire or the 24-h recall, the Working Group deemed it important to include bioassay protocols to address these nutrients. In addition, the Working Group identified a limited number of low-burden, validated screeners that are used to quantify dietary exposures. The selected measures will permit the research community to standardize data collection in nutrition, thus facilitating identification of gene–nutrient interactions in health promotion, disease prevention, and disease progression.
Table 2.
PhenX measures (Nutrition and dietary supplements domain)
| Breast-feeding |
| Calcium intake |
| (a) Calcium intake by adults (daily) |
| (b) Calcium intake by children |
| Caffeine intake |
| Dairy food intake (daily servings) |
| Dietary supplements use |
| Fiber intake |
| Fruits and vegetables intake |
| Percentage energy from fat |
| Selenium |
| Sugar intake (added) |
| Vitamin D |
| Total dietary intake |
Cardiovascular
Cardiovascular diseases (CVDs) have complex cause and are a prevalent cause of mortality and morbidity. The pathogenesis of CVD involves nearly all organ systems and metabolic pathways. Moreover, the evidence for multiple risk factors, both genetic and environmental, has been established, whereas other putative risk biomarkers are being investigated. Through observational and interventional epidemiologic studies the genetics of infrequent but high-impact effects of single genes, such as those for familial hypercholesterolemia [12], have been documented. However, much of the genetic risk for CVD is dependent on the relatively small effects of multiple genes and the interaction of these genes with environmental factors [13]. CVD provides a good example of the need to integrate a broad range of phenotypic measurements, both risk biomarkers and clinical outcomes, into GWAS.
The Cardiovascular Working Group developed a parsimonious list of measures that captures important components of both risk and disease expression, but with minimal personal and technical burden to both study subjects and investigators. The Working Group recognized that these considerations are critical to gaining widespread incorporation of the measures into studies that are not focused on CVD. This approach required compromises between technically sophisticated and expensive methods that could provide specific assessments and less burdensome, but less precise assessments drawn from numerous epidemiologic studies of cardiovascular conditions.
The PhenX Toolkit provides protocols for making robust measures of CVD risk markers, including environmental components (such as diet, obesity), and of clinical conditions and pharmacologic interventions. In addition, the Cardiovascular Working Group selected 14 measures (Table 3) that can assess clinical conditions and that are widely accepted and standard measures. For example, a measure for a lipid panel includes protocols for total cholesterol, low-density lipoprotein cholesterol, high-density lipoprotein cholesterol, and triglycerides. The lipid protocols are measured as continuous variables without classification (e.g. normal, borderline, or abnormal), facilitating statistical analysis for GWAS and avoiding the potential issues that can arise as categorical variables are reinterpreted or revised by expert committees seeking to refine clinical treatment goals.
Table 3.
PhenX measures (Cardiovascular domain)
| Family history of heart attack |
| Lipid profile |
| Blood pressure (adult/primary) |
| High blood pressure during pregnancy |
| Heart valve function |
| Angina |
| Sudden cardiac arrest |
| Myocardial infarction |
| Peripheral arterial disease |
| Abdominal aortic aneurysm |
| Arrhythmia (atrial and ventricular) |
| Deep venous thrombosis |
| Pulmonary embolism |
| Rheumatic fever/rheumatic heart disease |
The field of genetics has played an important role in the understanding of CVD. For example, the correlation between Lp(a) levels and coronary disease had been observed frequently but a causal relationship between the two was not supported. However, this causal relationship was recently elucidated by demonstrating a relationship between a genotype for increased Lp(a) and an increased risk of coronary disease [14•]. The association of polymorphisms at the LPA locus on 6q26–27 for Lp(a) explained the majority of variability for coronary disease and established this link to polymorphisms predicting coronary heart disease [14•].
Using PhenX measures: metabolic syndrome
Metabolic syndrome is a cluster of inter-related risk biomarkers for CVD and type 2 diabetes. This clustering reflects both environmental and genetic factors and their interaction. As such, metabolic syndrome provides a salient example of the utility of the PhenX Toolkit to investigators to test associations across different clusters of phenotypes and exposures to define complex conditions or syndromes. A harmonized definition of metabolic syndrome includes elevated blood pressure, dyslipidemia (elevated triglycerides and lower high-density lipoprotein cholesterol), raised fasting glucose, and central obesity [15••]. These factors for metabolic syndrome are strongly influenced by genetic factors and the molecular and physiological relationships require accurate phenotypic and genetic assessment to determine potential explanatory mechanisms [16••].
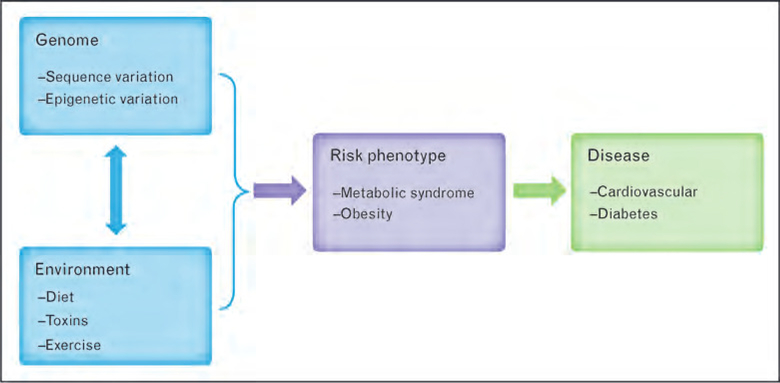
Figure 1 illustrates the interaction of genome and environment and the potential effect on risk phenotypes, highlighting the importance of multiple cross-cutting variables chosen from varying disciplines. In addition to the measures identified by the Cardiovascular and Nutrition Working Groups, the obesity risk phenotype may include waist circumference and body composition, measures identified by the Anthropometrics Working Group. The risk phenotype metabolic syndrome might further expand the list of variables of interest by including age (Demographics Working Group) and smoking (Alcohol, Tobacco, and Other Substances Working Group), in addition to those already included in obesity as well as those highlighted by both the Nutrition and Cardiovascular Working Groups.
Figure 1. Gene–environment interactions in the ontogeny of disease.
The diagram illustrates how an individual’s genome and environment interact and contribute to risk phenotypes which are associated with the development of disease.
Thus, PhenX provides a common platform on which to build an integrated approach for GWAS to define the interacting risk factors and exposures that underpin CVD, diabetes, and their common complications. More specifically, FTO was identified as a risk allele for metabolic syndrome through GWAS and has been implicated in hunger/satiation control, energy intake, and energy metabolism [17•]. FTO variants are associated with body mass index, hip circumference, and body weight [18] and have recently been shown to interact with the macronutrient composition of diet [19]. The use of PhenX measures will lead to the ability to analyze and reanalyze data from numerous studies and to systematically establish risk factors for obesity-related morbidity. GWAS of coronary artery disease risk have also identified unexpected associations with metabolic pathways whose function is dependent on essential nutrients, and thereby reveal new gene–nutrient interactions and the potential for novel nutritional interventions, including genes involved in mitochondrial folate-dependent one-carbon metabolism [13] and uric acid metabolism [20]. Likewise, investigations involving the influences of maternal diet on fetal genome programming leading to risk of metabolic syndrome in offspring are also amenable to association studies [21].
The PhenX Toolkit
The PhenX Toolkit presents the measures and protocols free of charge to the scientific community in a web-based resource (https://www.phenxtoolkit.org/). Within the toolkit, a brief description of each measure, its purpose, the rationale for its inclusion, the standard protocols for collecting the data, and references are provided. The requirements (e.g. training, personnel, and equipment needed to collect the data) are described. Toolkit users can search or browse for measures and protocols. Users select measures by adding them to a Cart (i.e. a specific collection of measures). Registered users can also save multiple collections of measures (Carts) and share their Carts with other registered users via a Toolkit Network. This enables investigators who are planning studies or expanding an existing study to work together to include a common set of PhenX measures for future analyses. Users can download a report that details their selections and can easily ‘cut and paste’ protocols to incorporate them into their study design.
Conclusion
The PhenX Toolkit is designed to assist investigators who are planning genomic studies and to promote the use of standard measures. The PhenX Toolkit makes it easier for researchers to expand their studies by incorporating measures of scientific interest that are outside of their primary research focus. As noted earlier, metabolic syndrome is just one example of an area of research that can benefit from the use of broadly accepted measures in two differing fields of study such as those included in the Cardiovascular and Nutrition and Dietary Supplements domains. As scientists seek to broaden their understanding of underlying determinants of complex diseases and effects of gene–environment interactions, the use of high-priority, standardized measures in fields which may not be their area of expertise will promote cross-study analysis, effectively increasing sample size and statistical power for validating prior study results and detecting relatively modest genetic associations. Increased statistical power will facilitate the identification of gene–gene and gene–environment interactions and lead to a better understanding of risk phenotypes. This knowledge should promote development of multi-faceted strategies for the prevention, management, and treatment of disease.
Acknowledgements
The authors thank Dr Erin M. Ramos for the critical review of the manuscript and Jennifer Drolet for editorial review. This work was supported by NHGRI, Award No. U01 HG004597–01.
References and recommended reading
Papers of particular interest, published within the annual period of review, have been highlighted as:
• of special interest
•• of outstanding interest
Additional references related to this topic can also be found in the Current World Literature section in this issue (p. 150).
- 1.Pennisi E. Breakthrough of the year: human genetic variation. Science 2007; 318:1842–1843. [DOI] [PubMed] [Google Scholar]
- 2.Pearson TA, Manolio TA. How to interpret a genome-wide association study. J Am Med Assoc 2008; 299:1335–1344. [DOI] [PubMed] [Google Scholar]
- 3.Hamilton CM, Strader LC, Pratt JG, et al. The PhenX Toolkit: get the most from your measures. Paper presented at the American Society of Human Genetics (ASHG) Annual Meeting, October 20–24, 2009; Honolulu, HI.• This presentation highlighted the PhenX project. The goal of the project is to facilitate cross-study comparisons by providing standard measures of phenotype and environmental exposures for use in genome-wide association studies (GWAS) and other large-scale genomic studies.
- 4.Smith CE, Ordovás JM. Fatty acid interactions with genetic polymorphisms for cardiovascular disease. Curr Opin Clin Nutr Metab Care 2009. [Epub ahead of print].•• This excellent review considers the limitations and challenges in investigating gene–diet associations.
- 5.Stover PJ, Caudill M. Genetic and epigenetic contributions to human nutrition and health: managing genome-diet interactions. J Am Diet Assoc 2008; 108:1480–1487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Williams CM, Ordovas JM, Lairon D, et al. The challenges for molecular nutrition research 1: linking genotype to healthy nutrition. Genes Nutr 2008; 3:41–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jenab M, Slimani N, Bictash M, et al. Biomarkers in nutritional epidemiology: applications, needs and new horizons. Hum Genet 2009; 125:507–525.•• This article considers the methodological challenges in quantifying nutritional status and intake for gene-diet-lifestyle-disease risk associations.
- 8.Stover P. Human nutrition and genetic variation. Food Nutr Bull 2007; 28:S101–S115. [DOI] [PubMed] [Google Scholar]
- 9.Solis C, Veenema K, Ivanov AA, et al. Folate intake at RDA levels is inadequate for Mexican American men with the methylenetetrahydrofolate reductase 677TT genotype. J Nutr 2008; 138:67–72.•• This article provides a salient example of how gene variants can determine the recommended dietary allowance of an essential nutrient in humans.
- 10.Trujillo E, Davis C, Milner J. Nutrigenomics, proteomics, metabolomics, and the practice of dietetics. J Am Diet Assoc 2006; 106:403–413. [DOI] [PubMed] [Google Scholar]
- 11.Thompson FE, Byers T. Dietary assessment resource manual. J Nutr 1994; 124:2245S–2317S. [DOI] [PubMed] [Google Scholar]
- 12.Goldstein JL, Brown MS. The LDL receptor defect in familial hypercholesterolemia. Implications for pathogenesis and therapy. Med Clin North Am 1982; 66:335–362. [DOI] [PubMed] [Google Scholar]
- 13.Samani N, Erdmann J, Hall A, et al. Genomewide association analysis of coronary artery disease. N Engl J Med 2007; 357:443–453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Clarke R, Peden JF, Hopewell JC, et al. Genetic variants associated with Lp(a) lipoprotein level and coronary disease. N Engl J Med 2009; 361:2518–2528.• This article discusses the use of genotyping to establish the causal relationship between genetic coding for Lp(a) and the development of coronary heart disease.
- 15.Alberti KG, Eckel RH, Grundy S, et al. Harmonizing the metabolic syndrome. Circulation 2009; 120:1640–1645.•• A consolidated statement from an international panel of the cluster of risk factors comprising the metabolic syndrome.
- 16.Lusis A, Attie A, Reue K. Metabolic syndrome: from epidemiology to systems biology. Nature Rev Genet 2008; 9:819–830.•• An integrated approach to investigating the genetic components of the metabolic syndrome and the interactions with the environment.
- 17.Olszewski PK, Fredriksson R, Olszewska AM, et al. Hypothalamic FTO is associated with the regulation of energy intake not feeding reward. BMC Neurosci 2009; 10:129.• This article highlights the need to understand the biological and physiological function of genes and gene variants when designing gene-diet-disease association studies.
- 18.Scuteri A, Sanna S, Chen WM, et al. Genome-wide association scan shows genetic variants in the FTO gene are associated with obesity-related traits. PLoS Genet 2007; 3:e115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Grau K, Hansen T, Holst C, et al. Macronutrient-specific effect of FTO rs9939609 in response to a 10-week randomized hypo-energetic diet among obese Europeans. Int J Obes (Lond) 2009; 33:1227–1234. [DOI] [PubMed] [Google Scholar]
- 20.Kolz M, Johnson T, Sanna S, et al. Meta-analysis of 28,141 individuals identifies common variants within five new loci that influence uric acid concentrations. PLoS Genet 2009; 5:e1000504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Waterland RA, Michels KB. Epigenetic epidemiology of the developmental origins hypothesis. Annu Rev Nutr 2007; 27:363–388. [DOI] [PubMed] [Google Scholar]