Abstract
Understanding the mechanisms of the Warburg shift to aerobic glycolysis is critical to defining the metabolic basis of cancer. Hereditary leiomyomatosis and renal cell carcinoma (HLRCC) is an aggressive cancer characterized by bi-allelic inactivation of the gene encoding the Krebs cycle enzyme fumarate hydratase, an early shift to aerobic glycolysis, and rapid metastasis. We observed impairment of the mitochondrial respiratory chain in tumors from HLRCC patients. Biochemical and transcriptomic analyses revealed that respiratory chain dysfunction in the tumors was due to loss of expression of mitochondrial DNA (mtDNA)-encoded subunits of respiratory chain complexes, caused by a marked decrease in mtDNA content and increased mtDNA mutations. We demonstrated that accumulation of fumarate in HLRCC tumors inactivated the core factors responsible for replication and proofreading of mtDNA, leading to loss of respiratory chain components, thereby promoting the shift to aerobic glycolysis and disease progression in this prototypic model of glucose-dependent human cancer.
One-Sentence Summary:
Kidney tumors lacking fumarate hydratase become aggressive due to a metabolic shift arising from altered mitochondrial DNA.
Editor’s Summary:
A metabolic shift from altered mitochondrial DNA
Deficiency in the metabolic enzyme fumarate hydratase distinguishes an aggressive and lethal form of kidney cancer called hereditary leiomyomatosis and renal cell carcinoma (HLRCC). Crooks et al. investigated the molecular basis for why HLRCC tumors rapidly grow and metastasize. Deficiency in fumarate hydratase led to the accumulation of the metabolite fumarate, resulting in the modification and inactivation of factors involved in mitochondrial DNA replication and proofreading. Subsequently, mitochondrial DNA mutations increased, leading to loss of mitochondria and a metabolic shift to aerobic glycolysis. Thus, lack of a crucial metabolic enzyme leads to mitochondrial dysfunction and metabolic rewiring that promote tumor progression and metastasis.
Introduction
Understanding the metabolic basis of cancer can provide the foundation for the development of effective forms of therapy for patients affected with localized as well as advanced forms of this disease (1). In the 1920s, Warburg demonstrated that many types of cancer showed greatly increased lactate fermentation even in the presence of oxygen, now termed aerobic glycolysis (2), and later suggested that mitochondrial dysfunction was responsible for the shift to aerobic glycolysis (3). Although several types of cancer exhibit aerobic glycolysis despite competent mitochondrial function, the identification of fundamental mechanisms underlying glucose dependence remains an unresolved question central to the genesis of cancer (1, 4-7).
Fumarate hydratase-deficient type 2 papillary renal cell carcinoma (RCC) exhibits characteristics consistent with the Warburg effect (8). This aggressive cancer is characterized by germline mutations of the gene encoding the Krebs cycle enzyme, fumarate hydratase (FH) (9). HLRCC-associated FH-deficient RCC is the most aggressive and lethal of the 14 known types of genetically defined heritable forms of RCC (10, 11). Whereas VHL-deficient clear cell RCC has been estimated to take up to 50 years to develop and metastasize (12), FH-deficient RCC is characterized by rapid growth with a propensity to metastasize when the primary tumor is very small (1/2 cm) within a short period of time (10). FH-deficient type 2 papillary renal cell carcinoma exhibits impaired oxidative phosphorylation, a metabolic shift to aerobic glycolysis, upregulation of the pentose phosphate pathway and a notable dependence on glucose for growth (13-16). Upregulation of the pentose phosphate pathway facilitates production of NADPH, providing cytosolic reducing equivalents for anabolic reactions including lipid biosynthesis.
FH-deficient tumors accumulate high levels of fumarate which facilitates the non-enzymatic formation of an S-(2-succinyl)-cysteine (2-SC) adduct that inactivates multiple proteins involved in tumor biology and metabolism (17). FH-deficient tumors are also characterized by enhanced glucose uptake, demonstrated in vivo by 18fluoro-2-deoxy-D-glucose positron emission tomography (18FDG-PET) imaging. In FH-deficient RCC, the shift to aerobic glycolysis is associated with a decrease in cellular oxygen consumption (16, 18-20). The origin of respiratory chain dysfunction in this prototypic model of the Warburg shift has remained elusive. Fumarate hydratase, which catalyzes the conversion of fumarate to malate in the Krebs cycle, is not known to interact directly with the mitochondrial respiratory chain and it does not directly catalyze oxygen consumption.
In the present study, we evaluated the metabolic and biochemical phenotype of FH-deficient renal cancer in human tumors as well as tumor-derived cell lines. Our results demonstrated a loss of respiratory chain components in HLRCC tumors driven by deleterious mutations in the mtDNA-encoded subunits of the respiratory complexes and by diminished mtDNA content in the tumors. Both the mutations as well as the loss of mtDNA in FH-deficient tumors were associated with fumarate-mediated covalent modification of the mtDNA replicative machinery, including POLG, the sole mitochondrial DNA polymerase which is responsible for replication and proofreading of mtDNA. Our findings demonstrate that a defining feature of the highly aggressive FH-deficient renal cancers is the inactivation of mitochondrial DNA maintenance and repair proteins, caused directly by elevated fumarate levels, which leads to loss of respiration and an early, irreversible shift to aerobic glycolysis.
Results
FH-deficient HLRCC tumors are characterized by mitochondrial dysfunction and loss of respiratory chain components
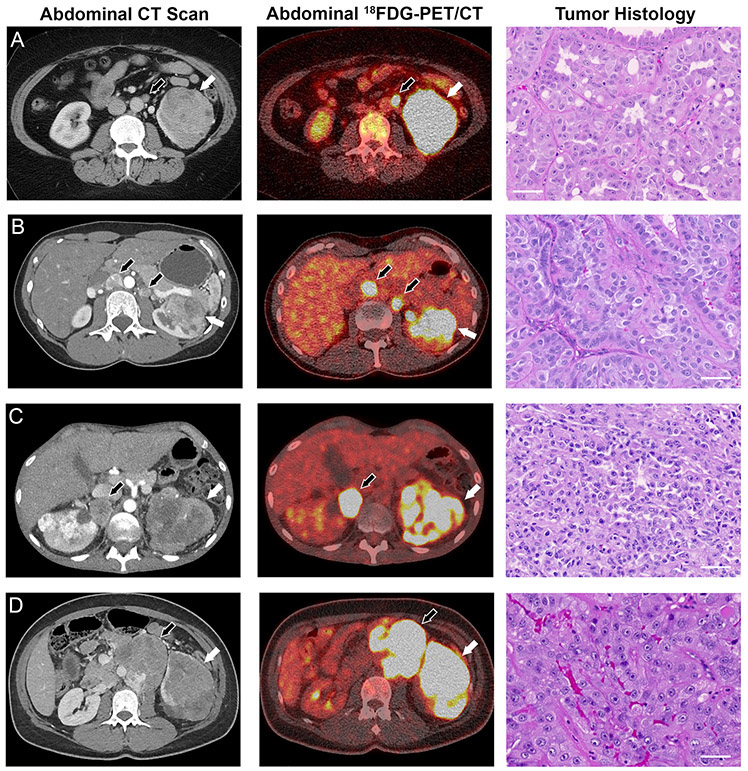
Tissue specimens from 25 patients with HLRCC-associated renal cell carcinoma with germline heterozygous pathogenic alterations in the FH gene (FH+/−) were evaluated (Table 1). Tumor specimens from 24 patients were removed during clinically indicated surgical procedures; specimens from one patient were obtained during rapid autopsy shortly after the patient’s death (Table 1). All patients gave informed consent and were managed on clinical protocols approved by the National Cancer Institute Institutional Review Board. The patients’ ages ranged from 21 to 70 years old (median 40). At the time of surgery, six patients were in their third decade and six were in their fourth decade of life. Three patients had localized disease, four had locally advanced, and eighteen had metastatic disease at the time of surgery/removal of tumors. When the present study was performed, four patients had no evidence of remaining disease, seven were alive with disease, and fourteen had died of metastatic HLRCC-associated type 2 papillary RCC. Tumors analyzed included both primary and metastatic type 2 papillary FH-deficient RCCs. Although other types of renal cell carcinoma, such as clear cell, are characterized by variable 18FDG-PET avidity in both metastatic as well as primary sites (21, 22), 20 out of 20 FH−/− metastatic lesions and seven of the eight primary FH−/− tumors evaluated by PET imaging were 18FDG-PET avid. Patients 1 and 2 and their tumor-derived FH-deficient cell lines have been reported previously (16, 18), and brief clinical histories (Supplementary Notes), abdominal CT scans, 18FDG-PET imaging, and tumor histology of patients 3-6 are presented in Figure 1A-D. In most cases in which complete nephrectomy was performed, paired non-cancerous FH+/− (heterozygous) renal cortical tissue was available for analysis. FH-deficient type 2 papillary renal cell carcinoma cell lines were derived from five HLRCC patients (Figures S1A, S1D, S1F, S1H, S1K and S2A-C).
Table 1:
Summary of patients evaluated in this study.
| Patient | M/F | Age |
FH Germline Mutation |
TNM Stage | AJCC Stage | Clinical Stage | Surgical Procedure† | Clinical Status | Age at Death |
|---|---|---|---|---|---|---|---|---|---|
| 1 | F | 49 | Q439P | III | T3a N1 M0 | Metastatic | RPLND | Dead of disease | 49 |
| 2 | F | 25 | H235D | I | T1a N0 M0 | Localized | Left partial nephrectomy | No evidence of disease | N/A |
| 3 | F | 50 | L244R | III | T3a N1 M0 | Metastatic | Left radical nephrectomy, adrenalectomy, and RPLND | Dead of disease | 54 |
| 4 | F | 40 | K371* | IV | T1b N1 M1 | Metastatic | RPLND | Alive with disease | N/A |
| 5 | M | 38 | V322G | IV | T2a N1 M1 | Metastatic | Right video-assisted thoracoscopic surgery (VATS) with parietal pleural mass biopsy | Dead of disease | 41 |
| 6 | F | 42 | R343X | IV | T3a N1 M1 | Metastatic | Cytoreductive left nephrectomy, adrenalectomy, and RPLND | Alive with disease | N/A |
| 7 | F | 24 | dupT,p.M266fs | IV | T4 N1 M1 | Metastatic | Distal pancreatectomy, splenectomy, left colectomy, RPLND and resection of retroperitoneal recurrence | Dead of disease | 26 |
| 8 | M | 27 | Q439P | III | T3b N1 M0 | Metastatic | Left radical nephrectomy with inferior vena cava thrombectomy | Dead of disease | 32 |
| 9 | M | 68 | A101* | III | T3a N0 M0 | Locally Advanced | Right radical nephrectomy and RPLND | No evidence of disease | N/A |
| 10 | M | 31 | Complete deletion | IV | T3a N1 M1 | Metastatic | Left partial nephrectomy and RPLND with bilateral pelvic LND | Dead of disease | 32 |
| 11 | M | 21 | G280D | IV | T3a N0 M1 | Metastatic | Right hepatectomy, partial left hepatectomy, and cholecystectomy | Dead of disease | 24 |
| 12 | M | 63 | Complete deletion | III | T3a N1 M0 | Metastatic | Right radical nephrectomy, RPLND, bilateral pelvic LND and cholecystectomy | Dead of disease | 63 |
| 13 | F | 56 | R233H | IV | T3a N1 M1 | Metastatic | RPLND, pelvic LND, omentectomy, left oopherectomy, splenectomy, left lower lobe pulmonary wedge resection, abdominal mass resection | Dead of disease | 61 |
| 14 | F | 43 | N340K | IV | T1a N1 M1 | Metastatic | RPLND | Alive with disease | N/A |
| 15 | M | 49 | dupT,p.M266fs | IV | T3a N1 M1 | Metastatic | RPLND | Alive with disease | N/A |
| 16 | M | 27 | T147A | IV | T3a N1 M1 | Metastatic | Right partial nephrectomy and RPLND | Dead of disease | 29 |
| 17 | F | 38 | S365G | I | T1a N0 M0 | Localized | Right partial nephrectomy | No evidence of disease | N/A |
| 18 | M | 52 | S365G | IV | T4 N1 M1 | Metastatic | Right lung lower lobectomy and mediastinal LND | Alive with disease | N/A |
| 19 | M | 70 | I81Dfs14 | IV | T4 N1 M1 | Metastatic | Left nephrectomy and RPLND | Dead of disease | 71 |
| 20 | M | 40 | Complete deletion | III | T1b N1 M0 | Locally Advanced | Left radical nephrectomy and RPLND | Alive with disease | N/A |
| 21 | M | 30 | Q439P | IV | T3b N1 M1 | Metastatic | RPLND | Dead of disease | 32 |
| 22 | M | 32 | delA p.E432fs | III | T1a N1 M0 | Locally Advanced | Left radical nephrectomy | Alive with disease | N/A |
| 23 | M | 38 | R233C | II | T2b N0 M0 | Localized | Left radical nephrectomy | Dead of disease | 48 |
| 24 | M | 23 | R233H | IV | T2b N1 M1 | Metastatic | Left radical nephrectomy (autopsy) | Dead of disease | 23 |
| 25 | M | 59 | N107T | III | T3a N1 M0 | Locally Advanced | Right partial nephrectomy, left partial adrenalectomy, and RPLND | No evidence of disease | N/A |
All surgical procedures were performed open, unless otherwise stated. RPLND: Retroperitoneal lymph node dissection; LND: Lymph node dissection; M: Male. F: Female.
Figure 1: Manifestations of FH−/− renal cell carcinoma in four HLRCC patients.
(A) A contrast-enhanced computed tomography (CT) scan revealed a 7 cm mass on the left kidney of patient 3. 18FDG-PET imaging demonstrated both intra- and extra-renal avidity indicative of locally advanced disease and lymph node metastasis, and hematoxylin and eosin (H&E) staining of tumor tissue revealed high grade type 2 papillary renal cell carcinoma. (B) A CT scan of patient 4 revealed a 6 cm left kidney tumor and enlarged retroperitoneal lymph nodes. 18FDG-PET suggested regional lymph node involvement as well, and H&E staining revealed high grade type 2 papillary renal cell carcinoma. (C) Patient 5 showed evidence of bilateral kidney lesions and retroperitoneal lymph node metastases. The kidney lesions and metastases showed avidity on 18FDG-PET imaging, and H&E staining of the tumor post mortem revealed high grade type 2 papillary renal cell carcinoma. (D) Patient 6 exhibited a mixed solid/cystic left kidney lesion with hilar adenopathy. 18FDG-PET imaging revealed PET-positive renal mass and extra-renal retroperitoneal metastases, and H&E staining revealed that the lung metastases from patient 6 were high grade type 2 papillary renal cell carcinoma. White arrows indicate primary tumors, black arrows indicate metastases. Scale bars = 50μm.
Loss of FH expression results in mitochondrial dysfunction in several model systems, including human tumor-derived cell lines (13, 16, 18, 23). We evaluated mitochondrial ultrastructure in FH−/− patient-derived tumors (Figures 2A and 2B) and in paired normal renal cortex tissue (Figure 2C). Electron microscopy analyses revealed aberrations in mitochondrial morphology in the tumor samples, which included swollen mitochondria containing fewer cristae structures that were frequently fragmented, as well as breach sites where the integrity of the inner and outer mitochondrial membranes appeared to be disrupted (Figures 2A and 2B). We also observed the presence of autophagic structures near or within some tumor mitochondria that may indicate ongoing mitochondrial autophagy and turnover (Figures 2A and 2B).
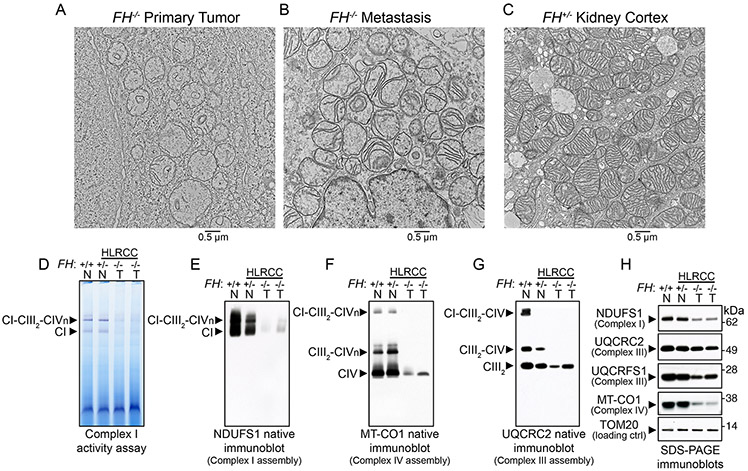
Figure 2. FH-deficient tumors exhibited altered mitochondrial ultrastructure and respiratory chain dysfunction.
(A-C) Electron microscopy of HLRCC primary tumor (A, FH−/−), metastasis (B, FH−/−), and non-tumor renal cortex from HLRCC patients (C, FH+/−). A total of 10 tissue specimens were obtained from three patients. 100-500 cells per specimen grid were evaluated before representative images were obtained. (D) In-gel assay of NADH dehydrogenase activity of Complex I (CI) in mitochondrial lysates from primary HLRCC tumors and renal cortex samples. (E) Native immunoblot (IB) for the nuclear-encoded Complex I subunit NDUFS1 in normal renal cortical and HLRCC tumor mitochondria (T, FH−/−). (F) Native IB for the mitochondrial-encoded Complex IV (CIV) subunit MT-CO1 in normal renal cortical and HLRCC tumor mitochondria to assess Complex IV levels and incorporation of Complex IV into supercomplexes containing Complex I, dimeric complex III (CIII2) and variable number of copies (from 1 to 4) of Complex IV (CIVn). (G) Native IB for the nuclear-encoded subunit of Complex III UQCRC2 in normal renal cortical and HLRCC tumor mitochondria to assess the assembly of Complex III-containing supercomplexes. (H) SDS-PAGE immunoblots for Complex I NDUFS1, Complex III UQCRC2 and Rieske protein UQCRFS1, and Complex IV MT-CO1 in normal renal cortical and tumor mitochondria. TOM20 served as the loading control for mitochondrial proteins. In (D) to (H), lane 1 consisted of renal cortex mitochondria from a non-HLRCC patient (FH+/+, N stands for normal), lane 2 was renal cortex mitochondria from HLRCC patient 10 (FH+/−, N stands for normal), and lanes 3 and 4 were primary tumor mitochondria from patients 4 and 3, respectively (FH−/− ,T stands for tumor).
Because oxidative phosphorylation (OXPHOS) complexes I-V (CI-CV) influence the architecture of the mitochondrial inner membrane, where they localize (24), we assessed respiratory chain assembly and function in FH-deficient HLRCC tumors in which the ultrastructural organization of the cristae appeared to be compromised (Figures 2A and 2B). We found that Complex I activity was decreased in HLRCC tumor mitochondria relative to renal cortex mitochondria (Figures 2D and S3A). The decreased Complex I activity was due to its defective assembly, as assessed by BN-PAGE immunoblot with an antibody against NDUFS1 (Figures 2E and S3B). Similarly, Complex IV assembly, as assessed by native immunoblot with an antibody against mitochondrially-encoded MT-CO1, was compromised in the tumor samples (FH−/−) compared to non-tumor HLRCC (FH+/−) and normal kidney cortex (FH+/+) samples (Figures 2F and S3C). Incorporation of Complex IV into supercomplexes, which are composed of Complex IV bound to Complex III and Complex I at varying stoichiometries, was lost in the tumor samples (Figures 2F and S3C). Complex III levels were also decreased and incorporation of Complex III into supercomplexes was absent in the tumor samples (Figures 2G, S3C). Complex V (F0-F1 ATPase) assembly was also impaired to varying degrees in tumor samples (Figure S3D). SDS-PAGE immunoblots on mitochondrial extracts from renal cortex and HLRCC tumor specimens revealed that levels of mtDNA-encoded subunits of Complex IV MT-CO1 (Figures 2H, S3E), MT-CO2 (Figure S3E), MT-CO3 (Figure S3E), and of CI MT-ND6 (Figure S3E) were decreased in the tumor samples. The levels of the nuclear encoded Complex I subunits NDUFS1 (Figure 2H) and NDUFB8 (Figure S3E) were also considerably lower in the tumor samples, whereas levels of the Complex III core subunit, UQCRC2, were only moderately decreased (Figure 2H). Together, these data demonstrate an impairment in the assembly of respiratory Complexes I, III and IV in HLRCC tumor mitochondria with a substantial loss of protein abundance of mitochondrially-encoded subunits.
HLRCC tumors exhibited downregulation of mitochondrial encoded mRNAs, which resulted from decreased mtDNA copy number
We performed RNAseq gene expression analysis in a cohort of primary HLRCC tumors, metastatic HLRCC tumors, and renal cortex samples obtained during surgery to assess expression levels of tricarboxylic acid (TCA) cycle enzymes and both nuclear and mitochondrial genes encoding Complexes I-V components. Unlike previous observations in clear cell RCC (25, 26), our gene expression analysis showed no evidence of coordinated down-regulation of nuclear-encoded TCA cycle or respiratory chain genes in FH−/− tumors (Figure 3A). In fact, some nuclear-encoded TCA cycle or respiratory chain genes were slightly up-regulated in HLRCC primary tumors and metastases as compared to renal cortex specimens (Figure 3A). In contrast, RNAseq analyses demonstrated a global decrease in mtDNA-encoded mRNAs in HLRCC primary tumors and metastases, with all seven Complex I genes (MT-ND1 to MT-ND6) showing from 2 to 8-fold down-regulation in HLRCC primary tumors and metastases (Figure 3A). The single mtDNA-encoded subunit of Complex III, MT-CYB, was decreased by 8-fold, and the three mtDNA-encoded subunits of Complex IV (MT-CO1 to MT-CO3) were decreased by 2-4 fold in tumors (Figure 3A). Finally, expression of the two mtDNA-encoded subunits of Complex V (MT-ATP6 and MT-ATP8) was decreased by 2-4 fold in HLRCC tumors (Figure 3A).
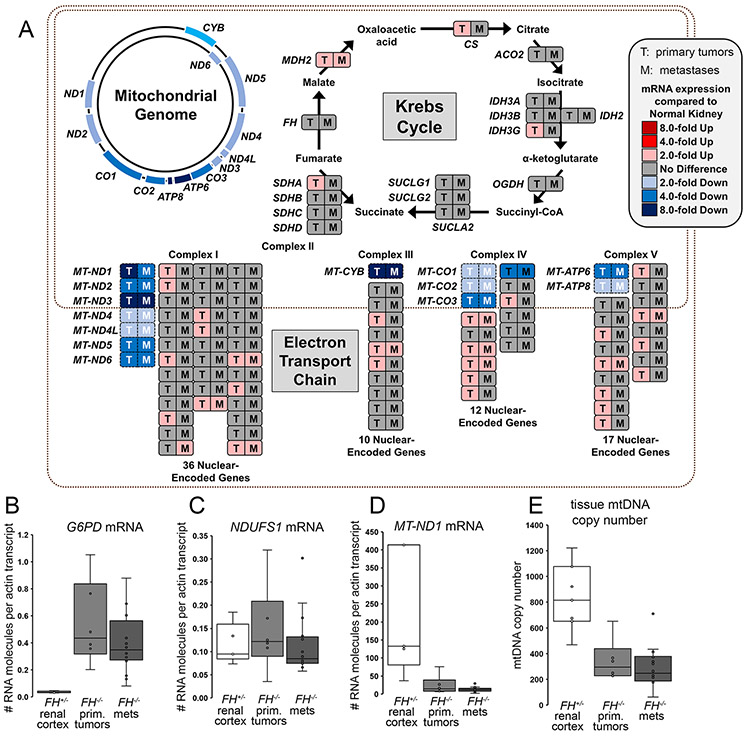
Figure 3. HLRCC FH−/− tumors exhibited down-regulation of mitochondrial encoded mRNAs, which resulted from decreased mtDNA copy number.
(A) RNAseq pathway analysis of nuclear-encoded and mtDNA-encoded genes in HLRCC tumor (T; n=5 patient samples) and metastases (M; n=16 patient samples) and paired renal cortex samples (n=5 patient samples), organized according to Krebs cycle and electron transport chain complex. (B) qPCR measurement of the mRNA expression of the NRF2 transcriptional target G6PD showed upregulation in FH−/− primary tumors (prim. tumors) and metastases (mets) versus FH+/− renal cortex (P=0.0010, cortex vs tumors, Wilcoxon Two-Sample Test). (C) qPCR revealed no significant change in mRNA expression of the nuclear-encoded NDUFS1 in FH+/− renal cortex versus FH−/− primary tumors and metastases (P=0.2522, cortex vs tumors). (D) qPCR measurement demonstrated decreased mRNA expression of MT-ND1 in FH−/− primary tumors and metastases versus FH+/− renal cortex (P=0.0020, cortex vs tumors). For (B) to (D), n=6 independent primary tumor samples, n=16 metastatic tumor samples obtained from seven patients, and n=5 independent FH+/− renal cortex samples. (E) qPCR measurement of mtDNA copy number relative to nuclear DNA content demonstrated significant (P=0.0012, cortex vs tumors) decrease in mtDNA content in FH−/− primary tumors (n=6 patient samples) and FH−/− metastases (n=16 metastatic tumor samples obtained from 14 patients) versus FH+/− renal cortex (n=7 patient samples).
Further validation of the RNA expression profiles in HLRCC primary tumors and metastases was obtained by qPCR assays. In contrast to the strong and significant induction of mRNA expression of the pentose phosphate pathway enzyme glucose-6-phosphate dehydrogenase (G6PD) in HLRCC tumors (Figure 3B) (14, 27), we observed no change in the mRNA levels for the nuclear-encoded Complex I subunit NDUFS1 (Figure 3C) and significant down-regulation of mtDNA-encoded subunits of respiratory complexes I, III, IV and V (Figures 3D and S4A-C), suggesting that the decreased NDUFS1 protein levels reported above (Figures 2E and S3B) resulted from posttranscriptional protein instability caused by impaired Complex I assembly in FH−/− tumors. RNAseq gene expression patterns in FH−/− tumors were consistent with strong upregulation of glycolysis, the pentose phosphate pathway, fatty acid biosynthesis, and glutamine uptake and utilization (Figure S5). Genes encoding enzymes that are transcriptional targets of NRF2 (nuclear factor (erythroid-derived 2)-like 2) that utilize cytosolic NADPH were also coordinately upregulated in FH−/− tumors (28) (Figures S6A-F).
Our transcriptome analyses indicated that the underlying cause of respiratory chain dysfunction in HLRCC FH−/− tumors was the selective loss of expression of mtDNA-encoded subunits of Complexes I, III, IV and V. We therefore asked whether these changes in gene expression in the tumors were mirrored by a decrease in mtDNA copy number. To address this question, we quantified mtDNA copy number in 11 primary FH−/− HLRCC tumors and 26 FH−/− metastases and compared these findings to those in normal FH+/− renal cortex DNA samples (Figure 3E). The number of mtDNA molecules decreased from an average of 833 copies per cell in FH+/− renal cortex specimens to an average of 344 and 294 copies per cell in primary and metastatic HLRCC tumors, respectively (Figure 3E), and the decreased mtDNA copy numbers in FH−/− tumors was statistically significant. These data suggest that decreased mRNA expression of genes encoded by mtDNA is a result of decreased mtDNA content in the tumors.
The activities and assembly of respiratory complexes were impaired or absent in FH−/− HLRCC tumor-derived cell lines.
Tumors are heterogeneous tissues that are composed of tumor cells as well as stromal and vascular tissues and infiltrates that contain lymphocytes and other non-cancerous cells. To evaluate the activities and assembly of the mitochondrial OXPHOS complexes in a homogeneous population of FH−/− cells, we analyzed five FH-deficient tumor cell lines derived from patients who underwent surgery at the NIH Clinical Center, including two previously-reported HLRCC cell lines UOK262 and UOK268 (16, 18) (Figures S1A-E), and the three novel tumor-derived FH−/− cell lines, UOK271, UOK348 and UOK350 (Figure S1F-L, S2A-C). For reference, we also evaluated respiratory chain function in a mtDNA-deficient clear cell RCC cell line UOK121rho0 (29) and its parental cell line, UOK121. Complex I activity and assembly were diminished in UOK262 cells as compared to cultured primary renal proximal tubule epithelial cells (RPTEC) and human renal cortical epithelial (HRCE) control cells (Figures 4A and 4B). UOK268, UOK271, UOK348 and UOK350 HLRCC cell lines showed more marked defects in CI assembly and activity (Figures 4A and 4B), specifically undetectable levels of the fully assembled CI (Figure 4B) and complete loss of CI activity (Figure 4A) to a degree comparable to that in UOK121rho0 cells which have no mtDNA and therefore cannot encode the seven Complex I subunits (Figures 4A and 4B). The protein levels of nuclear-encoded NDUFS1 were slightly decreased in all HLRCC cell lines as compared to controls (Figure 4C), whereas the levels of the mtDNA-encoded subunit MT-ND6 were unchanged in UOK262 and UOK268 cells, diminished in UOK348 cells, and undetectable in UOK271, UOK350 and UOK121rho0 cells (Figure 4C).
Figure 4. Activities and assembly of respiratory complexes were severely impaired or absent in FH−/− HLRCC tumor-derived cell lines.
(A) In-gel NADH-dehydrogenase activity assay to assess Complex I (CI) activity was performed in five FH−/− HLRCC tumor-derived cell lines, two control cell lines (RPTEC and HRCE), a mtDNA-deficient clear cell RCC cell line UOK121rho0 and its respiration-competent parental cell line, UOK121. (B) Mitochondrial lysates from the cell lines indicated in (A) were analyzed by native IB for the nuclear-encoded NDUFS1 to assess Complex I levels and assembly. CI-CIII2-CIVn designates Complex I-containing supercomplexes which include dimeric Complex III and 1-4 copies of Complex IV. (C) Mitochondrial lysates from the cell lines indicated in (A) were analyzed by SDS IB for the nuclear-encoded CI subunit NDUFS1. (D) Mitochondrial lysates from the cell lines indicated in (A) were analyzed by native IB for the core Complex III subunit UQCRC1. CIII2-CIV and CIII2-CIVn are supercomplexes containing dimeric Complex III and 1-4 copies of Complex IV. (E) Mitochondrial lysates from the cell lines indicated in (A) were analyzed by SDS IB for the Complex III nuclear encoded subunit UQCRC1 and the mtDNA-encoded Complex III subunit MT-CYB. (F) In-gel cytochrome c oxidase assay to assess Complex IV activity was performed in the cell lines indicated in (A). (G) Mitochondrial lysates from the cell lines indicated in (A) were analyzed by native IB for the mtDNA-encoded Complex IV subunit MT-CO1 to assess Complex IV assembly. (H) Mitochondrial lysates from the cell lines indicated in (A) were analyzed by SDS IB for the mtDNA-encoded Complex IV subunits MT-CO1, MT-CO2 and MT-CO3. (I) Mitochondrial lysates from the cell lines indicated in (A) were analyzed by native IB (top) for the nuclear-encoded Complex V subunit ATP5A to assess Complex V assembly (F1/F0 ATPase complex). SDS IB for ATP5A and MT-ATP6 proteins (lower panels) were performed on the same samples. Data shown in (A) to (I) are representative of three or more independent experiments. (J) NDUFS1 mRNA expression levels in FH−/− tumor cell lines and controls. (K) MT-ND1 mRNA expression levels in FH−/− tumor cell lines and controls. Data in (J) to (K) were obtained from three independent biological replicates per group and expression levels normalized to RPTEC cells, mean % control ± SD. (L) qPCR measurement of mtDNA copy number in FH−/− tumor cell lines and controls, n=3-5 biological replicates per group, means ± SD.
A slight defect in Complex III assembly was also detected in UOK262 cells and Complex III assembly was absent in UOK271, UOK348 and UOK350 cells (Figure 4D). The abundance of the nuclear DNA-encoded Complex III subunit UQCRC1 was decreased in UOK271, UOK348 and UOK350 cell lines (Figure 4E), whereas the mtDNA-encoded MT-CYB was absent in UOK271 and UOK350 cells and diminished in UOK348 cells (Figure 4E). Complex III assembly starts with the co-translational insertion of MT-CYB in the inner mitochondrial membrane, where it acts as the “seed” around which the remaining subunits coalesce (30, 31). Therefore, loss of expression of MT-CYB may explain the defective Complex III assembly in UOK271, UOK348 and UOK350 cell lines (Figures 4D and 4E). Defective Complex I assembly in UOK262 and UOK268 accounted for the loss of Complex III-containing supercomplexes (Figures 4B and 4D). Complex IV (cytochrome c oxidase) activity (Figure 4F) and assembly (Figure 4G) were impaired in UOK262 and UOK268 cells and absent in UOK271 cells, UOK348 cells and UOK350 cells, in which total levels of the mtDNA-encoded subunits MT-CO1, MT-CO2 and MT-CO3 were either undetectable (UOK271 and UOK350 cells, Figure 4H) or diminished (UOK348 cells, Figure 4H). MT-CO1, which contains a heme-a group and a binuclear heme a3/copper (CuB) center, is the core subunit around which CIV assembles (31, 32). Neither of the other two mtDNA-encoded Complex IV core subunits, MT-CO2, which ligates the other copper center (CuA), and MT-CO3, which does not play a direct role in catalysis (33), can initiate Complex IV assembly in the absence of MT-CO1 (32). Diminished or absent expression of the core subunits of Complex IV explains the reduced levels or complete loss of Complex IV in HLRCC cell lines.
Complex V (F0-F1 ATPase) assembly was impaired in UOK271 and UOK350 cells to levels comparable to those in UOK121rho0 cells (Figure 4I), in which only the nuclear-encoded F1 module formed. Defective assembly of Complex V in UOK271 and UOK350 FH-deficient cells was due to loss of expression of MT-ATP6 (Figure 4I, lower panel), which together with MT-ATP8, forms the F0 subcomplex that contains mtDNA-encoded subunits (Figure 4I). Consistent with the impaired assembly and loss of function of respiratory complexes I, III, IV and V in the cell lines (Figures 4A-I), oxygen consumption rates were markedly reduced in UOK262 cells and nearly undetectable in UOK268 cells (Figure S7), as previously reported (16, 18, 20). UOK271, UOK350, and UOK348 cells showed loss of respiration to levels comparable to that of UOK121rho0 cells (Figure S7), which lacks respiratory complexes.
mRNA expression of the nuclear DNA (nDNA)-encoded Complex I subunit NDUFS1 was similar across all cell lines (Figure 4J), indicating the absence of a coordinated transcriptional down-regulation of nuclear-encoded respiratory chain gene expression in the HLRCC cell lines. MT-ND1 mRNA expression levels were not decreased in UOK262, UOK268, or UOK348 cells compared to RPTEC or UOK121 ccRCC cells (Figure 4K), whereas UOK271 and UOK350 cells exhibited undetectable or nearly undetectable expression of MT-ND1 mRNA, respectively (Figure 4K). Similar patterns of expression were found in the FH−/− HLRCC cell lines for the other mitochondrial genes encoding subunits of Complex III-V (Figure S8A-C). mtDNA copy numbers in the cell lines ranged from slightly decreased in UOK262 to ~50% lower in UOK268 and UOK348 cells, to absent or nearly undetectable in UOK271 and UOK350 cells, respectively, compared to RPTEC control cells (Figure 4L). Thus, the deficiency of mtDNA and the resulting loss of mtDNA-encoded mRNAs of subunits of the respiratory complexes provided an explanation for the defective activity and assembly of the respiratory complexes in UOK271 and UOK350 cells. However, the respiratory chain defects of UOK262, UOK268 and UOK348 cells could not be explained by mtDNA deficiency, because these cell lines still contained mtDNA and showed expression of mtDNA-encoded mRNAs.
Re-expression of wild-type FH in HLRCC tumor-derived cell lines did not restore functionality of the respiratory chain
The data presented so far demonstrated that malfunction of respiratory chain complexes differed in the five FH−/− HLRCC cell lines (Figure 4A-I). Complex I inactivation in FH-deficient murine cells has been previously suggested to be caused by impaired biogenesis of its eight iron-sulfur clusters due to succination of the Fe-S biogenesis factor Nfu1 (20). To evaluate a potential causal association between accumulation of fumarate and respiratory chain disfunction, we restored FH expression in all five HLRCC cell lines. Robust FH activity was detected in all five HLRCC cell lines after polyclonal expression of HA-tagged FH (Figure 5A). The presence of the HA tag resulted in faster migration of the recombinant FH tetramers on native gels compared to HRCE and RPTEC cells (Figure 5A). Immunoblots confirmed high levels of expression of wild type HA-FH relative to untransfected cells (Figure 5B). However, restoration of FH expression in the HLRCC cell lines caused a minor increase in Complex I assembly in UOK262 cells and a partial restoration of Complex I activity and assembly in UOK350 cells, but did not affect Complex I activity or assembly in UOK268, UOK271, and UOK348 cells (Figures 5C and 5D). Similarly, restoration of FH activity led to a small recovery of levels and/or activities of respiratory complexes III, IV and V in UOK262 and UOK350 cells (Figures 5E-H), but it did not enhance respiratory chain activities in the other three cell lines. NDUFS1 protein abundance was moderately restored in UOK262 and UOK350 cells upon re-expression of WT FH, reflecting the partial restoration of Complex I assembly in these cell lines (Figure 5D). In contrast, UQCRC2 remained depleted in UOK271 and UOK348 cells (Figure 5I). Notably, MT-CO1 was undetectable in UOK271 cells even after restoration of FH activity (Figure 5I), whereas MT-CO1 protein levels were substantially increased in UOK350 cells upon re-expression of FH (Figure 5I).
Figure 5. Re-expression of wild type FH in HLRCC tumor-derived cell lines did not restore a functional respiratory chain.
(A) In-gel assay of FH activity in five HLRCC FH−/− cell lines and in the FH−/− cell lines after re-expression of wild type HA-tagged FH. HRCE and RPTEC cells were used as controls. The presence of the HA tag resulted in a faster mobility of the recombinant FH tetramers (FHWT-HA) on native gels compared to endogenous FH (FHWT) in HRCE and RPTEC cells. (B) Immunoblots for FH and HA-tag in the same cell lines as in (A). The cytoplasmic protein malate dehydrogenase (MDH1) was used as a loading control for total protein. (C) In-gel NADH dehydrogenase assay to assess Complex I (CI) activity in the same cell lines as in (A). CI-CIII2-CIVn indicates supercomplexes containing Complex I, dimeric Complex III and one to four copies of Complex IV. (D) Mitochondrial lysates from the cell lines indicated in (A) were analyzed by native IB for the nuclear-encoded NDUFS1 to assess Complex I levels and assembly. (E) Mitochondrial lysates from the cell lines indicated in (A) were analyzed by native IB for the nuclear-encoded Complex III subunit UQCRC2 to assess Complex III levels. (F) In-gel cytochrome c oxidase activity for Complex IV activity was performed in the same cell lines as in (A). (G) Mitochondrial lysates from the cell lines indicated in (A) were analyzed by native IB for the mtDNA-encoded MT-CO1 to assess Complex IV assembly. (H) Mitochondrial lysates from the cell lines indicated in (A) were analyzed by native IB for the nuclear-encoded ATP5A to assess Complex V assembly. (I) Mitochondrial lysates from the cell lines indicated in (A) were analyzed by SDS IB for the Complex I subunit NDUFS1, Complex III subunit UQCRC2, Complex IV subunit MT-CO1 and Complex V subunit before and after restoration of FH expression. Data shown in (A) to (I) are representative of three independent experiments.
Our observations indicate that loss of FH activity had a lasting impact on mitochondrial function even after FH activity was restored and fumarate levels were reduced in the tumor-derived cells. Together, these data demonstrated that restoration of FH activity in FH−/− HLRCC cells was not sufficient to fully normalize the multiple deficiencies in the respiratory chain to levels found in normal control RPTEC cells (Figure 5A-I). Accordingly, we sought changes in mitochondria that could be irreversible and performed sequencing of mtDNA from FH−/− tumor-derived cell lines and tumors.
Increased mutational loads in mtDNA-encoded genes associated with loss of FH in HLRCC cell lines and tumors
To gain insights into the nature of the mitochondrial respiratory dysfunction in HLRCC tumors and tumor-derived UOK262, UOK268 and UOK348 cell lines, we sequenced mtDNA using both Sanger and next generation sequencing platforms. We identified notable mutations in the mitochondrial DNA of all cell lines that still retained mtDNA. We found that UOK262 cells harbored a homoplasmic m.G14607A (p.P23S) mutation in the MT-ND6 subunit of Complex I that was predicted to be pathogenic by multiple databases (Figures 6A, S9A, Table S1). Notably, this mutation was also specifically found at homoplasmic levels in the tumor from which UOK262 cells were derived but was absent in the germline mtDNA from the blood of the patient from which UOK262 cells were derived (Figures 6A and S9A). We also detected a somatic mtDNA m.T11552C (p.S265P) missense mutation (of medium predicted pathogenicity) in the MT-ND4 subunit of Complex I in UOK262 that was present at 36% heteroplasmy in the cell line and 24% in the tumor from which the cell line was derived (Figures 6A and S9B), but absent from the germline. UOK268 cells harbored a homoplasmic m.T4891C (p.I141T) mutation of high predicted pathogenicity in the MT-ND2 subunit of Complex I, a substitution that was not present in the patient’s germline (Figures 6B and S9C). Due to loss of mtDNA, we were unable to amplify or sequence the mitochondrial genome of UOK271 or UOK350 cells (Figures 6C and 6D); however, sequencing data on the tumor from which UOK271 was derived revealed a heteroplasmic m.C13679T (p.P448L) mutation in the MT-ND5 subunit of Complex I of medium pathogenicity (Table S1). Notably, the site variability of these mtDNA positions in the HmtDB (Human Mitochondrial DataBase(34)) database is zero, suggesting that these mutations are not tolerated in normal human tissues.
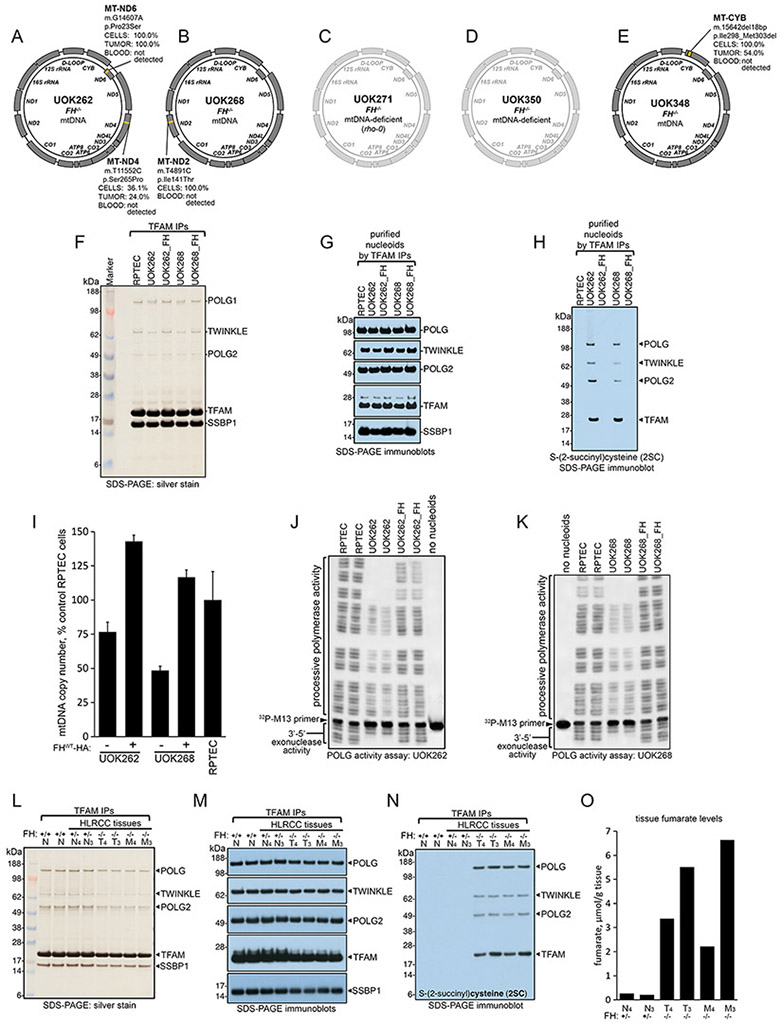
Figure 6. Fumarate covalently modified the core components of mitochondrial nucleoids in FH-deficient cell lines and primary tumors, leading to loss of mtDNA and increased mutational loads in mtDNA genes.
(A-E) Schematic representation of the mutations in the mtDNA genome identified in the five HLRCC FH−/− cell lines. In C and D, the mtDNA of UOK271 and UOK350 cells is shadowed because these cells have entirely or nearly entirely lost their mitochondrial genome, respectively. (F) Silver staining of nucleoid core components purified by immunoprecipitation of TFAM in control cells (RPTEC) and in the FH−/− HLRCC UOK262 and UOK268 cell lines before and after re-expression of FH (UOK262_FH; UOK268_FH). (G) Immunoblots to core nucleoid components POLG, POLG2, TWINKLE, TFAM and SSBP1 on nucleoids isolated by differential centrifugation and purified by TFAM immunoprecipitations. (H) Samples as in (F) were immunoblotted for 2SC. Data in F-H are representative of five independent experiments. (I) mtDNA copy number relative to control RPTEC cells in FH−/− HLRCC cell lines before and after re-expression of WT FH (n=3-5 biological replicates per cell line); this experiment was repeated three times. (J and K) POLG activity assay of processive and exonuclease activities of POLG present in purified nucleoids from UOK262 (J) or UOK268 (K) cells. Data are representative of four independent experiments. (L) Silver staining of nucleoid core components purified by differential centrifugation steps and TFAM immunoprecipitations in two FH+/+ normal kidney cortex samples, in two HLRCC normal kidney cortex samples FH+/− (N4 and N3), in two primary tumors FH−/− (T4 and T3), and in two metastatic tumors FH−/− (M4 and M3). (M) Immunoblots to the core nucleoid components POLG, POLG2, TWINKLE, TFAM and SSBP1 isolated from tissue samples as in (L). (N) Samples as in (L) were immunoblotted for 2SC. Data in L-N are representative of four independent experiments. (O) Fumarate levels in HLRCC FH+/− normal kidney cortex samples (N4, N3), in two HLRCC FH−/− primary tumors (T4 and T3) and in two metastases (M4 and M3).
Finally, we discovered that UOK348 cells harbored a homoplasmic 18 base pair deletion in the MT-CYB gene (m.15642del18bp, p.I298-M303del, Figures 6E and S10), which was also found in the tumor from which this cell line was derived (Figures 6E and S10, Table S1), but which was absent from the germline. A mitochondrial myopathy patient with severe exercise intolerance and multisystem disorders including deafness, retinal pigmentary dystrophy, and severe brain lactate accumulation was reported to harbor a nearly identical 18bp deletion of the MT-CYB gene at 50% heteroplasmy in her muscle tissue (35). Cybrids harboring this 18bp MT-CYB deletion failed to grow in the absence of glucose and demonstrated greatly impaired mitochondrial oxidative phosphorylation (35). The presence of these pathogenic mutations in our FH-deficient tumor cell lines is consistent with the observed persistent defect in respiratory chain function following restoration of FH activity. We conclude that the respiratory chain defects of UOK262, UOK268 and UOK348 cell lines are due to the acquisition of deleterious mtDNA mutations that impair assembly of a functional respiratory chain.
We also performed mtDNA sequencing analyses on 25 FH−/− samples obtained from primary and metastatic tumors (Table S1, Figure S14). 22 samples showed one or more somatic changes in mtDNA sequence (Table S1). Samples with mtDNA changes had on average 2.27 mutations per sample (range of 1-7 mtDNA changes; Figures S11A-C). Out of the 50 total somatic mtDNA changes identified, 1 change (2%) was present in a non-coding region, 11 changes (23.4%) were present in the non-coding D-loop region, 2 (4.3%) were present in tRNA-coding regions, 8 (17%) changes were in rRNA-coding regions, and 28 changes (53.2%) were in genes encoding proteins (Figure S11A, Table S1). Notably, synonymous protein coding mtDNA mutations were present only at low levels of heteroplasmy. The heteroplasmy levels clustered at either >70% (40% of samples) or <25% (56% of samples) (Figures S12A and B), which was not consistent with a normal or uniform distribution. 14 samples presented with at least one base change above 70% heteroplasmy, whereas six tumor samples had base changes below 25% mutation load (Figure S13). Considering the presence of varying amounts of stromal, vascular, and immune FH+/− cell infiltrates in the tumors we have analyzed in this study, the levels of heteroplasmy reported in tumor mtDNA mutations is likely an underestimate of that which is specific to the tumor cells only.
We further classified samples by the types of mtDNA mutations present, based on the mutational classification system developed by Ricketts et al. in the comprehensive TCGA analysis of renal cell carcinomas (36). According to this classification, 11 tumors (44%; Figure S15A) were classified as carrying pathogenic mutations (36% mutation category 4 and 8% mutation category 5; Figure S15B), whereas 14 tumors (56%) belonged to the “no pathogenic mutation” group (Figure S15A), carrying variants classified as category 1 (12%), 2 (20%), or 3 (24%) (Figure S15C), overall suggesting an increased pathogenic mutational load as compared to chromophobe (20%), papillary (33%), or clear cell RCC (13%) (36). Together, these data corroborate the accumulation of mtDNA variants in HLRCC tumor samples with our initial observation of mtDNA mutations in our FH−/− tumor-derived cell lines and suggest that a selective pressure for the accumulation of pathogenic mtDNA mutations in FH−/− tumors might exist.
Given the prevalence of mtDNA mutations that we identified in FH−/− HLRCC tumors, we assessed nuclear DNA mutational loads in five FH−/− tumors that were present in the TCGA analysis of RCC (36). Data from the five FH−/− tumors present in the TCGA analysis was not suggestive of a high number of somatic mutations in protein coding genes as compared to non-FH type 2 papillary RCC tumors, Type 1 papillary RCC tumors, and clear cell RCC tumors (Figure S16). Moreover, spectral karyotype analysis of our cell lines did not reveal frank aneuploidy or excessive chromosomal rearrangements indicative of genomic instability (Figure S17). These data are consistent with the notion that FH-deficient tumors grow rapidly as compared to other RCC tumor types such as ccRCC (11, 37), which can take decades to grow into a clinically-detectable tumor (12). To explore the origins of mtDNA mutations in FH-deficient tumors, we next evaluated the machinery responsible for the maintenance and replication of mtDNA.
Fumarate covalently modified the core components of mitochondrial nucleoids in FH-deficient cells and primary tumors
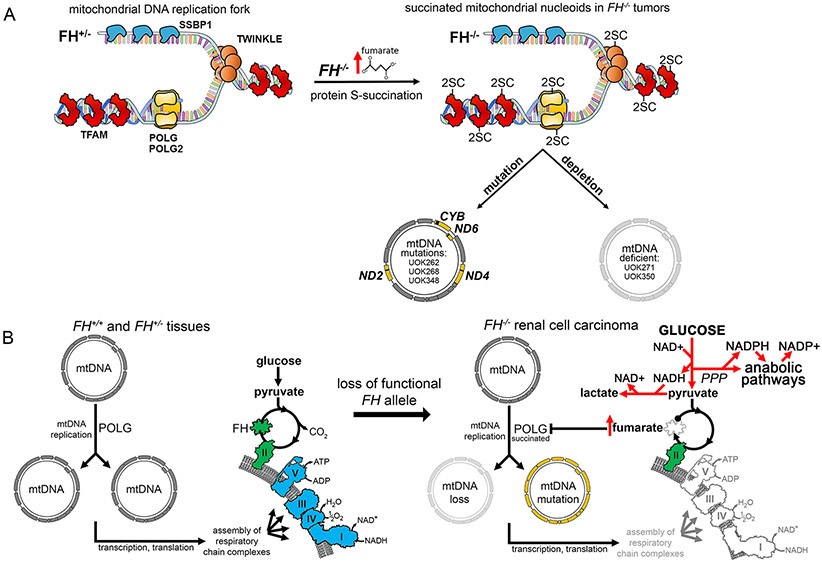
Although initially assumed to be naked and prone to damage, mtDNA is now known to be compacted into discrete nucleoprotein complexes known as nucleoids, which are composed of five core proteinaceous components (POLG, POLG2, TWINKLE, TFAM, and SSBP1) that are responsible for the maintenance and replication of mtDNA (38). Loss and mutations of mtDNA occur upon depletion or mutations of nucleoid components (39-42). The major protein constituent of mammalian nucleoids, the mitochondrial transcription factor A (TFAM), which is present at levels of about 1000 molecules per mtDNA molecule, has been reported to be covalently modified by fumarate at Cys246 in FH-deficient tumors and patient-derived cell lines (43). Succination of many diverse cytoplasmic and mitochondrial proteins has been reported to cause their inactivation. These include mitochondrial aconitase (44), the Fe-S biogenesis factors ISCU, NFU1, BOLA1 and BOLA3 (20), the glycolytic enzyme glyceraldehyde 3-phosphate dehydrogenase (GAPDH) (45), the Kelch-like ECH-associated protein 1 (KEAP1) [which loses its ability to repress the NRF2-mediated antioxidant response pathway (27, 46)], and α- and β-tubulin (47). TFAM mutations in patients and knockdown studies in mammalian cells have shown that this abundant nucleoid component is essential for maintaining mtDNA copy number (48-50). Our analyses on FH-deficient HLRCC tumors and cell lines revealed loss of mtDNA and increased mutational loads in the tumors, which, together with the reported succination of TFAM (43), prompted us to analyze the full complement of mtDNA maintenance and replication machinery in nucleoids isolated from HLRCC cell lines and tumors.
Silver staining of nucleoids purified by velocity sedimentation and TFAM immunoprecipitation confirmed the presence of all five main nucleoid components in UOK262 and UOK268 cells (Figure 6F). The identity of each of the nucleoid-associated proteins was confirmed by Western blot (Figure 6G) and mass spectrometry analysis (Table S2). Using an antibody raised against the 2SC covalent modification in control and FH−/− HLRCC cells, we found that four of the five core nucleoid components (POLG, TWINKLE, POLG2, and TFAM) were robustly succinated in UOK262 and UOK268 cells (Figure 6H). Succination of SSBP1 was not observed because this protein does not contain any cysteine residues. Restoration of WT FH activity in UOK262 and UOK268 cells resulted in loss of succination of the nucleoid components (Figure 6H), further demonstrating that succination of nucleoid components in HLRCC cells was driven by accumulation of fumarate due to loss of FH activity (Figure 6H). Notably, restoration of FH activity in UOK262 and UOK268 cells, which exhibited 25 to 45% reduction in mtDNA content compared to RPTEC control cells (Figure 4L), increased the mtDNA copy number level by 1.9-fold and 2.4-fold in UOK262 and UOK268 cells, respectively (Figure 6I). These data suggested that loss of FH expression and succination of the nucleoid components impaired mtDNA replication and maintenance in FH-deficient cells, resulting in progressive loss of mtDNA. We adapted an assay to assess the activity of POLG in nucleoids purified from FH-deficient cells (Figures 6J and 6K). We found that the POLG present in nucleoids obtained from UOK262 and UOK268 cells was deficient in both its processivity as well as its error-correcting 3’−5’ exonuclease activity, as observed by decreased laddering of radioactive 32P-labeled M13 primer derivatives in both the upward (processive) and downward (exonuclease) directions (Figures 6J and 6K). Re-expression of WT FH enzyme in UOK262 or UOK268 cells restored the processive and exonuclease activities of POLG (Figures 6J and 6K), demonstrating that fumarate-mediated succination was the cause of POLG impairment in UOK262 and UOK268 cells.
Widespread succination of mitochondrial proteins was specifically detected in FH−/− HLRCC primary tumors (Figure S18, input lanes), but not in control FH+/− renal cortex samples (Figure S18). Moreover, POLG immunoprecipitated from HLRCC tumors showed reactivity when immunoblotted for the 2SC modification (Figure S18), indicating that POLG was also succinated in primary tumors, consistent with the results in the cell lines. Isolation of nucleoids from FH−/− HLRCC primary and metastatic tumors (Figures 6L and 6M) revealed that POLG, POLG2, TWINKLE, and TFAM were succinated in tumors (Figure 6N), consistent with the results obtained in the cell lines and with the accumulation of fumarate in FH−/− HLRCC tumors (Figure 6O).
Mutations in POLG also cause error-prone mtDNA replication, leading to accumulation of point mutations over time as well as depletion of mtDNA (42). We propose that succination of POLG in primary tumors and FH−/− cells may lead to the same phenotype (Figure 7A), as demonstrated by the selective increase in mtDNA mutational loads in HLRCC tumors and cell lines. FH re-expression did not correct the mtDNA mutations in UOK262, UOK268 or UOK348 cells (Figure S9A-C, S10), indicating that the inability to fully restore respiratory chain activity in these cells upon re-expression of wild type FH was likely due to the deleterious nature of the mutations that they had acquired. The absence or near-absence of mtDNA in UOK271 and UOK350 cells, respectively, suggested that mtDNA loss occurred in these two cell lines likely as a result of fumarate-mediated succination of the replication machinery (Figure 7A), in line with the phenotypes observed upon inactivation of POLG or TFAM (42, 48-50).
Figure 7. Summary of findings in the study.
(A) Schematic illustrating the core proteinaceous nucleoid components bound to mtDNA at a replication fork. Nucleoid components are covalently modified at cysteine residues (to generate 2-succinocysteine modifications) in the presence of elevated fumarate concentrations in FH−/− tumor mitochondria. 2-succinocysteine modification of the DNA polymerase POLG leads to decreased and error-prone polymerase activity, which over time leads to loss and mutation of mtDNA in FH−/− tumor cells. Mutations in mtDNA were observed in three of the five patient-derived FH−/− tumor cell lines (UOK262, UOK268, UOK348), whereas depletion or complete absence of mtDNA was observed in all five FH−/− tumor cell lines. (B) Schematic illustrating a normally functioning TCA cycle and respiratory chain (left). Respiratory complexes that require mtDNA-encoded proteins for their function are indicated in blue, and FH and complex II which are encoded by nuclear genes only are colored in green. Following loss of the remaining functional FH allele, excess fumarate accumulation in FH−/− renal tumors results in the succination of POLG (right). Loss and mutations of mtDNA resulting from POLG succination lead to impaired respiratory chain assembly and activity, promoting aerobic glycolysis and pentose phosphate pathway (PPP) flux and driving anabolic pathways that require NADPH, thus facilitating tumor cell growth and progression (right). These mtDNA mutational events induce a permanent commitment of FH−/− tumor cells to aerobic glycolysis (the Warburg effect).
Discussion
FH-deficient kidney cancer is an early onset tumor characterized by a metabolic shift to aerobic glycolysis and a propensity to spread aggressively when the primary tumors are small (11). The underlying mechanisms that drive the progression of metabolic reprogramming in HLRCC tumors have thus far remained elusive. In this study, we showed that upon loss of FH, fumarate accumulation drove an irreversible loss of mitochondrial respiration in FH−/− cells mediated through inactivation of the core enzymes responsible for replication and proofreading of mtDNA, including POLG, POLG2, TFAM and TWINKLE, by covalent succination of reactive cysteine residues (Figure 7A). Covalent succination and associated inactivation of mitochondrial DNA metabolism proteins resulted in loss of mtDNA due to its ineffective replication, as well as in error-prone replication of mtDNA. Loss of respiratory chain activity obligates cells to compensate for energy loss by shifting to dependence on aerobic glycolysis as an energy source. We identified multiple deleterious mutations or deletions of mtDNA genes that encoded subunits of the electron transport chain complexes in nearly all of the HLRCC tumors analyzed. Fumarate-induced covalent modifications in FH-deficient tumors and cell lines were identified in four out of the five predominant core components of mitochondrial nucleoids (Figures 6H, 6N and 7A). Other cytoplasmic and mitochondrial proteins that are inactivated by succination include mitochondrial aconitase (ACO2) (44), KEAP1 (27, 46), and iron-sulfur assembly proteins such as NFU1, ISCU, BOLA1 and BOLA3 (20). Depletion of the core nucleoid components POLG, POLG2, TFAM and TWINKLE can lead to complete loss of mtDNA (50-53) and to point mutations and deletions (39-42), consistent with the phenotypes observed in HLRCC tumors and cell lines upon succination of nucleoid proteins.
In the HLRCC tumor-derived cell lines, which represent a homogeneous FH−/− model because the stromal and immune cells of the tumor are excluded, we found variable degrees of mtDNA aberrations, ranging from various missense mutations that disabled mitochondrial-encoded respiratory chain components, to the complete loss of the mitochondrial genome (Figures 6A-E and 7A). Given the deleterious nature of the mutations that had accumulated in the mtDNA at homoplasmic levels, loss of mitochondrial respiration in FH−/− cells was irreversible, because re-expression of FH did not restore respiration despite restoration of POLG activity in an environment that no longer contained elevated fumarate levels. Accordingly, restoration of FH enzyme activity allowed mtDNA copy numbers to normalize in FH−/− UOK262 and UOK268 tumor cell lines but did not correct the inactivating mutations in their mtDNA. Notably, benign renal oncocytomas harbor mtDNA mutations mainly in Complex I and high mtDNA copy numbers, but in the absence of increased fumarate (62). Irreversible loss of respiration forced FH-deficient tumors to engage in a complex metabolic adaptation that included a shift to dependence on aerobic glycolysis for energy production, which is sustained by elevated glucose uptake (Figures 7B and S5). Our data suggest that glucose was diverted to the pentose phosphate pathway (PPP), which is up-regulated in tumors and HLRCC RCC cell lines (Figure S5) (54). The PPP is the main source of cytosolic NADPH, a reduced cofactor that feeds NADPH-consuming anabolic and antioxidant reactions in the cytosol which thereby re-oxidize the NADP+ needed to maintain activity of the pentose phosphate shunt. Succination of the E3 ubiquitin ligase KEAP1 in FH−/− tumors prevents degradation of NRF2, allowing NRF2 to translocate to the nucleus and coordinately upregulate enzymes of the PPP (Figure S6) (27, 46). NRF2 concomitantly induces a potent antioxidant response mediated by genes encoding enzymes such as the NADPH dehydrogenase quinone 1 (NQO1), heme oxygenase 1 (HMOX1) and glutathione reductase (14, 27, 46), all of which consume cytosolic NADPH provided by the PPP to regenerate NADP+, and which were up-regulated at the mRNA level in our FH−/− tumors.
In rapidly growing cancers, such as HLRCC-associated RCC, cell proliferation depends on fatty acid synthesis which provides the building blocks for biogenesis of membranes (55). De novo fatty acid synthesis is an energetically demanding process which requires acetyl-CoA and large amounts of NADPH (56), thereby representing another NADPH-consuming pathway activated in FH-deficient tumors and enabling them to restore their redox balance and avoid saturation of the PPP. Acetyl CoA carboxylase (ACC), one of the main regulatory enzymes of de novo lipid biosynthesis, is up-regulated in FH−/− cells (13), and we showed here that the genes encoding fatty acid synthase (FASN) and ATP-citrate synthase (ACLY) were significantly up-regulated in HLRCC tumors (Figure S5), which would be expected to increase the generation of lipids needed to support membrane formation and cell division. The precursor molecule for fatty acid synthesis, citrate, is mainly derived from glutamine in cells with defects in mtDNA (19), such as FH-deficient RCC, by means of a linear series of reactions that could exclusively take place in the cytosol and mediated by the activity of enzymes that metabolize exogenously imported glutamine, including glutaminase (57), numerous transaminases, cytosolic isocitrate dehydrogenase (IDH1) consuming NADPH (58), and cytosolic aconitase, IRP1 (59) (Figure S5). Therefore, the metabolic compensations dictated by loss of mtDNA may result in features of the aggressive growth phenotype that characterizes HLRCC tumors, including increased fatty acid biosynthesis contributing to formation of new cell membranes, generation of the intermediates of the citric acid cycle that are utilized in anaplerotic reactions, and synthesis of other precursors necessary for biosynthesis of amino acids and nucleotides that are needed for rapid growth and invasiveness of this type of cancer (60, 61).
Our data suggest that loss of FH in HLRCC tumors led to a buildup of fumarate that covalently modified the thiol groups of cysteine residues in core enzymes required for replication and proofreading of mtDNA, including the sole DNA polymerase POLG, impairing its ability to effectively replicate the mitochondrial genome. Mitochondrial DNA replication and error correction capacity were thereby decreased, leading to loss of respiratory chain activity and a Warburg metabolic shift to aerobic glycolysis. Fumarate accumulated to millimolar levels in FH-deficient tumors (Fig. 6O) and because the initial fumarate accumulation likely occurred within the mitochondrial matrix, mitochondrial enzymes would be expected to be rapidly targeted and inactivated by succination. The modified cysteines may normally ligate crucial cofactors such as iron-sulfur clusters, as in the case of NFU1, ISCU, BOLA1 and BOLA3 (20) or, as has already been reported, for many enzymes involved in nuclear DNA metabolism (63). We propose that mitochondrial function is incapacitated at an early stage of tumor development in HLRCC due to loss of FH enzyme activity, resulting in fumarate accumulation in the mitochondrial matrix that leads to loss and mutation of mtDNA and to inactivation of several mitochondrial enzymes (Figure 7B). Although in the present work, we studied HLRCC patient tissues and patient-derived tumor cell lines, further investigation will be required to more completely characterize the mechanisms and kinetics of succination induced by loss of FH. These studies, as well as additional in-depth analyses of the earliest HLRCC renal lesions will hopefully provide more fundamental insight into the origins of FH-deficient tumorigenesis.
In summary, mitochondrial dysfunction forces metabolic remodeling in HLRCC tumors that favors anabolic pathways offering crucial bioenergetic and biosynthetic advantages for tumor growth and metastasis. Understanding that mitochondrial DNA mutations and deletions represent the fundamental cause for the shift to complete dependence on aerobic glycolysis in this aggressive form of inherited, genetically defined renal cell carcinoma will hopefully provide insights into the development of effective forms of therapy for patients with HLRCC and other related cancers.
Materials and Methods
Patients, Cell Lines and Tissue Culture
Patients with type 2 papillary renal cancers were evaluated and treated at the Clinical Center of the National Institutes of Health on a Urologic Oncology Branch, National Cancer Institute protocol (NCI-97-C-0147) approved by the NCI’s Institutional Review Board and written informed consent was given for participation in this study. Patents were confirmed to have germline mutations in the FH gene by Clinical Laboratory Improvement Amendments (CLIA) certified genetic testing. Clinical tumor staging was performed according to the current AJCC guidelines (64). UOK271 cells (Patient 3) were isolated from a large FH−/− primary renal tumor. UOK348 (Patient 7) and UOK350 (Patient 6) cells were derived from metastatic FH−/− RCC lesions of the retroperitoneum and lung, respectively. Briefly, tumor tissues were minced into small fragments and the explants were maintained on tissue culture dishes in a humidified incubator at 37 °C in the presence of 5% O2 and 5% CO2. Tumor explants were propagated in DMEM medium containing 25mM D-glucose, 4mM glutamine, 1mM pyruvate, 10% fetal bovine serum (FBS), 100 units/mL of penicillin and streptomycin, 0.25μg/mL amphotericin-B, and 1X non-essential amino acid mixture (all reagents from Gibco, Thermo Fisher Scientific Inc., MA, USA). Following 10 passages of continuous growth, cell lines were assessed for the absence of residual wild type FH alleles from contaminating stromal cells by Sanger DNA sequencing using established methods and FH activity assay and were subjected to STR and mycoplasma testing to verify purity and identity. Unless otherwise noted, all cell-based experiments in this work were conducted in DMEM medium containing 25mM D-glucose, 4mM glutamine, 1mM pyruvate and 10% fetal bovine serum (FBS) in humidified room air incubators at 5% CO2.
Clear native PAGE enzyme analysis
FH and malic NAD+-reductase enzyme activities were separated by native gel electrophoresis using pre-cast 1 mm 7% Tris-acetate polyacrylamide gels (Invitrogen, CA, USA). Briefly, whole cell lysates were prepared on ice by sonication in lysis buffer consisting of 10mM Tris, pH 8.0, 0.1% Triton X-100, 3mM KCl, 3mM MgCl2, 3mM sodium citrate, and protease inhibitors (Complete™ EDTA-free, Roche, IN, USA), followed by centrifugation at 21,000 x g for 30 min at 4 °C. Protein concentration of the recovered supernatants was measured by BCA assay and 10μg of total cellular protein was loaded into the gel, and the samples were subjected to electrophoresis at 4 °C for 2 hours at 125 V, with running buffer consisting of 25mM Tris, 192mM glycine. For measurement of FH activity, the gels were immersed in 10mL of a solution pre-warmed to 37 °C, consisting of 100mM Tris, pH 7.4, 1mg/mL 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT), 1mM NAD+, 0.184 mg/mL phenazine methosulfate (PMS), 150 units of porcine heart malic dehydrogenase (Sigma-Aldrich), and 5mM Na fumarate. The gels were gently rocked at 37 °C in the dark for 5-10 min, until FH enzyme activity bands were developed, at which point the gels were de-stained in the dark with sequential incubations in de-ionized water until the background was clear and scanned immediately. Malic NAD+-reductase enzyme activity was assessed by incubation of duplicate gels with 100mM Tris, pH 7.4, 1mg/mL iodonitrotetrazolium bromide (INT), 1mM NAD+, 0.184mg/mL phenazine methosulfate (PMS), and 5mM Na L-malate.
Polar metabolite analysis by IC-UHR-FTMS
Cells cultured to 80-90% confluence were washed 3 times in cold PBS, quenched in cold acetonitrile, and extracted in acetonitrile:water:chloroform (2:1.5:1) (65). The upper polar metabolite fractions were frozen in several aliquots each and lyophilized overnight. Metabolite analyses were performed on an Orbitrap Fusion Tribrid Mass Spectrometer following ion-pairing chromatography as previously described (66, 67). Data are expressed as nmol/mg protein.
Subcellular fractionation of cultured cells
Cellular fractionation into cytosol and intact mitochondria was done essentially as previously described (68, 69). All the steps were performed at 4 °C, on ice and in refrigerated centrifuges. Cells were washed with RHM buffer (210mM mannitol, 20mM sucrose and 4mM HEPES), and then permeabilized for 5 min in RHM buffer supplemented with 0.1% digitonin. The supernatant after centrifugation at 700xg for 5 min was subjected to centrifugation at 21,000xg for 20 min and the resulting supernatant was saved as soluble cytosolic fraction. The pellet after the centrifugation at 700xg for 5 min was subjected to centrifugation for 5 min at 1,600xg to generate the postnuclear supernatant, which contained a mixture of mitochondria, lysosomes, peroxisomes and ER. Mitochondria were purified from the postnuclear supernatant on a density gradient consisting of Optiprep density gradient medium (60% w/v; Sigma #D1556) diluted to the required final concentration in 5mM MOPS, pH 8.0, containing 1mM EDTA and 0.1% ethanol. The postnuclear supernatant was initially supplemented with Optiprep density gradient to a final concentration of 22.5% in a final volume of 450μL. 400μL of the diluted sample was layered over 200 μL of 27.5% Optiprep solution. The sample was then carefully overlayered with 200 μL of 20% Optiprep solution and subjected to centrifugation for 1.5h to 100,000xg. A ring formed at the 22.5-27.5% interface that contained purified mitochondria, which were further lysed in 1.25x buffer I (50 mM BisTris, 50 mM NaCl, 10% w/v Glycerol, 0.001% Ponceau S, 1.2% Lauryl maltoside, pH 7.2, protease and phosphatase inhibitors). The supernatant after centrifugation at 21,000xg for 15’ was saved as mitochondrial lysate.
Subcellular fractionation from tissue samples
Snap-frozen tissue specimens were ground to powder with mortar and pestle. Four volumes of extraction buffer (5mM MOPS, pH 7.65 supplemented with 250mM sucrose, 1mM EDTA, and 0.1% ethanol) were added per gram of tissue, and samples were extracted by two sets of dounce strokes (20 strokes) in a pre-chilled dounce homogenizer and were incubated on ice for 10 min. The homogenate was centrifuged at 1000xg for 10min. The supernatant was transferred to a new tube and the pellet containing nuclei was washed with dilution buffer (5mM MOPS, pH 8.0, supplemented with 1mM EDTA and 0.1% ethanol) and subsequently lysed in RIPA buffer and sonicated for the extraction of nuclear proteins. The supernatant after the first centrifugation step at 1000xg was spun at 2000xg for 10 min. The supernatant after the centrifugation at 2000xg was spun at 25,000xg for 20 min and the pellet, containing a mixture of mitochondria, lysosomes, peroxisomes and ER membranes, was subjected to further purification by density gradient as outlined above.
Immunoprecipitation (IP) experiments
Immunoprecipitations of SDH complex, POLG and TFAM were done by covalently coupling anti-SDHB antibody produced in mouse (Abcam #AB14714), anti-POLG antibody produced in rabbit (Millipore #ABC462), or anti-TFAM antibody (Proteintech #19998-1-AP) onto an amine-reactive resin (Pierce Co-IP kit, Cat. No. 26149), following the manufacturer’s instructions. Mitochondrial lysates were incubated with the beads overnight at 4 °C, and then washed extensively with lysis buffer I. Bound proteins were eluted with Tris-Glycine pH 2.8 for 10 min at 4 °C. Aliquots corresponding to 20-30% of mitochondrial lysates were run alongside the IP fractions onto the gels as inputs.
Native PAGE (BN-PAGE) and native immunoblots
The NativePAGE Novex Bis-Tris gel system (Thermo Fisher Scientific) was used to analyze native membrane protein complexes and native mitochondrial matrix complexes, with the following modifications: only the Light Blue Cathode Buffer was used; 20 μg of membrane protein extracts were loaded/well; the electrophoresis was performed at 150 V for 1 hour and 250 V for 2 hours. For the native immunoblot, PVDF was used as the blotting membrane. The transfer was performed at 25 V for 4 hours at 4 °C. After transfer, the membranes were washed with 8% acetic acid for 20 min to fix the proteins, and then rinsed with water before air-drying. The dried membranes were washed 5-6 times with methanol (to remove residual Coomassie Blue G-250), rinsed with water and then blocked for 2 hours at room temperature in 5% milk, before incubating with the desired antibodies diluted in 2.5% milk overnight at 4 °C. To avoid strip and reprobing of the same membrane, which might allow detection of signals from the previous immunoblots, samples were loaded and run in replicates on adjacent wells of the same gel and probed independently with different antibodies.
Complex I, Complex II and Complex IV in-gel activity assays
In-gel Complex I, Complex II and Complex IV activity assays were performed as previously described (68, 70, 71). Briefly, for Complex I activity, after resolution of the respiratory chain complexes by BN-PAGE, the gel was incubated with 0.1M TrisCl, pH 7.4, containing 1mg/ml nitrobluetetrazolium chloride (NBT) and 0.14mM NADH at room temperature for 30 min. For complex II, detection of succinate CoQ-reductase activity (SQR) (CoQ-mediated NBT reduction) was performed by incubating the gel for 30 min with 84mM succinate, 2mg/ml NBT, 4.5mM EDTA, 10mM KCN, 1mM sodium azide and 10μM ubiquinone in 50mM PBS, pH 7.4. For complex IV, the gel was incubated in 50mM phosphate buffer pH 7.4 containing 1mg/ml DAB (3,3’-diaminobenzidine) and 1mg/ml cytochrome c at room temperature for 30-45 min.
Nucleoid isolation
Nucleoid isolation was performed as previously described (72, 73) with some modifications. Briefly, mitochondria purified from 4x 150mm dishes were resuspended in 100 μL of MSH buffer (210mM mannitol, 70mM sucrose, 20mM HEPES, pH 8.0, 2mM EDTA) containing 1mM ADP, 5mM sodium pyruvate, 1mg/ml bovine serum albumin, 60mM KCl, 10mM MgCl2, 1mM K2HPO4, 100 units/ml Rnase-free Dnase I (Sigma type II) and 50 units/ml Benzonase nuclease HC (Millipore), and incubated at 37 °C for 15 min. The nuclease digestion was stopped by addition of 10μL of 250mM EDTA to chelate Mg2+. Mitochondria were layered over 0.8M sucrose, 20mM HEPES, 2mM EDTA, 2mM DTT and spun at 21,000xg for 15 min to remove the digested nuclear DNA fragments and to sediment pure mitochondria, which were washed once with MSH buffer and finally resuspended at approximately 7mg/ml protein in lysis buffer containing 30mM HEPES, pH 8, 5% glycerol, 2mM DTT, 1mM EDTA, 1.2% Triton X-100. The lysate was mixed on ice for 5 min and centrifuged at 3,000xg for 5 min. A total of 700 μL of the 1.2% Triton X-100 lysates after centrifugation was loaded on top of a step gradient prepared by layering 4.5 ml of 17% glycerol over 5 ml of 45% glycerol above a pad of 500 μL of 30% Nycodenz and 30% glycerol. All gradient layers contained 0.5% Triton X-100, 30mM HEPES, pH 8, 2mM EDTA, 2mM DTT, 500μL phenylmethylsulfonyl fluoride and 70 mM NaCl. Fractions were collected after centrifugation at 186,000xg for 1.5 hours. The fraction of nucleoids that sedimented very quickly and reached the pad, along with the fraction distributed in the lower half of the gradient, were subjected to immunoaffinity purification with anti-TFAM antibody covalently immobilized onto an amine-reactive resin (Pierce Co-IP kit, Cat. No. 26149), by following the manufacturer’s instructions.
RNAseq analysis
Total RNA was extracted from cell lines or cryo-pulverized tumor/normal tissues using Trizol Reagent (Invitrogen) following the manufacturer’s protocol and stored at −80 °C. RNA concentrations and purities were measured using a NanoDrop 2000 UV-Vis Spectrophotometer (Thermo Fisher Scientific) and 5μg of total RNA was sent to ACGT, Inc. (Germantown, MD, USA) for RNAseq analysis. All samples were processed with the TruSeq mRNA kit using oligo-dT beads to capture polyA tails and data was delivered as a Multiplex Fast-Q file with adaptors trimmed. 43-60 million read pairs per sample were obtained. The sequencing quality of the reads was assessed using FastQC (version 0.11.5), Preseq (version 2.0.3)(74), Picard tools (version 1.119), and RSeQC (version 2.6.4). Reads were trimmed using Trimmomatic (version 0.33) (75) to remove sequencing adapters, prior to mapping to the human reference genome, hg38, using STAR (version 2.5.2b) in two-pass mode (76). Across the samples, the median percentage of mapped read pairs was 76.8%. Expression levels were quantified using RSEM version 1.3.0 (77) with GENCODE annotation version 21 (78). The RSEM counts were then normalized using the variance stabilization transformation (79) from the DESeq2 R package (version 1.24.0) (80). These normalized values were used for visualizing mRNA expression levels and pathway enrichment.
qPCR
cDNA was synthesized for each tissue sample using the SuperScript® VILO™ cDNA Synthesis Kit (Invitrogen) with a starting amount of 1μg of total mRNA in a 20μl volume as described in the manufacturer’s protocol. Completed cDNA synthesis reactions were diluted 10-fold with 180μl of RNase-Free Water to produce a working stock. RT-PCR amplifications were performed in triplicate in 10μl reaction volumes consisting of 2μl of diluted cDNA, 5μl of Taqman® universal PCR master mix (Thermo Fisher Scientific), 0.5μl of the relevant Taqman® Gene Expression Assay probe (Thermo Fisher Scientific), and 2.5μl of RNase-Free Water. Reactions were run on an ABI ViiA7 real-time PCR system (Thermo Fisher Scientific) and expression levels were calculated using the ViiA7 software as comparative CT (ΔΔCT) values using the ACTB housekeeping gene (Hs99999903_m1) as a normal control. Taqman® Gene Expression Assay probes were used to assess the expression levels for the following genes: MT-ND1 (Hs02596873_s1), NDUFS1 (Hs00192297_m1), G6PD (Hs00166169_m1), MT-CYB (Hs02596867_s1), MT-CO1 (Hs02596864_g1), MT-ATP6 (Hs02596862_g1), ATP5B (Hs00969569_m1), UQCRB (Hs00559884_m1), and COX5A (Hs00362067_m1).
Electron Microscopy
Tissue samples were fixed with 2% glutaraldehyde and 4% formaldehyde in 0.1M Na Cacodylate buffer for an hour at room temperature for Transmission electron microscopy (TEM) processing. Each tissue sample was cut into five smaller pieces and transferred to a glass vial containing 0.1M Na Cacodylate buffer for EM processing. Tissue samples were washed with 0.1M Na Cacodylate buffer two times for 10 min. After the second buffer wash, the tissue samples were post fixed with 1% Osmium Tetroxide for an hour in the dark. Tissue samples were washed with 0.1M Na Cacodylate buffer twice for 10 min and washed once with 0.1N Na Acetate buffer and then stained with 0.5% Uranyl Acetate for an hour in the dark. After en block staining, tissue samples were washed with 0.1N Na Acetate buffer twice for 10 min. After the last buffer wash, tissue samples were subjected to gradual dehydration in the order of 35%, 50%, 70% and 95% Ethanol twice each step for 10 min and three times with 100% Ethanol for 10 min. After the last step of 100% Ethanol rinse, tissue samples were further dehydrated in Propylene oxide (PO) for 10 min for three times. After the last step in PO, tissue samples were infiltrated in 50:50 epoxy resin and PO overnight at room temperature.
The next day, after overnight infiltration, each tissue sample was removed from 50:50 epoxy resin and PO, blotted, and embedded in a plastic mold containing 100% pure epoxy resin and transferred to a 55 °C oven for 48 hours. Epoxy resin (PolyScience Resin) ingredients consisted of a mixture of Poly/Bed 812 embedding Media, Dodecenylsuccinic Anhydride (DDSA), Nadic Methyl Anhydride (NSA) and DMP-30 to solidify the resin. After 48 hours, tissue samples were taken out of the oven, and the best tissue embedded in resin mold was selected, then ultra-thin sectioned with a UC6 Leica Microtome. The ultra-thin sections were picked up on a 150 Copper mesh grid and were examined under a Hitachi H7600 Transmission electron microscope. The grids were post stained with 1:1 0.5% Uranyl Acetate in ddH2O and 70% Ethanol for 2 min and then rinsed with ddH2O for four times. Then cells stained with 1:1 Lead Citrate and H2O for 2 min and rinsed with ddH2O four times. The grids were carbon coated with a TedPella/Cressington Evaporator and imaged in a Hitachi H7600 transmission electron microscope at 80KeV.
Seahorse Respiration Analysis
Oxygen consumption rate (OCR) was calculated using XF Extracellular Flux Analyzer from Seahorse Bioscience (North Billerica, MA). 7,000 cells per well were cultured in 96-well plates for 36 hours in the presence of complete medium, washed, and incubated with complete medium lacking sodium bicarbonate and FBS at 37 °C for 1 hour in room air before respiration measurements were taken. 0.5μM antimycin A and 1μM rotenone were injected following three respiration measurements to inhibit mitochondrial respiration mediated by complexes I and III.
mtDNA Copy Number Analysis
DNA from tissue and cell samples was extracted using the Maxwell 16 Tissue and Cell extraction kits, respectively. Mitochondrial DNA copy number was evaluated using the NovaQUANT™ Human Mitochondria to nuclear DNA Ratio Kit (Millipore, MA, USA). This kit was used in accordance with the manufacturer’s protocol and provides Real-Time PCR primers for 2 mitochondria genome encoded genes, MT-ND1 and MT-ND6, and 2 nuclear genome encoded genes, BECN1 and NEB. Experiments were performed in triplicate using SYBR Green PCR Master Mix (Life Technologies, NY 14072, USA) and a ViiA™ 7 Real-Time PCR System (Life Technologies, NY 14072, USA). Mitochondrial copy number was calculated as a ratio of the mitochondrially encoded genes to the nuclear encoded genes assuming 2 copies of each nuclear encoded gene were present per cell.
Next generation mtDNA sequencing
DNA extracted from cells and tissues was assessed for quality and purity using an Agilent Tapestation analyzer. mtDNA sequencing was performed upon amplification of mitochondrial genomes from 1-10μl whole genomic DNA extracted from blood, cells, tumor tissues and surrounding normal kidney tissues using the QIAseq Targeted DNA Panel (DHS-105Z Human Mitochondria Panel; Qiagen) according to the manufacturer’s instructions. The resulting libraries were pair-end sequenced on a MiSeq instrument (Illumina). Read processing, alignment, and variant calling was carried out on the QIAseq Targeted Sequencing Data Analysis Portal with the Biomedical Genomics Analysis plugin (Qiagen).
Variants below 0.03 variant allele fraction (VMF) were filtered out and variant lists were compared between tumor and normal DNA samples (blood or normal kidney) to identify somatic changes. SIFT (81), Provean (82), PolyPhen-2 (83) and MutationAssessor (84) algorithms were used together to predict the consensus pathogenicity of missense mutations detected in FH-deficient cell lines and tumors.
Spectral Karyotyping (SKY)
Metaphase chromosomes were prepared by incubating cultured cells with 0.02 mg/ml Colcemid (Invitrogen) for 2 hours. The harvested cells were then incubated in 0.075M KCl then fixed with methanol/acetic acid (3:1). Metaphases were then prepared in a controlled humidity chamber (Thermotron, Holland, MI). SKY was performed and analyzed as previously described (85). Approximately 15 metaphases were imaged and analyzed using the hiSKY 7.2.7 Software (ASI, Carlsbad, CA) on a Leica DMRXA microscope.
Statistical Analysis
Statistical assessments were conducted using SAS, version 9.4 (SAS Institute, Inc, Cary, NC). Differences between expression of selected RNAseq, qRT-PCR, and mtDNA copy number data were assessed using the Wilcoxon rank sum test with exact p-values (PROC NPAR1WAY). To avoid overly optimistic assessment due to multiple samples obtained from the same patient that could be correlated, we selected the renal cortex whenever available and for all other tissue types (primary and metastasis) one sample per patient was randomly selected. To increase power, we combined data from primary and metastatic tumors into a single group for the comparisons. Statistical assessments of selected qRT-PCR gene expression data were conducted using five FH+/− independent renal cortex samples and nine independent FH−/− tumor samples. Analyses of RNAseq gene expression data were conducted using five FH+/− independent renal cortex samples and eight independent FH−/− tumor samples. Finally, statistical analysis of mtDNA copy number data were conducted using seven FH+/− independent renal cortex samples and 11 independent FH−/− tumor samples
Supplementary Material
Fig. S1. Images and genetic analysis of five FH−/− tumor cell lines.
Figure S2. FH in-gel activity assay and protein abundance analysis.
Figure S3. FH-deficient tumors exhibited a dysfunctional respiratory chain.
Figure S4: qPCR analysis of additional mtDNA-encoded genes in HLRCC tissues.
Figure S5. Up-regulation of expression of genes involved in glycolysis, glutaminolysis, pentose phosphate pathway, and fatty acid biosynthesis pathways in HLRCC FH−/− tumors.
Figure S6: RNAseq analysis of the expression of NADP-dependent NRF2 transcriptional target genes in HLRCC tissues.
Figure S7: Analysis of respiration rates in FH−/− HLRCC tumor-derived cell lines.
Figure S8: qPCR analysis of additional nuclear and mtDNA-encoded genes in FH−/− tumor cell lines.
Figure S9: Mitochondrial genome mutations present in UOK262 and UOK268 cells.
Figure S10: Mitochondrial genome mutation present in UOK348 cells.
Figure S11: Breakdown of mtDNA mutations found in FH−/− tumors and cell lines.
Figure S12: Heteroplasmy of somatic mtDNA mutations in FH−/− tumors.
Figure S13: Heteroplasmy of somatic mtDNA mutations found in each FH−/− tumor sample in the study.
Figure S14: Map of selected protein-coding mtDNA mutations found in tumor samples.
Figure S15: Classification of mtDNA mutation impact in tumor samples.
Figure S16: Comparative mutational analysis of nuclear genes in The Cancer Genome Atlas (TCGA) analysis of RCC.
Figure S17: Spectral karyotyping of HLRCC-derived cell lines.
Figure S18: Analysis of POLG immunoprecipitates from mitochondrial lysates isolated from FH−/− tumor tissue samples and control kidney cortex samples.
Table S1: Summary of mtDNA mutations in HLRCC tumors.
Table S2. Mass spectrometry analysis on nucleoid components isolated by TFAM immunoprecipitation in RPTEC and FH−/− UOK262 cell lines.
Acknowledgments:
The authors wish to thank Rabindra Gautam for assistance with clinical image assessment and processing and James Peterson for assistance with database management. We also thank Ashley Ferguson for assistance with laboratory analyses.
Funding: This work was supported by the Intramural Research Programs of NCI-CCR and NICHD. This project was funded in part with federal funds from the Frederick National Laboratory for Cancer Research, NIH, under contract HHSN26120080001E. Sanger sequencing was conducted at the CCR Genomics Core at the National Cancer Institute, NIH, Bethesda, MD 20892.
Footnotes
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: The RNAseq data were deposited in the GEO database, under accession GSE157256. The mass spectrometry data have been deposited in Peptide Atlas (URL: https://db.systemsbiology.net/sbeams/cgi/PeptideAtlas/PASS_View; Dataset identifier: PASS01618; Dataset Password: ZB3349ir). All other data needed to evaluate the conclusions in the paper are present in the paper or the Supplementary Materials. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the U.S. Government.
References and Notes
- 1.Kaelin WG Jr., Thompson CB, Cancer: clues from cell metabolism. Nature 465, 562–564 (2010). [DOI] [PubMed] [Google Scholar]
- 2.Warburg O, Posener K, Negelein E, Metabolism of the Carcinoma Cell. Biochem Z 152, 309–344 (1924). [Google Scholar]
- 3.Warburg O, On the origin of cancer cells. Science 123, 309–314 (1956). [DOI] [PubMed] [Google Scholar]
- 4.Thompson CB, Bielska AA, Growth factors stimulate anabolic metabolism by directing nutrient uptake. J Biol Chem 294, 17883–17888 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Vander Heiden MG, Cantley LC, Thompson CB, Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science 324, 1029–1033 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Koppenol WH, Bounds PL, Dang CV, Otto Warburg's contributions to current concepts of cancer metabolism. Nat Rev Cancer 11, 325–337 (2011). [DOI] [PubMed] [Google Scholar]
- 7.DeBerardinis RJ, Chandel NS, We need to talk about the Warburg effect. Nature Metabolism 2, 127–129 (2020). [DOI] [PubMed] [Google Scholar]
- 8.Warburg O, Wind F, Negelein E, The Metabolism of Tumors in the Body. J Gen Physiol 8, 519–530 (1927). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tomlinson IP et al. , Germline mutations in FH predispose to dominantly inherited uterine fibroids, skin leiomyomata and papillary renal cell cancer. Nat Genet 30, 406–410 (2002). [DOI] [PubMed] [Google Scholar]
- 10.Grubb RL et al. , Hereditary leiomyomatosis and renal cell cancer: a syndrome associated with an aggressive form of inherited renal cancer. J Urol 177, 2074–2080 (2007). [DOI] [PubMed] [Google Scholar]
- 11.Linehan WM et al. , The Metabolic Basis of Kidney Cancer. Cancer Discov, (2019). [DOI] [PubMed] [Google Scholar]
- 12.Mitchell TJ et al. , Timing the Landmark Events in the Evolution of Clear Cell Renal Cell Cancer: TRACERx Renal. Cell 173, 611–623 e617 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tong WH et al. , The glycolytic shift in fumarate-hydratase-deficient kidney cancer lowers AMPK levels, increases anabolic propensities and lowers cellular iron levels. Cancer Cell 20, 315–327 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sourbier C et al. , Targeting ABL1-mediated oxidative stress adaptation in fumarate hydratase-deficient cancer. Cancer Cell 26, 840–850 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Linehan WM, Ricketts CJ, The metabolic basis of kidney cancer. Semin Cancer Biol 23, 46–55 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yang Y et al. , UOK 262 cell line, fumarate hydratase deficient (FH-/FH-) hereditary leiomyomatosis renal cell carcinoma: in vitro and in vivo model of an aberrant energy metabolic pathway in human cancer. Cancer Genet Cytogenet 196, 45–55 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bardella C et al. , Aberrant succination of proteins in fumarate hydratase-deficient mice and HLRCC patients is a robust biomarker of mutation status. J Pathol 225, 4–11 (2011). [DOI] [PubMed] [Google Scholar]
- 18.Yang Y et al. , A novel fumarate hydratase-deficient HLRCC kidney cancer cell line, UOK268: a model of the Warburg effect in cancer. Cancer Genet 205, 377–390 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mullen AR et al. , Reductive carboxylation supports growth in tumour cells with defective mitochondria. Nature 481, 385–388 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tyrakis PA et al. , Fumarate Hydratase Loss Causes Combined Respiratory Chain Defects. Cell Rep 21, 1036–1047 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Noda Y et al. , 18-F fluorodeoxyglucose uptake in positron emission tomography as a pathological grade predictor for renal clear cell carcinomas. Eur Radiol 25, 3009–3016 (2015). [DOI] [PubMed] [Google Scholar]
- 22.Miyakita H et al. , Significance of 18F-fluorodeoxyglucose positron emission tomography (FDG-PET) for detection of renal cell carcinoma and immunohistochemical glucose transporter 1 (GLUT-1) expression in the cancer. Int J Urol 9, 15–18 (2002). [DOI] [PubMed] [Google Scholar]
- 23.Frezza C et al. , Haem oxygenase is synthetically lethal with the tumour suppressor fumarate hydratase. Nature 477, 225–228 (2011). [DOI] [PubMed] [Google Scholar]
- 24.Cogliati S, Enriquez JA, Scorrano L, Mitochondrial Cristae: Where Beauty Meets Functionality. Trends Biochem Sci 41, 261–273 (2016). [DOI] [PubMed] [Google Scholar]
- 25.Reznik E, Wang Q, La K, Schultz N, Sander C, Mitochondrial respiratory gene expression is suppressed in many cancers. Elife 6, (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.N. Cancer Genome Atlas, Comprehensive Molecular Characterization of Clear Cell Renal Cell Carcinoma. Nature 499, 43–49 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ooi A et al. , An antioxidant response phenotype shared between hereditary and sporadic type 2 papillary renal cell carcinoma. Cancer Cell 20, 511–523 (2011). [DOI] [PubMed] [Google Scholar]
- 28.Tonelli C, Chio IIC, Tuveson DA, Transcriptional Regulation by Nrf2. Antioxid Redox Signal 29, 1727–1745 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Greer YE et al. , ONC201 kills breast cancer cells in vitro by targeting mitochondria. Oncotarget 9, 18454–18479 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stoldt S et al. , Spatial orchestration of mitochondrial translation and OXPHOS complex assembly. Nat Cell Biol 20, 528–534 (2018). [DOI] [PubMed] [Google Scholar]
- 31.Signes A, Fernandez-Vizarra E, Assembly of mammalian oxidative phosphorylation complexes I-V and supercomplexes. Essays Biochem 62, 255–270 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nijtmans LG, Taanman JW, Muijsers AO, Speijer D, Van den Bogert C, Assembly of cytochrome-c oxidase in cultured human cells. Eur J Biochem 254, 389–394 (1998). [DOI] [PubMed] [Google Scholar]
- 33.Wikstrom M, Krab K, Sharma V, Oxygen Activation and Energy Conservation by Cytochrome c Oxidase. Chem Rev 118, 2469–2490 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Clima R et al. , HmtDB 2016: data update, a better performing query system and human mitochondrial DNA haplogroup predictor. Nucleic Acids Res 45, D698–D706 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Carossa V et al. , A novel in-frame 18-bp microdeletion in MT-CYB causes a multisystem disorder with prominent exercise intolerance. Hum Mutat 35, 954–958 (2014). [DOI] [PubMed] [Google Scholar]
- 36.Ricketts CJ et al. , The Cancer Genome Atlas Comprehensive Molecular Characterization of Renal Cell Carcinoma. Cell Rep 23, 313–326 e315 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ball MW et al. , Growth Rates of Genetically Defined Renal Tumors: Implications for Active Surveillance and Intervention. J Clin Oncol, JCO1902263 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bonekamp NA, Larsson NG, SnapShot: Mitochondrial Nucleoid. Cell 172, 388–388 e381 (2018). [DOI] [PubMed] [Google Scholar]
- 39.Spelbrink JN et al. , Human mitochondrial DNA deletions associated with mutations in the gene encoding Twinkle, a phage T7 gene 4-like protein localized in mitochondria. Nat Genet 28, 223–231 (2001). [DOI] [PubMed] [Google Scholar]
- 40.Longley MJ et al. , Mutant POLG2 disrupts DNA polymerase gamma subunits and causes progressive external ophthalmoplegia. Am J Hum Genet 78, 1026–1034 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Copeland WC, Defects of mitochondrial DNA replication. J Child Neurol 29, 1216–1224 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Spelbrink JN et al. , In vivo functional analysis of the human mitochondrial DNA polymerase POLG expressed in cultured human cells. J Biol Chem 275, 24818–24828 (2000). [DOI] [PubMed] [Google Scholar]
- 43.Yang M et al. , The Succinated Proteome of FH-Mutant Tumours. Metabolites 4, 640–654 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ternette N et al. , Inhibition of mitochondrial aconitase by succination in fumarate hydratase deficiency. Cell Rep 3, 689–700 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kornberg MD et al. , Dimethyl fumarate targets GAPDH and aerobic glycolysis to modulate immunity. Science 360, 449–453 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Adam J et al. , Renal cyst formation in Fh1-deficient mice is independent of the Hif/Phd pathway: roles for fumarate in KEAP1 succination and Nrf2 signaling. Cancer Cell 20, 524–537 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Piroli GG et al. , Identification of protein succination as a novel modification of tubulin. Biochem J 462, 231–245 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ekstrand MI et al. , Mitochondrial transcription factor A regulates mtDNA copy number in mammals. Hum Mol Genet 13, 935–944 (2004). [DOI] [PubMed] [Google Scholar]
- 49.Stiles AR et al. , Mutations in TFAM, encoding mitochondrial transcription factor A, cause neonatal liver failure associated with mtDNA depletion. Mol Genet Metab 119, 91–99 (2016). [DOI] [PubMed] [Google Scholar]
- 50.Larsson NG et al. , Mitochondrial transcription factor A is necessary for mtDNA maintenance and embryogenesis in mice. Nat Genet 18, 231–236 (1998). [DOI] [PubMed] [Google Scholar]
- 51.Hance N, Ekstrand MI, Trifunovic A, Mitochondrial DNA polymerase gamma is essential for mammalian embryogenesis. Hum Mol Genet 14, 1775–1783 (2005). [DOI] [PubMed] [Google Scholar]
- 52.Humble MM et al. , Polg2 is essential for mammalian embryogenesis and is required for mtDNA maintenance. Hum Mol Genet 22, 1017–1025 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Milenkovic D et al. , TWINKLE is an essential mitochondrial helicase required for synthesis of nascent D-loop strands and complete mtDNA replication. Hum Mol Genet 22, 1983–1993 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yang Y et al. , Metabolic reprogramming for producing energy and reducing power in fumarate hydratase null cells from hereditary leiomyomatosis renal cell carcinoma. PLoS One 8, e72179 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Currie E, Schulze A, Zechner R, Walther TC, Farese RV Jr., Cellular fatty acid metabolism and cancer. Cell Metab 18, 153–161 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wallace M, Metallo CM, Tracing insights into de novo lipogenesis in liver and adipose tissues. Semin Cell Dev Biol, (2020). [DOI] [PubMed] [Google Scholar]
- 57.Wang Y et al. , KAT2A coupled with the alpha-KGDH complex acts as a histone H3 succinyltransferase. Nature 552, 273–277 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lewis CA et al. , Tracing compartmentalized NADPH metabolism in the cytosol and mitochondria of mammalian cells. Mol Cell 55, 253–263 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Rouault TA, Maio N, Biogenesis and functions of mammalian iron-sulfur proteins in the regulation of iron homeostasis and pivotal metabolic pathways. J Biol Chem 292, 12744–12753 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Mullen AR et al. , Reductive carboxylation supports growth in tumour cells with defective mitochondria. Nature 481, 385–388 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Mullen AR et al. , Oxidation of alpha-ketoglutarate is required for reductive carboxylation in cancer cells with mitochondrial defects. Cell Rep 7, 1679–1690 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Gopal RK et al. , Early loss of mitochondrial complex I and rewiring of glutathione metabolism in renal oncocytoma. Proc Natl Acad Sci U S A 115, E6283–E6290 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Barton JK, Silva RMB, O'Brien E, Redox Chemistry in the Genome: Emergence of the [4Fe4S] Cofactor in Repair and Replication. Annu Rev Biochem 88, 163–190 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Amin MB, Edge SB, American Joint Committee on Cancer, AJCC cancer staging manual. (Springer, Switzerland, ed. Eighth edition., 2017), pp. xvii, 1024 pages. [Google Scholar]
- 65.Higashi RM, Fan TW, Lorkiewicz PK, Moseley HN, Lane AN, Stable isotope-labeled tracers for metabolic pathway elucidation by GC-MS and FT-MS. Methods Mol Biol 1198, 147–167 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Sun RC et al. , Noninvasive liquid diet delivery of stable isotopes into mouse models for deep metabolic network tracing. Nat Commun 8, 1646 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Fan TW et al. , Distinctly perturbed metabolic networks underlie differential tumor tissue damages induced by immune modulator beta-glucan in a two-case ex vivo non-small-cell lung cancer study. Cold Spring Harb Mol Case Stud 2, a000893 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Maio N et al. , Cochaperone binding to LYR motifs confers specificity of iron sulfur cluster delivery. Cell Metab 19, 445–457 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Maio N, Kim KS, Singh A, Rouault TA, A Single Adaptable Cochaperone-Scaffold Complex Delivers Nascent Iron-Sulfur Clusters to Mammalian Respiratory Chain Complexes I-III. Cell Metab 25, 945–953 e946 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Diaz F, Barrientos A, Fontanesi F, Evaluation of the mitochondrial respiratory chain and oxidative phosphorylation system using blue native gel electrophoresis. Curr Protoc Hum Genet Chapter 19, Unit19 14 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wittig I, Carrozzo R, Santorelli FM, Schagger H, Functional assays in high-resolution clear native gels to quantify mitochondrial complexes in human biopsies and cell lines. Electrophoresis 28, 3811–3820 (2007). [DOI] [PubMed] [Google Scholar]
- 72.Wang Y, Bogenhagen DF, Human mitochondrial DNA nucleoids are linked to protein folding machinery and metabolic enzymes at the mitochondrial inner membrane. J Biol Chem 281, 25791–25802 (2006). [DOI] [PubMed] [Google Scholar]
- 73.Bogenhagen DF, Biochemical isolation of mtDNA nucleoids from animal cells. Methods Mol Biol 554, 3–14 (2009). [DOI] [PubMed] [Google Scholar]
- 74.Daley T, Smith AD, Predicting the molecular complexity of sequencing libraries. Nat Methods 10, 325–327 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Bolger AM, Lohse M, Usadel B, Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Dobin A et al. , STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Li B, Dewey CN, RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics 12, 323 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Harrow J et al. , GENCODE: the reference human genome annotation for The ENCODE Project. Genome Res 22, 1760–1774 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Law CW, Chen Y, Shi W, Smyth GK, voom: Precision weights unlock linear model analysis tools for RNA-seq read counts. Genome Biol 15, R29 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Love MI, Huber W, Anders S, Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15, 550 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Vaser R, Adusumalli S, Leng SN, Sikic M, Ng PC, SIFT missense predictions for genomes. Nat Protoc 11, 1–9 (2016). [DOI] [PubMed] [Google Scholar]
- 82.Choi Y, Sims GE, Murphy S, Miller JR, Chan AP, Predicting the functional effect of amino acid substitutions and indels. PLoS One 7, e46688 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Adzhubei IA et al. , A method and server for predicting damaging missense mutations. Nat Methods 7, 248–249 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Reva B, Antipin Y, Sander C, Predicting the functional impact of protein mutations: application to cancer genomics. Nucleic Acids Res 39, e118 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Schrock E et al. , Multicolor spectral karyotyping of human chromosomes. Science 273, 494–497 (1996). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1. Images and genetic analysis of five FH−/− tumor cell lines.
Figure S2. FH in-gel activity assay and protein abundance analysis.
Figure S3. FH-deficient tumors exhibited a dysfunctional respiratory chain.
Figure S4: qPCR analysis of additional mtDNA-encoded genes in HLRCC tissues.
Figure S5. Up-regulation of expression of genes involved in glycolysis, glutaminolysis, pentose phosphate pathway, and fatty acid biosynthesis pathways in HLRCC FH−/− tumors.
Figure S6: RNAseq analysis of the expression of NADP-dependent NRF2 transcriptional target genes in HLRCC tissues.
Figure S7: Analysis of respiration rates in FH−/− HLRCC tumor-derived cell lines.
Figure S8: qPCR analysis of additional nuclear and mtDNA-encoded genes in FH−/− tumor cell lines.
Figure S9: Mitochondrial genome mutations present in UOK262 and UOK268 cells.
Figure S10: Mitochondrial genome mutation present in UOK348 cells.
Figure S11: Breakdown of mtDNA mutations found in FH−/− tumors and cell lines.
Figure S12: Heteroplasmy of somatic mtDNA mutations in FH−/− tumors.
Figure S13: Heteroplasmy of somatic mtDNA mutations found in each FH−/− tumor sample in the study.
Figure S14: Map of selected protein-coding mtDNA mutations found in tumor samples.
Figure S15: Classification of mtDNA mutation impact in tumor samples.
Figure S16: Comparative mutational analysis of nuclear genes in The Cancer Genome Atlas (TCGA) analysis of RCC.
Figure S17: Spectral karyotyping of HLRCC-derived cell lines.
Figure S18: Analysis of POLG immunoprecipitates from mitochondrial lysates isolated from FH−/− tumor tissue samples and control kidney cortex samples.
Table S1: Summary of mtDNA mutations in HLRCC tumors.
Table S2. Mass spectrometry analysis on nucleoid components isolated by TFAM immunoprecipitation in RPTEC and FH−/− UOK262 cell lines.