Abstract
Background
Lipid-metabolizing enzymes and their metabolites affect inflammation and fibrosis, but their roles in chronic kidney disease (CKD) have not been completely understood.
Methods
To clarify their role in CKD, we measured the mRNA levels of major lipid-metabolizing enzymes in 5/6 nephrectomized (Nx) kidneys of C57BL/6 J mice. Mediator lipidomics was performed to reveal lipid profiles of CKD kidneys.
Results
In 5/6 Nx kidneys, both mRNA and protein levels of Alox15 were higher when compared with those in sham kidneys. With respect to in situ hybridization, the mRNA level of Alox15 was higher in renal tubules of 5/6 Nx kidneys. To examine the role of Alox15 in CKD pathogenesis, we performed 5/6 Nx on Alox15−/− mice. Alox15−/− CKD mice exhibited better renal functions than wild-type mice. Interstitial fibrosis was also inhibited in Alox15−/− CKD mice. Mediator lipidomics revealed that Alox15−/− CKD mouse kidneys had significantly higher levels of PGD2 than the control. To investigate the effects of PGD2 on renal fibrosis, we administered PGD2 to TGF-β1-stimulated NRK-52E cells and HK-2 cells, which lead to a dose-dependent suppression of type I collagen and αSMA in both cell lines.
Conclusion
Increased PGD2 in Alox15−/− CKD mouse kidneys could inhibit fibrosis, thereby resulting in CKD improvement. Thus, Alox15 inhibition and PGD2 administration may be novel therapeutic targets for CKD.
Supplementary Information
The online version contains supplementary material available at 10.1007/s10157-021-02021-y.
Keywords: Chronic kidney disease, Lipoxygenase, ALOX15, Mediator lipidomics, Polyunsaturated fatty acids, Fibrosis
Introduction
Polyunsaturated fatty acids (PUFA) and their metabolites are linked to inflammation and its resolution in several organs [1]. Oxylipins, which are produced by the oxidation of PUFA, are important for PUFA biological activity as lipid mediators [1, 2]. Biosynthesis of oxylipins is mediated by several enzymes, such as lipoxygenase (LOX), cyclooxygenase (COX) and cytochrome P450 (CYP) [3]. These enzymes produce several lipid metabolites, such as prostaglandins, leukotrienes, and lipoxins, which are all heavily involved in the regulation of inflammation [1, 4]. For instance, ALOX15, a major subtype of LOX, has a dual aspect of proinflammatory and anti-inflammatory properties through its metabolites [5]: ALOX15 is highly expressed in eosinophils, bronchoalveolar epithelial cells and alveolar macrophages under nonpathological condition [5, 6], and promotes severity of asthma [7], lung injury [8], and heart failure [9], whereas it counteracts inflammation in arthritis [10] and ischemic brain [11]. Additionally, lipid mediators and their enzymes affect organ fibrosis as well as inflammation. Specific lipid mediators are involved in the pathogenesis of lung [12], liver [13], and heart [14] fibrosis. Similarly, the above-mentioned ALOX15 is also linked to the pathogenesis of fibrosis such as dermal fibrosis [15].
One of the common chronic diseases is chronic kidney disease (CKD), which affects approximately 8–16% of the general population in all stages combined [16]. Although CKD pathogenesis is complex and varies depending on the underlying disease, the kidney tissue generally becomes dysfunctional, leading to end-stage renal failure caused by chronic inflammation and subsequent fibrosis [17]. As mentioned, lipid mediators derived from PUFAs are strongly linked to inflammation and its resulting fibrosis. For instance, lipoxins and resolvins inhibit renal fibrosis [18, 19], but these effects are not yet examined in the CKD model with impaired kidney function. Moreover, comprehensive lipidomic profiles in CKD kidney tissues are still unreported. Thus, the role of lipid metabolic enzymes and their products in CKD pathogenesis has remained poorly understood.
This study aimed to elucidate the involvement of fatty acid metabolizing enzymes and their products in the renal impairment of 5/6 nephrectomized (Nx) CKD model mice. This study revealed that both of the transcription and protein expression levels of Alox15 were increased in CKD kidneys, and Alox15−/− mice demonstrated improved kidney dysfunction and fibrosis in the CKD model. Moreover, PGD2, which is the increased lipid metabolite in the CKD kidneys of Alox15−/− mice, inhibited the epithelial–mesenchymal transition (EMT) in proximal tubular cultured cells. Therefore, Alox15 inhibition and/or PGD2 administration could be a novel therapeutic target of CKD and fibrosis.
Materials and methods
Animals and experiments
This study used 8-week-old male C57BL/6 J mice (CLEA Japan), which were acclimatized for 1 week before all the experiments were performed. Moreover, Alox15−/− mice were generated in the C57BL/6 J background (Jackson Laboratory), and the 5/6 Nx model was established according to a previous study [20]. We collected blood samples for the evaluation of renal function and kidney tissue samples for immunoblotting and polymerase chain reaction at 8 weeks after 5/6 Nx, and for histological analysis at 40 weeks after 5/6 Nx. All experiments conformed to the guidelines for animal research of TMDU, and The Animal Care and Use Committee of TMDU approved our study protocol (approval number: A2019-117C4).
LC–MS/MS-based mediator lipidomics
We conducted LC–MS/MS analysis as described previously [21, 22]. Details of the procedure are described in Supplementary Methods.
Other experimental methods
We described other experimental methods in Supplementary Methods.
Results
mRNA level of Alox15 was increased in 5/6 Nx kidney
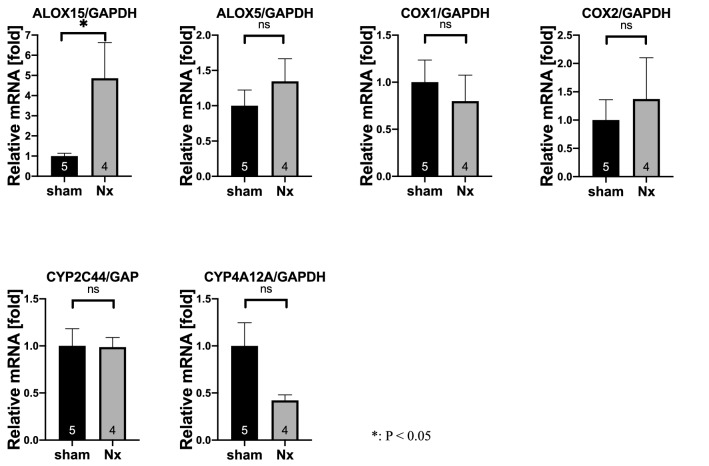
The quantitative changes of oxylipin enzymes in the CKD kidney were investigated by applying C57BL/6 mice to sham operation or 5/6 Nx as described previously [20]. To analyze the expression of major oxylipin enzymes expressed in the kidney (Alox15, Alox5, Ptgs1 (COX1), Ptgs2 (COX2), Cyp4a12 and Cyp2c44) [23, 24], we extracted kidney samples. Among the enzymes, Alox15 in 5/6 Nx kidneys had a significantly elevated mRNA level (P = 0.0004) compared with that in sham kidneys (Fig. 1). Conversely, the mRNA levels of the other major enzymes did not significantly change in this CKD model.
Fig. 1.
mRNA levels of major renal oxylipin enzymes in CKD kidney samples. Quantitative reverse transcription-polymerase chain reaction (qRT-PCR) analysis of major oxylipin enzymes expressed in the kidney (Alox15, Alox5, Ptgs1 [COX1], Ptgs2 [COX2], Cyp4a12 and Cyp2c44) was performed using sham and 5/6 nephrectomized (Nx) kidney samples. Compared with the sham kidneys, the 5/6 Nx kidneys had significantly increased mRNA level of Alox15 (P = 0.0004). The number of samples is shown at the bottom of the bar graph. Values are mean ± SEM. Unpaired Student’s t test, *P < 0.05
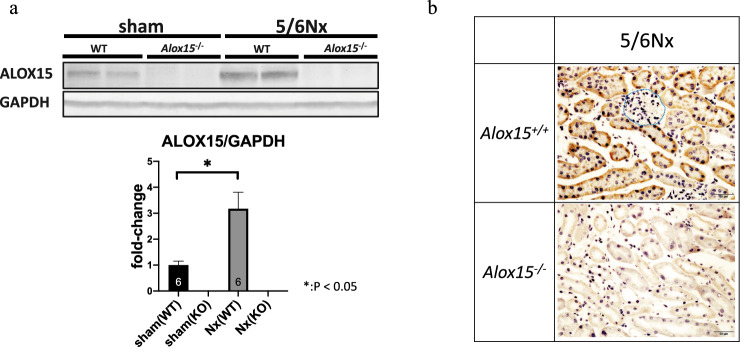
Protein level of Alox15 was increased in renal proximal tubular cells
In 5/6 Nx kidneys, the ALOX15 protein levels were also increased (P = 0.0078), as expected by increased transcriptional levels (Fig. 2a). To determine which cell types had increased Alox15 protein level in the CKD kidney, we examined the localization of Alox15 mRNA by in situ hybridization. In situ hybridization to mRNA revealed that the high expression level of Alox15 was localized at renal tubular cells in the 5/6 Nx model, but not in glomeruli (Fig. 2b). Histological features suggested that Alox15 mRNA was strongly expressed in the proximal tubules.
Fig. 2.
Alox15 expression in 5/6 Nx kidney at the protein level and localization of the mRNA of Alox15 in kidney tissues. a (upper) Representative immunoblots of ALOX15 of kidneys from sham-control and 5/6 nephrectomized (Nx) CKD mice. The band of ALOX15 was confirmed by the absence of ALOX15 band in Alox15−/− mice samples. (lower) Densitometry analysis of immunoblots. The ALOX15 in 5/6 Nx mouse kidneys was significantly increased compared with sham-control mouse kidneys (P = 0.0078). The number of samples is given at the bottom of the bar graph. Values are mean ± SEM. Unpaired Student’s t test, *P < 0.05. b in situ hybridization of kidneys from 5/6 Nx Alox15+/+ mice and 5/6 Nx Alox15−/− mice. In 5/6 Nx mouse kidneys, Alox15 mRNA was strongly expressed in the tubules, especially proximal tubules. The glomerulus had no mRNA expression of Alox15 (the blue border indicates a glomerulus)
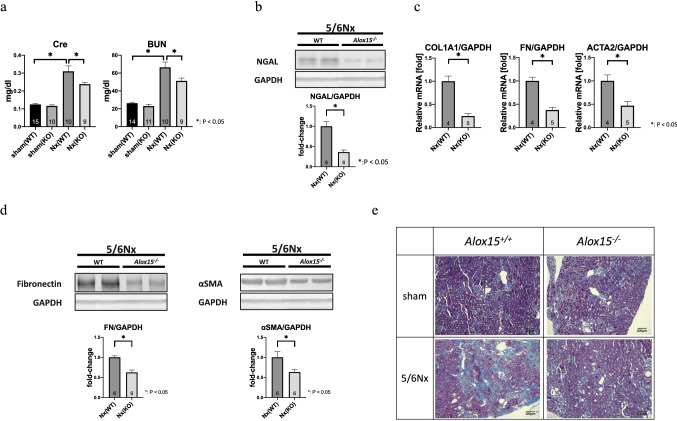
Alox15−/− mice were resistant to renal damage and fibrosis in the 5/6 Nx model
In 5/6 Nx CKD kidneys, the transcriptional and protein expression levels of ALOX15 were elevated. Therefore, we investigated whether and how ALOX15 plays a role in CKD in terms of kidney dysfunction and renal fibrosis. For this purpose, Alox15−/− mice were applied to the 5/6 Nx CKD model. Interestingly, serum Cre and blood urea nitrogen (BUN) levels were lower in Alox15−/− 5/6 Nx mice than those in wild-type (WT) mice (Fig. 3a), indicating that Alox15 depletion showed a protective effect in CKD pathogenesis. Accordingly, NGAL (a renal damage marker) protein expression was suppressed in Alox15−/− mice compared with that in WT mice (Fig. 3b). In addition, Alox15−/− mouse kidneys showed reduced Col1a1, Fn and Acta2 (αSMA) mRNA levels (Fig. 3c), and also showed decreased fibronectin and αSMA protein expression (Fig. 3d). Moreover, Alox15−/− mouse kidneys exhibited clearly suppressed interstitial fibrotic changes shown in Masson’s trichrome staining (Fig. 3e). Therefore, Alox15 deletion ameliorates kidney dysfunction and fibrosis in the CKD animal model.
Fig. 3.
Alox15−/− mice demonstrated kidney function and renal fibrosis amelioration in the CKD model. a Serological data from CKD mice at 8 weeks after 5/6 nephrectomy (Nx) showed that both serum creatinine and blood urea nitrogen levels of Alox15−/− CKD mice were significantly lower than those of WT CKD mice. The number of samples is shown at the bottom of the bar graph. Values are mean ± SEM. One-way analysis of variance was followed by Tukey’s multiple comparisons test, *P < 0.05. b Representative immunoblots of NGAL (a renal damage marker) of kidneys from 5/6 Nx mice. NGAL was suppressed in Alox15−/− CKD mouse kidneys compared with that in WT CKD mice. The number of samples is shown at the bottom of the bar graph. Values are mean ± SEM. Unpaired Student’s t test, *P < 0.05. c The mRNA of collagen type I and fibronectin (fibrosis markers), and Acta2 (αSMA; an EMT marker) were significantly suppressed in Alox15−/− mouse kidneys under 5/6 Nx condition compared with WT CKD mouse kidneys. The number of samples is shown at the bottom of the bar graph. Values are mean ± SEM. Unpaired Student’s t test, *P < 0.05. d Representative immunoblots of fibronectin and αSMA of kidneys from 5/6 Nx mice. Both of fibronectin and αSMA were suppressed in Alox15−/− CKD mouse kidneys compared with that in WT CKD mice. The number of samples is shown at the bottom of the bar graph. Values are mean ± SEM. Unpaired Student’s t test, *P < 0.05. e Kidney with Masson’s trichrome staining. Kidneys from Alox15+/+ and Alox15−/− mice under sham and 5/6 Nx conditions, respectively. Kidney tissues were obtained at 40 weeks after 5/6 Nx. Although interstitial fibrotic changes are evident in 5/6 Nx mouse kidneys, interstitial fibrosis in 5/6 Nx Alox15−/− mouse kidneys was significantly improved compared with that in 5/6 Nx Alox15+/+ mouse kidneys
Mediator lipidomics revealed altered PUFA metabolism in Alox15−/− CKD kidneys
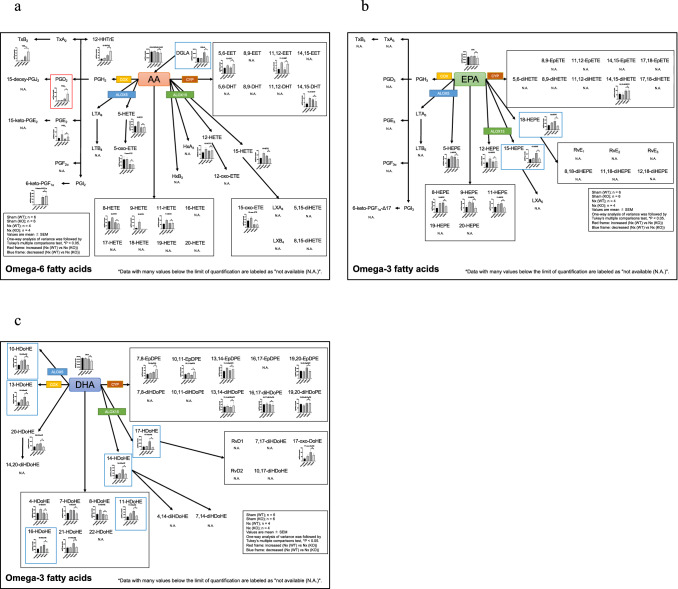
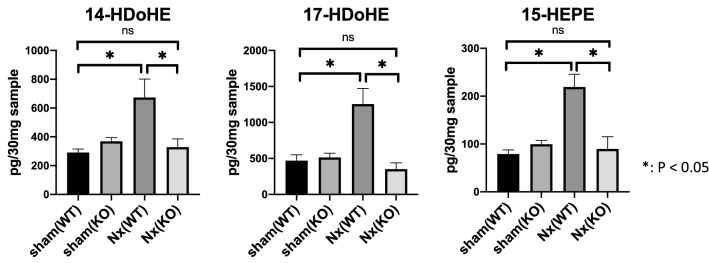
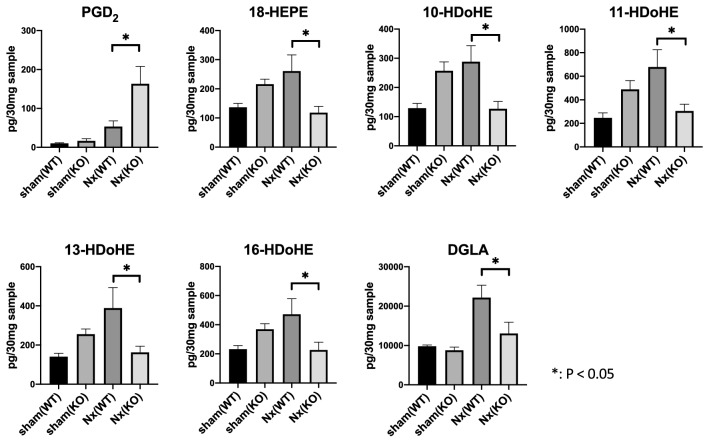
As mentioned in the Introduction section of this paper, the biological effects of Alox15 are related with the lipid mediators generated by Alox15. To elucidate the profile of lipid metabolites produced by Alox15 in the CKD kidney, we examined and compared the sham and 5/6 Nx kidney samples by LC–MS/MS-based mediator lipidomics to determine the lipid mediator profiles between WT mice and Alox15−/− CKD mice (Supplementary Table 1). Table 1 shows lipid metabolites which were significantly different between Alox15+/+ and Alox15−/− mice under 5/6 Nx condition. In addition, we made pathway maps of lipid mediators with classification by their substrates and their metabolizing enzymes (Fig. 4). A series of Alox15-derived PUFA metabolites such as 14-HDoHE, 17-HDoHE, and 15-HEPE [5, 25–27] was significantly increased in CKD models that correlated well to the increased level of ALOX15 expression in the kidneys (P = 0.0018, 0.0008, < 0.0001, respectively), and their elevations under CKD conditions were completely suppressed in Alox15−/− mice (Fig. 5). Besides 14-HDoHE, 17-HDoHE, and 15-HEPE, the levels of 18-HEPE, 10-HDoHE, 11-HDoHE, 13-HDoHE, 16-HDoHE and DGLA were also significantly decreased in Alox15−/− CKD kidneys compared to those in WT CKD kidneys, whereas only PGD2 was significantly increased in Alox15−/− CKD kidneys (Fig. 6).
Table 1.
List of lipid metabolites which were significantly different between Alox15+/+ and Alox15−/− mice under 5/6 Nx condition
| Sample name | Sham WT (n = 6) | Sham KO (n = 6) | Nx WT (n = 4) | Nx KO (n = 4) |
P value (Nx WT vs Nx KO) |
Increased or decreased in KO compared to WT | ||||
|---|---|---|---|---|---|---|---|---|---|---|
| Ave | SE | Ave | SE | Ave | SE | Ave | SE | |||
| PGD2 | 10.0 | 1.5 | 16.5 | 5.9 | 53.4 | 14.5 | 163.1 | 45.1 | 0.0093 | Increased |
| 15-HEPE | 78.8 | 8.7 | 99.4 | 8.1 | 219.2 | 26.7 | 89.5 | 25.9 | 0.0006 | Decreased |
| 18-HEPE | 136.9 | 13.3 | 215.7 | 17.6 | 260.8 | 55.7 | 118.3 | 21.7 | 0.0186 | Decreased |
| 10-HDoHE | 128.8 | 16.5 | 256.9 | 30.7 | 288.1 | 54.7 | 126.7 | 25.4 | 0.0246 | Decreased |
| 11-HDoHE | 245.9 | 43.2 | 488.0 | 74.5 | 678.4 | 146.6 | 305.5 | 57.8 | 0.0443 | Decreased |
| 13-HDoHE | 140.8 | 17.4 | 255.2 | 26.8 | 388.1 | 104.6 | 162.8 | 31.2 | 0.0341 | Decreased |
| 14-HDoHE | 289.5 | 25.1 | 368.6 | 26.2 | 673.2 | 128.3 | 328.0 | 57.6 | 0.0092 | Decreased |
| 16-HDoHE | 232.1 | 25.1 | 369.1 | 38.3 | 471.7 | 106.7 | 227.0 | 53.4 | 0.0497 | Decreased |
| 17-HDoHE | 468.5 | 82.0 | 513.1 | 59.0 | 1253.0 | 218.2 | 350.6 | 88.7 | 0.0005 | Decreased |
| DGLA | 9794.6 | 349.6 | 8797.8 | 791.8 | 22,167.8 | 3136.0 | 13,040.0 | 2866.7 | 0.0195 | Decreased |
By mediator lipidomics, the above fatty acid metabolites were detected in the kidney tissue (30 mg). The P values in the table were obtained by comparing Alox15+/+ and Alox15−/− mice under 5/6 Nx condition. The number of samples is as follows: sham (WT), n = 6, sham (KO), n = 6, Nx (WT), n = 4, Nx (KO), n = 4. One-way analysis of variance was followed by Tukey’s multiple comparisons test
Fig. 4.
Pathway maps of lipid mediators derived from omega-6 and omega-3 fatty acids. a Pathway map of lipid mediators derived from arachidonic acids (AA). b Pathway map of lipid mediators derived from eicosapentaenoic acids (EPA). c Pathway map of lipid mediators derived from docosahexaenoic acids (DHA)
Fig. 5.
Lipidomics confirmed that the production of ALOX15-dependent lipid metabolites was suppressed in Alox15−/− CKD kidneys. 14-HDoHE, 17-HDoHE, and 15-HEPE, which are ALOX15-dependent lipid metabolites, were significantly increased in WT CKD kidneys (P = 0.0018, 0.0008, < 0.0001 respectively). The increase was suppressed in Alox15−/− mice. The number of samples is as follows: sham (WT), n = 6, sham (KO), n = 6, Nx (WT), n = 4, Nx (KO), n = 4. Values are mean ± SEM. One-way analysis of variance was followed by Tukey’s multiple comparisons test, *P < 0.05
Fig. 6.
Lipid metabolites which were significantly different between Alox15+/+ and Alox15−/− mice under 5/6 Nx condition. Alox15−/− CKD kidneys had significantly increased levels of PGD2 compared with WT CKD kidneys (P = 0.0093). In contrast, Alox15−/− CKD kidneys had significantly decreased levels of 18-HEPE, 10-HDoHE, 11-HDoHE, 13-HDoHE, 16-HDoHE and DGLA compared with WT CKD kidneys (P = 0.0186, 0.0246, 0.0443, 0.0341, 0.0497, 0.0195 respectively). Additionally, 30 mg of each sample was analyzed with lipidomics. Sham (WT); n = 6, sham (KO); n = 6, Nx (WT); n = 4, Nx (KO); n = 4. Values are mean ± SEM. One-way analysis of variance was followed by Tukey’s multiple comparisons test, *P < 0.05
PGD2 suppressed EMT in cultured kidney cells
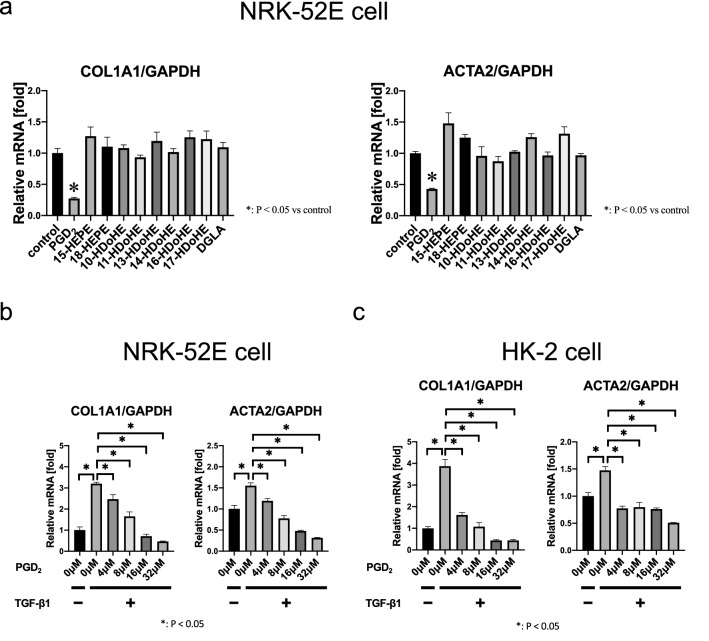
The effects of lipid metabolites which were significantly different between Alox15+/+ and Alox15−/− mice under 5/6 Nx condition were examined by administering these lipids to NRK-52E cells that were activated by pro-fibrotic cytokine TGF-β1. We assessed the mRNA expression of Cola1a and Acta2 (αSMA) in response to TGF-β1 to determine the effects of these lipids in culture (Fig. 7a). Among those, PGD2 conferred potent antifibrotic effect on NRK-52E cells in response to TGF-β1. The suppressive effect of PGD2 on COL1A1 and αSMA expression were dose dependent with the EC50 of 7.12 μM and 6.48 μM, respectively (Fig. 7b). Additionally, we conducted the same experiments in HK-2 cells, which are immortalized proximal tubule epithelial cells from normal adult human kidneys, and found a similar outcome, that is, COL1A1 and αSMA inhibition in a dose-dependent manner, in response to treatment with PGD2 (Fig. 7c). Therefore, increased levels of PGD2 in Alox15−/− CKD kidneys may contribute to the antifibrotic effects in CKD.
Fig. 7.
PGD2 suppressed EMT in cultured kidney cells. a Effects of lipid metabolites which were significantly different between Alox15+/+ and Alox15−/− mice under 5/6 Nx condition on the EMT of NRK-52E cells. These lipid metabolites were administered at 8 μM each to NRK-52E cells with TGF-β1 (5 ng/mL). PGD2 significantly suppressed Col1a1 and αSMA (Acta2) expression. None of the other fatty acid metabolites inhibited or promoted Col1a1 and αSMA. b Effects of PGD2 on the EMT of NRK-52E cells. PGD2 significantly suppressed Col1a1 and Acta2 expression induced with TGF-β1 (5 ng/mL) in NRK-52E cells dose-dependently. c Effects of PGD2 on the EMT of HK-2 cells. PGD2 also suppressed Col1a1 and Acta2 expression induced with TGF-β1 (5 ng/mL) in HK-2 cells dose-dependently. n = 3 for each group. Values are mean ± SEM. One-way analysis of variance was followed by Tukey’s multiple comparisons test, *P < 0.05
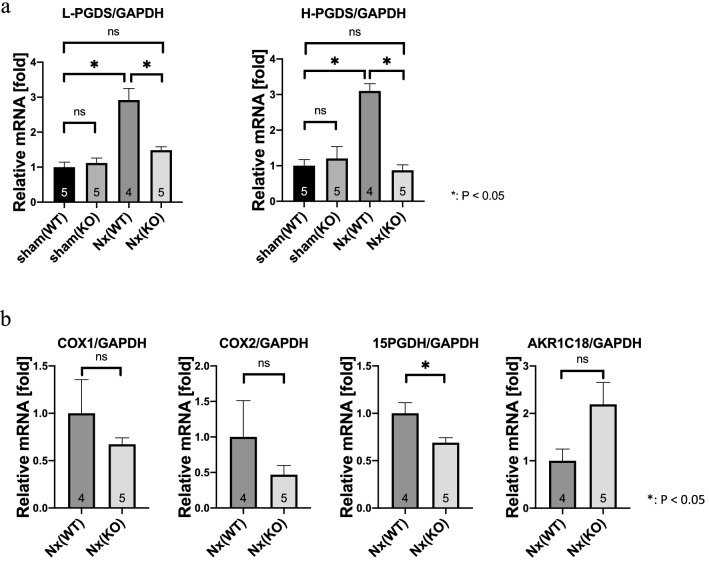
15-PGDH, a major PGD2-metabolizing enzyme, was reduced in Alox15−/− CKD kidneys
To clarify the mechanism of the increase in PGD2 in the kidneys of Alox15−/− CKD model mice, we measured the mRNA level of PGD2 synthase. PGD2 synthase (PGDS) has two isoforms: lipocalin PGDS (L-PGDS) and hematopoietic PGDS (H-PGDS) [28]. Both L-PGDS and H-PGDS levels were significantly increased under 5/6 Nx conditions when compared with those under sham conditions in the kidneys of WT mice (Fig. 8a, both P < 0.0001) indicating that increased PGD2 in CKD is due to increased PGD2 synthases. Conversely, the increase in L-PGDS and H-PGDS under 5/6 Nx conditions was significantly inhibited in Alox15−/− mice (Fig. 8a) indicating that the increase in PGD2 in Alox15−/− CKD model mice was not due to increased production by PGDS. Then, we examined mRNA levels of COX-1 and COX-2 as more upstream synthases of the prostaglandin-producing pathway. We also measured mRNA levels of 15-PGDH and AKR1C18, which are known to be major PGD2-metabolizing enzymes [29, 30]. While the mRNA levels of COX-1, COX-2, and AKR1C18 were unchanged, the mRNA level of 15-PGDH was significantly reduced in CKD kidneys of Alox15−/− mice when compared with those of WT mice (Fig. 8b, P = 0.0325), that potentially lead to the increase in PGD2 in CKD kidneys of Alox15−/− mice.
Fig. 8.
15-PGDH, a major PGD2-metabolizing enzyme, was reduced in Alox15−/− CKD kidneys. a Relative mRNA levels of each of the two PGDS synthase isoforms in the kidney tissue of sham or CKD model mice. In WT mice, both L-PGDS and H-PGDS levels were significantly increased under 5/6 Nx conditions when compared with those under sham conditions (both P < 0.0001). Conversely, the increase in L-PGDS and H-PGDS under 5/6 Nx conditions was significantly inhibited in Alox15−/− mice (P = 0.0004, < 0.0001, respectively), and their mRNA levels did not differ from those of sham WT mice. The number of samples is shown at the bottom of the bar graph. Values are mean ± SEM. One-way analysis of variance was followed by Tukey’s multiple comparisons test, *P < 0.05. b Relative mRNA levels of PGD2-related enzymes in the kidneys of CKD model mice. The mRNA level of 15-PGDH was significantly reduced in CKD kidneys of Alox15−/− mice when compared with those of WT mice (P = 0.0325). The mRNA levels of COX-1, COX-2 and AKR1C18 were not significantly changed between WT mice and Alox15−/− mice under 5/6 Nx conditions. The number of samples is shown at the bottom of the bar graph. Values are mean ± SEM. Unpaired Student’s t test, *P < 0.05
Discussion
This study demonstrated that in CKD kidney samples, both of the transcription and protein levels of Alox15 were increased, and kidney dysfunction and fibrosis were ameliorated in Alox15−/− mice. In addition, LC–MS/MS-based mediator lipidomics revealed that Alox15−/− CKD mouse kidneys had significantly increased level of PGD2 compared with WT mice. PGD2 inhibited the EMT of NRK-52E and HK-2 cells; hence, PGD2 increase may contribute to the resistance of Alox15−/− mice to renal injury and fibrosis.
The relationship between renal disease and lipid profiles using lipidomics on human serum or plasma have been extensively investigated [31–33], but not on kidney tissue. Similarly, although reports on lipidomics using CKD animal models are few [34–36], all of these studies performed lipidomics on plasma samples from a CKD animal model; however, no comprehensive lipidomics on kidney tissues from the CKD model have been reported yet. Direct lipidomics on tissue is also effective, as well as plasma samples, in identifying functional metabolites. Moreover, metabolite changes in tissues, as well as blood samples, are considerably different by organ [20]. In the present study, we thoroughly analyzed the profile of PUFA-derived lipid mediators in the kidney tissues of a CKD animal model.
This study focused on ALOX15, which is one of the major enzymes that metabolize PUFAs. In this study, both protein and mRNA expression levels of ALOX15 were clearly increased in CKD kidneys. We addressed the cell types responsible for the ALOX15 expression in CKD kidneys, and by in situ hybridization, we found an increased expression of ALOX15 mRNA in renal tubular cells in the 5/6 Nx model.
ALOX15 is involved in chronic diseases, such as atherosclerosis, and its deletion in animal disease models improves these diseases [5]. Regarding the association of ALOX15 with renal disease, proteinuria is decreased in Alox15−/− mice under glomerular injury by a streptozotocin-induced diabetic nephropathy model [37]. However, this diabetes mellitus mouse model does not show any decline in kidney function; thus, the mechanism on how ALOX15 is associated with impaired kidney function in CKD models has remained unclear. In the present study, using the 5/6 Nx mouse model, we found that ALOX15 deletion ameliorated kidney dysfunction and renal fibrosis in a CKD model.
To date, only IL-4 and IL-13 in monocytes are known to be regulatory factors that directly increase the expression of ALOX15 [5]. However, it remains unclear what increases ALOX15 in renal tubular cells. A variety of cytokines and uremic toxins are known to be increased in the plasma and kidney in CKD [38, 39]. Among them, there might be novel regulators of ALOX15. Further studies are needed to elucidate the major regulatory factors of ALOX15 in CKD.
In our studies to identify specific intervening oxylipins that link Alox15 deletion to its renoprotective effect in the 5/6 Nx model, we found that PGD2 may be involved in the resistance to renal damage caused by ALOX15 deletion. As a general understanding, to generate PGD2, COX-1 and COX-2 produce PGG2 from arachidonic acids, which is then converted into PGH2. When PGH2 is metabolized by PGD2 synthase (PGDS), PGD2 is produced [28]. PGDS has two genetically distinct isoforms, namely, lipocalin-type PGDS (L-PGDS) and hematopoietic-type PGDS (H-PGDS) [28]. Although PGD2 was increased in the Alox15−/− CKD mouse kidneys, neither L-PGDS nor H-PGDS was increased in the Alox15−/− CKD mouse kidneys, despite the large increase in PGDS in the WT CKD mouse kidneys (Fig. 8a). Furthermore, we also revealed that neither COX-1 nor COX-2 was increased in the Alox15−/− CKD mouse kidneys (Fig. 8b).
These results suggest that the increase in PGD2 in the Alox15−/− CKD mouse kidneys was not due to an increase in synthases but due to the increased substrate availability or an inhibition of degradation, and in fact, we have revealed a reduction in 15-PGDH, one of the major PGD2 metabolizing enzymes [29], in the Alox15−/− CKD mouse kidneys (Fig. 8b). Further investigation is needed to elucidate the reason why the deletion of Alox15 leads to decrease in 15-PGDH.
As mentioned above, PGD2 could be involved in the resistance to renal injury caused by ALOX15 loss. In this study, PGD2 inhibited EMT by TGF-β1 in NRK-52E and HK-2 cells, representing proximal tubular cells. PGD2 binds to two different G protein-coupled receptors, namely, DP1 and DP2, whose functions are different [28]. However, in our in vitro experiments, PGD2 was effective at concentrations ranging from 4 to 32 μM, which are relatively high, considering that the Ki values of DP1 and DP2 are 1.7 nM and 2.4 nM, respectively [28]. This result indicates that PGD2 may exert antifibrotic effects not via G protein-coupled receptors but via other pathways such as PPARγ, through which 15-d-PGJ2, a downstream PGD2 metabolite, is known to exert its effects [28].
In this study, which focuses on lipid metabolic enzymes and their metabolites, ALOX15 inhibition and/or PGD2 administration could be a promising therapeutic target for CKD. Unfortunately, no ALOX15 specific inhibitor that can be used in clinical practice has been developed [40, 41]. Therapeutic targeting of downstream functional metabolites, such as PGD2, rather than inhibition of fatty acid metabolizing enzymes, which affect various metabolites, could be a novel ideal CKD therapy.
Supplementary Information
Below is the link to the electronic supplementary material.
Acknowledgements
We thank all members of the Uchida Laboratory for their helpful discussions regarding this work, and Ms. Mie Honda for technical assistance in LC-MS analyses. This work was supported in part by Grants-in-Aid for Scientific Research (KAKENHI) from Japan Society of the Promotion of Science (grant numbers of S.U.: 25221306-00, 19H01049 and 18K19534; grant numbers of E.S.: 16H05314 and 19H03672; grant numbers of M.A.: 15H05897, 15H05898 and 20H00495), grants from Yukiko Ishibashi Foundation and from Salt Science Research Foundation (1925 and 2030).
Author contributions
NT, MA and ES designed the study; NT and HK carried out experiments; NT, HK, MA, ES, AU, TF, TF, TF, TY, YM, KA, KY, FA, KS, SM, TM, TR and SU analyzed the data; NT and ES made the figures; NT and ES drafted and revised the paper; all authors approved the final version of manuscript.
Data availability
All data are available from the corresponding author upon reasonable request.
Compliance with ethical standards
Conflict of interest
The authors declare no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Makoto Arita and Eisei Sohara equally contributed to this work.
Contributor Information
Makoto Arita, Email: makoto.arita@riken.jp.
Eisei Sohara, Email: esohara.kid@tmd.ac.jp.
References
- 1.Serhan CN. Pro-resolving lipid mediators are leads for resolution physiology. Nature. 2014;510(7503):92–101. doi: 10.1038/nature13479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Serhan CN, Chiang N. Resolution phase lipid mediators of inflammation: agonists of resolution. Curr Opin Pharmacol. 2013;13(4):632–640. doi: 10.1016/j.coph.2013.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gabbs M, Leng S, Devassy JG, Monirujjaman M, Aukema HM. Advances in our understanding of oxylipins derived from dietary PUFAs. Adv Nutr. 2015;6(5):513–540. doi: 10.3945/an.114.007732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dennis EA, Norris PC. Eicosanoid storm in infection and inflammation. Nat Rev Immunol. 2015;15(8):511–523. doi: 10.1038/nri3859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Singh NK, Rao GN. Emerging role of 12/15-Lipoxygenase (ALOX15) in human pathologies. Prog Lipid Res. 2019;73:28–45. doi: 10.1016/j.plipres.2018.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kuhn H, Banthiya S, van Leyen K. Mammalian lipoxygenases and their biological relevance. Biochim Biophys Acta. 2015;1851(4):308–330. doi: 10.1016/j.bbalip.2014.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bradding P, Redington AE, Djukanovic R, Conrad DJ, Holgate ST. 15-lipoxygenase immunoreactivity in normal and in asthmatic airways. Am J Respir Crit Care Med. 1995;151(4):1201–1204. doi: 10.1164/ajrccm/151.4.1201. [DOI] [PubMed] [Google Scholar]
- 8.Rossaint J, Nadler JL, Ley K, Zarbock A. Eliminating or blocking 12/15-lipoxygenase reduces neutrophil recruitment in mouse models of acute lung injury. Crit Care. 2012;16(5):R166. doi: 10.1186/cc11518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kayama Y, Minamino T, Toko H, Sakamoto M, Shimizu I, Takahashi H, Okada S, Tateno K, Moriya J, Yokoyama M, Nojima A, Yoshimura M, Egashira K, Aburatani H, Komuro I. Cardiac 12/15 lipoxygenase-induced inflammation is involved in heart failure. J Exp Med. 2009;206(7):1565–1574. doi: 10.1084/jem.20082596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kronke G, Katzenbeisser J, Uderhardt S, Zaiss MM, Scholtysek C, Schabbauer G, Zarbock A, Koenders MI, Axmann R, Zwerina J, Baenckler HW, van den Berg W, Voll RE, Kuhn H, Joosten LA, Schett G. 12/15-lipoxygenase counteracts inflammation and tissue damage in arthritis. J Immunol. 2009;183(5):3383–3389. doi: 10.4049/jimmunol.0900327. [DOI] [PubMed] [Google Scholar]
- 11.Sun L, Xu YW, Han J, Liang H, Wang N, Cheng Y. 12/15-Lipoxygenase metabolites of arachidonic acid activate PPARgamma: a possible neuroprotective effect in ischemic brain. J Lipid Res. 2015;56(3):502–514. doi: 10.1194/jlr.M053058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gouveia-Figueira S, Karimpour M, Bosson JA, Blomberg A, Unosson J, Pourazar J, Sandstrom T, Behndig AF, Nording ML. Mass spectrometry profiling of oxylipins, endocannabinoids, and N-acylethanolamines in human lung lavage fluids reveals responsiveness of prostaglandin E2 and associated lipid metabolites to biodiesel exhaust exposure. Anal Bioanal Chem. 2017;409(11):2967–2980. doi: 10.1007/s00216-017-0243-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bellanti F, Villani R, Facciorusso A, Vendemiale G, Serviddio G. Lipid oxidation products in the pathogenesis of non-alcoholic steatohepatitis. Free Radic Biol Med. 2017;111:173–185. doi: 10.1016/j.freeradbiomed.2017.01.023. [DOI] [PubMed] [Google Scholar]
- 14.Endo J, Sano M, Isobe Y, Fukuda K, Kang JX, Arai H, Arita M. 18-HEPE, an n-3 fatty acid metabolite released by macrophages, prevents pressure overload-induced maladaptive cardiac remodeling. J Exp Med. 2014;211(8):1673–1687. doi: 10.1084/jem.20132011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kronke G, Reich N, Scholtysek C, Akhmetshina A, Uderhardt S, Zerr P, Palumbo K, Lang V, Dees C, Distler O, Schett G, Distler JH. The 12/15-lipoxygenase pathway counteracts fibroblast activation and experimental fibrosis. Ann Rheum Dis. 2012;71(6):1081–1087. doi: 10.1136/annrheumdis-2011-200745. [DOI] [PubMed] [Google Scholar]
- 16.Chen TK, Knicely DH, Grams ME. Chronic kidney disease diagnosis and management: a review. JAMA. 2019;322(13):1294–1304. doi: 10.1001/jama.2019.14745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Romagnani P, Remuzzi G, Glassock R, Levin A, Jager KJ, Tonelli M, Massy Z, Wanner C, Anders HJ. Chronic kidney disease. Nat Rev Dis Primers. 2017;3:17088. doi: 10.1038/nrdp.2017.88. [DOI] [PubMed] [Google Scholar]
- 18.Borgeson E, Docherty NG, Murphy M, Rodgers K, Ryan A, O'Sullivan TP, Guiry PJ, Goldschmeding R, Higgins DF, Godson C. Lipoxin A(4) and benzo-lipoxin A(4) attenuate experimental renal fibrosis. FASEB J. 2011;25(9):2967–2979. doi: 10.1096/fj.11-185017. [DOI] [PubMed] [Google Scholar]
- 19.Qu X, Zhang X, Yao J, Song J, Nikolic-Paterson DJ, Li J. Resolvins E1 and D1 inhibit interstitial fibrosis in the obstructed kidney via inhibition of local fibroblast proliferation. J Pathol. 2012;228(4):506–519. doi: 10.1002/path.4050. [DOI] [PubMed] [Google Scholar]
- 20.Kikuchi H, Sasaki E, Nomura N, Mori T, Minamishima YA, Yoshizaki Y, Takahashi N, Furusho T, Arai Y, Mandai S, Yamashita T, Ando F, Maejima Y, Isobe K, Okado T, Rai T, Uchida S, Sohara E. Failure to sense energy depletion may be a novel therapeutic target in chronic kidney disease. Kidney Int. 2019;95(1):123–137. doi: 10.1016/j.kint.2018.08.030. [DOI] [PubMed] [Google Scholar]
- 21.Arita M. Mediator lipidomics in acute inflammation and resolution. J Biochem. 2012;152(4):313–319. doi: 10.1093/jb/mvs092. [DOI] [PubMed] [Google Scholar]
- 22.Isobe Y, Itagaki M, Ito Y, Naoe S, Kojima K, Ikeguchi M, Arita M. Comprehensive analysis of the mouse cytochrome P450 family responsible for omega-3 epoxidation of eicosapentaenoic acid. Sci Rep. 2018;8(1):7954–7964. doi: 10.1038/s41598-018-26325-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Imig JD, Khan MA. Cytochrome P450 and lipoxygenase metabolites on renal function. Compr Physiol. 2015;6(1):423–441. doi: 10.1002/cphy.c150009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Capdevila JH, Falck JR. The arachidonic acid monooxygenase: from biochemical curiosity to physiological/pathophysiological significance. J Lipid Res. 2018;59(11):2047–2062. doi: 10.1194/jlr.R087882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kuda O. Bioactive metabolites of docosahexaenoic acid. Biochimie. 2017;136:12–20. doi: 10.1016/j.biochi.2017.01.002. [DOI] [PubMed] [Google Scholar]
- 26.Archambault AS, Turcotte C, Martin C, Provost V, Larose MC, Laprise C, Chakir J, Bissonnette E, Laviolette M, Bosse Y, Flamand N. Comparison of eight 15-lipoxygenase (LO) inhibitors on the biosynthesis of 15-LO metabolites by human neutrophils and eosinophils. PLoS ONE. 2018;13(8):e0202424. doi: 10.1371/journal.pone.0202424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Biringer RG. The enzymology of human eicosanoid pathways: the lipoxygenase branches. Mol Biol Rep. 2020 doi: 10.1007/s11033-020-05698-8. [DOI] [PubMed] [Google Scholar]
- 28.Herlong JL, Scott TR. Positioning prostanoids of the D and J series in the immunopathogenic scheme. Immunol Lett. 2006;102(2):121–131. doi: 10.1016/j.imlet.2005.10.004. [DOI] [PubMed] [Google Scholar]
- 29.Tai HH, Ensor CM, Tong M, Zhou H, Yan F. Prostaglandin catabolizing enzymes. Prostaglandins Other Lipid Mediat. 2002;68–69:483–493. doi: 10.1016/S0090-6980(02)00050-3. [DOI] [PubMed] [Google Scholar]
- 30.Byrns MC, Duan L, Lee SH, Blair IA, Penning TM. Aldo-keto reductase 1C3 expression in MCF-7 cells reveals roles in steroid hormone and prostaglandin metabolism that may explain its over-expression in breast cancer. J Steroid Biochem Mol Biol. 2010;118(3):177–187. doi: 10.1016/j.jsbmb.2009.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhao YY, Vaziri ND, Lin RC. Lipidomics: new insight into kidney disease. Adv Clin Chem. 2015;68:153–175. doi: 10.1016/bs.acc.2014.11.002. [DOI] [PubMed] [Google Scholar]
- 32.Jia L, Wang C, Zhao S, Lu X, Xu G. Metabolomic identification of potential phospholipid biomarkers for chronic glomerulonephritis by using high performance liquid chromatography-mass spectrometry. J Chromatogr B Analyt Technol Biomed Life Sci. 2007;860(1):134–140. doi: 10.1016/j.jchromb.2007.10.033. [DOI] [PubMed] [Google Scholar]
- 33.Reis A, Rudnitskaya A, Chariyavilaskul P, Dhaun N, Melville V, Goddard J, Webb DJ, Pitt AR, Spickett CM. Top-down lipidomics of low density lipoprotein reveal altered lipid profiles in advanced chronic kidney disease. J Lipid Res. 2015;56(2):413–422. doi: 10.1194/jlr.M055624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dou F, Miao H, Wang JW, Chen L, Wang M, Chen H, Wen AD, Zhao YY. An integrated lipidomics and phenotype study reveals protective effect and biochemical mechanism of traditionally used Alisma orientale Juzepzuk in chronic kidney disease. Front Pharmacol. 2018;9:53. doi: 10.3389/fphar.2018.00053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang ZH, Vaziri ND, Wei F, Cheng XL, Bai X, Zhao YY. An integrated lipidomics and metabolomics reveal nephroprotective effect and biochemical mechanism of Rheum officinale in chronic renal failure. Sci Rep. 2016;6:22151. doi: 10.1038/srep22151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chen DQ, Chen H, Chen L, Vaziri ND, Wang M, Li XR, Zhao YY. The link between phenotype and fatty acid metabolism in advanced chronic kidney disease. Nephrol Dial Transplant. 2017;32(7):1154–1166. doi: 10.1093/ndt/gfw415. [DOI] [PubMed] [Google Scholar]
- 37.Faulkner J, Pye C, Al-Shabrawey M, Elmarakby AA. Inhibition of 12/15-lipoxygenase reduces renal inflammation and injury in streptozotocin-induced diabetic mice. J Diabetes Metab. 2015 doi: 10.4172/2155.6156.1000555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Castillo-Rodriguez E, Pizarro-Sanchez S, Sanz AB, Ramos AM, Sanchez-Nino MD, Martin-Cleary C, Fernandez-Fernandez B, Ortiz A. Inflammatory cytokines as uremic toxins: "Ni Son Todos Los Que Estan, Ni Estan Todos Los Que Son". Toxins (Basel) 2017;9(4):114. doi: 10.3390/toxins9040114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vanholder R, De Smet R, Glorieux G, Argiles A, Baurmeister U, Brunet P, Clark W, Cohen G, De Deyn PP, Deppisch R, Descamps-Latscha B, Henle T, Jorres A, Lemke HD, Massy ZA, Passlick-Deetjen J, Rodriguez M, Stegmayr B, Stenvinkel P, Tetta C, Wanner C, Zidek W, European Uremic Toxin Work Group (EUTox) Review on uremic toxins: classification, concentration, and interindividual variability. Kidney Int. 2003;63(5):1934–1943. doi: 10.1046/j.1523-1755.2003.00924.x. [DOI] [PubMed] [Google Scholar]
- 40.Sadeghian H, Jabbari A. 15-Lipoxygenase inhibitors: a patent review. Expert Opin Ther Pat. 2016;26(1):65–88. doi: 10.1517/13543776.2016.1113259. [DOI] [PubMed] [Google Scholar]
- 41.Orafaie A, Mousavian M, Orafai H, Sadeghian H. An overview of lipoxygenase inhibitors with approach of in vivo studies. Prostaglandins Other Lipid Mediat. 2020;148:106411. doi: 10.1016/j.prostaglandins.2020.106411. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data are available from the corresponding author upon reasonable request.